- 1Youjiang Medical University for Nationalities, Baise, Guangxi, China
- 2Guangxi Key Laboratory of basic and translational research of Bone and Joint Degenerative Diseases, Guangxi Biomedical Materials Engineering Research Center for Bone and Joint Degenerative Diseases, Department of Orthopedics, Affiliated Hospital of Youjiang Medical University for Nationalities, Baise, Guangxi, China
- 3Guangxi Botanical Garden of Medicinal Plants, Nanning, China
- 4Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, China
- 5School of Mechanical Engineering, Dongguan University of Technology, Dongguan, Guangdong, China
- 6Academy of Orthopedics, Guangdong Provincial Key Laboratory of Bone and Joint Degeneration Diseases, The Third Affiliated Hospital of Southern Medical University, Guangzhou, China
- 7State Key Laboratory of Metal Matrix Composites, School of Material Science and Engineering, Shanghai Jiao Tong University, Shanghai, China
Regenerative medicine is a discipline that studies how to use biological and engineering principles and operation methods to repair and regenerate damaged tissues and organs. Until now, regenerative medicine has focused mainly on the in-depth study of the pathological mechanism of diseases, the further development and application of new drugs, and tissue engineering technology strategies. The emergence of aptamers has supplemented the development methods and types of new drugs and enriched the application elements of tissue engineering technology, injecting new vitality into regenerative medicine. The role and application status of aptamers screened in recent years in various tissue regeneration and repair are reviewed, and the prospects and challenges of aptamer technology are discussed, providing a basis for the design and application of aptamers in long-term transformation.
Introduction
Regenerative medicine is closely related to the treatment of injury and aging. The methods and strategies of regenerative medicine involve mainly stem cell technology, tissue engineering technology, and physical and chemical intervention (Fu, 2018; Edgar et al., 2020). Although regenerative medicine based on these strategies has achieved partial success in tissue and organ repair and regeneration, it is still subject to many defects. These defects include difficulty in obtaining exogenous stem cells, immunogenicity, tumorigenicity, low survival rate of exogenous stem cells (Tang, 2019), and high production cost of traditional protein active molecules. In addition, the environmental conditions for their biological activity are harsh (Perwein et al., 2018), and the application elements of tissue engineering technology are outdated (Atala and Forgacs, 2019; Matai et al., 2020).
Aptamers are a new class of ligand molecules that are manually screened by SELEX (systematic evolution of ligands by exponential enrichment) technology (Saito, 2021). They are short single-stranded DNA or RNA that can be folded into specific spatial structures. Moreover, they can bind to specific target molecules with high affinity and specificity through spatial complementarity, electrostatic attraction, van der Waals force, hydrophobic interaction, π-π stacking, and hydrogen bonding (Cai et al., 2018; Plach and Schubert, 2020). Compared with antibodies, aptamers have lower production cost, higher thermal stability, stronger renaturation after denaturation, easier chemical modification and labeling, wider target range, lower immunogenicity and cytotoxicity, and higher tissue penetration ability (Byun, 2021; Ni et al., 2021). In view of this, they have been widely applied in the biomedical field for testing, diagnosis, and targeted cancer therapy in the form of detection probes and a targeting medium (Zhang et al., 2021a; Mandal et al., 2021; Ni et al., 2021; Riccardi et al., 2021; Ma et al., 2022).
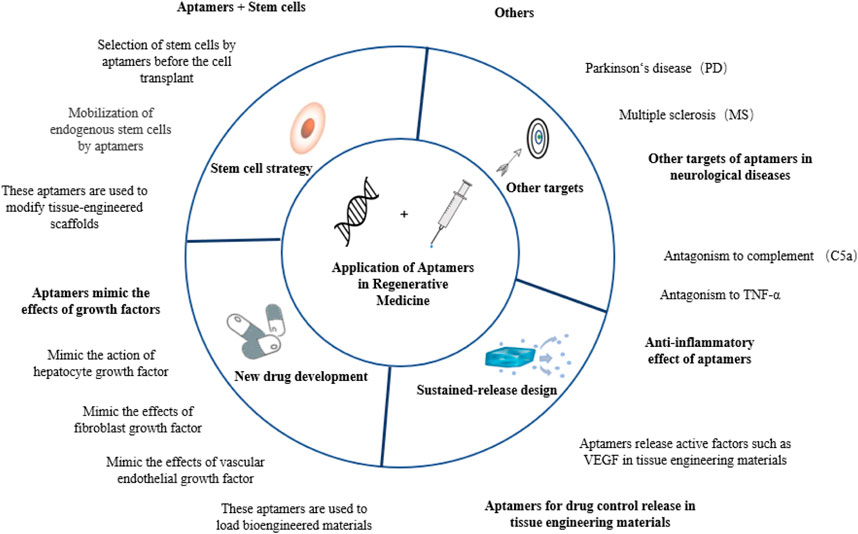
Aptamers have recently been introduced into various regenerative medicine strategies, showing high application value in tissue repair and disease treatment. In this review, aptamers are classified according to their different modes of action, and their applications in tissue regeneration are reviewed. See Figure 1 for a general preview.
Stem cell strategies combining aptamers in regenerative medicine
Stem cells can theoretically divide and proliferate infinitely. They exist widely in various tissues and can be derived and differentiated into various types of cells under specific environmental stimulation. Stem cells have become important resources in regenerative medicine because of their potency and growth capacity, especially for tissues with limited regenerative capacity, such as neural tissues (Gong et al., 2020) and muscle tissues (Roshanbinfar et al., 2021). Currently, stem cell strategies include mainly exogenous stem cell transplantation and endogenous stem cell mobilization and regulation. After arriving at the site of injury, stem cells from different sources play mainly a regenerative and repair role by integrating defects, secreting a variety of bioactive molecules, and partaking in immune regulation (Sarkar et al., 2021).
Aptamer-based stem cell sorting technology can be used to obtain and purify target cells during stem cell transplantation
For exogenous stem cell transplantation, the sorting and collection of various stem cells is the primary problem. Classical cell sorting methods include immunomagnetic bead sorting (Laghmouchi et al., 2020), flow cytometry (Manohar et al., 2021), and solid surface sorting strategy based on cell adhesion differences (Durand, 2021). The first two methods are based mainly on the immunological principle of antibodies. However, during the sorting operations, cell viability and biological characteristics are often affected by antibody binding, fluid shear force, treatment of related enzymes, temperature changes, and electrical and chemical stimulation. Finally, they become unstable (Yao et al., 2020). For example, the combination of protein antibodies with stem cells can lead to changes in cell Stemness, changing the initial state of the cell and too intense ambient temperature, potential of hydrogen (pH) and electric field intensity can even induce cell degeneration and apoptosis, etc.
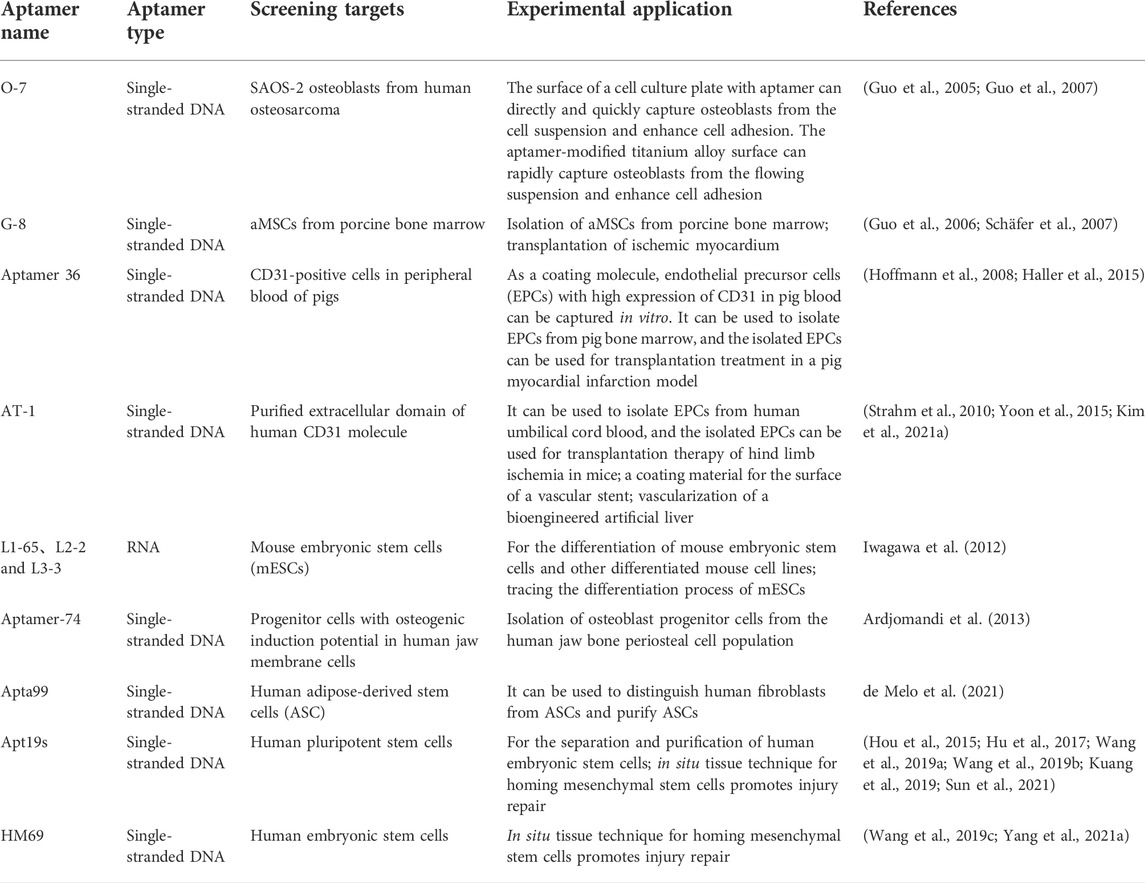
The emergence of SELEX technology based on cell or cell-specific markers has enabled aptamers to be used for sorting target cells. It has the advantages of low production cost, strong resistance to enzymatic hydrolysis, high purity, high efficiency, and little influence on cells. Over the past decade, a large number of aptamers with specificity and high affinity for various types of progenitor cells have been identified (Table 1). These include O-7 (a single-stranded DNA aptamer that binds human osteoblasts) (Guo et al., 2005; Guo et al., 2007), G-8 (an aptamer for binding adult mesenchymal stem cells (aMSCs) from pig bone marrow) (Guo et al., 2006; Schäfer et al., 2007), aptamer 36 (for CD31-positive cells in pig peripheral blood) (Hoffmann et al., 2008; Haller et al., 2015), specific binding aptamers of purified human CD31 extracellular domain (Strahm et al., 2010; Yoon et al., 2015; Kim et al., 2021a), specific RNA aptamers (L1-65, L2-2, and L3-3 binding to mouse embryonic stem cells (mESCs)) (Iwagawa et al., 2012), and DNA aptamers ((Aptamer-74) for osteoblast progenitors present in human jaw membrane cell population) (Ardjomandi et al., 2013). They have shown the ability to capture and enrich corresponding stem cells in bone marrow, whole blood, and other cell suspensions, suggesting their potential application in the collection of exogenous stem cells before transplantation and surface modification of biomaterials for tissue regeneration. Recently, de Melo et al. (2021) used adipose-derived stem cells (ASCs) from human adipose tissue and fibroblasts from skin tissue as screening targets and used Cell-SELEX and quantitative PCR technology to screen and identify an aptamer named Apta99. The aptamer can specifically recognize ASCs and shows a low affinity for fibroblasts, which is expected to be a powerful tool in ASC purification and therapeutic applications. Yao et al. (2020) also reported a cell capture technique beneficial to the survival of bone marrow mesenchymal stem cells (BMSCs). They introduced Apt19s (Hou et al., 2015), a single-stranded DNA aptamer with high specificity and affinity for BMSCs, into another cell-friendly DNA hydrogel system (Nam et al., 2021; Wu et al., 2021) to achieve a specific envelope and capture of stem cells during the formation of a hydrogel 3D DNA network. BMSCs showed uniform spatial distribution and good survival activity in this 3D network, and this system could achieve controlled release of captured cells under the digestion of nuclease. This principle is shown in Figure 2.
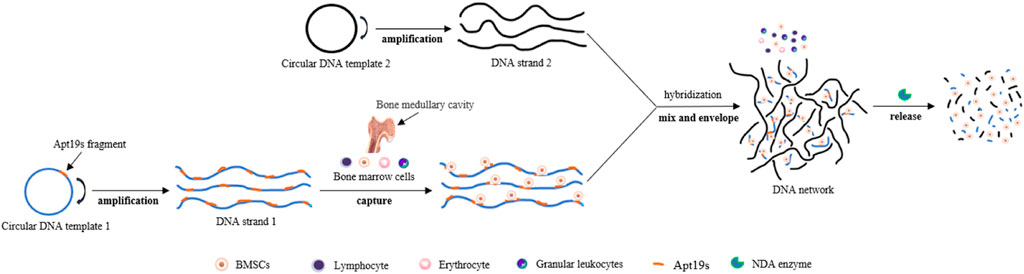
FIGURE 2. Schematic of a DNA cross-linking network designed for harvesting bone marrow mesenchymal stem cells. The process of capturing, enveloping, and releasing bone marrow stem cells (BMSCs). Capturing: DNA strand 1 was incubated with BMSCs, and cell capture was performed by aptamer Apt19s anchoring. Enveloping: Introducing DNA strand 2 into a cell solution containing DNA strand 1 triggers the formation of DNA networks. Releasing: The DNA network can be digested by DNA enzymes, which release BMSCs.
Aptamers can capture and enrich endogenous stem cells at the site of tissue injury to promote tissue regeneration and repair
When tissue is damaged, the stimuli can activate the silent stem cells residing in situ to divide and proliferate for self-repair. Compared with exogenous stem cells, endogenous stem cells do not have problems with immunogenicity and origin (Yamanaka, 2020). Activating endogenous stem cells facilitate tissue repair. Traditional chemoattractant types have been used to activate and migrate endogenous stem cells and have gained recognition in situ tissue engineering techniques (Andreas et al., 2014).
In recent years, aptamers with in vitro cell capture ability have also been applied to in situ enrichment of endogenous stem cells and promote tissue regeneration. Apt19s (Hou et al., 2015; Wang et al., 2019a) is a single-stranded DNA aptamer derived from the SELEX sequence database of human pluripotent stem cells (PSCs). It shows high specificity and affinity binding to multiple gene-derived progenitor cells in vitro. Apt19s can be used as a powerful tool for the isolation, purification, and enrichment of PSCs. The binding target was identified as alkaline phosphatase-ALPL, a membrane protein type highly expressed on the surface of PSCs and its derived progenitor cells. The possibility of applying Apt19s to tissue engineering in situ, which facilitates the homing of stem cells, has been explored. In these designs (Hu et al., 2017; Wang et al., 2019b; Sun et al., 2021), the authors chemically coupled amino-modified Apt19s with activated carboxyl groups in various tissue-engineered scaffold components through the dehydration condensation reaction. They expected that the tissue-engineered scaffolds functionalized by this novel “cell catcher” could achieve a better repair effect after implantation at the injured site. The results showed that Apt19s on these scaffolds could recruit bone marrow mesenchymal stem cells and promote injury healing in a rat osteochondral knee joint defect model, rabbit osteochondral knee joint defect model, and critical size rat skull bone defect model. Kuang et al. (2019) produced a functional hydrogel by the covalent combination of Apt19s, which was modified by Acrydite (acrylic phosphoramidite) at the 5′ end of the nucleotide chain with an injectable composite hydrogel whose matrix was mainly composed of dimethylaminoethyl methacrylate (DMAEMA) and 2-hydroxyethyl methacrylate (HEMA). This hydrogel was also found to have obvious endogenous MSC recruitment when implanted into a rat femur bone defect model. Wang et al. (2019c) recently developed a new MSC-targeting aptamer HM69 using whole cell SELEX technology and constructed functional nanoparticles based on this aptamer. Such nanoparticles are essentially oligonucleotides formed by the end-to-end connection of nucleotide chains of the aptamer as independent units. Both HM69 and HM69 functional nanoparticles showed high specificity and affinity binding to MSCs in vitro, and such binding properties were evaluated to be superior to Apt19s. They applied the novel nanoparticles to treat a femur bone defect in a rat model and found that nanoparticles reached the bone defect via the circulation and then mediated in situ recognition, capture, and enrichment of MSCs. Yang et al. (2021a) also designed a 3D bio-printing dual-function bionic scaffold for in-situ cartilage regeneration based on the directional recruitment of MSCs by the HM69 aptamer. MSC cartilage differentiation was enhanced by transforming growth factor, which achieved good results in a rabbit full-layer cartilage injury model of a knee joint. In addition, a large number of designs have been reported that use aptamers to recruit and enrich endogenous repair factors at tissue injury sites (e.g.,vascular endothelial growth factor (VEGF) (Son et al., 2019), fractalkine (FKN) (Enam et al., 2017), etc.) and these designs also take full advantage of the binding specificity and adhesion between aptamers and target molecules.
In conclusion, aptamers play a role as a new cell sorting tool and a new cell chemoattractant and trapping agent in regenerative medicine stem cell strategies. Compared with the traditional application elements represented by active protein molecules, aptamers have more efficient cell sorting efficiency and better adaptability in vivo, and are easier to modify and connect with tissue engineering scaffolds. However, there are no reports on human experimental applications.
Aptamer elements in a sustained-release hydrogel system
Aptamers with growth factor-like activity can be introduced into biomedical materials such as hydrogel to prolong the release time of aptamers, compared with using them alone. Moreover, aptamers can also be used as a controlled release device to control the release of other active molecules in sustained-release hydrogels.
Active aptamers in hydrogels
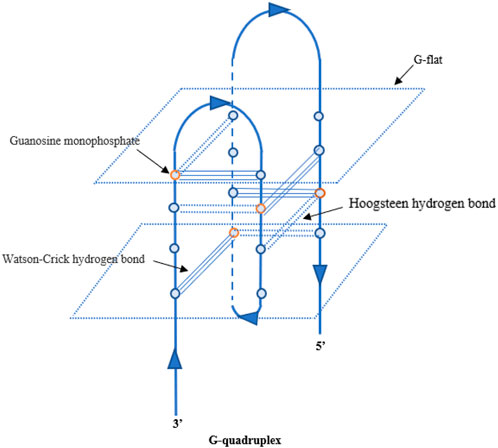
So far, some aptamers with growth factor-like activity have been screened to promote tissue regeneration. They have been combined with various hydrogel materials, showing a good application prospect in the field of tissue engineering. Ueki et al. (2016); Ueki et al. (2019); Ueki et al. (2020) obtained two aptamer dimers, ss-0 and TD0, that simulate the physiological effects of hepatocyte growth factor and basic fibroblast growth factor, respectively. The ss-0 is composed of two DNA aptamer monomers cross-linked by their complementary 5′-end sequences. These two monomers bind specifically and with high affinity to the receptor Met and have strong anti-nuclease stability. In vitro experiments have shown that ss-0 binds two Met receptor molecules to mediate intermolecular dimerization, including autophosphorylation of the receptor molecule, which stimulates cell migration and proliferation. After intravenous administration, the aptamer dimer showed ideal tissue distribution and pharmacokinetic characteristics, which are closely related to the G-quadruplex structure of the dimer sequence (see Figure 3). In a mouse model of outbreak hepatitis, ss-0 antagonizes the apoptosis of liver cells and alleviates the inflammatory response by specifically activating Met, proving its biosafety in vivo. TD0 specifically activates fibroblast growth factor receptor-1 (FGFR1). TD0 is directly linked by two SL38.2 DNA aptamers that target the extracellular domain of FGFR1 and can simulate the biological role of bFGF in vitro. This induces activation of FGFR1 dimerization and subsequent phosphorylation signal transduction pathways, supporting self-renewal and maintenance of stemness in induced pluripotent stem cells (iPSCs). It is noteworthy that SL38.2 also contains a guanine (G)-rich sequence that can form an antiparallel G-quadruplex structure in the physiological environment. This is considered to be key to its anti-nuclease stability and high-affinity binding to FGFR1. In 2015, Ramaswamy et al. (2015) reported a divalent aptamer (AptDivalent) assembled from two AptM80mer monomers, which specifically targets vascular endothelial growth factor receptor-2 (VEGFR2). Binding of AptDivalent to VEGFR2 on the surface of umbilical vein endothelial cells can activate the phosphorylation of receptors, activate the downstream Akt pathway, upregulate nitric oxide synthase content in endothelial cells, and ultimately promote endothelial cells to participate in the formation of new capillaries (all in vitro). James and Allen (2021) also reported the formation of a tubular phenotype in vitro in human umbilical vein endothelial cells (HUVECs) mediated by composite microfibers formed by the self-assembly of this aptamer dimer and collagen. Roy et al. (2021) introduced AptM80mer into a hyaluronic acid hydrogel and realized the functionalization of the hydrogel. They coupled the aptamer whose 5′ end of the nucleotide chain had been modified by an acrydite group to a thiolated-hyaluronic acid in the form of thioether bonds, which are formed under conditions of copolymerization. In vitro experiments based on HUVECs demonstrated that AptM80mer plays a similar role in endothelial cell activation and migration mediated by vascular endothelial growth factor (VEGF) in this hydrogel system and promoted angiogenesis. These studies indicate that the aptamer is expected to be an effective biochemical element for the vascularization design of engineering tissues (Figure 4).
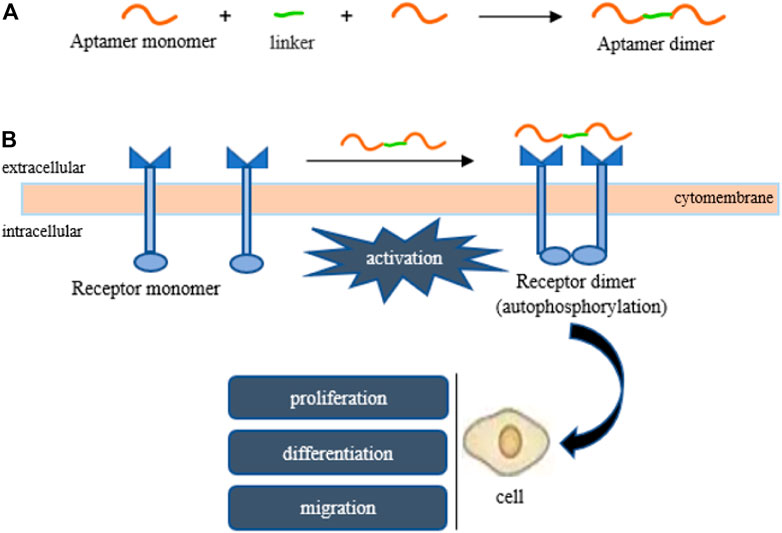
FIGURE 4. Aptamers mimic the activity of growth factors (HGF, bFGF, VEGF). (A) The formation of an aptamer dimer, (B) receptor-to-receptor dimerization and subsequent activation are both mediated by the aptamer dimer.
Aptamers can control the release of other active molecules in hydrogels
A novel sustained-release hydrogel system has been constructed by using aptamers as the affinity sites for proteins and bioactive oligonucleotides in hydrogels. This sustained-release hydrogel system has been proven beneficial to the long-term and flexible regulation of histopathological repair. In 2019, Abune et al. (2019) designed a double-aptamer-functionalized large-hole polyethylene glycol (PEG) hydrogel. They applied the same bonding method to introduce two DNA aptamers, modified by acrylate and targeting bFGF and VEGF, into the PEG network in the hydrogel system. This design also slowed the release rate of protein factors in the hydrogel and realized the long-term and stable release of active factors. The release mode of the two active factors mediated by this release platform synergistically promoted the migration of HUVECs and the regeneration of blood vessels in the chorioallantoic membrane (CAM) model in vitro. Zhao et al. (2019) also developed an aptamer-fibrin (Ap-Fn) hydrogel for controlling VEGF delivery, but the difference was the modification method of the aptamer. They first chemically coupled the mercaptosylated anti-VEGF aptamer with acrydite-modified natural fibrinogen in a copolymerization reaction between mercaptosylation and olefin to synthesize an aptamer-coupled fibrinogen macromolecule monomer. Compared with simple fibrinogen, the overall size and biological activity of this monomer hardly changed, and it could assemble and form a fibrin hydrogel under the action of thrombin and other clotting-related factors. It was proven that the anti-VEGF aptamer could significantly prolong the retention of VEGF in the Ap-Fn hydrogel, thus realizing the sustained release of VEGF. This improved VEGF release modality and promoted p-VEGFR2 expression and cell growth in HUVECs in vitro. Compared with the control hydrogels loaded with VEGF (hydrogels without aptamer modification or nonspecific aptamer modification), the Ap-Fn hydrogels loaded with VEGF could more effectively promote the generation of new blood vessels in the mouse skin trauma model and accelerate skin wound healing. The experiment by Juhl et al. (2019) also confirmed that the functional fibrinogen-aptamer did not lead to significant changes in gelation time, mechanical properties, or microstructure of the gel after gelation. However, the introduction of specific aptamers improved VEGF release kinetics in the aptamer-fibrin hydrogels (AFH), reducing the diffusion rate of VEGF compared with non-functional fibrin hydrogels (FH). The release time of VEGF was prolonged. In a mouse skull defect model of critical size, VEGF release mediated by the release platform functionalized by the aptamer increased the degree of vascularization in vivo and promoted osteogenesis more significantly than VEGF release mediated by the release platform without the introduction of the aptamer.
It can be seen from the above that aptamers could be used not only as a new bioactive ingredient to load hydrogels and other biological materials to promote tissue regeneration but also as a control device in biological materials to control the release process of other active ingredients. Screening more aptamer molecules for biological activity would be beneficial to regenerative medicine. In addition, we still need to pay more attention to the connection method between aptamers and biomaterial. We must also fully ensure that the biological activity of aptamers does not change after the connection, which is key to the future role of aptamers in the field of regenerative medicine.
Antagonistic aptamers against pro-inflammatory mediators (plays a blocking role in inflammatory mediators)
The immune-inflammatory response plays an important role in the repair of organ and tissue injury. A large number of therapeutic strategies targeting inflammatory factors or inflammatory factor receptors have been attempted for tissue regeneration. As a new type of ligand molecule, aptamers can influence the biological activities mediated by the target after binding with their targets. As a result, a number of aptamers directly involved in the regulation of inflammation have been developed. These aptamers can specifically bind inflammatory mediators, including interleukin-2 (IL-2), IL-6, IL-10, IL-11, IL-17, IL-32, transforming growth factor-β (TGF-β), tumor necrosis factor-α (TNF-α), interferon-γ (IFN-γ), chemokines CCL2, IP-10, and some related receptors (Boshtam et al., 2017), and even some inflammatory autoantigens (Mor-Vaknin et al., 2017; Cao et al., 2021), etc. Here, we focus on several aptamers with immunomodulatory activity developed in recent years. The difference is that they have been studied in vivo, and the types of aptamers that regulate complement activity have been developed.
Aptamers that bind TNF-α or TNF receptor
TNF-α is an important member of the TNF superfamily and is derived mainly from monocytes and macrophages. Other cells such as T and B lymphocytes, natural killer cells, mast cells and endothelial cells, neutrophils, smooth muscle cells, and cardiomyocytes can also release this cytokine. TNF-α is also one of the most potent cytokines found in mammals. Its pleiotropy is reflected mainly in its ability to bind both TNFR1 and TNFR2. The binding of TNF-α to TNFR1, one receptor type, leads to a negative inflammatory process or apoptosis. The binding of TNF-α to TNFR2 produces positive repair effects, such as maintaining cell survival and promoting tissue regeneration. These two opposite biological effects of TNF-α are closely related to the mitogen-activated protein kinase (MAPK) signaling pathway (Tseng et al., 2018; Zhang et al., 2020).
The inhibitors represented by TNF-α monoclonal antibodies are thought to effectively block the destructive effect of TNF-α (Mitoma et al., 2018). According to the same principle, a new class of aptamer-based TNF-α inhibitors has been developed. In 2013, Orava et al. (2013) identified a DNA aptamer VR11 that specifically recognizes human recombinant TNF-α. In vitro experiments showed that the aptamer could block the binding process between TNFα and TNFα receptor, thus inhibiting the cytotoxicity of TNF-α, which was similar to the action of TNF-α antibody. Kim et al. (2021b) introduced VR11 into a metal nanoparticle that is often used as a carrier material in biomedical applications. This not only enhanced the stability of aptamers but also played a role in capturing TNF-α at the inflammatory site in vivo and inhibiting the inflammatory response. Lai et al. (2019) developed a novel TNF-α targeting aptamer (aptTNF-α) and its PEG derivative (aptTNF-α-PEG), which showed good affinity for human/mouse TNF-α in vitro. In addition, targeted inhibition of TNF-α was also shown in acute lung injury and acute liver failure mouse models with high TNF-α expression. In vivo application of aptTNF-α/aptTNF-α-PEG reduced the degree of the acute inflammatory response in both models and promoted early regeneration of liver tissue. These studies indicate the potential of aptamers as non-immunogenic oligonucleotide inhibitors against TNFα.
Although TNF-α inhibitors inhibit the negative effects of TNF-α, they also eliminate the positive contribution of TNF-α to inflammation. To solve this problem, Zhang et al. (2020) screened specific aptamers for the inflammatory receptor TNFR1 downstream of the TNF-α signaling pathway, hoping to inhibit inflammation and improve the therapeutic effect by selectively inhibiting TNFR1. So far, they have obtained eight aptamers with high affinity for TNFR1 and two with low affinity for TNFR2. However, there are no relevant experimental data in cells and zoology.
Aptamers antagonizing complement (aptamers binding to complement)
Among these aptamers, NOX-D20 (Hoehlig et al., 2013), which binds and blocks complement C5a, has been studied the most. C5a is considered a potent anaphylactic toxin, and excessive C5a demonstrates a cascade of inflammatory destruction (Wood et al., 2018). NOX-D20 is a mirror aptamer synthesized from an unnatural L-oligonucleotide known as Spiegelmers. In vitro experiments have shown that NOX-D20 could block the release of elastase from polymorphonuclear leukocytes induced by C5a and the chemotaxis of CD88 expression cell lines. These two pathological events are closely related to pro-inflammation. In rodent sepsis models induced by cecal ligation and puncture, it showed therapeutic effects of reducing inflammation and organ damage, preventing the destruction of the vascular endothelial barrier, and improving the survival rate. The specific binding and inhibitory activity of NOX-D20 on C5a has been proven to be closely related to the G-quadruplex structure of its molecular structure (Yatime et al., 2015). In recent years, people have developed ways of using and treating NOX-D20. Li et al. (2019) combined a framework of nucleic acid (FNA) (Zhang et al., 2021b; Zhang et al., 2021c; Gao et al., 2022) in the form of a DNA origami nanostructure to improve the stability and targeting of NOX-D20 in vivo. They used bipyramidal FNA as a drug delivery platform to deliver NOX-D20 to treat cerebral ischemia-reperfusion injury and achieved good results. Zhang et al. (2021d) conjugated NOX-D20 with cerium dioxide nanoparticles (Ni et al., 2019) with active oxygen scavenging activity and formed the complex Ceria@Apt, which not only integrates the anti-inflammatory advantages of both but also makes full use of the liver absorption effect of nanomaterials. This composite material shows great application potential in the repair of liver ischemia-reperfusion injury.
In general, although various inhibitors against C5a have been extensively studied, the types of inhibitors against downstream c5A-related signaling molecules have not been reported.
Aptamer types that act on other pathological targets in neurological diseases
In recent years, studies on the types of aptamers involved in treating several neurological diseases (represented by multiple sclerosis and Parkinson’s disease) have become increasingly mature. These have provided ideas for developing new aptamer-based drugs, which are also introduced here.
Aptamers associated with multiple sclerosis
Inspired by the fact that a natural IgM antibody can promote myelin regeneration after binding to oligodendrocytes both in a mouse multiple sclerosis (MS) model infected with Theiler’s myeloencephilitis virus (TMEV) (Dubik et al., 2021) and in a mouse model of focal demyelination induced by lysophosphatidylcholine (Bieber et al., 2002), Nastasijevic et al. (2012) identified a small molecule single-stranded DNA aptamer 3064, or LJM-3064 (Wilbanks et al., 2019). This aptamer is smaller than the IgM monoclonal antibody by in vitro screening and targets the crude extract of myelin from SJL mice (the myelin preparation is a crude mixture of proteins and lipids). The aptamer contains a guanylate-rich domain and has shown high affinity and specific binding ability to myelin basic protein (MBP) subtypes in vitro. Previous studies (Bose, 2021) have shown that the sequence encoded by MBP exon 2 in the MBP subtype is an effective target for myelin regeneration. Their further study found that a streptavidin tetramer could mediate the coupling of four aptamers. This formed the tetramer aptamer complex Myaptavin-3064, which mimics the activity of polyvalent antibodies (Fereidan-Esfahani et al., 2020). The biological stability of the aptamer complex was enhanced compared with that of the aptamer monomer. Intraperitoneal injection of demyelinated mice infected with TMEV enhanced myelin regeneration in the mouse model, while aptamer monomers did not.
In later studies, Smestad and Maher (2013) verified that 26 nucleotides at the 5′ terminal of LJM-3064 (guanylate-rich region) were involved in forming the G-quadruplex through intramolecular folding. They also proved the univalent ion-dependent conformational switching of the G-tetrahedron structure in the physiological environment. Namely, in the absence of sodium and potassium ions, LJM-3064 adopts an antiparallel G-tetrahedron structure. When transferred into a buffer that mimics low potassium concentrations in tissue fluid and plasma, LJM-3064 rapidly transitions into a thermodynamically more stable parallel chain G-tetrahedron conformation. Their data also showed that the parallel G-quadruplex structure of the LJM-3064 monomer still existed in the tetravalent complex Myaptavin-3064. Perschbacher et al. (2015) used quantitative PCR to analyze the effect of the nucleotide sequence structure of LJM-3064 on its pharmacokinetics and the pharmacokinetic characteristics of various LJM-3064 derivatives produced by different modification methods. The results suggest that LJM-3064s tissue penetration and anti-enzymatic activity may be related to the G-rich sequence in its composition structure, and the polyvalent property of MyAPtavin-3064 may more affect the biological activity of myelin regeneration. Wilbanks et al. (2019) truncated and optimized a DNA aptamer LJM-5708 containing only 20 nucleotides based on LJM-3064, which retained the rich G sequence of the G-quadruplex structure formed by the original aptamer. The polyvalent aptamer complex formed by LJM-5708 showed enhanced myelin-binding properties compared with the LJM-5708 monomer, and also showed significant binding ability to human oligodendrocytes cultured in vitro. The study also pointed out that the absence of the G-quadruplex formation of the LJM-5708 aptamer results in the loss of myelin binding function of the polyvalent aptamer complex. Heider et al. (2018) designed a streptavidin-free aptamer polymer (named 3064-4WJ-LNA) on the basis of previous work, which is expected to improve immunogenicity.
In addition to studying the role of the LJM-3064 aptamer alone in MS myelin regeneration therapy, LJM-3064 was also conjugated to a mesenchymal stem cell-derived exosome with immunomodulatory functions. Two synergistic biological effects of myelin regeneration and anti-inflammation were achieved in the multiple sclerosis mouse model induced by a myelin oligodendrocyte glycoprotein, which expanded the application of the LJM-3064 aptamer (Hosseini et al., 2019).
In 2020, Fereidan-Esfahani et al. (2020) used flow cytometry and immunocytochemistry to determine the cell-binding properties of Myaptavin-3064 in vitro. Myaptavin-3064 shows relatively specific binding to the human oligodendroglioma (HOG) cell line. In particular, the differentiated HOG shows enhanced binding ability with no affinity for lung (L2) or kidney (BHK) cell lines. In addition to HOG cells, Myaptavin-3064 could bind to adult rat oligodendrocytes (OLs) but not to primary cortical cells derived from embryonic mice. The cell-binding properties of Myaptavin-3064 indicate that the target of Myaptavin-3064 exists in the more mature myelin sheath and also support the hypothesis that Myaptavin-3064 induces myelin regeneration through OL binding.
The migration of autoreactive immune cells across the BBB (blood-brain barrier) is considered to be a pathological feature of MS, and it plays a key role in the formation of MS demyelinating plaques (Goverman, 2009). The α4-integrin family mediates the adhesion, exudation, and migration of these inflammatory cells (Mitroulis et al., 2015). Based on the importance of α4-integrin in the pathogenesis of MS, a blocking antibody against α4-integrin, natalizumab, has been used for phase 3 clinical observation in patients with secondary progressive multiple sclerosis (Kapoor et al., 2018). According to a similar concept of action, Kouhpayeh et al. (2019) screened a specific single-stranded DNA aptamer with a high affinity for α4-integrin, which is expected to play a therapeutic effect similar to natalizumab on MS.
Aptamers associated with Parkinson’s disease
The α-synuclein (α-syn) is a small natural non-folding protein containing 140 amino acids, existing mainly in the presynaptic terminal in neurons (Burré et al., 2018). In a physiological state, α-syn is a kind of disordered protein in nature. Once stimulated by various injury factors, it tends to form oligomers and aggregates, which may produce toxic effects on neurons through mitochondrial injury, axon transport dysfunction, inflammation, and other mechanisms (Shahnawaz et al., 2020). More importantly, once the abnormal α-syn protein aggregates are absorbed by other normal cells, they can secondarily cause the same pathological process as other normal α-syn proteins in the recipient cells. This leads to intercellular infection and diffusion (Valera and Masliah, 2016), which are important in the formation and progression of PD. Zheng et al. (2018) attempted an aptamer strategy to regulate α-syn toxicity by screening two aptamers with high specificity and affinity for α-syn protein (named F5R1 and F5R2, respectively) using optimized SELEX. Both aptamers could effectively reduce α-syn accumulation in extracellular and intracellular domains, especially in cells. When delivered into human neuroblastoma cells and primary neurons by a carrier peptide, the targeted binding of the two aptamers to α-syn inhibits the formation of pathogenic aggregates and mediates intracellular degradation of α-syn, saving mitochondrial dysfunction and cell damage caused by α-syn overexpression. Ren et al. (2019) studied the in vivo effect of these aptamers on PD-related neuropathological defects. They used a neuron-targeting exosome (Alvarez-Erviti et al., 2011) as a loading vehicle for these therapeutic aptamers, achieving efficient delivery of aptamers to neurons in vivo and in vitro. When co-cultured with primary neurons, aptamer-loaded exosomes significantly reduced the formation of intracellular secondary α-syn insoluble aggregates induced by α-syn fibrils and saved synaptic protein loss and neuronal death. Intraperitoneal injection of this compound in a PD mouse model showed a significant reduction in the pathological aggregates of α-syn in the brain and a significant improvement in the associated motor dysfunction, further confirming the therapeutic ability of these aptamers in vivo.
Aptamers associated with Nogo-66 receptor
Myelin-derived inhibitors, represented by Nogo, myelin-associated glycoprotein (MAG), and oligodendrocyte myelin glycoprotein (OMgp), inhibit the growth of neuronal processes by binding to the Nogo-66 receptor (NgR). Using the NgR as the screening target, Wang et al. (2010) obtained a set of RNA aptamers. In vitro experiments showed that the binding properties of these aptamers to NgR did not affect neurite growth but could competitively block the inhibition of neurite growth by these three inhibitors. Agrawal et al. (2020) designed a sustained-release hyaluronic acid hydrogel modified by several antisense oligonucleotides, and realized the slow and sustained release of these highly therapeutic aptamers by using the complementary pairing relationship between antisense oligonucleotide sequence and NgR targeted aptamers to varying degrees. It had the same protective effect on the neuronal processes damaged in vitro.
Summary and prospects
Regenerative medicine makes full use of various biological techniques and engineering principles to provide better and faster repair of diseases. In the past, stem-cell strategies, tissue engineering techniques, and physicochemical techniques have been applied to regenerative medicine, which has achieved remarkable results. However, in the actual process of operation, people are still faced with many problems, such as difficulties in obtaining stem cells and limited means of regulating cell biological behavior. In addition, there is a lack of development mode and type of new chemical preparations with biological activity, as well as a scarcity of tissue engineering technology application factors.
Aptamers are a kind of ligand nucleic acid molecule with similar binding properties to antibodies. The binding targets are varied, including inorganic ions, amino acids, antibiotics, peptides, proteins, sugars, nucleotides, and their derivatives (Plach and Schubert, 2020). At the same time, these targets can be used as specific markers of bacteria (Yang et al., 2021b), viruses (Lu et al., 2021), cells (Lin et al., 2021), and other living organisms. Aptamers are widely used in various fields of biomedicine, especially in testing, diagnosis, and targeted therapy of cancer, due to their different binding effects with different targets. In a word, aptamers can be called “artificial high quality nucleic acids” because of their universality of application purposes, wide range of action targets, flexibility of action time and space, and diversity of action forms.
From the perspective of tissue regeneration, this article on aptamers in related research in recent years has continued the review and summary. The results show that aptamers can be used alone or crossed with each other through various technical means, such as stem cell technology, biological engineering materials such as hydrogels, nanomedicine, immunotherapy, and so on. Therefore, they show strong plasticity and a broad transformation prospect for applied research in regenerative medicine. At the same time, this paper also provides a reference for the long-term design and utilization of aptamers. Due to the limited amount of literature and the inability to categorize and report, we could not summarize the modification methods of aptamers for therapeutic application.
Many problems still need to be solved for the practical application of aptamers. These include further cost control, development of new aptamer types, improvement in screening efficiency, creation of modifications that improve pharmacokinetics and enhance biological activity in vivo, optimization of connection methods with other biomedical materials, and advancing zoological and human experiments. We believe that aptamers can better serve us after these problems have been solved.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
Study design: LW, JL, and YT. Study procedures and data collection: ZL, SMC, and JZ. Data analysis: ZL and CW. Drafting the manuscript: ZL, CW, and KL. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 32160209, 82160357, 82071361), Guangxi Key Laboratory of basic and translational research of Bone and Joint Degenerative Diseases (NO. 21-220-06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abune, L., Zhao, N., Lai, J., Peterson, B., Szczesny, S., and Wang, Y. (2019). Macroporous hydrogels for stable sequestration and sustained release of vascular endothelial growth factor and basic fibroblast growth factor using nucleic acid aptamers. ACS Biomater. Sci. Eng. 5 (5), 2382–2390. doi:10.1021/acsbiomaterials.9b00423
Agrawal, N. K., Allen, P., Song, Y. H., Wachs, R. A., Du, Y., Ellington, A. D., et al. (2020). Oligonucleotide-functionalized hydrogels for sustained release of small molecule (aptamer) therapeutics. Acta Biomater. 102, 315–325. doi:10.1016/j.actbio.2019.11.037
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., and Wood, M. J. A. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29 (4), 341–345. doi:10.1038/nbt.1807
Andreas, K., Sittinger, M., and Ringe, J. (2014). Toward in situ tissue engineering: Chemokine-guided stem cell recruitment. Trends Biotechnol. 32 (9), 483–492. doi:10.1016/j.tibtech.2014.06.008
Ardjomandi, N., Niederlaender, J., Aicher, W. K., Reinert, S., Schweizer, E., Wendel, H.-P., et al. (2013). Identification of an aptamer binding to human osteogenic-induced progenitor cells. Nucleic Acid. Ther. 23 (1), 44–61. doi:10.1089/nat.2012.0349
Atala, A., and Forgacs, G. (2019). Three-dimensional bioprinting in regenerative medicine: Reality, hype, and future. Stem Cells Transl. Med. 8 (8), 744–745. doi:10.1002/sctm.19-0089
Bieber, A. J., Warrington, A., Asakura, K., Ciric, B., Kaveri, S. V., Pease, L. R., et al. (2002). Human antibodies accelerate the rate of remyelination following lysolecithin-induced demyelination in mice. Glia 37 (3), 241–249. doi:10.1002/glia.10033
Bose, P. (2021). Central pontine myelinolysis and the osmotic demyelination syndromes: An open and shut case? Acta Neurol. Belg 121 (4), 849–858. doi:10.1007/s13760-021-01634-0
Boshtam, M., Asgary, S., Kouhpayeh, S., Shariati, L., and Khanahmad, H. (2017). Aptamers against pro- and anti-inflammatory cytokines: A review. Inflammation 40 (1), 340–349. doi:10.1007/s10753-016-0477-1
Burré, J., Sharma, M., and Südhof, T. C. (2018). Cell biology and pathophysiology of α-synuclein. Cold Spring Harb. Perspect. Med. 8 (3). doi:10.1101/cshperspect.a024091
Byun, J. (2021). Recent progress and opportunities for nucleic acid aptamers. Life (Basel) 11 (3). doi:10.3390/life11030193
Cai, S., Yan, J., Xiong, H., Liu, Y., Peng, D., and Liu, Z. (2018). Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 143 (22), 5317–5338. doi:10.1039/c8an01467a
Cao, J., Su, J., An, M., Yang, Y., Zhang, Y., Zuo, J., et al. (2021). Novel DEK-targeting aptamer delivered by a hydrogel microneedle attenuates collagen-induced arthritis. Mol. Pharm. 18 (1), 305–316. doi:10.1021/acs.molpharmaceut.0c00954
de Melo, M., da Silva Cunha, S. C. P., de Miranda, M. C., Barbosa, J. L., Faria, J. A. Q. A., Rodrigues, M. A., et al. (2021). Selection of DNA aptamers for differentiation of human adipose-derived mesenchymal stem cells from fibroblasts. Appl. Biochem. Biotechnol. 193 (11), 3704–3718. doi:10.1007/s12010-021-03618-5
Dubik, M., Marczynska, J., Mørch, M. T., Webster, G., Jensen, K. N., Wlodarczyk, A., et al. (2021). Innate signaling in the CNS prevents demyelination in a focal EAE model. Front. Neurosci. 15, 682451. doi:10.3389/fnins.2021.682451
Durand, M. (2021). Large-scale simulations of biological cell sorting driven by differential adhesion follow diffusion-limited domain coalescence regime. PLoS Comput. Biol. 17 (8), e1008576. doi:10.1371/journal.pcbi.1008576
Edgar, L., Pu, T., Porter, B., Aziz, J. M., La Pointe, C., Asthana, A., et al. (2020). Regenerative medicine, organ bioengineering and transplantation. Br. J. Surg. 107 (7), 793–800. doi:10.1002/bjs.11686
Enam, S. F., Krieger, J. R., Saxena, T., Watts, B. E., Olingy, C. E., Botchwey, E. A., et al. (2017). Enrichment of endogenous fractalkine and anti-inflammatory cells via aptamer-functionalized hydrogels. Biomaterials 142, 52–61. doi:10.1016/j.biomaterials.2017.07.013
Fereidan-Esfahani, M., Yue, W. Y., Wilbanks, B., Johnson, A. J., Warrington, A. E., Howe, C. L., et al. (2020). Remyelination-promoting DNA aptamer conjugate myaptavin-3064 binds to adult oligodendrocytes in vitro. Pharm. (Basel) 13 (11). doi:10.3390/ph13110403
Fu, X. (2018). Regenerative medicine in China: New advances and hopes. Sci. China Life Sci. 61 (10), 1135–1136. doi:10.1007/s11427-018-9394-9
Gao, Y., Chen, X., and Tian, T. (2022). A lysosome-activated tetrahedral nanobox for encapsulated siRNA delivery[J]. Adv. Mater 2022, e2201731.
Gong, Z., Xia, K., Xu, A., Yu, C., Wang, C., Zhu, J., et al. (2020). Stem cell transplantation: A promising therapy for spinal cord injury. Cscr 15 (4), 321–331. doi:10.2174/1574888x14666190823144424
Goverman, J. (2009). Autoimmune T cell responses in the central nervous system. Nat. Rev. Immunol. 9 (6), 393–407. doi:10.1038/nri2550
Guo, K. T., SchÄfer, R., Paul, A., Gerber, A., Ziemer, G., and Wendel, H. P. (2006). A new technique for the isolation and surface immobilization of mesenchymal stem cells from whole bone marrow using high‐specific DNA aptamers. Stem Cells 24 (10), 2220–2231. doi:10.1634/stemcells.2006-0015
Guo, K. T., Scharnweber, D., Schwenzer, B., Ziemer, G., and Wendel, H. P. (2007). The effect of electrochemical functionalization of Ti-alloy surfaces by aptamer-based capture molecules on cell adhesion. Biomaterials 28 (3), 468–474. doi:10.1016/j.biomaterials.2006.09.021
Guo, K., Wendel, H. P., Scheideler, L., Ziemer, G., and Scheule, A. M. (2005). Aptamer-based capture molecules as a novel coating strategy to promote cell adhesion. J. Cell. Mol. Med. 9 (3), 731–736. doi:10.1111/j.1582-4934.2005.tb00503.x
Haller, C., Sobolewska, B., Schibilsky, D., Avci-Adali, M., Schlensak, C., Wendel, H.-P., et al. (2015). One-staged aptamer-based isolation and application of endothelial progenitor cells in a porcine myocardial infarction model. Nucleic Acid. Ther. 25 (1), 20–26. doi:10.1089/nat.2014.0499
Heider, R. M., Smestad, J. A., Lemus, H. N., Wilbanks, B., Warrington, A. E., Peters, J. P., et al. (2018). An assay that predicts in vivo efficacy for DNA aptamers that stimulate remyelination in a mouse model of multiple sclerosis. Mol. Ther. - Methods & Clin. Dev. 9, 270–277. doi:10.1016/j.omtm.2018.03.005
Hoehlig, K., Maasch, C., Shushakova, N., Buchner, K., Huber-Lang, M., Purschke, W. G., et al. (2013). A novel C5a-neutralizing mirror-image (l-)Aptamer prevents organ failure and improves survival in experimental sepsis. Mol. Ther. 21 (12), 2236–2246. doi:10.1038/mt.2013.178
Hoffmann, J., Paul, A., Harwardt, M., Groll, J., Reeswinkel, T., Klee, D., et al. (2008). Immobilized DNA aptamers used as potent attractors for porcine endothelial precursor cells. J. Biomed. Mat. Res. 84A (3), 614–621. doi:10.1002/jbm.a.31309
Hosseini, S. F., Alibolandi, M., and Rafatpanah, H. (2019). Immunomodulatory properties of MSC-derived exosomes armed with high affinity aptamer toward mylein as a platform for reducing multiple sclerosis clinical score[J]. J. Control Release 299, 149–164.
Hou, Z., Meyer, S., Propson, N. E., Nie, J., Jiang, P., Stewart, R., et al. (2015). Characterization and target identification of a DNA aptamer that labels pluripotent stem cells. Cell Res. 25 (3), 390–393. doi:10.1038/cr.2015.7
Hu, X., Wang, Y., Tan, Y., Wang, J., Liu, H., Wang, Y., et al. (2017). A difunctional regeneration scaffold for knee repair based on aptamer-directed cell recruitment. Adv. Mater 29 (15). doi:10.1002/adma.201605235
Iwagawa, T., Ohuchi, S. P., Watanabe, S., and Nakamura, Y. (2012). Selection of RNA aptamers against mouse embryonic stem cells. Biochimie 94 (1), 250–257. doi:10.1016/j.biochi.2011.10.017
James, B. D., and Allen, J. B. (2021). Self-assembled VEGF-R2 targeting DNA aptamer-collagen fibers stimulate an angiogenic-like endothelial cell phenotype. Mater. Sci. Eng. C 120, 111683. doi:10.1016/j.msec.2020.111683
Juhl, O. T., Zhao, N., Merife, A. B., Cohen, D., Friedman, M., Zhang, Y., et al. (2019). Aptamer-functionalized fibrin hydrogel improves vascular endothelial growth factor release kinetics and enhances angiogenesis and osteogenesis in critically sized cranial defects. ACS Biomater. Sci. Eng. 5 (11), 6152–6160. doi:10.1021/acsbiomaterials.9b01175
Kapoor, R., Ho, P. R., Campbell, N., Chang, I., Deykin, A., Forrestal, F., et al. (2018). Effect of natalizumab on disease progression in secondary progressive multiple sclerosis (ASCEND): A phase 3, randomised, double-blind, placebo-controlled trial with an open-label extension. Lancet Neurol. 17 (5), 405–415. doi:10.1016/S1474-4422(18)30069-3
Kim, D. H., Ahn, J., Kang, H. K., Kim, M.-S., Kim, N.-G., Kook, M. G., et al. (2021). Development of highly functional bioengineered human liver with perfusable vasculature. Biomaterials 265, 120417. doi:10.1016/j.biomaterials.2020.120417
Kim, J., Park, H., Saravanakumar, G., and Kim, W. J. (2021). Polymer/aptamer-integrated gold nanoconstruct suppresses the inflammatory process by scavenging ROS and capturing pro-inflammatory cytokine TNF-α. ACS Appl. Mat. Interfaces 13 (8), 9390–9401. doi:10.1021/acsami.0c15727
Kouhpayeh, S., Hejazi, Z., Boshtam, M., Mirian, M., Rahimmanesh, I., Darzi, L., et al. (2019). Development of α4 integrin DNA aptamer as a potential therapeutic tool for multiple sclerosis. J Cell. Biochem. 120 (9), 16264–16272. doi:10.1002/jcb.28907
Kuang, L., Ma, X., Ma, Y., Yao, Y., Tariq, M., Yuan, Y., et al. (2019). Self-assembled injectable nanocomposite hydrogels coordinated by in situ generated CaP nanoparticles for bone regeneration. ACS Appl. Mat. Interfaces 11 (19), 17234–17246. doi:10.1021/acsami.9b03173
Laghmouchi, A., Hoogstraten, C., Falkenburg, J., and Jedema, I. (2020). Long-term in vitro persistence of magnetic properties after magnetic bead-based cell separation of T cells. Scand. J. Immunol. 92 (3), e12924. doi:10.1111/sji.12924
Lai, W. Y., Wang, J. W., Huang, B. T., Lin, E. P.-Y., and Yang, P.-C. (2019). A novel TNF-α-targeting aptamer for TNF-α-mediated acute lung injury and acute liver failure. Theranostics 9 (6), 1741–1751. doi:10.7150/thno.30972
Li, S., Jiang, D., Rosenkrans, Z. T., Barnhart, T. E., Ehlerding, E. B., Ni, D., et al. (2019). Aptamer-conjugated framework nucleic acids for the repair of cerebral ischemia-reperfusion injury. Nano Lett. 19 (10), 7334–7341. doi:10.1021/acs.nanolett.9b02958
Lin, N., Wu, L., Xu, X., Wu, Q., Wang, Y., Shen, H., et al. (2021). Aptamer generated by cell-SELEX for specific targeting of human glioma cells. ACS Appl. Mat. Interfaces 13 (8), 9306–9315. doi:10.1021/acsami.0c11878
Lu, T., Zhang, H., Zhou, J., Ma, Q., Yan, W., Zhao, L., et al. (2021). Aptamer-targeting of Aleutian mink disease virus (AMDV) can be an effective strategy to inhibit virus replication. Sci. Rep. 11 (1), 4649. doi:10.1038/s41598-021-84223-8
Ma, W., Yang, Y., and Zhu, J. (2022). Biomimetic nanoerythrosome-coated aptamer-DNA tetrahedron/maytansine conjugates: pH-responsive and targeted cytotoxicity for HER2-positive breast cancer[J]. Adv. Mater 2022, e2109609.
Mandal, M., Dutta, N., and Dutta, G. (2021). Aptamer-based biosensors and their implications in COVID-19 diagnosis. Anal. Methods 13 (45), 5400–5417. doi:10.1039/d1ay01519b
Manohar, S. M., Shah, P., and Nair, A. (2021). Flow cytometry: Principles, applications and recent advances. Bioanalysis 13 (3), 181–198. doi:10.4155/bio-2020-0267
Matai, I., Kaur, G., Seyedsalehi, A., McClinton, A., and Laurencin, C. T. (2020). Progress in 3D bioprinting technology for tissue/organ regenerative engineering. Biomaterials 226, 119536. doi:10.1016/j.biomaterials.2019.119536
Mitoma, H., Horiuchi, T., Tsukamoto, H., and Ueda, N. (2018). Molecular mechanisms of action of anti-TNF-α agents - comparison among therapeutic TNF-α antagonists. Cytokine 101, 56–63. doi:10.1016/j.cyto.2016.08.014
Mitroulis, I., Alexaki, V. I., Kourtzelis, I., Ziogas, A., Hajishengallis, G., and Chavakis, T. (2015). Leukocyte integrins: Role in leukocyte recruitment and as therapeutic targets in inflammatory disease. Pharmacol. Ther. 147, 123–135. doi:10.1016/j.pharmthera.2014.11.008
Mor-Vaknin, N., Saha, A., Legendre, M., Carmona-Rivera, C., Amin, M. A., Rabquer, B. J., et al. (2017). DEK-targeting DNA aptamers as therapeutics for inflammatory arthritis. Nat. Commun. 8, 14252. doi:10.1038/ncomms14252
Nam, K., Im, B. I., Kim, T., Kim, Y. M., and Roh, Y. H. (2021). Anisotropically functionalized aptamer-DNA nanostructures for enhanced cell proliferation and target-specific adhesion in 3D cell cultures. Biomacromolecules 22 (7), 3138–3147. doi:10.1021/acs.biomac.1c00619
Nastasijevic, B., Wright, B. R., Smestad, J., Warrington, A. E., Rodriguez, M., and Maher, L. J. (2012). Remyelination induced by a DNA aptamer in a mouse model of multiple sclerosis. PLoS One 7 (6), e39595. doi:10.1371/journal.pone.0039595
Ni, D., Wei, H., Chen, W., Bao, Q., Rosenkrans, Z. T., Barnhart, T. E., et al. (2019). Ceria nanoparticles meet hepatic ischemia-reperfusion injury: The perfect imperfection. Adv. Mater 31 (40), e1902956. doi:10.1002/adma.201902956
Ni, S., Zhuo, Z., Pan, Y., Yu, Y., Li, F., Liu, J., et al. (2021). Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl. Mat. Interfaces 13 (8), 9500–9519. doi:10.1021/acsami.0c05750
Orava, E. W., Jarvik, N., Shek, Y. L., Sidhu, S. S., and Gariépy, J. (2013). A short DNA aptamer that recognizes TNFα and blocks its activity in vitro. ACS Chem. Biol. 8 (1), 170–178. doi:10.1021/cb3003557
Perschbacher, K., Smestad, J. A., Peters, J. P., Standiford, M. M., Denic, A., Wootla, B., et al. (2015). Quantitative PCR analysis of DNA aptamer pharmacokinetics in mice. Nucleic Acid. Ther. 25 (1), 11–19. doi:10.1089/nat.2014.0515
Perwein, M. K., Smestad, J. A., Warrington, A. E., Heider, R. M., Kaczor, M. W., Maher, L. J., et al. (2018). A comparison of human natural monoclonal antibodies and aptamer conjugates for promotion of CNS remyelination: Where are we now and what comes next? Expert Opin. Biol. Ther. 18 (5), 545–560. doi:10.1080/14712598.2018.1441284
Plach, M., and Schubert, T. (2020). Biophysical characterization of aptamer-target interactions. Adv. Biochem. Eng. Biotechnol. 174, 1–15. doi:10.1007/10_2019_103
Ramaswamy, V., Monsalve, A., Sautina, L., Segal, M. S., Dobson, J., and Allen, J. B. (2015). DNA aptamer assembly as a vascular endothelial growth factor receptor agonist. Nucleic Acid. Ther. 25 (5), 227–234. doi:10.1089/nat.2014.0519
Ren, X., Zhao, Y., Xue, F., Zheng, Y., Huang, H., Wang, W., et al. (2019). Exosomal DNA aptamer targeting α-synuclein aggregates reduced neuropathological deficits in a mouse Parkinson's disease model. Mol. Ther. - Nucleic Acids 17, 726–740. doi:10.1016/j.omtn.2019.07.008
Riccardi, C., Napolitano, E., Platella, C., Musumeci, D., Melone, M. A. B., and Montesarchio, D. (2021). Anti‐VEGF DNA‐based aptamers in cancer therapeutics and diagnostics. Med. Res. Rev. 41 (1), 464–506. doi:10.1002/med.21737
Roshanbinfar, K., Esser, T. U., and Engel, F. B. (2021). Stem cells and their cardiac derivatives for cardiac tissue engineering and regenerative medicine. Antioxidants Redox Signal. 35 (3), 143–162. doi:10.1089/ars.2020.8193
Roy, T., James, B. D., and Allen, J. B. (2021). Anti-VEGF-R2 aptamer and RGD peptide synergize in a bifunctional hydrogel for enhanced angiogenic potential. Macromol. Biosci. 21 (2), e2000337. doi:10.1002/mabi.202000337
Saito, S. (2021). SELEX-based DNA aptamer selection: A perspective from the advancement of separation techniques. Anal. Sci. 37 (1), 17–26. doi:10.2116/analsci.20sar18
Sarkar, A., Saha, S., Paul, A., Maji, A., Roy, P., and Maity, T. K. (2021). Understanding stem cells and its pivotal role in regenerative medicine. Life Sci. 273, 119270. doi:10.1016/j.lfs.2021.119270
Schäfer, R., Wiskirchen, J., Guo, K., Neumann, B., Kehlbach, R., Pintaske, J., et al. (2007). Aptamer-based isolation and subsequent imaging of mesenchymal stem cells in ischemic myocard by magnetic resonance imaging. Fortschr Röntgenstr 179 (10), 1009–1015. doi:10.1055/s-2007-963409
Shahnawaz, M., Mukherjee, A., Pritzkow, S., Mendez, N., Rabadia, P., Liu, X., et al. (2020). Discriminating α-synuclein strains in Parkinson's disease and multiple system atrophy. Nature 578 (7794), 273–277. doi:10.1038/s41586-020-1984-7
Smestad, J., and Maher, L. R. (2013). Ion-dependent conformational switching by a DNA aptamer that induces remyelination in a mouse model of multiple sclerosis. Nucleic Acids Res. 41 (2), 1329–1342. doi:10.1093/nar/gks1093
Son, J., Kim, J., Lee, K., Hwang, J., Choi, Y., Seo, Y., et al. (2019). DNA aptamer immobilized hydroxyapatite for enhancing angiogenesis and bone regeneration. Acta Biomater. 99, 469–478. doi:10.1016/j.actbio.2019.08.047
Strahm, Y., Flueckiger, A., Billinger, M., Meier, P., Mettler, D., Weisser, S., et al. (2010). Endothelial-cell-binding aptamer for coating of intracoronary stents. J. Invasive Cardiol. 22 (10), 481–487.
Sun, T., Meng, C., Ding, Q., Yu, K., Zhang, X., Zhang, W., et al. (2021). In situ bone regeneration with sequential delivery of aptamer and BMP2 from an ECM-based scaffold fabricated by cryogenic free-form extrusion. Bioact. Mater. 6 (11), 4163–4175. doi:10.1016/j.bioactmat.2021.04.013
Tang, W. (2019). Challenges and advances in stem cell therapy. Bst 13 (4), 286. doi:10.5582/bst.2019.01241
Tseng, W. Y., Huang, Y. S., Lin, H. H., Luo, S.-F., McCann, F., McNamee, K., et al. (2018). TNFR signalling and its clinical implications. Cytokine 101, 19–25. doi:10.1016/j.cyto.2016.08.027
Ueki, R., Atsuta, S., Ueki, A., Hoshiyama, J., Li, J., Hayashi, Y., et al. (2019). DNA aptamer assemblies as fibroblast growth factor mimics and their application in stem cell culture. Chem. Commun. 55 (18), 2672–2675. doi:10.1039/c8cc08080a
Ueki, R., Uchida, S., Kanda, N., Yamada, N., Ueki, A., Akiyama, M., et al. (2020). A chemically unmodified agonistic DNA with growth factor functionality for in vivo therapeutic application. Sci. Adv. 6 (14), eaay2801. doi:10.1126/sciadv.aay2801
Ueki, R., Ueki, A., Kanda, N., and Sando, S. (2016). Oligonucleotide‐based mimetics of hepatocyte growth factor. Angew. Chem. Int. Ed. 55 (2), 579–582. doi:10.1002/anie.201508572
Valera, E., and Masliah, E. (2016). Combination therapies: The next logical Step for the treatment of synucleinopathies? Mov. Disord. 31 (2), 225–234. doi:10.1002/mds.26428
Wang, M., Wu, H., Li, Q., Yang, Y., Che, F., Wang, G., et al. (2019). Novel aptamer-functionalized nanoparticles enhances bone defect repair by improving stem cell recruitment. Ijn 14, 8707–8724. doi:10.2147/ijn.s223164
Wang, X., Song, X., Li, T., Chen, J., Cheng, G., Yang, L., et al. (2019). Aptamer-functionalized bioscaffold enhances cartilage repair by improving stem cell recruitment in osteochondral defects of rabbit knees. Am. J. Sports Med. 47 (10), 2316–2326. doi:10.1177/0363546519856355
Wang, X., Zheng, X., Duan, Y., Ma, L., and Gao, C. (2019). Defined substrate by aptamer modification with the balanced properties of selective capture and stemness maintenance of mesenchymal stem cells. ACS Appl. Mat. Interfaces 11 (16), 15170–15180. doi:10.1021/acsami.9b03333
Wang, Y., Khaing, Z. Z., Li, N., Hall, B., Schmidt, C. E., and Ellington, A. D. (2010). Aptamer antagonists of myelin-derived inhibitors promote axon growth. PLoS One 5 (3), e9726. doi:10.1371/journal.pone.0009726
Wilbanks, B., Smestad, J., Heider, R. M., Warrington, A. E., Rodriguez, M., and Maher, L. J. (2019). Optimization of a 40-mer antimyelin DNA aptamer identifies a 20-mer with enhanced properties for potential multiple sclerosis therapy. Nucleic Acid. Ther. 29 (3), 126–135. doi:10.1089/nat.2018.0776
Wood, A., Vassallo, A., Summers, C., Chilvers, E. R., and Conway-Morris, A. (2018). C5a anaphylatoxin and its role in critical illness-induced organ dysfunction. Eur. J. Clin. Invest. 48 (12), e13028. doi:10.1111/eci.13028
Wu, J., Liyarita, B. R., Zhu, H., Liu, M., Hu, X., and Shao, F. (2021). Self-assembly of dendritic DNA into a hydrogel: Application in three-dimensional cell culture. ACS Appl. Mat. Interfaces 13 (42), 49705–49712. doi:10.1021/acsami.1c14445
Yamanaka, S. (2020). Pluripotent stem cell-based cell therapy-promise and challenges. Cell Stem Cell 27 (4), 523–531. doi:10.1016/j.stem.2020.09.014
Yang, M., Chen, X., Zhu, L., Lin, S., Li, C., Li, X., et al. (2021). Aptamer-functionalized DNA-silver nanocluster nanofilm for visual detection and elimination of bacteria. ACS Appl. Mat. Interfaces 13 (32), 38647–38655. doi:10.1021/acsami.1c05751
Yang, Z., Zhao, T., Gao, C., Cao, F., Li, H., Liao, Z., et al. (2021). 3D-Bioprinted difunctional scaffold for in situ cartilage regeneration based on aptamer-directed cell recruitment and growth factor-enhanced cell chondrogenesis. ACS Appl. Mat. Interfaces 13 (20), 23369–23383. doi:10.1021/acsami.1c01844
Yao, C., Tang, H., Wu, W., Tang, J., Guo, W., Luo, D., et al. (2020). Double rolling circle amplification generates physically cross-linked DNA network for stem cell fishing. J. Am. Chem. Soc. 142 (7), 3422–3429. doi:10.1021/jacs.9b11001
Yatime, L., Maasch, C., Hoehlig, K., Klussmann, S., Andersen, G. R., and Vater, A. (2015). Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer. Nat. Commun. 6, 6481. doi:10.1038/ncomms7481
Yoon, J. W., Jang, I. H., Heo, S. C., Kwon, Y. W., Choi, E. J., Bae, K. H., et al. (2015). Isolation of foreign material-free endothelial progenitor cells using CD31 aptamer and therapeutic application for ischemic injury. PLoS One 10 (7), e0131785. doi:10.1371/journal.pone.0131785
Zhang, N., Wang, Z., and Zhao, Y. (2020). Selective inhibition of Tumor necrosis factor receptor-1 (TNFR1) for the treatment of autoimmune diseases. Cytokine & Growth Factor Rev. 55, 80–85. doi:10.1016/j.cytogfr.2020.03.002
Zhang, T., Tian, T., and Lin, Y. (2021). Functionalizing framework nucleic-acid-based nanostructures for biomedical application[J]. Adv. Mater 2021, e2107820.
Zhang, X., Hu, J., Becker, K. V., Engle, J. W., Ni, D., Cai, W., et al. (2021). Antioxidant and C5a-blocking strategy for hepatic ischemia-reperfusion injury repair. J. Nanobiotechnol 19 (1), 107. doi:10.1186/s12951-021-00858-9
Zhang, X., Liu, N., Zhou, M., Li, S., and Cai, X. (2021). The application of tetrahedral framework nucleic acids as a drug carrier in biomedicine fields. Cscr 16 (1), 48–56. doi:10.2174/1574888x15666200422103415
Zhang, Z., Pandey, R., Li, J., Gu, J., White, D., Stacey, H. D., et al. (2021). High‐affinity dimeric aptamers enable the rapid electrochemical detection of wild‐type and B.1.1.7 SARS‐CoV‐2 in unprocessed saliva. Angew. Chem. Intl Ed. 60 (45), 24266–24274. doi:10.1002/anie.202110819
Zhao, N., Coyne, J., Xu, M., Zhang, X., Suzuki, A., Shi, P., et al. (2019). Assembly of bifunctional aptamer-fibrinogen macromer for VEGF delivery and skin wound healing. Chem. Mat. 31 (3), 1006–1015. doi:10.1021/acs.chemmater.8b04486
Keywords: aptamer, tissue regeneration, regenerative medicine, repair of tissue injury, transformation and application
Citation: Luo Z, Chen S, Zhou J, Wang C, Li K, Liu J, Tang Y and Wang L (2022) Application of aptamers in regenerative medicine. Front. Bioeng. Biotechnol. 10:976960. doi: 10.3389/fbioe.2022.976960
Received: 23 June 2022; Accepted: 08 August 2022;
Published: 29 August 2022.
Edited by:
V. Prasad Shastri, University of Freiburg, GermanyReviewed by:
Xiaoxiao Cai, Sichuan University, ChinaSiu Hong Dexter Wong, Hong Kong Polytechnic University, Hong Kong SAR, China
Copyright © 2022 Luo, Chen, Zhou, Wang, Li, Liu, Tang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Li, bGs1MTY0MzM0MTVAc211LmVkdS5jbg==; Jia Liu, bGl1amlhMDExMUBsaXZlLmNu; Yujin Tang, dGFuZ3l1amluMTk2N0AxNjMuY29t
†These authors have contributed equally to this work
 Zhaohui Luo1,2†
Zhaohui Luo1,2† Chong Wang
Chong Wang Jia Liu
Jia Liu Yujin Tang
Yujin Tang Liqiang Wang
Liqiang Wang