
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 01 November 2022
Sec. Bioprocess Engineering
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.964396
Unspecific peroxygenases (UPOs, EC 1.11.2.1) are fungal enzymes that catalyze the oxyfunctionalization of non-activated hydrocarbons, making them valuable biocatalysts. Despite the increasing interest in UPOs that has led to the identification of thousands of putative UPO genes, only a few of these have been successfully expressed and characterized. There is currently no universal expression system in place to explore their full potential. Cell-free protein synthesis has proven to be a sophisticated technique for the synthesis of difficult-to-express proteins. In this work, we aimed to establish an insect-based cell-free protein synthesis (CFPS) platform to produce UPOs. CFPS relies on translationally active cell lysates rather than living cells. The system parameters can thus be directly manipulated without having to account for cell viability, thereby making it highly adaptable. The insect-based lysate contains translocationally active, ER-derived vesicles, called microsomes. These microsomes have been shown to allow efficient translocation of proteins into their lumen, promoting post-translational modifications such as disulfide bridge formation and N-glycosylations. In this study the ability of a redox optimized, vesicle-based, eukaryotic CFPS system to synthesize functional UPOs was explored. The influence of different reaction parameters as well as the influence of translocation on enzyme activity was evaluated for a short UPO from Marasmius rotula and a long UPO from Agrocybe aegerita. The capability of the CFPS system described here was demonstrated by the successful synthesis of a novel UPO from Podospora anserina, thus qualifying CFPS as a promising tool for the identification and evaluation of novel UPOs and variants thereof.
Unspecific peroxygenases (UPO, EC 1.11.2.1) are a recently discovered group of heme-containing oxidoreductases. Initially identified as a haloperoxidase in 2004, AaeUPO from Agrocybe aegerita (syn. Cyclocybe aegerita) was the first UPO described. It is a secreted glycoprotein with oxidizing activity towards several substrates (Ullrich et al., 2004). In the following years, UPOs were found to be secreted by other fungi as well (Anh et al., 2007; Gröbe et al., 2011), and further characterization revealed a broad catalytic spectrum with respect to both the substrate range and the numerous catalyzed reactions. Their catalytic repertoire includes one-electron oxidations (as for a classical peroxidase) as well as the activation of C-H (Peter et al., 2011; Babot et al., 2013) and C=C bonds (Peter et al., 2013), the cleavage of ethers (Kinne et al., 2009), and the oxygenation of heteroatoms (Ullrich 2009). The substrates of UPOs ranges from small aromatic (Kluge et al., 2009) or aliphatic (Babot et al., 2013) molecules to more sterically demanding substrates such as steroids (Babot et al., 2015). An overview of their catalytic diversity is given by Hofrichter et al. (Hofrichter et al., 2022). Because of this versatility, UPOs are promising candidates for the development of novel biosynthetic pathways. In this context, UPOs were successfully used to produce isophorone derivatives that are important precursors for the synthesis of pharmaceuticals (Aranda et al., 2019) and generate building blocks from renewable sources such as 5-hydroxymethylfurfural for the polymer industry (Carro et al., 2014). The ability of UPOs to generate pharmacologically relevant substrates is also of particular interest. The conversion of the antiplatelet drug clopidogrel to its active metabolite by different UPOs was also recently demonstrated (Kiebist et al., 2021).
Despite their vast capabilities, the full potential of UPOs remains untapped partly because of their limited availability. Through the extensive screening of genomic data, over 4,000 putative UPO genes have been identified and these can be roughly divided into long and short UPOs (Kinner et al., 2021). However, until today, only a little over 20 different UPOs have been expressed in an active form (Hofrichter et al., 2022). Two main strategies have been applied: homologous expression in the wild-type host and heterologous expression, mostly in Pichia pastoris (syn. Komagataella phaffii). The cultivation of wild-type UPO producers sometimes results in large amounts of a single UPO as reported for Marasmius rotula UPO (MroUPO) (Gröbe et al., 2011) yielding 445 mg/L. In addition to the time-consuming cultivation, a major drawback of producing UPOs in their original host is often the poor accessibility of the host to genetic engineering, which limits the production of UPOs to the unmodified wild-type enzyme. However, genetically engineered UPOs can outperform their wild-type equivalent in terms of secretion levels, catalytic activity (Molina-Espeja et al., 2014), substrate specificity (Gomez de Santos et al., 2019), and solvent stability (Martin-Diaz et al., 2021). In order to overcome these limitations, several classical production hosts have been examined for UPO expression. In principle, recombinant expression in E. coli is possible but typically leads to low amounts of active enzyme (Carro et al., 2019; Linde et al., 2020). The use of fungal production hosts such as Saccharomyces cerevisiae (Molina-Espeja et al., 2015) and Aspergillus oryzae (Babot et al., 2013) have proven to be more promising. In this context, P. pastoris has to be highlighted. In recent years, considerable progress has been made in establishing P. pastoris as a heterologous host for UPO production. The production of 735 g recombinant AaeUPO in a 2500 L fermentation was recently reported (Tonin et al., 2021). Several other UPOs, including newly characterized UPOs, were also produced in P. pastoris (Püllmann and Weissenborn, 2020; Bormann et al., 2021; Püllmann et al., 2021; Rotilio et al., 2021). However, not all UPOs can be easily produced in P. pastoris as recently demonstrated by Bormann et al. wherein only three out of 10 UPOs were expressed in an active form (Bormann et al., 2021). To improve UPO production in P. pastoris, extensive efforts were made to identify favorable promotors and signal peptides, which would allow an efficient UPO expression and secretion (Püllmann and Weissenborn 2020; Püllmann et al., 2021). Although these studies have shown promising results, including the successful expression of novel UPOs, there were still UPOs that could not be expressed in an active form such as GmaUPO and MweUPO (Püllmann et al., 2021), that can be expressed by and isolated from only its natural host Marasmius wettseinii (Ullrich et al., 2018). A universal platform for UPO production is thus yet to be shown.
A cell-free approach for the production of recombinant UPOs was recently published (Schramm et al., 2022). In a cell-free protein synthesis (CFPS), translationally active cell-lysate is used to carry out translation reactions. Because of its open nature, reaction conditions can be directly manipulated, thus making it a versatile tool for protein production (Gregorio et al., 2019). CFPS has proven valuable for the synthesis of numerous proteins (Gregorio et al., 2019; Dondapati et al., 2020), including heme-containing proteins such as horseradish peroxidase (HRP) (Zhu et al., 2015; Park and Kim, 2021) as well as bacterial cytochromes P450 (Kwon et al., 2013), cytochrome C oxidase (Katayama et al., 2010), and manganese peroxidase (Ninomiya et al., 2014). These examples all used E. coli lysate, which is by far the most commonly used CFPS system. E. coli-based CFPS is relatively inexpensive and scalable and has high productivity. However, it is only of limited use for the production of eukaryotic proteins for which post-translational modification (PTM) and correct folding might become a bottleneck. To address this problem, several eukaryotic CFPS systems, each having distinct features and thus qualifying them for special applications, were developed (Zemella et al., 2015). In this work, we focus on lysates containing endogenous microsomes. These microsomes are ER-derived vesicles that allow protein translocation and were reported for Chinese hamster ovary (CHO) (Brödel et al., 2015) and Sf21-based systems (Tarui et al., 2001). Microsomes enable posttranslational modifications such as N-glycosylations (Gurramkonda et al., 2018; Zemella et al., 2018), phosphorylation (Quast et al., 2016), lipidation (Shaklee et al., 2010), and signal peptide cleavage (Zemella et al., 2018). Furthermore, the microsomal lumen is much more favorable for the formation of disulfide bonds. This was shown by the example of antibody fragments synthesized in an insect-based CFPS reaction (Stech et al., 2014). Of those PTMs, N-glycosylation and disulfide bond formation are of particular interest when producing UPOs. Eukaryotic CFPS might thus prove itself a suitable platform for the synthesis of UPOs. This work aims to evaluate the capacity of the well-characterized Sf21 cell-free system to synthesize UPOs. The procedure for the cell-free protein production was shown for the model enzymes MroUPO and AaeUPO as representative for short and long UPOs, respectively. We also sought to demonstrate the capability of this cell-free system through the synthesis of the novel short PanUPO.
The templates for the Sf21-based synthesis of AaeUPO and MroUPO were manufactured by BioCat GmbH (Heidelberg, Germany). The construct design was based on the design reported earlier (Brödel et al., 2013b). The constructs were equipped with a T7 promotor. An internal ribosome entry site (IRES) from the Cricket-paralysis virus was used to enhance cap-independent translation initiation. The native signal peptide (and propeptide in case of AaeUPO) was removed, and constructs with and without melittin signal peptide were prepared. The mature proteins of AaeUPO and MroUPO were identified based on previous experimental data (Pecyna et al., 2009; Gröbe et al., 2011). PanUPO propeptide was identified based on homology. The last codon of the melittin signal peptide coding for the C-terminal Asp was omitted. pUC57-1.8 k was used as a plasmid backbone.
Sf21 lysate preparation and coupled CFPS have already been described and explained (Brödel et al., 2013b; Quast et al., 2015). Minor changes were made during lysate preparation. In all buffers used during the lysate preparation, DTT was omitted, and 0.05 mM GSH and 0.25 mM GSSG were added. Coupled transcription-translation reactions were performed in a batch mode format. Protein synthesis was performed in a thermomixer (Thermomixer comfort, Eppendorf, Hamburg, Germany) at 21°C (if not stated otherwise) and with gentle shaking at 500 rpm for 3 h. Reaction volumes of 50 μL were composed of 40% (v/v) Sf21 cell lysate, 100 µM of each canonical amino acid, nucleoside triphosphates (1.75 mM ATP, 0.30 mM CTP, 0.30 mM GTP, and 0.30 mM UTP), 100 ng/μL vector DNA, and 1 U/µL T7 RNA-polymerase (Agilent, Waldbronn, Germany), 0–75 µM porcine hemin (Alfa Aesar, Haverhill, USA) dissolved in NaOH. To monitor protein quality and quantity, reaction mixtures were supplemented with 14C-labeled leucine (Perkin Elmer, Inc.; Baesweiler, Germany). No template controls (NTC) were prepared in the same way as the samples except that the DNA template was replaced by water.
After incubation, the translation mix (TM) was centrifuged at 16,000 × g and 4°C for 10 min. The resulting supernatant (SN1) was collected, and the microsome harboring pellet was resuspended in phosphate-buffered saline (PBS) containing 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS). The resuspended pellet was incubated at 21°C and 750 rpm for 45 min in order to release translocated proteins out of the microsomes. The samples were then centrifuged again, and the supernatant (SN2) was collected.
Total protein yields of cell-free synthesized, 14C-labeled protein were determined by hot trichloroacetic acid (TCA) precipitation and subsequent liquid scintillation counting (Stech et al., 2012). In short, 3 µl of the fraction analyzed were mixed with 3 ml 10% TCA (Carl Roth GmbH & Co. KG; Karlsruhe, Germany) with 2% casein hydrolysate (Carl Roth GmbH & Co. KG; Karlsruhe, Germany) and incubated in a water bath at 80°C for 15 min followed by 30 min incubation on ice. The TCA solution was then transferred to membrane filters (VWR International GmbH, Darmstadt, Germany) using a vacuum filtration device (Hoefer, Inc., Holliston, USA). The filters were washed with 5% TCA and dried with acetone. The dry filters were transferred into scintillation vials (Sarstedt AG & Co KG) and incubated in 3 ml scintillation cocktail for 1 h. The samples were then measured using the Hidex 600 SL (Hidex; Turku, Finland). Protein yields were calculated based on the counted disintegrations per minute, the specific activity of 14C-leucine in the cell-free reaction, the molecular weight, and the number of leucines in the protein.
For qualitative analysis, 3 µl of the fraction analyzed was mixed with 47 µl water and precipitated in 150 µl cold acetone (Carl Roth GmbH & Co. KG; Karlsruhe, Germany) for at least 15 min. After centrifugation at 16,000 × g at 4°C for 10 min, the supernatant was removed, and the precipitate dried for 30 min at 45°C. The precipitate was resuspended in NuPage™ loading buffer (Fisher Scientific GmbH, Schwerte, Germany). Samples were incubated at 70°C for 10 min. Samples were loaded onto NuPage™ 10% Bis-Tris Gels (Fisher Scientific GmbH, Schwerte, Germany). The SeeBlue Pre-Stained marker (Fisher Scientific GmbH, Schwerte, Germany) was used as standard. Gels were run at 185 V for 35 min and then dried at 70°C (Unigeldryer 3545D, Uniequip, Planegg, Germany). The dry gels were placed on a phosphor screen (GE-Healthcare GmbH; Munich, Germany). After 1–2 days, the phosphor screen was scanned using an Amersham Typhoon RGB (GE-Healthcare GmbH; Munich, Germany) to visualize labeled proteins.
Protein N-glycosylation was investigated using PNGaseF (peptide N-glycosidase F, New England Biolabs GmbH, Frankfurt am Main, Germany) according to the manufacturer’s protocol. Proteins were synthesized in a cell-free manner in the presence of 14C-leucine. 3 µl of the protein sample were then used for the deglycosylation assay and precipitated in acetone for SDS-PAGE analysis as described above.
The activity of UPO was analyzed through the conversion of 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) (Merck KGaA, Darmstadt, Germany) to the ABTS radical cation (ε418 = 36,000 mM−1 cm−1), two molecules of 2,6-dimethoxyphenol (DMP) (Merck KGaA, Darmstadt, Germany) to coerulignone (ε469 = 49,600 mM−1 cm−1) and 5-nitro-1,3-benzodioxole (NBD) (Merck KGaA, Darmstadt, Darmstadt, Germany) to 4-nitrocatechol (ε425 = 9,700 mM−1 cm−1). For the activity assay, 5 μL–20 µL of sample were added to 85 μL–70 µL of assay buffer. The assay buffer compositions were as follows: ABTS-Assay: McIlvaine buffer pH 4.5, 0.3 mM ABTS, 10 mM GSSG; DMP-Assay: 0.1 M potassium phosphate buffer pH 6 (MroUPO) or pH 7 (AaeUPO), 6 mM DMP; NBD-Assay: McIlvaine buffer pH 7, 0.5 mM NBD. The reactions were started by adding 10 µL of 0.03% H2O2. Extinction measurements were performed using a microplate reader (Mithras2 LB 943 Multimode Plate Reade, Berthold Technologies GmbH & Co. KG).
The UPO activities towards the pharmaceuticals propranolol, diclofenac, clopidogrel, and desipramine were analyzed by LC-MS. For this purpose, 10 µl of UPO sample were incubated with a reaction mixture containing 1 mM substrate, 1 mM hydrogen peroxide, 5 mM ascorbate, and 20 mM potassium phosphate buffer (pH 7) at 25°C and 800 rpm on a thermal shaker (TurboShaker 3,500, Scienova GmbH, Jena, Germany) for 5 min and stopped by adding 100 µl of cold acetonitrile (−20°C). The samples were centrifuged at 14,000 × g for 10 min, and the supernatants were analyzed by LC-MS. Calibrations were performed with authentic standards 4-hydroxypropranolol, 4′-hydroxydiclofenac, 2-oxoclopidogrel, and 2-hydroxydesipramine.
Chromatographic separations in LC-MS experiments were performed on a Thermo Scientific Vanquish Flex Quaternary UHPLC system (Fisher Scientific GmbH, Waltham, MA, USA) using a Kinetex® column (C18, 2.6 µm, 100 Å, 150 × 2.1 mm, Phenomenex, Torrance, CA, USA). The injection volume was 1 μL, and the column was eluted at a flow rate of 0.5 ml min−1 and 40°C with two mobile phases: A (diH2O, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid) and the following gradient: 0 min, 10% B; 0.5 min, 10% B; 5 min, 80% B; 7 min, 80% B; 7.1 min, 10% B; 10 min, 10% B. MS spectra were obtained using the Thermo Scientific Q Exactive Focus quadrupole-Orbitrap mass spectrometer (Thermo Electron, Waltham, MA, USA) coupled with a heated electrospray ionization source in positive mode. The tune operating parameters were as follows: rate of sheath gas flow and auxiliary gas flow—40 and 15 (arbitrary units), respectively; spray voltage—4.0 kV; the temperature of capillary and sample heater—260°C and 400°C, respectively; high-resolution MS was operated at full scan positive mode with a mass range of m/z 150–1,500 at a resolution of 70,000 (m/z 200).
In order to obtain a universally applicable platform for UPO production, said platform must support glycosylation and disulfide bond formation. A Sf21-based lysate is thus used for the cell-free synthesis because of its high translocation and glycosylation efficiency (Batista et al., 2021). To achieve efficient translocation, the sequences coding for the native signal peptide and following residues that are processed during the maturation of AaeUPO and MroUPO were replaced by the melittin signal peptide, which was shown to enhance translocation as well as translation efficiency in Sf21 and CHO CFPS (Brödel et al., 2013b; Brödel et al., 2014). The melittin signal peptide was modified by removing the C-terminal Asp, which is known to remain at the N-terminus of the processed target protein after signal peptide cleavage. The lysate was also supplemented with glutathione and glutathione-disulfide (GSH/GSSG) during lysate preparation in order to promote the formation of disulfide bonds. The translation mix (TM) was supplemented with 14C-leucine, thereby enabling visualization through autoradiography after SDS-PAGE as well as protein quantification. After synthesis, the TM was fractionated by centrifugation, resulting in supernatant 1 (SN1), which contains all soluble components of the cell-free reaction, and the pelleted microsomal fraction (MF), which contains the microsomes including the translocated protein. The microsomes were then treated with 0.5% CHAPS in order to release translocated protein from the microsomal lumen. Soluble proteins (SN2) were separated from any remaining insoluble debris by an additional centrifugation step. Using the redox optimized lysate, AaeUPO and MroUPO were successfully synthesized (Figure 1A,B). For cell-free AaeUPO (cfAaeUPO), a band of about 38 kDa and for cfMroUPO, a band of about 28 kDa was detected. This is well in accordance with the theoretical masses of AaeUPO and MroUPO (38.4 kDa and 28 kDa, respectively). Also weaker bands of higher mass were observed for both UPOs in the TM, MF, and SN2; these bands are no longer present after PNGase F digestion, thereby indicating glycosylation. In the fractions TM, MF, and SN2 of cfMroUPO a pronounced band of approx. 62 kDa was detected; which was no longer visible under reducing conditions. The band most likely corresponds to the expected homodimer.
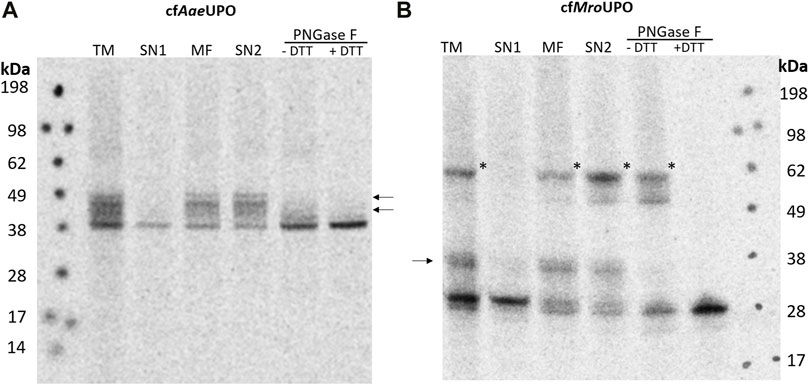
FIGURE 1. SDS-PAGE and autoradiography of cfAaeUPO and cfMroUPO. Samples of the translation mix (TM), supernatant 1 (SN1, containing non-translocated protein), the microsomal fraction (MF), and supernatant 2 (SN2, containing translocated protein recovered from the microsomes) were acetone precipitated, separated by SDS-PAGE, and analyzed through autoradiography. Two additional SN2 samples were collected and subjected to PNGase F digestion. The PNGase F digestion was performed under both native and denaturing/reducing conditions. All samples were treated with non-reducing denaturing sample buffer and heated before performing SDS-PAGE. The analysis was performed for cfAaeUPO (A) and cfMroUPO (B). N-glycosylation is indicated by arrows, a possible dimer band is labeled by asterisks.
One major advantage of CFPS is the open nature of the system, which allows the easy manipulation of reaction parameters and buffer conditions. Many general factors such as temperature and salt and lysate concentrations influence both protein yields and activity. Insect-based CFPS has a wide adjustment range, which was sufficiently covered in previous studies (Tarui et al., 2001; Groll et al., 2007; Brödel et al., 2013b), thereby providing a framework for further reaction design. In order to obtain active enzyme, the reaction temperature was reduced from 27°C to 21°C (Supplementary Figure S1). To our knowledge, no hemoprotein had been produced in an insect-based CFPS until now. In order to assess the influence of heme addition on protein synthesis, different hematin concentrations were applied. To determine the enzyme activity, the conversions of ABTS to its radical cation and DMP to coerulignone were analyzed spectrophotometrically. For both UPOs, no activity was observed when hematin was not supplemented to the reaction. The optimum for cfAaeUPO and cfMroUPO was 20 μM and 10 µM hematin, respectively (Figures 2A–C). Higher hematin concentrations decreased the UPO activity while increasing background activity. In order to account for background activity, a no-template-control (NTC) was analyzed for every sample. Data were corrected by the background activity. The autoradiograph of cfAaeUPO synthesized at different hematin concentrations showed no remarkable differences (Figure 2B). However, for cfMroUPO, the formation of dimers is drastically reduced when hematin is omitted (Figure 2D).
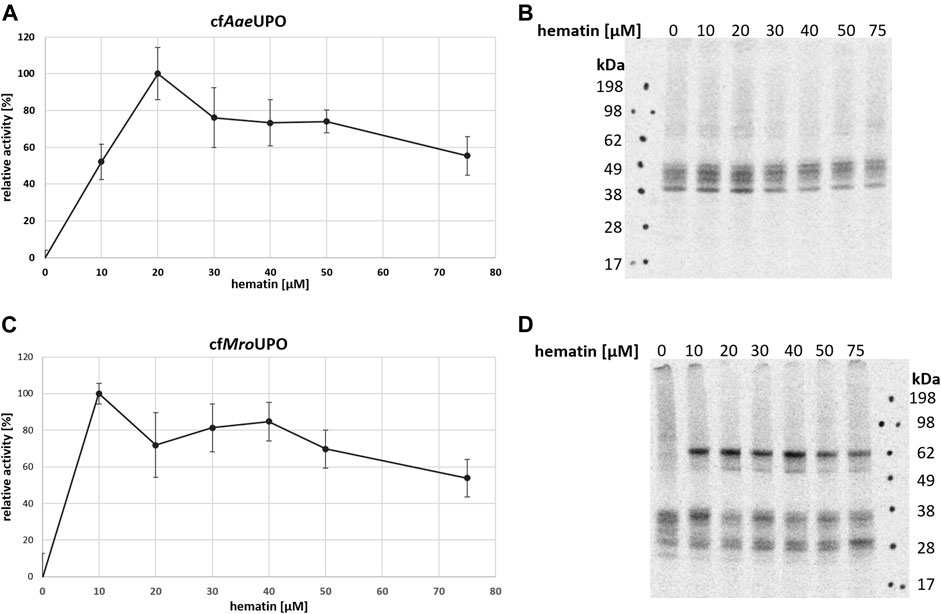
FIGURE 2. Influence of hematin supplementation on cfUPO activity. The activity of cfAaeUPO (A) and cfMroUPO (C) when synthesized in the presence of differing amounts of hematin was determined by ABTS-Assay (A) or DMP-assay (C). Measurements were performed in triplicate. For each sample, an NTC was analyzed and subtracted from the corresponding data points (B + D). SN2 of cfAaeUPO (B) and cfMroUPO (D) synthesized at different hematin concentrations was separated by SDS-PAGE and visualized through autoradiography.
In order to further elucidate the contribution of the melittin signal peptide (Mel) and protein translocation to the activity of cell-free synthesized UPO, additional UPO templates were designed. These constructs contain either the complete amino acid sequence of the unprocessed wild-type UPOs (including the native signal peptide and, if present, further residues that are cleaved during maturation) or only the mature UPO. In the case of cfAaeUPO, CFPS was successful only when the melittin signal peptide was used (Supplementary Figure S2). In contrast, all three cfMroUPO variants were successfully synthesized independently of any signal peptide. However, the cfMroUPO without signal peptide showed only a low translation efficiency. Using the native signal peptide led to an increase of synthesized cfMroUPO, that was located mainly in the SN1 and formed dimers with low efficiency. The introduction of a melittin signal peptide not only resulted in a higher overall protein yield but also led to translocated protein in the SN2 with glycosylation and dimerization as described above (Figure 3A).
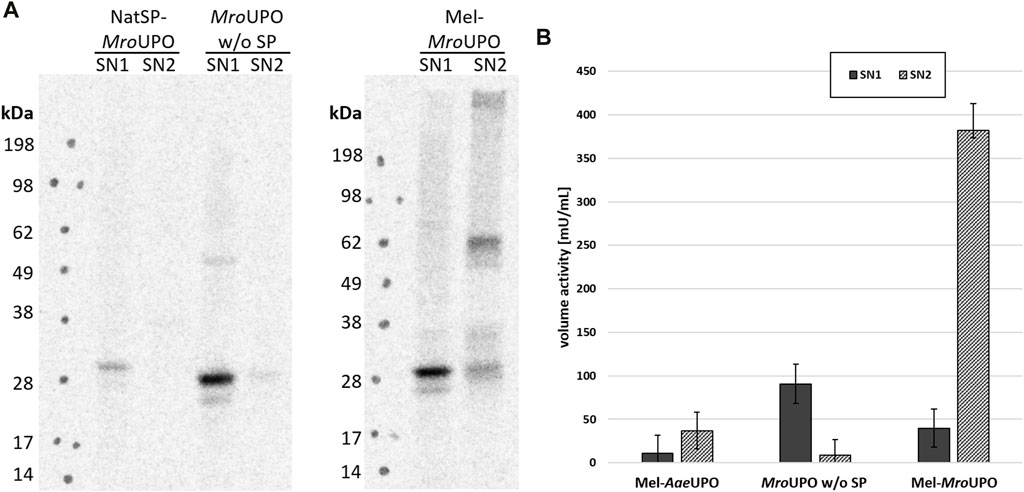
FIGURE 3. Influence of translocation on cfUPO activity. (A) Different MroUPO constructs either without signal peptide (MroUPO w/o SP), with native signal peptide (NatSP-MroUPO), or with melittin signal peptide (Mel-MroUPO) were synthesized, and supernatant 1 (SN1, containing non translocated protein) and supernatant 2 (SN2, containing translocated protein recovered from the microsomes) were analyzed by SDS-PAGE with subsequent autoradiography. (B) The conversion of DMP to coerulignone was analyzed spectrophotometrically for cfAaeUPO, cfMroUPO without SP, and Mel-MroUPO.
The activity of Mel-cfAaeUPO as well as of cfMroUPO with and without native signal peptide was analyzed by the conversion of DMP. By far, the highest volume activity of 382 mU/mL was observed for Mel-cfMroUPO in the SN2 compared with 40 mU/mL in the SN1. Omitting the signal peptide leads to increased activity in the SN1 (91 mU/mL). However, no activity was displayed in the SN2 without signal peptide. The volume activity of Mel-cfAaeUPO in the SN2 was 37 mU/mL while no activity was observed in the SN1 of Mel-cfAaeUPO. Because the translocation seems to play a pivotal role in terms of UPO activity, all further experiments were performed with the melittin constructs. Background activity detected in the NTC was determined to be 14 mU/mL and 22 mU/mL in the SN1 and SN2, respectively. In order to account for this background, for every sample an NTC was analyzed for every sample (Figure 3B).
ABTS was successfully converted by both UPOs obtained from SN2 (volume activities of 490 mU/mL and 29.5 mU/mL for cfAaeUPO and cfMroUPO, respectively (Supplementary Figure S3A), which corresponds to specific activities of 314 U/mg and 18.5 U/mg (Table 1). Applying the SN1 sample to the ABTS-assay buffer led to clouding regardless of the presence of UPO in the sample. Therefore, no colorimetric data was obtained for the SN1 by means of the ABTS assay.
The reaction products in the ABTS and DMP assay are the result of a one-electron oxidation, which is a classical peroxidase reaction. While peroxidase activity is expected from UPOs (Ullrich et al., 2004), their true potential lies in their peroxygenase activity. Therefore, cfAaeUPO and cfMroUPO were analyzed for the conversion of NBD to 4-nitrocatechol. The specific activities were 1.7 U/mg and 1.4 U/mg for cfAaeUPO and cfMroUPO, respectively. The volume activities were 4.7 mU/mL and 5.2 mU/mL for cfAaeUPO and cfMroUPO, respectively. This was more than 1.5-fold higher than the background activity of 2.8 mU/mL (Supplementary Figure S3B).
The conversion of chromogenic model substrates as shown above is a suitable tool for screening purposes. However, these substrates play a subordinate role when it comes to the application of UPOs. One of those applications is the biocatalytic synthesis of pharmaceuticals and their metabolites. In this work, we analyzed the conversion of four different pharmaceutically relevant substrates by LC-MS: β-adrenergic blocker propranolol to 5-hydroxypropanolol, anti-inflammatory diclofenac to 4′-hydroxydiclofenac, antithrombotic prodrug clopidogrel to 2-oxoclopidogrel, and antidepressant desipramine to its 2-hydroxy-derivate. cfAaeUPO showed volume activities in the mU/mL range for propranolol, diclofenac, and clopidogrel (Table 2). The conversion of desipramine by cfAaeUPO was also shown, albeit at much lower levels. In contrast, cfMroUPO samples exhibited a noticeably lower volume activity towards propranolol, diclofenac, and desipramine, whereas the volume activity towards clopidogrel exceeds that of cfAaeUPO by more than 2-fold.
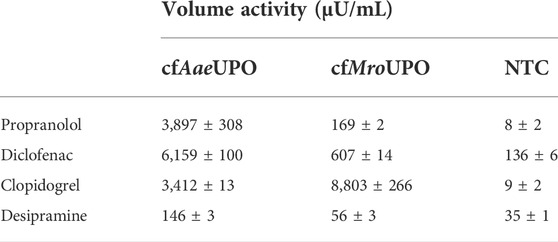
TABLE 2. Activity of cfAaeUPO and cfMroUPO for the conversion of different pharmaceutically relevant molecules.
We then sought to expand the scope of available UPOs with our new cell-free system. To do so, we prepared a template containing a putative UPO sequence from the filamentous ascomycete Podospora anserina. As for the other UPOs, the signal peptide was removed and exchanged for a modified melittin signal peptide as described above. The SDS-PAGE with subsequent autoradiography showed a band of about 28 kDa in all fractions; this is well in accordance with the theoretical size of 29.9 kDa (Figure 4A). Additional bands above the main band were observed in the TM fraction. In SN2, only a faint smear was present; which becomes visible as a distinct band after PNGase F digestion, thereby indicating heterogenic glycosylation. Subjecting SN1 and SN2 of cfPanUPO to the DMP assay revealed activity of 27.4 mU/mL in SN2 (Figure 4B). The PanUPOs conversion of the four pharmaceutical molecules investigated for AaeUPO and MroUPO was investigated through LC-MS. Considerable activity was observed only for clopidogrel (Table 3).
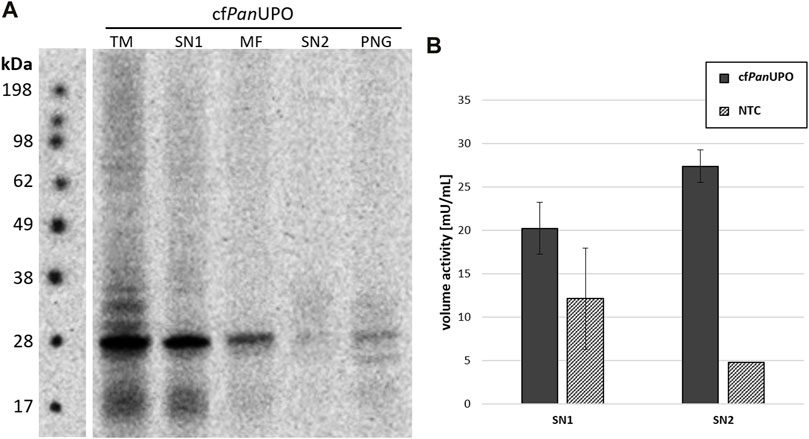
FIGURE 4. Cell-free synthesis of novel PanUPO. (A) SDS-PAGE and autoradiography of cfPanUPO. Samples of the translation mix (TM), supernatant 1 (SN1, containing non-translocated protein), the microsomal fraction (MF), and supernatant 2 (SN2, containing translocated protein recovered from the microsomes) were acetone precipitated, separated by SDS-PAGE, and analyzed by autoradiography. An additional SN2 sample was subjected to a native PNGase F digestion (PNG). All samples were treated with non-reducing denaturing sample buffer and heated prior to SDS-PAGE (B) DMP conversion of cfPanUPO from SN1 and SN2 compared with the background activity in those fractions.
Because of their broad catalytic spectrum, UPOs are highly attractive biocatalysts, especially for the generation of molecules of pharmaceutical relevance. For example, MroUPO is capable of cleaving corticosteroid side chains and can thus provide intermediates for the industrial synthesis of steroids (Ullrich et al., 2018). Another example would be the conversion of the anticancer drug cyclophosphamide using UPOs in order to circumvent the complex and inefficient chemical synthesis of its drug metabolite. The efficiency of the conversion of cyclophosphamide was dependent on the UPO that was used (Steinbrecht et al., 2020). Substantial differences in the catalytic activity of different UPOs were also reported for substrates like long chain terminal alkenes (Babot et al., 2022) and isophorone derivatives (Aranda et al., 2019). This underlines the need for a comprehensive UPO toolbox to take advantage of the full catalytic potential of UPOs. In order to expand this toolbox, we sought to establish a vesicle-based CFPS approach for the synthesis of long and short UPOs. We first aimed to produce the well-characterized UPOs AaeUPO and MroUPO because the data available on the activity of these UPOs would facilitate the evaluation of the performance of the Sf21-based cell-free system. In this work, we have shown that both AaeUPO and MroUPO were successfully synthesized in an Sf21-based cell-free reaction. For both UPOs, glycosylation was confirmed through PNGase F digestion. As expected, glycosylation was not observed in SN1 because it requires translocation into the microsomes. A downshift in the protein size of cfMroUPO from SN1 to the MF indicates the successful cleavage of the melittin signal peptide during the translocation. A similar slight downshift can also be observed for AaeUPO; however, this shift is much more subtle because of the resolution of the SDS-PAGE at higher molecular weights. Because the N-terminus of AaeUPO is rather sensitive and requires accurate processing (Püllmann et al., 2021), its activity supports the assumption of a correctly cleaved signal peptide. Both N-glycosylation and signal peptide cleavage has been reported for Sf21-based CFPS (Zemella et al., 2018). UPOs are N-glycosylated proteins, and studies on AaeUPO showed that their glycosylations are of the high mannose type (Pecyna et al., 2009; Piontek et al., 2010; Piontek et al., 2013). High mannose type glycosylation can also be expected for Sf21 CFPS (Zemella et al., 2018); however, the glycan structure might be less complex. The presence of multiple bands or smears of glycosylated protein indicates non-homogenous glycosylation, whereby not all glycosylation sites are addressed and/or are differently glycosylated (Zemella et al., 2018). Different glycoforms of AaeUPO and MroUPO are also observed when they are expressed in their wild-type host; however, the differences in glycosylation appear to be more subtle (Ullrich et al., 2009; Gröbe et al., 2011). In addition to the glycosylation, cfMroUPO displayed another band at about 62 kDa, which indicates dimer formation. This band was no longer detectable after applying reducing conditions during the deglycosylation assay. This is in accordance with the reported 3D structure of MroUPO (PDB 5FUJ), which forms dimers through intermolecular disulfide bridges. Although these results indicate dimerization through disulfide bonds, a definite proof (e.g., through mass spectrometry) is necessary. These potential dimers were predominantly observed in the SN2; this is not surprising because disulfide bonds are not expected to form in a cytosolic environment like the SN1. However, the possible presence of dimers in the SN1 could be a result of the redox optimization that was performed on the lysate used. Previous studies applied redox optimizations with varying results. While the redox supplementation of an insect-based cell-free reaction barely increased the activity of antibody fragments (Stech et al., 2014), the addition of not only GSH/GSSG but also protein disulfide isomerase to an insect-based cell-free reaction resulted in disulfide bridged alkaline phosphatase and human lysozyme (Ezure et al., 2007). A third approach for redox-optimization was utilized in this work; GSH/GSSG were added during lysate preparation, thereby resulting in a ready-to-use redox-optimized CFPS system that requires no further supplementation. A similarly optimized lysate was shown to promote the activity of ice structuring proteins (Brödel et al., 2013a); however, the reaction mix was also supplemented with GSH/GSSG in this case.
To the best of our knowledge, this is the first time a hemoprotein was synthesized in an insect-based CFPS reaction. We thus analyzed the effect of hematin on the cell-free synthesis and UPO activity. For that, two different assays (ABTS and DMP) were chosen to maximize the readout because both UPOs have differing activities towards these substrates (Ullrich et al., 2004; Gröbe et al., 2011). For both UPOs, no activity was observed without hematin supplementation, thereby indicating that no or at least an insufficient amount of heme is present in the unsupplemented lysate. Without the addition of hematin, no dimer formation of MroUPO was observed, thus indicating insufficient folding in the absence of heme. Increasing UPO activity was observed with rising hematin concentrations up to 10 μM–20 µM. No remarkable effect of the hematin or its solvent on the protein amount in SN2 was observed. For the synthesis of other hemoproteins in E. coli lysate, where heme was supplemented as hemin, an optimal hemin concentration in the range of 0.15 µM (Katayama et al., 2010) to 16.7 µM (Zhu et al., 2015) was reported. As an alternative to heme supplementation, the heme can be generated in situ from glucose and glycine in the presence of 5-aminolevulinic acid synthase as shown for bacterial CYP (Kwon et al., 2013) and HRP (Park and Kim, 2021). However, because hemin is inexpensive and seems to have no major adverse effects towards the CFPS itself, there currently is no need for following that approach in the Sf21-based CFPS system.
The contribution of the signal peptide and translocation to the UPO activity was then assessed. The introduction of the melittin signal peptide had a drastic effect on translation. Whereas no cfAaeUPO was synthesized without Mel, the cfMroUPO was synthesized independently of the use of the melittin signal peptide. However, the melittin signal peptide increased overall yields of cfMroUPO. This is well in accordance with studies that found a similar positive effect of melittin on translation efficiency (Brödel et al., 2013a; Stech et al., 2014). The conversion of DMP by cfMroUPO with and without melittin signal sequence was analyzed. cfMroUPO without melittin signal sequence showed activity only in the SN1. The activity observed was even higher than for Mel-cfMroUPO even though Mel-cfMroUPO had similar protein yields in SN1. This reduced activity results from the uncleaved signal peptide at the N-terminus, which is close to the active center of the protein. Deviations from the native N-terminus of the mature UPO were shown to have a negative effect on activity (Püllmann et al., 2021). The SN2 of Mel-cfMroUPO showed the highest activity by far. This underlines the importance of translocation for protein activity. The effect of the translocation on the enzyme activity can presumably be traced back to three factors: the formation of disulfide bonds, N-glycosylation, and protein folding in general. While analysis of the latter is difficult, the contribution of the other factors is more easily to assess. So far, there have been no studies of those factors on the activity of cell-free synthesized UPO. However, the effect of PTMs varies from UPO to UPO. For example, not all UPOs form disulfide bonds (Hofrichter et al., 2015), while glycosylations are present in all known UPOs; however, their impact on protein activity differs greatly depending on the individual UPO. AaeUPO was barely affected by deglycosylation, whereas MroUPO showed considerably lower activity when deglycosylated. Some UPOs activity is completely abolished upon deglycosylation (Püllmann et al., 2021). When further evaluating the contribution of PTMs to the activity of cfUPO, these factors have to be considered.
As expected, cfMroUPO exhibits a higher activity towards DMP than AaeUPO (Ullrich et al., 2004; Gröbe et al., 2011). For the substrate ABTS, the specific activity is in the range of wild-type AaeUPO (wtAaeUPO) with 295.7 U/mg (Ullrich et al., 2004), while the expression of an AaeUPO variant with an optimized signal peptide for P. pastoris expression achieved a specific activity of 233 U/mg (Molina-Espeja et al., 2014). When cfUPOs were compared with conventional prepared UPOs, the activity assays for the cfUPOs were performed using UPO directly after fractionation without any further purification. The cell-free synthesis of AaeUPO in a fungal lysate was recently shown (Schramm et al., 2022). The comparison shows that the activities of the cfAaeUPOs from both systems are in a comparable range. However, some different trends must be highlighted. While we achieved a higher volume activity of Sf21 cfAaeUPO towards ABTS, the cfAaeUPO synthesized in a fungal system shows a higher activity towards NBD. The conversion of pharmaceutical molecules also shows divergent trends. The fungal cfAaeUPO showed by far the highest activity towards propranolol; this was even higher than the wtAaeUPO. However, the Sf21 cfAaeUPO showed the highest activity towards diclofenac followed by propranolol and clopidogrel with about half the volume activity. These different catalytic profiles are surprising considering that they are both variants of the same UPO. Possible explanations include the presence of a His-tag at the C-terminus of the cfAaeUPO produced in fungal lysate, or differences in the glycosylation pattern and protein folding because of the presence of chaperones. In addition, buffer conditions such as salt concentrations and reaction components could affect UPO activity, especially in the cell-free approaches in which unpurified UPO was used for activity studies. Further investigation will be necessary to investigate the influence of these different parameters on catalytic activity. Taking advantage of these factors could provide a valuable approach to fine-tuning UPO activity. The data obtained analyzing the activity of both UPOs showed that the detection of UPO activity is tightly linked to the substrate applied. This underlines, that in pursuit of novel UPOs not only a versatile synthesis/expression platform but also a versatile analytical approach for testing different substrates is necessary. A promising step in this direction is the MISER (multiple injections in a single experimental run) approach (Knorrscheidt et al., 2020; Knorrscheidt et al., 2021), which allowed the analysis of multiple substrates and products at the same time in order to identify improved variants of the recently described Myceliophthora thermophila UPO (MthUPO) (Knorrscheidt et al., 2021). For the successful analysis of a high amount of UPOs, production strategy and analysis have to be well matched. As an easy-to-handle and versatile platform, CFPS is highly compatible and can support high-throughput approaches.
The true potential of CFPS lies in its flexibility. Yield-wise, CFPS will hardly compete with conventional expression strategies on an industrial scale. Yields reported for MroUPO produced in its wild-type host reach up to about 450 mg/L (Gröbe et al., 2011). While the expression of AaeUPO in its wild-type host yields only 10–30 mg/L (Ullrich et al., 2004; Pecyna et al., 2009), the recombinant expression of an AaeUPO variant, improved for secretion, yields over 200 mg/L (Molina-Espeja et al., 2015) with a recently reported pilot-scale production approach yielding 290 mg/L in a 2500 L fermentation (Tonin et al., 2021). The CFPS reaction shown here produced 1–2 mg/L of cfUPO. However, the scale of CFPS was much smaller. Reaction volumes typically are below 1 ml. Although 100 L CFPS reactions have been reported for E. coli CFPS (Zawada et al., 2011), we do not aim to present industrial-scale production levels. In this study, we sought to scale down the production to in order to achieve a high-throughput screening system. The downscaling of screening processes is an elementary feature of CFPS, which, on a larger scale, normally tends to have a higher cost per synthesized protein than common recombinant expression approaches (Thaore et al., 2020). While a higher price is acceptable for pharmaceutical applications (e.g., personalized medicine), the examples for industrial enzymes are less abundant and rely mainly on E. coli CFPS (Boyer et al., 2008; Li et al., 2016). Here, we sought for a fast and flexible screening platform in order to speed up engineering and functional characterization. Cell lysates and other components can be prepared in batches and stored over long periods. The CFPS reaction can then be prepared on demand. Depending on the assay applied, results can be obtained only 5 h after the reaction was started, thus allowing increased flexibility and handling. The capability of cell-free systems for those screening approaches was recently demonstrated by the synthesis and characterization of several novel azoreductases (Rolf et al., 2022).
We have shown that insect-based CFPS can be used to generate a novel short UPO from Podospora anserina. Initial activity assessment through DMP conversion revealed a lower activity of PanUPO compared with AaeUPO and MroUPO. Subsequent LC-MS analysis of the conversion of pharmaceutical substrates showed considerable activity only towards clopidogrel. This catalytic profile similar to that of MroUPO coincides with a previously reported tendency observed for short UPOs to catalyze the hydroxylation of benzene rings and related structures with lower efficiency than long UPOs (Hofrichter et al., 2022).
The example of PanUPO shows that the synthesis platform described is not only able to provide short and long UPOs but is also a promising approach to expanding the scope of available UPOs. As pointed out, some UPOs are still difficult to express in cell-based systems. The success depends not only on the expression itself but also on secretion and folding (Püllmann et al., 2021). CFPS might allow the decoupling of those factors. The secretion (or in this case translocation in the microsomes) uses a melittin signal peptide that leaves the N-terminus unaltered after cleavage. This approach worked well for AaeUPO, which was described to be quite sensitive to different signal peptides investigated for expression in yeast (Püllmann et al., 2021). An improvement in folding might be achieved by the supplementation of chaperones. The addition of eukaryotic chaperones to E. coli-based cell-free systems for the production of eukaryotic target proteins has been shown to have a beneficial effect on protein folding (Bonomo et al., 2010). Another approach to increase the activity of cell-free produced UPOs might be the use of a fungal system. This was recently shown for a novel fungal lysate (Schramm et al., 2022). While the use of fungal CFPS might be favorable because of its phylogenetic proximity to UPO producers and both host systems used do, in fact, produce UPOs themselves, there is yet much work to be put into further optimizing these lysates in order to harness their full potential. Because insect-based CFPS is well established, there are several methods available to address bottlenecks in protein yield and folding. These include methods to increase protein yields (e.g., dialysis-based approaches; (Quast et al., 2016); as well as repetitive synthesis approaches that can be used to increase the amount of translocated protein (Sachse et al., 2013). Another advantage is the flexible use of different templates. The mentioned fungal CFPS of UPOs so far relies on mRNA as a template. This requires in vitro transcription and purification of the mRNA before the translation reaction (Schramm et al., 2022). While decoupling transcription and translation reactions can have advantages, it is still cumbersome and hard to integrate into streamlined screening processes. Insect-based CFPS can be performed from either an RNA (Tarui et al., 2001) or a DNA template. Beyond that, the DNA templates can be either plasmids or linear DNA fragments (Ramm et al., 2022). Both approaches also apply to UPOs, thereby giving the insect-based CFPS higher flexibility for screening approaches. Overall, this makes insect-based CFPS a sophisticated screening platform for the identification and characterization of novel UPOs and variants thereof.
We successfully demonstrated that insect-based CFPS is well suited for the production of long and short UPOs using the example of the model enzymes MroUPO and AaeUPO as well as the novel PanUPO. The synthesis of active UPO was possible using translocationally active microsomes in the insect lysate. This work clearly depicts the importance of such vesicles and their capability to perform PTMs in order to obtain UPO. The open nature of CFPS in general and the well-established technologies for insect-based CFPS in particular make this platform a promising candidate for comprehensive UPO screening in order to identify novel UPOs and thus expand the biocatalytic toolbox for the production of pharmaceuticals and fine chemicals.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
RW: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing; AZ: conceptualization, methodology, reviewing, editing; MS: investigation, reviewing; JK: data curation, methodology, reviewing SK: conceptualization, funding acquisition, project administration, resources, supervision, validation, reviewing, editing.
This research was funded by the Ministry of Science, Research and Culture (MWFK, Brandenburg, Germany), project PZ-Syn (project number F241-03-FhG/005/001), and the Federal Ministry of Education and Research (BMBF, Germany), project CEFOX (031B0831).
For the Sf21 lysate preparation, the authors would like to thank D. Wenzel and Dipl. Ing. (FH) D. A. Wüstenhagen (Fraunhofer IZI-BB, Potsdam-Golm, Germany). The coding sequences for the wtUPOs were kindly provided by Prof. M. Hofrichter.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.964396/full#supplementary-material
AaeUPO, Agrocybe aegerita UPO; ABTS, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid; CFPS, cell-free protein synthesis; cfUPO, cell-free synthesized UPO; DMP, 2,6-dimethoxyphenol; Mel, melittin signal peptide; MF, microsomal fraction; MroUPO, Marasmius rotula UPO; NBD, 5-nitro-1,3-benzodioxole; PanUPO, Podospora anserina UPO; PTM, post-translational modification; SN1, supernatant 1; SN2, supernatant 2; TM, translation mix.
Anh, D. H., Ullrich, R., Benndorf, D., Svatos, A., Muck, A., and Hofrichter, M. (2007). The coprophilous mushroom Coprinus radians secretes a haloperoxidase that catalyzes aromatic peroxygenation. Appl. Environ. Microbiol. 73, 5477–5485. doi:10.1128/AEM.00026-07
Aranda, C., Municoy, M., Guallar, V., Kiebist, J., Scheibner, K., Ullrich, R., et al. (2019). Selective synthesis of 4-hydroxyisophorone and 4-ketoisophorone by fungal peroxygenases. Catal. Sci. Technol. 9, 1398–1405. doi:10.1039/C8CY02114G
Babot, E. D., Aranda, C., Kiebist, J., Scheibner, K., Ullrich, R., Hofrichter, M., et al. (2022). Enzymatic epoxidation of long-chain terminal alkenes by fungal peroxygenases. Antioxidants (Basel) 11, 522. doi:10.3390/antiox11030522
Babot, E. D., del Río, J. C., Cañellas, M., Sancho, F., Lucas, F., Guallar, V., et al. (2015). Steroid hydroxylation by basidiomycete peroxygenases: A combined experimental and computational study. Appl. Environ. Microbiol. 81, 4130–4142. doi:10.1128/AEM.00660-15
Babot, E. D., del Río, J. C., Kalum, L., Martínez, A. T., and Gutiérrez, A. (2013). Oxyfunctionalization of aliphatic compounds by a recombinant peroxygenase from Coprinopsis cinerea. Biotechnol. Bioeng. 110, 2323–2332. doi:10.1002/bit.24904
Batista, A. C., Soudier, P., Kushwaha, M., and Faulon, J.‐L. (2021). Optimising protein synthesis in cell‐free systems, a review. Eng. Biol. 5, 10–19. doi:10.1049/enb2.12004
Bonomo, J., Welsh, J. P., Manthiram, K., and Swartz, J. R. (2010). Comparing the functional properties of the Hsp70 chaperones, DnaK and BiP. Biophys. Chem. 149, 58–66. doi:10.1016/j.bpc.2010.04.001
Bormann, S., Hertweck, D., Schneider, S., Bloh, J. Z., Ulber, R., Spiess, A. C., et al. (2021). Modeling and simulation-based design of electroenzymatic batch processes catalyzed by unspecific peroxygenase from A. aegerita. Biotechnol. Bioeng. 118, 7–16. doi:10.1002/bit.27545
Boyer, M. E., Stapleton, J. A., Kuchenreuther, J. M., Wang, C.-W., and Swartz, J. R. (2008). Cell-free synthesis and maturation of FeFe hydrogenases. Biotechnol. Bioeng. 99, 59–67. doi:10.1002/bit.21511
Brödel, A. K., Raymond, J. A., Duman, J. G., Bier, F. F., and Kubick, S. (2013a). Functional evaluation of candidate ice structuring proteins using cell-free expression systems. J. Biotechnol. 163, 301–310. doi:10.1016/j.jbiotec.2012.11.001
Brödel, A. K., Sonnabend, A., and Kubick, S. (2014). Cell-free protein expression based on extracts from CHO cells. Biotechnol. Bioeng. 111, 25–36. doi:10.1002/bit.25013
Brödel, A. K., Sonnabend, A., Roberts, L. O., Stech, M., Wüstenhagen, D. A., and Kubick, S. (2013b). IRES-mediated translation of membrane proteins and glycoproteins in eukaryotic cell-free systems. PLoS One 8, e82234. doi:10.1371/journal.pone.0082234
Brödel, A. K., Wüstenhagen, D. A., and Kubick, S. (2015). Cell-free protein synthesis systems derived from cultured mammalian cells. Methods Mol. Biol. 1261, 129–140. doi:10.1007/978-1-4939-2230-7_7
Carro, J., Ferreira, P., Rodríguez, L., Prieto, A., Serrano, A., Balcells, B., et al. (2014). 5-hydroxymethylfurfural conversion by fungal aryl-alcohol oxidase and unspecific peroxygenase. FEBS J. 282, 3218–3229. doi:10.1111/febs.13177
Carro, J., González-Benjumea, A., Fernández-Fueyo, E., Aranda, C., Guallar, V., Gutiérrez, A., et al. (2019). Modulating fatty acid epoxidation vs hydroxylation in a fungal peroxygenase. ACS Catal. 9, 6234–6242. doi:10.1021/acscatal.9b01454
Dondapati, S. K., Stech, M., Zemella, A., and Kubick, S. (2020). Cell-free protein synthesis: A promising option for future drug development. BioDrugs 34, 327–348. doi:10.1007/s40259-020-00417-y
Ezure, T., Suzuki, T., Shikata, M., Ito, M., Ando, E., Nishimura, O., et al. (2007). Expression of proteins containing disulfide bonds in an insect cell-free system and confirmation of their arrangements by MALDI-TOF MS. Proteomics 7, 4424–4434. doi:10.1002/pmic.200700774
Gomez de Santos, P., Cervantes, F. V., Tieves, F., Plou, F. J., Hollmann, F., and Alcalde, M. (2019). Benchmarking of laboratory evolved unspecific peroxygenases for the synthesis of human drug metabolites. Tetrahedron 75, 1827–1831. doi:10.1016/j.tet.2019.02.013
Gregorio, N. E., Levine, M. Z., and Oza, J. P. (2019). A user's guide to cell-free protein synthesis. Methods Protoc. 2, 24. doi:10.3390/mps2010024
Gröbe, G., Ullrich, R., Pecyna, M. J., Kapturska, D., Friedrich, S., Hofrichter, M., et al. (2011). High-yield production of aromatic peroxygenase by the agaric fungus Marasmius rotula. Amb. Express 1, 31. doi:10.1186/2191-0855-1-31
Groll, U. von, Kubick, S., Merk, H., Stiege, W., and Schäfer, F. (2007). “Advances in insect-based cell-free protein expression,” in Cell-free protein expression. Editors Kudlicki, F. Katzen, R. P. Bennett, and R. P. Bennett (Austin, TX: Landes Bioscience), 37–48.
Gurramkonda, C., Rao, A., Borhani, S., Pilli, M., Deldari, S., Ge, X., et al. (2018). Improving the recombinant human erythropoietin glycosylation using microsome supplementation in CHO cell-free system. Biotechnol. Bioeng. 115, 1253–1264. doi:10.1002/bit.26554
Hofrichter, M., Kellner, H., Herzog, R., Karich, A., Kiebist, J., Scheibner, K., et al. (2022). Peroxide-Mediated oxygenation of organic compounds by fungal peroxygenases. Antioxidants (Basel) 11, 163. doi:10.3390/antiox11010163
Hofrichter, M., Kellner, H., Pecyna, M. J., and Ullrich, R. (2015). Fungal unspecific peroxygenases: Heme-thiolate proteins that combine peroxidase and cytochrome p450 properties. Adv. Exp. Med. Biol. 851, 341–368. doi:10.1007/978-3-319-16009-2_13
Katayama, Y., Shimokata, K., Suematsu, M., Ogura, T., Tsukihara, T., Yoshikawa, S., et al. (2010). Cell-free synthesis of cytochrome c oxidase, a multicomponent membrane protein. J. Bioenerg. Biomembr. 42, 235–240. doi:10.1007/s10863-010-9285-8
Kiebist, J., Schmidtke, K.-U., Schramm, M., König, R., Quint, S., Kohlmann, J., et al. (2021). Biocatalytic syntheses of antiplatelet metabolites of the thienopyridines clopidogrel and prasugrel using fungal peroxygenases. J. Fungi (Basel). 7, 752. doi:10.3390/jof7090752
Kinne, M., Poraj-Kobielska, M., Aranda, E., Ullrich, R., Hammel, K. E., Scheibner, K., et al. (2009). Regioselective preparation of 5-hydroxypropranolol and 4'-hydroxydiclofenac with a fungal peroxygenase. Bioorg. Med. Chem. Lett. 19, 3085–3087. doi:10.1016/j.bmcl.2009.04.015
Kinner, A., Rosenthal, K., and Lütz, S. (2021). Identification and expression of new unspecific peroxygenases - recent advances, challenges and opportunities. Front. Bioeng. Biotechnol. 9, 705630. doi:10.3389/fbioe.2021.705630
Kluge, M., Ullrich, R., Dolge, C., Scheibner, K., and Hofrichter, M. (2009). Hydroxylation of naphthalene by aromatic peroxygenase from Agrocybe aegerita proceeds via oxygen transfer from H2O2 and intermediary epoxidation. Appl. Microbiol. Biotechnol. 81, 1071–1076. doi:10.1007/s00253-008-1704-y
Knorrscheidt, A., Püllmann, P., Schell, E., Homann, D., Freier, E., and Weissenborn, M. J. (2020). Identification of novel unspecific peroxygenase chimeras and unusual YfeX axial heme ligand by a versatile high‐throughput GC‐MS approach. ChemCatChem 12, 4788–4795. doi:10.1002/cctc.202000618
Knorrscheidt, A., Soler, J., Hünecke, N., Püllmann, P., Garcia-Borràs, M., and Weissenborn, M. J. (2021). Simultaneous screening of multiple substrates with an unspecific peroxygenase enabled modified alkane and alkene oxyfunctionalisations. Catal. Sci. Technol. 11, 6058–6064. doi:10.1039/D0CY02457K
Kwon, Y.-C., Oh, I.-S., Lee, N., Lee, K.-H., Yoon, Y. J., Lee, E. Y., et al. (2013). Integrating cell-free biosyntheses of heme prosthetic group and apoenzyme for the synthesis of functional P450 monooxygenase. Biotechnol. Bioeng. 110, 1193–1200. doi:10.1002/bit.24785
Li, J., Lawton, T. J., Kostecki, J. S., Nisthal, A., Fang, J., Mayo, S. L., et al. (2016). Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J. 11, 212–218. doi:10.1002/biot.201500030
Linde, D., Olmedo, A., González-Benjumea, A., Estévez, M., Renau-Mínguez, C., Carro, J., et al. (2020). Two new unspecific peroxygenases from heterologous expression of fungal genes in Escherichia coli. Appl. Environ. Microbiol. 86, 028999–e2919. doi:10.1128/AEM.02899-19
Martin-Diaz, J., Molina-Espeja, P., Hofrichter, M., Hollmann, F., and Alcalde, M. (2021). Directed evolution of unspecific peroxygenase in organic solvents. Biotechnol. Bioeng. 118, 3002–3014. doi:10.1002/bit.27810
Molina-Espeja, P., Garcia-Ruiz, E., Gonzalez-Perez, D., Ullrich, R., Hofrichter, M., and Alcalde, M. (2014). Directed evolution of unspecific peroxygenase from Agrocybe aegerita. Appl. Environ. Microbiol. 80, 3496–3507. doi:10.1128/AEM.00490-14
Molina-Espeja, P., Ma, S., Mate, D. M., Ludwig, R., and Alcalde, M. (2015). Tandem-yeast expression system for engineering and producing unspecific peroxygenase. Enzyme Microb. Technol. 73-74, 29–33. doi:10.1016/j.enzmictec.2015.03.004
Ninomiya, R., Zhu, B., Kojima, T., Iwasaki, Y., and Nakano, H. (2014). Role of disulfide bond isomerase DsbC, calcium ions, and hemin in cell-free protein synthesis of active manganese peroxidase isolated from Phanerochaete chrysosporium. J. Biosci. Bioeng. 117, 652–657. doi:10.1016/j.jbiosc.2013.11.003
Park, Y.-J., and Kim, D.-M. (2021). Production of recombinant horseradish peroxidase in an engineered cell-free protein synthesis system. Front. Bioeng. Biotechnol. 9, 778496. doi:10.3389/fbioe.2021.778496
Pecyna, M. J., Ullrich, R., Bittner, B., Clemens, A., Scheibner, K., Schubert, R., et al. (2009). Molecular characterization of aromatic peroxygenase from Agrocybe aegerita. Appl. Microbiol. Biotechnol. 84, 885–897. doi:10.1007/s00253-009-2000-1
Peter, S., Kinne, M., Ullrich, R., Kayser, G., and Hofrichter, M. (2013). Epoxidation of linear, branched and cyclic alkenes catalyzed by unspecific peroxygenase. Enzyme Microb. Technol. 52, 370–376. doi:10.1016/j.enzmictec.2013.02.013
Peter, S., Kinne, M., Wang, X., Ullrich, R., Kayser, G., Groves, J. T., et al. (2011). Selective hydroxylation of alkanes by an extracellular fungal peroxygenase. FEBS J. 278, 3667–3675. doi:10.1111/j.1742-4658.2011.08285.x
Piontek, K., Strittmatter, E., Ullrich, R., Gröbe, G., Pecyna, M. J., Kluge, M., et al. (2013). Structural basis of substrate conversion in a new aromatic peroxygenase: Cytochrome P450 functionality with benefits. J. Biol. Chem. 288, 34767–34776. doi:10.1074/jbc.M113.514521
Piontek, K., Ullrich, R., Liers, C., Diederichs, K., Plattner, D. A., and Hofrichter, M. (2010). Crystallization of a 45 kDa peroxygenase/peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 66, 693–698. doi:10.1107/S1744309110013515
Püllmann, P., Knorrscheidt, A., Münch, J., Palme, P. R., Hoehenwarter, W., Marillonnet, S., et al. (2021). A modular two yeast species secretion system for the production and preparative application of unspecific peroxygenases. Commun. Biol. 4, 562. doi:10.1038/s42003-021-02076-3
Püllmann, P., and Weissenborn, M. J. (2020). Improving the heterologous production of fungal peroxygenases through an episomal Pichia pastoris promoter and signal peptide shuffling system. ACS Synth. Biol. 10, 1360–1372. doi:10.1021/acssynbio.0c00641
Quast, R. B., Kortt, O., Henkel, J., Dondapati, S. K., Wüstenhagen, D. A., Stech, M., et al. (2015). Automated production of functional membrane proteins using eukaryotic cell-free translation systems. J. Biotechnol. 203, 45–53. doi:10.1016/j.jbiotec.2015.03.015
Quast, R. B., Sonnabend, A., Stech, M., Wüstenhagen, D. A., and Kubick, S. (2016). High-yield cell-free synthesis of human EGFR by IRES-mediated protein translation in a continuous exchange cell-free reaction format. Sci. Rep. 6, 30399. doi:10.1038/srep30399
Ramm, F., Jack, L., Kaser, D., Schloßhauer, J. L., Zemella, A., and Kubick, S. (2022). Cell-free systems enable the production of AB5 toxins for diagnostic applications. Toxins (Basel) 14, 233. doi:10.3390/toxins14040233
Rolf, J., Ngo, A. C. R., Lütz, S., Tischler, D., and Rosenthal, K. (2022). Cell-free protein synthesis for the screening of novel azoreductases and their preferred electron donor. Chembiochem. 23, e202200121. doi:10.1002/cbic.202200121
Rotilio, L., Swoboda, A., Ebner, K., Rinnofner, C., Glieder, A., Kroutil, W., et al. (2021). Structural and biochemical studies enlighten the unspecific peroxygenase from Hypoxylon sp. EC38 as an efficient oxidative biocatalyst. ACS Catal. 11, 11511–11525. doi:10.1021/acscatal.1c03065
Sachse, R., Wüstenhagen, D., Šamalíková, M., Gerrits, M., Bier, F. F., and Kubick, S. (2013). Synthesis of membrane proteins in eukaryotic cell‐free systems. Eng. Life Sci. 13, 39–48. doi:10.1002/elsc.201100235
Schramm, M., Friedrich, S., Schmidtke, K.-U., Kiebist, J., Panzer, P., Kellner, H., et al. (2022). Cell-free protein synthesis with fungal lysates for the rapid production of unspecific peroxygenases. Antioxidants (Basel) 11, 284. doi:10.3390/antiox11020284
Shaklee, P. M., Semrau, S., Malkus, M., Kubick, S., Dogterom, M., and Schmidt, T. (2010). Protein incorporation in giant lipid vesicles under physiological conditions. Chembiochem 11, 175–179. doi:10.1002/cbic.200900669
Stech, M., Hust, M., Schulze, C., Dübel, S., and Kubick, S. (2014). Cell-free eukaryotic systems for the production, engineering, and modification of scFv antibody fragments. Eng. Life Sci. 14, 387–398. doi:10.1002/elsc.201400036
Stech, M., Merk, H., Schenk, J. A., Stöcklein, W. F. M., Wüstenhagen, D. A., Micheel, B., et al. (2012). Production of functional antibody fragments in a vesicle-based eukaryotic cell-free translation system. J. Biotechnol. 164, 220–231. doi:10.1016/j.jbiotec.2012.08.020
Steinbrecht, S., Kiebist, J., König, R., Thiessen, M., Schmidtke, K.-U., Kammerer, S., et al. (2020). Synthesis of cyclophosphamide metabolites by a peroxygenase from Marasmius rotula for toxicological studies on human cancer cells. Amb. Express 10, 128. doi:10.1186/s13568-020-01064-w
Tarui, H., Murata, M., Tani, I., Imanishi, S., Nishikawa, S., and Hara, T. (2001). Establishment and characterization of cell-free translation/glycosylation in insect cell (Spodoptera frugiperda 21) extract prepared with high pressure treatment. Appl. Microbiol. Biotechnol. 55, 446–453. doi:10.1007/s002530000534
Thaore, V., Tsourapas, D., Shah, N., and Kontoravdi, C. (2020). Techno-economic assessment of cell-free synthesis of monoclonal antibodies using CHO cell extracts. Processes 8, 454. doi:10.3390/pr8040454
Tonin, F., Tieves, F., Willot, S., van Troost, A., van Oosten, R., Breestraat, S., et al. (2021). Pilot-scale production of peroxygenase from Agrocybe aegerita. Org. Process Res. Dev. 25, 1414–1418. doi:10.1021/acs.oprd.1c00116
Ullrich, R., Liers, C., Schimpke, S., and Hofrichter, M. (2009). Purification of homogeneous forms of fungal peroxygenase. Biotechnol. J. 4, 1619–1626. doi:10.1002/biot.200900076
Ullrich, R., Nüske, J., Scheibner, K., Spantzel, J., and Hofrichter, M. (2004). Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 70, 4575–4581. doi:10.1128/AEM.70.8.4575-4581.2004
Ullrich, R., Poraj-Kobielska, M., Scholze, S., Halbout, C., Sandvoss, M., Pecyna, M. J., et al. (2018). Side chain removal from corticosteroids by unspecific peroxygenase. J. Inorg. Biochem. 183, 84–93. doi:10.1016/j.jinorgbio.2018.03.011
Zawada, J. F., Yin, G., Steiner, A. R., Yang, J., Naresh, A., Roy, S. M., et al. (2011). Microscale to manufacturing scale-up of cell-free cytokine production a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 108, 1570–1578. doi:10.1002/bit.23103
Zemella, A., Thoring, L., Hoffmeister, C., and Kubick, S. (2015). Cell-free protein synthesis: Pros and cons of prokaryotic and eukaryotic systems. Chembiochem 16, 2420–2431. doi:10.1002/cbic.201500340
Zemella, A., Thoring, L., Hoffmeister, C., Šamalíková, M., Ehren, P., Wüstenhagen, D. A., et al. (2018). Cell-free protein synthesis as a novel tool for directed glycoengineering of active erythropoietin. Sci. Rep. 8, 8514. doi:10.1038/s41598-018-26936-x
Keywords: cell-free protein synthesis, in vitro transcription/translation, enzymes, unspecific peroxygenases, insect cell lysate
Citation: Walter RM, Zemella A, Schramm M, Kiebist J and Kubick S (2022) Vesicle-based cell-free synthesis of short and long unspecific peroxygenases. Front. Bioeng. Biotechnol. 10:964396. doi: 10.3389/fbioe.2022.964396
Received: 08 June 2022; Accepted: 23 September 2022;
Published: 01 November 2022.
Edited by:
Tajalli Keshavarz, University of Westminster, United KingdomReviewed by:
Jiangang Yang, Tianjin Institute of Industrial Biotechnology (CAS), ChinaCopyright © 2022 Walter, Zemella, Schramm, Kiebist and Kubick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Kubick, c3RlZmFuLmt1Ymlja0BpemktYmIuZnJhdW5ob2Zlci5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.