
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 16 September 2022
Sec. Biosafety and Biosecurity
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.956271
This article is part of the Research Topic Forestry Biosafety View all 6 articles
Plant nuclear factor-Y (NF-Y) transcription factors (TFs) are key regulators of growth and stress resistance. However, the role of NF-Y TFs in poplar in response to biotic stress is still unclear. In this study, we cloned 26 PdbNF-Y encoding genes in the hybrid poplar P. davidiana × P. bollena, including 12 PdbNF-YAs, six PdbNF-YBs, and eight PdbNF-YCs. Their physical and chemical parameters, conserved domains, and phylogeny were subsequently analyzed. The protein–protein interaction (PPI) network showed that the three PdbNF-Y subunits may interact with NF-Y proteins belonging to two other subfamilies and other TFs. Tissue expression analysis revealed that PdbNF-Ys exhibited three distinct expression patterns in three tissues. Cis-elements related to stress-responsiveness were found in the promoters of PdbNF-Ys, and most PdbNF-Ys were shown to be differentially expressed under Alternaria alternata and hormone treatments. Compared with the PdbNF-YB and PdbNF-YC subfamilies, more PdbNF-YAs were significantly induced under the two treatments. Moreover, loss- and gain-of-function analyses showed that PdbNF-YA11 plays a positive role in poplar resistance to A. alternata. Additionally, RT‒qPCR analyses showed that overexpression and silencing PdbNF-YA11 altered the transcript levels of JA-related genes, including LOX, AOS, AOC, COI, JAZ, ORCA, and MYC, suggesting that PdbNF-YA11-mediated disease resistance is related to activation of the JA pathway. Our findings will contribute to functional analysis of NF-Y genes in woody plants, especially their roles in response to biotic stress.
To eliminate pathogens, plants have developed a complex defense system, of which phytohormones act as signals to trigger and mediate a diverse array of defense responses. Studies have shown that jasmonic acid (JA) triggers plant resistance by regulating the accumulation of ROS, callose, lignin and the expression of defense-related genes (Behr et al., 2018; Block et al., 2018; Moosa et al., 2019; Varsani et al., 2019). Salicylic acid (SA) is known to play a crucial role in systemic-acquired resistance (SAR) and plant innate immunity (Peng et al., 2021). Recently, transcription factors (TFs) were shown to be involved in plant disease resistance by regulating the hormone signaling pathway. For example, MYC2 and its downstream MYC2-targeted TFs (MTFs) form a transcription module that may directly regulate the JA-induced transcription of late defense genes during tomato resistance to B. cinerea (Du et al., 2017). PeTGA1 from Populus euphratica was shown to play a key role in plant immunity by positively regulating both the SA biosynthesis pathway and SA signaling during Colletotrichum gloeosporioides infection (Yang et al., 2022). WRKY transcription factors, including AtWRKY75, AtWRKY28, WRKY50, WRKY51, WRKY70, WRKY62, WRKY3, WRKY4, WRKY33, WRKY18, WRKY40, WRKY48, WRKY60, and WRKY41, in Arabidopsis (Wang et al., 2006; Zheng et al., 2006; Knoth et al., 2007; Lai et al., 2008; Pandey and Somssich 2009; Chen et al., 2013; Chen et al., 2020), CaWRKY27 in Capsicum annum (Dang et al., 2014), and PtrWRKY89 and PtrWRKY73 in poplar (Duan et al., 2015; Jiang et al., 2016) were shown to play roles in disease resistance by regulating the SA- and JA/ET-mediated signaling pathways. However, compared with studies on other TFs, little is known about the roles of nuclear factor Y (NF-Y) in plant disease resistance.
Nuclear factor Y (NF-Y), also called heme activator proteins (HAPs) or CCAAT-binding factors (CBFs), is widespread and evolutionarily conserved in plants, fungi, and animals (Kim et al., 1996). Unlike the other transcription factors (TFs), NF-Y is a heterotrimeric complex consisting of three distinct subunits NF-YA (HAP2/CBF-B), NF-YB (HAP3/CBF-A), and NF-YC (HAP5/CBF-C). The subfamilies (NF-YA, NF-YB, and NF-YC) are characterized by their conserved domains and sequence lengths. Generally, NF-YA sequences with a core region composed of 53 amino acids that contains two helices (α1 and α2) are longer than those of NF-YB and NF-YC (Petroni et al., 2012; Laloum et al., 2013). Of the two helices, α1 is required for subunit interactions in the N-terminal side and α2 is located in the C-terminal for sequence specificity to bind DNA at the CCAAT boxes. NF-YB and NF-YC, which contain three α-helices (α1, α2, and α3) and another α-helix domain in the C-terminus (αC), share histone-fold domains (HFDs) with the core histones H2B and H2A, respectively (Romier et al., 2003; Hackenberg et al., 2012). When NF-Y regulates downstream target genes, NF-YB and NF-YC form a dimer in the cytoplasm first, and then this dimer is transferred into the nucleus, where it interacts with NF-YA to complete the assembly of the heterotrimeric complex, binding to the CCAAT box and finally regulating the expression of downstream target genes (Petroni et al., 2012; Laloum et al., 2013).
In plants, NF-Y has been validated to be involved in multiple physiological and developmental processes, such as embryogenesis, seed germination, flowering time, root growth, and fruit ripening. In Arabidopsis thaliana, AtNF-YC3, -4, and -9 have been proven to control flowering time (Kumimoto et al., 2010); AtNF-YB2 was shown to positively regulate primary root elongation (Ballif et al., 2011); NF-YB9 has been shown to be a pivotal regulator in embryogenesis (Siefers et al., 2009); and NF-YCs were shown to integrate the GA and ABA signaling pathways by interacting with RGL2 during seed germination (Liu et al., 2016). In Chrysanthemum, CmNFYB8 was shown to be involved in flowering time by directly regulating the expression of cmo-MIR156 (Wei et al., 2017). In Triticum monococcum, the NF-Y regulates flowering initiation by integrating vernalization and photoperiod seasonal signals (Li et al., 2011). Overexpression of PtoNF-YC6 and PtoNF-YC8 from Populus tomentosa showed early flowering (Li et al., 2020), and PdNF-YB21 in poplar was shown to positively regulate root growth (Han et al., 2021). Moreover, plant NF-Ys are also involved in abiotic stress responses, especially salt or drought stress. For example, members of the NF-YA subfamily were proven to participate in drought resistance, such as AtNF-YA2, AtNF-YA3, and AtNF-YA5 in Arabidopsis (Leyva-González et al., 2012). StNF-YC9 played an important role in drought tolerance by increasing photosynthesis rate, antioxidant enzyme activity, and proline accumulation coupled to lowered malondialdehyde accumulation in potato (Li et al., 2021). Moreover, overexpression of OsNF-YA7 in transgenic rice plants increased drought tolerance by modulating gene regulation in an ABA-independent manner (Dong-Keun et al., 2015). Heterologous expression of both CdtNF-YC1 from Bermuda grass and TaNF-YA10–1 from wheat resulted in enhanced drought and salinity tolerance (Chen et al., 2015; Ma et al., 2015). The accumulation of GmNFYA in soybean may lead to derepression and maintenance of histone acetylation for activation of salt-responsive genes, which ultimately confer salt tolerance (Lu et al., 2021). In addition, several studies have also shown that NF-Ys play roles in plant interactions with pathogenic microorganisms. For example, the deregulation of NF-YA enhanced resistance to Ralstonia solanacearum in Arabidopsis (Sorin et al., 2014), and MtNF-YA1 appears to be a repressor of responses triggered in Medicago truncatula by Aphanomyces euteiches invasion (Rey et al., 2016). Both ZmNF-YA3 and ZmNF-YA8 in maize were upregulated after infection by the pathogens F. moniliforme, S. reiliana, and C. graminicola (Zhang et al., 2016). Moreover, the overexpression of OsHAP2E conferred resistance to pathogens of transgenic rice (Alam et al., 2015), and overexpression of GmNF-YC4-2 in soybean increased broad disease resistance for bacterial, viral, and fungal infections in the field (O’Conner et al., 2021). In cassava, the NF-Y transcription factor complex (MeNF-YA1/3, MeNF-YB11/16, and MeNF-YC11/12) was shown to play a role in the transcriptional activation of MePR promoters, contributing to the plant defense response (He et al., 2020).
Populus spp. are important landscape tree species around the world, that also have great economic and ecological value. However, abiotic stresses, such as cold injury and salinity, and biotic stresses, such as Alternaria alternata, Melampsora larici-populina, and Dothiorella gregaria, could cause damage to poplars and reduce their production. However, to date, few studies of the NF-Y family concerning biotic stress resistance have been performed in Populus. As such, to obtain a better understanding of the NF-Y family and boost future studies of this family with regard to resistance to biotic stress in Populus, we identified and characterized the NF-Y gene family in P. davidiana × P. bollena.
In this study, 26 NF-Y candidate genes were cloned and subjected to comprehensive analysis, including phylogenetic analysis, conserved motifs, promoter analysis, protein–protein interaction (PPI) network, and expression pattern analysis under biotic treatments. To further study this possibility, the function of PdbNF-YA11 was characterized in transgenic poplar. The results of PdbNF-Y analysis provided a basis for further studies on the functions of the PdbNF-Y family in poplar disease resistance.
Amino acid sequences of Populus trichocarpa PtNF-Ys were obtained from PlantTFDB (https://planttfdb.cbi.pku.edu.cn/blast.php). The sequences were used for a local BLAST search of P. davidiana × P. bollena transcriptomes. Additionally, the hidden Markov model (HMM) profile (PF02045 and PF00808) of the NF-Y conserved domain was used to search the protein sequences from poplar transcriptomes. The results of the two methods were merged to obtain candidate PdbNF-Y members and then submitted to the CCD (https://www.ncbi.nlm.nih.gov/cdd/) and SMARAT (https://smart.embl-heidelberg.de/) programs to verify the complete NF-Y domains.
Physical and chemical parameters of the PdbNF-Y proteins were predicted using the ExPASy-ProtParam tool (https://web.expasy.org/protparam/). Multiple sequence alignment of PdbNF-Ys conserved domain sequences and the formation of sequence logos by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and WebLogo program (https://weblogo.berkeley.edu/logo.cgi) were performed (Crooks et al., 2004). A neighbor-joining (NJ) tree was constructed by MEGA 5.0 software (https://www.megasoftware.net/index.php) with 1000 bootstrap replicates. The MEME program (https://meme-suite.org/tools/meme) was used to detect the conserved motifs in PdbNF-Y proteins with the following parameters: the maximum number of motifs was 10 and the length range was 6–100 amino acids. Exon–intron structure visualization was performed by comparing cDNA sequences with their corresponding full-length genomic DNA sequences using the online tool Gene Structure Display Server version 2.0 (gsds.cbi.pku.edu.cn/).
The promoter sequences (length, 2 kb) of PdbNF-Ys were collected from the P. davidiana × P. bollena genome database (not made public). The cis-elements were analyzed in the PlantCARE program (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
The interaction networks between PdbNF-Ys and other TFs were created by the online software STRING (https://string-db.org/) and visualized by Cytoscape. The intersection database of P. trichocarpa was selected as a reference.
Hybrid poplar seedlings were grown in a greenhouse under 16-h light/8-h dark conditions at 25°C for 15 days. The plant hormone treatments of poplar were carried out as follows: for methyl jasmonate (MeJA) treatment, the seedlings were sprayed with a freshly prepared 0.001% (w/v) MeJA solution; for the salicylic acid (SA) treatment, a new working solution of 5 mM SA was sprayed on the leaves. Leaves from plants were collected at 0, 2, 6, 12, 24, and 48 h after treatment, frozen in liquid nitrogen, and stored at −80°C for RNA extraction. Three independent biological replicates were performed.
The pathogenic fungus Alternaria alternata was cultured for 7 days on potato dextrose agar (PDA) medium in the dark and then diluted to 1 × 106 spores/ml with sterile water. Leaves were collected from three-month-old seedlings and inoculated with 10 µl of spore suspension and then placed in a transparent square petri dish with high humidity at 25°C with a 16-h light/8-h dark cycle. Leaves inoculated with sterile water only served as mock controls. The leaves were then sampled at 12, 24, 48, and 72 hpi after inoculation. Three biological replicates, each containing ten parallel leaves from ten different plants, were used for each treatment time point or control. The samples were then rapidly placed in liquid nitrogen and stored at −80°C.
Total RNA was extracted using the EZNA Plant RNA Kit (R6827, OMEGA, America), and cDNA was synthesized for quantitative real-time PCR (RT‒qPCR) by the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (AT311, TransGen Biotech, Beijing, China) according to the manufacturer’s method. RT‒qPCR was carried out using SYBR® Green real-time PCR Master Mix (QPK-201, TOYOBO, OSAKA, Japan) in a CFX96 TouchTM real-time PCR detection system (Bio-Rad, Hercules, CA, USA). The housekeeping genes PdbEF and Pdbactin were selected as internal controls, and the primers used for RT‒qPCR analysis are listed in Supplementary Table S1. Relative gene expression levels were measured using the 2−ΔΔCt method (Livaka and Schmittgen, 2001). Genes with changes in the expression level greater than 2-fold were considered to be significantly regulated.
The complete ORF of PdbNF-YA11 was cloned into the KpnI and XbaI restriction enzyme sites of pROKII under the control of the CaMV 35S promoter and NOS terminator to generate the overexpression construct pROKII-PdbNF-YA11 (PdbNF-YA11-OE). A 228-bp truncated inverted repeat CDS of PdbNF-YA11 was inserted into the RNAi vector pFGC5941 (pFGC::PdbNF-YA11) to silence the expression of PdbNF-YA11 (PdbNF-YA11-IE). The primers used are listed in Supplementary Table S1. Then, the construct was introduced into the Agrobacterium strain EHA105, which was then transformed into P. davidiana × P. bollena via Agrobacterium-mediated leaf-disk transformation (Ji et al., 2016). The transgenic plants were selected using kanamycin (50 mg/L) or glyphosate (1 mg/L), and all transgenic plants were verified via genomic PCR and RT‒qPCR.
Leaves collected from 1-month-old transgenic plants and wild-type plants were inoculated with 1 × 106/ml A. alternata spores and then placed in a transparent square petri dish under the conditions described above. The leaves were observed and sampled 4 d after inoculation, and the leaf lesion areas were measured by using ImageJ software (Geng et al., 2020). The infection ratings were assigned and calculated according to the method of Pandey et al. (2003). Three biological replicates, each containing ten parallel leaves from ten different plants, were used for each treatment time point or control. The sampled leaves were then used to determine the JA contents and the expression of JA-related genes. The JA contents were determined via enzyme-linked immunosorbent assay (ELISA) kits (Shanghai, China RJ-21830). The RT‒qPCR followed the method described above, and the primers used for analysis are listed in Supplementary Table S1. For each assay, three biological replicates were analyzed.
Statistical analyses were carried out using SPSS v.20 software. The data were compared using Student’s t test. Differences between the two groups of data were considered statistically significant at p < 0.05. * indicates a significant difference.
A total of 26 PdbNF-Y genes (12 PdbNF-YAs, six PdbNF-YBs, and eight PdbNF-YCs) were identified and cloned after the local BLAST and HMMER searches (Table 1, Supplementary Figure S1). The coding sequences (CDSs) of PdbNF-Ys ranged in size from 453 (PdbNF-YB5) to 1101 (PdbNF-YA1) bp with deduced proteins of 217–366 amino acids, showing a wide distribution of PdbNF-Y lengths. Among the three subfamilies of PdbNF-Y, the PdbNF-YA lengths (average length 326.25 AA) were the longest, the PdbNF-YC lengths (average length 251.2 AA) were shorter, and the PdbNF-YB lengths (average length 193.5 AA) were the shortest. The isoelectric point (pI) of PdbNF-Ys ranged from 5.04 (PdbNF-YB3) to 9.65 (PdbNF-YC4). All PdbNF-Y proteins received negative GRAVY scores, indicating that they are all hydrophilic proteins. Subcellular localization analysis revealed that most of the NF-Y proteins were predicted to be located in the nucleus, while a few were targeted to the cytoplasm or nucleus.
Multiple alignment analysis revealed that the amino acid sequences of NF-Ys contain an interaction domain for interacting with other NF-Y subunits and a DNA binding domain for recognizing the CCAAT binding sites. The conserved domain of PdbNF-YAs consists of two subdomains involved in the NF-YB/C interaction and DNA binding (Figure 1A). The conserved domain of PdbNF-YBs is composed of a DNA-binding domain and NF-YA/NF-YC interaction domains (Figure 1B). The conserved domain of PdbNF-YCs is characterized by a subdomain (distributed in two separate regions) responsible for NF-YA interaction (Figure 1C) and a DNA-binding domain consisting of only two residues in series “A” and “R.” Furthermore, the logo of PdbNF-Y conserved domain sequences suggested that PdbNF-YBs were more evolutionarily conserved than other types of PdbNF- Y subunits.
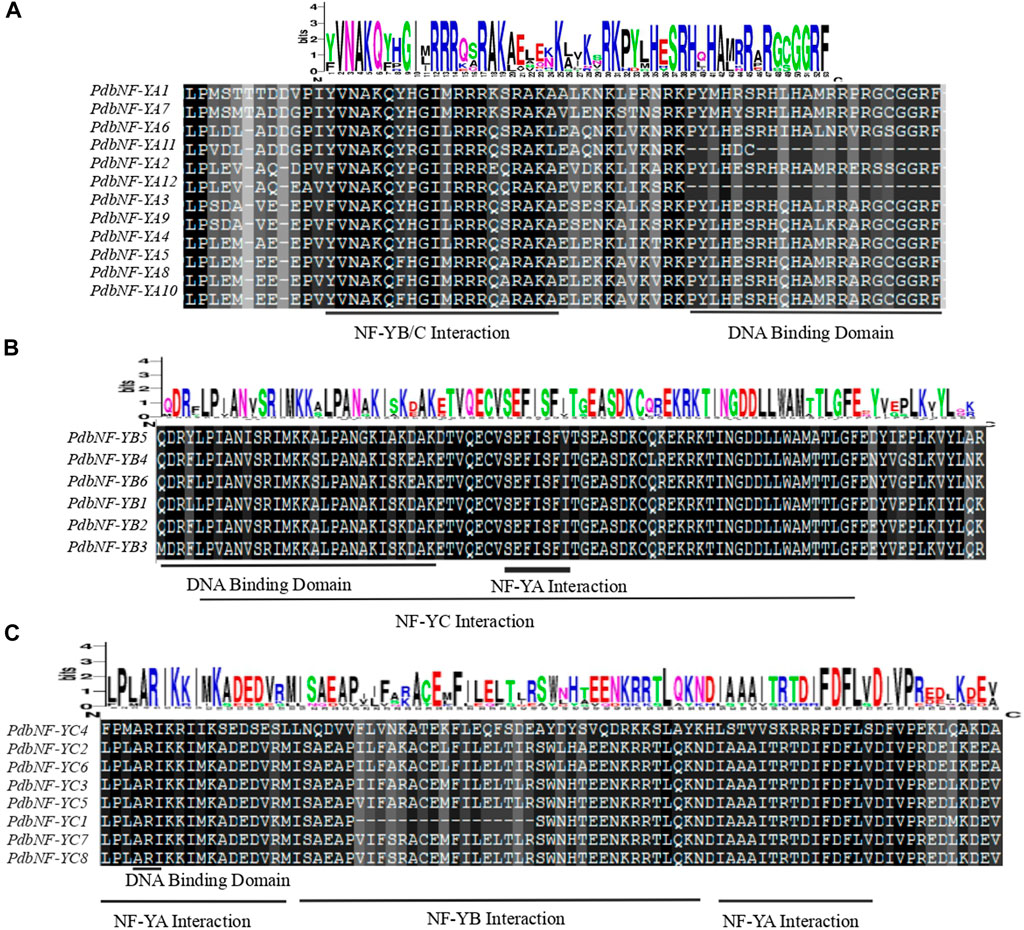
FIGURE 1. Multiple alignments of the PdbNF-Y family. (A) Multiple alignments of the PdbNF-YA conserved domains. (B) Multiple alignments of the PdbNF-YB conserved domains. (C) Multiple alignments of the PdbNF-YC conserved domains. The logo of the conserved domains was visualized in WebLogo.
To further elucidate the evolutionary relationships of the NF-Y family members between poplar and Arabidopsis, a phylogenetic tree was constructed with the NF-Y protein sequences of poplar and Arabidopsis (Figure 2). The NF-Y proteins were clustered into three clades (NF-YA, NF-YB, and NF-YC). Both the NF-YA and NF-YC groups contained one pair of paralogous NF-Y proteins, while the NF-YB group contained four pairs. The phylogenetic tree showed close relationships between the NF-Y members within each of the three subunits and a close relationship of the NF-Y family members between the two poplars and Arabidopsis, which indicated that the NF-Y protein family in poplar may have a structure and function similar to those in Arabidopsis.
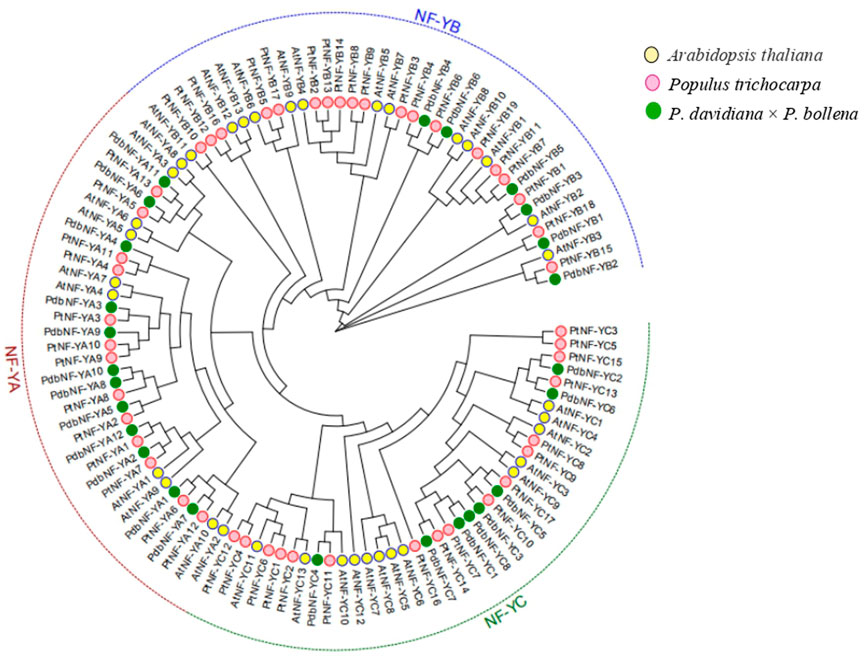
FIGURE 2. Phylogenetic analysis of the NF-Y TFs in poplar and Arabidopsis. All NF-Y TFs were clustered into three clades (NF-YA, NF-YB, and NF-YC).
Phylogenetic analysis was performed for PdbNF-Ys, and most of the PdbNF-Ys were clustered in pairs, except PdbNF-YA4, PdbNF-YA5, PdbNF-YB1, PdbNF-YB5, PdbNF-YC4, and PdbNF-YC7 (Figure 3A). These paired genes also shared similar gene structures, including the numbers and positions of both introns and exons (Figure 3B). Among these, the PdbNF-YA subfamily contained a larger number of introns (from four to six); one or three introns were found in the PdbNF-YC, while most of the PdbNF-YBs contained no introns, except PdbNF-YB5 (Figure 3B). The conserved motifs in PdbNF-Y proteins were analyzed by MEME, and a total of 10 motifs were identified. The results showed that all the PdbNF-Ys contained motifs involved in interacting with the other two NF-Y subfamilies and DNA binding. In addition, motif 9 was identified in most of the PdbNF-YAs, except PdbNF-YA1 and PdbNF-YA7, and motifs 8 and 10 were observed only in PdbNF-YA5/8/10 and PdbNF-YA8/10, respectively (Figure 3C).
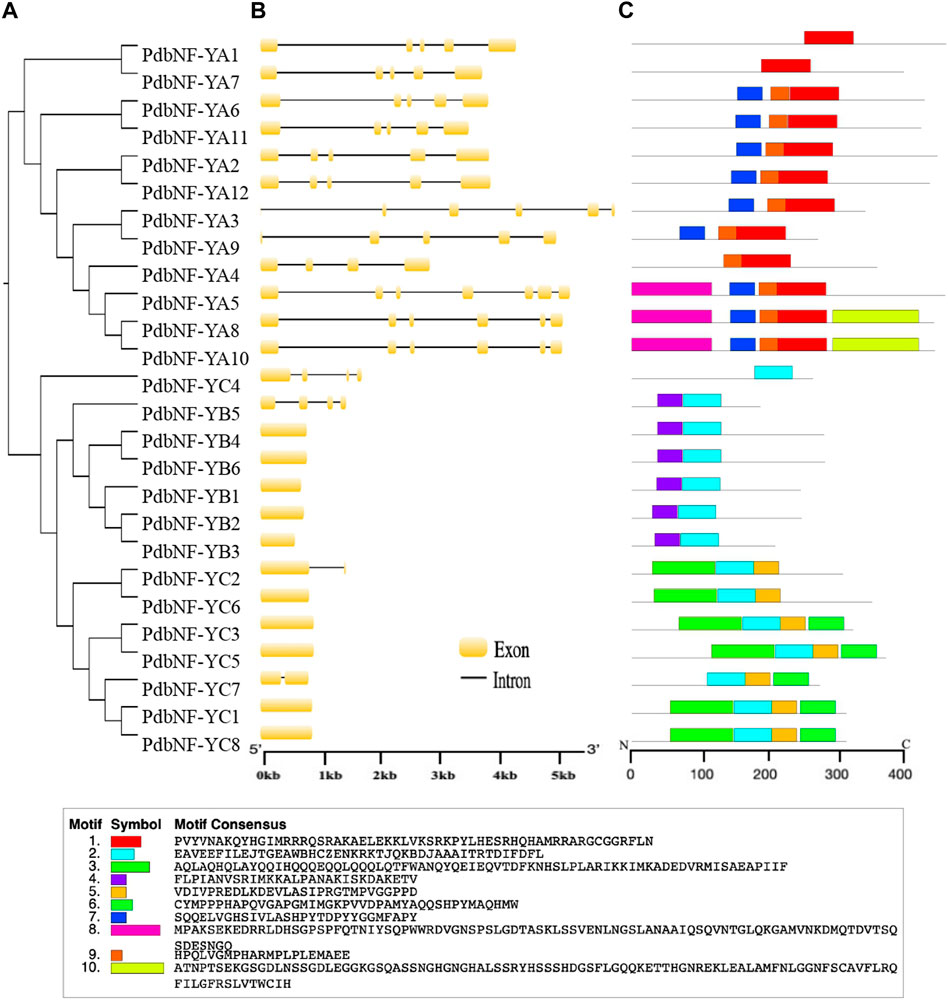
FIGURE 3. Phylogenetic relationships, gene structures, and motif compositions of the PdbNF-Y genes. (A) Unrooted NJ tree. (B) Exon/intron organization. Exons are shown as yellow boxes, and introns are shown as black lines. (C) Schematic representation of conserved MEME motifs of full-length NF-Y proteins. Colored boxes indicate different conserved motifs, while the black lines represent sequences with no MEME motifs detected.
To explore the potential function and regulation of PdbNF-Y genes, the 2000 bp upstream sequences of their promoters were analyzed. A total of 39 cis-elements were found, including 12 light responsive, five phytohormone-responsive, five plant growth-responsive, and 17 stress-responsive cis-elements (Figure 4). Among the cis-elements, stress-responsive elements accounted for the largest proportion. Several elements related to disease resistance were found among the stress-responsive elements. For example, MYC is related to JA signaling; W-box can bind to WRKY, which plays roles in plant disease resistance; and MYB is also related to disease response. Analysis of the phytohormone-responsive elements indicated that the TGA element is involved in auxin responsiveness, GARE motif is involved in gibberellin responsiveness, and TGACG-motif and CGTCA-motif are involved in MeJA responsiveness. In summary, the aforementioned results indicate the potential role of PdbNF-Y genes in response to the regulation of a variety of hormones and disease resistance.
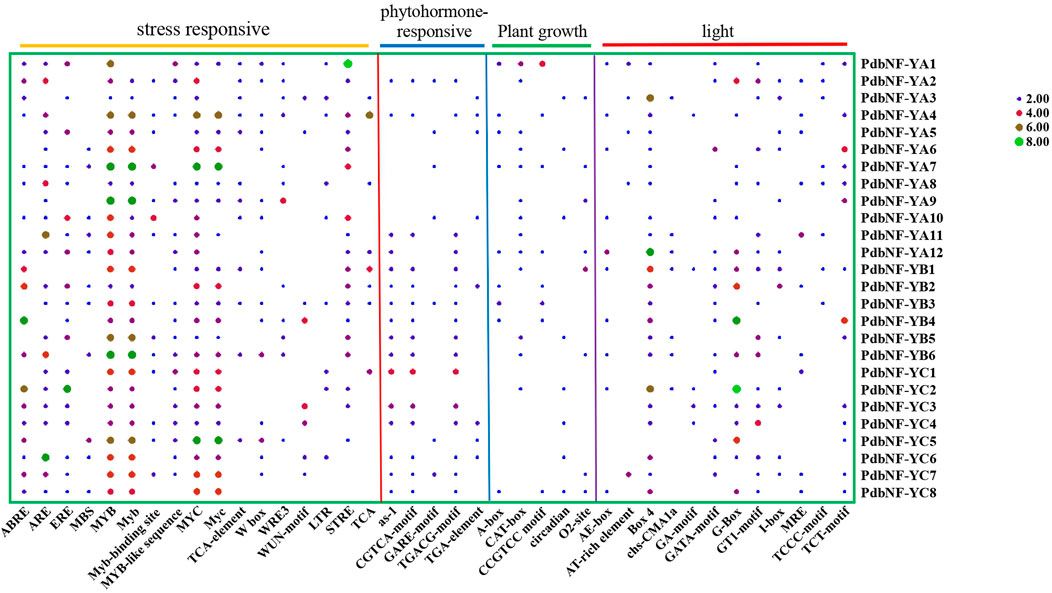
FIGURE 4. Heatmap of cis-elements in the promoter regions of the PdbNF-Ys. Colored bars indicate the numbers of cis-elements.
To obtain a more complete overview of the transcriptional regulation function of PdbNF-Ys, a protein–protein interaction (PPI) network was constructed. The results showed that the three PdbNF-Y subunits may interact with the NF-Y proteins belonging to the other two subfamilies, among which the interactions in PdbNF-YB2/PdbNF-YCs (PdbNF-YC3–5, PdbNF-YC7, and PdbNF-YC8), PdbNF-YB2/PdbNF-YAs (PdbNF-YA6 and PdbNF-YA11), and PdbNF-YC4/PdbNF-YB3 had combined scores higher than 0.95 (Figure 5, Supplementary Table S2). Moreover, these PdbNF-Ys can also interact with other TFs. For example, PdbNF-YAs may interact with TFs, such as GATA, GRF, MYB, bHLH, and bZIP; PdbNF-YBs may interact with bHLH and bZIP; and PdbNF-YCs may interact with TFs, including ERF, MYB, WRKY, bHLH, and bZIP (Figure 5). In addition, GO enrichment showed that the aforementioned TFs were primarily involved in “regulation of transcription,” “regulation of molecular function,” “DNA binding,” “CCAAT-binding factor complex,” and so on (Supplementary Table S3).
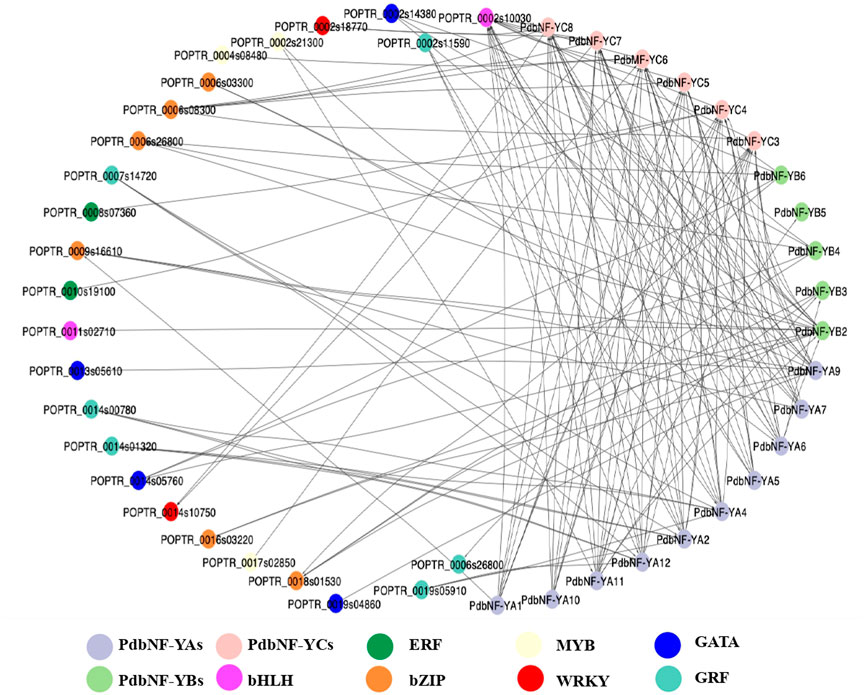
FIGURE 5. Protein–protein interaction analysis of PdbNF-Ys and other transcription factors. Protein–protein interaction network of PdbNF-Ys and other transcription factors. Colored circles indicate different transcription factors (TFs).
Since studies have already shown that NF-Ys play roles in regulating plant growth, the expression patterns of PdbNF-Ys in vegetative (root, stem, and leaf) tissues were analyzed by RT‒qPCR (Figure 6). As the expression of PdbNF-YA3 in roots was lower than that of the other genes in all three tissues, we chose its expression as a control for normalization of gene expression in these tissues. For the PdbNF-YAs, six of them (PdbNF-YA2, PdbNF-YA4–7, and PdbNF-YA11) had higher transcript levels in roots than in both stems and leaves, and five of them (PdbNF-YA1, PdbNF-YA8–10, and PdbNF-YA12) had higher transcript levels in leaves than in stems and roots, while the expression of PdbNF-YA3 was higher in stem than in roots and leaves. Among them, PdbNF-YA2 had the highest transcript level in roots, PdbNF-YA9 had the highest transcript level in leaves, and PdbNF-YA6 had the highest transcript level in stems. For the PdbNF-YBs, four of them (PdbNF-YB1, PdbNF-YB3, PdbNF-YB4, and PdbNF-YB6) had higher transcript levels in stems than in roots and leaves, especially PdbNF-YB3, which also had high expression in roots and leaves. However, the expression of PdbNF-YB2 in leaves was higher than that in roots and leaves, and PdbNF-YB5 had high expression in roots. For the PdbNF-YCs, three of them (PdbNF-YC1, PdbNF-YC5, and PdbNF-YC8) had higher transcript levels in roots than in stems and leaves, while the expression levels of the other PdbNF-YCs were higher in leaves than in stems and roots. These results showed tissue-specific PdbNF-Ys expression.
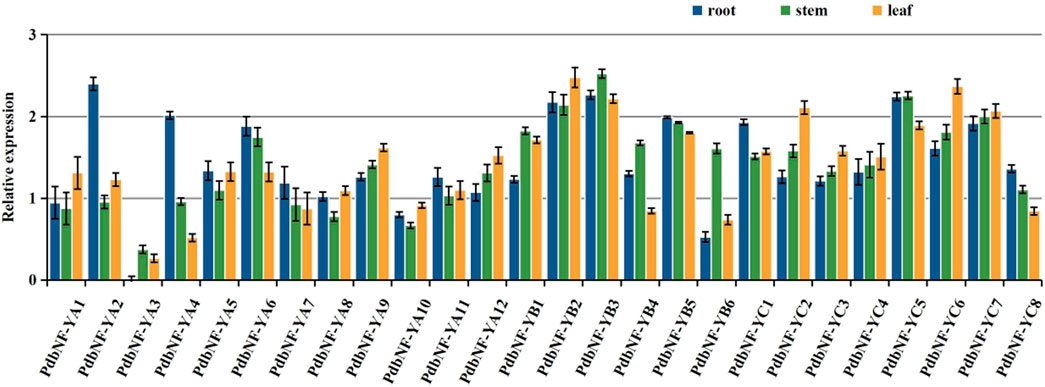
FIGURE 6. Expression profiles of PdbNF-Ys in three poplar tissues. X-axis: different PdbNF-Ys and Y-axis: expression level = log2 (fold change in expression).
To investigate the transcription of PdbNF-Ys in response to plant hormones, RT‒qPCR was performed (Figure 7). Genes with changes in the expression level greater than 2-fold were considered to be significantly regulated. The transcription of PdbNF-Ys was induced by exogenous MeJA at different time points in leaves. For the PdbNF-YAs, three (PdbNFYA2–4) were induced soon after the treatment, but their expressions were subsequently downregulated, while four (PdbNFYA6–9) were inhibited after treatment, but their expressions were subsequently upregulated. PdbNF-YA5 and PdbNF-YA11 were continuously upregulated, with peak transcription levels of 3.46 fold at 24 h and 4.44 fold at 12 h, respectively. For the PdbNF-YBs, two (PdbNF-YB2 and PdbNF-YB3) were continuously downregulated after treatment, while PdbNF-YB5 was continuously upregulated, with peak transcription levels of 3.56 fold at 6 h. For the PdbNF-YCs, three (PdbNF-YC1–3) were inhibited after treatment, while three (PdbNF-YC5-7) were induced but then downregulated. PdbNF-YC8 was upregulated and peaked at 6 h.

FIGURE 7. Heatmap of PdbNF-Ys expression under biotic stresses. The log2 fold change was colored using Cluster 3.0 (red for upregulated and green for downregulated). Each horizontal row represents a gene, and the vertical columns represent time points.
The transcription levels of the PdbNF-Ys were also induced by exogenous SA at different time points in leaves, and most of them were downregulated at 48 h, except PdbNF-YA4 and PdbNF-YB6. For the PdbNF-YAs, seven (PdbNF-YA1, PdbNF-YA2, PdbNF-YA6, PdbNF-YA7, PdbNF-YA9, PdbNF-YA11, and PdbNF-YA12) were induced soon after the treatment, but their expressions were subsequently downregulated, while the others were upregulated at 24 h after treatment. For the PdbNF-YBs, PdbNF-YB5 was continuously upregulated after treatment, with peak transcription levels of 5.03 fold at 6 h, while PdbNF-YB2 was inhibited under SA treatment. For the other PdbNF-YBs, PdbNF-YB1, PdbNF-YB3, and PdbNF-YB4 had the same expression patterns. Among the PdbNF-YCs, PdbNF-YC6 was inhibited after treatment, while four (PdbNF-YC1, PdbNF-YC4, PdbNF-YC5, and PdbNF-YC7) were upregulated after 6 h (Figure 7).
The expression of PdbNF-Ys after inoculation with the pathogen A. alternata was also determined (Figure 7). The RT‒qPCR showed that most of the PdbNF-YAs were induced after treatment, except PdbNF-YA2. Among these PdbNF-YAs, PdbNF-YA11 was significantly expressed during the treatment, while PdbNF-YA1,3–5 and PdbNF-YA9,12 had high transcript levels at 12 hpi, but their expressions were subsequently downregulated. For the other PdbNF-YAs, their expressions were repressed or not obviously induced at different time points. The PdbNF-YBs, including PdbNF-YB1-2 and PdbNF-YB6, were induced at 12 hpi, and their expression levels were downregulated later, while the expression of PdbNF-YB5 was upregulated during the treatments. For the PdbNF-YCs, PdbNF-YC1 was induced after inoculation, and the expression levels of PdbNF-YC2 and PdbNF-YC6 were upregulated at early stages of treatment (12 and 24 hpi), while two PdbNF-YCs (PdbNF-YC3 and PdbNF-YC8) were inhibited at 12 and 24 hpi. These results indicate that PdbNF-Ys may be involved in different stages of progression during poplar interaction with A. alternata. Moreover, JA and SA, the main hormones related to plant disease resistance, are considered the most important signaling molecules in plant defense responses. The upregulation of marker genes involved in the two pathways indicates the activation of these pathways. The expression profiles of marker genes belonging to the JA and SA pathway were also determined by RT‒qPCR, and gene cluster analysis was performed to illustrate the putative molecular mechanism by which the PdbNF-Ys respond to pathogen infection (Supplementary Figure S2). The result showed that 15 PdbNF-Ys, including nine PdbNF-YAs, four PdbNF-YBs, and two PdbNF-YCs, had the same expression profiles as the genes involved in the JA signaling, while the others had the same expression profiles as the genes involved in the SA signaling.
In brief, the transcription of the PdbNF-Ys was affected by all the treatments. Both plant hormone and plant pathogenic fungus treatments could trigger PdbNF-Ys transcription.
Based on the expression analysis aforementioned, PdbNF-YA11 was chosen for further study of its role in poplar disease resistance. The PdbNF-YA11-overexpressing (PdbNF-YA11-OE) and RNAi-silenced PdbNF-YA11 (PdbNF-YA11-IE) plants were obtained via Agrobacterium infection (Figure 8B). To examine whether PdbNF-YA11 affects leaf rot during A. alternata infection, intact leaves obtained from the transgenic lines and the corresponding wild-type (WT) parental lines were spot-inoculated with conidia. The results showed that the severity of tissue damage upon A. alternata infection was significantly lower in the leaves of overexpression poplars than in their WT siblings at 4 d post inoculation (dpi), while the damage severity was higher in RNAi poplars, further confirming the percentage disease index results (Figures 8A–D). In addition, as the expression of PdbNF-YA11 was induced under MeJA treatment, the expression profiles of genes related to JA synthesis and signaling were analyzed in transgenic plants by RT‒qPCR, and the JA contents were also analyzed during infection. These genes showed significantly higher relative expression levels than those in the WT during infection in PdbNF-YA11-OE plants, while the expression levels of these genes were downregulated in PdbNF-YA11-IE plants (Figures 9B–I). Meanwhile, the JA contents in the overexpression lines were higher than those in the WT during infection but were lower in the PdbNF-YA11-IE lines than the WT (Figure 9A). These results implied that PdbNF-YA11 is involved in the resistance of poplar to A. alternata infection by regulating the JA synthesis and signaling pathways.
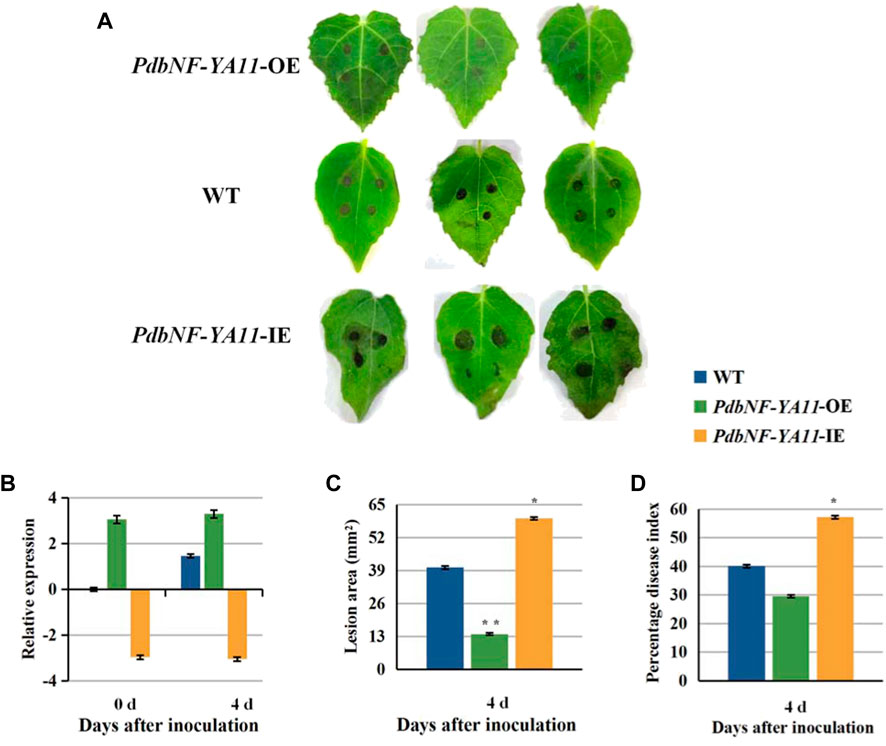
FIGURE 8. PdbNF-YA11 enhances resistance to A. alternata in poplar. (A) Leaves of WT and transgenic poplar inoculated with A. alternata plugs for 4 dpi. (B) Relative expression levels of PdbNF-YA11 during infection in WT and transgenic plants. (C) Lesion areas of leaves. (D) Percentage disease index at 4 dpi. The error bars represent the standard deviation of triplicate assays. Asterisks show significant differences between inoculated and noninoculated leaves (**p < 0.01 and *p < 0.05).
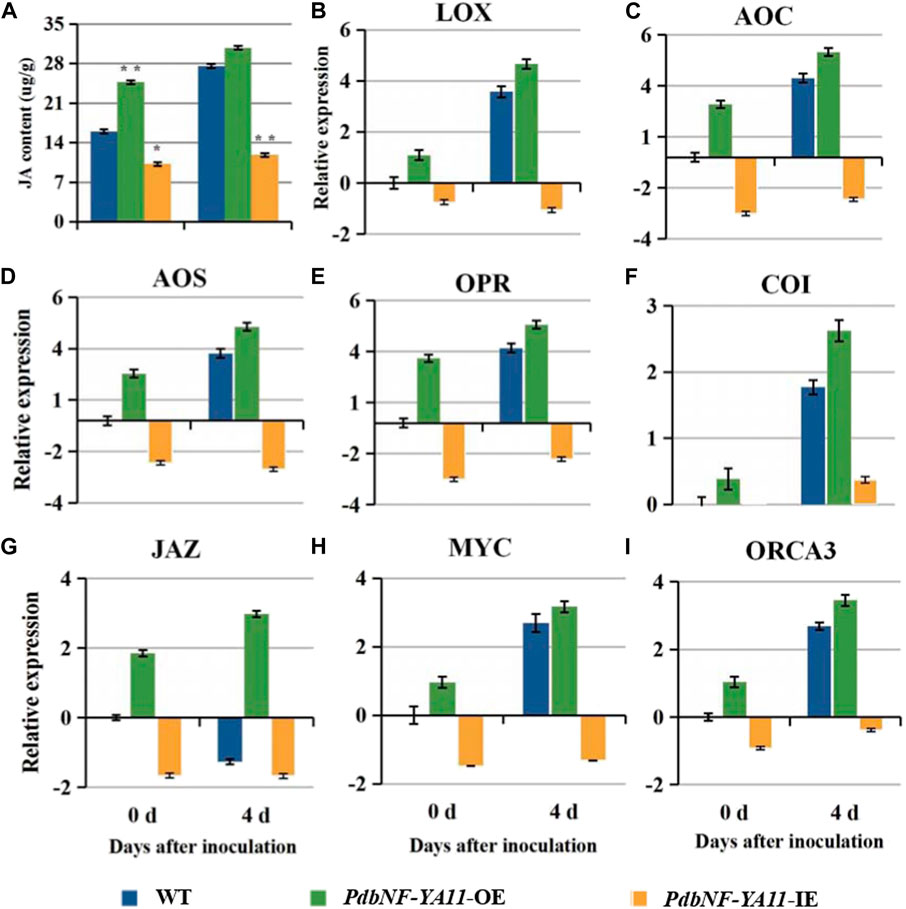
FIGURE 9. PdbNF-YA11 regulates JA signaling. (A) JA contents in WT and transformed plants. (B)–(I) Relative expression levels of the JA biosynthetic and JA signaling pathway genes. The error bars represent the standard deviation of triplicate assays. Asterisks show significant differences between inoculated and noninoculated leaves (**p < 0.01 and *p < 0.05).
As a transcription factor (TF) that regulates plant growth and stress resistance, the NF-Y has been identified and studied in multiple plants. There were 36 NF-Ys in A. thaliana (Siefers et al., 2009), 25 NF-Ys in Ricinus communi (Wang et al., 2018), 59 NF-Ys in Solanum lycopersicum (Li et al., 2016), 32 NF-Ys in Vitis vinifera (Ren et al., 2016), 33 NF-Ys in Juglans regia (Quan et al., 2018), 24 NF-Ys in peach (Li et al., 2019), 28 NF-Ys in Pinus tabuliformis (Guo et al., 2020), 52 NF-Ys in P. trichocarpa (Liu et al., 2021), and 46 NF-Ys in P. tomentosa (Li et al., 2022). In this study, a total of 26 NF-Y members were cloned in P. davidiana × P. bollena, including 12 PdbNF-YAs, six PdbNF-YBs, and eight PdbNF-YCs (Table 1, Supplementary Figure S1). The reason for the low numbers of NF-Y genes identified in P. davidiana × P. bollena may be associated with the lack of a high-quality genome assembly, which may have resulted in some NF-Y family genes not being successfully cloned. The NF-Ys are well known to be involved in plant growth and physiology. However, few studies of the NF-Y family concerning biotic stress resistance have been performed in poplar. As such, we further analyzed the function of PdbNF-Ys involved in biotic stress response, providing a valuable resource for further functional research on NF-Y genes in Populus.
In Arabidopsis and P. trichocarpa, the biological functions of some NF-Ys have been identified. For example, PtNF-YA9 was shown to be involved in abiotic stress and to alter leaf size by mediating the expression of auxin-related genes (Lian et al., 2018). Both AtNF-YB2 and AtNF-YB3 were shown to be involved in root development and flowering time regulation (Kumimoto et al., 2008). AtNF-YC3, -4, and -9 have been proven to control flowering time (Kumimoto et al., 2010), and NF-YB9 has been shown to be a pivotal regulator in embryogenesis (Siefers et al., 2009). In this study, phylogenetic relationships between Arabidopsis and poplar were examined, and the high sequence identity between them can be used to infer similar functions. The results showed that PdbNF-YA9 is homologous to PtNF-YA9 (Figure 2) and had a high transcript level in leaves (Figure 6), suggesting that it may be involved in regulation of leaf development in P. davidiana × P. bollena. Moreover, PdbNF-YB3 and PdbNF-YB2 showed homology to AtNF-YB2 and AtNF-YB3, respectively (Figure 2), which both have high transcript levels in three tissues (Figure 6) and may have similar roles in poplar.
Recently, NF-Ys have been shown to play key roles in plants in response to stress resistance. For example, NF-YAs from Arabidopsis, StNF-YC9 from potato, and OsNF-YA7 from rice were proven to participate in drought resistance (Leyva-González et al., 2012; Dong-Keun et al., 2015; Li et al., 2021). Both CdtNF-YC1 from B. grass and TaNF-YA10–1 from wheat play roles in drought and salinity tolerance (Chen et al., 2015; Ma et al., 2015). GmNFYA may regulate salt tolerance in soybean (Lu et al., 2021). ZmNF-YA3 and ZmNF-YA8 in maize were upregulated after infection by the leaf pathogens, S. reiliana and C. graminicola (Zhang et al., 2016). OsHAP2E conferred resistance to M. oryzae and X. oryzae pv. oryzae in rice by regulating defense- and signaling-related genes (Alam et al., 2015). The NF-Y in cassava may play roles in the plant defense response by regulating MePR (He et al., 2020). In poplar, PtNF-YA9 and PtNF-YB7 were proven to regulate resistance to abiotic stress in transgenic Arabidopsis (Han et al., 2013; Lian et al., 2018), but little is known about the roles of NF-Ys during disease resistance. In this study, cis-element analysis showed that the PdbNF-Y promoters contain elements related to disease resistance, such as MYC, W-box, and MYB (Figure 4), and RT-qPCR showed that most of the PdbNF-Ys were significantly changed during the A. alternata infection (Figure 7). Further analysis also showed that these PdbNF-Ys have the same expression profiles as the genes involved in SA/JA signaling, which are known to be involved in disease resistance (Supplementary Figure S2). These results proved that PdbNF-Ys may be involved in poplar disease resistance.
Phytohormones, especially jasmonates (JAs) act as signals to trigger and mediate a diverse array of defense responses. Transcription factors (TFs) such as MYC2, ERF, and WRKY have been shown to be involved in plant disease resistance by regulating the hormone signaling pathway. For example, MYC2 and its downstream MYC2-targeted TFs (MTFs) form a transcription module that may directly regulate the JA-induced transcription of late defense genes during tomato resistance to B. cinerea (Du et al., 2017). WRKY75 could also positively regulate the JA-mediated plant defense against B. cinerea and A. brassicicola by regulating genes such as ORA59 and PDF1.2 in Arabidopsis (Chen et al., 2020). In Pinus tabuliformis, the NF-Ys were shown to respond to SA and JA treatments, which suggests their roles in affecting disease resistance by dynamically regulating SA and JA signaling (Guo et al., 2020). In this study, the PdbNF-Y promoters contain elements related to hormone response (Figure 4), and the protein–protein interaction (PPI) network showed that the PdbNF-Ys may interact with TFs involved in plant hormone signal transduction, such as ERF, MYB, WRKY, and bHLH (Figure 5, Supplementary Table S3). Importantly, the expression levels of PdbNF-Ys were significantly changed under phytohormone treatments (Figure 7), which indicates that these PdbNF-Ys may be involved in hormone-mediated regulation of defense responses.
Based on the results aforementioned, PdbNF-YA11 contains multiple types of disease-responsive cis-elements in its promoter, and its expression under pathogen infection and hormone treatments was significantly induced. As such, PdbNF-YA11 was chosen for further study of its role during plant‒pathogen interactions in leaves. Both the PdbNF-YA11-overexpressing (OE) and PdbNF-YA11 RNAi-silenced (IE) poplars were generated by Agrobacterium-mediated leaf-disk transformation, and the leaves were observed after inoculation with A. alternata. The PdbNF-YA11-OE plants exhibited significantly higher resistance to A. alternata infection than WT plants, while the PdbNFYA11-IE lines were more susceptible to A. alternata than WT plants (Figure 8). Moreover, JA is commonly known to be involved in plant defense mechanisms against necrotrophic pathogens, such as Alternaria, Botrytis cinerea, Sclerotinia sclerotiorum, Plectosphaerella cucumerina, Fusarium oxysporum, and Pythium (Glazebrook et al., 2005). In this study, the expression levels of JA-related gene and JA accumulation were increased in the PdbNF-YA11-OE plants, but the opposite effects were observed in the PdbNF-YA11-IE plants during infection (Figure 9). Overall, these results proved that PdbNFYA11 may be involved in poplar resistance to pathogen infection by positively regulating the JA-related pathway.
In this study, 26 PdbNF-Ys were cloned from the hybrid poplar P. davidiana × P. bollena. Specific information on the protein architecture was determined, and phylogenetic analysis was performed. Then, the expression profiles of the genes were detected in diverse poplar tissues and seedlings under different treatments. Additionally, PdbNF-YA11 was shown to be involved in the resistance of poplar to A. alternata infection by regulating JA synthesis and signaling pathways by genetic engineering. These results suggested that PdbNF-Ys may be essential not only for plant development but also for the response to biotic stress and provide a basis for further studies on the functions of the PdbNF-Y family.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
CW designed the experiments. YH and HM performed the experiments. XW, TC, and GH analyzed the data. YH and CW drafted and revised the manuscript.
This work was supported by the National Natural Science Foundation of China (No. 31870665) and the Innovation Project of the State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) (No. 2014B03).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.956271/full#supplementary-material
Alam, M. M., Tanaka, T., Nakamura, H., Ichikawa, H., Kobayashi, K., Yaeno, T., et al. (2015). Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 13, 85–96. doi:10.1111/pbi.12239 | |
Ballif, J., Endo, S., Kotani, M., MacAdam, J., and Wu, Y. (2011). Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol. biochem. 49, 579–583. doi:10.1016/j.plaphy.2011.01.013 | |
Behr, M., Lutts, S., Hausman, J. F., and Guerriero, G. (2018). Jasmonic acid to boost secondary growth in hemp hypocotyl. Planta 248, 1029–1036. doi:10.1007/s00425-018-2951-5 | |
Block, A., Christensen, S. A., Hunter, C. T., and Alborn, H. T. (2018). Herbivore-derived fatty-acid amides elicit reactive oxygen species burst in plants. J. Exp. Bot. 69, 1235–1245. doi:10.1093/jxb/erx449 | |
Chen, L., Zhang, L., Xiang, S., Chen, Y., Zhang, H., and Yu, D. (2020). The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 72, 1473–1489. doi:10.1093/jxb/eraa529 | |
Chen, M., Zhao, Y., Zhuo, C., Lu, S., and Guo, Z. (2015). Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 13, 482–491. doi:10.1111/pbi.12270 | |
Chen, X., Liu, J., Lin, G., Wang, A., Wang, Z., and Lu, G. (2013). Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell. Rep. 32, 1589–1599. doi:10.1007/s00299-013-1469-3 | |
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: A sequence logo generator: Figure 1. Genome Res. 14, 1188–1190. doi:10.1101/gr.849004 | |
Dang, F., Wang, Y., She, J., Lei, Y., Liu, Z., Eulgem, T., et al. (2014). Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 150, 397–411. doi:10.1111/ppl.12093 | |
Dong-Keun, L., Kim, H. I., Jang, G., Chung, P. J., Jeong, J. S., Kim, Y. S., et al. (2015). The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 241, 199–210. doi:10.1016/j.plantsci.2015.10.006 | |
Du, M., Zhao, J., Tzeng, D. T., Liu, Y., Deng, L., Yang, T., et al. (2017). Myc2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell. 29, 1883–1906. doi:10.1105/tpc.16.00953 | |
Duan, Y., Jiang, Y., Ye, S., Karim, A., Ling, Z., He, Y., et al. (2015). PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell. Rep. 34, 831–841. doi:10.1007/s00299-015-1745-5 | |
Geng, D., Shen, X., Xie, Y., Yang, Y., Bian, R., Gao, Y., et al. (2020). Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Hortic. Res. 7, 102. doi:10.1038/s41438-020-0324-2 | |
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi:10.1146/annurev.phyto.43.040204.135923 | |
Guo, Y., Niu, S., El‐Kassaby, Y. A., and Li, W. (2020). Transcriptome ‐ wide isolation and expression of NF‐Y gene family in male cone development and hormonal treatment of Pinus tabuliformis. Physiol. Plant. 171, 34–47. doi:10.1111/ppl.13183 | |
Hackenberg, D., Wu, Y., Voigt, A., Adams, R., Schramm, P., and Grimm, B. (2012). Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 5, 876–888. doi:10.1093/mp/ssr107 | |
Han, X., Lin, S., Zhou, Y., Wen, J., Kang, X., et al. (2021). Pdnf-yb21 positively regulated root lignin structure in poplar. Ind. Crops Prod. 168, 113609. doi:10.1016/j.indcrop.2021.113609 |
Han, X., Tang, S., An, Y., Zheng, D. C., Xia, X. L., and Yin, W. L. (2013). Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 64, 4589–4601. doi:10.1093/jxb/ert262 | |
He, X., Liu, G., Li, B., Xie, Y., Wei, Y., Shang, S., et al. (2020). Functional analysis of the heterotrimeric NF-Y transcription factor complex in cassava disease resistance. Ann. Bot. 124, 1185–1197. doi:10.1093/aob/mcz115 | |
Ji, S. D., Wang, Z. Y., Fan, H. J., Zhang, R. S., Yu, Z. Y., Wang, J. J., et al. (2016). Heterologous expression of the Hsp24 from Trichoderma asperellum im-proves antifungal ability of Populus transformant Pdpap-Hsp24 s to Cytospora chrysosperma and Alternaria alternate. J. Plant Res. 129, 921–933. doi:10.1007/s10265-016-0829-9 | |
Jiang, Y., Guo, L., Liu, R., Jiao, B., Zhao, X., Ling, Z., et al. (2016). Overexpression of poplar PtrWRKY89 in transgenic Arabidopsis leads to a reduction of disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. PLoS One 11, e0149137. doi:10.1371/journal.pone.0149137 | |
Kim, I. S., Sinha, S., de Crombrugghe, B., and Maity, S. N. (1996). Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 16, 4003–4013. doi:10.1128/mcb.16.8.4003 | |
Knoth, C., Ringler, J., Dangl, J. L., and Eulgem, T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant. Microbe. Interact. 20, 120–128. doi:10.1094/mpmi-20-2-0120 | |
Kumimoto, R, Adam, L., Hymus, G. J., Repetti, P. P., Reuber, T. L., Marion, C. M., et al. (2008). The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 5, 709–723. doi:10.1007/s00425-008-0773-6 | |
Kumimoto, R. W., Zhang, Y., Siefers, N., and Holt, B. F. (2010). NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63, 379–391. doi:10.1111/j.1365-313x.2010.04247.x | |
Lai, Z., Vinod, K., Zheng, Z., Fan, B., and Chen, Z. (2008). Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 8, 68. doi:10.1186/1471-2229-8-68 | |
Laloum, T., De Mita, S., Gamas, P., Baudin, M., and Niebel, A. (2013). CCAAT-Box binding transcription factors in plants: Y so many? Trends Plant Sci. 18, 157–166. doi:10.1016/j.tplants.2012.07.004 | |
Leyva-González, M. A., Ibarra-Laclette, E., Cruz-Ramirez, A., and Herrera-Estrella, L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One 7, e48138. doi:10.1371/journal.pone.0048138 | |
Li, C., Distelfeld, A., Comis, A., and Dubcovsky, J. (2011). Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 67, 763–773. doi:10.1111/j.1365-313x.2011.04630.x | |
Li, J., Gao, K., Khan, W. U., Yang, X., Yang, X., Zhao, T., et al. (2020). Genome-wide analysis of the poplar NF-Y gene family and its expression in floral bud development of Populus tomentosa. Trees 34, 285–296. doi:10.1007/s00468-019-01917-3 |
Li, J., Gao, K., Yang, X., Guo, B., Xue, Y., Miao, D., et al. (2022). Comprehensive analyses of four PtoNF-YC genes from populus tomentosa and impacts on flowering timing. Int. J. Mol. Sci. 23, 3116. doi:10.3390/ijms23063116 | |
Li, M., Li, G., Liu, W., Dong, X., and Zhang, A. (2019). Genome-wide analysis of the NF-Y gene family in peach (Prunus persica L.) BMC Genomics 20, 612–627. doi:10.1186/s12864-019-5968-7 | |
Li, S., Li, K., Ju, Z., Cao, D., Fu, D., Zhu, H., et al. (2016). Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics 17, 36–51. doi:10.1186/s12864-015-2334-2 | |
Li, S., Zhang, N., Zhu, X., Ma, R., Liu, S., Wang, X., et al. (2021). Genome-wide analysis of NF-Y genes in potato and functional identification of stNF-YC9 in drought tolerance. Front. Plant Sci. 12, 749688. doi:10.3389/fpls.2021.749688 | |
Lian, C., Li, Q., Yao, K., Zhang, Y., Meng, S., Yin, W., et al. (2018). Populus trichocarpa PtNF-YA9, A multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis. Front. Plant Sci. 9, 954–968. doi:10.3389/fpls.2018.00954 | |
Liu, R., Wu, M., Liu, H., Gao, Y., Chen, J., Yan, H., et al. (2021). Genome-wide identification and expression analysis of the nf-y transcription factor family in populus. Physiol. Plant. 171, 309–327. doi:10.1111/ppl.13084 | |
Liu, X., Hu, P., Huang, M., Tang, Y., Li, Y., Li, L., et al. (2016). The NF-YC-RGL2 module integrates GA and ABA signalling to reg ulate seed germination in Arabidopsis. Nat. Commun. 7, 12768. doi:10.1038/ncomms12768 | |
Livaka, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262 | |
Lu, L., Wei, W., Tao, J., Lu, X., Bian, X., Hu, Y., et al. (2021). Nuclear factor Y subunit GmNFYA competes with GmHDA13 for interaction with GmFVE to positively regulate salt tolerance in soybean. Plant Biotechnol. J. 19, 2362–2379. doi:10.1111/pbi.13668 | |
Ma, X., Li, C., and Wang, M. (2015). Wheat NF-YA10 functions independently in salinity and drought stress. Bioengineered 6, 245–247. doi:10.1080/21655979.2015.1054085 | |
Moosa, A., Sahi, S. T., Khan, S. A., and Malik, A. U. (2019). Salicylic acid and jasmonic acid can suppress green and blue moulds of citrus fruit and induce the activity of polyphenol oxidase and peroxidase. Folia Hortic. 31, 195–204. doi:10.2478/fhort-2019-0014 |
O’Conner, S., Zheng, W., Qi, M., Kandel, Y., Fuller, R., Whitham, S. A., et al. (2021). GmNF-YC4-2 increases protein, exhibits broad disease resistance and expedites maturity in soybean. Int. J. Mol. Sci. 22, 3586. doi:10.3390/ijms22073586 | |
Pandey, K. K., Pandey, P. K., Kalloo, G., and Banerjee, M. K. (2003). Resistance to early blight of tomato with respect to various parameters of disease epidemics. J. Gen. Plant Pathol. 69, 364–371. doi:10.1007/s10327-003-0074-7 |
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi:10.1104/pp.109.138990 | |
Peng, Y., Yang, J., Li, X., and Zhang, Y. (2021). Salicylic acid: Biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791. doi:10.1146/annurev-arplant-081320-092855 | |
Petroni, K., Kumimoto, R. W., Gnesutta, N., Calvenzani, V., Fornari, M., Tonelli, C., et al. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 24, 4777–4792. doi:10.1105/tpc.112.105734 | |
Quan, S., Niu, J., Zhou, L., Xu, H., Ma, L., and Qin, Y. (2018). Identification and characterization of NF-Y gene family in walnut (Juglans regia L.) BMC Plant Biol. 18, 255–272. doi:10.1186/s12870-018-1459-2 | |
Ren, C., Zhang, Z., Wang, Y., Li, S., and Liang, Z. (2016). Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.) BMC Genomics 17, 605–608. doi:10.1186/s12864-016-2989-3 | |
Rey, T., Laporte, P., Bonhomme, M., Jardinaud, M. F., Huguet, S., Balzergue, S., et al. (2016). MtNF-YA1, a central transcriptional regulator of symbiotic nodule development, is also a determinant of Medicago truncatula susceptibility toward a root pathogen. Front. Plant Sci. 7, 1837. doi:10.3389/fpls.2016.01837 | |
Romier, C., Cocchiarella, F., Mantovani, R., and Moras, D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278, 1336–1345. doi:10.1074/jbc.m209635200 | |
Siefers, N., Dang, K. K., Kumimoto, R. W., Bynum, W. E., Tayrose, G., and Holt, B. F. (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149, 625–641. doi:10.1104/pp.108.130591 | |
Sorin, C., Declerck, M., Christ, A., Blein, T., Ma, L., Lelandais‐Briere, C., et al. (2014). A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 202, 1197–1211. doi:10.1111/nph.12735 | |
Varsani, S., Grover, S., Zhou, S., Koch, K. G., Huang, P. C., Kolomiets, M. V., et al. (2019). 12-Oxo-phytodienoic acid acts as a regulator of maize defense against corn leaf aphid. Plant Physiol. 179, 1402–1415. doi:10.1104/pp.18.01472 | |
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, e123. doi:10.1371/journal.ppat.0020123 | |
Wang, Y., Xu, W., Chen, Z., Han, B., Haque, M. E., and Liu, A. (2018). Gene structure, expression pattern and interaction of nuclear factor-Y family in castor bean (Ricinus communis). Planta 247, 559–572. doi:10.1007/s00425-017-2809-2 | |
Wei, Q., Ma, C., Xu, Y., Wang, T., Chen, Y., Lu, J., et al. (2017). Control of chrysanthemum flowering through integration with an aging pathway. Nat. Commun. 8, 829. doi:10.1038/s41467-017-00812-0 | |
Yang, Y., Li, H. G., Liu, M., Wang, H. L., Yang, Q., Yan, D. H., et al. (2022). PeTGA1 enhances disease resistance against Colletotrichum gloeosporioides through directly regulating PeSARD1 in poplar. Int. J. Biol. Macromol. 214, 672–684. doi:10.1016/j.ijbiomac.2022.06.099 | |
Zhang, Z., Li, X., Zhang, C., Zou, H., and Wu, Z. (2016). Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 478, 752–758. doi:10.1016/j.bbrc.2016.08.020 | |
Keywords: poplar, NF-Y transcription factor, expression pattern, JA signal, disease resistance
Citation: Huang Y, Ma H, Wang X, Cui T, Han G, Zhang Y and Wang C (2022) Expression patterns of the poplar NF-Y gene family in response to Alternaria alternata and hormone treatment and the role of PdbNF-YA11 in disease resistance. Front. Bioeng. Biotechnol. 10:956271. doi: 10.3389/fbioe.2022.956271
Received: 30 May 2022; Accepted: 16 August 2022;
Published: 16 September 2022.
Edited by:
Mulualem Tigabu, Swedish University of Agricultural Sciences, SwedenReviewed by:
Roberto Mantovani, University of Milan, ItalyCopyright © 2022 Huang, Ma, Wang, Cui, Han, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Wang, Y2hhb3dhbmdAbmVmdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.