- 1Shenzhen Key Laboratory of Musculoskeletal Tissue Reconstruction and Function Restoration, Department of Orthopaedic Surgery, Shenzhen People’s Hospital (The Second Clinical Medical College of Jinan University), Shenzhen, China
- 2Department of Orthopedics, Shenzhen Second People’s Hospital (First Affiliated Hospital of Shenzhen University, Health Science Center), Shenzhen, China
- 3Tsinghua University Shenzhen International Graduate School, Shenzhen, China
- 4Department of Chemistry, The Chinese University of Hong Kong, Hong Kong, China
- 5Department of Child and Adolescent Psychiatry, Shenzhen Kangning Hospital, Shenzhen Mental Health Center, Shenzhen, China
- 6Affiliated Hospital of Jining Medical University, Jining Medical University, Jining, China
Imaging of extracellular vesicles (EVs) will facilitate a better understanding of their biological functions and their potential as therapeutics and drug delivery vehicles. In order to clarify EV-mediated cellular communication in vitro and to track the bio-distribution of EV in vivo, various strategies have been developed to label and image EVs. In this review, we summarized recent advances in the tracking of EVs, demonstrating the methods for labeling and imaging of EVs, in which the labeling methods include direct and indirect labeling and the imaging modalities include fluorescent imaging, bioluminescent imaging, nuclear imaging, and nanoparticle-assisted imaging. These techniques help us better understand the mechanism of uptake, the bio-distribution, and the function of EVs. More importantly, we can evaluate the pharmacokinetic properties of EVs, which will help promote their further clinical application.
1 Introduction
Extracellular vesicles (EVs) are lipid-bound vesicles that are secreted to the extracellular space by cells, which contain proteins and nucleic acids in the lumen, and are wrapped with the lipid membrane (Raposo and Stoorvogel, 2013). EVs were discovered decades ago and considered as waste products from cells at first (Johnstone et al., 1987). Only recently, they were found to mediate cell-to-cell communication by transferring molecules from donor cells to recipient cells, thus regulating the signal pathway in target cells (Valadi et al., 2007). Almost all types of cells could release EVs, and they could be detected in all body fluids, such as blood (Deatherage and Cookson, 2012). Moreover, EVs are excellent deliver vehicles for small molecules, proteins, nucleic acids, and nanoparticles, as they are biocompatible, could cross the biological barrier, and protect the inclusions during circulation at the same time (Alvarez-Erviti et al., 2011; El-Andaloussi et al., 2012).
Compared to the bulky studies of applying EVs in therapy development and diagnosis, the study of their behavior in cells and in vivo is insufficient due to the lack of related techniques, which should be addressed before the application of EVs as therapeutics (Svenson, 2013; Duan et al., 2021). It is of great challenge to track EVs as they are nanometer-sized and have a complex membrane structure (Mulcahy et al., 2014). Currently, lipophilic dyes are most frequently used for EV staining and fluorescent tracking (Liang et al., 2020a; Xu et al., 2020; Nakazaki et al., 2021). Lipophilic dyes, including PKH26 and DIR, can easily label EVs by intercalating into the lipid membrane. However, these lipophilic dyes form aggregates with similar size as EVs and bring false-positive results (Dehghani et al., 2020). In addition, when tracking with lipophilic-stained EV in vivo, the resident cells could also be labeled by the diffused dye (Progatzky et al., 2013). Moreover, the dye has long half-life, so its signal cannot represent EVs in the end (Kim et al., 2019). Therefore, techniques with high efficiency and great tracking capability are in need.
Here, we summarized the arisen tracking methods reported these years in the present study, including fluorescent imaging, bioluminescent imaging, and radio-labeling-assisted imaging, as well as techniques employing nanoparticles for imaging.
2 Extracellular vesicles
There are three main types of EVs, namely, exosomes, microvesicles (MVs), and apoptotic body, in which the first two are frequently studied (Todorova et al., 2017). Different EVs have different biogenesis pathways, size, and contents. Exosomes are first generated as intraluminal membrane vesicles in the multivesicular body (MVB) within the endocytic system and then released upon the fusion of the MVB with the plasma membrane (Hessvik and Llorente, 2018). MVs are secreted directly through cell membrane budding (Lv et al., 2019). The size of exosomes ranges from 30–150 nm and MVs from 100–1,000 nm. Large MVs could be isolated by differentiation ultracentrifugation. Small MVs and exosomes are often collected together due to size overlap and the limitation of current purification techniques, which are termed exosomes or extracellular vesicles by different researchers (Théry et al., 2006; Thery et al., 2018). The contents of exosomes are different from the cytosolic contents of the donor cells, and some proteins are sorted into exosomes through the endosomal sorting complex required for transport (ESCRT) machinery (Wei et al., 2021). For MVs, their membrane structure is similar to the plasma membrane of the donor cells, and their inclusions are a portion of cytosol (McDaniel et al., 2013).
EVs mediate cell-to-cell communication through the transfer of signaling molecules from donor cells to recipient cells, which enables the modulation of various processes in the target cells (Figure 1) (Valadi et al., 2007). Thus, understanding the mechanism of uptake is important as it is the critical step of cell-to-cell communication. However, the clear mechanism of EVs’ uptake is unknown (Mulcahy et al., 2014). Various studies proposed that EV internalization occurs through endocytosis or membrane fusion (Mulcahy et al., 2014). Endocytosis includes a range of uptake pathways including clathrin-dependent endocytosis, caveolin-dependent endocytosis, micropinocytosis, phagocytosis, lipid-raft-involved endocytosis, and receptor-mediated endocytosis (Figure 1). Among these ways, the receptor-mediated endocytosis endows EVs with the ability of selective uptake by specific cell types (Marlin and Springer, 1987; Hoen et al., 2009). It has been reported that EVs with integrin α6β4 could be specifically internalized by lung fibroblasts, explaining the lung tropism of breast cancer metastasis (Hoshino et al., 2015). In addition to endocytosis, researchers observed that some of the EVs were taken up by target cells through membrane fusion by employing the fluorescent lipid dequenching method (Tian et al., 2013). Overall, EVs could be internalized by virtually all cells tested, and multiple pathways may be involved (Mulcahy et al., 2014). However, for different cell types, the uptake pathways may be different (Mulcahy et al., 2014).
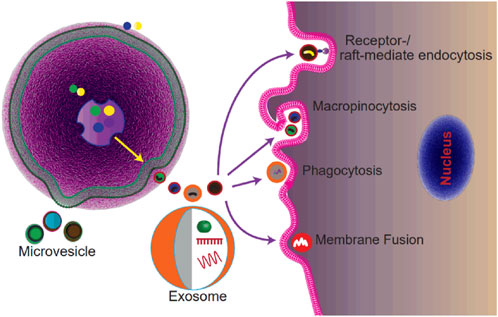
FIGURE 1. Schematic of the biogenesis and uptake of extracellular vesicles. Exosomes are generated as ILVs within MVBs and secreted after MVBs fusing with the plasma membrane. MVs are generated by the outward budding of the plasma membrane. EVs are internalized by recipient cells via micropinocytosis, or phagocytosis, or receptor-/lipid raft-mediated endocytosis, or membrane fusion.
3 Labeling of EVs
The efficient labeling of EVs is the essential step for tracking. Labeling methods for EVs include directly labeling EVs and labeling the donor cells in an indirect manner. In the following sections, we summarized the labeling methods from these two categories.
3.1 Direct labeling
Direct labeling means directly modifying the isolated EVs, which have membranes to bind to signal molecules and have lumen to encapsulate tracer elements.
3.1.1 Anchoring tracer elements on the EV membrane
To anchor the signal molecules on the EV membrane, the potential binding sites can be the lipid bilayer membrane, the transmembrane protein, and the reactive function group on the membrane.
3.1.1.1 Binding to the lipid bilayer mambrane
The lipid bilayer membrane of EVs facilitates the hydrophobic binding of lipophilic dyes and lipid-tagged molecules and also the electrostatic bonding between the positively charged signal molecules and the negatively charged membrane.
The lipophilic dyes, such as PKH26, can bind to EVs by intercalating into the lipid bilayer membrane, which is the most frequently used method for EV labeling currently (Elmahalaway et al., 2018; Wang et al., 2020; Liu et al., 2021). Moreover, molecules with a lipid tail could also bind to the membrane of EVs due to the hydrophobic binding between the lipid tail and lipid bilayer membrane (Salunkhe et al., 2020). He et al. (2019) synthesized the cholesterol-tagged fluorescent probe complex with two complementary single-stranded nucleic acid chains (A and S). The S chain included the sequence of the aptamer of ATP and had the Cy3 modification at the 3’ end. The A chain included the complementary sequence of the ATP aptamer, a BHQ-1 dye at the 5’ end, and a FAM dye at the 3’ end. They designed this device to monitor the uptake pathway of EVs by cells. When EVs are internalized via membrane fusion, the cell membrane showed the green fluorescence of FAM, while if EVs were taken up by endocytosis, the free ATP molecules in the cytoplasm could bind to the aptamer and freed the S chain and showed the red fluorescence of Cy3.
In addition to facilitating the hydrophobic binding, the negatively charged membrane of EVs also allows the attachment of the positively charged dyes by electrostatic bonding (Kim et al., 2016; Feng et al., 2021). Cao et al. (2019) synthesized an aggregation-induced emission luminogen, DPA-SCP, which was positively charged and could attach to the EV membrane for labeling. This method was simple, as it only needed to incubate EVs together with the labeling agents and yet showed superior labeling efficiency.
The labeling methods by anchoring tracer elements to the membrane are often convenient and efficient, which makes their usage frequent. However, the non-covalent binding is not robust and specific, which results in unspecific labeling.
3.1.1.2 Binding to the membrane proteins
Several proteins present on the EV membrane, including EV-specific membrane proteins and some membrane proteins inherent from the donor cells (Hu et al., 2020). Antibodies or aptamers specific to these membrane proteins can be used to EV labeling (Theodoraki et al., 2020; Zhu et al., 2021). Jiang et al. (2018) used gold-carbon dots to label tumor cell-derived EVs by ligating the tumor-specific antibodies to the surface of dots.
The affinity-based binding is tough and specific, but the introduction of antibodies or aptamers may be expensive and laborious. Gao et al. (2018) discovered a peptide, CP05, by phage display, which could bind to EVs specifically. Therefore, EVs could be specifically labeled by dye-tagged CP05. It is well known that the synthesis and modification of peptides are much easier than the production and modification of antibodies or aptamers, and thus this strategy can greatly simplify the labeling.
3.1.1.3 Ligating to the reactive group on EV membrane
There are abundant reactive groups on the surface of EVs, which can react with the tracer molecules (González et al., 2021). However, the reaction should proceed under mild conditions, as EVs cannot tolerate harsh conditions. In this regard, bio-orthogonal reactions are suitable for labeling EVs (Wang et al., 2015). Zhang et al. (2020) employed 4-formylbenzoate to 6-hydrozinonicotinate acetone hydrazine click chemistry to label EVs with quantum dots (QDs) by reacting with the –NH2 group on the membrane. This covalent labeling was solid, but it could also react with the free proteins collected together with EVs as current purification methods can not get rid of all the protein contaminates.
3.1.2 Encapsulating the tracer elements in the lumen of EVs
EVs are lipid-bound vesicles. Loading tracers into the lumen can label EVs and protect the tracer elements during circulation at the same time. As EVs are frequently used as delivery vehicles for therapy development, methods used for cargo loading for delivery purpose can be used for labeling as well (Duan et al., 2021).
3.1.2.1 Incubation
Several tracers can be loaded into EVs by a simple procedure of incubation. During incubation, the molecules penetrate the bilayer membrane by physical diffusion or active uptake.
Molavipordanjani et al. (2020) obtained radiolabeled EVs by incubating the fac-[99mTc(CO)3(H2O)3]+ synthon with EVs. Then, the bio-distribution of EVs in mouse was imaged using the gamma camera. This labeling process occurs via physical diffusion as small molecules can penetrate the membrane spontaneously. Large materials, such as gold nanoparticles, can also be loaded into EVs through incubation. Betzer et al. (2017) reported that glucose-modified nanoparticles could be taken up into EVs via an active, energy-dependent manner as EVs could actively uptake glucose.
3.1.2.2 Electroporation
Electroporation is broadly used for cargo loading as the electrical field could generate micro-pores on the membrane, which allows molecules or nanoparticles enter into the vehicle (Xi et al., 2021). Spab et al. (2020) employed electroporation for loading the tracer element, PMA/Au-BSA@Ce6, into EVs and generated the passion fruit-like EV for tracking and treatment at the same time.
3.1.2.3 Sonication
The integrity of the EV membrane could be disrupted by sonication, which allows the co-incubated substances to load into EVs simultaneously upon the reassemble of the membrane (Fu et al., 2020). In addition to the loaded cargo, the processed EVs may have altered potency as the inner contents leak out during sonication, while this outcome may benefit the study under certain circumstances. Jang et al. (2020) utilized electroporation to load the chlorin e6 photosensitizer into tumor cell-derived EVs for photoacoustic imaging and photodynamic therapy. EVs from tumor cells could promote tumor cell proliferation, which might counteract the effects of the photodynamic therapy. However, after sonication, EVs showed no effect on tumor cell proliferation even with a very high dose (50 μg/ml), which possibly resulted from the significantly decreased contents of protein and RNA after sonication.
3.1.2.4 Extrusion
Nanovesicles produced via extrusion of cells are another type of attractive delivery vehicles as they have comparable advantages as EVs. Ding et al. (2019) decorated the tracer element, GQDzyme/ABTS, with cell membrane by extruding the erythrocyte, which built a barrier for the nanozyme from circulation, and provided the nanozyme with excellent biocompatibility and stealth ability for long blood circulation. Extrusion of erythrocytes for nanovesicle production shows great potential for therapy development, as erythrocytes are easily obtained from patients and the yield is high (Jo et al., 2014; Kuo et al., 2017).
3.1.2.5 Fusion
Fusion was employed to coat amorphous mesoporous silica nanoparticles with the liposome membrane by Liu et al. (2009) to retain and protect the loaded drugs for further in vivo circulation. The fusion occurs spontaneously upon electrostatic bonding between the positively charged silica nanoparticles and negatively charged liposomes. Illes et al. first employed this method to coat the metal–organic framework (MOF) with EVs (Illes et al., 2017; Xhy et al., 2021). The EV coating could seal the MOF from leakage, facilitate the endosomal escape of the drugs, protect them from the circulation, and endow the biocompatibility to the MOF. Compared with other cargo loading methods, the fusion methods retained the membrane integrity to a great extent and thus reserved the advantageous properties of the EV as much as possible.
3.2 Indirect labeling
Modifying the donor cells to produce labeled EVs is what called the indirect labeling method. To achieve fine labeling efficiency, the modification should be sorted into the secreted EVs as much as possible.
3.2.1 Genetic engineering
Genetic engineering is the most frequently used method for EV labeling. The tracer elements were expressed either as recombinant proteins on the EV membrane or as cargo in the EV lumen (Figure 2). CD63, the exosomal marker protein, is often edited for labeling (Logozzi et al., 2009). Fluorescent or luminescent tags are engineered either to the terminus or to the extracellular loop of CD63, which are tagged either inside or outside of EVs, respectively, such as pHluorin, NLuc, and Antares2 (Sung et al., 2015; Hikita et al., 2018; Verweij et al., 2018; Hikita et al., 2020). The M-shaped topology of CD63 possesses restrictions for engineering as both the N- and C-terminus are distributed inside the vesicles. Curley et al. (2020) designed several CD63 truncates for tagging and found a distinct sequence of CD63 for flexible tagging. In addition to CD63, other exosomal proteins employed for tagging tracer proteins are Lamp-2B, lactadherin, and PDGFR (Alvarez-Erviti et al., 2011; Ohno et al., 2013; Takahashi et al., 2013; Liang et al., 2020b; Liang et al., 2021). However, engineering a single protein for labeling may result in bias as it would only label one subtype of EVs. Gupta et al. (2021) systematically screened the exosomal proteins for loading and found that the combination of two exosomal proteins greatly increases the efficiency for therapy development.
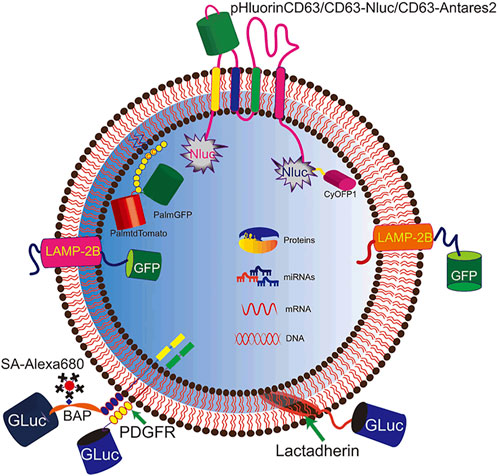
FIGURE 2. Genetic engineering for EV labeling. Fluorescent and luminescent proteins are tagged to EVs for labeling. Signal molecules (pHluorin, NLuc, Antares2, GFP, and GLuc) are introduced to EVs by tagging to membrane proteins (CD63, Lamp2B, and PDGFR), membrane-binding protein (lactadherin), and lipid tail (PalmGFP).
In addition to tagging to the exosomal proteins, fluorescent proteins can also be engineered to express in the lumen of EVs. Lu et al. engineered the donor cells to express PalmGFP, which could attach to the EV membrane through the lipid tail, and thus increased the exosomal loading of GFP (Lai et al., 2015; Lu et al., 2021). Other than fluorescent or luminescent elements, nucleic acids can be used as tracer elements as well. Bala et al. (2015) introduced miR-155 to the lumen of EVs through engineering the donor cells, and then they analyzed the bio-distribution of EVs in mouse via quantitative PCR because of the absence of miR-155 in mouse cells. Very recently, Ferreira et al. (2022) found that proteins containing the KFERQ motif pentapeptide could be loaded into EVs via the membrane protein Lamp2A. The recombinant protein of mCherry tagged with the KFERQ-like sequences was enriched in EVs, which provided a powerful method for EV labeling.
3.2.2 Metabolic labeling
During cell metabolism, cells uptake nutrition from the environment, such as H2O and glucose. The cells will be metabolically labeled when cultured in a medium with labeled nutrient substances and produce labeled EVs. Horgan et al. (2020) obtained deuterium-labeled EVs by culturing cells in a medium containing deuterium oxide, deuterated choline chloride, or deuterated d-glucose. The labeled EVs acted as a bio-orthogonal Raman-active tag for direct Raman identification of EVs during cell internalization. Lee et al. (2018) also metabolically incorporated the active function group, -azido, into EVs by incubating the donor cell with azido-sugar. Then, EVs were further modified by click chemistry. This type of labeling methods provides a simple and convenient manner for introducing tags to EVs, which has a minimum impact on the properties of the produced EVs.
3.2.3 Lipid exchange
Since MVs are generated from direct membrane budding, they have similar membrane structure as the parental cells (Lv et al., 2019). Introducing tags to the cell membrane of the donor cells can produce tagged MVs spontaneously. Chen et al. incubated the donor cells with biotin-functionalized phosphatidylethanolamine to produce biotin-labeled MVs (Valadi et al., 2007; Chen et al., 2015). These MVs were further labeled with streptavidin-functionalized quantum dots (QDs) for imaging (Valadi et al., 2007; Chen et al., 2015).
3.2.4 Labeling donor cells
Stained cells can also produce stained EVs. Monopoli et al. (2018) used an amphiphilic dye to stain the donor cells. The cells could produce NIR-labeled EVs for imaging. The labeling processes included steps of uptake of dyes by cells first and second the secretion of labeled-EVs, which was a route similar to the exocytosis of the anti-cancer drugs from tumor cells as a drug-resistance defense of the cells.
4 Imaging methods
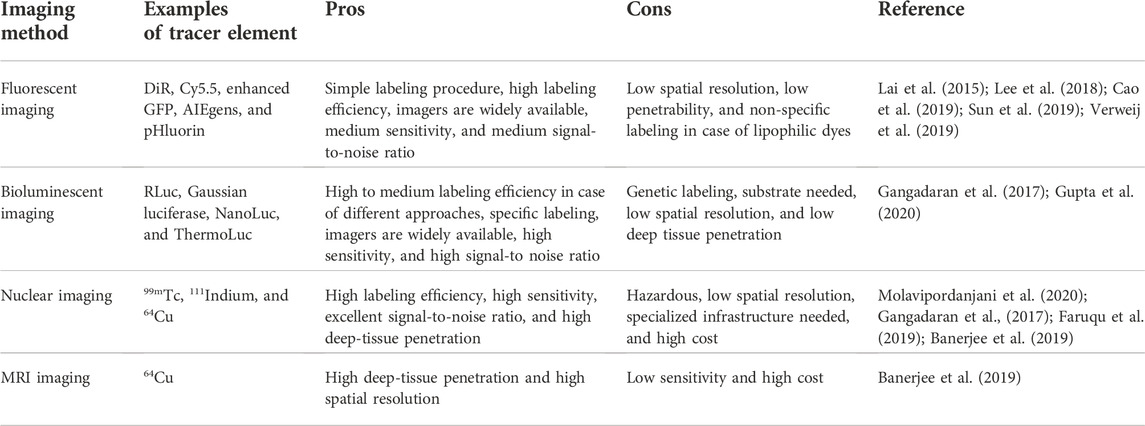
Several imaging modalities are employed for EV imaging till now. We categorize them into four types: fluorescent imaging, bioluminescent imaging, nuclear imaging, and nanoparticle-based imaging. Table 1 contains the list of the advantages and disadvantages of the principal imaging approaches.
4.1 Fluorescent imaging
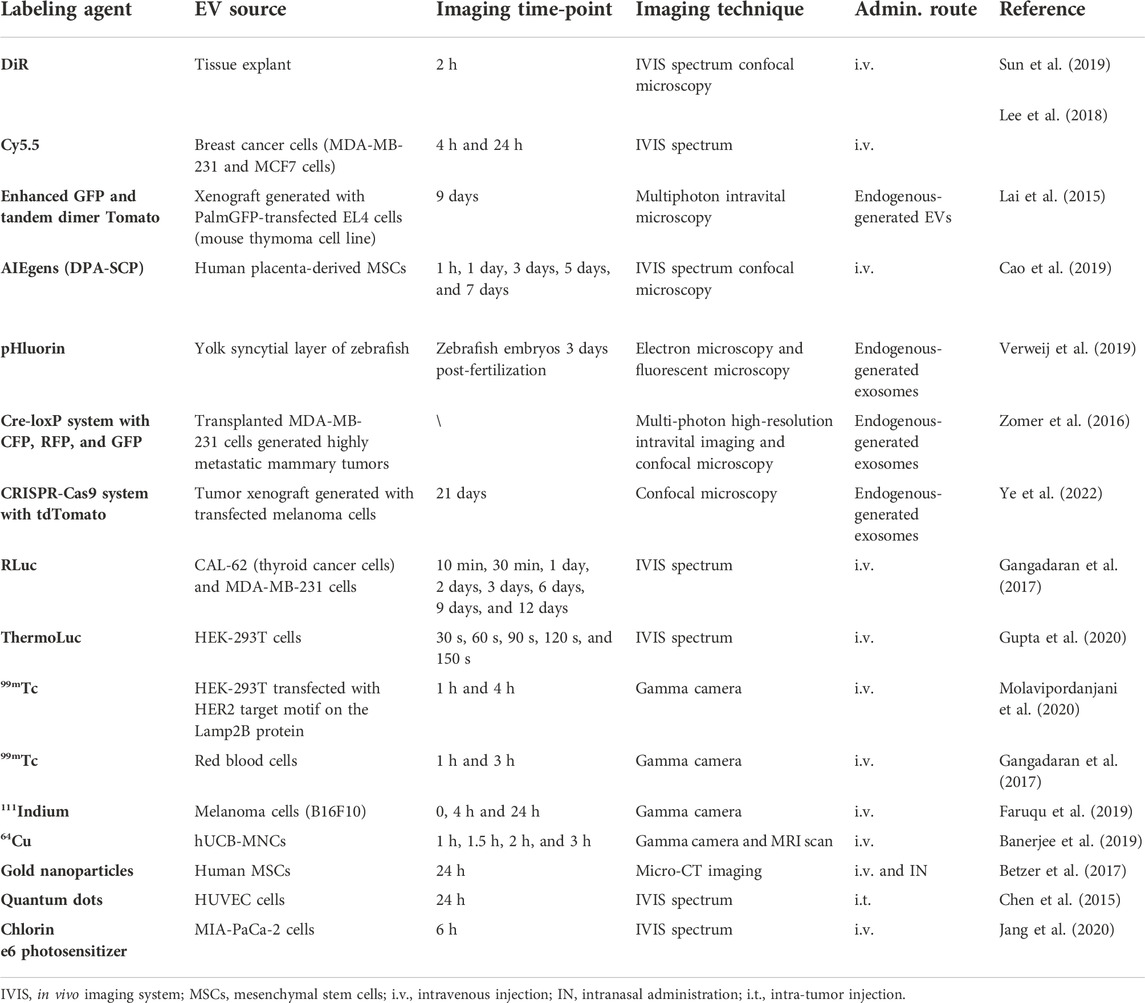
Fluorescent imaging is a strategy extensively used in molecular and cellular biology (Lu et al., 2019).Fluorescent microscopy is easy to operate and could provide information in real time and noninvasively. Chemical dyes, fluorescent proteins, and aggregation-induced emission luminogens (AIEgens) are well-used indicators for fluorescent imaging. However, there are several disadvantages for in vivo fluorescent imaging, as the fluorescent signal has low penetration capability and high background due to the auto-fluorescence of the tissue, which makes it more suitable for in vitro imaging (Ntziachristos et al., 2003). Plenty of studies tracked labeled EVs via fluorescent imaging, in which the signal could keep constant or altered upon environment change. Representative examples are listed in Table 2.
4.1.1 With constant fluorescent
The tracking of the constant signals can reveal the distribution of labeled EVs in cells or in vivo. For this purpose, chemical dyes, fluorescent proteins, and AIEgens with the conventional design can provide satisfactory results.
Chemical dyes are widely used for tracking EVs as they have stable fluorescent signals, minimal impact on EV property, and simple labeling procedures. Sun et al. (2019) employed the lipophilic dye, DIR, for EV tracking. They analyzed the bio-distribution of the labeled EVs using the confocal laser scanning microscope (CLSM) with the tissue sections. By comparing the bio-distribution of EVs with or without ultrasound-targeted microbubble destruction (UTMD), they concluded that UTMD provided an excellent strategy for the targeted delivery of EVs. Lipophilic dyes are convenient and efficient for EV labeling. However, aggregate formation, possible staining of resident cells, and long half-life limit their tracking efficiency (Dehghani et al., 2019). To achieve better performance, the purification step for the stained EVs should be cautiously performed. Pužar Dominkuš et al. (2018) reported that only 10% of particles collected by ultracentrifugation after staining with lipophilic dyes were labeled EVs, and they suggested to use the sucrose gradient to improve the purity. In addition to lipophilic dyes, cyanine dyes are broadly used for labeling in cell research studies (Shi et al., 2016). Usually, they label biomacromolecules via covalent ligation, and thus several functionalized derivatives are commercially available for this purpose. Lee et al. (2018) labeled EVs with Cy5.5 via click chemistry-assisted ligation and tracked the labeled EVs in vivo by CLSM with the tissue sections. They found that EVs derived from highly metastatic cancer cells had selective organ distribution. Chemical dyes are stable, easy to use, and sensitive, but they have limited penetration depth, which hinders the application in deep body structure imaging.
Fluorescent protein is another well-used imaging indicator for cell research. Lai et al. (2015) labeled EVs with PalmGFP to monitor the communication among tumor cells and between tumors cells and the environment via EV exchange by multiphoton intravital microscopy. They found that EVs released by tumor cells distributed intracellularly, or associated with the cytoplasmic membrane, or outside of the tumor cells. They also had the time-lapse recordings, which showed that the EVs could transport individually in small, fast-moving clusters. This indicated that EVs from tumor cells could be internalized by surrounding cells, such as the tumor-infiltrating immune cells.
Another popular fluorescent indicator, AIEgens, is luminogens with aggregation-induced emission properties (Huang et al., 2020). AIEgens show excellent results for biomedical imaging with the advantages of excellent biocompatibility, super resistance to photobleaching, and high signal-to-noise ratios. Cao et al. (2019) employed the positively charged AIEgens, DPA-SCP, for labeling EVs through electrostatic bonding, which required only a simple labeling procedure but exerted superior labeling efficiency. Furthermore, they found that DPA-SCP could track EVs in vivo noninvasively, precisely, and quantitatively without impacting their therapeutic effects. So DPA-SCP supported the convenient, precise, noninvasive, and real-time tracking of EVs, which was superior over other fluorescent dyes.
4.1.2 With changeable fluorescent signal
Tracking with the changeable fluorescent signal can provide more information than tracking with the constant fluorescence signal, such as EV secretion and cellular uptake. Moreover, it can be used to monitor the journey and fate of endogenous EVs, but not pre-isolated EVs, which reveals the in situ communication between cells via EV exchange.
pHluorin is a pH-sensitive green fluorescent protein and represents a powerful tool for monitoring EV secretion and the fusion of MVB with the plasma membrane (Sung et al., 2015; Bebelman et al., 2020; Sung et al., 2020). Verweij et al. (2019) applied the CD63-pHluorin-engineered zebrafish embryos for live-tracking of the biogenesis, journey, and internalization of individual endogenous EVs in vivo. They found that EVs released from the yolk syncytial layer were first captured, endocytosed, and degraded by the patrolling macrophages and endothelial cells during the blood circulation. The CD63-pHluorin zebrafish embryos represented a powerful tool to study the function of endogenous EVs. However, pHluorin has shortcomings, including low expression level in target cells and fast photobleaching. In addition, the small penetration depth of the fluorescent signal further limits the application in clinical translation.
Apart from fluorescent proteins with changeable signals upon environment change, gene editing tools can also change the fluorescent signal following activation. The Cre-loxP technology allowed gene ablation in cells, and thus it was applied to monitor EV transfer between highly metastatic human MDA-MB-231 tumor cells and less malignant human T47D tumor cells in mouse in real time (Zomer et al., 2015; Zomer et al., 2016). For this purpose, MDA-MB-231 cells were engineered to express the Cre recombinase and CFP, while T47D cells were edited to express floxed-DsRed-floxed-eGFP. When T47D cells internalized EVs from MDA-MB-231 cells, their fluorescent signals turned from red to green. High-resolution intravital imaging observed that EVs derived from malignant cells were internalized by less malignant tumors, both located near and distant places. However, the efficiency of tracking might be low, as only mRNAs of Cre recombinase but not the proteins were detected in EVs derived from the engineered donor cells in this study. The CRISPR-Cas9 system is another popular gene editing tool in molecular and cellular biology (Wang et al., 2016). Ye et al. employed this strategy for EV tracking in cells and in vivo (Ye et al., 2020; Ye et al., 2022). The author loaded the single-guide RNA, Cas9 ribonucleoprotein complex, into EVs by editing the donor cells to express the complex tagged with GFP nanobody and CD63-GFP at the same time, resulting in a high loading efficiency. Then, the genetically engineered melanoma cells were implanted subcutaneously in a mouse model that ubiquitously expressed STOP-tdTomato. By analyzing the tissue sections, they found that EVs from the tumor xenografts preferentially targeted the brain and liver as indicated by the fluorescent signal of tdTomato. The method for cargo loading in this study was efficient, and the in vivo tracking was noninvasive and real-time, making it a powerful method.
Apart from the aforementioned tools, monitoring EVs by the changeable fluorescence signal could be toggled by a fine design. He et al. (2019) developed a novel device to track the EV internalization pathway by using the ATP aptamer and its complementary sequence. The unit included the ATP aptamer with Cy3 modification at the 3’ end, its complementary sequence with a BHQ-1 dye at the 5’ end, and a FAM dye at the 3’ end. The whole unit was anchored on EVs by lipid insertion. Recipient cell membranes exhibited green in case of membrane fusion, and its cytosol demonstrated red in case of endocytosis. Thus, the EV uptake pathway by distinct cells was clearly visible by fluorescent imaging. This method was efficient on monitoring the EV uptake pathway and possessed great potential for EV tracking in vivo.
4.2 Bioluminescent imaging
Bioluminescent imaging is the most widely used in vivo imaging technique (Yi et al., 2020). The bioluminescence has advantages of high sensitivity, low background, and credible results, as the labeling is intrinsic. However, it requires genetic modification and substrate injections. Moreover, bioluminescence tracking is limited by the facts that the signal fades quickly and the penetration ability and spatial resolution are low. Representative examples are listed in Table 2.
Several kinds of luciferases were employed for EV labeling and tracking. Gangadaran et al. (2017) employed the Renilla luciferase (RLuc) for EV tracking in vivo by transducing RLuc into the donor cells. Then, EVs derived from different cells having different bio-distribution patterns were revealed by in vivo bioluminescence imaging. EVs isolated from thyroid cancer cells emitted strong signals at the lungs followed by the liver, spleen, and kidney, while EVs derived from breast cancer cells showed strong signals at the liver, followed by the lung, spleen, and kidney. In this study, luciferases were introduced into EVs by merely overexpressing RLuc in donor cells without any EV-guiding module. Thus, it was very likely that the loading efficiency was unsatisfactory, and free luciferases might be released in the cell culture media and be co-isolated with EVs by ultracentrifugation, leading to fake signals.
Compared with the regular luciferases, including Gaussia luciferase (GLuc) and RLuc, NanoLuc has a larger dynamic range and high signal intensity, making it more suitable for in vivo tracking (Gupta et al., 2020). Gupta et al. (2020) performed the in vivo tracking of EVs with NanoLuc, which was genetically ligated to CD63 to improve the loading efficiency. Nevertheless, free NanoLuc was detected in the cell culture medium. For this reason, they examined the tracking results of other luciferases and identified ThermoLuc as the best luciferase for the in vivo imaging of EVs (Gupta et al., 2020). Tracking with ThermoLuc indicated that EV bio-distribution was different according to different injection routes. Moreover, they tracked EVs from different subtypes by tagging ThermoLuc to CD63 or CD9 and concluded that different subtypes had different bio-distribution patterns.
4.3 Nuclear imaging
Nuclear imaging is broadly used in cellular biology and clinical diagnosis. The labeling is durable, and the imaging is highly sensitive and has high penetrability. However, it is expensive, time-consuming, and unavailable for most laboratories. Representative examples are listed in Table 2.
For radiolabeling, the radioactive substances are easily introduced into EVs by incubation. The in vivo imaging was then obtained by positron emission tomography (PET) or single-photon emission computed tomography (SPECT) (Khan, 2021). Gangadaran et al. (2018) produced 99mTc-labeled nanovesicles by extruding the red blood cells, and the in vivo imaging indicated that the nanovesicles were highly distributed in the liver and spleen but not in the thyroid. Molavipordanjani et al. (2020) also labeled EVs with 99mTc and found that tumor cells derived EVs, which were genetically modified to have a ligand of HER2 on the membrane surface, showed a high accumulation in xenograft tumor generated with high-HER2-expression tumor cells.
Radioactive substances can also be engineered on the EV surface by chemical ligation. Faruqu et al. (2019) compared the 111indium labeling efficiency of intraluminal labeling by incubation and membrane labeling by chemical ligation and concluded that membrane-labeled melanoma cell-derived EVs had a superior radiolabeling efficiency and radiochemistry stability. Then, the in vivo distribution of the membrane-labeled EVs was imaged by the SPECT scan in melanoma-bearing mice. The labeled EVs accumulated primarily in the liver and spleen, followed by the kidneys. Similarly, Banerjee et al. (2019) labeled small EVs with 64Cu by decorating the metal chelator, 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA), on the membrane surface via thiol–maleimide conjugation. By magnetic resonance imaging (MRI) and PET, they observed the accumulation of small EVs derived from human umbilical cord blood mononuclear cells (hUCB-MNCs) in mice at 1 h, 1.5 h, 2 h, and 3 h after intravenous injection.
4.4 Nanoparticle-assisted imaging
Nanoparticles have been extensively employed for biomedical imaging and have great potential as probes for disease diagnosis. Among them, gold nanoparticles (GNPs), quantum dots (QDs), and metal–organic framework (MOF) are powerful tools for EV tracking. Representative examples are listed in Table 2.
4.4.1 Gold nanoparticles (GNPs)
GNPs can be used for cellular biomedical imaging due to their unique plasmonic properties. Other than the optical modalities, the signal of GNPs can penetrate deep inside the tissues, which benefits the imaging of the deep body structure. Betzer et al. (2017) employed glucose-coated GNPs for tracking EVs in the deep brain structure. EVs were labeled through active uptake of glucose-coated GNPs and in vivo images were taken by computed tomography, indicating that the intranasal administration resulted in the superior brain accumulation of EVs compared with intravenous injection. They further noninvasively tracked the intranasally administered EVs in the mouse model with focal brain ischemia and found that EVs had an increased accumulation at the focal site over 24 h.
4.4.2 Quantum dots (QDs)
QDs are nanoscale crystals that can emit fluorescence of various wavelengths upon being excited by UV light. Compared with the conventional fluorophore, QDs have brighter emission, a higher signal-to-noise ratio, and less photobleaching, making them attractive for imaging. Chen et al. (2015) employed QDs for noninvasively tracking MVs in vivo. MVs were labeled with QDs via binding between biotin and streptavidin. Then, with a mouse model bearing melanoma xenografts, they found that MVs derived from malignant melanoma cells administered intratumorally could target tumor cells but not transfer to the liver, in which injected-free QDs were distributed.
4.4.3 Metal–organic framework (MOF)
MOFs, composed of metal ions and the coordinated organic ligands, have attracted extensive interests for biomedical research, such as biological imaging and delivery. Illes et al. (2017) coated the iron-based MOFs, MIL-88A NPs, with EVs by the fusion method. In this study, they loaded calcein to the MOFs for imaging. In addition, the iron-based MOFs are capable of MRI imaging, so it is worth expecting in their future work.
4.5 Other tracing methods
4.5.1 Photoacoustic imaging
Photoacoustic imaging (PAI), developed based on the photoacoustic effect, is an attractive imaging modality as it allows noninvasive and real-time imaging with the advantages of deep tissue penetration and high spatial resolution. Proper contrast agents are needed for PAI, which absorb the laser irradiation energy and convert it into the ultrasound signal. Apart from the contrast agents, PAI is similar to ultrasound imaging, which is cheaper than CT and MRI, and has no risk of radiation exposure. Moreover, the contrast agents, which could emit optical signals, allow optical imaging with the advantage of high contrast at the same time. Representative examples are listed in Table 2.
PAI is employed for EV imaging in several studies. Ding et al. (2019) encapsulated the H2O2-sensitive PAI contrast agent into the erythrocyte membrane by extrusion. The generated nanoparticles could image the nasopharyngeal carcinoma cells, which produces H2O2 following laser radiation. The strategy combines the excellent biocompatibility and stability of the encapsulated nanoparticles, the sensitive and specific reactions between the nanoparticles and cancer cells, and the high spatial resolution of PAI, present as an ideal platform for tumor diagnosis. In addition to the application in diagnosis, Jang et al. (2020) developed the photodynamic and immune therapy assisted by PAI. The photosensitizer, chlorin e6, was loaded into the isolated tumor cell-derived EVs by sonication. The generated nanoparticles could be visualized via PAI in vivo and produce cytotoxic reactive oxygen species (ROS) inside the tumor cells under the tissue-specific laser irradiation. Tumor cells were damaged by tumor cell-derived EV-stimulated ROS and cytokines from immune cells.
4.5.2 Raman spectrum for imaging
Raman spectroscopy is an analytical technique developed based on the inelastic scattering of the molecular vibration and widely applied in many research fields, as it can reveal the fingerprint spectrum of the samples in a non-destructive and label-free manner. Raman spectroscopy is applied to analyze the compositional characterization of purified EVs. EVs from different subtypes and different cells could be distinguished by Raman spectroscopy, and the latter had great potential in developing diagnostic approaches for early-stage cancer (Park et al., 2017).
However, to trace EVs in cells or in tissues, signal of EVs can be confounded by the cells’ signal as the similar chemical bond they possess. The introduction of Raman tags, such as alkyne and carbon–deuterium bonds, which vibrates in the region different from the endogenous biological molecules, can circumvent this issue. Horgan et al. (2020) introduced the carbon–deuterium bonds to EVs by metabolic incorporation. Then, by using the confocal spontaneous Raman microspectroscopy system, they obtained a high-resolution image of EV uptake by cells in vitro, both in 2D and 3D.
4.5.3 Quantitative analysis of EVs distributed in vivo with YRNA-based amplification
In addition to the involvement of different imaging modalities, the quantitative measurements of EVs distributed in different tissues can be obtained by nucleic acid-based amplification. YRNAs, first discovered as the RNA component of Ro RNA particle in 1981 and having a direct role in DNA replication, are particularly plentiful in EVs (Kowalski and Krude, 2015). Among all the YRNAs, NT4, a 24-nuleotide 5’ fragment of human YRNA 4, was highly enriched in human CDC-derived EVs but absent in mouse, which could act as the tracer of EVs in mouse (Ciullo et al., 2022). Then, by using the quantitative PCR approach, the distribution of CDC-derived EVs in the mouse and mouse cells was measured.
4.5.4 Multimodal imaging
Multimodal imaging refers to the production of two or more sets of signals at the same time, such as the combination of optical and magnetic imaging. It is attractive as it can provide multiplex information for one sample, especially when the involved modalities have complementary advantages. Several studies tracked EVs by multimodal imaging. Zhao et al. (2016) encapsulated Mn-magnetofunctionalized Ag2Se QDs (Ag2Se@Mn QDs) in MVs by sonication, which had great near-infrared fluorescence and magnetic resonance imaging capabilities. Further, they carried out the continuous dual-modal tracking of MVs in nude mice after the intraperitoneal injection and realized the long-term, noninvasive, whole-body, high-resolution, in situ quantitative, and dual-mode tracking of MVs in vivo.
5 Conclusion
Developments in the field of EVs offer very promising approaches for the diagnosis and treatment of diseases. However, the current understanding of these EVs, especially their in vivo behavior and distribution, is still insufficient. Current methods for in vivo imaging and tracking of EVs, including fluorescent imaging, bioluminescent imaging, photoacoustic imaging, nuclear imaging, and MRI have greatly aided our understanding of the uptake mechanism of EVs, bio-distribution patterns, migration, and functions of EVs and more importantly, pharmacokinetic properties can be assessed and dosage can be optimized by these tracking methods. The development of a reliable, non-invasive, and stable EV labeling technology and in vivo imaging technology to reveal their EVs in disease diagnosis, drug delivery, and barrier penetration will promote further clinical applications of EVs.
Tracking EVs in cells or in vivo provides information involved in cellular uptake and cell–cell communication. Various uptake mechanisms of EVs have been reported. He et al. (2019) visualized different uptake pathways using a fine-designed fluorescent device. The results indicated that the occurrence of uptake pathways was different with different EV origins and recipient cell types. Specific cell–cell communication in vivo could be monitored by EV tracking as well. Lai et al. (2015) visualized the communication between tumor cells and surrounding cells via EV exchange through fluorescent labeling. Also, Wiklander et al. (2015) reported that EV in vivo bio-distribution was determined by cell source, route of administration, and targeting. In addition to the mechanism study, EV tracking has an instructive function for laboratorial and clinical research studies. Betzer et al. (2017) found that intranasally administered EVs had enhanced brain accumulation compared to intravenously injected EVs.
Despite promising applications, several important aspects remain to be explored in future research studies. First, the labeling of EVs with fluorescent dyes or genetic engineering techniques is limited by the inherent background generated by natural biomolecules such as hemoglobin and lipoproteins, which can significantly interfere with the staining process of EVs and lipophilic dyes, resulting in high contamination during sample isolation, thereby affecting the in vivo distribution. The evaluation of in vivo administration routes based on optical imaging is limited to scientific research, while the conversion of optical imaging to humans is largely hindered by the limited penetration of light. Also, EVs lack standard procedures. For example, the number of injected EVs or the labeling efficiency of EVs is not uniform in various literatures. Therefore, dose normalization is recommended for future studies in order to obtain comparisons between studies. Currently, there is almost a lack of imaging platforms dedicated to exosomal dynamic tracking. Due to the lack of an EV-specific imaging platform system, it is necessary to perform multimodal imaging methods to reveal the true fate of EVs, such as using MRI and radionuclide imaging platforms (PET or SPECT) to improve its high sensitivity. Moreover, these multimodal imaging instruments are readily available in the clinic and can be easily extended from research to clinics. It is believed that elucidating the unclear behavior and function of EVs in vivo may facilitate its translation into clinical applications. As tagging methods and tracking techniques improve, the mysteries of EVs will be unraveled.
Author contributions
QL and YL conceived and designed the manuscript. YL drew the figures. QL wrote the paper. JH, JX, and GL provided feedback on the manuscript. All authors contributed to data analysis, provided critical feedback, and proofread the final manuscript.
Funding
This work was funded by the Science and Technology Innovation Committee of Shenzhen (Nos. JCYJ20210324113401003, GJHZ20190820115203714, and JSGG20191129094218565), the Guangdong Basic and Applied Basic Research Foundation (2021A1515010985 and 2021A1515220134), the Sanming Project of Medicine (No. SZSM201612079), the Shenzhen Fund for Guangdong Provincial High Level Clinical Key Specialties (No. SZGSP013), and the Shenzhen Key Medical Discipline Construction Fund (No. SZXK042) and Natural Science Foundation of China (No. 82172463).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., and Wood, M. J. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29 (4), 341–345. doi:10.1038/nbt.1807
Bala, S., Csak, T., Momen-Heravi, F., Lippai, D., Kodys, K., Catalano, D., et al. (2015). Biodistribution and function of extracellular miRNA-155 in mice. Sci. Rep. 5, 10721. doi:10.1038/srep10721
Banerjee, A., Alves, V., Rondão, T., Sereno, J., Neves, Â., Lino, M., et al. (2019). A positron-emission tomography (PET)/magnetic resonance imaging (MRI) platform to track in vivo small extracellular vesicles. Nanoscale 11 (28), 13243–13248. doi:10.1039/c9nr02512j
Bebelman, M. P., Bun, P., Huveneers, S., van Niel, G., Pegtel, D. M., and Verweij, F. J. (2020). Real-time imaging of multivesicular body-plasma membrane fusion to quantify exosome release from single cells. Nat. Protoc. 15 (1), 102–121. doi:10.1038/s41596-019-0245-4
Betzer, O., Perets, N., Angel, A., Motiei, M., Popovtzer, R., Yadid, G., et al. (2017). In vivo neuroimaging of exosomes using gold nanoparticles. ACS Nano 11 (11), 10883–10893. doi:10.1021/acsnano.7b04495
Cao, H., Yue, Z., Gao, H., Chen, C., Cui, K., Zhang, K., et al. (2019). In vivo real-time imaging of extracellular vesicles in liver regeneration via aggregation-induced emission luminogens. ACS Nano 13 (3), 3522–3533. doi:10.1021/acsnano.8b09776
Chen, G., Zhu, J. Y., Zhang, Z. L., Zhang, W., Ren, J. G., Wu, M., et al. (2015). Transformation of cell-derived microparticles into quantum-dot-labeled nanovectors for antitumor siRNA delivery. Angew. Chem. Int. Ed. Engl. 54 (3), 1050–1054. doi:10.1002/ange.201410223
Ciullo, A., Li, C., Li, L., Ungerleider, K. C., Peck, K., Marbán, E., et al. (2022). Biodistribution of unmodified cardiosphere-derived cell extracellular vesicles using single RNA tracing. J. Extracell. Vesicles 11 (1), e12178. doi:10.1002/jev2.12178
Curley, N., Levy, D., Do, M. A., Brown, A., Stickney, Z., Marriott, G., et al. (2020). Sequential deletion of CD63 identifies topologically distinct scaffolds for surface engineering of exosomes in living human cells. Nanoscale 12 (22), 12014–12026. doi:10.1039/d0nr00362j
Deatherage, B. L., and Cookson, B. T. (2012). Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 80 (6), 1948–1957. doi:10.1128/iai.06014-11
Dehghani, M., Gulvin, S. M., Flax, J., and Gaborski, T. R. (2019). Exosome labeling by lipophilic dye PKH26 results in significant increase in vesicle size. BioRxivAvailable from: https://www.biorxiv.org/content/10.1101/532028v2.article-info (Accessed January 27, 2019).532028, doi:10.1101/532028
Dehghani, M., Gulvin, S. M., Flax, J., and Gaborski, T. R. (2020). Systematic evaluation of PKH labelling on extracellular vesicle size by nanoparticle tracking analysis. Sci. Rep. 10, 9533. doi:10.1038/s41598-020-66434-7
Ding, H., Cai, Y., Gao, L., Liang, M., Miao, B., Wu, H., et al. (2019). Exosome-like nanozyme vesicles for H 2 O 2 -responsive catalytic photoacoustic imaging of xenograft nasopharyngeal carcinoma. Nano Lett. 19 (1), 203–209. doi:10.1021/acs.nanolett.8b03709
Duan, L., Xu, L., Xu, X., Qin, Z., Zhou, X., Xiao, Y., et al. (2021). Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale 13 (3), 1387–1397. doi:10.1039/d0nr07622h
El-Andaloussi, S., Lee, Y., Lakhal-Littleton, S., Li, J., Seow, Y., Gardiner, C., et al. (2012). Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 7 (12), 2112–2126. doi:10.1038/nprot.2012.131
Elmahalaway, A. M., Elazab, E. E., Abdrabbo, M., Said, O. M., Sabry, D., and Sibaie, M. M. E. (2018). Comparative light and electron microscopic study on the therapeutic efficacy of adipose derived stem cells versus exosomes for experimentally induced acute corneal injuries in rats. J. Stem Cell. Res. Ther. 8, 6.
Faruqu, F. N., Wang, J. T., Xu, L., McNickle, L., Chong, E. M., Walters, A., et al. (2019). Membrane radiolabelling of exosomes for comparative biodistribution analysis in immunocompetent and immunodeficient mice - a novel and universal approach. Theranostics 9 (6), 1666–1682. doi:10.7150/thno.27891
Feng, K., Xie, X., Yuan, J., Gong, L., Zhu, Z., Zhang, J., et al. (2021). Reversing the surface charge of MSC-derived small extracellular vesicles by εPL-PEG-DSPE for enhanced osteoarthritis treatment. J. Extracell. Vesicles 10 (13), e12160. doi:10.1002/jev2.12160
Ferreira, J. V., da Rosa Soares, A., Ramalho, J., Máximo Carvalho, C., Cardoso, M. H., Pintado, P., et al. (2022). LAMP2A regulates the loading of proteins into exosomes. Sci. Adv. 8 (12), eabm1140. doi:10.1126/sciadv.abm1140
Fu, S., Wang, Y., Xia, X., and Zheng, J. C. (2020). Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 20, 100261. doi:10.1016/j.impact.2020.100261
Gangadaran, P., Hong, C. M., Oh, J. M., Rajendran, R. L., Kalimuthu, S., Son, S. H., et al. (2018). In vivo non-invasive imaging of radio-labeled exosome-mimetics derived from red blood cells in mice. Front. Pharmacol. 9, 817. doi:10.3389/fphar.2018.00817
Gangadaran, P., Li, X. J., Lee, H. W., Oh, J. M., Kalimuthu, S., Rajendran, R. L., et al. (2017). A new bioluminescent reporter system to study the biodistribution of systematically injected tumor-derived bioluminescent extracellular vesicles in mice. Oncotarget 8 (66), 109894–109914. doi:10.18632/oncotarget.22493
Gao, X., Ning, R., Dong, X., Zuo, B., Rong, Y., Zhou, Q., et al. (2018). Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci. Transl. Med. 10 (444), eaat0195. doi:10.1126/scitranslmed.aat0195
González, M., González-Arjona, M., Santos-Coquillat, A., Vaquero, J., Salinas, B., de Molina, A., et al. (2021). Covalently labeled fluorescent exosomes for in vitro and in vivo applications. Biomedicines 9 (1), 81. doi:10.3390/biomedicines9010081
Gupta, D., Liang, X., Pavlova, S., Wiklander, O. P. B., Corso, G., Zhao, Y., et al. (2020). Quantification of extracellular vesicles in vitro and in vivo using sensitive bioluminescence imaging. J. Extracell. Vesicles 9 (1), 1800222. doi:10.1080/20013078.2020.1800222
Gupta, D., Wiklander, O. P. B., Görgens, A., Conceição, M., Corso, G., Liang, X., et al. (2021). Amelioration of systemic inflammation via the display of two different decoy protein receptors on extracellular vesicles. Nat. Biomed. Eng. 5 (9), 1084–1098. doi:10.1038/s41551-021-00792-z
He, F., Ye, Z. Y., Zhao, L. D., Yin, B. C., and Ye, B. C. (2019). Probing exosome internalization pathways through confocal microscopy imaging. Chem. Commun. 55 (93), 14015–14018. doi:10.1039/c9cc07491k
Hessvik, N. P., and Llorente, A. (2018). Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 75 (2), 193–208. doi:10.1007/s00018-017-2595-9
Hikita, T., Miyata, M., Watanabe, R., and Oneyama, C. (2020). In vivo imaging of long-term accumulation of cancer-derived exosomes using a BRET-based reporter. Sci. Rep. 10 (1), 16616. doi:10.1038/s41598-020-73580-5
Hikita, T., Miyata, M., Watanabe, R., and Oneyama, C. (2018). Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci. Rep. 8 (1), 14035. doi:10.1038/s41598-018-32535-7
Hoen, E. N. M. N-t., Buschow, S. I., Anderton, S. M., Stoorvogel, W., and Wauben, M. H. M. (2009). Activated T-cells recruit exosomes secreted by dendritic cells via LFA-1. Blood 113 (9), 1977–1981. doi:10.1182/blood-2008-08-174094
Horgan, C. C., Nagelkerke, A., Whittaker, T. E., Nele, V., Massi, L., Kauscher, U., et al. (2020). Molecular imaging of extracellular vesicles in vitro via Raman metabolic labelling. J. Mat. Chem. B 8 (20), 4447–4459. doi:10.1039/d0tb00620c
Hoshino, A., Costa-Silva, B., Shen, T. L., Rodrigues, G., Hashimoto, A., Tesic Mark, M., et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527 (7578), 329–335. doi:10.1038/nature15756
Hu, Q., Su, H., Li, J., Christopher, L., Tang, W., Wan, M., et al. (2020). Clinical applications of exosome membrane proteins. Precis. Clin. Med. 3 (1), 54–66. doi:10.1093/pcmedi/pbaa007
Huang, X., Guo, Q., Zhang, R., Zhao, Z., Leng, Y., Lam, J. W. Y., et al. (2020). AIEgens: An emerging fluorescent sensing tool to aid food safety and quality control. Compr. Rev. food Sci. food Saf. 19 (4), 2297–2329. doi:10.1111/1541-4337.12591
Illes, B., Hirschle, P., Barnert, S., Cauda, V., Wuttke, S., and Engelke, H. (2017). Exosome-coated metal-organic framework nanoparticles: An efficient drug delivery platform. Chem. Mat. 29, 8042–8046. acs.chemmater.7b02358. doi:10.1021/acs.chemmater.7b02358
Jang, Y., Kim, H., Yoon, S., Lee, H., Kim, H., Jung, J., et al. (2020). Exosome-based photoacoustic imaging guided photodynamic and immunotherapy for the treatment of pancreatic cancer. J. Control. Release 330, 293–304. doi:10.1016/j.jconrel.2020.12.039
Jiang, X., Zong, S., Chen, Z., Yizhi, W., Cui, Z., and Cui, Y. (2018). Gold-carbon dots for the intracellular imaging of cancer-derived exosomes. Nanotechnology 29, 175701. doi:10.1088/1361-6528/aaaf14
Jo, W., Kim, J., Yoon, J., Jeong, D., Cho, S., Jeong, H., et al. (2014). Large-scale generation of cell-derived nanovesicles. Nanoscale 6 (20), 12056–12064. doi:10.1039/c4nr02391a
Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L., and Turbide, C. (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262 (19), 9412–9420. doi:10.1016/s0021-9258(18)48095-7
Khan, A. A., and , de Rosales, T. M. (2021). Radiolabelling of extracellular vesicles for PET and SPECT imaging. Nanotheranostics 5 (3), 256–274. doi:10.7150/ntno.51676
Kim, D. H., Kothandan, V. K., Kim, H. W., Kim, K. S., Kim, J. Y., Cho, H. J., et al. (2019). Noninvasive assessment of exosome pharmacokinetics in vivo: A review. Pharmaceutics 11 (12), 649. doi:10.3390/pharmaceutics11120649
Kim, D. K., Nishida, H., An, S. Y., Shetty, A. K., Bartosh, T. J., and Prockop, D. J. (2016). Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc. Natl. Acad. Sci. U. S. A. 113 (1), 170–175. doi:10.1073/pnas.1522297113
Kowalski, M. P., and Krude, T. (2015). Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell. Biol. 66, 20–29. doi:10.1016/j.biocel.2015.07.003
Kuo, W. P., Tigges, J. C., Toxavidis, V., and Ghiran, I. (2017). Red blood cells: A source of extracellular vesicles. Methods Mol. Biol. 1660, 15–22. doi:10.1007/978-1-4939-7253-1_2
Lai, C. P., Kim, E. Y., Badr, C. E., Weissleder, R., Mempel, T. R., Tannous, B. A., et al. (2015). Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 6, 7029. doi:10.1038/ncomms8029
Lee, T. S., Kim, Y., Zhang, W., Song, I. H., and Tung, C. H. (2018). Facile metabolic glycan labeling strategy for exosome tracking. Biochimica Biophysica Acta - General Subj. 1862 (5), 1091–1100. doi:10.1016/j.bbagen.2018.02.001
Liang, Y., Duan, L., Lu, J., and Xia, J. (2021). Engineering exosomes for targeted drug delivery. Theranostics 11 (7), 3183–3195. doi:10.7150/thno.52570
Liang, Y., Duan, L., Xu, X., Li, X., Liu, M., Chen, H., et al. (2020). Mesenchymal stem cell-derived exosomes for treatment of autism spectrum disorder. ACS Appl. Bio Mat. 3 (9), 6384–6393. doi:10.1021/acsabm.0c00831
Liang, Y., Xu, X., Li, X., Xiong, J., Li, B., Duan, L., et al. (2020). Chondrocyte-targeted MicroRNA delivery by engineered exosomes toward a cell-free osteoarthritis therapy. ACS Appl. Mat. Interfaces 12 (33), 36938–36947. doi:10.1021/acsami.0c10458
Liu, J., Jiang, X., Ashley, C., and Brinker, C. J. (2009). Electrostatically mediated liposome fusion and lipid exchange with a nanoparticle-supported bilayer for control of surface charge, drug containment, and delivery. J. Am. Chem. Soc. 131 (22), 7567–7569. doi:10.1021/ja902039y
Liu, J., Wu, X., Lu, J., Huang, G., Zhang, G., Zhang, H., et al. (2021). Exosomal transfer of osteoclast-derived miRNAs to chondrocytes contributes to osteoarthritis progression. Nat. Aging 1 (4), 368–384. doi:10.1038/s43587-021-00050-6
Logozzi, M., Milito, A. D., Lugini, L., Borghi, M., Calabrò, L., Spada, M., et al. (2009). High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. Plos One 4 (4), e5219–e. doi:10.1371/journal.pone.0005219
Lu, K., Vu, C. Q., Matsuda, T., and Nagai, T. (2019). Fluorescent protein-based indicators for functional super-resolution imaging of biomolecular activities in living cells. Int. J. Mol. Sci. 20 (22), 5784. doi:10.3390/ijms20225784
Lu, Y., Eguchi, T., Sogawa, C., Taha, E. A., Tran, M. T., Nara, T., et al. (2021). Exosome-based molecular transfer activity of macrophage-like cells involves viability of oral carcinoma cells: Size exclusion chromatography and concentration filter method. Cells 10 (6), 1328. doi:10.3390/cells10061328
Lv, Y., Tan, J., Miao, Y., and Zhang, Q. (2019). The role of microvesicles and its active molecules in regulating cellular biology. J. Cell. Mol. Med. 23 (12), 7894–7904. doi:10.1111/jcmm.14667
Marlin, S. D., and Springer, T. A. (1987). Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1). Cell. 51 (5), 813–819. doi:10.1016/0092-8674(87)90104-8
McDaniel, K., Correa, R., Zhou, T., Johnson, C., Francis, H., Glaser, S., et al. (2013). Functional role of microvesicles in gastrointestinal malignancies. Ann. Transl. Med. 1 (1), 4. doi:10.3978/j.issn.2305-5839.2012.10.01
Molavipordanjani, S., Khodashenas, S., Abedi, S. M., Moghadam, M. F., and Hosseinimehr, S. J. (2020). 99mTc-radiolabeled HER2 targeted exosome for tumor imaging. Eur. J. Pharm. Sci. 148, 105312. doi:10.1016/j.ejps.2020.105312
Monopoli, M. P., Zendrini, A., Wu, D., Cheung, S., Sampedro, G., Ffrench, B., et al. (2018). Endogenous exosome labelling with an amphiphilic NIR-fluorescent probe. Chem. Commun. 54 (52), 7219–7222. doi:10.1039/c8cc02135j
Mulcahy, L. A., Pink, R. C., and Carter, D. (2014). Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3 (1), 24641. doi:10.3402/jev.v3.24641
Nakazaki, M., Morita, T., Lankford, K. L., Askenase, P. W., and Kocsis, J. D. (2021). Small extracellular vesicles released by infused mesenchymal stromal cells target M2 macrophages and promote TGF-β upregulation, microvascular stabilization and functional recovery in a rodent model of severe spinal cord injury. J. Extracell. Vesicles 10 (11), e12137. doi:10.1002/jev2.12137
Ntziachristos, V., Bremer, C., and Weissleder, R. (2003). Fluorescence imaging with near-infrared light: New technological advances that enable in vivo molecular imaging. Eur. Radiol. 13 (1), 195–208. doi:10.1007/s00330-002-1524-x
Ohno, S., Takanashi, M., Sudo, K., Ueda, S., Ishikawa, A., Matsuyama, N., et al. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 21 (1), 185–191. doi:10.1038/mt.2012.180
Park, J., Hwang, M., Choi, B., Jeong, H., Jung, J. H., Kim, H. K., et al. (2017). Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal. Chem. 89 (12), 6695–6701. doi:10.1021/acs.analchem.7b00911
Progatzky, F., Dallman, M. J., and Lo Celso, C. (2013). From seeing to believing: Labelling strategies for in vivo cell-tracking experiments. Interface Focus 3 (3), 20130001. doi:10.1098/rsfs.2013.0001
Pužar Dominkuš, P., Stenovec, M., Sitar, S., Lasič, E., Zorec, R., Plemenitaš, A., et al. (2018). PKH26 labeling of extracellular vesicles: Characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochimica Biophysica Acta - Biomembr. 1860 (6), 1350–1361. doi:10.1016/j.bbamem.2018.03.013
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell. Biol. 200 (4), 373–383. doi:10.1083/jcb.201211138
Salunkhe, S., Dheeraj, B. M., Chitkara, D., and Mittal, A. (2020). Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. J. Control. Release 326 (10), 599–614. doi:10.1016/j.jconrel.2020.07.042
Shi, C., Wu, J. B., and Pan, D. (2016). Review on near-infrared heptamethine cyanine dyes as theranostic agents for tumor imaging, targeting, and photodynamic therapy. J. Biomed. Opt. 21 (5), 050901. doi:10.1117/1.jbo.21.5.050901
Pan, S., Pei, L., Zhang, A., Zhang, Y., Zhang, C., Huang, M., et al. (2020). Passion fruit-like exosome-PMA/Au-BSA@Ce6 nanovehicles for real-time fluorescence imaging and enhanced targeted photodynamic therapy with deep penetration and superior retention behavior in tumor. Biomaterials 230, 119606. doi:10.1016/j.biomaterials.2019.119606
Sun, W., Li, Z., Zhou, X., Yang, G., and Yuan, L. (2019). Efficient exosome delivery in refractory tissues assisted by ultrasound-targeted microbubble destruction. Drug Deliv. 26 (1), 45–50. doi:10.1080/10717544.2018.1534898
Sung, B. H., Ketova, T., Hoshino, D., Zijlstra, A., and Weaver, A. M. (2015). Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 6, 7164. doi:10.1038/ncomms8164
Sung, B. H., von Lersner, A., Guerrero, J., Krystofiak, E. S., Inman, D., Pelletier, R., et al. (2020). A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun. 11 (1), 2092. doi:10.1038/s41467-020-15747-2
Svenson, S. (2013). Theranostics: Are we there yet? Mol. Pharm. 10 (3), 848–856. doi:10.1021/mp300644n
Takahashi, Y., Nishikawa, M., Shinotsuka, H., Matsui, Y., Ohara, S., Imai, T., et al. (2013). Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol. 165 (2), 77–84. doi:10.1016/j.jbiotec.2013.03.013
Theodoraki, M., Hong, C., Donnenberg, V. S., Donnenberg, A. D., and Whiteside, T. L. (2020). Evaluation of exosome proteins by on-bead flow cytometry. Cytom. A 99 (4), 372–381. doi:10.1002/cyto.a.24193
Théry, C., Amigorena, S., Raposo, G., and Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell. Biol. 30 (1), 322. doi:10.1002/0471143030.cb0322s30
Thery, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7 (1), 1461450. doi:10.1080/20013078.2018.1461450
Tian, T., Zhu, Y. L., Hu, F. H., Wang, Y. Y., Huang, N. P., and Xiao, Z. D. (2013). Dynamics of exosome internalization and trafficking. J. Cell. Physiol. 228 (7), 1487–1495. doi:10.1002/jcp.24304
Todorova, D., Simoncini, S., Lacroix, R., Sabatier, F., and Dignat-George, F. (2017). Extracellular vesicles in angiogenesis. Circ. Res. 120 (10), 1658–1673. doi:10.1161/circresaha.117.309681
Valadi, H., EkstroM, K., Bossios, A., SjoStrand, M., Lee, J. J., and LoTvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 9 (6), 654–659. doi:10.1038/ncb1596
Verweij, F. J., Bebelman, M. P., Jimenez, C. R., Garcia-Vallejo, J. J., Janssen, H., Neefjes, J., et al. (2018). Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J. Cell. Biol. 217 (3), 1129–1142. doi:10.1083/jcb.201703206
Verweij, F. J., Revenu, C., Arras, G., Dingli, F., Loew, D., Pegtel, D. M., et al. (2019). Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev. Cell. 48 (4), 573–589.e4. e4. doi:10.1016/j.devcel.2019.01.004
Wang, H., La Russa, M., and Qi, L. S. (2016). CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264. doi:10.1146/annurev-biochem-060815-014607
Wang, M., Altinoglu, S., Takeda, Y. S., and Xu, Q. (2015). Integrating protein engineering and bioorthogonal click conjugation for extracellular vesicle modulation and intracellular delivery. Plos One 10 (11), e0141860. doi:10.1371/journal.pone.0141860
Wang, X., Yao, X., Xie, T., Chang, Z., Guo, Y., and Ni, H. (2020). Exosome-derived uterine miR-218 isolated from cows with endometritis regulates the release of cytokines and chemokines. Microb. Biotechnol. 13 (4), 1103–1117. doi:10.1111/1751-7915.13565
Wei, H., Chen, Q., Lin, L., Sha, C., Zhu, X., Liu, Y., et al. (2021). Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 17 (1), 163–177. doi:10.7150/ijbs.53671
Wiklander, O. P., Nordin, J. Z., O'Loughlin, A., Gustafsson, Y., Corso, G., Mäger, I., et al. (2015). Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 4, 26316. doi:10.3402/jev.v4.26316
Xhy, A., Hj, B., Ccw, A., Yang, L. A., Yhl, A., Chen, Z. A., et al. (2021). Photocatalysis-activated SR-AOP over PDINH/MIL-88A(Fe) composites for boosted chloroquine phosphate degradation: Performance, mechanism, pathway and DFT calculations. Appl. Catal. B Environ. 293, 120229. doi:10.1016/j.apcatb.2021.120229
Xi, X. M., Xia, S. J., and Lu, R. (2021). Drug loading techniques for exosome-based drug delivery systems. Pharmazie 76 (2), 61–67. doi:10.1691/ph.2021.0128
Xu, R., Bai, Y., Min, S., Xu, X., and Ju, S. (2020). <p>in vivo monitoring and assessment of exogenous mesenchymal stem cell-derived exosomes in mice with ischemic stroke by molecular imaging</p>. Int. J. Nanomedicine 15, 9011–9023. doi:10.2147/ijn.s271519
Ye, Y., Shi, Q., Yang, T., Xie, F., Zhang, X., Xu, B., et al. (2022). In vivo visualized tracking of tumor-derived extracellular vesicles using CRISPR-cas9 system. Technol. Cancer Res. Treat. 21, 153303382210853. doi:10.1177/15330338221085370
Ye, Y., Zhang, X., Xie, F., Xu, B., Xie, P., Yang, T., et al. (2020). An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 8 (10), 2966–2976. doi:10.1039/d0bm00427h
Yi, Y. W., Lee, J. H., Kim, S. Y., Pack, C. G., Ha, D. H., Park, S. R., et al. (2020). Advances in analysis of biodistribution of exosomes by molecular imaging. Int. J. Mol. Sci. 21 (2), 665. doi:10.3390/ijms21020665
Zhang, M., Vojtech, L., Ye, Z., Hladik, F., and Nance, E. (2020). Quantum dot labeling and visualization of extracellular vesicles. ACS Appl. Nano Mat. 3 (7), 7211–7222. doi:10.1021/acsanm.0c01553
Zhao, J. Y., Chen, G., Gu, Y. P., Cui, R., Zhang, Z. L., Yu, Z. L., et al. (2016). Ultrasmall magnetically engineered Ag2Se quantum dots for instant efficient labeling and whole-body high-resolution multimodal real-time tracking of cell-derived microvesicles. J. Am. Chem. Soc. 138 (6), 1893–1903. doi:10.1021/jacs.5b10340
Zhu, L., Xu, Y., Wei, X., Lin, H., Huang, M., Lin, B., et al. (2021). Coupling aptamer-based protein tagging with metabolic glycan labeling for in situ visualization and biological function study of exosomal protein-specific glycosylation. Angew. Chem. Int. Ed. Engl. 60 (33), 18259–18263. doi:10.1002/ange.202103696
Zomer, A., Maynard, C., Verweij, F. J., Kamermans, A., Schäfer, R., Beerling, E., et al. (2015). In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 161 (5), 1046–1057. doi:10.1016/j.cell.2015.04.042
Keywords: exosomes, extracellular vesicles, exosome labeling, exosome tracking, in vivo imaging
Citation: Liu Q, Huang J, Xia J, Liang Y and Li G (2022) Tracking tools of extracellular vesicles for biomedical research. Front. Bioeng. Biotechnol. 10:943712. doi: 10.3389/fbioe.2022.943712
Received: 14 May 2022; Accepted: 03 November 2022;
Published: 18 November 2022.
Edited by:
Principia Dardano, Consiglio Nazionale delle Ricerche, Napoli, ItalyReviewed by:
Annarita Falanga, Huazhong University of Science and Technology, Wuhan, ChinaDawei Jiang, Huazhong University of Science and Technology, Wuhan, China
Copyright © 2022 Liu, Huang, Xia, Liang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujie Liang, bGlhbmd5amllQDEyNi5jb20=; Guangheng Li, bGlndWFuZ2hlbmdAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Qisong Liu
Qisong Liu Jianghong Huang2,3†
Jianghong Huang2,3† Jiang Xia
Jiang Xia Yujie Liang
Yujie Liang Guangheng Li
Guangheng Li