- 1Department of Chemical, Biological and Environmental Engineering, Universitat Autònoma de Barcelona, Bellaterra (Cerdanyola del Vallès), Catalonia, Spain
- 2TBI, Université de Toulouse, CNRS, INRAE, INSA, Toulouse, France
Production of 3-hydroxypropionic acid (3-HP) in Pichia pastoris (syn. Komagataella phaffii) via the malonyl-CoA pathway has been recently demonstrated using glycerol as a carbon source, but the reported metrics were not commercially relevant. The flux through the heterologous pathway from malonyl-CoA to 3-HP was hypothesized as the main bottleneck. In the present study, different metabolic engineering approaches have been combined to improve the productivity of the original 3-HP producing strains. To do so, an additional copy of the gene encoding for the potential rate-limiting step of the pathway, i.e., the C-terminal domain of the malonyl-CoA reductase, was introduced. In addition, a variant of the endogenous acetyl-CoA carboxylase (ACC1S1132A) was overexpressed with the aim to increase the delivery of malonyl-CoA. Furthermore, the genes encoding for the pyruvate decarboxylase, aldehyde dehydrogenase and acetyl-CoA synthase, respectively, were overexpressed to enhance conversion of pyruvate into cytosolic acetyl-CoA, and the main gene responsible for the production of the by-product D-arabitol was deleted. Three different screening conditions were used to classify the performance of the different strains: 24-deep-well plates batch cultures, small-scale cultures in falcon tubes using FeedBeads® (i.e., slow release of glycerol over time), and mini bioreactor batch cultures. The best two strains from the FeedBeads® screening, PpHP8 and PpHP18, were tested in bioreactor fed-batch cultures using a pre-fixed exponentially increasing feeding rate. The strain PpHP18 produced up to 37.05 g L−1 of 3-HP at 0.712 g L−1 h−1 with a final product yield on glycerol of 0.194 Cmol−1 in fed-batch cultures. Remarkably, PpHP18 did not rank among the 2-top producer strains in small scale batch cultivations in deep-well plates and mini bioreactors, highlighting the importance of multiplexed screening conditions for adequate assessment of metabolic engineering strategies. These results represent a 50% increase in the product yield and final concentration, as well as over 30% increase in volumetric productivity compared to the previously obtained metrics for P. pastoris. Overall, the combination of glycerol as carbon source and a metabolically engineered P. pastoris strain resulted in the highest 3-HP concentration and productivity reported so far in yeast.
Introduction
Microbial production of platform chemicals is an increasingly attractive alternative to petrochemical-derived products. One such chemical, 3-hydroxypropionic acid (3-HP), is an organic acid that can be chemically converted to acrylic acid, 1,3-propanediol, and malonic acid, among others (della Pina et al., 2011), offering a great potential for bioproduction of a wide range of polymers. In fact, 3-HP has been included in the list of the top platform chemicals that can be obtained from biomass (Werpy and Petersen, 2004).
3-HP can be naturally produced by several bacteria through different biosynthetic routes. Among them, the glycerol-dependant pathway, and the pathways via malonyl-CoA and β-alanine intermediates have been the most extensively investigated (Kumar et al., 2013; de Fouchécour et al., 2018). Multiple microorganisms have been engineered to produce 3-HP, including bacteria (Jers et al., 2019) and yeast (Ji et al., 2018). The highest 3-HP concentrations and productivities achieved so far have been obtained in Escherichia coli [76.2 g L−1 of 3-HP and 1.89 g L−1 h−1, respectively (Kim et al., 2020)] and Klebsiella pneumoniae (Zhao et al., 2019). Both bacterial species were engineered to produce 3-HP through the glycerol-dependent pathway. This facilitated the use of glycerol as a carbon source and, ultimately, of crude glycerol, which is an abundant, inexpensive and renewable feedstock produced as a main by-product of the conventional biodiesel production process. Morevover, glycerol is very attractive for production of organic acids such as 3-HP due to its higher degree of reduction compared to glucose.
Production of 3-HP in yeast has been mainly pursued by introducing the malonyl-CoA or β-alanine pathways, e.g., reaching a titer of about 13.7 g L−1 and with a 0.14 ± 0.0 C-mol−1 yield on glucose in Saccharomyces cerevisiae fed-batch cultures (Borodina et al., 2015). The methylotrophic yeast Pichia pastoris (syn. Komagataella phaffii) is able to grow efficiently on glycerol. Furthermore, crude glycerol may contain from 1% to 25% w/w methanol (Luo et al., 2016), making this feedstock ideal for a methylotrophic microorganism such as P. pastoris. In addition, this yeast can grow at a pH as low as 3 (Cregg et al., 1993; Werten et al., 1999), which allows performing the fermentation process at a pH below the pKa of 3-HP (4.51). This would enable the use of in situ product recovery systems, thereby avoiding 3-HP accumulation to toxic levels and facilitating 3-HP export from cells (van Maris et al., 2004). The 3-HP extraction yield using a membrane-assisted in situ product recovery system was increased from 5% to 74% when the pH of the medium was decreased from 5 to 3.2 (Chemarin et al., 2019).
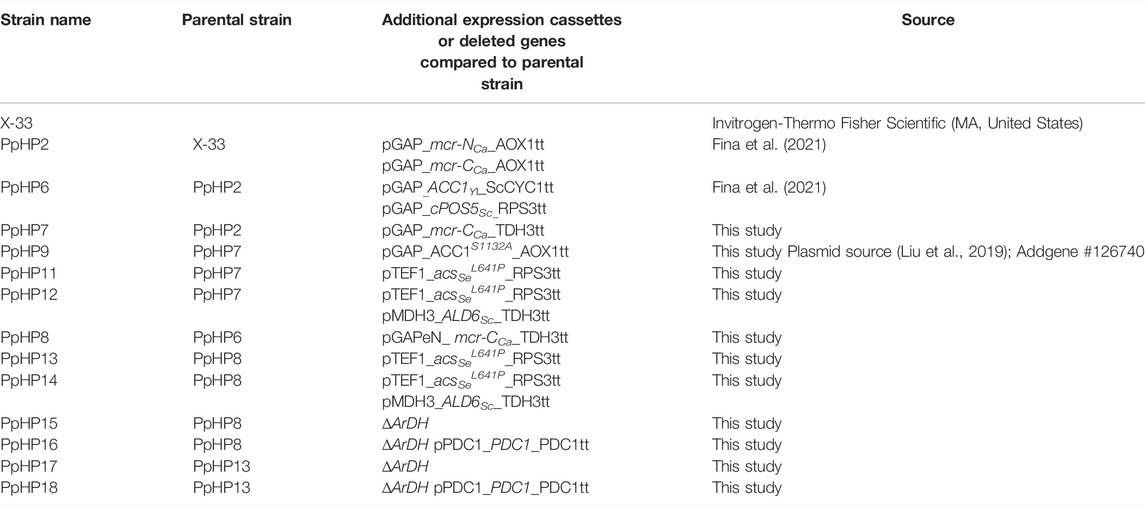
In a recent study, we addressed the use of P. pastoris to produce 3-HP from glycerol (Fina et al., 2021). This yeast was metabolically engineered to express the malonyl-CoA reductase pathway, which consists of two consecutive steps reducing malonyl-CoA into 3-HP, while consuming two NADPH molecules (Figure 1). Specifically, we expressed the gene encoding for the bi-functional enzyme malonyl-CoA reductase (MCR) from Chloroflexus aurantiacus. This pathway provides the simplest way to produce 3-HP from glycerol in P. pastoris, as the glycerol-dependant pathway is coenzyme B12-dependant. Since P. pastoris does not naturally produce coenzyme B-12, the introduction of the glycerol-dependant pathway for 3-HP biosynthesis pathway in this cell factory would require either the addition of such expensive cofactor to the fermentation medium or the co-expression of the multistep B-12 biosynthetic pathway (Fang et al., 2018).
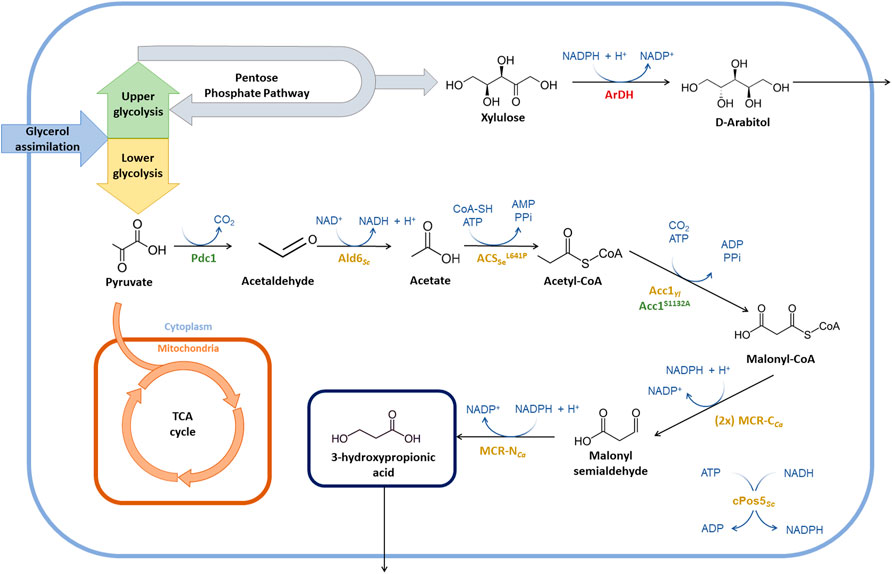
FIGURE 1. Summary of metabolic engineering strategies for the optimisation of 3-HP production in P. pastoris. Strain engineering started from the 3-HP producing strain PpHP6 containing the malonyl-CoA to 3-HP pathway (Fina et al., 2021). Heterologous enzymes are depicted in yellow, endogenous enzymes being overexpressed are shown in green, and the enzymes catalyzing the reactions of genes being deleted are shown in red. The (2x) indicates that a second copy of the MCR-CCa gene was introduced. Enzyme abbreviations: Pdc1, pyruvate decarboxylase; Ald6Sc, aldehyde dehydrogenase from S. cerevisiae; ACSSeL641P, acetyl-CoA synthase from S. enterica with the mutation L641P; Acc1Yl, acetyl-CoA carboxylase from Y. lipolytica; Acc1S1132A, acetyl-CoA carboxylase from P. pastoris with the mutation S1132A; MCR-CCa, C-terminal domain of the malonyl-CoA reductase from C. aurantiacus; MCR-NCa, N-terminal domain of the malonyl-CoA reductase from C. aurantiacus; cPos5Sc, NADH kinase from S. cerevisiae relocated into the cytoplasm; ArDH, D-arabitol dehydrogenase.
In addition, several improvements were made to the original strain. First, the independent expression of the two MCR subunits was implemented, leading to a 12.5-fold increase in 3-HP production. Second, the resulting strain was further engineered to increase the availability of NADPH and malonyl-CoA, which are the two substrates of this route. To this end, the acetyl-CoA carboxylase gene from Yarrowia lipolytica (ACC1Yl) and a NADH kinase from Saccharomyces cerevisiae (cPOS5Sc) were expressed in the cytosol. However, this strategy resulted in rather modest improvements in terms of product yield, pointing to the delivery of malonyl-CoA as a major bottleneck to further improve 3-HP production. Notably, this strain produced up to 24.75 g L−1 of 3-HP in 45.5 h in a fed-batch culture using glycerol as a carbon source, which is, to our knowledge, the highest productivity reported so far in yeast (0.54 g L−1 h−1) (Fina et al., 2021).
Despite providing the first demonstration of the potential of P. pastoris for 3-HP production from glycerol, 3-HP production metrics from our previous study are still far from being industrially-relevant, as a minimum productivity of 2.5 g L−1 h−1, 50∼100 g L−1 and >0.5 g/g yield are generally required to achieve an economically feasible bioprocess for carboxylic acid production (Werpy and Petersen, 2004; Mazzoli, 2021).
Here, further metabolic engineering of the previously developed 3-HP-producing strains (Fina et al., 2021) was performed to increase the production metrics. In particular, we aimed at improving both the biosynthetic pathway for 3-HP from its metabolic precursor (acetyl-CoA), as well as improving the metabolic flux towards acetyl-CoA synthesis and reducing by-product (arabitol) formation. Notably, we used three different small-scale cultivation systems for better characterisation of the impact of genetic modifications on cell growth and product yields and subsequent transfer to bioreactor-scale cultivations.
Materials and Methods
Molecular Biology and Strains
All the strains used in this study are listed in Table 1. A detailed description of the molecular biology workflow to generate each strain can be found in the Supplementary Material S1. The modular cloning vectors GoldenPics [(Prielhofer et al., 2017); Addgene Kit #1000000133] and the vectors for CRISPR-Cas9 of the CrisPi kit (Gassler et al., 2019); Addgene Kit #1000000136) were used.
The strains generated using the GoldenPics plasmids were generated by integration of the heterologous DNA at the loci described in the Supplementary Material S1.
For the new strains generated by CRISPR-Cas9, the integration cassette was amplified from P. pastoris genome using a high-fidelity Q5 polymerase (New England Biolabs, MA, United States) and the recommended protocol (Gassler et al., 2019). The integration loci are also indicated in the Supplementary Material S1. The integrity of the inserted cassette was verified by Sanger sequencing at the Genomics and Bioinformatics Service of the Universitat Autònoma de Barcelona.
Copy Number Determination by Droplet Digital PCR
The number of copies of the gene mcr-CCa of the highest producing clones of the strains PpHP7 and PpHP8 was tested with droplet digital PCR (ddPCR) following a previously reported method (Cámara et al., 2016), except that the QX200TM ddPCRTM EvaGreen Supermix (Biorad, CA, United States) was used instead of Taqman probes. First, the genomic DNA of the highest producing clones was purified using the Wizard® Genomic DNA Purification Kit from Promega (WI, United States). Afterwards, 0.5 µg of genomic DNA were restricted using the restriction enzymes BamHI-FD and EcoRI-FD from Thermo Fisher Scientific (MA, United States). Subsequently, the genomic DNA was diluted to a concentration of 1 ng μL−1.
Second, a master mix of 22.5 µL was prepared with the forward primer at 0.4 µM, the reverse primer at 0.2 µM, and the restricted genomic DNA at 0.08 ng μL−1. Afterwards, the master mix was mixed with 22.5 µL of EvaGreen 2X master solution and was thoroughly mixed by vortexing. Following droplet generation using a Droplet generator (Biorad, CA, United States), the droplets were transferred to a 96-tubes rack and subsequently sealed with a PCR Plate Heat Seal. The PCR was carried using a C1000 Touch Thermal Cycler (Biorad, CA, United States). The annealing temperature was set to 60°C. Finally, the ddPCR results were analysed using a QX200 Droplet Digital PCR system (Biorad, CA, United States).
The primers used for ddPCR are listed in Table 2. The primer pairs ddPCR_mcrCCa_FW and ddPCR_mcrCCa_RV (Table 2) were used to amplify the mcr-CCa gene. Primers ddPCR_act_FW and ddPCR_act_RV were used to amplify the β-actin (ACT1) endogenous gene, which was used as reference of a single-copy gene.
24-Deep-Well Plates Screening
P. pastoris strains were inoculated into 50 mL falcon tubes containing 5 mL of YPG (1% yeast extract, 2% peptone and 1% v/v glycerol) supplemented with 100 μg mL−1 zeocin (InvivoGen, CA, United States). The cells were grown overnight at 30°C and 200 rpm in an incubator shaker Multitron Standard (Infors HT, Bottmingen, Switzerland) with a 2.5 cm orbit. 50 μL of overnight-grown cultures were used to inoculate each well of a 24 deep-well plate containing 2 mL of Buffered Minimal Glycerol (BMG) medium, containing 100 mM potassium phosphate buffer pH 6, 1.34% yeast nitrogen base (YNB), 1% v/v glycerol, and 0.4 mg L−1 biotin. The cultures were incubated at 28°C and 220 rpm in the same incubator shaker using a platform with a slope of 20° to improve the aeration. The cultures were grown for 48 h to ensure the full consumption of the substrate. Thereafter, culture samples were centrifuged at 12,000 g for 5 min and filtered through a 0.20 µm pore size syringe filter (SLLGX13NK, Merck Millipore, CA, United States). The 3-HP was quantified using HPLC.
The parental clones PpHP2 and PpHP6 were screened in triplicates. Six to eight clones were screened in triplicates for the strains generated by single-homology integration (PpHP7, PpHP8, and PpHP9). One single clone was screened in triplicates for the strains generated using CRISPR-Cas9.
Small-Scale Screening in Falcon Tubes Using FeedBeads®
The inoculum was prepared following the same protocol described for the deep-well plates screenings. Afterwards, 50-mL falcon tubes were filled with 5 mL of Buffer Minimal medium (BM; 100 mM potassium phosphate buffer pH 6, 1.34% YNB and 0.4 mg L−1 biotin), supplemented with one Glycerol FeedBeads® (SMFB12001, Kuhner Shaker GmbH, Germany). This FeedBead® releases 40 mg of glycerol in 48 h. The cultures were inoculated with 50 µL of the overnight saturated cultures. The falcon tubes were incubated in an incubator shaker at 200 rpm and 30°C for 48 h. Each clone was tested in triplicate. A triplicate control was performed by adding one FeedBead® to a falcon with 5 mL of BM medium. These controls were used to determine the actual release of glycerol under the tested conditions.
Culture samples were centrifuged at 12,000 g for 5 min and filtered through a 0.20 µm pore size syringe filter (SLLGX13NK, Merck Millipore). The 3-HP was quantified using HPLC as described below.
Mini Bioreactors Screening
The automated cultivation and sampling platform described elsewhere was used (Heux et al., 2014).
The bioreactor medium contained 2.5 g L−1 glycerol, 1.8 g L−1 citric acid, 0.02 g L−1 CaCl2 · 2 H2O, 12.6 g L−1 (NH4)2HPO4, 0.5 g L−1 MgSO4 · 7 H2O, 0.9 g L−1 KCl, 50 μL antifoam Glanapon 2000 kz (Bussetti and Co., GmbH, Vienna, Austria), 0.4 mg L−1 biotin and 4.6 mL L−1 of PTM1 trace salts (Maurer et al., 2006). The pH was set to 5 using 1 M HCl. The medium was autoclaved without the trace salts and the biotin, which were filter sterilized and added to the medium under sterile conditions. Each mini bioreactor was filled with 15 mL of medium. The pre-inoculum was prepared as described for the other two screening methods (deep-well plates and falcon tubes with FeedBeads®). The overnight-saturated cultures were used to inoculate 250 mL shake flasks with 25 mL of YPG at a starting OD600 of 0.5–1.5. The cells were grown for 8 h at 28°C and 160 rpm in a shaker incubator to ensure the cells were at the exponential phase. Each bioreactor was inoculated at a starting OD600 of 0.025. Each strain was tested in triplicate.
The temperature of the bioreactors was set to 28°C. The dissolved oxygen (DO) and the pH were monitored throughout the cell culture. Samples were automatically withdrawn from the bioreactors every 2 h for the first 16 h of culture to monitor the OD600. After 16 h, samples were withdrawn hourly during 8 h for both OD600 monitoring and supernatant analysis. The 250 µL samples for culture supernatant analysis were automatically placed on 96-well plates with a 0.45 µm pore size filter bottom. The samples were filtered by applying vacuum. Glycerol, 3-HP, and D-arabitol were quantified using NMR as described below.
The growth rate (µmax), the specific substrate consumption rate (qSGlyc), and the specific 3-HP and D-arabitol production rates (qP3HP and qPAbt) were calculated during the exponential growth phase using the R program PhysioFit (Peiro et al., 2019).
Fed-Batch Cultures in Bioreactors
The cultures were carried out using a DASGIP Parallel Bioreactor System (Eppendorf, Germany). The starting volume of each 1.3 L reactor vessel was 400 mL. The Batch Medium described for the mini bioreactors with 40 g L−1 of glycerol was used. All the medium components except the biotin and the trace salts were mixed, placed into the reactor, and autoclaved. Biotin and trace salts were added through the septum port after autoclaving the reactor. The pH was controlled at 5 using 15% ammonia (only base addition was used). Aeration was set to 1 vvm (0.4 L min−1). The DO was set to 30%, and it was automatically controlled using the following cascade: 1) Increasing the stirring rate from 400 to 1,200 rpm; 2) Compressed air was mixed with pure oxygen to increase the percentage of oxygen of the inlet gas. The reactors were inoculated at a starting OD600 of 1. The inocula were prepared as described elsewhere (Garcia-Ortega et al., 2013).
The feeding medium composition was 400 g L−1 glycerol, 10 g L−1 KCl, 6.45 g L−1 MgSO4 · 7 H2O, 0.35 g L−1 CaCl2 · 2 H2O, 0.2 mL L−1 antifoam Glanapon 2,000 kz, 1.2 mg L−1 biotin and 15 mL L−1 PTM1 trace salts. All the feeding medium components except the biotin and the trace salts were mixed and autoclaved. Biotin and trace salts were filter-sterilized and added to the feeding medium under sterile conditions. Feeding medium was added to the bioreactor using the exponentially increasing feed-rate described by Eq. 1, where F(t) is the feeding rate at every time, t0 is the time where the feeding starts (end of the batch phase), X (t0) is the biomass concentration at the end of the batch phase, V (t0) is the starting volume of the reactor (the volume for the batch phase),
Therefore, the flow rate of the feeding medium was set using Eq. 3.
The feeding started once the DO increased above 60%, indicating that all the substrate of the batch phase had been consumed. Samples were collected for biomass cell dry weight (CDW) determination and supernatant analysis. The CDW was quantified in triplicates. From 0.5 to 2 mL of culture were filtered through pre-weighted glass microfiber filters (APFF04700, Merck Millipore). The filters were then washed with 10 mL of distilled water with 9 g L−1 NaCl and dried overnight at 105°C. The filters containing the dry biomass were weighted to calculate the CDW.
Culture samples were centrifuged 5 min at 12,000 g and the supernatant was filtered with a 0.2 µm pore size syringe filter (SLLGX13NK, Merck Millipore). The filtered supernatant was analysed with HPLC for glycerol, 3-HP, and D-arabitol quantification.
HPLC Analysis
HPLC was used to quantify the glycerol, the D-arabitol, and the 3-HP from the supernatant of the deep-well plates, the FeedBeads® screening, and the fed-batch bioreactor samples. A previously described HPLC protocol was used (Fina et al., 2021). The 3-HP was quantified from the UV spectrum. Glycerol and D-arabitol were quantified from the Refractive Index (RI) spectrum. As 3-HP and glycerol have the same retention time and 3-HP is also detected on the RI detector, the area corresponding to the 3-HP (previously quantified from the UV signal) was subtracted to latter calculate the actual glycerol concentration.
NMR Analysis
Glycerol, D-arabitol, and 3-HP were quantified from the culture supernatants of the mini bioreactor cultures using 1D-1H Nuclear Magnetic Resonance (NMR) on a Bruker Advance III 800 MHz spectrometer (Bruker BioSpin, Germany). Prior to the analyses, 180 µL of filtered culture supernatant samples were mixed with 20 µL of 10 mM TSP (3-(trimethylsilyl)-[2,2,3,3-2H4]-propionic acid sodium salt), which was used as an internal standard. The NMR spectrometer was coupled to a 5 mm CQPI cryoprobe, which was set to 280 K. A 30° presaturation pulse was recorded, followed by a relaxation delay of 7 s. The software TopSpin 3.6.4 (Bruker BioSpin, Germany) was used for the integration of the peaks.
Results and Discussion
Increasing mcr-CCa Copy Number Leads to Higher 3-HP Production
Previous results showed that the co-overexpression of ACC1 and cPOS5 in the initial 3HP-producing strain PpHP2 (Table 1) led to a small but significant increase in the 3-HP yield on glycerol in strain PpHP6 (from 0.131 Cmol Cmol−1 to 0.146 Cmol Cmol−1) (Fina et al., 2021). However, the overexpression of either ACC or cPOS5 did not result into any further significant impact on 3-HP production, highlighting the malonyl-CoA reductase step as a major bottleneck for 3-HP production, rather than the supply of malonyl-CoA and NADPH availability.
Thereby, a second copy of the C-terminal domain of the malonyl-CoA reductase gene (mcr-CCa) under the control of the strong and constitutive GAP promoter was added to the PpHP2 and PpHP6 strains, generating strains PpHP7 and PpHP8, respectively. For each strain, six independently isolated clones were tested in triplicate using deep-well plates and BMG medium. The highest producer clone for strains PpHP7 and PpHP8 produced 1.81 ± 0.02 g L−1 and 2.29 ± 0.01 g L−1 of 3-HP (i.e., 0.143 ± 0.001 and 0.180 ± 0.001 Cmol−1 yield on glycerol, respectively). The gene copy analysis showed that both clones contained 2 copies of mcr-CCa. The 3-HP yield achieved by the strains PpHP2 and PpHP6 was 0.130 ± 0.005 and 0.146 ± 0.004 Cmol−1, respectively. Therefore, the addition of a second copy of mcr-CCa to PpHP2 and PpHP6 led to a 1.38 and 1.23-fold improvement, respectively (p-values of 0.01 and 0.0001), as shown in Figure 2.
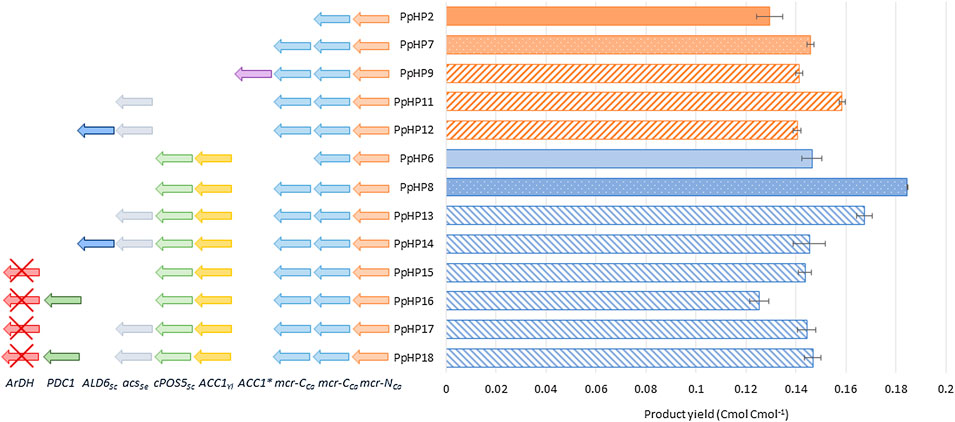
FIGURE 2. 3-HP production yield of the recombinant P. pastoris strains tested in deep-well plates. The genetic modifications performed to each strain are shown at the left side of the graph. The product yield in Cmol−1 is shown at the right side of the graph.
These results are consistent with previous studies with other yeasts such as S. cerevisiae or Schizosaccharomyces pombe. The use of a multicopy integrative vector containing the mcrCa and the ACC1 gene from S. cerevisiae led to a 3-fold increase in 3-HP production in S. cerevisiae (Kildegaard et al., 2016). In S. pombe, the addition of a second copy of mcr-CCa led to an almost 3-fold improvement in the 3-HP yield. Addition of a third copy of mcr-CCa led to a further 2-fold improvement in 3-HP production, while the addition of a fourth copy of the same gene, or the addition of a second copy mcr-NCa did not have an impact on the final 3-HP titre (Takayama et al., 2018).
Nevertheless, the increase in 3-HP production achieved in P. pastoris when additional copies of the mcr-CCa gene were introduced was significantly lower than the one achieved in other yeasts. Notably, the MCR specific activity of cell extracts of P. pastoris expressing mcrCa under the control of pGAP was more than one order of magnitude higher than the same gene expressed under the control of pTEF1 in S. cerevisiae (Chen et al., 2014; Fina et al., 2021). Additional bottleneck(s) upstream of the malonyl-CoA reductase reaction steps are probably limiting the 3-HP production, so no further copies of mcr-CCa were introduced.
Expression of the de-regulated ACC1S1132A Mutant Does Not Improve 3-HP Production
The acetyl-CoA carboxylase (Acc) catalyses the conversion of acetyl-CoA into malonyl-CoA. The enzyme Acc1S1132A from P. pastoris contains a site mutation (S1132A) that prevents the post-translational inactivation of the enzyme by phosphorylation when glucose is depleted (Choi and da Silva, 2014). Expression of this endogenous ACC1 variant under the control of the GAP promoter showed a positive impact on the production of acetyl-CoA derived drugs in P. pastoris (Liu et al., 2019). The same approach was used in PpHP7, but the new strain (PpHP9) produced less 3-HP than the parental strain (0.138 ± 0.001 Cmol−1, Figure 2).
Acc1 is organized as a homodimer. The phosphorylation of one of the two monomers inactivates the enzymatic complex (Hunkeler et al., 2016). Therefore, it is plausible that the co-existence of the heterologous mutated version of Acc1 and the endogenous non-mutated versions leads to the formation of inactive heterodimers, hindering the effect of Acc1S1132A overexpression. This hypothesis is supported by previous studies showing that replacement of the endogenous ACC1 sequence with ACC1S1132A was required to ensure that Acc1 actvity would only be controlled by Acc1S1132A expression levels (Wen et al., 2020). Deletion of the endogenous ACC1 gene in strains PpHP9 may thus had a positive effect on 3-HP production. Moreover, it is well reported that Acc1 is a highly regulated enzyme (Hunkeler et al., 2016; Chen et al., 2018). In addition to its post-translational regulation, this enzyme is also regulated by allosteric inhibition (Goodridge, 1972; Qiao et al., 2015). Differences in the kinetic parameters between the two Acc1 variants might also explain the differences in the final 3-HP yield.
In contrast, the overexpression of ACC1 from Y. lipolytica in P. pastoris led to a small increase in 3-HP production (Fina et al., 2021). The yield in PpHP8 (harboring Acc1Yl) was around 1.25-fold higher than in PpHP7. These results are consistent with recent studies where the heterologous expression of ACC1 from Y. lipolytica yielded a higher accumulation of malonyl-CoA than the overexpression of the endogenous S. cerevisiae ACC1 (Pereira et al., 2022).
Metabolic Engineering for Increased Acetyl-CoA Supply and Minimization of By-Product Formation
The endogenous cytosolic acetyl-CoA biosynthesis pathway was overexpressed to pull the conversion of pyruvate into acetyl-CoA. To this end, a modified Acetyl-CoA synthase from Salmonella enterica was used (acsSeL641P). The gene harbours a mutation (L641P) to suppress the post-translational inhibition of the enzyme by acetylation (Shiba et al., 2007). Expression of acsSeL641P was tested with or without the co-expression of the aldehyde dehydrogenase gene ALD6 from S. cerevisiae. The two genes were expressed under the control of the TEF1 and MDH3 promoters, respectively, which are two mid-to-high expression constitutive promoters (Prielhofer et al., 2017). Use of pGAP was dismissed to avoid the dilution of the expression of the other genes, as there are already 5 heterologous genes under the control of pGAP (Marx et al., 2009; Aw and Polizzi, 2013).
Expression of acsSe in strains PpHP7 and PpHP8, generating strains PpHP11 and PpHP13, led to opposite effects. While PpHP11 produced significantly more 3-HP (0.153 ± 0.002 Cmol−1 yield) than PpHP7 (p-value of 0.0003), its expression in PpHP13 led to a reduction of the yield (p-value 0.0006), as shown in Figure 2.
The expression of the aldehyde dehydrogenase ALD6 in both PpHP11 and PpHP13 (PpHP12 and PpHP14, respectively) led to a decrease in the 3-HP yield for both strains (Figure 2). Interestingly, the deletion of ALD6 in S. cerevisiae led to a reduced susceptibility to 3-HP, as Ald6 might catalyse the conversion of 3-HP into the toxic compound 3-hydroxypropanaldehyde (3-HPA) (Kildegaard et al., 2014). Thus, it is plausible that ALD6 overexpression in 3-HP producing P. pastoris strains leads to an increased accumulation of 3-HPA.
Finally, the overexpression of endogenous pyruvate decarboxylase (PDC1) was tested. To this end, an additional copy of PDC1 under the control of its endogenous promoter was added to the genomes of the strains PpHP8 and PpHP13. Addition of a second copy of PDC1 was coupled to the deletion of the ArDH gene, which is reported as the main responsible of the D-arabitol production in P. pastoris (Melo et al., 2020). To distinguish the effect of the deletion of ArDH from the overexpression of PDC1, strains PpHP15 and PpHP17 were generated by deleting ArDH from strains PpHP8 and PpHP13, respectively (without an additional copy of PDC1). The strains PpHP16 and PpHP18 were also obtained from PpHP8 and PpHP13, and they harbour both the deletion of ArDH and a second copy of the endogenous PDC1. None of the new strains produced more 3-HP than their parental strains (Figure 2).
Screening Under Substrate-Limiting Conditions Yields Different Ranking of Strains Compared to Conventional Substrate-Excess Screening Strategies
Previous results obtained in deep-well plates showed that the overexpression of the cytosolic acetyl-CoA production pathway did not result in a significant increase in the final 3-HP production. Moreover, accumulation of D-arabitol was not observed in none of the strains, as was the case for PpHP6 strain in fed-batch cultivations (Fina et al., 2021). Thus, the effect of ArDH deletion could not be properly assessed at small scale. Therefore, a representative subset of the strains constructed in this study were further cultured in two additional systems: 1) Mini bioreactors, favouring fully aerobic conditions and easy withdrawal of multiple samples. 2) Falcon tubes containing FeedBeads®, which release glycerol at a low and constant rate (40 mg in 48 h), thereby permitting cell growth at a low rate, i.e., under substrate-limiting conditions resembling a fed-batch process.
The mini bioreactors were sampled during the exponential phase, which allowed to accurately determine the µmax and the q-rates of the consumption of glycerol and the production of 3-HP and D-arabitol of the different strains, whereas endpoint samples (at 48 h) were taken from cultivations in falcon tubes containing FeedBeads®.
Notably, comparison of the results from these alternative screening conditions with the deep-well plate cultivations (Figure 3) reveals that the ranking of the strains based on their product yield varies depending on the screening condition. PpHP8 exhibited the highest product yield for the three screening conditions. However, the combined effect of ACS and Pdc1 overexpression and ArDH deletion (strain PpHP18) was clearly growth condition dependent. Indeed, the 3-HP production of the two strains, PpHP8 and PpHP18 was very similar at a low growth rate (i.e., using FeedBeads®), but differed significantly at µmax (i.e., in the mini bioreactor experiment). Interestingly, 3-HP yields were generally higher in deep-well plates and falcon tubes, where oxygen transfer is usually considered as rate-limiting during cultivation, than in fully aerobic mini bioreactor cultures. This is consistent with the fact that the pGAP promoter is upregulated in P. pastoris cells growing on glucose under hypoxic conditions (Baumann et al., 2011).
FIGURE 3. Comparison of the most relevant 3-HP producing P. pastoris strains under three different screening conditions. (A) 3-HP production yield in deep-well plates (grey solid bars), FeedBeads® (blue dotted bars), and mini bioreactors (orange striped bars). (B) Growth rate (yellow vertically striped bars) and arabitol production yield (green horizontally striped bars) for each strain grown in mini bioreactors.
Moreover, strains PpHP13, PpHP15, PpHP17, and PpHP18 showed a much lower 3-HP product yield than PpHP8 when the cells were grown in mini bioreactors. Conversely, the µmax of strain PpHP8 was the lowest of this subset of stains. An inverse correlation between µmax and 3-HP product yield was observed. Despite lower growth rates would be generally expected for the strains with a larger number of heterologously overexpressed genes, overexpression of ACS and Pdc1 and the deletion of ArDH led to a higher µmax. Acc1 overexpression has been reported to cause growth impairment in yeast (Shi et al., 2014; Kildegaard et al., 2016). This reduction in the growth rate was attributed to a metabolic burden. The fact the cell growth improves when the cytosolic acetyl-CoA pathway is overexpressed suggests that the lower growth rate of PpHP8 might be caused by a limitation in acetyl-CoA supply for the biosynthetic reactions. Such hypothesis is supported by the fact that the product yield of strains PpHP8 and PpHP18 are almost identical when the cells are grown at a low growth rate (i.e., using FeedBeads®). This is also reinforced by the differential D-arabitol production of strains PpHP8 and PpHP13. The presence of arabitol denotes a redox imbalance (Fina et al., 2021). When the delivery of acetyl-CoA is improved by overexpressing ACS (PpHP13), the D-arabitol production decreases.
Notably, the mini bioreactors data reveals that the deletion of ArDH from strain PpHP8 (strain PpHP15) resulted in a decrease in the D-arabitol yield and an improvement of the µmax. Production of D-arabitol from glycerol results in net ATP consumption. Thus, the reduction in the D-arabitol production leads to higher energetic yield from the substrate, as well as a better carbon conservation (as less by-product D-arabitol is produced), thereby potentially increasing acetyl-CoA supply and supporting higher growth rates. However, this might result in reduced Acetyl-CoA availability for 3-HP production.
Altogether, the observed results indicate the supply of cytosolic acetyl-CoA as a major limiting factor to further increase the 3-HP yield.
Fed-Batch Cultures
The use of FeedBeads® mimics the conditions at a fed-batch culture, where cells grow under substrate-limiting conditions, i.e., at growth rates below the µmax. The strains PpHP8 and PpHP18, which showed the highest 3-HP yields in FeedBeads® cultures, were thus further tested in a fed-batch bioreactor culture using a pre-programmed exponential glycerol feeding strategy to maintain the growth rate at 0.075 h−1, which is approximately half the value of the µmax of the two strains (0.133 h−1 and 0.165 h−1, respectively). The feeding phase finished when the equivalent to 195 g L−1 of glycerol were added to the reactor, to compare the results with our previous experiments where the same overall amount of substrate was used (Fina et al., 2021). Thereafter, the cultivation was finalised when the DO signal increased, indicating the depletion of any residual glycerol that might have been accumulated during the late phase of the fed-batch culture. The cultivation profiles for both strains are shown in Figure 4.
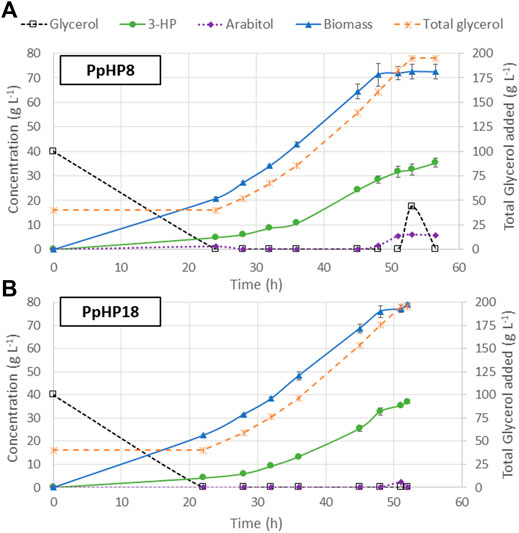
FIGURE 4. Profiles of fed-batch cultivations of the PpHP8 (A) and PpHP18 (B) strains. Concentration of glycerol, biomass, 3-HP, and D-arabitol are represented using the left-side y-axis. The total amount of glycerol added to the reactor, normalized by the actual volume of the reactor at every time is represented using the y-axis at the right side. The average result of two independent cultivations is shown. The error bars correspond to the standard deviation for the duplicate.
At the end of the batch phase, PpHP8 produced 4.92 ± 0.17 g L−1 3-HP, while the strain PpHP18 produced 4.11 ± 0.17 g L−1 3-HP. These results agree with the observations from the mini bioreactors, where the yield of PpHP8 was higher than the yield of PpHP18. However, 3-HP concentration at the end of the fed-batch culture of PpHP18 reached 37.05 ± 0.73 g L−1, while the PpHP8 strain reached 35.40 ± 1.85 g L−1. Thus, the yield during the feeding phase was higher for PpHP18 than PpHP8. Moreover, the fact PpHP18 µmax is higher than the one of PpHP8 resulted in a shorter batch phase (22 h compared to 24 h). Previous studies revealed that P. pastoris cannot sustain the growth rate in fed-batch cultivations at a pre-set µ of 0.10 h−1 when 3-HP concentrations reached concentrations above 15 g L−1, leading to the accumulation of glycerol and D-arabitol in the reactor medium (Fina et al., 2021). This phenomenon is still observed in PpHP8 strain growing at 0.075 h−1, where 17.41 g L−1 of glycerol and 6.01 g L−1 of D-arabitol accumulated into the reactor, while no glycerol nor D-arabitol accumulation were observed for PpHP18. For this reason, the PpHP18 cultivation was finalised just after stopping the addition of the feeding. Overall, the fed-batch for PpHP18 lasted 52 h, while the fed-batch of PpHP8 lasted 56.5 h. Thus, the volumetric productivity of PpHP18 was significantly higher than the volumetric productivity of PpHP8 (0.712 ± 0.010 g L−1 h−1 compared to 0.627 ± 0.033 g L−1 h−1). The overall product yield on glycerol was 0.194 ± 0.004 Cmol−1 for PpHP18 and 0.186 ± 0.010 Cmol−1 for PpHP8.
The productivity obtained for the strain PpHP18 is the highest productivity reported in yeast, and, to our knowledge, it is also the highest productivity reported using the malonyl-CoA pathway to produce 3-HP in any microorganism. Furthermore, 37.05 g L−1 is the highest 3-HP concentration reported in yeast, highlighting the potential of the combined use of the P. pastoris cell factory and glycerol as feedstock for 3-HP production. Notably, the metabolically engineered PpHP18 strain showed a 50% increase in the final 3-HP titre and yield, and a 31.9% increase in volumetric productivity compared to the initial strain (PpHP6).
Conclusion
Production of 3-HP in P. pastoris through the malonyl-CoA reductase has been reported. The aim of this work was to improve its production by several metabolic engineering strategies. Improvement of the 3HP yield was obtained by adding a second copy of mcr-CCa . However, 1) increasing the availability of malonyl-CoA by overexpressing a post-translationally unregulated Acc1 from P. pastoris; 2) overexpressing endogenous acetyl-CoA pathway (ACS, Ald6, and Pdc1) or 3) limiting the production of arabitol by deleting ArDH did not improve 3-HP production in small-scale cultures. Nevertheless, deletion of ArDH improved the 3-HP production in fed-batch bioreactor cultures.
The work of this study demonstrates that the system and the mode of cultivation clearly affected the phenotype of the strains. For instance, the strain PpHP18 showed the highest production of 3-HP in fed-batch cultivation mode but not in deep-well plates and mini bioreactors operated in batch. This provides clear evidence on the importance to implement robust and reliable small-scale cultivation methods allowing for the mid/high throughput characterisation of strain performance under bioprocess-like conditions. However, the results obtained with the FeedBeads® were similar to the ones obtained in fed-bath cultures, thereby validating this methodology as a reliable method for strain screening/ranking and fast translation to bioreactor-scale.
Overall, our study showed that the multiplexed screening methodologies allowed to improve the information content, thereby supporting the formulation of hypotheses of previously unidentified metabolic bottlenecks such as that the supply of cytosolic acetyl-CoA may be limiting 3-HP production. Nonetheless, further studies, e.g., using 13C-based metabolic flux analysis and metabolomics are needed to design novel metabolic engineering strategies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AF, JA, and PF conceived and designed the research project. AF constructed all strains, performed the deep-well plates, the FeedBeads® and the fed-batch experiments. AF and SH performed the mini bioreactors screening. AF drafted the manuscript. All authors critically reviewed the manuscript.
Funding
This work was supported by project PID2019-104666GB-100 from the Spanish Ministry of Science and Innovation; 2017-SGR-1462 from the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) of the Catalan Government. AF was supported with a FPU scholarship (FPU17/05434) from the Spanish Ministry of Science and Innovation and a Short-Term Fellowship (8853) from the European Molecular Biology Organization (EMBO).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.942304/full#supplementary-material
References
Aw, R., and Polizzi, K. M. (2013). Can Too Many Copies Spoil the Broth? Microb. Cell Fact. 12, 128. doi:10.1186/1475-2859-12-128
Baumann, K., Dato, L., Graf, A. B., Frascotti, G., Dragosits, M., Porro, D., et al. (2011). The Impact of Oxygen on the Transcriptome of Recombinant S. cerevisiae and P. pastoris - A Comparative Analysis. BMC Genomics 12, 218. doi:10.1186/1471-2164-12-218
Borodina, I., Kildegaard, K. R., Jensen, N. B., Blicher, T. H., Maury, J., Sherstyk, S., et al. (2015). Establishing a Synthetic Pathway for High-Level Production of 3-hydroxypropionic Acid in Saccharomyces cerevisiae via β-alanine. Metab. Eng. 27, 57–64. doi:10.1016/j.ymben.2014.10.003
Cámara, E., Albiol, J., and Ferrer, P. (2016). Droplet Digital PCR-Aided Screening and Characterization of Pichia pastoris multiple Gene Copy Strains. Biotechnol. Bioeng. 113, 1542–1551. doi:10.1002/bit.25916
Chemarin, F., Athès, V., Bedu, M., Loty, T., Allais, F., Trelea, I. C., et al. (2019). Towards an In Situ Product Recovery of Bio-Based 3-hydroxypropionic Acid: Influence of Bioconversion Broth Components on Membrane-Assisted Reactive Extraction. J. Chem. Technol. Biotechnol. 94, 964–972. doi:10.1002/jctb.5845
Chen, X., Yang, X., Shen, Y., Hou, J., and Bao, X. (2018). Screening Phosphorylation Site Mutations in Yeast Acetyl-CoA Carboxylase Using Malonyl-CoA Sensor to Improve Malonyl-CoA-Derived Product. Front. Microbiol. 9, 47. doi:10.3389/fmicb.2018.00047
Chen, Y., Bao, J., Kim, I.-K., Siewers, V., and Nielsen, J. (2014). Coupled Incremental Precursor and Co-factor Supply Improves 3-hydroxypropionic Acid Production in Saccharomyces cerevisiae. Metab. Eng. 22, 104–109. doi:10.1016/j.ymben.2014.01.005
Choi, J. W., and da Silva, N. A. (2014). Improving Polyketide and Fatty Acid Synthesis by Engineering of the Yeast Acetyl-CoA Carboxylase. J. Biotechnol. 187, 56–59. doi:10.1016/j.jbiotec.2014.07.430
Cregg, J. M., Vedvick, T. S., and Raschke, W. C. (1993). Recent Advances in the Expression of Foreign Genes in Pichia pastoris. Nat. Biotechnol. 11, 905–910. doi:10.1038/nbt0893-905
de Fouchécour, F., Sánchez-Castañeda, A.-K., Saulou-Bérion, C., and Spinnler, H. É. (2018). Process Engineering for Microbial Production of 3-Hydroxypropionic Acid. Biotechnol. Adv. 36, 1207–1222. doi:10.1016/j.biotechadv.2018.03.020
della Pina, C., Falletta, E., and Rossi, M. (2011). A Green Approach to Chemical Building Blocks. The Case of 3-Hydroxypropanoic Acid. Green Chem. 13, 1624–1632. doi:10.1039/c1gc15052a
Fang, H., Li, D., Kang, J., Jiang, P., Sun, J., and Zhang, D. (2018). Metabolic Engineering of Escherichia coli for De Novo Biosynthesis of Vitamin B12. Nat. Commun. 9, 4917. doi:10.1038/s41467-018-07412-6
Fina, A., Brêda, G. C., Pérez‐Trujillo, M., Freire, D. M. G., Almeida, R. V., Albiol, J., et al. (2021). Benchmarking Recombinant Pichia pastoris For 3‐Hydroxypropionic Acid Production from Glycerol. Microb. Biotechnol. 14, 1671–1682. doi:10.1111/1751-7915.13833
Garcia-Ortega, X., Ferrer, P., Montesinos, J. L., and Valero, F. (2013). Fed-Batch Operational Strategies for Recombinant fab Production with Pichia pastoris Using the Constitutive GAP Promoter. Biochem. Eng. J. 79, 172–181. doi:10.1016/j.bej.2013.07.013
Gassler, T., Heistinger, L., Mattanovich, D., Gasser, B., and Prielhofer, R. (2019). CRISPR/Cas9-Mediated Homology-Directed Genome Editing in Pichia pastoris. Methods Mol. Biol. 1923, 211–225. doi:10.1007/978-1-4939-9024-5_9
Goodridge, A. G. (1972). Regulation of The Activity of Acetyl Coenzyme A Carboxylase By Palmitoyl Coenzyme A and Citrate. J. Biol. Chem. 247, 6946–6952. doi:10.1016/s0021-9258(19)44677-2
Heux, S., Poinot, J., Massou, S., Sokol, S., and Portais, J.-C. (2014). A Novel Platform for Automated High-Throughput Fluxome Profiling of Metabolic Variants. Metab. Eng. 25, 8–19. doi:10.1016/j.ymben.2014.06.001
Hunkeler, M., Stuttfeld, E., Hagmann, A., Imseng, S., and Maier, T. (2016). The Dynamic Organization of Fungal Acetyl-Coa Carboxylase. Nat. Commun. 7, 11196. doi:10.1038/ncomms11196
Jers, C., Kalantari, A., Garg, A., and Mijakovic, I. (2019). Production of 3-Hydroxypropanoic Acid from Glycerol by Metabolically Engineered Bacteria. Front. Bioeng. Biotechnol. 7, 124. doi:10.3389/fbioe.2019.00124
Ji, R.-Y., Ding, Y., Shi, T.-Q., Lin, L., Huang, H., Gao, Z., et al. (2018). Metabolic Engineering of Yeast for the Production of 3-Hydroxypropionic Acid. Front. Microbiol. 9, 2185. doi:10.3389/fmicb.2018.02185
Kildegaard, K. R., Hallström, B. M., Blicher, T. H., Sonnenschein, N., Jensen, N. B., Sherstyk, S., et al. (2014). Evolution Reveals A Glutathione-Dependent Mechanism of 3-Hydroxypropionic Acid Tolerance. Metab. Eng. 26, 57–66. doi:10.1016/j.ymben.2014.09.004
Kildegaard, K. R., Jensen, N. B., Schneider, K., Czarnotta, E., Özdemir, E., Klein, T., et al. (2016). Engineering and Systems-Level Analysis of Saccharomyces cerevisiae For Production of 3-Hydroxypropionic Acid Via Malonyl-Coa Reductase-Dependent Pathway. Microb. Cell Fact. 15, 53. doi:10.1186/s12934-016-0451-5
Kim, J. W., Ko, Y. S., Chae, T. U., and Lee, S. Y. (2020). High‐Level Production of 3‐Hydroxypropionic Acid From Glycerol as a Sole Carbon Source Using Metabolically Engineered Escherichia coli. Biotechnol. Bioeng. 117, 2139–2152. doi:10.1002/bit.27344
Kumar, V., Ashok, S., and Park, S. (2013). Recent Advances in Biological Production of 3-Hydroxypropionic Acid. Biotechnol. Adv. 31, 945–961. doi:10.1016/j.biotechadv.2013.02.008
Liu, Y., Bai, C., Liu, Q., Xu, Q., Qian, Z., Peng, Q., et al. (2019). Engineered Ethanol-Driven Biosynthetic System for Improving Production of Acetyl-CoA Derived Drugs in Crabtree-Negative Yeast. Metab Eng. 54, 275–284. doi:10.1016/j.ymben.2019.05.001
Luo, X., Ge, X., Cui, S., and Li, Y. (2016). Value-Added Processing of Crude Glycerol into Chemicals and Polymers. Bioresour. Technol. 215, 144–154. doi:10.1016/j.biortech.2016.03.042
Maris, A. J. A. v., Konings, W. N., Dijken, J. P. v., and Pronk, J. T. (2004). Microbial Export of Lactic and 3-Hydroxypropanoic Acid: Implications for Industrial Fermentation Processes. Metab. Eng. 6, 245–255. doi:10.1016/j.ymben.2004.05.001
Marx, H., Mecklenbräuker, A., Gasser, B., Sauer, M., and Mattanovich, D. (2009). Directed Gene Copy Number Amplification in Pichia pastoris by Vector Integration into the Ribosomal DNA Locus. FEMS Yeast Res. 9, 1260–1270. doi:10.1111/j.1567-1364.2009.00561.x
Maurer, M., Kühleitner, M., Gasser, B., and Mattanovich, D. (2006). Versatile Modeling and Optimization of Fed Batch Processes for the Production of Secreted Heterologous Proteins with Pichia pastoris. Microb. Cell Fact. 5, 37. doi:10.1186/1475-2859-5-37
Mazzoli, R. (2021). Current Progress in Production of Building-Block Organic Acids by Consolidated Bioprocessing of Lignocellulose. Fermentation 7, 248. doi:10.3390/fermentation7040248
Melo, N. T. M., Pontes, G. C., Procópio, D. P., de Gois e Cunha, G. C., Eliodório, K. P., Costa Paes, H., et al. (2020). Evaluation of Product Distribution in Chemostat and Batch Fermentation in Lactic Acid-Producing Komagataella phaffii Strains Utilizing Glycerol as Substrate. Microorganisms 8, 781. doi:10.3390/microorganisms8050781
Peiro, C., Millard, P., de Simone, A., Cahoreau, E., Peyriga, L., Enjalbert, B., et al. (2019). Chemical and Metabolic Controls on Dihydroxyacetone Metabolism Lead To Suboptimal Growth of Escherichia coli. Appl. Environ. Microbiol. 85, e00768. doi:10.1128/AEM.00768-19
Pereira, H., Azevedo, F., Domingues, L., and Johansson, B. (2022). Expression of Yarrowia lipolytica Acetyl-CoA Carboxylase in Saccharomyces cerevisiae and its Effect on In-Vivo Accumulation of Malonyl-CoA. Comput. Struct. Biotechnol. J. 20, 779–787. doi:10.1016/j.csbj.2022.01.020
Prielhofer, R., Barrero, J. J., Steuer, S., Gassler, T., Zahrl, R., Baumann, K., et al. (2017). GoldenPiCS: A Golden Gate-Derived Modular Cloning System for Applied Synthetic Biology in the Yeast Pichia pastoris. BMC Syst. Biol. 11, 123. doi:10.1186/s12918-017-0492-3
Qiao, K., Imam Abidi, S. H., Liu, H., Zhang, H., Chakraborty, S., Watson, N., et al. (2015). Engineering Lipid Overproduction in the Oleaginous Yeast Yarrowia lipolytica. Metab. Eng. 29, 56–65. doi:10.1016/j.ymben.2015.02.005
Shi, S., Chen, Y., Siewers, V., and Nielsen, J. (2014). Improving Production of Malonyl Coenzyme A-Derived Metabolites by Abolishing Snf1-Dependent Regulation of Acc1. mBio 5, e01130. doi:10.1128/mBio.01130-14
Shiba, Y., Paradise, E. M., Kirby, J., Ro, D.-K., and Keasling, J. D. (2007). Engineering of the Pyruvate Dehydrogenase Bypass in Saccharomyces cerevisiae For High-Level Production of Isoprenoids. Metab. Eng. 9, 160–168. doi:10.1016/j.ymben.2006.10.005
Takayama, S., Ozaki, A., Konishi, R., Otomo, C., Kishida, M., Hirata, Y., et al. (2018). Enhancing 3-Hydroxypropionic Acid Production in Combination With Sugar Supply Engineering by Cell Surface-Display and Metabolic Engineering of Schizosaccharomyces pombe. Microb. Cell Fact. 17, 176. doi:10.1186/s12934-018-1025-5
Wen, J., Tian, L., Xu, M., Zhou, X., Zhang, Y., and Cai, M. (2020). A Synthetic Malonyl-Coa Metabolic Oscillator in Komagataella phaffii. ACS Synth. Biol. 9, 1059–1068. doi:10.1021/acssynbio.9b00378
Werpy, T., and Petersen, G. (2004). Top Value Added Chemicals from Biomass Volume I - Results of Screening for Potential Candidates from Sugars and Synthesis Gas. Golden, CO, USA: U.S. Department of Energy. Available at: http://www.osti.gov/bridge.
Werten, M. W. T., van den Bosch, T. J., Wind, R. D., Mooibroek, H., and de Wolf, F. A. (1999). High-Yield Secretion of Recombinant Gelatins By Pichia pastoris. Yeast 15, 1087–1096. doi:10.1002/(sici)1097-0061(199908)15:11<1087::aid-yea436>3.0.co;2-f
Keywords: 3-hydroxypropionic acid, Pichia pastoris, glycerol, malonyl-CoA, acetyl-CoA, metabolic engineering
Citation: Fina A, Heux S, Albiol J and Ferrer P (2022) Combining Metabolic Engineering and Multiplexed Screening Methods for 3-Hydroxypropionic Acid Production in Pichia pastoris. Front. Bioeng. Biotechnol. 10:942304. doi: 10.3389/fbioe.2022.942304
Received: 12 May 2022; Accepted: 22 June 2022;
Published: 22 July 2022.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Yongjin Zhou, Dalian Institute of Chemical Physics (CAS), ChinaYang Gu, Nanjing Normal University, China
Copyright © 2022 Fina, Heux, Albiol and Ferrer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pau Ferrer, UGF1LkZlcnJlckB1YWIuY2F0
 Albert Fina
Albert Fina Stephanie Heux2
Stephanie Heux2 Joan Albiol
Joan Albiol Pau Ferrer
Pau Ferrer