
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 01 July 2022
Sec. Bioprocess Engineering
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.925311
This article is part of the Research TopicMetabolic Engineering for Bioresources and Bioenergies Production from MicroalgaeView all 9 articles
 Yu Dan1,2,3†
Yu Dan1,2,3† Jiahui Sun3,4,5,6†
Jiahui Sun3,4,5,6† Shanshan Zhang3,4,5,6
Shanshan Zhang3,4,5,6 Yannan Wu1,2,3
Yannan Wu1,2,3 Shaoming Mao1,2*
Shaoming Mao1,2* Guodong Luan3,4,5,6*
Guodong Luan3,4,5,6* Xuefeng Lu3,4,5,6,7*
Xuefeng Lu3,4,5,6,7*Cyanobacteria are a promising photosynthetic chassis to produce biofuels, biochemicals, and pharmaceuticals at the expense of CO2 and light energy. Glycogen accumulation represents a universal carbon sink mechanism among cyanobacteria, storing excess carbon and energy from photosynthesis and may compete with product synthesis. Therefore, the glycogen synthesis pathway is often targeted to increase cyanobacterial production of desired carbon-based products. However, these manipulations caused severe physiological and metabolic impairments and often failed to optimize the overall performance of photosynthetic production. Here, in this work, we explored to mobilize the glycogen storage by strengthening glycogen degradation activities. In Synechococcus elongatus PCC 7942, we manipulated the abundances of glycogen phosphorylase (GlgP) with a theophylline dose-responsive riboswitch approach, which holds control over the cyanobacterial glycogen degradation process and successfully regulated the glycogen contents in the recombinant strain. Taking sucrose synthesis as a model, we explored the effects of enhanced glycogen degradation on sucrose production and glycogen storage. It is confirmed that under non-hypersaline conditions, the overexpressed glgP facilitated the effective mobilization of glycogen storage and resulted in increased secretory sucrose production. The findings in this work provided fresh insights into the area of cyanobacteria glycogen metabolism engineering and would inspire the development of novel metabolic engineering approaches for efficient photosynthetic biosynthesis.
Cyanobacteria are important prokaryotic microorganisms performing oxygenic photosynthesis and playing essential roles in global carbon and nitrogen cycles on Earth (Waterbury et al., 1979; Flombaum et al., 2013; Rousseaux and Gregg, 2014). Being abundant and widely distributed among diverse types of habitats, cyanobacteria provide 20–30% of the primary organic carbon in the biosphere, with their efficient oxygenic photosynthesis systems converting solar energy and carbon dioxide into organic compounds (Flombaum et al., 2013; Rousseaux and Gregg, 2014). In cyanobacteria cells, the surplus carbon flow beyond the requirements of cellular growth and maintenance would be stored as a carbon sink, supporting materials and energy for the cells to survive in dark and stressful conditions (Ball and Morell, 2003; Nakamura et al., 2005; Damrow et al., 2016). Glycogen is the most essential carbon sink compound in cyanobacteria, accounting for up to 50% of the total cellular biomass in specific species or environments (Aikawa et al., 2014; Song et al., 2016).
As promising photosynthetic microbial platforms for biotechnological and industrial applications (Angermayr et al., 2009; Lu, 2010; Oliver and Atsumi, 2014; Zhou et al., 2016; Luan and Lu, 2018), diverse strategies and tools have been developed to manipulate carbon flow in cyanobacteria, and glycogen metabolism has been generally recognized as a promising target (Carrieri et al., 2012; Melis, 2012; Zhou et al., 2016; Luan et al., 2019). Metabolic pathways for glycogen synthesis and degradation have been clearly deciphered in cyanobacteria (Figure 1A) and inhibition of the key enzymes for glycogen synthesis, and GlgC (ADP-glucose pyrophosphorylase, catalyzing ADP-glucose formation with glucose-1-phosphate) and GlgA (glycogen synthase, incorporating glucose monomers into the growing 1-4 α-linked glucose polymer) have successfully facilitated effective regulation of glycogen storages (Carrieri et al., 2012; Melis, 2012; Zhou et al., 2016) and intracellular carbon distribution (Xu et al., 2013; Hendry et al., 2017). However, impaired glycogen synthesis usually causes severe disturbance to cell physiology, including reduced photosynthesis, growth, respiration, and robustness toward environmental stresses (Miao et al., 2003; Suzuki et al., 2010; Grundel et al., 2012; Guerra et al., 2013; Hickman et al., 2013). In many cases, blocking glycogen synthesis decreased rather than increasing the productivity of heterologous pathways in engineered cyanobacterial strains, which might be resulted from the combined effect of physiological damage and metabolic rebalance (Davies et al., 2014; Jacobsen and Frigaard, 2014; Li et al., 2014; van der Woude et al., 2014; Work et al., 2015).
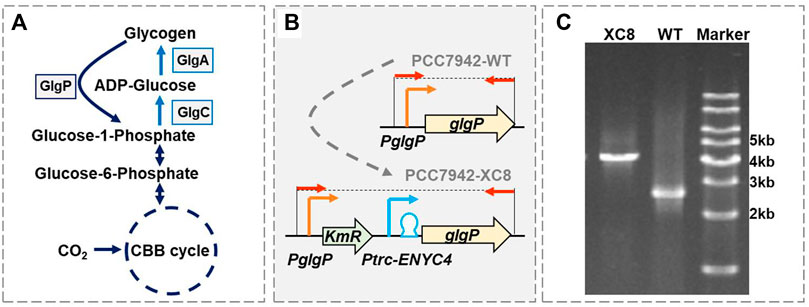
FIGURE 1. Construction of a theophylline-responsive glgP expression system in Synechococcus elongatus PCC 7942. (A) Glycogen metabolism pathways in cyanobacteria. GlgP, glycogen phosphorylase; GlgA, glycogen synthase; GlgC, glucose-1-phosphate adenylyltransferase or ADP-glucose pyrophosphorylase; CBB cycle, Calvin–Benson–Bassham cycle. (B) Strategy for constructing the theophylline-responsive riboswitch control system for glgP expression in PCC 7942. A kanamycin resistance gene (KmR) and theophylline dose-regulated expression cassette (Ptrc-ENYC4) were inserted between the native promoter of glgP (PglgP) and the glgP CDS, and the obtained mutant was termed as PCC 7942 XC8. The red arrows represented the primer pairs used for checking the genotypes of PCC 7942 wildtype (PCC7942-WT) and XC8 mutant (PCC7942-XC8) strains. (C) Identification of the genotypes of PCC 7942 wildtype (WT) and XC8 mutant by PCR with the primers (shown as red arrows in B).
Since elimination of the glycogen synthesis flux failed to optimize the photosynthetic biosynthesis performances of the cyanobacterial cell factories, strengthening the glycogen degradation pathways to mobilize the carbon sink into central metabolism could undoubtedly be an alternative strategy worth trying (Luan et al., 2019). However, approaches and effects to manipulate glycogen degradation metabolism in cyanobacteria have been relatively less reported. As shown in Figure 1A, glycogen phosphorylase (encoded by glgP), catalyzing the glycogen digestion into glucose-1-phosphate (G-1-P), is mainly responsible for glycogen degradation in cyanobacteria (Fu and Xu, 2006). In 2012, Ducat et al. overexpressed glgP in an engineered strain of Synechococcus elongatus PCC 7942 for secretory sucrose production and reported that the additional cassette for glgP expression caused a 10% reduction in sucrose productivity (Ducat et al., 2012). In general, the possibilities to engineer and accelerate glycogen degradation and the subsequent effects on cellular physiology and metabolism in cyanobacteria are yet to be explored.
In the present study, we aimed to manipulate glycogen metabolism by regulating the expression of glgP and the activities of glycogen degradation. Adopting a theophylline dose-responsive riboswitch, GlgP abundances, glycogen phosphorylase activities, and glycogen storage in a model cyanobacterium Synechococcus elongatus PCC 7942 (hereafter PCC 7942 for short) were successfully regulated, and the relative effects on cellular physiology and metabolism were explored. Sucrose is another important type of carbohydrate in PCC 7942 to be accumulated as osmoprotectants in response to extracellular hypersaline stress (Hagemann, 2011). In the past years, interactions between sucrose synthesis and glycogen metabolism have been explored in different cyanobacteria species (Suzuki et al., 2010; Guerra et al., 2013; Xu et al., 2013), and several glycogen metabolism engineering strategies have been adopted to improve the production of sucrose and other metabolic products in engineered cyanobacterial cell factories (Ducat et al., 2012; Qiao et al., 2018; Luan et al., 2019). Thus, sucrose synthesis was also taken as a model to explore whether the strategy of accelerating glycogen degradation could enhance the performance of photosynthetic biosynthesis. Our findings provided fresh insights into the area of cyanobacteria glycogen metabolism engineering and would inspire the development of novel metabolic engineering approaches for efficient photosynthetic biosynthesis.
As shown in Figure 1A, glycogen phosphorylase (GlgP) plays an essential role in catalyzing the glycogen degradation process by breaking the α-1,4-glycosidic bond on the glycogen chain and removing glucose monomers in the form of glucose-1-phosphate (G-1-P). It has been confirmed that the knockout of two glgP genes in Synechocystis sp. PCC 6803 would increase the glycogen contents by about 7% with continuous illumination and 250%–450% in day–night cycles (Shimakawa et al., 2014). However, so far, there have not been reports about quantitatively evaluating the effects of enhanced glgP expression on glycogen storage in cyanobacteria. We designed to regulate the expression of glgP and glycogen degradation activities adopting a theophylline-dose responsive riboswitch system ENYC4 in PCC 7942. Previously the Ptrc-ENYC4 system (Ptrc promoter combined with the ENYC4 riboswitch sequence) had been successfully utilized to regulate the expression of glgC and facilitated dynamic regulation of GlgC abundances and glycogen contents (Qiao et al., 2018; Chi et al., 2019). In this work, we adopted a similar strategy by replacing the native glgP promoter sequence with a Ptrc-ENYC4 sequence (Figures 1B,C). As designed, the expression of glgP on the chromosome of PCC 7942 would be regulated by the dose of theophylline.
To evaluate the effects of the theophylline-responsive riboswitch on regulating glgP expression, concentrations of theophylline (0,100, and 1,000 μM) were supplemented into the culture broth of XC8, and the abundances of GlgP were evaluated by immunoblotting. As shown in Figure 2A; Supplementary Figure S1, supplementation of theophylline with all of the three concentrations into the culture medium caused minor influence on growths of the XC8 strain, showing similar growth patterns with the wildtype control. During the process, the GlgP abundances were significantly regulated by the addition of theophylline (Figure 2B). Samples were collected from three time points of the cultivation process, and Western blot with GlgP-specific antibodies was performed. When no theophylline was added, the GlgP abundances were significantly reduced to an undetectable level compared with those of the control. The addition of theophylline (100 and 1,000 μM) as an inducer would elevate the expressions of GlgP in XC8. In the medium containing 100 μM theophylline, GlgP abundances would significantly exceed those of the wildtype, and increasing the theophylline concentration to 1,000 μM would further improve the GlgP abundances. The results indicated that in PCC 7942 recombinant strain XC8 carrying the Ptrc-ENYC4-glgP cassette, the abundances of GlgP could be effectively regulated with theophylline dose added in the culture broth. In addition, the change in GlgP abundances also brought in consistent changes in glycogen phosphorylase activities in the XC8 strain with increasing theophylline inductions (Figure 2C). With 100 μM theophylline induction, glycogen phosphorylase activities were increased to nearly 4-fold higher than those of the wildtype control.
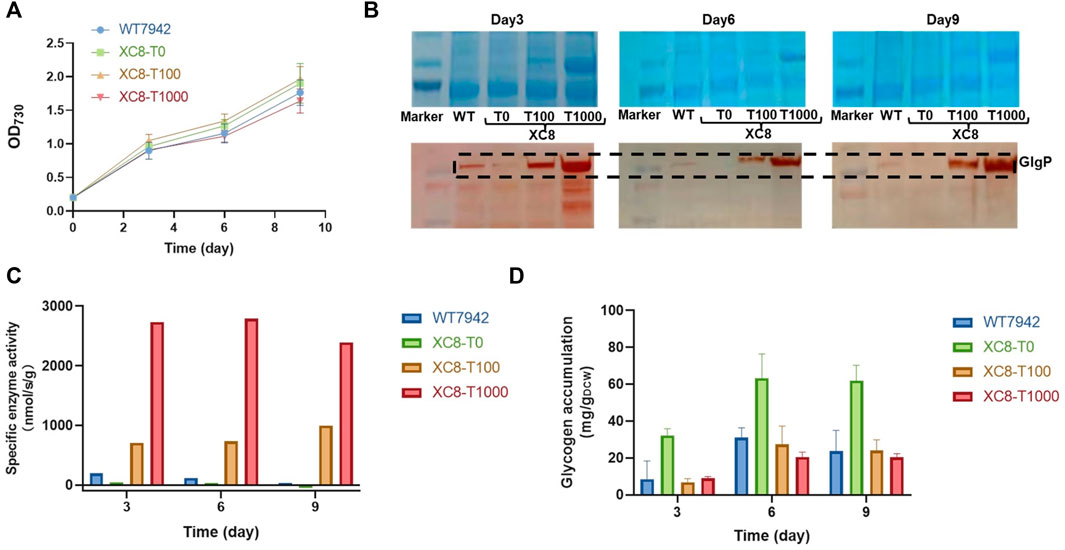
FIGURE 2. Theophylline-regulated glgP expression and glycogen storage in PCC 7942. (A) Cell growths, (B) GlgP abundances, (C) glycogen phosphorylase activities, and (D) glycogen contents of XC8 strain induced with different concentrations of theophylline during the 10 days of cultivation. To confirm the effects of theophylline induction on glgP expression regulation, different concentrations of theophylline (0 μM, T0; 100 μM, T100; 1,000 μM, T1000) were supplemented into culture broth of XC8 strain in flasks. The cells would be sampled on Day 3, Day 6, and Day 9 for GlgP Western blot assays, glycogen phosphorylase activity calculations, and glycogen content determination.
It has been confirmed that the GlgP abundances and activities could be artificially regulated through theophylline dose in the XC8 strain; thus, we further determined the influence of artificially regulated glycogen phosphorylase activities on glycogen storage in Synechococcus cells. Intracellular glycogen contents of XC8 and wildtype control were calculated on Day 3, Day 6, and Day 9 of the flask cultivation process (Figure 2D). As expected, intracellular glycogen contents of the recombinant strain showed a significantly negative relationship with the theophylline dose and GlgP abundances. Glycogen contents in the wildtype PCC 7942 cells were maintained in the range between 10 mg/gDCW and 30 mg/gDCW. While for the XC8 recombinant strain, when no theophylline was added (meaning that the translation of glgP transcripts was still inhibited), the intracellular contents were increased by 2–3 folds compared with those of the wildtype control. When theophylline was supplemented in the culture broth, glycogen storage in XC8 cells would be correspondingly reduced. However, it is also noteworthy that although the GlgP abundances and activities could be increased by theophylline supplementation to levels much higher than those of the wildtype control, the glycogen contents would still be maintained on a normal physiological level similar to the wildtype control.
With relatively weak illumination (50 μmol photons/m2/s) and limited carbon supply for the cells cultivated in flasks, the glycogen synthesis and storage in PCC 7942 cells were maintained at a relatively low level; thus, the effects of enhanced glgP expression and glycogen degradation activities might not be significant enough. We supposed that under conditions facilitating rapid glycogen accumulation, the effects of enhanced glycogen degradation activities might be more obvious; thus, we further evaluated the performance of the XC8 cells in column photobioreactors with bubbled 3% carbon-air gas. The illumination strengths were also elevated to 150 μmol photons/m2/s. As shown in Figure 3A, with enhanced illumination and carbon supply, both the wildtype and the XC8 strains grow significantly faster, with or without theophylline inductions, while the theophylline induction significantly reduced the final cell densities of XC8. During the cultivation process, intracellular glycogen contents of the wildtype were gradually increased, reaching from about 40 mg/gDCW on Day 3 to about 120 mg/gDCW on Day 6 and then were maintained on a stable level in the stationary phase; the addition of theophylline caused no influence on glycogen contents as well as cell growths (Figure 3B). As for XC8, when no theophylline was added, the glycogen contents would be maintained on a much higher level than those of the wildtype control, although the difference was declining with the prolonged cultivation process (about 5-fold on Day 3 and 1.75-fold on Day 6). When 500 μM theophylline was added, the glycogen contents would be reduced by about 70% to 60 mg/gDCW after 6 days of induction and cultivation, which is about 50% of the wildtype level on Day 6. The growth of the XC8 was reduced by the theophylline-induced glgP overexpression (Figure 3A). The final cell density was about 30% lower than that of the controls, indicating that the elevated glycogen turn-over rates might cause a non-significant cycle between glucose-1-phosphate and glycogen, leading to waste or inefficient utilization of the photosynthesis-derived energy and material flow.
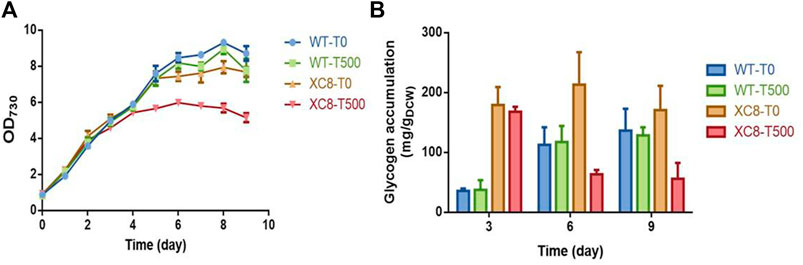
FIGURE 3. Effects of theophylline-regulated glgP expression on PCC 7942 cell growths and glycogen contents in column photobioreactors. (A) Cell growth and (B) glycogen contents of the wildtype and XC8 strains when cultivated in column bioreactors. The cultivation would be bubbled with CO2 (3% in air) and supplied with elevated illuminations (150 μmol photons/m2/s). 0 μM (T0) and 500 μM (T500) theophylline were added to the culture broth.
As the most important carbon sink mechanism in cyanobacteria, glycogen metabolism regulates and buffers the intracellular carbon flow distribution derived from photosynthesis. When treated with salt stress, carbon distribution in cyanobacteria cells would be rewired and the osmolyte synthesis would take up a significant portion of intracellular carbon, indicating potential interactions and competition with the glycogen metabolism network. Sucrose is the primary osmolyte synthesized in PCC 7942 to resist extracellular hypersaline conditions, and previously it has been reported that the blocking of the glycogen synthesis pathway would reduce the capacity to accumulate sucrose in PCC 7942 under hypersaline conditions and inhibit the salt tolerances (Miao et al., 2003; Suzuki et al., 2010). To explore the effects of enhanced glgP expression on sucrose accumulation of PCC 7942 facing salt stress, we imposed salt stress on PCC 7942 wildtype and XC8 strains supplemented with concentrations of theophylline. As shown in Figure 4A, when cultivating in hypersaline conditions, growths of XC8 were not influenced by the addition of theophylline to regulate GlgP abundances and glycogen storage. Although glgP expression in XC8 was significantly inhibited when no theophylline was added and the glycogen would be accumulated to a 2-fold higher level than that of the wildtype control, glycogen storage would still be dramatically reduced with the accumulation of intracellular sucrose under hypersaline conditions (Figure 4B). During the cultivation process with salt stress, glycogen contents in XC8 were maintained at the same level as that of wildtype control, with or without theophylline induction. However, the theophylline induction (1,000 μM) of glgP expression failed to elevate sucrose accumulation in XC8 cells. After 12 days of salt stressed cultivation, although intracellular sucrose concentration of XC8 cells was slightly higher than that of the wildtype control, no significant difference could be detected between the samples with or without theophylline induction (Figure 4C). Under hypersaline conditions, sucrose would be synthesized and accumulated in Synechococcus cells to resist the hyperosmotic stress and would be maintained at a favorable concentration rather than unlimited accumulation, which might be a possible explanation for the phenomenon that the reduction of glycogen contents failed to bring in elevated sucrose yield.
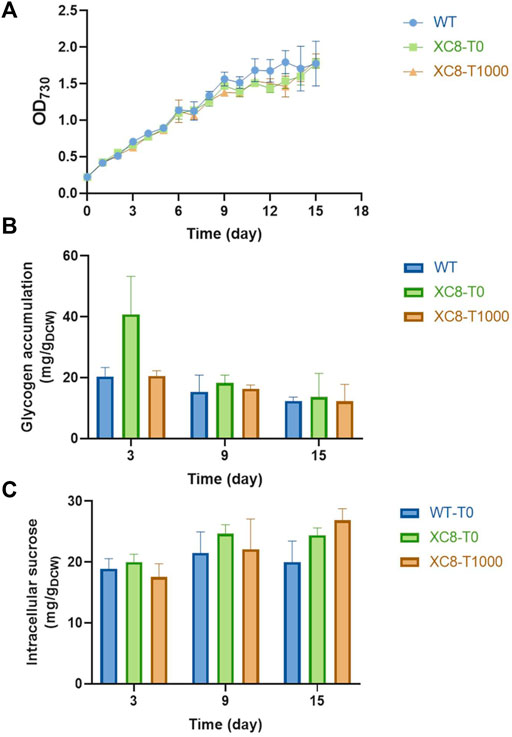
FIGURE 4. Effects of theophylline-regulated glgP expression on salt stress tolerance and intracellular sucrose storage in PCC 7942. (A) Growths of PCC 7942 wildtype and XC8 mutant strains facing 150 mM salt stress imposed on Day 3 with different doses of theophylline. (B) Glycogen storage of PCC 7942 wildtype and XC8 mutant strains facing 150 mM salt stress imposed on Day 3 with different doses of theophylline. (C) Intracellular sucrose concentrations of PCC 7942 wildtype and XC8 mutant strains facing 150 mM salt stress imposed on Day 3 with different doses of theophylline.
Previously, it has been confirmed that the introduction of the E. coli–sourced cscB sucrose permease could facilitate the secretory synthesis of sucrose, which removes the potential intracellular over-accumulation effects and significantly improved sucrose production titers in cyanobacteria (Ducat et al., 2012; Song et al., 2016). Inspired by this, we explored whether the GlgP overexpression strategy could be used for enhancing secretory sucrose production in engineered cell factories.
With a previously developed engineered strain FL130 carrying a heterologous cscB gene and an overexpressed native sps gene (encoding the sucrose-6-phosphate synthase, the rate-limiting enzyme catalyzing sucrose synthesis in PCC 7942), we utilized the Ptrc-ENYC4 system to replace the native glgP promoter sequence and obtained the recombinant strain XC14 (Figure 5A). As shown in Figures 5B,C, when 1,000 μM theophylline was added to induce the overexpression of glgP, growth of XC14 would be inhibited, and the glycogen contents were reduced to a similar level to those of the FL130 control. After 150 mM NaCl was added to activate Sps and sucrose synthesis, glycogen storage in all the sets would be continuously decreased, indicating that a significantly rewired intracellular carbon distribution leads to enhanced sucrose production. After 3 days of cultivation under salt stress, glycogen contents in the XC14 strain without theophylline induction were still about 2-folds higher than those of the FL130 control and the XC14 cells with induction of 1,000 μM theophylline (Figure 5C). However, the difference in glycogen storage and glgP expressions caused minor effects on specific productivities of secreted sucrose (Figure 5D) on the per cell levels, and the final sucrose titer of the theophylline-induced XC14 strain was lower than that of the control due to the reduced cell growth. Comparing the wildtype and XC8 strains cultivated in flasks and column photobioreactors, the theophylline-induced glgP overexpression failed to improve the actual performances of the sucrose synthesis cell factories stressed by 150 mM NaCl. A possible explanation would be the low contribution ratio of glycogen storage in the salt-induced sucrose synthesis process. Due to the introduction of sucrose permease, the limitation of intracellular sucrose accumulation was removed, and a much larger portion of the carbon source would be rewired into sucrose synthesis, which would be then secreted to the extracellular environments. The carbon source from mobilized glycogen storage would take a much lower ratio for sucrose synthesis than that in the non-secretory mode. Thus, enhanced glycogen degradation could only bring in negligible contribution to carbon flow distribution, while the burden on protein overexpression and theophylline further caused the weakened performance of cell growth and sucrose titers, which is in accordance with the previously reported phenomenon by Ducat et al. (2012).
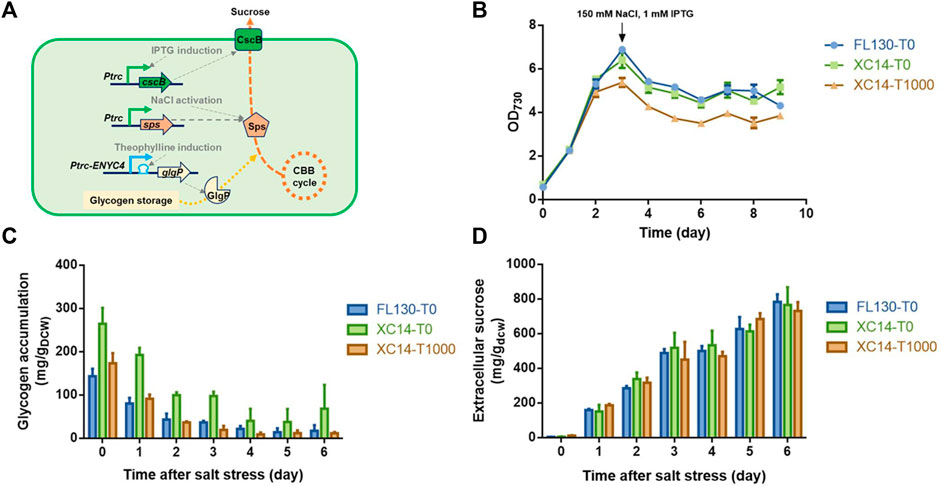
FIGURE 5. Effects of theophylline-induced glgP overexpression on salt-induced sucrose synthesis and secretion in PCC 7942 cell factories. (A) Design and working mode of the theophylline-regulated glgP expression system in a previously constructed sucrose-synthesizing strain FL130. The expression of the cscB gene would be induced by the addition of IPTG, facilitating sucrose secretion out of the cells. An additional copy of the native sucrose-6-phosphate synthase gene would be expressed by a constitutive strong promoter Ptrc, and when salt stress was imposed, the intracellular Sps would be activated by increased concentrations of NaCl, leading to enhanced sucrose synthesis with the carbon source from CBB cycle and glycogen storage. The promoter sequence of the native glgP gene would be replaced with the Ptrc-ENYC4 system, and the addition of theophylline would lead to overexpression of glgP and enhanced glycogen degradation. The strain derived from FL130 was termed XC14. (B) Cell growths, (C) glycogen storage, and (D) extracellular sucrose production of FL130 and XC14 (with or without theophylline induction) strains before and after salt stress on Day 3. T0 and T1000 represented theophylline concentrations of 0 and 1,000 μM, respectively.
Based on the effects of theophylline-induced glgP expression on glycogen storage and sucrose synthesis in XC14 facing salt stress, we supposed that this strategy might more possibly be valuable in non-hypersaline conditions to link the glycogen degradation metabolism and synthesis of desired metabolites. As for PCC 7942, sucrose synthesis would be activated through salt ion–induced activation of sucrose phosphate synthase and inhibition of invertase (Liang et al., 2020), while it has been confirmed that Sps from PCC 6803 (hereafter termed as Sps6803 for short) was not a salt-activated enzyme, and introduction of the Sps6803 in the another Synechococcus strain UTEX 2973 successfully resulted in a secretory synthesis of sucrose without the need for salt stress induction (Lin et al., 2020). We took the same strategy to construct a PCC 7942–derived cell factory producing sucrose under non-hypersaline conditions by overexpressing Sps6803 in the FL92 strain which was previously developed and carried the same cscB-expression cassette as FL130 (Figure 6A), and the strain was termed as JS28 (Ptrc-cscB; PcpcB-sps6803). As shown in Figure 6B, growth patterns of the FL130, JS28, and the two respective glgP-overexpressing strains (JS33 and JS34) were quite similar under non-hypersaline conditions, while the glgP-overexpression caused a slight decrease of cell densities. As shown in Figure 6D, JS28 could synthesize about 500 mg/L sucrose, about 2.5 folds higher than that of the FL130 (Ptrc-cscB; Ptrc-sps) under the same non-hypersaline conditions. Glycogen storage in JS28 was also 50% lower than that of FL130, indicating that the secretory sucrose synthesis deprived a large portion of carbon flow from glycogen storage under non-hypersaline conditions (Figure 6C).
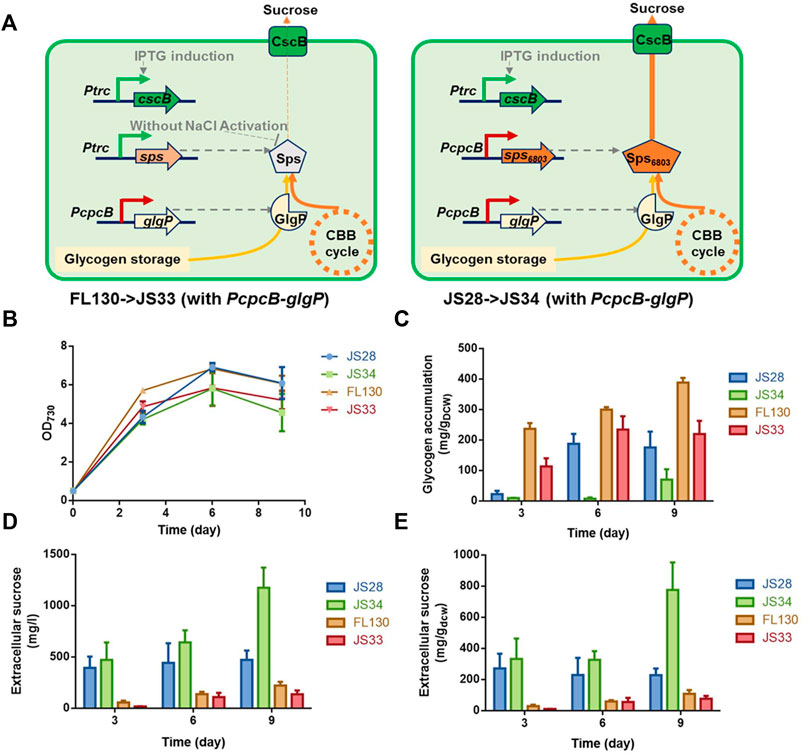
FIGURE 6. Effects of glgP overexpression on sucrose synthesis of PCC 7942 cell factories under non-hypersaline conditions. (A) To facilitate sucrose synthesis under non-hypersaline conditions, the PCC 6803 sourced sps gene sps6803 was put under the control of the PcpcB promoter to replace the Ptrc expressed native sps gene in FL130, and the obtained strain was termed as JS28. To enhance glgP expression, a PcpcB promoter–driven glgP expression cassette was introduced into FL130 and JS28, generating JS33 and JS34, respectively. Without salt stress induction, the Sps in FL130 and JS33 would be inactivated, and the sucrose synthesis would be inhibited. In JS28 and JS34, the NaCl-activation–independent Sps6803 could normally catalyze the sucrose synthesis process. Cultivated in column photobioreactors in standard conditions without salt stress, (B) cell growth, (C) glycogen contents, and (D,E) sucrose production of FL130, JS33, JS28, and JS34 cells were calculated in 9 days.
To explore the effects of enhanced glycogen degradation, we expressed glgP with a strong constitutive promoter PcpcB in both JS28 and FL130 (Figure 6A) and obtained the strains JS34 (Ptrc-cscB; PcpcB-sps6803; PcpcB-glgP) and JS33 (Ptrc-cscB; Ptrc-sps; PcpcB-glgP) respectively. As expected, glycogen storage in both of the two strains was reduced. In JS34, sucrose synthesis was correspondingly increased by about 2.4-folds compared with that of JS28 (with no glgP overexpression), reaching about 1,200 mg/L (accounting for up to about 40% of the fixed carbon, Figure 6E), while in JS33 (FL130 with PcpcB expressed glgP), the sucrose productivities and sucrose titers were not significantly increased compared with those of FL130, indicating that as for FL130 cells under non-hypersaline conditions, carbon flux was not the rate-limiting factor restricting sucrose synthesis.
Previously Ducat et al. have reported that combined overexpression of glgP and cscB could not improve sucrose productivities while causing a 5%–10% decrease (Ducat et al., 2012). In this work, we also evaluated the performances of similar strains JS33 (Ptrc-cscB; Ptrc-sps; PcpcB-glgP) cultivated under hypersaline conditions and got similar results when compared with the FL130 control (Ptrc-cscB; Ptrc-sps). As shown in Supplementary Figure S2, when cultivated under hypersaline conditions, expression of glgP reduced the sucrose titer from 2 g/L to 1.5 g/L after 9 days of cultivation. As mentioned above, the sharply reduced glycogen storage of salt-stress–induced sucrose-synthesizing cells provides a reasonable explanation for that phenomenon. As for FL130 strain facing continuous salt stress, a large portion of the carbon source (including the glycogen storage) has been motivated for sucrose synthesis; thus, the enhanced glgP expression and glycogen degradation activities could only bring in minor contributions but just consume more energy and resources on meaningless protein (GlgP) synthesis, which in turn causes impairment on the final sucrose yields.
The salt stress–responsive sucrose synthesis serves as a special case for cyanobacteria-based photosynthetic production, and the naturally evolved condition–induced metabolism shift spontaneously plays a role in regulating glycogen metabolism, and thus the artificially implemented glycogen motivation strategy could not bring in necessary effects. However, as for a majority of cyanobacteria metabolic engineering cases, the carbon sink in glycogen storage (Davies et al., 2014; van der Woude et al., 2014; David et al., 2018) and how to rewire the natural carbon sink into artificially assembled heterologous pathway was still an important issue to be solved. With the salt stress–independent sucrose synthesizing cell factories as a model, we have confirmed the feasibility of regulating the glycogen degradation pathway to engineer the glycogen metabolism and production capacities of the desired product.
Unless noted otherwise, all reagents were purchased from Sigma-Aldrich (United States). Taq DNA polymerase and all restriction enzymes were purchased from Fermentas (Canada) or Takara (Japan). The kits used for molecular cloning were obtained from Omega (United States) or Takara (Japan). Oligonucleotides were synthesized, and DNA-sequencing was performed by Genewiz (Suzhou, China).
All the cyanobacterial strains utilized in this work are listed in Table 1. To achieve theophylline dose–regulated glgP expression in PCC 7942 and FL130, a 155-bp DNA fragment, containing the Ptrc promoter and the theophylline-dependent riboswitch ENYC4, was synthesized according to the reported sequence (Nakahira et al., 2013). A KmR (kanamycin resistance gene) fragment, a 1.3-kb upstream fragment of the glgP gene (Synpcc7942_0244, according to the sequence from KEGG), the 155-bp Ptrc-ENYC4 fragment, and a 1.0-kb glgP ORF (open reading frame) fragment were ligated in sequence (5′->3′) through infusion PCR, and the obtained fragment would be cloned into pMD18T (TaKaRa, Dalian, China) to get the pDY150 plasmid. The plasmid pDY150 would be transformed into PCC 7942 wildtype strain for construction of XC8 and into FL130 for the construction of XC14.
To achieve sucrose synthesis under non-hypersaline conditions in PCC 7942, the PCC 6803-sourced sps gene (sll0045, according to the sequence from KEGG, termed as sps6803) would be cloned and introduced into FL92. The backbone of a previously constructed plasmid pQL225 (Qiao et al., 2018), containing upstream and downstream homologous fragments of neutral site 2 (NS2) on the chromosome of PCC 7942 and the PcpcB promoter sequence, would be amplified and fused with the sps6803 fragment and the spectinomycin-resistance gene (SpR) fragment using the Seamless Assembly Cloning Kit (CloneSmarter, C5891) to generate the plasmid pJS13. The pJS13 plasmid would be transformed into FL92 for construction of JS28.
To enhance glgP expression in FL130 and JS28, the native glgP gene from PCC 7942 would be amplified. The backbone of a previously constructed plasmid pQL225 (Qiao et al., 2018) containing upstream and downstream homologous fragments of neutral site 2 (NS2) on the chromosome of PCC 7942, the PcpcB promoter sequence, and the gentamicin resistance gene (GmR) fragment would be amplified and fused with the glgP fragment to generate the plasmid pJS23. The backbone of another previously constructed plasmid pFL20n (Duan et al., 2016) containing the upstream and downstream homologous fragments of neutral site 1 (NS1) on the chromosome of PCC 7942 would be amplified and ligated with the PcpcB-glgP-GmR fragment from the pJS23, and the generated plasmid was termed as pJS22. Plasmids pJS22 and pJS23 were transformed to the strains JS28 and FL130, respectively, for the construction of JS34 and JS33.
Escherichia coli DH5α was used as the host for the construction of plasmids. The respective plasmids would be transformed into Synechococcus cells, and the antibiotic-resistant transformants were usually obtained after 7–10 days of cultivation on selective BG11 agar plates. The genotypes of the transformants were verified by PCR and DNA-sequencing.
The wildtype and engineered Synechococcus strains were cultivated with BG11 medium (Rippka et al., 1979) in 250-ml flasks or 350-ml column photobioreactors (580 mm by 30 mm). The cultivation with flasks would be performed in a horizontal rotary shaker with 130 rpm under moderate intensity (30–50 μmol photons/m2/s) provided by white-light illumination lamps. When the column photobioreactors were utilized, 3% CO2 (V/V in the air) would be bubbled to provide a carbon source, and the illuminations would be elevated to about 150 μmol photons/m2/s. As for the recombinant strains, concentrations of antibiotics (20 μg/ml spectinomycin, 20 μg/ml kanamycin, 10 μg/ml chloramphenicol, and 2 μg/ml gentamicin) would be added to BG11 medium when required. When multiple antibiotics are used at the same time, the concentration of each antibiotic would be reduced. During the cultivation process, cell growths were calculated by measuring the optical density at a wavelength of 730 nm (OD730) and converted to dry cell weight (DCW) with a pre-established calibration between the OD730 and DCW of PCC 7942 cultures (1.0 OD730 unit equals approximately 0.34 gDCW/L). To regulate glycogen expression or to induce sucrose synthesis, theophylline, IPTG, and NaCl would be supplemented as necessary. Concentrations of theophylline were selected based on previous experience with the Ptrc4-ENYC4 element (Qiao et al., 2018; Chi et al., 2019). At least three biological replicates were performed for each experiment to ensure repeatabilities.
Synechococcus cells were harvested by centrifugation at 4°C. The resuspended cells (in 50 mM Tris-HCl buffer, pH 8.0) were disrupted with 100-mm glass beads (Sigma) in an ice bath. After removing the cell debris and glass beads by 4°C centrifugation, the supernatants were collected. Protein concentrations of the cell-free extracts were also measured with the Bradford method. The protein samples were analyzed on 12% SDS-PAGE with a standard procedure and blotted onto PVDF membranes, sealed with 5% nonfat milk-TBST buffer (TBS added with 0.05% Tween-20) at 4°C overnight. First, the membrane was incubated with polyclonal antibodies to GlgC (1:1000) (Hangzhou HuaAn Biotechnology Co., Ltd., Hangzhou, China) for 3 h and washed three times with TBST (15 min each time). Second, the membrane was incubated with an alkaline phosphatase–linked secondary antibody (goat anti-rabbit, Invitrogen, Shanghai, China) for 1 h and washed three times with TBST (15 min each time). Finally, the membrane was developed with BCIP/NBT (Sigma).
To evaluate the sucrose production of the strains constructed in this work, the culture broth would be sampled and centrifuged at 10,000 g. To determine the extracellular sucrose amounts, the supernatants would be analyzed with the Sucrose/D-glucose Assay Kit (Megazyme, K-SUCGL). To calculate the intracellular sucrose concentrations, the pellet would be resuspended with 80% (V / V) ethanol and heated at 65°C for 4 h. After being centrifuged at 15,000 g for 5 min, the supernatant would be carefully collected and completely dried by blowing of N2 stream at 55°C. Finally, the dried samples would be resuspended with ddH2O and assayed with the Sucrose/D-glucose Assay Kit.
Glycogen phosphorylase activities of the Synechococcus strains were calculated following the previously introduced protocols with modifications (Fu and Xu, 2006). The cells were resuspended and disrupted in buffer A (containing 18 mM KH2PO4, 27 mM Na2HPO4, 15 mM MgCl2, 100 μM EDTA, pH 6.8). A 200-μl reaction system for assay contained 30 μl crude enzyme, 18 mM KH2PO4, 27 mM Na2HPO4, 15 mM MgCl2, 100 μM EDTA, 340 μM Na2NADP (Amresco), 4 μM glucose-1,6-biphosphate (Santa Cruz), six units ml-1 glucose-6-phosphate dehydrogenase (Sigma), 0.8 units ml-1 phosphoglucomutase (Sigma), and 2 mg ml-1 glycogen (Sigma). The same system lacking glycogen was used as a control. GlgP activities were measured by calculating the generation speeds of NADPH with a change in absorbance at 340 nm. The calculations of Abs340 were performed with 96-well plates by a microplate reader. The reactions were initiated by adding the crude enzyme.
Intracellular glycogen contents would be calculated as previously described (Grundel et al., 2012), with minor modifications. Synechococcus cells would be collected by centrifugation at 10,000 g for 15 min. The pellet would be washed three times with ddH2O, resuspended with 30% (W/V) KOH, heated at 95°C for 2 h, and finally, ice-cooled ethanol would be added to a final concentration of 70%–75% (V/V). The mixture would be cooled at -20°C overnight and then centrifuged at 15,000 g for 15 min to collect the glycogen precipitation. The glycogen pellet would be washed twice with 70% ethanol (V/V) and 98% ethanol (V/V) successively and then dried by vacuum centrifugation. The glycogen finally obtained was suspended in 100 mM sodium acetate and digested by amyloglucosidase (Novozymes). The glucose contents generated in the glycogen solution were determined with the Sucrose/D-glucose Assay Kit.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
YD, JS, SZ, and YW, performed the research project. SM, GL, and XL supervised the research project and guided the design of experiments. YD, JS, SZ, SM, GL, and XL. drafted and revised the manuscript. All authors read the manuscript and agree to submit to Frontiers in Bioengineering and Biotechnology.
This work was supported by the National Key Research and Development Program of China (Grant number 2021YFA0909700), the National Science Foundation of China (Grant numbers 31872624, 32070084, and 31770092), the DNL Cooperation Fund, CAS (DNL202014), and the Youth Innovation Promotion Association CAS (to GL) the Shandong Taishan Scholarship (to XL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.925311/full#supplementary-material
Aikawa, S., Nishida, A., Ho, S.-H., Chang, J.-S., Hasunuma, T., and Kondo, A. (2014). Glycogen Production for Biofuels by the Euryhaline Cyanobacteria Synechococcus Sp. Strain PCC 7002 from an Oceanic Environment. Biotechnol. Biofuels 7, 88. doi:10.1186/1754-6834-7-88
Angermayr, S. A., Hellingwerf, K. J., Lindblad, P., and Teixeira de Mattos, M. J. (2009). Energy Biotechnology with Cyanobacteria. Curr. Opin. Biotechnol. 20, 257–263. doi:10.1016/j.copbio.2009.05.011
Ball, S. G., and Morell, M. K. (2003). From Bacterial Glycogen to Starch: Understanding the Biogenesis of the Plant Starch Granule. Annu. Rev. Plant Biol. 54, 207–233. doi:10.1146/annurev.arplant.54.031902.134927
Carrieri, D., Paddock, T., Maness, P.-C., Seibert, M., and Yu, J. (2012). Photo-catalytic Conversion of Carbon Dioxide to Organic Acids by a Recombinant Cyanobacterium Incapable of Glycogen Storage. Energy Environ. Sci. 5, 9457–9461. doi:10.1039/c2ee23181f
Chi, X., Zhang, S., Sun, H., Duan, Y., Qiao, C., Luan, G., et al. (2019). Adopting a Theophylline-Responsive Riboswitch for Flexible Regulation and Understanding of Glycogen Metabolism in Synechococcus Elongatus PCC7942. Front. Microbiol. 10, 551. doi:10.3389/fmicb.2019.00551
Damrow, R., Maldener, I., and Zilliges, Y. (2016). The Multiple Functions of Common Microbial Carbon Polymers, Glycogen and PHB, during Stress Responses in the Non-diazotrophic Cyanobacterium Synechocystis Sp. PCC 6803. Front. Microbiol. 7, 966. doi:10.3389/fmicb.2016.00966
David, C., Schmid, A., Adrian, L., Wilde, A., and Bühler, K. (2018). Production of 1,2-propanediol in photoautotrophicSynechocystisis Linked to Glycogen Turn-Over. Biotechnol. Bioeng. 115, 300–311. doi:10.1002/bit.26468
Davies, F. K., Work, V. H., Beliaev, A. S., and Posewitz, M. C. (2014). Engineering Limonene and Bisabolene Production in Wild Type and a Glycogen-Deficient Mutant of Synechococcus Sp. PCC 7002. Front. Bioeng. Biotechnol. 2, 21. doi:10.3389/fbioe.2014.00021
Duan, Y., Luo, Q., Liang, F., and Lu, X. (2016). Sucrose Secreted by the Engineered Cyanobacterium and its Fermentability. J. Ocean. Univ. China 15, 890–896. doi:10.1007/s11802-016-3007-8
Ducat, D. C., Avelar-Rivas, J. A., Way, J. C., and Silver, P. A. (2012). Rerouting Carbon Flux to Enhance Photosynthetic Productivity. Appl. Environ. Microbiol. 78, 2660–2668. doi:10.1128/aem.07901-11
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincón, J., Zabala, L. L., Jiao, N., et al. (2013). Present and Future Global Distributions of the Marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. doi:10.1073/pnas.1307701110
Fu, J., and Xu, X. (2006). The Functional Divergence of Two glgP Homologues in Synechocystis Sp. PCC 6803. Fems Microbiol. Lett. 260, 201–209. doi:10.1111/j.1574-6968.2006.00312.x
Gründel, M., Scheunemann, R., Lockau, W., and Zilliges, Y. (2012). Impaired Glycogen Synthesis Causes Metabolic Overflow Reactions and Affects Stress Responses in the Cyanobacterium Synechocystis Sp. PCC 6803. Microbiology-Sgm 158, 3032–3043. doi:10.1099/mic.0.062950-0
Guerra, L. T., Xu, Y., Bennette, N., Mcneely, K., Bryant, D. A., and Dismukes, G. C. (2013). Natural Osmolytes Are Much Less Effective Substrates Than Glycogen for Catabolic Energy Production in the Marine Cyanobacterium Synechococcus Sp. Strain PCC 7002. J. Biotechnol. 166, 65–75. doi:10.1016/j.jbiotec.2013.04.005
Hagemann, M. (2011). Molecular Biology of Cyanobacterial Salt Acclimation. Fems Microbiol. Rev. 35, 87–123. doi:10.1111/j.1574-6976.2010.00234.x
Hendry, J. I., Prasannan, C., Ma, F., Möllers, K. B., Jaiswal, D., Digmurti, M., et al. (2017). Rerouting of Carbon Flux in a Glycogen Mutant of Cyanobacteria Assessed via Isotopically Non-stationary 13 C Metabolic Flux Analysis. Biotechnol. Bioeng. 114, 2298–2308. doi:10.1002/bit.26350
Hickman, J. W., Kotovic, K. M., Miller, C., Warrener, P., Kaiser, B., Jurista, T., et al. (2013). Glycogen Synthesis Is a Required Component of the Nitrogen Stress Response in Synechococcus Elongatus PCC 7942. Algal Res. 2, 98–106. doi:10.1016/j.algal.2013.01.008
Jacobsen, J. H., and Frigaard, N.-U. (2014). Engineering of Photosynthetic Mannitol Biosynthesis from CO2 in a Cyanobacterium. Metab. Eng. 21, 60–70. doi:10.1016/j.ymben.2013.11.004
Li, X., Shen, C. R., and Liao, J. C. (2014). Isobutanol Production as an Alternative Metabolic Sink to Rescue the Growth Deficiency of the Glycogen Mutant of Synechococcus Elongatus PCC 7942. Photosynth. Res. 120, 301–310. doi:10.1007/s11120-014-9987-6
Liang, Y., Zhang, M., Wang, M., Zhang, W., Qiao, C., Luo, Q., et al. (2020). Freshwater Cyanobacterium Synechococcus Elongatus PCC 7942 Adapts to an Environment with Salt Stress via Ion-Induced Enzymatic Balance of Compatible Solutes. Appl. Environ. Microbiol. 86, e02904–02919. doi:10.1128/AEM.02904-19
Lin, P.-C., Zhang, F., and Pakrasi, H. B. (2020). Enhanced Production of Sucrose in the Fast-Growing Cyanobacterium Synechococcus Elongatus UTEX 2973. Sci. Rep. 10, 390. doi:10.1038/s41598-019-57319-5
Lu, X. (2010). A Perspective: Photosynthetic Production of Fatty Acid-Based Biofuels in Genetically Engineered Cyanobacteria. Biotechnol. Adv. 28, 742–746. doi:10.1016/j.biotechadv.2010.05.021
Luan, G., and Lu, X. (2018). Tailoring Cyanobacterial Cell Factory for Improved Industrial Properties. Biotechnol. Adv. 36, 430–442. doi:10.1016/j.biotechadv.2018.01.005
Luan, G., Zhang, S., Wang, M., and Lu, X. (2019). Progress and Perspective on Cyanobacterial Glycogen Metabolism Engineering. Biotechnol. Adv. 37, 771–786. doi:10.1016/j.biotechadv.2019.04.005
Melis, A. (2012). Photosynthesis-to-fuels: from Sunlight to Hydrogen, Isoprene, and Botryococcene Production. Energy Environ. Sci. 5, 5531–5539. doi:10.1039/c1ee02514g
Miao, X., Wu, Q., Wu, G., and Zhao, N. (2003). Sucrose Accumulation in Salt-Stressed Cells Ofagpgene Deletion-Mutant in cyanobacteriumSynechocystissp. PCC 6803. Fems Microbiol. Lett. 218, 71–77. doi:10.1111/j.1574-6968.2003.tb11500.x
Nakahira, Y., Ogawa, A., Asano, H., Oyama, T., and Tozawa, Y. (2013). Theophylline-dependent Riboswitch as a Novel Genetic Tool for Strict Regulation of Protein Expression in Cyanobacterium Synechococcus Elongatus PCC 7942. Plant Cell Physiol. 54, 1724–1735. doi:10.1093/pcp/pct115
Nakamura, Y., Takahashi, J.-i., Sakurai, A., Inaba, Y., Suzuki, E., Nihei, S., et al. (2005). Some Cyanobacteria Synthesize Semi-amylopectin Type α-Polyglucans Instead of Glycogen. Plant Cell Physiol. 46, 539–545. doi:10.1093/pcp/pci045
Oliver, J. W. K., and Atsumi, S. (2014). Metabolic Design for Cyanobacterial Chemical Synthesis. Photosynth. Res. 120, 249–261. doi:10.1007/s11120-014-9997-4
Qiao, C., Duan, Y., Zhang, M., Hagemann, M., Luo, Q., and Lu, X. (2018). Effects of Reduced and Enhanced Glycogen Pools on Salt-Induced Sucrose Production in a Sucrose-Secreting Strain of Synechococcus Elongatus PCC 7942. Appl. Environ. Microbiol. 84, e02023–02017. doi:10.1128/AEM.02023-17
Rippka, R., Stanier, R. Y., Deruelles, J., Herdman, M., and Waterbury, J. B. (1979). Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. J. General Microbiol. 111, 1–61. doi:10.1099/00221287-111-1-1
Rousseaux, C. S., and Gregg, W. W. (2014). Interannual Variation in Phytoplankton Primary Production at a Global Scale. Remote Sens. (Basel) 6, 1–19. doi:10.3390/rs6010001
Shimakawa, G., Hasunuma, T., Kondo, A., Matsuda, M., Makino, A., and Miyake, C. (2014). Respiration Accumulates Calvin Cycle Intermediates for the Rapid Start of Photosynthesis in Synechocystis Sp. PCC 6803. Biosci. Biotechnol. Biochem. 78, 1997–2007. doi:10.1080/09168451.2014.943648
Song, K., Tan, X., Liang, Y., and Lu, X. (2016). The Potential of Synechococcus Elongatus UTEX 2973 for Sugar Feedstock Production. Appl. Microbiol. Biotechnol. 100 (18), 7865–7875. doi:10.1007/s00253-016-7510-z
Suzuki, E., Ohkawa, H., Moriya, K., Matsubara, T., Nagaike, Y., Iwasaki, I., et al. (2010). Carbohydrate Metabolism in Mutants of the Cyanobacterium Synechococcus Elongatus PCC 7942 Defective in Glycogen Synthesis. Appl. Environ. Microbiol. 76, 3153–3159. doi:10.1128/aem.00397-08
Van Der Woude, A. D., Angermayr, S. A., Puthan Veetil, V., Osnato, A., and Hellingwerf, K. J. (2014). Carbon Sink Removal: Increased Photosynthetic Production of Lactic Acid by Synechocystis Sp. PCC6803 in a Glycogen Storage Mutant. J. Biotechnol. 184, 100–102. doi:10.1016/j.jbiotec.2014.04.029
Waterbury, J. B., Watson, S. W., Guillard, R. R. L., and Brand, L. E. (1979). Widespread Occurrence of a Unicellular, Marine, Planktonic, Cyanobacterium. Nature 277, 293–294. doi:10.1038/277293a0
Work, V. H., Melnicki, M. R., Hill, E. A., Davies, F. K., Kucek, L. A., Beliaev, A. S., et al. (2015). Lauric Acid Production in a Glycogen-Less Strain of Synechococcus Sp. PCC 7002. Front. Bioeng. Biotechnol. 3, 48. doi:10.3389/fbioe.2015.00048
Xu, Y., Tiago Guerra, L., Li, Z., Ludwig, M., Charles Dismukes, G., and Bryant, D. A. (2013). Altered Carbohydrate Metabolism in Glycogen Synthase Mutants of Synechococcus Sp. Strain PCC 7002: Cell Factories for Soluble Sugars. Metab. Eng. 16, 56–67. doi:10.1016/j.ymben.2012.12.002
Keywords: cyanobacteria, glycogen, sucrose, glycogen phosphorylase, Synechococcus elongatus PCC7942
Citation: Dan Y, Sun J, Zhang S, Wu Y, Mao S, Luan G and Lu X (2022) Manipulating the Expression of Glycogen Phosphorylase in Synechococcus elongatus PCC 7942 to Mobilize Glycogen Storage for Sucrose Synthesis. Front. Bioeng. Biotechnol. 10:925311. doi: 10.3389/fbioe.2022.925311
Received: 21 April 2022; Accepted: 11 May 2022;
Published: 01 July 2022.
Edited by:
Fantao Kong, Dalian University of Technology, ChinaReviewed by:
Jaruswan Warakanont, Kasetsart University, ThailandCopyright © 2022 Dan, Sun, Zhang, Wu, Mao, Luan and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoming Mao, bXNtNTI2QDE2My5jb20=; Guodong Luan, bHVhbmdkQHFpYmVidC5hYy5jbg==; Xuefeng Lu, bHZ4ZkBxaWJlYnQuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.