
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol., 05 September 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.920639
This article is part of the Research TopicInsights in Synthetic Biology 2021: Novel Developments, Current Challenges, and Future PerspectivesView all 16 articles
Biomining is a biotechnological approach where microorganisms are used to recover metals from ores and waste materials. While biomining applications are motivated by critical issues related to the climate crisis (e.g., habitat destruction due to mine effluent pollution, metal supply chains, increasing demands for cleantech-critical metals), its drawbacks hinder its widespread commercial applications: lengthy processing times, low recovery, and metal selectivity. Advances in synthetic biology provide an opportunity to engineer iron/sulfur-oxidizing microbes to address these limitations. In this forum, we review recent progress in synthetic biology-enhanced biomining with iron/sulfur-oxidizing microbes and delineate future research avenues.
Biomining is a technique that utilizes microorganisms to recover target metals from ores and waste materials, and potentially capture them for recovery and downstream purification. Chemolithotrophic iron/sulfur (Fe/S)-oxidizing microbes have attracted attention in biomining research since they thrive in extremely acidic metal-rich environments, produce a bioleaching lixiviant containing Fe3+ and H+ by oxidizing ferrous iron and/or reduced inorganic sulfur compounds (RISCs) as an energy source, and fix CO2 from the atmosphere as the carbon source (Chen et al., 2022a). Considerable efforts have been devoted to applying synthetic biology to improve the biomining performance of Fe/S-oxidizing microbes, and significant progress has been made in recent years. This mini review presents the up-to-date developments of synthetic biology in biomining with Fe/S-oxidizing microbes and provides perspectives on this research area.
A typical biomining microbial community consists of autotrophic Fe/S-oxidizers, mixotrophs and heterotrophs (Li and Wen, 2021) (Figure 1). Neutrophilic Fe-oxidizers (eg. Rhizobiales spp. and Ralstonia spp.) may play a role to kick start the mineral oxidation at circumneutral pH (Percak-Dennett et al., 2017). The acidophilic Fe-oxidizers (e.g., Leptospirillum spp. and Ferroplasma spp.) obtain electrons from dissolved Fe2+ for energy generation, thus generating Fe3+ that can attack the mineral’s lattice structure to release additional iron, target metals and sulfur compounds. However, S0 or jarosite (KFe3(SO4)2(OH)6) can accumulate to form a passivation layer covering the mineral surface thus inhibiting further leaching. S-oxidizers (e.g., Acidianus spp., some species in Acidithiobacillus and Sulfolobus) can oxidize sulfur compounds like S0 to sulfuric acid. This process generates additional H+ that assists Fe3+ in further degrading the mineral lattice structure to release the target metal. Combined, Fe-oxidizers and S-oxidizers maintain extremely acidic conditions with a high oxidation-redox potential (ORP) to leach target metals from the mineral without precipitating them. Moreover, mixotrophs and heterotrophs found in genera such as Acidiphilium, Metallosphaera, Sulfobacillus and Sulfolobus can also degrade organic compounds mitigating potential toxicity issues to autotrophic Fe/S-oxidizers.
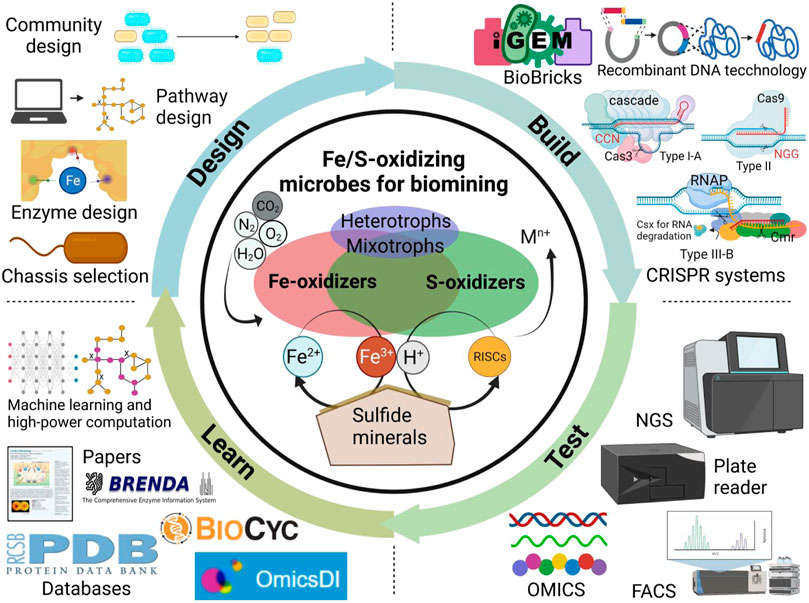
FIGURE 1. Harnessing synthetic biology for sustainable biomining with Fe/S-oxidizing microbes. Technology advances accelerate the design-build-test-learn cycle of synthetic biology-assisted biomining. NGS, next-generation sequencing; FACS, fluorescence-activated cell sorting, OMICS refers to genomics, proteomics, transcriptomics, metabolomics, and fluxomics.
Several species in the genera Acidithiobacillus are capable of oxidizing both Fe2+ and RISCs. In particular, Acidithiobacillus ferrooxidans is the most widely studied chassis for biomining applications. Mixed cultures of A. ferrooxidans with other Fe-oxidizing and S-oxidizing microbes, mixotrophs and heterotrophs have been shown to be effective for bioleaching (Wang et al., 2020). Hence, A. ferrooxidans with its metabolic versatility and ability to fix CO2 is a prime target for synthetic biology efforts to enhance biomining kinetics, to produce value-added products from CO2 and to confer advanced functionality like selective metal recovery. In addition, new Fe/S-oxidizing microbes found in extreme environments will also contribute valuable genetic diversity, such as the novel archaeal group found in Yellowstone National Park and Zetaproteobacteria from marine hydrothermal vents (Kozubal et al., 2013; McAllister et al., 2019).
A hallmark of synthetic biology is the application of principles outlined in the design-build-test-learn (DBTL) cycle commonly found in traditional engineering disciplines to the field of biology. This DBTL cycle integrates rational design, genetic engineering, results testing, and data analysis to learn and resolve bottlenecks in biomanufacturing. Such a DBTL cycle could be specifically applied to engineering Fe/S-oxidizing microbes for biomining applications (Figure 1). For Design, computational modelling tools combined with biological databases (e.g., genome sequences, OMICS datasets) enable the rational design of enzymes, metabolic pathways, and communities of biomining microbes. A genome-scale metabolic model (GEM) of A. ferrooxidans (iMC507) is available for in silico analysis and design, and has been used to predict gene deletions for growth-coupled extracellular polymeric substance (EPS) production (Campodonico et al., 2016).
For Build, there are genetic engineering tools to engineer Fe/S-oxidizing microbes with desirable biomining features. Some conjugative/mobilizable plasmids and viruses have been reported in species of Acidianus, Acidiphilium, Leptospirillum, Metallosphaera and Sulfobacillus to allow for heterologous gene expression (Beard et al., 2021). However, significant advances have been made in Acidithiobacillus and archaea Sulfolobus since prior efforts were mostly devoted to studying these model organisms for acidophiles. For Sulfolobus spp., the endogenous type I-A and type III-B CRISPR/Cas systems have been harnessed for efficient targeted genome editing and gene silencing (Peng et al., 2017). Comparatively, genetic engineering of Acidithiobacillus spp. is time-consuming due to its generally slower growth rate, thus making it harder to engineer. Nonetheless, studies have shown it is still possible to use conventional genome editing methods like transposon-based gene interruption or suicide plasmid-based gene deletion (Chen et al., 2022a). Recently, Moya-Beltrán et al. investigated type IV CRISPR-Cas systems in the class Acidithiobacillia, which may enable genome editing like Sulfolobus spp. (Moya-Beltrán et al., 2021). Recently, there was a report of a successful gene knockdown in A. ferrooxidans using the CRISPR-dCas9 system derived from S. pyogenes (Yamada et al., 2022). At the same time, our group first achieved seamless genome editing using the CRISPR-Cas9 system in Acidithiobacillus ferridurans, which significantly accelerates the genetic engineering process (Chen et al., 2022b). However, additional studies are needed to apply CRISPR systems in Acidithiobacillus strains and other Fe/S-oxidizing microbes for biomining applications.
The Test and Learn phases of the DBTL involve validating and analyzing data generated from engineered strains for subsequent engineering. These components of the DBTL cycle are most underdeveloped for biomining microbes. The development of high-throughput technologies such as next-generation sequencing (NGS), automated liquid handling/plate reader workflows, fluorescence-activated cell sorting (FACS), and microfluidics have facilitated the screening of large strain libraries (Sarnaik et al., 2020). These big datasets combined with existing databases like OmicsDI, BioCyc, BRENDA, and PDB can enable machine learning methods to generate critical insights to inform design iterations. While for example high-throughput sequencing of 16S rRNA gene has been used in biomining studies to monitor microbial community dynamics, efforts are still needed in high-throughput engineering, culturing, testing, screening of Fe/S-oxidizing microbes with desirable biomining traits.
While a comprehensive DBTL methodology is yet to be applied for engineering biomining, there are several examples where synthetic biology has been deployed in this context. Gumulya et al. (2018) reviewed the manipulation of genes responsible for acid tolerance, metal tolerance, osmotolerance, thermotolerance, Fe/S-oxidation, and carbon fixation to improve microbe’s suitability for industrial use. In Acidithiobacillus strains, overexpression of rus and cyc2 genes was found to improve Fe2+ oxidation activity, whereas overexpression of the quorum sensing (QS) operon afeI-orf-afeR improved sulfur oxidation rates and promoted EPS synthesis. Glutathione biosynthesis gene gshB overexpression significantly increased intracellular reactive oxygen species (ROS) levels and expanded the halotolerance of the cells. Other genes related to Fe/S-metabolisms (e.g., fur, tetH, sdo1, sdo2, rsrR, rsrS, and sor), metal resistance (e.g., cueO, ars and mer), c-di-GMP signaling (e.g., dgc and pelD) and quorum sensing (e.g., aar, ado and act) were also investigated (Jung et al., 2021).
A deeper understanding of Fe/S-oxidizing microbes’ genes is nonetheless still needed for more detailed metabolic engineering towards desirable biomining properties, compared to the model chassis Escherichia coli and Saccharomyces cerevisiae. Schmitz et al. (2021) harnessed high-throughput genome editing and sequencing approaches in the study of heterotrophic bacterium Gluconobacter oxydans to produce organic acids for rare earth elements biomining. They created a library of single-gene transposon mutants in G. oxydans and found the bioleaching rates of rare earth elements increased up to 18% when phosphate-specific transport systems genes were disrupted. Therefore, high-throughput engineering, culture, sequencing, screening processes may facilitate faster development of engineered Fe/S-oxidizing microbes.
Biomining processes are commercially applied around the world in two broad categories: irrigation-type and stirred tank-type. Irrigation-type examples include heap bioleaching processes carried out by companies like Newmont Mining and Phelps Dodge, which are suitable for metal recovery from low-grade ores because these systems are easier and cheaper to construct, although the process can be slow with inefficient recoveries (Petersen, 2016). In contrast, the more widely used processes are of stirred tank-type, such as BIOX™ technology from Metso-Outotec. The control of operating parameters contribute significantly to the fast and efficient metal recovery, but the operating cost is higher (Rawlings et al., 2003). Recently, Kremser et al. (2020) compared the potential of heap and stirred-tank bioreactors for metal recovery from shredder-light-fractions. Pure and co-culture of A. ferrooxidans and L. ferrooxidans were used for Cu, Zn and Ni recovery in batch and up-scale experiments, which highlights the potential for future commercial biomining applications with engineered Fe/S-oxidizing microbes. However, for large-scale applications, both heap and stirred-tank biomining provide an open, non-sterile environment. This open environment can lead to difficulties in optimizing a stable microbial consortia that can outcompete the native consortia and match the scale and complex nature of the ore feedstocks (Rawlings and Johnson, 2007). The design and construction of synthetic and mixed microbial consortia is an emerging area in synthetic biology, and may help to address these issues for better industrial biomining applications (Brune and Bayer, 2012; McCarty and Ledesma-Amaro, 2019).
Synthetic biology-enhanced biomining can be a sustainable, eco-friendly, and cost-effective technology with applications in leaching and recovering base metals, precious metals, and rare earth elements from a variety of feedstocks like low-grade ores, mine wastes, and electronic wastes. Though engineering Fe/S-oxidizing microbes is still at its earliest stage, we believe that the following developments can accelerate synthetic biology-enhanced biomining research:
(1) Fast and efficient genome editing methods and tools are needed for Fe/S-oxidizing microbes (i.e., CRISPR/Cas9 system). Genetic tools are available for Sulfolobus strains that are adapted to high temperature (80oC) and acidic conditions (pH < 3), highlighting their suitability for engineering (Lewis et al., 2021). We summarized the diverse genetic tools that are feasible for Acidithiobacillus species (Chen et al., 2022a). Different CRISPR systems have been successfully used for genome editing in Sulfolobus and Acidithiobacillus species. We believe genome editing methods for other Fe/S-oxidizing microbes can offer us a larger solution space for designing synthetic biomining consortia with desirable leaching properties.
(2) Development of high-throughput technologies to culture and screen of Fe/S-oxidizing microbes will accelerate the engineering of strains with more desirable industrial properties such as heavy metal tolerance, robustness under large bioreactor conditions. Example studies could include designing synthetic histidine kinases that can sense heavy-metals (Sarnaik et al., 2020), creating a whole-genome single-gene mutant library to enable screens for desired properties (Schmitz et al., 2021), or using CRISPRi system as an efficient platform for rapid identification of target genes (Wu et al., 2020).
(3) The leaching of metals from the mineral body is typically not a selective process as all metals contained in the mineral lattice are released into solution. Such a complex leachate complicates downstream processing. To mitigate this complexity, metal-sensing regulators (e.g., riboswitches) could be used to construct circuits that enable selective and sequential metal recovery by Fe/S-oxidizing microbes by regulating pathways responsible for specific metal uptake, storage, and efflux during biomining process (Diep et al., 2018).
(4) Engineering microbial communities using synthetic biology tools, such as CRISPR/Cas systems, riboswitches, quorum sensing or syntrophic exchanges (McCarty and Ledesma-Amaro, 2019) may promote synergistic effects among Fe-oxidizers, S-oxidizers and heterotrophs and further enhance biomining efficiency, especially in heap and stirred-tank biomining that carried out in the non-sterile environment.
(5) Rather than directly genetically engineering Fe/S-oxidizing bacteria, directed evolution may also be a good research direction since stakeholders are more likely to accept industrial bacteria with more natural origins. For example, the EvolvR system can help evolve industrial microbes with desired phenotypes, such as fast growth rate, high tolerance to complex metals, acidity, salinity, and more (Halperin et al., 2018).
JC, YL, and RM conceived and designed the structure of the minireview; JC and YL wrote the first draft; PD contributed to illustration and re-writing the manuscript; all authors ratified the submitted and revised versions.
This work was funded by Elements of Biomining Grant from the Province of Ontario through the ORF Research Excellence funding program, and the Postdoctoral fellowship of 2022-2023 Climate Positive Energy Rising Stars in Clean Energy from University of Toronto.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Beard, S., Ossandon, F. J., Rawlings, D. E., and Quatrini, R. (2021). The flexible genome of acidophilic prokaryotes. Curr. Issues Mol. Biol. 40, 231–266. doi:10.21775/cimb.040.231
Brune, K. D., and Bayer, T. (2012). Engineering microbial consortia to enhance biomining and bioremediation. Front. Microbiol. 3, 203. doi:10.3389/fmicb.2012.00203
Campodonico, M. A., Vaisman, D., Castro, J. F., Razmilic, V., Mercado, F., Andrews, B. A., et al. (2016). Acidithiobacillus ferrooxidans's comprehensive model driven analysis of the electron transfer metabolism and synthetic strain design for biomining applications. Metab. Eng. Commun. 3, 84–96. doi:10.1016/j.meteno.2016.03.003
Chen, J., Liu, Y., Diep, P., and Mahadevan, R. (2022a). Genetic engineering of extremely acidophilic Acidithiobacillus species for biomining: Progress and perspectives. J. Hazard. Mat. 438, 129456. doi:10.1016/j.jhazmat.2022.129456
Chen, J., Liu, Y., and Mahadevan, R. (2022b). Genetic engineering of Acidithiobacillus ferridurans with CRISPR-Cas9/dCas9 systems. bioRxiv.
Diep, P., Mahadevan, R., and Yakunin, A. F. (2018). Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front. Bioeng. Biotechnol. 6, 157. doi:10.3389/fbioe.2018.00157
Gumulya, Y., Boxall, N. J., Khaleque, H. N., Santala, V., Carlson, R. P., and Kaksonen, A. H. (2018). In a quest for engineering acidophiles for biomining applications: Challenges and opportunities. Genes 9, 116. doi:10.3390/genes9020116
Halperin, S. O., Tou, C. J., Wong, E. B., Modavi, C., Schaffer, D. V., and Dueber, J. E. (2018). CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248–252. doi:10.1038/s41586-018-0384-8
Jung, H., Inaba, Y., and Banta, S. (2021). Genetic engineering of the acidophilic chemolithoautotroph Acidithiobacillus ferrooxidans. Trends Biotechnol. 40, 677–692. doi:10.1016/j.tibtech.2021.10.004
Kozubal, M. A., Romine, M., Jennings, R. D., Jay, Z. J., Tringe, S. G., Rusch, D. B., et al. (2013). Geoarchaeota: A new candidate phylum in the archaea from high-temperature acidic iron mats in Yellowstone national Park. ISME J. 7, 622–634. doi:10.1038/ismej.2012.132
Kremser, K., Thallner, S., Schoen, H., Weiss, S., Hemmelmair, C., Schnitzhofer, W., et al. (2020). Stirred-tank and heap-bioleaching of shredder-light-fractions (SLF) by acidophilic bacteria. Hydrometallurgy 193, 105315. doi:10.1016/j.hydromet.2020.105315
Lewis, A. M., Recalde, A., Bräsen, C., Counts, J. A., Nussbaum, P., Bost, J., et al. (2021). The biology of thermoacidophilic archaea from the order Sulfolobales. FEMS Microbiol. Rev. 45, fuaa063. fuaa063. doi:10.1093/femsre/fuaa063
Li, M., and Wen, J. (2021). Recent progress in the application of omics technologies in the study of bio-mining microorganisms from extreme environments. Microb. Cell Fact. 20, 178. doi:10.1186/s12934-021-01671-7
Mcallister, S. M., Moore, R. M., Gartman, A., Luther, G. W., Emerson, D., and Chan, C. S. (2019). The Fe (II)-oxidizing Zetaproteobacteria: Historical, ecological and genomic perspectives. FEMS Microbiol. Ecol. 95, fiz015. fiz015. doi:10.1093/femsec/fiz015
Mccarty, N. S., and Ledesma-Amaro, R. (2019). Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 37, 181–197. doi:10.1016/j.tibtech.2018.11.002
Moya-Beltrán, A., Makarova, K. S., Acuña, L. G., Wolf, Y. I., Covarrubias, P. C., Shmakov, S. A., et al. (2021). Evolution of type IV CRISPR-cas systems: Insights from CRISPR loci in integrative conjugative elements of Acidithiobacillia. CRISPR J. 4, 656–672. doi:10.1089/crispr.2021.0051
Peng, N., Han, W., Li, Y., Liang, Y., and She, Q. (2017). Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Sci. China Life Sci. 60, 370–385. doi:10.1007/s11427-016-0355-8
Percak‐Dennett, E., He, S., Converse, B., Konishi, H., Xu, H., Corcoran, A., et al. (2017). Microbial acceleration of aerobic pyrite oxidation at circumneutral pH. Geobiology 15, 690–703. doi:10.1111/gbi.12241
Petersen, J. (2016). Heap leaching as a key technology for recovery of values from low-grade ores–A brief overview. Hydrometallurgy 165, 206–212. doi:10.1016/j.hydromet.2015.09.001
Rawlings, D. E., Dew, D., and Du Plessis, C. (2003). Biomineralization of metal-containing ores and concentrates. Trends Biotechnol. 21, 38–44. doi:10.1016/s0167-7799(02)00004-5
Rawlings, D. E., and Johnson, D. B. (2007). The microbiology of biomining: Development and optimization of mineral-oxidizing microbial consortia. Microbiology 153, 315–324. doi:10.1099/mic.0.2006/001206-0
Sarnaik, A., Liu, A., Nielsen, D., and Varman, A. M. (2020). High-throughput screening for efficient microbial biotechnology. Curr. Opin. Biotechnol. 64, 141–150. doi:10.1016/j.copbio.2020.02.019
Schmitz, A. M., Pian, B., Medin, S., Reid, M. C., Wu, M., Gazel, E., et al. (2021). Generation of a Gluconobacter oxydans knockout collection for improved extraction of rare earth elements. Nat. Commun. 12, 6693–6711. doi:10.1038/s41467-021-27047-4
Wang, L., Yin, S., Wu, A., and Chen, W. (2020). Synergetic bioleaching of copper sulfides using mixed microorganisms and its community structure succession. J. Clean. Prod. 245, 118689. doi:10.1016/j.jclepro.2019.118689
Wu, J., Cheng, Z., Min, D., Cheng, L., He, R., Liu, D., et al. (2020). CRISPRi system as an efficient, simple platform for rapid identification of genes involved in pollutant transformation by Aeromonas hydrophila. Environ. Sci. Technol. 54, 3306–3315. doi:10.1021/acs.est.9b07191
Keywords: synthetic biology, Fe/S-oxidizing microbes, biomining, design-build-test-learn (DBTL) cycle, CRISPR
Citation: Chen J, Liu Y, Diep P and Mahadevan R (2022) Harnessing synthetic biology for sustainable biomining with Fe/S-oxidizing microbes. Front. Bioeng. Biotechnol. 10:920639. doi: 10.3389/fbioe.2022.920639
Received: 14 April 2022; Accepted: 08 August 2022;
Published: 05 September 2022.
Edited by:
Jean Marie François, Institut Biotechnologique de Toulouse (INSA), FranceReviewed by:
Danielle Maass, Federal University of São Paulo, BrazilCopyright © 2022 Chen, Liu, Diep and Mahadevan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radhakrishnan Mahadevan, a3Jpc2huYS5tYWhhZGV2YW5AdXRvcm9udG8uY2FjYQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.