
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Bioeng. Biotechnol. , 23 June 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.919969
This article is part of the Research Topic Advances and Trends in Microbial Production of Biopolymers and Their Building Blocks View all 11 articles
Polylactic acid (PLA), a homopolymer of lactic acid (LA), is a bio-derived, biocompatible, and biodegradable polyester. The evolved class II PHA synthase (PhaC1Ps6-19) was commonly utilized in the de novo biosynthesis of PLA from biomass. This study tested alternative class I PHA synthase (PhaCCs) from Chromobacterium sp. USM2 in engineered Escherichia coli for the de novo biosynthesis of PLA from glucose. The results indicated that PhaCCs had better performance in PLA production than that of class II synthase PhaC1Ps6-19. In addition, the sulA gene was engineered in PLA-producing strains for morphological engineering. The morphologically engineered strains present increased PLA production. This study also tested fused propionyl-CoA transferase and lactate dehydrogenase A (fused PctCp/LdhA) in engineered E. coli and found that fused PctCp/LdhA did not apparently improve the PLA production. After systematic engineering, the highest PLA production was achieved by E. coli MS6 (with PhaCCs and sulA), which could produce up to 955.0 mg/L of PLA in fed-batch fermentation with the cell dry weights of 2.23%, and the average molecular weight of produced PLA could reach 21,000 Da.
Biopolyesters have been developed in recent years as alternatives to petroleum-based synthetic polyesters (Nduko and Taguchi, 2021). As a commercialized biopolyester, polylactic acid (PLA) can be prepared from renewable biomass with promising physical performance, biocompatibility, and biodegradability (Taguchi et al., 2008; Yang et al., 2010; Paço et al., 2019). The industrial preparation of PLA is accomplished through several steps: first, the monomer of lactic acid (LA) is prepared from biomass through fermentation; then LA is converted to lactide followed by the ring-opening polymerization of lactide to PLA (Maharana et al., 2009).
With the fast development of biotechnology, PLA homopolymer and LA-containing copolymers can be de novo biosynthesized from renewable biomass by engineered strains (Taguchi et al., 2008; Yang et al., 2010; Choi et al., 2016; Zou et al., 2021). However, compared with the efficient bioproduction of LA-containing copolymers, the microbial production of PLA homopolymers is still challenging with low productivity. One of the barriers in the biosynthesis of the PLA homopolymer is that the PHA synthases involved exhibit higher activities toward other substrates than LA monomer (Park et al., 2012).
Based on the primary structure, subunit compositions, and substrate specificity, four classes of PHA synthases have been found in nature (Yang et al., 2010; Zou et al., 2017; Chek et al., 2019). Class I, III, and IV PHA synthases prefer short-chain length (SCL) monomers, whereas class II PHA synthases exhibit higher activities toward medium-chain length (MCL) monomers (Rehm, 2003; Tsuge et al., 2015; Zou et al., 2017). Class II synthase from Pseudomonas sp. was engineered (E130D, S325T, S477G, and Q481K) to gain the ability to synthesize PLA and LA-containing copolymers. Although the engineered strains (with engineered class II synthase) can efficiently produce LA-containing copolymers (Jung et al., 2010; Yang et al., 2010; Choi et al., 2016; Li et al., 2016), PLA homopolymer productivity was as low as 0.5 wt% of dry cell weight (Yang et al., 2010; Park et al., 2012). The low productivity of the PLA homopolymer was presumably due to the low substrate specificity of PHA synthase (Park et al., 2012), low mobility of the generated PLA, and low concentration of LA-CoA in the engineered strains (Matsumoto et al., 2018). Robust PHA synthases with higher substrate specificity toward LA monomer need to be discovered or engineered for the enhancement of the biosynthesis of PLA homopolymers (Taguchi and Matsumoto, 2021).
Morphological engineering is another trend in the biosynthesis of biopolymers, which aims at larger cell space for the storage of the produced biopolymers in vivo. FtsZ, a bacterial tubulin homolog, is one of the targets in the morphological engineering of bacteria strains (Erickson et al., 2010; Chen et al., 2012; Wang et al., 2014). The inhibition of FtsZ can affect the formation of the Z ring during bacteria division and hence enlarge cell space (Dajkovic et al., 2008; Adams and Errington, 2009; Chen et al., 2012). Higher expression of SulA can inhibit FtsZ and reduce the cell division rate (Dajkovic et al., 2008; Chen et al., 2012). For example, morphologically engineered E. coli JM109 exhibits the increased production of poly (3-hydroxybutyrate) (PHB) (from 8 to 9 g/L) and poly (3HB-co-4HB) (from 8.2 to 9.2 g/L) (Wang et al., 2014; Wu et al., 2016).
In this study, to screen robust PHA synthase in PLA biosynthesis, we selected the class I PHA synthase from Chromobacterium sp. USM2, which has been used in the polymerization of 3-hydroxypropionic acid (3HP, an isomer of LA) (Linares-Pastén et al., 2015), and evaluated its performance in PLA homopolymer production by engineered E. coli. In addition, we additionally expressed the sulA gene and evaluated the PLA production of morphologically engineered strains.
All the information on strains and plasmids is listed in Table 1. The information of key plasmids is provided in Supplemental Figure S1. The primers are summarized in Supplemental Table S1. Engineered strains were constructed using the following methods.
The suicide plasmid–mediated genome editing method (Gao et al., 2014) was utilized in the deletion of the ackA gene from the genome of E. coli BL21 (DE3). The homolog arms (600 bp upstream and downstream of the ackA gene) were cloned into the suicide plasmid of pRE112. The constructed pRE112-△ackA was transformed into E. coli χ7213 to construct the donor strain of E. coli χ7213/pRE112-△ackA. Then, the conjugation occurred between the donor strain and the receptor strain of E. coli BL21 (DE3) for the deletion of the ackA gene. The strain of E. coli MS1 (Table 1) was constructed after knocking off ackA.
Based on the chassis strain of E. coli MS1, other engineered strains (MS2–MS7) were constructed using the following methods. As shown in Figure 1, the biosynthetic pathway of PLA was engineered in PLA-producing strains; in addition, the sulA gene was expressed in strains for larger cell space.
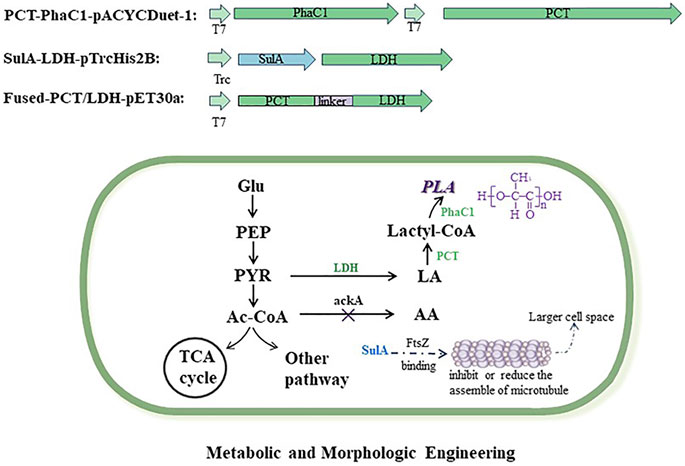
FIGURE 1. Metabolic and morphologic engineering of E. coli strains for polylactic acid production. Exogenous vectors expressed the genes of key enzymes which are involved in the biosynthesis pathway of polylactic acid. The gene of ackA was deleted in the engineered strains to reduce the competitive by-product of acetic acid. The gene of sulA was expressed in engineered stains for larger cell space. Full names of abbreviations are as follows: PLA, polylactic acid; PhaC, PHA synthase; PCT, propionyl-CoA transferase; LDH, lactate dehydrogenase; Glu, glucose; PEP, phosphoenolpyruvate; PYR, pyruvate; Ac-CoA, acetyl co-enzyme A; AA, acetic acid; LA, lactic acid.
From E. coli MS1, E. coli MS2 additionally expressed the ldhA gene from E. coli BL21 (lactate dehydrogenase, NCBI No. NC_012892.2). From E. coli MS2, E. coli MS3 additionally expressed the evolved PctCp gene (propionyl-CoA transferase, NCBI No. CAB77207.1, with mutation of V193A) from Clostridium propionicum DSM 1682 and the evolved PhaC1Ps6-19 gene (class II PHA synthase, NCBI No. ACM68707.1, with mutations of E130D, S325T, S477G, and Q481K) from Pseudomonas sp. MBEL 6-19. From E. coli MS3, E. coli MS4 additionally expressed the sulA gene from E. coli str. K-12 substr. MG1655 (NCBI No. NC_000913.3).
From E. coli MS2, E. coli MS5 additionally expressed the evolved PctCp gene from C. propionicum and the PhaCCs gene (class I PHA synthase, NCBI No. ADL70203.1) from Chromobacterium sp. USM2. From E. coli MS5, E. coli MS6 additionally expressed the sulA gene from E. coli str. K-12.
From E. coli MS1, E. coli MS7 additionally expressed the gene of the fused enzyme of PctCp/LdhA (Supplemental Figure S2). A flexible linker (Gly–Gly–Gly–Gly–Ser)3 was inserted between PctCp and LdhA; in addition, E. coli MS7 expressed the evolved PhaC1Ps6-19 gene from C. propionicum and the sulA gene from E. coli str. K-12.
All strains utilized in this study are cultivated in the Luria–Bertani (LB) medium (flask-level) or M9 medium (fermenter-level). The LB medium contains (per liter) 10 g tryptone, 5 g yeast extract, and 10 g NaCl. The M9 medium consists of (per liter) 1 g (NH4)2SO4, 3 g K2HPO4.3H2O, 1.9 g KCl, 5 g yeast extract, 1 g sodium citrate, 1 g citric acid, 1 g glycine betaine, 0.24 g MgSO4, and 1 ml of the stored solution of trace element. The stored solution of trace element consists of (per liter) 3.7 g (NH4)6Mo7O24·4H2O, 2.9 g ZnSO4 7H2O, 24.7 g H3BO3, 2.5 g CuSO4 5H2O, and 15.8 g MnCl2 4H2O. Variable antibiotics were supplemented in the cultivation medium for different strains (Table 1). The concentration of antibiotics in the medium is as follows: ampicillin 48 mg/L, chloramphenicol 24 mg/L, and kanamycin 45 mg/L. The sucrose-containing (10% w/w) LB medium was utilized to screen the strain of E. coli MS1.
For fermentation of PLA, individual strains were cultivated in 10 ml LB medium at 37°C overnight in a rotary shaker at 220 rpm. Then, 5% (V/V) seed cultures were added into 100 ml LB medium (in a flask) and cultivated at 37°C overnight in a rotary shaker at 220 rpm. The secondary seed cultures were inoculated into a 5-L fermenter (Bailun Inc., China) containing 2 L M9 medium, and 20 g/L glucose was added as a starting carbon source. Fed-batch fermentation starts at 37°C. During the fermentation process, ammonium hydroxide solution (6M) was automatically added to maintain the pH at 7. The dissolved oxygen concentration (DOC) was maintained at 10–20% by changing the agitation speed and ventilatory capacity (VC). Ten hours after inoculation, 0.5 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) was added in the fermenter, and then the cultivation temperature was decreased to 30°C. Then, 100 ml of 50% glucose (w/v) was supplemented every 12 h. The cell growth value of OD600 was monitored using the spectrophotometer at 600 nm. The duration time of fermentation was 72 h.
The solvent extraction method (Jung et al., 2010) was utilized for the purification of PLA products from the cells. After fermentation, the cells were harvested by centrifugation at 4000 rpm for 20 min. The harvested cells were washed twice with absolute ethanol and distilled water before lyophilization. Then, the cells were lyophilized for 24 h and the cell dry weights (CDW) of different samples were measured and recorded. PLA was extracted from dried cells by chloroform in the Soxhlet apparatus at 80°C for 16 h. Excessive chloroform was removed using a rotary evaporator, and cell debris was removed by passing through a PTFE filter. PLA was precipitated by adding five-fold ice-cold methanol. Weights of purified and dried PLA was measured and recorded.
To qualitatively determine the polymer structure, the samples were analyzed by 1H and 13C nuclear magnetic resonance (NMR) spectra using a Bruker AM-500 MHz spectrometer at 500 and 125 MHz, respectively. The sample was solved in CDCl3 with tetramethylsilane (TMS) as an internal chemical shift standard. The number-average molecular weight (Mn) and the weight-average molecular weight (Mw) of PLA were determined by gel permeation chromatography (GPC) equipped with TSKgel SuperMutipore HZ-M*2 column and GPC data processing software. The PLA sample (1 mg/ml) was eluted using tetrahydrofuran (THF) before injection (20 μl), and GPC was operated at a flow rate of 0.35 ml/min at 40°C. Polystyrene with a narrow range of polydispersity (1.03-1.05) was used for calibration.
The morphological form of engineered E. coli was characterized using a scanning electron microscope (SEM) on a Hitachi S-4800 instrument. The cells were harvested by centrifugation at 5000 rpm for 5 min and subsequently washed with phosphate-buffered saline (PBS) (pH 7.4) three times. Then, the washed cells were fixed with 2.5% (V/V) glutaraldehyde overnight at 4°C. The fixed cells were washed again with phosphate-buffered saline (PBS) (pH 7.4) three times (30 min each). Afterward, ethanol gradients of 30%, 50%, 70%, 80%, 90%, and 95% (V/V) solutions were used to dehydrate the fixed cells in a sequential way. The cell samples were further dehydrated with 100% ethanol three times. After that, tertiary butyl alcohol was mixed with ethanol in a ratio of 1:1 and pure tertiary butyl alcohol was used to achieve metathesis of ethanol in the cells (Wang et al., 2014). At last, the cells were mixed in with tertiary butyl alcohol and lyophilized for imaging.
The PLA granules in E. coli cells were characterized using a transmission electron microscope (TEM) on a Hitachi H-7650 instrument. The cells were harvested by centrifugation at 5000 rpm for 5 min and subsequently washed with phosphate-buffered saline (PBS) (pH 7.4) three times. Then, the washed cells were fixed with 2.5% (V/V) glutaraldehyde overnight at 4°C. The fixed cells were washed again with phosphate-buffered saline (PBS) (pH 7.4) three times (30 min each), and then 1% (V/V) osmic acid was added to fix the cell for 1 h. The fixed cells were washed again with PBS (pH 7.4) three times (30 min each). Afterward, acetone gradient solutions of 30%, 50%, 70%, 80%, 90%, and 95% (V/V) were used to dehydrate the fixed cells in a sequential way. The resulting cell samples were further dehydrated with 100% acetone three times. Finally, the cell samples are embedded with Spurr resin for imaging.
The significance of differences between the mean values of testing samples was compared using Student’s t-test. Differences were considered statistically obvious if p < 0.05 and significant if p < 0.01.
The qualitative 1H and 13C assay of polymer product (Supplemental Figure S3) confirmed that PLA homopolymer was de novo–biosynthesized from glucose by the engineered strains of E. coli, and the 1H assay also indicated the presence of oligomeric PLA or LA monomer indicated the hydrolysis of PLA during analysis processing. The quantitative comparison of PLA production by different strains (Figure 2) showed that E. coli MS5 (with PhaCcs) could produce up to 323.4 mg/L of PLA (0.85% of CDW), obviously higher (p < 0.05) than the PLA production (189.5 mg/L, 0.54% of CDW) by the control strain of E. coli MS3 (with evolved PhaC1Ps6-19). The results indicated that class I PHA synthase (from Chromobacterium sp. USM2) exhibited better performance in the biosynthesis of the PLA homopolymer. This study further compared the Mw of the produced PLA with the strains of MS5 and MS3 (Table 2). Although the PLA products with two strains present a similar average Mw (around 21,000 Da), the lower Mw/Mn value of MS5 indicated that the PLA produced with MS5 has more centralized molecular weight distribution than that of MS3.
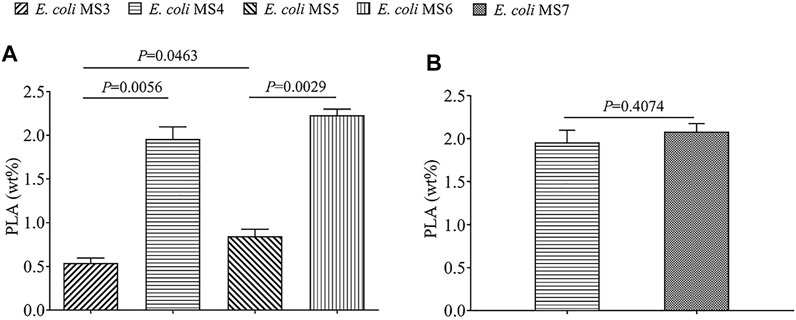
FIGURE 2. Comparison of the PLA production by five different engineered strains. The PLA production was recorded by wt% (weight of PLA/dry cell weight). The data shown are expressed as means of three repeated experiments, and the error bars present their standard deviation. Statistical difference (p value) between different groups is also present. (A) Strains which express the gene of PhaC1Cs (class I PHA synthase) and sulA (for morphological engineering) have higher PLA production. (B) Strain which expresses the fused enzyme of PctCp/LdhA has similar PLA production with the control strain.
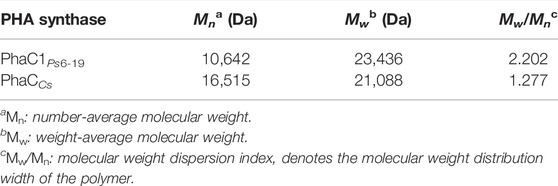
TABLE 2. Gel permeation chromatography (GPC) of PLA synthesized by strains with different PHA synthases.
As described in earlier studies, only engineered class II PHA synthases were utilized in the biosynthesis of PLA homopolymer and LA-containing copolymers, and the mutation of several residues (E130D, S325T, S477G, and Q481K) can improve their substrate specificities toward LA monomers (Taguchi et al., 2008; Yamada et al., 2009; Jung et al., 2010; Yang et al., 2010; Choi et al., 2016). Class I PHA synthases present higher substrate specificity toward SCL monomers (Zou et al., 2017), but have not been utilized in the biosynthesis of the PLA homopolymer and LA-containing copolymers before. The results of this study confirmed that class I PHA synthase could also be utilized in PLA production (Figure 2). Moreover, the same synthase (PhaCCs) could catalyze the polymerization of 3-hydroxypropionic acid (3HP, an isomer of LA), and the CDW of the produced P (3HP) can reach 40% (Linares-Pastén et al., 2015), indicating that class I PhaCCs shows promising substrate specificity toward the SCL hydroxypropionic acids.
At the time point of 36 h after inoculation, the cells of E. coli MS4 (morphological engineering via sulA) and MS3 (control strain without sulA) were harvested for SEM/TEM analysis. The SEM results revealed that E. coli MS4 has an elongated rod cell shape (Figure 3A), which presents a longer cell shape than the control strain of E. coli MS3 (Figure 3B). Moreover, TEM results showed that the intracellular PLA granules in E. coli MS4 (Figure 3C) occupied larger cell space than in the control strain of E. coli MS3 (Figure 3D), indicating the increased PLA production in E. coli MS4. The results of fed-batch fermentation (Figure 2) confirmed that E. coli MS4 significantly (p < 0.01) had higher production of PLA (CDW of 1.96%) than E. coli MS3 (CDW of 0.54%). The highest PLA production was achieved by E. coli MS6 (contains both PhaCCs and sulA), which can produce up to 955.0 mg/L of PLA in fed-batch fermentation with the CDW of 2.23%.
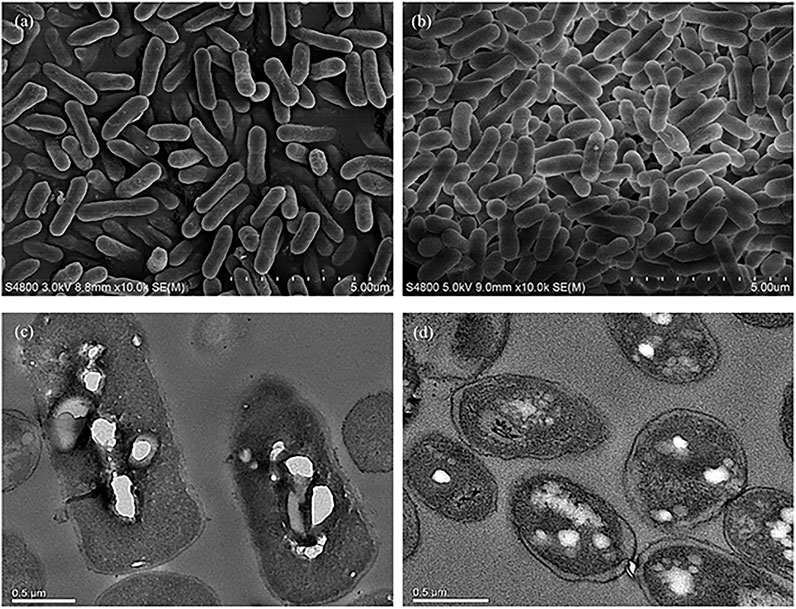
FIGURE 3. Morphological comparison of strains with or without morphological engineering using SEM and TEM assays. (A) SEM assay of cells from the morphologically engineered strain (with the expression of sulA). (B) SEM assay of cells from the control strain without expressing sulA. (C) TEM assay of cells from the morphologically engineered strain (with the expression of sulA). (D) TEM assay of cells from the control strain without expressing sulA.
The significantly increased PLA production in E. coli MS4 and MS6 than their control strains of E. coli MS3 and MS5 (Figure 2) indicated that the overexpression of the sulA gene in E. coli strains could morphologically affect the strains to achieve improved production of intracellular polymers. Similar to this study, E. coli strains have been morphologically engineered to achieve the increased production of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) (Wang et al., 2014). It has been demonstrated that SulA could bind the tubulin of FtsZ to inhibit the formation of Z loop in the cell division of E. coli strains (Pichoff and Lutkenhaus, 2002, 2005; Chen et al., 2012). The SulA/FtsZ interaction was also found in Pseudomonas (Chen et al., 2012), indicating that this strategy can be more broadly applied in the biosynthesis of biopolymers by variable chassis strains.
As shown in Figure 1, LdhA and PctCp are two key enzymes in the biosynthetic pathway of lactyl-CoA. In order to supply the increased level of lactyl-CoA for PLA biosynthesis, the fused PctCp/LdhA enzyme was engineered in E. coli MS7 (Supplemental Figure S2). Compared with the PLA production of the control strain of E. coli MS4 (CDW of 1.96%), the PLA production of E. coli MS7 (CDW of 2.08%) was not apparently increased (p > 0.05). In addition, E. coli MS7 (containing PctCp and LdhA fusion enzyme) had a lower dry cell weight (28.9 g/L, 72 h) than E. coli MS4 (31.6 g/L, 72 h), which indicated that the presence of fused enzyme exhibits growth stress toward the engineered strain of MS7.
Similar to other fused proteins and enzymes (Yu et al., 2015), the tandem fusion of PctCp and LdhA can spatially restrain multiple catalytic domains in one fused enzyme. The results of this study indicated that fused PctCp/LdhA enzyme retained the biocatalytic functions of individual PctCp and LdhA. However, fused PctCp/LdhA did not apparently improve the general biosynthesis of PLA, which indicated that the final step (polymerization of lactyl-CoA into PLA by PHA synthase) could be the bottleneck step in the biosynthesis of the PLA homopolymer, as reported by earlier studies (Park et al., 2012; Matsumoto et al., 2018).
The present study demonstrated that class I PHA synthase from Chromobacterium sp. USM2 (PhaCCs) is feasible to catalyze the polymerization of the PLA homopolymer from lactyl-CoA. In de novo PLA fermentation from glucose, engineered E. coli strains having PhaCCs present improved PLA production than control strains, which expressed the evolved class II PHA synthase PhaC1Ps6-19. In addition, sulA-mediated morphological engineering could enlarge the cell space and further improve PLA production of the engineered E. coli strains.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
HZ, MS, and LY conceived and presented the idea. MS, ML, and AY performed the experiment. MS, HZ, and JP wrote the manuscript in consultation with others. All authors read and approved the final manuscript.
The present study was supported by the Talent Foundation and the Province and Ministry Co-construction Collaborative Innovation Center of Eco-chemical Engineering (STHGYX2221).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.919969/full#supplementary-material
Adams, D. W., and Errington, J. (2009). Bacterial Cell Division: Assembly, Maintenance and Disassembly of the Z Ring. Nat. Rev. Microbiol. 7, 642–653. doi:10.1038/nrmicro2198
Chek, M. F., Hiroe, A., Hakoshima, T., Sudesh, K., and Taguchi, S. (2019). PHA Synthase (Phac): Interpreting the Functions of Bioplastic-Producing Enzyme from a Structural Perspective. Appl. Microbiol. Biotechnol. 103, 1131–1141. doi:10.1007/s00253-018-9538-8
Chen, Y., Milam, S. L., and Erickson, H. P. (2012). SulA Inhibits Assembly of FtsZ by a Simple Sequestration Mechanism. Biochemistry 51, 3100–3109. doi:10.1021/bi201669d
Choi, S. Y., Park, S. J., Kim, W. J., Yang, J. E., Lee, H., Shin, J., et al. (2016). One-Step Fermentative Production of Poly(Lactate-Co-Glycolate) from Carbohydrates in Escherichia coli. Nat. Biotechnol. 34, 435–440. doi:10.1038/nbt.3485
Dajkovic, A., Mukherjee, A., and Lutkenhaus, J. (2008). Investigation of Regulation of FtsZ Assembly by SulA and Development of a Model for FtsZ Polymerization. J. Bacteriol. 190, 2513–2526. doi:10.1128/JB.01612-07
Erickson, H. P., Anderson, D. E., and Osawa, M. (2010). FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol. Mol. Biol. Rev. 74, 504–528. doi:10.1128/mmbr.00021-10
Gao, Y., Liu, C., Ding, Y., Sun, C., Zhang, R., Xian, M., et al. (2014). Development of Genetically Stable Escherichia coli Strains for Poly(3-Hydroxypropionate) Production. PLoS One 9, e97845. doi:10.1371/journal.pone.0097845
Jung, Y. K., Kim, T. Y., Park, S. J., and Lee, S. Y. (2010). Metabolic Engineering of Escherichia Coli for the Production of Polylactic Acid and its Copolymers. Biotechnol. Bioeng. 105, 161–171. doi:10.1002/bit.22548
Li, Z.-J., Qiao, K., Shi, W., Pereira, B., Zhang, H., Olsen, B. D., et al. (2016). Biosynthesis of Poly(Glycolate-Co-Lactate-Co-3-Hydroxybutyrate) from Glucose by Metabolically Engineered Escherichia coli. Metab. Eng. 35, 1–8. doi:10.1016/j.ymben.2016.01.004
Linares-Pastén, J. A., Sabet-Azad, R., Pessina, L., Sardari, R. R. R., Ibrahim, M. H. A., and Hatti-Kaul, R. (2015). Efficient Poly(3-Hydroxypropionate) Production From Glycerol Using Lactobacillus Reuteri And Recombinant Escherichia Coli Harboring L. Reuteri Propionaldehyde Dehydrogenase And Chromobacterium Sp. PHA Synthase Genes. Bioresour. Technol. 180, 172–176. doi:10.1016/j.biortech.2014.12.099
Maharana, T., Mohanty, B., and Negi, Y. S. (2009). Melt-Solid Polycondensation of Lactic Acid and its Biodegradability. Prog. Polym. Sci. 34, 99–124. doi:10.1016/j.progpolymsci.2008.10.001
Matsumoto, K. i., Hori, C., Fujii, R., Takaya, M., Ooba, T., Ooi, T., et al. (2018). Dynamic Changes of Intracellular Monomer Levels Regulate Block Sequence of Polyhydroxyalkanoates in Engineered Escherichia coli. Biomacromolecules 19, 662–671. doi:10.1021/acs.biomac.7b01768
Nduko, J. M., and Taguchi, S. (2021). Microbial Production of Biodegradable Lactate-Based Polymers and Oligomeric Building Blocks from Renewable and Waste Resources. Front. Bioeng. Biotechnol. 8, 1–18. doi:10.3389/fbioe.2020.618077
Paço, A., Jacinto, J., da Costa, J. P., Santos, P. S. M., Vitorino, R., Duarte, A. C., et al. (2019). Biotechnological Tools for the Effective Management of Plastics in the Environment. Crit. Rev. Environ. Sci. Technol. 49, 410–441. doi:10.1080/10643389.2018.1548862
Park, S. J., Lee, S. Y., Kim, T. W., Jung, Y. K., and Yang, T. H. (2012). Biosynthesis of Lactate-Containing Polyesters by Metabolically Engineered Bacteria. Biotechnol. J. 7, 199–212. doi:10.1002/biot.201100070
Pichoff, S., and Lutkenhaus, J. (2002). Unique and Overlapping Roles for ZipA and FtsA in Septal Ring Assembly in Escherichia coli. EMBO J. 21, 685–693. doi:10.1093/emboj/21.4.685
Pichoff, S., and Lutkenhaus, J. (2005). Tethering the Z Ring to the Membrane through a Conserved Membrane Targeting Sequence in FtsA. Mol. Microbiol. 55, 1722–1734. doi:10.1111/j.1365-2958.2005.04522.x
Rehm, B. H. A. (2003). Polyester Synthases: Natural Catalysts for Plastics. Biochem. J. 376, 15–33. doi:10.1042/BJ20031254
Taguchi, S., and Matsumoto, K. i. (2021). Evolution of Polyhydroxyalkanoate Synthesizing Systems toward a Sustainable Plastic Industry. Polym. J. 53, 67–79. doi:10.1038/s41428-020-00420-8
Taguchi, S., Yamada, M., Matsumoto, K. i., Tajima, K., Satoh, Y., Munekata, M., et al. (2008). A Microbial Factory for Lactate-Based Polyesters Using a Lactate-Polymerizing Enzyme. Proc. Natl. Acad. Sci. U.S.A. 105, 17323–17327. doi:10.1073/pnas.0805653105
Tsuge, T., Hyakutake, M., and Mizuno, K. (2015). Class IV Polyhydroxyalkanoate (PHA) Synthases and PHA-Producing Bacillus. Appl. Microbiol. Biotechnol. 99, 6231–6240. doi:10.1007/s00253-015-6777-9
Wang, Y., Wu, H., Jiang, X., and Chen, G.-Q. (2014). Engineering Escherichia coli for Enhanced Production of Poly(3-Hydroxybutyrate-Co-4-Hydroxybutyrate) in Larger Cellular Space. Metab. Eng. 25, 183–193. doi:10.1016/j.ymben.2014.07.010
Wu, H., Chen, J., and Chen, G.-Q. (2016). Engineering the Growth Pattern and Cell Morphology for Enhanced PHB Production by Escherichia coli. Appl. Microbiol. Biotechnol. 100, 9907–9916. doi:10.1007/s00253-016-7715-1
Yamada, M., Matsumoto, K. i., Nakai, T., and Taguchi, S. (2009). Microbial Production of Lactate-Enriched Poly[(R)-lactate-co-(R)-3-hydroxybutyrate] with Novel Thermal Properties. Biomacromolecules 10, 677–681. doi:10.1021/bm8013846
Yang, T. H., Kim, T. W., Kang, H. O., Lee, S.-H., Lee, E. J., Lim, S.-C., et al. (2010). Biosynthesis of Polylactic Acid and its Copolymers Using Evolved Propionate CoA Transferase and PHA Synthase. Biotechnol. Bioeng. 105, 150–160. doi:10.1002/bit.22547
Yu, K., Liu, C., Kim, B.-G., and Lee, D.-Y. (2015). Synthetic Fusion Protein Design and Applications. Biotechnol. Adv. 33, 155–164. doi:10.1016/j.biotechadv.2014.11.005
Zou, H., Shi, M., Zhang, T., Li, L., Li, L., and Xian, M. (2017). Natural and Engineered Polyhydroxyalkanoate (PHA) Synthase: Key Enzyme in Biopolyester Production. Appl. Microbiol. Biotechnol. 101, 7417–7426. doi:10.1007/s00253-017-8485-0
Keywords: class I polyhydroxyalkanoate synthase, engineered E. coli, polylactic acid, biopolyester, degradable polymer
Citation: Shi M, Li M, Yang A, Miao X, Yang L, Pandhal J and Zou H (2022) Class I Polyhydroxyalkanoate (PHA) Synthase Increased Polylactic Acid Production in Engineered Escherichia Coli. Front. Bioeng. Biotechnol. 10:919969. doi: 10.3389/fbioe.2022.919969
Received: 14 April 2022; Accepted: 09 May 2022;
Published: 23 June 2022.
Edited by:
Xinglin Jiang, Technical University of Denmark, DenmarkReviewed by:
Liangzhi Li, Suzhou University of Science and Technology, ChinaCopyright © 2022 Shi, Li, Yang, Miao, Yang, Pandhal and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huibin Zou, em91aGJAcWliZWJ0LmFjLmNu, aHVpYmluem91QGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.