
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 13 May 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.908804
This article is part of the Research Topic Genomic Strategies for Efficient Microbial Cell Factories View all 5 articles
 Jiwei Chen1†
Jiwei Chen1† Yifan Huang1†
Yifan Huang1† Yuexuan Shu1†
Yuexuan Shu1† Xiaoyue Hu2,3
Xiaoyue Hu2,3 Di Wu1
Di Wu1 Hangjin Jiang2
Hangjin Jiang2 Kui Wang1
Kui Wang1 Weihua Liu3
Weihua Liu3 Weiqi Fu1,4*
Weiqi Fu1,4*Microalgae have drawn much attention for their potential applications as a sustainable source for developing bioactive compounds, functional foods, feeds, and biofuels. Diatoms, as one major group of microalgae with high yields and strong adaptability to the environment, have shown advantages in developing photosynthetic cell factories to produce value-added compounds, including heterologous bioactive products. However, the commercialization of diatoms has encountered several obstacles that limit the potential mass production, such as the limitation of algal productivity and low photosynthetic efficiency. In recent years, systems and synthetic biology have dramatically improved the efficiency of diatom cell factories. In this review, we discussed first the genome sequencing and genome-scale metabolic models (GEMs) of diatoms. Then, approaches to optimizing photosynthetic efficiency are introduced with a focus on the enhancement of biomass productivity in diatoms. We also reviewed genome engineering technologies, including CRISPR (clustered regularly interspaced short palindromic repeats) gene-editing to produce bioactive compounds in diatoms. Finally, we summarized the recent progress on the diatom cell factory for producing heterologous compounds through genome engineering to introduce foreign genes into host diatoms. This review also pinpointed the bottlenecks in algal engineering development and provided critical insights into the future direction of algal production.
Diatoms are natural cell factories that synthesize various value-added compounds, such as polyunsaturated fatty acids, pigments, terpenes, and sterols (Yang et al., 2020). Recent years have witnessed tremendous progress in genome sequencing and editing of diatoms. The genome sequencing of two model diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana, has offered the genetic basis for the biotechnology development of diatoms. The reconstruction of genome-scale metabolic models (GEMs) in diatoms has promoted metabolic engineering strategies to improve algal productivity. Genome engineering tools such as CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR associated system), transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), microRNA (miRNA), and small interfering RNA (siRNA) are in rapid development to harness the genomic potential. For instance, the genome of diatom P. tricornutum could be efficiently edited by CRISPR/Cas9 system (Ng et al., 2017) and the TALEN-mediated system (Serif et al., 2017). Thus, diatoms have broad prospects in the production of value-added compounds with systems and synthetic biology tools enabling the characterization of many biosynthetic pathways and related genes. Nevertheless, the bottleneck of commercializing commodities like biomaterials (such as polyhydroxyalkanoates) and biofuels by microalgal cell factories lies in the biomass productivity in high-density culture. Optimizing photosynthetic efficiency (PE) may achieve high overall productivity as microalgae are considered efficient solar energy converters with metabolic flexibility. For compound production in the high-density culture of diatoms, it is feasible to utilize a two-phase culture, which possesses high biomass productivity under optimal conditions in the first stage and then accumulates value-added compounds such as EPA and fucoxanthin under stress conditions in the second stage, showing the potential for industrial applications (Yang and Wei, 2020; Parkes et al., 2021).
As a group of oil-producing algae, diatoms can be used as a source to produce biofuels and bioactive compounds such as fatty acids and carotenoids. The omega-3 polyunsaturated fatty acids such as docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) from diatoms have critical biological functions for fetal growth and development based on their roles in maintaining the healthy function of the brain and retina (Marella and Tiwari, 2020). In addition, diatoms can produce non-native value-added compounds through heterologous expression systems, such as plant triterpenoids, monoclonal antibodies, and bioplastics (Butler et al., 2020; Lu et al., 2021). The traditional heterologous expression platforms like Escherichia coli and Saccharomyces cerevisiae have already been widely used. However, these classical expression systems require external organic carbon sources, limiting their carbon neutrality contributions (Hempel and Maier, 2012). Compared to other cell factories, such as bacteria, yeast, plants, and mammals (Table 1), diatoms have shown promise to become an ideal natural expression system due to their advantages of high efficiency, low cost, and the ability to synthesize complex value-added compounds. Green microalgae, such as the model species Chlamydomonas reinhardtii, have also been reported for the heterologous synthesis of value-added compounds. For comparison, diatoms could achieve stable genetic modifications with an available genetic toolbox and have a number of features; for example, P. tricornutum does have a free intracellular pool of GPP, while C. reinhardtii doesn’t accumulate GPP naturally (Fabris et al., 2020). Further, the rapid progress of diatom sequencing also provides new insights into diatom systems and synthetic biology.
In this review, we specifically focus on 1) diatom systems biology, including genome sequencing and GEMs, 2) recent progress in PE optimization, and 3) native and non-native bioactive compounds and their biosynthetic pathways in diatoms manipulated by synthetic biology. Finally, we highlight challenges and discuss possible directions for developing algae-based value-added products.
P. tricornutum (Bowler et al., 2008) and T. pseudonana (Armbrust et al., 2004) have been used as two model diatoms for biological research due to their rapid growth and relatively small genome sizes. Their genome sequencing has incredibly advanced system biology development. Recently, more diatoms have been sequenced, with the complete genomes available, including Fistulifera solaris (Tanaka et al., 2015), Skeletonema costatum (Ogura et al., 2018), Seminavis robusta (Osuna-Cruz et al., 2020), and Cyclotella cryptica (Traller et al., 2016). Twenty-four whole-genome sequences from 21 diatom species are available to the public, according to the data retrieved from NCBI (the National Center for Biotechnology Information, United States) (as shown in Table 2). The genome sequencing results indicate that many species-specific genes are present in the diatom genome, and some diatom genes are obtained from bacteria by horizontal gene transfer (Falciatore et al., 2020; Bhattacharjya et al., 2021).
GEMs are the bottom-up description of cell metabolism based on the annotated genome and known metabolic reactions (Levering et al., 2016; Broddrick et al., 2019). They offer a whole-cell view of metabolic fluxes, act as powerful tools to discover the exceptional metabolic capabilities of cells and predict the internal metabolism of the organism under different conditions. Based on the whole life cycle, GEM development is usually divided into 4 phases: inception (the GEM model is firstly constructed based on an existing reference database), maturation (GEM is continuously updated as new information/knowledge becomes available), specialization (existing high-quality GEMs are tailored to specific strain cell lines), and amalgamation (several high-quality GEMs are wired together to form a multicellular model) (Seif and Palsson, 2021). The first diatom GEM was reconstructed for P. tricornutum (iLB1027_lipid), with 2,172 metabolites distributed in 6 compartments and 1,027 genes for 4,456 reactions, focusing on lipid metabolism (Levering et al., 2016). Later, the GEMs of T. pseudonana CCMP 1335 (iThaps987 and iTps1432) and polar diatom Fragilariopsis cylindrus were reported (Ahmad et al., 2020; Lavoie et al., 2020; van Tol and Armbrust, 2021). Previous studies also developed several strategies to improve the reconstruction efficiency of GEMs. For example, a standard sub-model chloroplast template (iGR774) with 774 genes, 788 reactions, and 764 metabolites could be incorporated into chloroplast-containing organisms with a minor modification (Bjerkelund Rokke et al., 2020). A novel software pipeline, Rapid Annotation of Photosynthetic Systems (RAPS), has been developed to create the first draft GEM of new species by using manual curation efforts of published models. The models produced by RAPS could capture more genes, showing more excellent capabilities to predict experimentally determined growth rates (Metcalf et al., 2020). By applying the GEMs, the physiological performance of diatoms can be modeled and predicted. After elevating CO2 levels, 137 reactions in eight different metabolic groups of P. tricornutum have fluxes rising, indicating pathways related to biomass production enhancement (Levering et al., 2017). However, this phenomenon was not observed under nitrogen limitation (Levering et al., 2017). The intercompartment reductant shuttles of P. tricornutum to transfer and consume additional light energy were deciphered under the combination of photo-physiology data and GEMs, supporting the prediction of photoprotective pathways for diatoms under different light intensities (Broddrick et al., 2019). A compartmentalized GEM for microalgae was reconstructed to reveal the metabolic pathway for chitosan and rhamnose synthesis, enabling metabolic engineering with synthetic biology tools (Juneja et al., 2016). Therefore, GEMs can provide insights into the metabolic information of biomass allocation in diatoms and build the foundation for synthetic biology development.
However, only three diatoms’ GEMs have been reported so far. A small proportion of genes are used in modeling (9.9% for P. tricornutum, 8.6% for T. pseudonana, and 3.9% for F. cylindrus). The reconstruction of diatoms’ GEMs is still in its early stages. Two bottlenecks of GEM reconstruction are maturation (Parts of the model need to be improved based on future research, and genes for newly discovered metabolic pathways also need to be added to the model) and content removal (remove incorrect or redundant parts).
To tackle the reduction of photosynthetic efficiency at high-density culture, strain engineering for PE is needed to accumulate more value-added compounds. More than half of the energy is lost when the electron transport chain (ETC) is saturated due to the fast rate of photon capture, which may be 100 folds greater than the downstream rate of photosynthetic electron flow (Perrine et al., 2012). Therefore, the gap between currently achievable and maximum thermodynamic efficiency for photosynthesis is substantial. Accordingly, physical and biotechnological methods could reduce energy loss during photosynthesis in some microalgae, such as diatoms.
In photonic fields, a recent study showed that an ideal arrangement of timing in photosynthetic pathways achieved by pulsed light is the key to increasing algal PE (Sforza et al., 2012; Zarmi et al., 2020). Apart from that, the interaction between ocean acidification and different light environments presents another insight for PE enhancement (Briggs and Carpenter, 2019). For diatoms, the photonic structures of their frustules have also inspired photonics and nanotechnology studies. The role of frustules in trapping light was studied and applied in dye-sensitized solar cells (DSSCs) (Toster et al., 2013). The cellular light field could be altered by the optical properties of the nanostructure of frustule valves. Thus, in centric diatoms Coscinodiscus granii, PE was enhanced by photon trapping and scattering (Goessling et al., 2018). Therefore, developing biotechniques to synchronize photonics with biological timescales and modifying optical properties of frustules could be promising for PE enhancement.
In biotechnological fields, in addition to conventional culture condition optimization, such as a study of pH effects on microalgae (Costache et al., 2013), synthetic biology has emerged for PE enhancement through comparative phylogenomic analyses, proteomic studies, and transcriptomic analyses. In addition, transcriptome sequencing and annotation give clues to target genes for PE and biomass productivity enhancement (Xi et al., 2021). However, photosynthetic genes and relevant pathways in many microalgae are underexplored or restrained to certain model species like C. reinhardtii, which possesses a genome-wide library of mutants (Li et al., 2019), as half of the protein-coding genes for photosynthetic organisms remain uncharacterized (Krishnakumar et al., 2015). That dilemma mainly lies in its genetic diversity and variability (Fu et al., 2019; Butler et al., 2020). However, the recent developments in PE are still encouraging, as PE in diatoms has been studied at the cellular and subcellular level (Yan et al., 2018; Uwizeye et al., 2021).
One study on photosystem (PS) structure in diatoms suggested that light utilization could be improved via interconnected subunits to ensure the equilibration of electron carriers (Flori et al., 2017). Over the years, many studies focused on diatom PS II-light-harvesting antenna complex (PSII-FCPII) composing of PS and fucoxanthin chlorophyll a/c proteins (FCP) as light-harvesting antenna complex (LHC) for its structural basis (Nagao et al., 2019), light-harvesting ability (Nagao et al., 2014a), and energy transfer mechanism (Nagao et al., 2014b). With the elucidation of the PSII-FCPII supercomplex, the structural basis for detailed mechanisms regulating energy transfer and quenching in diatoms was deciphered (Wang W. et al., 2020). The structure of the PSI-FCPI supercomplex was also reported in diatom Chaetoceros gracilis consisting of cores, antenna subunits, and pigments (Xu et al., 2020). These results revealed the structural basis of PE and provided a foundation for further study of PE by reconstructing or replacing PS subunits. The process of exploring PE has always been accompanied by the study of the inhibition of photosynthesis. One research on allelochemicals from the toxic dinoflagellate revealed the negative effect of membrane damage to the PE of diatom Chaetoceros muelleri (Long et al., 2021). Similarly, recent studies on diatom P. tricornutum have shown that plasma membrane-type aquaporins (AQPs) were related to the efflux of NH3, which promoted non-photochemical quenching (NPQ) (Matsui et al., 2018). Moreover, the research on Synedra sp. showed PE decreased under the structural damage of thylakoids (Yao et al., 2019). Once the connections between these factors and the inhibition of PE are clear, one can engineer a more efficient apparatus for microalgal photosynthesis.
Currently, optimization of inorganic carbon utilization has also shown promise for PE enhancement. The carbon concentrating mechanisms (CCMs) play a vital role in concentrating CO2 to ribulose-1, 5-bisphosphate carboxylase oxygenase (Rubisco). It consists of Ci transportation, carboxysomes, and CAs (Barati et al., 2021). A recent study has demonstrated that related genes such as HLA3 (Yamano et al., 2015) and LCI1 (Kono et al., 2020) are essential for CCM functioning. It is also feasible to genetically modify CCM (Gao et al., 2015). Carbonic anhydrases (CAs), one of the critical components in CCMs, have received extensive studies. In 2014, δ-class carbonic anhydrase (TweCA) and a novel CA called CDCA1 were discovered in the diatom Thalassiosira weissflogii, demonstrating the relationship between photosynthesis and metal ions in water and the potential use of metalloenzyme to capture CO2 (Del Prete et al., 2014; Alterio et al., 2015). The genome sequencing of two model diatoms, i.e., P. tricornutum and T. pseudonana, enabled the elucidation of genes related to their CCMs, indicating that CA played a crucial role in CCM for inorganic carbon acquisition (Hopkinson et al., 2016). More CAs were discovered, such as Pt43233. It was a novel θ-type CA essential for PE in P. tricornutum, which is sited in the lumen of thylakoid, demonstrating direct usage of the pH gradient to provide CO2 to the Calvin cycle (Kikutani et al., 2016). CCMs in some diatoms involved an external carbonic anhydrase (CA (ext)) that was influenced by external CO2 concentration (Smith-Harding et al., 2017). CCMs exhibited great diversity and were regulated mainly by the LysR family of transcriptional regulators, while CCMs in diatoms were related to pyrenoids (Tomar et al., 2017). Research on the accumulation of purple acid phosphatases (PAPs) showed that PAP1 promoted growth and photosynthesis in P. tricornutum by modifying carbon flux and phosphate acquisition (Wang et al., 2021). One recent study identified distinct carbon flux patterns supporting higher PE in microalgae than in C3 and C4 plants (Treves et al., 2022).
Photosynthetic pigments, consisting of three major classes, are responsible for harvesting light energy from photosynthetically active radiation (PAR) ranging from 400 to 700 nm in green plants and most microalgae, while broader absorption spectra could be achieved by varied chlorophyll species that were discovered naturally in several bacteria with more remarkable light-harvesting ability (Naduthodi et al., 2021). Although the detailed genes involved remain unclarified, it’s feasible to enhance PE by expanding the light absorption spectrum with non-native pigments. An approach for PE enhancement termed intracellular spectrum recompositioning (ISR) in diatoms also showed promising results with a 50% increase in PE and biomass productivity (Fu et al., 2017). In the ISR study, a green fluorescent protein (GFP) was expressed and engineered in the model diatom P. tricornutum to shift excessive blue light to green light intracellularly, which optimized light distribution and absorption by heterologous counterparts (Fu et al., 2017). One recent study aimed at non-genetic tailoring light-harvesting by adding a molecular dye (Cy5) to fix the orange gap of absorption spectrum in the diatom Thalassiosira weissflogii, demonstrating an enhancement of PE and biofuel generation (Leone et al., 2021). The study on the diatom Corethron hystrix under high UV stress found the gene expression of PSII-related proteins D1, CP43, and CP47 was suppressed, indicating a genetic engineering clue for PE enhancement under high UV threat through a unique protective mechanism against UV exposure (Read et al., 2019). NPQ, termed non-photochemical quenching, represents the light energy lost in photosynthesis due to inefficiency caused by the wrong size of the LHC related to chlorophyll pigments. Antenna size optimization was achieved via several approaches, such as RNAi technology (Barry et al., 2015) and mutagenesis attained by UV and ethyl methanesulfonate (EMS) mediation (Cazzaniga et al., 2014; Shin et al., 2016). Recent studies have shown the possibility to adjust LHC sizes dynamically by proteins such as chloroplast signal recognition particle (CpSRP) developed for antenna size reduction (Naduthodi et al., 2021).
The key enzymes in Calvin–Benson–Bassham (CBB) cycle play a vital role in rate-limiting steps. The shortcomings of Rubisco, an enzyme allocating the carbon flux in the CBB cycle, serve as an engineering target in genetic modifications for PE. Over-expression of this enzyme or its activase (Ng et al., 2017) is a possible way to enhance its catalytic ability. In recent years, protein engineering and the genome editing toolbox have boosted the crucial progress on Rubisco engineering for better performance by point mutations and genetic modification (Sharwood, 2017). In addition, the native Rubisco enzyme could be replaced with heterologous expression of superior variants identified in systems biology studies (Satagopan et al., 2019). For example, diatoms evolved with Rhodophyte Form 1D Rubisco, which is more suitable in CCM to achieve high carboxylation rates under high O2/CO2 ratios (Rickaby and Eason Hubbard, 2019). However, these variants are usually only available with a specific supplement of activates or chaperone proteins as the heterologous proteins may also disrupt subcompartments with unique binding motifs (Meyer et al., 2020). Compared to engineered Rubisco enzyme variants, the heterologous expression for homodimeric Rubisco may be an ideal option in microalgae. Following this approach, other enzymes of the CBB cycle were considered. For instance, the overexpression of sedoheptulose bisphosphatase (SBP) (Yang et al., 2017), fructose 1,6-bisphosphate aldolase (FBA) (Ogawa et al., 2015; Yang et al., 2017), and ribulose-5-phosphate kinase (PRKase) (Ng et al., 2017) activated intermediate regeneration and energy reallocation in CBB cycle. The efficiency of the CBB cycle relies largely on CCM. A study integrated the individual components of C. reinhardtii CCM into the chloroplast of tobacco plants (C3 plant) by inserting cDNAs encoding the protein (CAH3 and LCIA), demonstrating increased PS II efficiency (up to 18%) as well as more accumulation of biomass in the newly generated transgenic strains (Nolke et al., 2019). A further study proved that when C. reinhardtii LCIA (CrLCIA) gene was heterologously expressed in Nannochloropsis salina CCMP1776, the activity of Ci and CA were enhanced, resulting in an increase in biomass productivity up to two folds (Vikramathithan et al., 2020). These studies on green microalgae indicate that the heterologous expression of CCM components is the potential for PE enhancement in diatoms. But due to the complexity of the steps and pathways involved, the engineering scheme for this mechanism is still in the early stage and needs further study.
Some green microalgae, such as C. reinhardtii, are model organisms for physiological study, but the difficulty and inefficiency in heterologous gene expression limit the research development. In that case, diatoms, as one of the most successful photosynthetic groups (Hippmann et al., 2022), have caught great attention from the science community in recent years for their genetic modification potential. Figure 1 summarizes the development of green microalgae and diatoms in PE enhancement. Great possibilities still lie in improving PE and biomass productivity in diatoms. The exploration of photosynthesis raises insight into fields beyond. By testing carbon dioxide- or bicarbonate-capturing efficiencies in open ponds, microalgae demonstrated their strong ability to fix bicarbonate via photosynthesis, thus thought to be well suited for carbon capture and sequestration (Sayre, 2010). Bioenergy production combined with carbon sequestration (BECCS) is considered a key strategy to deal with the global threat of climate change caused by greenhouse gases in the context of sustainable development goals (SDGs) (Barry et al., 2015). Meanwhile, the enhancement of photosynthetic efficiency also indicates the potential for microalgae in advanced wastewater treatment (Qi et al., 2021)and photovoltaic devices (Cho et al., 2019). To overcome the drawbacks of natural and artificial photosynthesis, a semi-artificial photosynthetic system was developed to efficiently generate solar fuels and chemicals (Kornienko et al., 2018).
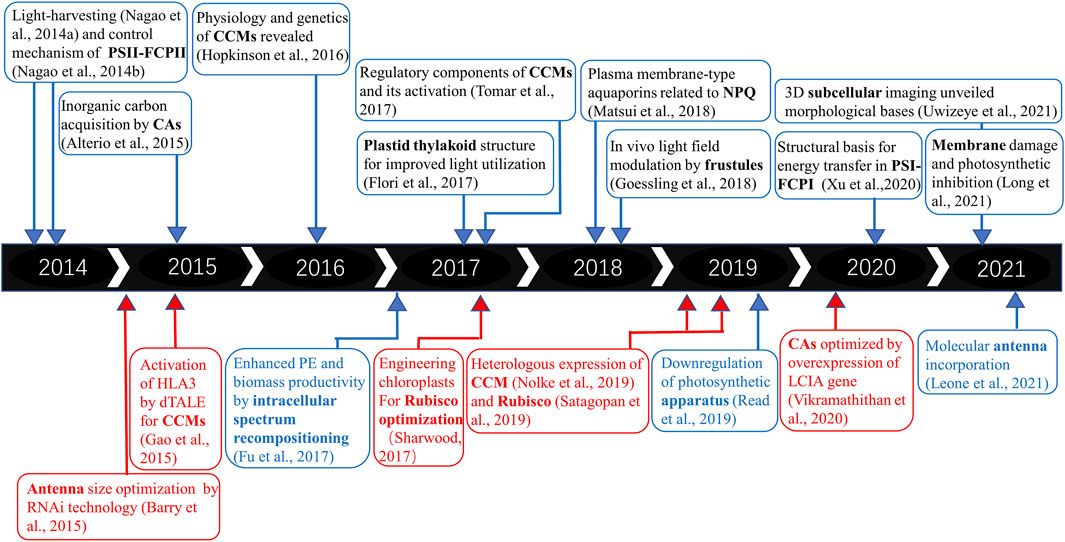
FIGURE 1. Timeline and basic and applied research trends on microalgal PE and biomass productivity. The upper panel refers to fundamental research on PE in diatoms, and the lower panel refers to practical applications of synthetic biology tools. The red part represents green microalgae, while the blue region represents diatoms.
Fucoxanthin (Fx) is an essential marine carotenoid that widely exists in microalgae, functioning as an anticancer, antibacterial, and hypoglycemic agent (Wang J. et al., 2019; Karpiński and Adamczak, 2019). Currently, Phaeophyta (brown algae) is the primary source of Fx. But due to its low yield, low quality, and high cost, it is difficult to meet market demand. Alternatively, the marine diatom P. tricornutum has more than 100 times the Fx content of Phaeophyta. Thus it has been considered a promising Fx commercial producer (Yang and Wei, 2020). Fx with biological activity plays a vital role in diatom growth. It exists together with chlorophylls as fucoxanthin–chlorophyll protein (FCP), which is crucial for the light-harvesting complex (Gelzinis et al., 2015; Wang W. et al., 2019). FCP enables diatoms to absorb light in a broad spectrum, contributing to energy transfer and dissipation in photosynthesis (Wang W. et al., 2019).
The biosynthesis of Fx in diatoms has been extensively studied but not fully understood (Figure 2A). It is generally agreed that the synthesis of Fx starts from isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) that are catalyzed by geranylgeranyl pyrophosphate synthase (GGPPS) to generate geranylgeranyl diphosphate (GGPP) (Fraser Paul et al., 2002; Cazzonelli and Pogson, 2010). Then the GGPP is converted to phytoene by phytoene synthase (PSY), which is the rate-determining step (Cazzonelli and Pogson, 2010). With phytoene desaturase (PDS), ζ-carotene desaturase (ZDS), and carotenoid isomerase (CRISO), phytoene is converted to lycopene, which is further catalyzed by lycopene β-cyclase (LCYB) to produce β-carotene. Cytochrome P450-type carotene hydroxylases (CYP97A/B/C) are believed to be involved in synthesizing the subsequent zeaxanthin (Zx) in microalgae. However, only the gene encoding CYP97B has been identified in P. tricornutum (Cui et al., 2019). Then, Zx is catalyzed to generate violaxanthin (Vx) by zeaxanthin epoxidase (ZEP1, ZEP2, and ZEP3) through intermediate metabolites antheraxanthin (Ax), which is further converted to Zx by violaxanthin de-epoxidase (VDE), forming the violaxanthin cycle (Eilers et al., 2016b). However, the next step to generate Fx is not fully understood with two hypothetical paths: 1) Vx is catalyzed by violaxanthin de-epoxidase–like (VDL) proteins to generate neoxanthin (Nx) as the precursor of Fx and diadinoxanthin (Ddx) (Dautermann et al., 2020), 2) Vx transfers to Ddx as the precursor of Fx and diatoxanthin (Dtx) (Bertrand, 2010; Dambek et al., 2012).
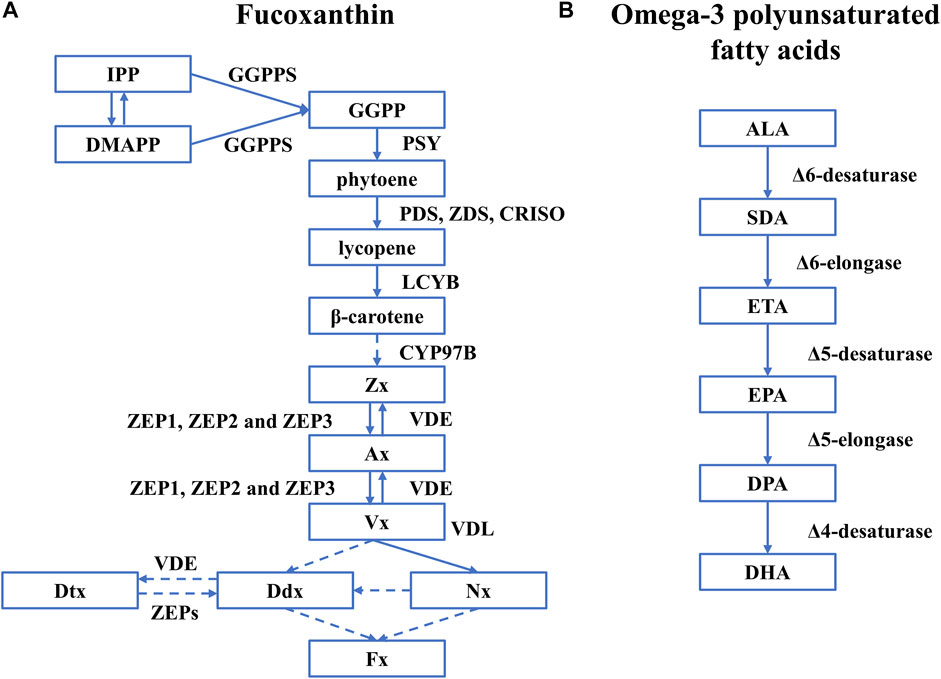
FIGURE 2. The biosynthetic pathway of Fx and omega-3 polyunsaturated fatty acids in diatoms. Rectangles represent intermediates, and arrows represent biosynthetic pathways and active enzymes. (A) Fucoxanthin. The figure was adapted from a previous publication (Manfellotto et al., 2020). A dashed line represents the unknown biosynthetic process. (B) omega-3 polyunsaturated fatty acids. The figure was adapted from a previous publication (Sayanova et al., 2017) and only covers the pathway of omega-3 polyunsaturated fatty acids. Abbreviations: IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl diphosphate; PSY, phytoene synthase; GGPPS, geranylgeranyl pyrophosphate synthase; GGPP, geranylgeranyl diphosphate; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRISO, carotenoid isomerase; LCYB, lycopene β-cyclase; Zx, zeaxanthin; ZEP1, ZEP2, and ZEP3, three types of Zea epoxidase; VDE, violaxanthin de-epoxidase; Vx, violaxanthin; VDL, violaxanthin de-epoxidase-like proteins; Nx, neoxanthin; Ddx, diadinoxanthin; Dtx, diatoxanthin.
It has been found that 1-deoxy-D-xylulose 5-phosphate synthase is necessary to generate IPP for initiating Fx biosynthesis. With the overexpression of the gene that encodes 1-deoxy-D-xylulose 5-phosphate synthase, the content of Fx increased up to 2.4 folds (Eilers et al., 2016a). Phytoene synthase (PSY) is one of the most important enzymes involved in the rate-determining step. The PSY gene fused with the enhanced green fluorescent protein (eGFP) gene was successfully introduced into P. tricornutum and overexpressed, resulting in an increase of Fx content by about 1.45 folds in the stationary phase and about 1.19 folds in total compared with the wild type (WT) (Kadono et al., 2015). A new technique for high-throughput screening of high-Fx-content diatom mutants using flow cytometry was invented and effectively separated the strains with higher Fx content, which was 25.5% higher than that in the WT (Fan et al., 2021). The simultaneous overexpression of three genes, violaxanthin de-epoxidase (Vde), Vde-related (Vdr), and zeaxanthin epoxidase 3 (Zep3) genes, resulted in a four-fold increase in Fx content in P. tricornutum (Manfellotto et al., 2020).
Diatoms can produce a variety of lipids, including triacylglycerol (TAG) and other neutral lipids, which makes them a potential source for biofuels. In addition, diatoms can produce biologically active omega-3 polyunsaturated fatty acids such as EPA and DHA that support the growth of organisms (Li-Beisson et al., 2019). They can be used to promote the learning and memory ability of the brain, inhibit the proliferation of some tumor cells, and improve glucose and lipid metabolism disorders (Zhang et al., 2019). EPA and DHA are mainly extracted from marine fish that can quickly accumulate harmful substances. Given that the ecological crisis caused by the overfishing of marine animals is also grievous, diatoms are considered a healthy and sustainable substitute (Fu et al., 2022).
The synthesis of omega-3 polyunsaturated fatty acids starts from α-linolenic (ALA, C18:3) obtained by the saturated fatty acids (C18:0) with the Δ9-, Δ12-, and Δ15-desaturases. EPA and DHA are synthesized with a series of desaturases and elongases, including Δ6-, Δ5- and Δ4-desaturases, and Δ6- and Δ5-elongases (Sayanova et al., 2017). The specific synthetic route is as follows (Figure 2): 1) As a precursor of omega-3 LC-PUFAs, α-linolenic (ALA, C18:3) is used to generate stearic acid (SDA, C18:4) by the action of Δ6-desaturase, and then synthesize eicosatetraenoic acid (ETA, C20:4) under the catalysis of Δ6-elongase. 2) ETA is converted to eicosapentaenoic acid (EPA, C20: 5) by Δ5-desaturase. 3) EPA is first elongated to docosapentaenoic acid (DPA, C22:5) by Δ5-elongase and then desaturated by a Δ4-desaturase to produce docosahexaenoic acid (DHA, C22:6) (Zulu et al., 2018).
Abiotic stress and genetic modification approaches have improved diatoms’ productivity for value-added compounds. Abiotic stresses, such as light, salinity, pH, and temperature stress, can increase the content of metabolites but reduce the biomass productivity of diatoms, leading to a decrease in overall yield. For example, under low salinity conditions, the EPA content in the arctic diatom Attheya septentrinalis increased, while its growth rate dropped significantly compared with that under the natural seawater condition (Steinrücken et al., 2018). In addition, the lipid content in P. tricornutum increased under high pH stress, while its carotenoid content increased under low pH stress, although their growth rates decreased under both pH stress conditions (Mus et al., 2013). On the contrary, genetic engineering can improve the content of bioactive compounds without reducing biomass production, which is beneficial to the productivity of target products. By inserting an additional endogenous Δ6 fatty acid desaturase gene (PtD6) into the P. tricornutum genome, the overexpression of PtD6 resulted in an increase in total lipids and EPA (Zhu et al., 2017). Similarly, overexpression of the Δ5-desaturase gene (PtD5b) in P. tricornutum increased the EPA content by 58% and also produced 0.44 mg/g DHA, which was not observed in the wild type (Peng et al., 2014). Under the synergistic effect with Δ5-desaturase (PtD5b) and malonyl CoA-acyl carrier protein transacylase (MCAT), the total lipid content was increased by 2.61 folds without impairing the growth of P. tricornutum, and the content of EPA and DHA also increased at the same time (Wang et al., 2017). One diacylglycerol acyltransferases (DGAT2B) gene was introduced and co-expressed with the Δ5-elongase (OtElo5) gene in P. tricornutum, leading to a higher content of DHA (3.8–8.4% of dry biomass) compared with the WT (1.8% of dry biomass) (Haslam et al., 2020).
Meganucleases (MNs) and transcription activator-like effector nucleases (TALENs) are practical tools for gene editing, such as gene knockout and gene knock-in. Using these technologies to disrupt the carbohydrate storage gene UDP-glucose pyrophosphorylase (UGPase) in P. tricornutum resulted in a 45-fold increase in TAG content in nutrient replete culture and a 3-fold increase in nitrogen-depleted culture (Daboussi et al., 2014). With the development of gene-editing technology, CRISPR/Cas9 has been applied to diatoms, which can realize gene editing quickly and efficiently at a low cost but with high accuracy (Nymark et al., 2017). With the aid of this technique, up to five genes in P. tricornutum were edited successfully, increasing total lipid content dramatically (Sharma et al., 2021).
Sterols, a type of triterpenoid, are the most abundant isoprenoids produced by diatoms, effective for cholesterol-lowering and anti-inflammation (Aldini et al., 2014). Sterols function as regulators of cell membrane dynamics in diatoms, but their synthetic pathways are not fully deciphered. Overexpression of HMGR (3-hydroxy-3-methyl glutaryl CoA reductase) in P. tricornutum increased the levels of sterol intermediates such as squalene, cycloartenol, and obtusifoliol, but the total sterol content did not change, indicating that there is a mechanism controlling the overall content in diatoms (Jaramillo-Madrid et al., 2020). Recently, independent genes distinct from other diatom species have been found in the marine diatom Rhizosolenia setigera, which encode two isopentenyl diphosphate isomerases (RsIDI1 and RsIDI2) and squalene synthase (RsSQS) that catalyze the synthesis of isoprenes. The sequence analyses suggested that these genes might come from a secondary gene fission event (Ferriols et al., 2017).
Diatoms have also been developed and engineered as photosynthetic cell factories to produce heterologous value-added bioactive compounds (as shown in Figure 3). When diatoms express heterologous compounds, the native product in diatoms (such as TAGs) can be extracted simultaneously to reduce costs further (D'Adamo et al., 2019). Heterologous compounds expressed in diatoms were summarized in Table 3, including plasmid design and transformation methods.
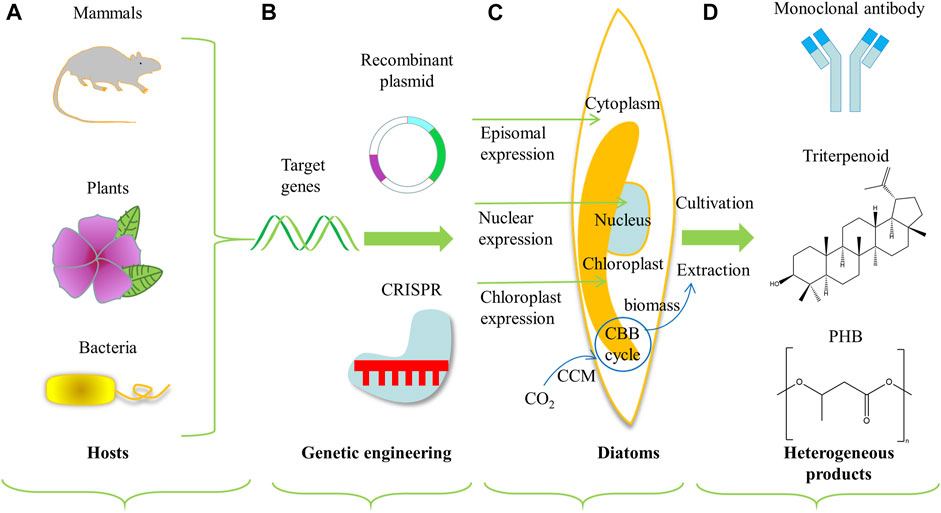
FIGURE 3. Schematic diagram of engineering diatoms as cell factories for heterologous production of value-added compounds. Heterologous expression system includes (A) the selection of target source genes, (B) conventional genetic engineering or genome engineering technologies like CRISPR, (C) DNA transformation and expression locus in diatoms, and (D) production of value-added compounds (D'Adamo et al., 2019). The part with blue cycle and arrows represents the framework for inorganic carbon utilization. Abbreviations: CBB, Calvin–Benson–Bassham cycle; CCM, carbon concentrating mechanism; PHB, Poly-β-hydroxybutyrate.
Due to specificity in targeting and modulating disease-related pathways, recombinant biologics have been widely used to treat various diseases (Dehghani et al., 2020). The vast majority of recombinant medical proteins are glycoproteins, implying that complex N-glycosylation and disulfide bond formation are necessary for adequately folding glycoprotein structures and achieving glycoprotein function. It is challenging to produce safe and qualified recombinant therapeutic protein products with prokaryotic cells by conventional heterologous expression. In addition, the high cost of mammalian cells limits their applications in developing and less-developed countries. With the natural advantages of photosynthetic autotrophy, such as fast reproduction and low cost, diatoms also have a complete post-translational expression pathway, which can directly secrete the hetero-expressed proteins into the medium. This feature reduces the complexity and cost of subsequent separation and purification in diatoms (Specht and Mayfield, 2014). Heterologous expression of recombinant proteins and antimicrobial peptides has also been reported in green microalgae. A single chain antibody that recognizes B-cell surface molecule CD22 and a chimeric immunotoxin gene contained the hinge and CH2 and CH3 domains of a human IgG1 were expressed in chloroplasts of C. reinhardtii (Tran et al., 2013). Antimicrobial peptide piscidin-4 gene with optimized codon was expressed in chloroplasts of the freshwater green microalga Haematococcus pluvialis (Wang K. et al., 2020).
Recombinant pharmaceutical proteins synthesized by microalgae mainly include monoclonal antibodies (mAbs), immunotoxins, human cytokines, recombinant hormones, and recombinant vaccines. The first recombinant proteins reported are the monoclonal IgG1/kappa antibody CL4mAb and the Hepatitis B surface antigen (HBsAg) subtype adr expressed in the P. tricornutum, resulting in 8.7% CL4mAb and 0.7% HBsAg of the total soluble protein yielded in the endoplasmic reticulum (ER). Antibody accumulation does not impair the growth rate and cell viability of P. tricornutum, and antibody expression levels do not decline over 1 year of testing, which is critical for large-scale industrial production (Hempel et al., 2011b). Similarly, two studies successfully expressed a human IgG antibody against the Hepatitis B Virus surface protein and monoclonal IgG antibodies against the nucleoprotein of Marburg virus in P. tricornutum (Hempel and Maier, 2012; Hempel et al., 2017).
Terpenoids can be divided into monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), disesquiterpenes (C25), triterpenes (C30), and tetraterpenes (C40) according to the number of isoprene units in the molecule (Ma et al., 2021). Terpenoids are natural compounds widely used in the pharmaceutical industry, such as anticancer and antimalarial applications. But many terpenoids, such as triterpenes, are found in low concentrations in living organisms, increasing production costs due to purification complexity and limiting their commercial application (Ma et al., 2021). E. coli and S. cerevisiae are usually selected as the heterologous hosts for terpenoid production based on their rapid growth and clear metabolic pathways. However, diatoms are more closely related to plants than bacteria or yeast, meaning that cell environments of diatoms are more favorable to the synthesis of plant-derived terpenoids. Diatoms contain both the cytosolic mevalonate pathway (MVA) and the chloroplastic non-mevalonate pathway (MEP) to develop IPP, which is an essential precursor of the sterol metabolic pathway (Fabris et al., 2020). Moreover, the flux towards isoprenoids in microalgae may be higher than in other organisms (Lauersen, 2019). However, the unknown regulation and underlying mechanism of the isoprenoid pathway in diatoms affect further investigations of heterologous terpenoid production.
Intron-mediated enhancement is a phenomenon in many eukaryotes, increasing transcription speed, mRNA stability, and protein translation efficiency when introns are present (Shaul, 2017). By introducing this strategy, the native glyceraldehyde 3-phosphate dehydrogenase (GAPDH) intron was inserted into the coding sequence of Abies grandis bisabolene (a representative sesquiterpene) synthase gene to realize heterologous expression in the diatom Fistulifera solaris (Tanaka et al., 2021).
Geraniol is a plant monoterpenoid, the main component of rose essential oil. It is formed by the direct conversion of geranyl diphosphate (GPP) by the enzyme geraniol synthase. Unlike the cell factories that naturally do not accumulate GPP, such as yeast, E. coli, and C. reinhardtii, the marine diatom P. tricornutum has an intracellular pool of GPP but has no genes for terpene synthases (Jaramillo-Madrid et al., 2019; Fabris et al., 2020). These properties indicate no endogenous monoterpenoid biosynthetic pathways to compete for heterologous production of monoterpenoids. A full-length coding sequence of Catharanthus roseus geraniol synthase was introduced into P. tricornutum using an episomal expression strategy with episomes delivered into the diatom nuclei by bacterial conjugation (Fabris et al., 2020). The target gene was expressed in the cytoplasm, and the production of monoterpenoid geraniol reached 0.309 mg/L, a similar level as that achieved by yeast engineering (Fabris et al., 2020).
Lupeol is a plant triterpenoid with antioxidant, anti-inflammatory, and tumor-suppressive effects. It can be synthesized from 2,3-oxidosqualene precursors under the catalysis of oxidosqualene cyclases. In the plant sapogenin biosynthetic pathway, lupeol can be oxidized into betulinic acid under the catalysis of cytochrome P450 and NADPH reductase. Two lupeol synthase coding sequences from Lotus japonicus and Arabidopsis thaliana were successfully integrated into the P. tricornutum nuclear genome. Later, Medicago truncatula CYP716A12, a cytochrome P450-dependent monooxygenase, and the Medicago truncatula cytochrome P450 reductases (MtCPR) were expressed in P. tricornutum to synthesize betulinic acid (D'Adamo et al., 2019). Lupeol is harmless to host cells and may be secreted into the medium, which has inherent advantages in downstream extraction at the industrial level (D'Adamo et al., 2019). This study is more complex than other reports of modified diatoms, involving relatively complex material reactions and pathways. The stability of heterologous genes and their impact on the original metabolism should be considered when the complexity of strains increases (Ng et al., 2020).
The vast majority of plastic made from petroleum is challenging to degrade and lacks a complete recycling process. Most plastics are discharged into the sea after use and become microplastics gradually, which have detrimental effects on human health and the environment. Biodegradable bioplastics can effectively reduce plastic pollution and carbon emissions (Park and Lee, 2022). However, bioplastics made from land crops require large amounts of arable land, fertilizers, and freshwater. Among many bioplastics, polyhydroxybutyrate (PHB) produced in bacteria and cyanobacteria has attracted much attention due to its good mechanical, heat resistance, and biocompatibility properties (Hempel et al., 2011a; Sirohi et al., 2021). The entire PHB pathway of Ralstonia eutropha H16, which has three enzymes, PhaA (ketothiolase), PhaB (acetoacetyl CoA reductase), and PhaC (PHB synthase), has been expressed in the cytoplasm of P. tricornutum. However, the study of PHB-producing microalgae is still in its infant stage, and the metabolic pathway of PHB production in diatoms is not fully understood (Sirohi et al., 2021). Although diatoms synthesize PHB at a rate of 100 times faster than plant expression systems, they still cannot compete with the soil bacterium R. eutropha, which has been commercially used as the PHB producer for many years (Hempel et al., 2011a).
This review summarizes the current systems and synthetic biology progress for diatom cell factories toward biotechnology applications (Figure 4). The systems biology approaches, including the GEMs, have facilitated metabolic prediction and rapid development of strain engineering and metabolic engineering in diatoms. Unlike GEMs of well-studied model organisms like E. coli, the establishment of diatom GEM is still in its early stage. The lack of high-quality and high-quantity genome annotations impedes the development of diatoms’ system biology (Vavitsas et al., 2021). PE and biomass productivity enhancements are essential foundations and boosters for cost-effective algal production. There are also considerable gaps between diatoms and some model green microalgae in the photosynthetic field, such as the modification on CCMs and Rubisco. Although more studies have to be done on diatoms, it is clear that the long-awaited breakthrough in PE will be realized under the recent progress in synthetic biology, therefore boosting biomass productivity to a brand-new level in diatoms.
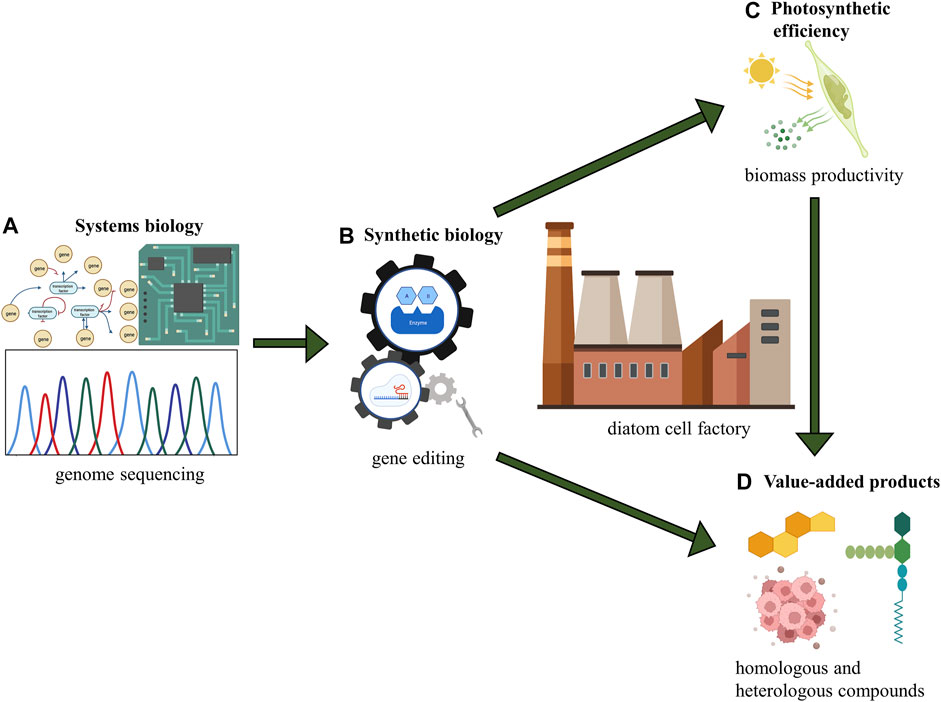
FIGURE 4. Conceptual scheme for diatom cell factory. (A) systems biology: understanding the whole picture of metabolism in diatoms; (B) synthetic biology: genetic manipulation and genome engineering; (C) photosynthetic efficiency: fundamental principle of energy conversion and biomass production; (D) value-added products: output from diatom cell factory including homologous and heterologous compounds.
Although the knowledge of the biosynthetic steps of bioactive compounds and genetic engineering help improve the yield and productivity of value-added compounds, diatom synthetic biology is still in its infancy. The essential enzymes and genes of interest, together with their regulation, are not fully understood. The diversity of the genome also poses a significant challenge to developing efficient, convenient, and stable transgenic tools for practical application since there has been no report of using the popular CRISPR technology for heterologous expression in diatoms yet. However, with the continuous development of gene-editing technology, new tools such as TALENs and CRISPR/Cas9 play a critical role in producing value-added compounds in diatoms. The combination of synthetic and systems biology will also allow us to understand better the compound conversion pathways and energy flows in diatom cell factories. These new technologies will facilitate the discovery of new compounds and the improvement of their yield, making diatoms one of the best candidates for cell factories. In the foreseeable future, we can anticipate that more compounds for bioenergy, food, feed, and pharmaceutical industries will be developed through algal cell factories, which could play an essential role in the green economy.
In short, diatom cell factories may provide solutions to global challenges like rising CO2 and energy crises. The diversity of diatom species offers us tremendous possibilities for digging potential value-added products. More discoveries and technological breakthroughs are needed to drive diatom research from the laboratory to commercialization. Moreover, this progress requires support from government policymakers, investors, educators, and other stakeholders. We believe a sustainable diatom cell factory is promising and achievable under continuous development of systems and synthetic biology to combine PE enhancement and compound yield improvement for biosustainability and carbon neutrality.
JC, YH, YS, and WF co-wrote the draft of the manuscript, and WF conceived and developed the idea of the review. All authors reviewed, edited, and approved the final manuscript.
This work was supported by the Hundred Talents Program of Zhejiang University and the Icelandic Technology Development Fund (Rannis).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, A., Tiwari, A., and Srivastava, S. (2020). A Genome-Scale Metabolic Model of Thalassiosira Pseudonana CCMP 1335 for a Systems-Level Understanding of its Metabolism and Biotechnological Potential. Microorganisms 8 (9), 1396. doi:10.3390/microorganisms8091396
Aldini, R., Micucci, M., Cevenini, M., Fato, R., Bergamini, C., Nanni, C., et al. (2014). Antiinflammatory Effect of Phytosterols in Experimental Murine Colitis Model: Prevention, Induction, Remission Study. PLoS One 9 (9), e108112. doi:10.1371/journal.pone.0108112
Alterio, V., Langella, E., De Simone, G., and Monti, S. (2015). Cadmium-Containing Carbonic Anhydrase CDCA1 in Marine Diatom Thalassiosira Weissflogii. Mar. Drugs 13 (4), 1688–1697. doi:10.3390/md13041688
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The Genome of the Diatom Thalassiosira Pseudonana : Ecology, Evolution, and Metabolism. Science 306 (5693), 79–86. doi:10.1126/science.1101156
Barati, B., Zeng, K., Baeyens, J., Wang, S., Addy, M., Gan, S.-Y., et al. (2021). Recent Progress in Genetically Modified Microalgae for Enhanced Carbon Dioxide Sequestration. Biomass Bioenergy 145, 105927. doi:10.1016/j.biombioe.2020.105927
Barry, A. N., Starkenburg, S. R., and Sayre, R. T. (2015). Strategies for Optimizing Algal Biology for Enhanced Biomass Production. Front. Energy Res. 3. doi:10.3389/fenrg.2015.00001
Bertrand, M. (2010). Carotenoid Biosynthesis in Diatoms. Photosynth Res. 106 (1), 89–102. doi:10.1007/s11120-010-9589-x
Bhattacharjya, R., Tiwari, A., Marella, T. K., Bansal, H., and Srivastava, S. (2021). New Paradigm in Diatom Omics and Genetic Manipulation. Bioresour. Technol. 325, 124708. doi:10.1016/j.biortech.2021.124708
Bjerkelund Røkke, G., Hohmann-Marriott, M. F., and Almaas, E. (2020). An Adjustable Algal Chloroplast Plug-And-Play Model for Genome-Scale Metabolic Models. PLoS One 15 (2), e0229408. doi:10.1371/journal.pone.0229408
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. nature 456 (7219), 239–244. doi:10.1038/nature07410
Briggs, A. A., and Carpenter, R. C. (2019). Contrasting Responses of Photosynthesis and Photochemical Efficiency to Ocean Acidification under Different Light Environments in a Calcifying Alga. Sci. Rep. 9 (1), 3986. doi:10.1038/s41598-019-40620-8
Broddrick, J. T., Du, N., Smith, S. R., Tsuji, Y., Jallet, D., Ware, M. A., et al. (2019). Cross‐compartment Metabolic Coupling Enables Flexible Photoprotective Mechanisms in the Diatom Phaeodactylum Tricornutum. New Phytol. 222 (3), 1364–1379. doi:10.1111/nph.15685
Butler, T., Kapoore, R. V., and Vaidyanathan, S. (2020). Phaeodactylum Tricornutum: A Diatom Cell Factory. Trends Biotechnol. 38 (6), 606–622. doi:10.1016/j.tibtech.2019.12.023
Cazzaniga, S., Dall'Osto, L., Szaub, J., Scibilia, L., Ballottari, M., Purton, S., et al. (2014). Domestication of the Green Alga Chlorella Sorokiniana: Reduction of Antenna Size Improves Light-Use Efficiency in a Photobioreactor. Biotechnol. Biofuels 7 (1), 157. doi:10.1186/s13068-014-0157-z
Cazzonelli, C. I., and Pogson, B. J. (2010). Source to Sink: Regulation of Carotenoid Biosynthesis in Plants. Trends Plant Sci. 15 (5), 266–274. doi:10.1016/j.tplants.2010.02.003
Cho, C., Nam, K., Kim, G.-Y., Seo, Y. H., Hwang, T. G., Seo, J.-W., et al. (2019). Multi-bandgap Solar Energy Conversion via Combination of Microalgal Photosynthesis and Spectrally Selective Photovoltaic Cell. Sci. Rep. 9 (1), 18999. doi:10.1038/s41598-019-55358-6
Costache, T. A., Acién Fernández, F. G., Morales, M. M., Fernández-Sevilla, J. M., Stamatin, I., and Molina, E. (2013). Comprehensive Model of Microalgae Photosynthesis Rate as a Function of Culture Conditions in Photobioreactors. Appl. Microbiol. Biotechnol. 97 (17), 7627–7637. doi:10.1007/s00253-013-5035-2
Cui, H., Ma, H., Cui, Y., Zhu, X., Qin, S., and Li, R. (2019). Cloning, Identification and Functional Characterization of Two Cytochrome P450 Carotenoids Hydroxylases from the Diatom Phaeodactylum Tricornutum. J. Biosci. Bioeng. 128 (6), 755–765. doi:10.1016/j.jbiosc.2019.06.008
D'Adamo, S., Schiano di Visconte, G., Lowe, G., Szaub-Newton, J., Beacham, T., Landels, A., et al. (2019). Engineering the Unicellular Alga Phaeodactylum Tricornutum for High-Value Plant Triterpenoid Production. Plant Biotechnol. J. 17 (1), 75–87. doi:10.1111/pbi.12948
Daboussi, F., Leduc, S., Maréchal, A., Dubois, G., Guyot, V., Perez-Michaut, C., et al. (2014). Genome Engineering Empowers the Diatom Phaeodactylum Tricornutum for Biotechnology. Nat. Commun. 5 (1), 3831. doi:10.1038/ncomms4831
Dambek, M., Eilers, U., Breitenbach, J., Steiger, S., Buchel, C., and Sandmann, G. (2012). Biosynthesis of Fucoxanthin and Diadinoxanthin and Function of Initial Pathway Genes in Phaeodactylum Tricornutum. J. Exp. Bot. 63 (15), 5607–5612. doi:10.1093/jxb/ers211
Dautermann, O., Lyska, D., Andersen-Ranberg, J., Becker, M., Fröhlich-Nowoisky, J., Gartmann, H., et al. (2020). An Algal Enzyme Required for Biosynthesis of the Most Abundant Marine Carotenoids. Sci. Adv. 6 (10), eaaw9183. doi:10.1126/sciadv.aaw9183
Dehghani, J., Adibkia, K., Movafeghi, A., Maleki-Kakelar, H., Saeedi, N., and Omidi, Y. (2020). Towards a New Avenue for Producing Therapeutic Proteins: Microalgae as a Tempting Green Biofactory. Biotechnol. Adv. 40, 107499. doi:10.1016/j.biotechadv.2019.107499
Del Prete, S., Vullo, D., Scozzafava, A., Capasso, C., and Supuran, C. T. (2014). Cloning, Characterization and Anion Inhibition Study of the δ-class Carbonic Anhydrase (TweCA) from the Marine Diatom Thalassiosira Weissflogii. Bioorg. Med. Chem. 22 (1), 531–537. doi:10.1016/j.bmc.2013.10.045
Eilers, U., Bikoulis, A., Breitenbach, J., Büchel, C., and Sandmann, G. (2016a). Limitations in the Biosynthesis of Fucoxanthin as Targets for Genetic Engineering in Phaeodactylum Tricornutum. J. Appl. Phycol. 28 (1), 123–129. doi:10.1007/s10811-015-0583-8
Eilers, U., Dietzel, L., Breitenbach, J., Büchel, C., and Sandmann, G. (2016b). Identification of Genes Coding for Functional Zeaxanthin Epoxidases in the Diatom Phaeodactylum Tricornutum. J. Plant Physiology 192, 64–70. doi:10.1016/j.jplph.2016.01.006
Fabris, M., George, J., Kuzhiumparambil, U., Lawson, C. A., Jaramillo-Madrid, A. C., Abbriano, R. M., et al. (2020). Extrachromosomal Genetic Engineering of the Marine Diatom Phaeodactylum Tricornutum Enables the Heterologous Production of Monoterpenoids. ACS Synth. Biol. 9 (3), 598–612. doi:10.1021/acssynbio.9b00455
Falciatore, A., Jaubert, M., Bouly, J.-P., Bailleul, B., and Mock, T. (2020). Diatom Molecular Research Comes of Age: Model Species for Studying Phytoplankton Biology and Diversity. Plant Cell 32 (3), 547–572. doi:10.1105/tpc.19.00158
Fan, Y., Ding, X.-T., Wang, L.-J., Jiang, E.-Y., Van, P. N., and Li, F.-L. (2021). Rapid Sorting of Fucoxanthin-Producing Phaeodactylum Tricornutum Mutants by Flow Cytometry. Mar. Drugs 19 (4), 228. doi:10.3390/md19040228
Ferriols, V. M. E. N., Yaginuma-Suzuki, R., Fukunaga, K., Kadono, T., Adachi, M., Matsunaga, S., et al. (2017). An Exception Among Diatoms: Unique Organization of Genes Involved in Isoprenoid Biosynthesis in Rhizosolenia Setigera CCMP 1694. Plant J. 92 (5), 822–833. doi:10.1111/tpj.13719
Flori, S., Jouneau, P.-H., Bailleul, B., Gallet, B., Estrozi, L. F., Moriscot, C., et al. (2017). Plastid Thylakoid Architecture Optimizes Photosynthesis in Diatoms. Nat. Commun. 8 (1), 15885. doi:10.1038/ncomms15885
Fu, W., Chaiboonchoe, A., Khraiwesh, B., Sultana, M., Jaiswal, A., Jijakli, K., et al. (2017). Intracellular Spectral Recompositioning of Light Enhances Algal Photosynthetic Efficiency. Sci. Adv. 3(9), e1603096. doi:10.1126/sciadv.1603096
Fu, W., Nelson, D. R., Mystikou, A., Daakour, S., and Salehi-Ashtiani, K. (2019). Advances in Microalgal Research and Engineering Development. Curr. Opin. Biotechnol. 59, 157–164. doi:10.1016/j.copbio.2019.05.013
Fu, W., Shu, Y., Yi, Z., Su, Y., Pan, Y., Zhang, F., et al. (2022). Diatom Morphology and Adaptation: Current Progress and Potentials for Sustainable Development. Sustainable Horizons 2, 100015. doi:10.1016/j.horiz.2022.100015
Gao, H., Wang, Y., Fei, X., Wright, D. A., and Spalding, M. H. (2015). Expression Activation and Functional Analysis of HLA 3, a Putative Inorganic Carbon Transporter in C Hlamydomonas Reinhardtii. Plant J. 82 (1), 1–11. doi:10.1111/tpj.12788
Gelzinis, A., Butkus, V., Songaila, E., Augulis, R., Gall, A., Büchel, C., et al. (2015). Mapping Energy Transfer Channels in Fucoxanthin-Chlorophyll Protein Complex. Biochimica Biophysica Acta (BBA) - Bioenergetics 1847 (2), 241–247. doi:10.1016/j.bbabio.2014.11.004
Goessling, J. W., Su, Y., Cartaxana, P., Maibohm, C., Rickelt, L. F., Trampe, E. C. L., et al. (2018). Structure-based Optics of Centric Diatom Frustules: Modulation of the In Vivo Light Field for Efficient Diatom Photosynthesis. New Phytol. 219 (1), 122–134. doi:10.1111/nph.15149
Grabowski, G. A., Golembo, M., and Shaaltiel, Y. (2014). Taliglucerase Alfa: an Enzyme Replacement Therapy Using Plant Cell Expression Technology. Mol. Genet. Metabolism 112 (1), 1–8. doi:10.1016/j.ymgme.2014.02.011
Haslam, R. P., Hamilton, M. L., Economou, C. K., Smith, R., Hassall, K. L., Napier, J. A., et al. (2020). Overexpression of an Endogenous Type 2 Diacylglycerol Acyltransferase in the Marine Diatom Phaeodactylum Tricornutum Enhances Lipid Production and Omega-3 Long-Chain Polyunsaturated Fatty Acid Content. Biotechnol. Biofuels 13, 87. doi:10.1186/s13068-020-01726-8
Hempel, F., Bozarth, A. S., Lindenkamp, N., Klingl, A., Zauner, S., Linne, U., et al. (2011a). Microalgae as Bioreactors for Bioplastic Production. Microb. Cell Fact. 10, 81. doi:10.1186/1475-2859-10-81
Hempel, F., Lau, J., Klingl, A., and Maier, U. G. (2011b). Algae as Protein Factories: Expression of a Human Antibody and the Respective Antigen in the Diatom Phaeodactylum Tricornutum. PLoS One 6 (12), e28424. doi:10.1371/journal.pone.0028424
Hempel, F., and Maier, U. G. (2012). An Engineered Diatom Acting like a Plasma Cell Secreting Human IgG Antibodies with High Efficiency. Microb. Cell Fact. 11 (1), 126. doi:10.1186/1475-2859-11-126
Hempel, F., Maurer, M., Brockmann, B., Mayer, C., Biedenkopf, N., Kelterbaum, A., et al. (2017). From Hybridomas to a Robust Microalgal-Based Production Platform: Molecular Design of a Diatom Secreting Monoclonal Antibodies Directed against the Marburg Virus Nucleoprotein. Microb. Cell Fact. 16 (1), 131. doi:10.1186/s12934-017-0745-2
Hippmann, A. A., Schuback, N., Moon, K. M., McCrow, J. P., Allen, A. E., Foster, L. F., et al. (2022). Proteomic Analysis of Metabolic Pathways Supports Chloroplast-Mitochondria Cross‐talk in a Cu‐limited Diatom. Plant Direct 6 (1), e376. doi:10.1002/pld3.376
Hopkinson, B. M., Dupont, C. L., and Matsuda, Y. (2016). The Physiology and Genetics of CO2 Concentrating Mechanisms in Model Diatoms. Curr. Opin. Plant Biol. 31, 51–57. doi:10.1016/j.pbi.2016.03.013
Jaramillo-Madrid, A. C., Abbriano, R., Ashworth, J., Fabris, M., Pernice, M., and Ralph, P. J. (2020). Overexpression of Key Sterol Pathway Enzymes in Two Model Marine Diatoms Alters Sterol Profiles in Phaeodactylum Tricornutum. Pharmaceuticals 13 (12), 481. doi:10.3390/ph13120481
Jaramillo-Madrid, A. C., Ashworth, J., Fabris, M., and Ralph, P. J. (2019). Phytosterol Biosynthesis and Production by Diatoms (Bacillariophyceae). Phytochemistry 163, 46–57. doi:10.1016/j.phytochem.2019.03.018
Juneja, A., Chaplen, F. W. R., and Murthy, G. S. (2016). Genome Scale Metabolic Reconstruction of Chlorella Variabilis for Exploring its Metabolic Potential for Biofuels. Bioresour. Technol. 213, 103–110. doi:10.1016/j.biortech.2016.02.118
Kadono, T., Kira, N., Suzuki, K., Iwata, O., Ohama, T., Okada, S., et al. (2015). Effect of an Introduced Phytoene Synthase Gene Expression on Carotenoid Biosynthesis in the Marine Diatom Phaeodactylum Tricornutum. Mar. Drugs 13 (8), 5334–5357. doi:10.3390/md13085334
Karpiński, T. M., and Adamczak, A. (2019). Fucoxanthin-An Antibacterial Carotenoid. Antioxidants 8 (8), 239. doi:10.3390/antiox8080239
Kikutani, S., Nakajima, K., Nagasato, C., Tsuji, Y., Miyatake, A., and Matsuda, Y. (2016). Thylakoid Luminal θ-carbonic Anhydrase Critical for Growth and Photosynthesis in the Marine Diatom Phaeodactylum Tricornutum. Proc. Natl. Acad. Sci. U.S.A. 113 (35), 9828–9833. doi:10.1073/pnas.1603112113
Kono, A., Chou, T. H., Radhakrishnan, A., Bolla, J. R., Sankar, K., Shome, S., et al. (2020). Structure and Function of LCI1: a Plasma Membrane CO 2 Channel in the Chlamydomonas CO 2 Concentrating Mechanism. Plant J. 102 (6), 1107–1126. doi:10.1111/tpj.14745
Kornienko, N., Zhang, J. Z., Sakimoto, K. K., Yang, P., and Reisner, E. (2018). Interfacing Nature's Catalytic Machinery with Synthetic Materials for Semi-artificial Photosynthesis. Nat. Nanotech 13 (10), 890–899. doi:10.1038/s41565-018-0251-7
Krishnakumar, V., Hanlon, M. R., Contrino, S., Ferlanti, E. S., Karamycheva, S., Kim, M., et al. (2015). Araport: the Arabidopsis Information Portal. Nucleic Acids Res. 43 (Database issue), D1003–D1009. doi:10.1093/nar/gku1200
Lauersen, K. J. (2019). Eukaryotic Microalgae as Hosts for Light-Driven Heterologous Isoprenoid Production. PLANTA 249 (1), 155–180. doi:10.1007/s00425-018-3048-x
Lavoie, M., Saint-Béat, B., Strauss, J., Guérin, S., Allard, A., V. Hardy, S., et al. (2020). Genome-Scale Metabolic Reconstruction and In Silico Perturbation Analysis of the Polar Diatom Fragilariopsis Cylindrus Predicts High Metabolic Robustness. Biology 9 (2), 30. doi:10.3390/biology9020030
Leone, G., De la Cruz Valbuena, G., Cicco, S. R., Vona, D., Altamura, E., Ragni, R., et al. (2021). Incorporating a Molecular Antenna in Diatom Microalgae Cells Enhances Photosynthesis. Sci. Rep. 11 (1), 5209. doi:10.1038/s41598-021-84690-z
Levering, J., Broddrick, J., Dupont, C. L., Peers, G., Beeri, K., Mayers, J., et al. (2016). Genome-Scale Model Reveals Metabolic Basis of Biomass Partitioning in a Model Diatom. PLoS One 11 (5), e0155038. doi:10.1371/journal.pone.0155038
Levering, J., Dupont, C. L., Allen, A. E., Palsson, B. O., and Zengler, K. (2017). Integrated Regulatory and Metabolic Networks of the Marine Diatom Phaeodactylum Tricornutum Predict the Response to Rising CO 2 Levels. MSYSTEMS 2 (1). doi:10.1128/mSystems.00142-16
Li, X., Patena, W., Fauser, F., Jinkerson, R. E., Saroussi, S., Meyer, M. T., et al. (2019). A Genome-wide Algal Mutant Library and Functional Screen Identifies Genes Required for Eukaryotic Photosynthesis. Nat. Genet. 51 (4), 627–635. doi:10.1038/s41588-019-0370-6
Li-Beisson, Y., Thelen, J. J., Fedosejevs, E., and Harwood, J. L. (2019). The Lipid Biochemistry of Eukaryotic Algae. Prog. Lipid Res. 74, 31–68. doi:10.1016/j.plipres.2019.01.003
Liu, C.-P., Tsai, T.-I., Cheng, T., Shivatare, V. S., Wu, C.-Y., Wu, C.-Y., et al. (2018). Glycoengineering of Antibody (Herceptin) through Yeast Expression and In Vitro Enzymatic Glycosylation. Proc. Natl. Acad. Sci. U.S.A. 115 (4), 720–725. doi:10.1073/pnas.1718172115
Long, M., Peltekis, A., González-Fernández, C., Hégaret, H., and Bailleul, B. (2021). Allelochemicals of Alexandrium Minutum: Kinetics of Membrane Disruption and Photosynthesis Inhibition in a Co-occurring Diatom. Harmful Algae 103, 101997. doi:10.1016/j.hal.2021.101997
Lu, Y., Zhang, X., Gu, X., Lin, H., and Melis, A. (2021). Engineering Microalgae: Transition from Empirical Design to Programmable Cells. Crit. Rev. Biotechnol. 41 (8), 1233–1256. doi:10.1080/07388551.2021.1917507
Ma, C., Zhang, K., Zhang, X., Liu, G., Zhu, T., Che, Q., et al. (2021). Heterologous Expression and Metabolic Engineering Tools for Improving Terpenoids Production. Curr. Opin. Biotechnol. 69, 281–289. doi:10.1016/j.copbio.2021.02.008
Manfellotto, F., Stella, G. R., Falciatore, A., Brunet, C., and Ferrante, M. I. (2020). Engineering the Unicellular Alga Phaeodactylum Tricornutum for Enhancing Carotenoid Production. Antioxidants 9 (8), 757. doi:10.3390/antiox9080757
Marella, T. K., and Tiwari, A. (2020). Marine Diatom Thalassiosira Weissflogii Based Biorefinery for Co-production of Eicosapentaenoic Acid and Fucoxanthin. Bioresour. Technol. 307, 123245. doi:10.1016/j.biortech.2020.123245
Matsui, H., Hopkinson, B., Nakajima, K., and Matsuda, Y. (2018). Plasma-membrane-type Aquaporins from Marine Diatoms Function as CO2/NH3 Channels and Provide Photoprotection. Plant Physiol. 178 (1), 00453–357. doi:10.1104/pp.18.00453
Metcalf, A. J., Nagygyor, A., and Boyle, N. R. (2020). Rapid Annotation of Photosynthetic Systems (RAPS): Automated Algorithm to Generate Genome-Scale Metabolic Networks from Algal Genomes. Algal Res. 50, 101967. doi:10.1016/j.algal.2020.101967
Meyer, M. T., Itakura, A. K., Patena, W., Wang, L., He, S., Emrich-Mills, T., et al. (2020). Assembly of the Algal CO 2 -fixing Organelle, the Pyrenoid, Is Guided by a Rubisco-Binding Motif. Sci. Adv. 6(46), eabd2408. doi:10.1126/sciadv.abd2408
Mus, F., Toussaint, J.-P., Cooksey, K. E., Fields, M. W., Gerlach, R., Peyton, B. M., et al. (2013). Physiological and Molecular Analysis of Carbon Source Supplementation and pH Stress-Induced Lipid Accumulation in the Marine Diatom Phaeodactylum Tricornutum. Appl. Microbiol. Biotechnol. 97 (8), 3625–3642. doi:10.1007/s00253-013-4747-7
Naduthodi, M. I. S., Claassens, N. J., D’Adamo, S., van der Oost, J., and Barbosa, M. J. (2021). Synthetic Biology Approaches to Enhance Microalgal Productivity. Trends Biotechnol. 39 (10), 1019–1036. doi:10.1016/j.tibtech.2020.12.010
Nagao, R., Kato, K., Suzuki, T., Ifuku, K., Uchiyama, I., Kashino, Y., et al. (2019). Structural Basis for Energy Harvesting and Dissipation in a Diatom PSII-FCPII Supercomplex. Nat. Plants 5 (8), 890–901. doi:10.1038/s41477-019-0477-x
Nagao, R., Yokono, M., Teshigahara, A., Akimoto, S., and Tomo, T. (2014a). Light-Harvesting Ability of the Fucoxanthin Chlorophyll A/C-Binding Protein Associated with Photosystem II from the Diatom Chaetoceros Gracilis as Revealed by Picosecond Time-Resolved Fluorescence Spectroscopy. J. Phys. Chem. B 118 (19), 5093–5100. doi:10.1021/jp502035y
Nagao, R., Yokono, M., Tomo, T., and Akimoto, S. (2014b). Control Mechanism of Excitation Energy Transfer in a Complex Consisting of Photosystem II and Fucoxanthin Chlorophyll A/c-Binding Protein. J. Phys. Chem. Lett. 5 (17), 2983–2987. doi:10.1021/jz501496p
Ng, I. S., Keskin, B. B., and Tan, S. I. (2020). A Critical Review of Genome Editing and Synthetic Biology Applications in Metabolic Engineering of Microalgae and Cyanobacteria. Biotechnol. J. 15 (8), 1900228. doi:10.1002/biot.201900228
Ng, I. S., Tan, S. I., Kao, P. H., Chang, Y. K., and Chang, J. S. (2017). Recent Developments on Genetic Engineering of Microalgae for Biofuels and Bio‐Based Chemicals. Biotechnol. J. 12 (10), 1600644. doi:10.1002/biot.201600644
Nölke, G., Barsoum, M., Houdelet, M., Arcalís, E., Kreuzaler, F., Fischer, R., et al. (2019). The Integration of Algal Carbon Concentration Mechanism Components into Tobacco Chloroplasts Increases Photosynthetic Efficiency and Biomass. Biotechnol. J. 14 (3), 1800170. doi:10.1002/biot.201800170
Nymark, M., Sharma, A., Hafskjold, M., Sparstad, T., Bones, A., and Winge, P. (2017). CRISPR/Cas9 Gene Editing in the Marine Diatom Phaeodactylum Tricornutum. Bio-protocol 7 (15), e2442. doi:10.21769/BioProtoc.2442
Ogawa, T., Tamoi, M., Kimura, A., Mine, A., Sakuyama, H., Yoshida, E., et al. (2015). Enhancement of Photosynthetic Capacity in Euglena Gracilis by Expression of Cyanobacterial Fructose-1,6-/Sedoheptulose-1,7-Bisphosphatase Leads to Increases in Biomass and Wax Ester Production. Biotechnol. Biofuels 8, 80. doi:10.1186/s13068-015-0264-5
Ogura, A., Akizuki, Y., Imoda, H., Mineta, K., Gojobori, T., and Nagai, S. (2018). Comparative Genome and Transcriptome Analysis of Diatom, Skeletonema Costatum, Reveals Evolution of Genes for Harmful Algal Bloom. BMC Genomics 19, 12. doi:10.1186/s12864-018-5144-5
Osuna-Cruz, C. M., Bilcke, G., Vancaester, E., De Decker, S., Bones, A. M., Winge, P., et al. (2020). The Seminavis Robusta Genome Provides Insights into the Evolutionary Adaptations of Benthic Diatoms. Nat. Commun. 11 (1), 13. doi:10.1038/s41467-020-17191-8
Park, Y.-K., and Lee, J. (2022). Achievements in the Production of Bioplastics from Microalgae. Phytochem. Rev. doi:10.1007/s11101-021-09788-8
Parkes, R., Archer, L., Gee, D. M., Smyth, T. J., Gillespie, E., and Touzet, N. (2021). Differential Responses in EPA and Fucoxanthin Production by the Marine Diatom Stauroneis Sp. Under Varying Cultivation Conditions. Biotechnol. Prog. 37 (6), e3197. doi:10.1002/btpr.3197
Peng, K.-T., Zheng, C.-N., Xue, J., Chen, X.-Y., Yang, W.-D., Liu, J.-S., et al. (2014). Delta 5 Fatty Acid Desaturase Upregulates the Synthesis of Polyunsaturated Fatty Acids in the Marine Diatom Phaeodactylum Tricornutum. J. Agric. Food Chem. 62 (35), 8773–8776. doi:10.1021/jf5031086
Perrine, Z., Negi, S., and Sayre, R. T. (2012). Optimization of Photosynthetic Light Energy Utilization by Microalgae. Algal Res. 1 (2), 134–142. doi:10.1016/j.algal.2012.07.002
Qi, F., Jia, Y., Mu, R., Ma, G., Guo, Q., Meng, Q., et al. (2021). Convergent Community Structure of Algal-Bacterial Consortia and its Effects on Advanced Wastewater Treatment and Biomass Production. Sci. Rep. 11 (1), 21118. doi:10.1038/s41598-021-00517-x
Read, R. W., Vuono, D. C., Neveux, I., Staub, C., and Grzymski, J. J. (2019). Coordinated Downregulation of the Photosynthetic Apparatus as a Protective Mechanism against UV Exposure in the Diatom Corethron hystrix. Appl. Microbiol. Biotechnol. 103 (4), 1837–1850. doi:10.1007/s00253-018-9544-x
Rickaby, R. E. M., and Eason Hubbard, M. R. (2019). Upper Ocean Oxygenation, Evolution of RuBisCO and the Phanerozoic Succession of Phytoplankton. Free Radic. Biol. Med. 140, 295–304. doi:10.1016/j.freeradbiomed.2019.05.006
Satagopan, S., Huening, K. A., and Tabita, F. R. (2019). Selection of Cyanobacterial ( Synechococcus Sp. Strain PCC 6301) RubisCO Variants with Improved Functional Properties that Confer Enhanced CO 2 -Dependent Growth of Rhodobacter Capsulatus, a Photosynthetic Bacterium. MBIO 10 (4). doi:10.1128/mBio.01537-19
Sayanova, O., Mimouni, V., Ulmann, L., Morant-Manceau, A., Pasquet, V., Schoefs, B., et al. (2017). Modulation of Lipid Biosynthesis by Stress in Diatoms. Phil. Trans. R. Soc. B 372 (1728), 20160407. doi:10.1098/rstb.2016.0407
Sayre, R. (2010). Microalgae: The Potential for Carbon Capture. Biosci. Biotechnol. Biochem. 60 (9), 722–727. doi:10.1525/bio.2010.60.9.9
Seif, Y., and Palsson, B. Ø. (2021). Path to Improving the Life Cycle and Quality of Genome-Scale Models of Metabolism. Cell Syst. 12 (9), 842–859. doi:10.1016/j.cels.2021.06.005
Serif, M., Lepetit, B., Weissert, K., Kroth, P. G., and Rio Bartulos, C. (2017). A Fast and Reliable Strategy to Generate TALEN-Mediated Gene Knockouts in the Diatom Phaeodactylum Tricornutum. Algal Res. 23, 186–195. doi:10.1016/j.algal.2017.02.005
Sforza, E., Simionato, D., Giacometti, G. M., Bertucco, A., and Morosinotto, T. (2012). Adjusted Light and Dark Cycles Can Optimize Photosynthetic Efficiency in Algae Growing in Photobioreactors. PLoS One 7 (6), e38975. doi:10.1371/journal.pone.0038975
Sharma, A. K., Nymark, M., Flo, S., Sparstad, T., Bones, A. M., and Winge, P. (2021). Simultaneous Knockout of Multiple LHCF Genes Using Single sgRNAs and Engineering of a High‐fidelity Cas9 for Precise Genome Editing in Marine Algae. Plant Biotechnol. J. 19 (8), 1658–1669. doi:10.1111/pbi.13582
Sharwood, R. E. (2017). Engineering Chloroplasts to Improve Rubisco Catalysis: Prospects for Translating Improvements into Food and Fiber Crops. New Phytol. 213 (2), 494–510. doi:10.1111/nph.14351
Shaul, O. (2017). How Introns Enhance Gene Expression. Int. J. Biochem. Cell Biol. 91 (Pt B), 145–155. doi:10.1016/j.biocel.2017.06.016
Shin, W.-S., Lee, B., Jeong, B.-r., Chang, Y. K., and Kwon, J.-H. (2016). Truncated Light-Harvesting Chlorophyll Antenna Size in Chlorella Vulgaris Improves Biomass Productivity. J. Appl. Phycol. 28 (6), 3193–3202. doi:10.1007/s10811-016-0874-8
Sirohi, R., Lee, J. S., Yu, B. S., Roh, H., and Sim, S. J. (2021). Sustainable Production of Polyhydroxybutyrate from Autotrophs Using CO2 as Feedstock: Challenges and Opportunities. Bioresour. Technol. 341, 125751. doi:10.1016/j.biortech.2021.125751
Smith‐Harding, T. J., Beardall, J., and Mitchell, J. G. (2017). The Role of External Carbonic Anhydrase in Photosynthesis during Growth of the Marine Diatom Chaetoceros Muelleri. J. Phycol. 53 (6), 1159–1170. doi:10.1111/jpy.12572
Specht, E. A., and Mayfield, S. P. (2014). Algae-based Oral Recombinant Vaccines. Front. Microbiol. 5, 60. doi:10.3389/fmicb.2014.00060
Steinrücken, P., Mjøs, S. A., Prestegard, S. K., and Erga, S. R. (2018). Enhancing EPA Content in an Arctic Diatom: A Factorial Design Study to Evaluate Interactive Effects of Growth Factors. Front. Plant Sci. 9, 491. doi:10.3389/fpls.2018.00491
Tanaka, T., Maeda, Y., Suhaimi, N., Tsuneoka, C., Nonoyama, T., Yoshino, T., et al. (2021). Intron-Mediated Enhancement of Transgene Expression in the Oleaginous Diatom Fistulifera Solaris towards Bisabolene Production. Algal Res. 57, 102345. doi:10.1016/j.algal.2021.102345
Tanaka, T., Maeda, Y., Veluchamy, A., Tanaka, M., Abida, H., Maréchal, E., et al. (2015). Oil Accumulation by the Oleaginous Diatom Fistulifera Solaris as Revealed by the Genome and Transcriptome. Plant Cell 27 (1), 162–176. doi:10.1105/tpc.114.135194
Tomar, V., Sidhu, G. K., Nogia, P., Mehrotra, R., and Mehrotra, S. (2017). Regulatory Components of Carbon Concentrating Mechanisms in Aquatic Unicellular Photosynthetic Organisms. Plant Cell Rep. 36 (11), 1671–1688. doi:10.1007/s00299-017-2191-3
Toster, J., Iyer, K. S., Xiang, W., Rosei, F., Spiccia, L., and Raston, C. L. (2013). Diatom Frustules as Light Traps Enhance DSSC Efficiency. NANOSCALE 5 (3), 873–876. doi:10.1039/c2nr32716c
Traller, J. C., Cokus, S. J., Lopez, D. A., Gaidarenko, O., Smith, S. R., McCrow, J. P., et al. (2016). Genome and Methylome of the Oleaginous Diatom Cyclotella Cryptica Reveal Genetic Flexibility toward a High Lipid Phenotype. Biotechnol. Biofuels 9, 20. doi:10.1186/s13068-016-0670-3
Tran, M., Van, C., Barrera, D. J., Pettersson, P. L., Peinado, C. D., Bui, J., et al. (2013). Production of Unique Immunotoxin Cancer Therapeutics in Algal Chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 110 (1), E15–E22. doi:10.1073/pnas.1214638110
Uwizeye, C., Decelle, J., Jouneau, P.-H., Flori, S., Gallet, B., Keck, J.-B., et al. (2021). Morphological Bases of Phytoplankton Energy Management and Physiological Responses Unveiled by 3D Subcellular Imaging. Nat. Commun. 12 (1), 1049. doi:10.1038/s41467-021-21314-0
van Tol, H. M., and Armbrust, E. V. (2021). Genome-Scale Metabolic Model of the Diatom Thalassiosira Pseudonana Highlights the Importance of Nitrogen and Sulfur Metabolism in Redox Balance. PLoS One 16 (3), e0241960. doi:10.1371/journal.pone.0241960
van Veen, H. A., Koiter, J., Vogelezang, C. J. M., van Wessel, N., van Dam, T., Velterop, I., et al. (2012). Characterization of Recombinant Human C1 Inhibitor Secreted in Milk of Transgenic Rabbits. J. Biotechnol. 162 (2-3), 319–326. doi:10.1016/j.jbiotec.2012.09.005
Vavitsas, K., Kugler, A., Satta, A., Hatzinikolaou, D. G., Lindblad, P., Fewer, D. P., et al. (2021). Doing Synthetic Biology with Photosynthetic Microorganisms. Physiol. Plant. 173 (2), 624–638. doi:10.1111/ppl.13455
Vikramathithan, J., Hwangbo, K., Lim, J.-M., Lim, K.-M., Kang, D. Y., Park, Y.-I., et al. (2020). Overexpression of Chlamydomonas Reinhardtii LCIA (CrLCIA) Gene Increases Growth of Nannochloropsis Salina CCMP1776. Algal Res. 46, 101807. doi:10.1016/j.algal.2020.101807
Walsh, G. (2018). Biopharmaceutical Benchmarks 2018. Nat. Biotechnol. 36 (12), 1136–1145. doi:10.1038/nbt.4305
Wang, J., Ma, Y., Yang, J., Jin, L., Gao, Z., Xue, L., et al. (2019a). Fucoxanthin Inhibits Tumour-Related Lymphangiogenesis and Growth of Breast Cancer. J. Cell Mol. Med. 23 (3), 2219–2229. doi:10.1111/jcmm.14151
Wang, K., Cui, Y., Wang, Y., Gao, Z., Liu, T., Meng, C., et al. (2020a). Chloroplast Genetic Engineering of a Unicellular Green Alga Haematococcus pluvialis with Expression of an Antimicrobial Peptide. Mar. Biotechnol. 22 (4), 572–580. doi:10.1007/s10126-020-09978-z
Wang, W., Yu, L.-J., Xu, C., Tomizaki, T., Zhao, S., Umena, Y., et al. (2019b). Structural Basis for Blue-Green Light Harvesting and Energy Dissipation in Diatoms. Science 363 (6427). eaav0365. doi:10.1126/science.aav0365
Wang, W., Zhao, S., Pi, X., Kuang, T., Sui, S. F., and Shen, J. R. (2020b). Structural Features of the Diatom Photosystem II-Light‐harvesting Antenna Complex. FEBS J. 287 (11), 2191–2200. doi:10.1111/febs.15183
Wang, X., Balamurugan, S., Liu, S.-F., Ji, C.-Y., Liu, Y.-H., Yang, W.-D., et al. (2021). Hydrolysis of Organophosphorus by Diatom Purple Acid Phosphatase and Sequential Regulation of Cell Metabolism. J. Exp. Bot. 72 (8), 2918–2932. doi:10.1093/jxb/erab026
Wang, X., Liu, Y.-H., Wei, W., Zhou, X., Yuan, W., Balamurugan, S., et al. (2017). Enrichment of Long-Chain Polyunsaturated Fatty Acids by Coordinated Expression of Multiple Metabolic Nodes in the Oleaginous Microalga Phaeodactylum Tricornutum. J. Agric. Food Chem. 65 (35), 7713–7720. doi:10.1021/acs.jafc.7b02397
Xi, Y., Yin, L., Chi, Z. y., and Luo, G. (2021). Characterization and RNA-Seq Transcriptomic Analysis of a Scenedesmus Obliqnus Mutant with Enhanced Photosynthesis Efficiency and Lipid Productivity. Sci. Rep. 11 (1), 11795. doi:10.1038/s41598-021-88954-6
Xu, C., Pi, X., Huang, Y., Han, G., Chen, X., Qin, X., et al. (2020). Structural Basis for Energy Transfer in a Huge Diatom PSI-FCPI Supercomplex. Nat. Commun. 11 (1), 5081. doi:10.1038/s41467-020-18867-x
Yamano, T., Sato, E., Iguchi, H., Fukuda, Y., and Fukuzawa, H. (2015). Characterization of Cooperative Bicarbonate Uptake into Chloroplast Stroma in the Green Alga Chlamydomonas Reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 112 (23), 7315–7320. doi:10.1073/pnas.1501659112
Yan, D., Beardall, J., and Gao, K. (2018). Variation in Cell Size of the Diatom Coscinodiscus Granii Influences Photosynthetic Performance and Growth. Photosynth Res. 137 (1), 41–52. doi:10.1007/s11120-017-0476-6
Yang, B., Liu, J., Ma, X., Guo, B., Liu, B., Wu, T., et al. (2017). Genetic Engineering of the Calvin Cycle toward Enhanced Photosynthetic CO2 Fixation in Microalgae. Biotechnol. Biofuels 10, 229. doi:10.1186/s13068-017-0916-8
Yang, R., and Wei, D. (2020). Improving Fucoxanthin Production in Mixotrophic Culture of Marine Diatom Phaeodactylum Tricornutum by LED Light Shift and Nitrogen Supplementation. Front. Bioeng. Biotechnol. 8. doi:10.3389/fbioe.2020.00820
Yang, R., Wei, D., and Xie, J. (2020). Diatoms as Cell Factories for High-Value Products: Chrysolaminarin, Eicosapentaenoic Acid, and Fucoxanthin. Crit. Rev. Biotechnol. 40 (7), 993–1009. doi:10.1080/07388551.2020.1805402
Yao, J., Chen, X., Zhang, M., Zhang, Y., Zhang, Z., Xian, X., et al. (2019). Inhibition of the Photosynthetic Activity of Synedra Sp. By Sonication: Performance and Mechanism. J. Environ. Manag. 233, 54–62. doi:10.1016/j.jenvman.2018.12.029
Zarmi, Y., Gordon, J. M., Mahulkar, A., Khopkar, A. R., Patil, S. D., Banerjee, A., et al. (2020). Enhanced Algal Photosynthetic Photon Efficiency by Pulsed Light. ISCIENCE 23 (5), 101115. doi:10.1016/j.isci.2020.101115
Zhang, T.-T., Xu, J., Wang, Y.-M., and Xue, C.-H. (2019). Health Benefits of Dietary Marine DHA/EPA-enriched Glycerophospholipids. Prog. Lipid Res. 75, 100997. doi:10.1016/j.plipres.2019.100997
Zhu, B.-H., Tu, C.-C., Shi, H.-P., Yang, G.-P., and Pan, K.-H. (2017). Overexpression of Endogenous Delta-6 Fatty Acid Desaturase Gene Enhances Eicosapentaenoic Acid Accumulation in Phaeodactylum Tricornutum. Process Biochem. 57, 43–49. doi:10.1016/j.procbio.2017.03.013
Keywords: microalgae, diatoms, synthetic biology, systems biology, photosynthesis, biomass productivity
Citation: Chen J, Huang Y, Shu Y, Hu X, Wu D, Jiang H, Wang K, Liu W and Fu W (2022) Recent Progress on Systems and Synthetic Biology of Diatoms for Improving Algal Productivity. Front. Bioeng. Biotechnol. 10:908804. doi: 10.3389/fbioe.2022.908804
Received: 31 March 2022; Accepted: 29 April 2022;
Published: 13 May 2022.
Edited by:
Eugene Fletcher, Escarpment Laboratories Inc., CanadaReviewed by:
Zhengquan Gao, Shandong University of Technology, ChinaCopyright © 2022 Chen, Huang, Shu, Hu, Wu, Jiang, Wang, Liu and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiqi Fu, d2VpcWlmdUB6anUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.