
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 21 October 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.904636
 Qingguo Liu1,2†
Qingguo Liu1,2† Jiaqing Zhu1†
Jiaqing Zhu1† Na Liu1
Na Liu1 Wenjun Sun1
Wenjun Sun1 Bin Yu1
Bin Yu1 Huanqing Niu1
Huanqing Niu1 Dong Liu1
Dong Liu1 Pingkai Ouyang1
Pingkai Ouyang1 Hanjie Ying1
Hanjie Ying1 Yong Chen1
Yong Chen1 Gulin Zhao1*
Gulin Zhao1* Tianpeng Chen1*
Tianpeng Chen1*The biofilm (BF) provides favorable growth conditions to cells, which has been exploited in the field of industrial biotechnology. Based on our previous research works on type I fimbriae for the biosynthesis of L-threonine (LT) in Escherichia coli, in this study, a fimA-overexpressing strain was engineered, which improved BF formation under industrial fermentation conditions. The morphological observation and characterization of BF formation were conducted to verify the function of the subunit FimA. However, it was not suitable for repeated-batch immobilized fermentation as the LT titer was not elevated significantly. The underlying molecular mechanisms of BF formation and the LT carbon flux were explored by transcriptomic analysis. The results showed that fimA regulated E. coli BF formation but affected LT carbon distribution. This study will stimulate thoughts about how the fimbriae gene regulated biofilms and amino acid excretion and will bring some consideration and provide a reference for the development of BF-based biomanufacturing processes in E. coli.
L-threonine (LT) is an essential amino acid with a variety of functions in pharmaceutical, chemical, and food industries, as well as feed additives (Leuchtenberger et al., 2005). Thus, its demand is sharply increasing. At present, microbial fermentation is generally used for LT synthesis in the industry due to its low raw material cost, high fermentation yield, and easy large-scale production. Moreover, Escherichia coli (E. coli) has become the best candidate strain in industrial production with the characteristics of easy culture and a clear genetic background (Lee et al., 2006; Dong et al., 2011). However, the preparation of the seed culture was required every time in free-cell batch LT fermentation, which led to the unavailability for the continuous catalytic process (Beloin et al., 2004; Rajkumar et al., 2013). In addition, the increasing products and salt concentrations of E. coli might affect the membrane osmotic pressure for cell tolerance, and then, the imbalance of cell homeostasis could result in a low reaction intensity for LT production.
Immobilized fermentation based on a continuous catalytic process is one of the most important ways to realize the continuous production of chemicals, which has gained worldwide attention. Hence, establishing a new immobilized fermentation system can achieve continuous and efficient production of LT in E. coli. The biofilm (BF) is a complex cell community closely related to biotic or abiotic surfaces (Sauer, 2003). Cells can employ these structures to withstand shear forces and obtain nutrients. E. coli covered by BFs can tolerate extreme conditions, including high cell density, hypoxia restriction, and high osmotic pressure, which are essential for the continuous fermentation process.
BF-based immobilized fermentation is considered a solution instead of free-cell fermentation, owing to its unique advantages (e.g., BF matrix protection, repeated cells, and improve metabolic activities) compared to free-cell fermentation (Zhao et al., 2015). The microbial BFs, such as Aspergillus niger, Saccharomyces cerevisiae, Clostridium acetobutylicum, and E. coli, have been effectively used for immobilized fermentation (Chen et al., 2013; Liu et al., 2013; Yu et al., 2018; Chen et al., 2019).
Type I fimbriae, about 1–2 μm in length and about 6.9 nm in width, are one of the most critical structures for BF biosynthesis in prokaryotic bacteria, including E. coli, which contain a large number of fimbrial gene clusters belonging to the chaperone/usher assembly class (Tripathi et al., 2013). Each operon encodes type I fimbriae structural subunits that are assembled into a fimbrial filament on the cell surface by a periplasmic chaperone and outer membrane usher protein (Hung and Hultgren, 1998). Previous research has confirmed that the overexpression of fimH could improve the efficiency of LT production by enhancement of BF formation and immobilization intensity (Chen et al., 2019). Moreover, it has been reported that FimA is the main structural unit of type I fimbriae, which consists of 500–3000 FimA proteins, accounting for 95%. Meanwhile, fimbriated bacteria can express 4200 fimbrial filaments on their surface, which plays a key role in the fimbriae assembly and BF formation (Berne et al., 2015). Thus, upon expression of a fimbrial gene cluster, the respective major structural subunit could become one of the most abundant proteins in the bacterial cell.
In the present study, a fimA-overexpressing E. coli strain was constructed for enhancing BF formation. A BF-based fermentation system was developed with polyurethane as a carrier for BF enrichment. After adhesion to the carrier surfaces, the cells formed a mass of BFs to withstand high shear force. The morphological observation was performed for the characterization analysis of BFs. In addition, the effects of fimA disturbance on the microbial physiological process of E. coli were explored, in combination with the detection of LT yield. Subsequently, transcriptomic analysis was conducted to explore to the role of fimA disturbance in cell physiological metabolism.
E. coli W1688 (CCTCC M2015233), an LT-producing strain, was derived from E. coli MG1655 by introducing mutations and molecular modifications of the LT metabolic pathway, and it could form BFs. All detailed characteristics of plasmids and strains in this study are presented in Table 1. Amplification of the fimA gene was performed using the E. coli W1688 genomic DNA. The plasmid pET28a and the fimA gene were ligated with the restriction enzymes Xba I and Nco I using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). Red homologous recombination was used in the fimA deletion strain of E. coli W1688 (Datsenko and Wanner, 2000; Madyagol et al., 2011). Briefly, the gene of the kanamycin resistance sequence and homologous regions (about 50 bp) were combined by PCR. Subsequently, the One Step Cloning Kit (Vazyme, Nanjing, China) was used in the cloning of the plasmid pKD4 and the resistance fragment. The knockout of the gene fimA was performed by electroporation (Bio-Rad, United States) with 2.0 kV, 25 mF, and a 200 Ω pulse with the recombinant plasmid transforming into the E. coli W1688 strain. The specific operation processes were adopted from the study by Chen et al. (2019).
The wild-type and recombinant strains of E. coli were cultured in an LB medium containing NaCl (10 g/L), tryptone (10 g/L), and yeast extract (5 g/L). The fermentation medium contained glucose (30 g/L), (NH4)2SO4 (20 g/L), CaCO3 (15 g/L), yeast extract (2 g/L), KH2PO4 (1 g/L), MgSO4·7H2O (0.8 g/L), MnSO4·5H2O (0.2 g/L), and FeSO4·7H2O (0.2 g/L). The initial pH was adjusted to 7.2 using acetic acid. All culture media were sterilized at 115°C for 20 min, and 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added when necessary. The shake flask fermentation was operated at 37°C and 200–220 rpm.
Briefly, 5% seed culture with final OD600 0.05 was inoculated in free-cell fermentation with flasks. Sampling was performed, followed by centrifugation (8000 rpm, 5 min, 4°C), and the supernatants were subjected to the quantitative analysis of LT and residual glucose. Immobilized fermentation was conducted with the carrier (30 g/L), according to the aforementioned conditions. After the first batch, 10% of the residual broth with the carrier covered by BFs was added into the fresh culture medium, followed by the second batch of culture. The subsequent operations were the same as mentioned earlier.
The concentrations of LT were determined by HPLC (high-performance liquid chromatography) with a UV detector at 338 nm, according to its maximum UV absorption wavelength, 1 ml/min, and 36°C. The mobile phase consisted of 80% acetonitrile and 0.1 M sodium acetate. The concentrations of glucose were detected using an RI detector with the Aminex HPX-87H column at 1 ml/min and 55°C. The mobile phase consisted of 5 mM H2SO4.
Total biomass staining was performed for the semi-quantitative analysis of BFs using the basic dye crystal violet (Li et al., 2003). After 12-h cultivation at 37°C, the strains were diluted (1:2000) and cultured again in a 96-well plate (200 μl/well) at 37°C. PBS (1%) was used to wash the wells twice for removing free cells. The BFs were fixed with methanol at 4°C for 15 min. Subsequently, the BFs were stained with crystal violet (1%) for 15 min, and PBS was used to wash the wells repeatedly 3–4 times. Then, 200 μl of acetic acid (33%) was added to dissolve crystal violet. Finally, the absorbance was measured at 570 nm using a Thermo Scientific microplate reader (Multiskan SkyHigh, Thermo Fisher Scientific, United States).
After 30-h cultivation, the coverslip was gently removed from the fermentation broth for the observation of BFs through microscopy. The BFs were fixed in paraformaldehyde (4%) and dehydrated using a vacuum freeze dryer. Transmission electron microscopy (TEM) (HT7820, Hitachi, Japan), TM3000 electron microscopy (Hitachi, Japan), and scanning electron microscopy (SEM) (SEM 4800, Hitachi, Japan) were carried out.
The adhesion properties of bacteria and distribution of cells in the BFs were examined using a fluorescence microscope. Briefly, 4% paraformaldehyde was used for BF fixing as described previously. Then, the fixed BFs were stained with 0.2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 30 min.
The pretreatment steps of the three strains before transcriptomic analysis were adopted from the study by Chen et al. (2019). Frasergen Inc. (http://www.frasergen.com/) provided sequencing platforms and technical assistance in data analyses. Briefly, the samples of immobilized fermentation were centrifuged (8000 rpm, 5 min, 4°C) to remove the liquid supernatant. Three biological replicates were applied for parallel analysis. The differential expression level of each gene was assessed according to the absolute value of Log2Ratio ≥ 1 and a false discovery rate (FDR) of ≤ 0.05.
After harvesting E. coli recombinant strains and wild-type strains at the exponential phase, the RNAprep Pure Cell/bacteria Kit was used to isolate total RNA, and RNase-Free DNase was used to digest the residual DNA. cDNA was prepared for RT-qPCR after reverse transcription. The primer pairs were designed by Primer Express software with avoidance of choosing hairpin structures (Table 2). RT-qPCR assays were conducted on a StepOnePlus Real-Time PCR System using the SYBR Green PCR Master Mix. The relative expression levels of target genes were measured using the 2-ΔΔCt method. One negative control and three technical replicates were used for each sample.
After spread plate cultivation, negative control and false positive test were carried out simultaneously. Then, 549 bp of the whole fimA fragment was deleted by the replacement of kanamycin resistance with the results of PCR and sequencing verification, which was named E. coli W1688-ΔfimA. For the fimA overexpression strain, E. coli W1688-fimA* was also successfully constructed. The different abilities of BF formation were revealed by the 96-well plate experiments. The crystal violet staining OD570 of W1688-fimA* in the LB medium was remarkably elevated by 96% compared to that of the wild-type strain (1.34 versus 2.62, respectively), which could be benefited by fimA overexpression (Figure 1A). By contrast, the knockout of fimA gene markedly attenuated BF formation in the W1688-ΔfimA strain compared to the wild-type strain (1.34 versus 1.25, respectively). Consistent results were also found in the fermentation medium. Actually, after the construction was completed, the difference between the vector control alone with pET28a and the wild-type strain was less than 5% in the aspect of the microbial growth, biofilm formation, and amino acid production. Therefore, these data were omitted. In addition, the fluorescent dye DAPI was used to observe the living cells, which preliminarily verified the effect of fimA on BF formation.
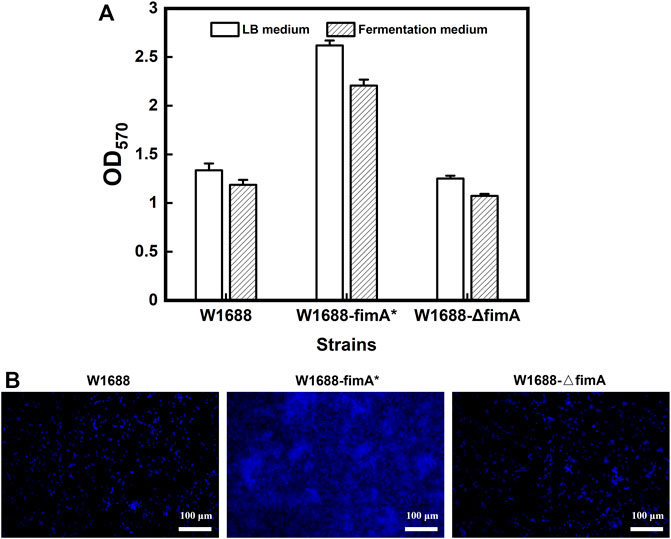
FIGURE 1. Semi-quantitative analysis of BF formation in the three strains. (A) Crystal violet staining in the two media. (B) Fluorescence microscopy of the BFs stained by DAPI.
TM3000 electron microscopy indicated that cell adhesion and BF formation were more noticeable in E. coli W1688-fimA* at different magnifications than those in the wild-type strain (Figure 2). Cells were encased in BFs by extracellular secretions, which were in the form of aggregation with each other. On the contrary, BF formation was obviously decreased and presented as a sparse bacterial distribution in E. coli W1688-ΔfimA. In addition, SEM was also used for the observation of BF formation (Figure 3A). The physiological function of fimA, as the subunit of type I fimbriae, was investigated. Based on the results of TEM, tiny type I fimbriae were seen in the wild-type strain (Figure 3B1). As expected, the deletion of the fimA gene resulted in the absence of fimbriae with a smooth appearance (Figure 3B3). This may be due to the fact that FimA is the main structural unit in the fimbrial assembly, which account for 95%. In contrast, the overexpression of fimA not only contributed to an increase in fimbriae which assembled on the cell surface evenly but also led to the accumulation of flagella with the filamentous substance attaching to bacteria, which intertwined cells with each other (Figure 3B2).
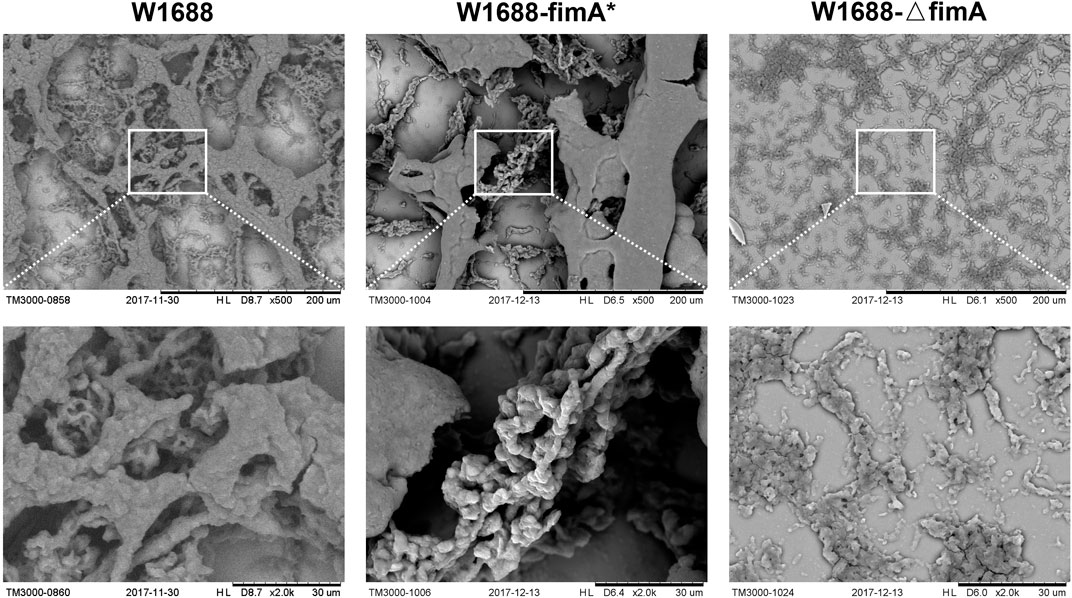
FIGURE 2. TM3000 electron microscopy of cell adhesion and BF formation at various magnification levels. First column: W1688; second column: W1688-fimA*; third column: W1688-△fimA. Cell adhesion and BF formation were noticeable in E. coli W1688-fimA*. Cells were encased in BFs by extracellular secretions with the form of aggregation with each other. BF formation was decreased and presented as a sparse distribution in E. coli W1688-ΔfimA.
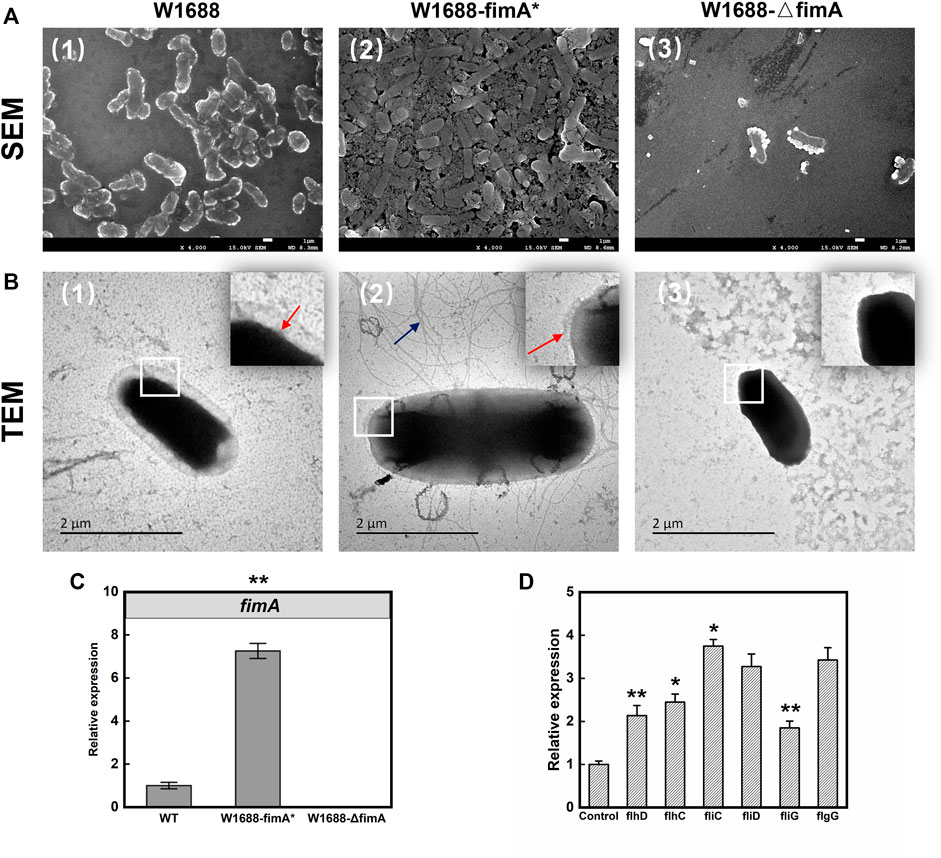
FIGURE 3. SEM (A) and TEM (B) images of BF formation and cell adhesion. First row: SEM; second row: TEM. First column: W1688; second column: W1688-fimA*; third column: W1688-△fimA. (C) RT-qPCR validation of the fimA expression. (D) RT-qPCR validation of flagellar gene expression in WT and the fimA* strain. Mean ± SD (n = 3). Two-tailed t-test. ***p < 0.001, **p < 0.01, and *p < 0.05. The results of SEM for the observation of BF formation were similar to crystal violet staining of the BF. Type I fimbriae were tiny on the cell surface (Figure 3B1, the red arrow). The absence of fimbriae with smooth appearance was observed in W1688-△fimA (Figure 3B3). An increase was observed in the extracellular surface fimbriae in W1688-fimA* (Figure 3B2, the red arrow), and the accumulation of the flagellum assembly with the filamentous substance attached to bacteria was observed in W1688-fimA*, which intertwined cells with each other (Figure 3B2, the blue arrow).
RT-PCR test results showed the relative expression of fimA in the fimA* strain was seven times more than that in the wild-type strain (Figure 3C), which indicated that due to the high efficiency of the T7 promoter, the fimA gene could still be activated efficiently when IPTG was added at the beginning of the batch fermentation. To investigate the increased flagellum in the fimA* strain, RT-PCR was used to detect the expression of some representative genes in the flagella. The results showed that the transcriptional activators of the flagellar regulon (FlhDC), flagellar filament structure protein (FliC), flagellar filament capping protein (FliD), flagellar motor switch protein (FliG), and flagellar basal-body rod protein (FlgG) significantly increased with varying degrees (Figure 3D), suggesting that the overexpression of fimA gene may enhance the assembly of the flagellum structure. Here, we describe the phenomenon that overexpression of fimA could result in an increase in flagella. The expression of flagella could represent two meanings: one was the expression of flagellum composition and structure, which led to the increase in the flagellum assembly. The other one was the expression of flagellum-mediated motility with energy expenditure, which is represented by the flagellar motor gene motA/B. The overexpression of the fimA gene might enhance the assembly of the flagellum structure, and RT-PCR results for relative expression of motA/B had not shown a significant change (data not shown), which indicated fimA overexpression might be not associated with the activation of bacterial motility directly. It was similar that Lane et al. (2007) showed that the overexpression of type 1 fimbriae dramatically downregulated motility and flagellum expression in E. coli. Altogether, these data provided further insight into the complex interplay between type I fimbriae expression and the flagellum-mediated assembly and motility.
These findings demonstrated that fimA overexpression could facilitate cell adhesion and contribute to the aggregation effects of E. coli during BF formation. However, fimA deletion exerted a negative impact on BF formation. Hence, the fimA gene can serve as an important regulatory factor for BF formation in E. coli.
Batch fermentation of the recombinant and wild-type strains was investigated for the capabilities of BF formation. However, LT production was different from that in E. coli W1688-fimH* (Chen et al., 2019). As shown in Figure 4, the LT titer was elevated by 7% in the W1688-fimA* strain compared to the wild-type strain (11.3 g/L versus 10.5 g/L, respectively). In addition, the fermentation cycle was reduced from 36 h to 32 h. Moreover, the level of LT in E. coli W1688-ΔfimA was decreased compared to the wild-type strain (8.1 g/L versus 10.5 g/L, respectively) accompanied by the reduced glucose consumption at the first stage of fermentation. Different from the effect of fimH on LT production that some enzymes were involved in LT biosynthesis, the significant increase in production for the fimA gene had not been observed due to the subtle effect on LT synthesis and metabolism. Instead, the efficiency of BF-based immobilization could be improved, along with the shortened fermentation period of 4 h, thus leading to an increase in BF formation. All these observations indicated that the overexpression of fimA might influence the titer and productivity of LT in the E. coli W1688-fimA* strain.
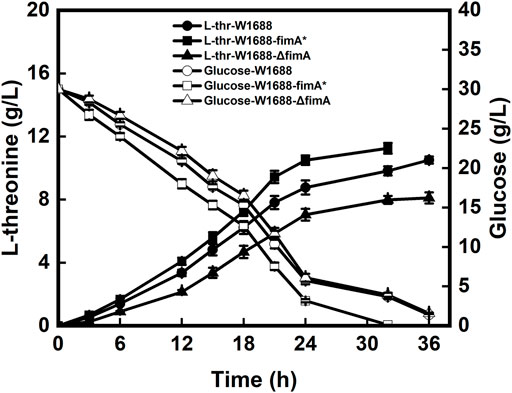
FIGURE 4. Levels of LT and glucose produced and consumed by the three strains during batch fermentation, respectively.
A BF-based immobilized fermentation strategy was applied to further improve the efficiency of fermentation. The polyurethane carrier was used to support BFs, which could impose a beneficial effect on cell aggregation. During the immobilized fermentation, the LT titer in the E. coli W1688-fimA* strain was almost similar to that in the wild-type strain (data not shown). As a result, BF on the carrier increased significantly, as revealed by SEM observation, and the efficiency of BF immobilization was improved during the continuous LT fermentation of the E. coli W1688-fimA* strain. However, the improvement of the LT titer and productivity was still negligible compared to that in the wild-type strain, indicating that the disturbance of fimA might be associated with the carbon flux distribution of LT.
To further confirm the carbon flow in the E. coli W1688-fimA* strain under the industrial fermentation conditions, dry cell weight (DCW) experiments were performed in the engineered strains. The data on OD570 for BF and DCW are shown in Figure 5. The overexpression of the fimA gene could lead to enhanced BF formation. In addition, more than 26% of DCW was related to fimA overexpression (1.18 g/L versus 1.49 g/L), which was consistent with the tendency of results in OD570 for BF formation. Moreover, fimA overexpression could lead to the accumulation of extracellular surface fimbriae and filamentous flagella (Figure 3B2).
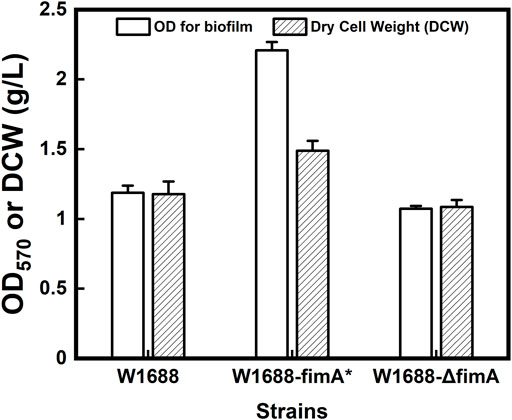
FIGURE 5. Quantitative analysis of crystal violet staining for BFs and dry cell weight in the three different strains.
To explore the mechanisms underlying improved BF formation, transcriptomic analysis was conducted on the recombinant and wild-type strains. The expression ratios of genes related to BF formation were significantly different among the three strains (Figure 6), which could be classified into several distinct clusters. The E. coli fimbrial gene cluster is proposed to be composed of a seven-gene operon encoding the structural and assembly components (fimAICDFGH), which shares the same promoter in its 5′ region. The fimA gene (encodes for the type Ⅰ fimbriae subunit) and other fim family genes guided the assembly of fimbriae to the cell surface (Reisner et al., 2003; Schembri et al., 2003) with high expression in the 1688-fimA* strain. FimI is a putative fimbrial protein and has been shown to encode a 16.4-kDa noncytoplasmic protein product. FimC and FimD are the chaperone and usher proteins used to assemble fimbriae on the surface of the cell, and a number of well-characterized fimbrial systems are assembled using the chaperone/usher pathway (Nuccio and Baumler, 2007). FimC (the “chaperone”) accelerates protein folding and delivers the subunits to the pore-forming protein FimD (“the usher”) in the outer membrane. Subsequently, the subunits are translocated and incorporated into the growing fimbriae (Costello et al., 2012).
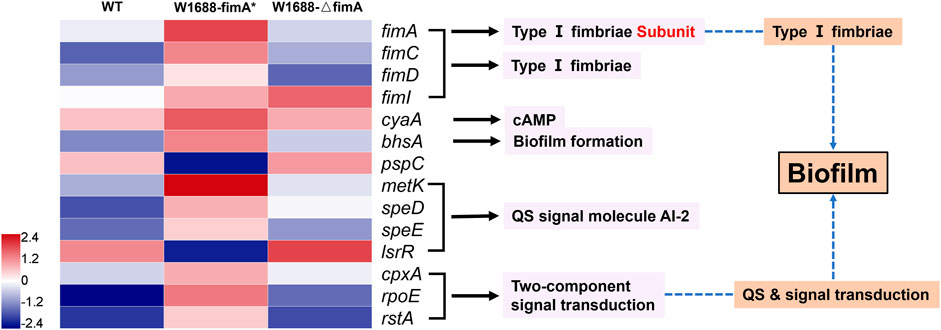
FIGURE 6. Transcriptomic analysis of genes responsible for the BF formation pathway in the three strains. Blue and red denote downregulated and upregulated genes, respectively.
In E. coli, the cAMP level is primarily regulated by the activity of adenylate cyclase encoded by cyaA (Notley-McRobb et al., 1997; Jeong et al., 2021). It catalyzes the production of cAMP from ATP and is a ubiquitous regulatory factor for gene expression and metabolic pathways (Gancedo, 2013; Bassler et al., 2018). BhsA is a stress resistance protein, which is directly related to BF formation under extreme conditions. metK, speD, and speE are associated with the synthesis of quorum-sensing signal molecule autoinducer-2 (AI-2), which in turn can promote BF formation (Vendeville et al., 2005). LsrR is a transcriptional repressor, and its downregulation can accelerate the transport and internalization of AI-2 for more BFs (Schauder et al., 2001; Barrios et al., 2006). In fact, metK, which catalyzes the reaction to AI-2, is commonly expressed in prokaryotic bacteria. cpxA, rpoE, and rstA belong to a two-component signal transduction module including signal secretion, reception of the quorum-sensing signal molecule, phosphorylation, and dephosphorylation. They are involved in histidine kinase-mediated cell adhesion, transcriptional control, and gene regulation.
All these genes were modulated to various degrees in the fimA-overexpressed and fimA-knockout strains compared to the wild-type strain. Our findings indicated that fimA overexpression could regulate many other genes to improve BF formation in the fimA* strain.
The overexpression of fimA led to the over-assembly of type I fimbriae subunit protein, which was beneficial for cell aggregation. However, BF-based immobilized fermentation was not applicable for increasing LT production. It was preliminarily inferred that fimA overexpression could disturb the carbon flow of LT. Hence, transcriptomic analysis was carried out to understand whether the LT titer could regulate biosynthetic pathway genes and clarify the potential mechanisms of carbon distribution in the LT pathway. As expected, fimA overexpression not only led to the formation of BFs but also regulated the genes in the LT biosynthetic pathway (Figures 7A,B). For instance, asd, metL, lysC, and thrA that convert L-aspartate into L-homoserine, as well as thrB and thrC that convert L-homoserine into LT (Lee et al., 2003), were all upregulated in E. coli W1688-fimA*. In spite of this, the LT transporter-encoding gene rhtA was downregulated, thereby decelerating the production of LT (Livshits et al., 2003).
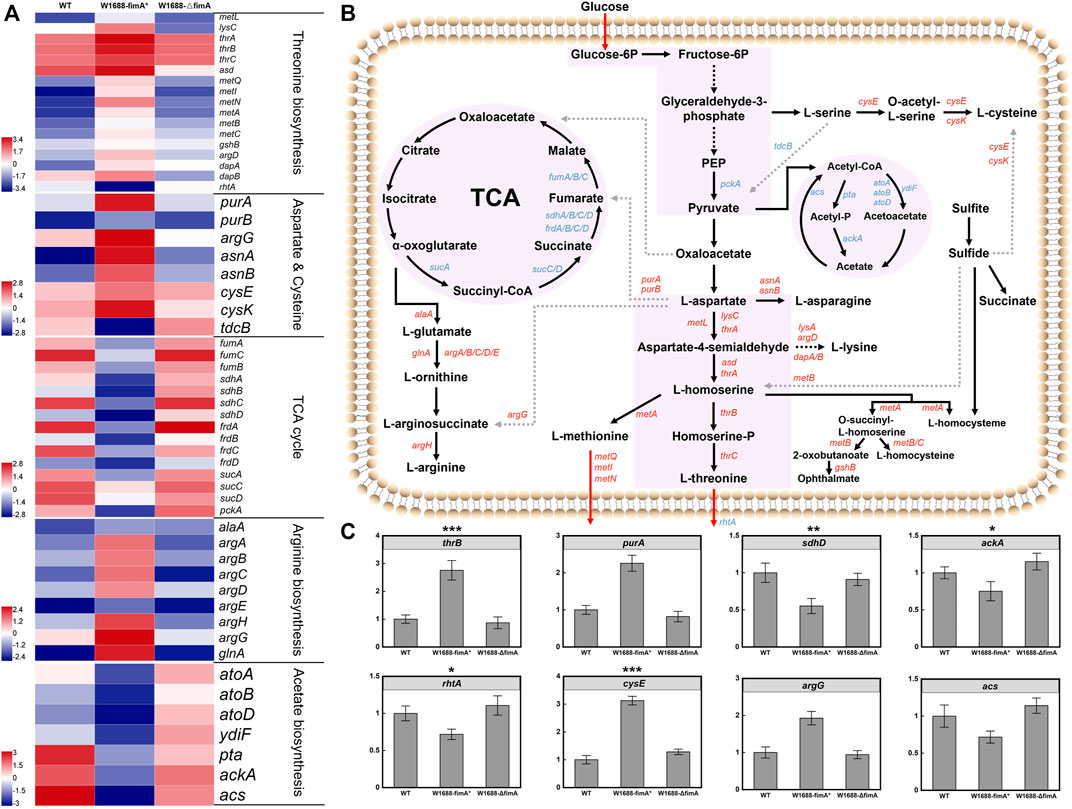
FIGURE 7. Transcriptomic profiles of LT biosynthetic pathways in three different strains. (A) Transcriptomic analysis of the three different strains. Blue and red denote downregulated and upregulated genes, respectively. From the top to the bottom panels: genes in the biosynthesis of LT, L-aspartate, L-cysteine, L-arginine, and acetate biosynthesis and the TCA cycle, respectively; (B) LT biosynthetic pathways in E. coli. Blue and red arrows indicate downregulated and upregulated genes, respectively. (C) RT-qPCR validation of the genes associated with LT biosynthesis and transportation. Mean ± SD (n = 3). Two-tailed t-test. ***p < 0.001, **p < 0.01, and *p < 0.05.
Before the formation of pyruvate, the expression levels of the L-serine and L-cysteine branches were upregulated, which could partly shunt the accumulation of target products. With regard to the L-aspartate branch, purA/B, argG, and asnA/B shared the central carbon metabolism flux of LT, resulting in the accumulation of fumarate, L-arginine, and L-asparagine. Meanwhile, benefiting from the upregulation of met family genes, AI-2 biosynthesis was increased, which led to the enhancement of BF formation and L-methionine. The genes of the whole branch from α-oxoglutarate to L-arginine were also upregulated, and they further shunt the carbon flux. In addition, more metabolic precursors of central carbon metabolism could divert to the by-path. Although previous findings have demonstrated that the improved BF formation facilitated the fermentation efficiency of LT, other by-paths also excessively occupied the central carbon flux due to the overexpression of fimA.
Indeed, purA/B encoding proteins to synthesize target chemical products of the TCA cycle often bring a competitive effect on the carbon flux. However, the weakened TCA cycle may reduce the deoxidization and energy supply to various other biochemical pathways such as NADH and ATP, thus leading to a reduction in the LT titer. Generally, the higher specific growth rates would lead to better production yields. However, it is inevitable that aerobic E. coli cultivation will generate acetate, which is caused by an imbalance between the glucose uptake and TCA cycle capacity, resulting in pyruvate and acetyl-CoA accumulation (Lee et al., 2006; Nahku et al., 2010). Transcriptomic analysis showed that the expression levels of ackA, pta, and acs which were about acetate production were noticeably downregulated. Actually, acetate accumulation with the dramatic decrease in pH could reduce the maximum growth rate and inhibit protein production. Meanwhile, it diverts carbon from biomass formation and exhibits harmful effects on BF and cells such as the inhibition of the enzyme activity, changes in the cell membrane permeability and osmotic pressure, and disaggregation of the biofilm. The expression levels of genes in the acetate pathway were downregulated, which might facilitate LT production. Thus, it was confirmed that the glucose consumed by E. coli W1688-fimA* in free fermentation was faster than the other strains. The genes in the branched metabolic pathway were differentially expressed, and the genes associated with central carbon metabolism were remarkably upregulated in E. coli W1688-fimA* (Figure 7C). Taken altogether, these findings demonstrated that fimA overexpression improved the metabolic flux of LT but could also affect other carbon distribution mechanisms.
In this work, the immobilized fermentation system was proposed for LT production in E. coli, together with the benefits of BF formation. The fimA-overexpressing strain could promote BF formation under industrial fermentation conditions but is not suitable for continuous immobilized fermentation. The LT titer was only elevated from 10.5 g/L to 11.3 g/L in E. coli W1688-fimA*. Transcriptomic profiling indicated that BF formation was improved by modulation of BF-related genes, while LT biosynthesis was affected by genes in the carbon metabolism pathway. In conclusion, the type I fimbriae subunit fimA regulated E. coli BF formation but affected LT carbon distribution. This study will stimulate thoughts about how the type 1 fimbriae gene regulated cell adhesion and excretion of amino acids and bring some consideration and provide a reference for the development of BF-based fermentation in E. coli.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The reads and the HiSeq transcriptomic reads generated for E. coli W1688, E. coli W1688-fimA* and E. coli W1688-ΔfimA, respectively, have been submitted to the BioProject database of National Center for Biotechnology Information (NCBI) under accession number SRR8335002, SRR8335003 and SRR8335000, respectively.
QL and JZ conceived of the study, designed the experiments, performed the laboratory work, analyzed and interpreted the data, and drafted the manuscript. NL constructed the plasmids and strains, participated in the fermentation experiments, performed electron microscopy, analyzed the metabolic products, and performed the statistical analysis. WS, HN, and DL performed the transcriptomic analysis of different strains and revised the manuscript. BY supplied the carriers. PO, HY, and YC critically revised the manuscript. GZ and TC contributed to experimental design and data interpretation and also critically revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Key R&D Program of China (Grant No. 2021YFC2101100), the National Natural Science Foundation of China (22178176), the National Key Research and Development Program of China (Grant No. 2018YFB1501705), the key program of the National Natural Science Foundation of China (Grant No. 21636003), the Outstanding Youth Foundation of China (Grant No. SBK2017010373), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R28), the Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Key Research and Development Program of Nanjing Jiangbei New Area (ZDYF20200220).
The authors thank Prof. Sheng Yang for providing strain E. coli W1688.
The author QL is employed by Nanjing Hi-Tech Biological Technology Research Institute Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Barrios, A. F. G., Zuo, R., Hashimoto, Y., Yang, L., Bentley, W. E., and Wood, T. K. (2006). Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188, 305–316. doi:10.1128/JB.188.1.305-316.2006
Bassler, J., Schultz, J. E., and Lupas, A. N. (2018). Adenylate cyclases: Receivers, transducers, and generators of signals. Cell. Signal. 46, 135–144. doi:10.1016/j.cellsig.2018.03.002
Beloin, C., Valle, J., Latour‐Lambert, P., Faure, P., Kzreminski, M., Balestrino, D., et al. (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51, 659–674. doi:10.1046/j.1365-2958.2003.03865.x
Berne, C., Ducret, A., Hardy, G. G., and Brun, Y. V. (2015). Adhesins involved in attachment to abiotic surfaces by Gram‐negative bacteria. Microb. Biofilms, 163–199. doi:10.1128/9781555817466.ch9
Chen, T., Liu, N., Ren, P., Xi, X., Yang, L., Sun, W., et al. (2019). Efficient biofilm-based fermentation strategies for L-threonine production by Escherichia coli. Front. Microbiol. 10, 1773. doi:10.3389/fmicb.2019.01773
Chen, Y., Liu, Q., Zhou, T., Li, B., Yao, S., Li, A., et al. (2013). Ethanol production by repeated batch and continuous fermentations by Saccharomyces cerevisiae immobilized in a fibrous bed bioreactor. J. Microbiol. Biotechnol. 23 (4), 511–517. doi:10.4014/jmb.1209.09066
Costello, C. M., Yeung, C. L., Rawson, F. J., and Mendes, P. M. (2012). Application of nanotechnology to control bacterial adhesion and patterning on material surfaces. J. Exp. Nanosci. 7 (6), 634–651. doi:10.1080/17458080.2012.740640
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97 (12), 6640–6645. doi:10.1073/pnas.120163297
Dong, X., Quinn, P. J., and Wang, X. (2011). Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of L-threonine. Biotechnol. Adv. 29, 11–23. doi:10.1016/j.biotechadv.2010.07.009
Gancedo, J. M. (2013). Biological roles of cAMP: Variations on a theme in the different kingdoms of life. Biol. Rev. 88 (3), 645–668. doi:10.1111/brv.12020
Hung, D. L., and Hultgren, S. J. (1998). Pilus biogenesis via the chaperone/usher pathway: An integration of structure and function. J. Struct. Biol. X. 124, 201–220. doi:10.1006/jsbi.1998.4049
Jeong, S.-H., Park, J.-B., Wang, Y., Kim, G.-H., Zhang, G., Wei, G., et al. (2021). Regulatory molecule cAMP changes cell fitness of the engineered Escherichia coli for terpenoids production. Metab. Eng. 65, 178–184. doi:10.1016/j.ymben.2020.11.009
Lane, M. C., Simms, A. N., and Mobley, H. L. (2007). Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J. Bacteriol. 189 (15), 5523–5533. doi:10.1128/JB.00434-07
Lee, J.-H., Lee, D.-E., Lee, B.-U., and Kim, H.-S. (2003). Global analyses of transcriptomes and proteomes of a parent strain and an L-threonine-overproducing mutant strain. J. Bacteriol. 185, 5442–5451. doi:10.1128/JB.185.18.5442-5451.2003
Lee, M.-H., Lee, H.-W., Park, J.-H., Ahn, J.-O., Jung, J.-K., and Hwang, Y.-I. (2006). Improved L-threonine production of Escherichia coli mutant by optimization of culture conditions. J. Biosci. Bioeng. 101, 127–130. doi:10.1263/jbb.101.127
Leuchtenberger, W., Huthmacher, K., and Drauz, K. (2005). Biotechnological production of amino acids and derivatives: Current status and prospects. Appl. Microbiol. Biotechnol. 69, 1–8. doi:10.1007/s00253-005-0155-y
Li, X., Yan, Z., and Xu, J. (2003). Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 149, 353–362. doi:10.1099/mic.0.25932-0
Liu, D., Chen, Y., Li, A., Ding, F., Zhou, T., He, Y., et al. (2013). Enhanced butanol production by modulation of electron flow in Clostridium acetobutylicum B3 immobilized by surface adsorption. Bioresour. Technol. 129, 321–328. doi:10.1016/j.biortech.2012.11.090
Livshits, V. A., Zakataeva, N. P., Aleshin, V. V., and Vitushkina, M. V. (2003). Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 154, 123–135. doi:10.1016/S0923-2508(03)00036-6
Madyagol, M., Al-Alami, H., Levarski, Z., Drahovská, H., Turňa, J., and Stuchlík, S. (2011). Gene replacement techniques for Escherichia coli genome modification. Folia Microbiol. (Praha). 56, 253–263. doi:10.1007/s12223-011-0035-z
Nahku, R., Valgepea, K., Lahtvee, P.-J., Erm, S., Abner, K., Adamberg, K., et al. (2010). Specific growth rate dependent transcriptome profiling of Escherichia coli K12 MG1655 in accelerostat cultures. J. Biotechnol. 145, 60–65. doi:10.1016/j.jbiotec.2009.10.007
Notley-McRobb, L., Death, A., and Ferenci, T. (1997). The relationship between external glucose concentration and cAMP levels inside Escherichia coli: Implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology 143 (6), 1909–1918. doi:10.1099/00221287-143-6-1909
Nuccio, S.-P., and Baumler, A. J. (2007). Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71 (4), 551–575. doi:10.1128/MMBR.00014-07
Rajkumar, R., Yaakob, Z., and Takriff, M. S. (2013). Potential of micro and macro algae for biofuel production: A brief review. Bioresources 9, 1606–1633. doi:10.15376/biores.9.1.1606-1633
Reisner, A., Haagensen, J. A., Schembri, M. A., Zechner, E. L., and Molin, S. (2003). Development and maturation of Escherichia coli K‐12 biofilms. Mol. Microbiol. 48, 933–946. doi:10.1046/j.1365-2958.2003.03490.x
Sauer, K. (2003). The genomics and proteomics of biofilm formation. Genome Biol. 4, 219. doi:10.1186/gb-2003-4-6-219
Schauder, S., Shokat, K., Surette, M. G., and Bassler, B. L. (2001). The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum‐sensing signal molecule. Mol. Microbiol. 41, 463–476. doi:10.1046/j.1365-2958.2001.02532.x
Schembri, M. A., Kjærgaard, K., and Klemm, P. (2003). Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48, 253–267. doi:10.1046/j.1365-2958.2003.03432.x
Tripathi, P., Beaussart, A., Alsteens, D., Dupres, V., Claes, I., Von Ossowski, I., et al. (2013). Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano 7, 3685–3697. doi:10.1021/nn400705u
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., and Hardie, K. R. (2005). Making'sense'of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. doi:10.1038/nrmicro1146
Yu, B., Zhang, X., Sun, W., Xi, X., Zhao, N., Huang, Z., et al. (2018). Continuous citric acid production in repeated-fed batch fermentation by Aspergillus niger immobilized on a new porous foam. J. Biotechnol. 276, 1–9. doi:10.1016/j.jbiotec.2018.03.015
Keywords: Escherichia coli, fimA gene, biofilm, L-threonine carbon distribution, transcriptomic analysis
Citation: Liu Q, Zhu J, Liu N, Sun W, Yu B, Niu H, Liu D, Ouyang P, Ying H, Chen Y, Zhao G and Chen T (2022) Type I fimbriae subunit fimA enhances Escherichia coli biofilm formation but affects L-threonine carbon distribution. Front. Bioeng. Biotechnol. 10:904636. doi: 10.3389/fbioe.2022.904636
Received: 14 June 2022; Accepted: 03 October 2022;
Published: 21 October 2022.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Kuang-Sheng Yeh, National Taiwan University, TaiwanCopyright © 2022 Liu, Zhu, Liu, Sun, Yu, Niu, Liu, Ouyang, Ying, Chen, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianpeng Chen, Y2hlbnRpYW5wZW5nQG5qdGVjaC5lZHUuY24=; Gulin Zhao, emhhb2d1bGluQG5qdGVjaC5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.