- 1College of Life Sciences, Key Laboratory for Cell and Gene Engineering of Zhejiang Province, Zhejiang University, Hangzhou, China
- 2National Engineering Laboratory for Cereal Fermentation Technology, Jiangnan University, Wuxi, China
- 3Zhejiang Esigma Biotechnology Company Limited, Haining, China
- 4Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Anthocyanins are natural pigments found in various plants. As multifunctional natural compounds, anthocyanins are widely used in food, pharmaceuticals, health products, and cosmetics. At present, the anthocyanins are heterologously biosynthesized in prokaryotes from flavan-3-ols, which is rather expensive. This study aimed to metabolically engineer Saccharomyces cerevisiae for anthocyanin production. Anthocyanin production has been extensively studied to understand the metabolic pathway enzymes in their natural hosts, including CHS (chalcone synthase); FLS (flavonol synthase); CHI (chalcone isomerase); F3H (flavanone 3-hydroxylase); F3′H (flavonoid 3′-hydroxylase); F3′5′H (flavonoid 3′,5′-hydroxylase); DFR (dihydroflavonol 4-reductase); ANS (anthocyanidin synthase); LAR (leucoanthocyanidin reductase); and UFGT (flavonoid 3-O-glucosyltransferase). The anthocyanin transporter MdGSTF6 was first introduced and proven to be indispensable for the biosynthesis of anthocyanins. By expressing MdGSTF6, FaDFR, PhANS0, and Dc3GT and disrupting EXG1 (the main anthocyanin-degrading enzyme), the BA-22 strain produced 261.6 mg/L (254.5 mg/L cyanidin-3-O-glucoside and 7.1 mg/L delphinidin-3-O-glucoside) anthocyanins from 2.0 g/L dihydroflavonols, which was known to be the highest titer in eukaryotes. Finally, 15.1 mg/L anthocyanins was obtained from glucose by expressing the de novo biosynthesis pathway in S. cerevisiae, which is known to be the highest de novo production. It is the first study to show that through the introduction of a plant anthocyanin transporter and knockout of a yeast endogenous anthocyanin degrading enzyme, the anthocyanin titer has been increased by more than 100 times.
Introduction
Anthocyanins are water-soluble pigments belonging to the class of flavonoids existing in a plethora of plants, which endow nature with rich colors (Khoo et al., 2017). They are mainly stored in the vacuoles of plant cells (Liu et al., 2018). More than 700 anthocyanins have been verified (Salehi et al., 2020), among which pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin are the most common types (Khoo et al., 2017). They play important roles in plant growth by protecting against pathogens and reducing the harmful effects of ultraviolet radiation (Jiang et al., 2019). A large number of studies have demonstrated that anthocyanins have multiple physiological activities within the human body, such as improvement in vision (Nomi et al., 2019), prevention of brain disorders (Henriques et al., 2020), intervention against obesity (Yamei et al., 2020), and treatment of hyperglycemia and hyperuricemia (Yang et al., 2020). As natural multifunctional additives, anthocyanins are approved as food colorants by many countries (Belwal et al., 2020). Given that the market demand for anthocyanins is growing, more diverse strategies for improving the efficiencies of anthocyanin production are yet to be explored. However, issues such as the “browning effect,” (Oszmianski and Lee, 1990), unstable supply, and low efficiency remain to be resolved.
Multiple genes are involved in the biosynthesis of anthocyanins (Ichino et al., 2014). In previous studies, synthesis commenced from naringenin (NAR) and proceeded through a series of hydroxylation reactions catalyzed by flavonoid 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), and flavonoid 3′5′-hydroxylase (F3′5′H) (Figure 1). These dihydroflavonols were reduced to unstable intermediates; leucoanthocyanidins by dihydroflavonol 4-reductase (DFR) (Belwal et al., 2020; Saigo et al., 2020). These were converted into anthocyanidins in the form of aglycone by anthocyanidin synthase (ANS) and then subsequently converted into anthocyanin in the form of glycoside by UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT/3GT). The biosynthesis of anthocyanin in plants has been extensively studied (Passeri et al., 2016) and is strictly regulated by the MBW (MYB-bHLH-WDR) protein complex in plant cells (Xu et al., 2015). After synthesis in the cytoplasm, the anthocyanins are known to be transported into the vacuole and restored (Chanoca et al., 2016; Olejnik et al., 2016; Jiang et al., 2019).
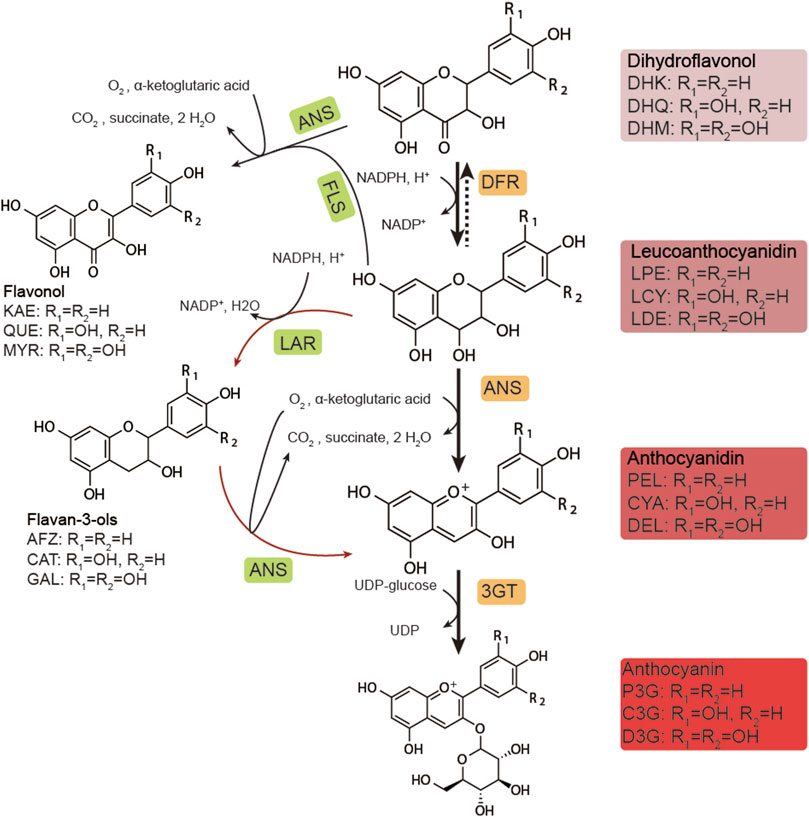
FIGURE 1. Biosynthetic pathway of anthocyanins. DHK, dihydrokaempferol; DHQ, dihydroquercetin; DHM, dihydromyricetin; LPE, leucopelargonidin; LCY, leucocyanidin; LDE: leucodelphinidin; PEL, pelargonidin; CYA, cyanidin; DEL, delphinidin; P3G, pelargonidin-3-O-glucoside; C3G, cyanidin-3-O-glucoside; D3G, delphinidin-3-O-glucoside; KAE, kaempferol; QUE, quercetin; MYR, myricetin; AFZ, afzelechin; CAT, catechin; GAL, gallocatechin; DFR, dihydroflavonol-4-reductase; ANS, anthocyanidin synthase; 3GT, UDP-glucose flavonoid 3-O-glucosyltransferase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase.
Like most high-value natural compounds, the acquisition of anthocyanins still relies significantly on extractions from plants. The slow growth of plants and their dependence on arable land, as well as the long-winded, destructive extraction methods, makes high-value chemicals so expensive (Belwal et al., 2020; Marchev et al., 2020). Using microbes is an alternative and benign approach for anthocyanin production (Yan et al., 2005). The heterologous biosynthesis of anthocyanins was first reported in Escherichia coli, and titers of up to 439 mg/L of cyanidin-3-O-glucoside (C3G) from catechin have been obtained in this microbial host (Yan et al., 2008; Lim et al., 2015; Shrestha et al., 2019). Alternative prokaryotic hosts such as Corynebacterium glutamicum (Zha et al., 2018) and Lactococcus lactis (Solopova et al., 2019) have also proven themselves as capable hosts for anthocyanin production, though in these particular cases, flavan-3-ols (afzelechin and catechin) were used as the initial starting point, which was rather expensive. However, inefficient conversion of dihydroflavonols to anthocyanins remains unresolved. In this work, Saccharomyces cerevisiae was engineered to biosynthesize anthocyanins by introducing an anthocyanin transporter, screening of optimal genes, and identifying as well as knock out of endogenous anthocyanin endogenous degrading enzymes. Finally, 261.6 mg/L and 15.1 mg/L anthocyanins were obtained from dihydroflavonols and glucose, respectively.
Materials and methods
Genes, plasmids, strains, and chemicals
All the genes, plasmids, and strains used in this study are listed in Supplementary Tables S1, S2 and Table 1. The genes were synthesized by Sangon (Shanghai, China), which were optimized for S. cerevisiae expression, except for GhANS0, PhANS0, and At3GT0. The pY26-TEF-GPD plasmid (Li et al., 2008) was used for gene expression, and the pRS426-PTEF1-Cas9-PSNR52-sgRNA plasmid was used for genome editing of S. cerevisiae. E. coli JM109 was used for plasmid propagation.
Cyanidin, cyanidin-3-O-glucoside, delphinidin, and delphinidin-3-O-glucoside were purchased from Herbest (Baoji, China). Catechin, protocatechuic acid, and phloroglucinol aldehyde were purchased from Yuanye (Shanghai, China). The BM-31 strain was used for dihydroflavonol production in a 5-L bioreactor following the protocol of culture conditions described in Section 2.5. The substrate used in this study was obtained through methanol extraction and concentration using an RE100-Pro rotary evaporator (DLAB Scientific, Beijing, China). The substrate contained 69.3% of dihydroquercetin, 24.0% of dihydromyricetin, and 6.7% of other intermediate products.
Genetic procedures
All the DNA fragments were amplified using 2 × Phanta Max Master Mix (Dye Plus) (Vazyme, Nanjing, China). A ready-to-use seamless cloning kit (Sangon, Shanghai, China) was used for plasmid construction; the length of the homologous arm was not less than 25 bp between the fragments. All the recombinant plasmids were determined by DNA sequencing. The knockout and integration of genes were performed using CRISPR-Cas9 systems (Dicarlo et al., 2013). The CRISPRdirect tool (http://crispr.dbcls.jp) was used for selecting a 20-nt guide sequence of the single-guide RNA (sgRNA). All the 20-nt guide sequences used in this study are listed in Supplementary Table S3. The gene deletion and expression cassettes (donor DNA) for genome editing were obtained by overlap extension polymerase chain reaction, in which the homologous arms were amplified from the genome and were not less than 300 bp. A Frozen-EZ Yeast Transformation II Kit (Zymo Research, CA, United States) was used for the transformation of S. cerevisiae following the manufacturer’s protocol.
Ammonium sulfate precipitation
The strain CENPK2-1D was grown on the YPD medium (10 g/L yeast extract, 20 g/L tryptone, and 20 g/L glucose) plate for 2–3 days at 30°C. Five to seven colonies were inoculated into a 250-ml shaking flask containing 20 ml of the YPD medium for 16 h and cultivated at 220 rpm and 30°C. Four 2-L shaking flasks containing 500 ml of YPD medium were inoculated with 5 ml of preculture for 24-h cultivation at 220 rpm and 30°C. Then, the culture was centrifuged at 4°C and 4000 g for 10 min using an Avanti J-26S XP Centrifuge (Beckman Coulter, CA, United States). Approximately 2 L of the supernatant was collected in a 4-L centrifuge bucket and placed on a magnetic stirrer in an ice bath. Then, 328 g (NH4)2SO4 was added slowly to the supernatant with stirring, which made the concentration of ammonium sulfate in the solution reach 30% (g/g), followed by centrifugation at 4°C and 4,000 g for 20 min. The supernatant was collected into a centrifuge bucket again; 10 ml 20 mmol/L PB (phosphate buffer, pH = 5.0, 4°C) was used to dissolve the protein precipitate, and 0–30% protein component was obtained. Then, 234 g (NH4)2SO4 was slowly added to the supernatant mentioned above with stirring, which made the concentration of ammonium sulfate in the solution reach 50% (g/g), followed by centrifugation at 4°C and 4,000 g for 20 min; the supernatant was collected into the centrifuge bucket again, 10 ml of 20 mmol/L PB was used to dissolve the protein precipitate, and 3–50% protein component was obtained. Finally, 696 g (NH4)2SO4 was added slowly to the supernatant with stirring, which made the concentration of ammonium sulfate in the solution reach 100% (g/g), followed by centrifugation at 4°C and 4,000 g for 20 min, the supernatant was removed, and 10 ml of 20 mmol/L PB was used to dissolve the protein precipitate, and 50–100% of the protein component was obtained.
Identification of anthocyanin-degrading enzymes
The protein component was filtered through a 0.45-μm filter membrane before purification. ÄKTA Avant 25 (GE Healthcare, WA, United States) equipped with HiLoad 26/600 Superdex 200 pg (GE Healthcare) was used for size-exclusion chromatography. The protein component was loaded at the rate of 5 ml/min, and 20 mm Tris, pH 5.0 was used for elution at the rate of 3 ml/min. The eluates corresponding to different signal peaks were collected. Amicon Ultra Centrifugal Filters of 10k MWCO (Merck, NJ, United States) were used for the protein concentration at 3000 g and 4°C until the volume was concentrated to 200 μl. SDS-PAGE was used for separating the protein concentrates using a NuPAGE 10% Bis-Tris Gel (Thermo Fisher Scientific, MA, United States). The protein bands were excised from the gel. The protein profiles provided by Sangon (Shanghai, China) were used for identifying the protein bands.
Preparation of dihydroflavonols
Dihydroflavonols were produced from glucose by the strain of BM-31 (Li G. et al., 2020) through the fed-batch fermentation as described below with a modification that supplemented 10 g/L CaCO3 in the medium. The fermentation broth was placed in a 4-L centrifuge bucket and then centrifuged for 15 min at 4°C and 1,500 g using an Avanti J-26S XP (Beckman Coulter, CA, United States). The supernatant and cell pellet were treated separately: the water from the supernatant was removed by using a rotary evaporator, an equal volume of ethanol was added to the solid, and suspension A was obtained; equal volume of ethanol was added to the cell pellet, and suspension B was obtained. Suspensions A and B were mixed, followed by magnetic stirring for 24 h to extract flavonoids, then placed in a Buchner funnel for suction filtration, and washed with an appropriate amount of ethanol. The filtrate was evaporated to dryness using a rotary evaporator; finally, a small amount of ethanol was used to dissolve precipitated flavonoids. The components of dihydroflavonols in the extract were 95%, of which DHQ (dihydroquercetin) and DHM (dihydromyricetin) account for 72 and 23%, respectively, as per the analysis by HPLC.
Culture conditions
All the recombinant yeasts were grown on the auxotrophic SC medium (1.7 g/L yeast nitrogen base, 5.0 g/L ammonium sulfate, 20 g/L glucose, and appropriate amino acids: 50 mg/L leucine, 50 mg/L histidine, 50 mg/L tryptophan, and 50 mg/L uracil) plate for 2–4 days at 30°C. A single colony was inoculated into a 12-ml sterile culture tube containing 2 ml of SC medium for 24-h growth at 220 rpm and 30°C. Then, 0.5 ml of preculture was inoculated into a 250-ml shake flask containing 50 ml of SC medium at 220 rpm and 30°C. After 24-h growth, the culture was centrifuged at 4°C and 3000 g for 5 min using an Allegra X-15R Centrifuge (Beckman Coulter, CA, United States). Then, the strains were resuspended in 10 ml of YPD medium supplemented with 500 mg/L dihydroflavonols for 24-h cultivation.
The fed-batch fermentation was performed, and dissolved oxygen was maintained at 10% (degree of saturation) with the agitation ranging from 250 to 600 rpm. A 5-L bioreactor (T&J Bioengineering, Shanghai, China) contained 2.5 L of YPD medium for de novo biosynthesis of anthocyanins. Medium A (400 g/L glucose, 18 g/L KH2PO4, 15 g/L yeast extract, 10.24 g/L MgSO47H2O, 7 g/L K2SO4, 0.56 g/L Na2SO4, 20 ml/L trace metal solution, and 24 ml/L vitamin solution) was as described in the previous study (Li G. et al., 2020); it fed for 15 h at the rate of 5 ml/h and stopped at 60 h. After 24 h, 0.05 mmol/L α-ketoglutaric acid, 0.5 mmol/L ascorbic acid, and 0.5 mmol/L FeSO4 were added individually to the bioreactor.
Analytical methods
Synergy H1 (BioTek, VT, United States) was used for measuring OD600. Equal volumes of fermentation broth and acidified methanol (1% HCL, v/v) were mixed for cell disruption using FastPrep-24 5G (MP Biomedicals, Shanghai, China). Then, the mixed liquid was centrifuged and filtered using a 0.22-μm filter membrane. A high-performance liquid chromatography system (Shimadzu Corporation, Kyoto, Japan) equipped with a diode array detector was used for flavonoid analysis (Li G. et al., 2020). Cyanidin, cyanidin-3-O-glucoside, delphinidin, delphinidin-3-O-glucoside, and catechin were detected at 527 nm, 517 nm, 532 nm, 524 nm, and 278 nm, respectively. Dihydroquercetin, dihydromyricetin, protocatechuic acid, and phloroglucinol aldehyde were detected at 290 nm.
Results
Introduction of an anthocyanin vacuolar transporter
As a eukaryote, S. cerevisiae has been proven to be a suitable host for the biosynthesis of many natural products, such as artemisinic acid (Paddon et al., 2013). It has organelles similar to plant cells, especially the vacuole that is also the storehouse in plant cells. Some transporters of anthocyanin from plant sources were reported to promote vacuolar absorption of anthocyanins when expressed in S. cerevisiae (Zhao and Dixon, 2009). Therefore, three transporters, AtTT12 (Zhao and Dixon, 2009), AtGFS9 (Ichino et al., 2014), and MdGSTF6 (Jiang et al., 2019), were synthesized and co-expressed with FaDFR, MdANS, and At3GT (Supplementary Table S1), respectively. Supplemented with 500 mg/L dihydroflavonols for 24-h cultivation, the results are shown in Figure 2A When FaDFR, MdANS, and At3GT were expressed in S. cerevisiae, only 1.8 mg/L anthocyanins (1.6 mg/L C3G and 0.2 mg/L D3G) were obtained. However, 22.1 mg/L anthocyanins (19.6 mg/L C3G and 2.5 mg/L D3G) were obtained when MdGSTF6 was co-expressed in the aforementioned strain, which was greater than that of AtTT12 and AtGFS9. Meanwhile, ANS belonging to the 2-oxoglutarate-dependent dioxygenase (2ODD), has properties of another 2ODD flavanol synthase (FLS) that catalyzes the conversion of dihydroflavonols into flavonols (Wilmouth et al., 2002); then, it can be glycosylated by 3GT (Lyu et al., 2020). Therefore, quercetin-3-O-glucoside (Q3G) and myricetin-3-O-glucoside (M3G) were detected in this study. The introduction of MdGSTF6, on the contrary, significantly reduced the occurrence of side reactions; the contents of flavonol glycosides were decreased to 4.9 mg/L (4.1 mg/L Q3G and 0.8 mg/L M3G).
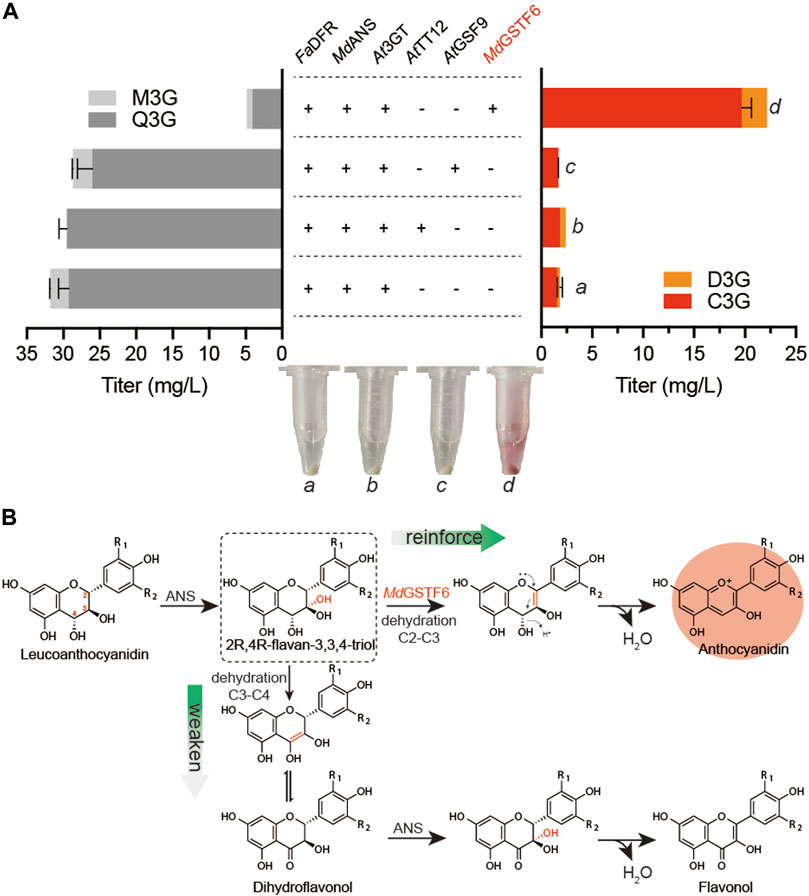
FIGURE 2. Metabolic engineering strategies for anthocyanin production (A) AtTT12, AtGSF9, and MdGSTF6 promoted by PINO1 co-expressed with FaDFR and MdANS promoted by PTDH1 and PGAL10 in the BA-01 strain with the pY26-TEF-GPD. The “a,” “b,” “c,” and “d” are the fermentation broth (equal volume of acidified methanol was added) of different groups (B) Possible mechanism of MdGSTF6 for anthocyanin production in S. cerevisiae.
Screening of high-efficiency anthocyanidin synthase and UDP-glucose flavonoid 3-O-glucosyltransferase
In the anthocyanin biosynthetic process, the unstable aglycone form (anthocyanidin) can be converted into a stable glycoside form (anthocyanin) by UDP-glucose flavonoid 3-O-glucosyltransferase (3GT). Another three 3GTs, namely, At3GT0, Fa3GT, and Dc3GT were integrated into the GAL80 site of CENPK2-1D, resulting in BA-02, BA-03, and BA-04 (Table 1). The pY26-PTDH1-FaDFR-TGAA1-PGAL10-MdANS-TALT1-PINO1-MdGSTF6-TCYC1 plasmid was transformed into the strains of BA-01, BA-02, BA-03, and BA-04. After 24-h cultivation, the Dc3GT was optimal and 27.4 mg/L anthocyanins (25.1 mg/L C3G and 2.3 mg/L D3G) were obtained when supplemented with 500 mg/L dihydroflavonols (Figure 3A). Anthocyanidin synthase (ANS) is a vital enzyme for biosynthesis of anthocyanin. Other 11 ANS, namely, DpANS, GhANS, GhANS0, LrANS, TsANS, VvANS, BoANS, CsANS, GmANS, MiANS, and PhANS0 were tested in the strain BA-04. The result (Figure 3B) showed PhANS0 had the highest activity and 38.2 mg/L anthocyanins (35.8 mg/L C3G and 2.4 mg/L D3G) were obtained. After “were obtained.”, please add a sentence “Two 3GT (At3GT0, Dc3GT) and three ANS (PhANS0, LrANS, GhANS0) were paired into six groups that Dc3GT and PhANS0 were found to be the best catalytic combination (Figure 3C), when supplemented with 2 g/L dihydroflavonol, 83.7 mg/L (81.4 mg/L C3G and 2.3 mg/L D3G) anthocyanins were obtained at 48-h fermentation (Figure 3D).” It is worth noting that the catalytic ability of At3GT and At3GT0, GhANS, and GhANS0 was higher than that of their codon-optimized counterparts. These results indicate that S. cerevisiae can be used as a host for the expression of enzymes from plant sources.
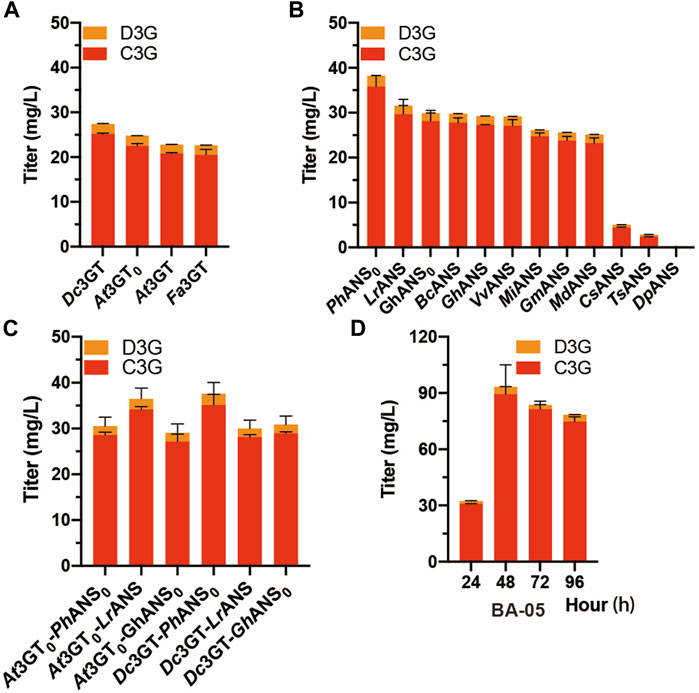
FIGURE 3. Screening of ANS and 3 GT (A) At3GT, At3GT0, Fa3GT, and Dc3GT represent BA-01, BA-02, BA-03, and BA-04, respectively, harboring the pY26-PTDH1-FaDFR-TGAA1-PGAL10-MdANS-TALT1-PINO1-MdGSTF6- TCYC1 plasmid for 24-h cultivation supplemented with 500 mg/L dihydroflavonols (B) BA-04 harboring 12 different pY26-PTDH1-FaDFR-TGAA1-PGAL10-ANSs-TALT1-PINO1-MdGSTF6-TCYC1 plasmids (ANSs indicate 12 different types of ANS) for 24-h cultivation supplemented with 500 mg/L dihydroflavonols (C) Two strains BA-02 and BA-04 and three plasmids pY26-PTDH1-FaDFR-TGAA1-PGAL10-ANSs-TALT1-PINO1-MdGSTF6-TCYC1 plasmid (ANSs indicate PhANS0, LrANS, and GhANS0) composed of six combinations for 24-h cultivation supplemented with 500 mg/L dihydroflavonols (D) 96-h fermentation of the BA-05 strain; 500 mg/L dihydroflavonols were added after 0 h, 1,000 mg/L dihydroflavonols were added after 24 h, and 500 mg/L dihydroflavonols were added after 48 h.
Identification of endogenous anthocyanin-degrading enzymes
To further investigate the phenomenon of anthocyanin degradation, 100 mg/L C3G was added to the SC and YPD medium using CENPK2-1D. The C3G degraded much faster in nutrient-rich YPD medium than in SC medium (Supplementary Figure S1A), suggesting that the anthocyanins underwent significant enzymatic degradation in S. cerevisiae. The 24-h fermentation broth (with YPD medium) of CENPK2-1D was centrifuged to collect the extracellular proteins, and the cells were fragmented to collect the intracellular proteins (with 20 mmol/L PB, pH = 5.0 buffer). Furthermore, 100 mg/L C3G was added to extracellular and intracellular proteins for 12 h (220 rpm, 30°C). The results showed that the degradative protein mainly existed extracellularly (Figure 4A). According to the protocol of ammonium sulfate precipitation, the ammonium sulfate precipitation protein components of 0%–30%, 30%–50%, and 50–100% were obtained, and the 100 mg/L C3G was added for 12-h incubation at 220 rpm and 30°C. The result showed that the degradative protein mainly existed in 30–50% of the protein components (Figure 4B).
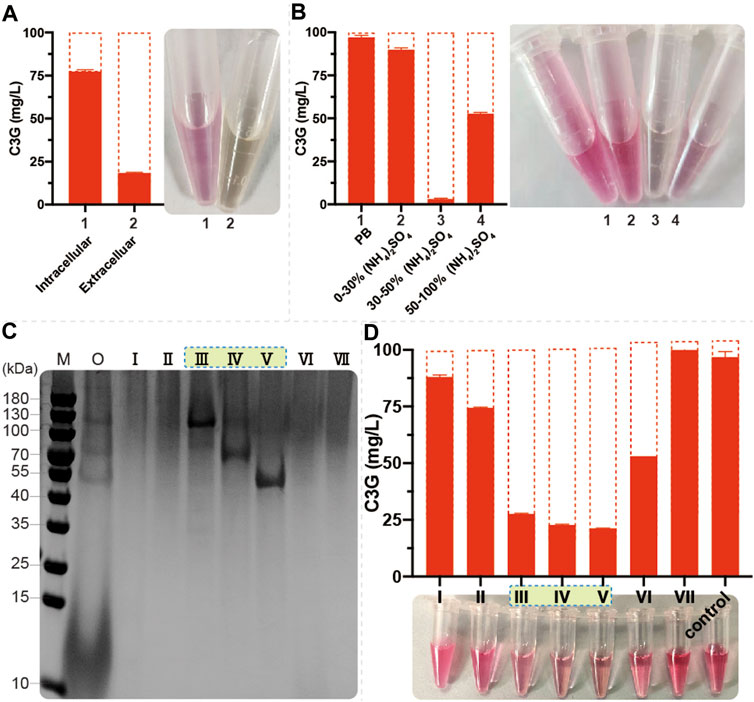
FIGURE 4. Identification of endogenous anthocyanin-degrading enzymes (A) 1 ml of fermentation broth (with YPD medium) of CENPK2-1D was centrifuged at 6000g and 30°C for 1 min; the supernatant was extracellular proteins. The cell pellet was washed twice with 2 ml of 20 mmol/L PB (pH = 5.0, 4°C). The intracellular proteins were obtained by cell disruption with 1 ml of 20 mm PB and 0.1-mm glass beads in FastPrep-24 5G. Then, 100 mg/L C3G was added to extracellular and intracellular proteins at 220 rpm and 30°C for 12-h incubation. An equal volume of acidified methanol as the chromogenic reagent was added to the sample (B) Then, 0%–30%, 30%–50%, and 50–100% protein components were obtained during ammonium sulfate precipitation; 100 mg/L C3G was added for 12-h incubation at 220 rpm and 30°C. An equal volume of acidified methanol as the chromogenic reagent was added to the sample (C) M: marker; O: 30–50% protein component; Ⅰ–Ⅶ: seven elution signal peaks (D) 100 mg/L C3G was added to Ⅰ–Ⅶ for 12-h incubation at 220 rpm and 30°C. An equal volume of acidified methanol as the chromogenic reagent was added to the sample.
Size-exclusion chromatography was performed to separate 0–50% of protein components, and seven elution signal peaks were collected. The proteins corresponding to Ⅲ, Ⅳ, and Ⅴ protein bands had a noticeable degradation effect on anthocyanins (Figures 4C,D). The anthocyanin degradation ratio of Ⅲ, Ⅳ, and Ⅴ was 72.4, 77.3, and 78.7%, respectively. Then, using the protein profiles, four potential anthocyanin-degrading enzymes, namely, SCW2, SIM1, SCW4, and EXG1 (Table 2) were selected by protein profile identification. Moreover, SPR1 and YIR007W were reported to be efficient β-glucosidases (Schmidt et al., 2011) considered to be potential anthocyanin-degrading enzymes.
Knockout of anthocyanin-degrading enzymes
The six potential anthocyanin-degrading enzymes (SCW2, SIM1, SCW4, EXG1, SPR1, and YIR007W) were knocked out in the strain CENPK2-1D, resulting in strains BA-10, BA-11, BA-12, BA-13, BA-14, BA-15, and BA-16. The anthocyanin degradation ratio of BA-13 showed that disrupting EXG1 significantly reduced the degradation rate from 88.0 to 17.3%. Thus, higher anthocyanin production could be achieved by knocking out EXG1 (Figure 5A). At the lowest anthocyanin degradation ratio (12.2%), the BA-20 strain was integrated with Dc3GT in the GAL80 site, resulting in BA-21. As shown in Figure 5B, although the anthocyanin-degrading enzymes knocked out the BA-21 strain, anthocyanins could not be synthesized effectively; the titer of anthocyanins was 2.9 mg/L (2.4 mg/L C3G and 0.5 mg/L D3G). The BA-21 strain further expressed MdGSTF6 (the crucial enzyme for anthocyanin biosynthesis in S. cerevisiae), and 74.7 mg/L anthocyanins (72.3 mg/L C3G and 2.4 mg/L D3G) were obtained.
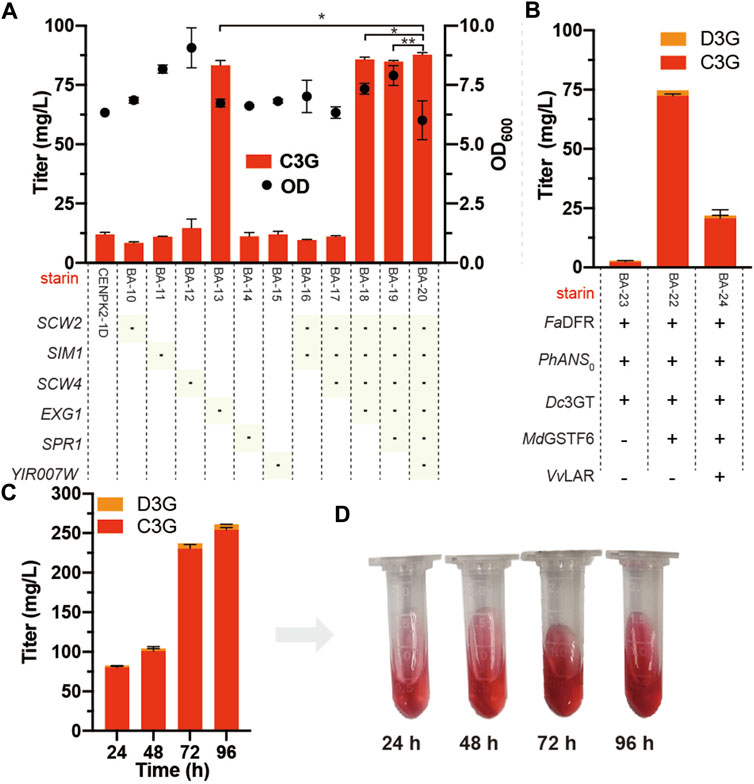
FIGURE 5. Knockout of anthocyanin-degrading enzymes (A) Marker of “-” indicates that the corresponding gene was knocked out in the CENPK2-1D. Student’s t test (one-tailed; two-sample unequal variance; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) was performed by Origin 2018 (B) FaDFR, PhANS0, and MdGSTF6 were promoted by PTDH1, PGAL10, and PINO1 in the BA-21 strain with the pY26-TEF-GPD supplemented with 500 mg/L dihydroflavonols for 24-h cultivation, respectively. VvLAR was connected to the N-terminal of FaDFR with a flexible (GGGGS)2 linker (CTACCGCCACCGCCGCTACCGCCACCGCC) and promoted by PTDH1 (C) 96-h fermentation of the BA-22 strain; 500 mg/L dihydroflavonols were added after 0 h, 1,000 mg/L dihydroflavonols were added after 24 h, and 500 mg/L dihydroflavonols were added after 48 h (D) Fermentation broth corresponding to different times. Equal volume of acidified methanol as the chromogenic reagent was added to the sample.
In addition, flavan-3-ols are the precursors of anthocyanins obtained by the introduction of leucoanthocyanidin reductase (LAR) into the anthocyanin biosynthetic pathway (Figure 1). According to our previous study (Sun et al., 2021), VvLAR was connected to the FaDFR N-terminal with a flexible (GGGGS) 2 linker (CTACCGCCACCGCCGCTACCGCCACCGCC), and the output of flavan-3-ols increased sevenfold. However, the introduction of VvLAR reduced the production of anthocyanins; out of our expectation, only 21.9 mg/L anthocyanins (20.8 mg/L C3G and 1.1 mg/L D3G) were obtained (Figure 5B). When supplemented with 2.0 g/L dihydroflavonols, the accumulation of anthocyanins reached 261.6 mg/L (254.5 mg/L C3G and 7.1 mg/L D3G) by BA-22 (BA-21 harboring the pY26-PTDH1-FaDFR-TGAA1-PGAL10-PhANS0-TALT1-PINO1-MdGSTF6-TCYC1 plasmid) (Figures 5C,D).
De novo biosynthesis of anthocyanins
At present, some progress has been achieved in the heterologous biosynthesis of dihydroflavonols in S. cerevisiae (Li G. et al., 2020; Gao et al., 2020), laying the foundation for de novo biosynthesis of anthocyanins. As EXG1 was proven to be the main anthocyanin-degrading enzyme, Dc3GT was integrated into the EXG1 site of BM-31 (Li G. et al., 2020), resulting in BA-31. Then, the pY26-PTDH1-FaDFR-TGAA1-PGAL10-PhANS0-TALT1-PINO1-MdGSTF6-TCYC1 plasmid was transformed into BA-31, resulting in BA-32. Red colonies grew on the SC medium plate because of successful expression of the anthocyanin de novo biosynthesis pathway in S. cerevisiae (Figure 6A). Fed-batch fermentation was performed in the 5-L bioreactor for 72 h. The results are shown in Figure 6B. A large amount of dihydroflavonols accumulated, containing 11.3 mg/L dihydrokaempferol, 138.5 mg/L dihydroquercetin, and 37.1 mg/L dihydromyricetin. The highest titer of anthocyanins was obtained in 60 h (15.1 mg/L, containing 8.0 mg/L C3G, 3.0 mg/L CYA, 3.5 mg/L D3G, and 0.7 mg/L DEL), after which the titer of anthocyanins decreased gradually.
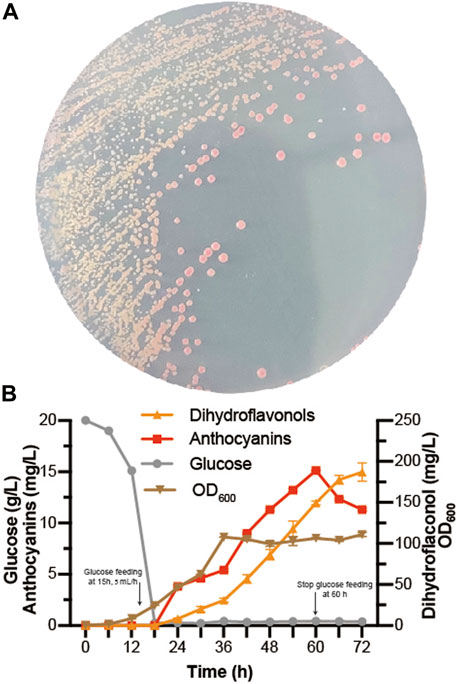
FIGURE 6. De novo biosynthesis of anthocyanins (A) BA-32 strain was grown for 4 days in the SC medium plate (B) Dihydroflavonols, including dihydrokaempferol, dihydroquercetin, and dihydromyricetin; anthocyanins, including pelargonidin-3-O-glucoside, cyanidin-3-O-glucoside, delphinidin-3-O-glucoside, pelargonidin, cyaniding, and delphinidin. The fed-batch fermentation was performed with a 5-L bioreactor containing 2.5 L of YPD. The dissolved oxygen was maintained at 10% (degree of saturation) with the agitation ranging from 250 to 600 rpm. The glucose was fed after 15 h at the rate of 5 ml/h and stopped at 60 h. After 24 h, 0.05 mmol/L α-ketoglutaric acid, 0.5 mmol/L ascorbic acid, and 0.5 mmol/L FeSO4 were added individually to the bioreactor.
Discussion
The production of anthocyanins with microorganisms by metabolic engineering is meaningful for sustainable development, whereas the large-scale production of anthocyanins is still difficult (Li J. et al., 2020). The expensive flavan-3-ols (afzelechin and catechin) are usually the substrates for producing anthocyanins by ANS and 3 GT in prokaryotes. In this study, anthocyanins were successfully obtained from dihydroflavonols. A high titer of anthocyanins was harvested, reaching 261.6 mg/L, through the introduction of MdGSTF6 and the knockout of anthocyanin-degrading enzymes. Finally, by assembling the anthocyanin de novo biosynthetic pathway, 15.1 mg/L anthocyanins were obtained.
In plant cells, the stable storage of anthocyanins is inseparable from the transportation of anthocyanins; however, the mechanism is unclear (Behrens et al., 2019). MdGSTF6 is an anthocyanin transporter involved in transporting anthocyanins from the cytoplasm to the vacuole in the plant cell (Jiang et al., 2019). However, it did not transport anthocyanins to the vacuole of S. cerevisiae because more than 90% of anthocyanins were transported to the extracellular environment. Actually, as polyphenols, many flavonoids, including anthocyanins, would be toxic to cells during intracellular accumulation. In E. coli, the production of anthocyanins could be improved by enhancing the secretion of anthocyanins (Lim et al., 2015). The transport of target products is of positive significance for their accumulation. In this aspect, S. cerevisiae has obvious advantages over E. coli for anthocyanin production. In microorganisms, 2R,4R-flavan-3,3,4-triol, in the production of leucoanthocyanidin by ANS, prefers dehydration to form double bonds at C3–C4 to C2–C3 (Zhang et al., 2019). The resulting production leads to the formation of flavonols after a series of spontaneous reactions and the catalysis of ANS. (Figure 2B). However, under the action of MdGSTF6, the 2R,4R-flavan 3,3,4-triol was likely to form double bonds at C2–C3, which was beneficial to the formation of anthocyanins.
Although the biosynthesis of anthocyanins has been extensively studied (Morita et al., 2014), the mechanism of anthocyanin degradation by S. cerevisiae remains unclear (Pojer et al., 2013). EXG1 was found to be the main anthocyanin-degrading enzyme in this study. EXG1 was reported to be the flavonoid glycoside hydrolase with significant hydrolysis of anthocyanidins (Schmidt et al., 2011) and C-7 flavonoid glycosides (Lyu et al., 2020). Through the knockout of EXG1, the titer of anthocyanins was approximately increased by 100% in the case of co-expressing MdGSTF6. However, an improvement in anthocyanin production by deleting EXG1 was not detected in the absence of MdGSTF6 (Figure 5B). EXG1 did not have a strong hydrolytic effect on the 3-position of flavonoid glycoside because trace amounts of quercetin (QUE) were detected during the co-culture of Q3G with S. cerevisiae (Supplementary Figure S1). Aromatic compound lyases exist in many microorganisms (Patel et al., 2017; Barbosa et al., 2019). Kallscheuer et al. prevented the degradation of phenylpropanoids in Corynebacterium glutamicum by deleting related gene clusters for constructing a platform strain of overproducing stilbenes and (2S)-flavanones (Kallscheuer et al., 2016). The degradation of C3G might be due to the cleavage of EXG1. In fact, the speculative C3G degradation products were reported previously (Barbosa et al., 2019), and protocatechuic acid and phloroglucinol aldehyde were detected by LC-MS (Supplementary Figure S2).
ANS is the key as well as the multifunctional enzyme in the anthocyanin biosynthetic pathway. As mentioned earlier, ANS has the same function as FLS that can convert dihydroflavonols into flavonols. In addition, ANS also had the function of F3H belonging to 2ODD, which could also convert dihydroflavonoids into corresponding dihydroflavonols. When naringenin was supplemented to the S. cerevisiae expressing ANS, dihydrokaempferol and kaempferol were detected simultaneously. The promiscuity of ANS could lead to the accumulation of the by-products flavonol-3-O-glucosides, such as Q3G and M3G. Fortunately, in the presence of MdGSTF6, the titer of anthocyanins was increased by 13-fold, while the flavonol-3-O-glucoside was decreased by sevenfold. In contrast to the prokaryotes (E. coli, C. glutamicum, and L. lactis), S. cerevisiae hardly produced anthocyanins with the flavan-3-ols as the substrate when expressing ANS and 3GT in the EXG1 knockout strain, which was consistent with a previous study (Eichenberger et al., 2018). When co-expressed with MdGSTF6 and supplemented with 500 mg/L catechin (Supplementary Figure S5), 2.3 mg/L C3G was detected, which was increased 15-fold compared to the control group (without MdGSTF6). This suggests that MdGSTF6 also has the potential ability to assist the conversion of flavan-3-ols to anthocyanins.
To further increase the production of anthocyanins, some problems should be resolved in the future. The anthocyanins tend to degrade due to the instability of their molecular structure (Coutinho et al., 2015). pH is one of the main factors affecting the stabilization of anthocyanins. Generally, anthocyanins are prone to degradation in a neutral or alkaline environment (Coutinho et al., 2015), which limits their application in complex environments. The stability of anthocyanins could be improved by some modifications, such as introducing acetyl and methyl groups (Zhao et al., 2017). The supply of UDP-glucose was an important factor in anthocyanin production (Lim et al., 2015; Zha et al., 2018). However, it was not suitable to add it to the medium as a substrate, owing to its excessive values. Rewiring UDP-glucose metabolism in S. cerevisiae should be an effective method (Feng et al., 2020). In addition, directed evolution is an important technology to improve the catalytic efficiency of key enzymes. Also, the intensity of red colonies caused by anthocyanins is a promising high-throughput screening indicator, which makes it effective to select high-catalytic activity mutants.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFA0905500).
Conflict of interest
Authors SX and GC were employed by the company Zhejiang Esigma Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.899182/full#supplementary-material
References
Barbosa, P., Araujo, P., Oliveira, J., Fraga, I., Pissarra, J., Amaral, C., et al. (2019). Metabolic pathways of degradation of malvidin-3-O-monoglucoside by Candida oleophila. Int. Biodeterior. Biodegrad. 144, 104768. doi:10.1016/j.ibiod.2019.104768
Behrens, C. E., Smith, K. E., Iancu, C. V., Choe, J. Y., and Dean, J. V. (2019). Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-binding cassette transporter AtABCC2. Sci. Rep. 9, 437. doi:10.1038/s41598-018-37504-8
Belwal, T., Singh, G., Jeandet, P., Pandey, A., Giri, L., Ramola, S., et al. (2020). Anthocyanins, multi-functional natural products of industrial relevance: Recent biotechnological advances. Biotechnol. Adv. 43, 107600. doi:10.1016/j.biotechadv.2020.107600
Chanoca, A., Burkel, B., Kovinich, N., Grotewold, E., Eliceiri, K. W., Otegui, M. S., et al. (2016). Using fluorescence lifetime microscopy to study the subcellular localization of anthocyanins. Plant J. 88, 895–903. doi:10.1111/tpj.13297
Coutinho, I. B., Freitas, A., Maçanita, A. L., and Lima, J. C. (2015). Effect of water content on the acid–base equilibrium of cyanidin-3-glucoside. Food Chem. x. 172, 476–480. doi:10.1016/j.foodchem.2014.09.060
Dicarlo, J. E., Norville, J. E., Mali, P., Rios, X., Aach, J., Church, G. M., et al. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. doi:10.1093/nar/gkt135
Eichenberger, M., Hansson, A., Fischer, D., Durr, L., and Naesby, M. (2018). De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 18, 13. doi:10.1093/femsyr/foy046
Feng, Y., Yao, M., Wang, Y., Ding, M., Zha, J., Xiao, W., et al. (2020). Advances in engineering UDP-sugar supply for recombinant biosynthesis of glycosides in microbes. Biotechnol. Adv. 41, 107538. doi:10.1016/j.biotechadv.2020.107538
Gao, S., Xu, X., Zeng, W., Xu, S., Lyv, Y., Feng, Y., et al. (2020). Efficient biosynthesis of (2S)-eriodictyol from (2S)-naringenin in Saccharomyces cerevisiae through a combination of promoter adjustment and directed evolution. ACS Synth. Biol. 9, 3288–3297. doi:10.1021/acssynbio.0c00346
Henriques, J. E., Serra, D., Dinis, T. C. P., and Almeida, L. M. (2020). The anti-neuroinflammatory role of anthocyanins and their metabolites for the prevention and treatment of brain disorders. Int. J. Mol. Sci. 21, 8653. doi:10.3390/ijms21228653
Ichino, T., Fuji, K., Ueda, H., Takahashi, H., Koumoto, Y., Takagi, J., et al. (2014). GFS9/TT9 contributes to intracellular membrane trafficking and flavonoid accumulation in Arabidopsis thaliana. Plant J. 80, 410–423. doi:10.1111/tpj.12637
Jiang, S. H., Chen, M., He, N. B., Chen, X. L., Wang, N., Sun, Q. G., et al. (2019). MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 6, 40. doi:10.1038/s41438-019-0118-6
Kallscheuer, N., Vogt, M., Stenzel, A., Gatgens, J., Bott, M., Marienhagen, J., et al. (2016). Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 38, 47–55. doi:10.1016/j.ymben.2016.06.003
Khoo, H. E., Azlan, A., Tang, S. T., and Lim, S. M. (2017). Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 61, 1361779. doi:10.1080/16546628.2017.1361779
Li, A., Liu, Z., Li, Q., Yu, L., Wang, D., Deng, X., et al. (2008). Construction and characterization of bidirectional expression vectors in Saccharomyces cerevisiae. FEMS Yeast Res. 8, 6–9. doi:10.1111/j.1567-1364.2007.00335.x
Li, G., Li, H., Lyu, Y., Zeng, W., and Zhou, J. (2020a). Enhanced biosynthesis of dihydromyricetin in Saccharomyces cerevisiae by coexpression of multiple hydroxylases. J. Agric. Food Chem. 68, 14221–14229. doi:10.1021/acs.jafc.0c05261
Li, J., Zhu, K., and Zhao, H. (2020b). Transcriptome analysis reveals the protection mechanism of proanthocyanidins for Saccharomyces cerevisiae during wine fermentation. Sci. Rep. 10, 6676. doi:10.1038/s41598-020-63631-2
Lim, C. G., Wong, L., Bhan, N., Dvora, H., Xu, P., Venkiteswaran, S., et al. (2015). Development of a recombinant Escherichia coli strain for overproduction of the plant pigment anthocyanin. Appl. Environ. Microbiol. 81, 6276–6284. doi:10.1128/aem.01448-15
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L. F. M., Visser, R. G. F., Bovy, A., et al. (2018). Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 6, 52. doi:10.3389/fchem.2018.00052
Lyu, Y., Liu, S., Gao, S., and Zhou, J. (2020). Identification and characterization of three flavonoid 3-O-glycosyltransferases from Epimedium koreanum Nakai. Biochem. Eng. J. 163, 107759. doi:10.1016/j.bej.2020.107759
Marchev, A. S., Yordanova, Z. P., and Georgiev, M. I. (2020). Green (cell) factories for advanced production of plant secondary metabolites. Crit. Rev. Biotechnol. 40, 443–458. doi:10.1080/07388551.2020.1731414
Morita, Y., Takagi, K., Fukuchi-Mizutani, M., Ishiguro, K., Tanaka, Y., Nitasaka, E., et al. (2014). A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 78, 294–304. doi:10.1111/tpj.12469
Nomi, Y., Iwasaki-Kurashige, K., and Matsumoto, H. (2019). Therapeutic effects of anthocyanins for vision and eye health. Molecules 24, 3311. doi:10.3390/molecules24183311
Olejnik, A., Rychlik, J., Kidoń, M., Czapski, J., Kowalska, K., Juzwa, W., et al. (2016). Antioxidant effects of gastrointestinal digested purple carrot extract on the human cells of colonic mucosa. Food Chem. x. 190, 1069–1077. doi:10.1016/j.foodchem.2015.06.080
Oszmianski, J., and Lee, C. Y. (1990). Inhibition of polyphenol oxidase activity and browning by honey. J. Agric. Food Chem. 38, 1892–1895. doi:10.1021/jf00100a002
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., Mcphee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi:10.1038/nature12051
Passeri, V., Koes, R., and Quattrocchio, F. M. (2016). New challenges for the design of high value plant products: Stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 7, 153. doi:10.3389/fpls.2016.00153
Patel, A., Sartaj, K., Arora, N., Pruthi, V., and Pruthi, P. A. (2017). Biodegradation of phenol via meta cleavage pathway triggers de novo TAG biosynthesis pathway in oleaginous yeast. J. Hazard. Mat. 340, 47–56. doi:10.1016/j.jhazmat.2017.07.013
Pojer, E., Mattivi, F., Johnson, D., and Stockley, C. S. (2013). The case for anthocyanin consumption to promote human health: A review. Compr. Rev. Food Sci. Food Saf. 12, 483–508. doi:10.1111/1541-4337.12024
Saigo, T., Wang, T., Watanabe, M., and Tohge, T. (2020). Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 55, 93–99. doi:10.1016/j.pbi.2020.04.001
Salehi, B., Sharifi-Rad, J., Cappellini, F., Reiner, Z., Zorzan, D., Imran, M., et al. (2020). The therapeutic potential of anthocyanins: Current approaches based on their molecular mechanism of action. Front. Pharmacol. 11, 1300. doi:10.3389/fphar.2020.01300
Schmidt, S., Rainieri, S., Witte, S., Matern, U., and Martens, S. (2011). Identification of a Saccharomyces cerevisiae glucosidase that hydrolyzes flavonoid glucosides. Appl. Environ. Microbiol. 77, 1751–1757. doi:10.1128/aem.01125-10
Shrestha, B., Pandey, R. P., Darsandhari, S., Parajuli, P., and Sohng, J. K. (2019). Combinatorial approach for improved cyanidin 3-O-glucoside production in Escherichia coli. Microb. Cell. Fact. 18, 7. doi:10.1186/s12934-019-1056-6
Solopova, A., Van Tilburg, A. Y., Foito, A., Allwood, J. W., Stewart, D., Kulakauskas, S., et al. (2019). Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate. Metab. Eng. 54, 160–169. doi:10.1016/j.ymben.2019.04.002
Sun, C. C., Li, G. J., Li, H. B., Lyu, Y. B., Yu, S. Q., Zhou, J. W., et al. (2021). Enhancing flavan-3-ol biosynthesis in Saccharomyces cerevisiae. J. Agric. Food Chem. 69, 12763–12772. doi:10.1021/acs.jafc.1c04489
Wilmouth, R. C., Turnbull, J. J., Welford, R. W. D., Clifton, I. J., Prescott, A. G., Schofield, C. J., et al. (2002). Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10, 93–103. doi:10.1016/s0969-2126(01)00695-5
Xu, W. J., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi:10.1016/j.tplants.2014.12.001
Yamei, Y., Danping, F., Xiaoyan, C., Jia, M., Lu, L., Linwu, R., et al. (2020). Review of obesity intervention of anthocyanins from Lycium ruthenicum Murr. J. Food Sci. Technol. 38, 21–26. doi:10.3969/j.issn.2095-6002.2020.04.003
Yan, Y. J., Chemler, J., Huang, L. X., Martens, S., and Koffas, M. a. G. (2005). Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 71, 3617–3623. doi:10.1128/aem.71.7.3617-3623.2005
Yan, Y. J., Li, Z., and Koffas, M. a. G. (2008). High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol. Bioeng. 100, 126–140. doi:10.1002/bit.21721
Yang, Y., Zhang, J.-L., and Zhou, Q. (2020). Targets and mechanisms of dietary anthocyanins to combat hyperglycemia and hyperuricemia: A comprehensive review. Crit. Rev. Food Sci. Nutr. 62, 1119–1143. doi:10.1080/10408398.2020.1835819
Zha, J., Zang, Y., Mattozzi, M., Plassmeier, J., Gupta, M., Wu, X., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for anthocyanin production. Microb. Cell. Fact. 17, 143. doi:10.1186/s12934-018-0990-z
Zhang, J. R., Trossat-Magnin, C., Bathany, K., Delrot, S., and Chaudiere, J. (2019). Oxidative transformation of leucocyanidin by anthocyanidin synthase from Vitis vinifera leads only to quercetin. J. Agric. Food Chem. 67, 3595–3604. doi:10.1021/acs.jafc.8b06968
Zhao, C. L., Yu, Y. Q., Chen, Z. J., Wen, G. S., Wei, F. G., Zheng, Q., et al. (2017). Stability-increasing effects of anthocyanin glycosyl acylation. Food Chem. x. 214, 119–128. doi:10.1016/j.foodchem.2016.07.073
Keywords: anthocyanins, degrading enzyme, Saccharomyces cerevisiae, transporter, de novo biosynthesis
Citation: Xu S, Li G, Zhou J, Chen G and Shao J (2022) Efficient production of anthocyanins in Saccharomyces cerevisiae by introducing anthocyanin transporter and knocking out endogenous degrading enzymes. Front. Bioeng. Biotechnol. 10:899182. doi: 10.3389/fbioe.2022.899182
Received: 24 March 2022; Accepted: 13 July 2022;
Published: 19 August 2022.
Edited by:
M. Kalim Akhtar, United Arab Emirates University, United Arab EmiratesReviewed by:
Jian Zha, Shaanxi University of Science and Technology, ChinaHuadong Peng, Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain), Denmark
Copyright © 2022 Xu, Li, Zhou, Chen and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guicai Chen, Z3VpY2FpY2hlbjg4QDEyNi5jb20=; Jianzhong Shao, c2hhb2p6QHpqdS5lZHUuY24=
†These authors have contributed equally to this work
 Sha Xu
Sha Xu Guangjian Li2†
Guangjian Li2† Jingwen Zhou
Jingwen Zhou Guicai Chen
Guicai Chen Jianzhong Shao
Jianzhong Shao
