- 1School of Food Science and Technology, Jiangnan University, Wuxi, China
- 2The Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi, China
- 3The Science Center for Future Foods, Jiangnan University, Wuxi, China
Hyaluronidases are a group of glycosidases catalyzing the degradation of hyaluronic acid (HA). Because of the advantages of effectively hydrolyzing the HA-rich matrix and low immunogenicity, human hyaluronidase PH20 (hPH20) is widely used in the medical field. Here, we realized the active expression of recombinant hPH20 by Pichia pastoris under a methanol-induced promoter PAOX1. By optimizing the composition of the C-terminal domain and fusing protein tags, we constructed a fusion mutant AP2-△491C with the extracellular hyaluronidase activity of 258.1 U·L−1 in a 3-L bioreactor, the highest expression level of recombinant hPH20 produced by microbes. Furthermore, we found recombinant hPH20 hydrolyzed the β-1,4 glycosidic bonds sequentially from the reducing end of o-HAs, with HA6NA as the smallest substrate. The result will provide important theoretical guidance for the directed evolution of the enzyme to prepare multifunctional o-HAs with specific molecular weights.
Introduction
Hyaluronidases (Hyals) are a large class of glycosidases that predominately catalyze the degradation of hyaluronic acid (HA) (Fronza et al., 2016; Khan et al., 2018). Due to their functions of anesthetic assistance, reducing intraocular pressure, facilitating drug absorption, and resistance to tumor signaling, Hyals have been widely used in medical fields (Locke et al., 2019; Feng et al., 2021; and Knowles et al., 2021). Based on the differences in their catalytic mechanisms and end products, Hyals are classified into three families (Bookbinder et al., 2006; El-Safory et al., 2010; and Kang et al., 2018): hyaluronate 4-glycanohydrolases (EC 3.2.1.35, mammalian hyaluronidases, hydrolyzing the β-1,4 glycosidic bonds of HA, and furnishing tetrasaccharide as the main product), hyaluronate 3-glycanohydrolases (EC 3.2.1.36, leech hyaluronidases, hydrolyzing the β-1,3 glycosidic bonds of HA, and generating tetra- and hexasaccharide end products), hyaluronate lyases (EC 4.2.2.1, bacterial lyases, degrading HA by a β-elimination reaction, and yielding unsaturated disaccharides as the main products). To date, Hyals from animal tissues, leeches, venoms, and various microorganisms have been studied (Tan and Ponnudurai, 1992; Oettl et al., 2003; Rigden and Jedrzejas, 2003; Yang and Lee, 2006; Jin et al., 2014).
Human hyaluronidase PH20 (hPH20), a glycosylphosphatidylinositol (GPI)-anchored membrane protein in mammalian sperm, is a widely recognized hyaluronidase (Cherr et al., 2001). hPH20 could effectively hydrolyze the HA-rich matrix of the oocyte to facilitate the penetration of the sperm. Thus, hPH20 has great potential applications in medical fields (Hong et al., 2019; Usmani et al., 2019; Connor et al., 2020). In past years, the successful expression of recombinant hPH20 in insect or Chinese hamster ovary (CHO) cells has been reported (Frost, 2007; Hofinger et al., 2007); however, the disadvantages such as complex operation, long culture cycle, and high cost restrict its applications. By contrast, Pichia pastoris gives higher expression levels of a wide variety of recombinant enzymes and is generally regarded as easier, faster, and less expensive than insect and mammalian expression systems (Cereghino and Cregg, 2000). In a previous study, Chen et al. have realized the constitutive expression of recombinant hPH20 in P. pastoris, and even the activity was merely 2 U·L−1 (Chen et al., 2016). To meet the industrial requirement, the expression of recombinant hPH20 needs a considerable improvement. Furthermore, to date, the hydrolysis pattern of recombinant hPH20 on HA still remains unclear.
In this study, we analyzed the hPH20 sequence and achieved the comparatively high expression of recombinant hPH20 in P. pastoris with the methanol-induced promoter PAOX1. Through the optimization of the C-terminal domain and application of protein fusion tags, we successfully constructed a fusion mutant AP2-△491C with the extracellular hyaluronidase activity of 258.1 U·L−1 in a 3-L bioreactor, which was the highest value from microbes to date. Moreover, hydrolysis pattern analysis results demonstrate that recombinant hPH20 hydrolyzes the β-1,4 glycosidic bonds sequentially from the reducing end of o-HAs with HA6NA as the smallest substrate.
Materials and Methods
Strains, Plasmids, and Reagents
E. coli JM109 and plasmid pPIC9K were used for gene cloning and propagation procedures. P. pastoris GS115 was used as a host for the expression of recombinant hPH20 and its mutants. PrimeSTAR Max DNA Polymerase, T4 DNA ligase, T4 polynucleotide kinase, restriction endonucleases, and protein markers were purchased from TaKaRa (Dalian, China). Ampicillin, G418 sulfate, and plasmid Mini Prep Kit were purchased from Sangon Biotech (Shanghai, China). The gel extraction kit and 10% Bis-Tris protein gel were purchased from Thermo Scientific (Shanghai, China). Hyaluronidase from bovine testicular tissue (BTH) was obtained from Sigma-Aldrich (St Louis, MO, United States). Other chemicals were obtained commercially and were of reagent grade.
Medium
Luria–Bertani (LB) medium (10 g L−1 tryptone, 10 g L−1 NaCl, 5 g L−1 yeast extract, and pH 7.0) was used for the cloning experiment. The MD plate [20 g L−1 glucose, 13.4 g L−1 yeast nitrogen base (YNB), and 20 g L−1 agar] was used for screening P. pastoris GS115 recombinants. YPD medium (10 g L−1 yeast extract, 20 g L−1 tryptone, and 20 g L−1 glucose), BMGY medium (10 g L−1 yeast extract, 20 g L−1 tryptone, 100 mM potassium phosphate, 13.4 g L−1 YNB, 4 × 10–4 g L−1 D-biotin, and 10 g L−1 glycerol), and BMMY medium [10 g L−1 yeast extract, 20 g L−1 tryptone, 100 mM potassium phosphate, 13.4 g L−1 YNB, 4 × 10–4 g L−1 D-biotin, and 1% (v/v) methanol] were used for flask cultures. Trace metal solution (PTM1, 3 g L−1 MnSO4·H2O, 6 g L−1 CuSO4·5H2O, 0.2 g L−1 MoNa2O4·2H2O, 65 g L−1 FeSO4·7H2O, 0.5 g L−1 CoCl2, 20 g L−1 ZnCl2, 0.09 g L−1 KI, 0.02 g L−1 H3BO3, 0.2 g L−1 D-biotin, and 5 ml L−1 H2SO4) and BSM (18 g L−1 K2SO4, 14.9 g L−1 MgSO4·7H2O, 4.13 g L−1 KOH, 40 g L−1 glycerol, 27 ml L−1 H3PO4, and 0.93 g L−1 CaSO4 with 4.4 ml L−1 PTM1) were used for fed-batch fermentation.
Genetic Operations
According to the codon bias of P. pastoris, we optimized the codons of the hPH20 gene carrying the His × 6-tag coding sequence. The codon-optimized DNA fragment was cloned to restriction sites EcoR I and Not I in pPIC9K to obtain plasmid pPIC9K-hPH20. The primers (Supplementary Table S1) were used for cloning the C-terminal truncated mutants. The PCR products were extracted using the gel extraction kit. The extracted product, T4 DNA ligase, T4 polynucleotide kinase, and T4 DNA ligase buffer were added to a PCR tube at a ratio of 7:1:1:1 (v/v), incubated at 16°C for 12 h, and then transformed into E. coli JM109 for sequencing. Plasmids with the correct sequences were linearized by Sal I and transformed into competent P. pastoris GS115 cells via electroporation (voltage: 2000 V, capacitance: 25 μF, resistance: 200 Ω, and cuvette: 2 mm) using a GenePulser Xcell™ apparatus purchased from Bio-Rad (Hercules, United States). The transformants were screened on MD plates, and the selection of multi-copy cells was performed on YPD plates with 4 g L−1 G418 sulfate. The gene copy numbers of all recombinant strains were determined by real-time PCR, as described previously (Kang et al., 2016). Clones harboring nine copies of the target gene of each construct were selected.
We had three protein fusion tags (AP2, HL28, and Sumo-tag) synthesized before the N terminal of the truncated mutant △491C to obtain three recombinant expression plasmids: pPIC9K-ap2-△491C, pPIC9K-hl28-△491C, and pPIC9K-sumo-△491C. Then, we transformed and screened the P. pastoris GS115 recombinants as before.
Shake Flask Culture and Fed-Batch Fermentation
P. pastoris GS115 strains carrying recombinant hPH20 were inoculated in 250-ml flasks supplied with 50 ml YPD medium and grown at 30°C and 220 rpm for 24 h. Then, the yeast strains were transferred to a BMGY medium with a 10% (v/v) inoculum and cultivated at 30°C and 220 rpm until the OD600 value reached 6. The yeast strains were collected and washed with 0.9% NaCl three times, then transferred to 50 ml BMMY medium to induce the expression at 30°C and 220 rpm. All the flasks were added with 1% (v/v) methanol every 24 h, and experiments were carried out in triplicate.
Fed-batch fermentation was performed in a 3-L bioreactor (BXBIO, Shanghai, China) using a 900 ml BSM. A 10% (v/v) inoculum was obtained after cultivating the strains at 30°C and 220 rpm for 24 h. The initial fermentation parameters were set at 30°C, pH 5.0, 2.0 vvm, and 220 rpm. After glycerol exhaustion indicated by a sudden increase in dissolved oxygen, 50% (w/v) glycerol supplemented with 1.2% (v/v) PTM1 was fed at a constant rate of 25 ml L−1 h−1 for 10 h. Methanol supplemented with 1.2% (v/v) PTM1 was fed at a constant rate of 7 ml L−1 h−1 to induce the expression of recombinant hPH20. The culture broth was collected every 12 h for analysis of cell growth and hyaluronidase activity.
Western Blot Analysis of Recombinant hPH20 Expression
For the detection of recombinant hPH20 expression, samples were harvested by centrifugation (9,000 × g for 10 min at 4°C). The supernatant was mixed with 4 × Protein Native PAGE Loading Buffer (TaKaRa, Dalian, China) and heated at 100°C for 10 min. SDS-PAGE was performed by 10% Bis-Tris Protein Gels in MES running buffer at 100 V. Then, SDS-PAGE gel was electroblotted to PVDF transfer membrane (PerkinElmer, Shanghai, China) using a transfer buffer containing 20% methanol, 15.1 g L−1 glycine, and 3.0 g L−1 Tris. Blotted membrane was blocked with QuickBlock™ Blocking Buffer (Beyotime Biotechnology, Shanghai, China) for 1 h. For immunodetection, His-tag Mouse Monoclonal Antibody (Beyotime Biotechnology, Shanghai, China) diluted 1: 1,000 with TBS (2.4 g L−1, pH 7.6 Tris buffer containing 8.0 g L−1 NaCl) was used as a primary antibody. After washing with TBST (2.4 g L−1, pH 7.6 Tris buffer containing 8.0 g L−1 NaCl and 0.1% Tween-20) for 30 min, a 1:1,000 dilution of the secondary antibody, HRP-labeled Goat Anti-Mouse lgG (H + L) (Beyotime Biotechnology, Shanghai, China) was added and incubated at 4°C with gentle shaking for 1 h. After washing with TBST for 30 min, DAB Horseradish Peroxidase Color Development Kit (Beyotime Biotechnology, Shanghai, China) was used to develop the color on the membrane.
Determination of Recombinant hPH20 Activity
Recombinant hPH20 activity was determined using the 3,5-dinitrosalicylic acid (DNS) method to measure the reducing sugar released from hyaluronic acid (Asteriou et al., 2001). In this study, 100 μl of 1.25 mg ml−1 hyaluronic acid in 100 mM, pH 5.0 acetate buffer was mixed with 100 μl of enzyme sample and incubated at 37°C for 30 min. Thereafter, 200 μl of DNS was added to the standard reaction system to terminate the reaction. The reaction was boiled in a water bath at 100°C for 6 min and then cooled on ice immediately. The reaction system with inactivated enzyme was used as a control. Enzyme activity (U) was defined as the amount of enzyme that released 1 μmol of reducing sugar [equivalent to N-acetyl-glucosamine (NAG)] per minute under specified assay conditions. All values were expressed as the mean ± standard deviation (SD ≤ 5%) of three independent experiments.
Hydrolysis of Recombinant hPH20 on Different Glycosaminoglycan Substrates
An aliquot of 100 μl of 1.25 mg ml−1 different glycosaminoglycan substrates [HA, chondroitin sulfate type A (CSA), chondroitin sulfate type C (CSC), and heparin (HP)] in 100 mM, pH 5.0 acetate buffer was mixed with 100 μl of enzyme sample and incubated at 37°C for 30 min. Thereafter, 200 μl of DNS was added to the standard reaction system to terminate the reaction. Recombinant hPH20 activity was quantified as described earlier.
Preparation of HA2nNA o-HAs
The enzymatic properties and hydrolysis process of LHyal have been studied (Jin et al., 2014). To prepare HA2nNA o-HAs (HA4NA, HA6NA, and HA8NA), 1 g of HA was fully dissolved in 100 ml of distilled water and pre-incubated at 37°C. Afterward, 3000 U·ml−1 of LHyal was mixed with the aforementioned HA solution and incubated at 37°C for 20 h, then LHyal was inactivated by boiling for 5 min. After high-speed centrifugation (9,000 × g for 30 min at 4°C) to remove inactivated enzyme, a HiPrep Q HP 16/10 column (column volume: 20 ml, GE Healthcare) was used to separate HA2nNA o-HAs from the supernatant. The column was balanced by buffer A (0.02 M, pH 8.0 PBS) before eluting by a linear gradient of 0–100 mM NaCl (pH 8.0) for 3 CV under 3 ml min−1 flow rate.
Each fraction corresponding to a chromatographic peak was desalted using a Superdex 30 Increase 10/300 G L column and lyophilized to get the final products. Each HA2nNA o-HA fraction was qualitatively analyzed by high-performance liquid chromatography–mass spectrometry (HPLC-MS) to confirm purity, as described previously (He et al., 2020).
Hydrolysis of HA2nNA o-HAs and Analysis of Hydrolysates
Recombinant hPH20 was purified using a Ni-NTA column (GE Healthcare, Shanghai, China). An aliquot of 100 μl of recombinant hPH20 was mixed with 80 μl of purified o-HAs (HA4NA, HA6NA, and HA8NA), respectively, and incubated at 37°C for 8 h; then recombinant hPH20 was inactivated by boiling for 5 min. To remove the salt, 180 μl of methanol was added to the reaction system, mixed gently, and then centrifuged at 12,000 rpm for 10 min. Electrospray ionization-mass spectrometry (ESI-MS) spectra were acquired using a Quattro Premier XE system (Waters). Additionally, HPLC separation was performed using a Zorbax NH2 column (4.6 × 250 mm) at 40°C. The products were eluted with acetonitrile/water (75:25) at a flow rate of 1.0 ml min−1. The eluent was monitored by measuring the total ions in the mass range of m/z 100–600 in the negative mode.
Results and Discussion
Structure and Catalytic Domain Analysis of hPH20
The hPH20 gene (GenBank accession number: NP_003,108.2) cloned from human sperm encodes a polypeptide of 511 amino acids. The molecular weight and pI value of hPH20 were estimated to be 58.4 kDa and 7.0, respectively. The conserved amino acids in hPH20 structural domains associated with substrate catalysis and glycosylation were deduced by the NCBI online protein blast tool: the active site Asp146 and Glu148, and the glycosylation site Asn82, Asn166, Asn235, Asn254, Asn368, and Asn393. Homology analysis revealed that hPH20 shows 86.1% identity with hyaluronidase from Sapajus apella, 75.1% with hyaluronidase from Carlito syrichta, and 70.0% with hyaluronidase from Otolemur garnettii, and 65.4% with hyaluronidase from Ictidomys tridecemlineatus.
hPH20 was predicted to contain four structural domains by Protein Domain Analysis Tool (http://smart.embl-heidelberg.de). As shown in Figure 1, the domain compositions were signal peptide coding region (positions 1–35): found in the wild-type sequence of hPH20, related to transport and secretion; Glyco_hydro_56 (positions 42–369): N-terminal catalytic domain of hPH20, commonly found in GH56 protein; zona pellucida recognition domain (∼110 aa linked after Glyco_hydro_56): recognized by the zona pellucida of the oocyte, related to the binding of sperm and egg cells in fertilization; and GPI anchoring region (positions 484–511): a short hydrophobic structure in the C-terminal of hPH20, associated with the binding of protein to the cell membrane. Moreover, a protease cleavage site was predicted between Ser490 and Ala491 of the GPI anchoring region.
Active Expression of Recombinant hPH20 With Its Truncated Variants
In consideration of deletion of functionally unnecessary domains could simplify the whole structure and improve the secretion efficiency and production of an enzyme (Kim et al., 2011; Duan and Wu, 2015), recombinant hPH20 was truncated and expressed with the P. pastoris expression system for characterization. Through the analysis of structural domains of recombinant hPH20, we found that the GPI anchoring region (positions 484–511) played a role in the binding of recombinant hPH20 and cell membrane, not related to the catalysis according to the description of domain function. Furthermore, a protease cleavage site existed between positions 490 and 491, indicating the amino acid sequence after position 490 may not affect the enzymatic properties of recombinant hPH20. Therefore, as shown in Figure 2A, we designed and constructed two C-terminal truncated mutants (hPH20△484C and hPH20△491C).
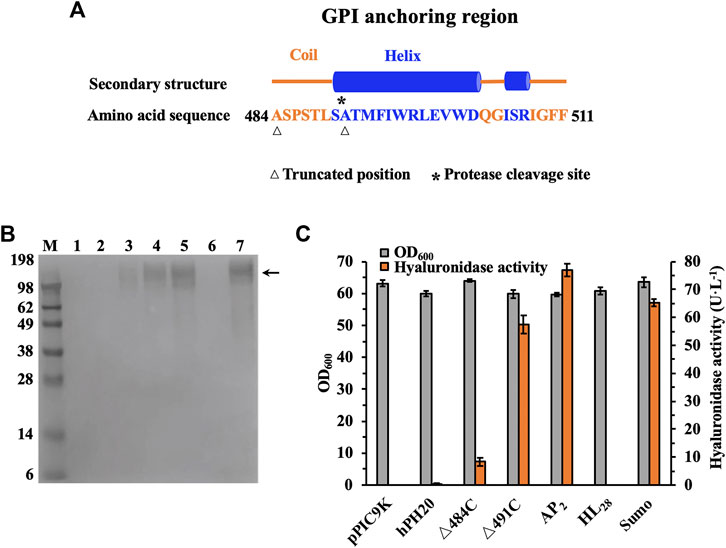
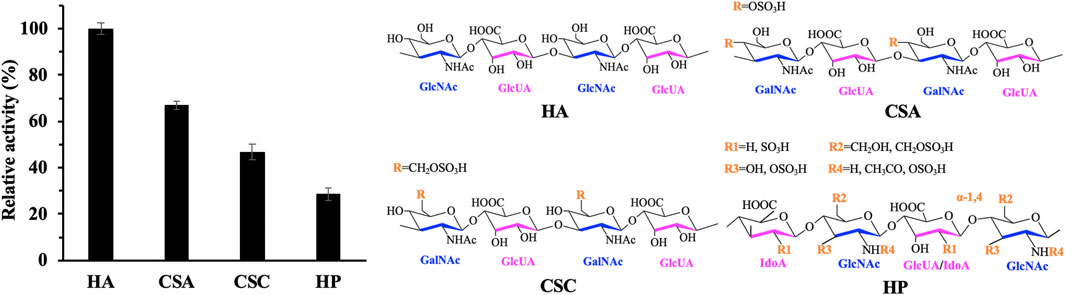
FIGURE 2. Expression and activity of recombinant hPH20, truncated mutants (△484C △491C), and △491C with different tags in the culture broth. (A) Secondary structure and sequence analysis of the GPI anchoring region. (B) Western blot analysis of the recombinant hPH20, truncated mutants (△484C △491C), and △491C with different tags. M: standard protein marker; 1: control; 2: recombinant hPH20; 3: △484C; 4: △491C; 5: AP2; 6: HL28; 7: Sumo. The truncated mutants (△484C △491C) and △491C with different tags are shown by the black arrow. (C) Hyaluronidase activity of recombinant hPH20, truncated mutants (△484C △491C), and △491C with different tags.
The molecular weights of secreted recombinant hPH20, hPH20△484C, and hPH20△491C were estimated to be 55.4, 52.2, and 52.8 kDa, respectively. However, we were not sure whether the protein bands of recombinant hPH20, hPH20△484C, and hPH20△491C in culture supernatants were present or not by SDS-PAGE analysis (Supplementary Figure S1). To confirm the expression of these variants, Western blot analysis with sensitive protein detection was carried out. As shown in Figure 2B, hPH20△484C and hPH20△491C in the culture supernatants were successfully detected with a very broad molecular mass of 62–198 kDa, which were higher than the calculated values. This may be due to the glycosylation that occurred in the endoplasmic reticulum. By contrast, the recombinant hPH20 in the supernatant was not detected. To identify the reason, we mixed the whole cell of P. pastoris GS115-pPIC9K-hPH20 with HA and determined whether HA could be hydrolyzed. As shown in Supplementary Figure S2, the whole-cell hydrolyzed HA, indicating the recombinant hPH20 fixed on the cell membrane with the GPI anchoring region and its N-terminal catalytic domain, was exposed to the outside of the cell membrane, which accounted for not detecting recombinant hPH20 in the supernatant.
In parallel, the hyaluronidase activities of recombinant hPH20, hPH20△484C, and hPH20△491C in the culture broth were determined by using the DNS method. As shown in Figure 2C, the extracellular hyaluronidase activity of hPH20△491C reached 57.5 U·L−1, which was 6.8 times higher than that of hPH20△484C (8.4 U·L−1), indicating that the structure between 490 and 484 was indispensable for maintaining a high catalytic activity of recombinant hPH20. On the contrary, we did not observe any extracellular hyaluronidase activity of recombinant hPH20, indicating the C-terminal structure between 511 and 491 played a key role in the anchoring of recombinant hPH20 and cell membrane. The results of hyaluronidase activity determination were consistent with those of Western blot analysis.
To further improve the secretion efficiency and production, we optimized the composition of the C-terminal structure and constructed truncated mutants hPH20△507C, hPH20△502C, hPH20△497C, hPH20△489C, hPH20△487C, and hPH20△485C, comparing extracellular hyaluronidase activities of these mutants with that of hPH20△491C. As shown in Supplementary Figure S3, these mutants displayed lower hyaluronidase activity and expression levels than those of hPH20△491C. Therefore, we used hPH20△491C in the following experiments.
Introduction of Protein Fusion Tags to Improve Recombinant hPH20 Expression
Protein fusion tags functioned to facilitate the expression of exogenous protein and increase the secretion efficiency of soluble protein (Moua et al., 2016; and Wang et al., 2019). For instance, Huang et al. achieved the extracellular high-level expression of LHyal in P. pastoris by fusing different N-terminal tags, the soluble expression of leech hyaluronidase (Huang et al., 2020). The secretion efficiency of Candida antarctica lipase B (Kim et al., 2015) and the (+)-zizaene synthase from Chrysopogon zizanioides (Hartwig et al., 2015) were also improved. Therefore, three commonly used protein fusion tags AP2, HL28, and Sumo-tag were fused to the N terminal of the variant hPH20△491C to assist the expression and secretion.
As shown in Figure 2B, fusion mutants AP2-△491C and Sumo-△491C in the culture broth were both detected by Western blot analysis, indicating the successful expression and secretion. By contrast, no HL28-△491C was documented, which might be due to the inhibitory effect of the protein tag HL28 on the expression of △491C. The hyaluronidase activities of AP2-△491C and Sumo-△491C were also comparatively analyzed. As shown in Figure 2C, the extracellular hyaluronidase activities of AP2-△491C and Sumo-△491C reached 76.9 U·L−1 and 65.4 U·L−1, respectively, 1.3 and 1.1 times higher than that of △491C, indicating the protein tags AP2 and Sumo could facilitate the expression and secretion of △491C. Significantly, the extracellular hyaluronidase activity of fusion mutant AP2-△491C in this study was 38.5 times higher than that of the previous report (Chen et al., 2016), suggesting the stronger promoter strength of PAOX1 than that of the constitutive promoter PGAP.
Production of Recombinant hPH20 With High-Cell Density Fermentation
To evaluate the hyaluronidase-producing ability of recombinant P. pastoris GS115-pPIC9K-ap2-△491C, a high-cell fermentation was performed in a 3-L bioreactor. As shown in Figure 3B, the extracellular hyaluronidase activity of AP2-△491C reached 258.1 U·L−1 at 96 h, 3.4 times higher than that of flask cultures. Consistently, the Western blot analysis of the extracellular culture supernatant indicated the inducible expression and secretion of AP2-△491C during the whole process of fermentation (Figure 3A). To our knowledge, we achieved the highest expression level of recombinant hPH20 in studies of producing recombinant hPH20 by the microbial expression system. This result would lay a foundation for the large-scale preparation of recombinant hPH20 in future studies.
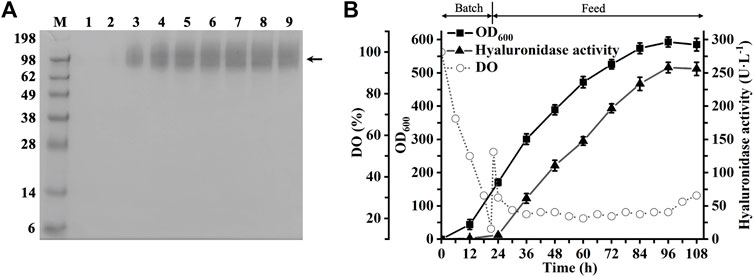
FIGURE 3. Fed-batch fermentation of recombinant strain P. pastoris GS115-pPIC9K-ap2-△491C. (A) Western blot analysis of the culture supernatants. M: standard protein marker; 1-9, samples were taken at 12, 24, 36, 48, 60, 72, 84, 96, and 108 h, respectively. The target proteins are shown by the black arrow. (B) Time course of cell growth, dissolved oxygen, and hyaluronidase activity in a 3-L bioreactor.
Substrate Specificity of Recombinant hPH20
To analyze the substrate specificity of recombinant hPH20, we chose different glycosaminoglycans (HA, CSA, CSC, and HP) as substrates to measure the hydrolytic activity of recombinant hPH20. As shown in Figure 4, HA, CSA, CSC, and HP could be hydrolyzed. Recombinant hPH20 showed the highest hydrolytic capacity toward substrate HA, 1.5, 2.1, and 3.5 times higher than those of substrates CSA, CSC, and HP. Compared with LHyal and BTH (Jin et al., 2014), recombinant hPH20 exhibited a broader substrate spectrum.
Hydrolysis Pattern of Recombinant hPH20 on Substrate HA
In a previous study, it has been reported that recombinant hPH20 hydrolyzes the β-1,4 glycosidic bonds of the HA chain producing o-HAs with different molecular weights (Arming et al., 1997), but the hydrolysis pattern remains unknown. To solve this problem, we prepared HA2nNA o-HAs (HA4NA, HA6NA, and HA8NA shown in Supplementary Figure S4) by LHyal-catalyzed HA hydrolysis and analyzed the hydrolysates of o-HAs cleaved by recombinant hPH20.
As shown in Figure 5, recombinant hPH20 hydrolyzed HA6NA producing HA5NN as the sole detectable hydrolysate, indicating the enzyme hydrolyzed the β-1,4 glycosidic bond at the reducing end of HA6NA, while HA5NN could not be further cleaved. When using HA8NA as the substrate, HA5NN and HA2 were the main products, while HA3AA and HA7NN were not detected (Figures 6A,B). The result suggested that recombinant hPH20 hydrolyzed the β-1,4 glycosidic bonds sequentially from the reducing end of HA8NA, and the hydrolysis capacity toward the penultimate β-1,4 glycosidic bond from the reducing end was higher than that of the β-1,4 glycosidic bond at the reducing end (Figure 6C). In addition, it was noteworthy that recombinant hPH20 could not degrade the β-1,4 glycosidic bond of HA4NA (data not shown). From the aforementioned results, we could conclude that recombinant hPH20 hydrolyzed the β-1,4 glycosidic bonds sequentially from the reducing end of o-HAs with HA6NA as the smallest substrate. The hydrolysis pattern of recombinant hPH20 differed from BTH, which hydrolyzed the β-1,4 glycosidic bonds from the nonreducing end of o-HAs (Takagaki et al., 1994). A study on the hydrolysis pattern of recombinant hPH20 was of great significance for the directed evolution of the enzyme to prepare a variety of multifunctional o-HAs with specific molecular weights in the future.
FIGURE 5. Analysis of the end products of HA6NA hydrolyzed by recombinant hPH20. (A,B) Extracted ion chromatogram/mass spectra of end products of HA6NA by HPLC-MS. (C) Schematic diagram of the hydrolysis process of HA6NA.
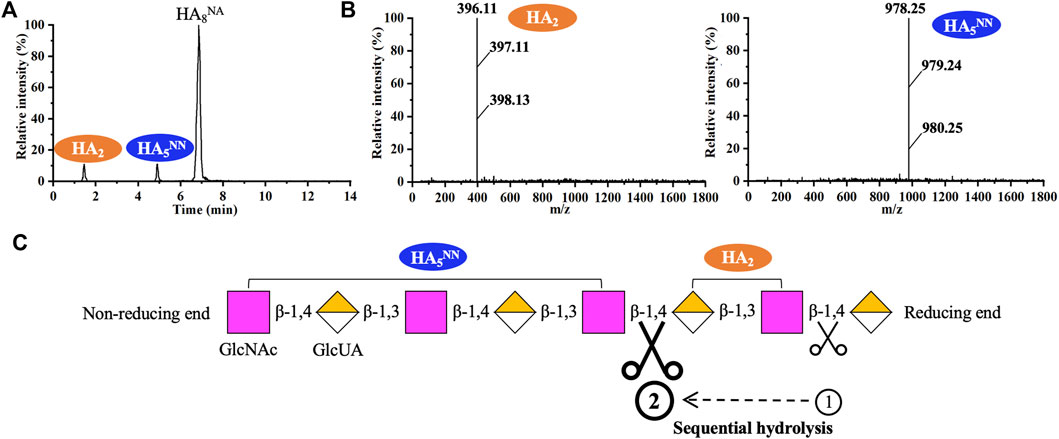
FIGURE 6. Analysis of the end products of HA8NA hydrolyzed by recombinant hPH20. (A,B) Extracted ion chromatogram/mass spectra of end products of HA8NA by HPLC-MS. (C) Schematic diagram of the hydrolysis process of HA8NA.
Conclusion
In the present study, human hyaluronidase PH20 was engineered and expressed with high activity in P. pastoris. By simplifying the C-terminal domain and introducing protein fusion tags, a fusion mutant AP2-△491C was constructed, which showed the extracellular hyaluronidase activity of 258.1 U·L−1 in a 3-L bioreactor. To the best of our knowledge, this was the highest reported value of human hyaluronidase PH20. Moreover, our results found that recombinant hPH20 hydrolyzes the β-1,4 glycosidic bonds sequentially from the reducing end of the hyaluronan polysaccharide chain. The smallest substrate for recognition and digestion was HA6NA.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
BP conceived the project and designed the experiments, JH assisted with material preparation, WZ constructed the recombinants, HH carried out cell culture and enzyme activity tests, YW helped with data processing, and MW, GD, and ZK contributed to writing—review and editing.
Funding
This work was financially supported by the National Natural Science Foundation of China (32100057, 32000058), Jiangsu Province Natural Science Fund for Distinguished Young Scholars (BK20200025), the China Postdoctoral Science Foundation (2021M691286), and a grant from the Key Technologies R&D Program of Jiangsu Province (BE2019630).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.885888/full#supplementary-material
References
Arming, S., Strobl, B., Wechselberger, C., and Kreil, G. (1997). In Vitro mutagenesis of PH-20 Hyaluronidase from Human Sperm. Eur. J. Biochem. 247 (3), 810–814. doi:10.1111/j.1432-1033.1997.t01-1-00810.x
Asteriou, T., Deschrevel, B., Delpech, B., Bertrand, P., Bultelle, F., Merai, C., et al. (2001). An Improved Assay for the N-Acetyl-Gglucosamine Reducing Ends of Polysaccharides in the Presence of Proteins. Anal. Biochem. 293 (1), 53–59. doi:10.1006/abio.2001.5068
Bookbinder, L. H., Hofer, A., Haller, M. F., Zepeda, M. L., Keller, G.-A., Lim, J. E., et al. (2006). A Recombinant Human Enzyme for Enhanced Interstitial Transport of Therapeutics. J. Controlled Release 114 (2), 230–241. doi:10.1016/j.jconrel.2006.05.027
Cereghino, J. L., and Cregg, J. M. (2000). Heterologous Protein Expression in the Methylotrophic yeastPichia Pastoris. FEMS Microbiol. Rev. 24 (1), 45–66. doi:10.1111/j.1574-6976.2000.tb00532.x
Chen, K.-J., Sabrina, S., El-Safory, N. S., Lee, G.-C., and Lee, C.-K. (2016). Constitutive Expression of Recombinant Human Hyaluronidase PH20 by Pichia pastoris. J. Biosci. BioengineeringJ. Biosci. Bioeng. 122 (6), 673–678. doi:10.1016/j.jbiosc.2016.06.007
Cherr, G. N., Yudin, A. I., and Overstreet, J. W. (2001). The Dual Functions of GPI-Anchored PH-20: Hyaluronidase and Intracellular Signaling. Matrix Biol. 20 (8), 515–525. doi:10.1016/s0945-053x(01)00171-8
Connor, R. J., Taverna, D. M., Thrall, K., LaBarre, M. J., and Kang, D. W. (2020). Use of Computed Tomography to Assess Subcutaneous Drug Dispersion with Recombinant Human Hyaluronidase PH20 in a Swine Model. J. Pharmacol. Toxicol. Methods 106, 106936. doi:10.1016/j.vascn.2020.106936
Duan, X., and Wu, J. (2015). Enhancing the Secretion Efficiency and Thermostability of a Bacillusderamificans Pullulanase Mutant (D437H/D503Y) by N-Terminal Domain Truncation. Appl. Environ. Microbiol. 81 (6), 1926–1931. doi:10.1128/AEM.03714-14
El-Safory, N. S., Fazary, A. E., and Lee, C.-K. (2010). Hyaluronidases, a Group of Glycosidases: Current and Future Perspectives. Carbohydr. Polym. 81 (2), 165–181. doi:10.1016/j.carbpol.2010.02.047
Feng, C., Xiong, Z., Wang, C., Xiao, W., Xiao, H., Xie, K., et al. (2021). Folic Acid-Modified Exosome-PH20 Enhances the Efficiency of Therapy via Modulation of the Tumor Microenvironment and Directly Inhibits Tumor Cell Metastasis. Bioactive Mater. 6 (4), 963–974. doi:10.1016/j.bioactmat.2020.09.014
Fronza, M., Muhr, C., da Silveira, D. S. C., Sorgi, C. A., Rodrigues, S. F. d. P., Farsky, S. H. P., et al. (2016). Hyaluronidase Decreases Neutrophils Infiltration to the Inflammatory Site. Inflamm. Res. 65 (7), 533–542. doi:10.1007/s00011-016-0935-0
Frost, G. I. (2007). Recombinant Human Hyaluronidase (rHuPH20): an Enabling Platform for Subcutaneous Drug and Fluid Administration. Expert Opin. Drug Deliv. 4 (4), 427–440. doi:10.1517/17425247.4.4.427
Hartwig, S., Frister, T., Alemdar, S., Li, Z., Scheper, T., and Beutel, S. (2015). SUMO-fusion, Purification, and Characterization of a (+)-zizaene Synthase from Chrysopogon Zizanioides. Biochem. Biophysical Res. Commun. 458 (4), 883–889. doi:10.1016/j.bbrc.2015.02.053
He, J., Huang, H., Zou, X., Wang, Y., Du, G., and Kang, Z. (2020). Construction of Saturated Odd- and Even-Numbered Hyaluronan Oligosaccharide Building Block Library. Carbohydr. Polym. 231, 115700. doi:10.1016/j.carbpol.2019.115700
Hofinger, E. S. A., Bernhardt, G., and Buschauer, A. (2007). Kinetics of Hyal-1 and PH-20 Hyaluronidases: Comparison of Minimal Substrates and Analysis of the Transglycosylation Reaction. Glycobiology 17 (9), 963–971. doi:10.1093/glycob/cwm070
Hong, Y., Kim, Y. K., Kim, G. B., Nam, G.-H., Kim, S. A., Park, Y., et al. (2019). Degradation of Tumour Stromal Hyaluronan by Small Extracellular Vesicle-PH20 Stimulates CD103+ Dendritic Cells and in Combination with PD-L1 Blockade Boosts Anti-tumour Immunity. J. Extracellular Vesicles 8 (1), 1670893. doi:10.1080/20013078.2019.1670893
Huang, H., Liang, Q., Wang, Y., Chen, J., and Kang, Z. (2020). High-level Constitutive Expression of Leech Hyaluronidase with Combined Strategies in Recombinant Pichia pastoris. Appl. Microbiol. Biotechnol. 104 (4), 1621–1632. doi:10.1007/s00253-019-10282-7
Jin, P., Kang, Z., Zhang, N., Du, G., and Chen, J. (2014). High-yield Novel Leech Hyaluronidase to Expedite the Preparation of Specific Hyaluronan Oligomers. Sci. Rep. 4, 4471. doi:10.1038/srep04471
Kang, Z., Zhang, N., and Zhang, Y. (2016). Enhanced Production of Leech Hyaluronidase by Optimizing Secretion and Cultivation in Pichia pastoris. Appl. Microbiol. Biotechnol. 100 (2), 707–717. doi:10.1007/s00253-015-7056-5
Kang, Z., Zhou, Z., Wang, Y., Huang, H., Du, G., and Chen, J. (2018). Bio-based Strategies for Producing Glycosaminoglycans and Their Oligosaccharides. Trends Biotechnol. 36 (8), 806–818. doi:10.1016/j.tibtech.2018.03.010
Khan, N., Niazi, Z. R., Rehman, F. U., Akhtar, A., Khan, M. M., Khan, S., et al. (2018). Hyaluronidases: A Therapeutic Enzyme. Ppl 25 (7), 663–676. doi:10.2174/0929866525666180629121823
Kim, S.-K., Park, Y.-C., Lee, H. H., Jeon, S. T., Min, W.-K., and Seo, J.-H. (2015). Simple Amino Acid Tags Improve Both Expression and Secretion ofCandida Antarcticalipase B in recombinantEscherichia Coli. Biotechnol. Bioeng. 112 (2), 346–355. doi:10.1002/bit.25361
Kim, Y.-M., Shimizu, R., Nakai, H., Mori, H., Okuyama, M., Kang, M.-S., et al. (2011). Truncation of N- and C-Terminal Regions of Streptococcus Mutans Dextranase Enhances Catalytic Activity. Appl. Microbiol. Biotechnol. 91 (2), 329–339. doi:10.1007/s00253-011-3201-y
Knowles, S. P., Printz, M. A., Kang, D. W., LaBarre, M. J., and Tannenbaum, R. P. (2021). Safety of Recombinant Human Hyaluronidase PH20 for Subcutaneous Drug Delivery. Expert Opin. Drug Deliv. 18 (11), 1673–1685. doi:10.1080/17425247.2021.1981286
Locke, K. W., Maneval, D. C., and LaBarre, M. J. (2019). Correction. Drug Deliv. 26 (1), 1300. doi:10.1080/10717544.2019.1594569
Moua, P. S., Gonzalez, A., Oshiro, K. T., Tam, V., Li, Z. H., Chang, J., et al. (2016). Differential Secretion Pathways of Proteins Fused to the Escherichia coli Maltose Binding Protein (MBP) in Pichia pastoris. Protein Expr. Purif. 124, 1–9. doi:10.1016/j.pep.2016.04.005
Oettl, M., Hoechstetter, J., Asen, I., Bernhardt, G., and Buschauer, A. (2003). Comparative Characterization of Bovine Testicular Hyaluronidase and a Hyaluronate Lyase from Streptococcus Agalactiae in Pharmaceutical Preparations. Eur. J. Pharm. Sci. 18 (3-4), 267–277. doi:10.1016/s0928-0987(03)00022-8
Rigden, D. J., and Jedrzejas, M. J. (2003). Structures of Streptococcus Pneumoniae Hyaluronate Lyase in Complex with Chondroitin and Chondroitin Sulfate Disaccharides. J. Biol. Chem. 278 (50), 50596–50606. doi:10.1074/jbc.M307596200
Takagaki, K., Nakamura, T., Izumi, J., Saitoh, H., Endo, M., Kojima, K., et al. (1994). Characterization of Hydrolysis and Transglycosylation by Testicular Hyaluronidase Using Ion-spray Mass Spectrometry. Biochemistry 33 (21), 6503–6507. doi:10.1021/bi00187a017
Tan, N. H., and Ponnudurai, G. (1992). Comparative Study of the Enzymatic, Hemorrhagic, Procoagulant and Anticoagulant Activities of Some Animal Venoms. Comp. Biochem. Physiol. C Comp. Pharmacol. Toxicol. 103 (2), 299–302. doi:10.1016/0742-8413(92)90011-U
Usmani, S. Z., Nahi, H., Mateos, M.-V., van de Donk, N. W. C. J., Chari, A., Kaufman, J. L., et al. (2019). Subcutaneous Delivery of Daratumumab in Relapsed or Refractory Multiple Myeloma. Blood 134 (8), 668–677. doi:10.1182/blood.2019000667
Wang, X., Chen, Y., Nie, Y., and Xu, Y. (2019). Improvement of Extracellular Secretion Efficiency of Bacillus Naganoensis Pullulanase from Recombinant Escherichia coli: Peptide Fusion and Cell wall Modification. Protein Expr. Purif. 155, 72–77. doi:10.1016/j.pep.2018.09.011
Yang, P.-F., and Lee, C.-K. (2006). Purification of Recombinant Hyaluronan Lyase of Streptococcus Pyogenes Bacteriophage H4489A Expressed in Escherichia coli and its Application for the Specific Determination of Hyaluronan Concentration. Carbohydr. Polym. 65 (2), 159–164. doi:10.1016/j.carbpol.2005.12.037
Keywords: Pichia pastoris, human hyaluronidase, PH20, domain truncation, heterologous expression, hydrolysis pattern
Citation: Pang B, He J, Zhang W, Huang H, Wang Y, Wang M, Du G and Kang Z (2022) Active Expression of Human Hyaluronidase PH20 and Characterization of Its Hydrolysis Pattern. Front. Bioeng. Biotechnol. 10:885888. doi: 10.3389/fbioe.2022.885888
Received: 28 February 2022; Accepted: 12 April 2022;
Published: 13 May 2022.
Edited by:
Hao Song, Tianjin University, ChinaReviewed by:
Hui Li, Nanjing Tech University, ChinaSenthilkumar Sivaprakasam, Indian Institute of Technology Guwahati, India
Copyright © 2022 Pang, He, Zhang, Huang, Wang, Wang, Du and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Kang, emthbmdAamlhbmduYW4uZWR1LmNu
 Bo Pang
Bo Pang Jing He2,3
Jing He2,3 Yang Wang
Yang Wang Guocheng Du
Guocheng Du Zhen Kang
Zhen Kang