- 1College of Food Engineering and Biotechnology, Hanshan Normal University, Chaozhou, China
- 2Shenzhen Key Laboratory of Marine Bioresource and Eco-Environmental Science, Shenzhen Engineering Laboratory for Marine Algal Biotechnology, Guangdong Provincial Key Laboratory for Plant Epigenetics, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 3Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province, College of Optoelectronic Engineering, Shenzhen University, Shenzhen, China
- 4Shenzhen-Hong Kong Institute of Brain Science, Shenzhen, China
Euglena is a genus of single-celled eukaryotes that show both plant- and animal-like characteristics. Euglena gracilis, a model species, is of great academic interest for studying endosymbiosis and chloroplast development. As an industrial species, E. gracilis is also of primary biotechnological and economic importance as high value-added food, medicine, and cosmetic and high-quality feedstock for jet-fuel production because of its cells containing many high-value products, such as vitamins, amino acids, pigments, unsaturated fatty acids, and carbohydrate paramylon, as metabolites. For more than half a century, E. gracilis has been used as an industrial biotechnology platform for fundamental biology research, mainly exploring relevant physiological and biochemical method studies. Although many researchers focused on genetic engineering tools for E. gracilis in recent years, little progress has been achieved because of the lack of high-quality genome information and efficient techniques for genetic operation. This article reviewed the progress of the genetic transformation of E. gracilis, including methods for the delivery of exogenous materials and other advanced biotechnological tools for E. gracilis, such as CRISPR and RNA interference. We hope to provide a reference to improve the research in functional genomics and synthetic biology of Euglena.
Introduction
Euglena gracilis (E. gracilis) is a single-celled eukaryotic alga without a cell wall but has flagella on the top of the cell. It majorly lives in freshwater, is distributed worldwide, and could bloom in ponds, rivers, wastewater, etc. E. gracilis exhibits both animal and plant features with various modes of nutrition, including autotrophy, heterotrophy, and mixotrophy (Torihara and Kishimoto, 2015). As a model microorganism, E. gracilis has been studied extensively in many aspects, such as the development of secondary endosymbiotic chloroplasts, bioremediation of environmental pollutions, and decreasing carbon dioxide emissions, especially the improved harvest of high-value products that people in the market favor (Klinthong et al., 2015; Gissibl et al., 2019; Tahira et al., 2019). E. gracilis cells contain linear polysaccharide paramylon, wax esters, vitamins, and amino acids and have immense commercial importance (Gissibl et al., 2019). For example, paramylon, with a specific molecular structure (β-1, 3-glucan), can be used as a functional food ingredient. The content of paramylon in E. gracilis is much higher than that in other species such as fungi. Yasuda et al. (2018) reported that the maximum dry weight of the cell in E. gracilis can reach up to 70%. E. gracilis can not only be valid for the production of nutraceuticals and cosmeceuticals but also be used in biofuels. The glucan crystals produced from E. gracilis can be transformed into wax esters under anerobic conditions. They have relatively lower freezing points and are appropriate for being a feedstock of carbon chemical- origin used for biofuel (Hiroshi et al., 2017). Moreover, paramylon can be processed into plastics produced with succinic and lactic acids, the ideal bio-ingredients from E. gracilis cells under anerobic conditions. This is an environmentally friendly application of E. gracilis (Tomita et al., 2016).
Presently, the genetic development of E. gracilis for metabolic engineering is limited. Researchers developed traditional strain improvement by transferring DNA vectors for overexpression of genes that encoded the desired proteins. Its performance did not seem ideal based on the historical development of the subject, which could be attributed to both transformation efficiency and lack of complete genome information of E. gracilis (Doron et al., 2016). In 2019, Ebenezer et al. (2019) had published the draft nuclear genome of E. gracilis with an estimated haploid genome size of about 500 Mb, and a total of 1,266,288 contigs were assembled with a total length of 1.4 Gb, including 1,459 contigs with a length longer than 10 Kb. The longest contig was 16 Kb, and the N50 was only 955 bp. Compared with the previous estimation genome size of E. gracilis (1–4 Gb), the new version of the genome size was smaller, and the data can only be used for fundamental analysis. Even the prediction of complete ORF (open reading frame) information about E. gracilis was not easy to be provided (Ebenezer et al., 2017). The short reads make it tough to build a complete genome. Other problems such as gene duplication, repetitive sequences, and high complexity of its genome lead to failure in obtaining the full potential of E. gracilis in synthesis biology.
To date, microalgae have a remarkable ability to synthesize many value-added natural products that are becoming more significant for academics or industries globally. E. gracilis, as a model species, is one of the potential selected microalgae among other industrial Chlamydomonas reinhardtii, Chlorella sp., and Haematococcus pluvialis species , under the increasingly fierce competition with the development of synthetic biology. However, with the genetic studies that focused on analyzing metabolic pathways for the efficient synthesis of high-value products of interest, it is imperative to develop proven bioengineering tools for E. gracilis as an industrial biotechnology platform. Thus, we reviewed the progress of the genetic transformation of E. gracilis, such as CRISPR and RNA interference, and new ideas for delivery methods of exogenous materials, such as single-cell microinjection and electroporation for algae.
Harnessing Bioengineering Tools for the Development of E. gracilis
The Progress of the Genetic Transformation of E. gracilis
Studies related to the transformation of the nuclear genome of E. gracilis were insufficient. The earlier reports of the genetic transformation of E. gracilis were focused on the chloroplast genome. Biolistic bombardments coupled with selection biomarkers (i.e., streptomycin, spectinomycin, and neomycin phosphotransferase II) have been used for plastid transformation (Doetsch et al., 2001; Ogawa et al., 2015b). Krajcovic et al. (2011) first reported the successful nuclear genome transformation of E. gracilis with a zeocin resistance transformation cassette by electroporation. The Ble gene was cloned into the gene of LHCP (light-harvesting chlorophyll a/b binding protein of photosystem) II between the 5′- and 3′- terminal of it so that it could be driven and expressed by the LHCPII promoter. The mutants were detected with Ble gene-positive by PCR screening and can be stable for more than a year. Khatiwada et al. (2019) explored the genetic engineering tools to obtain stable E. gracilis nuclear transformants via constructing plasmid vectors with the selection marker of hygromycin by Agrobacterium-mediated transformation, biolistic bombardment, and electroporation techniques. The results showed that only Agrobacterium-mediated transformation could produce stable nuclear transformants while the others lost their property after repeated rounds of cultivation. Recently, another Agrobacterium-mediated nuclear transformation of E. gracilis has been successfully conducted with the selection markers of hygromycin and zeocin. The transformation efficiency was up to 8.26 ± 4.9% after the optimization of co-cultivation parameters (Becker et al., 2021). Although successful nuclear transformation reports were in a small number, those cases showed bright prospects in bioengineering. They provided the genetic tools to produce stable nuclear transformation for E. gracilis.
Antibiotic Resistance and Selection Markers of E. gracilis
Antibiotic resistance is critical for transformant screening and selection. However, there is limited study on the sensitivity of Euglena to antibiotics. In 2019, Khatiwada et al. introduced vectors with the hygromycin resistance gene hptⅡ (encoding hygromycin phosphotransferase Ⅱ) into E. gracilis by electroporation, Agrobacterium-mediated transformation, and biolistic bombardment. In 2021, Becker et al. explored the sensitivity of E. gracilis to a range of antibiotics in liquids and agar plates with different concentrations. The results showed that these antibiotics caused a 55–70% reduction in the growth rate at a concentration of 10 μg/ml of hygromycin and 15 μg/ml of zeocin in liquid culture. The growth was completely inhibited when the concentration was more than 30 μg/ml of hygromycin and zeocin. Under the tested concentrations, the antibiotic kanamycin did not affect E. gracilis growth (0, 20, 50, 100, or 200 μg/ml). They also assessed the susceptibility of E. gracilis on agar plates with varying concentrations of antibiotics. The lethal concentration of the antibiotics hygromycin and zeocin was 30 μg/ml. No colonies were observed on agar plates more than 30 μg/ml. Those selected markers mentioned previously could facilitate the screening and selection of E. gracilis.
The Delivery Methods of Exogenous Materials for E. gracilis
Several exogenous materials, such as DNA, RNA, and RNPs (ribonucleoproteins), are the standard delivery objects for bioengineering. An excellent review on the delivery mode of E. gracilis, including protoplast transformation, electroporation, biolistic bombardment, and Agrobacterium-mediated transformation, has been summarized elsewhere and will not be dwelt on here (Khatiwada et al., 2020). It is proposed that single-cell microinjection and electroporation could be the efficient molecule delivery ways for E. gracilis.
Microinjection is a physical method that uses a glass capillary needle to deliver a small volume of substances such as plasmids and proteins into cells. The materials can be accurately delivered to specific locations of cells, such as the cytoplasm and nucleus. Under the microscope, the manipulated cells can be observed all through the injection procedure, which is helpful for the real-time tracking of injected materials. The microinjection method has been successfully used in mice, zebrafish, rabbits, and much larger livestock such as cattle, sheep, pigs, and other animals as a result of its high efficiency and low cell death rates (Gordon et al., 1980; Hammer et al., 1985; Yuan and Sun, 2009). However, it is rarely conducted in microalgae cells. In 1986, Neuhaus et al. (1986) delivered SV40 DNA and pSV2neo into the nuclei of Acetabularia sp. by using the microinjection technique. This single-cell alga could grow vertically to a length of 5 cm. The injected nuclei were implanted into anucleate cell fragments of the same species. The treated cells not only survived but also formed progeny. In 1994, Meindl et al. (1994) had injected phalloidin into the cells of the green alga Micrasterias denticulata (the cell size is larger than 100 μm) and observed two types of actin filament systems in the cells, indicating that actin plays a key role in the cell morphogenetic process. However, it is difficult to perform microalgae injection when the size of the cells is less than 20 μm, such as the cells of E. gracilis in sphere form. In 1978, Nichols and Rikmenspoel (1978) injected EDTA, EGTA, Zn2+, and Mn2+ into E. gracilis and C. reinhardtii, respectively, and explored the relationship between microalgae flagellar movement and divalent cations. Flagellar motility stopped when E. gracilis and C. reinhardtii were injected with 7 × 10−14 and 2 × 10−14 L 0.02 M EDTA, respectively. Since then, no delivery of active molecules was reported by microinjection into Euglena and the other microalgae with the cell size less than 100 μm.
Single-cell electroporation has been developed based on the single-cell analysis technique, allowing the delivery of ions or molecules into the cell. As we know, millions of cells in a bath mode are analyzed with a low efficiency rate by conventional electroporation. Recently, developed nanofabricated electrodes were able to achieve single-cell electroporation and deliver different types of molecules with high transfection efficiency and high cell survival rate compared to the conventional one (Santra and Tseng, 2016). A modified single-cell electroporation method has been developed for molecular delivery into E. gracilis cells (Ohmachi et al., 2016). A variety of molecules, including GFP, Alexa Fluor 488, and exciton-controlled hybridization-sensitive fluorescent oligonucleotide (ECHO) RNA probes have been successfully introduced into living E. gracilis cells. This new method realized high transfection efficiency and viability rate after electroporation. Thus, single-cell electroporation techniques of E. gracilis will open up a new window for bioengineering manipulation at the single-cell level.
Gene Editing of E. gracilis Meditated by CRISPR-Cas9
In 2013, Zhang’s and Church’s team used the type II system (CRISPR/Cas9) derived from Streptococcus to successfully perform genome editing in mammalian cells (Cong et al., 2013; Mali et al., 2013). Since then, CRISPR/Cas9-based genome editing technology has been extensively researched and applied, and many Cas9-based application tools have been developed.
CRISPR/Cas9 is a system based on the endonuclease activity of Cas9 protein and the specific localization function of gRNA for the precise manipulation of foreign genomes. It can be used in many genetic engineering organisms, including animal cells, plant cells, and microalgae cells. The researchers introduced RNPs into C. reinhardtii cells by electroporation and successfully obtained CRISPR-/Cas9-induced mutations at the targeted sites of MAA7, CpSRP43, and ChlM genes (Shin et al., 2016). Compared with the first reported mutations induced by transgenic Cas9, the efficiency of delivered Cas9 RNPs has increased by 100 times, significantly improving gene knockout efficiency (Jiang et al., 2014). Researchers have successfully implemented CRISPR/Cas9 gene-editing technology in microalgae such as Nannochloropsis spp, Phaeodactylum tricornutum, and Thalassiosira pseudonana in the past 10 years (Hopes et al., 2016; Nymark et al., 2016; Wang et al., 2016). Recently, Nomura et al. (2019), Nomura et al. (2020) had successfully introduced Cas9 RNPs into E. gracilis through electroporation. Through a non-homologous end-repair mechanism, the mutation efficiency of the targeted gene EgGSL2 was as high as 90.1%. These two are the only research articles on the successful gene editing of E. gracilis using the CRISPR/Cas9 system, and no successful reports have followed yet.
Application of RNA Interference Technology in Euglena Cells
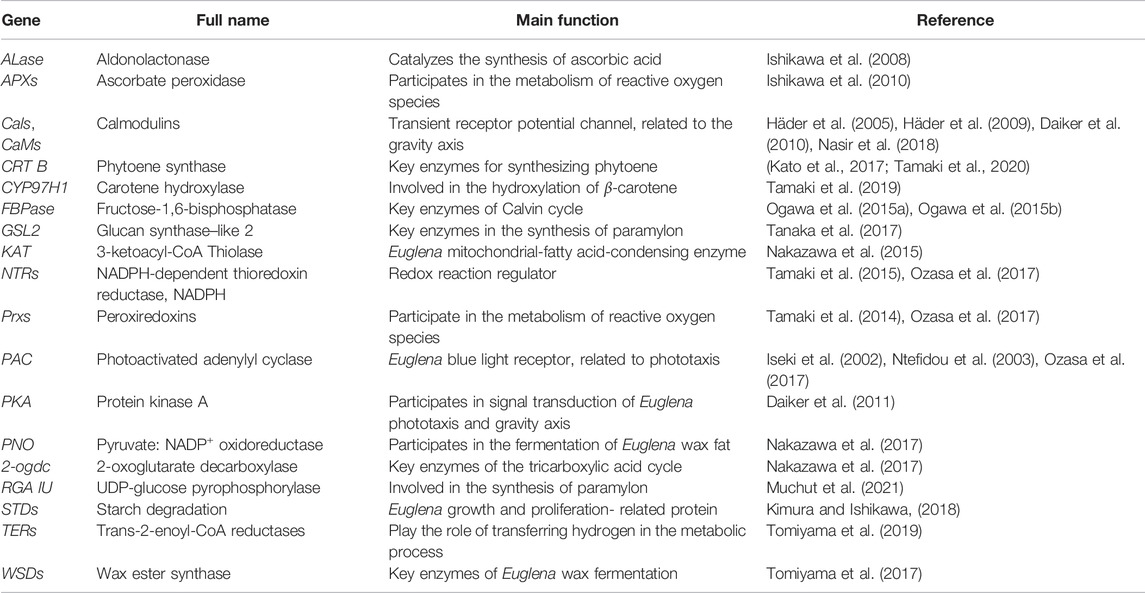
RNA interference (RNAi) is a phenomenon of the specific degradation of homologous mRNA induced by double-stranded RNA (dsRNA). It is a conservative self-defense mechanism to resist transgenes and foreign viruses in the evolutionary process. The homologous dsRNA of the target gene is artificially constructed and introduced into the cell to silence the expression of the target gene. In 2002, RNAi technology was used to study the function of a nuclear gene (blue light receptor-photosensitized adenylate cyclase, PAC) in E. gracilis, resulting in significant reduction of PAC content in the cells (Iseki et al., 2002). Subsequently, RNAi technology has been frequently used in the gene function verification of E. gracilis. For instance, RNAi was conducted to silence calmodulin-related genes to explore the influence of calmodulin-related genes on the gravity axis of E. gracilis and its movement mode (Häder et al., 2005; Daiker et al., 2010; Nasir et al., 2018). Nasir focused on the flagellin EgPCDUF4201 and its interaction with calmodulin CaM.2 (Nasir et al., 2018). In subsequent research, the focus shifted to the basic metabolic processes of E. gracilis, especially the metabolites such as wax and paramylons. Since 2014, more than ten researchers have used RNAi technology to explore a variety of basic metabolic processes, including photosynthetic pigments, redox processes, wax metabolism pathways, and paramylon metabolism processes, and search for the connection of genes and metabolic processes. So far, many studies about gene function have been carried out in E. gracilis by RNAi technology (Table 1). These genes are involved in E. gracilis’ primary metabolism, light response, and metabolite synthesis. The methods of introducing DNA and gene cloning used in these documents can be optimized to study the genetic transformation of E. gracilis.
Discussion
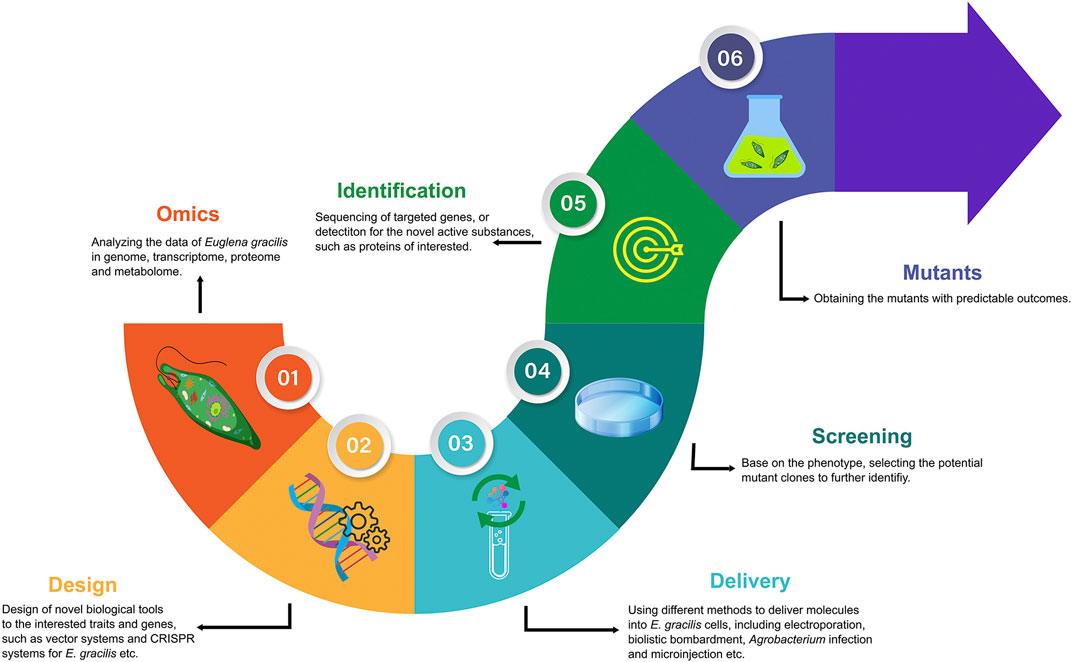
Presently, synthetic biology on microalgae has followed the workflow of analyzing the omics data, designing the transformation systems, delivering exogenous molecules, and screening, identification, and obtaining mutants with predictable outcome (Figure 1). The excavation of genetic information to assist metabolic engineering of E. gracilis is essential to improve the subsequent design of novel biological tools. The design of promoters, targeted sites of interested genes, transcription terminators and regulators, plasmids, etc., is mainly based on genome integrity and quality when performing bioengineering works. However, the number of nuclear transformation reports for E. gracilis is minimal because of lack of complete genome information. To some extent, the draft genome of E. gracilis has adequately promoted the promoters or other elements to develop. For the gene function identifying of E. gracilis, the tools of RNAi that can silence the gene function were recommended. Because the number of the relevant cases was much greater than those of the nuclear transformation, it means a high efficiency rate but will not cause permanent phenotype (Table 1). The CRISPR technique offers the opportunity to obtain mutants with predictable outcomes permanently. However, there are only two research articles on E. gracilis regarding CRISPR/Cas9 so far [Nomura et al. (2019), Nomura et al. (2020)]. The efficiency rate was as high as approximately 70–90% using electroporation without a knock-in selected marker gene, which was hardly reproducible by other teams, including ours, since 2020. The reasons may be difficulty in optimizing the parameters by electroporation or lack of the knock-in selected marker for mutant screening. For example, Shamoto et al. (2018) have obtained a 55% gene editing efficiency rate of fap70 in C. reinhardtii by using a knock-in paromomycin-resistance gene as a selected marker, which was much higher than that in the report of CRISPR/Cas9 in C. reinhardtii without selected markers by delivering RNPs using electroporation (Baek et al., 2016). Agrobacterium-mediated transformation may be the most efficient way among electroporation and biolistic bombardment for obtaining stable mutants in bulk cells of E. gracilis based on Khatiwada et al. (2019), and also can be supported by Becker et al. (2021). Nevertheless, with the rise of single-cell analysis technologies, microinjection and single-cell electroporation will be powerful delivery tools to introduce exogenous molecules into microalgae cells with high transformation efficiency and high cell viability. We conducted the CRISPR/Cas9 genome editing on E. gracilis and successfully knocked out the Crtp1 gene by microinjection with a high efficiency rate (16.7%) (Chen et al., 2021). However, the following problems still limit its wide use: expensive equipment, experienced operators, and time consumption. For screening, reducing the workload of screening was determined by designed steps at the beginning. A significant phenotype would be helpful in picking up the transformants conveniently and quickly. Finally, the sequencing of targeted genes can be used to identify mutants further, and other methods such as Western blot or HPLC (high-performance liquid chromatography) are the ideal means to analyze the quality and quantity for the designed active substances, such as the proteins, lipids, and pigments.
In summary, E. gracilis, with its special biological characters, has excellent potential for academic and economic values. However, stable nuclear transformation is still a problem today. We are happy to see the significant contributions of researchers from all over the world when the “1st Annual International Congress on Euglenoids” has been successfully held and closed in London in 2021, and we believe that the genetic conversion of Euglena has the following research directions: 1) to complete the genome sequencing of Euglena; 2) to elucidate the replication mechanism of DNA, internal and external DNA; 3) to find out high-expression promoters, codon optimization, and intron embedding; 4) to construct more efficient Euglena selection biomarkers; and 5) to develop CRISPR-based genome editing systems for RNAi and other genetic engineering tools that Euglena has not reported.
Recently, gene engineering targeted genes mainly focusing on commercial bioproducts, with single gene/enzyme manipulations: 1) synthesis or degradation of bioactive compounds and high valued-added ingredients such as paramylon, wax, fatty acids, and carotenoids; 2) reactive oxygen species scavengers; and 3) receptors related to phototaxis and gravity axis. In the near future, E. gracilis might become a model synthetic microbial chassis for exploring photosynthesis; retrograde pathways as the essential communication between the nucleus and the two DNA-containing organelles, synthetic chloroplast; flagellar elongation and shortening; and epigenetic components and mechanisms, including DNA methylation, histone modifications, and microRNAs. Thus, with the improvement of bioengineering tools, the synthetic biology research of Euglena will achieve enormous progress, and genetic engineering transformation will significantly promote the industrial application and scientific progress of these unique protists.
Author Contributions
ZC and JW conceived and wrote the review. JZ, MD and ZC helped collect materials and finish the manuscript. AL, JW, QL and HZ offered help to this project and/or revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was partially supported by China’s National Key R&D Programs (2018YFA0902500; 2020YFA0908703; 2021YFA0910800) and the National Natural Science Foundation of China (41876188).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Baek, K., Kim, D. H., Jeong, J., Sim, S. J., Melis, A., Kim, J.-S., et al. (2016). DNA-free Two-Gene Knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 Ribonucleoproteins. Sci. Rep. 6 (1), 1–7. doi:10.1038/srep30620
Becker, I., Prasad, B., Ntefidou, M., Daiker, V., Richter, P., and Lebert, M. (2021). Agrobacterium Tumefaciens-Mediated Nuclear Transformation of a Biotechnologically Important Microalga-Euglena Gracilis. Ijms 22 (12), 6299. doi:10.3390/ijms22126299
Chen, Z., Zhu, J., Chen, Z., Li, Q., Zhu, H., Lei, A., et al. (2021). “A High-Efficient CRISPR/Cas9 Mediated a Successful Genome Editing on Euglena gracilis CrtP1 Gene by Microinjection,” in Program of Event 1st Annual International Congress on Euglenoids (London. (Abstract) https://drive.google.com/drive/folders/1RQRcqUpUedGxsXDmjKNe0Wa5dVANYkij.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819–823. doi:10.1126/science.1231143
Daiker, V., Häder, D.-P., Richter, P. R., and Lebert, M. (2011). The Involvement of a Protein Kinase in Phototaxis and Gravitaxis of Euglena gracilis. Planta 233 (5), 1055–1062. doi:10.1007/s00425-011-1364-5
Daiker, V., Lebert, M., Richter, P., and Häder, D.-P. (2010). Molecular Characterization of a Calmodulin Involved in the Signal Transduction Chain of Gravitaxis in Euglena gracilis. Planta 231 (5), 1229–1236. doi:10.1007/s00425-010-1126-9
Doetsch, N. A., Favreau, M. R., Kuscuoglu, N., Thompson, M. D., and Hallick, R. B. (2001). Chloroplast Transformation in Euglena gracilis : Splicing of a Group III Twintron Transcribed from a Transgenic psbK Operon. Curr. Genet. 39 (1), 49–60. doi:10.1007/s002940000174
Doron, L., Segal, N. a., and Shapira, M. (2016). Transgene Expression in Microalgae-From Tools to Applications. Front. Plant Sci. 7, 505. doi:10.3389/fpls.2016.00505
Ebenezer, T. E., Carrington, M., Lebert, M., Kelly, S., and Field, M. C. (2017). “Euglena gracilis Genome and Transcriptome: Organelles, Nuclear Genome Assembly Strategies and Initial Features,” in Euglena: Biochemistry, Cell and Molecular Biology. Editors S. D. Schwartzbach, and S. Shigeoka (Switzerland: Springer), 125–140. doi:10.1007/978-3-319-54910-1_7
Ebenezer, T. E., Zoltner, M., Burrell, A., Nenarokova, A., Novák Vanclová, A. M. G., Prasad, B., et al. (2019). Transcriptome, Proteome and Draft Genome of Euglena gracilis. BMC Biol. 17 (1), 1–23. doi:10.1186/s12915-019-0626-8
Gissibl, A., Sun, A., Care, A., Nevalainen, H., and Sunna, A. (2019). Bioproducts from Euglena gracilis: Synthesis and Applications. Front. Bioeng. Biotechnol. 7, 108. doi:10.3389/fbioe.2019.00108
Gordon, J. W., Scangos, G. A., Plotkin, D. J., Barbosa, J. A., and Ruddle, F. H. (1980). Genetic Transformation of Mouse Embryos by Microinjection of Purified DNA. Proc. Natl. Acad. Sci. U.S.A. 77 (12), 7380–7384. doi:10.1073/pnas.77.12.7380
Häder, D.-P., Richter, P., Ntefidou, M., and Lebert, M. (2005). Gravitational Sensory Transduction Chain in Flagellates. Adv. Space Res. 36 (7), 1182–1188. doi:10.1016/j.asr.2005.03.081
Häder, D.-P., Richter, P. R., Schuster, M., Daiker, V., and Lebert, M. (2009). Molecular Analysis of the Graviperception Signal Transduction in the Flagellate Euglena gracilis: Involvement of a Transient Receptor Potential-like Channel and a Calmodulin. Adv. Space Res. 43 (8), 1179–1184. doi:10.1016/j.asr.2009.01.029
Hammer, R. E., Pursel, V. G., Rexroad, C. E., Wall, R. J., Bolt, D. J., Ebert, K. M., et al. (1985). Production of Transgenic Rabbits, Sheep and Pigs by Microinjection. Nature 315 (6021), 680–683. doi:10.1038/315680a0
Hopes, A., Nekrasov, V., Kamoun, S., and Mock, T. (2016). Editing of the Urease Gene by CRISPR-Cas in the Diatom Thalassiosira pseudonana. Plant Methods 12 (1), 1–12. doi:10.1186/s13007-016-0148-0
Inui, H., Ishikawa, T., and Tamoi, M. (2017). “Wax Ester Fermentation and its Application for Biofuel Production,” in Euglena: Biochemistry, Cell and Molecular Biology. Editors S. D. Schwartzbach, and S. Shigeoka (Switzerland: Springer), 269–283. doi:10.1007/978-3-319-54910-1_13
Iseki, M., Matsunaga, S., Murakami, A., Ohno, K., Shiga, K., Yoshida, K., et al. (2002). A Blue-Light-Activated Adenylyl Cyclase Mediates Photoavoidance in Euglena gracilis. Nature 415 (6875), 1047–1051. doi:10.1038/4151047a
Ishikawa, T., Nishikawa, H., Gao, Y., Sawa, Y., Shibata, H., Yabuta, Y., et al. (2008). The Pathway via D-Galacturonate/L-Galactonate Is Significant for Ascorbate Biosynthesis in Euglena gracilis. J. Biol. Chem. 283 (45), 31133–31141. doi:10.1074/jbc.M803930200
Ishikawa, T., Tajima, N., Nishikawa, H., Gao, Y., Rapolu, M., Shibata, H., et al. (2010). Euglena gracilis Ascorbate Peroxidase Forms an Intramolecular Dimeric Structure: its Unique Molecular Characterization. Biochem. J. 426 (2), 125–134. doi:10.1042/bj20091406
Jiang, W., Brueggeman, A. J., Horken, K. M., Plucinak, T. M., and Weeks, D. P. (2014). Successful Transient Expression of Cas9 and Single Guide RNA Genes in Chlamydomonas reinhardtii. Eukaryot. Cel. 13 (11), 1465–1469. doi:10.1128/EC.00213-14
Kato, S., Soshino, M., Takaichi, S., Ishikawa, T., Nagata, N., Asahina, M., et al. (2017). Suppression of the Phytoene Synthase Gene (EgcrtB) Alters Carotenoid Content and Intracellular Structure of Euglena gracilis. BMC Plant Biol. 17 (1), 125. doi:10.1186/s12870-017-1066-7
Khatiwada, B., Kautto, L., Sunna, A., Sun, A., and Nevalainen, H. (2019). Nuclear Transformation of the Versatile Microalga Euglena gracilis. Algal. Research 37, 178–185. doi:10.1016/j.algal.2018.11.022
Khatiwada, B., Sunna, A., and Nevalainen, H. (2020). Molecular Tools and Applications of Euglena gracilis: From Biorefineries to Bioremediation. Biotechnol. Bioeng. 117 (12), 3952–3967. doi:10.1002/bit.27516
Kimura, M., and Ishikawa, T. (2018). Suppression of DYRK Ortholog Expression Affects Wax Ester Fermentation in Euglena gracilis. J. Appl. Phycol. 30 (1), 367–373. doi:10.1007/s10811-017-1235-y
Klinthong, W., Yang, Y.-H., Huang, C.-H., and Tan, C.-S. (2015). A Review: Microalgae and Their Applications in CO2 Capture and Renewable Energy. Aerosol Air Qual. Res. 15 (2), 712–742. doi:10.4209/aaqr.2014.11.0299
Krajcovic, J., Vejerla, V. K., Vacula, R., Dobáková, E., Gavurníková, G., and Schwartzbach, S. D. (2011). Development of an Effective Transformation System for the Nuclear Genome of the Flagellate Euglena gracilis. Curr. Opin. Biotechnol. 22 (22), S45. doi:10.1016/j.copbio.2011.05.115
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided Human Genome Engineering via Cas9. Science 339 (6121), 823–826. doi:10.1126/science.1232033
Meindl, U., Zhang, D., and Hepler, P. K. (1994). Actin Microfilaments Are Associated with the Migrating Nucleus and the Cell Cortex in the Green Alga Micrasterias. Studies on Living Cells. J. Cel Sci. 107 (7), 1929–1934. doi:10.1242/jcs.107.7.1929
Muchut, R. J., Calloni, R. D., Arias, D. G., Arce, A. L., Iglesias, A. A., and Guerrero, S. A. (2021). Elucidating Carbohydrate Metabolism in Euglena gracilis: Reverse Genetics-Based Evaluation of Genes Coding for Enzymes Linked to Paramylon Accumulation. Biochimie 184, 125–131. doi:10.1016/j.biochi.2021.02.016
Nakazawa, M., Andoh, H., Koyama, K., Watanabe, Y., Nakai, T., Ueda, M., et al. (2015). Alteration of Wax Ester Content and Composition in Euglena gracilis with Gene Silencing of 3-Ketoacyl-CoA Thiolase Isozymes. Lipids 50 (5), 483–492. doi:10.1007/s11745-015-4010-3
Nakazawa, M., Hayashi, R., Takenaka, S., Inui, H., Ishikawa, T., Ueda, M., et al. (2017). Physiological Functions of pyruvate:NADP+ Oxidoreductase and 2-oxoglutarate Decarboxylase in Euglena gracilis under Aerobic and Anaerobic Conditions. Biosci. Biotechnol. Biochem. 81 (7), 1386–1393. doi:10.1080/09168451.2017.1318696
Nasir, A., Le Bail, A., Daiker, V., Klima, J., Richter, P., and Lebert, M. (2018). Identification of a Flagellar Protein Implicated in the Gravitaxis in the Flagellate Euglena gracilis. Sci. Rep. 8 (1), 7605. doi:10.1038/s41598-018-26046-8
Neuhaus, G., Neuhaus-Url, G., de Groot, E. J., and Schweiger, H.-G. (1986). High Yield and Stable Transformation of the Unicellular Green Alga Acetabularia by Microinjection of SV40 DNA and pSV2neo. EMBO J. 5 (7), 1437–1444. doi:10.1002/j.1460-2075.1986.tb04380.x
Nichols, K. M., and Rikmenspoel, R. (1978). Control of Flagellar Motility in Euglena and Chlamydomonas. Exp. Cel Res. 116 (2), 333–340. doi:10.1016/0014-4827(78)90456-1
Nomura, T., Inoue, K., Uehara‐Yamaguchi, Y., Yamada, K., Iwata, O., Suzuki, K., et al. (2019). Highly Efficient Transgene‐free Targeted Mutagenesis and Single‐stranded Oligodeoxynucleotide‐mediated Precise Knock‐in in the Industrial Microalga Euglena gracilis Using Cas9 Ribonucleoproteins. Plant Biotechnol. J. 17 (11), 2032–2034. doi:10.1111/pbi.13174
Nomura, T., Yoshikawa, M., Suzuki, K., and Mochida, K. (2020). Highly Efficient CRISPR-Associated Protein 9 Ribonucleoprotein-Based Genome Editing in Euglena gracilis. STAR Protoc. 1 (1), 100023. doi:10.1016/j.xpro.2020.100023
Ntefidou, M., Iseki, M., Watanabe, M., Lebert, M., and Häder, D.-P. (2003). Photoactivated Adenylyl Cyclase Controls Phototaxis in the Flagellate Euglena gracilis. Plant Physiol. 133 (4), 1517–1521. doi:10.1104/pp.103.034223
Nymark, M., Sharma, A. K., Sparstad, T., Bones, A. M., and Winge, P. (2016). A CRISPR/Cas9 System Adapted for Gene Editing in Marine Algae. Sci. Rep. 6 (1), 1–6. doi:10.1038/srep24951
Ogawa, T., Kimura, A., Sakuyama, H., Tamoi, M., Ishikawa, T., and Shigeoka, S. (2015a). Characterization and Physiological Role of Two Types of Chloroplastic Fructose-1,6-Bisphosphatases in Euglena gracilis. Arch. Biochem. Biophys. 575, 61–68. doi:10.1016/j.abb.2015.04.002
Ogawa, T., Kimura, A., Sakuyama, H., Tamoi, M., Ishikawa, T., and Shigeoka, S. (2015b). Identification and Characterization of Cytosolic Fructose-1,6-Bisphosphatase in Euglena gracilis. Biosci. Biotechnol. Biochem. 79 (12), 1957–1964. doi:10.1080/09168451.2015.1069694
Ohmachi, M., Fujiwara, Y., Muramatsu, S., Yamada, K., Iwata, O., Suzuki, K., et al. (2016). A Modified Single-Cell Electroporation Method for Molecule Delivery into a Motile Protist, Euglena gracilis. J. Microbiol. Methods 130, 106–111. doi:10.1016/j.mimet.2016.08.018
Ozasa, K., Won, J., Song, S., Tamaki, S., Ishikawa, T., and Maeda, M. (2017). Temporal Change of Photophobic Step-Up Responses of Euglena gracilis Investigated through Motion Analysis. PLoS One 12 (2), e0172813. doi:10.1371/journal.pone.0172813
Santra, T. S., and Tseng, F.-G. (2016). “Electroporation for Single-Cell Analysis,” in Essentials of Single-Cell Analysis (Springer), 55–83. doi:10.1007/978-3-662-49118-8_3
Shamoto, N., Narita, K., Kubo, T., Oda, T., and Takeda, S. (2018). CFAP70 Is a Novel Axoneme-Binding Protein that Localizes at the Base of the Outer Dynein Arm and Regulates Ciliary Motility. Cells 7 (9), 124. doi:10.3390/cells7090124
Shin, S.-E., Lim, J.-M., Koh, H. G., Kim, E. K., Kang, N. K., Jeon, S., et al. (2016). CRISPR/Cas9-induced Knockout and Knock-In Mutations in Chlamydomonas reinhardtii. Sci. Rep. 6 (1), 1–15. doi:10.1038/srep27810
Tahira, S., Khan, S., Samrana, S., Shahi, L., Ali, I., Murad, W., et al. (2019). Bio-assessment and Remediation of Arsenic (Arsenite As-III) in Water by Euglena gracilis. J. Appl. Phycol. 31 (1), 423–433. doi:10.1007/s10811-018-1593-0
Tamaki, S., Kato, S., Shinomura, T., Ishikawa, T., and Imaishi, H. (2019). Physiological Role of β-carotene Monohydroxylase (CYP97H1) in Carotenoid Biosynthesis in Euglena gracilis. Plant Sci. 278, 80–87. doi:10.1016/j.plantsci.2018.10.017
Tamaki, S., Maruta, T., Sawa, Y., Shigeoka, S., and Ishikawa, T. (2015). Biochemical and Physiological Analyses of NADPH-dependent Thioredoxin Reductase Isozymes in Euglena gracilis. Plant Sci. 236, 29–36. doi:10.1016/j.plantsci.2015.03.016
Tamaki, S., Maruta, T., Sawa, Y., Shigeoka, S., and Ishikawa, T. (2014). Identification and Functional Analysis of Peroxiredoxin Isoforms in Euglena gracilis. Biosci. Biotechnol. Biochem. 78 (4), 593–601. doi:10.1080/09168451.2014.89003710.1080/09168451.2014.890037
Tamaki, S., Tanno, Y., Kato, S., Ozasa, K., Wakazaki, M., Sato, M., et al. (2020). Carotenoid Accumulation in the Eyespot Apparatus Required for Phototaxis Is Independent of Chloroplast Development in Euglena gracilis. Plant Sci. 298, 110564. doi:10.1016/j.plantsci.2020.110564
Tanaka, Y., Ogawa, T., Maruta, T., Yoshida, Y., Arakawa, K., and Ishikawa, T. (2017). Glucan Synthase‐like 2 Is Indispensable for Paramylon Synthesis in Euglena gracilis. FEBS Lett. 591 (10), 1360–1370. doi:10.1002/1873-3468.12659
Tomita, Y., Yoshioka, K., Iijima, H., Nakashima, A., Iwata, O., Suzuki, K., et al. (2016). Succinate and Lactate Production from Euglena gracilis during Dark, Anaerobic Conditions. Front. Microbiol. 7, 2050. doi:10.3389/fmicb.2016.02050
Tomiyama, T., Goto, K., Tanaka, Y., Maruta, T., Ogawa, T., Sawa, Y., et al. (2019). A Major Isoform of Mitochondrial Trans-2-enoyl-CoA Reductase Is Dispensable for Wax Ester Production in Euglena gracilis under Anaerobic Conditions. PLoS One 14 (1), e0210755. doi:10.1371/journal.pone.0210755
Tomiyama, T., Kurihara, K., Ogawa, T., Maruta, T., Ogawa, T., Ohta, D., et al. (2017). Wax Ester Synthase/Diacylglycerol Acyltransferase Isoenzymes Play a Pivotal Role in Wax Ester Biosynthesis in Euglena gracilis. Sci. Rep. 7 (1), 13504. doi:10.1038/s41598-017-14077-6
Torihara, K., and Kishimoto, N. (2015). Evaluation of Growth Characteristics of Euglena gracilis for Microalgal Biomass Production Using Wastewater. J. Wat. Envir. Tech. 13 (3), 195–205. doi:10.2965/jwet.2015.195
Wang, Q., Lu, Y., Xin, Y., Wei, L., Huang, S., and Xu, J. (2016). Genome Editing of Model Oleaginous Microalgae Nannochloropsis Spp. By CRISPR/Cas9. Plant J. 88 (6), 1071–1081. doi:10.1111/tpj.13307
Yasuda, K., Ogushi, M., Nakashima, A., Nakano, Y., and Suzuki, K. (2018). Accelerated Wound Healing on the Skin Using a Film Dressing with β-Glucan Paramylon. In Vivo 32 (4), 799–805. doi:10.21873/invivo.11310
Keywords: Euglena gracilis, genetic transformation, biotechnology, CRISPR, RNAi
Citation: Chen Z, Zhu J, Du M, Chen Z, Liu Q, Zhu H, Lei A and Wang J (2022) A Synthetic Biology Perspective on the Bioengineering Tools for an Industrial Microalga: Euglena gracilis. Front. Bioeng. Biotechnol. 10:882391. doi: 10.3389/fbioe.2022.882391
Received: 23 February 2022; Accepted: 11 March 2022;
Published: 06 April 2022.
Edited by:
Xupeng Cao, Dalian Institute of Chemical Physics (CAS), ChinaReviewed by:
Zhengquan Gao, Shandong University of Technology, ChinaHongli Zheng, Nanchang University, China
Copyright © 2022 Chen, Zhu, Du, Chen, Liu, Zhu, Lei and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangxin Wang, anh3YW5nQHN6dS5lZHUuY24=
 Zhenfan Chen
Zhenfan Chen Jiayi Zhu
Jiayi Zhu Ming Du
Ming Du Zixi Chen
Zixi Chen Qiong Liu
Qiong Liu Hui Zhu1
Hui Zhu1 Anping Lei
Anping Lei Jiangxin Wang
Jiangxin Wang