- 1School of Life Science and Technology, Xinxiang Medical University, Xinxiang, China
- 2International Joint Research Laboratory for Recombinant Pharmaceutical Protein Expression System of Henan, Xinxiang, China
- 3Institutes of Health Central Plain, Xinxiang Medical University, Xinxiang, China
- 4Department of Biochemistry and Molecular Biology, Xinxiang Medical University, Xinxiang, China
Recombinant therapeutic proteins (RTPs) are important parts of biopharmaceuticals. Chinese hamster ovary cells (CHO) have become the main cell hosts for the production of most RTPs approved for marketing because of their high-density suspension growth characteristics, and similar human post-translational modification patterns et al. In recent years, many studies have been performed on CHO cell expression systems, and the yields and quality of recombinant protein expression have been greatly improved. However, the expression levels of some proteins are still low or even difficult-to express in CHO cells. It is urgent further to increase the yields and to express successfully the “difficult-to express” protein in CHO cells. The process of recombinant protein expression of is a complex, involving multiple steps such as transcription, translation, folding processing and secretion. In addition, the inherent characteristics of molecular will also affect the production of protein. Here, we reviewed the factors affecting the expression of recombinant protein and improvement strategies in CHO cells.
Introduction
In recent years, the demand for biopharmaceuticals has been increasing and innovative and improved bioprocessing technologies are essential to meet the demand for industrial production of products. As an important part of biological drugs, recombinant therapeutic proteins (RTPs), especially RTPs represented by recombinant antibodies, have achieved good efficacy and great economic benefits and have received more and more attention. Mammalian cells have become the main host cells for the production of RTPs because of their ability to perform post-translational modifications (PTMs) such as correct folding and glycosylation of recombinant proteins. Since the production of tissue type plasminogen activator (tPA) in Chinese hamster ovary (CHO) cells in 1986, CHO cells have become the production host of most approved recombinant proteins and recombinant antibody drugs due to their high density suspension growth, PTMs similar to humanized cells, easy gene operation and process scale-up. This expression system has been used to produce 84% of the antibodies approved during 2015–2018, accounting for more than half of all approved antibodies during this period (Walsh, 2018). By 2019, all six of the top ten best-selling drugs were produced in CHO cells (Urquhart, 2020). RTPs are the fastest growing part of the biopharmaceutical industry, with the US Food and Drug Administration (FDA) approving an average of more than 25 new approvals per year from 2014 to 2020. It is predicted that the global biopharmaceutical market value will reach $389 million by 2024 (O'Flaherty et al., 2020).
With the advancement of CHO cell line development and process optimization, the yields of recombinant mAbs (monoclonal antibodies, mAbs) of CHO cells have achieved as high as 5 g/L, or even more than 10 g/L (Alves and Dobrowsky, 2017; Handlogten et al., 2018). Although CHO cell platforms have been widely used to produce RTPs, there are still inherent limitations in the synthesis and secretion of many complex RTPs, such as low productivity, growth restriction and expression instability, low resistance to culture-related stresses and high costs of production. The reasons of those are that intracellular protein processing can be affected by many factors, such as cellular secretory capacity, protein aggregation or degradation, and protein folding (Kaneyoshi et al., 2019), which results in unsatisfactory expression or difficult-to-express of RTSs. It has become urgent challenges to express specific protein in specific cell line with good quantity and quality using strategies involving genetic modification, optimization of expression vector, cell line modification and process optimization.
Recombinant Protein Expression Process in Chinese Hamster Ovary Cell
From gene structure to correctly folded protein products, a series of procedure are involved, such as transcription, translation, PTM, protein folding, and secretion et al. The nucleotide sequence of its coding region determines the amino acid sequence of the corresponding protein. Transcription process can control protein production in CHO cell, and the stability of mRNA is directly related to the number of translation products mRNA produced by transcription is prone to degradation, so the degradation rate of mRNA directly affects the expression level of protein (Narula et al., 2019). Mature mRNAs need to be processed into the translation process by capping, splicing and polyadenylation, and can also affect the translation process with the structure of the composition of (eukaryotic initiation factors eIFs) and ribosomal proteins, and mRNAs can be degraded by co-translation. Studies have found that codon selection can affect the rate of ribosome elongation, which in turn affects the rate of mRNA degradation (Bicknell and Ricci, 2017), Others have found that mRNA levels and expression levels are not consistent (Reinhart et al., 2014). After transcription in the nucleus, mRNA is transported to the cytoplasm and recognized and translated by ribosomes (Baek et al., 2017), and peptides of secreted proteins are translocated into the ER in a co-translational manner, followed by protein folding and maturation in the ER lumen tube, transport of correctly folded proteins into the Golgi apparatus for PTMs, and finally secretion into the extracellular environment in the form of secretory vesicles to function. Unfolded or terminally misfolded proteins can be removed by ER-associated protein degradation (ERAD) (Ruggiano et al., 2014; Wu and Rapoport, 2018; Mathias et al., 2020). Accumulation of unfolded or misfolded proteins in the ER lumen beyond the transport capacity impairs ER function, triggering the unfolded protein response (UPR) ultimately leading to apoptosis and affecting yield (Hetz, 2012; Hetz and Papa, 2018; Mathias et al., 2020).
When proteins are synthesized on ribosomes, signal recognition particle (SRP) in the cytoplasm recognizes signal peptides that newly generate 5∼30 amino acids at the N-terminus of proteins and function to transport proteins into the correct subcellular compartment (Saraogi and Shan, 2011). Next, SRP transfers SRP and the signal recognition particle-ribosome nascent chain (SRP-RNC) combinatorial complex to receptors on the ER membrane, which is finally transferred to the lumen of the ER, and then the signal peptide is cleaved by signal peptidase. Therefore, the transport of protein to the endoplasmic reticulum is the rate-limiting step of the secretory pathway, and the signal peptide sequence can promote the intracellular transport of recombinant protein products to secretory organelles and promote expression. If the nascent polypeptide chain is inefficiently translocated to the endoplasmic reticulum, SRP complex function may lead to miscleavage of the signal peptide, resulting in inefficient assembly and low expression of the antibody light chain (Reinhart et al., 2014).
Polypeptide chains with complete primary structure can have normal biological activity only when they fold into the correct three-dimensional spatial structure. If the folding process is wrong and a misplaced spatial structure is formed, they will lose their biological function and may even cause diseases. Typically correctly folded proteins form small vesicles that leave the ER, which move toward the Golgi and fuse with its membrane. Transport inside the Golgi and transport of proteins from the Golgi to the plasma membrane are also mediated by vesicles. If the high concentration of heterologous protein exceeds the secretory capacity of CHO cells, it will lead to inefficient secretion, protein degradation, intracellular and extracellular aggregation, etc., which in turn affects the yield of protein (Delic et al., 2014). The inherent properties of other molecules may also contribute to the susceptibility of proteins to degradation, aggregation, and other adverse protein-protein interactions that may lead to cytotoxicity. The cells themselves may produce toxicity, one is intracellular toxicity, involved in cellular regulation, and the other is extracellular toxicity, involved in negative feedback regulation. Usually most polypeptide chains can fold correctly, but a small proportion of proteins are still difficult to fold correctly.
PTMs, usually in two ways, through proteolytic cleavage and covalent modification through amino acids, may lead to changes in protein characteristics and function (Spahr et al., 2018; Bryan et al., 2021). When the protein itself is improperly designed or the DNA coding sequence is inappropriate, it will lead to the lack of PTM (Unzueta et al., 2015), insufficient supply of chaperones, difficulty in the formation of interchain and intra-chain disulfide bonds, and ineffective vesicle transport (Delic et al., 2014) or other adverse forms of protein-protein interactions, which in turn will affect the expression of proteins and affect the yield and quality of recombinant proteins.
Overcoming Difficulty in Expressing Recombinant Protein in Chinese Hamster Ovary Cells and Improving Strategies
Although the technology of producing recombinant protein by CHO cells has been more mature and a large number of recombinant proteins have been successfully produced, there are still some proteins that are DTE and proteins with low expression levels. To solve this problem, the main strategies mainly include ar the increasing the cell viability, viable cell density, and cell specific production rate (Qp) through some molecular and cell methods, using genetically engineered cell lines.
Genetic Modification
Ribosomes have traditionally been considered deciphers of the 64-word genetic code translated into complex proteins, while ribosomes can also shape the transcriptome by controlling mRNA stability. In recombinant protein production, the expression rate, yield, and final product quality are affected by the codon bias of the relevant gene and the codon bias of the expression system (Quax et al., 2015). Although all codons affect the translation rate, some codons will be rapidly deciphered, while others will cause ribosome pausing due to the relative cognate tRNA concentration. Due to the species preference of codons, codon optimization can be used to overcome the difficulty in protein expression or improve the expression level of recombinant proteins in recombinant protein expression (Ang et al., 2016; Chung et al., 2013). It is generally accepted that codon bias can improve translation efficiency by modulating the elongation rate of the process (Quax et al., 2015). Heterologous expression of recombinant human interferon beta (rhIFN-β) gene in suspension-adapted Chinese hamster ovary (CHO-s) cells with optimized codons to increase the GC content at the third position of each codon increased the expression level by 2.8-fold (Shayesteh et al., 2020). Overexpression of transcription factors such as ZFP-TF, ATF4, or GADD34 significantly increased the yield of various cellular recombinant proteins compared with parental cells, up to 10-fold (Kwon et al., 2006; Omasa et al., 2008). Due to the low frequency of availability of certain transport RNA (tRNA) molecules, codons associated with them are slowly translated. Codons with higher tRNA abundance will therefore translate more accurately (Shah and Gilchrist, 2010). Another key protein that plays an important role in protein biosynthesis is mammalian target of rapamycin (mTOR), which enhances the ectopic expression of mTOR gene and can also significantly improve the cell growth (increase cell size and protein content), proliferation (increase cell cycle progression), viability (decrease apoptosis), robustness (decreased sensitivity to suboptimal growth factors and oxygen supply), and specific productivity of secreted human glycoproteins of recombinant CHO cells. Culturing an mTOR transgenic CHO-derived cell line for the secretion of therapeutic immunoglobulins with an antibody of 50 pg/cell/day increased fourfold compared to the parental production cell line (Dreesen and Fussenegger, 2011).
Gene sequence optimization can also adjust the GC content of genes, avoid base duplications, and eliminat restriction enzyme recognition sites, and also avoid similarity to important RNA motifs located in open reading frames that may interfere with mRNA processing and translational function, because they may cause ribosome pausing, in addition to factors such as CpG content and TATA boxes. Another 5′ untranslated region (5′ UTR) insertion into the RNA hairpin structure, the “regulatory element (RGE)”, also improves expression (Eisenhut et al., 2020). In addition to gene sequence optimization, sequence motifs such as complementarity determining region three influences the rate of LC-HC dimerization during MAb synthesis, results in product-specific difference of yield (Pybus et al., 2014).
Optimization of Expression Vector
In mammalian cells, a highly complex process controlled by multiple DNA signaling elements and corresponding trans-acting factors is called transcription. In this cascade, promoters play a key role in integration and transcriptional signal processing, therefore, selecting the correct promoter, optimizing the combination of the promoter with different regulatory elements, and preventing promoter methylation can improve the expression and stability of recombinant proteins (Kim et al., 2011; Ho et al., 2015). In CHO cells, human cytomegalovirus immediate-early enhancer/promoter (hCMVp), simian virus 40 early promoter (SV40E), CMV enhancer/chicken β-actin promoter (CAG), human elongation-1α promoter (HEF-1α) and Chinese hamster elongation factor-1α promoter (CHEF-1α) are commonly used to express GOI, and CMV promoter has the highest transgene expression level and the highest transfection efficiency, and SV40 promoter can maintain transgene expression more stably during long-term culture (Wang et al., 2016). Generally, selectable marker genes are driven by weak promoters (Nguyen et al., 2019), hypomethylation of DNA in promoter regions can improve gene transcriptional activity, and acetylation of acetylated histones (Dong et al., 2019) can also play a role in the transcriptional control of active genes.
Due to the structure of heavy chain (HC) and light chain (LC), the matching of HC and LC is need to be considered in the construction of expression vector. When free LC does not bind to HC, HC is misfolded, assembled and accumulated. LC peptides are very important for the efficient folding and assembly of Mabs. IgG antibody sequences with high aggregation/aggregation tendency are more likely to form Russell bodies in the ER lumen. LC and HC cannot be completely and correctly folded and assemble to form heterodimers in the endoplasmic reticulum and Golgi apparatus, which will cause inefficient assembly and affect antibody yield (Kaneyoshi et al., 2019) and overall Qp of cells (Ishii et al., 2014), so the LC expression level is high and the secretion rate is high. Compared with HC, LC excess is conducive to antibody production, and the ratio of HC to LC can be optimized by optimizing the number of transfected DNA or vector design. If a two-plasmid system is utilized, the HC and LC are located on different vectors and different plasmid ratios are used for transfection. In addition, single-plasmid systems can also be used to control HC to LC expression with different ratios using IRES (internal ribosome entry site, IRES) mediated vectors up to 1.15 g/L, double or tricistronic vectors (Li et al., 2020). The polycistronic vector contains IRES and Furin-2A (F2A), and the expression of multiple cistrons on a single vector is achieved by IRES and F2A (Ho et al., 2013) to optimize the expression of recombinant antibodies.
It has been found that random transgene insertions may be associated with position effects. Inefficient transgene expression is mainly due to transcriptional silencing, involving chromatin condensation, histone deacetylation and methylation at CpG sequences. By changing the structure of chromatin through specific chromosomal elements and maintaining chromatin in an open state, the transcription level of target genes can be increased. For example, ubiquitous chromatin opening element (UCOE) (Neville et al., 2017), as well as matrix attachment region (MAR), upstream chromatin opening element and locus control region, can prevent transgene silencing and are used to improve transgene expression (Li et al., 2018). MARs are DNA sequences that can interact with the nuclear matrix in eukaryotic genomes. Through the binding of MARs to the nuclear matrix, chromatin can form independent ring structures that regulate the transcription and expression of genes and reduce transgene silencing due to position effects. It has been found that MARs increase transgene expression levels in mammalian cell expression systems and reduce differences in expression levels between transgenic individuals, thereby inhibiting transgene silencing (Chang et al., 2014). MARs can also improve transgene expression levels in CHO cells, and there is a distance effect in MAR-regulated transgene expression (Zhang et al., 2014; Zhang et al., 2020).
Cell Line Modification
The entire “CHO cell system” includes a variety of different cell lines, which are likely to come from the first isolation of clone, naturally immortalized CHO in 1956 (Puck et al., 1958). The productivity of dihydrofolate reductase (DHFR) deficient CHO cell can increase with gene copy number amplification (Yeo et al., 2017), which is commonly used for RTPs production.
In general, the factors that affect product quality depend on the type of RTPs and the rCHO cell line used. In a mathematical modeling of recombinant Mab manufacture in seven GS-CHO cell lines, the improving of Mab production through cell line modification should be cell line specific (O'Callaghan et al., 2010). The difference in integration site of GOI might influence the production of Mab in the strategies of cell line modification. Efficient expression of the transgene during development of an rCHO cell line based on random integration of the transgene is highly dependent on the number of gene copies integrated into the genome as well as the location of the integration. The latter is greatly influenced by changes in position effects, which are influenced by local permeability of the site and proximity and interaction with local and distal enhancers. It has been shown that transgenes with high copy number are not always consistent with high cell productivity (Reisinger et al., 2008), but the number of integrated transgenes will affect the expression of certain cell lines (Chusainow et al., 2009). Integration sites in different structural genomes and regulatory genomes as well as vector regulatory elements and their structural rearrangements can lead to clonal variation (Grav et al., 2018). Site specific integration has been used for CHO cell line development and engineering (Igarashi and Kashiwagi, 2021), the site is important for the production of GOI.
It is crucial to randomly integrate the transgene into CHO host cells and select the appropriate cell line to produce the protein of interest (POI). Generally, the most commonly used methods to engineer cell lines include gene overexpression and gene knockout, and the former method is to introduce the GOI into the host cell to realize the overexpression of POI. Knockout is more specific to delete a gene from the genome by chemically and radiation-induced random mutation or gene editing directed mutation, or to shut down its function and target genetic engineering, which is superior to random mutation. In this regard, the most advanced technologies mainly include zinc finger nucleases (ZFNs), transcription activator-like effector nuclease transcription activator-like effector nuclease (TALENs) and regularly clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-related protein 9 (Cas9) systems (Dong et al., 2019; Carver et al., 2020), which are able to delete or introduce proteins in CHO cells while improving the productivity of recombinant proteins, which in turn promotes product quality (Grav et al., 2018). In addition, the production of rCHO cell lines is allowed by multi-copy number targeted integration to support the horizontal production of RTPs by their industrial production (Sergeeva and Lee, 2020) (Figure 1).
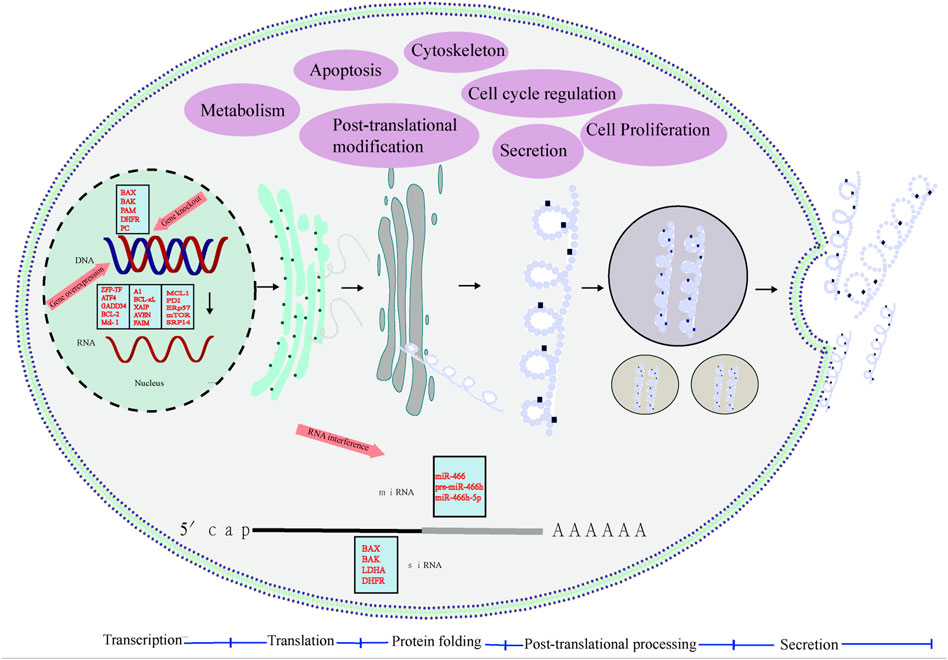
FIGURE 1. Process of protein synthesis. mRNA is transcribed in the nucleus, transported to the cytoplasm and translated by the ribosome, and then the protein binds to the rough endoplasmic reticulum, folding and processing before entering the Golgi apparatus. The soluble protein is PTMs and then secreted through the secretory pathway. Through cell engineering strategies such as anti-apoptotic engineering, metabolic engineering, cell cycle engineering, and protein PTMs engineering, overexpression of dominant genes, gene knockout, and siRNA interference inhibit unfavorable gene expression, transform mammalian cell lines, and improve cell lines Performance, effectively improve cell viability and increase the production of recombinant protein.
Mammalian cell lines are engineered by cell engineering strategies, such as anti-apoptotic engineering, metabolic engineering, cell cycle engineering, and protein PTM engineering, overexpression of dominant genes, gene knockout, and siRNA interference to inhibit unfavorable gene expression to improve the performance of cell lines, effectively improve cell viability, and improve the yield of recombinant proteins. In addition, non-coding RNAs technology can also improve and improve cell lines in growth, apoptosis inhibition, metabolism, yield, and the ability to express glycosylated recombinant proteins (Table 1), with great potential (Raab et al., 2021).
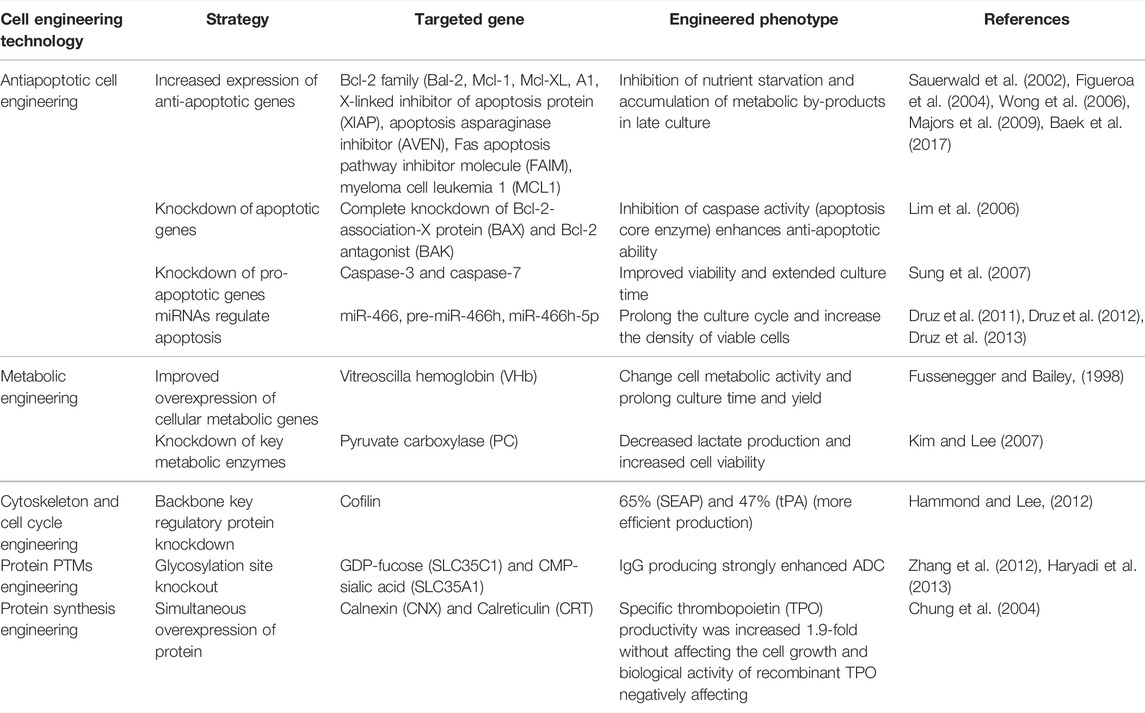
TABLE 1. Overcoming the difficulty in expressing recombinant protein in CHO cells and improving cell line modification strategies.
Cell apoptosis refers to the autonomous and orderly death of cells controlled by genes to maintain the stability of the internal environment, which can reduce apoptosis and prolong culture time during bioprocessing, thereby increasing volume product yield (Henry et al., 2020). Overexpression of anti-apoptotic genes can facilitate genetic engineering of CHO-producing cells. Apoptotic proteins include Bc1-2 family (Bcl-2, Mcl- 1, Bcl-XL, A1), X-linked inhibitor of apoptosis protein (XIAP), inhibitor of apoptosis asparaginase (AVEN), Fas apoptotic pathway inhibitor molecule (FAIM), myeloma cell leukemia 1 (MCL1), etc., which inhibit nutrient loss and metabolic byproduct accumulation in the late stage of culture, induce the occurrence of apoptosis and then prolong the culture time (Figueroa et al., 2004; Wong et al., 2006; Majors et al., 2009; Raab et al., 2021), and another strategy is to knock down bcl-2 associated X protein (BAX) and bcl-2 antagonist/killer (BAK) in weakly promote CHO cells and prolong the culture time of CHO cells in nutrient-poor or high-osmolarity media (Lim et al., 2006).
MicroRNAs are defined as small molecules of 22–25 nucleotides that can regulate approximately 100 mRNA targets involved in one or more regulatory networks simultaneously. Studies have demonstrated that up-regulation or down-regulation of microRNAs in host cells can change productivity levels by regulating cellular processes, such as apoptosis, cell cycle, histone methylation, and cell growth (Klanert et al., 2016; Muluhngwi et al., 2017; Amadi et al., 2020) and other non-coding RNAs technologies can also enable optimization of cell lines. Reduced expression levels of miR-23 in CHO cells can triple the specific expression level of secreted proteins from this cell without affecting cell growth (Kelly et al., 2015). In CHO cells, small interfering RNA (siRNA) is widely used for specific gene silencing to improve apoptosis resistance, glycosylation, metabolism, or specific productivity (Sarkar and Tran, 2021). Using siRNA pairs of caspase-3 (CASP3) and -7 (CASP7), both genes were silenced simultaneously, resulting in improved performance to 5 g/L in CHO cells treated with sodium butyrate (NaBu, a non-specific inhibitor of histone deacetylases (HDACs) (Sung et al., 2007). It has been found that miRNAs induce apoptosis in CHO cells in nutrient-poor medium, in which miR-466h induces apoptosis in CHO cells (Druz et al., 2011; Druz et al., 2012). Pro-apoptotic knockdown of pre-miR-466h in recombinant CHO-SEAP cells mediated by short hairpin RNA (shRNA) prolonged culture for more than 35 h and increased viable cell density by more than 40%. The above results demonstrate the potential of this novel approach to create more production cell lines by stably manipulating specific miRNA expression. Knockdown of anti-apoptotic genes (BCL2L2, DAD1, BIRC6, STAT5A, and SMO) by pre-miR-466h was up-regulated 2.1- to 12.5-fold, demonstrating that these genes are direct targets of miR-466h-5p in CHO cells (Druz et al., 2013) and directly affect protein yield.
Most metabolic engineering can reduce the accumulation of toxic by-products and improve protein production, which can regulate many biological process-related traits through CHO cell metabolic engineering, such as glycosylation (Richelle and Lewis, 2017). Changing of the metabolic activity of CHO by overexpressing specific genes can affect cell metabolism, and prolong the culture time and yield. Enhanced expression of Vitreoscilla hemoglobin (VHb) in CHO cells has been shown to increase tPA production 40%–100% (Fussenegger and Bailey, 1998) (Fussenegger and Bailey, 1998). During cell culture, the culture medium is gradually acidified with the accumulation of lactate, and lactate dehydrogenase (LDH) converts pyruvate to produce lactate. The decreasing in pH resulting from the increase in lactic acid ultimately resulted in the inhibition of cell growth. It can inhibit LDH by avoiding the oxidation of pyruvate to lactate, reduce lactate dehydrogenase A (LDHA) to enzyme activity, and down-regulate lactate levels without affecting cell proliferation and yield (Zhou et al., 2011). In addition, the simultaneous decreasing in LDH and pyruvate dehydrogenase kinase (PDHK) activities can lead to a decrease in lactate concentration and increase the antibody yield per unit volume (Fu et al., 2016).
Alterations in cytoskeletal protein expression are thought to affect cellular processes associated with recombinant protein production, including transcription, cell cycle progression, metabolism, and secretory vesicle trafficking. Differential expression of cytoskeletal proteins has been observed in modified or gene-amplified cell lines (Kuystermans et al., 2010), and both small interfering RNA (siRNA) and shRNA can increase the Qp of recombinant CHO cell lines (Hammond and Lee, 2012). Cell cycle detection key kinases control cell cycle and proliferation, and overexpression of cell cycle inhibitors such as p21cip1 or p27Kip1 leads to cell cycle arrest, but can increase cell Qp in CHO cells (Astley and Al-Rubeai, 2008).
Optimization of Cell Culture Process
The substances and energy required for biochemical reactions such as new cell synthesis and cell metabolism are obtained from the culture medium. It was found that amino acids as the backbone in the medium component had a significant effect on product formation and critical quality attributes of mAbs. Recombinant protein production requires high amino acids to support not only host cell growth and heterologous recombinant protein synthesis, but also provide raw materials as carbon and nitrogen sources for nucleotide, lipid, ATP, and NADPH synthesis (Duarte et al., 2014). Nutrients in the medium are necessary for the cell growth environment. Among these components, amino acids constitute the constituent recombinant proteins and native CHO proteins, accounting for approximately 70% of the stem cell mass (Carrillo-Cocom et al., 2015). Mammalian cells can only produce some of the amino acids required for protein synthesis, while others are essential amino acids that need to be replenished from the outside. The consumption of extracellular amino acids has a negative impact on cell growth, culture time, specific productivity (qP), and monoclonal antibody glycosylation (Fomina-Yadlin et al., 2014). Amino acids are important sources of nitrogen and potential carbon for many intermediates in a large number of metabolic pathways when other sources are limited. Therefore, CHO cells must be supplemented with a sufficient number of essential amino acids to maintain continued survival, growth and proliferation, and with non-essential amino acids. In the process of cell proliferation, amino acids account for the majority of cell biomass consumption (Hosios et al., 2016). Studies have confirmed that post-translational modification of amino acid residues affects the diversity and quality of Mabs. Glutamine (Gln) has also been shown to restore or stimulate mTORC1 (mechanistic target of rapamycin complex 1) activity upon amino acid starvation (Tan et al., 2017) and can also affect protein yield through glutamine hydrolysis (Durán et al., 2012).
During the optimization of production process, the effects of culture medium on cell viability, metabolism, and product quality should be considered. In the early stage, necessary nutrients were provided by the serum of animal tissues or blood. Due to the regulatory limitations of mAb production, the serum-free medium formulation was invented, thus avoiding the use of serum in cell culture (Song et al., 2013). There are now many formulations available for different cell lines and different growth levels, including CD CHO (Gibco), Excell CD CHO (SAFC), etc., During culture, cells may experience a decrease in recombinant protein yield during prolonged culture, even during extension of the final bioreactor scale (Tharmalingam et al., 2018). Modern industrial culture of CHO cells must support high viable cell density, promote the synthesis and extracellular transport of biological products, and optimize nutrients and formulations of the medium is very important for protein quality and yield improvement (Ritacco et al., 2018).
Using CHO mAb cell line, the fed batch culture was increased by 12-fold compared with batch culture, while the optimal choline chloride to glucose ratio of batch culture CHO cells could be optimized, the culture time could be shortened, and the protein yield could be improved (Kuwae et al., 2018). It was found that during perfusion cell culture, the use of concentrated nutrient diet decreased the medium consumption by 1.8-fold, increased the volume expression by 1.67-fold, and also increased the cell density (Bielser et al., 2020). Chemical additives such as sodium butyrate (NaBu) and dimethyl sulfoxide (DMSO) can increase Qp in rCHO cells in a dose-dependent manner by inducing cell cycle arrest (Ha and Lee, 2014), decreasing oxidative stress (Ha et al., 2018), and inhibiting endoplasmic reticulum stress (Ha et al., 2019). However, these chemical additives are cytotoxic, can induce apoptosis, and have a negative impact on product quality in rCHO cell culture. Therefore, harmless and effective chemical additives are needed. Driven by high volumetric titers and a small footprint, perfusion-based bioprocess research has regained an interest from academia and industry. The future of perfusion bioprocesses is bright and that the fields of media optimization and continuous processing hold the greatest potential (Igarashi and Kashiwagi, 2021).
To better maintain product quality during culture, it is important to minimize the accumulation of host cell proteins in the culture medium, otherwise it will damage product quality. In the process of rCHO cell culture, there are thousands of host cell proteins (HCPs) in the culture supernatant, and the total number and number of different types of HCPs depend on the host cell type, culture method and culture time. Due to higher cell concentrations and longer culture cycles, the extracellular accumulation level of HCPs in fed batch cultures is much higher than that in batch cultures (Park et al., 2017). Removal of unnecessary HCPs by multiple secretome engineering of CHO cells can reduce the total amount and number of different types of HCPs in the culture medium (Kol et al., 2020). In addition, if specific HCPs that damage POIs are identified, the genes encoding these HCPs can be knocked out in CHO cells, thereby eliminating their accumulation in the culture medium. By using CRISPR/cas9-mediated methods, heavy sialylated glycoproteins such as EPO can be produced in batches by knocking out sialidase and pro-apoptotic genes in rCHO cells (Ha et al., 2020).
Cell culture parameters for Qp include partial pressure of carbon dioxide, osmolality, temperature, dissolved oxygen, and pH, and improvements in cell secretion, folding, and glycosylation are important ways to optimize mammalian cell production systems (Omasa et al., 2010). The optimal temperature for CHO cell growth is 37°C, and the temperature was controlled lower than 37°C during protein production (Sou et al., 2015; Torres et al., 2018), which can prolong the time of antibodies and other RTPs and improve the final product yields Long and low temperature two-phase cell culture process can improve product quality and purification yield (Gomez et al., 2018). pH value of culture medium is another important environmental parameter, can affect cell growth, cell metabolism, and the yield and quality of recombinant proteins in rCHO cells (Xie et al., 2016; Paul et al., 2019). CHO cells grow well at neutral pH (Michl and Park, 2019). The intracellular pH is not uniform, ranging from 4.7 to 8.0 in lysosomesand mitochondria (Jaworska et al., 2021). Every cellular process that occurs in intracellular organelles has an optimal pH, which shows optimal function when the pH value is reached. When the optimal pH deviates from a specific cellular process, it can cause changes in conformational enzymes and impair function. The osmotic pressure of the medium can also affect cell growth and recombinant protein production, and the osmotic pressure of CHO cells was 280∼320 mOsm. When osmolality deviates from this standard range, rCHO cells showed a decrease in Qp, but generally show an increase in Qp (Qin et al., 2019). High osmolality has a positive effect on the product quality of protein aggregation, but a negative effect on protein glycosylation. In addition, in rCHO cell culture, the parameters of CO2 and O2 dissolved oxygen should also be coordinated.
Promote Correct Folding and Processing of Proteins
Molecular chaperones are a family of proteins whose role is to assist in the folding and targeting of proteins in normal and stressed cells. Insufficient supply of chaperones limits correct protein folding in recombinant protein production. Intramolecular chaperones can help protein folding and assist protein subunits to correctly fold with multiple subunits to assemble into intact proteins (O'Rourke et al., 2019) (Fatima et al., 2021). Following protein synthesis and translocation into the ER lumen, secreted proteins must be properly folded and modified with the help of chaperones and folding enzymes. Chaperone proteins in CHO cells can improve the productivity of difficult-to-express proteins, such as chaperone heat shock proteins (HSPs), which help protein folding, assembly, translocation, and degradation under cellular stress. It also prevents the aggregation of newly synthesized polypeptide chains into defective proteins. Protein disulfide isomerase (PDI) catalyzes the formation and cleavage of disulfide bonds and helps protein folding improve the productivity of Mabs (Borth et al., 2005), and its isomer ERp57 can increase protein yield up to 2.1-fold without reducing cell growth (Hwang et al., 2003). It should be pointed out that this would only be an effective strategy where this was the only limiting factor. Engineering of protein folding and assembly in the ER is inherently protein specific, and possibly Mab specific. Mabs, even of the same subclass, likely differ in their kinetics of folding and assembly and thus may benefit to a variable extent from overexpression of different ER molecular chaperones or foldases. PDI has a certain inhibitory effect on the formation of disulfide bridges in antibody LC. When PDI recognizes less antibody light chain (LC), the formation of disulfide bridges in antibody light chain (LC) is blocked, and secretion is blocked due to incorrect folding (Mathias et al., 2020). Paracycline B interacts with other proteins in the ER to form a partner complex that promotes protein folding. Chemical chaperones are compounds that improve ER folding capacity, promote protein folding in the ER, and enhance protein secretion. Expression levels were improved by optimizing chaperones, UPR transactivators, chemical chaperones, maximizing cell-specific yield and eliminating protein aggregation during transient expression (Johari et al., 2015). The introduction of promoted genes in protein folding, such as simultaneous overexpression of calnexin (CNX) and calreticulin (CRT), increased specific thrombopoietin (TPO) productivity 1.9-fold, did not affect the cell growth and biological activity of recombinant TPO, increased protein synthesis and secretion, and then increased the yield of recombinant protein in CHO cells (Chung et al., 2004).
Signal peptides transport proteins to the correct subcellular compartment, but natural signal peptides are not necessarily optimal (Kober et al., 2013). Moreover, signal peptides can be interchanged between different species (Knappskog et al., 2007). Therefore, the selection of appropriate signal peptides is conducive to the correct processing and secretion of proteins (Haryadi et al., 2015) Optimizing different signal peptides can improve recombinant protein expression (Cecconi et al., 2021).
Toxic Protein Expression Strategy
Another factor causing insufficient protein production is the inherent toxicity of proteins. Avoiding exposure of cells to high concentrations of protein, or optimizing cell lines to resist toxic proteins, can promote growth and then increase yield. Protein toxicity can be mitigated by a variety of methods. One approach is to change the protein itself to reduce its cytotoxicity modify the protein to produce a more stable, less toxic form, and the other option is to change the promoter in the expression vector rather than the POI. The inherent characteristics of molecules can also lead to easy degradation, aggregation and other adverse protein-protein interactions of proteins, which leads to cytotoxicity. If the GOI is determined to be toxic, then one option to increase yield is to adapt host cells and reduce growth process by-products (Knappskog et al., 2007). Decreased sensitivity, protein toxicity, or degradation of cell lines cultured in the presence of low concentrations of protein can also be alleviated by reducing protein contact with production cultures. Avoiding the burden of recombinant protein metabolism during cell growth produces potential toxicity and inducible expression vectors can be applied (Ong et al., 2019). It can also be achieved by chemical supplementation, in which toxic proteins competitively inhibit interactions with cells through antagonists. Another approach is to use a perfusion growth system in which the culture supernatant is continuously removed by filtration and the cell mass is retained, which can improve cell density and volume productivity (Junne and Neubauer, 2018).
Summary and Prospect
To improve the protein production capacity of recombinant proteins in CHO cells, the bottlenecks in transcription, translation, PTMs, protein folding and secretion during the production process must be recognized, and solutions must be developed for each rate-limiting step. Cell engineering technology can precisely control the relative expression of multiple functional gene components, which in turn affects protein yield. New therapeutic applications are opened up using new protein forms such as bispecific antibodies and fusion proteins. An understanding of the critical quality attributes of RTPs during CHO cell culture and their contributing factors is critical to the production of high quality RTPs. The proper assembly of these new RTPs may be more complex, which may affect productivity and potency. The solution strategy can start from the molecular design of proteins, combined with appropriate cell line, vector engineering and process optimization, based on the understanding of folding mechanism and the understanding of potential bottlenecks, optimize protein secretion and prevent aggregation, maximize the yield of recombinant proteins, reduce large-scale production costs, and promote the development of biopharmaceuticals.
Author Contributions
Z-ML: manuscript preparation and revision; Z-LF: Proof Read and manuscript revison; X-YW: manuscript revison; T-YW: manuscript design and revison.
Funding
This work was supported by the Grants from the Natural Science Foundation of Henan Province (No.202300410320);the Key Scientific Research Project of Higher Education of Henan Province, China (No.22A310009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alves, C. S., and Dobrowsky, T. M. (2017). Strategies and Considerations for Improving Expression of "Difficult to Express" Proteins in CHO Cells. Methods Mol. Biol. 1603, 1–23. doi:10.1007/978-1-4939-6972-2_1
Amadi, I. M., Agrawal, V., Christianson, T., Bardliving, C., Shamlou, P., and LeBowitz, J. H. (2020). Inhibition of Endogenous Mir-23a/Mir-377 in CHO Cells Enhances Difficult-to-Express Recombinant Lysosomal Sulfatase Activity. Biotechnol. Prog. 36 (3), e2974. doi:10.1002/btpr.2974
Ang, K. S., Kyriakopoulos, S., Li, W., and Lee, D. Y. (2016). Multi-Omics Data Driven Analysis Establishes Reference Codon Biases for Synthetic Gene Design in Microbial and Mammalian Cells. Methods 102, 26–35. doi:10.1016/j.ymeth.2016.01.016
Astley, K., and Al-Rubeai, M. (2008). The Role of Bcl-2 and its Combined Effect with p21CIP1 in Adaptation of CHO Cells to Suspension and Protein-free Culture. Appl. Microbiol. Biotechnol. 78, 391–399. doi:10.1007/s00253-007-1320-2
Baek, E., Noh, S. M., and Lee, G. M. (2017). Anti-Apoptosis Engineering for Improved Protein Production from CHO Cells. Methods Mol. Biol. 1603, 71–85. doi:10.1007/978-1-4939-6972-2_5
Bicknell, A. A., and Ricci, E. P. (2017). When mRNA Translation Meets Decay. Biochem. Soc. Trans. 45, 339–351. doi:10.1042/bst20160243
Bielser, J. M., Kraus, L., Burgos-Morales, O., Broly, H., and Souquet, J. (2020). Reduction of Medium Consumption in Perfusion Mammalian Cell Cultures Using a Perfusion Rate Equivalent Concentrated Nutrient Feed. Biotechnol. Prog. 36, e3026. doi:10.1002/btpr.3026
Borth, N., Mattanovich, D., Kunert, R., and Katinger, H. (2005). Effect of Increased Expression of Protein Disulfide Isomerase and Heavy Chain Binding Protein on Antibody Secretion in a Recombinant CHO Cell Line. Biotechnol. Prog. 21 (1), 106–111. doi:10.1021/bp0498241
Bryan, L., Clynes, M., and Meleady, P. (2021). The Emerging Role of Cellular Post-translational Modifications in Modulating Growth and Productivity of Recombinant Chinese Hamster Ovary Cells. Biotechnol. Adv. 49, 107757. doi:10.1016/j.biotechadv.2021.107757
Carrillo-Cocom, L. M., Genel-Rey, T., Araíz-Hernández, D., López-Pacheco, F., López-Meza, J., Rocha-Pizaña, M. R., et al. (2015). Amino Acid Consumption in Naïve And Recombinant CHO Cell Cultures: Producers of a Monoclonal Antibody. Cytotechnology 67 (5), 809–820. doi:10.1007/s10616-014-9720-5
Carver, J., Ng, D., Zhou, M., Ko, P., Zhan, D., Yim, M., et al. (2020). Maximizing Antibody Production in a Targeted Integration Host by Optimization of Subunit Gene Dosage and Position. Biotechnol. Prog. 36, e2967. doi:10.1002/btpr.2967
Cecconi, D., Di Carlo, C., and Brandi, J. (2021). Protein Secretion Prediction Tools and Extracellular Vesicles Databases. Methods Mol. Biol. 2361, 213–227. doi:10.1007/978-1-0716-1641-3_13
Chang, M., Liu, R., Jin, Q., Liu, Y., and Wang, X. (2014). Scaffold/Matrix Attachment Regions From CHO Cell Chromosome Enhanced The Stable Transfection Efficiency and the Expression of Transgene in CHO Cells. Biotechnol. Appl. Biochem. 61 (5), 510–516. doi:10.1002/bab.1204
Chung, B. K., Yusufi, F. N., Mariati, , Yang, Y., and Lee, D. Y. (2013). Enhanced Expression of Codon Optimized Interferon Gamma in CHO Cells. J. Biotechnol. 167(3), 326–333. doi:10.1016/j.jbiotec.2013.07.011
Chung, J. Y., Lim, S. W., Hong, Y. J., Hwang, S. O., and Lee, G. M. (2004). Effect of Doxycycline-Regulated Calnexin and Calreticulin Expression on Specific Thrombopoietin Productivity of Recombinant Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 85, 539–546. doi:10.1002/bit.10919
Chusainow, J., Yang, Y. S., Yeo, J. H., Toh, P. C., Asvadi, P., Wong, N. S., et al. (2009). A Study of Monoclonal Antibody-Producing CHO Cell Lines: What Makes a Stable High Producer? Biotechnol. Bioeng. 102 (4), 1182–1196. doi:10.1002/bit.22158
Delic, M., Göngrich, R., Mattanovich, D., and Gasser, B. (2014). Engineering of Protein Folding and Secretion-Strategies to Overcome Bottlenecks for Efficient Production of Recombinant Proteins. Antioxidants Redox Signal. 21, 414–437. doi:10.1089/ars.2014.5844
Dong, W., Li, C., Yang, Y., Wang, T., and Wang, F. (2019). Increasing Transgenic Expression in Recombinant Chinese Hamster Ovary Cells Using Introns in Different Directions. Sheng Wu Gong Cheng Xue Bao 35, 1071–1078. doi:10.13345/j.cjb.180516
Dreesen, I. A., and Fussenegger, M. (2011). Ectopic Expression of Human Mtor Increases Viability, Robustness, Cell Size, Proliferation, and Antibody Production of Chinese Hamster Ovary Cells. Biotechnol. Bioeng. 108 (4), 853–866. doi:10.1002/bit.22990
Druz, A., Betenbaugh, M., and Shiloach, J. (2012). Glucose Depletion Activates Mmu-Mir-466h-5p Expression Through Oxidative Stress and Inhibition of Histone Deacetylation. Nucl. Acids Res. 40 (15), 7291–7302. doi:10.1093/nar/gks452
Druz, A., Chu, C., Majors, B., Santuary, R., Betenbaugh, M., and Shiloach, J. (2011). A Novel Microrna Mmu-Mir-466h Affects Apoptosis Regulation in Mammalian Cells. Biotechnol. Bioeng. 108 (7), 1651–1661. doi:10.1002/bit.23092
Druz, A., Son, Y. J., Betenbaugh, M., and Shiloach, J. (2013). Stable Inhibition of Mmu-Mir-466h-5p Improves Apoptosis Resistance and Protein Production in CHO Cells. Metab. Eng. 16, 87–94. doi:10.1016/j.ymben.2012.12.004
Duarte, T. M., Carinhas, N., Barreiro, L. C., Carrondo, M. J., Alves, P. M., and Teixeira, A. P. (2014). Metabolic Responses of CHO Cells to Limitation of Key Amino Acids. Biotechnol. Bioeng. 111 (10), 2095–2106. doi:10.1002/bit.25266
Durán, R. V., Oppliger, W., Robitaille, A. M., Heiserich, L., Skendaj, R., Gottlieb, E., et al. (2012). Glutaminolysis Activates Rag-Mtorc1 Signaling. Mol. Cell. 47 (3), 349–358. doi:10.1016/j.molcel.2012.05.043
Eisenhut, P., Mebrahtu, A., Moradi Barzadd, M., Thalén, N., Klanert, G., Weinguny, M., et al. (2020). Systematic Use of Synthetic 5′-UTR RNA Structures to Tune Protein Translation Improves Yield and Quality of Complex Proteins in Mammalian Cell Factories. Nucleic Acids Res. 48, e119. doi:10.1093/nar/gkaa847
Figueroa, B., Chen, S., Oyler, G. A., Hardwick, J. M., and Betenbaugh, M. J. (2004). Aven and Bcl-xL Enhance Protection against Apoptosis for Mammalian Cells Exposed to Various Culture Conditions. Biotechnol. Bioeng. 85, 589–600. doi:10.1002/bit.10913
Fatima, K., Naqvi, F., and Younas, H. (2021). A Review: Molecular Chaperone-mediated Folding, Unfolding and Disaggregation of Expressed Recombinant Proteins. 79(2), 153–174. doi:10.1007/s12013-021-00970-5
Fomina-Yadlin, D., Gosink, J. J., McCoy, R., Follstad, B., Morris, A., Russell, C. B., et al. (2014). Cellular Responses to Individual Amino-Acid Depletion in Antibody-Expressing and Parental CHO Cell Lines. Biotechnol. Bioeng. 111, 965–979.
Fu, T., Zhang, C., Jing, Y., Jiang, C., Li, Z., Wang, S., et al. (2016). Regulation of Cell Growth and Apoptosis through Lactate Dehydrogenase C Over-expression in Chinese Hamster Ovary Cells. Appl. Microbiol. Biotechnol. 100, 5007–5016. doi:10.1007/s00253-016-7348-4
Fussenegger, M., and Bailey, J. E. (1998). Molecular Regulation of Cell-Cycle Progression and Apoptosis in Mammalian Cells: Implications for Biotechnology. Biotechnol. Prog. 14, 807–833. doi:10.1021/bp9800891
Gomez, N., Wieczorek, A., Lu, F., Bruno, R., Diaz, L., Agrawal, N. J., et al. (2018). Culture Temperature Modulates Half Antibody and Aggregate Formation in a Chinese Hamster Ovary Cell Line Expressing a Bispecific Antibody. Biotechnol. Bioeng. 115, 2930–2940. doi:10.1002/bit.26803
Grav, L. M., Sergeeva, D., Lee, J. S., Marin de Mas, I., Lewis, N. E., Andersen, M. R., et al. (2018). Minimizing Clonal Variation during Mammalian Cell Line Engineering for Improved Systems Biology Data Generation. ACS Synth. Biol. 7, 2148–2159. doi:10.1021/acssynbio.8b00140
Ha, T. K., Hansen, A. H., Kol, S., Kildegaard, H. F., and Lee, G. M. (2018). Baicalein Reduces Oxidative Stress in CHO Cell Cultures and Improves Recombinant Antibody Productivity. Biotechnol. J. 13, e1700425. doi:10.1002/biot.201700425
Ha, T. K., Hansen, A. H., Kildegaard, H. F., and Lee, G. M. (2019). BiP Inducer X: An ER Stress Inhibitor for Enhancing Recombinant Antibody Production in CHO Cell Culture. Biotechnol. J. 14, e1900130. doi:10.1002/biot.201900130
Ha, T. K., Hansen, A. H., Kildegaard, H. F., and Lee, G. M. (2020). Knockout of Sialidase and Pro-apoptotic Genes in Chinese Hamster Ovary Cells Enables the Production of Recombinant Human Erythropoietin in Fed-Batch Cultures. Metab. Eng. 57, 182–192. doi:10.1016/j.ymben.2019.11.008
Ha, T. K., and Lee, G. M. (2014). Effect of Glutamine Substitution by TCA Cycle Intermediates on the Production and Sialylation of Fc-Fusion Protein in Chinese Hamster Ovary Cell Culture. J. Biotechnol. 180, 23–29. doi:10.1016/j.jbiotec.2014.04.002
Hammond, S., and Lee, K. H. (2012). RNA Interference of Cofilin in Chinese Hamster Ovary Cells Improves Recombinant Protein Productivity. Biotechnol. Bioeng. 109, 528–535. doi:10.1002/bit.23322
Handlogten, M. W., Lee-O'Brien, A., Roy, G., Levitskaya, S. V., Venkat, R., Singh, S., et al. (2018). Intracellular Response to Process Optimization and Impact on Productivity and Product Aggregates for a High-Titer CHO Cell Process. Biotechnol. Bioeng. 115, 126–138. doi:10.1002/bit.26460
Henry, M. N., MacDonald, M. A., Orellana, C. A., Gray, P. P., Gillard, M., Baker, K., et al. (2020). Attenuating Apoptosis in Chinese Hamster Ovary Cells for Improved Biopharmaceutical Production. Biotechnol. Bioeng. 117, 1187–1203. doi:10.1002/bit.27269
Haryadi, R., Zhang, P., Chan, K. F., and Song, Z. (2013). CHO-gmt5, a Novel CHO Glycosylation Mutant for Producing Afucosylated and Asialylated Recombinant Antibodies. Bioengineered 4 (2), 90–94. doi:10.4161/bioe.22262
Hetz, C., and Papa, F. R. (2018). The Unfolded Protein Response and Cell Fate Control. Mol. Cell. 69, 169–181. doi:10.1016/j.molcel.2017.06.017
Hetz, C. (2012). The Unfolded Protein Response: Controlling Cell Fate Decisions under ER Stress and beyond. Nat. Rev. Mol. Cell. Biol. 13, 89–102. doi:10.1038/nrm3270
Ho, S. C. L., Mariati, fnm., Yeo, J. H. M., Fang, S. G., and Yang, Y. (2015). Impact of Using Different Promoters and Matrix Attachment Regions on Recombinant Protein Expression Level and Stability in Stably Transfected CHO Cells. Mol. Biotechnol. 57, 138–144. doi:10.1007/s12033-014-9809-2
Ho, S. C., Bardor, M., Li, B., Lee, J. J., Song, Z., Tong, Y. W., et al. (2013). Comparison of Internal Ribosome Entry Site (IRES) and Furin-2A (F2A) for Monoclonal Antibody Expression Level and Quality in CHO Cells. PLoS One 8 (5), e63247. doi:10.1371/journal.pone.0063247
Hosios, A. M., Hecht, V. C., Danai, L. V., Johnson, M. O., Rathmell, J. C., Steinhauser, M. L., et al. (2016). Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell 36 (5), 540–549. doi:10.1016/j.devcel.2016.02.012
Hwang, S. O., Chung, J. Y., and Lee, G. M. (2003). Effect of Doxycycline-Regulated Erp57 Expression on Specific Thrombopoietin Productivity of Recombinant CHO Cells. Biotechnol. Prog. 19 (1), 179–184. doi:10.1021/bp025578m
Igarashi, K., and Kashiwagi, K. (2021). Functional Roles of Polyamines and Their Metabolite Acrolein in Eukaryotic Cells. Amino Acids 53, 1473–1492. doi:10.1007/s00726-021-03073-w
Ishii, Y., Murakami, J., Sasaki, K., Tsukahara, M., and Wakamatsu, K. (2014). Efficient Folding/Assembly In Chinese Hamster Ovary Cells is Critical for High Quality (Low Aggregate Content) of Secreted Trastuzumab as Well as for High Production: Stepwise Multivariate Regression Analyses. J. Biosci. Bioeng. 118 (2), 223–230. doi:10.1016/j.jbiosc.2014.01.013
Jaworska, A., Malek, K., and Kudelski, A. (2021). Intracellular pH - Advantages and Pitfalls of Surface-Enhanced Raman Scattering and Fluorescence Microscopy - A Review. Spectrochimica Acta Part A Mol. Biomol. Spectrosc. 251, 119410. doi:10.1016/j.saa.2020.119410
Junne, S., and Neubauer, P. (2018). How Scalable and Suitable Are Single-Use Bioreactors? Curr. Opin. Biotechnol. 53, 240–247. doi:10.1016/j.copbio.2018.04.003
Johari, Y. B., Estes, S. D., Alves, C. S., Sinacore, M. S., and James, D. C. (2015). Integrated Cell and Process Engineering for Improved Transient Production of a "Difficult-to-Express" Fusion Protein by CHO Cells. Biotechnol. Bioeng. 112 (12), 2527–2542. doi:10.1002/bit.25687
Kaneyoshi, K., Kuroda, K., Uchiyama, K., Onitsuka, M., Yamano-Adachi, N., Koga, Y., et al. (2019). Secretion Analysis of Intracellular "Difficult-To-Express" Immunoglobulin G (IgG) in Chinese Hamster Ovary (CHO) Cells. Cytotechnology 71, 305–316. doi:10.1007/s10616-018-0286-5
Kelly, P. S., Breen, L., Gallagher, C., Kelly, S., Henry, M., Lao, N. T., et al. (2015). Re-Programming CHO Cell Metabolism Using Mir-23 Tips The Balance Towards a Highly Productive Phenotype. Biotechnol. J. 10 (7), 1029–1040. doi:10.1002/biot.201500101
Kim, S. H., and Lee, G. M. (2007). Down-Regulation of Lactate Dehydrogenase-A by Sirnas for Reduced Lactic Acid Formation of Chinese Hamster Ovary Cells Producing Thrombopoietin. Appl. Microbiol. Biotechnol. 74 (1), 152–159. doi:10.1007/s00253-006-0654-5
Kim, M., O'Callaghan, P. M., Droms, K. A., and James, D. C. (2011). A Mechanistic Understanding of Production Instability in CHO Cell Lines Expressing Recombinant Monoclonal Antibodies. Biotechnol. Bioeng. 108, 2434–2446. doi:10.1002/bit.23189
Klanert, G., Jadhav, V., Shanmukam, V., Diendorfer, A., Karbiener, M., Scheideler, M., et al. (2016). A Signature of 12 microRNAs is Robustly Associated With Growth Rate in a Variety of CHO Cell Lines. J. Biotechnol. 235, 150–161. doi:10.1016/j.jbiotec.2016.03.022
Knappskog, S., Ravneberg, H., Gjerdrum, C., Tröβe, C., Stern, B., and Pryme, I. F. (2007). The Level of Synthesis and Secretion of Gaussia Princeps Luciferase in Transfected CHO Cells Is Heavily Dependent on the Choice of Signal Peptide. J. Biotechnol. 128, 705–715. doi:10.1016/j.jbiotec.2006.11.026
Kober, L., Zehe, C., and Bode, J. (2013). Optimized Signal Peptides for the Development of High Expressing CHO Cell Lines. Biotechnol. Bioeng. 110, 1164–1173. doi:10.1002/bit.24776
Kol, S., Ley, D., Wulff, T., Decker, M., Arnsdorf, J., Schoffelen, S., et al. (2020). Multiplex Secretome Engineering Enhances Recombinant Protein Production and Purity. Nat. Commun. 11, 1908. doi:10.1038/s41467-020-15866-w
Kuwae, S., Miyakawa, I., and Doi, T. (2018). Development of a Chemically Defined Platform Fed-Batch Culture Media for Monoclonal Antibody-Producing CHO Cell Lines with Optimized Choline Content. Cytotechnology 70, 939–948. doi:10.1007/s10616-017-0185-1
Kuystermans, D., Dunn, M. J., and Al-Rubeai, M. (2010). A Proteomic Study of cMyc Improvement of CHO Culture. BMC Biotechnol. 10, 25. doi:10.1186/1472-6750-10-25
Kwon, R. J., Kim, S. K., Lee, S. I., Hwang, S. J., Lee, G. M., Kim, J. S., et al. (2006). Artificial Transcription Factors Increase Production of Recombinant Antibodies in Chinese Hamster Ovary Cells. Biotechnol. Lett. 28 (1), 9–15. doi:10.1007/s10529-005-4680-7
Li, Q., Wang, X. Y., Zhao, C. P., Tian, Z. W., Xu, D. H., Wang, T. Y., et al. (2018). Effects of Different Promoters and MAR Combinations on Transgene Expression of Recombinant CHO Cells. Sichuan Da Xue Xue Bao Yi Xue Ban 49 (1), 18–23.
Li, Y. M., Wang, M., Wang, T. Y., Wei, Y. G., Guo, X., Mi, C. L., et al. (2020). Effects of Different 2A Peptides on Transgene Expression Mediated by Tricistronic Vectors in Transfected CHO Cells. Mol. Biol. Rep. 47 (1), 469–475. doi:10.1007/s11033-019-05153-3
Lim, S. F., Chuan, K. H., Liu, S., Loh, S. O. H., Chung, B. Y. F., Ong, C. C., et al. (2006). RNAi Suppression of Bax and Bak Enhances Viability in Fed-Batch Cultures of CHO Cells. Metab. Eng. 8, 509–522. doi:10.1016/j.ymben.2006.05.005
Majors, B. S., Betenbaugh, M. J., Pederson, N. E., and Chiang, G. G. (2009). Mcl-1 Overexpression Leads to Higher Viabilities and Increased Production of Humanized Monoclonal Antibody in Chinese Hamster Ovary Cells. Biotechnol. Prog. 25, 1161–1168. doi:10.1002/btpr.192
Mathias, S., Wippermann, A., Raab, N., Zeh, N., Handrick, R., Gorr, I., et al. (2020). Unraveling what Makes a Monoclonal Antibody Difficult‐to‐express: From Intracellular Accumulation to Incomplete Folding and Degradation via ERAD. Biotechnol. Bioeng. 117, 5–16. doi:10.1002/bit.27196
Michl, J., Park, K. C., and Swietach, P. (2019). Evidence-based Guidelines for Controlling pH in Mammalian Live-Cell Culture Systems. Commun. Biol. 2, 144. doi:10.1038/s42003-019-0393-7
Muluhngwi, P., Richardson, K., Napier, J., Rouchka, E. C., Richardson, K. C., Mott, J. L., and Klinge, C. M. (2017). Regulation of miR-29b-1/a Transcription and Identification of Target mRNAs in CHO-K1 Cells. Mol. Cell Endocrinol. 444, 38–47. doi:10.1016/j.mce.2017.01.044
Narula, A., Ellis, J., Taliaferro, J. M., and Rissland, O. S. (2019). Coding Regions Affect mRNA Stability in Human Cells. Rna 25, 1751–1764. doi:10.1261/rna.073239.119
Neville, J. J., Orlando, J., Mann, K., McCloskey, B., and Antoniou, M. N. (2017). Ubiquitous Chromatin-Opening Elements (UCOEs): Applications in Biomanufacturing and Gene Therapy. Biotechnol. Adv. 35 (5), 557–564. doi:10.1016/j.biotechadv.2017.05.004
Nguyen, L. N., Baumann, M., Dhiman, H., Marx, N., Schmieder, V., Hussein, M., et al. (2019). Novel Promoters Derived from Chinese Hamster Ovary Cells via In Silico and In Vitro Analysis. Biotechnol. J. 14, e1900125. doi:10.1002/biot.201900125
O'Callaghan, P. M., McLeod, J., Pybus, L. P., Lovelady, C. S., Wilkinson, S. J., Racher, A. J., et al. (2010). Cell Line-Specific Control of Recombinant Monoclonal Antibody Production by CHO Cells. Biotechnol. Bioeng. 106 (6), 938–951. doi:10.1002/bit.22769
O'Flaherty, R., Bergin, A., Flampouri, E., Mota, L. M., Obaidi, I., Quigley, A., et al. (2020). Mammalian Cell Culture For Production of Recombinant Proteins: A Review of the Critical Steps in Their Biomanufacturing. Biotechnol. Adv. 43, 107552. doi:10.1016/j.biotechadv.2020.107552
Omasa, T., Takami, T., Ohya, T., Kiyama, E., Hayashi, T., Nishii, H., et al. (2008). Overexpression of GADD34 Enhances Production of Recombinant Human Antithrombin III in Chinese Hamster Ovary Cells. J. Biosci. Bioeng. 106 (6), 568–573. doi:10.1263/jbb.106.568
Omasa, T., Onitsuka, M., and Kim, W.-D. (2010). Cell Engineering and Cultivation of Chinese Hamster Ovary (CHO) Cells. Cpb 11, 233–240. doi:10.2174/138920110791111960
Ong, E. C., Smidt, P., and McGrew, J. T. (2019). Limiting the Metabolic Burden of Recombinant Protein Expression during Selection Yields Pools with Higher Expression Levels. Biotechnol. Prog. 35, e2839. doi:10.1002/btpr.2839
O'Rourke, S. M., Morozov, G. I., Roberts, J. T., Barb, A. W., and Sgourakis, A. W. (2019). Production of Soluble pMHC-I Molecules in Mammalian Cells Using the Molecular Chaperone TAPBPR. Protein Eng. Des. Sel. 32 (12), 525–532. doi:10.1093/protein/gzaa015
Park, J. H., Jin, J. H., Lim, M. S., An, H. J., Kim, J. W., and Lee, G. M. (2017). Proteomic Analysis of Host Cell Protein Dynamics in the Culture Supernatants of Antibody-Producing CHO Cells. Sci. Rep. 7, 44246. doi:10.1038/srep44246
Paul, K., Rajamanickam, V., and Herwig, C. (2019). Model-based Optimization of Temperature and pH Shift to Increase Volumetric Productivity of a Chinese Hamster Ovary Fed-Batch Process. J. Biosci. Bioeng. 128, 710–715. doi:10.1016/j.jbiosc.2019.06.004
Puck, T. T., Cieciura, S. J., and Robinson, A. (1958). Genetics of Somatic Mammalian Cells. J. Exp. Med. 108, 945–956. doi:10.1084/jem.108.6.945
Pybus, L. P., James, D. C., Dean, G., Slidel, T., Hardman, C., Smith, A., et al. (2014). Predicting the Expression of Recombinant Monoclonal Antibodies in Chinese Hamster Ovary Cells Based on Sequence Features of the CDR3 Domain. Biotechnol. Prog. 30, 188–197. doi:10.1002/btpr.1839
Qin, J., Wu, X., Xia, Z., Huang, Z., Zhang, Y., Wang, Y., et al. (2019). The Effect of Hyperosmolality Application Time on Production, Quality, and Biopotency of Monoclonal Antibodies Produced in CHO Cell Fed-Batch and Perfusion Cultures. Appl. Microbiol. Biotechnol. 103, 1217–1229. doi:10.1007/s00253-018-9555-7
Quax, T. E., Claassens, N. J., Söll, D., and van der Oost, J. (2015). Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cell. 59 (2), 149–161. doi:10.1016/j.molcel.2015.05.035
Raab, N., Zeh, N., Schlossbauer, P., Mathias, S., Lindner, B., Stadermann, A., et al. (2022). A Blueprint from Nature: miRNome Comparison of Plasma Cells and CHO Cells to Optimize Therapeutic Antibody Production. N. Biotechnol. 66, 79–88. doi:10.1016/j.nbt.2021.10.005
Reinhart, D., Sommeregger, W., Debreczeny, M., Gludovacz, E., and Kunert, R. (2014). In Search of Expression Bottlenecks in Recombinant CHO Cell Lines-A Case Study. Appl. Microbiol. Biotechnol. 98, 5959–5965. doi:10.1007/s00253-014-5584-z
Reisinger, H., Steinfellner, W., Stern, B., Katinger, H., and Kunert, R. (2008). The Absence of Effect of Gene Copy Number and Mrna Level on the Amount of Mab Secretion From Mammalian Cells. Appl. Microbiol. Biotechnol. 81 (4), 701–710. doi:10.1007/s00253-008-1701-1
Richelle, A., and Lewis, N. E. (2017). Improvements in Protein Production in Mammalian Cells from Targeted Metabolic Engineering. Curr. Opin. Syst. Biol. 6, 1–6. doi:10.1016/j.coisb.2017.05.019
Ritacco, F. V., Wu, Y., and Khetan, A. (2018). Cell Culture Media for Recombinant Protein Expression in Chinese Hamster Ovary (CHO) Cells: History, Key Components, and Optimization Strategies. Biotechnol. Prog. 34, 1407–1426. doi:10.1002/btpr.2706
Ruggiano, A., Foresti, O., and Carvalho, P. (2014). ER-associated Degradation: Protein Quality Control and beyond. J. Cell. Biol. 204, 869–879. doi:10.1083/jcb.201312042
Saraogi, I., and Shan, S.-o. (2011). Molecular Mechanism of Co-translational Protein Targeting by the Signal Recognition Particle. Traffic 12, 535–542. doi:10.1111/j.1600-0854.2011.01171.x
Sarkar, S., and Tran, N. (2021). Cuboplex-Mediated Nonviral Delivery of Functional siRNA to Chinese Hamster Ovary (CHO) Cells. 13 (2), 2336–2345. doi:10.1021/acsami.0c20956
Sauerwald, T. M., Betenbaugh, M. J., and Oyler, G. A. (2002). Inhibiting Apoptosis in Mammalian Cell Culture Using the Caspase Inhibitor XIAP and Deletion Mutants. Biotechnol. Bioeng. 77 (6), 704–716. doi:10.1002/bit.10154
Sergeeva, D., Lee, G. M., Nielsen, L. K., and Grav, L. M. (2020). Multicopy Targeted Integration for Accelerated Development of High-Producing Chinese Hamster Ovary Cells. ACS Synth. Biol. 9, 2546–2561. doi:10.1021/acssynbio.0c00322
Shah, P., and Gilchrist, M. A. (2010). Effect of Correlated Trna Abundances on Translation Errors and Evolution of Codon Usage Bias. PLoS Genet. 6 (9), e1001128. doi:10.1371/journal.pgen.1001128
Shayesteh, M., Ghasemi, F., Tabandeh, F., Yakhchali, B., and Shakibaie, M. (2020). Design, Construction, and Expression of Recombinant Human Interferon Beta Gene in CHO-S Cell Line Using EBV-Based Expression System. Res. Pharm. Sci. 15 (2), 144–153. doi:10.4103/1735-5362.283814
Sung, Y. H., Lee, J. S., Park, S. H., Koo, J., and Lee, G. M. (2007). Influence of Co-Down-Regulation of Caspase-3 and Caspase-7 by Sirnas on Sodium Butyrate-Induced Apoptotic Cell Death of Chinese Hamster Ovary Cells Producing Thrombopoietin. Metab. Eng. 9 (5–6), 452–464. doi:10.1016/j.ymben.2007.08.001
Song, S.-W., Lee, S.-J., Kim, C.-Y., Han, B., and Oh, J.-W. (2013). Rapid Establishment of CHO Cell Lines Producing the Anti-hepatocyte Growth Factor Antibody SFN68. J. Microbiol. Biotechnol. 23, 1176–1184. doi:10.4014/jmb.1305.05056
Sou, S. N., Sellick, C., Lee, K., Mason, A., Kyriakopoulos, S., Polizzi, K. M., et al. (2015). How Does Mild Hypothermia Affect Monoclonal Antibody Glycosylation? Biotechnol. Bioeng. 112, 1165–1176. doi:10.1002/bit.25524
Spahr, C. S., Daris, M. E., Graham, K. C., Soriano, B. D., Stevens, J. L., and Shi, S. D.-H. (2018). Discovery, Characterization, and Remediation of a C-Terminal Fc-Extension in Proteins Expressed in CHO Cells. MAbs 10, 1291–1300. doi:10.1080/19420862.2018.1511197
Tan, H. W. S., Sim, A. Y. L., and Long, Y. C. (2017). Glutamine Metabolism Regulates Autophagy-Dependent Mtorc1 Reactivation During Amino Acid Starvation. Nat. Commun. 8 (1), 338. doi:10.1038/s41467-017-00369-y
Tharmalingam, T., Barkhordarian, H., Tejeda, N., Daris, K., Yaghmour, S., Yam, P., et al. (2018). Characterization of Phenotypic and Genotypic Diversity in Subclones Derived from a Clonal Cell Line. Biotechnol. Prog. 34, 613–623. doi:10.1002/btpr.2666
Torres, M., Zúñiga, R., Gutierrez, M., Vergara, M., Collazo, N., Reyes, J., et al. (2018). Mild Hypothermia Upregulates Myc and Xbp1s Expression and Improves Anti-tnfα Production in CHO Cells. PLoS ONE 13, e0194510. doi:10.1371/journal.pone.0194510
Unzueta, U., Vázquez, F., Accardi, G., Mendoza, R., Toledo-Rubio, V., Giuliani, M., et al. (2015). Strategies for the Production of Difficult-To-Express Full-Length Eukaryotic Proteins Using Microbial Cell Factories: Production of Human Alpha-Galactosidase A. Appl. Microbiol. Biotechnol. 99, 5863–5874. doi:10.1007/s00253-014-6328-9
Urquhart, L. (2020). Top Companies and Drugs by Sales in 2019. Nat. Rev. Drug. Discov. 19(4), 228. doi:10.1038/d41573-020-00047-7
Walsh, G. (2018). Biopharmaceutical Benchmarks 2018. Nat. Biotechnol. 36, 1136–1145. doi:10.1038/nbt.4305
Wang, X.-Y., Zhang, J.-H., Zhang, X., Sun, Q.-L., Zhao, C.-P., and Wang, T.-Y. (2016). Impact of Different Promoters on Episomal Vectors Harbouring Characteristic Motifs of Matrix Attachment Regions. Sci. Rep. 6, 26446. doi:10.1038/srep26446
Wong, D. C. F., Wong, K. T. K., Nissom, P. M., Heng, C. K., and Yap, M. G. S. (2006). Targeting Early Apoptotic Genes in Batch and Fed-Batch CHO Cell Cultures. Biotechnol. Bioeng. 95, 350–361. doi:10.1002/bit.20871
Wu, X., and Rapoport, T. A. (2018). Mechanistic Insights into ER-Associated Protein Degradation. Curr. Opin. Cell. Biol. 53, 22–28. doi:10.1016/j.ceb.2018.04.004
Xie, P., Niu, H., Chen, X., Zhang, X., Miao, S., Deng, X., et al. (2016). Elucidating the Effects of pH Shift on IgG1 Monoclonal Antibody Acidic Charge Variant Levels in Chinese Hamster Ovary Cell Cultures. Appl. Microbiol. Biotechnol. 100, 10343–10353. doi:10.1007/s00253-016-7749-4
Yeo, J. H. M., Ho, S. C. L., Mariati, M., Koh, E., Tay, S. J., Woen, S., et al. (2017). Optimized Selection Marker and CHO Host Cell Combinations for Generating High Monoclonal Antibody Producing Cell Lines. Biotechnol. J. 12 (12). doi:10.1002/biot.201700175
Zhang, J. H., Wang, X. Y., Wang, T. Y., Wang, F., Dong, W. H., Wang, L., et al. (2014). Distance Effect of Matrix Attachment Regions on Transgene Expression in Stably Transfected Chinese Hamster Ovary Cells. Biotechnol. Lett. 36(10), 1937–1943. doi:10.1007/s10529-014-1563-9
Zhang, J. H., Zhang, J. H., Wang, X. Y., Xu, D. H., and Wang, T. Y. (2020). Distance Effect Characteristic of the Matrix Attachment Region Increases Recombinant Protein Expression in Chinese Hamster Ovary Cells. Biotechnol. Lett. 42 (2), 187–196. doi:10.1007/s10529-019-02775-2
Zhang, P., Haryadi, R., Chan, K. F., Teo, G., Goh, J., Pereira, N. A., et al. (2012). Identification of Functional Elements of the GDP-Fucose Transporter SLC35C1 Using a Novel Chinese Hamster Ovary Mutant. Glycobiology. 22 (7), 897–911. doi:10.1093/glycob/cws064
Zhou, M., Crawford, Y., Ng, D., Tung, J., Pynn, A. F., Meier, A., et al. (2011). Decreasing Lactate Level and Increasing Antibody Production in Chinese Hamster Ovary Cells (CHO) by Reducing the Expression of Lactate Dehydrogenase and Pyruvate Dehydrogenase Kinases. J. Biotechnol. 153, 27–34. doi:10.1016/j.jbiotec.2011.03.003
Keywords: recombinant protein, CHO, difficult to express, expression vector, recombinant therapeutic proteins
Citation: Li Z-M, Fan Z-L, Wang X-Y and Wang T-Y (2022) Factors Affecting the Expression of Recombinant Protein and Improvement Strategies in Chinese Hamster Ovary Cells. Front. Bioeng. Biotechnol. 10:880155. doi: 10.3389/fbioe.2022.880155
Received: 21 February 2022; Accepted: 01 June 2022;
Published: 04 July 2022.
Edited by:
Dong-Yup Lee, Sungkyunkwan University, South KoreaReviewed by:
Muriel Bardor, Université de Rouen, FranceKelvin H. Lee, University of Delaware, United States
Copyright © 2022 Li, Fan, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian-Yun Wang, d3RpYW55dW5jbkAxMjYuY29t
 Zheng-Mei Li
Zheng-Mei Li Zhen-Lin Fan
Zhen-Lin Fan Xiao-Yin Wang2,4
Xiao-Yin Wang2,4 Tian-Yun Wang
Tian-Yun Wang