
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Bioeng. Biotechnol. , 14 April 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.878795
Streptomyces mobaraensis transglutaminase (TGase) is extracellularly expressed as a zymogen and then activated by TGase-activating protease (TAP). In this study, we reported the strategy for improving TGase production via the regulation of TAP activity in S. mobaraensis. First, we analyzed the effects of three inorganic nitrogen sources on TGase production. With 30 mM nitrogen content, the time to the peak of TGase activity induced by (NH4)2SO4 or NH4Cl was 72 h, 12 h earlier than that of the fermentation without adding NH4+. SDS-PAGE analysis indicated that NH4+ accelerated the TGase activation in S. mobaraensis. Then, we examined the effect of NH4+ on TAP biosynthesis using a TGase-deficient S. mobaraensis strain. It showed that NH4+ enhanced the TAP activity at the early stage of the fermentation, which was dependent on the concentration and time of NH4+ addition. Last, the yield and productivity of S. mobaraensis TGase were increased by 1.18-fold and 2.1-fold, respectively, when optimal NH4+ addition (60 mM and 12 h) was used. The fermentation period was shortened from 84 to 48 h. The NH4+ addition also increased the storage stability of crude enzyme at room temperature. These findings will benefit the TGase production and its activation mechanism in S. mobaraensis.
Transglutaminase (TGase, EC 2.3.2.13) belongs to the transferases family that introduces covalent cross-links in proteins between glutamine residues and primary amines through an acyl-transfer reaction (Akbari et al., 2021). Due to the unique catalytic reaction, TGase has been exploited to improve the texture properties of protein-based foods (Miwa, 2020). Recently, it has also exhibited application potential in pharmacological production, textile industry, and leather processing (Zhu and Tramper, 2008; Duarte et al., 2019; Doti et al., 2020). In contrast to the enzymes extracted from animals and plants, Streptomyces mobaraensis TGase is a Ca2+-independent enzyme and easier to be produced on a large scale (Ando et al., 1989). Although Streptomyces TGases have been expressed in a variety of heterologous hosts, only S. mobaraensis TGase is approved as generally recognized as safe (GRAS) (Yin et al., 2021). These advantages benefit the TGase application and make S. mobaraensis fermentation become the main source of commercial TGase products (Santhi et al., 2017). Therefore, it is desirable to improve the TGase production by S. mobaraensis.
Since the discovery of Streptomyces TGase in 1989, isolating novel TGase-producing strains (Ceresino et al., 2018) and screening high-yielding mutant strains have long been used for enhancing TGase production (Jiang et al., 2017; Yin et al., 2021). On the other hand, the media composition and fermentation process were critical for TGase biosynthesis (Akbari et al., 2021). Transcription analysis indicated that protease, CTAB, and MgCl2 were proven to promote TGase expression in S. mobaraensis (Fatima and Khare, 2021). To improve the economic efficiency, TGase fermentation was conducted using agricultural wastes, such as wheat bran and non-commercial potatoes (Guerra-Rodriguez and Vazquez, 2014; Fatima et al., 2019). Compared to genetic modification, these “non-genetic” strategies are much easier to be accepted for food industries. To date, the highest TGase activity (19.7 U/mL) in S. mobaraensis was achieved by random mutagenesis based on atmospheric and room-temperature plasma mutagenesis and flow cytometry technology (Yin et al., 2021). However, the fermentation period of Streptomyces TGase usually reached 72–96 h or even over 10 days (Akbari et al., 2021). Thus, reducing the fermentation period is crucial for enhancing TGase productivity in S. mobaraensis.
In S. mobaraensis, TGase is exported in the form of inactive zymogen (pro-TGase) and then fully activated into a mature form by its endogenous metalloprotease (TAMP) and AP-specific tri/tetrapeptidyl aminopeptidase within the next 2 days (Zotzel et al., 2003a; Zotzel et al., 2003b). This activation process is regulated by a Streptomyces subtilisin inhibitor (SSTI), which could inhibit TAMP activity (Juettner et al., 2020). Therefore, improving the activation process is an important strategy to reduce the fermentation period. It has been demonstrated that in vitro protease addition reduced the fermentation period of the Streptomyces hygroscopicus TGase by 18% (Zhang et al., 2010). By inducing the overexpression of total protease, metalloprotease, and serine protease, MgCl2 can also accelerate the activation of pro-TGase (Zhang et al., 2012). However, protease addition is not cost-effective, while excessive MgCl2 is detrimental to cell growth (Zhang et al., 2012). This activation was effective in the context of a small amount of zymogen. In addition, cetyltrimethylammonium bromide is predicted to inactivate the protease inhibitor, resulting in improved activation (Zhang et al., 2008). To be noted, all the protease-mediated activations were investigated using the Streptomyces strains with relatively low TGase production.
In this study, we first investigated the effects of three inorganic nitrogen sources on the fermentation period of smY 2019 (a robust variant of S. mobaraensis DSM40587) (Yin et al., 2021), determining that NH4+ could improve TGase activation. Based on a reliable measure for TAP activity using a TGase-deficient S. mobaraensis strain smY2019∆tg, the changes of TAP activity during fermentation were precisely characterized. It was shown that NH4+ enhanced TAP activity, dependent on the concentration and time of NH4+ added. Finally, TGase productivity was significantly improved by regulating zymogen activation in smY 2019.
S. mobaraensis smY2109 and smY2019∆tg were used to study TGase and TAP, respectively (Yin et al., 2021). The plasmid pET-22b (+) and Escherichia coli BL21 (DE3) were used for expressing pro-TGase.
The spore culture on the GYM agar medium and seed culture in shake flasks were performed as described in the previous study (Yin et al., 2021). The composition of the basal fermentation medium was as follows: 2% glycerol, 2% peptone, 0.5% yeast extract, 2% soya flour, 0.4% K2HPO4, 0.2% KH2PO4, and 0.2% MgSO4. To study the influence of different inorganic nitrogen sources on the TGase fermentation period, NaNO3, (NH4)2SO4, and NH4Cl were added to the basal fermentation medium in the same total nitrogen content (30 mM final concentration).
The gene fragment of pro-TGase was amplified from the S. mobaraensis smY2019 genome by PrimeSTAR GXL DNA Polymerase (TaKaRa, Dalian, China) using the primer pair ptg-F (CATGCCATGGGCAGCGGCACCGGGGAAGAGAAGAG)/ptg-R (CCGCTCGAGCGGCCAGCCCTGTGTCACCTTGTCG) and cloned into the Nco I-Xho I sites of pET-22b (+), generating the pro-TGase expression plasmid pET-22b/ptg. The plasmid pET-22b/ptg was introduced into E. coli BL21 (DE3). The recombinant E. coli strain was inoculated into a Luria–Bertani medium containing 100 μg/ml ampicillin for seed culture at 37°C for 12 h. Then, 1-ml seed cultures were transferred into a 50-ml terrific broth (TB) medium containing the same amount of antibiotics and further cultivated at 37°C. At an OD600 of 0.8, the cells were induced by adding the inducer isopropyl beta-D-1-thiogalactopyranoside (400 μM, final concentration). Growth was continued at 20°C for up to 40 h. The culture supernatant was subjected to affinity purification using the His-Trap column (GE Healthcare, NY, United States). The pro-TGase was eluted with elution buffer (50 mM Tris-HCl, 50 mM NaCl, and 150 mM imidazole, pH 8.0) and dialyzed against dialysis buffer (50 mM Tris-HCl, pH 8.0). Protein concentration of purified pro-TGase was determined by using the BCA protein assay kit (Beyotime, Shanghai, China). The samples were diluted to 0.5 mg/ml of protein concentrations and used as the substrate for TAP activity measurement.
For detecting the protease activity, the activation reaction was initiated by mixing the purified pro-TGase (0.5 mg/ml) with an equal volume of the culture supernatant of smY2019∆tg. One unit of the TAP toward the pro-TGase was defined as the amount of enzyme needed to generate one unit of mature TGase per hour at 30°C.
The biomass of S. mobaraensis was measured by using the dry cell weight (DCW) method. S. mobaraensis cells were harvested by centrifugation (5,000 × g, 15 min) from 10 ml fermentation broth. After washing with sterile water three times, the cell pellets were dried at 105°C until they had a constant weight.
According to the previous report (Yin et al., 2021), the colorimetric method was conducted to measure TGase activity using N-CBZ-Gln-Gly (Sigma-Aldrich, Shanghai, China) as the substrate. One unit of TGase activity is defined as the amount of enzyme needed to generate 1 μmol of hydroxamate per min at 37°C.
SDS-PAGE analysis was performed to separate proteins on a 10% running gel, which was visualized after staining with Coomassie Brilliant Blue R250.
The logistic function was used to fit the curve of the specific production rate by OriginLab 2018 software (OriginLab Corporation, Northampton, United States). First-order kinetics was applied to calculate the specific production rate as follows:
where X is DCW (g/L); t is culture time (h); and P is TGase activity (U/mL).
All experiments were carried out in triplicate at least.
Nitrogen source is critical for cell growth and product biosynthesis. The ectoine production was improved by optimizing the type and quantity of the nitrogen sources (Zhang et al., 2022). However, previously, there were very few reports on the biosynthesis of TGase using inorganic nitrogen sources. In the present study, based on the basal fermentation medium, we analyzed the effects of three inorganic nitrogen sources at a constant concentration (30 mM) on the production of TGase by the S. mobaraensis smY 2019. As shown in Figure 1A, the cultivation without the inorganic nitrogen sources rapidly accumulated the TGase after 36 h, and the maximal enzyme activity was at 84 h. In the case of (NH4)2SO4 and NH4Cl, the rapid TGase production started at 24 h, and the peak value of the enzyme activity occurred at 72 h, 12 h earlier than the control. In contrast, NaNO3 did not affect the TGase biosynthesis (Figure 1A). The cell growth of smY2019 was not significantly affected by adding the inorganic nitrogen sources (Figure 1B), suggesting that the reduced TGase production period is not due to cell growth changes. As we all know, TGase was secreted as inactive pro-TGase and then transformed into active mature TGase (Zotzel et al., 2003a). To understand the reason for the accelerated TGase biosynthesis, the culture supernatant of each condition was taken at 36 h and 72 h and subjected to SDS-PAGE analysis. For the samples taken at 36 h, both pro-TGase (43 kDa) and TGase (38 kDa) bands could be seen in all cases (Figure 1D). The sample with NaNO3 added had similar protein bands with the control, with thick bands of pro-TGase and thin bands of TGase, while the addition of (NH4)2SO4 or NH4Cl had more thick bands of TGase. When fermented in the medium with (NH4)2SO4 or NH4Cl for 72 h, the pro-TGase bands were completely converted into TGase bands. However, in addition to the TGase bands, thin pro-TGase bands were still detected at 72 h in the case of control and NaNO3 (Figure 1D). After the in vitro activation with dispase, all the samples at 36 h and 72 h shared similar TGase activities (18.6–21.1 U/mL) (Figure 1C). These findings indicated that the addition of (NH4)2SO4 and NH4Cl significantly influences S. mobaraensis TGase activation instead of pro-TGase expression. However, Na2SO4 and NaCl did not improve the TGase activation (data not shown), suggesting the critical role of NH4+.
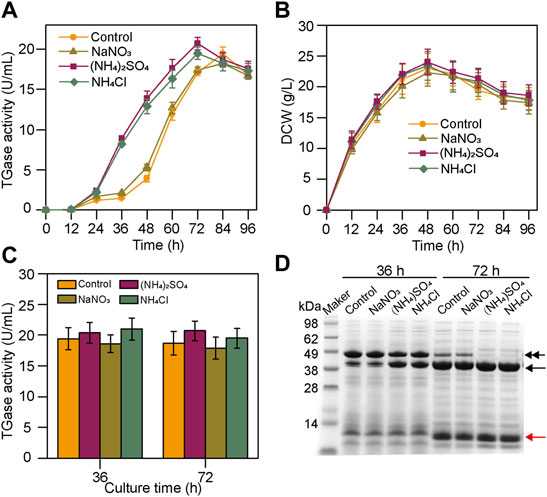
FIGURE 1. Effects of an inorganic nitrogen source on TGase production by smY 2019. (A) Time course of TGase production with the addition of different inorganic nitrogen sources. (B) Time course of biomass. (C) TGase activity assay after the activation by dispase. (D) SDS-PAGE analysis of the culture supernatants. The pro-TGase and TGase bands are indicated with black double and single arrows, respectively. The SSTI bands were identified using MALDI-TOF/MS (data not shown) and are indicated with red arrows. Each fermentation process was performed in a 250-ml flask containing 30 ml of the fermentation medium added with 30 mM of inorganic nitrogen at 30°C and 220 rpm. In the control experiment, no inorganic nitrogen source was added.
It has shown that the biosynthesis of pro-TGase is simultaneous with its activation at the first half of S. mobaraensis fermentation, and the inactivation of the mature TGase could be seen in the later stage of the fermentation (Figure 1A and Supplementary Figure S1). Therefore, it is hard to characterize TAP activity using S. mobaraensis with TGase production. It is essential to establish a method to measure the TAP activity accurately. The pro-TGase from S. mobaraensis was expressed in E. coli BL21 (DE3) and purified (Supplementary Figure S2A). The TAP activity was measured using the purified pro-TGase as a substrate and indicated by TGase activity. Meanwhile, the previously constructed TGase-deficient strain smY2019∆tg was used as the research host for analyzing TAP activity during fermentation (Yin et al., 2021). smY2019∆tg did not produce TGase under the same cultivation condition, eliminating the interference of its own TGase activity on TAP activity measurement (Figure 2A). Compared with the previous method, this method exclusively reflects the activity of protease that can activate pro-TGase, which was more sensitive and reliable (Zhang et al., 2012).
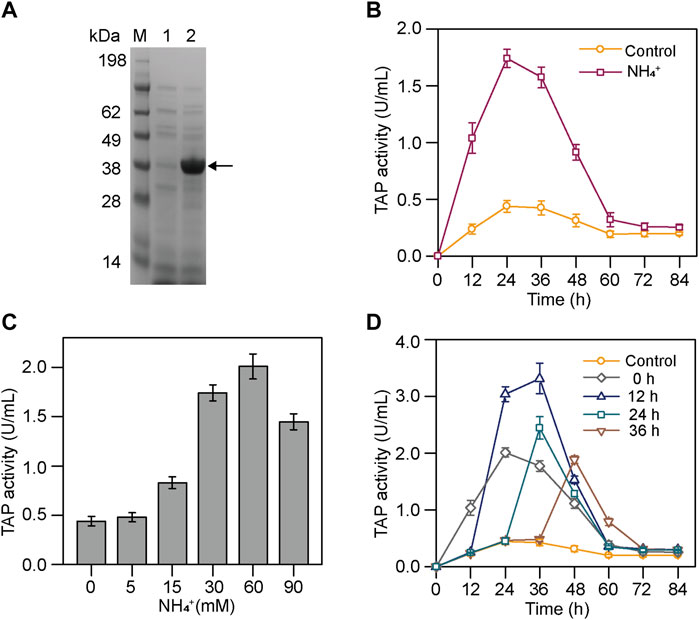
FIGURE 2. Effects of NH4+ on the TAP activity in smY2019∆tg. (A) SDS-PAGE analysis of culture supernatant after cultivation for 84 h. Lane 1: smY2019∆tg; lane 2: smY 2019. The TGase band is indicated with single arrows. (B) Time course of TAP activity in smY2019∆tg with or without 30 mM NH4+ addition. (C) Effect of NH4+ concentration on the TAP activity in smY2019∆tg. The culture supernatant from the cultivation for 24 h was used for TAP activity determination. (D) Effect of NH4+ addition time on TAP activity during the culture process at a constant NH4+ concentration of 60 mM. NH4+ was added in the form of (NH4)2SO4.
We here analyzed the TAP activities of smY2019∆tg in the absence and presence of (NH4)2SO4. The TAP activity in smY2019∆tg with 30 mM NH4+ addition grew much faster than that of the cultivation without NH4+ at the initial 24 h, and the peak value achieved the former reaching 1.74 U/mL, four times higher than that of the latter. Within the next 36 h, the TAP activities under both conditions declined gradually and maintained a very low level (Figure 2B). To further compare the TAP activities, the in vitro activation process of samples taken at 24 h was analyzed. The cultivation with NH4+ completely activated the 0.5 mg/ml pro-TGase within 9 h, while that without NH4+ did not even activate even after 18 h (Supplementary Figures S2B,C). These results confirmed that NH4+ increased the TAP activity at the early stage of fermentation. TAMP (purified from surface colonies on plates) was considered to be involved in TGase activation and regulated by SSTI in S. mobaraensis (Zotzel et al., 2003a; Juettner et al., 2018; Fuchsbauer, 2021). However, the transcript levels of TAMP and SSTI at 24 h were not changed in the presence and absence of NH4+ (data not shown). Streptomyces are prodigious producers of proteases. (Chater et al., 2010). Streptomyces coelicolor, a model organism for the study of Streptomyces, contains 56 genes encoding protease, including eight metalloproteinase genes (Bentley et al., 2002). Gene expression often differs when the growth conditions were changed. Adding NH4+ may induce the expression of a novel metalloproteinase or relieve the inhibition of this activating protease in the early stage of fermentation.
Then, the concentration and time of NH4+ addition were optimized to further improve the TGase activation. When (NH4)2SO4 was added at the beginning of the fermentation, the TAP activity at 24 h increased with the concentration of NH4+ from 0 to 60 mM, while further increase in the NH4+ concentration (90 mM) reduced the protease activity (Figure 2C). The effect of NH4+ addition time on TAP activity was investigated at a constant NH4+ concentration (60 mM) during the culture process. After the NH4+ addition, the TAP activity showed an initial increase followed by a drop in all addition cases (Figure 2D). When NH4+ was added at 0 or 12 h, the TAP activity increased continuously for 24 h. In contrast, the increase phase was reduced to 12 h in the case of the addition at 24 h or 36 h. Finally, the NH4+ addition at 12 h achieved the highest TAP activity among the tested addition time. To be noted, SSTI bands at 72 h were smaller than those at 36 h (Figure 1D). Researchers have demonstrated that SSTI is secreted into the fermentation medium in an early cultivation stage and partially degraded by tripeptidyl aminopeptidase in the later stage (Juettner et al., 2020). Generally, partial degradation endows SSTI with full TAP inhibitory activity (Juettner et al., 2020). Thus, SSTI might undergo a similar processing, which may account for the rapid decrease in TAP activity in the later phase of fermentation.
To improve TGase productivity, NH4+ (60 mM) was added at 12 h during the fermentation of smY 2019. As shown in Figure 3A, the fermentation with NH4+ addition achieved the peak value of TGase activity at 48 h, 36 h earlier than the fermentation without NH4+ addition. Accordingly, the peak of the specific production rate also shifted forward when NH4+ was added (Figure 3B). As indicated by SDS-PAGE analysis, the pro-TGase band was completely converted into the mature TGase band at 48 h in the case of the NH4+ addition, and this band conversion ended at 84 h without NH4+ addition (Figure 3C). Moreover, the TGase yield of the former was 18% higher than that of the latter (Figure 3A). Finally, through the NH4+ addition, TGase productivity was increased from 0.23 U/(mLh) to 0.48 U/(mLh). In a previous study, MgCl2 had been shown to have a positive effect on the activation of pro-TGase (Zhang et al., 2012). It is noteworthy that this activation was investigated in the context of a small amount of zymogen. For smY 2019, a high-yielding pro-TGase mutant, the activation effect was not improved after optimizing the amount of MgCl2 (Supplementary Figure S3). Excessive addition of MgCl2 could even be deleterious for the growth of S. mobaraensis (Zhang et al., 2012). As a nitrogen source, NH4+ was harmless to cell growth and was a more appropriate activator of pro-TGase.
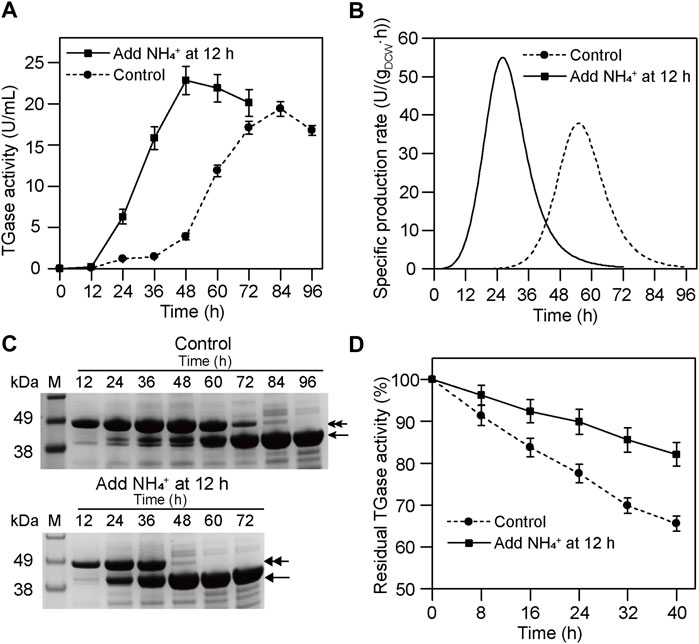
FIGURE 3. Effects of NH4+ on TGase production by smY 2019. (A) Time course of TGase production. (B) Specific production rate curve of TGase. (C) SDS-PAGE analysis of the culture supernatant. The pro-TGase and TGase bands are indicated with double and single arrows, respectively. (D) Storage stability of the crude TGase solution at room temperature. The crude TGase solution referred to 48-h culture supernatant with 60 mM NH4+ addition. The 84-h culture supernatant without addition of NH4+ was used as a control. NH4+ was added in the form of (NH4)2SO4.
To be noted, NH4+ addition improved the storage stability of the crude TGase solution of smY 2019. When treated at room temperature for 40 h, the TGase activity of the culture supernatant from the 48-h culture broth with NH4+ addition retained 82% of initial activity, while that from 84-h culture broth without NH4+ only obtained 65% residual activity (Figure 3D). This is probably due to the fact that a lot of proteases were produced at the later stage of S. mobaraensis, resulting in proteolytic degradation of TGase (Fuchsbauer, 2021). Therefore, the reduced fermentation period could not only increase the economy of the TGase but also its storage stability.
This study was the first to demonstrate that NH4+ addition was capable to enhance TAP activity in S. mobaraensis. After optimizing the amount and time of NH4+ added, pro-TGase activation was considerably improved as TAP activity was significantly enhanced. Hence, the TGase productivity was increased 2.1 times relative to that without NH4+ addition, and maximum production was obtained in 43% less time. Our study makes the production of TGase more cost-effective and enhances the storage stability of crude enzyme solutions. Future work is to explore the key protease by transcriptomic comparison and regulate its expression at the gene level to efficiently activate pro-TGase using an inexpensive medium.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
XY: methodology, writing—original draft, formal analysis, and investigation. SR: methodology and investigation. JZ: investigation and formal analysis. GD: validation and project administration. JC: data curation and project administration. SL: writing—review and editing, supervision, and validation.
This work was supported by the National Key Research and Development Program of China (No. 2021YFC2101400) and the National Natural Science Foundation of China (No. 32071474 and 31771913).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.878795/full#supplementary-material
Akbari, M., Razavi, S. H., and Kieliszek, M. (2021). Recent Advances in Microbial Transglutaminase Biosynthesis and its Application in the Food Industry. Trends Food Sci. Technol. 110, 458–469. doi:10.1016/j.tifs.2021.02.036
Ando, H., Adachi, M., Umeda, K., Matsuura, A., Nonaka, M., Uchio, R., et al. (1989). Purification and Characteristics of a Novel Transglutaminase Derived from Microorganisms. Agric. Biol. Chem. 53 (10), 2613–2617. doi:10.1080/00021369.1989.10869735
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A.-M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete Genome Sequence of the Model Actinomycete Streptomyces Coelicolor A3(2). Nature 417 (6885), 141–147. doi:10.1038/417141a
Ceresino, E. B., de Melo, R. R., Kuktaite, R., Hedenqvist, M. S., Zucchi, T. D., Johansson, E., et al. (2018). Transglutaminase from Newly Isolated Streptomyces Sp. CBMAI 1617: Production Optimization, Characterization and Evaluation in Wheat Protein and Dough Systems. Food Chem. 241, 403–410. doi:10.1016/j.foodchem.2017.09.010
Chater, K. F., Biró, S., Lee, K. J., Palmer, T., and Schrempf, H. (2010). The Complex Extracellular Biology ofStreptomyces. FEMS Microbiol. Rev. 34 (2), 171–198. doi:10.1111/j.1574-6976.2009.00206.x
Doti, N., Caporale, A., Monti, A., Sandomenico, A., Selis, F., and Ruvo, M. (2020). A Recent Update on the Use of Microbial Transglutaminase for the Generation of Biotherapeutics. World J. Microbiol. Biotechnol. 36 (4), 53. doi:10.1007/s11274-020-02829-y
Duarte, L., Matte, C. R., Bizarro, C. V., and Ayub, M. A. Z. (2019). Review Transglutaminases: Part II-Industrial Applications in Food, Biotechnology, Textiles and Leather Products. World J. Microbiol. Biotechnol. 36 (1), 11–20. doi:10.1007/s11274-019-2792-9
Fatima, S. W., and Khare, S. K. (2021). Effect of Key Regulators in Augmenting Transcriptional Expression of Transglutaminase in Streptomyces Mobaraensis. Bioresour. Technol. 340, 125627. doi:10.1016/j.biortech.2021.125627
Fatima, S. W., Tiwari, R., and Khare, S. K. (2019). Utilization of Agro-Industrial Waste for Production of Transglutaminase from Streptomyces Mobaraensis. Bioresour. Technol. 287, 121391. doi:10.1016/j.biortech.2019.121391
Fuchsbauer, H. L. (2021). Approaching Transglutaminase from Streptomyces Bacteria over Three Decades. FEBS J. doi:10.1111/febs.16060
Guerra-Rodríguez, E., and Vázquez, M. (2014). Evaluation of a Novel Low-Cost Culture Medium Containing Exclusively Milk, Potato and Glycerol for Microbial Transglutaminase Production by Streptomyces Mobaraensis. Chem. Eng. Res. Des. 92 (4), 784–791. doi:10.1016/j.cherd.2013.06.027
Jiang, Y., Shang, Y. P., Li, H., Zhang, C., Pan, J., Bai, Y. P., et al. (2017). Enhancing Transglutaminase Production of Streptomyces Mobaraensis by Iterative Mutagenesis Breeding with Atmospheric and Room-Temperature Plasma (ARTP). Bioresour. Bioproc. 4, 37. doi:10.1186/s40643-017-0168-2
Juettner, N. E., Classen, M., Colin, F., Hoffmann, S. B., Meyners, C., Pfeifer, F., et al. (2018). Features of the Transglutaminase-Activating Metalloprotease from Streptomyces Mobaraensis DSM 40847 Produced in Escherichia coli. J. Biotechnol. 281, 115–122. doi:10.1016/j.jbiotec.2018.07.004
Juettner, N. E., Schmelz, S., Anderl, A., Colin, F., Classen, M., Pfeifer, F., et al. (2020). The N‐terminal Peptide of the Transglutaminase‐activating Metalloprotease Inhibitor from Streptomyces Mobaraensis Accommodates Both Inhibition and Glutamine Cross‐linking Sites. FEBS J. 287 (4), 708–720. doi:10.1111/febs.15044
Miwa, N. (2020). Innovation in the Food Industry Using Microbial Transglutaminase: Keys to success and Future Prospects. Anal. Biochem. 597, 113638. doi:10.1016/j.ab.2020.113638
Santhi, D., Kalaikannan, A., Malairaj, P., and Arun Prabhu, S. (2017). Application of Microbial Transglutaminase in Meat Foods: A Review. Crit. Rev. Food Sci. Nutr. 57 (10), 2071–2076. doi:10.1080/10408398.2014.945990
Yin, X., Li, Y., Zhou, J., Rao, S., Du, G., Chen, J., et al. (2021). Enhanced Production of Transglutaminase in Streptomyces Mobaraensis through Random Mutagenesis and Site-Directed Genetic Modification. J. Agric. Food Chem. 69 (10), 3144–3153. doi:10.1021/acs.jafc.1c00645
Zhang, D., Wang, M., Du, G., Zhao, Q., Wu, J., and Chen, J. (2008). Surfactant Protein of the Streptomyces Subtilisin Inhibitor Family Inhibits Transglutaminase Activation in Streptomyces Hygroscopicus. J. Agric. Food Chem. 56 (9), 3403–3408. doi:10.1021/jf703567t
Zhang, D., Zhu, Y., and Chen, J. (2010). Microbial Transglutaminase Production: Understanding the Mechanism. Biotechnol. Genet. Eng. Rev. 26, 205–222. doi:10.5661/bger-26-205
Zhang, L., Zhang, L., Han, X., Du, M., Zhang, Y., Feng, Z., et al. (2012). Enhancement of Transglutaminase Production in Streptomyces Mobaraensis as Achieved by Treatment with Excessive MgCl2. Appl. Microbiol. Biotechnol. 93 (6), 2335–2343. doi:10.1007/s00253-011-3790-5
Zhang, H., Liang, Z., Zhao, M., Ma, Y. Q., Luo, Z. S., Li, S., et al. (2022). Metabolic Engineering of Escherichia coli for Ectoine Production with a Fermentation Strategy of Supplementing the Amino Donor. Front. Bioeng. Biotechnol. 10, 824859. doi:10.3389/fbioe.2022.824859
Zhu, Y., and Tramper, J. (2008). Novel Applications for Microbial Transglutaminase beyond Food Processing. Trends Biotechnol. 26 (10), 559–565. doi:10.1016/j.tibtech.2008.06.006
Zotzel, J., Keller, P., and Fuchsbauer, H.-L. (2003a). Transglutaminase from Streptomyces Mobaraensis Is Activated by an Endogenous Metalloprotease. Eur. J. Biochem. 270 (15), 3214–3222. doi:10.1046/j.1432-1033.2003.03703.x
Keywords: transglutaminase, Streptomyces mobaraensis, NH4+, zymogen activation, productivity
Citation: Yin X, Rao S, Zhou J, Du G, Chen J and Liu S (2022) Improved Productivity of Streptomyces mobaraensis Transglutaminase by Regulating Zymogen Activation. Front. Bioeng. Biotechnol. 10:878795. doi: 10.3389/fbioe.2022.878795
Received: 18 February 2022; Accepted: 21 March 2022;
Published: 14 April 2022.
Edited by:
Fengjie Cui, Jiangsu University, ChinaReviewed by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaCopyright © 2022 Yin, Rao, Zhou, Du, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song Liu, bGl1c29uZ0BqaWFuZ25hbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.