
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 05 April 2022
Sec. Bioprocess Engineering
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.870672
This article is part of the Research Topic From Biomass to Bio-energy and Bio-chemicals: Pretreatment, Thermochemical Conversion, Biochemical Conversion and Its Bio-Based Applications View all 20 articles
Dehydrins (DHNs) belong to group II of late embryogenesis-abundant (LEA) proteins, which are up-regulated in most plants during cold, drought, heat, or salinity stress. Despite the importance of dehydrins for the plants to resist abiotic stresses, it is necessary to obtain plant-derived dehydrins from different biomass. Generally, dehydrin PicW1 from Picea wilsonii is involved in Kn-type dehydrin with five K-segments, which has a variety of biological activities. In this work, Picea wilsonii dehydrin PicW1 was expressed in Escherichia coli and purified by chitin-affinity chromatography and size-exclusion chromatography, which showed as a single band by SDS-PAGE. A cold-sensitive enzyme of lactate dehydrogenase (LDH) is used to explore the protective activities of other proteins. Temperature stress assays showed that PicW1 had an effective protective effect on LDH activity, which was better than that of bovine serum albumin (BSA). This study provides insights into the purification and protective activity of K5 DHNs for the advancement of dehydrin structure and function from biomass.
Biomass refers to the organic matter produced by all organisms and growth. Generally, biomass is the abundant renewable resource that can not only be transformed into many high value-added products of chemicals, biofuels, and advanced materials (Dong et al., 2020; Liu et al., 2021; Wang et al., 2021; Xu et al., 2021; Hu et al., 2022) but also contains highly abundant stress-resistant gene resources (Ning et al., 2021). Obtaining proteins from biomass tissues requires extracting genomic DNA and RNA, reverse transcription to obtain cDNA, and then designing specific primers to amplify a specific segment of genomic DNA according to the known similar biomass genes. PCR primers were designed with the recombinant plasmid containing the target gene as the template. The amplified products were recovered and connected to the cloning vector, transformed into E. coli and extracted the plasmid, and then the appropriate expression vector was selected. The target protein was successfully obtained through a variety of separation and purification techniques (Zhou et al., 2006). An area of considerable research interest is the plant’s ability to resist a variety of abiotic stresses such as drought, cold, high temperature, and high salinity (Hara et al., 2003; Saavedra et al., 2006; Hundertmark and Hincha, 2008; Hirayama and Shinozaki, 2010). All of these result in cell dehydration (Galau et al., 1986). One family of proteins that is expressed during cell dehydration has been named dehydration proteins (dehydrins, or DHNs) (Close, 1996; Graether and Boddington, 2014). DHNs are highly hydrophilic, and their structural analysis implies that they are intrinsically disordered proteins (IDPs), which provide a flexible property to interact with metal ions and biomolecules (Dunker et al., 2001; Halder et al., 2018; Lv et al., 2018; Gupta et al., 2019). DHNs belong to LEA-DII (late embryogenesis-abundant proteins) family and their types have been classified using the segments K, Y, and S. The K-segment is an approximately 15-amino-acid-long conserved lysine-rich motif, and it tends to adopt an amphipathic α-helix structure according to computational prediction (Lv et al., 2018). The Y-segment is a relatively short segment which is named for the conserved tyrosine residues (Hara et al., 2010). The S-segment is a serine-rich region that can be phosphorylated, and it may play a role in the protein delivery to the nucleus (Goday, 1994). In addition to these segments, Φ-segments (G- and polar amino acid-rich sequences) (Close, 1997), F-segments (Strimbeck, 2017; Wei et al., 2019; Ohkubo et al., 2020), and ChP-segments (Graether and Boddington, 2014) have also been described.
In previous studies, four different proteins from various Spruce species, namely, PicW1, PicW2, PicM, and PicK, were identified and expressed in E. coli. It had been proved that PicW genes were an effective antifreeze resource by thermal hysteresis activity and E. coli antifreeze tests (Zhao et al., 2017; Zhang et al., 2018). In this study, we report the purification of a protective protein PicW1 from Picea wilsonii. PicW1 belongs to the K5DHN family, because the protein has five highly conservative QKA segments, and each segment contains 2–5 Lys amino acids. PicW1 was cloned from Picea wilsonii and expressed in E. coli, and the protein was separated and purified by chitin-affinity chromatography and gel filtration chromatography. In addition, the activity of LDH is commonly used as a marker for the ability of dehydrins to rescue protein function during stress. We found that PicW1 can protect the enzyme activity of LDH which is better than BSA (a known protective protein) under freeze-thaw stress. However, in the high-temperature tests, the protective effect of PicW1 is similar to that of BSA and better than the negative control lysozyme (LZM).
The chitin column (New England Biolabs, United States) and AKTA protein purification system were used for two-step protein purification. An ultrafiltration tube (Amicon ultra, United States) of 10 kDa millipore was used for protein concentration in the purification process. Liquid nitrogen was purchased from the Institute of Semiconductors, Chinese Academy of Sciences. The DU730 ultraviolet spectrophotometer (BECKMAN COULTER, United States) was used to measure the absorbance in the enzyme activity reaction. The protein concentration was measured using NanoDrop 2000c (Thermo Fisher Scientific, United States).
The Picea wilsonii dehydrin gene pTWIN1-PicW1 was given by Professor Jichen Xu (National Engineering Laboratory of Tree Breeding, Beijing Forestry University), and the recombinant plasmid, pRSF-Duet-LDH5, was given by Professor Fei Xiao (Peking University Fifth Clinical College of Geriatrics Key Laboratory). Both E. coli DH5α strain and E. coli BL21 (DE3) were purchased from TaKaRa Company.
E. coli BL21 (DE3) competent cells were transformed with the recombinant construct (Figure 1A). Ten milliliters of LB medium pH 7.0 containing ampicillin (100 μg/ml) were inoculated with a positive BL21 colony and then incubated for 12 h at 37°C with shaking at 220 rpm. One liter fresh LB medium containing ampicillin (100 μg/ml) was inoculated with the aforementioned cells and grown until the OD600 nm reached 0.6–0.8 absorbance units (mid-log phase). Upon reaching the mid-log phase, protein expression was induced with isopropyl β-D-thiogalactoside (IPTG) at a concentration of 0.5 mM for 20 h at 16°C and 180 rpm. Then, the cells were harvested by centrifugation at 4,000 rpm for 30 min at 4°C. The pellet was saved and the supernatant was discarded.
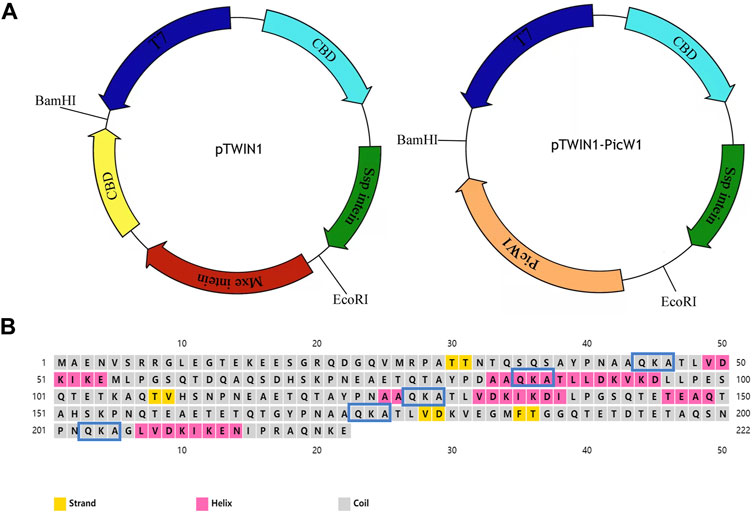
FIGURE 1. Prokaryotic expression vector pTWIN1 and secondary structures of PicW1. (A) CBD, chitin-binding domain; Mxe intein, Mycobacterium xenopi GyrA intein; Ssp intein, Synechocystis sp. DnaB intein gene; PicW1, the coding region of PicW1 gene; T7, T7promoter. (B) PicW1 has 222 amino acids, of which 5 K-segments start from the amino acid sequence QKA. Yellow squares represent the strand, pink squares represent the helix, and gray squares represent the coil.
The pellet was collected from 300 ml culture, lysed in 25 ml lysis buffer A containing 20 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM EDTA, and 1 mM phenylmethylsulfonnmkyl fluoride (PMSF). The cells were sonicated on ice for 4 s 70–100 times spaced by about 6 s time interval to disrupt cell membranes. Then, the lysates were centrifuged for 30 min at 13,000 rpm at 4°C to separate soluble proteins from other cellular components. The clarified lysate was used for further purification.
Chitin-affinity chromatography was equilibrated with buffer A1 (20 mM Tris-HCl pH 8.0, 500 mM NaCl, and 1 mM EDTA). The clarified lysate (supernatant) was run onto a 3 ml chitin column which was followed by washing with 20-bed volumes of washing buffer B (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM EDTA, 0.1% Triton-X100) to thoroughly remove non-specific proteins (O'Shea et al., 2015). The column was supplemented with three column volumes of cleavage buffer C (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM EDTA, and 120 mM DTT) and placed on a rotary mixer. Then, the flow was stopped and the column was placed on a rotary mixer at 4°C for 48 h for the on-column cleavage of PicW1 protein. DTT was added every 6 h. The samples were collected in 6, 12, 18, 24, 30, 36, and 48 h and analyzed by 15% polyacrylamide gel electrophoresis on sodium dodecyl sulfate (SDS-PAGE). When the cutting was completed, the chitin matrix naturally settled through gravity sedimentation. The protein that flowed down was concentrated to within 2 ml in a 10 kDa concentration tube at 4,000 rpm, filtered through a 0.22 ultrafiltration membrane to remove impurities, and then prepared for the next purification operation.
The preliminarily purified protein solution was applied onto a Superdex 200 pg 10/300 GL column which was equilibrated with buffer D (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 5 mM DTT) (O'Shea et al., 2015). Size exclusion was performed at 0.4 ml/min. The peaks of the target proteins were collected and concentrated to 10 mg/ml. The purified proteins were collected and tested through 18% SDS-PAGE. The LDH5 was prepared as previously described (Liu et al., 2017). The purified PicW1 and LDH5 were temporarily stored at 4°C. If long-term storage is required, 50% glycerol can be added to the protein and stored at −80°C.
The LDH protection assay was carried out in order to determine the protective role of the PicW1 protein. The activity of LDH was measured using the modified technique of Lin and Thomashow (1992). The negative control was LZM and the positive control was bovine serum albumin (BSA). All the proteins and reagents were dissolved in buffer E (10 mM phosphate buffer, pH 7.5). Different protein samples and LDH were mixed at the molar ratio of 0:1, 5:1, 10:1, and 20:1 into a 60 μl solution; the final concentration of LDH was 0.13 μM. The enzyme solution was divided equally into six tubes. For the cryoprotective assay, three tubes were subjected to freeze-thaw stress treatment and the other three tubes were left untreated. Freezing stress was imparted on the enzyme by five cycles of freezing in liquid nitrogen for 1 min and subsequent thawing in a water bath at 25°C for 5 min in the presence and absence of the PicW1 protein.
For the high-temperature stress assay, we set four temperatures which were 43, 49, 55, and 61°C, respectively, and prepared 60 μl enzyme solutions with the molar ratio (5:1) of different protein samples and LDH. The protein mixture solution was equally divided into six tubes; three copies were not processed, and the others were treated in the specific temperature water bath for 2 min, and then it was placed in the 25°C water bath for 5 min renaturation; the process was repeated three times.
Then, 5 μl enzyme solution was added to 2 ml of substrate (10 mM phosphate buffer pH 7.5, 0.1 mM NADH, and 7.5 mM pyruvate). The reduction of A340 in the reaction system was determined using a DU730 ultraviolet spectrophotometer within 3 min. The results were expressed as the LDH5 residual enzyme activity (%) relative to the activity prior to treatment. The untreated LDH activity was considered to be 100% at 25°C.
Here, ∆A340a is the difference of A340 before the test sample is processed and ∆A340b is the difference of A340 after the test sample is processed.
Three independent assays were performed, and SE was included. Values were the mean ± SD (n = 3). Student’s t-test analysis was performed between each sample of enzyme solution low-temperature protection experiment and 61°C high-temperature stress experiment.
In previous research, the full-length DHN gene PicW1 was isolated from P. wilsonii (Liu et al., 2014). Protein sequence analysis shows that PicW1 has a total length of 669 bp and encodes 222 amino acids. It is mainly composed of hydrophilic amino acids without cysteine(C) and tryptophan (W), with a high proportion of charged and polar amino acids and a low proportion of non-polar and hydrophobic residues (Table 1). According to Figure 1B, PicW1 is mainly composed of coil, which is normal for an IDP. DHNs contain at least one K-segment and as many as eleven (Jean-Marie et al., 2008), while PicW1 has five K-segments. Four K-segments are in the helical region predicted by the secondary structure, but only the second K-segment and the fourth K-segment form a relatively complete helical structure. The K-segment could form α-helices under abiotic stress to help stabilize membranes and proteins (Eriksson et al., 2011; Matthew et al., 2015; Mouillon et al., 2008). Some studies showed that the K-segment played a major role in the protection of enzymes at low temperatures, and prevented harmful changes in the secondary and tertiary structure of proteins (Yang et al., 2015). The truncated K-segment experiment showed that ERD10, RcDHN 5, and TaDHN -5 decreased the low-temperature protection of LDH (Peng et al., 2010; Cedeno et al., 2017). In addition, the lipid vesicle binding analysis of the three K-segments deletion derivatives of maize DHN1 proved that the α-helical conformation of the K-segment was essential for the binding of DHN to anionic phospholipid vesicles (Hara et al., 2011). The K-segment of wheat DHN-5 (YSK2) protected E. coli exposed to various stresses by preventing protein aggregation (Clarke et al., 2015).
The constructs used in the study are shown in Figure 1A. We selected E. coli as the heterologous expression system of Picea dehydrin gene, and the expressed protein can play its protective function (Yin et al., 2007). Some studies have shown that wheat dehydrin DHN-5, Ammopiptanthus mongolicus dehydrin AmCIP, and Arabidopsis thaliana dehydrins RAB18, LTI29, LTI30, and COR47 were heterologously expressed in the prokaryotic expression system, and the protein size and activity are correct (Svensson et al., 2000; Brini et al., 2010; Shi et al., 2016). The presence of the expressed protein in the supernatant of lysates from the transformant cells was examined by SDS-PAGE gel electrophoresis. Figure 2A indicated that induced cells produced a band with a predicted molecular weight of CBD-intein-PicW1 at around 55 kDa. The recombinant protein mainly existed in the supernatant after cell disruption and a small amount in insoluble components. We purified the PicW1 protein by affinity chromatography via intein-mediated purification using an affinity chitin-binding tag system; a novel protein purification system which utilizes the inducible self-cleavage activity of protein splicing elements (termed inteins) to separate the target protein from the affinity tag. PicW1-intein-CBD can specifically bind to chitin beads. Intein could self-cleavage in the presence of the reducing agent DTT, so that the target protein PicW1 and intein-CBD will be separated. Intein-CBD was adsorbed on the chitin beads, and the target protein PicW1 existed in the cutting buffer and was finally eluted by the gravity flow. As shown in Figure 2B, most of the target proteins were completely cleaved at 48 h.
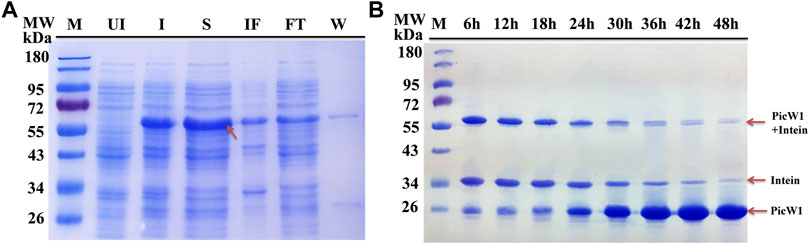
FIGURE 2. Purification of recombinant PicW1 by chitin-affinity chromatography. (A) SDS-PAGE analysis of the purification of PicW1-intein expressed in E. coli BL21 (DE3) cells. Lane 1, Marker = M; Lane 2, Un-induced BL21 cells lysate = UI; Lane 3, IPTG-induced BL21 cells lysate = I; Lane 4, Cell supernatant = S; Lane 5, Insoluble fraction = IF; Lane 6, Flow through = FT; Lane 7, wash with buffer B =W. Protein extraction along with intein tag at a range of about 55 kDa. (B) SDS-PAGE of PicW1 purification process using the chitin column. Lane1, marker; Lane 2 to Lane 9: 6, 12, 18, 24, 30, 36, 42, and 48 h represents the purification time.
PicW1 was further purified using the AKTA protein purification system of the Surperdex 200 pg 10/300 GL column, with a major and symmetrical peak at 17 ml. The gel filtration curve of the Superdex 200 preparative column showed that PicW1 was a monomer that matched with the molecular weight value calculated from its primary sequence (Figure 3A). The PicW1 components were collected from the AKTA system for crystallization and subsequent functional tests. As shown in Figure 3B, the target protein at the peak of the curve was identified using 18% SDS-PAGE, with a molecular weight of about 24 kDa. The final yield was ∼20 mg PicW1 per liter of culture medium.
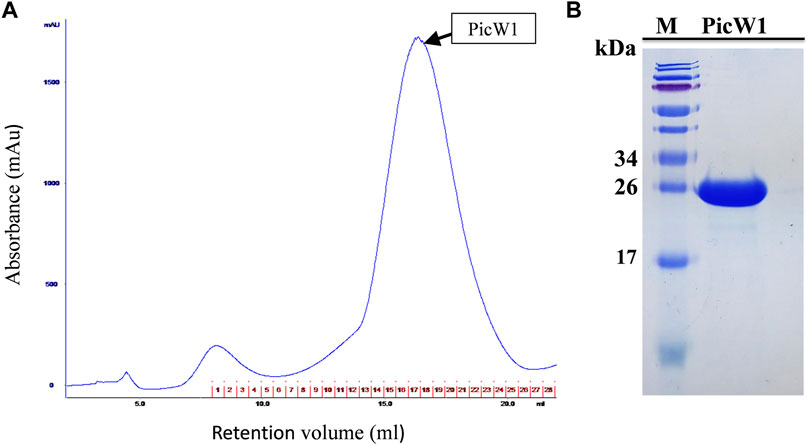
FIGURE 3. Purification of PicW1 by size-exclusion chromatography. (A) Gel filtration chromatography of PicW1 on a Superdex 200 pg 10/300 GL column. Elution was monitored at 280 nm at a flow rate of 0.4 ml/min. The red numbers represent the tubes of protein components collected using the AKTA collector. (B) SDS-PAGE analysis of “PicW1” purified using the Superdex 200 pg 10/300 GL column. Lane 1, Marker = M; Lane 2, protein components at peak value of gel filtration curve.
Because LDH loses its activity during freezing and high-temperature stress, we select this enzyme to evaluate the protective effect of PicW1. The activity of LDH before the experiment was considered as 100%. The enzyme LDH lost its activity completely upon repeated 1 min freezing and 5 min thawing at -196°C for five times (Figure 4A). In contrast, LDH activity retained to nearly 84% in the presence of 0.65 µM (5:1) PicW1, which was higher than that with BSA (71%; Figure 4A). We also found approximately 92% LDH activity was retained when 1.30 µM (10:1) and 2.6 µM (20:1) PicW1 were added, which was higher than that with BSA (84%; Figure 4B, Figure 4C). The increased concentration of purified PicW1 increased the percentage of LDH activity retained after freeze-thaw assay (Figure 4).
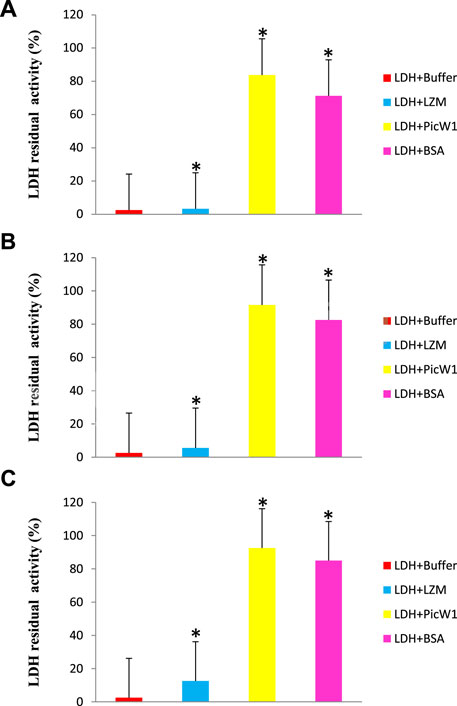
FIGURE 4. PicW1 protected LDH activity from freeze-thaw stress. Different molar ratios of LDH solutions in the presence of BSA, LZM, and purified recombinant proteins PicW1 were subjected to freeze-thaw stress for the specified times (A), 5:1; (B), 10:1; (C), 20:1 represent the molar ratio of added protein to LDH; the buffer group only has LDH. LDH activity was measured at each particular time point. The results are expressed as the LDH activity (%) relative to the activity before treatment. Three independent assays were performed, and SE was included. Values are the mean ± SD (n = 3). Significant differences in the LDH enzyme activity retention are indicated as *p < 0.05, which were evaluated with a t-test (t-test was performed on the protective effect of every two proteins of LZM, PicW1, and BSA on LDH).
Many cold sensitive enzymes are oligomers, including LDH, β-glucosidase (bglG), and catalase. Low-temperature stress makes subunit denaturation unable to combine correctly, and then irreversibly reduces the enzyme activity (Goyal et al., 2005). Brini et al. showed that wheat DHN-5 increased the activity and thermal stability of fungal bglG and glucose oxidase (GOD/POD) (Brini et al., 2010). However, PicW1 contains five K-segments and is different from the typical K-segment sequence. It has been reported that the deletion of K-segment of wheat dehydrin WZY2 leads to the reduction of low-temperature protection, indicating that these segments are involved in low-temperature protection (Yang et al., 2015). It is speculated that the hydrophobic side of picw1 may interact with the lipid components of the biofilm or combine with the hydrophobic region of the denatured part of the protein to form a protective layer to prevent the denatured macromolecular substances from further denaturation and polymerization (Rahman et al., 2010). Cor15 may form a random curl in the low-temperature protection experiment, which means that except for the K-segment, the irregular curl improves the structural flexibility and may play an important role in the low-temperature protection of cold-sensitive enzymes (Ingram and Bartels, 1996). Picw1 protein contains a large number of irregular curls, which may play a positive role in freeze-thaw stress. In conclusion, PicW1 has cryoprotective activity on LDH and has a more significant protective effect than BSA (a known protective protein) from the deleterious effects of freeze-thaw treatment. These findings serve to not only further define the molecular characteristics and possible functions of PicW1, but also add to the pool of evidence that supports a role for DHN proteins in cold tolerance.
The effect of PicW1 in protecting LDH activity during high-temperature stress was also evaluated. Similarly, BSA was selected as a positive control, and LZM was used as a negative control. As shown in Figure 5A, the enzyme activity of LDH decreased with the increase of stress treatment temperature. The LDH enzyme activity in the presence of PicW1 was higher than that in the LZM experimental group and blank control group at each temperature, but it was only slightly better than that in the BSA group at 43–55°C. The LDH enzyme almost completely lost its activity after three heat stress cycles at 61°C, only 7%. A minimum concentration of 0.65 µM PicW1 was found to be sufficient to retain 14% of the enzymatic activity under high-temperature stress (61°C) than the 9% of 0.65 µM BSA. The student t-test of 61°C data (Figure 5B) showed that the protective effect of PicW1 on LDH was better than that of other experimental groups, including the BSA group. The cryoprotection of dehydrin on LDH has been widely studied. The dehydrated protein is more effective than small molecules (such as sucrose) or other proteins (such as BSA) in protecting LDH activity from freeze-thaw damage (Hughes and Graether, 2011). The protective effect of BSA is generally considered as non-specific ones of high protein concentrations (Abriata et al., 2016; Huang et al., 2022). Although the protective effect of dehydrins on other proteins is mainly observed at low temperature, there are relatively few studies on high-temperature stress. Previously, LEA protein COR15 could protect the activity of LDH, but the temperature study was limited to 43°C (Nakayama et al., 2008). In addition, DHN-5 could significantly improve the activity and thermal stability of bglG at 50 and 70°C (Brini et al., 2010). Here, we propose a heat stress assay at 43–61°C, which shows that PicW1 can effectively protect LDH under high-temperature treatment up to 61°C.
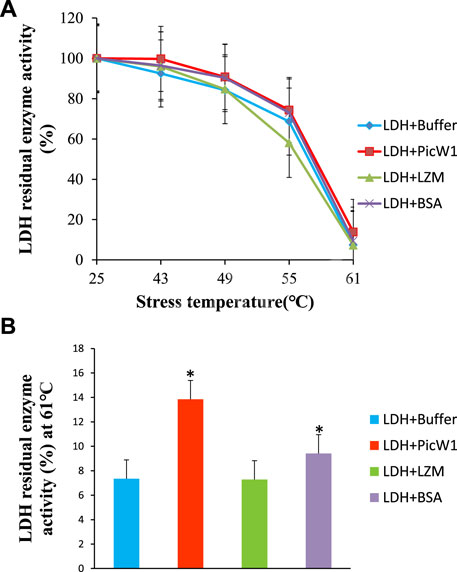
FIGURE 5. Protective effect of PicW1 protein on LDH activity under high-temperature stress. (A) LDH solution was cultured in high-temperature water bath (43, 49, 55, and 61°C) for a specific time in the presence of PicW1, BSA, and LZM. The results were expressed as LDH residual enzyme activity (%) relative to the activity before stress. Error bar represents ±SEM from at least three replicates. (B) Heat stress treatment of LDH in the presence and absence of PicW1 at 61°C. Significant differences in the LDH enzyme activity retention are indicated as *p < 0.05, which were evaluated with a t-test (t-test was performed on the protective effect of every two proteins of LZM, PicW1, and BSA on LDH).
Dehydrin interacts with the lipid components of biofilm or combines with the hydrophobic region of protein denatured part to form a protective layer, prevent the further denaturation and polymerization of denatured macromolecules, and help plants stabilize cell membrane and proteins under stress. Immunolocalization studies showed that dehydrin accumulated more in the plasma membrane or areas rich in the membrane structure, around lipids and proteins. For example, the dehydrin lti29 gene was overexpressed in Arabidopsis. During cold acclimation, the LTI29 of transgenic plants was transferred from cytoplasm to near membrane, and the survival rate of plants under freezing stress was improved (Puhakainen et al., 2004). DHN can act as a partner for other proteins to help them fold correctly and prevent them from aggregating under heat or cold stress (Hara et al., 2001). However, the chaperone activity of traditional proteins can also form specific complexes with target proteins through the interaction of hydrophobic plaques (Ellis, 2004). Therefore, DHN is more like a barrier between molecules, preventing the aggregation of denatured proteins. We have compared the activity of the PicW1 protein with another known protectant like BSA. The results showed that PicW1 protein could protect the LDH activity better than BSA under freeze-thaw stress. In addition, the higher the concentration of PicW1, the greater the protective effect on LDH. It may be imagined that PicW1 is like a “wall” that prevents the denaturation and aggregation of LDH. The more the “walls”, the more LDH activity is retained (Figure 6).
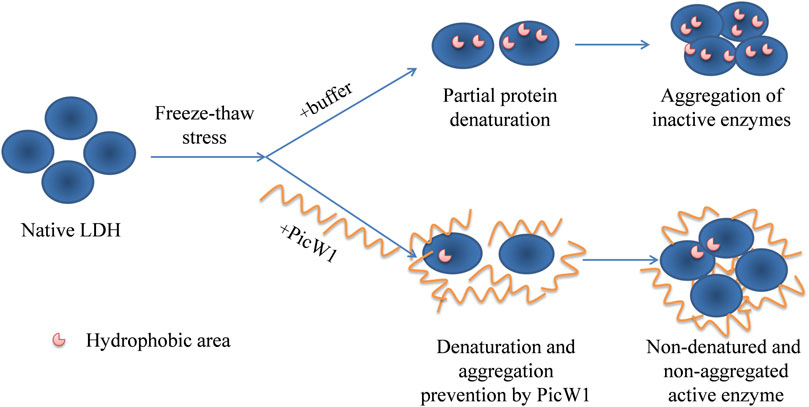
FIGURE 6. Putative mechanisms for cryoprotective activity of PicW1. When LDH is frozen and thawed, hydrophobic patches appear on the surface of the proteins, and then hydrophobic interaction is generated between the patches. PicW1 acts as a shielding molecule to interfere with the hydrophobic interaction between LDHs. Finally, LDH maintains a more natural state.
Among the group of LEA proteins, dehydrins play a major role in protection during abiotic stress. DHNs are known as stress-responsive proteins that are up-regulated in plants during stress such as drought, cold, and salinity (Hara et al., 2005). In this work, we have highly expressed cold acclimation protein PicW1 from Picea wilsonii in E. coli. We also proved that PicW1 has superior low-temperature protection on LDH. These approaches should be applicable to other plant dehydrins. The success in PicW1 expression and purification will facilitate future structural studies to understand how it works in stress resistance. In the future, we will explore the mechanism of PicW1 in depth.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
JuL performed the experiment of dehydrin protein expression and purification and the experiment of enzyme activity protection under temperature stress, and wrote the manuscript. MD did the expression and purification of LDH protein. JiL, YZ, and YR carried out image processing. JX provides the dehydrin gene. WG and SG proposed this idea and revised the manuscript.
We greatly acknowledge the National Key R&D Program of China (2019YFD1001604); the Fundamental Research Funds for the Central Universities (grant number 2018ZY18); and the National Natural Science Foundation of China (grant number 31070651).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abriata, L. A., Spiga, E., and Peraro, M. D. (2016). Molecular Effects of Concentrated Solutes on Protein Hydration, Dynamics, and Electrostatics. Biophysical J. 111 (4), 743–755. doi:10.1016/j.bpj.2016.07.011
Brini, F., Saibi, W., Amara, I., Gargouri, A., Masmoudi, K., and Hanin, M. (2010). Wheat Dehydrin DHN-5 Exerts a Heat-Protective Effect on β-Glucosidase and Glucose Oxidase Activities. Biosci. Biotechnol. Biochem. 74 (5), 1050–1054. doi:10.1271/bbb.90949
Cedeño, C., Żerko, S., Tompa, P., and Koźmiński, W. (2017). 1H, 15N, 13C Resonance Assignment of Plant Dehydrin Early Response to Dehydration 10 (ERD10). Biomol. NMR Assign 11 (2), 127–131. doi:10.1007/s12104-017-9732-0
Clarke, M. W., Boddington, K. F., Warnica, J. M., Atkinson, J., McKenna, S., Madge, J., et al. (2015). Structural and Functional Insights into the Cryoprotection of Membranes by the Intrinsically Disordered Dehydrins. J. Biol. Chem. 290 (45), 26900–26913. doi:10.1074/jbc.M115.678219
Close, T. J. (1997). Dehydrins: A Commonalty in the Response of Plants to Dehydration and Low Temperature. Physiol. Plant 100 (2), 291–296. doi:10.1111/j.1399-3054.1997.tb04785.x
Close, T. J. (1996). Dehydrins: Emergence of a Biochemical Role of a Family of Plant Dehydration Proteins. Physiol. Plant 97 (4), 795–803. doi:10.1111/j.1399-3054.1996.tb00546.x
Dong, H., Zheng, L., Yu, P., Jiang, Q., Wu, Y., Huang, C., et al. (2020). Characterization and Application of Lignin-Carbohydrate Complexes from Lignocellulosic Materials as Antioxidants for Scavenging In Vitro and In Vivo Reactive Oxygen Species. ACS Sust. Chem. Eng. 8 (1), 256–266. doi:10.1021/acssuschemeng.9b05290
Dunker, A. K., Lawson, J. D., Brown, C. J., Williams, R. M., Romero, P., Oh, J. S., et al. (2001). Intrinsically Disordered Protein. J. Mol. Graphics Model. 19 (1), 26–59. doi:10.1016/s1093-3263(00)00138-8
Eriksson, S. K., Kutzer, M., Procek, J., Gröbner, G., and Harryson, P. (2011). Tunable Membrane Binding of the Intrinsically Disordered Dehydrin Lti30, a Cold-Induced Plant Stress Protein. Plant Cell 23, 2391–2404. doi:10.1105/tpc.111.085183
Galau, G. A., Hughes, D. W., and Dure, L. (1986). Abscisic Acid Induction of Cloned Cotton Late Embryogenesis-Abundant (Lea) mRNAs. Plant Mol. Biol. 7 (3), 155–170. doi:10.1007/bf00021327
Goday, A., Jensen, A. B., Culiáñez-Macià, F. A., Mar Albà, M., Figueras, M., Serratosa, J., et al. (1994). The maize Abscisic Acid-Responsive Protein Rab17 Is Located in the Nucleus and Interacts with Nuclear Localization Signals. Plant Cell 6, 351–360. doi:10.1105/tpc.6.3.351
Goyal, K., Walton, L. J., and Tunnacliffe, A. (2005). LEA Proteins Prevent Protein Aggregation Due to Water Stress. Biochem. J. 388 (Pt 1), 151–157. doi:10.1042/BJ20041931
Graether, S. P., and Boddington, K. F. (2014). Disorder and Function: a Review of the Dehydrin Protein Family. Front. Plant Sci. 5, 576. doi:10.3389/fpls.2014.00576
Gupta, A., Marzinek, J. K., Jefferies, D., Bond, P. J., Harryson, P., and Wohland, T. (2019). The Disordered Plant Dehydrin Lti30 Protects the Membrane during Water-Related Stress by Cross-Linking Lipids. J. Biol. Chem. 294 (29416), 6468–6482. doi:10.1074/jbc.RA118.007163
Halder, T., Upadhyaya, G., Basak, C., Das, A., Chakraborty, C., and Ray, S. (2018). Dehydrins Impart Protection against Oxidative Stress in Transgenic Tobacco Plants. Front. Plant Sci. 9 (9), 136. doi:10.3389/fpls.2018.00136
Hara, M., Shinoda, Y., Tanaka, Y., and Kuboi, T. (2010). DNA Binding of Citrus Dehydrin Promoted by Zinc Ion. Plant Cel Environ 32 (5), 532–541. doi:10.1111/j.1365-3040.2009.01947.x
Hara, M., Fujinaga, M., and Kuboi, T. (2005). Metal Binding by Citrus Dehydrin with Histidine-Rich Domains. J. Exp. Bot. 56 (420), 2695–2703. doi:10.1093/jxb/eri262
Hara, M., Shinoda, Y., Kubo, M., Kashima, D., Takahashi, I., Kato, T., et al. (2011). Biochemical Characterization of the Arabidopsis KS-type Dehydrin Protein, Whose Gene Expression Is Constitutively Abundant rather Than Stress Dependent. Acta Physiol. Plant 33 (6), 2103–2116. doi:10.1007/s11738-011-0749-1
Hara, M., Terashima, S., Fukaya, T., and Kuboi, T. (2003). Enhancement of Cold Tolerance and Inhibition of Lipid Peroxidation by Citrus Dehydrin in Transgenic Tobacco. Planta 217 (2), 290–298. doi:10.1007/s00425-003-0986-7
Hara, M., Terashima, S., and Kuboi, T. (2001). Characterization and Cryoprotective Activity of Cold-Responsive Dehydrin from Citrus Unshiu. J. Plant Physiol. 158 (10), 1333–1339. doi:10.1078/0176-1617-00600
Hirayama, T., and Shinozaki, K. (2010). Research on Plant Abiotic Stress Responses in the post-genome Era: Past, Present and Future. Plant J. 61 (6), 1041–1052. doi:10.1111/j.1365-313x.2010.04124.x
Hu, Y., Yan, B., Stephen Chen, Z., Wang, L., and Tang and Caoxing Huang, W. (2022). Recent Technologies for the Extraction and Separation of Polyphenols in Different Plants: A Review. J. Renew. Mater. 10 (6), 1471–1490. doi:10.32604/jrm.2022.018811
Huang, C., Jiang, X., Shen, X., Hu, J., Tang, W., Wu, X., et al. (2022). Lignin-enzyme Interaction: A Roadblock for Efficient Enzymatic Hydrolysis of Lignocellulosics. Renew. Sust. Energ. Rev. 154, 111822. doi:10.1016/j.rser.2021.111822
Hughes, S., and Graether, S. P. (2011). Cryoprotective Mechanism of a Small Intrinsically Disordered Dehydrin Protein. Protein Sci. 20 (1), 42–50. doi:10.1002/pro.534
Hundertmark, M., and Hincha, D. K. (2008). LEA (Late Embryogenesis Abundant) Proteins and Their Encoding Genes in Arabidopsis thaliana. Bmc Genomics 9 (1), 118. doi:10.1186/1471-2164-9-118
Ingram, J., and Bartels, D. (1996). The Molecular Basis of Dehydration Tolerance in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 (1), 377–403. doi:10.1146/annurev.arplant.47.1.377
John Ellis, R. (2004). From Chloroplasts to Chaperones: How One Thing Led to Another. Photosynth Res. 80 (1-3), 333–343. doi:10.1023/B:PRES.0000030439.62331.d0
Lin, C., and Thomashow, M. F. (1992). A Cold-Regulated Arabidopsis Gene Encodes A Polypeptide Having Potent Cryoprotective Activity. Biochem. Biophysical Res. Commun. 183 (3), 1103–1108. doi:10.1016/s0006-291x(05)80304-3
Liu, C., Wang, Y., Wei, H., Meng, W., Zhang, E., and Jian, G. (2017). Expression and Purification of Recombinant Human LDH5 in Escherichia coli. Biotechnology 27 (2), 174–179.
Liu, H., Du, H., Zheng, T., Liu, K., Ji, X., Xu, T., et al. (2021). Cellulose Based Composite Foams and Aerogels for Advanced Energy Storage Devices. Chem. Eng. J. 426, 130817. doi:10.1016/j.cej.2021.130817
Liu, J., Xu, X., Xu, Q., Wang, S., and Xu, J. (2014). Transgenic Tobacco Plants Expressing PicW Gene from Picea Wilsonii Exhibit Enhanced Freezing Tolerance. Plant Cel Tiss Organ. Cult 118 (3), 391–400. doi:10.1007/s11240-014-0491-7
Lv, A., Su, L., Liu, X., Xing, Q., Huang, B., An, Y., et al. (2018). Characterization of Dehydrin Protein, CdDHN4-L and CdDHN4-S, and Their Differential Protective Roles against Abiotic Stress In Vitro. BMC Plant Biol. 18 (18), 299. doi:10.1186/s12870-018-1511-2
Matthew, W., Clarke, , and Kelly, F. (2015). Structural and Functional Insights into the Cryoprotection of Membranes by the Intrinsically Disordered Dehydrins. The Journal of biological chemistry 290 (45), 26900–26913.
Mouillon, J.-M., Eriksson, S. K., and Harryson, P. (2008). Mimicking the Plant Cell Interior under Water Stress by Macromolecular Crowding: Disordered Dehydrin Proteins Are Highly Resistant to Structural Collapse. Plant Physiol. 148 (4), 1925–1937. doi:10.1104/pp.108.124099
Nakayama, K., Okawa, K., Kakizaki, T., and Inaba, T. (2008). Evaluation of the Protective Activities of a Late Embryogenesis Abundant (LEA) Related Protein, Cor15am, during Various Stressesin Vitro. Biosci. Biotechnol. Biochem. 72 (6), 1642–1645. doi:10.1271/bbb.80214
Ning, P., Yang, G., Hu, L., Sun, J., Shi, L., Zhou, Y., et al. (2021). Recent Advances in the Valorization of Plant Biomass. Biotechnol. Biofuels 14 (1), 102. doi:10.1186/s13068-021-01949-3
O'Shea, C., Kryger, M., Stender, E. G., Kragelund, B. B., Willemoës, M., and Skriver, K. (2015). Protein Intrinsic Disorder in Arabidopsis NAC Transcription Factors: Transcriptional Activation by ANAC013 and ANAC046 and Their Interactions with RCD1. Biochem. J. 465 (465), 281–294. doi:10.1042/BJ20141045
Ohkubo, T., Kameyama, A., Kamiya, K., Kondo, M., and Hara, M. (2020). F-segments of Arabidopsis Dehydrins Show Cryoprotective Activities for Lactate Dehydrogenase Depending on the Hydrophobic Residues. Phytochemistry 173, 112300. doi:10.1016/j.phytochem.2020.112300
Peng, Y., Reyes, J. L., Wei, H., Yang, Y., Karlson, D., Covarrubias, A. A., et al. (2010). RcDhn5, a Cold Acclimation-Responsive Dehydrin from Rhododendron Catawbiense Rescues Enzyme Activity from Dehydration Effects In Vitro and Enhances Freezing Tolerance in RcDhn5-Overexpressing Arabidopsis Plants. Physiol. Plant 134, 583–597. doi:10.1111/j.1399-3054.2008.01164.x
Puhakainen, T., Hess, M. W., Mäkelä, P., Svensson, J., Heino, P., and Palva, E. T. (2004). Overexpression of Multiple Dehydrin Genes Enhances Tolerance to Freezing Stress in Arabidopsis. Plant Mol. Biol. 54 (5), 743–753. doi:10.1023/b:plan.0000040903.66496.a4
Rahman, L. N., Chen, L., Nazim, S., Bamm, V. V., Yaish, M. W., Moffatt, B. A., et al. (2010). Interactions of Intrinsically Disordered Thellungiella Salsuginea Dehydrins TsDHN-1 and TsDHN-2 with Membranes - Synergistic Effects of Lipid Composition and Temperature on Secondary Structure. Biochem. Cel Biol. 88 (5), 791–807. doi:10.1139/o10-026
Richard Strimbeck, G. (2017). Hiding in plain Sight: the F Segment and Other Conserved Features of Seed Plant SKn Dehydrins. Planta 245 (5), 1061–1066. doi:10.1007/s00425-017-2679-7
Saavedra, L., Svensson, J., Carballo, V., Izmendi, D., Welin, B., and Vidal, S. (2006). A Dehydrin Gene inPhyscomitrella Patensis Required for Salt and Osmotic Stress Tolerance. Plant J. 45 (2), 237–249. doi:10.1111/j.1365-313x.2005.02603.x
Shi, J., Liu, M., Chen, Y., Wang, J., and Lu, C. (2016). Heterologous Expression of the Dehydrin-like Protein Gene AmCIP from Ammopiptanthus Mongolicus Enhances Viability of Escherichia coli and Tobacco under Cold Stress. Plant Growth Regul. 79 (1), 71–80. doi:10.1007/s10725-015-0112-4
Svensson, J., Palva, E. T., and Welin, B. (2000). Purification of Recombinant Arabidopsis thaliana Dehydrins by Metal Ion Affinity Chromatography. Protein Expr. Purif. 20 (2), 169–178. doi:10.1006/prep.2000.1297
Wang, X., Tang, S., Chai, S., Wang, P., Qin, J., Pei, W., et al. (2021). Preparing Printable Bacterial Cellulose Based Gelatin Gel to Promote In Vivo Bone Regeneration. Carbohydr. Polym. 270 (11), 118342. doi:10.1016/j.carbpol.2021.118342
Wei, H., Yang, Y., Himmel, M. E., Tucker, M. P., Ding, S.-Y., Yang, S., et al. (2019). Identification and Characterization of Five Cold Stress-Related Rhododendron Dehydrin Genes: Spotlight on a FSK-type Dehydrin with Multiple F-Segments. Front. Bioeng. Biotechnol. 7, 30. doi:10.3389/fbioe.2019.00030
Xu, T., Du, H., Liu, H., Liu, W., Zhang, X., Si, C., et al. (2021). Advanced Nanocellulose‐Based Composites for Flexible Functional Energy Storage Devices. Adv. Mater. 33 (48), 2101368. doi:10.1002/adma.202101368
Yang, W., Zhang, L., Lv, H., Li, H., Zhang, Y., Xu, Y., et al. (2015). The K-Segments of Wheat Dehydrin WZY2 Are Essential for its Protective Functions under Temperature Stress. Front. Plant Sci. 6, 406. doi:10.3389/fpls.2015.00406
Yin, J., Li, G., Ren, X., and Herrler, G. (2007). Select what You Need: a Comparative Evaluation of the Advantages and Limitations of Frequently Used Expression Systems for Foreign Genes. J. Biotechnol. 127 (3), 335–347. doi:10.1016/j.jbiotec.2006.07.012
Zhang, B., Guo, G., Lu, F., Song, Y., Liu, Y., Xu, J., et al. (2018). PicW2 from Picea Wilsonii: Preparation, Purification, Crystallization and X-ray Diffraction Analysis. Acta Crystallogr. F Struct. Biol. Commun. 74 (Pt 6), 363–366. doi:10.1107/S2053230X18007537
Zhao, Q. H., Liu, J., Xu, X., Xu, Q., Gao, W., Xu, J. C., et al. (2017). PicW Orthologs from spruce with Differential Freezing Tolerance Expressed in Escherichia coli. Int. J. Biol. Macromolecules 101, 595–602.
Keywords: dehydrin, expression, purification, lactate dehydrogenase, protective effects
Citation: Liu J, Dai M, Li J, Zhang Y, Ren Y, Xu J, Gao W and Guo S (2022) Expression, Purification, and Preliminary Protection Study of Dehydrin PicW1 From the Biomass of Picea wilsonii. Front. Bioeng. Biotechnol. 10:870672. doi: 10.3389/fbioe.2022.870672
Received: 07 February 2022; Accepted: 07 March 2022;
Published: 05 April 2022.
Edited by:
Caoxing Huang, Nanjing Forestry University, ChinaReviewed by:
Chuan-Ling Si, Tianjin University of Science and Technology, ChinaCopyright © 2022 Liu, Dai, Li, Zhang, Ren, Xu, Gao and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Gao, d19nYW9AYmpmdS5lZHUuY24=; Sujuan Guo, Z3dhbmd6c0AyNjMubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.