
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 12 April 2022
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.863690
This article is part of the Research Topic Bioproduction of Valuable Compounds from Food By-Products, Residues and Algae Biomass as Growth Substrates View all 7 articles
The increasing global demand for food and energy production encourages the development of new production strategies focused on sustainability. Often, microbial bioprocesses rely on food or feed competitive feedstocks; hence, there is a trending need for green substrates. Here, we have proven the potential of brown seaweed biomass as microbial feedstock on account of its content of mannitol and the glucose polymer laminarin. Our host, Corynebacterium glutamicum, was engineered to enable access to mannitol as a carbon source through the heterologous expression of the mannitol-specific phosphotransferase system and the mannitol-1-phosphate-5-dehydrogenase from Bacillus subtilis. Overproduction of riboflavin was coupled with mannitol and glucose consumption via constitutive overexpression of the biosynthetic riboflavin operon ribGCAH from C. glutamicum. Brown seaweed extract and brown seaweed hydrolysate from Laminaria hyperborea, containing mannitol and glucose, were used as a carbon source for flask and bioreactor fermentations. In a seaweed-based fed-batch fermentation, the riboflavin final titer, yield, and volumetric productivity values of 1,291.2 mg L−1, 66.1 mg g−1, and 17.2 mg L−1 h−1, respectively, were achieved.
Nowadays, there is a continuous and fast increment of the world´s population and, therefore, an increase in the demand for food and energy production, which is commonly associated with environmental, social, and ethical issues. To avoid the dependence on fossil-based fuels and chemicals, the use of new and renewable sources of energy and industry feedstocks is in the spotlight (Barbot et al., 2016). Brown seaweed is a promising feedstock for microbial bioprocesses due to its high growth rates, great biomass production yields, abundance of fermentable carbohydrates, lack of arable land needed, and no freshwater requirement for cultivation. Brown macroalgal biomass, specifically from Laminaria, contains up to 60% of fermentable carbohydrates from dry weight (Barbot et al., 2016), including the polysaccharide of glucose, laminarin, and the sugar alcohol mannitol. The bacterium Corynebacterium glutamicum appears very suitable to be engineered for the utilization of macroalgae biomass for several reasons. First, C. glutamicum is used in amino acid fermentation industry at a million-ton-scale (Becker et al., 2018; Pérez-García and Wendisch, 2018). Moreover, C. glutamicum has successfully been engineered for the utilization of non-native carbon sources such as lignocellulosic sugars, glycerol as a by-product of the biodiesel process, or amino sugars from the fishing industry wastes (Wendisch et al., 2016; Becker et al., 2018; Zhang et al., 2020). Finally, C. glutamicum lacks a complex carbon catabolite repression system, hence it can consume many substrates in parallel, unlike other biotechnology workhorses like Escherichia coli or Bacillus subtilis (Görke and Stülke, 2008). Based on previous studies, C. glutamicum can utilize mannitol under specific conditions (Laslo et al., 2012). C. glutamicum possesses an arabitol/mannitol catabolic operon (Laslo et al., 2012), which comprises three genes (mtlTDR). In spite of this, wild-type C. glutamicum lacks the ability to utilize mannitol as the sole carbon source. Deletion of the transcriptional repressor gene mtlR or the overexpression of the mannitol 2-dehydrogenase (mtlD) and the arabitol/mannitol permease (mtlT) genes can enable consumption of mannitol. Through this pathway mannitol, is ingested and converted into fructose, which is non-optimal since C. glutamicum cannot utilize unphosphorylated fructose. Therefore, fructose has to be released to the medium and then has to be assimilated and phosphorylated through the fructose-specific phosphotransferase system (PTS) (Laslo et al., 2012). On the other hand, the molecule laminarin is a storage linear polysaccharide of glucose made up of β (1→3)-glucan with β (1→6) branches (Becker et al., 2020). While some organisms like Vibrio spp. can hydrolysate laminarin releasing glucoses (Alderkamp et al., 2007), C. glutamicum lacks the enzymatic machinery to hydrolyze laminarin and, hence, cannot use it as carbon source. However, pretreatment of dissolved laminarin with commercial enzymes containing cellulases, ß-glucosidases, and hemicellulose can release glucose units (Hakvåg et al., 2020). Therefore, it is required to develop tools to enable the utilization of brown seaweed-derived carbohydrates by C. glutamicum and, ultimately, the utilization of carbohydrates in seaweed hydrolysate and seaweed extract.
Traditionally used for the million-ton-scale production of amino acids, C. glutamicum has become a flexible and efficient production host for various chemicals like non-proteogenic amino acids, diamines, carotenoids, or biofuels (Becker et al., 2018; Pérez-García and Wendisch, 2018). Riboflavin or vitamin B2 is an essential nutrient for higher animals which is involved, for instance, in lipid and energy metabolism and needs to be obtained from the diet (Pinto and Zempleni, 2016; Revuelta et al., 2017). Currently, B. subtilis and the filamentous fungus Ashbya gossypii are two of the main hosts to produce riboflavin at large scales (Averianova et al., 2020). Such organisms have been extensively engineered to enhance riboflavin production (Schwechheimer et al., 2016; Averianova et al., 2020), although not from alternative and renewable feedstocks. Production of riboflavin by C. glutamicum was enabled via overexpression of the sigma factor gene sigH (Taniguchi and Wendisch, 2015; Pérez-García et al., 2021). However, the productivity values were not competitive in comparison to those in the current large-scale producers (Averianova et al., 2020).
Hence, we explored the use of mannitol-specific PTSs from mannitol natural microbial consumers in C. glutamicum. In combination with a laminarin pretreatment to release glucose, two of the most abundant carbohydrates from brown seaweed were accessible for use by C. glutamicum as carbon source. Besides, competitive riboflavin production was achieved and coupled with the synthetic consumption of seaweed substrates in C. glutamicum (Figure 1). Finally, scaling-up was achieved in lab-scale bioreactors proving the feasibility of newly constructed strains.
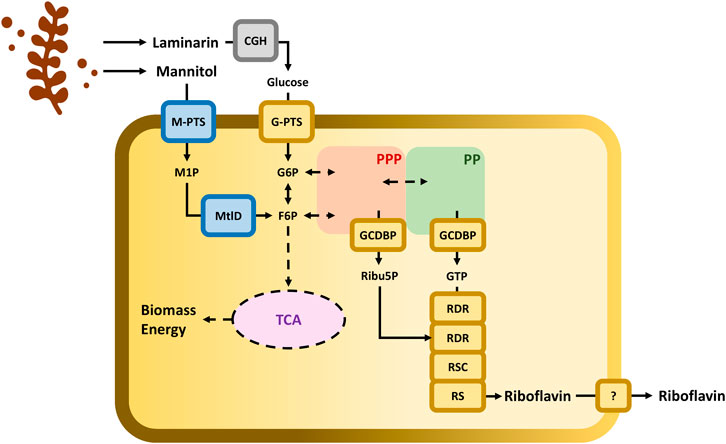
FIGURE 1. Schematic representation of C. glutamicum cell factory with the metabolic connection between the utilization of brown seaweed-derived carbohydrates and the riboflavin biosynthetic pathway. Pathways: The pentose phosphate pathway (PPP) is depicted with a red-shadowed shape; the purine pathway (PP) is depicted with a green-shadowed shape; and tricarboxylic acid (TCA) cycle is depicted with a purple shadowed shape. Enzymes: Endogenous enzymatic steps are depicted in yellow boxes. G-PTS, glucose-specific PTS coded by ptsG; GCDBP, GTP cyclohydrolase II/3,4-dihydroxy-2-butanone-4-phosphatesynthase coded by ribA; RDR, bifunctional riboflavin-specific deaminase/reductase coded by ribG; RSC, riboflavin synthase beta chain coded by ribH; and RS, riboflavin synthase by ribC. Enzymatic steps implemented in C. glutamicum are depicted in blue boxes. M-PTS, mannitol-specific PTS coded by mtlA or mtlAF; MtlD, mannitol-1-phosphate-5-dehydrogenase coded by mtlD. Pretreatment of laminarin with commercial enzymes containing cellulases, ß-glucosidases, and hemicellulose (CGH) is depicted in grey box. Metabolites: M1P, mannitol 1-phosphate; G6P, glucose 6-phosphase; F6P, fructose 6-phosphate; Ribu5P, ribulose 5-phosphate; and GTP, Guanosine 5-triphosphate.
All plasmids and bacterial strains used in this work are listed in Table 1 and Table 2, respectively. E. coli was cultivated in lysogeny broth (LB) or on LB agar plates at 37°C. Pre-cultures of C. glutamicum strains were cultivated in brain heart infusion (BHI) medium or BHI agar plates at 30°C. The main cultures of C. glutamicum were done in CGXII minimal medium (Eggeling et al., 2005), inoculated to an optical density at 600 nm (OD600) of one approx. Optical densities were measured via the spectrophotometer WPA CO 8000 Biowave Cell Density Meter from Biochrom Ltd. All biomass calculations were done by using the correlation factor 1 g L−1 biomass = 0.343 OD600. When necessary, kanamycin 25 μg ml−1, spectinomycin 100 μg ml−1, and/or tetracycline 5 μg ml−1 were supplemented to the medium. For the induction of genes cloned into the vectors pVWEx6 (Henke et al., 2021) and pEKEx3 (Stansen et al., 2005), the medium was supplemented with different concentrations of isopropyl-β-D-1-thiogalactopyranoside (IPTG), which is specified in Results. All chemicals were purchased from Merck/Sigma. As carbon source, the sugars glucose, mannitol, seaweed extract (SWE), or seaweed hydrolysate (SWH) were added at the concentrations indicated in Results. For the determination of the sugars, organic acids, and riboflavin, samples were withdrawn from the cultures, centrifuged at 16,200 × G and 4°C to remove the cells and stored at −20°C till used. All carbon sources, antibiotics, and biotin were sterilized by filtration, while the rest of the components were sterilized by autoclavation.
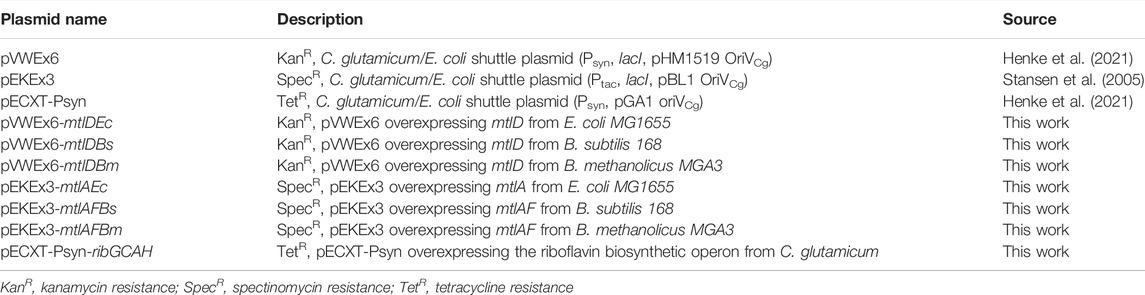
TABLE 1. List of plasmids used in this work. KanR, kanamycin resistance; SpecR, spectinomycin resistance; TetR, tetracycline resistance.
Standard molecular genetics techniques were performed as described elsewhere (Green and Sambrook, 2012). E. coli cells were transformed by heat shock (Green and Sambrook, 2012) and C. glutamicum cells were transformed by electroporation with a single pulse at 2.5 kV, 200 Ω, and 25 µF (Eggeling et al., 2005). CloneAmp™ HiFi PCR Premix Protocol (Takara Bio Inc.) was used for cloning PCR. GoTaq® DNA polymerase Protocol (Promega) was used for colony PCR. The genes mtlDEc and mtlAEc were amplified from genomic DNA from E. coli MG1655 with the primers mtlD-Ec-Fw, mtlD-Ec-Rv, mtlA-Ec-Fw, and mtlA-Ec-Rv. The genes mtlDBs and mtlAFBs were amplified from genomic DNA from B. subtilis str. 168 with the primers mtlD-Bs-Fw, mtlD-Bs-Rv, mtlAF-Bs-Fw, and mtlAF-Bs-Rv. The genes mtlDBm, mtlABm, and mtlFBm were amplified from genomic DNA from Bacillus methanolicus MGA3 with the primers mtlD-Bm-Fw, mtlD-Bm-Rv, mtlA-Bs-Fw, mtlA-Bs-Rv, mtlF-Bs-Fw, and mtlF-Bs-Rv. The whole riboflavin biosynthetic operon (ribGCAH) from C. glutamicum was amplified from genomic DNA from C. glutamicum ATCC13021 with the primers RibCg-Fw and RibCg-Rv. Colony PCRs for the pEKEx3-based cloning were carried out with the primers X3Fw and X3Rv. Colony PCRs for the pVWEx6-based cloning were carried out with the primers X6Fw and X6Rv. Colony PCRs for the pECXT-Psyn-based cloning were carried out with the primers SYFw and SYRv. All primers were purchased from Sigma, and all primer sequences are listed in Table 3. The plasmids pVWEx6 (Henke et al., 2021), pEKEx3 (Stansen et al., 2005), and pECXT-Psyn (Henke et al., 2021) were restricted with BamHI (New England Biolabs) and were combined with the amplified PCR products in Gibson Assembly (Gibson et al., 2009) to construct the plasmids listed in Table 1.
As study material for this work, brown seaweed biomass and brown SWH from Laminaria hyperborea was kindly provided by the company Alginor in Norway. For the preparation of SWE, frozen and milled Laminaria hyperborea leaf biomass was thawed overnight at 4°C. Hot water (70°C approx.) was added to the biomass with the proportion of 1:1 (grams biomass: grams water). The pH was then adjusted to 3.5 by the addition of sulfuric acid. Next, the mixture was incubated at 70°C for 2 to 3 h. When still hot, the mixture was centrifuged at 47,800 × G for 20 min. The supernatant was collected, and the biomass was discarded. For the enzymatic digestion of laminarin, the pH was adjusted to 5.0, and 1 ml of the enzyme blend Cellic CTect2 (Merck/Sigma) was added per 100 ml of supernatant, followed by incubation at 50°C and the mixture was shaken at 200 rpm for 24 h. The pH of the enzyme-treated supernatant was adjusted to 7.0 and the substrate was sterile-filtered before use. The L. hyperborea hydrolysate (SWH) provided by the company Alginor was also treated with the enzyme blend Cellic CTect2 as described previously (Hakvåg et al., 2020).
For the quantification of extracellular sugars, furans (furfural and 5-hydroxymethylfurfural), and riboflavin, a high-pressure liquid chromatography (HPLC) system was used (Waters Alliance e2695 Separations Module). Supernatants of culture broth were diluted in a proportion of 1:10 before analyzing and storing at −20°C. The quantification of sugars and furans was performed using a 300 mm × 7.7 mm Hi-Plex H column (Agilent Technologies) prewarmed at 45°C and detected by a refractive index detector (2414 RI Detector, Waters). Sulfuric acid of 5 mM was used as mobile phase at a flow rate of 0.6 ml min−1 . The quantification of riboflavin was performed using a 75 mm × 4.6 mm Symmetry C18 Column 3.5 µm column (Waters) prewarmed at 25°C, and detected by a fluorescence detector (2475 FLR Detector, Waters) with the excitation and emission wavelengths of 370 and 520 nm, respectively. Ammonium acetate of 5 mM with a pH of 6.0 and methanol (3:1) was used as mobile phase at a flow rate of 0.8 ml min−1 . Sample preparation for riboflavin quantification was carried out as described in a previous study (Petteys and Frank, 2011).
Baffled glass autoclavable bioreactors with a total volume of 2 L (working volume of 1.7 L) were used (Applikon Biotechnology). Two four-bladed Rushton impellers with outer diameters of 45 mm were placed in the stirrer axis with a distance from the bottom of the reactor of 6 and 12 cm. The relative dissolved oxygen saturation (rDOS) was automatically controlled and maintained via the stirring rpm. The rDOS levels were monitored with 12 mm polarographic DO2 sensors (AppliSens). A pH of 7.0 was established and controlled by the automatic addition of phosphoric acid (10 % (w/w)) and potassium hydroxide (4 M). The pH values were monitored with 12 mm pH sensors (AppliSens). Antifoam 204 (Sigma) was added manually when needed. The cultivation temperature was maintained at a constant temperature of 30°C using a heating jacket via a bioreactor wall. A constant aeration rate of 0.75 vvm of air was applied from the bottom of the bioreactor with a L-type sparger.
The initial working volume of 0.7 L was inoculated to an OD600 of five from the overnight shake flask pre-culture in a complex medium 2 YT (16 g of tryptone 10 g yeast extract and 5 g of NaCl per liter). Samples were collected manually using Super Safe Samplers (Infors HT) and used for OD600 measurements and for the preparation of HPLC samples. Per liter of batch medium, it contained: 10 g (NH4)2SO4, 5 g Urea, 0.5 g KH2PO4, 0.5 g K2HPO4, 0.01325 g CaCl2 x 2H2O, 0.25 g MgSO4 x 7H2O, 0.2 mg biotin, and the trace elements 1 mg FeSO4 x 7H2O, 1 mg MnSO4 x H2O, 0.1 mg ZnSO4 x 7H2O, 0.02 mg CuSO4, and 0.002 mg NiCl2 x 6H2O. The batch medium contained 80% (v/v) of SWE or SWH. The feed medium contained 100% (v/v) of SWE or SWH. Linear feeding rates of 2.5 ml min−1 were manually initiated when the rDOS values increased from 30% to 65% for the first time. The processes were completed, which was indicated by the increase of the rDOS values from 30 to 65 % during the feeding phase.
Mannitol, a type of sugar alcohol, is one of the most abundant energy and carbon storage molecules in nature, which is also present in brown seaweed biomass (Song and Vieille, 2009; Barbot et al., 2016). However, mannitol is not a natural substrate for C. glutamicum. When growing C. glutamicum (pVWEx6) (pEKEx3) (named CgSW9) in minimal medium supplemented with 1% of glucose along with gradually increasing concentrations of mannitol (Figure 2), we observed that the growth rates of C. glutamicum slightly increased with mannitol concentrations from 0 to 1%, having a negative impact in growth rate and biomass formation at 4% of mannitol. HPLC data indicated that only glucose was depleted. C. glutamicum can utilize mannitol under certain conditions (Laslo et al., 2012). However, the utilization of mannitol by C. glutamicum via mannitol-specific PTS remains unexplored. C. glutamicum lacks a mannitol-specific PTS (Laslo et al., 2012), hence we assessed mannitol-specific PTS from different microorganisms that were able to consume mannitol through a PTS. The genes for the mannitol-1-phosphate-5-dehydrogenase (mtlD) and the mannitol-specific PTS genes mtlAF from E. coli, B. subtilis, and B. methanolicus were tested and evaluated in C. glutamicum wild-type. While MtlD is a cytosolic protein that converts mannitol-1-phosphate in fructose 1-phosphate, MtlAF are membrane proteins that participate in the uptake of mannitol and its phosphorylation to mannitol 1-phosphate. The mtlD genes were cloned into the IPTG-inducible expression vector pVWEx6 [Henke et al., 2021). Transformation of C. glutamicum with the mtlD carrying vectors and the empty vector yielded the strains CgSW1 (C. glutamicum (pVWEx6)], CgSW2 [C. glutamicum (pVWEx6-mtlDEc)], CgSW3 [C. glutamicum (pVWEx6-mtlDBs)], and CgSW4 [C. glutamicum (pVWEx6-mtlDBm)] (Table 2). On the other hand, the mtlAF genes were cloned into the IPTG-inducible expression vector pEKEx3 (Stansen et al., 2005). Transformation of C. glutamicum with the mtlAF carrying vectors and the empty vector yielded the strains CgSW5 [C. glutamicum (pEKEx3)], CgSW6 [C. glutamicum (pEKEx3-mtlAEc)], CgSW7 [C. glutamicum (pEKEx3-mtlAFBs)], and CgSW8 [C. glutamicum (pEKEx3-mtlAFBm)] (Table 2). Finally, the combination of the pVWEx6- and pEKEx3-derived vectors in C. glutamicum yielded the strains CgSW9 [C. glutamicum (pVWEx6) (pEKEx3)], CgSW10 [C. glutamicum (pVWEx6-mtlDEc) (pEKEx3-mtlAEc)], CgSW11 [C. glutamicum (pVWEx6-mtlDBs) (pEKEx3-mtlAFBs)], and CgSW12 [C. glutamicum (pVWEx6-mtlDBm) (pEKEx3-mtlAFBm)] (Table 2). All the strains were tested under fermentation conditions for 48 h with minimal medium inoculated from the BHI pre-cultures to an OD600 of around one and supplemented with 0.5% of mannitol as sole carbon source and different IPTG concentrations (ranging from 0 to 600 µM). However, the single vector strains did not grow and, hence, native mannitol-1-phosphate-5-dehydrogenase and mannitol-specific PTS activities were not detected under the conditions tested (data not shown). Instead, the double vector strains carrying the genes from B. subtilis and B. methanolicus showed growth (Figures 3A,C). Under the conditions assayed, the growth rate of CgSW11 and CgSW12 peaked at 0.07 ± 0.00 h−1 and 0.06 ± 0.00 h−1 when using 100 µM of IPTG. Higher IPTG concentrations showed negative impact in the growth rates of CgSW11 and CgSW12 (Figure 3A). The control strain CgSW10 and the strain overexpressing the PTS genes from E. coli did not grow. As for the biomass formed, CgSW11 and CgSW12 were able to reach concentrations of about 1.6 g L−1. A negative impact in the biomass formation was observed from 200 µM of IPTG for CgSW11 and from 400 µM of IPTG for CgSW12 (Figure 3C). The genes from B. subtilis are more harmful than those from B. methanolicus for C. glutamicum since high induction via IPTG causes an important drop in the growth rate and biomass formation (Figures 3A,C).
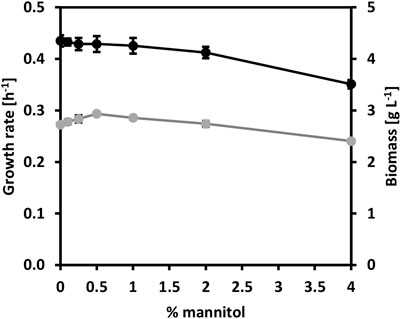
FIGURE 2. Growth behavior of strain CgSW9 in the presence of mannitol. Glucose-based minimal medium was supplemented with gradually increasing concentrations of mannitol. Growth rate values are depicted in grey. Biomass values are depicted in black. Means and standard deviations of triplicates are shown.
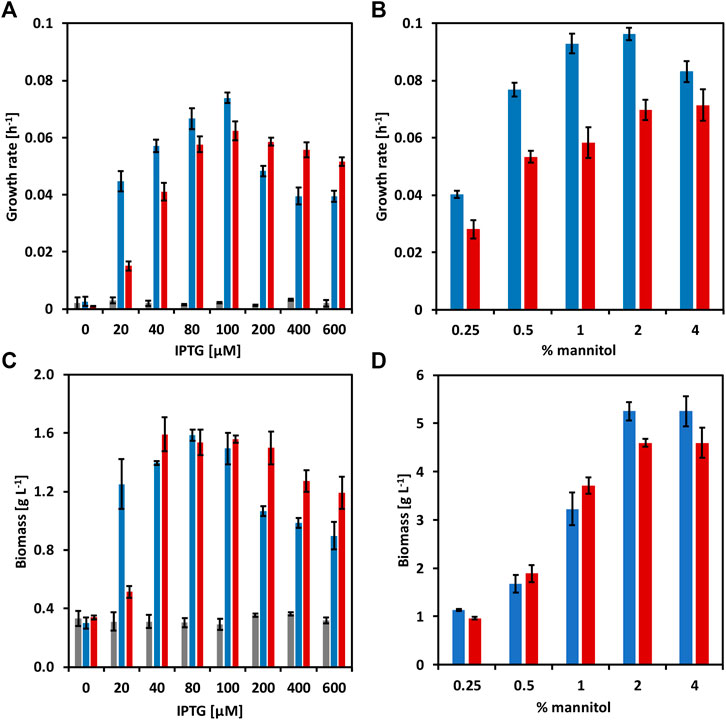
FIGURE 3. Growth behavior of the strains CgSW10, CgSW11, and CgSW12 with mannitol as sole carbon source. (A) Growth rates of CgSW10 (grey columns), CgSW11 (blue columns), and CgSW12 (red columns) strains grown in 0.5% mannitol minimal medium supplemented with different concentrations of IPTG. (B) Growth rates of CgSW11 and CgSW12 strains grown in minimal medium supplemented with different concentrations mannitol and 100 µM of IPTG. (C) Final biomass of CgSW10, CgSW11, and CgSW12 strains grown in 0.5% mannitol minimal medium supplemented with different concentrations of IPTG. (D) Final biomass of CgSW11 and CgSW12 strains grown in minimal medium supplemented with 100 µM of IPTG and different concentrations of mannitol. Means and standard deviations of triplicates are shown.
CgSW11 and CgSW12 strains were further tested in minimal medium supplemented with increasing concentrations of mannitol as sole carbon source and 100 µM of IPTG (Figures 3B,D). Under those conditions, CgSW11 grew faster at 1 and 2 % of mannitol with growth rates of 0.09 ± 0.00 h−1and 0.10 ± 0.00 h−1, respectively. CgSW12 grew faster at 2 and 4% of mannitol with growth rates of 0.07 ± 0.00 h−1 and 0.07 ± 0.01 h−1, respectively (Figure 3B). When using 2 and 4% of mannitol, CgSW11 reached a biomass of 5.2 g L−1, while under the same mannitol concentrations, CgSW12 reached a slightly lower biomass of 4.6 g L−1 (Figure 3D). The strain CgSW11 seems to perform better at lower mannitol concentrations, although it grew faster than CgSW12 under all mannitol concentrations tested here (Figures 3B,D).
Apart from mannitol, brown seaweed also contains large reserves of laminarin (Barbot et al., 2016). C. glutamicum lacks the laminarin-degrading enzyme systems and hence, cannot utilize laminarin as carbon source. Therefore, in this work, laminarin was enzymatically predigested to release glucose.
The strains CgSW9, CgSW11, and CgSW12 were tested under fermentation for 48 h with minimal medium with glucose, mannitol, and glucose along with mannitol as sole carbon source. Data for the growth rate, biomass formation, and biomass yield were collected and depicted in Table 4. The control strain CgSW9 only grew when glucose was supplemented in the medium, independently of the presence of mannitol. The growth rate of CgSW9 with only glucose was 0.25 ± 0.02 h−1 and with glucose plus mannitol was 0.26 ± 0.00 h−1. The strains CgSW11 and CgSW12 could grow with mannitol, glucose, and with the combination of both sugars (Table 4). The growth rate, biomass, and biomass yield values for both strains with glucose as sole carbon source were similar to those of the control strain CgSW9 (Table 4). As shown previously (Figures 3A,B), CgSW11 could grow faster on mannitol than CgSW12 (Table 4). The main differences observed were from the cultures with the combined carbon sources glucose and mannitol. CgSW11 grew slightly at a slower rate (0.23 ± 0.01 h−1) combined with both sugars together than only with glucose (0.26 ± 0.00 h−1). However, CgSW12 showed a more dramatic drop in the growth rate when growing on mannitol and glucose (0.18 ± 0.01 h−1) as compared to its growth only with glucose (0.25 ± 0.01 h−1) (Table 4). The biomass and yield values from the CgSW12 as compared to those from CgSW9 and CgSW11 indicated the possibility that not all sugars were fully depleted. This was confirmed via HPLC analysis, since more than 9 g L−1 of mannitol were detected in the supernatants of CgSW12 when the growth was completed. The fact that the proteins from B. methanolicus showed some sort of competition for glucose and mannitol made the strain CgSW12 not suitable for the purpose of this study. Therefore, the CgSW11 strain was chosen for further steps.
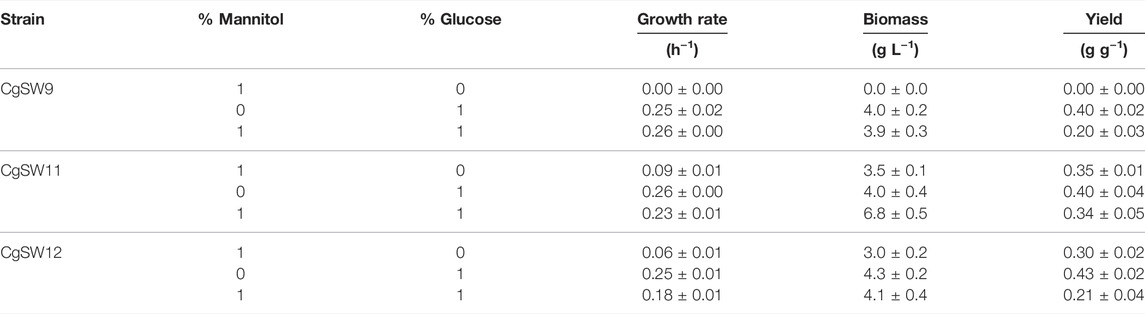
TABLE 4. Growth values of the strains CgSW9, SgSW11, and CgSW12 when using mannitol, glucose, or mannitol plus glucose as sole carbon sources.
For laminarin hydrolysis, the enzyme blend Cellic CTect2, which contains cellulases, ß-glucosidases, and hemicellulose was applied to the SWE and the SWH. The content of mannitol and glucose in the SWE and SWH was quantified via HPLC before and after the laminarin hydrolysis treatment, and the results can be seen in Table 5.
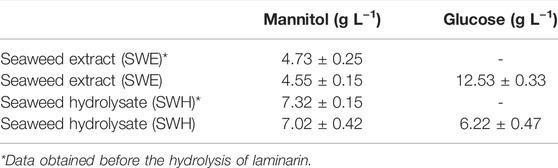
TABLE 5. Mannitol and glucose concentrations in the seaweed extract (SWE) and seaweed hydrolysate (SWH) used in this work.
Next, the strains CgSW9 and CgSW11 were grown on minimal medium supplemented with different % of SWE or SWH as carbon source (Figure 4). Both strains were able to grow using SWH and SWE. While CgSW9 and CgSW11 grew faster with SWE than with SWH (Figures 4A,D), the biomass yields were higher with SWH than with SWE (Figures 4B,E). Hence, even though SWH may contain additional substrates apart from glucose and mannitol, SWE seems less harmful for the cells. When measuring furans as growth inhibitors via HPLC, 0.42 ± 0.04 mM of furfural was detected only in SWH. A growth rate of 0.16 ± 0.01 h−1 and a biomass of 5.8 0.1 g L−1 were achieved when using the strain CgSW11 and 80% of SWE as carbon source; the highest growth rate and biomass values were attained under the conditions tested here (Figures 4D,E). In addition, CgSW9 and CgSW11 were also tested with 100 % of SWE or SWH, with and without the addition of trace elements and biotin. However, the growth rate and biomass values were, in general, lower in comparison with the values obtained from the cultivations with SWE or SWH combined with minimal medium CGXII, especially with SWE and when no trace elements and biotin were supplemented (Figures 4A–E). Hence, SWH seems to cope with (Figures 4A–E) the lack of micronutrients needed for the cultivation of C. glutamicum strains to a certain extent.
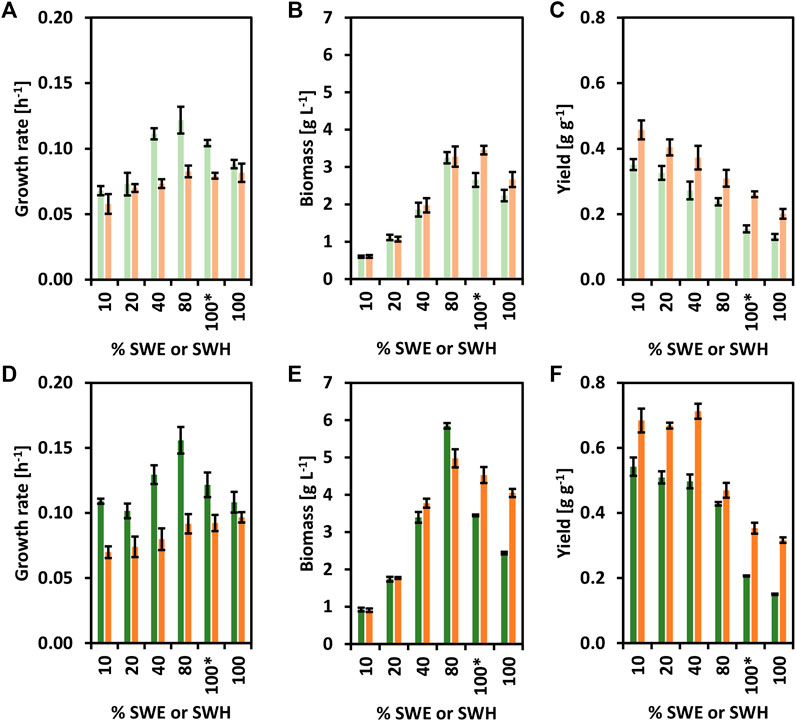
FIGURE 4. Growth of the strains CgSW9 and CgSW11 in brown seaweed-based substrates. Concentrations of SWE or SWH that ranged from 10 to 100% were used as carbon source. The condition 100* means 100% seaweed-based substrate supplemented with trace elements and biotin. (A) Growth rates (h−1) of CgSW9. (B) Final biomasses formed (g L−1) of CgSW9. (C) Biomass yields (g g−1) of CgSW9. (D) Growth rates (h−1) of CgSW11. (E) Final biomasses formed (g L−1) of CgSW11. (F) Biomass yields (g g−1) of CgSW11. Values from CgSW9 grown on SWE are depicted with light green columns. Values from CgSW9 grown on SWH are depicted with light orange columns. Values from CgSW11 grown on SWE are depicted with dark green columns. Values from CgSW11 grown on SWH are depicted with dark orange columns. Means and standard deviations of triplicates are shown.
Concluding this block, CgSW11 proved to be a suitable strain for the consumption of brown seaweed-based substrates, better growth values can be achieved via a combination of minimal medium together with SWE or SWH. It is also noteworthy that SWH is a richer medium than SWE, but more harmful for C. glutamicum.
To reinforce the concept of circular bioeconomy, a newly added value chain from brown seaweed was created here. Riboflavin and its active forms have been extensively used in the food, feed, and pharmaceutical industries (Liu et al., 2020). To overproduce riboflavin with C. glutamicum, the whole riboflavin biosynthetic operon (ribGCAH, Figure 1) from C. glutamicum was cloned into the vector pECXT-Psyn under the control of the constitutive promoter Psyn (Henke et al., 2021). The vectors pECXT-Psyn and pECXT-Psyn-ribGCAH were transferred to C. glutamicum wild-type yielding the strains CgRibo1 and CgRibo2 (Table 2). Growth and production of CgRibo1 and CgRibo2 were tested in minimal medium supplemented with different concentrations of glucose as sole carbon source (Figure 5). The highest riboflavin titer achieved by the strain CgRibo2 was 944.6 ± 87.1 mg L−1 using 4% of glucose and within 50 h, meaning a volumetric productivity of 18.9 ± 1.7 mg L−1 h−1 (Figures 5D–F). However, biomass and riboflavin yields were affected when supplementing 2 and 4% of glucose (Figures 5C–E). Growth rates of CgRibo2 were also impaired as compared to those from CgRibo1 when the strain was fed with high glucose concentrations (Figure 5A). Nevertheless, the vector pECXT-Psyn-ribGCAH proved to be an excellent tool to establish riboflavin production in C. glutamicum strains. Therefore, the vector pECXT-Psyn was transferred to the strain CgSW9 and the vector pECXT-Psyn-ribGCAH was transferred to the strain CgSW11, yielding the strains CgRibo3 and CgRibo4, respectively (Table 2). Afterward, growth and production of CgRibo3 and CgRibo4 were tested in minimal medium supplemented with 1% mannitol, 1% glucose, or 1% mannitol and 1% glucose (Figure 6). In general, CgRibo4 grew at a slower rate and reached lower biomass yields than CgRibo3 under all conditions tested here. Particularly, when using glucose and mannitol as carbon source, the biomass formation was drastically lower for CgRibo4 than for CgRibo3 (Figures 6A–C). The riboflavin titer of CgRibo4 with 1% glucose as sole carbon source was 387.1 ± 32.7 mg L−1, which is 41.6% lower titer as compared with the riboflavin titer achieved by CgRibo2 with 1% glucose as sole carbon source. Hence, the riboflavin production is impaired by the metabolic modifications implemented in CgRibo4. When using 1% mannitol as sole carbon source, CgRibo4 produced 467.3 ± 45.7 mg L−1 of riboflavin, which is17.2% higher titer than the one obtained from 1% glucose (Figure 6D). Finally, when using 1% glucose together with 1% mannitol as carbon source, the CgRibo4 strain produced 607.3 ± 33.1 mg L−1 of riboflavin, implying a riboflavin yield of 30.4 ± 3.3 mg g−1 (Figures 6D–F).
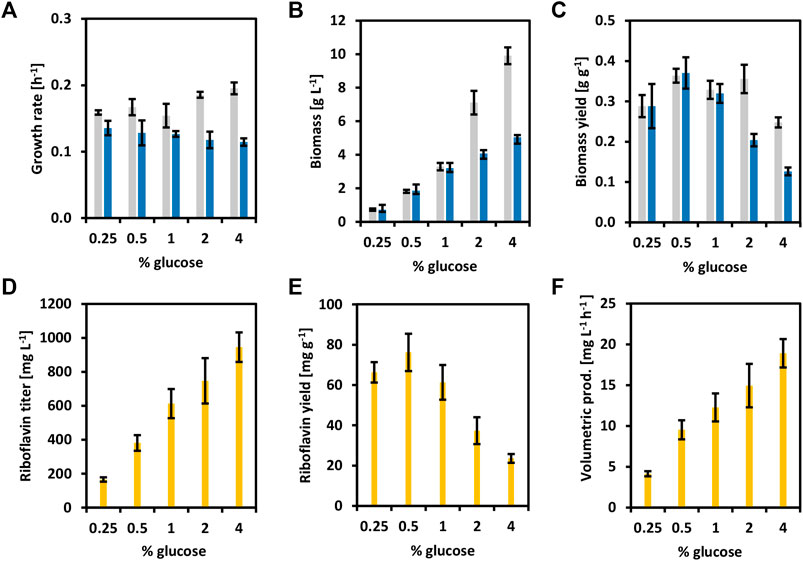
FIGURE 5. Growth and riboflavin production from glucose of the strains CgRibo1 and CgRibo2. Concentrations of glucose ranged from 0.25 to 4 g L−1 were tested. Growth rates (h−1) (A), final biomasses formed (g L−1) (B), and biomass yields (g g−1) (C) of CgRibo1 (grey) and CgRibo2 (blue) are depicted. Production values of final riboflavin titers (mg L−1) (D), riboflavin yields (mg g−1) (E), and riboflavin volumetric productivities (mg L−1 h−1) (F) of CgRibo2 are depicted with yellow columns. Means and standard deviations of triplicates are shown.
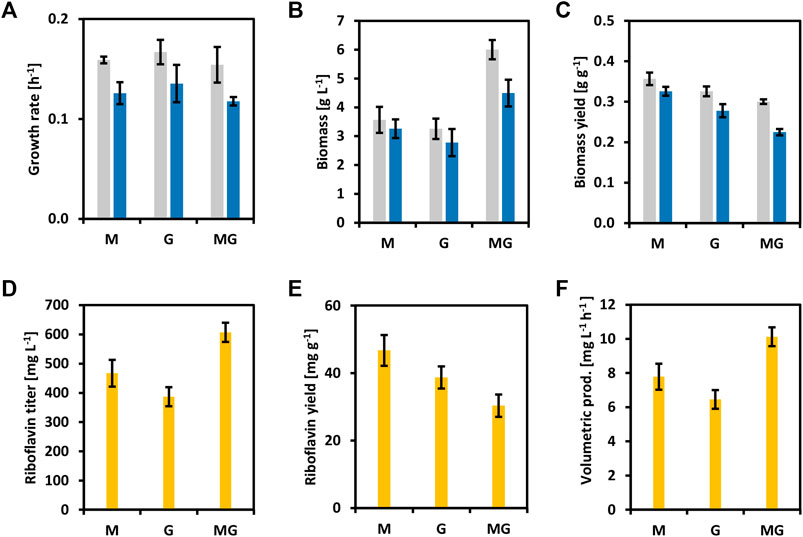
FIGURE 6. Growth and riboflavin production from brown seaweed sugars of the strains CgRibo3 and CgRibo4. 1% of glucose, 1% of mannitol, and 1% of glucose and 1% of mannitol were assayed. (A) Growth rates (h−1) of CgRibo3 and CgRibo4. (B) Final biomasses of formed (g L−1) CgRibo3 and CgRibo4. (C) Biomass yields (g g−1) of CgRibo3 and CgRibo4. (D) Final riboflavin titers (mg L−1) of CgRibo4. (E) Riboflavin yields (mg g−1) of CgRibo4. (F) Riboflavin volumetric productivities (mg L−1 h−1) of CgRibo4. Growth values of CgRibo3 are depicted with grey columns. Growth values of CgRibo4 are depicted with blue columns. Production values of CgRibo4 are depicted with yellow columns. Means and standard deviations of triplicates are shown.
The riboflavin producer CgRibo4 was also cultivated with SWE and SWH as carbon sources to prove the establishment of a newly added value chain from seaweed-based substrates. The concentrations of 80 and 100% of SWE or SWH were chosen as the most relevant conditions for the purpose of this study (Figure 7). As previously, when using 80% of SWE or SWH the medium contained all the components of the minimal medium as well. Similar to earlier results, the CgRibo4 strain grew faster on SWE, but the biomass yields were higher with SWH, especially when 100% SWH was used (Figures 7A–C). With regard to riboflavin, the best productivity values were achieved with 80% SWE. Under such conditions, a riboflavin titer of 635.0 ± 40.0 mg L−1 peaked after 40 h, indicating a volumetric productivity of 15.9 ± 1.0 mg L−1 h−1. However, the best riboflavin yield of 57.8 ± 6.6 mg g−1 was achieved from 80% SWH (Figures 7D–F). Mannitol and glucose were fully depleted by the end of the growth experiment.
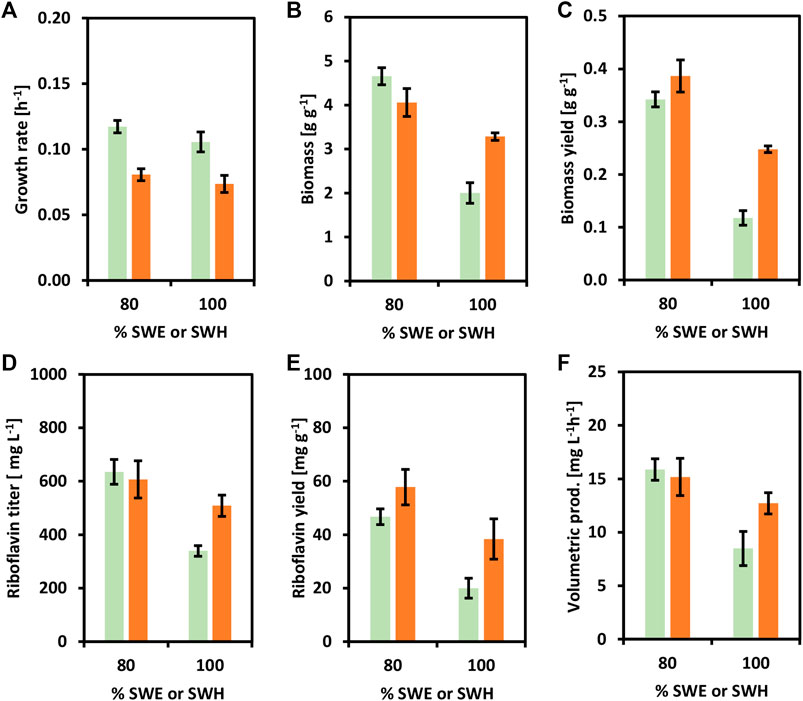
FIGURE 7. Growth and riboflavin production from brown seaweed-based substrates of the strain CgRibo4. SWE (green columns) or SWH (orange columns) were assayed in the concentrations of 80 and 100%. (A) Growth rates (h−1) of CgRibo4. (B) Final biomasses of formed (g L−1) CgRibo4. (C) Biomass yields (g g−1) of CgRibo4. (D) Final riboflavin titers (mg L−1) of CgRibo4. (E) Riboflavin yields (mg g−1) of CgRibo4. (F) Riboflavin volumetric productivities (mg L−1 h−1) of CgRibo4. Means and standard deviations of triplicates are shown.
Here, we have successfully established competitive riboflavin production in C. glutamicum strains from glucose and seaweed-based substrates. Production values up to this point are summarized in Table 6. Subsequently, the feasibility of using seaweed-based substrates as alternative microbial substrate was evaluated in bioreactor fermentations.
To fully test the performance of CgRibo4, such strain was grown in lab-scale bioreactors following a fed-batch fermentation approach. SWE or SWH were used as carbon source in the concentrations of 80 and 100% for the batch and feed phases, respectively. According to HPLC measurements, the SWE-based fermentation media contained 3.5 g L−1 of mannitol and 10.6 g L−1 of glucose in the batch medium and 4.2 g L−1 of mannitol and 12.5 g L−1 of glucose in the feed medium. While the SWH-based fermentation contained 5.9 g L−1 of mannitol and 5.3 g L−1 of glucose in the batch medium and 6.9 g L−1 of mannitol and 6.2 g L−1 of glucose in the feed medium. A linear feeding rate of 2.5 ml min−1 was applied in both fermentations when the relative dissolved oxygen saturation (rDOS) value increased from 30% to 65% and above (after 30 h). The fermentation processes were considered completed when the rDOS value increased from 30% to 65% and above, during the feeding phase (after 75 h). By the end of the SWE-based fermentation, 0.1 g L−1 of mannitol was left, and 0.8 g L−1 of mannitol and 0.2 g L−1 of glucose were not used in the SWH-based fermentation. The riboflavin final titer, yield, and volumetric values of 1,291.2 mg L−1, 66.1 mg g−1, and 17.2 mg L−1 h−1, respectively, were achieved when using SWE (Figure 8A). On the other hand, 1,108.9 mg L−1, 75.8 mg g−1, and 14.8 mg L−1 h−1 values were achieved when using SWH (Figure 8B). While the highest riboflavin titer and volumetric productivity were achieved with SWE as carbon source, the highest riboflavin yield was reached when using SWH as carbon source. Differences were also observed in the new biomasses formed, since 4.8 g L−1 of biomass was produced from the SWE sugars while 5.5 g L−1 were produced from the SWH sugars, exhibiting biomass yields of 0.25 g g−1 and 0.38 g g−1, respectively. Interestingly, in both fermentations, the biomass reached steady state during the feeding phases, but riboflavin continued to be accumulated until the end of the processes.
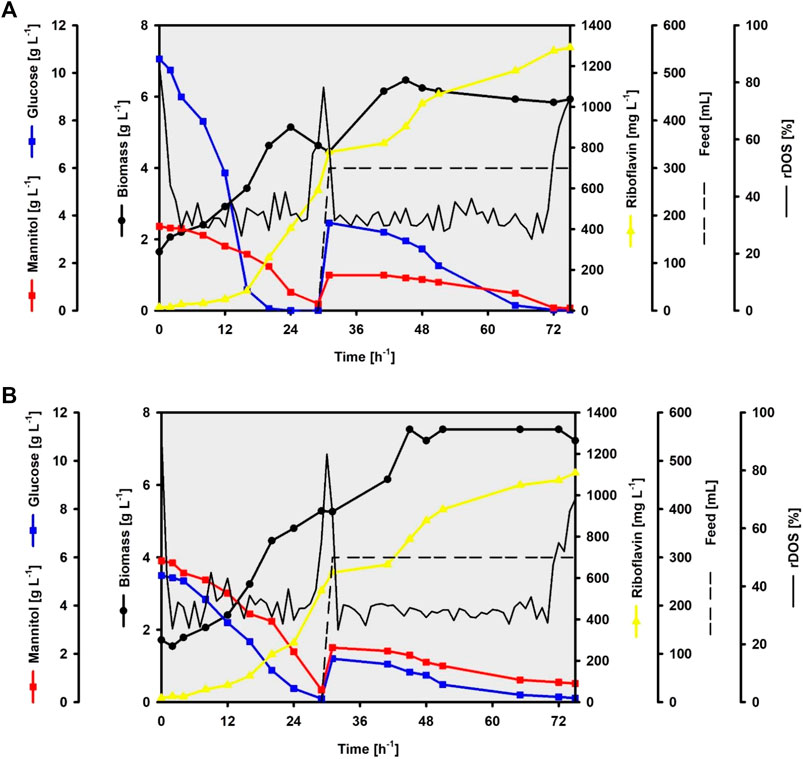
FIGURE 8. Fed-batch fermentations of the strain CgRibo4 using brown seaweed-based substrates. Glucose and mannitol concentrations are depicted with blue and red lines, respectively. Biomass concentrations are depicted with black circles. Riboflavin concentrations are depicted with yellow lines. The rDOS levels are depicted with black lines. Feed supplementations are depicted with dashed black lines. (A) SWE-based process. (B) SWH-based process.
Our bioreactor fermentations proved that the production of riboflavin can be based on seaweed substrates up to 100%, being also scalable to higher working volumes.
Nowadays, alternative feedstocks like wood-related residual biomass, crop residues, wastewater, or agri-food co-products are in the spotlight (Javourez et al., 2021). Biomass is a key sustainable feedstock for the transition toward circular and low fossil carbon economies. In particular, seaweed biomass is a promising candidate due to the lack of need for arable land, freshwater requirement for cultivation, and the presence of fermentable carbohydrates in their composition (Barbot et al., 2016). The total carbohydrate content among seaweed species varies from 24% to 65% in some brown seaweeds like Laminaria digitata, being comparable with the carbohydrate content of 65–75% in lignocellulosic biomass (Olsson et al., 2020). Mannitol, laminarin, alginate, and fucoidan in brown seaweeds are promising substrates for the utilization in biorefinery processes. In comparison, red seaweed varieties consist of different typical carbohydrates kinds including water-soluble sulfur-containing galactans like agar and carrageenan (Jiménez-Escrig and Sánchez-Muniz, 2000). Some seaweed species have been investigated as fermentation substrate previously. For instance, the red seaweed Gracilaria sp. was used as substrate for Lactobacillus acidophilus and Lactobacillus plantarum for lactic acid production (Monteiro et al., 2021). Regarding brown seaweeds, the species Saccharina japonica was fermented by Monascus purpureus to produce lovastatin (Suraiya et al., 2018). In another example, the S. japonica extracts containing mannitol and glucose were used to produce value-added chemicals like cadaverine and C30 terpenoids by B. methanolicus (Hakvåg et al., 2020). In this study, the brown seaweed L. hyperborea from the Norwegian coasts was shown to be a promising alternative microbial substrate for bioprocesses with the microbial workhorse C. glutamicum. Seaweed extract and hydrolysate contained mannitol and glucose from laminarin, which were targeted as fermentable sugars here. For most of the common biotechnological workhorses like E. coli, B. subtilis, or C. glutamicum, glucose is a native carbon source (Fujita, 2009; Lindner et al., 2011; Bren et al., 2016) and many efforts have been taken to fully understand and improve glucose utilization by these microbial workhorses (Fujita, 2009; Pérez-García et al., 2016; Alva et al., 2020). Regarding mannitol, some microorganisms have specific systems for the uptake and phosphorylation of mannitol, as it is the case for E. coli, B. subtilis, and B. methanolicus, which possess mannitol-specific PTSs (Figge et al., 1994; Heravi and Altenbuchner, 2014; López et al., 2019). The wild-type C. glutamicum grows on arabitol, but not on other sugar alcohols (Laslo et al., 2012). However, growth of C. glutamicum in the presence of arabitol allows subsequent growth on mannitol. Utilization of mannitol by C. glutamicum can be triggered by the overexpression of the arabitol/mannitol catabolic operon mtlTD (Peng et al., 2011).In addition, the deletion of the transcriptional repressor gene mtlR also led to the overexpression of mtlTD and, hence allowing mannitol utilization by C. glutamicum (Laslo et al., 2012). These approaches have been used to establish C. glutamicum mannitol-consuming strains (Laslo et al., 2012; Hoffmann et al., 2018). However, mannitol is not directly included in the central metabolism, but it is rather oxidized to fructose, which escapes the cells to be further assimilated and phosphorylated via the fructose-specific PTS (Laslo et al., 2012). In this study, the mannitol-specific PTSs from E. coli, B. subtilis, and B. methanolicus were tested in C. glutamicum. Mannitol-based growth was achieved by using the systems from B. subtilis and B. methanolicus. However, the system from E. coli could not enable mannitol-based growth in C. glutamicum. In a PTS system, the mannitol-specific components (EII) include three domains, the mannitol permease IIC (EIIC) component, the mannitol-specific phosphotransferase enzyme IIB (EIIB) component, and the mannitol-specific phosphotransferase enzyme IIA (EIIA) component (Clore and Venditti, 2013). While in E. coli, the EIICBA components are coded by mtlA, and in B. subtilis and B. methanolicus, the EIICB components are coded by mtlA and the EIIA component is coded by mtlF as independent protein (Heravi and Altenbuchner, 2014; López et al., 2019). This gene distribution may affect the interaction between the PTS components, and ultimately may disturb the final function of those proteins. The EIIA component is the direct phosphoryl group acceptor which is donated by the phosphocarrier HPr protein (Lindner et al., 2011). C. glutamicum ATCC 13032 has PTS genes encoding four different EII permeases, specific for glucose (PtsG), fructose (PtsF), sucrose (PtsS), and one unknown substrate (Moon et al., 2007). The C. glutamicum deletion mutant lacking the general PTS component HPr is unable to use the PTS systems for growth (Lindner et al., 2011). Therefore, it is important that HPr can interact with the EIIA component. Aligning protein sequences via BLASTP (Altschul et al., 1990), we observed that the EIIA components from E. coli, B. subtilis, and B. methanolicus present similarities in their protein sequences with PtsF from C. glutamicum. Query covers and identities for E. coli’s EIIA component were, 68 and 29% , 88 and 27% for B. subtilis’s EIIA component, and 72 and 36% for B. methanolicus’s EIIA component, respectively. According to these values, the EIIA component sequence from B. subtilis and B. methanolicus are closer to the sequence of PtsF from C. glutamicum. B. methanolicus is a thermotolerant microorganism with an optimum growth temperature of 50—53°C (Irla et al., 2014), hence the folding of MtlAF and MtlD may have been negatively affected when using 30°C during growth experiments with C. glutamicum, which supports the fact that the MtlAFD from B. subtilis performed best in C. glutamicum, since both are mesophilic microorganism (Droffner and Yamamoto, 1985; Eggeling et al., 2005). Nevertheless, C. glutamicum growth was shown from the seaweed-based substrates SWE and SWH. According to our results, SWE seems to be a less harmful substrate for the C. glutamicum cells in comparison to SWH. The presence of the furfural only in SWH might have contributed to growth inhibition even though C. glutamicum showed recalcitrance against growth inhibitors from some hydrolysates, such as methylfurfural and hydroxymethylfurfural (Gopinath et al., 2011; Mindt et al., 2019).
Industrial riboflavin production is traditionally carried out using the organisms B. subtilis and the filamentous fungus Ashbya gossypii (Schwechheimer et al., 2016; Averianova et al., 2020) reaching values up to 26.8 g L−1 of riboflavin B. subtilis strain KCCM 10445 (Lee et al., 2004). Some of the genetic modifications used for the development of riboflavin-producing strains include the overexpression of the riboflavin biosynthetic genes, increasing the metabolic flux towards the pentose phosphate pathway, or activation of the glycine and the purine pathways (Averianova et al., 2020). In C. glutamicum, a differential gene expression analysis revealed that the overexpression of sigma factor gene sigH upregulates the riboflavin biosynthesis genes ribH, ribA, and ribC, increasing the formation of riboflavin (Taniguchi and Wendisch, 2015). By overexpressing sigH and following a dynamic co-cultivation approach in bioreactors with a mixture of sugars, a riboflavin titer of 27 mg L−1 was achieved (Pérez-García et al., 2021). In this work, the riboflavin biosynthetic genes from C. glutamicum were constitutively overexpressed under the control of the synthetic Psyn promoter (Henke et al., 2021). The best production values were obtained with the strain CgRibo2 when using 4% of glucose as sole carbon source, peaking at 944.6 ± 87.1 mg L−1 of riboflavin in flask fermentations, showing a 35-fold higher titer than the previously reported by Pérez-García et al., in 2021. In a similar approach, a B. subtilis strain containing multiple copies of the endogenous riboflavin biosynthetic operon expressed constitutively from strong phage promoters produced 14 g L−1 after 48 h in fed-batch operating bioreactors (Perkins et al., 1999). However, the production values of the strain CgRibo4 decreased significantly compared to those of CgRibo2 using glucose. This may be explained by the metabolic burden of carrying three vectors in CgRibo4 instead of one vector in CgRibo2 (Wein et al., 2019). Yet, riboflavin production was also shown from seaweed-based substrates for the first time here. The best riboflavin titer of 1,291.2 mg L−1 was achieved after 75 h using SWE as sole carbon source in a fed-batch fermentation. The fed-batch method is a common approach that has been applied to riboflavin industrial strains (Averianova et al., 2020). For instance, production of riboflavin with A. gossypii is carried out in fed-batch fermentations with aerobic conditions during 6–8 days (Stahmann et al., 2000). In another example, the process of fermentation for riboflavin production with the B. subtilis strain RB 50 is carried out on carbon-limited fed-batch cultivations during 48–56 h (Bretzel et al., 1999). Nevertheless, the feasibility of the newly generated riboflavin producer was successfully tested in seaweed-based fed-batch fermentations proving to be a promising process. In this regard, de novo production via microbial fermentations offers the flexibility for using broad natural substrates or non-natural carbon sources prior microbial engineering (Wendisch et al., 2016). Moreover, alternative renewable feedstocks or carbon containing waste streams have the potential to become more attractive within these processes (Gopinath et al., 2011; Buschke et al., 2013; Meiswinkel et al., 2013; Mindt et al., 2019).
In conclusion, we have established C. glutamicum as a promising riboflavin production host that utilizes brown seaweed-derived carbohydrates as feedstock. Heterologous expression of the mannitol-specific PTS together with mannitol-1-phosphate 5-dehydrogenase from B. subtilis enabled efficient utilization of mannitol by C. glutamicum. Combined with the constitutive overexpression of the endogenous riboflavin biosynthetic operon from C. glutamicum, we generated the C. glutamicum strain with the highest riboflavin production values regarding titer, yield, and productivity from glucose. Moreover, we have shown the production of riboflavin from seaweed-based substrates in flask and bioreactor fermentations.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
FP-G: conceptualization, investigation, methodology, supervision, writing—original draft, writing—review and editing, and project administration. VJK: investigation, methodology, and writing—review and editing. LFB: investigation, methodology, and writing—review and editing. TB: supervision, writing—review and editing, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
FP-G and VJK were funded by the Department of Biotechnology and Food Science at the Norwegian University of Science and Technology. LFB was funded by The Research Council of Norway within ERA CoBioTech, project number 327216.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank Georg Kopplin, Research & Development Manager of the company Alginor ASA, for kindly providing biomass and hydrolysate from the brown seaweed Laminaria hyperborea.
Abe, S., Takayama, K.-I., and Kinoshita, S. (1967). Taxonomical Studies on Glutamic Acid-Producing Bacteria. J. Gen. Appl. Microbiol. 13, 279–301. doi:10.2323/jgam.13.279
Alderkamp, A.-C., Van Rijssel, M., and Bolhuis, H. (2007). Characterization of marine Bacteria and the Activity of Their Enzyme Systems Involved in Degradation of the Algal Storage Glucan Laminarin. FEMS Microbiol. Ecol. 59, 108–117. doi:10.1111/j.1574-6941.2006.00219.x
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Alva, A., Sabido-Ramos, A., Escalante, A., and Bolívar, F. (2020). New Insights into Transport Capability of Sugars and its Impact on Growth from Novel Mutants of Escherichia coli. Appl. Microbiol. Biotechnol. 104, 1463–1479. doi:10.1007/s00253-019-10335-x
Averianova, L. A., Balabanova, L. A., Son, O. M., Podvolotskaya, A. B., and Tekutyeva, L. A. (2020). Production of Vitamin B2 (Riboflavin) by Microorganisms: An Overview. Front. Bioeng. Biotechnol. 8, 1172. doi:10.3389/fbioe.2020.570828
Barbot, Y., Al-Ghaili, H., and Benz, R. (2016). A Review on the Valorization of Macroalgal Wastes for Biomethane Production. Mar. Drugs 14, 120. doi:10.3390/md14060120
Becker, J., Rohles, C. M., and Wittmann, C. (2018). Metabolically Engineered Corynebacterium Glutamicum for Bio-Based Production of Chemicals, Fuels, Materials, and Healthcare Products. Metab. Eng. 50, 122–141. doi:10.1016/j.ymben.2018.07.008
Becker, S., Tebben, J., Coffinet, S., Wiltshire, K., Iversen, M. H., Harder, T., et al. (2020). Laminarin Is a Major Molecule in the marine Carbon Cycle. Proc. Natl. Acad. Sci. U.S.A. 117, 6599–6607. doi:10.1073/pnas.1917001117
Bren, A., Park, J. O., Towbin, B. D., Dekel, E., Rabinowitz, J. D., and Alon, U. (2016). Glucose Becomes One of the Worst Carbon Sources for E.Coli on Poor Nitrogen Sources Due to Suboptimal Levels of cAMP. Sci. Rep. 6, 24834. doi:10.1038/srep24834
Bretzel, W., Schurter, W., Ludwig, B., Kupfer, E., Doswald, S., Pfister, M., et al. (1999). Commercial Riboflavin Production by Recombinant Bacillus Subtilis : Down-Stream Processing and Comparison of the Composition of Riboflavin Produced by Fermentation or Chemical Synthesis. J. Ind. Microbiol. Biotechnol. 22, 19–26. doi:10.1038/sj.jim.2900604
Buschke, N., Schäfer, R., Becker, J., and Wittmann, C. (2013). Metabolic Engineering of Industrial Platform Microorganisms for Biorefinery Applications - Optimization of Substrate Spectrum and Process Robustness by Rational and Evolutive Strategies. Bioresour. Tech. 135, 544–554. doi:10.1016/j.biortech.2012.11.047
Clore, G. M., and Venditti, V. (2013). Structure, Dynamics and Biophysics of the Cytoplasmic Protein-Protein Complexes of the Bacterial Phosphoenolpyruvate: Sugar Phosphotransferase System. Trends Biochem. Sci. 38, 515–530. doi:10.1016/j.tibs.2013.08.00310.1016/j.tibs.2013.08.003
Droffner, M. L., and Yamamoto, N. (1985). Isolation of Thermophilic Mutants of Bacillus Subtilis and Bacillus Pumilus and Transformation of the Thermophilic Trait to Mesophilic Strains. Microbiology 131, 2789–2794. doi:10.1099/00221287-131-10-2789
Eggeling, L., Bott, M., and Bott, M. (2005). Handbook of Corynebacterium Glutamicum. CRC Press. doi:10.1201/9781420039696
Figge, R. M., Ramseier, T. M., and Saier, M. H. (1994). The Mannitol Repressor (MtlR) of Escherichia coli. J. Bacteriol. 176, 840–847. doi:10.1128/jb.176.3.840-847.1994
Fujita, Y. (2009). Carbon Catabolite Control of the Metabolic Network inBacillus Subtilis. Biosci. Biotechnol. Biochem. 73, 245–259. doi:10.1271/bbb.80479
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 6, 343–345. doi:10.1038/nmeth.1318
Gopinath, V., Meiswinkel, T. M., Wendisch, V. F., and Nampoothiri, K. M. (2011). Amino Acid Production from rice Straw and Wheat Bran Hydrolysates by Recombinant Pentose-Utilizing Corynebacterium Glutamicum. Appl. Microbiol. Biotechnol. 92, 985–996. doi:10.1007/s00253-011-3478-x
Görke, B., and Stülke, J. (2008). Carbon Catabolite Repression in Bacteria: many Ways to Make the Most Out of Nutrients. Nat. Rev. Microbiol. 6, 613–624. doi:10.1038/nrmicro1932
Green, M. R., and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press.
Hakvåg, S., Nærdal, I., Heggeset, T. M. B., Kristiansen, K. A., Aasen, I. M., and Brautaset, T. (2020). Production of Value-Added Chemicals by Bacillus Methanolicus Strains Cultivated on Mannitol and Extracts of Seaweed Saccharina Latissima at 50°C. Front. Microbiol. 11, 680. doi:10.3389/fmicb.2020.00680
Hanahan, D. (1983). Studies on Transformation of Escherichia coli with Plasmids. J. Mol. Biol. 166, 557–580. doi:10.1016/S0022-2836(83)80284-8
Henke, N. A., Krahn, I., and Wendisch, V. F. (2021). Improved Plasmid-Based Inducible and Constitutive Gene Expression in Corynebacterium Glutamicum. Microorganisms 9, 204. doi:10.3390/microorganisms9010204
Heravi, K. M., and Altenbuchner, J. (2014). Regulation of the Bacillus Subtilis Mannitol Utilization Genes: Promoter Structure and Transcriptional Activation by the Wild-type Regulator (MtlR) and its Mutants. Microbiology (Reading) 160, 91–101. doi:10.1099/mic.0.071233-0
Hoffmann, S. L., Jungmann, L., Schiefelbein, S., Peyriga, L., Cahoreau, E., Portais, J.-C., et al. (2018). Lysine Production from the Sugar Alcohol Mannitol: Design of the Cell Factory Corynebacterium Glutamicum SEA-3 through Integrated Analysis and Engineering of Metabolic Pathway Fluxes. Metab. Eng. 47, 475–487. doi:10.1016/j.ymben.2018.04.019
Irla, M., Neshat, A., Winkler, A., Albersmeier, A., Heggeset, T. M. B., Brautaset, T., et al. (2014). Complete Genome Sequence of Bacillus Methanolicus MGA3, a Thermotolerant Amino Acid Producing Methylotroph. J. Biotechnol. 188, 110–111. doi:10.1016/j.jbiotec.2014.08.013
Javourez, U., O’Donohue, M., and Hamelin, L. (2021). Waste-to-nutrition: a Review of Current and Emerging Conversion Pathways. Biotechnol. Adv. 53, 107857. doi:10.1016/j.biotechadv.2021.107857
Jiménez-Escrig, A., and Sánchez-Muniz, F. J. (2000). Dietary Fibre from Edible Seaweeds: Chemical Structure, Physicochemical Properties and Effects on Cholesterol Metabolism. Nutr. Res. 20, 585–598. doi:10.1016/S0271-5317(00)00149-4
Laslo, T., von Zaluskowski, P., Gabris, C., Lodd, E., Ruckert, C., Dangel, P., et al. (2012). Arabitol Metabolism of Corynebacterium Glutamicum and its Regulation by AtlR. J. Bacteriol. 194, 941–955. doi:10.1128/JB.06064-11
Lee, K. H., Choi, H., Han, J. K., Park, Y. H., and Park, J. H. (2004). Microorganisms and Process for the Production of Riboflavin by Fermentation, 809–1401. Available at: https://patents.google.com/patent/EP1426450A1/en (Accessed December 5, 2021).
Lindner, S. N., Seibold, G. M., Henrich, A., Krämer, R., and Wendisch, V. F. (2011). Phosphotransferase System-independent Glucose Utilization in Corynebacterium Glutamicum by Inositol Permeases and Glucokinases. Appl. Environ. Microbiol. 77, 3571–3581. doi:10.1128/AEM.02713-10
Liu, S., Hu, W., Wang, Z., and Chen, T. (2020). Production of Riboflavin and Related Cofactors by Biotechnological Processes. Microb. Cel Fact. 19, 31. doi:10.1186/s12934-020-01302-7
López, M. G., Irla, M., Brito, L. F., and Wendisch, V. F. (2019). Characterization of D-Arabitol as Newly Discovered Carbon Source of Bacillus Methanolicus. Front. Microbiol. 10. doi:10.3389/fmicb.2019.01725
Meiswinkel, T. M., Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2013). Crude Glycerol-Based Production of Amino Acids and Putrescine by Corynebacterium Glutamicum. Bioresour. Tech. 145, 254–258. doi:10.1016/j.biortech.2013.02.053
Mindt, M., Hannibal, S., Heuser, M., Risse, J. M., Sasikumar, K., Nampoothiri, K. M., et al. (2019). Fermentative Production of N-Alkylated Glycine Derivatives by Recombinant Corynebacterium Glutamicum Using a Mutant of Imine Reductase DpkA from Pseudomonas Putida. Front. Bioeng. Biotechnol. 7, 232. doi:10.3389/fbioe.2019.00232
Monteiro, P., Lomartire, S., Cotas, J., Pacheco, D., Marques, J. C., Pereira, L., et al. (2021). Seaweeds as a Fermentation Substrate: A Challenge for the Food Processing Industry. Processes 9, 1953. doi:10.3390/pr9111953
Moon, M.-W., Park, S.-Y., Choi, S.-K., and Lee, J.-K. (2007). The Phosphotransferase System of Corynebacterium Glutamicum: Features of Sugar Transport and Carbon Regulation. J. Mol. Microbiol. Biotechnol. 12, 43–50. doi:10.1159/000096458
Olsson, J., Toth, G. B., and Albers, E. (2020). Biochemical Composition of Red, green and Brown Seaweeds on the Swedish West Coast. J. Appl. Phycol 32, 3305–3317. doi:10.1007/s10811-020-02145-w
Peng, X., Okai, N., Vertès, A. A., Inatomi, K.-i., Inui, M., and Yukawa, H. (2011). Characterization of the Mannitol Catabolic Operon of Corynebacterium Glutamicum. Appl. Microbiol. Biotechnol. 91, 1375–1387. doi:10.1007/s00253-011-3352-x
Pérez-García, F., Burgardt, A., Kallman, D. R., Wendisch, V. F., and Bar, N. (2021). Dynamic Co-cultivation Process of Corynebacterium Glutamicum Strains for the Fermentative Production of Riboflavin. Fermentation 7, 11. doi:10.3390/fermentation7010011
Pérez-García, F., Peters-Wendisch, P., and Wendisch, V. F. (2016). Engineering Corynebacterium Glutamicum for Fast Production of L-Lysine and L-Pipecolic Acid. Appl. Microbiol. Biotechnol. 100, 8075–8090. doi:10.1007/s00253-016-7682-6
Pérez-García, F., and Wendisch, V. F. (2018). Transport and Metabolic Engineering of the Cell Factory Corynebacterium Glutamicum. FEMS Microbiol. Lett. 365. doi:10.1093/femsle/fny166
Perkins, J. B., Sloma, A., Hermann, T., Theriault, K., Zachgo, E., Erdenberger, T., et al. (1999). Genetic Engineering of Bacillus Subtilis for the Commercial Production of Riboflavin. J. Ind. Microbiol. Biotechnol. 22, 8–18. doi:10.1038/sj.jim.2900587
Petteys, B. J., and Frank, E. L. (2011). Rapid Determination of Vitamin B2 (Riboflavin) in Plasma by HPLC. Clinica Chim. Acta 412, 38–43. doi:10.1016/j.cca.2010.08.037
Revuelta, J. L., Ledesma-Amaro, R., Lozano-Martinez, P., Díaz-Fernández, D., Buey, R. M., and Jiménez, A. (2017). Bioproduction of Riboflavin: a Bright Yellow History. J. Ind. Microbiol. Biotechnol. 44, 659–665. doi:10.1007/s10295-016-1842-7
Schwechheimer, S. K., Park, E. Y., Revuelta, J. L., Becker, J., and Wittmann, C. (2016). Biotechnology of Riboflavin. Appl. Microbiol. Biotechnol. 100, 2107–2119. doi:10.1007/s00253-015-7256-z
Song, S. H., and Vieille, C. (2009). Recent Advances in the Biological Production of Mannitol. Appl. Microbiol. Biotechnol. 84, 55–62. doi:10.1007/s00253-009-2086-5
Stahmann, K.-P., Revuelta, J. L., and Seulberger, H. (2000). Three Biotechnical Processes Using Ashbya Gossypii, Candida Famata, or Bacillus Subtilis Compete with Chemical Riboflavin Production. Appl. Microbiol. Biotechnol. 53, 509–516. doi:10.1007/s002530051649
Stansen, C., Uy, D., Delaunay, S., Eggeling, L., Goergen, J.-L., and Wendisch, V. F. (2005). Characterization of a Corynebacterium Glutamicum Lactate Utilization Operon Induced during Temperature-Triggered Glutamate Production. Appl. Environ. Microbiol. 71, 5920–5928. doi:10.1128/AEM.71.10.5920-5928.2005
Suraiya, S., Kim, J.-H., Tak, J. Y., Siddique, M. P., Young, C. J., Kim, J. K., et al. (2018). Influences of Fermentation Parameters on Lovastatin Production by Monascus purpureus Using Saccharina Japonica as Solid Fermented Substrate. LWT 92, 1–9. doi:10.1016/j.lwt.2018.02.013
Taniguchi, H., and Wendisch, V. F. (2015). Exploring the Role of Sigma Factor Gene Expression on Production by Corynebacterium Glutamicum: Sigma Factor H and FMN as Example. Front. Microbiol. 6, 740. doi:10.3389/fmicb.2015.00740
Wein, T., Hülter, N. F., Mizrahi, I., and Dagan, T. (2019). Emergence of Plasmid Stability under Non-selective Conditions Maintains Antibiotic Resistance. Nat. Commun. 10, 2595. doi:10.1038/s41467-019-10600-7
Wendisch, V. F., Brito, L. F., Gil Lopez, M., Hennig, G., Pfeifenschneider, J., Sgobba, E., et al. (2016). The Flexible Feedstock Concept in Industrial Biotechnology: Metabolic Engineering of Escherichia coli, Corynebacterium Glutamicum, Pseudomonas, Bacillus and Yeast Strains for Access to Alternative Carbon Sources. J. Biotechnol. 234, 139–157. doi:10.1016/j.jbiotec.2016.07.022
Keywords: seaweed, mannitol, phosphotransferase system, Corynebacterium glutamicum, riboflavin, fed-batch fermentation
Citation: Pérez-García F, Klein VJ, Brito LF and Brautaset T (2022) From Brown Seaweed to a Sustainable Microbial Feedstock for the Production of Riboflavin. Front. Bioeng. Biotechnol. 10:863690. doi: 10.3389/fbioe.2022.863690
Received: 27 January 2022; Accepted: 09 March 2022;
Published: 12 April 2022.
Edited by:
Ana Blandino, University of Cádiz, SpainReviewed by:
Dae-Hee Lee, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaCopyright © 2022 Pérez-García, Klein, Brito and Brautaset. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernando Pérez-García, ZmVybmFuZG8ucGVyZXotZ2FyY2lhQG50bnUubm8=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.