
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 14 February 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.843843
This article is part of the Research Topic Synthetic Biology for the Sustainable Production of Biochemicals in Engineered Microbes View all 17 articles
The demand for raspberry ketone (RK) as a plant-based natural flavoring agent is high, but natural RK is one of the most expensive flavor compounds due to its limited content in plants. Here, we produced RK de novo from simple carbon sources in Escherichia coli. We genetically engineered E. coli metabolism to overproduce the metabolic precursors tyrosine and p-coumaric acid and increase RK production. The engineered E. coli produced 19.3- and 1.9 g/L of tyrosine and p-coumaric acid from glucose, respectively. The p-coumaric acid CoA ligase from Agrobacterium tumefaciens and amino acid substituted benzalacetone synthase of Rhemu palmatum (Chinese rhubarb) were overexpressed in E. coli overproducing p-coumaric acid. The overexpression of fabF, encoding β-ketoacyl-acyl carrier protein synthetase II increased intracellular malonyl-CoA, the precursor of benzalacetone synthase for RK biosynthesis, and improved RK production. Fed-batch cultures given glucose as a carbon source produced 62 mg/L of RK under optimized conditions. Our production system is inexpensive and does not rely on plant extraction; thus, it should significantly contribute to the flavor and fragrance industries.
The raspberry phenylbutanoid ketone 4-(4-hydroxyphenyl)butan-2-one (RK) is a natural flavor in plants such as raspberries, grapes, peaches, and rhubarb. The berry flavor of RK with a low odor threshold is used as a food additive to create various aromas such as cherry, strawberry, kiwi and other fruits (Beekwilder et al., 2007; Bredsdorff et al., 2015; Sun et al., 2021). The alleged health benefits of RK include weight reduction (Morimoto et al., 2005; Wang et al., 2012) and skin lightening (Harada et al., 2008) and it is in high demand in the supplements and cosmetics industries (Milke et al., 2020; Vandamme and Soetaert, 2002). As the demand for RK is second only to vanillin, the potential of RK in the natural flavor market RK is 6–10 million Euros (Feron and Wache 2005). However, RK is difficult to cost-effectively produce from plants due to low contents. For example, raspberries contain only 1 to 4 mg/kg of RK (Larsen et al., 1991; Beekwilder et al., 2007), and extraction costs are high (Böker et al., 2001). Thus, the market for naturally occurring RK as a flavoring agent is United States $3,000–$20,000/kg (Milke et al., 2020). Chemically synthesized RK (Malkar and Yadav 2019) is not regarded as a natural flavor by United States and EU regulations, and it is essentially unacceptable to some consumers. However, microbial fermentation is an alternative strategy that allows inexpensive mass production of RK without the need for extraction from plants.
The RK biosynthesis pathways and mechanisms in raspberries and rhubarb have been investigated (Borejsza-Wysocki and Hrazdina 1994; Abe et al., 2001). The pathway starts from p-coumaroyl-CoA (Figure 1), which is a ubiquitous intermediate of the plant lignin biosynthetic pathway. During RK synthesis, the non-oxidative deamination of phenylalanine is catalyzed by phenylalanine ammonia lyase (PAL) and followed by hydroxylation and CoA-activation catalyzed by cinnamate-4-hydroxylase (C4H) and p-coumarate CoA ligase (CL) to produce p-coumaroyl-CoA. Thereafter, p-coumaroyl-CoA is condensed with malonyl-CoA to generate p-hydroxybenzalacetone by the activity of benzalacetone synthase (BAS), which is a type III polyketide synthase. The resulting p-hydroxybenzalacetone is reduced by the NADPH-dependent benzalacetone reductase (BAR) to yield RK (Figure 1).
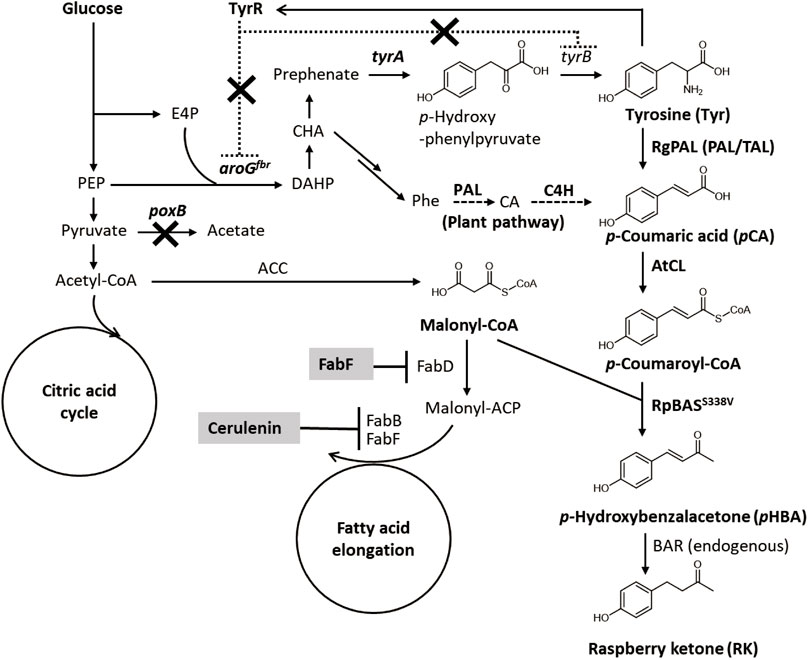
FIGURE 1. Metabolic network for RK biosynthesis from glucose in engineered E. coli. ACC, acetyl-CoA carboxylase; ACP, acyl carrier protein; aroGfbr, feedback resistant isozyme of DAHP synthase; BAR, benzalacetone reductase; BAS, benzalacetone synthase; CA, cinnamic acid; CL, 4-coumarate CoA ligase; C4H, cinnamate-4-hydroxylase; DAHP, 3-deoxy-D-arabino-heptulosonate 7-phosphate; E4P, erythrose 4-phosphate; FabB, β-ketoacyl-ACP synthase I; FabD, malonyl-CoA-ACP transacylase; FabF, β-ketoacyl-ACP synthase II; PAL, phenylalanine ammonia lyase; PEP, phosphoenolpyruvate; Phe, phenylalanine; poxB, pyruvate oxidase; TAL, tyrosine ammonia lyase; tyrA, chorismate mutase/prephenate dehydrogenase; tyrB, tyrosine aminotransferase; TyrR, transcriptional repressor of aromatic amino acid biosynthesis genes.
Microbial RK has been produced using genetically manipulated microorganisms such as yeast, Escherichia coli and other bacteria (Lee at al. 2016; Wang et al., 2019; Milke et al., 2020). The common approach converts p-coumaric acid as a starting material to RK in host cells producing heterogenous CL, BAS and BAR that originate from plants. Host E. coli and Corynebacterium glutamicum cells convert p-coumaric acid to RK with titers of 91.0 and 99.8 mg/l, respectively (Wang et al., 2019; Milke et al., 2020). However, few efforts have been made to produce RK de novo using renewable carbon sources. One exception is a wine yeast that generates plant PAL, C4H, and the synthetic CL and BAS fusion enzyme, and this yeast produces 3.5 mg/l of RK in grape juice medium (Lee at al. 2016), which is far lower than that yielded using p-coumaric acid as the raw material. The low titer could be due to the low availability of p-coumaroyl-CoA and malonyl-CoA that are substrates of BAS, the key enzyme of RK biosynthesis. Here, we showed that improving p-coumaric acid and malonyl-CoA availability significantly increased the productivity of microbial de novo RK synthesis. We constructed E. coli that generated abundant p-coumaric acid and malonyl-CoA by metabolic engineering and chemical stimulation. After gene selection and stepwise culture optimization, our recombinant E. coli produced RK biosynthesis enzymes and fermented glucose to produce 62 mg/L of RK.
Supplementary Table S1 lists the strains used in this study. We produced RK and constructed plasmids using E. coli BL21 (DE3) and E. coli JM109 (Novagen, Madison, WI, United States), respectively. We purchased 4-coumaroyl-CoA from Sigma–Aldrich (St. Louis, MO, United States) and l-tyrosine, p-coumaric acid, and p-hydroxybenzalacetone from Wako Chemicals (Tokyo, Japan). Plasmids were constructed using KOD One PCR Master Mix (Toyobo, Osaka, Japan), restriction enzymes (Takara Bio Inc., Shiga, Japan), Ligation high Ver. 2 (Toyobo) and NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs Inc., Ipswich, MA, United States). Metabolites were analyzed by high-performance liquid chromatography (HPLC) using a 1200 infinity photodiode array detector (Agilent Technologies Inc., Santa Clara, CA, United States), and by LC-ESI-MS/MS and GC-MS using LCMS-8045 and QP-2010 mass spectrometers (both from Shimadzu, Kyoto, Japan).
Supplementary Table S2 lists the plasmids used in this study. Nucleotide fragments of the tyrA gene were amplified by PCR using E. coli MG1655 genomic DNA and primers (Supplementary Table S3), digested with NcoI and BamHI, and cloned into pETduet-1 (Novagen) that was also digested with these enzymes to generate pET-tyrA. Plasmid pET-FevV for producing TAL (fevV from Streptomyces sp. WK-5344), was provided by Dr. Kawaguchi (Kawaguchi et al., 2017). Nucleotide sequences of the PAL genes of Camellia sinensis (Matsumoto et al., 1994) and Lithospermum erythrorhizon (Yazaki et al., 1995) were optimized according to E. coli codon usage (accession numbers; MZ439822 and MZ439823, Cspal and Lepal), synthesized and cloned into pUC57 (Genscript Biotech Corp., Piscataway, NJ, United States) to generate pUC-Cspal and pUC-Lepal. These plasmids were digested with NdeI and EcoRI and the resulting PAL gene fragments were cloned into pET-28b (Novagen) to obtain pET-Cspal and pET-Lepal, respectively. Plasmid pET28a-pal (Zhu et al., 2013) was digested with NdeI and XhoI to obtain Rhodotorula glutinis PAL gene fragments, and these were cloned into pRSFduet-1 (Novagen) to generate pRSF-Rgpal. The Agrobacterium tumefaciens 4-coumarate CoA ligase gene (Atu1416) was amplified by PCR using A. tumefaciens C58 genomic DNA and primers (Supplementary Table S3), digested with EcoRI and SalI, and cloned into pCDFduet-1 (Novagen) to generate pCDF-AtCL. The Rubus idaeus and Rhemu palmatum BAS genes were codon optimized (accession numbers; MZ439820 and MZ439821), synthesized, and cloned into pEX-A2 to obtain pEX-RiBAS and pEX-RpBAS (Eurofins Genomics Inc., Tokyo, Japan), respectively. We mutated BAS genes using QuickChange Site-directed Mutagenesis Kits (Agilent Technologies) and primers (Supplementary Table S3) to generate pEX-RiBASS338V pEX-RpBASS331V. The DNA fragments of RiBAS and RpBAS genes were digested with NdeI and XhoI and cloned into pCDF-AtCL to generate pCDF-AtCL-RiBAS, pCDF-AtuCL-RpBAS, pCDF-AtCL-RiBASS338V and pCDF-AtuCL-RpBASS331V, respectively. We amplified fabF by PCR using E. coli MG1655 genomic DNA and primers (Supplementary Table S3), then assembled it with pET28b that was digested with NcoI and XhoI to generate pET-fabF. Fragments of DNA containing the T7lac promoter, fabF, and T7 terminator sequences were amplified by PCR using pET-fabF and primers (Supplementary Table S3), then assembled with pCDF-AtCL-RpBAS that was digested with PacI to generate pCDF-AtCL-RpBAS-fabF. The tyrR and poxB genes of BL21 (DE3) were disrupted using the Red/ET recombination system (Gene Bridges, Heidelberg, Germany) with the described primers (Supplementary Table S3) to generate ΔtyrR and ΔpoxB strains (Masuo et al., 2016).
Recombinant E. coli BL21 (DE3) strains were cultured in 3 ml of LB medium, then 2 ml portions were inoculated into 500-ml conical flasks containing 100 ml of fermentation medium (10 g glucose, 10 g tryptone, 5 g yeast extract, 24 g Na2HPO4, 12 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 0.5 g MgSO4 7H2O, 15 mg CaCl2, 50 mg thiamine-HCl and 2 ml of trace element solution/L; Fujita et al., 2013) or modified fermentation medium (10 g glucose, 10 g tryptone, 5 g yeast extract, 12 g Na2HPO4, 6 g KH2PO4, 42 g MOPS, 0.5 g NaCl, 10 g (NH4)2SO4, 0.5 g MgSO4 7H2O, 15 mg CaCl2, 50 mg thiamine-HCl and 2 ml of trace element solution/L). The flasks were rotary-shaken at 120 rpm and 30°C for 3 h under aerobic conditions unless otherwise stated. When the OD600 reached 0.6, 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) was added, then the cells were further incubated for the indicated amounts of time. Fed-batch cultures in a 1.0-L BMJ-01 fermenter (Biott, Tokyo, Japan) containing 0.5 L of fermentation or modified fermentation medium were agitated at 550 rpm, 30°C, and aerated at 1.0 L/min. When the OD600 reached 0.6, 0.1 mM IPTG was added. Peristaltic pumps fed the cultures with 500 g/L of glucose when the glucose concentration dipped below 1.5 g/L. The pH was monitored using an electrode and maintained between 7.0 and 7.1 by adding 10% NH4OH.
Escherichia coli BL21 (DE3) harboring either pET-FevV, pET-28a-pal, pET-Cspal or pET-Lepal was cultured in 3 ml of LB medium, then 2 ml portions were inoculated into 100 ml of LB medium and rotary-shaken at 120 rpm at 30°C under aerobic conditions. When the OD600 of the cultures reached 0.6, the cultures were incubated for 18 h with 0.5 mM IPTG, then the cells were sedimented by centrifugation at 3,000 × g for 10 min. The cells were washed with 50 mM Tris-HCl (pH 7.0), suspended in 50 mM Tris-HCl (pH 8.0) containing 1.8 g/L of tyrosine and incubated at 30°C with agitation at 120 rpm.
Culture supernatants were acidified with formic acid and extracted twice with equal amounts ethyl acetate. The ethyl acetate was evaporated, then the extracts were dissolved in methanol and analyzed by GC-MS equipped with a ZB-5MS capillary column (30 m × 0.32 mm internal diameter × 0.25-μm film thickness: Phenomenex, Torrance, CA, United States). The injection and ion-source temperatures were 250 and 200°C, respectively. The linear velocity of the carrier helium gas was 45 cm/s. The oven temperature was controlled at 40°C for 4 min, increased to 250°C at a rate of 12°C/min, then maintained at 250°C for 7 min.
Tyrosine and p-coumaric acid were quantified using HPLC and a TSKgel® ODS-100V column (4.6 mm × 25 cm, particle size 3 μm, Tosoh, Tokyo, Japan). The initial mobile phase comprised 95%:5% 10 mM ammonium formate (pH 7.0): acetonitrile for 8 min, followed by an increase to 50% acetonitrile for 6 min and maintained for 2 min. The flow rate of 0.8 ml min−1 and the column temperature of 30°C were maintained throughout the analysis.
We quantified RK and intermediates using the multiple-reaction monitoring mode on the LC-ESI-MS/MS under the following conditions: capillary voltage, 4.5 kV; desolvation line, 250°C; heat block, 400°C; nebulizer nitrogen gas 3 L/min; drying gas, 10 L/min. Compounds of interest were separated by the LC system equipped with a 150 × 2.1 mm ACQUITY UPLC CSH™ C18 2.1 × 150 mm (Waters, Corp., Milford, MA United States) with particle and pore sizes of 1.7 μm and 130
We initially generated E. coli that overproduced tyrosine, the precursor of p-coumaric acid to construct a microbial de novo RK synthesis system (Figure 1). A feedback resistant mutant of 3-deoxy-d-arabinoheptulosonate-7-phosphate (DAHP) synthase (aroGfbr) and the chorismate mutase/prephenate dehydrogenase gene (tyrA) were overexpressed in E. coli BL21 (DE3) with the native aroG and T7 promoters, respectively (Figure 1). The resulting strain produced 0.4 ± 0.1 g/L of tyrosine in the modified M9 medium, whereas the parental E. coli BL21 (DE3) almost did not (Supplementary Figure S1). Another potential parental strain, E. coli NST37 (DE3)/ΔpheLA, which has an enhanced shikimate pathway (Masuo et al., 2016), produced less tyrosine after the same genetic modification (Supplementary Figure S1B). The poxB gene deletion mutant (ΔpoxB) harboring aroGfbr and tyrA expression plasmids produced 0.9 ± 0.2 g/L of tyrosine in the 72-h flask culture (Figure 2A and Table 1). Deletion of poxB encoding the pyruvate oxidase that synthesizes acetate from pyruvate (Causey et al., 2004), led to a positive effect on tyrosine production probably due to the altered carbon flux including acetate, phosphoenolpyruvate, and erythrose-4-phosphate generation (Figure 1).
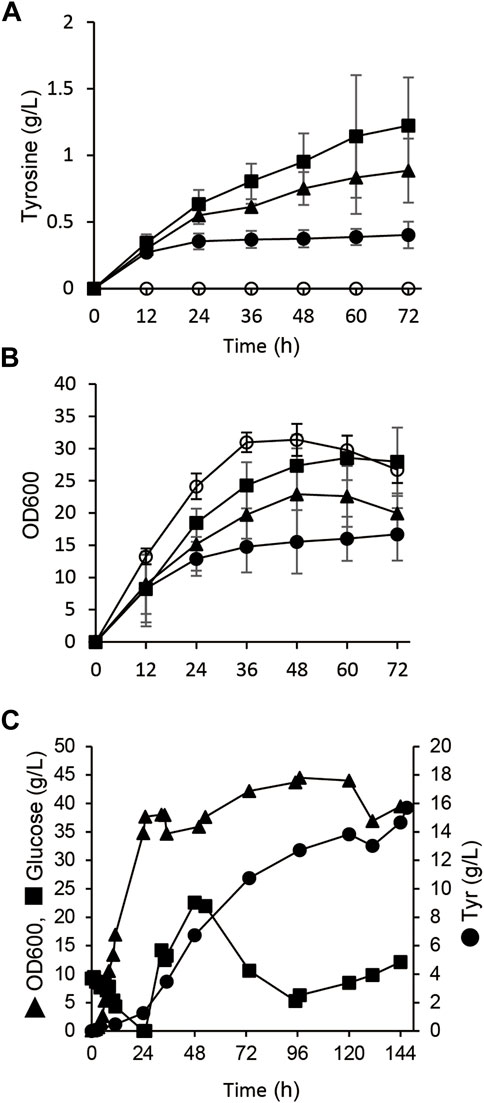
FIGURE 2. Tyrosine production from glucose by E. coli with engineered metabolism. (A,B) Time-dependent tyrosine production (A) and growth (B) of E. coli BL21 (DE3) (closed circles), ΔpoxB (closed triangles) and ΔtyrR (closed squares) harboring pET-tyrA and pACYC-aroGfbr in fermentation medium at 30°C. E. coli BL21 (DE3) harboring pET and pACYC empty plasmids was analyzed as a control (open squares). (C) Fed-batch cultures of ΔtyrR harboring pET-tyrA and pACYC-aroGfbr in jar fermenters containing 0.5 L of fermentation medium at 30°C and pH 7.1 (maintained by 10% ammonium) were incubated for 24 h, then fed with glucose (0.8 g/L/h). Error bars indicate standard deviation (n = 3).
A gene deletion mutant of tyrR (ΔtyrR), which encodes a transcriptional repressor of aromatic amino acid biosynthesis genes (Pittard et al., 2005), harboring the aroGfbr and tyrA expression plasmids (strain AT1) produced 1.2 ± 0.3 g/L of tyrosine in flask cultures. The production titer of AT1 was 3.0-fold higher than that of parental BL21 (DE3) strain (Figure 2A and Table 1). Growth defect caused by aroGfbr and tyrA overexpression was restored by gene disruption of tyrR (Figures 2A,B). Fed-batch cultures of AT1 with stepwise additions of glucose avoided excessive glucose accumulation in the jar fermenters. The culture generated 15.7 g/L tyrosine with a production yield of 7.4% vs glucose (Figure 2C). Precipitates that appeared on the inner walls of fermentation vessels (Supplementary Figure S2B), contained 1.9 g of tyrosine, and the total amount produced was 19.3 g/L. The culture generated low levels of acetate (<1.0 g L−1; Supplementary Figure S2A), indicating decreased metabolic flow of glucose to acetate, which resulted in increased tyrosine production.
To convert the produced tyrosine to pCA, we employed tyrosine ammonia lyase (TAL) or bifunctional PAL that deaminate phenylalanine and tyrosine. We cloned TAL or PAL genes derived from yeast, plant or bacteria into the pRSFduet1 vector under the control of the T7 promoter and the resulting plasmids were introduced into BL21 (DE3). These strains converted tyrosine into pCA (Table 2). The E. coli producing Rodotorura glutinis PAL (RgPAL) had the highest yield, conversion rate and efficiency, and was deemed adequate for pCA synthesis de novo. We introduced the RgPAL expression plasmid into the AT1 strain and the resulting AT2 strain produced 1.9 g/L of pCA from glucose in a fed-batch culture under optimized conditions over a period of 60 h (Figure 3). The culture accumulated >1.0 g/L tyrosine, indicating that PAL reaction limits pCA production.
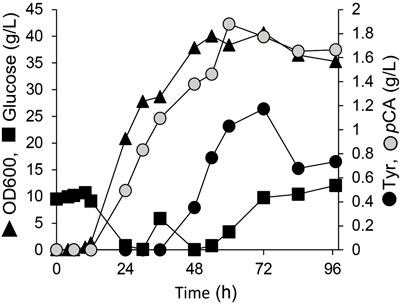
FIGURE 3. Production of p-coumaric acid by E. coli. Fed-batch cultures of AT2 strain in jar fermenters containing 0.5 L of fermentation medium at 30°C and pH 7.1 (maintained by 10% ammonium) were incubated for 24 h, then fed with glucose (0.8 g/L/h).
We used AT2 as a heterologous expression host to construct the RK pathway. We expressed both the genes for AtCL and RiBAS, derived from Agrobacterium tumefaciens C58 (Campillo et al., 2014) and Rubus idaeus BAS (Zheng and Hrazdina, 2008) in the AT2 strain under the control of T7lac promoter. The resulting AT2Ri strain was cultured in modified M9 medium for 169 h and its metabolites were extracted with ethyl acetate and analyzed by GC-MS (Figure 4A). The ion peaks had the same retention time and fragmentation ion pattern as standard RK in the metabolites of AT2Ri, but not AT2 (Figures 4A,B). The AT2Ri strain produced 0.44 mg L−1 of RK from glucose for 76 h, whereas is considerable amount of pCA remaining in the culture supernatant (Table 3), implied insufficient BAS activities in the cells. Substituting serine 331 with valine in BAS from Rhemu palmatum (RpBAS) increases the catalytic activity (Abe et al., 2007). Alignment of the amino acid sequences showed that this serine residue was conserved in RiBAS (Supplementary Figure S3). The corresponding mutants of RiBAS (RiBASS338V), RpBAS, and mutated RpBASS331V, together with AtCL, were introduced into AT2, respectively. The resulting AT2Rp and AT2RpSV strains, expressing RpBAS and RpBASS331V, produced 28 and 29 mg/L of RK from glucose as the raw material after 76 h, respectively. These yields were 60-fold higher than that of AT2Ri (Table 3). The AT2Rp and AT2RpSV strains accumulated less pCA than AT2Ri, indicating that conversion of pCA to RK is more efficient with RpBAS and RpBASS331V. Our study showed that the mutations in RpBAS and RiBAS had little effect on cellular RK production (Table 3), probably due to the pH dependence of BAS activity (Abe et al., 2007). Less p-hydroxybenzalacetone accumulated in cultured AT2RpSV (Table 3), implying sufficient BAR activities in this strain under appropriate culture conditions.
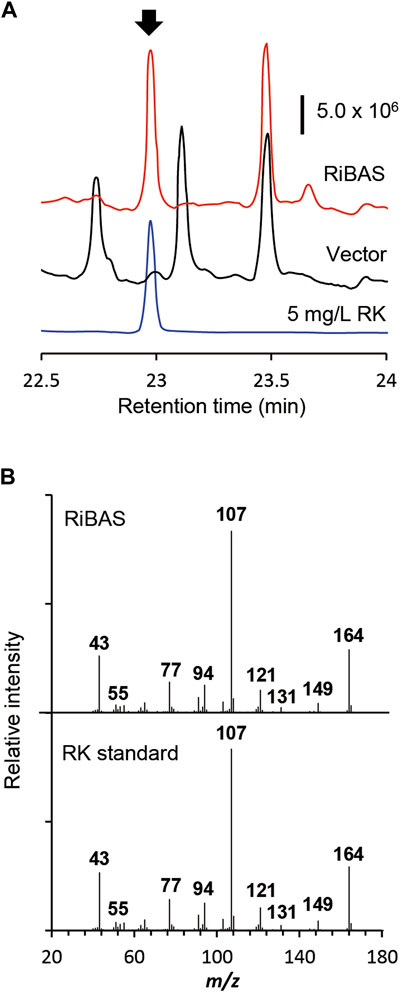
FIGURE 4. Production of RK from glucose by E. coli expressing 4CL and BAS. (A) GC-MS profile of extracts of cultured E. coli AT2 harboring pCDF-Atu4CL-RiBAS (RiBAS) or pCDFduet-1 (Vector). Arrow, peak of standard RK. (B) Gas chromatography-mass spectrometry fragmentation patterns of peaks at 23 min (arrow in (A)).
The overexpression of fabF, which encodes β-ketoacyl carrier protein synthase II, inhibits FabD activity and hence fatty acid elongation (Figure 1), and resulted in cellular accumulation of malonyl-CoA in E. coli (Subrahmanyam and Cronan 1998; Kassab et al., 2019). The AT3 strain that was the AT2 strain overexpressing in under the control of T7 lac promoter, accumulated 2.2-fold more malonyl-CoA than AT2 (Figure 5A). We constructed a plasmid to overexpress fabF along with the AtCL and RpBASS331V genes and introduced it into AT2 to generate AT3RpSV, which produced 24 mg/L of RK when cultured for 60 h in modified fermentation medium (Figure 5B). The production titer of AT3RpSV was 1.4-fold higher than that of AT2RpSV, indicating that the increased cellular malonyl-CoA levels improved the amount of malonyl-CoA available for RK production. We optimized aeration conditions, culture media, and IPTG concentrations for RK production by AT3RpSV (Supplementary Figure S4). Fed-batch cultured AT3RpSV under controlled glucose addition produced 41 mg/L of RK with a production yield of 0.08% (vs glucose, Figure 5C).
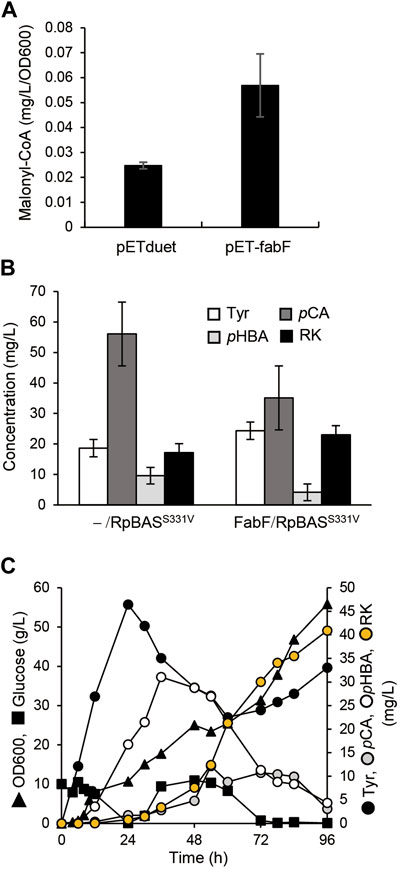
FIGURE 5. Overexpression of FabF increased intracellular malonyl-CoA and RK production. (A) Intracellular malonyl-CoA concentration in ΔtyrR strain harboring pETduet-1 or pET-fabF incubated in modified fermentation medium at 30°C for 60 h. (B) Concentrations of RK and intermediates in cultured AT2 strain harboring pCDF-Atu4CL-RpBASS331V (-/RpBASS331V) or pCDF-Atu4CL-RpBASS331V-fabF (FabF/RpBASS331V (AT3RpSV strain). Strains were incubated in modified fermentation medium at 30°C for 60 h. (C) Fed-batch cultures of AT3RpSV in jar fermenters containing 0.5 L of fermentation medium were incubated for 24 h at 30°C and pH 7.1 (maintained by 10% ammonium), then fed with glucose (0.8 g/L/h). Error bars indicate standard deviation (n = 3).
Cerulenin binds to ACP synthase to block its interaction with malonyl-CoA and inhibits fatty acid biosynthesis. Adding cerulenin to bacterial cultures thus accumulates high levels of intracellular malonyl-CoA (Davis et al., 2000). We simultaneously added IPTG and various concentrations of cerulenin to AT3RpSV cultures. The AT3RpSV strain produced 1.6-fold RK (54 mg/L) when cultured with 0.1 mM cerulenin than those without cerulenin (Figure 6A). Fed-batch cultures of AT3RpSV with 0.1 mM cerulenin in 1-L jar fermenters produced 62 mg/L of RK under optimized culture conditions, which increased the yield (Figure 6B) up to 0.12% vs glucose.
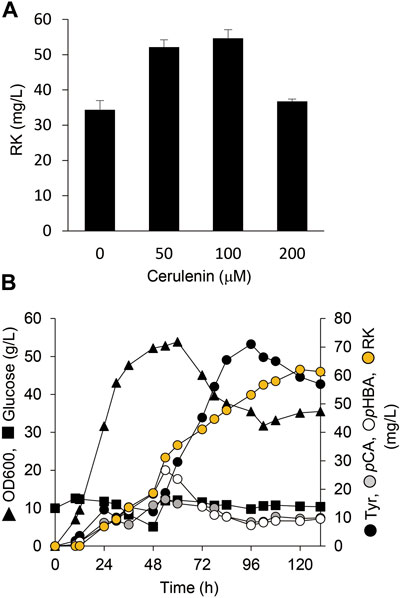
FIGURE 6. Cerulenin addition increased E. coli RK production. (A) Production of RK by AT3RpSV cultured in modified fermentation medium containing various concentrations of cerulenin at 30°C for 60 h. (B) Fed-batch cultures of AT3RpSV were incubated in jar fermenters containing 0.5 L of fermentation medium at 30°C and pH 7.1 (maintained by 10% ammonium). Cerulenin (100 mM) was added after incubation for 10 h and the cultures were fed with glucose (0.8 g/L/h) after incubation for 24 h. Error bars indicate standard deviation (n = 3).
We constructed a microbial platform that produced RK from glucose. Genetically engineering E. coli metabolism and subsequent PAL optimization resulted in the respective production of 19.3 and 1.9 g/L of Tyr and pCA from glucose (Figures 2, 3). A strain producing pCA harbored plasmids to express CL and BAS generated RK from glucose (Figure 4). Increasing cellular malonyl-CoA by genetic and chemical manipulation improved the RK yield (Figures 5, 6). Finally, fed-batch culture under optimal conditions fermented 62 mg/L of RK from glucose (Figure 6). Currently, the use of cerulenin is a disadvantage for cost-saving RK production. Notably, our platform produces 41 mg/L of RK without the cerulenin, which is 12-times more than the previous fermenting process (Lee et al., 2016).
During biosynthesis, BAS conjugates p-coumaroyl-CoA and malonyl-CoA, which are respectively synthesized via the phenylpropanoid and fatty acid synthesis pathways (Figure 1). The efficient production of RK from glucose in a heterologous host requires the optimal production of these substrates. Due to difficulties achieving this, published reports describing microbial RK production are limited to those in which phenylpropanoid pCA was the raw material (Lee at al. 2016; Wang et al., 2019; Milke et al., 2020). The present study engineered the phenylpropanoid- and fatty acid synthesis pathways using genetic and chemical approaches, as well as fermentation, and optimized pCA levels and malonyl-CoA supplies in E. coli.
The engineered AT2 strain fermented glucose to produce 1.9 g/L of pCA during culture for 60 h (Figure 3). This amount exceeded that produced by any previous E. coli system (Flourat et al., 2021). Not only is pCA important as an aromatic precursor of bioactive substances such as stilbenoids, flavonoids and curcuminoids (Katsuyama et al., 2008; Trantas et al., 2009), but it is also a raw material for thermo-tolerant plastics (Kaneko et al., 2006). Our platform could thus facilitate the production of these valuable compounds.
The culture supernatant of the AT2 strain still contained a large amount of tyrosine (>1.0 g/L), and this strain yielded 10-fold less pCA than AT1 (Figures 2, 3). This implied insufficient conversion of tyrosine to pCA, and that enhancing RgPAL production improves the generation of pCA and RK. The tyrosine conversion rate of RgPAL to pCA reached 0.8 g of pCA/L/h under resting-cell reactions (Table 2). This was much higher than the calculated maximum rate of 0.07 g of pCA/L/h generated by fermenting AT2 cells (Figure 3). This indicates that fine-tuning cellular metabolic flux further improves productivity.
The availability of intracellular malonyl-CoA was increased by FabF overexpression and cerulenin, which improved RK production de novo (Figures 5, 6). This implied that increasing the intracellular flux for malonyl-CoA synthesis would improve RK production. Another strategy might be to overproduce acetyl-CoA carboxylase that converts intracellular acetyl-CoA to malonyl-CoA (Figure 1), which is supported by the following findings. Excessive acetyl-CoA carboxylase production results in a 100-fold increase in intracellular malonyl-CoA levels (Davis et al., 2000). Overexpressed acetyl-CoA carboxylase combined with engineering the metabolism of the glycolytic system and the TCA cycle, results in 4- and 5.6-fold increases, respectively, in the amounts of intracellular malonyl-CoA and naringenin, a plant-specific secondary metabolite derived from malonyl-CoA (Xu et al., 2011).
This study used multiple plasmids for expressing heterologous genes. Although this approach allows rapid construction of recombinant E. coli, several issues should be addressed before applied to large-scale fermentation that includes stability of plasmids, cost of antibiotics, and stable enzyme production. Genomic integration of the introduced genes should be a possible approach to solve these issues. CRISPR/Cas9 or λ-Red recombination systems enable scarless genome engineering in E. coli (Pyne et al., 2015; Bayer et al., 2021; Vo et al., 2021) and must be powerful tools for this purpose. These approaches improve our RK fermentation system to a more stable and economically friendly one.
In conclusion, this study established a microbial system that produced useful amounts of RK at low cost, and within a reasonable time frame. The present supply of RK derived from plants is limited and extraction is costly. Our straightforward batch fermentation system offers an inexpensive and efficient alternative to extracting RK from plants based on a simple carbon source that should significantly contribute to the flavor and fragrance industries.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SM and NT conceived and designed research. SM, CS, KU, YS and YK conducted experiments. SM, CS, KU and YS analyzed data. SM and NT wrote the manuscript. All authors read and approved the manuscript.
This study was supported by the Japan Society for the Promotion of Science KAKENHI Grants-in-Aid for Scientific Research (17K15257), Japan Science and Technology Agency CREST (JPMJCR13B) and a Japan Science and Technology Agency ERATO Project Grant (JPMJER1502).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Norma Foster for critical reading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.843843/full#supplementary-material
Abe, I., Takahashi, Y., Morita, H., and Noguchi, H. (2001) Benzalacetone Synthase. Eur. J. Biochem. 268(11):3354–3359. doi:10.1046/j.1432-1327.2001.02255.x
Abe, T., Morita, H., Noma, H., Kohno, T., Noguchi, H., and Abe, I. (2007) Structure Function Analysis of Benzalacetone Synthase from Rheum Palmatum. Bioorg. Med. Chem. Lett. 17(11):3161–3166. doi:10.1016/j.bmcl.2007.03.029
Bayer, C. N., Rennig, M., Ehrmann, A. K., and Nørholm, M. H. H. (2021) A Standardized Genome Architecture for Bacterial Synthetic Biology (SEGA). Nat. Commun. 12(1):5876. doi:10.1038/s41467-021-26155-5
Beekwilder, J., van der Meer, I. M., Sibbesen, O., Broekgaarden, M., Qvist, I., Mikkelsen, J. D., et al. (2007) Microbial Production of Natural Raspberry Ketone. Biotechnol. J. 2:1270–1279. doi:10.1002/biot.200700076
Boker, A., Fischer, M., and Berger, R. G. (2001) Raspberry Ketone from Submerged Cultured Cells of the Basidiomycete Nidula Niveo-Tomentosa Biotechnol. Prog. 17(3):568–572. doi:10.1021/bp010011b
Borejsza-Wysocki, W., Goers, S. K., McArdle, R. N., and Hrazdina, G. (1992) (p-Hydroxyphenyl)butan-2-one Levels in Raspberries Determined by Chromatographic and Organoleptic Methods. J. Agric. Food Chem. 40: 1176–1177. doi:10.1021/jf00019a018
Borejsza-Wysocki, W., and Hrazdina, G. (1994) Biosynthesis of P-Hydroxyphenylbutan-2-One in Raspberry Fruits and Tissue Cultures. Phytochemistry 35(3):623–628. doi:10.1016/S0031-9422(00)90575-2
Bredsdorff, L., Wedebye, E. B., Nikolov, N. G., Hallas-Møller, T., and Pilegaard, K. (2015) Raspberry Ketone in Food Supplements - High Intake, Few Toxicity Data - A Cause for Safety Concern? Regul. Toxicol. Pharmacol. 73(1):196–200. doi:10.1016/j.yrtph.2015.06.022
Campillo, T., Renoud, S., Kerzaon, I., Vial, L., Baude, J., Gaillard, V., et al. (2014) Analysis of Hydroxycinnamic Acid Degradation in Agrobacterium Fabrum Reveals a Coenzyme A-dependent, Beta-Oxidative Deacetylation Pathway. Appl. Environ. Microbiol. 80(11):3341–3349. doi:10.1128/AEM.00475-14
Causey, T. B., Shanmugam, K. T., Yomano, L. P., and Ingram, L. O. (2004) Engineering Escherichia coli for Efficient Conversion of Glucose to Pyruvate. Proc. Natl. Acad. Sci. 101(8):2235–2240. doi:10.1073/pnas.0308171100
Davis, M. S., Solbiati, J., and Cronan, J. E. (2000) Overproduction of Acetyl-CoA Carboxylase Activity Increases the Rate of Fatty Acid Biosynthesis in Escherichia coli J. Biol. Chem. 275(37):28593–28598. doi:10.1074/jbc.M004756200
Feron, G., Mauvais, G., Martin, F., Sémon, E., and Blin-Perrin, C. 2007, Microbial Production of 4-hydroxybenzylidene Acetone, the Direct Precursor of Raspberry Ketone, Lett. Appl. Microbiol., 45) “Microbial Production of Food Flavour,” in Food Biotech, eds K. Shetty, A. L. Pometto, and G. Paliyath (New York, NY: M. Dekker Inc.), 29–35. doi:10.1111/j.1472-765X.2007.02147.x
Flourat, A. L., Combes, J., Bailly‐Maitre‐Grand, C., Magnien, K., Haudrechy, A., Renault, J. H., et al. (2021) Accessing P ‐Hydroxycinnamic Acids: Chemical Synthesis, Biomass Recovery, or Engineered Microbial Production? ChemSusChem 14(1):118–129. doi:10.1002/cssc.202002141
Fujita, T., Nguyen, H. D., Ito, T., Zhou, S., Osada, L., Tateyama, S., et al. (2013) Microbial Monomers Custom-Synthesized to Build True Bio-Derived Aromatic Polymers. Appl. Microbiol. Biotechnol. 97(20):8887–8894. doi:10.1007/s00253-013-5078-4
Harada, N., Okajima, K., Narimatsu, N., Kurihara, H., and Nakagata, N. (2008) Effect of Topical Application of Raspberry Ketone on Dermal Production of Insulin-like Growth Factor-I in Mice and on Hair Growth and Skin Elasticity in Humans. Growth Horm. IGF Res. 18:335–344. doi:10.1016/j.ghir.2008.01.005
Kaneko, T., Thi, T. H., Shi, D. J., and Akashi, M. (2006) Environmentally Degradable, High-Performance Thermoplastics from Phenolic Phytomonomers. Nat. Mater 5(12):966–970. doi:10.1038/nmat1778
Kassab, E., Fuchs, M., Haack, M., Mehlmer, N., and Brueck, T. B. (2019) Engineering Escherichia coli FAB System Using Synthetic Plant Genes for the Production of Long Chain Fatty Acids. Microb. Cel Fact. 18(1):163. doi:10.1186/s12934-019-1217-7
Katsuyama, Y., Matsuzawa, M., Funa, N., and Horinouchi, S. (2008) Production of Curcuminoids by Escherichia coli Carrying an Artificial Biosynthesis Pathway. Microbiology (Reading) 154(9):2620–2628. doi:10.1099/mic.0.2008/018721-0
Kawaguchi, H., Katsuyama, Y., Danyao, D., Kahar, P., Nakamura-Tsuruta, S., Teramura, H., et al. (2017) Caffeic Acid Production by Simultaneous Saccharification and Fermentation of Kraft Pulp Using Recombinant Escherichia coli Appl. Microbiol. Biotechnol. 101(13):5279–5290. doi:10.1007/s00253-017-8270-0
Kim, M., Baek, H. S., Lee, M., Park, H., Shin, S. S., Choi, D. W., et al. (2016) Rhododenol and Raspberry Ketone Impair the normal Proliferation of Melanocytes through Reactive Oxygen Species-dependent Activation of GADD45. Toxicol. Vitro 32:339–346. doi:10.1016/j.tiv.2016.02.003
Larsen, M., Poll, L., Callesen, O., and Lewis, M. (1991) Relations between the Content of Aroma Compounds and the Sensory Evaluation of 10 Raspberry Varieties (Rubus idaeusL). Acta Agriculturae Scand. 41:447–454. doi:10.1080/00015129109439927
Lee, D., Lloyd, N. D. R., Pretorius, I. S., and Borneman, A. R. (2016) Heterologous Production of Raspberry Ketone in the Wine Yeast Saccharomyces cerevisiae via Pathway Engineering and Synthetic Enzyme Fusion. Microb. Cel Fact 15:49. doi:10.1186/s12934-016-0446-2
Malkar, R. S., and Yadav, G. D. (2019) Selectivity Engineering in One Pot Synthesis of Raspberry Ketone: Crossed Aldol Condensation of P -Hydroxybenzaldehyde and Acetone and Hydrogenation over Novel Ni/Zn-La Mixed Oxide. ChemistrySelect 4:2140–2152. doi:10.1002/slct.201804060
Masuo, S., Zhou, S., Kaneko, T., and Takaya, N. (2016) Bacterial Fermentation Platform for Producing Artificial Aromatic Amines. Sci. Rep. 6:25764 doi:10.1038/srep25764
Matsumoto, S., Takeuchi, A., Hayatsu, M., and Kondo, S. (1994) Molecular Cloning of Phenylalanine Ammonia-Lyase cDNA and Classification of Varieties and Cultivars of tea Plants (Camellia Sinensis) Using the tea PAL cDNA Probe. Theoret. Appl. Genet. 89(6):671–675. doi:10.1007/BF00223703
Milke, L., Mutz, M., and Marienhagen, J. (2020) Synthesis of the Character Impact Compound Raspberry Ketone and Additional Flavoring Phenylbutanoids of Biotechnological Interest with Corynebacterium Glutamicum. Microb. Cel Fact 19:92. doi:10.1186/s12934-020-01351-y
Morimoto, C., Satoh, Y., Hara, M., Inoue, S., Tsujita, T., and Okuda, H. (2005) Anti-obese Action of Raspberry Ketone. Life Sci. 77:194–204. doi:10.1016/j.lfs.2004.12.029
Pittard, J., Camakaris, H., and Yang, J. (2005) The TyrR Regulon. Mol. Microbiol. 55(1):16–26. doi:10.1111/j.1365-2958.2004.04385.x
Pyne, M. E., Moo-Young, M., Chung, D. A., and Chou, C. P. (2015) Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli. Appl. Environ. Microbiol. 81(15):5103–5114. doi:10.1128/AEM.01248-15
Subrahmanyam, S., and Cronan, J. E. (1998) Overproduction of a Functional Fatty Acid Biosynthetic Enzyme Blocks Fatty Acid Synthesis in Escherichia coli J. Bacteriol. 180(17):4596–4602. doi:10.1128/JB.180.17.4596-4602.1998
Sun, L., Xin, F., and Alper, H. S. (2021) Bio-synthesis of Food Additives and Colorants-A Growing Trend in Future Food. Biotechnol. Adv. 47:107694. doi:10.1016/j.biotechadv.2020.107694
Trantas, E., Panopoulos, N., and Ververidis, F. (2009) Metabolic Engineering of the Complete Pathway Leading to Heterologous Biosynthesis of Various Flavonoids and Stilbenoids in Saccharomyces cerevisiae. Metab. Eng. 11(6):355–366. doi:10.1016/j.ymben.2009.07.004
Vandamme, E. J., and Soetaert, W. (2002) Bioflavours and Fragrances via Fermentation and Biocatalysis. J. Chem. Technol. Biotechnol. 77:1323–1332. doi:10.1002/jctb.722
Vo, P. L. H., Ronda, C., Klompe, S. E., Chen, E. E., Acree, C., Wang, H. H., et al. (2021) CRISPR RNA-Guided Integrases for High-Efficiency, Multiplexed Bacterial Genome Engineering. Nat. Biotechnol. 39(4):480–489. doi:10.1038/s41587-020-00745-y
Wang, C., Zheng, P., and Chen, P. (2019) Construction of Synthetic Pathways for Raspberry Ketone Production in Engineered Escherichia coli. Appl. Microbiol. Biotechnol. 103(9):3715–3725. doi:10.1007/s00253-019-09748-5
Wang, L., Meng, X., and Zhang, F. (2012) Raspberry Ketone Protects Rats Fed High-Fat Diets against Nonalcoholic Steatohepatitis. J. Med. Food 15:495–503. doi:10.1089/jmf.2011.1717
Xu, P., Ranganathan, S., Fowler, Z. L., Maranas, C. D., and Koffas, M. A. G. (2011) Genome-scale Metabolic Network Modeling Results in Minimal Interventions that Cooperatively Force Carbon Flux towards Malonyl-CoA. Metab. Eng. 13(5):578–587. doi:10.1016/j.ymben.2011.06.008
Yazaki, K., Kataoka, M., Honda, G., Severin, K., and Heide, L. (1997) cDNA Cloning and Gene Expression of Phenylalanine Ammonia-Lyase inLithospermum Erythrorhizon. Biosci. Biotechnol. Biochem. 61(12):1995–2003. doi:10.1271/bbb.61.1995
Zheng, D., and Hrazdina, G. (2008) Molecular and Biochemical Characterization of Benzalacetone Synthase and Chalcone Synthase Genes and Their Proteins from Raspberry (Rubus Idaeus L.). Arch. Biochem. Biophys. 470(2):139–145. doi:10.1016/j.abb.2007.11.013
Keywords: raspberry ketone, flavor agent, microbial production, plant secondary metabolite, metabolic engieering
Citation: Masuo S, Saga C, Usui K, Sasakura Y, Kawasaki Y and Takaya N (2022) Glucose-Derived Raspberry Ketone Produced via Engineered Escherichia coli Metabolism. Front. Bioeng. Biotechnol. 10:843843. doi: 10.3389/fbioe.2022.843843
Received: 27 December 2021; Accepted: 18 January 2022;
Published: 14 February 2022.
Edited by:
Jingyu Wang, Westlake Institute for Advanced Study (WIAS), ChinaReviewed by:
Jifeng Yuan, Xiamen University, ChinaCopyright © 2022 Masuo, Saga, Usui, Sasakura, Kawasaki and Takaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naoki Takaya, dGFrYXlhLm5hb2tpLmdlQHUudHN1a3ViYS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.