- 1School of Basic Medicine, Xinxiang Medical University, Xinxiang, China
- 2International Joint Research Laboratory for Recombinant Pharmaceutical Protein Expression System of Henan, Xinxiang Medical University, Xinxiang, China
Chinese hamster ovary (CHO) cells are currently the most widely used host cells for recombinant therapeutic protein (RTP) production. Currently, the RTP yields need to increase further to meet the market needs and reduce costs. In this study, three stabilizing and anti-repressor (SAR) elements from the human genome were selected, including human SAR7, SAR40, and SAR44 elements. SAR elements were cloned upstream of the promoter in the eukaryotic vector, followed by transfection into CHO cells, and were screened under G418 pressure. Flow cytometry was used to detect enhanced green fluorescent protein (eGFP) expression levels. The gene copy numbers and mRNA expression levels were determined through quantitative real-time PCR. Furthermore, the effect of the stronger SAR elements on adalimumab was investigated. The results showed that transgene expression levels in the SAR-containing vectors were higher than that of the control vector, and SAR7 and SAR40 significantly increased and maintained the long-term expression of the transgene in CHO cells. In addition, the transgene expression level increase was related with gene copy numbers and mRNA expression levels. Collectively, SAR elements can enhance the transgene expression and maintain the long-term expression of a transgene in transfected CHO cells, which may be used to increase recombinant protein production in CHO cells.
Introduction
Recombinant therapeutic proteins (RTPs, antibodies) are an important part of the biotechnological drugs produced (Overton, 2014; Rodgers and Chou, 2016). The mammalian cell line is the most used biopharmaceutical production platform, which is due to the ability to produce proteins with post-translation modifications that are similar to human natural proteins (Zhu, 2012; Dalton and Barton, 2014; Portolano et al., 2014). Chinese hamster ovary (CHO) cells are the most used mammalian expression system, which has become the most important expression system for the production of RTPs and vaccines (Betts and Dickson, 2017; Kuo et al., 2018; Valente et al., 2018). The establishment of transfected cell lines with high-level expression of the gene of interest (GOI) is often an arduous process due to the heterogeneity of transgene expression. To obtain stable and highly transfected GOI cell lines, cell line engineering, growth medium and production process modification, and expression vector design were performed (Jia et al., 2018; Guo et al., 2020; Wang et al., 2022). Among these strategies, the expression vector design is an effective method to increase GOI expression level and stability.
In previous studies, some epigenetic regulatory elements were found to improve the GOI yield and stability of clones in CHO cells. The most common epigenetic elements are the locus control regions, matrix attachment regions (MARs), insulators, ubiquitously acting chromatin opening elements (UCOEs), and stabilizing and anti-repressor elements (SARs) (Buceta et al., 2011; Pfaff et al., 2013; Harraghy et al., 2015; Guo et al., 2020). These elements can be cloned into different sites of the expression vector, such as the flank of the expression box, upstream of the promoter, or downstream of poly A, which can improve the transcription efficiency and stability, to significantly reduce the time and cost required to produce many recombinant proteins. The MAR, UCOE, promoter, intron, insulator, selective marker, and poly A can be used to increase GOI expression levels in CHO cells (Mariati Yeo et al., 2014; Chen et al., 2017; Zhao et al., 2017; Tian et al., 2018; Xu et al., 2018; Li et al., 2019; Wang et al., 2022). MAR can increase the expression level in CHO cells and reduce the variation of transgene expression among transformants (Wang et al., 2008; Wang et al., 2010; Wang et al., 2012; Li et al., 2013; Zhao et al., 2017; Li et al., 2018; Tian et al., 2018; Li et al., 2019).
The ability of epigenetic elements to protect transgenes from epigenetic changes caused by changes in the cell culture environment is an important feature of these elements because it contributes to the long-term, stability, and high-level expression of proteins. Inserting epigenetic regulatory elements into expression vectors can improve the production level of recombinant proteins and the stability of mammalian cell lines.
SARs are regulatory elements that can block the repressor proteins and are highly conserved between the human and mouse, either at the functional or sequence level. Kwaks et al. (2003) screened SAR elements from the human genome library and flanked them in transgene genes and transfected them into mammal cells. They found that SARs can achieve more efficient GOI production (Kwaks et al., 2003). Although several SARs have been shown to increase transgene expression and expression stability in CHO cells (Otte et al., 2007; van Blokland et al., 2007), there are no systematic studies about the effect of SARs on transgene expression level and stability and the mechanism of function is unclear. In this study, three SAR elements were cloned upstream of the promoter of the expression vector, and their effect and mechanism on the transgene on CHO cells were investigated.
Materials and Methods
Vector Construction
pIRES-neo vector (Clontech, Mountain View, CA, United States), a bicistronic vector driven by the CMV promoter, was used for the backbone vector. The enhanced green fluorescent protein (eGFP) from pEGFP-C1 (Clontech) was subcloned into pIRES-neo to generate the pIRES-EGFP vector. The human stabilizing and anti-repressor element 7 (SAR7) (GenBank no: AY190751.1, position 1-2101), stabilizing and anti-repressor element 40 (SAR40) (GenBank no: AY190756.1, position 1-1031), and stabilizing and anti-repressor element 44 (SAR44) (GenBank no: AP000525.1, position 8205-9790) were artificially synthesized by General Biosystems (Chuzhou, China). All SARs elements were digested by NruI/BstBI enzymes, and sub-cloned into the upstream of the CMV promoter in the pIRES-EGFP vector according to the molecular standard operation, thereby producing pIRES-SAR7, pIRES-SAR40, and pIRES-SAR44 (Figure 1). The resultant vectors were verified through enzyme digestion and sequencing. To study the effect of SAR elements on the production of recombinant proteins, an expression vector containing the adalimumab heavy chain (HC) and light chain (LC) was constructed. To obtain the stably transfected mAb cell colonies, the LC and HC were coded on two separate plasmids, both carrying the different antibiotic resistance gene (Figure 1). Briefly, the eGFP gene was replaced with the LC and HC of adalimumab gene upstream of the promoters in pIRES-EGFP, pIRES-SAR7, and pIRES-SAR40 to construct the expression vectors pIRES-LC, pIRES-HC, pIRES-LC-SAR7, pIRES-HC-SAR7, pIRES-LC-SAR40, and pIRES-HC-SAR40.
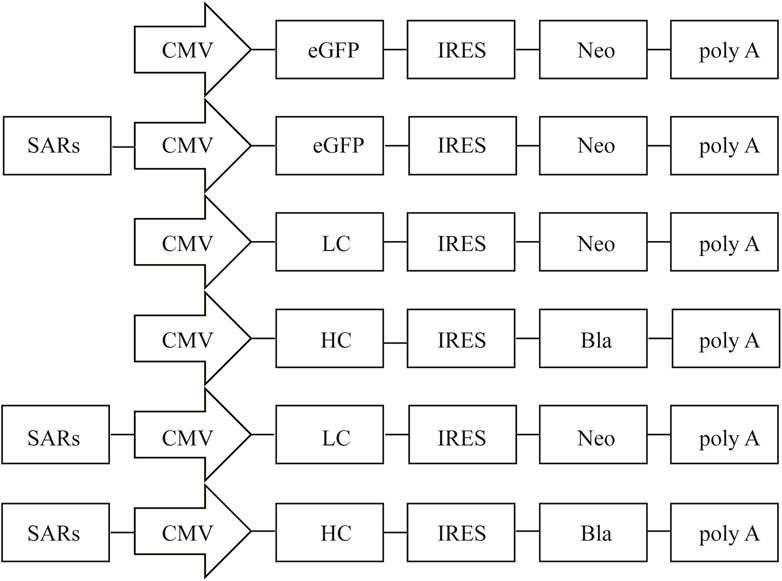
FIGURE 1. Representation of the vectors containing different SARs. Schematic illustration of the expression vector containing different SARs which were inserted upstream of CMV. CMV, human cytomegalovirus IE gene promoter; eGFP, enhanced green fluorescence protein; IRES, internal ribosome entry site; poly A, polyadenylation signal. Neo, neomycin; Bla, blasticidin.
Cell Culture and Transfection
CHO cells (#A11557-01; Life Technologies, Carlsbad, CA, United States) were cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient mixture F12 (Gibco, Carlsbad, CA, United States) with 10% fetal bovine serum (FBS; Gibco) at 37°C with 5% CO2. The cells were counted and inoculated in 24-well plates at a density of 4 × 105 cells/well. After reaching 80–90% confluence, the cells were transfected with pIRES-SAR7, pIRES-SAR40, or pIRES-SAR44; pIRES-EGFP was used as a control vector. Transfections were performed using the Lipofectamine® 2000 reagent (Invitrogen, Waltham, MA, United States) according to the manufacturer’s manual. All transfections were performed in triplicates.
Transient Expression
At 48-h post transfection, the transfection efficiency of each expression vector was observed under a fluorescence microscope (ECLIPSE Ti; Nikon, Tokyo, Japan), and mean fluorescence intensity (MFI) was detected by flow cytometry as previously described (Wang et al., 2019; Wang et al., 2022). Briefly, a total of 100,000 fluorescent events were obtained using a 530/15 bandpass filter for the green fluorescent signal, which was acquired with an emission wave length of 530 nm. Data acquisition and analyses were performed using Flow Jo software 7.6 (Tree Star, Ashland, OR).
Screening of Stably Transfected Cells
At 48-h after transfection, cells were cultured in the presence of 800 μg/ml G418 (Invitrogen) for approximately 14 days until the non-transfected control cells were eliminated. Next, transfected cells were seeded (4 × 105 cells/mL) in 6-well plates with 400 μg/ml G418 to produce the stably screened cells for determination of the fluorescence intensity. The eGFP expression was evaluated through flow cytometry using 1 × 106 CHO cells, and non-transfected cells were used as the negative control. Three stably transfected pools were generated for each vector. To analyze the expression long-term stability of the transgene, the stable cell lines were cultured in the absence of antibiotics selection pressure for 120 generations. At generations 30, 60, 90, and 120 after transfection, cells were collected and the eGFP expression level was analyzed through flow cytometry.
Quantitative Real-Time Polymerase Chain Reaction Analysis
Genomic DNA was extracted from 5 × 106 cells according to the manufacturer’s manual (Beyotime, Shanghai, China). The DNA concentration and purity were determined. Total RNA was extracted using the TRIzol reagent (Sigma, San Francisco, CA, United States) following the manufacturer’s protocols. The isolated RNA was treated with RNase-free DNaseI (Thermo Scientific, Waltham, MA, United States) to eliminate DNA contamination. cDNA synthesis was performed according to the manufacturer’s protocol (Cwbio, Jiangsu, China). Quantitative real-time PCR (qRT-PCR) was performed to determine the eGFP mRNA levels, and gene copy numbers using the SYBR Premix Ex Taq (Takara Bio, Beijing, China). The reaction mixture (10 μL) consisted of 5 μL SYBR Green (TAKARA, Dalian, China), 4 μL template DNA (0.05 μg/μL), 0.3 μL each of the forward and reverse primers (10 nmol/ml), and 0.4 μL deionized water. The glyceraldehyde phosphate dehydrogenase (GAPDH) gene was used as an endogenous control. The eGFP and GAPDH primers were designed (Table 1) and qRT-PCR was performed using an ABI 7500 Fast real-time PCR instrument (Applied Biosystems, Foster City, CA, United States) and the results were analyzed by 7500 Fast System SDS Software. The cycling parameters were as follows: 95°C for 3 min, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. The relative eGFP expression and eGFP copy number were determined with the 2-∆∆Ct method (Livak and Schmittgen, 2001). The experiment was repeated three times.
Expression of Adalimumab
The adalimumab LC and HC constructs were co-transfected at a 2:1 ratio, and a 4 µL Lipofectamine 2000 transfection reagent per 2 µg plasmid DNA was transfected into each well. After co-transfection, cells were selected in the presence of G418 as described above and Blasticidin (15 μg/ml) to establish stable CHO cell colonies. Then, the transfected CHO cells were suspension-cultured in a serum-free medium (ProteinEasy Biological Co., Ltd., Xinxiang, China) using a working volume of 30 ml in 125-ml Corning shake flasks at 37°C with 5% CO2. After 7 days, supernatants were collected and the expression level of adalimumab was analyzed through Western blot.
The cell supernatant transfected with the expression vector was collected and adalimumab expression was determined through western blot. The cell supernatant containing adalimumab and 5× buffer were mixed and boiled at 100°C for 1 min. First, the resolved proteins were transferred onto a PVDF membrane at 25 V for 90 min. The membrane was blocked with non-fat dry milk (NFDM) in TBS with 0.05% Tween-80 (TBS-T) for 1 h, and incubated with the secondary antibody at 1:8000 dilution (dilution buffer, 1% NFDM in TBS-T) (Catalogue no: E031610-01, Goat Anti-Mouse IgG, EarthOx, San Francisco, CA, United States) for 1 h, and washed three times with TBST. The membrane was soaked in ECL chemiluminescent solution and developed under a gel imaging system. At the same time, adalimumab expression was quantified with ELISA according to the provided protocol (ELK Biotechnology Co., Ltd., Wuhan, China).
Statistical Analysis
Data were analyzed with the SPSS 18.0 software (SPSS Inc., Chicago, IL, United States) using independent sample t-tests and reported as the mean ± standard deviation. p < 0.05 was considered statistically significant.
Results
Transfection Efficiency and Transient Expression
We first evaluated the effect of different SARs on the transfection efficiency using the eGFP reporter gene. The transfection efficiency and transient eGFP expression levels in CHO cells were observed through a fluorescence microscope 48 h after transfection. Our results showed that the eGFP expression in the vectors containing SAR7 and SAR40 were significantly higher than that of the control vector, while the eGFP expression mediated by SAR44 was weaker than that of the control vector (Figure 2A). The transfection efficiencies of SAR7, SAR40, and SAR44 were approximately 95, 92, and 70%, respectively (p < 0.05; Figure 2B), which were consistent with the transgene expression level. Compared with the control, SAR7, SAR40 increased the eGFP transient expression 1.32 and 1.19-fold, but SAR44 did not (Figure 2C).
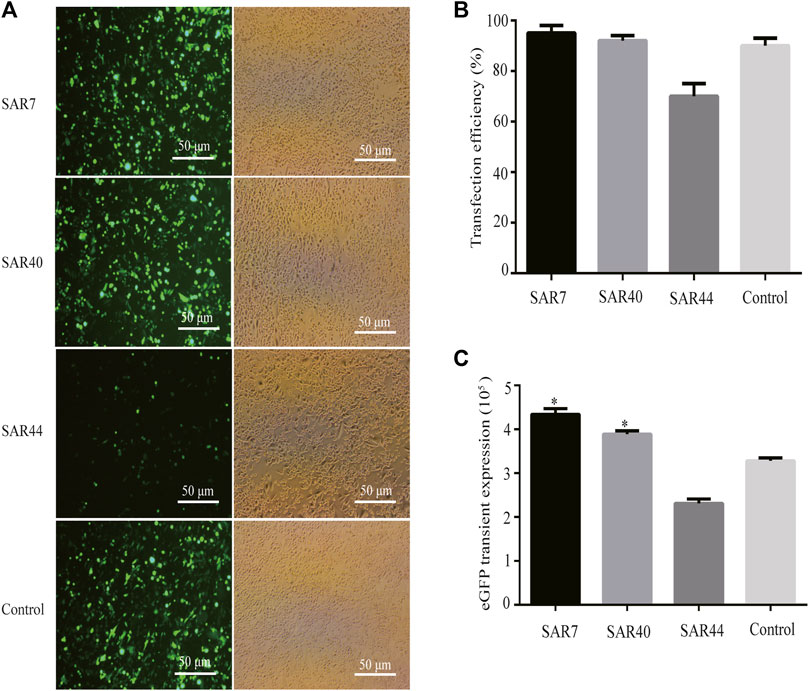
FIGURE 2. Transient expression of recombinant protein in transfected pools of different SARs. Three vectors were transfected into CHO cells, 48 h after transfection, and the transfection efficiencies were estimated. (A) The representative photos were observed by fluorescence microscopy. (B) Analysis of the transfection efficiency. (C) Analysis of the eGFP transient expression. The results are the mean values obtained for three independent experiments, and the standard deviation is indicated (*p < 0.05).
Transgene Expression Levels in Stably Transfected Cells
To establish stable transfectants, CHO cells were transfected with the respective vectors and subjected to G418 selection; the eGFP expression was further measured through flow cytometry (Figure 3A). Compared with the control vector, the mean fluorescence intensity of eGFP containing SAR7, SAR40, and SAR44 was 3.74-, 3.02- and 2.15-fold higher, respectively (Figure 3B), suggesting SARs can improve transgene expression levels in stably transfected cells.
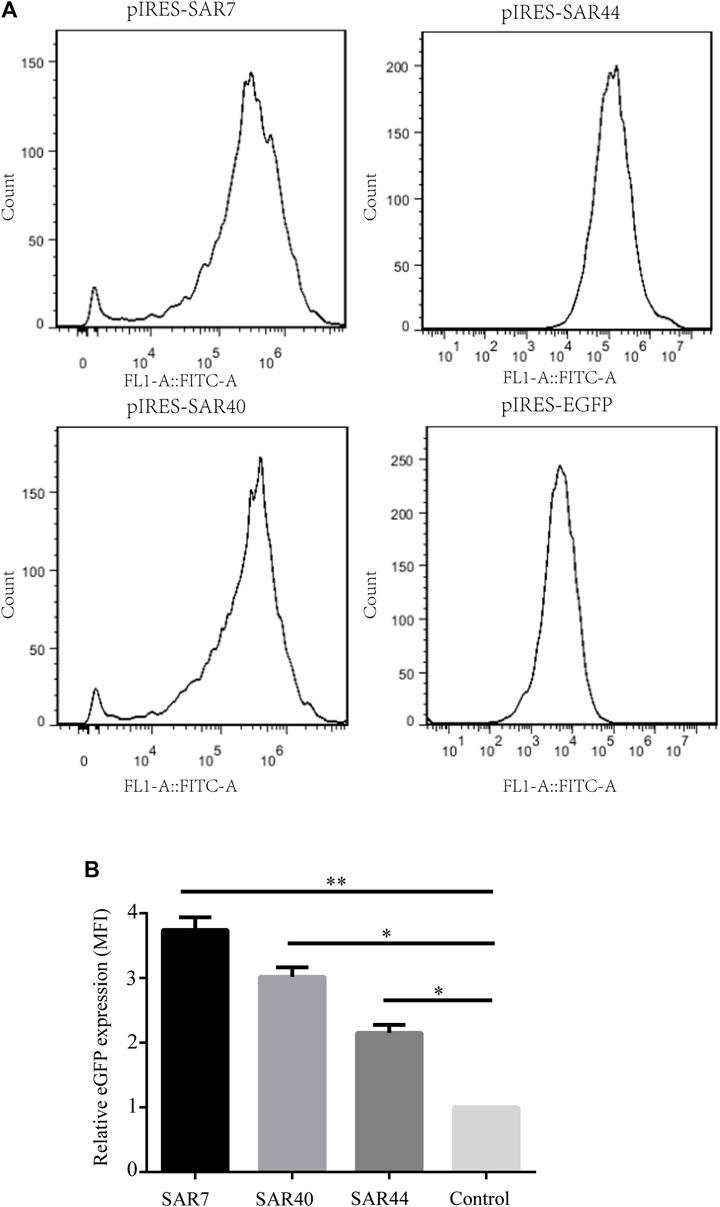
FIGURE 3. Analysis of MFI in stably transfected CHO cell line. (A) The MFI of eGFP of the different construct was measured by flow cytometry on day 20 under G418 selection. (B) Fold statistical analysis results of eGFP expression levels in the different constructs. The results are the mean values obtained for three independent experiments, and the standard deviation is indicated (*p < 0.05; **p < 0.05).
Subsequently, we performed qRT-PCR with cDNAs prepared from cells transfected with SARs and found that SARs increased the eGFP mRNA level compared with the control vector. SAR7, SAR40, and SAR44 increased the eGFP mRNA expression by approximately 8.5-, 4.8-, and 1.2-fold, respectively (Figure 4A).
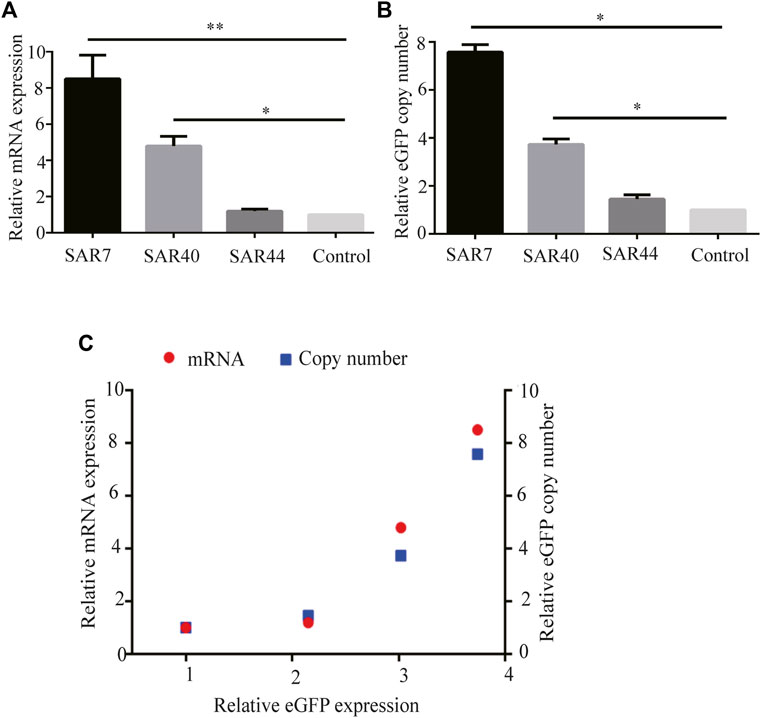
FIGURE 4. Analysis of the relative eGFP mRNA levels and eGFP copy number in stably transfected CHO cells. After screening under G418, the stable transfected CHO cell pools were acquired. (A) Total RNA was isolated and cDNA synthesis was performed, then the mRNA levels were detected using qRT-PCR. (B) Relative eGFP gene copy numbers were detected by qRT-PCR. (C) Correlation analysis between the relative eGFP copy (blue square), mRNA expression (red) and relative eGFP expression in long-term stable transfected CHO cells. Results were obtained from three independent experiments. The results are the mean values obtained for three independent experiments, and standard deviation is indicated (*p < 0.05; **p < 0.05).
Gene Copy Number Analysis
To investigate the relationship between the transgene expression level and gene copy number, we performed qRT-PCR using genomic DNA. The results showed that gene copy numbers in cells transfected with SAR7, SAR40, and SAR44 expression vectors were 7.58-, 3.73- and 1.45-fold higher than that of the control vector (Figure 4B), suggesting that the increase in eGFP expression levels was related to the increase in gene copy numbers (Figure 4C), which was consistent with previous studies (van Blokland et al., 2007).
Analysis of Long-Term Stability of Transgene Expression
As it is very important to maintain the stability of transgene expression in CHO cells for a long time, we investigated the long-term stability at generation 120 after transfection. The cells were collected and the eGFP expression was analyzed at generations 30, 60, 90, and 120 post-transfection. As shown in Figure 5, SAR40 showed the highest eGFP expression, followed by SAR7 (Figure 5A), and the transgenic expression maintenance rate of SAR40 was the highest followed by that of SAR7 (Figure 5B), suggesting that SAR can help to maintain the stability of transgene expression.
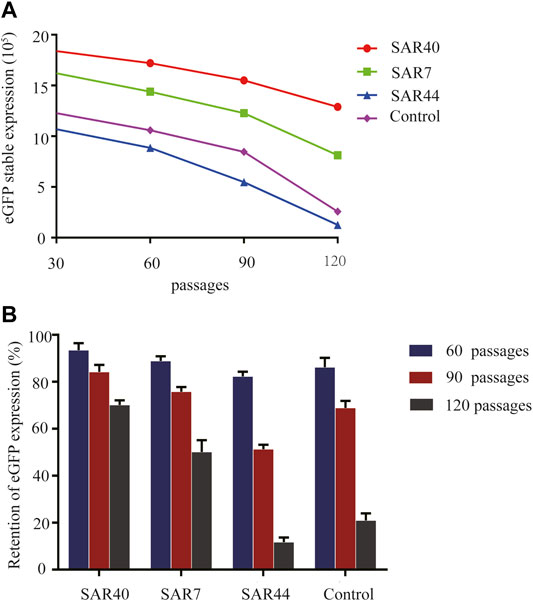
FIGURE 5. Analysis of long-term stability of the transgene expression in transfected CHO cells. Cells were collected, and FACSCalibur estimated the eGFP fluorescence profile at generation 30, 60,90, and 120 after transfection. (A) The eGFP expression (MFI) on different days after transfection. (B) The retention of eGFP expression of different constructs. The results are the mean values obtained for three independent experiments, and the standard deviation is indicated.
Analysis of Adalimumab Expression
We assessed the effect of SAR on secretory protein expression by analyzing the level of Adalimumab in stably transfected CHO cells cultured in serum-free suspension for 7 days. The cell supernatants were collected and the expression level of adalimumab was analyzed through Western blot. The results showed that adalimumab can be expressed in all vectors (Figure 6A). SAR40 and SAR7 can increase the Adalimumab expression levels 1.850 and 1.429 compared with the control in CHO cells (Figures 6B,C).
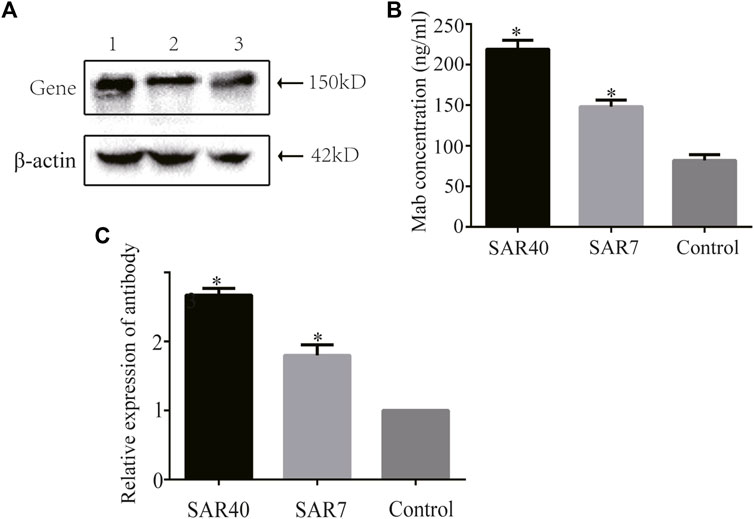
FIGURE 6. Analysis of adalimumab antibody expression of different constructs in CHO cells. The vectors containing adalimumab antibody gene were transformed into CHO cells, respectively. Cells supernatant was collected for analysis. (A) Adalimumab antibody expression was detected by Western blot. Lane 1, pIRES-mAb-SAR40; Lane 2, pIRES-mAb-SAR7; Lane 3, pIRES-mAb. (B) Expression of the adalimumab antibody was detected by ELISA. (C) Relative expression of adalimumab antibody. The results are the mean values obtained for three independent experiments, and the standard deviation is indicated (*p < 0.05).
Discussion
Recombinant therapeutic proteins have gained considerable attention in the biopharmaceutical industry. The large-scale industrial production of high-value therapeutic proteins is mainly achieved in mammalian cell lines, but this process is often hindered by low production and unstable expression (Kwaks and Otte, 2006; Jazayeri et al., 2018). Therefore, maintaining a high level of gene expression is a key factor affecting the production of recombinant proteins. The use of cis-epigenetic regulatory elements in the expression vector can result in more cell clones expressing proteins, as well as higher and more stable protein expression (Harraghy et al., 2015). In this study, we investigated the effect of SARs on transgenic expression and found that SARs can enhance the transgenic expression and maintain the long term expression of transgenes in transfected CHO cells, which may be used in the recombinant protein production in CHO cells.
SARs were discovered using a genomic screening strategy for elements that increase transgene expression, and are evolutionarily highly conserved at the sequence level, genome location, and function (Kwaks et al., 2003; Sautter and Enenkel, 2005). SAR is a blocker and not an enhancer of chromatin-related inhibitors. SAR can promote more cell clones expressing proteins, and confer a higher and more stable protein expression (Kwaks et al., 2003; van Blokland et al., 2007; Otte et al., 2007). Therefore, SAR has great significance in the application of recombinant protein production. In the present study, SARs were cloned upstream of the CMV promoter in the bicistronic expression vector, and the effects of three SARs on transgene expression were evaluated. eGFP is often used as a reporter gene to explore the regulatory effect of various cis-acting elements on gene expression (Seo et al., 2010; Wang et al., 2018); therefore, we first compared the transfection efficiency of different SARs using eGFP. We found that SAR7 and SAR40 showed the highest transfection efficiency, and their transient expression was stronger than that of SAR44 and the control vector, suggesting that the transfection efficiency is affected by SARs with different sequences. Moreover, the eGFP expression in the vectors containing SARs was indeed higher than that without SAR vectors, among which the eGFP expression for SAR7 and SAR40 was stronger than that for SAR44. Figure 2A is the representative photos observed by fluorescence microscopy and bright field image. The transfection efficiency of SAR44 is lower than that of the control, which is related to the vector structure. However, SAR44 has higher MFI than the control under the stably transfected state (Figure 3A), indicating that SAR44 can increase stably transgene in CHO cells. The increase in eGFP expression level correlated well with mRNA levels for the SAR-containing vectors and their gene copy numbers in a copy number-dependent manner, which was consistent with previous studies (Kwaks et al., 2003; Otte et al., 2007). However, SAR40-induced adalimumab expression was higher than that of SAR7. The effects of SAR on eGFP and mAb SAR are inconsistent, which may be due to eGFP is in one plasmid, but the mAb is coded by two plasmids. It is very important that the correct association of LC and HC during mAb production. The unbalanced proportion of LC to HC can cause low yields and aggregation. Another reason may be related to the secretory characteristics of the adalimumab antibody, and the exact mechanism needs further study. We hypothesize that the incorporation of SAR7 or SAR40 in the vectors may result in more colonies that produce higher gene expression levels. Moreover, SAR can interfere with the deacetylation or methylation of the surrounding chromatin, thus blocking the inhibitory effect of chromatin on the expression of exogenous genes at the molecular level (Kwaks et al., 2003).
During cell culture, the cell growth conditions can impact transgene expression. Cells change their gene expression mode according to different conditions such as nutrient consumption and shaking table speed (Sieck et al., 2014). Environmental stress has also been shown to cause epigenetic changes (Gim et al., 2015; Seo et al., 2015). Therefore, cell culture conditions may affect the epigenetic changes of cells, thus affecting the production of recombinant proteins.
In conclusion, in this study we explored the effects of SARs on transgene expression in CHO cells and found that SARs increase the transgenic expression in stably transfected CHO cells. SAR7 had the most remarkable effect, followed by SAR40. This effect may be caused by the increased gene copy numbers and their mRNA expression, but other factors need to be further explored. However, this study only compared three SARs in CHO cell line using eGFP and adalimumab. More SARs and transgenes should be used to investigate the effect of SAR elements on recombinant protein expression, for a successful industrial application.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, AY190751.1, AY190756.1.
Author Contributions
T-YW: designed research and manuscript revision. QL: original draft writing and manuscript revision. QL, R-FY, and Y-XY: conducted experiments. C-LM: contributed new reagents or analytical tools. Y-LJ: analyzed data. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No.32071468) and the National Natural Science Foundation of Henan Province, China (No. 212102310318).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Betts, Z., and Dickson, A. J. (2017). Improved CHO Cell Line Stability and Recombinant Protein Expression during Long-Term Culture. Methods Mol. Biol. 1603, 119–141. doi:10.1007/978-1-4939-6972-2_8
Buceta, M., Galbete, J. L., Kostic, C., Arsenijevic, Y., and Mermod, N. (2011). Use of Human MAR Elements to Improve Retroviral Vector Production. Gene Ther. 18, 7–13. doi:10.1038/gt.2010.115
Chen, S.-j., Wang, W., Zhang, F.-y., Jia, Y.-l., Wang, X.-y., Guo, X., et al. (2017). A Chimeric HS4 Insulator-Scaffold Attachment Region Enhances Transgene Expression in Transfected Chinese Hamster Ovary Cells. FEBS Open Bio 7, 2021–2030. doi:10.1002/2211-5463.12335
Dalton, A. C., and Barton, W. A. (2014). Over‐expression of Secreted Proteins from Mammalian Cell Lines. Protein Sci. 23, 517–525. doi:10.1002/pro.2439
Gim, J.-A., Hong, C. P., Kim, D.-S., Moon, J.-W., Choi, Y., Eo, J., et al. (2015). Genome-wide Analysis of DNA Methylation Before-And after Exercise in the Thoroughbred Horse with MeDIP-Seq. Mol. Cells 38, 210–220. doi:10.14348/molcells.2015.2138
Guo, X., Wang, C., and Wang, T.-Y. (2020). Chromatin-modifying Elements for Recombinant Protein Production in Mammalian Cell Systems. Crit. Rev. Biotechnol. 40 (7), 1035–1043. doi:10.1080/07388551.2020.1805401
Harraghy, N., Calabrese, D., Fisch, I., Girod, P.-A., LeFourn, V., Regamey, A., et al. (2015). Epigenetic Regulatory Elements: Recent Advances in Understanding Their Mode of Action and Use for Recombinant Protein Production in Mammalian Cells. Biotechnol. J. 10, 967–978. doi:10.1002/biot.201400649
Jazayeri, S. H., Amiri-Yekta, A., Bahrami, S., Gourabi, H., Sanati, M. H., and Khorramizadeh, M. R. (2018). Vector and Cell Line Engineering Technologies toward Recombinant Protein Expression in Mammalian Cell Lines. Appl. Biochem. Biotechnol. 185, 986–1003. doi:10.1007/s12010-017-2689-8
Jia, Y. L., Guo, X., Lu, J. T., Wang, X. Y., Qiu, L. L., and Wang, T. Y. (2018). CRISPR/Cas9-mediated Gene Knockout for DNA Methyltransferase Dnmt3a in CHO Cells Displays Enhanced Transgenic Expression and Long-Term Stability. J. Cel. Mol. Med. 22, 4106–4116. doi:10.1111/jcmm.13687
Kuo, C.-C., Chiang, A. W., Shamie, I., Samoudi, M., Gutierrez, J. M., and Lewis, N. E. (2018). The Emerging Role of Systems Biology for Engineering Protein Production in CHO Cells. Curr. Opin. Biotechnol. 51, 64–69. doi:10.1016/j.copbio.2017.11.015
Kwaks, T. H. J., Barnett, P., Hemrika, W., Siersma, T., Sewalt, R. G. A. B., Satijn, D. P. E., et al. (2003). Identification of Anti-repressor Elements that Confer High and Stable Protein Production in Mammalian Cells. Nat. Biotechnol. 21, 553–558. doi:10.1038/nbt814
Kwaks, T. H. J., and Otte, A. P. (2006). Employing Epigenetics to Augment the Expression of Therapeutic Proteins in Mammalian Cells. Trends Biotechnol. 24, 137–142. doi:10.1016/j.tibtech.2006.01.007
Li, Q., Wang, X. Y., Zhao, C. P., Tian, Z. W., Xu, D. H., Wang, T. Y., et al. (2018). Effects of Different Promoters and MAR Combinations on Transgene Expression of Recombinant CHO Cells. Sichuan Da Xue Xue Bao Yi Xue Ban. 49, 18–23. (In Chinese). doi:10.13464/j.scuxbyxb.2018.01.004
Li, Q., Dong, W., Wang, T., Liu, Z., Wang, F., Wang, X., et al. (2013). Effect of β-globin MAR Characteristic Elements on Transgene Expression. Mol. Med. Rep. 7, 1871–1874. doi:10.3892/mmr.2013.1424
Li, Q., Zhao, C.-p., Lin, Y., Song, C., Wang, F., and Wang, T.-y. (2019). Two Human MARs Effectively Increase Transgene Expression in Transfected CHO Cells. J. Cell. Mol. Med. 23, 1613–1616. doi:10.1111/jcmm.14018
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Mariati Yeo, J. H. M., Koh, E. Y. C., Ho, S. C. L., and Yang, Y. (2014). Insertion of Core CpG Island Element into Human CMV Promoter for Enhancing Recombinant Protein Expression Stability in CHO Cells. Biotechnol. Prog. 30, 523–534. doi:10.1002/btpr.1919
Otte, A. P., Kwaks, T. H. J., van Blokland, R. J. M., Sewalt, R. G. A. B., Verhees, J., Klaren, V. N. A., et al. (2007). Various Expression-Augmenting DNA Elements Benefit from STAR-Select, a Novel High Stringency Selection System for Protein Expression. Biotechnol. Prog. 23, 801–807. doi:10.1021/bp070107r10.1002/bp070107r
Overton, T. W. (2014). Recombinant Protein Production in Bacterial Hosts. Drug Discov. Today 19, 590–601. doi:10.1016/j.drudis.2013.11.008
Pfaff, N., Lachmann, N., Ackermann, M., Kohlscheen, S., Brendel, C., Maetzig, T., et al. (2013). A Ubiquitous Chromatin Opening Element Prevents Transgene Silencing in Pluripotent Stem Cells and Their Differentiated Progeny. Stem Cells 31, 488–499. doi:10.1002/stem.1316
Portolano, N., Watson, P. J., Fairall, L., Millard, C. J., Milano, C. P., Song, Y., et al. (2014). Recombinant Protein Expression for Structural Biology in HEK 293F Suspension Cells: a Novel and Accessible Approach. JoVE 16, e51897. doi:10.3791/51897
Rodgers, K. R., and Chou, R. C. (2016). Therapeutic Monoclonal Antibodies and Derivatives: Historical Perspectives and Future Directions. Biotechnol. Adv. 34, 1149–1158. doi:10.1016/j.biotechadv.2016.07.004
Sautter, K., and Enenkel, B. (2005). Selection of High-Producing CHO Cells Using NPT Selection Marker with Reduced Enzyme Activity. Biotechnol. Bioeng. 89, 530–538. doi:10.1002/bit.20374
Seo, H. W., Kim, T. M., Choi, J. W., Han, B. K., Song, G., and Han, J. Y. (2010). Evaluation of Combinatorial Cis-Regulatory Elements for Stable Gene Expression in Chicken Cells. BMC Biotechnol. 10, 69. doi:10.1186/1472-6750-10-69
Seo, J.-Y., Park, Y.-J., Yi, Y.-A., Hwang, J.-Y., Lee, I.-B., Cho, B.-H., et al. (2015). Epigenetics: General Characteristics and Implications for Oral Health. Restor. Dent. Endod. 40, 14–22. doi:10.5395/rde.2015.40.1.14
Sieck, J. B., Budach, W. E., Suemeghy, Z., Leist, C., Villiger, T. K., Morbidelli, M., et al. (2014). Adaptation for Survival: Phenotype and Transcriptome Response of CHO Cells to Elevated Stress Induced by Agitation and Sparging. J. Biotechnol. 189, 94–103. doi:10.1016/j.jbiotec.2014.08.042
Tian, Z.-W., Xu, D.-H., Wang, T.-Y., Wang, X.-Y., Xu, H.-Y., Zhao, C.-P., et al. (2018). Identification of a Potent MAR Element from the Human Genome and Assessment of its Activity in Stably Transfected CHO Cells. J. Cell. Mol. Med. 22, 1095–1102. doi:10.1111/jcmm.13361
Valente, K. N., Levy, N. E., Lee, K. H., and Lenhoff, A. M. (2018). Applications of Proteomic Methods for CHO Host Cell Protein Characterization in Biopharmaceutical Manufacturing. Curr. Opin. Biotechnol. 53, 144–150. doi:10.1016/j.copbio.2018.01.004
Vanblokland, H., Kwaks, T., Sewalt, R., Verhees, J., Klaren, V., Siersma, T., et al. (2007). A Novel, High Stringency Selection System Allows Screening of Few Clones for High Protein Expression. J. Biotechnol. 128, 237–245. doi:10.1016/j.jbiotec.2006.09.023
Wang, F., Wang, T.-Y., Tang, Y.-Y., Zhang, J.-H., and Yang, X.-J. (2012). Different Matrix Attachment Regions Flanking a Transgene Effectively Enhance Gene Expression in Stably Transfected Chinese Hamster Ovary Cells. Gene 500, 59–62. doi:10.1016/j.gene.2012.03.049
Wang, T. Y., Yang, R., Qin, C., Wang, L., and Yang, X. (2008). Enhanced Expression of Transgene in CHO Cells Using Matrix Attachment Region. Cell Biol. Int. 32, 1279–1283. doi:10.1016/j.cellbi.2008.07.014
Wang, T. Y., Zhang, J. H., Jing, C. Q., Yang, X. J., and Lin, J. T. (2010). Positional Effects of the Matrix Attachment Region on Transgene Expression in Stably Transfected CHO Cells. Cell. Biol. Int. 34, 141–145. doi:10.1042/CBI20090017
Wang, W., Guo, X., Li, Y. M., Wang, X. Y., Yang, X. J., Wang, Y. F., et al. (2018). Enhanced Transgene Expression Using Cis-Acting Elements Combined with the EF1 Promoter in a Mammalian Expression System. Eur. J. Pharm. Sci. 123, 539–545. doi:10.1016/j.ejps.2018.08.016
Wang, X. Y., Du, Q,J., Zhang, W. L., Xu, D. H., Zhang, X., Jia, Y. L., et al. (2022). Enhanced Transgene Expression by Optimization of Poly A in Transfected CHO Cells. Front. Bioeng. Biotech. 10, 722722. doi:10.3389/fbioe.2022.722722
Wang, X. Y., Yi, D. D., Wang, T. Y., Wu, Y. F., Chai, Y. R., Xu, D. H., et al. (2019). Enhancing Expression Level and Stability of Transgene Mediated by Episomal Vector via Buffering DNA Methyltransferase in Transfected CHO Cells. J. Cell Biochem. 120, 15661–15670. doi:10.1002/jcb.28835
Xu, D. h., Wang, X. y., Jia, Y. l., Wang, T. y., Tian, Z. w., Feng, X., et al. (2018). SV40 Intron, a Potent Strong Intron Element that Effectively Increases Transgene Expression in Transfected Chinese Hamster Ovary Cells. J. Cell. Mol. Med. 22, 2231–2239. doi:10.1111/jcmm.13504
Zhao, C.-P., Guo, X., Chen, S.-J., Li, C.-Z., Yang, Y., Zhang, J.-H., et al. (2017). Matrix Attachment Region Combinations Increase Transgene Expression in Transfected Chinese Hamster Ovary Cells. Sci. Rep. 7, 42805. doi:10.1038/srep42805
Keywords: Chinese hamster ovary cell, stabilizing and anti-repressor elements, transgene expression, epigenetic regulatory elements, green fluorescent protein
Citation: Li Q, Yan R-F, Yang Y-X, Mi C-l, Jia Y-l and Wang T-Y (2022) Stabilizing and Anti-Repressor Elements Effectively Increases Transgene Expression in Transfected CHO Cells. Front. Bioeng. Biotechnol. 10:840600. doi: 10.3389/fbioe.2022.840600
Received: 22 December 2021; Accepted: 21 April 2022;
Published: 26 May 2022.
Edited by:
Elizabeth R. Balmayor, University Hospital RWTH Aachen, GermanyReviewed by:
Mario Andrea Marchisio, Tianjin University, ChinaMartina Anton, Klinikum rechts der Isar der TUM, Germany
Copyright © 2022 Li, Yan, Yang, Mi, Jia and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian-Yun Wang, d3RpYW55dW5jbkAxMjYuY29t
 Qin Li
Qin Li Rui-Fang Yan1
Rui-Fang Yan1 Tian-Yun Wang
Tian-Yun Wang