- 1College of Ocean Food and Biology Engineering, Jimei University, Xiame, China
- 2Fujian Provincial Key Laboratory of Food Microbiology and Enzyme Engineering Technology, Xiamen, China
- 3Research Center of Food Biotechnology of Xiamen City, Xiamen, China
- 4Key Laboratory for Ecology of Tropical Islands, College of Life Sciences, Ministry of Education, Hainan Normal University, Haikou, China
The bacterial strain of Microbulbifer sp. ALW1 has demonstrated visible ability of degrading the cell wall of Laminaria japonica, and biochemical characterization has been performed on some individual enzymes to elucidate its genetic basis. However, it still remains elusive how strain ALW1 successfully breaks down the major cell wall component alginate polysaccharide and colonizes on its marine host. In this study, a mass spectrometry-based quantitative analysis of the extracellular and intracellular proteomes was introduced to elucidate the alginate degradation pathway in ALW1 strain. Mass spectrometry and biochemical assays indicated that strain ALW1 could effectively degrade alginate polysaccharide into disaccharides and trisaccharides within 12 h. Proteome analysis identified 156 and 1,047 proteins exclusively localized in extracellular and intracellular compartments, respectively, with 1,086 protein identities of dual localization. Functional annotation of the identified proteins suggested the involvement of diverse catalytic enzymes and non-catalytic molecules for the cleavage and metabolism of alginate polysaccharide. A simplified pathway was constructed to demonstrate the extracellular digestion, active transport, and intracellular conversion of alginate polysaccharide and its fragmented oligosaccharides, casting a picture of genetic loci controlling alginate catabolism by ALW1 strain. This study aims to provide a guide for utilization and genetic manipulation of the bacterial strain ALW1 for efficient alginate oligosaccharides production by fermentation.
Introduction
Seaweeds and their constituent compounds are utilized as food, fertilizer, industrial products, medical products, and cosmetics and for producing biofuel (Wargacki et al., 2012; Kawai and Murata, 2016; Jesumani et al., 2019; Ścieszka and Klewicka, 2019; Lopez-Santamarina et al., 2020). Alginates as natural polysaccharides are major structural components of the cell wall of seaweed (Li et al., 2021). The potential applications of alginate are restricted because of its large molecular weight, low water solubility, and high solution viscosity when significant concentrations are needed. In contrast, alginate oligosaccharides (AOS), oligomers containing 2 to 25 monomers, have better bioavailability because of their improved water solubility and low solution viscosity. AOS have shown immunomodulatory, antimicrobial, antioxidant, prebiotic, antihypertensive, antidiabetic, antitumor, anticoagulant, and other activities (Liu et al., 2019; Jiang et al., 2021). With the development of marine resources, AOS have potential applications in food, cosmetics, medicine, and other fields (Roohinejad et al., 2017).
Seaweed polysaccharides can be processed into oligosaccharides through different approaches (Liu et al., 2019), including physical and chemical methods, enzymatic method (Xu et al., 2018), and fermentation (Li et al., 2017). The chemical and physical methods normally have relatively low yield of oligosaccharides, violent reactions, and certain pollutions. The enzymatic method is difficult to manipulate in industry as a result of the poor thermal tolerance and sensitivity to mechanical disturbance. Alginate depolymerization with specific microorganisms serves as an effective and indispensable method for AOS production. Alginate-degrading bacteria have been isolated and utilized for AOS production such as Flavobacterium sp. LXA (An et al., 2008), Gracilibacillus A7 (Tang et al., 2009), Pseudoalteromonas agarovorans CHO-12 (Choi et al., 2009), Bacillus subtilis KCTC 11782BP (Bang et al., 2015), and Bacillus litoralis M3 (Wang et al., 2016).
Proteomics is emerging as a potent tool to dissect the complex biological processes, providing direct reflection of the gene expression profiles of the organism under defined environmental conditions. Paradendryphiella salina has been reported to be a microorganism capable of parasitizing and degrading algal cell walls. The enzymes of polysaccharide lyase family 7 (PL7) and 8 (PL8) were identified by proteomic analysis in P. salina, functioning as the forefront weapon to invade and degrade alginate in cell wall to facilitate its colonization on marine host (Pilgaard et al., 2019). The two-dimensional gel electrophoresis assay uncovered the proteomic dynamics of rumen bacteria Butyrivibrio proteoclasticus in response to polysaccharide carbon supplies, highlighting the implication of polysaccharidases, carbohydrate-binding proteins (CBPs), ATP-binding cassette (ABC) transporter, and substrate-binding proteins (SBPs) in polysaccharide degradation and oligosaccharide uptake (Dunne et al., 2012). Proteomics has been applied to investigate the mechanism of polysaccharide degradation by microbes (Sichert et al., 2020; de Figueiredo et al., 2021; Jensen et al., 2021).
A marine strain designated Microbulbifer sp. ALW1 has been isolated from rotten Laminaria japonica, and some alginate lyases have been characterized in our previous studies (Zhu et al., 2016; Jiang et al., 2019). To have a comprehensive understanding of the biological processes of strain ALW1 breaking down alginate, the quantitative proteomics using isobaric tags for relative and absolute quantitation of the label-free strategy was employed to identify the key proteins in polysaccharide metabolism extracellularly and intracellularly. Through revealing the metabolism process of the alginate by Microbulbifer sp. ALW1, the informative clues obtained could facilitate the use of brown algae or its waste to produce functional materials such as AOS using microbial method, and a genetic source of polysaccharide-degrading enzymes.
Materials and Methods
Cell Culture
Microbulbifer sp. ALW1 from our laboratory stock was refreshed in 50 ml of medium A [0.5% (w/v) kelp, 3.0% (w/v) NaCl, 0.2% (w/v) K2HPO4, 0.1% (w/v) MgSO4•7H2O, 0.001% (w/v) FeSO4•7H2O] with agitation (180 rpm) at 25°C for 24 h. After the strain revival, 1 ml of the suspension was subcultured into 50 ml of medium B [0.5% (w/v) (NH4)2SO4, 3.0% (w/v) NaCl, 0.2% (w/v) K2HPO4, 0.1% (w/v) MgSO4•7H2O, 0.001% (w/v) FeSO4•7H2O, pH 7.5] plus 0.5% (w/v) peptone with agitation (180 rpm) at 25°C for 12 h, with the OD600 value of about 1.2. Subsequently, the suspension was further subcultured with 50× dilution in 1,000 ml of medium B with the addition of 0.2% (w/v) sodium alginate instead of peptone at 25°C. The cell growth was monitored by measuring the OD600 value. The cell metabolite reducing sugar was determined by 3, 5-dinitrosalicylic acid (DNS) (Miller, 1959). The amount of reducing sugar produced was detected by measuring the absorbance value at 540 nm. Phenol-sulfuric acid assay for total carbohydrate determination to quantify total sugars in cellular metabolites (Montgomery, 1961). The identified protein was expressed in Escherichia coli BL21 (DE3) according to our previous study (Zhu et al., 2020).
HPLC-MS Analysis
Preparation of the hydrolysates from alginate lyase-treated sodium alginate was carried out according to the method described previously (Jiang et al., 2019). The samples of alginate metabolites and enzymatic hydrolysates were determined using an ultrahigh-performance liquid chromatography (Ultimate 3000, ThermoFisher Scientific, Rockford, United States) with hypersil Gold C18 column (ThermoFisher Scientific, Rockford, United States) and matrix-assisted laser desorption synaptquan ionization-time of flight mass spectrometer MS/MS (TSQ TQU04549, Thermo Fisher Scientific, Rockford, United States). The samples were determined from 150 to 800 m/z by mass spectrometry (MS) under positive ion mode.
Total Protein Extraction
The cells grown for 30 h in medium B containing alginate were pelleted by centrifugation at 12000 rpm for 10 min at 4°C. The supernatant was collected and applied on a Millipore centrifugal filter 3K device (3,000 nominal molecular weight limit, Millipore, United States). The concentrated supernatant was obtained and used as the extracellular sample. The cell pellet was milled into fine powder in liquid nitrogen to obtain the intracellular sample. Then the extracellular and intracellular proteins were extracted following the modified phenol protocol as described, and air-dried pellets are dissolved in lysis buffer (7 M urea, 2 M thiourea, 2% CHAPS, 13 mM DTT) (Wang et al., 2012). Protein concentration was determined by the Bradford assay with bovine serum albumin as the standard (Bradford, 1976). The total protein extracts were stored at −20°C after lyophilization for later analysis.
HPLC-ESI-MS/MS for Proteomics
Extracellular and intracellular proteins (300 μg each) were used for proteomic analysis, respectively. The lyophilized proteins were treated with dithiothreitol and iodoacetamide for reduction and alkylation, respectively, and then digested by trypsin following the standard procedures. Mass spectra of the peptides were acquired on an AB 5800 MALDI-TOF/TOF mass spectrometry (MS) instrument (AB SCIEX, Foster City, United States) as described (Yi et al., 2014).
Protein Identification
Protein identification was performed with Protein Pilot™ v4.5 software (AB SCIEX, United States) against the Malus × domestica database (http://www.rosaceae.org) using the Paragon Algorithm and searching against the genome database of Microbulbifer sp. ALW1 (Shilov et al., 2007). Up to two missed cleavages were permitted for fully tryptic peptides. Carboxyamidomethyl cysteine and oxidized methionine were set as fixed and variable modifications, respectively. The false discovery rate (FDR) was determined by using a target-decoy search strategy (Elias and Gygi, 2007). Mass tolerance for peptide and fragment ion was 40 ppm. The acceptance of peptide assignment and protein identification was the unused ProtScore greater than 1.3 with at least two matched peptides. FDR assessment was 0.01. Three biological replicates of proteins from extracellular and intracellular samples of strain ALW1 cells cultured with alginate as the carbon source were normalized to the intensity of many qualitatively matched proteins (or peptides). All protein hits were identified with a confidence of 95%. A Venn diagram was used to identify the proteins present in all three biological replicates.
Bioinformatics Analyses
Functional annotations of the proteins identified through label-free quantitation analysis were performed. Clusters of orthologous groups (COG) analysis of the proteins was performed for functional classification via searches in a database (http://eggnogdb.embl.de/). Gene ontology (GO) analysis was performed using the Blast2GO program (https://www.blast2go.com/) and further performed online using WEGO software (http://wego.genomics.org.cn). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using the OmicShare tools, a free online platform for data analysis (http://www.omicshare.com/tools). Searches for the polysaccharide related metabolic enzymes, including glycoside hydrolases (GH), glycosyltransferases (GT), carbohydrate esterases (CE), polysaccharide lyases (PL), and auxiliary activities (AA) were performed manually on the basis of the BLASTP and the CAZy database (Lombard et al., 2014).
Following the annotation and annotation augmentation steps, the targeted proteins were blasted against KEGG GENES to retrieve the KEGG Orthology (KO) and subsequently mapped to pathways in KEGG. These metabolic pathways of alginate polysaccharide were deduced on the basis of such enzymes and the KEGG database.
Statistical Analysis
All experiments were conducted with three technical replicates and three biological replicates. The statistical significance (p values) in mean values was determined with unpaired two-tailed Student’s t-test (SPSS 13.0), with p < 0.05 as statistically significant.
Results and Discussion
Growth of Strain ALW1
The growth dynamics of strain ALW1 cells in the medium containing alginate as the sole carbon source was recorded during the time course. The biomass was relatively stable in the first 16 h, and the OD600 reached the plateau of 1.36 at 30 h (Figure 1A). The total sugar content showed a downward trend and reached the lowest at about 30 h (Figure 1B). The released reducing sugar accumulated to the highest at 12 h and declined to the lowest afterward at 24 h (Figure 1C). These observations indicated that strain ALW1 could effectively break down the alginate polysaccharide to oligosaccharides and then take advantage of the degradation products. In order to coalesce the stationary phase, the cell culture at the time point of 30 h was selected for proteomic analysis.
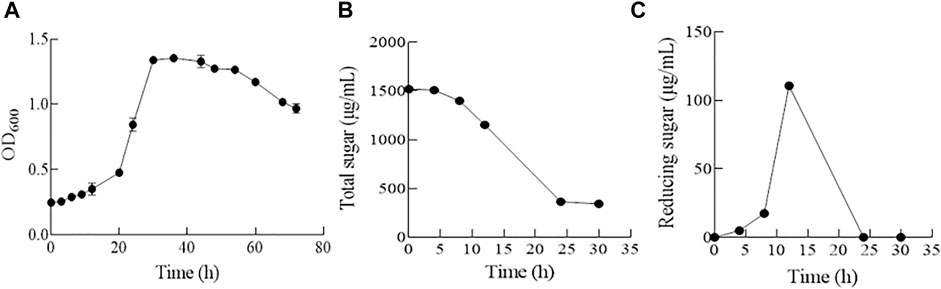
FIGURE 1. Biological growth of strain ALW1 cultured under alginate. (A) OD600, (B) total sugar, and (C) reducing sugar.
Analyses of Alginate Metabolites
To have a better knowledge of the alginate degradation patterns of strain ALW1, mass spectrometry was performed to analyze the alginate metabolites over the time course. Under the positive ion mode, the degrees of polymerization (DPs) with mass-to-charge ratios (m/z) of the products are presumed to be two at 374 m/z and three at 550 m/z. The initial alginate polysaccharide was not detectable in this assay due to its high molecular weight (Figure 2A). After 4 h of incubation, the metabolites of disaccharides and trisaccharides showed some accumulation (Figure 2B). According to their signal intensity, the contents of disaccharides and trisaccharides reached the maximum at 12 h with the passage of time (Figure 2B−D), and showed a significant decrease at the time points of 24 and 30 h (Figures 2E,F). The dynamics presented by mass spectra was in harmony with the observation of total reducing sugar quantification, suggesting the oligosaccharides were absorbed and further metabolized by strain ALW1 cells after liberation from alginate polysaccharide. The digestion patterns are consistent with those of the previous reports (Takeshita and Oda, 2016; Tang et al., 2020). Taken together, the aforementioned observations indicated that strain ALW1 had the ability to degrade the alginate polysaccharide to produce disaccharides and trisaccharides within 12 h.
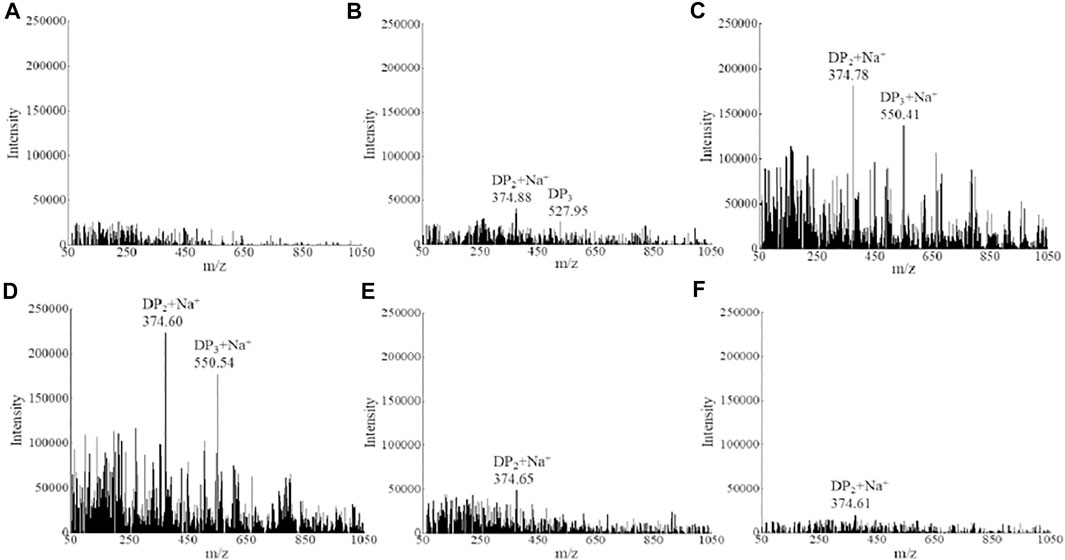
FIGURE 2. Mass spectrometry analysis of alginate metabolism. (A) 0 h, (B) 4 h, (C) 8 h, (D) 12 h, (E) 24 h, and (F) 30 h.
Identification of Intracellular and Extracellular Proteins
Based on the mass spectrometry and database alignment results, the proteins were initially screened for each sample according to the rules of peptides greater than or equal to two and Unused ProtScore >1.3 (p value <0.05, and confidence >95%). The Venn diagram was constructed to show the distribution of the identified proteins. Specifically, 2,133 and 1,242 protein identities were shared among three biological replicates as intracellular and extracellular proteins, respectively (Supplementary Figure S1). Moreover, only 156 and 1,047 proteins were identified as exclusively extracellular and intracellular proteins within their respective groups (Supplementary Figure S2), indicating that most of extracellular proteins were secreted from intracellular synthesis. It is worth mentioning that the intracellular involvement of alginate metabolism was significantly more than that of the extracellular counterpart, as the intercellular proteins far outnumbered the extracellular proteins. It is presumable that the intracellular environment provides a more favorable niche for enzyme activities, implying the more vigorous intracellular metabolic activities of strain ALW1.
Protein Functional Classification
To identify the significantly enriched GO functional groups of the intracellular and extracellular proteins of strain ALW1 cells, WEGO software was used to perform a GO analysis and confirm their classifications. According to the GO annotations, the subcategories of the proteins were listed under different terms, including biological process, cellular component, and molecular function. The results of GO analysis showed that strain ALW1 metabolized alginate polysaccharide mainly concentrated in catalytic activity (GO:0003824), metabolic process (GO:0008152), cellular process (GO: 0009987), binding (GO: 0005488), cell part (GO: 0044464), and cell (GO: 0005623) (Figure 3A). Notably, the proteins under catalytic activity and metabolic processes were predominant, especially for the intracellular proteins, substantiating the biological relevance of strain ALW1 in alginate decomposition. In addition, a significant portion of proteins was related to binding for the intracellular and extracellular proteins. As polysaccharides may serve as the main carbon source for many marine bacteria (Alderkamp et al., 2007; Kabisch et al., 2014; Sichert et al., 2020), proteins encoded in operons known as polysaccharide utilization loci (PULs) are often enriched to counter the physical barrier of a wide variety of polysaccharides, functioning as carbohydrate-degrading enzymes and binding proteins (Adams et al., 2021). The proteins related to cellular process (GO: 0009987) may participate in material transportation, information transmission, and the maintenance of basic life activities.
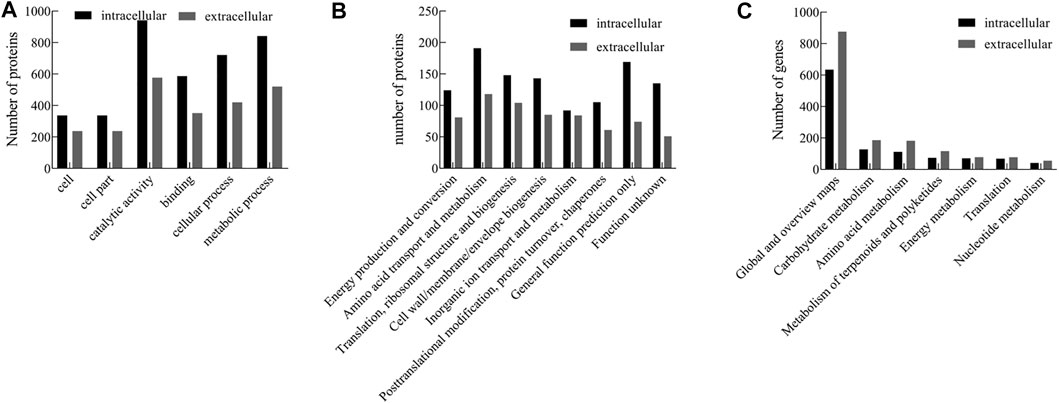
FIGURE 3. GO (A), COG (B), and KEGG (C) pathway analysis of intracellular and extracellular proteins from Microbulbifer sp. ALW1.
The intracellular and extracellular proteins of Microbulbifer sp. ALW1 were further grouped according to their main biological activities, as defined by the functional catalog of COG. The functional categories of intracellular proteins mainly included amino acid transport and metabolism, general function prediction only, translation, ribosomal structure and biogenesis, cell wall/membrane/envelope biogenesis, and function unknown (Figure 3B). Meanwhile, the extracellular counterparts were mainly amino acid transport and metabolism, translation, ribosomal structure, biogenesis, cell wall/membrane/envelope biogenesis, and inorganic ion transport and metabolism. In addition, there was a minority of intracellular and extracellular proteins under the category of posttranslational modification, protein turnover, and chaperones, which normally played essential roles in regulating protein fate and modulating protein functions (Latko et al., 2019).
To further investigate the biological functions of these proteins, KEGG pathway analysis was performed on the identified proteins that were mapped to the reference canonical pathways in the KEGG database. The results revealed that the extracellular and intracellular proteins of strain ALW1 were mainly mapped in global and overview maps, with low protein enrichment in the pathways such as carbohydrate metabolism, amino acid metabolism, metabolism of terpenoids and polyketides, energy metabolism, translation, and nucleotide metabolism (Figure 3C). The protein enrichment patterns indicated that the majority of the proteins were presenting the overall picture of cell metabolism.
Identification of CAZymes
CAZymes represent the carbohydrate-active enzymes to digest diverse sugar bonds in glycans (Kunath et al., 2017). To determine the key enzymes involved in alginate degradation by strain ALW1, the CAZymes analysis was performed for the intracellular and extracellular proteins of this strain. The distribution map of CAZymes indicated that except for glycosyltransferases (GT) and carbohydrate esterases (CE), the numbers of the other types including the non-catalytic protein carbohydrate-binding modules (CBMs) of extracellular proteins were greater than those of intracellular proteins (Figure 4), indicating the robust extracellular enzymatic activities of breaking down the alginate polysaccharide. In addition, among these carbohydrate-active enzymes, the GH were predominant over the other major types of CAZymes, and the PL were the least abundant (Figure 4). GH process the common carbohydrate of motif alpha-linked glycoside in nature (McGuire et al., 2020). PL are mainly responsible for the breakdown of polysaccharides into oligosaccharides and monomers through exolytic and endolytic cleavages (Li et al., 2021), and the resulting oligosaccharides can be transported into the bacterial cells via the transport systems. CBM is carbohydrate-active protein with a discrete fold having carbohydrate-binding activity (Shoseyov et al., 2006). There was a greater number of GH, PL, and CBM in the extracellular secretion than in the intracellular compartments (Figure 4), reflecting the active digestion of alginate polysaccharide extracellularly before the further actions intracellularly upon the uptake of their metabolites.
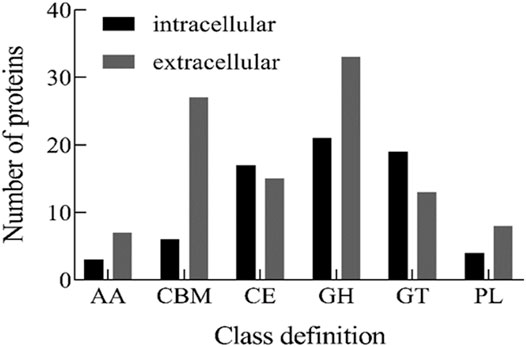
FIGURE 4. CAZy classification of intracellular and extracellular proteins from Microbulbifer sp. ALW1 cultured with alginate. AA means auxiliary activity. CBM means carbohydrate-binding module. CE means carbohydrate esterase. GH means glycoside hydrolases. GT means glycosyltransferases. PL means polysaccharide lyase.
Alginate Polysaccharide Metabolism Pathway in Strain ALW1
Based on KEGG pathway analysis, some proteins with key signatures to play roles in alginate polysaccharide metabolism were summarized to decipher the potential mechanism of alginate degradation and adsorption in strain ALW1 (Table 1). The secreted enzymes included alginate lyase (protein ID: 1_423, 1_424 and 1_430), arabinan endo-1,5-alpha-l-arabinosidase (protein ID: 1_1147), alpha-glucosidase (protein ID: 1_3641), oligosaccharide 4-alpha-d-glucosyltransferase (protein ID: 1_3668), and beta-glucosidase (protein ID: 1_2974), which are reported to function in degrading seaweed polysaccharides into oligosaccharide fragments or monosaccharide molecules (Zehavi et al., 1992; Wong et al., 2000; Zhu et al., 2019; Tang et al., 2020; Li et al., 2021). In addition, some transporters were identified, including ABC transporter (protein ID: 1_960 and 1_1141), major facilitator superfamily (MFS) transporter (protein ID: 1_426 and 1_2094), and TonB system transport protein (protein ID: 1_1049 and 1_1051), which may function as the vehicles (Matthews et al., 2000; Hollenstein et al., 2007; Yan, 2013; Maruyama et al., 2015; Reintjes et al., 2017) for the uptake of primary metabolites of alginate polysaccharide by strain ALW. After adsorption by strain ALW1 cells with the assistance of the transporters, the oligosaccharides and monomers were further degraded through different intracellular alginate lyases and glycolysis pathway.
To further verify the function of alginate lyases, the putative alginate lyase of protein ID 1_430 was expressed in E. coli BL21 (DE3) and applied to degrade sodium alginate. Under the positive ion mode, the mass spectra revealed that alginate lyase 1_430-mediated degradation of alginate generated a mixture of DP2 (disaccharides), DP3 (trisaccharides), and DP4 (tetrasaccharides) (Supplementary Figure S3). The molecular weight of the alginate lyase was 71.5 kDa, and the specific activity of enzyme could reach 26.98 U/mg. In addition, the other alginate lyases have been obtained from Microbulbifer sp. ALW1 in our previous study. These results further verified the ability of strain ALW1 to metabolize alginate polysaccharide to disaccharides and trisaccharides, while the monosaccharide was predicted to be absorbed into the cell and further utilized and was not detected extracellularly (Figure 2).
After integration of the molecular players and their potential nodes in the signaling pathways, the alginate polysaccharide metabolism pathway in strain ALW1 was sketched (Figure 5). Generally, upon the feeding of alginate polysaccharide, Microbulbifer sp. ALW1 produces some extracellularly localized enzymes collectively named alginate lyases and the related hydrolases and transferases to break down the polymers into smaller fragments and monomers. The oligosaccharide and monosaccharide are further actively transported into the periplasmic space of strain ALW1 cells. The monosaccharide is non-enzymatically converted and further reduced to 2-keto-3-deoxy-d-gluconic acid (KDG) subsequently entering into the Entner–Doudoroff (ED) pathway (Takase et al., 2014; Hobbs et al., 2016). In this pathway, KDG is utilized by a two-step catalytic reaction under the sequential actions of 2-keto-3-deoxygluconate aldolase (KdgA) and kinase (KdgK) to produce glyceraldehyde-3-phosphate and pyruvate (Hobbs et al., 2016; Zhang et al., 2021), which are metabolized through tricarboxylic acid (TCA) cycle to support cell growth.
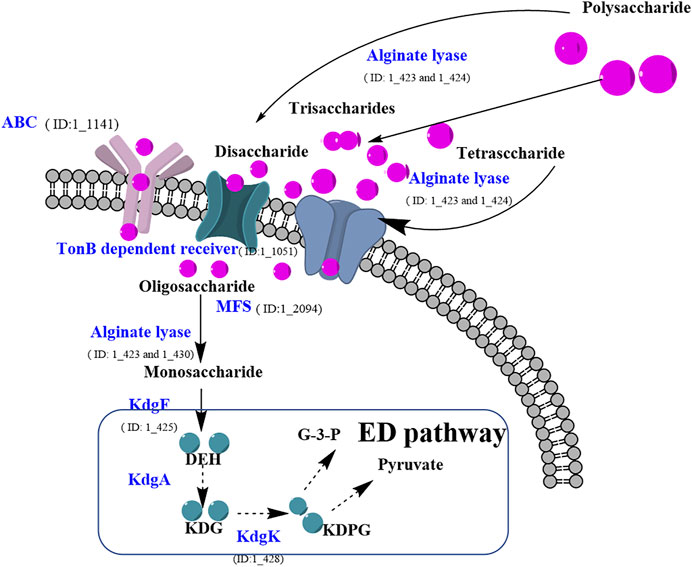
FIGURE 5. Prediction of alginate polysaccharide metabolic pathway of Microbulbifer sp. ALW1. MFS means major facilitator super family. DEH means 4-deoxy-l-erythro-5-hexoseulose urinate. KDPG means 2-keto-3-deoxy-6-phosphate-gluconic acid. KDG means 2-keto-3-deoxy-d-gluconic acid. KdgK means 2-keto-3-deoxygluconate kinase. G-3-P means glyceraldehyde-3-phosphate.
Conclusion
The dynamic patterns of the metabolite accumulation of alginate after digestion by Microbulbifer sp. ALW1 demonstrated its ability to fragment polysaccharide into oligosaccharides. The intracellular and extracellular proteome of strain ALW1 fed with an alginate carbon source presented the overall picture of the orchestration of polysaccharide breakdown, transport, and cellular metabolism. The identification of diverse enzymes and other molecular players involved in the catabolic pathway accentuates the complex actions of alginate degradation to produce oligosaccharides and to support cell growth. This study provides references for genetic engineering of the bacterial strain to enhance its performance in industrial fermentation to produce the bioactive AOS.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
ZPL and YBZ designed the experiments. ZPL, XCW, YBZ, YXH, XPD, HN, CHZ, and LY conceived of the study. ZPL, XCW, and YBZ drafted the manuscript. YXG and XYH carried out fermentation experiments. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (22178142, 22038012, and 32001672) and Natural Science Foundation of Fujian Province (2020J01678).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.829428/full#supplementary-material
References
Adams, A. N. D., Azam, M. S., Costliow, Z. A., Ma, X., Degnan, P. H., and Vanderpool, C. K. (2021). A Novel Family of RNA-Binding Proteins Regulate Polysaccharide Metabolism in Bacteroides Thetaiotaomicron. J. Bacteriol. 203 (21). doi:10.1128/jb.00217-21
Alderkamp, A.-C., Van Rijssel, M., and Bolhuis, H. (2007). Characterization of marine Bacteria and the Activity of Their Enzyme Systems Involved in Degradation of the Algal Storage Glucan Laminarin. FEMS Microbiol. Ecol. 59, 108–117. doi:10.1111/j.1574-6941.2006.00219.x
An, Q.-D., Zhang, G.-L., Wu, H.-T., Zhang, Z.-C., Zheng, G.-S., Zhang, Y.-L., et al. (2008). Properties of an Alginate-Degrading Flavobacterium Sp. Strain LXA Isolated from Rotting Algae from Coastal China. Can. J. Microbiol. 54, 314–320. doi:10.1139/W08-013
Bang, M.-A., Seo, J.-H., Seo, J.-W., Jo, G. H., Jung, S. K., Yu, R., et al. (2015). Correction: Bacillus Subtilis KCTC 11782BP-Produced Alginate Oligosaccharide Effectively Suppresses Asthma via T-Helper Cell Type 2-related Cytokines. PLoS One 10, e0130510–9. doi:10.1371/journal.pone.0130510
Bradford, M. M. (1976). A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 72 (1-2), 248–254. doi:10.1016/0003-2697(76)90527-310.1006/abio.1976.9999
Choi, D., Piao, Y. L., Shin, W.-S., and Cho, H. (2009). Production of Oligosaccharide from Alginate Using Pseudoalteromonas Agarovorans. Appl. Biochem. Biotechnol. 159, 438–452. doi:10.1007/s12010-008-8514-7
de Figueiredo, F. L., de Oliveira, A. C. P., Terrasan, C. R. F., Gonçalves, T. A., Gerhardt, J. A., Tomazetto, G., et al. (2021). Multi-omics Analysis Provides Insights into Lignocellulosic Biomass Degradation by Laetiporus Sulphureus ATCC 52600. Biotechnol. Biofuels 14, 1–17. doi:10.1186/s13068-021-01945-7
Dunne, J. C., Li, D., Kelly, W. J., Leahy, S. C., Bond, J. J., Attwood, G. T., et al. (2012). Extracellular Polysaccharide-Degrading Proteome of Butyrivibrio Proteoclasticus. J. Proteome Res. 11, 131–142. doi:10.1021/pr200864j
Elias, J. E., and Gygi, S. P. (2007). Target-decoy Search Strategy for Increased Confidence in Large-Scale Protein Identifications by Mass Spectrometry. Nat. Methods 4, 207–214. doi:10.1038/nmeth1019
Hobbs, J. K., Lee, S. M., Robb, M., Hof, F., Barr, C., Abe, K. T., et al. (2016). KdgF, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate. Proc. Natl. Acad. Sci. USA 113, 6188–6193. doi:10.1073/pnas.1524214113
Hollenstein, K., Dawson, R. J., and Locher, K. P. (2007). Structure and Mechanism of ABC Transporter Proteins. Curr. Opin. Struct. Biol. 17, 412–418. doi:10.1016/j.sbi.2007.07.003
Jensen, S., Frank, J. A., Arntzen, M. Ø., Duperron, S., Vaaje-Kolstad, G., and Hovland, M. (2021). Endozoicomonadaceae Symbiont in Gills of Acesta Clam Encodes Genes for Essential Nutrients and Polysaccharide Degradation. FEMS Microbiol. Ecol. 97 (6), fiab070. doi:10.1093/femsec/fiab070)
Jesumani, V., Du, H., Aslam, M., Pei, P., and Huang, N. (2019). Potential Use of Seaweed Bioactive Compounds in Skincare-A Review. Mar. Drugs 17, 688–719. doi:10.3390/md17120688
Jiang, Z., Guo, Y., Wang, X., Li, H., Ni, H., Li, L., et al. (2019). Molecular Cloning and Characterization of AlgL17, a New Exo-Oligoalginate Lyase from Microbulbifer Sp. ALW1. Protein Expr. Purif. 161, 17–27. doi:10.1016/j.pep.2019.03.015
Jiang, Z., Zhang, X., Wu, L., Li, H., Chen, Y., Li, L., et al. (2021). Exolytic Products of Alginate by the Immobilized Alginate Lyase Confer Antioxidant and Antiapoptotic Bioactivities in Human Umbilical Vein Endothelial Cells. Carbohydr. Polym. 251, 116976. doi:10.1016/j.carbpol.2020.116976
Kabisch, A., Otto, A., König, S., Becher, D., Albrecht, D., Schüler, M., et al. (2014). Functional Characterization of Polysaccharide Utilization Loci in the marine Bacteroidetes 'Gramella Forsetii' KT0803. ISME J. 8, 1492–1502. doi:10.1038/ismej.2014.4
Kawai, S., and Murata, K. (2016). Biofuel Production Based on Carbohydrates from Both Brown and Red Macroalgae: Recent Developments in Key Biotechnologies. Ijms 17, 145. doi:10.3390/ijms17020145
Kunath, B. J., Bremges, A., Weimann, A., McHardy, A. C., and Pope, P. B. (2017). Metagenomics and CAZyme Discovery. Methods Mol. Biol. 1588, 255–277. doi:10.1007/978-1-4939-6899-2_20
Latko, M., Czyrek, A., Porębska, N., Kucińska, M., Otlewski, J., Zakrzewska, M., et al. (2019). Cross-talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells 8 (5), 455–483. doi:10.3390/cells8050455
Li, M., Shang, Q., Li, G., Wang, X., and Yu, G. (2017). Degradation of marine Algae-Derived Carbohydrates by Bacteroidetes Isolated from Human Gut Microbiota. Mar. Drugs 15, 92–104. doi:10.3390/md15040092
Li, Q., Zheng, L., Guo, Z., Tang, T., and Zhu, B. (2021). Alginate Degrading Enzymes: an Updated Comprehensive Review of the Structure, Catalytic Mechanism, Modification Method and Applications of Alginate Lyases. Crit. Rev. Biotechnol. 41, 953–968. doi:10.1080/07388551.2021.1898330
Liu, J., Yang, S., Li, X., Yan, Q., Reaney, M. J. T., and Jiang, Z. (2019). Alginate Oligosaccharides: Production, Biological Activities, and Potential Applications. Compr. Rev. Food Sci. Food Saf. 18, 1859–1881. doi:10.1111/1541-4337.12494
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucl. Acids Res. 42, D490–D495. doi:10.1093/nar/gkt1178
Lopez-Santamarina, A., Miranda, J. M., Mondragon, A. d. C., Lamas, A., Cardelle-Cobas, A., Franco, C. M., et al. (2020). Potential Use of marine Seaweeds as Prebiotics: A Review. Molecules 25, 1004–1026. doi:10.3390/molecules25041004
Maruyama, Y., Itoh, T., Kaneko, A., Nishitani, Y., Mikami, B., Hashimoto, W., et al. (2015). Structure of a Bacterial ABC Transporter Involved in the Import of an Acidic Polysaccharide Alginate. Structure 23, 1643–1654. doi:10.1016/j.str.2015.06.021
Matthews, S. A., Iglesias, T., Rozengurt, E., and Cantrell, D. (2000). Spatial and Temporal Regulation of Protein Kinase D (PKD). EMBO J. 19, 2935–2945. doi:10.1093/emboj/19.12.2935
McGuire, B. E., Hettle, A. G., Vickers, C., King, D. T., Vocadlo, D. J., and Boraston, A. B. (2020). The Structure of a Family 110 Glycoside Hydrolase Provides Insight into the Hydrolysis of α-1,3-galactosidic Linkages in λ-carrageenan and Blood Group Antigens. J. Biol. Chem. 295, 18426–18435. doi:10.1074/jbc.RA120.015776
Miller, G. L. (1959). Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 31, 426–428. doi:10.1021/ac60147a030
Montgomery, R. (1961). Further Studies of the Phenol-Sulfuric Acid Reagent for Carbohydrates. Biochim. Biophys. Acta 48, 591–593. doi:10.1016/0006-3002(61)90059-2
Pilgaard, B., Wilkens, C., Herbst, F.-A., Vuillemin, M., Rhein-Knudsen, N., Meyer, A. S., et al. (2019). Proteomic Enzyme Analysis of the marine Fungus Paradendryphiella salina Reveals Alginate Lyase as a Minimal Adaptation Strategy for Brown Algae Degradation. Sci. Rep. 9, 1–13. doi:10.1038/s41598-019-48823-9
Reintjes, G., Arnosti, C., Fuchs, B. M., and Amann, R. (2017). An Alternative Polysaccharide Uptake Mechanism of marine Bacteria. ISME J. 11, 1640–1650. doi:10.1038/ismej.2017.26
Roohinejad, S., Koubaa, M., Barba, F. J., Saljoughian, S., Amid, M., and Greiner, R. (2017). Application of Seaweeds to Develop New Food Products with Enhanced Shelf-Life, Quality and Health-Related Beneficial Properties. Food Res. Int. 99, 1066–1083. doi:10.1016/j.foodres.2016.08.016
Ścieszka, S., and Klewicka, E. (2019). Algae in Food: a General Review. Crit. Rev. Food Sci. Nutr. 59, 3538–3547. doi:10.1080/10408398.2018.1496319
Shilov, I. V., Seymour, S. L., Patel, A. A., Loboda, A., Tang, W. H., Keating, S. P., et al. (2007). The Paragon Algorithm, a Next Generation Search Engine that Uses Sequence Temperature Values and Feature Probabilities to Identify Peptides from Tandem Mass Spectra. Mol. Cell Proteomics 6 (9), 1638–1655. doi:10.1074/mcp.T600050-MCP200
Shoseyov, O., Shani, Z., and Levy, I. (2006). Carbohydrate Binding Modules: Biochemical Properties and Novel Applications. Microbiol. Mol. Biol. Rev. 70, 283–295. doi:10.1128/mmbr.00028-05
Sichert, A., Corzett, C. H., Schechter, M. S., Unfried, F., Markert, S., Becher, D., et al. (2020). Verrucomicrobia Use Hundreds of Enzymes to Digest the Algal Polysaccharide Fucoidan. Nat. Microbiol. 5, 1026–1039. doi:10.1038/s41564-020-0720-2
Takase, R., Mikami, B., Kawai, S., Murata, K., and Hashimoto, W. (2014). Structure-based Conversion of the Coenzyme Requirement of a Short-Chain Dehydrogenase/reductase Involved in Bacterial Alginate Metabolism. J. Biol. Chem. 289, 33198–33214. doi:10.1074/jbc.M114.585661
Takeshita, S., and Oda, T. (2016). Usefulness of Alginate Lyases Derived from marine Organisms for the Preparation of Alginate Oligomers with Various Bioactivities. Adv. Food Nutr. Res. 79, 137–160. doi:10.1016/bs.afnr.2016.07.003
Tang, J.-C., Taniguchi, H., Chu, H., Zhou, Q., and Nagata, S. (2009). Isolation and Characterization of Alginate-Degrading Bacteria for Disposal of Seaweed Wastes. Lett. Appl. Microbiol. 48, 38–43. doi:10.1111/j.1472-765X.2008.02481.x
Tang, L., Wang, Y., Gao, S., Wu, H., Wang, D., Yu, W., et al. (2020). Biochemical Characteristics and Molecular Mechanism of an Exo-type Alginate Lyase VxAly7D and its Use for the Preparation of Unsaturated Monosaccharides. Biotechnol. Biofuels 13, 1–17. doi:10.1186/s13068-020-01738-4
Wang, M., Chen, L., Liu, Z., Zhang, Z., Qin, S., and Yan, P. (2016). Isolation of a Novel Alginate Lyase‐producing Bacillus Litoralis Strain and its Potential to Ferment Sargassum Horneri for Biofertilizer. Microbiologyopen 5, 1038–1049. doi:10.1002/mbo3.387
Wang, X., Wang, D., Wang, D., Wang, H., Chang, L., Yi, X., et al. (2012). Systematic Comparison of Technical Details in CBB Methods and Development of a Sensitive GAP Stain for Comparative Proteomic Analysis. Electrophoresis 33, 296–306. doi:10.1002/elps.201100300
Wargacki, A. J., Leonard, E., Win, M. N., Regitsky, D. D., Santos, C. N. S., Kim, P. B., et al. (2012). An Engineered Microbial Platform for Direct Biofuel Production from Brown Macroalgae. Science 335 (6066), 308–313. doi:10.1126/science.1214547
Wong, T. Y., Preston, L. A., and Schiller, N. L. (2000). Alginate Lyase: Review of Major Sources and Enzyme Characteristics, Structure-Function Analysis, Biological Roles, and Applications. Annu. Rev. Microbiol. 54, 289–340. doi:10.1146/annurev.micro.54.1.289
Xu, F., Wang, P., Zhang, Y.-Z., and Chen, X.-L. (2018). Diversity of Three-Dimensional Structures and Catalytic Mechanisms of Alginate Lyases. Appl. Environ. Microbiol. 84, 02040. doi:10.1128/AEM.02040-17
Yan, N. (2013). Structural Advances for the Major Facilitator Superfamily (MFS) Transporters. Trends Biochem. Sci. 38, 151–159. doi:10.1016/j.tibs.2013.01.003
Yi, X., Sun, Y., Yang, Q., Guo, A., Chang, L., Wang, D., et al. (2014). Quantitative Proteomics of Sesuvium portulacastrum Leaves Revealed that Ion Transportation by V-ATPase and Sugar Accumulation in Chloroplast Played Crucial Roles in Halophyte Salt Tolerance. J. Proteomics 99, 84–100. doi:10.1016/j.jprot.2014.01.017
Zehavi, U., Herchman, M., and Köpper, S. (1992). Polymers Having (1 → 4)- and (1 → 6)-linked α-d-glucopyranosyl Groups as Acceptors in the Glycogen Synthase Reaction,+. Carbohydr. Res. 228, 255–263. doi:10.1016/s0008-6215(00)90563-9
Zhang, L., Li, X., Zhang, X., Li, Y., and Wang, L. (2021). Bacterial Alginate Metabolism: an Important Pathway for Bioconversion of Brown Algae. Biotechnol. Biofuels 14, 1–18. doi:10.1186/s13068-021-02007-8
Zhu, B., Ni, F., Sun, Y., Ning, L., and Yao, Z. (2019). Elucidation of Degrading Pattern and Substrate Recognition of a Novel Bifunctional Alginate Lyase from Flammeovirga Sp. NJ-04 and its Use for Preparation Alginate Oligosaccharides. Biotechnol. Biofuels 12, 1–13. doi:10.1186/s13068-019-1352-8
Zhu, Y. B., Wu, L. Y., Chen, Y. H., Ni, H., Xiao, A. F., and Cai, H. N. (2016). Characterization of Alginate Lyase from Microbulbifer Sp. ALW5 and Antioxidant Activity of its Hydrolysates. J. Chin. Inst. Food Sci. Technol. 16, 79–87. doi:10.16429/j.1009-7848.2016.06.012
Keywords: alginate catabolism, proteomics, Microbulbifer sp. ALW1, oligosaccharides, polysaccharides
Citation: Li Z, Huang X, Guo Y, Zhang C, Yang L, Du X, Ni H, Wang X and Zhu Y (2022) Toward Understanding the Alginate Catabolism in Microbulbifer sp. ALW1 by Proteomics Profiling. Front. Bioeng. Biotechnol. 10:829428. doi: 10.3389/fbioe.2022.829428
Received: 05 December 2021; Accepted: 31 January 2022;
Published: 16 March 2022.
Edited by:
Haijin Mou, Ocean University of China, ChinaReviewed by:
Daochen Zhu, Jiangsu University, ChinaLianli Chi, Shandong University, China
Fu-Li Li, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2022 Li, Huang, Guo, Zhang, Yang, Du, Ni, Wang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuchu Wang, eGNod2FuZ0BoYWlubnUuZWR1LmNu; Yanbing Zhu, eWFuYmluZ3podUBqbXUuZWR1LmNu
 Zhipeng Li1,2,3
Zhipeng Li1,2,3 Hui Ni
Hui Ni Xuchu Wang
Xuchu Wang Yanbing Zhu
Yanbing Zhu