- 1Clean Energy Research Center, Korea Institute of Science and Technology (KIST), Seoul, South Korea
- 2Department of Biotechnology, Korea University, Seoul, South Korea
- 3Division of Energy and Environment Technology, University of Science and Technology (UST), Daejeon, South Korea
- 4Green School, Korea University, Seoul, South Korea
Efficient xylose catabolism in engineered Saccharomyces cerevisiae enables more economical lignocellulosic biorefinery with improved production yields per unit of biomass. Yet, the product profile of glucose/xylose co-fermenting S. cerevisiae is mainly limited to bioethanol and a few other chemicals. Here, we introduced an n-butanol-biosynthesis pathway into a glucose/xylose co-fermenting S. cerevisiae strain (XUSEA) to evaluate its potential on the production of acetyl-CoA derived products. Higher n-butanol production of glucose/xylose co-fermenting strain was explained by the transcriptomic landscape, which revealed strongly increased acetyl-CoA and NADPH pools when compared to a glucose fermenting wild-type strain. The acetate supplementation expected to support acetyl-CoA pool further increased n-butanol production, which was also validated during the fermentation of lignocellulosic hydrolysates containing acetate. Our findings imply the feasibility of lignocellulosic biorefinery for producing fuels and chemicals derived from a key intermediate of acetyl-CoA through glucose/xylose co-fermentation.
Introduction
Lignocellulosic biomass offers a sustainable and environmentally friendly source of raw materials for producing fuels and chemicals (Ko and Lee 2018). Commercial bioethanol production has been achieved using the yeast Saccharomyces cerevisiae and sugar- or starch-based biomass. To improve the economic feasibility of lignocellulosic biorefinery, S. cerevisiae strains have been engineered to co-ferment glucose and xylose, the most abundant hexose and pentose sugars in lignocellulosic hydrolysates, respectively, resulting in significantly improved lignocellulosic bioethanol yields, titers, and productivity (Hoang Nguyen Tran et al., 2020). In addition, increasing efforts have also been devoted to expanding the product profile of S. cerevisiae to include advanced fuels and chemicals demonstrating the great potential of microbial cell factories for biorefinery (Ekas et al., 2019). Yet, the production of non-native products using both glucose and xylose derived from lignocellulosic hydrolysates has been less explored. Thus, it is necessary to evaluate the feasibility of sustainable-biorefinery concepts based on lignocellulosic biomass, which is the most abundant and sustainable resource.
Efficient glucose/xylose co-fermentation enables complete utilization of all available sugars in lignocellulosic biomass, which increases the overall conversion yield in lignocellulosic biorefinery. By incorporating xylose into the substrate used by S. cerevisiae, the overall conversion yield could be increased by up to 84% during lignocellulosic bioethanol production (Ko et al., 2018). In addition, xylose metabolism supported the diversion of metabolic flux towards non-ethanol products in an ethanol-producing S. cerevisiae yeast strain (Li et al., 2019). Recently, xylose-utilizing strains have been suggested as promising microbial cell factories for producing acetyl-CoA-derived products (Kwak and Jin 2017), due to their altered cellular metabolism. Therefore, it is of interest to confirm the advantages of glucose/xylose co-fermentation in producing fuels and chemicals derived from acetyl-CoA at the phenotypic and transcriptional levels.
As a core intermediate in central carbon metabolism, acetyl-CoA can be converted into various fuels and chemicals. Yeast strains have been suggested to be better hosts for producing acetyl-CoA-derived products than other microbial hosts (Sun and Alper 2020). With limited metabolic flux through cytosolic acetyl-CoA, however, the successful production of fuels and chemicals derived from acetyl-CoA has been rarely demonstrated in S. cerevisiae. Specifically, the production of acetyl-CoA-derived n-butanol, a short-chain alcohol similar to ethanol, has been limited even with higher butanol tolerance of S. cerevisiae when compared to other non-native bacterial hosts (Knoshaug and Zhang 2009; Hong and Nielsen 2012; González-Ramos et al., 2013). Engineering S. cerevisiae for n-butanol production has been based mainly on the reverse-β-oxidation pathway of Clostridium sp., which naturally produce butanol. Therefore, successful n-butanol production would also enable successful incorporation of the reverse-β-oxidation pathway, an economic route for medium-chain fatty acids and alcohols with numerous industrial applications, into the cellular network of S. cerevisiae.
Here, we sought to evaluate the potential of a glucose/xylose co-fermenting strain of S. cerevisiae as a production host for acetyl-CoA-derived n-butanol in lignocellulosic biorefinery concept. To this end, an n-butanol-biosynthesis pathway was introduced in XUSEA, a previously developed S. cerevisiae strain that simultaneously ferments glucose and xylose (Tran Nguyen Hoang et al., 2018; Hoang Nguyen Tran et al., 2020), generating XUSEA-B. The transcription profile of the XUSEA strain was compared to that of the wild-type strain to understand the modified cellular network caused by the introduction of a heterologous route for bioconverting xylose into butanol. n-Butanol production in the glucose/xylose co-fermenting strain was further improved by supplementation with acetic acid, a major by-product generated during lignocellulosic biomass-pretreatment process. Finally, we successfully demonstrated lignocellulosic n-butanol production by using a glucose/xylose co-fermenting strain. Consequently, the results of this study show the potential of a glucose/xylose co-fermenting strain in the production of acetyl-CoA-derived n-butanol from lignocellulosic biomass.
Materials and Methods
Strains and Culture Conditions
All strains used in this study are shown in Table 1. The yeast strains used in this study were isogenic with respect to S. cerevisiae S288C BY4741. The yeast strains were routinely cultivated at 30 °C in yeast synthetic complete (YSC) medium composed of xylose (and/or glucose), 6.7 g/L of yeast nitrogen base (Difco, Detroit, MI, United States), and CSM-HIS-LEU-URA (MP Biomedicals, Solon, Ohio, United States). Escherichia coli DH10β cells were employed for DNA manipulation and were cultured at 37°C in LB medium supplemented with 100 μg/ml ampicillin.
Plasmid Construction
All plasmids used in this study are shown in Table 2. The homologous genes used in this study were ERG10 and ETR1, and the heterologous genes were EutE from E. coli, Kr and Htd from Yarrowia lipolytica, and Hbd, Crt, AdhE2, and BdhB from Clostridium acetobutylicum. The start codon of BdhB was changed from GTG to ATG, and the mitochondria-targeting sequences of the Kr, Htd, and Etr1 proteins were predicted and excluded using TargetP software version 1.1 (http://www.cbs.dtu.dk/services/TargetP/) and the MITOPROT online tools (https://ihg.gsf.de/ihg/mitoprot.html) according to the reference by Lian and Zhao (2015). Each gene cassette was cloned into an expression vector using the Gibson Assembly method and introduced in S. cerevisiae using the Frozen-EZ Yeast Transformation II Kit (Zymo Research), according to the manufacturer’s instructions.
Predicting Mitochondria-Targeting Sequences
The online MITOPROT tool was used to predict and exclude mitochondria-targeting signal peptide sequences, which were further validated using TargetP, version 1.1 (Lian and Zhao 2015). The nucleotide sequences encoding the excluded signal peptides are as follows: TTCCGACTCACCACTGCCCGAATTGCTTCTGTGCGAGGCTTCTCCACCTCCGCCAGCCTGTCC (for Kr), CGAAGCCTATATATAAACGTT CCGGGTCTTTTTCCTTCCACCTCTCTAGCACGAGAA (for Htd), and CTTCCCA CATTCAAACGTTACATG (for Etr1). Additionally, an intron sequence of Htd was excluded along with mitochondria-targeting sequence.
Fermentation
For seed cultures, yeast cells from a glycerol stock were inoculated in YSC medium containing 2% glucose. The yeast cells were then transferred to fresh YSC medium containing 2% glucose, 2% xylose, or 2% glucose plus 2% xylose with an inoculum size of 5%, and grown aerobically in flasks for 1.5–2 days. Subsequently, the yeast cells were harvested and finally transferred to fresh YSC medium. The pH of the fermentation medium was maintained by adding 70 mM phthalate buffer (pH 5.0) or 80 mM phosphate buffer (pH 6.5). Micro-aerobic fermentation was carried out in 125 ml serum bottles with a final working volume of 40 ml at a low cell density (initial optical density [OD] of 0.2) and a high cell density (initial OD of 15). The serum bottles were capped with rubber stoppers, with a needle for carbon dioxide release during fermentation. Lignocellulosic-biomass hydrolysates (Miscanthus sacchariflorus Goedae-Uksae 1), pretreated with dilute acid, were purchased from Sugaren Co., Ltd (Korea). The hydrolysates contained 30.6 g/L of glucose, 15.1 g/L of xylose, 1.3 g/L of arabinose, 0.15 g/L of formic acid, 0.05 g/L of acetic acid, 0.08 g/L of levulinic acid, 0.06 g/L of 5-HMF, and 0.07 g/L of furfural.
Analytical Methods
Cell growth was analyzed by measuring the OD at 600 nm with a Cary 60 Bio UV–Vis spectrometer (Agilent Technologies, United States). Glucose and xylose concentrations were analyzed using a high-performance liquid chromatography system (HPLC 1260 Infinity, Agilent Technologies, CA, United States) equipped with a refractive index detector, using a Hi-Plex H column (Agilent Technologies, Palo Alto, CA, United States). The system was operated using 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml/min, and the column temperature was maintained at 65 °C. Ethanol, n-butanol, and acetate concentrations were analyzed using a gas chromatograph instrument (Agilent Technologies, CA, United States) equipped with a flame ionization detector, using an HP-INNOWax polyethylene glycol column (30 m × 0.32 µm × 0.25 µm).
Transcriptomic Analysis
For transcriptomic analysis, cells were cultured and harvested at exponential phase during glucose fermentation and glucose/xylose co-fermentation. Cell pellets were collected by centrifugation at 1,000 g for 5 min. Total RNA extraction was performed using Trizol reagent (Invitrogen, CA, United States) according to the manufacturer’s protocol provided by Ebiogen (Seoul, Republic of Korea). Each of the total RNA samples was evaluated for RNA quality control based on the 28S/18S ratio and RIN measured on the 2,100 Bioanalyzer system (Agilent Technologies, Waldbronn, Germany). The cDNA library was constructed using the Clontech SMARTer Stranded RNA-Seq kit (Clontech, Mountain View, CA, United States). High-throughput sequencing was performed on an Illumina HiSeq 2,500 system (Illumina, Inc, San Diego, CA, United States).
Results
Introduction of a n-Butanol Production Pathway in a Glucose/Xylose Co-fermenting Strain
To develop an S. cerevisiae strain capable of n-butanol-production, genes that were previously reported to support butanol biosynthesis were introduced in an efficient glucose/xylose co-fermenting strain, XUSEA (Hoang Nguyen Tran et al., 2020), with various combinations. Specifically, Hbd and Crt originated from C. acetobutylicum (a representative n-butanol-producing bacterium), and the orthologous genes (Kr and Htd, respectively) originated from Y. lipolytica, a yeast with an efficient β-oxidation pathway. These genes were heterologously expressed along with the homologous genes, ERG10 and cytoETR1, to construct the reverse-β-oxidation pathway, thereby enabling the conversion of acetyl-CoA into butyryl-CoA (Dellomonaco et al., 2011; Lian and Zhao 2015). To generate a complete butanol-production pathway, Adhe2 or BdhB from C. acetobutylicum were heterologously expressed with or without EutE from E. coli (Figure 1A). Heterologous expression of these C. acetobutylicum genes increased butanol production more than heterologous expression of the indicated Y. lipolytica genes, even without codon optimization. When expressing heterologous genes, it is often expected that genes originating from yeasts or other eukaryotic cells will be expressed at higher levels in eukaryotic cells (such as S. cerevisiae). However, the distance in a phylogenetic tree does not seem to guarantee better performance, when expressed in a heterologous host.
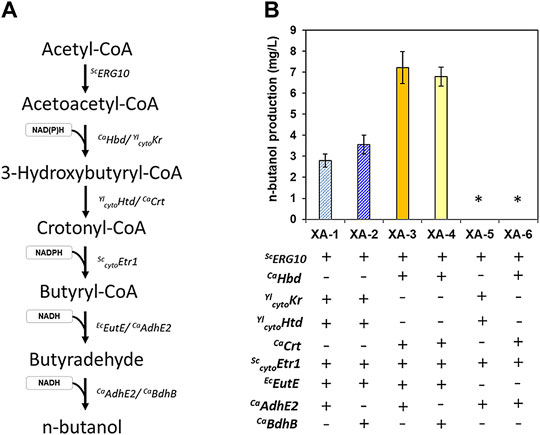
FIGURE 1. n-butanol production in the glucose/xylose co-fermenting S. cerevisiae strains expressing various gene sets used for n-butanol biosynthesis. The genes were sourced from S. cerevisiae (Sc), Y. lipolytica (Yl), E. coli (Ec), and C. acetobutyricum (Ca). Low-cell density fermentations (initial OD of 0.2) were conducted with 20 g/L xylose as the sole carbon source. (A) n-butanol pathway showing the genes used in this study. (B) butanol titer of the strains expressing six combinations of genes used to construct various n-butanol-production pathways. The error bars represent the standard deviations obtained using biological triplicates. *: below the detection limit of 2.5 mg/L.
The choice of the alcohol dehydrogenase used, either Adhe2 or BdhB, did not significantly affect the butanol titer. Although AdhE2 is a bi-functional enzyme that converts butyryl-CoA into butyraldehyde and then to n-butanol, it cannot fully support butanol production without EutE expression, suggesting that sufficient enzyme levels are required for each step in n-butanol synthesis. Of the strains expressing heterologous gene combinations, the XA-3 strain harboring the set comprised of Hdb, Crt, EutE, and AdhE2 resulted in the highest butanol titer of 7.2 mg/L, followed by the XA-4 strain harboring the set comprised of Hdb, Crt, EutE, and BdhB (6.8 mg/L) (Figure 1B). Based on the similar butanol titer with a half size of BdhB compared to Adhe2, the butanol production pathway genes in the XA-4 strain was selected as the combination of choice for further experiments throughout this study.
The n-butanol production pathway with the selected genes were introduced into both a xylose utilizing strain of XUSEA and a wild-type strain of S. cerevisiae BY4741, generating XUSEA-B and WT-B, respectively. Then, the n-butanol production performance of XUSEA-B and WT-B was compared during glucose fermentation, xylose fermentation, and glucose/xylose co-fermentation (Figure 2). During glucose fermentation, the XUSEA-B strain showed 2.1-fold higher n-butanol production than the WT-B strain (14.2 mg/L and 6.6 mg/L, respectively; Figure 2A). The XUSEA-B strain produced the same amount of n-butanol during xylose fermentation, whereas the WT-B strain produced no n-butanol because it could not utilize xylose. During glucose/xylose co-fermentation, XUSEA-B produced 26.3 mg/L of n-butanol within 72 h, which was 3.9-fold higher than that produced by WT-B (6.7 mg/L; Figures 2C,D), suggesting that the xylose-utilizing strain could serve as a promising host for producing n-butanol, an acetyl-CoA-derived product.
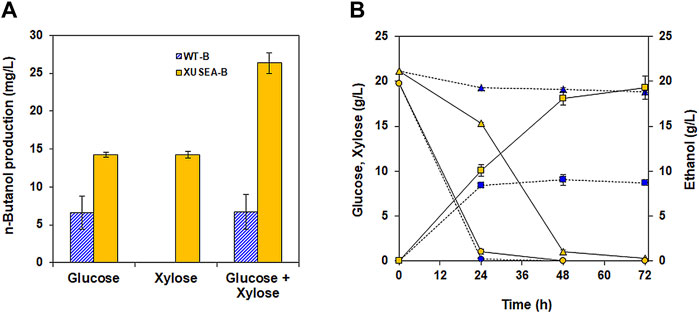
FIGURE 2. Comparison of n-butanol production between the WT-B (blue) and XUSEA-B (yellow) strains during glucose and/or xylose fermentation. Fermentation was conducted with an initial OD of 0.2. (A) n-butanol titer after 72 h of fermentation. (B) glucose (circles)/xylose (triangles) consumption and ethanol production (squares) during co-fermentation. The error bars represent the standard deviations obtained using biological triplicates.
Transcriptomic Landscape Revealed the Redirected Carbon Flux for Improved n-Butanol Production in a Xylose-Utilizing Strain
To understand the mechanism underlying efficient n-butanol production by the xylose-utilizing strain, we analyzed the global transcript profiles of XUSEA-B and WT-B during glucose fermentation and glucose/xylose co-fermentation. Compared with WT-B, the XUSEA-B strain showed marked differences in the gene-expression landscape (170 upregulated and 84 downregulated genes with glucose fermentation versus 172 upregulated and 82 downregulated genes with mixed-sugar fermentation) (Figure 3). Transcriptional changes in genes associated with core carbon metabolism and linked with the butanol-production pathway were similar between both fermentation conditions, despite the use of different carbon sources. The XUSEA-B strain appeared to show elevated central carbon flux through the n-butanol-production pathway, resulting from pathway upregulation due to increased cofactor generation and reduced use of the acetyl-CoA precursor. The ZWF1, SOL3, and GND1 genes (involved in the oxidative pentose phosphate pathway, from which the factor NADPH is generated), were upregulated in XUSEA-B compared to WT-B by 2.81-, 4.11-, and 7.82-fold during glucose fermentation and by 3.74-, 5.54-, and 9.93-fold during glucose/xylose co-fermentation, respectively (Figure 3, Table 3).
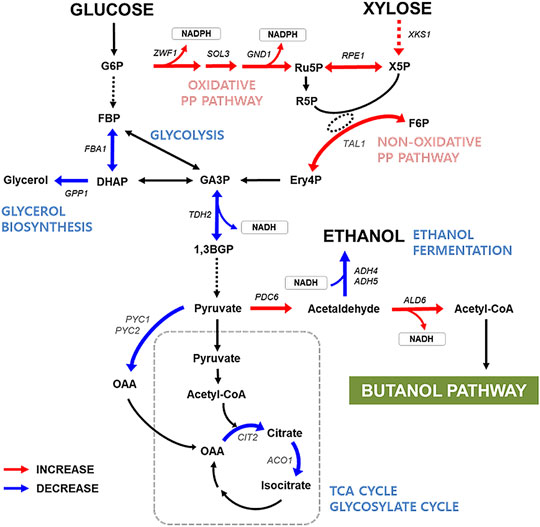
FIGURE 3. Transcription levels of the genes involved in central carbon metabolism in the XUSEA-B strain during glucose or glucose/xylose co-fermentation. Red/blue arrows indicate the enzymatic steps with more than 2-fold increased/decreased transcription levels in XUSEA-B compared to those in WT-B strain. The cells were grown on glucose medium (20 g/L glucose) or a mixed-sugar medium (20 g/L glucose and 20 g/L xylose) under micro-aerobic conditions. Sampling was conducted during the exponential phase of cell growth. The gene symbols were referenced from the Saccharomyces Genome Database.
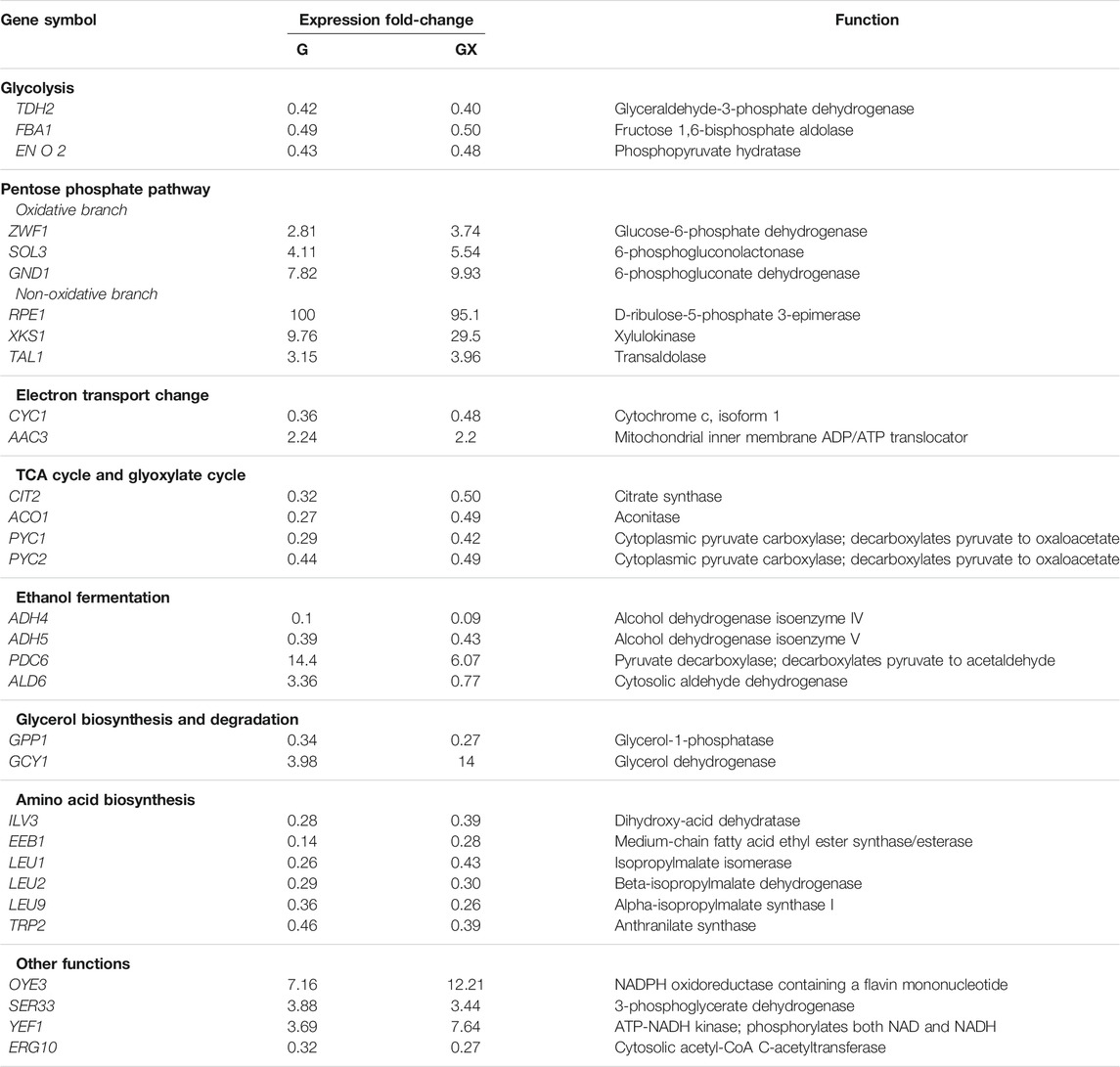
TABLE 3. Fold-changes in the expression levels of genes involved in metabolic pathways in XUSEA-B strain, versus the WT-B strain. G: glucose, GX: glucose and xylose.
It should be noted that genes involved in the non-oxidative pentose phosphate pathway (such as RPE1 and TAL1) and XKS1 were highly upregulated in the XUSEA-B strain, compared to WT-B (Figure 3, Table 3). These differences were mainly due to the genetic background XUSEA (the parental strain of XUSEA-B), in which the above-mentioned genes were overexpressed (Tran Nguyen Hoang et al., 2018; Hoang Nguyen Tran et al., 2020). However, it is noteworthy that RPE1 mRNA expression was upregulated by approximately 100-fold in the XUSEA-B strain. This finding suggests that carbon flux was enhanced through glycerate-3-phosphate, an interconnection point between the oxidative and non-oxidative pentose phosphate pathways. The transcription of PDC6, which is involved in converting pyruvate to acetaldehyde, was also strongly upregulated suggesting that the acetyl-CoA was increased in XUSEA-B. In contrast, the FBA1 and TDH2 genes (involved in glycolysis), were down-regulated more than 2-fold in XUSEA-B, leading to a low carbon flux through central carbon metabolism via glycolysis.
Supplementation With Acetate Improved n-Butanol Production by S. cerevisiae
After confirming the positive effect of increased acetyl-CoA availability in XUSEA-B, based on transcriptomic analysis, the effect of acetate supplementation on n-butanol production was investigated as a strategy for supplying additional acetyl-CoA. To this end, n-butanol production by XUSEA-B was evaluated during glucose and/or xylose fermentation supplemented with various concentrations of acetate (0 g/L, 1 g/L, or 2 g/L). During glucose fermentation, acetate supplementation increased n-butanol production by XUSEA-B (Figure 4). XUSEA-B strains produced 40% or 28% more n-butanol in the presence of one or 2 g/L acetate, respectively. In contrast, acetate supplementation did not result in increased n-butanol production during xylose fermentation. The production of n-butanol decreased by 56% in the presence of 1 g/L acetate during 48 h of xylose fermentation. No n-butanol was produced in the presence of 2 g/L acetate in the medium because the cell growth was significantly inhibited (data not shown). Interestingly, the negative effect of acetate supplementation on n-butanol production during xylose fermentation was compensated in the presence of glucose. During glucose/xylose co-fermentation, n-butanol production was reduced by 23% in the presence of 1 g/L acetate (Figure 4B).
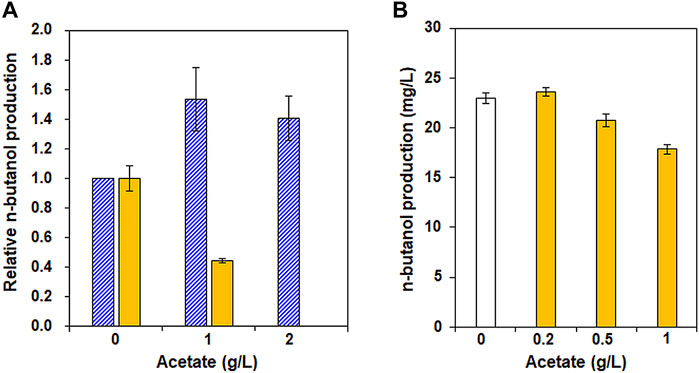
FIGURE 4. Effect of acetate supplementation on n-butanol production by XUSEA-B strains during glucose and/or xylose fermentation. Fermentation was conducted with an initial OD of 0.2 and various concentrations of acetate (0, 1, or 2 g/L). (A) n-butanol production during glucose (pattern, blue) or xylose (solid, yellow) fermentation. (B) n-butanol production during glucose/xylose co-fermentation without (white) and with (yellow) acetate supplementation (0.2 g/L, 0.5 g/L, or 1 g/L). The error bars represent standard deviations obtained using biological triplicates.
Given the both positive and negative effects of acetate supplementation on n-butanol production, we introduced the n-butanol-production pathway into an acetate-tolerant glucose/xylose co-fermenting strain of XUSAE-57 (Ko et al., 2019). To promote the conversion of acetate into acetyl-CoA, a heterologous acetate-utilization pathway was additionally integrated by overexpressing a mutant version of ACSL641P from Salmonella enterica and AADH from E. coli, thereby generating XUSAEA-B. The XUSAEA-B, an acetate tolerant and utilizing strain, showed dramatically improved n-butanol production resulting in an n-butanol titer of 46.5 mg/L with acetate supplementation (1 g/L) during glucose/xylose fermentation at pH 6.5, under which condition the inhibitory effect of acetate has been shown to decrease (Figure 5).
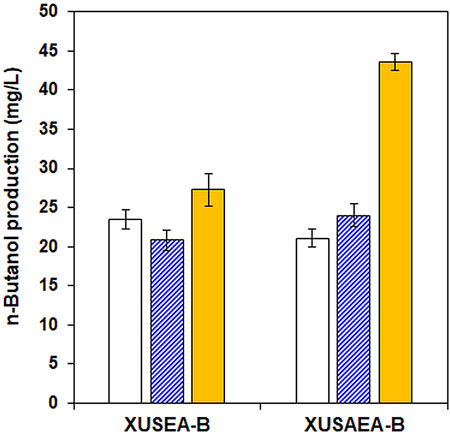
FIGURE 5. Comparison of n-butanol production by the XUSEA-B strain and the acetate-tolerant strain of XUSAEA-B during glucose/xylose co-fermentation. Fermentation was conducted using an initial OD of 0.2 with 0 g/L (solid, white) and 1 g/L (pattern, blue) of acetate supplementation at pH 5.0. n-butanol production at pH 6.5 was also evaluated with 1 g/L of acetate supplementation (solid, yellow). The error bars represent standard deviations obtained using biological triplicates.
Cellulosic n-Butanol Production Using Glucose/Xylose Co-fermenting S. cerevisiae
Encouraged by the increased n-butanol production during glucose/xylose co-fermentation in the presence of acetate, we evaluated the n-butanol-production performance of XUSAEA-B from lignocellulosic hydrolysates of Miscanthus sacchariflorus Goedae-Uksae, which were prepared through a H2SO4-catalyzed hydrothermal process (Figure 6). During a 21 h fermentation, XUSAEA-B completely utilized glucose and xylose, and produced 60.1 mg/L of n-butanol (Figures 6A,C). Interestingly, n-butanol production from lignocellulosic hydrolysates was 14% higher than that from YSC medium which was supplemented with same glucose and xylose concentrations as Micanthus hydrolysate (60.1 mg/L and 52.8 mg/L for lignocellulosic hydrolysates and YSC medium, respectively) (Figure 6C). To our knowledge, this study demonstrates n-butanol production from lignocellulosic hydrolysate by using a glucose/xylose co-fermenting S. cerevisiae for the first time whereas other previous studies mainly showed n-butanol production from synthetic glucose media (Lian et al., 2014; Sakuragi et al., 2015; Swidah et al., 2015; Schadeweg and Boles 2016a; Schadeweg and Boles, 2016b).
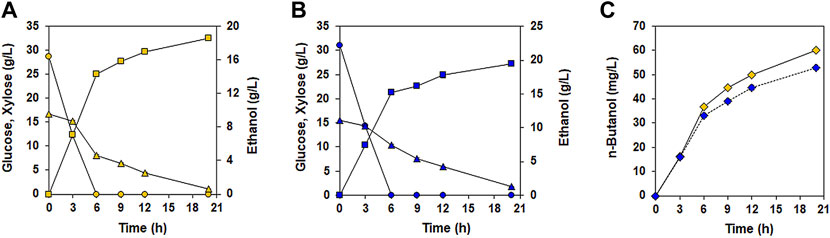
FIGURE 6. n-butanol fermentation by the XUSAEA-B strain using lignocellulosic hydrolysates. Fermentation was conducted with Miscanthus hydrolysates pretreated with diluted acids. Fermentation with YSC medium composed of the same concentration of sugars (30 g/L of glucose and 15 g/L of xylose) was also conducted under the same conditions. The initial OD and pH were 15 and 6.5, respectively. (A) Fermentation performance using lignocellulosic hydrolysates (yellow). (B) Fermentation performance using YSC medium (blue). (C) n-butanol production during fermentation using lignocellulosic hydrolysates (solid line, yellow diamond) and synthetic medium (dash line, blue diamond). Ethanol production (squares) and glucose (circles) and xylose (triangles) consumptions were measured during fermentation. The error bars represent standard deviations obtained using biological triplicates.
Discussion
Yeast engineering for lignocellulosic biorefinery has primarily focused on either the expansion of substrate ranges or product profiles through individual approaches. However, the product profiles of S. cerevisiae capable of xylose utilization are limited to only a few compounds, including isobutanol, carotene, and 1-hexadecanol (Brat and Boles, 2013; Guo et al., 2016; Sun et al., 2019). Recent reports highlighting the prospect of engineering a xylose-utilizing strain as a promising host for the production of acetyl-coA derived products (Kwak and Jin, 2017) offer easier diversion of product profiles from ethanol to other products, for a glucose/xylose co-utilizing S. cerevisiae.
In this study, we demonstrated the expansion of product profiles of a glucose/xylose co-fermenting S. cerevisiae to include the acetyl-coA derived product, n-butanol, which is a short chain alcohol similar to ethanol, however, has only been produced by S. cerevisiae from glucose with limited success. Despite the numerous attempts to develop n-butanol producing S. cerevisiae, the production titer remained suboptimal even with extensive metabolic engineering. There are several benefits associated with using a glucose/xylose co-fermenting strain for n-butanol production. For instance, it has a higher metabolic flux through acetyl-CoA compared to strains capable of only utilizing glucose (Kwak and Jin, 2017; Li et al., 2019). Additionally, it has an associated higher conversion yield per unit of biomass. These benefits were demonstrated in the current study. Specifically, the glucose/xylose co-fermenting S. cerevisiae strain, XUSEA-B, showed 3.9-fold higher n-butanol titer compared to the wild-type strain capable of only utilizing glucose during glucose/xylose co-fermentation. Given that the amount of xylose present was nearly half that of glucose, the additional n-butanol produced by XUSEA-B demonstrates a synergistic effect with an additional carbon source (xylose) in hydrolysates and an increased acetyl-CoA availability, supported by xylose catabolism in XUSEA-B.
In the study, we pointed out the increased acetyl-CoA pool of a glucose/xylose co-fermenting strain through analysis of the transcriptomic landscape in XUSEA-B, in which the increased metabolic flux was expected toward pyruvate, acetaldehyde, acetate, and cytosolic acetyl-CoA by re-directing the carbon flux through competing pathways such as the tricarboxylic acid (TCA) cycle, glyoxylate cycle, amino acid biosynthesis, and ethanol production (Figure 3, Table 1). The metabolic flux through pyruvate, specifically, could be directed toward acetaldehyde based on the highly upregulated expression of PDC6 accompanied by downregulation of genes associated with pyruvate carboxylation, and those involved in the TCA cycle, glyoxylate cycle and amino acid biosynthesis employing pyruvate as a precursor. Moreover, the fate of acetaldehyde through the ethanol-fermentation pathway also appeared to be redirected toward acetate formation, based on the decreased expression of alcohol dehydrogenases encoded by ADH4 and ADH5 and increased expression of aldehyde dehydrogenase encoded by ALD6 (Figure 3, Table 3) in XUSEA-B. Meanwhile, the high availability of the cytosolic acetyl-CoA contributed substantially to the improved n-butanol production in XUSEA-B during glucose and glucose/xylose fermentation, thus demonstrating the advantage of employing xylose-utilizing S. cerevisiae for the production of acetyl-CoA-derived biofuels and biochemicals from lignocellulosic biomass.
In XUSEA-B, the oxidative pentose phosphate pathway, a major route for NADPH production (Stincone et al., 2015), appeared to be actively regulated via upregulation of ZWF1, SOL3, and GND1, possibly supporting the notion that improved NADPH availability improves metabolic flux through the NADPH-dependent rate-limiting step in the n-butanol production pathway. Indeed, the n-butanol pathway in the glucose/xylose co-fermenting strain, containing an NADPH-preferring cytoETR1 protein, was better supported by the sufficient NADPH cofactor present in the XUSEA-B strain.
In terms of glycerol-metabolism, expression of the glycerol-1-phosphatase gene (GPP1), which is important for glycerol biosynthesis, decreased (Figure 3, Table 3), whereas transcription of the glycerol dehydrogenase gene (GCY1), which participates in glycerol catabolism under micro-aerobic conditions, was increased. Similarly, suppressed glycerol synthesis combined with disruption of the ethanol-fermentation pathway reportedly stimulates n-butanol production (Lian et al., 2014; Swidah et al., 2015; Schadeweg and Boles, 2016b) by increasing the abundance of NADH, a driving force of the n-butanol pathway (Kim et al., 2015; Schadeweg and Boles, 2016a). Furthermore, the upregulation of genes related to cofactor regeneration, such as OYE3, SER33, and YEF1, suggest that XUSEA-B maintained an optimal balance by varying redox cofactors, not only for efficient n-butanol production. Understanding the transcriptomic characteristics of XUSEA-B strain not only help to explain the improved butanol production during glucose/xylose fermentation, but also could offer engineering strategies to further improve the butanol titer in this minimally engineered strain of XUSEA-B. These engineering targets could also be used for rewiring biosynthetic routes to produce acetyl-CoA-derived chemicals in the context of lignocellulosic biorefinery.
Previously, acetate supplementation increased n-butanol production in a native producer of Clostridium sp. by upregulating the expression levels of CoA-transferase genes, thereby increasing the availability of acetyl-CoA and butyryl-CoA, two main precursors of the n-butanol synthesis pathway (Chen and Blaschek, 1999). During lignocellulosic fermentation by S. cerevisiae, however, acetate is regarded as a major inhibitory compound impeding cell growth and sugar utilization rate, and thus limits the fermentation performance of S. cerevisiae (Helle et al., 2003; Jönsson et al., 2013), particularly during xylose fermentation (Ko et al., 2015). S. cerevisiae endogenously expresses ACS1 and ACS2, which encode acetyl-coA synthetases to convert acetate into acetyl-CoA using ATP (van den Berg et al., 1996). Therefore, acetate supplementation could be expected to support additional carbon flux through the n-butanol biosynthetic pathway by enhancing acetyl-CoA, a crucial precursor for n-butanol production. As expected, in the current study n-butanol production increased during glucose fermentation in the presence of acetate. This could be explained due to the increased acetyl-CoA pool through the conversion of acetate by native ACS genes. The sufficient intracellular ATP supplied from glucose could also have alleviated the inhibitory effect of acetate (Zhang et al., 2016). During xylose fermentation, however, acetate supplementation did not positively affect n-butanol production, possibly due to insufficient detoxification and limited ATP generation by xylose (Casey et al., 2010). In fact, increased acetate concentration caused a decrease in the n-butanol titer during xylose fermentation (Figure 4A). Several strategies have been proposed to overcome the toxic effects of acetic acid while maintaining fermentation performance in S. cerevisiae. Ko et al. improved acetate tolerance of a xylose fermenting S. cerevisiae through adaptive laboratory evolution (Ko et al., 2019). Zhang et al. have coupled an acetate reduction pathway with xylose catabolism during cellulosic fermentation to improve sugar-to-product conversion yield while converting acetate into less inhibitory products (Zhang et al., 2016). When an acetate-tolerant glucose/xylose co-fermenting strain harboring an acetate conversion pathway, XUSAEA-B, was used, the beneficial effects of acetate supplementation were recovered resulting in significantly improved n-butanol production (Figure 5). Moreover, during lignocellulosic fermentation, XUSAEA-B produced 60.1 mg/L of n-butanol, accounting for a 14% higher yield compared to that obtained during fermentation using synthetic media with the same concentrations of sugars (52.8 mg/L), possibly due to the positive effect of acetate on elevating acetyl-CoA for the n-butanol pathway in lignocellulosic hydrolysates.
Conclusion
In this study, we investigated the production of the acetyl-coA-derived product, n-butanol, by a glucose/xylose co-fermenting S. cerevisiae strain. The transcription profiles of S. cerevisiae with efficient xylose catabolism revealed a modified cellular network that better supported the generation of acetyl-CoA and cofactors required for n-butanol fermentation. Incorporating acetate catabolism further improved n-butanol production from lignocellulosic hydrolysates. Consequently, the results of this study show the potential of using a glucose/xylose co-fermenting strain and lignocellulosic biomass as more attractive production host and resource for biorefinery.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Authors Contributions
YL: investigation, writing—original draft, writing—review and editing. PH: writing—original draft, writing—review and editing. JK: supervision, writing—review and editing. GG, YU, and SH: formal analysis, writing—review and editing. S-ML: conceptualization, formal analysis, writing—review and editing.
Funding
This research was supported by the Korea Institute of Science and Technology (KIST) Institutional Program (grant number. 2E31853) and the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (Information and Communication Technology) (grant number. NRF-2020M1A2A2080847).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.826787/full#supplementary-material
References
Brat, D., and Boles, E. (2013). Isobutanol Production Fromd-Xylose by recombinant Saccharomyces cerevisiae. FEMS Yeast Res. 13, 241–244. doi:10.1111/1567-1364.12028
Casey, E., Sedlak, M., Ho, N. W. Y., and Mosier, N. S. (2010). Effect of Acetic Acid and pH on the Cofermentation of Glucose and Xylose to Ethanol by a Genetically Engineered Strain of Saccharomyces cerevisiae. FEMS Yeast Res. 10, 385–393. doi:10.1111/j.1567-1364.2010.00623.x
Chen, C.-K., and Blaschek, H. P. (1999). Effect of Acetate on Molecular and Physiological Aspects of Clostridium Beijerinckii NCIMB 8052 Solvent Production and Strain Degeneration. Appl. Environ. Microbiol. 65, 499–505. doi:10.1128/AEM.65.2.499-505.1999
Dellomonaco, C., Clomburg, J. M., Miller, E. N., and Gonzalez, R. (2011). Engineered Reversal of the β-oxidation Cycle for the Synthesis of Fuels and Chemicals. Nature 476, 355–359. doi:10.1038/nature10333
Ekas, H., Deaner, M., and Alper, H. S. (2019). Recent Advancements in Fungal-Derived Fuel and Chemical Production and Commercialization. Curr. Opin. Biotechnol. 57, 1–9. doi:10.1016/j.copbio.2018.08.014
González-Ramos, D., van den Broek, M., van Maris, A. J., Pronk, J. T., and Daran, J.-M. G. (2013). Genome-scale Analyses of Butanol Tolerance in Saccharomyces cerevisiae Reveal an Essential Role of Protein Degradation. Biotechnol. Biofuels 6, 48. doi:10.1186/1754-6834-6-48
Guo, W., Sheng, J., Zhao, H., and Feng, X. (2016). Metabolic Engineering of Saccharomyces cerevisiae to Produce 1-hexadecanol from Xylose. Microb. Cel Fact 15, 24. doi:10.1186/s12934-016-0423-9
Helle, S., Cameron, D., Lam, J., White, B., and Duff, S. (2003). Effect of Inhibitory Compounds Found in Biomass Hydrolysates on Growth and Xylose Fermentation by a Genetically Engineered Strain of S. cerevisiae. Enzyme Microb. Technol. 33, 786–792. doi:10.1016/S0141-0229(03)00214-X
Hoang Nguyen Tran, P., Ko, J. K., Gong, G., Um, Y., and Lee, S.-M. (2020). Improved Simultaneous Co-fermentation of Glucose and Xylose by Saccharomyces cerevisiae for Efficient Lignocellulosic Biorefinery. Biotechnol. Biofuels 13, 12. doi:10.1186/s13068-019-1641-2
Hong, K.-K., and Nielsen, J. (2012). Metabolic Engineering of Saccharomyces cerevisiae: a Key Cell Factory Platform for Future Biorefineries. Cell. Mol. Life Sci. 69, 2671–2690. doi:10.1007/s00018-012-0945-1
Jönsson, L. J., Alriksson, B., and Nilvebrant, N.-O. (2013). Bioconversion of Lignocellulose: Inhibitors and Detoxification. Biotechnol. Biofuels 6, 16. doi:10.1186/1754-6834-6-16
Kim, S., and Hahn, J.-S. (2015). Efficient Production of 2,3-butanediol in Saccharomyces cerevisiae by Eliminating Ethanol and Glycerol Production and Redox Rebalancing. Metab. Eng. 31, 94–101. doi:10.1016/j.ymben.2015.07.006
Knoshaug, E. P., and Zhang, M. (2009). Butanol Tolerance in a Selection of Microorganisms. Appl. Biochem. Biotechnol. 153, 13–20. doi:10.1007/s12010-008-8460-4
Ko, J. K., Enkh‐Amgalan, T., Gong, G., Um, Y., and Lee, S. M. (2019). Improved Bioconversion of Lignocellulosic Biomass bySaccharomyces Cerevisiaeengineered for Tolerance to Acetic Acid. GCB Bioenergy 12, 90–100. doi:10.1111/gcbb.12656
Ko, J. K., Jung, J. H., Altpeter, F., Kannan, B., Kim, H. E., Kim, K. H., et al. (2018). Largely Enhanced Bioethanol Production through the Combined Use of Lignin-Modified Sugarcane and Xylose Fermenting Yeast Strain. Bioresour. Technol. 256, 312–320. doi:10.1016/j.biortech.2018.01.123
Ko, J. K., and Lee, S.-M. (2018). Advances in Cellulosic Conversion to Fuels: Engineering Yeasts for Cellulosic Bioethanol and Biodiesel Production. Curr. Opin. Biotechnol. 50, 72–80. doi:10.1016/j.copbio.2017.11.007
Ko, J. K., Um, Y., Park, Y.-C., Seo, J.-H., and Kim, K. H. (2015). Compounds Inhibiting the Bioconversion of Hydrothermally Pretreated Lignocellulose. Appl. Microbiol. Biotechnol. 99, 4201–4212. doi:10.1007/s00253-015-6595-0
Kwak, S., and Jin, Y.-S. (2017). Production of Fuels and Chemicals from Xylose by Engineered Saccharomyces cerevisiae: a Review and Perspective. Microb. Cel Fact 16, 82. doi:10.1186/s12934-017-0694-9
Li, X., Chen, Y., and Nielsen, J. (2019). Harnessing Xylose Pathways for Biofuels Production. Curr. Opin. Biotechnol. 57, 56–65. doi:10.1016/j.copbio.2019.01.006
Lian, J., Si, T., Nair, N. U., and Zhao, H. (2014). Design and Construction of Acetyl-CoA Overproducing Saccharomyces cerevisiae Strains. Metab. Eng. 24, 139–149. doi:10.1016/j.ymben.2014.05.010
Lian, J., and Zhao, H. (2015). Reversal of the β-Oxidation Cycle in Saccharomyces cerevisiae for Production of Fuels and Chemicals. ACS Synth. Biol. 4, 332–341. doi:10.1021/sb500243c
Sakuragi, H., Morisaka, H., Kuroda, K., and Ueda, M. (2015). Enhanced Butanol Production by Eukaryotic Saccharomyces cerevisiae Engineered to Contain an Improved Pathway. Biosci. Biotechnol. Biochem. 79, 314–320. doi:10.1080/09168451.2014.972330
Schadeweg, V., and Boles, E. (2016a). Increasing N-Butanol Production with Saccharomyces cerevisiae by Optimizing Acetyl-CoA Synthesis, NADH Levels and Trans-2-enoyl-CoA Reductase Expression. Biotechnol. Biofuels 9, 257. doi:10.1186/s13068-016-0673-0
Schadeweg, V., and Boles, E. (2016b). n-Butanol Production in Saccharomyces cerevisiae Is Limited by the Availability of Coenzyme A and Cytosolic Acetyl-CoA. Biotechnol. Biofuels 9, 44. doi:10.1186/s13068-016-0456-7
Stincone, A., Prigione, A., Cramer, T., Wamelink, M. M. C., Campbell, K., Cheung, E., et al. (2015). The Return of Metabolism: Biochemistry and Physiology of the Pentose Phosphate Pathway. Biol. Rev. 90, 927–963. doi:10.1111/brv.12140
Sun, L., and Alper, H. S. (2020). Non-conventional Hosts for the Production of Fuels and Chemicals. Curr. Opin. Chem. Biol. 59, 15–22. doi:10.1016/j.cbpa.2020.03.004
Sun, L., Kwak, S., and Jin, Y.-S. (2019). Vitamin A Production by Engineered Saccharomyces cerevisiae from Xylose via Two-phase In Situ Extraction. ACS Synth. Biol. 8, 2131–2140. doi:10.1021/acssynbio.9b00217
Swidah, R., Wang, H., Reid, P. J., Ahmed, H. Z., Pisanelli, A. M., Persaud, K. C., et al. (2015). Butanol Production in S. cerevisiae via a Synthetic ABE Pathway Is Enhanced by Specific Metabolic Engineering and Butanol Resistance. Biotechnol. Biofuels 8, 97. doi:10.1186/s13068-015-0281-4
Tran Nguyen Hoang, P., Ko, J. K., Gong, G., Um, Y., and Lee, S.-M. (2018). Genomic and Phenotypic Characterization of a Refactored Xylose-Utilizing Saccharomyces cerevisiae Strain for Lignocellulosic Biofuel Production. Biotechnol. Biofuels 11, 268. doi:10.1186/s13068-018-1269-7
van den Berg, M. A., de Jong-Gubbels, P., Kortland, C. J., van Dijken, J. P., Pronk, J. T., Steensma, H. Y., et al. (1996). The Two Acetyl-Coenzyme A Synthetases of Saccharomyces cerevisiae Differ with Respect to Kinetic Properties and Transcriptional Regulation. J. Biol. Chem. 71, 28953–28959. doi:10.1074/jbc.271.46.28953
Keywords: Saccharomyces cerevisiae, glucose/xylose co-fermentation, n-butanol, acetyl-CoA, acetate, lignocellulosic biomass
Citation: Lee Y-J, Hoang Nguyen Tran P, Ko JK, Gong G, Um Y, Han SO and Lee S-M (2022) Glucose/Xylose Co-Fermenting Saccharomyces cerevisiae Increases the Production of Acetyl-CoA Derived n-Butanol From Lignocellulosic Biomass. Front. Bioeng. Biotechnol. 10:826787. doi: 10.3389/fbioe.2022.826787
Received: 01 December 2021; Accepted: 27 January 2022;
Published: 16 February 2022.
Edited by:
Wenming Zhang, Nanjing Tech University, ChinaReviewed by:
Mingfeng Cao, Xiamen University, ChinaSoo Rin Kim, Kyungpook National University, South Korea
Copyright © 2022 Lee, Hoang Nguyen Tran, Ko, Gong, Um, Han and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sun-Mi Lee, c21sZWVAa2lzdC5yZS5rcg==
†Present address: Yeon-Jung Lee, Seegene Institute of Life Sciences, Seoul, South Korea
‡These authors have contributed equally to this work and share first authorship
 Yeon-Jung Lee
Yeon-Jung Lee Phuong Hoang Nguyen Tran1,3‡
Phuong Hoang Nguyen Tran1,3‡ Ja Kyong Ko
Ja Kyong Ko Youngsoon Um
Youngsoon Um Sun-Mi Lee
Sun-Mi Lee