- 1Department of Orthopaedics, Xijing Hospital, The Fourth Military Medical University, Xi’an, China
- 2Department of Orthopaedics, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
- 3Tangdu Hospital, The Fourth Military Medical University, Xi’an, China
Excessive chondrocyte degeneration and inflammation are the pathological features of osteoarthritis (OA), and altered miR-212-5p may contribute to meniscus and cartilage degeneration. Whether exosomes derived from miR-212-5p overexpressed synovial mesenchymal stem cells (SMSC-212-5p-Exos) could be utilized to treat degenerative chondrocytes is investigated in this study. Down-regulated miR-212-5p and up-regulated E74 Like ETS Transcription Factor 3 (ELF3) expression were detected in OA synovial tissues, which showed a negative correlation (r = −0.55, p = 0.002). miR-212-5p directly targeted ELF3 and regulated the relative expression of ELF3 in SMSCs as indicated by luciferase reporter assay and RT-PCR. The relative expression of ELF3, chondrocyte degeneration-related molecules, matrix metalloproteinase, and inflammatory molecules were detected in chondrocytes stimulated with interleukin (IL)-1β or co-incubated with SMSC-212-5p-Exos or SMSCs-derived exosomes (SMSC-Exos). IL-1β induced up-regulation of ELF3, down-regulation of degeneration molecules (Collagen II, Aggrecan, and Sox9), up-regulation of matrix metalloproteinase (MMP-1, MMP-3, and MMP-13), and up-regulation of inflammatory molecules (IL-6, MCP-1, TNF-α, COX-2, and iNOS) could be inhibited by SMSC-212-5p-Exos or SMSC-Exos administration. When compared with the SMSC-Exos, SMSC-212-5p-Exos showed more treatment benefits. All of these indicate that SMSC-212-5p-Exos could suppress chondrocyte degeneration and inflammation by targeting ELF3, which can be considered as a disease-modifying strategy.
Introduction
Osteoarthritis (OA) is a disease that comprises progressive cartilage degenerative and synovial membrane inflammation, having an increased frequency of osteophyte formation and subchondral bone sclerosis in the aging population (Hunter and Bierma-Zeinstra, 2019; Katz et al., 2021; Sharma, 2021). No curative treatment has been applied in the clinic, and joint replacement remains the most commonly applied and effective therapy for the disability. Chondrocyte degeneration and inflammation are the principal cause of OA, for chondrocytes are the only residents in the avascular articular cartilage to maintain the specialized structure. It is also reported that chondrocytes size could be potentially used to assess disease progression (Gratal et al., 2019). Therefore, the molecular mechanism underlying chondrocyte degeneration and inflammation is vital for OA treatment.
Exosomes are small size (30–100 nm), single-membrane organelles released by various cells to shunt loading bioactive molecules to target cells (Kalluri and LeBleu, 2020; O'Brien et al., 2020). Research has shown that exosomes derived from synovial mesenchymal stem cells (SMSC-Exos) have the potential to palliate the severity of interleukin (IL)-1β induced osteoarthritis (Wang et al., 2020; Qiu et al., 2021). Further research is urgent to confirm the effectiveness and feasibility of exosome-based therapy.
MicroRNAs (miRNAs) can post-transcriptionally regulate OA-associated genes in chondrocytes (Oliviero et al., 2019; Huang et al., 2021). Exosomal miRNAs differ significantly from those of the parent cell, and following uptake, exosomal miRNAs can mediate target gene repression and reprogramme the cellular response (Heidarzadeh et al., 2021; Moghadasi et al., 2021). In rheumatoid arthritis, miR-212-3p can reduce proliferation and promote apoptosis of fibroblast-like synoviocytes (Liu et al., 2018). On the other hand, altered miR-212-5p expression can lead to the meniscus and cartilage degenerative process in OA (Jiang et al., 2021). All of these indicate the treatment benefit of miR-212-5p delivery in OA and whether exosomes derived from miR-212-5p overexpressed SMSCs (SMSC-212-5p-Exos) could be utilized to treat OA is investigated in this study.
Methods and Materials
Patient Enrollment
Based on the criteria of the American College of Rheumatology, thirty OA patients (18 females and 12 males; 50–74 years old; mean age of 62.3 ± 6.4 years) underwent total knee arthroplasty were enrolled. Meanwhile, the synovial tissues were collected from OA patients and 20 donors with accidental deaths (excluding those with OA-related diseases) or normal knee-joint synovium who were subjected to lower limb amputation after acute trauma or open reduction and internal fixation after fracturing of the tibial plateau (13 females and 7 males; 50–75 years old; mean age of 61.87 ± 7.44 years). The study was approved by the Xijing Hospital, and informed written consent was derived from each participant or close relatives.
Synovial Mesenchymal Stem Cells Differentiation and Transfection
Synovial membrane specimens were digested with 0.2% type I collagenase (Thermo Fisher) at 37°C overnight, which were further collected by centrifugation and seeded in a high-glucose DMEM medium (Thermo Fisher, Waltham, MA, United States) supplemented with 10% FBS for 4 days to allow cell attachment. The medium was refreshed every 3 days, and at day 14, SMSCs were obtained. After blocking with human BD Fc Block™, SMSCs were stained with the following antibodies (Becton Dickinson): anti-CD34, anti-CD44, anti-CD45, anti-Sca-1, and anti-CD105 antibodies to confirm the phenotype using Guava® easyCyte™ flow cytometer (Merck-Millipore, Billerica, MA, United States). At passage 3, SMSCs were switched to osteogenic differentiation medium (Sigma-Aldrich, St. Louis, MO, United States) for 2 weeks or StemPro Adipogenesis Differentiation Kit (Gibco) for 4 weeks. The miR-212-5p mimic or mimic-negative control (NC) (Sigma-Aldrich) were transfected with Lipofectamine® 3,000 (Thermo Fisher) at the concentration of 100 nM according to the manufacturer’s instructions. The following sequences were used: miR-212-5p mimic (sense, 5′-ACCUUGGCUCUAGACUGCUUACU-3'; and antisense, 5′-UAAGCAGUCUAGAGCCAAGGUUU-3′), mimic-negative control miRNA (sense, 5′-UUCUCCGAACGUGUCACGUTT-3'; and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′).
Exosomes Isolation and Characterization
Conditioned medium of SMSCs or miR-212-5p overexpressing SMSCs was filtered with 0.22 μM filters (Merck-Millipore) and centrifuged (4,000×g) to concentrate the volume into approximately 200 μL, which was further ultracentrifuged (100,000×g, 1 h, 4°C) to obtain the exosomal particles. A transmission electron microscope (TEM) was utilized to observe the morphology of exosomes, and the size and distribution of exosomes were measured using dynamic light scattering (DLS) analysis.
Collection and Culture of Primary Chondrocyte
Primary chondrocytes were detached from cartilages dissected from the subchondral bone with 0.25 mg/ml collagenase P and 4 mg/ml protease, and the isolated chondrocytes (1×107) were seeded into the 6-well plates 1 day before treatment. When cell confluence reached 80%, 2 µg exosomes (EXO miR-NC and EXO-miR-212-5p) were introduced into the chondrocytes culture medium supplemented with 10 ng/ml IL-1β. Chondrocytes treated with PBS were regarded as the blank controls. After 48 h of treatment, cells were collected for subsequent use.
RNA Isolation and Quantitation
TRIzol (Invitrogen) was utilized to extract total RNA from chondrocytes or SMSCs. Total Exosome RNA & Protein Isolation Kit (Invitrogen, Waltham, MA, United States) was used to extract RNA and protein from exosomes for further analysis. TaqMan™ Advanced miRNA cDNA Synthesis Kit (Thermofisher) was utilized to reverse-transcript miRNA into cDNA, and RNU6B was utilized as an internal reference. Transscript® All-in-One-First-Strand cDNA synthesis supermix was utilized to reverse-transcript mRNA into cDNA, and GAPDH was utilized as an internal reference. SYBR Green Master Mix (Roche, Penzberg, Upper Bavaria, Germany) was applied to detect the amplification. The primers were listed as following: ELF3, forward 5′- CATGACCTACGAGAAGCTGAGC-3′, reverse 5′- GACTCTGGAGAACCTCTTCCTC-3’; miR-212-5p, forward 5′- CAGTCTCCAGTCACGG-3′, reverse 5′- GAACATGTCTGCGTATCTC-3’; Collagen II (COL2A1), forward 5′- CCTGGCAAAGATGGTGAGACAG-3′, reverse 5′- CCTGGTTTTCCACCTTCACCTG-3’; Aggrecan, forward 5′-CAGGCTATGAGCAGTGTGATGC-3′, reverse 5′- GCTGCTGTCTTTGTCACCCACA-3’; Sox9, forward 5′- AGGAAGCTCGCGGACCAGTAC-3′, reverse 5′- GGTGGTCCTTCTTGTGCTGCAC-3’; MMP-1, forward 5′- ATGAAGCAGCCCAGATGTGGAG -3′, 5′- TGGTCCACATCTGCTCTTGGCA -3’; MMP-3, forward 5′- CACTCACAGACCTGACTCGGTT-3′, reverse 5′- AAGCAGGATCACAGTTGGCTGG-3’; MMP-13, forward 5′- CCTTGATGCCATTACCAGTCTCC-3′, reverse 5′- AAACAGCTCCGCATCAACCTGC-3’; IL-6, forward 5′ AGACAGCCACTCACCTCTTCAG-3′, reverse 5′- TTCTGCCAGTGCCTCTTTGCTG-3; COX-2, forward 5′- CGGTGAAACTCTGGCTAGACAG-3′, reverse 5′- GCAAACCGTAGATGCTCAGGGA-3’; iNOS, forward 5′- GCTCTACACCTCCAATGTGACC-3′, reverse 5′- CTGCCGAGATTTGAGCCTCATG-3’; GAPDH, forward 5′- CTGTGCCGTTGAATTTGCCG-3′, forward 5′- CGGGTTCCTATAAATACGGACTG-3’.
Luciferase Assay
The QuikChange Lightning Site-Directed Mutagenesis kit (Agilent, Santa Clara, CA, United States) was utilized to mutate the binding site of ELF3 with miR-212-5p. 3′-UTR fragment of ELF3 mRNA (mutant or wide type) was subcloned into pGL3 luciferase vector (Promega, Madison, WI, United States), which was further co-transfected with miR-212-5p mimic or mimic-NC into HEK-293T cells for 36 h with Lipofectamine 3,000. Subsequently, the luciferase activity was assayed with Luciferase Assay System (Promega).
Immunoblot Assay
The cellular lysates were quantified using the BCA protein concentration kit (20201ES76, Yeasen Company, Shanghai, China), and 20 µg lysates were separated with 10% SDS-PAGE electrophoresis and transferred to PVDF membranes, which were further blocked in Tris-buffered saline with Tween 20 (TBST) containing 0.1% Tween 20 and 5% skimmed milk powder and followed by incubation with the primary antibodies (Santa Cruz, Dallas, TX, United States) specific for CD63, CD9, Alix, ELF3, and GAPDH, and then incubated in peroxidase-conjugated secondary antibody (Sigma-Aldrich) at room temperature for 1 h. The dilution used for the primary antibodies against CD63, CD9, Alix, and ELF3 was 1:1,000, the dilution used for the primary antibody against GAPDH was 1:2000, and the dilution used for the second antibody was 1:2000. The signal was developed with an ECL system (GE Healthcare), and the relative intensity was normalized with GAPDH expression with NIH-Image J1.51p 22.
Elisa
According to the manufacturer’s instructions, the concentrations of IL-6, MCP-1, MMP-3, MMP-13, and TNF-α were detected with commercial ELISA kits (eBiosciences, San Diego, CA, United States). Separated stock standard solutions and samples were measured with a SpectraMax M5 microplate reader at a wavelength of 450 nm.
Statistical Analysis
Statistical analysis were performed with Graph-Pad Prism 6.0. Data were presented as means ± SD. The Mann-Whitney test was performed to determine the statistical significance between any two groups, and Dunn’s multiple comparisons test was used to compare multiple groups. p-value < 0.05 was considered statistically significant.
Results
Down-Regulated miR-212-5p and Up-Regulated ELF3 Expression in OA Synovial Tissues
The relative miR-212-5p expression in OA synovial tissues was down-regulated compared with healthy control (Figure 1A, p < 0.001), and up-regulated ELF3 mRNA expression (Figure 1B, p < 0.001) and increased protein expression of IL-1β (Figure 1C, p < 0.001) were also observed. The expression correlation analysis indicated that both the correlation between ELF3 and miR-212-5p (Figure 1D, r = −0.55, p = 0.002) and the correlation between IL-1β and miR-212-5p (Figure 1E, r = −0.47, p = 0.009) were significantly negatively correlated. As expected, the correlation between ELF3 and IL-1β was positively correlated (Figure 1F, r = 0.41, p = 0.024).
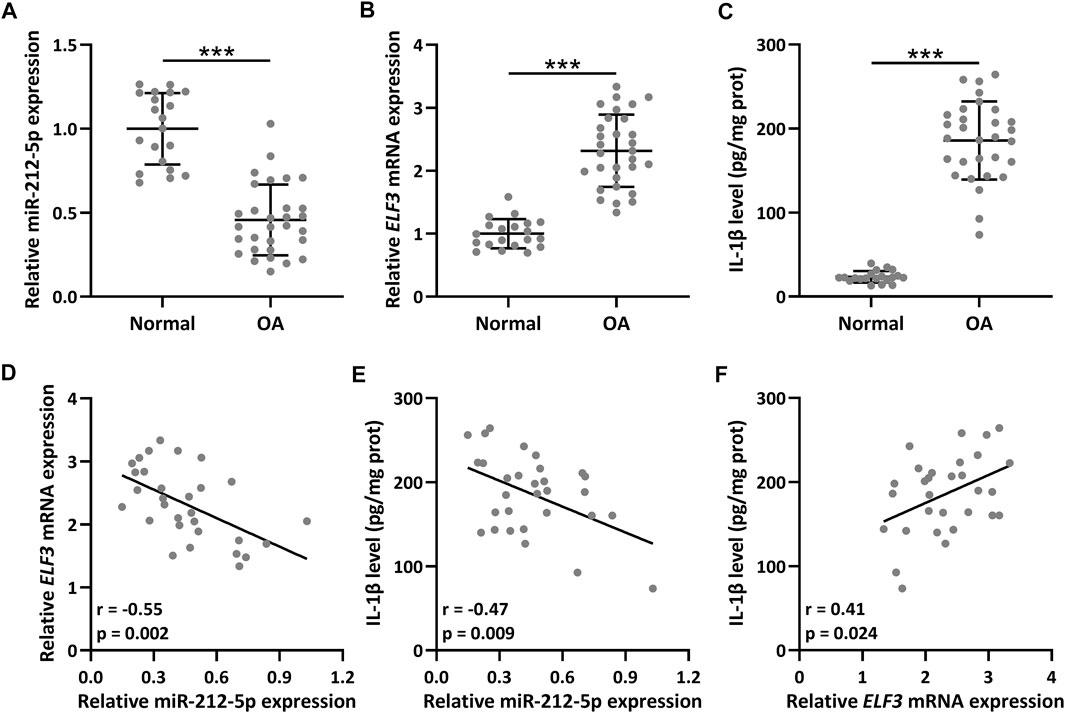
FIGURE 1. The expression of ELF3 and miR-212-5p in OA (n = 30) and normal synovial tissues (n = 20). qRT-PCR was utilized to detect the relative expressions of miR-212-5p (A) and ELF3 (B). ELISA was used to measure the IL-1β level (C). Pearson correlation coefficient of miR-212-5p and ELF3 mRNA (D), miR-212-5p and IL-1β level (E), ELF3 mRNA and IL-1β level (F) in OA synovial tissues. n = 30. Mean ± SD. ***p < 0.001.
SMSC-Derived Exosomes Isolation and Confirmation
Spindle-like SMSCs were observed at passage 3 (P3) (Figure 2A), which could be induced into osteogenic cells confirmed by Alizarin Red S staining of calcium mineral deposits (Figure 2B) and induced into adipogenic cells confirmed by Oil Red O staining of small cytoplasmic lipid droplets (Figure 2C). Flow cytometry analysis confirmed the phenotype of SMSCs, which were positive for CD105, CD44, and SCA-1, and negative for CD34 and CD45 (Figure 2D). TEM showed the single membrane structure of SMSC-Exos (Figure 2E), and dynamic light scattering assayed that most exosomes ranged from 30 to 150 nm in size (Figure 2F). The relative protein expression of exosome markers (CD63, CD9, and Alix) was significantly up-regulated as detected by Western blot (Figure 2G). All these results confirmed the success of SMSC-Exos isolation.
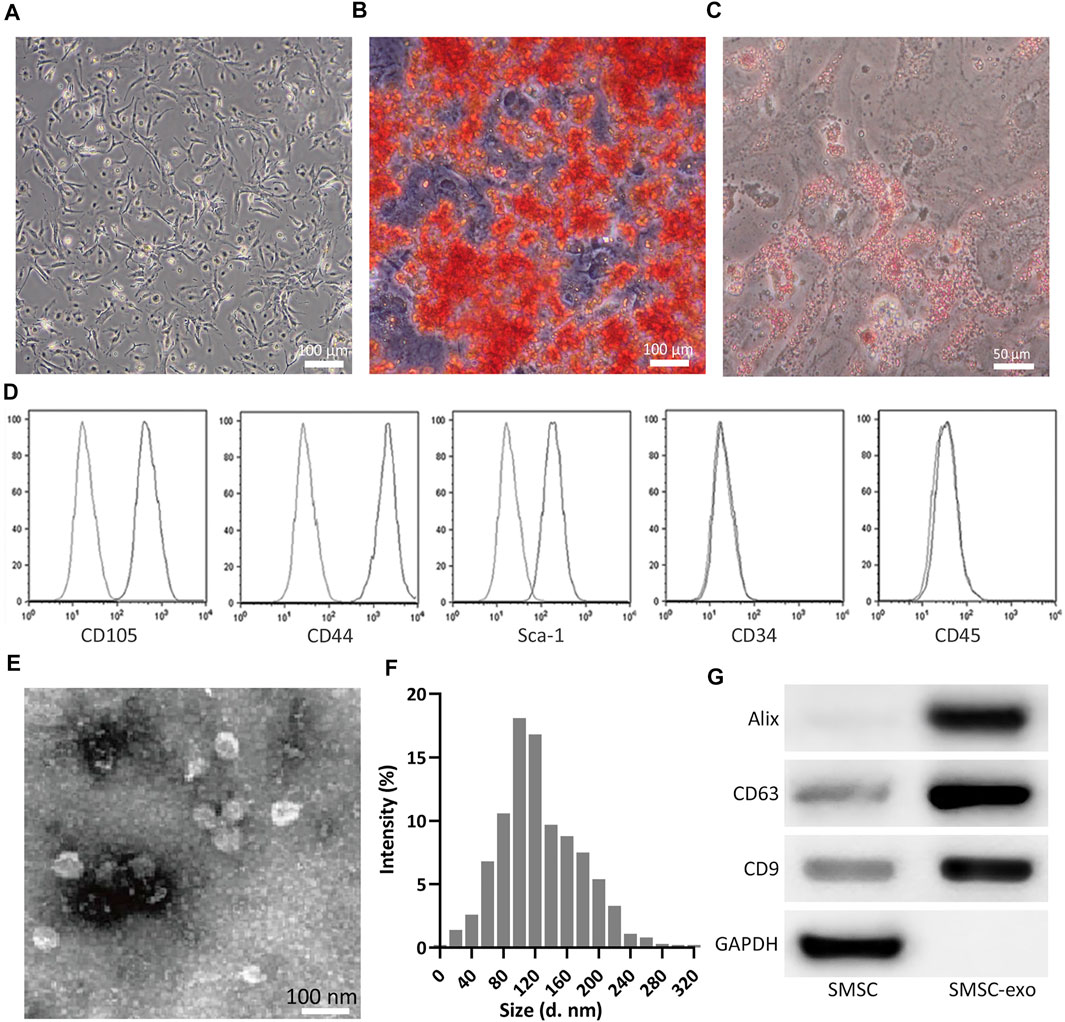
FIGURE 2. Isolation of SMSCs and SMSC-derived exosomes (SMSC-Exos). (A) Observation of SMSCs morphology at passage 3. (B) After 14 days of osteogenic differentiation, matrix mineralization of SMSCs was stained with Alizarin Red S. (C) after 14 days of adipogenesis induction, SMSCs were stained with Oil Red O. (D) Fluorescence Activated Cell Sorting was performed on SMSCs for CD105, CD44, Sca-1, CD34, and CD45 at passage 3. (E) Identification of exosomes by TEM. (F) Nanoparticle Tracking Analysis of the isolated exosome. (G) Detection of CD9, CD63, and Alix expression by Western blot analysis between SMSCs and SMSC-derived exosomes (SMSC-Exos).
miR-212-5p Directly Targets ELF3 in SMSCs
TargetScan (Agarwal et al., 2015) and miRBase (Ambros et al., 2003) database were utilized to predict the possible targets of miR-212-5p. ELF3 was among the top 10 molecules that could be binding with miR-212-5p. 3′-UTR region of ELF3, containing the predicted miR-212-5p binding sequences (Figure 3A), was cloned into the luciferase vector. MiR-212-5p overexpression could significantly decrease the luciferase activity of ELF3 (Figure 3B). In addition, up-regulation of miR-212-5p was observed in miR-212-5p overexpressed SMSCs (Figure 3C), which could diminish the relative mRNA (Figure 3D) and protein (Figures 3E,F) expression of ELF3. These results indicated that miR-212-5p directly targeted ELF3 and regulated the expression of ELF3 in SMSCs.
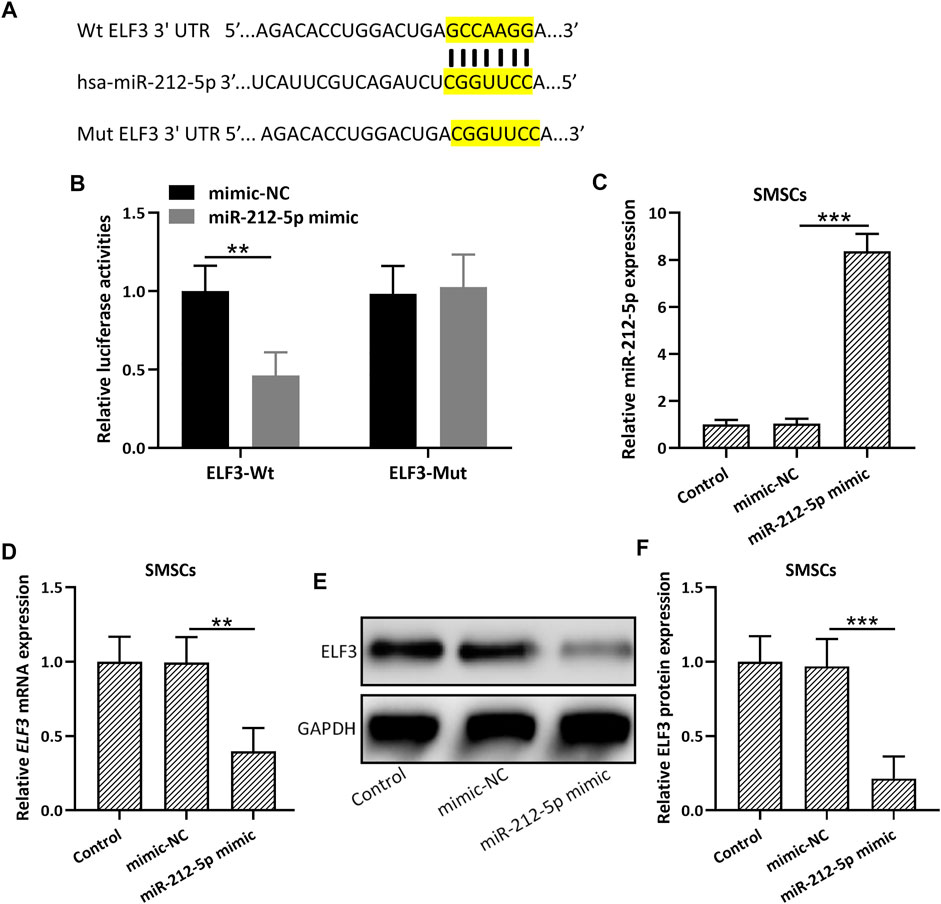
FIGURE 3. miR-212-5p targeted ELF3 and regulated the expressions of ELF3 in SMSCs. (A) The predicted binding sequences of miR-212-5p with ELF3 mRNA were shown. (B) HEK-293T cells were co-transfected with luciferase reporters containing ELF3 3′-UTR (wide type or mutant) and miR-212-5p mimics or negative control. After 48 h of incubation, the relative luciferase activity was measured. n = 3. Mean ± SD. **p < 0.01, Two-way ANOVA followed Turkey’s multiple comparisons test. (C) qRT-PCR was used to assay the expressions of miR-212-5p in SMSCs and miR-212-5p overexpressed SMSCs. (D–F), SMSCs were transfected with miR-212-5p mimics or negative control for 48 h qRT-PCR and Western blotting were used to analyze the mRNA and proteins expressions of ELF3. N = 3 for each group. Mean ± SD. **p < 0.01, ***p < 0.001, One-way ANOVA followed Dunn’s multiple comparisons test.
Exosomes Derived From miR-212-5p Overexpressed SMSCs (SMSC-212-5p-Exos) Inhibit IL-1β Induced ELF3 Expression in Chondrocytes
Up-regulated miR-212-5p expression could be detected in SMSC-212-5p-Exos (Figure 4A), indicating that SMSCs exosomes can carry miR-212-5p to target cells. SMSC-212-5p-Exos showed no difference in size and membrane structure when compared with SMSC-Exos (data not shown). Different concentrations of IL-1β were used to stimulate chondrocytes, and it was found that decreased cell viability was observed in accordance with the increased IL-1β concentration (Figure 4B). SMSC-Exos could reverse the diminished cell viability induced by IL-1β in chondrocytes, and SMSC-212-5p-Exos showed more treatment benefit when compared with SMSC-Exos (Figure 4C). On the other hand, IL-1β incubated chondrocytes showed up-regulated ELF3 expression in both transcription (Figure 4D) and protein levels (Figures 4E,F), which can be reversed by the co-incubation with SMSC-Exos or SMSC-212-5p-Exos, while SMSC-212-5p-Exos showed the better treatment benefit. All of these results indicated that exosomal miR-212-5p inhibited up-regulated ELF3 expression in chondrocytes induced by IL-1β.
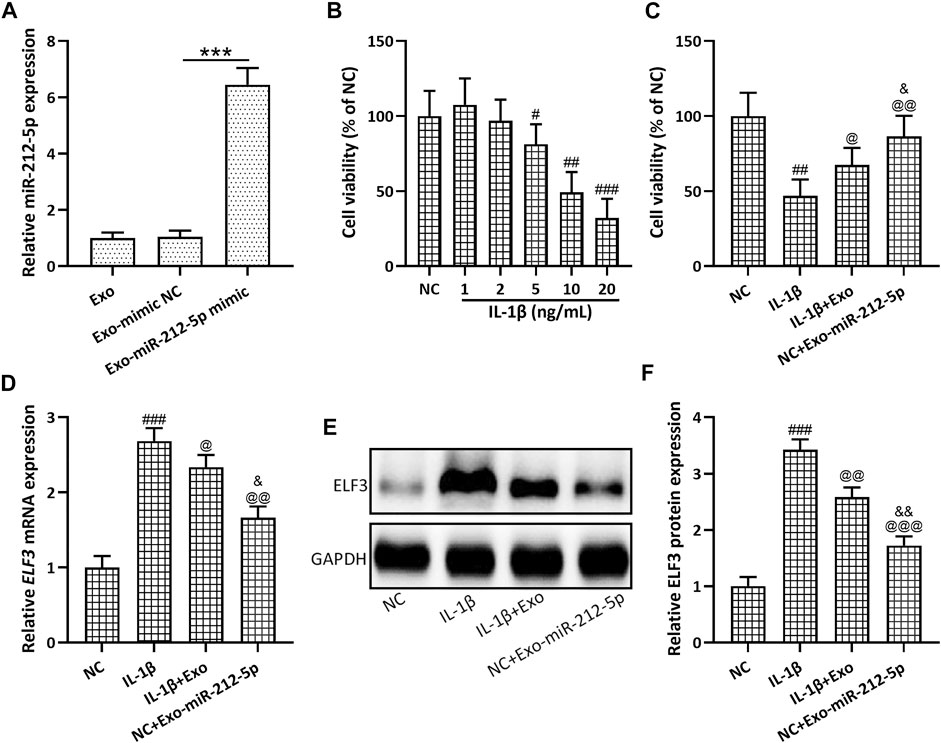
FIGURE 4. Exosomes derived from miR-212-5p overexpressed SMSCs (SMSC-212-5p-Exos) inhibited up-regulated ELF3 expression in chondrocytes induced by IL-1β. (A) qRT-PCR was utilized to analyze the relative expressions of miR-212-5p from SMSC-Exos or SMSC-212-5p-Exos. (B) Cell viability of chondrocytes after individual treatment with different concentrations of IL-1β for 48 h. (C) Cell viability of chondrocytes after co-treatment with IL-1β and SMSC-Exos, or SMSC-212-5p-Exos for 48 h. (D–F) IL-1β induced chondrocytes were treated with SMSC-Exos or SMSC-212-5p-Exos for 48 h qRT-PCR and western blotting were used to analyze the mRNA and proteins expressions of ELF3. n = 6 for each group in cell viability analysis, n = 3 for each group in qRT-PCR and western blotting tests. #p < 0.05, ##p < 0.01, ###p < 0.001 compared to NC. @p < 0.05, @@p < 0.01, @@@p < 0.001 compared to IL-1β. &p < 0.05, &&p < 0.01 compared to IL-1β+Exo. One-way ANOVA followed Dunn’s multiple comparisons test.
SMSC-212-5p-Exos Attenuate IL-1β Induced Chondrocyte Degeneration and Degradation
The relative mRNA expression of Collagen II (Figure 5A), Aggrecan (Figure 5B), and SOX9 (Figure 5C) in chondrocytes was decreased after the stimulation with IL-1β, while SMSC-Exos, especially SMSC-212-5p-Exos, could reverse such reduced expression of chondrocyte degeneration related molecules. The relative content of MMP-1 (Figure 6A), MMP-3 (Figure 6B), and MMP-13 (Figure 6C) in the culture supernatant of chondrocytes was increased after the stimulation with IL-1β, while SMSC-Exos or SMSC-212-5p-Exos could inhibit the secretion of the relevant molecules. On the other hand, the relative expression of MMP-1 (Figure 6D), MMP-3 (Figure 6E), and MMP-13 (Figure 6F) in chondrocytes was increased after the treatment with IL-1β, while SMSC-Exos or SMSC-212-5p-Exos could reverse such increased expression. When compared with SMSC-Exos treatment, SMSC-212-5p-Exos treatment could show more treatment benefit. These results demonstrated that exosomal miR-212-5p could attenuate IL-1β induced chondrocyte degeneration and degradation.
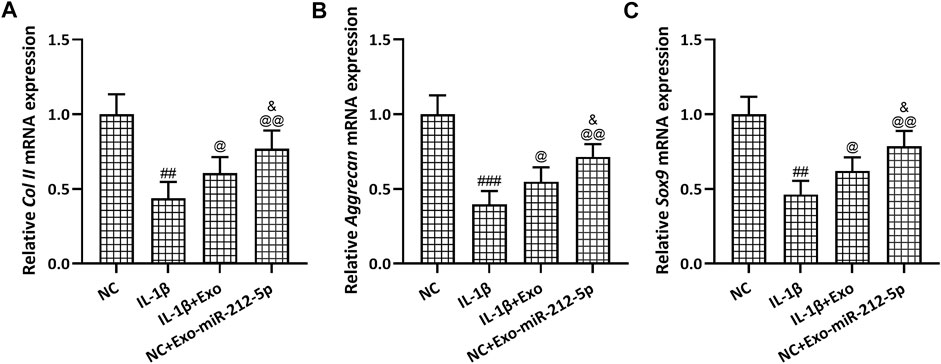
FIGURE 5. SMSC-212-5p-Exos attenuated IL-1β induced chondrocyte degeneration. IL-1β induced chondrocytes were treated with SMSC-Exos or SMSC-212-5p-Exos for 48 h qRT-PCR was utilized to analyze the mRNA expressions of Collagen II (A), Aggrecan (B) and Sox9 (C). n = 3 for each group. ##p < 0.01, ###p < 0.001 compared to NC. @p < 0.05, @@p < 0.01 compared to IL-1β. &p < 0.05 compared to IL-1β+Exo. One-way ANOVA followed Dunn’s multiple comparisons test.
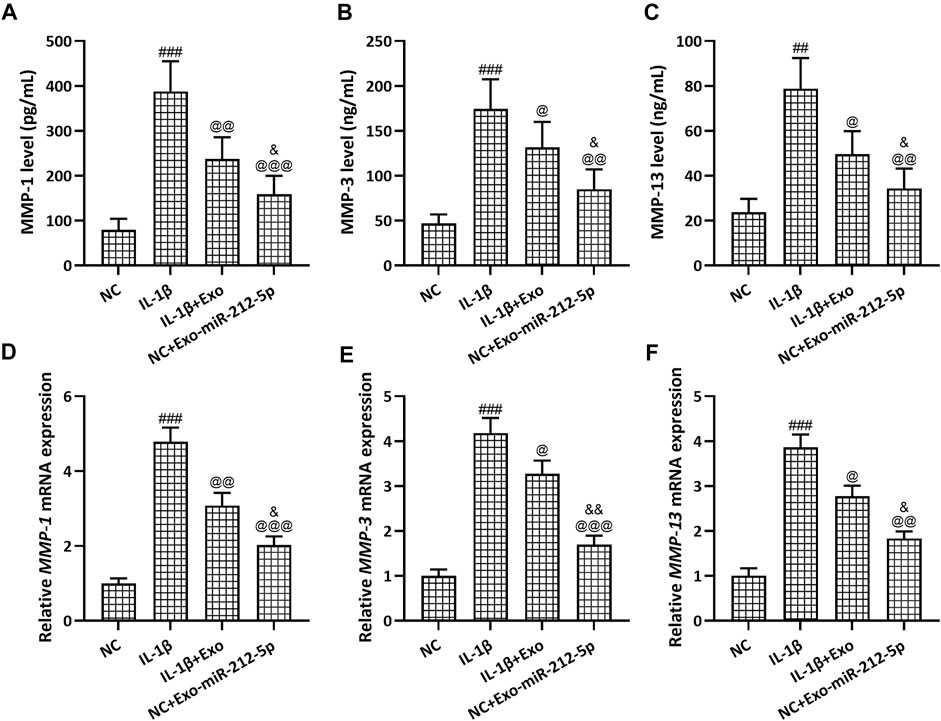
FIGURE 6. SMSC-212-5p-Exos attenuated IL-1β induced MMP-1, MMP-3, and MMP-13 expression in chondrocytes. IL-1β induced chondrocytes were treated with SMSC-Exos or SMSC-212-5p-Exos for 48 h. MMP-1, MMP-3, and MMP-13 production in cellular supernatant were measured by ELISA (A–C). mRNA expression of MMP-1, MMP-3, and MMP-13 from cell lysis were tested by qRT-PCR (D–F). n = 6 for each group in ELISA analysis, n = 3 for each group in qRT-PCR test. ##p < 0.01, ###p < 0.001 compared to NC. @p < 0.05, @@p < 0.01, @@@p < 0.001 compared to IL-1β. &p < 0.05, &&p < 0.01 compared to IL-1β+Exo. One-way ANOVA followed Dunn’s multiple comparisons test.
SMSC-212-5p-Exos Attenuate IL-1β Induced Inflammatory Responses in Chondrocytes
The relative content of IL-6 (Figure 7A), MCP1 (Figure 7B), and TNFα (Figure 7C) in the culture supernatant of chondrocytes was increased after the treatment with IL-1β, while SMSC-Exos or SMSC-212-5p-Exos could reverse such increased expression. On the other hand, the relative mRNA expression of IL-6 (Figure 7D), COX-2 (Figure 7E), and iNOS (Figure 7F) in chondrocytes was increased after the treatment with IL-1β, while SMSC-Exos or SMSC-212-5p-Exos could reverse such increased expression. It was worth noting that when compared with SMSC-Exos treatment, SMSC-212-5p-Exos treatment could decrease the levels of inflammatory molecules.
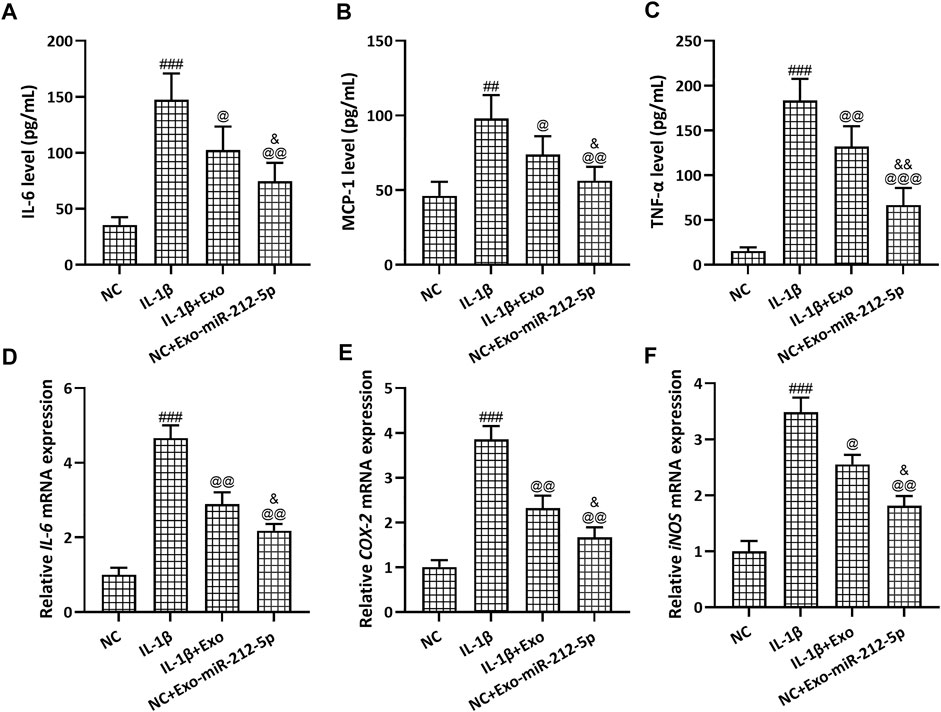
FIGURE 7. SMSC-212-5p-Exos attenuated IL-1β induced inflammatory responses in chondrocytes. IL-1β induced chondrocytes were treated with SMSC-Exos or SMSC-212-5p-Exos for 48 h. IL-6, MCP-1, and TNF-α production in cellular supernatant were measured by ELISA (A–C). mRNA levels of IL-6, COX-2, and iNOS from cell lysis were tested by qRT-PCR (D–F). n = 6 for each group in ELISA analysis, n = 3 for each group in qRT-PCR test. ##p < 0.01, ###p < 0.001 compared to NC. @p < 0.05, @@p < 0.01, @@@p < 0.001 compared to IL-1β. &p < 0.05, &&p < 0.01 compared to IL-1β+Exo. One-way ANOVA followed Dunn’s multiple comparisons test.
Discussion
In addition to inflammation, dysregulation between the synthesis and degradation of extracellular matrix mainly mediated by type-II collagen, aggrecan, and matrix metalloproteinases contributes to the articular cartilage degradation and loss process (Martel-Pelletier et al., 2016). In this study, we utilize IL-1β induced chondrocytes to mimic the pathology of OA. We found that SMSC-Exos showed the enrichment of miR-212-5p, which could target chondrocytes and further inhibit the ELF3 mediated chondrocyte degeneration, degradation, and inflammation process. This study highlights the significance and the need for further development of SMSC-212-5p-Exos on the clinical therapy of OA.
miR-132/212 clusters are maintained on down-regulation in mesenchymal stem cells at four different stages of transforming growth factor-β3-induced chondrogenesis differentiation (Yang et al., 2011). On the other hand, miR-132/212 clusters regulate hematopoietic stem cell maintenance and survival with age by buffering forkhead box O-3 (FoxO3) expression (Mehta et al., 2015). While no research has been performed to decipher the role of miR-212-5p in SMSCs. In our study, miR-212-5p is identified to contribute to the regulation of chondrocytes mediated by SMSC-Exos or SMSC-212-5p-Exos. All of these results indicate the universal utilization of miR-212-5p based treatment.
As transcription factors belonging to the ETS family, ELF3 participates in autoimmune and tumor neogenesis (Jedlicka and Gutierrez-Hartmann, 2008; Luk et al., 2018). The previous study demonstrates that ELF3 could modulate type II collagen transcription by prohibiting SOX9-CBP/p300-driven histone acetyltransferase activity in chondrocytes (Otero et al., 2017). On the other hand, ELF3 co-localizes with MMP-13 protein to regulate the transcription activity in human osteoarthritic cartilage (Otero et al., 2012). In accordance with the up-regulated SOX9 and MMP-13 expression testified in our study, all of these results define a pro-catabolic role for ELF3 to regulate MMP-13 and SOX9 mediated cartilage remodeling and degradation. Elevated ELF3 expression in OA cartilage tissues can act as a mediator of inflammatory and catabolism to destruct cartilage, which indicates the potential target of ELF3 to alleviate OA (Conde et al., 2018).
ELF3 is identified to direct target miR-212 to suppress nasopharyngeal carcinoma cells proliferation (Kang et al., 2020). In this research, miR-212-5p could target ELF3 to alleviate IL-1β induced chondrocytes degradation and inflammation. There are some limitations that should be indicated here. Microenvironment can alter the composition of exosomes (Skuratovskaia et al., 2020), and in vivo models should be constructed to demonstrate the therapeutic benefit of SMSC-212-5p-Exos. Whether other molecules, except miR-212-5p, could contribute to the treatment advantage in SMSC-212-5p-Exos compared with SMSC-Exos need further detailed analysis.
Accumulating investigations have indicated that exosomes will pave a new therapeutic paradigm for osteoarthritis treatment. Our study testifies that SMSC-212-5p-Exos could alleviate IL-1β induced chondrocytes degradation, degradation, and inflammation process involved in OA. It is worth noting that only in vitro model is utilized in the study, and the OA in vivo model should be constructed to demonstrate the treatment benefit of SMSC-212-5p-Exos. All in all, this study indicates that SMSC-212-5p-Exos could target ELF3-mediated OA pathology in chondrocytes, which can be considered as a future treatment option.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Xijing Hospital. The patients/participants provided their written informed consent to participate in this study. Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
Concept or design: TZ and ML. Acquisition of data: TZ, YL, XZ, JX, and ML. Analysis or interpretation of data: TZ, YL, and ML. Drafting of the manuscript: TZ, YL, XZ, JX, and ML. Critical revision of the manuscript for important intellectual content: All authors. All authors had full access to the data, contributed to the study, approved the final version for publication, and took responsibility for its accuracy and integrity.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agarwal, V., Bell, G. W., Nam, J.-W., and Bartel, D. P. (2015). Predicting Effective microRNA Target Sites in Mammalian mRNAs. Elife 4, e05005. doi:10.7554/eLife.05005
Ambros, V., Bartel, B., Bartel, D. P., Burge, C. B., Carrington, J. C., Chen, X., et al. (2003). A Uniform System for microRNA Annotation. RNA 9 (3), 277–279. doi:10.1261/rna.2183803
Conde, J., Otero, M., Scotece, M., Abella, V., Gómez, R., López, V., et al. (2018). E74-Like Factor (ELF3) and Leptin, a Novel Loop between Obesity and Inflammation Perpetuating a Pro-Catabolic State in Cartilage. Cell Physiol Biochem 45 (6), 2401–2410. doi:10.1159/000488227
Gratal, P., Mediero, A., Sánchez-Pernaute, O., Prieto-Potin, I., Lamuedra, A., Herrero-Beaumont, G., et al. (2019). Chondrocyte Enlargement Is a Marker of Osteoarthritis Severity. Osteoarthritis and Cartilage 27 (8), 1229–1234. doi:10.1016/j.joca.2019.04.013
Heidarzadeh, M., Gürsoy-Özdemir, Y., Kaya, M., Eslami Abriz, A., Zarebkohan, A., Rahbarghazi, R., et al. (2021). Exosomal Delivery of Therapeutic Modulators through the Blood-Brain Barrier; Promise and Pitfalls. Cell Biosci 11 (1), 142. doi:10.1186/s13578-021-00650-0
Huang, P.-y., Wu, J.-g., Gu, J., Zhang, T.-q., Li, L.-f., Wang, S.-q., et al. (2021). Bioinformatics Analysis of miRNA and mRNA Expression Profiles to Reveal the Key miRNAs and Genes in Osteoarthritis. J. Orthop. Surg. Res. 16 (1), 63. doi:10.1186/s13018-021-02201-2
Hunter, D. J., and Bierma-Zeinstra, S. (2019). Osteoarthritis. The Lancet 393 (10182), 1745–1759. doi:10.1016/s0140-6736(19)30417-9
Jedlicka, P., and Gutierrez-Hartmann, A. (2008). Ets Transcription Factors in Intestinal Morphogenesis, Homeostasis and Disease. Histol. Histopathol 23 (11), 1417–1424. doi:10.14670/hh-23.1417
Jiang, Z., Du, X., Wen, X., Li, H., Zeng, A., Sun, H., et al. (2021). Whole-Transcriptome Sequence of Degenerative Meniscus Cells Unveiling Diagnostic Markers and Therapeutic Targets for Osteoarthritis. Front. Genet. 12, 754421. doi:10.3389/fgene.2021.754421
Kalluri, R., and LeBleu, V. S. (2020). The Biology , Function , and Biomedical Applications of Exosomes. Science 367 (6478), eaau6977. doi:10.1126/science.aau6977
Kang, Y., Cui, Y., and Tan, M. (2020). MicroRNA-212 Suppresses Cell Proliferation in Nasopharyngeal Carcinoma by Targeting ELF3. Oncol. Lett. 19 (4), 2902–2908. doi:10.3892/ol.2020.11401
Katz, J. N., Arant, K. R., and Loeser, R. F. (2021). Diagnosis and Treatment of Hip and Knee Osteoarthritis. Jama 325 (6), 568–578. doi:10.1001/jama.2020.22171
Liu, Y., Zhang, X. L., Li, X. F., Tang, Y. C., and Zhao, X. (2018). miR-212-3p Reduced Proliferation, and Promoted Apoptosis of Fibroblast-Like Synoviocytes via Down-Regulating SOX5 in Rheumatoid Arthritis. Eur. Rev. Med. Pharmacol. Sci. 22 (2), 461–471. doi:10.26355/eurrev_201801_14196
Luk, I., Reehorst, C., and Mariadason, J. (2018). ELF3, ELF5, EHF and SPDEF Transcription Factors in Tissue Homeostasis and Cancer. Molecules 23 (9), 2191. doi:10.3390/molecules23092191
Martel-Pelletier, J., Barr, A. J., Cicuttini, F. M., Conaghan, P. G., Cooper, C., Goldring, M. B., et al. (2016). Osteoarthritis. Nat. Rev. Dis. Primers 2, 16072. doi:10.1038/nrdp.2016.72
Mehta, A., Zhao, J. L., Sinha, N., Marinov, G. K., Mann, M., Kowalczyk, M. S., et al. (2015). The MicroRNA-132 and MicroRNA-212 Cluster Regulates Hematopoietic Stem Cell Maintenance and Survival with Age by Buffering FOXO3 Expression. Immunity 42 (6), 1021–1032. doi:10.1016/j.immuni.2015.05.017
Moghadasi, S., Elveny, M., Rahman, H. S., Suksatan, W., Jalil, A. T., Abdelbasset, W. K., et al. (2021). A Paradigm Shift in Cell-Free Approach: The Emerging Role of MSCs-Derived Exosomes in Regenerative Medicine. J. Transl Med. 19 (1), 302. doi:10.1186/s12967-021-02980-6
O’Brien, K., Breyne, K., Ughetto, S., Laurent, L. C., and Breakefield, X. O. (2020). RNA Delivery by Extracellular Vesicles in Mammalian Cells and its Applications. Nat. Rev. Mol. Cel Biol 21 (10), 585–606. doi:10.1038/s41580-020-0251-y
Oliviero, A., Della Porta, G., Peretti, G. M., and Maffulli, N. (2019). MicroRNA in Osteoarthritis: Physiopathology, Diagnosis and Therapeutic challenge. Br. Med. Bull. 130 (1), 137–147. doi:10.1093/bmb/ldz015
Otero, M., Peng, H., Hachem, K. E., Culley, K. L., Wondimu, E. B., Quinn, J., et al. (2017). ELF3 Modulates Type II Collagen Gene (COL2A1) Transcription in Chondrocytes by Inhibiting SOX9-CBP/p300-Driven Histone Acetyltransferase Activity. Connect. Tissue Res. 58 (1), 15–26. doi:10.1080/03008207.2016.1200566
Otero, M., Plumb, D. A., Tsuchimochi, K., Dragomir, C. L., Hashimoto, K., Peng, H., et al. (2012). E74-Like Factor 3 (ELF3) Impacts on Matrix Metalloproteinase 13 (MMP13) Transcriptional Control in Articular Chondrocytes under Proinflammatory Stress. J. Biol. Chem. 287 (5), 3559–3572. doi:10.1074/jbc.M111.265744
Qiu, M., Liu, D., and Fu, Q. (2021). MiR-129-5p Shuttled by Human Synovial Mesenchymal Stem Cell-Derived Exosomes Relieves IL-1β Induced Osteoarthritis via Targeting HMGB1. Life Sci. 269, 118987. doi:10.1016/j.lfs.2020.118987
Sharma, L. (2021). Osteoarthritis of the Knee. N. Engl. J. Med. 384 (1), 51–59. doi:10.1056/NEJMcp1903768
Skuratovskaia, D., Vulf, M., Khaziakhmatova, O., Malashchenko, V., Komar, A., Shunkin, E., et al. (2020). Exosome Limitations in the Treatment of Inflammatory Diseases. Curr. Pharm. Des. 27, 3105–3121. doi:10.2174/1381612826666201210120444
Wang, K., Li, F., Yuan, Y., Shan, L., Cui, Y., Qu, J., et al. (2020). Synovial Mesenchymal Stem Cell-Derived EV-Packaged miR-31 Downregulates Histone Demethylase KDM2A to Prevent Knee Osteoarthritis. Mol. Ther. - Nucleic Acids 22, 1078–1091. doi:10.1016/j.omtn.2020.09.014
Keywords: miR-212-5p, synovial mesenchymal stem cells, ELF3, chondrocyte, SMSCs, osteoarthritis
Citation: Zheng T, Li Y, Zhang X, Xu J and Luo M (2022) Exosomes Derived From miR-212-5p Overexpressed Human Synovial Mesenchymal Stem Cells Suppress Chondrocyte Degeneration and Inflammation by Targeting ELF3. Front. Bioeng. Biotechnol. 10:816209. doi: 10.3389/fbioe.2022.816209
Received: 30 November 2021; Accepted: 28 January 2022;
Published: 24 February 2022.
Edited by:
Elizabeth R. Balmayor, Maastricht University, NetherlandsReviewed by:
Rald Groven, Maastricht University, NetherlandsVirginie Joris, Maastricht University, Netherlands
Copyright © 2022 Zheng, Li, Zhang, Xu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Luo, TGl6dzA4MEBmbW11LmVkdS5jbg==
†These authors have contributed equally to this work
 Tianlei Zheng
Tianlei Zheng Yan Li1†
Yan Li1†