- Department of Neurology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
The lack of regenerative capacity of neurons leads to poor prognoses for some neurological disorders. The use of small molecules to directly reprogram somatic cells into neurons provides a new therapeutic strategy for neurological diseases. In this review, the mechanisms of action of different small molecules, the approaches to screening small molecule cocktails, and the methods employed to detect their reprogramming efficiency are discussed, and the studies, focusing on neuronal reprogramming using small molecules in neurological disease models, are collected. Future research efforts are needed to investigate the in vivo mechanisms of small molecule-mediated neuronal reprogramming under pathophysiological states, optimize screening cocktails and dosing regimens, and identify safe and effective delivery routes to promote neural regeneration in different neurological diseases.
1 Introduction
The regenerative repair of neurons in acute injuries and neurodegeneration diseases has been clinically challenging (Sivandzade and Cucullo, 2021). The main reason for this is the extremely limited neuron-regenerative capacity in the adult mammalian Central Nervous System (CNS), which contributes to the poor prognosis of patients with CNS injuries (Goldman, 2016; Tedeschi et al., 2016; McMurran et al., 2018; Vignoles et al., 2019; Stricker and Gotz, 2021). In the last decades, stem cell transplantation therapy has been proposed to be an efficient method of replacing lost neurons in several neurological disorders (Albert et al., 2021; Lu et al., 2021; Salikhova et al., 2021). However, some existing problems, such as teratogenic effect, differentiation abnormality, ethical issues, and survival limitation, prevent its clinical appilications (Barker and de Beaufort, 2013; Qin et al., 2017; Zeng et al., 2021). Recently, an emerging reprogramming technique that transforms non-neuronal cells into induced neurons (iNs) can be regarded as a promising potential therapeutic tool in regenerative medicine (Ma et al., 2019). Instead of exogenous cells’ transplantation, this technique makes it possible to directly reprogram endogenous starting cells into target cells in vivo (Srivastava and DeWitt, 2016).
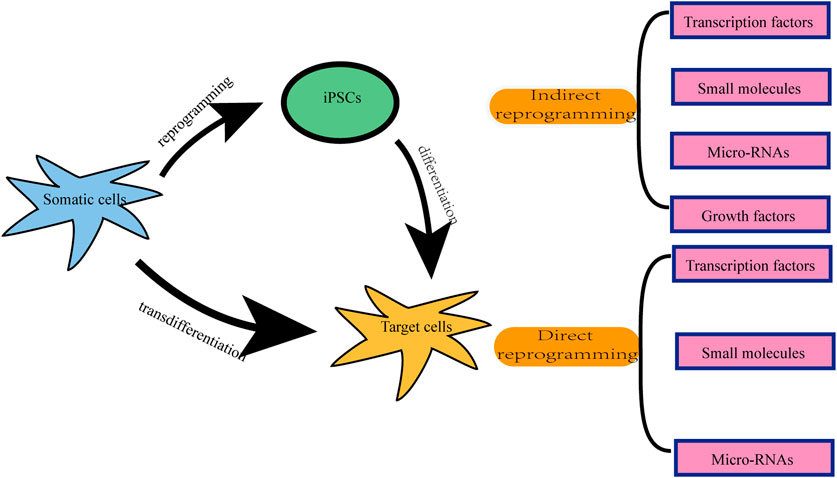
There are two types of neuronal reprogramming: indirect reprogramming and direct reprogramming (Figure 1) (Sato et al., 2019). The former is a two-step process, in which differentiated somatic cells are first reprogrammed into intermediated states such as induced pluripotent stem cells (iPSCs) or multipotent neural stem cells (iNSCs), followed by differentiation into neurons (Kim et al., 2011; Ladewig et al., 2013; Liu et al., 2016; Yuan et al., 2020). The latter is the direct transformation of terminally differentiated cells into target mature cells without passing through the stages of pluripotent or multipotent cells, which is also known as transdifferentiation (Kim et al., 2011; Ladewig et al., 2013; Liu et al., 2016; Mollinari et al., 2018; Yang et al., 2020a; Yuan et al., 2020; Wang et al., 2021a). Since direct lineage reprogramming does not go through an intermediate stem cell state, it possesses the appealing features of a low likelihood of tumor formation, no age-resetting, a high conversion speed, and an efficient cell differentiation (Yang et al., 2011; Li and Chen, 2016; Ma et al., 2019; Mollinari and Merlo, 2021).
There are mainly several approaches to achieving reprogramming, each with its advantages and disadvantages. One involves reprogramming mediated by the ectopic gene expression, such as the ectopic expression of neuronal transcription factors (TFs) via virus vectors (Vierbuchen et al., 2010; Meng et al., 2012). As for endogenous gene manipulation, an emerging and innovative technology, clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system can easily edit and modulate deoxyribonucleic acid (DNA) sequences within the endogenous genome (Hsu et al., 2014). It was reported that the application of CRISPR/Cas9 can directly convert fibroblasts into neuronal cells via silencing endogenous non-neuronal genes (Rubio et al., 2016). Another different method is associated with the use of small-molecule compounds during the process of reprogramming (Dai et al., 2015; Li et al., 2015). Besides, microRNAs are included in cellular conversion formulations to aid the inhibition of alternative cell fates (Ambasudhan et al., 2011; Yoo et al., 2011; Xue et al., 2013; Zhou et al., 2015). For the last decade, as one of the small non-coding RNAs, microRNAs were combined with TFs and small molecules to kickstart the successful conversion of iNs (Ambasudhan et al., 2011; Smith et al., 2016; Habekost et al., 2020; Nemoto et al., 2020), which suppress gene expression by promoting mRNA degradation or by blocking translation (Bushati and Cohen, 2007; Krol et al., 2010). Furthermore, studies have shown that growth factors promote the process of reprogramming (He et al., 2017; Yi et al., 2021).
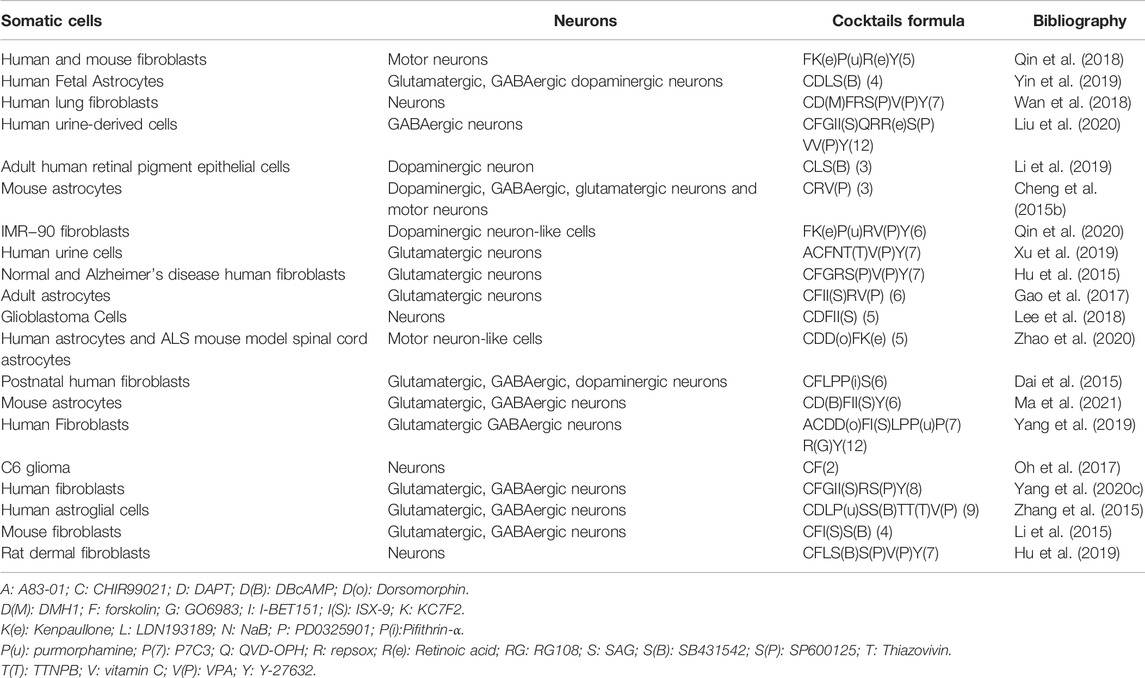
Reviewing the published research, the two most widely used methods in the field of reprogramming are the ectopic expression of TFs and the administration of small molecule cocktails. The overexpression of TFs using viral vectors has been reported to be efficient in mediating reprogramming (Yang et al., 2020b; Puls et al., 2020). The lineage-specific viral vectors are targeted (Brulet et al., 2017; Liang et al., 2018). The virus vector genomes can insert target cell genomes (lentiviruses and retroviruses) or form stable episomes (adeno-associated viruses) during the reprogramming, which cause TFs to sustain expression with long-term induction effects (Duan et al., 1998; Grande et al., 2013; Brulet et al., 2017; Chan et al., 2017; Morabito et al., 2017; Liang et al., 2018; Man et al., 2018; Haridhasapavalan et al., 2019). However, when integrating vectors, e.g., lentiviruses and retroviruses are used to deliver the TFs (Naldini et al., 1996; Lesbats et al., 2016), this approach may also carry the risk of causing mutations in the host cell genome (Li et al., 2002; Modlich et al., 2006; Medvedev et al., 2010); thus, raising safety issues, such as tumor formation (Oh et al., 2017; Pu et al., 2019). While the non-integrating vectors, such as adeno-associated viruses, are involved, the efficiency of reprogramming is low and the delivery capacity of TFs is limited (Shao and Wu, 2010; Hirsch et al., 2016). The other universally used approach is to induce somatic cells’ transdifferentiation using small molecules (Cheng et al., 2015a; Hu et al., 2015; Zhang et al., 2016a; Cao et al., 2016; Zeng et al., 2021). We summarized the most common small molecules during the transdifferentiation of somatic cells into neurons (Table 1). Since no exogenous genes are introduced and there is no risk of genetic manipulation, this approach seems to be a safer way of reprogramming (Cheng et al., 2015b; Hu et al., 2015; Zhang et al., 2016a; Cao et al., 2016; Zeng et al., 2021). In addition, compared with the viral vector-mediated reprogramming method, small molecules therapy has the advantages of being cost-effective, easy to obtain, reversible and controllable, strongly permeable, and lacking immunogenicity issues (Xu et al., 2008; Hou et al., 2013; Xu et al., 2015; Cao et al., 2016; Ye et al., 2016; Qin et al., 2017; Xie et al., 2017). Therefore, small molecules-mediated reprogramming has a great potential for clinical translation and is a promising therapeutic strategy for the treatment of neurological diseases. However, compared with TF-induced reprogramming, which is only required for ectopic expression of fate-determining TFs of related cells, in most cases, small molecules mediating cell reprogramming are not as specific as this latter methods (Yang et al., 2019). In general, single small molecules cannot induce somatic reprogramming, while TFs can (Yin et al., 2019). It has been demonstrated that small molecule compounds can directly convert terminally differentiated cells into neurons (Hou et al., 2013; Cheng et al., 2015b; Li et al., 2015; Zhang et al., 2015; Gao et al., 2017). Using a mixture of small molecules, somatic cells such as fibroblasts (Hu et al., 2015; Li et al., 2015; Qin et al., 2018; Wan et al., 2018; Yang et al., 2019; Xu et al., 2020), astrocytes (Zhang et al., 2015; Gao et al., 2017; Ma et al., 2019), human urine cells (Xu et al., 2019; Liu et al., 2020), peripheral blood T cells (Tanabe et al., 2018), and even glioma cells can be directly induced into neurons (Oh et al., 2017; Lee et al., 2018). In this review, we will mainly discuss small molecules-mediated direct reprogramming of the CNS neurons.
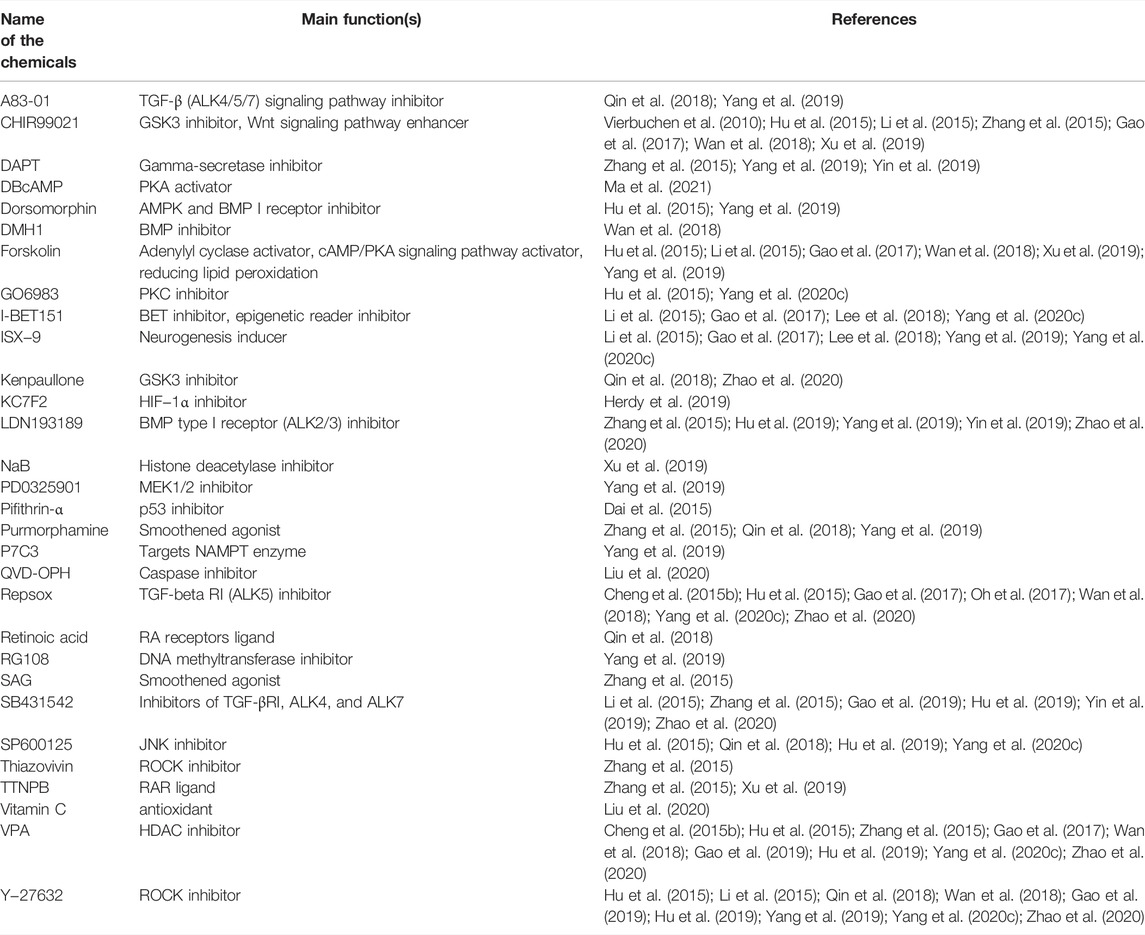
TABLE 1. The most common small molecules during the transdifferentiation of somatic cells into neurons.
2 Mechanisms of Small Molecules Induced Reprogramming
The patterns of small-molecule cocktails in neuronal reprogramming are decided on the types of starting cells and whether it is under pathological conditions (Hu et al., 2015; Li et al., 2019; Qin et al., 2020). However, the synergistic mechanisms of the used drug cocktails, remain unclear (Vasan et al., 2021). It has been reported that several combinations of small molecules, such as DFICBY (DBcAMP, Forskolin, ISX9, CHIR99021, I-BET151, Y-27632) (Ma et al., 2021), CAYTFVB (CHIR99021, A8301, Y27632, TTNPB, Forskolin, Valproic acid (VPA), NaB) (Xu et al., 2019), FRSCGYIB (Forskolin, RepSox, SP600125, CHIR99021, Go6983, Y27632, IXS9, I-BET151) (Yang et al., 2020c), and VCRFSGYIBRTQ (VPA, CHIR99021, Repsox, Forskolin, SP600625, GO6983, Y27632, ISX9, I-BET151, RA, Vit C, QVD-OPH) (Liu et al., 2020), are used to modulate signaling pathways, epigenetics, and the metabolisms of reprogramming, to effectively transform somatic cells into neurons, while these researchers did not illuminate the comprehensive mechanisms. Noteworthy, challenges exist in exploring the best combinations of small molecules in converting neurons from other differentiated cells.
2.1 Signaling Pathways Involved in Small Molecules Induced Reprogramming
2.1.1 The Wnt/GSK-3 Signaling Pathway
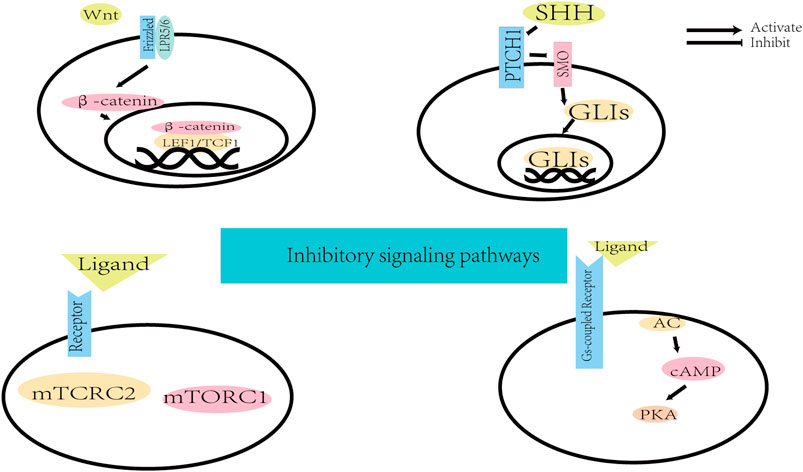
Small molecules can promote direct neuronal reprogramming by regulating several cellular signaling pathways (Figure 2) (Yin et al., 2019). The background information of these pathways is demonstrated in Figure 3 and Figure 4. Signaling by the Wingless/integrated (Wnt) family of secreted ligands via the transcriptional coactivator β-catenin controls embryonic development and disease processes by stimulating distinct intracellular signaling pathways (Angers and Moon, 2009; MacDonald et al., 2009). When activated by Wnt, Frizzled receptors recruit the cytoplasmic protein complex to the plasma membrane, which activates different signaling cascades (Inestrosa and Varela-Nallar, 2015). In addition, the Wnt ligands also interact with the co-receptor low-density lipoprotein receptor-related protein 5 (LRP5) or LRP6 to induce the stabilization of β-catenin by preventing its phosphorylation via glycogen synthase kinase-3 (GSK3) (Toledo and Inestrosa, 2010; Valvezan and Klein, 2012). Then β-catenin accumulates and moves to the nucleus where it interacts with members of the T-cell factor/lymphoid enhancer factor 1 (TCF/LEF1) family of transcription factors to modulate the expression of Wnt target genes (Angers and Moon, 2009; Arrázola et al., 2009; MacDonald et al., 2009; Clevers and Nusse, 2012).
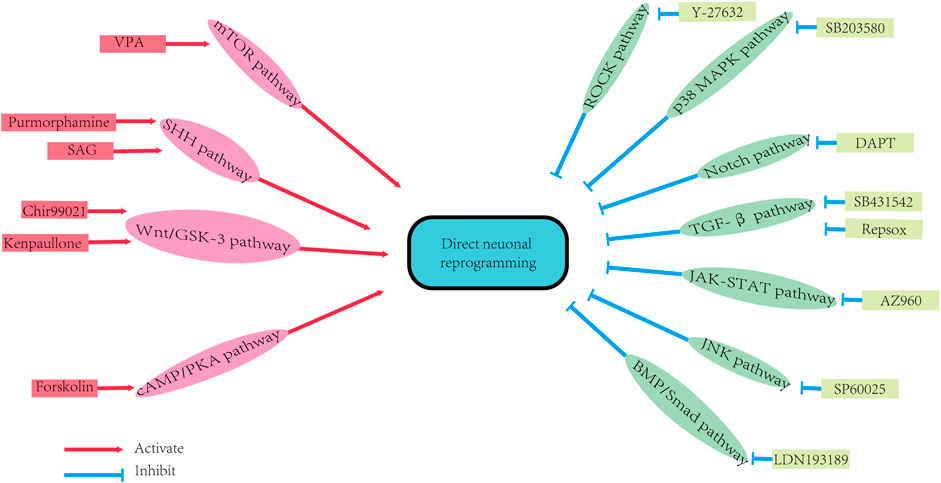
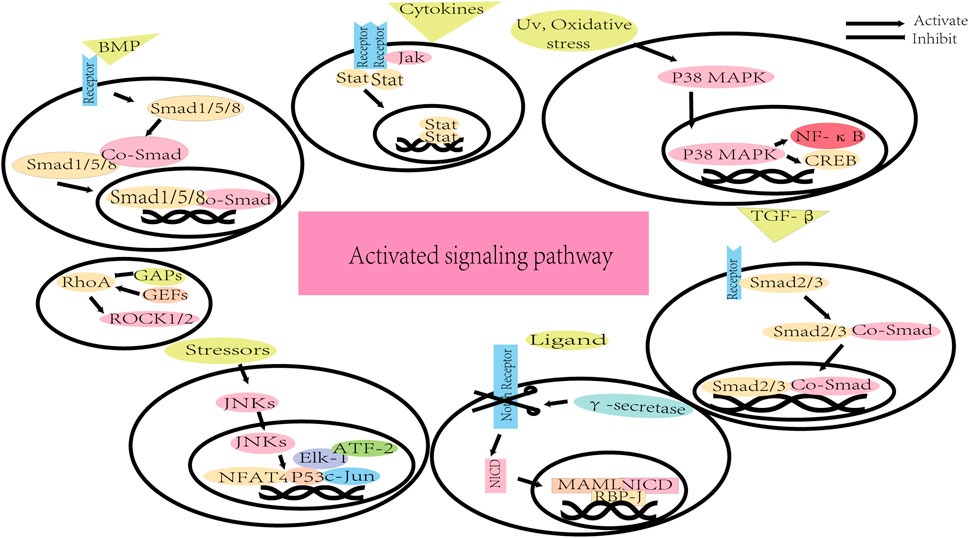
FIGURE 2. The signaling pathways of direct neuron reprogramming by small molecules. DAPT: N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; SAG: SHH activator smoothened agonist; VPA: Valproic acid. BMP/Smad pathway: bone morphogenetic protein/Smad pathway; cAMP/PKA pathway: cyclic adenosine monophosphate/protein kinase A system pathway; JAK-STAT pathway: Janus kinase-signal transducer and activator of transcription pathway; JNK pathway: Jun N-terminal Kinase pathway; mTOR pathway: Mammalian Target of Rapamycin pathway; P38 MAPK pathway: P38 mitogen-activated protein kinase pathway; ROCK pathway: Rho-associated protein kinase pathway; SHH pathway: Sonic Hedgehog pathway; TGF-β pathway: transforming growth factor-beta pathway; Wnt/GSK-3 pathway: (Wingless/integrated)/glycogen synthase kinase 3 pathway.
The Wnt/GSK-3 signaling pathway has been shown to regulate postnatal and adult neurogenesis (Lie et al., 2005; Li et al., 2009; Zhang et al., 2011), and is also involved in the regulation of cellular self-renewal, pluripotency and promotes neuronal differentiation (Rivetti di Val Cervo et al., 2017; Oproescu et al., 2021). It has been reported that Wnt’s activation promotes the process of reprogramming (Cheng et al., 2015b). Whereas the inhibition of the GSK-3 signaling promotes neural progenitor cells’ homeostasis and induction (Hur and Zhou, 2010; Li et al., 2012). Selective GSK-3 inhibitors, such as CHIR99021 and kenpaullone (Cheng et al., 2015b), have been shown to indirectly activate the Wnt pathway in human and mouse stem cells (Silva et al., 2008; Li et al., 2009; Qin et al., 2017; Ma et al., 2019). Moreover, the combination of CHIR99021 with other small molecules has been shown to promote the conversion of astrocytes (Yang et al., 2019; Yin et al., 2019; Ma et al., 2021), fibroblasts (Qin et al., 2018), and human bone marrow mesenchymal stromal cells (MSCs)-derived neural progenitor cells (NPCs) into neurons (George et al., 2018).
2.1.2 The JAK-STAT Signaling Pathway
The Janus family tyrosine kinase–signal transducer and activator of transcription (JAK-STAT) signaling pathway is broadly regulated by cytokines in CNS (Campbell, 2005). The binding of a cytokine with its unique cell membrane receptor triggers activation of the JAKs and tyrosine phosphorylation (Nabavi et al., 2019). Then the receptor phosphotyrosine sites interact with Src homology 2 (SH2) domains on STATs, and STATs also become phosphorylated by JAKs (Banerjee et al., 2017). These activated STATs form dimers that translocate to the nucleus and bind with DNA target sequences modulating gene expression (Campbell, 2005; Stark and Darnell, 2012; Au-Yeung et al., 2013).
The JAK-STAT signaling pathway is involved in cell proliferation, differentiation, apoptosis, and immune system regulation (Xin et al., 2020; Herrera and Bach, 2019; Villarino et al., 2017; O'Shea et al., 2015). Unlike neurons, which have a non-dividing property, fibroblasts and astrocytes can divide and proliferate. However, exiting the cell cycle is required for the fibroblasts and astrocytes transformation into neurons (Liu et al., 2013; Jiang et al., 2015; Tang, 2017). Interestingly, the inhibition of the JAK-STAT pathway can promote neuronal reprogramming via inducing fibroblasts to exit the cell cycle (Herdy et al., 2019). It has been demonstrated that the JAK2 inhibitor, AZ960, can reduce fibroblasts division while mediating the conversion of fibroblasts to neurons (Herdy et al., 2019).
Epithelial-mesenchymal transition (EMT) refers to a process in which epithelial cells lose their apical-basal polarity and cell-cell junctions to transdifferentiate into mesenchymal cells that have invasive properties (Amack, 2021). Mesenchymal-to-Epithelial Transition (MET) is the opposite process of EMT and has been reported to play an important role in the direct conversion of fibroblasts into neurons, and therefore, MET may be used to improve the process of reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010; He et al., 2017). However, the p-STAT3 dimer directly promotes the expression of genes related to EMT (Wendt et al., 2014). The JAK2 inhibitor, AZ960, has been reported to promote MET via indirect inhibition of STAT3, which leads to an enhanced conversion of fibroblasts into neurons (Herdy et al., 2019). In addition, the JAK/STAT pathway inhibitor, ruxolitinib, has been shown to improve the efficiency of cortical astroglia conversion into neurons in rats (Smith et al., 2016).
2.1.3 The SHH Signaling Pathway
The founding member of the Sonic Hedgehog (SHH) family of secreted proteins three decades ago opened a field that has encompassed embryonic development, stem cell biology, and tissue homeostasis (Briscoe and Thérond, 2013). The SHH signaling has involved in axonal guidance and stem cell maintenance in the nervous system (Fuccillo et al., 2006). The SHH, released by the cell producing the ligands, relieves the constitutive repression of the receptor smoothened (SMO) by the receptor patched 1 (PTCH1) (Tukachinsky et al., 2016; Hill et al., 2021). Once SMO activation, the zinc finger transcription factors known as GLIs travel to the nucleus and initiate transcription of downstream SHH target genes (Bai et al., 2004; Pan et al., 2006; Hill et al., 2021).
Apart from regulating neural development (De Luca et al., 2016; Patel et al., 2017; Hill et al., 2021), the SHH signaling pathway can also promote the proliferation of reactive astrocytes (Sirko et al., 2013; Guo et al., 2014). Embryonic stem cells or induced pluripotent stem cells can differentiate into motor neurons after treating with the SHH/Purmorphamine (an activator of the SHH signaling pathway), indicating that activation of the SHH signaling facilitates motor neuron generation (Qin et al., 2018). The activation of the SHH pathway has been shown to promote neuronal differentiation in the transformation process of somatic cells (Masai et al., 2005). The SHH activator, smoothened agonist (SAG), can promote the in vitro reprogramming of human fetal astrocytes into functional neurons (Zhang et al., 2015).
2.1.4 The BMP/Smad Signaling Pathway
The bone morphogenetic protein (BMP) belongs to the transforming growth factor-beta (TGF-β) superfamily and it has been demonstrated that BMPs are involved in the regulation of cell proliferation, survival, differentiation and apoptosis (Shi and Massagué, 2003; Xiao et al., 2007). When BMP ligands bind to the BMP receptors, the canonical BMP signaling pathway is initiated through activating Smads 1, 5, and 8 (Zhong and Zou, 2014; Kashima and Hata, 2018). Then the receptor-activated Smad proteins (R-Smads) form the complex with the common mediator-Smad (co-Smad, Smad4), followed by traveling to the nucleus and modulating gene expression (Heldin et al., 1997; Xiao et al., 2007).
The BMP/Smad signaling pathway regulates synaptic development and astroglial differentiation (Gross et al., 1996; Politano et al., 2019; Vuilleumier et al., 2019). Blocking BMP signaling has been shown to further promote iNs maturation (Vignoles et al., 2019). The BMP inhibitor, LDN193189, which was important for astroglial differentiation, promoted the reprogramming of human astrocytes into neurons (Gross et al., 1996; Zhang et al., 2015; Ma et al., 2019; Yin et al., 2019). For higher reprogramming efficiency, LDN193189 has been combined with SB431542 (a TGF-β inhibitor) to assist in neural transition (Lee et al., 2015; Mirakhori et al., 2015; Park et al., 2017). In addition, another BMP pathway blocker dorsomorphin and SB431542, improve the efficiency of the induction of the human adult peripheral blood T cells to iNs (Tanabe et al., 2018).
2.1.5 The TGF-β Signaling Pathway
The TGF-β pathway is involved in various biological processes during embryogenesis and in adult tissue homeostases, such as cell growth, differentiation, migration, and apoptosis (Massagué et al., 2000; Derynck and Akhurst, 2007; Kashima and Hata, 2018). Ligands bind to TGF-β ligand-specific receptors with serine/threonine kinases and then receptors are activated, which activate Smad proteins 2 and 3 (Massagué et al., 2000; Dijke and Hill, 2004). The activated Smads bind co-Smad to form the complex translocates to the nucleus and modulates gene transcription by binding to DNA-binding transcription factors (Dijke and Hill, 2004; Kashima and Hata, 2018).
It has been reported that the activation of the TGF-β signaling pathway can inhibit neuronal fate and promote glial fate (Rodriguez-Martinez and Velasco, 2012). The TGF-β signaling pathway has been shown to induce EMT, and its inhibition can facilitate reprogramming by promoting MET (Aguirre et al., 2010; Qin et al., 2017). SB431542 (a TGF-β inhibitor) is involved in inhibiting glial fate and promoting neuronal fate, which not only boosts reprogramming during MET (Ichida et al., 2009; Li et al., 2010; Rodriguez-Martinez and Velasco, 2012; Zhang et al., 2015), but also promotes the cell cycle exit and conversion of human astrocytes into neurons (Rodriguez-Martinez and Velasco, 2012; Ma et al., 2019; Yin et al., 2019). In addition, SB431542 can also inhibit Smad signaling by regulating the phosphorylation of ALK (Activin-receptor-like kinase) receptors (Dijke and Hill, 2004). Recently, another common TGF-β signaling pathway inhibitor, RepSox, showed similar promoting functions in neuronal reprogramming (Tu et al., 2019; Roudnicky et al., 2020; Zhang et al., 2020; Chen et al., 2021).
2.1.6 The Notch Signaling Pathway
Notch is not only a key regulator of adult neural stem cells but also plays important role in the regulation of migration, morphology, synaptic plasticity, and survival of neurons (Ables et al., 2011). Mammals possess four Notch receptors (Notch1, Notch2, Notch3 and Notch4) and many ligands, mainly containing jagged 1 (JAG1) and jagged 2 (JAG2, homologs of serrate), and delta-like proteins (Lindsell et al., 1996; Ables et al., 2011). Upon ligand binding, the transmembrane domain of the Notch receptor is cleaved by γ-secretase, and the Notch intracellular domain (NICD) is released into the cytoplasm (Ables et al., 2011). Then NICD translocates to the nucleus and is combined with mastermind-like protein 1 (MAML1), MAML2 or MAML3, which converts the recombining binding protein suppressor of hairless (RBPJ) complex from a transcriptional inhibitor to an activator to modulate Notch target genes expression (Ables et al., 2011).
The Notch signaling pathway is a highly conserved morphogenetic pathway, which plays an important role in regulating neuronal self-renewal and differentiation (Breunig et al., 2007; Roese-Koerner et al., 2017), survival (Saura et al., 2004), and neuronal plasticity (de Bivort et al., 2009; Zhang et al., 2014). The activation of this pathway maintains cells in an undifferentiated state, while its inhibition helps promote neuronal fate determination (Kageyama et al., 2005). DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester) inhibits Notch signaling pathway by targeting γ-secretase (Urbán and Guillemot, 2014; Borghese et al., 2010; Hitoshi et al., 2004), and has been used to promote the neuronal differentiation of embryonic stem cells (Borghese et al., 2010; Qi et al., 2017). Besides, DAPT facilitates the conversion of human astrocytes into neurons in vitro (Zhang et al., 2015; Yin et al., 2019). In a mouse stroke model, the inhibition of Notch signaling is required for the entry of mouse striatal astrocytes into neurogenesis (Magnusson et al., 2014), suggesting that the conversion of reactive astrocytes into neurons is associated with Notch signaling (Smith et al., 2016).
2.1.7 The p38 MAPK Signaling Pathway
MAPKs are a family of highly conserved proteins and are involved in different cellular processes including cell survival, proliferation, differentiation, and migration (Zarubin and Han, 2005; Ijomone et al., 2021). There are three major subfamilies of MAPK proteins: extracellular signal-regulated kinases (ERK), Jun N-terminal Kinase (JNK), and mitogen-activated protein kinase (p38 MAPK) (Zarubin and Han, 2005; Koga et al., 2019; Aluko et al., 2021). The p38 pathway comprises p38α, p38β, p38γ, and p38δ, which is activated by stress and regulates immune response, cell survival, and differentiation (Nebreda and Porras, 2000; Ono and Han, 2000; Cuenda and Rousseau, 2007). The MAPK pathways consist of three continuously activated protein kinases: MAPK, MAPK1, and MAPK2 (Morrison, 2012). The specific extracellular stimuli activate MAPK2 and MAPK1 successively and then initiate the p38 MAPK cascade (Ijomone et al., 2021). After activation, p38 translocates from the cytoplasm to the nucleus to modulate factors like cyclic AMP-responsive element-binding protein (CREB) and nuclear factor kappa B (NF-κB), or accumulates in the cytoplasm to regulate proteins (Raingeaud et al., 1995; Ben-Levy et al., 1998).
The activation of the p38 MAPK signaling pathway has been shown to promote astrocytes activation and glial scar formation during reactive gliosis (Cheon et al., 2016; Li et al., 2020). Its inhibition improved neuronal induction by inhibiting the proliferation of responsive astrocytes. SB203580 is a common p38 inhibitor that has been applied to induce the conversion of reactive astrocytes to neurons in vitro (Smith et al., 2016).
2.1.8 The mTOR Signaling Pathway
The mechanism of the Mammalian Target of Rapamycin (mTOR) signaling pathway is a crucial cellular signaling hub, involving growth control, protein synthesis, gene expression, and metabolic regulation (Lipton and Sahin, 2014). The mTOR is a large, highly conserved, serine/threonine kinase, which is a member of the phosphoinositide 3-kinase-related kinase family and is widely expressed in eukaryotic cell types, including neuronal cells (Sabatini et al., 1999). Extracellular activators of the mTOR pathway bind to membrane receptors, and then trigger intracellular signal transduction via mTOR complex 1 (mTORC1) and mTOR complex 1 (mTORC2) (Lipton and Sahin, 2014; O' Neill, 2013). The activation of the mTOR pathway regulates protein syntheses and enhances gene expressions involved in the regulation of cell proliferation and survival (Russo et al., 2012).
Studies have reported that the mTOR signaling pathway is a critical regulator of cell growth and proliferation (Endo et al., 2009; Kaeberlein, 2013; Qin et al., 2016). The reprogramming efficiency and neuronal differentiation may be enhanced by the VPA (an activator of the mTOR pathway)-activated mTOR signaling pathway (Duan et al., 2019).
2.1.9 The cAMP/PKA Signaling Pathway
The cAMP is the archetypal second messenger discovered in 1956 (Sutherland and Rall, 1958). A high level of cyclic adenosine monophosphate (cAMP) is connected with axonal growth during the development of the embryonic CNS (Cai et al., 2001). When an extracellular ligand binds to its specific Gs-protein coupled receptor and induces a structural change resulting in the Gs-protein complex dissociating, the cAMP signaling pathway is initiated (Billington and Penn, 2003). Gsα subunit from Gs-protein complex drives the classical signaling pathway via activation of adenylyl cyclase (AC) (Billington and Hall, 2012). Then AC catalyzes the conversion of intracellular ATP to cAMP. As the downstream of cAMP, the protein kinase A system (PKA) is activated and modulates protein phosphorylation and gene expression (Billington and Hall, 2012; Yang, 2018).
The cAMP is a ubiquitous second messenger in the regulation of many biological processes, such as cell migration, differentiation, proliferation, and apoptosis (Zhang et al., 2016b). Forskolin is a cAMP/PKA activator that stimulates the transformation of fibroblasts into neurons (Liu et al., 2013; Hu et al., 2019; Cardon et al., 2020; Yang et al., 2020c). Furthermore, the increased intracellular levels of cAMP by Forskolin provide neurotrophic effects, reduce oxidant stress injury, and improve cellular survivability (Xu et al., 2019). It has also been found that small molecules that target the cAMP/PKA pathway can overcome the interference of in vivo environment on transdifferentiation (Yuan et al., 2020).
2.1.10 The ROCK Signaling Pathway
The Rho-associated protein kinase (ROCK) is a serine-threonine kinase with two isoforms (ROCK1 and ROCK2), the inhibition of which causes several biological events such as promotion of neurite outgrowth, axonal regeneration, and activation of prosurvival Akt (Nakagawa et al., 1996; Chong et al., 2017). As the upstream of the ROCK, RhoA (a small GTPase protein) is regulated by guanine nucleotide-exchange factors (GEFs) and GTPase-activating proteins (GAPs) to shift its activation or inactivation (Nakagawa et al., 1996; Cherfils and Zeghouf, 2013; Duman et al., 2015). The activated RhoA activates the ROCK1 or ROCK2, followed by driving actin cytoskeletal remodeling, cell contractility, and cell death (Leung et al., 1995; Matsui et al., 1996; Kimura et al., 2021).
The ROCK signaling pathway is closely associated with the pathogenesis of various CNS disorders through its involvement in many aspects of neuronal functions, including neurite outgrowth and retraction (Koch et al., 2018). The inhibitor of ROCK, Thiazovivin, has been shown to improve cell survival and promote reprogramming efficiency (Watanabe et al., 2007; Lin et al., 2009). Another inhibitor of the ROCK pathway, Y-27632, helps induce fibroblasts into neuron-like cells (Hu et al., 2015). In addition, a study showed that Y-27632 could also effectively reduce reactive oxygen species (ROS) levels in fibroblasts during the process of motor neuron regeneration after injury (Kang et al., 2017).
2.1.11 The JNK Signaling Pathway
The JNK is one of the subfamilies of MAPK, accumulating evidence indicating that this family is also important for neuronal migration during brain development (Sun et al., 2007). Following exposure stressors, the JNKs translocate from the cytoplasm to the nucleus (Zhang et al., 1998). In the nucleus, JNKs phosphorylate several transcription factors, such as c-Jun, ATF-2, Elk-1, p53, and NFAT4, which then trigger specific cell stress responses (Tournier et al., 2000; Sun et al., 2007).
The JNK pathway is an evolutionarily conserved kinase cascade, which plays an important role in stress-induced apoptosis and tumor progression (Herrera and Bach, 2021). It is widely confirmed that there is oxidative stress during direct reprogramming (Gascón et al., 2016; Suzuki and Shults, 2019), and therefore, the inhibition of the JNK pathway can improve the survival of iNs. In this case, the JNK inhibitor, SP600125, is combined with other small molecules to drive the transdifferentiation of somatic cells into neurons (Hu et al., 2015; Hu et al., 2019; Liu et al., 2020).
Since each small molecule activates or inhibits different signaling pathways, the use of different drug cocktails results in variable neuronal properties and conversion efficiencies. It is challenging to elucidate the exact role of each chemical compound reprogramming. Furthermore, only pan-signaling pathways that are regulated by small molecules, are proposed. The specific targets of the chemical during transdifferentiation remain unclear and need further investigations. The review of published studies indicates that various combinations of small molecules are used for neuronal reprogramming. Notably, despite the analogs that act on the same pathway, the individual effect of each small molecule may not be consistent, resulting in different conversion efficiencies. For example, a study demonstrated that when replacing the cocktails, SLCD, consisting of SB431542 (S), LDN193189 (L), CHIR99021 (C), and DAPT (D) by their functional analogs in astrocytes conversion into neurons, the conversion efficiency becomes lower than that of the original SLCD combination (Yang et al., 2020c). Besides, the molecular pathways involved in the transformation process vary, due to the different characteristics of the starting cells, and the complexity of the pathways’ network. Therefore, it is complicated to interpret the specific mechanisms of these molecular combinations.
2.2 The Modulation of Epigenetics by Small Molecules
Some small molecules can regulate the efficiency of reprogramming by modulating epigenetics (Aguirre-Vázquez et al., 2021). These epigenetic modifications are heritable differences that include “Tags” such as DNA methylation and diverse histone modifications that can affect DNA accessibility and chromatin structure (Ebrahimi et al., 2019). DNA methylation and histone modifications are the main mechanisms responsible for the epigenetic regulation of gene expression during cell development and differentiation (Li et al., 2007; Cedar and Bergman, 2009). In general, DNA methylation is associated with gene silencing (Bird, 2002). DNA methyltransferases (DNMTs) are a family of enzymes responsible for DNA methylation (Tessier et al., 2021). DNMTs inhibitors, such as 5-aza-29-deoxycytosine (5-aza-dC), and RG108, have been shown to promote neuronal reprogramming by increasing DNA methylation of non-neuronal gene promoters (Zhang et al., 2015; Zeng et al., 2021). Zhang et al. revealed a significant increase of DNA methylation in the promoter region of the GFAP (glial fibrillary acidic protein) gene via epigenetic analyses during the chemical reprogramming, consistent with previous studies on epigenetic alteration of astrocytes during the neuronal reprogramming (Condorelli et al., 1994; Cheng et al., 2011; Zhang et al., 2015). Histones can be post-translationally modified via methylation and acetylation. Histone methylation contributes to gene activation or repression, while histone acetylation is likely to correlate with gene activation (Struhl, 1998; Boyer et al., 2006; Barski et al., 2007; Kouzarides, 2007; Smith et al., 2016). Histone acetylation is beneficial to open chromatin structures through its disruption of nucleosomal interactions and histone tails’ release from the linker DNA (Kretsovali et al., 2012), which is the basis for the binding of chromatin by neural TFs that facilitate the activation of neuronal programs (Smith et al., 2016). For example, H3K9 (histone H3 at lysine 9) acetylation is one of the most momentous epigenetic markers. This acetylation process can promote gene expression in transdifferentiation when it is enriched in the promoter region of genes (Liang et al., 2004; Bernstein et al., 2005). H3K27 (histone H3 at lysine 27) acetylation is perceived as a super-enhancer that promotes gene expression (Hnisz et al., 2013). Enrichment in H3K27 acetylation has been observed at TF binding sites in reprogrammed fibroblasts into neurons (Smith et al., 2016). The increased histone acetylation level by the HDACs (histone deacetylases) inhibitor, VPA, has been confirmed to increase the level of H3K4 methylation in the promoter region and decrease the level of H3K27 methylation at the transcription start site (TSS) of the NEUROG2 and NEUROD1, which leads to an improved reprogramming efficiency (Phiel et al., 2001; Huangfu et al., 2008; Shi et al., 2008; Hirabayashi et al., 2009; Zhang et al., 2015; Qin et al., 2017; Yin et al., 2019). However, VPA did not appear to serve a notable role or even decreased the reprogramming efficiency in several chemical cocktails (Yin et al., 2019; Yang et al., 2020c). An exciting finding showed that more cell death was observed after longer exposure to VPA because of its cytotoxicity (Zhang et al., 2015). In addition, all-trans retinoic acid (ATRA) can activate histone acetyltransferases (HAT) by binding retinoic acid (RA) receptors to improve the conversion of neuronal transdifferentiation (Han et al., 2010). TTNPB (an agonist of RA receptors) was found to play an important role in neural differentiation, while did not have a significant effect on the astrocyte-neuron reprogramming (Zhang et al., 2015). The lack of contribution of TTNPB indicated that RA may not be a necessary element in reprogramming astrocytes into neurons. Administration time and dosage of epigenetic regulators should be explored when incorporated into chemical cocktail formulas. Moreover, some small molecules, such as forskolin and dorsomorphin (inhibitor of AMP-activated protein kinase and BMP type 1 receptors), have been shown to induce reprogramming, by modulating the epigenetic state, improving chromatin accessibility, and promoting H3K27 acetylation (Liu et al., 2013).
At present, research on the regulation of epigenetics by small molecules is a novel and promising direction in the mechanistic studies of reprogramming. The modulation of epigenetics is temporal and reversible to the unchangeable DNA sequence of the host cells, which is retained during the disease state and age information of the starting cells. Some new techniques, such as high-throughput sequencing analysis, can be utilized to screen pivotal regulators of epigenetics (Headley et al., 2019; Vandana et al., 2021).
2.3 Modulation of Cellular Metabolism by Small Molecules
The metabolic state of cells is affected by intracellular and extracellular factors, such as specific cell cycle phases and cell functions, the oxygenation status, and metabolic demands of tissues under different physiological or pathological conditions (Ito and Suda, 2014). In general, proliferating cells, such as astrocytes and fibroblasts, mainly use anaerobic glycolysis and beta-oxidation for energy metabolism (Lunt and Vander Heiden, 2011; Ge et al., 2012; Gascón et al., 2017; Qin et al., 2017), whereas neurons are more dependent on oxidative phosphorylation (OXPHOS) (Tsacopoulos and Magistretti, 1996; Hülsmann et al., 2000; Bélanger et al., 2011; Shyh-Chang et al., 2013; Magistretti and Allaman, 2015; Zheng et al., 2016), which increase is a characteristic of neuronal identity. Some studies reported that anaerobic glycolysis limits neuronal reprogramming (Agostini et al., 2016; Gascón et al., 2016; Zheng et al., 2016). During the conversion of astrocytes into neurons, a downregulation of glycolysis-related genes and an increase in OXPHOS-related genes have been observed (Magistretti and Allaman, 2015; Masserdotti et al., 2016; Gascón et al., 2017; Ma et al., 2019). Hypoxia-inducible factor 1-alpha (HIF-1α) is a major inhibitor of OXPHOS and an activating mediator of glycolysis. The HIF-1α inhibitor, KC7F2, can improve the induction of neuronal conversion by promoting OXPHOS and its metabolic shift which are necessary for successful induction of neuronal conversion (Herdy et al., 2019). In addition to metabolic state, the metabolic factor ROS can also influence the regulation of cell fate (Maryanovich and Gross, 2013). During neuronal reprogramming, metabolic shift causes oxidative stress, which is a major obstacle to successful conversion (Gascón et al., 2016). It was also reported that there is an increase in lipid peroxidation during the reprogramming of astrocytes into neurons, and that antioxidants, such as vitamin E and forskolin, can promote iNs generation by reducing lipid peroxidation (Liu et al., 2013; Gascón et al., 2016).
In summary, the process of direct neuronal reprogramming is often accompanied by a metabolic shift that leads to an increase in oxidative products, which may inhibit the reprogramming process, and lead to cell death. Therefore, small molecules that can promote metabolic shift and inhibit oxidative stress may be needed in improving the efficiency of reprogramming.
3 Methods to Screen Small Molecule Cocktails
There are several approaches to screening small molecule compounds in the process of reprogramming. Yang et al. screened small molecules based on three major criteria of selection: 1) the disruption of somatic cell-specific programs, such as I-BET151 (Li et al., 2015); 2) the activation of neural reprogramming related signaling pathways, the promotion of neuronal gene expression, and the improvement of reprogramming efficiency, such as VPA, Forskolin, CHIR99021, GO6983, SP600125, ISX9, purmorphamine and dorsomorphin (Huangfu et al., 2008; Ladewig et al., 2012; Hu et al., 2015; Li et al., 2015; Madhu et al., 2016); and 3) the potential to promote neuronal survival and maturation, such as Repsox and Y-27632 (Ladewig et al., 2012; Lamas et al., 2014; Li et al., 2015). In addition, a team identified compounds that predominantly target epigenetic modifications, metabolic transformations, and signaling pathways involved in neuronal fate patterning (Yang et al., 2019). Moreover, Li et al. have used a stepwise protocol in which they screened for small molecules that improved the efficiency of transcription-factor-mediated reprogramming and determined if a combination of the resulting molecules could completely replace the involved TFs (Vierbuchen et al., 2010; Li et al., 2015). The authors found that combining small molecules which drive the conversion of somatic cells into neural progenitor cells, with small molecules that boost the differentiation of the neural progenitors to neurons, directly induces neuronal reprogramming (Hu et al., 2015). In 2016, another study showed that an unbiased screening assay can be used to screen small molecules to effectively increase the efficiency of reprogramming fibroblasts into neurons (Yin et al., 2019). Although numerous, current studies lack effective and standard methods to screen optimal small-molecule cocktail formulas. As we all know, there are plentiful small molecules with similar functions, which makes it challenging to choose the best formulas under different conditions. Theoretically, the above methods may be feasible for directly neuronal reprogramming, but extensive repetitions of experiments are still needed to illustrate these protocols.
When somatic cells are cultured in mediums until they reach a 90% confluence, treatment with small-molecule cocktails can initiate reprogramming (Zhang et al., 2015; Qin et al., 2018; Yin et al., 2019). Direct small molecules-mediated reprogramming is generally divided into two steps: One is associated with the induction culture, which is mainly mediated by small molecules; and the other is the differentiation or maturation culture, which is simply maintained by neurotrophic factors or fewer small molecules (Li et al., 2015; Gao et al., 2017; Ma et al., 2019; Yang et al., 2019). Sometimes, the newly generated neurons are also co-cultured with primary glial cells to support the maturation of induced neuronal electrophysiology and synaptic activities, indicating that glial-derived factors may contribute to the reprogramming (Kashani et al., 2011; Gao et al., 2017; An et al., 2018).
4 Chemical Cocktail Formulas in the Transdifferentiation of Somatic Cells Into Neurons
Small molecule cocktail formulas in the direct transformation of somatic cells into neurons were collected in Table 2. Furthermore, several common transformation protocols and conversion efficiency were summarized in Figure 5. Reviewing the published research, we speculate that the chemicals should be administrated in order. Generally, small molecules were added to induce neuronal conversion for one or 2 weeks firstly, then followed by administrating neurotrophic factors. It is hard to say which molecule is essential or optional in chemical cocktails. Various studies reached diverse conclusions due to different starting cells. Several studies reported that a combination of small chemicals, VCRF (VPA, CHIR99021, Repsox, Forskolin), had important effects on the neural differentiation and survival of mouse and human somatic cells (Cheng et al., 2015a; Hu et al., 2015; Li et al., 2015). Therefore, we speculate that VCRF can be used as a basic formula for direct neuronal reprogramming. However, it is a pity that there is no powerful evidence to confirm this. There are two most common types of somatic cells used for neuronal reprogramming, fibroblasts and astrocytes. Therefore, we will mainly talk about the small molecule cocktail formulas in the process of transdifferentiation starting from these two types of cells.
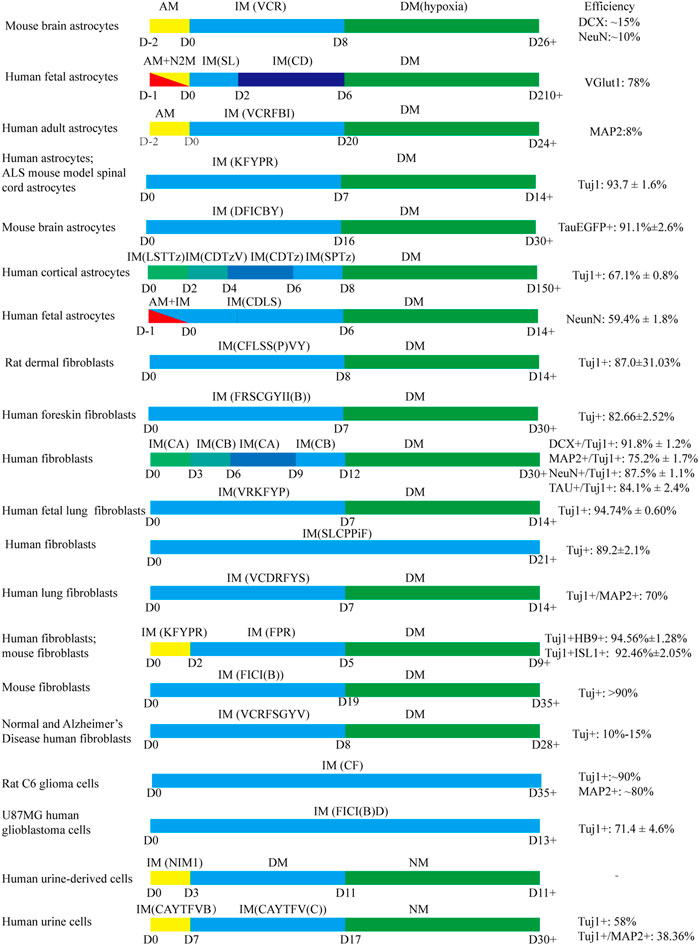
FIGURE 5. The common protocols use small molecules to generate iNs. AM: astrocyte medium; IM: induced medium; DM: differentiation medium; N2M: N2 medium; NM: neuron medium VCR: VPA, Chir99021, Repsox; SL: SB431542, LDN193189; CD: CHIR99021, DAPT; VCRFBI: VPA, Chir99021, Repsox, Forskolin, i-Bet151, ISX-9; KFYPR: Kenpaullone, Forskolin, Y-27632, Purmorphaine, Retinoic acid; DFICBY: DBcAMP, Forskolin, ISX9, CHIR99021, IBET151, Y-27632; LSTTz: LDN193189, SB431542, TTNPB, Tzv; CDTzV: CHIR99021, DAPT, Tzv, VPA; CDTz: CHIR99021, DAPT, Tzv; SPTz: SAG, Purmorphamine, Tzv; CDLS: CHIR99021, DAPT, LDN193189, SB431542; CFLSS(P)VY: CHIR99021, Forskolin, LDN193189, SB431542, SP600125, VPA, Y27632; FRSCGYII(B): Forskolin, RepSox, SP600125, CHIR99021, GO6983, Y-27632, ISX-9, I-BET151; CA: CHIR99021, LDN193189, RG108, Dorsomorphin, P7C3-A20, A83-01, ISX9; CB: Forskolin, Y27632, DAPT, PD0325901, A83-01, purmorphamine, P7C3-A20; VRKFYP: VPA, Repsox, Kenpaullone, Forskolin, Y-27632, Purmorphamine; SLCPPiF: SB431542, LDN-193189, CHIR99021, PD0325901, Pifithrin-α, Forskolin; VCDRFYS: VPA, CHIR99021, DMH1, Repsox, Forskolin, Y-27632, SP600125; KFYPR: KenpaulloneKenpaullone, Forskolin, Y27632, Purmorphamine, RA; FPR: Forskolin, Purmorphamine, RA; FICI(B): Forskolin, ISX9, CHIR99021, I-BET151; VCRFSGYV: VPA, CHIR99021, Repsox, Forskolin, SP600625, GO6983, Y-27632; D, Dorsomorphin; CF: CHIR99021, Forskolin; FICI(B)D: Forskolin, ISX9, CHIR99021, I-BET151, DAPT; NIM1: cAMP-Na, VPA, CHIR99021, Repsox, Forskolin, SP600125, GO6983, Y-27632, IBET151, ISX-9, RA, QVDOPh, vitamin C; CAYTFVB: CHIR99021, A8301, Y-27632, TTNPB, Forskolin, VPA, NaB; CAYTFV(C): CHIR99021, A8301, Y-27632, TTNPB, Forskolin, vitamin C.
Fibroblasts are the most common somatic cells of neuronal reprogramming. Generally, the induction of fibroblasts derived from different species and sites or in vivo and in vitro into neurons needs diverse cocktail formulas (Hu et al., 2015; Li et al., 2015; Wan et al., 2018). However, human and mouse fibroblasts, even rat fibroblasts can be efficiently and directly converted into neurons by the same small molecule cocktails (Qin et al., 2018; Hu et al., 2019). In 2015, Li et al. found that in the cocktail, FICSB (Forskolin, ISX9, CHIR99021, SB431542, and I-BET151), the neurogenesis inducer ISX9 was necessary to activate neuron-specific genes, and SB431542 was dispensable for generating neurons, although it enhanced the survival and neurite outgrowth of the iNs. In addition, I-BET151 (a BET family bromodomain inhibitor) disrupted the fibroblast-specific programs in early-stage reprogramming, dramatically enhancing the reprogramming rate and neurite outgrowth of the iNs (Li et al., 2015). Small molecules, Repsox, Y-27632, CHIR99021, Dorsomorphin, Forskolin, Kenpaullone, LDN193189, P7C3-A20 and SP600125 have been reported to promote neuronal survival, differentiation and maturation (Rawal et al., 2007; Hotta et al., 2009; Ladewig et al., 2012; Liu et al., 2013; Lamas et al., 2014; Hu et al., 2015; Qin et al., 2018; Wan et al., 2018; Yang et al., 2019). Yang et al. claimed that RG108, PD0325901 and A83-01 supplementation boosted the neuronal reprogramming efficiency significantly (Yang et al., 2019). Interestingly, the absence of either PD03259 or A83-01 had a slight effect on the conversion rate (Yang et al., 2019). We noticed that Forskolin was present in almost all small-molecule cocktails of the existing transdifferentiation of fibroblasts into neurons, the reason for which may be due to it promoting reprogramming as a PKA activator and antioxidant, or the synergy with other chemicals.
As for astrocytes, another somatic cell widely used in the field of neuronal reprogramming, are shared the same neuroectodermal linage and the same progenitor radial glia with neurons, which may partially underlie astrocyte-to-neuron conversion (Rao et al., 2021). Cheng et al. declared that VPA alone was able to induce astrocytes into neuroblasts with low efficiency, while the elimination of VPA significantly abolished the generation of neuroblasts from postnatal mouse astrocytes (Cheng et al., 2015b). Consistent with fibroblast-to-neuron reprogramming, ISX-9 activated neuronal genes while I-BET151 suppressed astrocyte genes in the direct generation of neuronal cells from adult astrocytes (Gao et al., 2017). Kenpaullone was shown to play an important role in the neuronal morphological changes in a chemical cocktail (Zhao et al., 2020). DAPT, CHIR99021, SB431542, LDN193189, DBcAMP and Y-27632 significantly enhanced the neuronal conversion efficiency of mouse astrocytes, and DAPT was the most (Ma et al., 2021).
In addition to fibroblasts and astrocytes, human urine-derived cells, which are easily obtained without invasive injury, were converted into neuron-like cells with the help of VPA, CHIR99021, Repsox, Forskolin, SP600625, GO6983, Y27632, ISX9, I-BET151, RA, VitC, QVD-OPH and CHIR99021, A8301, Y27632, TTNPB, Forskolin, VPA, NaB (Xu et al., 2019; Liu et al., 2020). More promisingly, glioblastoma cells and glioma cells can be transdifferentiated into fully differentiated neurons by chemical cocktails hence losing their malignant characteristics (Oh et al., 2017; Lee et al., 2018).
The comprehensive roles and synergetic effects of these small molecules in neuronal conversion remain to be further explored. It is hard to say the pros and cons of different chemical cocktails. On the one hand, the neuronal transdifferentiation was inefficient and ineffective when fewer small molecules were involved (Cheng et al., 2015b). On the other hand, it has been shown that the more small molecules were contained in the formulations, the greater the toxic effect and was difficult for clinical applications (Zhang et al., 2015; Yin et al., 2019).
5 Detection of the Efficiency of the Reprogramming Conversion
The transformation efficiency is one of the most momentous focuses in neuronal reprogramming. There are various existing detection techniques to evaluate the conversion efficiency of neuronal reprogramming. The most significant conversion of somatic cells into neurons usually occurs within the first 24 h of treatment with small molecules (Li et al., 2015; Ma et al., 2019). Therefore, it is necessary to observe the morphology change frequently on 1 day after the administration of chemicals. An easy and direct detection method is to microscopically observe the typical morphology of the reprogrammed cells. The iNs usually have smaller, more compact, and more shining cell bodies, with bipolar or multipolar shapes, and the appearance of secondary and tertiary branches (Xu et al., 2019; Yang et al., 2019; Ma et al., 2021). The neurite length can be captured by confocal microscopy (Gao et al., 2017). In addition, the conversion of astrocytes to neurons can be dynamically monitored by time-lapse live-cell imaging or continuous photo taking in vitro (Zhang et al., 2015; Gascón et al., 2016; Yin et al., 2019; Rao et al., 2021); however, these methods will be technically and expensively prohibitive for most research labs (Wang et al., 2021b). Besides, immunofluorescence staining is one of the most common methods used for the detection of reprogramming efficiency. Generally, successfully induced neurons express neuronal markers, such as β-tubulin III (TUJ1), microtubule association protein-2 (MAP2), neuronal nuclei (NeuN), and doublecortin (DCX) (Rao et al., 2021). The origin of iNs can be determined by co-labeled fluorescence markers of initiating cells and neurons, which can be observed by epifluorescent microscopy or confocal microscopy in vitro (Xu et al., 2019; Yin et al., 2019; Ma et al., 2021). Moreover, immunostaining with synapsin I highlight the presence of strong iNs synaptic puncta along dendrites, indicating the formation of the well-established synapses (Hu et al., 2015; Yang et al., 2019).
However, the observations of morphology and immunofluorescence staining cannot identify the lineage origins of iNs which is vital for neuronal reprogramming. In 1981, Sternberg and Hamilton were the first to describe Cre, a recombinase enzyme isolated from the P1 bacteriophage, that catalyzes a directional DNA recombination between two 34-bp recognition elements called loxP sites (Sternberg and Hamilton, 1981). The Cre-loxP lineage tracing system is a common conditional lineage tracing technology that specifically labels the original cells in vivo (Ma et al., 2021). In this regard, the Aldehyde dehydrogenase 1 family member L1-Cre (Aldh1l1-Cre) Mice were intracranially injected with a flexed EGFP (Enhanced Green Fluorescent Protein)-adeno-associated virus (AAV-FLEX-EGFP), that specifically labels Aldehyde dehydrogenase 1 family member L1 (ALDH1L1)-expressing astrocytes (Ma et al., 2021). Another tracing technology is mediated by a virus vector that labels the starting cells with a green fluorescent protein (GFP) (Zhang et al., 2015; Gao et al., 2017; Yin et al., 2019), which is more specific and precise than fluorescence staining. For example, Gao and his colleagues traced the cultured astrocytes with retrovirus expressing GFP from human GFAP promoter (GFAP::GFP) (Gao et al., 2017). These starting cells’ labeling techniques can be utod in co-label with neuron-specific markers to identify the origin of iNs. In addition, a common and achievable method is to detect the expression of neuronal-specific proteins using western blot (Hu et al., 2015; Oh et al., 2017). Similarly, RT-qPCR (real-time quantitative polymerase chain reaction) can be used to monitor the expression of neuronal-specific genes (Qin et al., 2018; Yang et al., 2019).
The successful reprogramming is characterized by the iNs and endogenous neurons sharing similar transcriptional patterns. When investigating the transcriptional mechanisms of reprogramming, RT-qPCR can also be applied to quantitatively detect the differentially expressed genes (DEGs) (Qin et al., 2018; Yang et al., 2019; Yin et al., 2019). The RNA-sequencing (RNA-seq) technology is also used to investigate the transcriptome changes during the somatic cell-to-neuron conversion process (Gao et al., 2017; Ma et al., 2019; Yin et al., 2019). Comparing single-cell RNA-sequencing data from endogenous neurons and iNs a more efficient reprogramming can be achieved by correcting the differential gene expression using the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technique (Götz and Bocchi, 2021). The results of Gene Ontology (GO) analysis can also be used to identify similarities between iNs and endogenous neurons by determining the DGEs involved in different cell fates (Yang et al., 2019; Ma et al., 2021).
Apart from the change in cell morphology and gene expression, it is equally important that the iNs possess mature electrophysiological functions. Electrophysiological properties of iNs can usually be examined by the whole-cell patch-clamp technique that records action potentials, inward sodium currents, and outward potassium currents in vitro (Ma et al., 2019; Yang et al., 2019; Ma et al., 2021). The detection of typical spontaneous postsynaptic currents (sPSCs) indicates the formation of synaptic connections among neurons (Hu et al., 2015; Zhang et al., 2015; Gao et al., 2017).
Taken together, the identification of converted neurons should be validated from various aspects, such as the expression of neuron-specific genes and protein markers, and neuronal electrophysiological function, which is the basis for the application of the chemical-induced neurons (CiNs) in clinical translation.
Moreover, most important is employing multiple stringent lineage-tracing methods to identify the cell origin for the iNs (Wang et al., 2021b; Rao et al., 2021). Due to the wide variety of cells in vivo, compared with the pure culture cells in vitro, the origin of the cells in vivo should be identified by stringent lineage tracing. Of note, wang et al. found that the AAV-mediated co-expression of NeuroD1 and a reporter efficiently induced reporter-labeled endogenous neurons because NeuroD1 likely cis-regulated the specificity of human GFAP (hGFAP) promoter for its expression in neurons (Wang et al., 2021b). There will be more explorations to figure out complex cross networks among the virus vectors, TFs, and reporters in various cells. They concluded that the tamoxifen-inducible Aldh1l1-CreERT2; R26R-YFP line seems preferable with minimal labeling of endogenous neurons and the Aldh1l1-CreERT2; R26R-tdTomato line also is generally specific by comparing several tracing methods in tracing the lineage of astrocytes (Wang et al., 2021b). At the same time, Rao et al. declared that NeuroD1 and other TFs (PAX6, ASCL1, SOX2, PTBP1) cannot induce microglia-to-neuron conversion via rigid lineage tracing (Rao et al., 2021). It is necessary to employ rigorous lineage tracing in the controversial TFs-mediated cross-lineage reprogramming. Noteworthy, whether there are mislabeling neurons is also needed further investigation during chemical reprogramming. It is misleading to gain exciting results by mislabeling the endogenous neurons (Wang et al., 2021b).
6 Administration of Small Molecules in vivo Neuronal Reprogramming
Since the microenvironment is complex in vivo, it is vital to pursue the optimal delivery strategies of the chemicals. There are several methods for the delivery of the small molecules into the body, such as single intracranial injection, intraperitoneal (i.p.) administration, and constant intracranial injection by an osmotic mini-pumping system, slow-release via biomaterials (Cheng et al., 2015c; Yin et al., 2019; Ma et al., 2021). However, which delivery route is best is a conundrum.
One of the biggest challenges for in vivo reprogramming studies is the maintenance of a constant concentration of small molecules inside the brain, and without causing serious damages (Yin et al., 2019). Although core drugs can be sent to the target sites of the brain through stereotaxic intracranial injection, they might cause invasive damage to the brain tissue. In addition, the one-time injection might limit the efficacy of reprogramming due to the inability of maintaining a constant concentration and the limited treatment time. Using intraperitoneal (i.p.) administration, researchers have confirmed that small molecules, such as DAPT, CHIR99021, SB431542, and LDN193189, can pass through the blood-brain barrier (BBB) to regulate adult neurogenesis (Yin et al., 2019). Compared with intracranial injection, intraperitoneal injection is more suitable for repeated administration and has less traumatic effects on rodents. However, repeat high-dose administration of the drug will cause high cost, which might restrain wide application from the clinical translation aspect. Besides, after passing through the BBB, the concentration of small molecules in the intravascular is extremely low, which may affect the in vivo efficiency of reprogramming. Therefore, the dosage of small molecules administered by i. p. injection would be much higher than that for intracranial treatment. Furthermore, the in vivo dosage of small molecules should usually be higher than that used in vitro (Ma et al., 2021), due to part of the drugs permeating the non-target cells in vivo. Recently, it has been reported that chemical compounds can be administrated into animal brains (striatum or cortex) at a constant rate via a mini-osmotic pumping system that maintains a constant concentration (Gao et al., 2017; Yang et al., 2020c). This method has the advantage of providing a stable and continuous release of the drugs. However, the prolonged presence of mini-osmotic pumps in the brain may cause infection and additional invasive injury to the rodents. In addition, some researchers have proposed to use biomaterial that encapsulates small molecules to achieve a targeted delivery to the brain (Cheng et al., 2015c). Besides, prodrugs could be used to improve the selectivity of the small molecules in the future (Ma et al., 2021). However, the attempt failed since the small molecules did not stay for a long time in the brain, which may be due to the small size and the unsuitability of biomaterial that has been selected for such small molecules.
In summary, no perfect delivery routes have been found for the delivery of small molecules into the brain. An ideal method should be explored in the future to maintain a constant concentration of small molecules in the brain without causing severe invasive damage to the brain, as well as lower off-target effects.
7 Direct Neuron Reprogramming in Neurological Disorders
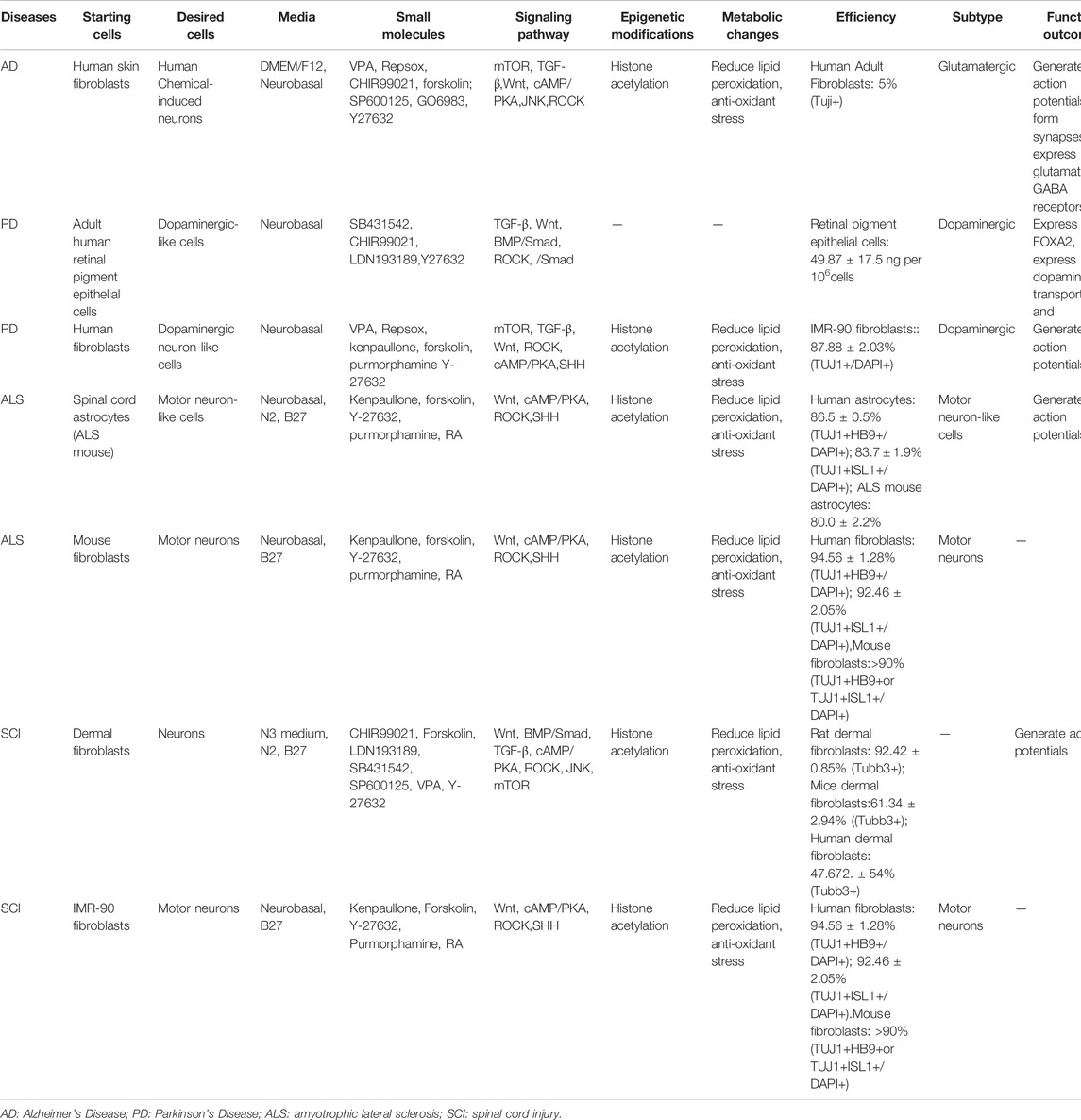
Some studies reported that the conversion of reactive dividing cells, that are found around the lesion sites, into various subtypes of neurons may be a promising strategy for some neurological diseases (An et al., 2018; Mollinari and Merlo, 2021). For example, induced motor neurons can promote recovery of amyotrophic lateral sclerosis (ALS) and spinal cord injury (SCI), and an induced dopaminergic neurons supplement is beneficial to patients with Parkinson’s Disease (PD). Here, we summarize current studies that focused on small molecules-mediated direct neuronal reprogramming in different neurological disease models (Table 3).
Alzheimer’s disease (AD) is a devastating condition of the aging population, characterized by learning and memory deficits due to plaques’ formation in the hippocampus (Kashani et al., 2011). A study has shown that fibroblasts deriving from skins of patients with familial Alzheimer’s disease (FAD) can be directly converted into human chemical-induced neurons (hCiNs) using the chemical cocktail, VCRFSGY, that includes VPA, CHIR99021, Repsox, Forskolin, SP600125, GO6983, Y27632. However, most of the hCiNs are glutamatergic neurons in their induction system (Hu et al., 2015). Regrettably, researchers didn’t conduct chemical transdifferentiation or the transplantation of hCiNs in vivo (Hu et al., 2015). What is expected is these hCiNs could be transplanted into the individual patient but don’t cause the risk of tumorigenicity and transplantation rejection, which helps to achieve personalized medicine.
PD is a neurodegenerative disease characterized by the progressive loss of dopaminergic neurons in the midbrain substantia nigra pars compacta (Dawson and Dawson, 2003; Dexter and Jenner, 2013). Motor and nonmotor symptoms are displayed in PD patients, including resting tremors, bradykinesia, postural instability, and rigidity (Hoehn and Yahr, 1967). Li’s team found that adult human retinal pigment epithelial cells can transdifferentiate into dopaminergic-like cells in vitro following induction by a combination of SB431542, CHIR99021, and LDN193189. After transplanting the inducted cells into the brain, recipient PD monkeys showed significant improvement in clinical conditions (Li et al., 2019). Another study showed that a cocktail consisting of VPA, Repsox, kenpaullone (another inhibitor of GSK-3β), forskolin, purmorphamine, and Y-27632 can convert human fibroblasts into dopaminergic neuron-like cells in vitro (Qin et al., 2020). The transplantation of the induced dopaminergic-like neurons can be utilized to ameliorate Parkinson’s disease in the future. In addition, the direct induction of responsive glial cells in the stratum by small molecules for the generation of dopaminergic-like neurons is another promising research direction.
ALS is a lethal neurodegenerative disorder that affects lower and upper motor neurons (Amico and Antel, 1981). Zhao et al. have found that the drug cocktails KFYPR (kenpaullone, forskolin, Y-27632, purmorphamine, and RA), can induce the transdifferentiation of spinal cord astrocytes, from an ALS mice model carrying a SOD1 mutation, into motor neuron-like cells in vivo (Zhao et al., 2020). However, the CiNs exhibited a decrease in cell survival and an increase in oxidative stress compared to wild-type motor neurons derived from healthy mice (Zhao et al., 2020). Another study reported that human fibroblasts can be directly reprogrammed into motor neurons using the same five small molecules in vitro (Qin et al., 2018). Moreover, it has been shown that ALS patients derived fibroblasts can be directly converted into motor neurons by introducing the TFs NEUROG2, SOX11, ISL1, and LHX3, and the morphological and survival deficits of ALS human-induced motor neurons (hiMNs) can be ameliorated by the small molecule kenpaullone (Liu et al., 2016).
SCI results in damage to motor, sensory and autonomic functions (Gris et al., 2004; Hou and Rabchevsky, 2014; Smith et al., 2016; Ghasemi-Kasman et al., 2018; Zheng et al., 2021). In 2019, Hu and his colleagues found that after transplantation of neural scaffolds consisting of dermal fibroblasts-reprogrammed neurons that were induced by a small-molecule cocktail CFLSSVY (CHIR99021, Forskolin, LDN193189, SB431542, SP600125, VPA, and Y27632), the damaged tissue was repaired obviously and the hindlimb movements and motor-nerve conductivities of SCI-treated rats were significantly improved (Hu et al., 2019). In addition, human IMR-90 fibroblasts have been directly converted into motor neurons with the small molecules, forskolin, kenpaullone, purmorphamine, RA, and Y-27632 (Qin et al., 2018).
To sum up, most of the current studies focused on the efficiency of different inductive chemical formulas in neuronal reprogramming in vitro. Only a few studies observed the in vivo transdifferentiation of reactive glial cells into neurons under pathophysiological states. More studies are required to explore the safety and efficacy of endogenous neuronal reprogramming in various neurological disorders from the perspective of future clinical translation.
8 Discussion
In the present review, we mainly discussed the mechanisms of different small molecules, the applied approaches in screening small molecule cocktails, the chemical cocktail formulas in neuronal transdifferentiation, the detection methods used to assess the conversion efficiency, and the administration of small molecules in vivo. We also summarized current studies that focused on direct small molecules-mediated reprogramming of somatic cells into neurons in different neurological disease models. For the molecular mechanistic studies, we reviewed some pathways involved in chemical reprogramming, such as the Wnt, JAK-STAT, SHH, BMP/Smad, TGF-β, Notch, p38 MAPK, mTOR, cAMP/PKA, JNK, and ROCK pathway. In addition, epigenetic modifications and metabolic changes during the chemical-induced reprogramming process have also been summarized. Some recent scientific tools such as single-cell sequencing and CRISPR-based genome-wide screening will help probe new chemical cocktails and elaborate the underlying induction mechanisms (Wen and Tang, 2016; Joung et al., 2017). At present, the studies about direct neuron reprogramming by small molecules in disease models are rare, with only a few cases in neurodegenerative diseases and SCI. More efforts are needed to explore the application of chemical transdifferentiation in other neurological diseases.
Existing issues and challenges need overcoming in future studies. Firstly, the optimal concentration of each small molecule in the cocktails is difficult to determine. In a study, the researchers tried hundreds of different cocktails tests before finding the optimal concentrations of the nine assessed small molecules (LDN193189, SB431542, TTNPB, thiazovivin, CHIR99021, VPA, DAPT, SAG, and purmorphamine) in converting astrocytes into neurons, which is time-wasting and costly (Zhang et al., 2015). Secondly, the length of the induction time is difficult to determine, as longer times can have toxic effects on the cells, while shorter times might not be effective in inducing neuronal transformation. Thirdly, it is a big challenge to seek an optimal formula to achieve acceptable conversion efficiency. From the clinical translation aspect, the composition of small molecules should be as simple as possible on the premise of acceptable conversion efficiency (Smith et al., 2016; Yin et al., 2019). We summarized the transformation efficiencies from somatic cells to neurons in this review, of which the higher efficiencies were 94.74 ± 0.60% (Tuj+/DAPI+), 94.56 ± 1.28% (Tuj1+HB9+/DAPI+) and 93.7 ± 1.6% (Tuj1+/DAPI+) (Qin et al., 2018; Qin et al., 2020; Zhao et al., 2020). Nevertheless, statistical indicators, cell types and induced protocols of each study were heterogeneous, so it was difficult to clarify the best protocols. More research is needed to explore the most efficient protocols in the future. The in vitro microenvironment of induction is relatively simple and easy to monitor. However, there are many unresolved questions vis-à-vis the in vivo application of small molecules-mediated programming. Specifically speaking, how the in vivo complex microenvironment interacts with small molecules and whether it changes the effect of small molecules, are questions that require further investigations. Moreover, since small molecules lack cellular specificity to enter the brain, it is hard to judge whether other cell types, apart from the intended starting cell type (Man et al., 2018), are affected. In addition, some of the chemicals may have some unwanted toxic effects on normal brain cells (Man et al., 2018). Another major concern is whether newly transformed neurons can survive for a long time and efficiently integrate into local neural circuits. Some researchers have adopted retrograde a mono-transsynaptic tracing technique using a pseudotyped rabies virus to determine whether CiNs can form synaptic connections with host neurons in vivo (Ma et al., 2021). Another challenge for in vivo chemical transdifferentiation is the development of an ideal administration method that preserves a constant concentration of small molecule compounds inside the brain without causing severe invasive damage. A micro-injection pump for continuous drug delivery and biological material for drugs’ slow-release are attractive tools for the delivery of the drugs (Yin et al., 2019; Ma et al., 2021). To achieve the cellular specificity of small molecules, nanoparticles containing specific signals which can recognize target cells can be utilized in the future (Prapainop et al., 2012). For the treatment of some neurological disorders, subtype-specific neurons should be induced in appropriate regions of the brain (Smith et al., 2016). Recently, it has been proposed that some in vitro chemical-induced stable neurons could be constructed for brains by a three-dimensional (3D) bioprinting technology, which prints biodegradable materials with cells as 3D tissue (Ho and Hsu, 2018; Yuan et al., 2020). In 2021, a lab bioprinted MSC-derived neural tissues successfully using a fibrin-based bioink and the RX1 bioprinter with the aid of SB431542, LDN-193189, purmorphamine, fibroblast growth factor 8 (FGF8), fibroblast growth factor-basic (bFGF), and brain-derived neurotrophic factor (BDNF) (Restan Perez et al., 2021). Even more interesting is that Ho et al. achieved cell reprogramming via 3D bioprinting of human fibroblasts in polyurethane hydrogel for the manufacture of neural-like constructs with Forkhead box D3 (FoxD3), a transcription factor (Ho and Hsu, 2018).
Since in vivo reprogramming of glial cells into neurons is an excellent research avenue for the treatment of neurological disorders, a better understanding of the mechanisms of small molecule cocktails-induced neuronal reprogramming is critical. However, there are currently limited studies that were published on the endogenous transdifferentiation from somatic cells into neurons and in different disease models. Future research efforts are needed to screen the optimal induction formula, attain cellular specificity entry of small molecules, optimize the routes of administration and improve transdifferentiation efficiency.
Author Contributions
ZT and GL contributed to the conception and revised the review paper. JyW and SC collected the related research articles. JyW wrote the manuscript, drew the figures, and summarized the tables. CP corrected pictures and tables. All the authors read and approved the final article.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82071330, No. 81901219).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Dr. Jiahui Wang and Dr. Danyang Chen for revising the manuscript.
References
Ables, J. L., Breunig, J. J., and Eisch, A. J. (2011). Not(ch) Just Development: Notch Signalling in the Adult Brain. Nat. Rev. Neurosci. 12, 269–283. doi:10.1038/nrn3024
Agostini, M., Romeo, F., Inoue, S., Niklison-Chirou, M. V., Elia, A. J., Dinsdale, D., et al. (2016). Metabolic Reprogramming during Neuronal Differentiation. Cell Death Differ. 23, 1502–1514. doi:10.1038/cdd.2016.36
Aguirre, A., Rubio, M. E., and Gallo, V. (2010). Notch and EGFR Pathway Interaction Regulates Neural Stem Cell Number and Self-Renewal. Nature 467, 323–327. doi:10.1038/nature09347
Aguirre-Vázquez, A., Salazar-Olivo, L. A., Flores-Ponce, X., Arriaga-Guerrero, A. L., Garza-Rodríguez, D., Camacho-Moll, M. E., et al. (2021). 5-Aza-2′-Deoxycytidine and Valproic Acid in Combination with CHIR99021 and A83-01 Induce Pluripotency Genes Expression in Human Adult Somatic Cells. Molecules 26, 1909. doi:10.3390/molecules26071909
Albert, K., Niskanen, J., Kälvälä, S., and Lehtonen, Š. (2021). Utilising Induced Pluripotent Stem Cells in Neurodegenerative Disease Research: Focus on Glia. Ijms 22, 4334. doi:10.3390/ijms22094334
Aluko, O. M., Lawal, S. A., Ijomone, O. M., and Aschner, M. (2021). Perturbed MAPK Signaling in ASD: Impact of Metal Neurotoxicity. Curr. Opin. Toxicol. 26, 1–7. doi:10.1016/j.cotox.2021.03.009
Amack, J. D. (2021). Cellular Dynamics of EMT: Lessons from Live In Vivo Imaging of Embryonic Development. Cell Commun. Signal 19, 79. doi:10.1186/s12964-021-00761-8
Ambasudhan, R., Talantova, M., Coleman, R., Yuan, X., Zhu, S., Lipton, S. A., et al. (2011). Direct Reprogramming of Adult Human Fibroblasts to Functional Neurons under Defined Conditions. Cell Stem Cell 9, 113–118. doi:10.1016/j.stem.2011.07.002
Amico, L. L., and Antel, J. P. (1981). Amyotrophic Lateral Sclerosis. Postgrad. Med. 70, 50–61. doi:10.1080/00325481.1981.11715822
An, N., Xu, H., and Gao, W.-Q. (2018). Direct Conversion of Somatic Cells into Induced Neurons. Mol. Neurobiol. 55, 642–651. doi:10.1007/s12035-016-0350-0
Angers, S., and Moon, R. T. (2009). Proximal Events in Wnt Signal Transduction. Nat. Rev. Mol. Cell Biol. 10, 468–477. doi:10.1038/nrm2717
Arrázola, M. S., Varela-Nallar, L., Colombres, M., Toledo, E. M., Cruzat, F., Pavez, L., et al. (2009). Calcium/calmodulin-dependent Protein Kinase Type IV Is a Target Gene of the Wnt/β-Catenin Signaling Pathway. J. Cell. Physiol. 221, 658–667. doi:10.1002/jcp.21902
Au-Yeung, N., Mandhana, R., and Horvath, C. M. (2013). Transcriptional Regulation by STAT1 and STAT2 in the Interferon JAK-STAT Pathway. JAK-STAT 2, e23931. doi:10.4161/jkst.23931
Bai, C. B., Stephen, D., and Joyner, A. L. (2004). All Mouse Ventral Spinal Cord Patterning by Hedgehog Is Gli Dependent and Involves an Activator Function of Gli3. Dev. Cell 6, 103–115. doi:10.1016/s1534-5807(03)00394-0
Banerjee, S., Biehl, A., Gadina, M., Hasni, S., and Schwartz, D. M. (2017). JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs 77, 521–546. doi:10.1007/s40265-017-0701-9
Barker, R. A., and de Beaufort, I. (2013). Scientific and Ethical Issues Related to Stem Cell Research and Interventions in Neurodegenerative Disorders of the Brain. Prog. Neurobiol. 110, 63–73. doi:10.1016/j.pneurobio.2013.04.003
Barski, A., Cuddapah, S., Cui, K., Roh, T.-Y., Schones, D. E., Wang, Z., et al. (2007). High-resolution Profiling of Histone Methylations in the Human Genome. Cell 129, 823–837. doi:10.1016/j.cell.2007.05.009
Bélanger, M., Allaman, I., and Magistretti, P. J. (2011). Brain Energy Metabolism: Focus on Astrocyte-Neuron Metabolic Cooperation. Cell Metab. 14, 724–738. doi:10.1016/j.cmet.2011.08.016
Ben-Levy, R., Hooper, S., and Wilson, R. (1998). Nuclear Export of the Stress-Activated Protein Kinase P38 Mediated by its Substrate MAPKAP Kinase-2. Curr. Biol. 8, 1049–1057. doi:10.1016/s0960-9822(98)70442-7
Bernstein, B. E., Kamal, M., and Lindblad-Toh, K. (2005). Genomic Maps and Comparative Analysis of Histone Modifications in Human and Mouse. Cell 120, 169–181. doi:10.1016/j.cell.2005.01.001
Billington, C. K., and Hall, I. P. (2012). Novel cAMP Signalling Paradigms: Therapeutic Implications for Airway Disease. Br. J. Pharmacol. 166, 401–410. doi:10.1111/j.1476-5381.2011.01719.x
Billington, C. K., and Penn, R. B. (2003). Signaling and Regulation of G Protein-Coupled Receptors in Airway Smooth Muscle. Respir. Res. 4, 2. doi:10.1186/rr195
Bird, A. (2002). DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 16, 6–21. doi:10.1101/gad.947102
Borghese, L., Dolezalova, D., Opitz, T., Haupt, S., Leinhaas, A., Steinfarz, B., et al. (2010). Inhibition of Notch Signaling in Human Embryonic Stem Cell-Derived Neural Stem Cells Delays G1/S Phase Transition and Accelerates Neuronal Differentiation In Vitro and In Vivo. Stem Cells 28, 955–964. doi:10.1002/stem.408
Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., et al. (2006). Polycomb Complexes Repress Developmental Regulators in Murine Embryonic Stem Cells. Nature 441, 349–353. doi:10.1038/nature04733
Breunig, J. J., Silbereis, J., Vaccarino, F. M., Šestan, N., and Rakic, P. (2007). Notch Regulates Cell Fate and Dendrite Morphology of Newborn Neurons in the Postnatal Dentate Gyrus. Proc. Natl. Acad. Sci. U.S.A. 104, 20558–20563. doi:10.1073/pnas.0710156104
Briscoe, J., and Thérond, P. P. (2013). The Mechanisms of Hedgehog Signalling and its Roles in Development and Disease. Nat. Rev. Mol. Cell Biol. 14, 416–429. doi:10.1038/nrm3598
Brulet, R., Matsuda, T., Zhang, L., Miranda, C., Giacca, M., Kaspar, B. K., et al. (2017). NEUROD1 Instructs Neuronal Conversion in Non-reactive Astrocytes. Stem Cell Rep. 8, 1506–1515. doi:10.1016/j.stemcr.2017.04.013
Bushati, N., and Cohen, S. M. (2007). microRNA Functions. Annu. Rev. Cell Dev. Biol. 23, 175–205. doi:10.1146/annurev.cellbio.23.090506.123406
Cai, D., Qiu, J., Cao, Z., McAtee, M., Bregman, B. S., and Filbin, M. T. (2001). Neuronal Cyclic AMP Controls the Developmental Loss in Ability of Axons to Regenerate. J. Neurosci. 21, 4731–4739. doi:10.1523/jneurosci.21-13-04731.2001
Campbell, I. L. (2005). Cytokine-mediated Inflammation, Tumorigenesis, and Disease-Associated JAK/STAT/SOCS Signaling Circuits in the CNS. Brain Res. Rev. 48, 166–177. doi:10.1016/j.brainresrev.2004.12.006
Cao, N., Huang, Y., Zheng, J., Spencer, C. I., Zhang, Y., Fu, J.-D., et al. (2016). Conversion of Human Fibroblasts into Functional Cardiomyocytes by Small Molecules. Science 352, 1216–1220. doi:10.1126/science.aaf1502
Cardon, T., Franck, J., Coyaud, E., Laurent, E. M. N., Damato, M., Maffia, M., et al. (2020). Alternative Proteins Are Functional Regulators in Cell Reprogramming by PKA Activation. Nucleic Acids Res. 48, 7864–7882. doi:10.1093/nar/gkaa277
Cedar, H., and Bergman, Y. (2009). Linking DNA Methylation and Histone Modification: Patterns and Paradigms. Nat. Rev. Genet. 10, 295–304. doi:10.1038/nrg2540
Chan, K. Y., Jang, M. J., Yoo, B. B., Greenbaum, A., Ravi, N., Wu, W.-L., et al. (2017). Engineered AAVs for Efficient Noninvasive Gene Delivery to the Central and Peripheral Nervous Systems. Nat. Neurosci. 20, 1172–1179. doi:10.1038/nn.4593
Chen, Y., Wu, B., He, J. F., Chen, J., Kang, Z. W., Liu, D., et al. (2021). Effectively Intervening Epithelial-Mesenchymal Transition of Retinal Pigment Epithelial Cells with a Combination of ROCK and TGF-β Signaling Inhibitors. Invest. Ophthalmol. Vis. Sci. 62, 21. doi:10.1167/iovs.62.4.21
Cheng, C. J., Tietjen, G. T., Saucier-Sawyer, J. K., and Saltzman, W. M. (2015). A Holistic Approach to Targeting Disease with Polymeric Nanoparticles. Nat. Rev. Drug Discov. 14, 239–247. doi:10.1038/nrd4503
Cheng, L., Gao, L., Guan, W., Mao, J., Hu, W., Qiu, B., et al. (2015). Direct Conversion of Astrocytes into Neuronal Cells by Drug Cocktail. Cell Res. 25, 1269–1272. doi:10.1038/cr.2015.120
Cheng, L., Hu, W., Qiu, B., Zhao, J., Yu, Y., Guan, W., et al. (2015). Erratum: Generation of Neural Progenitor Cells by Chemical Cocktails and Hypoxia. Cell Res. 25, 645–646. doi:10.1038/cr.2015.55
Cheng, P.-Y., Lin, Y.-P., Chen, Y.-L., Lee, Y.-C., Tai, C.-C., Wang, Y.-T., et al. (2011). Interplay between SIN3A and STAT3 Mediates Chromatin Conformational Changes and GFAP Expression during Cellular Differentiation. Plos One 6, e22018. doi:10.1371/journal.pone.0022018
Cheon, S. Y., Cho, K. J., Song, J., and Kim, G. W. (2016). Knockdown of Apoptosis Signal-Regulating Kinase 1 Affects Ischaemia-Induced Astrocyte Activation and Glial Scar Formation. Eur. J. Neurosci. 43, 912–922. doi:10.1111/ejn.13175
Cherfils, J., and Zeghouf, M. (2013). Regulation of Small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309. doi:10.1152/physrev.00003.2012
Chong, C.-M., Ai, N., and Lee, S. (2017). ROCK in CNS: Different Roles of Isoforms and Therapeutic Target for Neurodegenerative Disorders. Cdt 18, 455–462. doi:10.2174/1389450117666160401123825
Clevers, H., and Nusse, R. (2012). Wnt/β-Catenin Signaling and Disease. Cell 149, 1192–1205. doi:10.1016/j.cell.2012.05.012
Condorelli, D. F., Nicoletti, V. G., Barresi, V., Caruso, A., Conticello, S., de Vellis, J., et al. (1994). Tissue-specific DNA Methylation Patterns of the Rat Glial Fibrillary Acidic Protein Gene. J. Neurosci. Res. 39, 694–707. doi:10.1002/jnr.490390610
Cuenda, A., and Rousseau, S. (2007). p38 MAP-Kinases Pathway Regulation, Function and Role in Human Diseases. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1773, 1358–1375. doi:10.1016/j.bbamcr.2007.03.010
Dai, P., Harada, Y., and Takamatsu, T. (2015). Highly Efficient Direct Conversion of Human Fibroblasts to Neuronal Cells by Chemical Compounds. J. Clin. Biochem. Nutr. 56, 166–170. doi:10.3164/jcbn.15-39
Dawson, T. M., and Dawson, V. L. (2003). Molecular Pathways of Neurodegeneration in Parkinson's Disease. Science 302, 819–822. doi:10.1126/science.1087753
de Bivort, B. L., Guo, H.-F., and Zhong, Y. (2009). Notch Signaling Is Required for Activity-dependent Synaptic Plasticity at theDrosophilaNeuromuscular Junction. J. Neurogenetics 23, 395–404. doi:10.3109/01677060902878481
De Luca, A., Cerrato, V., and Fucà, E. (2016). Sonic Hedgehog Patterning during Cerebellar Development. Cell. Mol. Life Sci. 73, 291–303. doi:10.1007/s00018-015-2065-1
Derynck, R., and Akhurst, R. J. (2007). Differentiation Plasticity Regulated by TGF-β Family Proteins in Development and Disease. Nat. Cell Biol. 9, 1000–1004. doi:10.1038/ncb434
Dexter, D. T., and Jenner, P. (2013). Parkinson Disease: from Pathology to Molecular Disease Mechanisms. Free Radic. Biol. Med. 62, 132–144. doi:10.1016/j.freeradbiomed.2013.01.018
Dijke, P. t., and Hill, C. S. (2004). New Insights into TGF-β-Smad Signalling. Trends Biochem. Sci. 29, 265–273. doi:10.1016/j.tibs.2004.03.008
Duan, D., Sharma, P., Yang, J., Yue, Y., Dudus, L., Zhang, Y., et al. (1998). Circular Intermediates of Recombinant Adeno-Associated Virus Have Defined Structural Characteristics Responsible for Long-Term Episomal Persistence in Muscle Tissue. J. Virol. 72, 8568–8577. doi:10.1128/jvi.72.11.8568-8577.1998
Duan, Q., Li, S., Wen, X., Sunnassee, G., Chen, J., Tan, S., et al. (2019). Valproic Acid Enhances Reprogramming Efficiency and Neuronal Differentiation on Small Molecules Staged-Induction Neural Stem Cells: Suggested Role of mTOR Signaling. Front. Neurosci. 13, 867. doi:10.3389/fnins.2019.00867
Duman, J. G., Mulherkar, S., Tu, Y.-K., X. Cheng, J., and Tolias, K. F. (2015). Mechanisms for Spatiotemporal Regulation of Rho-GTPase Signaling at Synapses. Neurosci. Lett. 601, 4–10. doi:10.1016/j.neulet.2015.05.034
Ebrahimi, A., Keske, E., Mehdipour, A., Ebrahimi-Kalan, A., and Ghorbani, M. (2019). Somatic Cell Reprogramming as a Tool for Neurodegenerative Diseases. Biomed. Pharmacother. 112, 108663. doi:10.1016/j.biopha.2019.108663
Endo, M., Antonyak, M. A., and Cerione, R. A. (2009). Cdc42-mTOR Signaling Pathway Controls Hes5 and Pax6 Expression in Retinoic Acid-dependent Neural Differentiation. J. Biol. Chem. 284, 5107–5118. doi:10.1074/jbc.M807745200
Fuccillo, M., Joyner, A. L., and Fishell, G. (2006). Morphogen to Mitogen: the Multiple Roles of Hedgehog Signalling in Vertebrate Neural Development. Nat. Rev. Neurosci. 7, 772–783. doi:10.1038/nrn1990
Gao, L., Guan, W., Wang, M., Wang, H., Yu, J., Liu, Q., et al. (2017). Direct Generation of Human Neuronal Cells from Adult Astrocytes by Small Molecules. Stem Cell Rep. 8, 538–547. doi:10.1016/j.stemcr.2017.01.014
Gao, L., Huang, S., Zhang, H., Hua, W., Xin, S., Cheng, L., et al. (2019). Suppression of Glioblastoma by a Drug Cocktail Reprogramming Tumor Cells into Neuronal like Cells. Sci. Rep. 9, 3462. doi:10.1038/s41598-019-39852-5
Gascón, S., Masserdotti, G., Russo, G. L., and Götz, M. (2017). Direct Neuronal Reprogramming: Achievements, Hurdles, and New Roads to Success. Cell Stem Cell 21, 18–34. doi:10.1016/j.stem.2017.06.011
Gascón, S., Murenu, E., Masserdotti, G., Ortega, F., Russo, G. L., Petrik, D., et al. (2016). Identification and Successful Negotiation of a Metabolic Checkpoint in Direct Neuronal Reprogramming. Cell Stem Cell 18, 396–409. doi:10.1016/j.stem.2015.12.003
Ge, W.-P., Miyawaki, A., Gage, F. H., Jan, Y. N., and Jan, L. Y. (2012). Local Generation of Glia Is a Major Astrocyte Source in Postnatal Cortex. Nature 484, 376–380. doi:10.1038/nature10959
George, S., Hamblin, M. R., and Abrahamse, H. (2018). Current and Future Trends in Adipose Stem Cell Differentiation into Neuroglia. Photomed. Laser Surg. 36, 230–240. doi:10.1089/pho.2017.4411
Ghasemi-Kasman, M., Shojaei, A., Gol, M., Moghadamnia, A. A., Baharvand, H., and Javan, M. (2018). miR-302/367-induced Neurons Reduce Behavioral Impairment in an Experimental Model of Alzheimer's Disease. Mol. Cell. Neurosci. 86, 50–57. doi:10.1016/j.mcn.2017.11.012
Goldman, S. A. (2016). Stem and Progenitor Cell-Based Therapy of the Central Nervous System: Hopes, Hype, and Wishful Thinking. Cell Stem Cell 18, 174–188. doi:10.1016/j.stem.2016.01.012
Götz, M., and Bocchi, R. (2021). Neuronal Replacement: Concepts, Achievements, and Call for Caution. Curr. Opin. Neurobiol. 69, 185–192. doi:10.1016/j.conb.2021.03.014
Grande, A., Sumiyoshi, K., and López-Juárez, A. (2013). Environmental Impact on Direct Neuronal Reprogramming In Vivo in the Adult Brain. Nat. Commun. 4, 2373. doi:10.1038/ncomms3373
Gris, D., Marsh, D. R., and Oatway, M. A. (2004). Transient Blockade of the CD11d/CD18 Integrin Reduces Secondary Damage after Spinal Cord Injury, Improving Sensory, Autonomic, and Motor Function. J. Neurosci. 24, 4043–4051. doi:10.1523/jneurosci.5343-03.2004
Gross, R. E., Mehler, M. F., and Mabie, P. C. (1996). Bone Morphogenetic Proteins Promote Astroglial Lineage Commitment by Mammalian Subventricular Zone Progenitor Cells. Neuron 17, 595–606. doi:10.1016/s0896-6273(00)80193-2
Guo, Z., Zhang, L., Wu, Z., Chen, Y., Wang, F., and Chen, G. (2014). In Vivo direct Reprogramming of Reactive Glial Cells into Functional Neurons after Brain Injury and in an Alzheimer's Disease Model. Cell Stem Cell 14, 188–202. doi:10.1016/j.stem.2013.12.001
Habekost, M., Jørgensen, A. L., Qvist, P., and Denham, M. (2020). MicroRNAs and Ascl1 Facilitate Direct Conversion of Porcine Fibroblasts into Induced Neurons. Stem Cell Res. 48, 101984. doi:10.1016/j.scr.2020.101984
Han, J., Sachdev, P. S., and Sidhu, K. S. (2010). A Combined Epigenetic and Non-genetic Approach for Reprogramming Human Somatic Cells. Plos One 5, e12297. doi:10.1371/journal.pone.0012297
Haridhasapavalan, K. K., Borgohain, M. P., Dey, C., Saha, B., Narayan, G., Kumar, S., et al. (2019). An Insight into Non-integrative Gene Delivery Approaches to Generate Transgene-free Induced Pluripotent Stem Cells. Gene 686, 146–159. doi:10.1016/j.gene.2018.11.069
He, S., Chen, J., Zhang, Y., Zhang, M., Yang, X., Li, Y., et al. (2017). Sequential EMT-MET Induces Neuronal Conversion through Sox2. Cell Discov. 3, 17017. doi:10.1038/celldisc.2017.17
Headley, K. M., Kedziora, K. M., and Alejo, A. (2019). Chemical Screen for Epigenetic Barriers to Single Allele Activation of Oct4. Stem Cell Res. 38, 101470. doi:10.1016/j.scr.2019.101470
Heldin, C.-H., Miyazono, K., and ten Dijke, P. (1997). TGF-β Signalling from Cell Membrane to Nucleus through SMAD Proteins. Nature 390, 465–471. doi:10.1038/37284
Herdy, J., Schafer, S., and Kim, Y. (2019). Chemical Modulation of Transcriptionally Enriched Signaling Pathways to Optimize the Conversion of Fibroblasts into Neurons. Elife 8. doi:10.7554/eLife.41356
Herrera, S. C., and Bach, E. A. (2019). JAK/STAT Signaling in Stem Cells and Regeneration: from Drosophila to Vertebrates. Development 146, 146. doi:10.1242/dev.167643
Herrera, S. C., and Bach, E. A. (2021). The Emerging Roles of JNK Signaling in Drosophila Stem Cell Homeostasis. Ijms 22, 5519. doi:10.3390/ijms22115519
Hill, S. A., Fu, M., and Garcia, A. D. R. (2021). Sonic Hedgehog Signaling in Astrocytes. Cell. Mol. Life Sci. 78, 1393–1403. doi:10.1007/s00018-020-03668-8
Hirabayashi, Y., Suzki, N., Tsuboi, M., Endo, T. A., Toyoda, T., Shinga, J., et al. (2009). Polycomb Limits the Neurogenic Competence of Neural Precursor Cells to Promote Astrogenic Fate Transition. Neuron 63, 600–613. doi:10.1016/j.neuron.2009.08.021
Hirsch, M. L., Wolf, S. J., and Samulski, R. J. (2016). Delivering Transgenic DNA Exceeding the Carrying Capacity of AAV Vectors. Methods Mol. Biol. 1382, 21–39. doi:10.1007/978-1-4939-3271-9_2
Hitoshi, S., Seaberg, R. M., Koscik, C., Alexson, T., Kusunoki, S., Kanazawa, I., et al. (2004). Primitive Neural Stem Cells from the Mammalian Epiblast Differentiate to Definitive Neural Stem Cells under the Control of Notch Signaling. Genes Dev. 18, 1806–1811. doi:10.1101/gad.1208404
Hnisz, D., Abraham, B. J., Lee, T. I., Lau, A., Saint-André, V., Sigova, A. A., et al. (2013). Super-enhancers in the Control of Cell Identity and Disease. Cell 155, 934–947. doi:10.1016/j.cell.2013.09.053
Ho, L., and Hsu, S.-h. (2018). Cell Reprogramming by 3D Bioprinting of Human Fibroblasts in Polyurethane Hydrogel for Fabrication of Neural-like Constructs. Acta Biomater. 70, 57–70. doi:10.1016/j.actbio.2018.01.044
Hoehn, M. M., and Yahr, M. D. (1967). Parkinsonism: Onset, Progression, and Mortality. Neurology 17, 427. doi:10.1212/wnl.17.5.427
Hotta, R., Pepdjonovic, L., Anderson, R. B., Zhang, D., Bergner, A. J., Leung, J., et al. (2009). Small-molecule Induction of Neural Crest-like Cells Derived from Human Neural Progenitors. Stem Cells 27, 2896–2905. doi:10.1002/stem.208
Hou, P., Li, Y., Zhang, X., Liu, C., Guan, J., Li, H., et al. (2013). Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 341, 651–654. doi:10.1126/science.1239278
Hou, S., and Rabchevsky, A. G. (2014). Autonomic Consequences of Spinal Cord Injury. Compr. Physiol. 4, 1419–1453. doi:10.1002/cphy.c130045
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278. doi:10.1016/j.cell.2014.05.010
Hu, W., Qiu, B., Guan, W., Wang, Q., Wang, M., Li, W., et al. (2015). Direct Conversion of Normal and Alzheimer's Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell 17, 204–212. doi:10.1016/j.stem.2015.07.006
Hu, Y. n., Zhang, F., Zhong, W., Liu, Y. n., He, Q., Yang, M., et al. (2019). Transplantation of Neural Scaffolds Consisting of Dermal Fibroblast-Reprogrammed Neurons and 3D Silk Fibrous Materials Promotes the Repair of Spinal Cord Injury. J. Mat. Chem. B 7, 7525–7539. doi:10.1039/c9tb01929d
Huangfu, D., Maehr, R., Guo, W., Eijkelenboom, A., Snitow, M., Chen, A. E., et al. (2008). Induction of Pluripotent Stem Cells by Defined Factors Is Greatly Improved by Small-Molecule Compounds. Nat. Biotechnol. 26, 795–797. doi:10.1038/nbt1418
Hülsmann, S., Oku, Y., Zhang, W., and Richter, D. W. (2000). Metabolic Coupling between Glia and Neurons Is Necessary for Maintaining Respiratory Activity in Transverse Medullary Slices of Neonatal Mouse. Eur. J. Neurosci. 12, 856–862. doi:10.1046/j.1460-9568.2000.00973.x
Hur, E.-M., and Zhou, F.-Q. (2010). GSK3 Signalling in Neural Development. Nat. Rev. Neurosci. 11, 539–551. doi:10.1038/nrn2870
Ichida, J. K., Blanchard, J., Lam, K., Son, E. Y., Chung, J. E., Egli, D., et al. (2009). A Small-Molecule Inhibitor of Tgf-β Signaling Replaces Sox2 in Reprogramming by Inducing Nanog. Cell Stem Cell 5, 491–503. doi:10.1016/j.stem.2009.09.012
Ijomone, O. M., Iroegbu, J. D., and Aschner, M. (2021). Impact of Environmental Toxicants on P38- and ERK-MAPK Signaling Pathways in the Central Nervous System. Neurotoxicology 86, 166–171. doi:10.1016/j.neuro.2021.08.005
Inestrosa, N. C., and Varela-Nallar, L. (2015). Wnt Signalling in Neuronal Differentiation and Development. Cell Tissue Res. 359, 215–223. doi:10.1007/s00441-014-1996-4
Ito, K., and Suda, T. (2014). Metabolic Requirements for the Maintenance of Self-Renewing Stem Cells. Nat. Rev. Mol. Cell Biol. 15, 243–256. doi:10.1038/nrm3772
Jiang, H., Xu, Z., and Zhong, P. (2015). Cell Cycle and P53 Gate the Direct Conversion of Human Fibroblasts to Dopaminergic Neurons. Nat. Commun. 6, 10100. doi:10.1038/ncomms10100
Joung, J., Konermann, S., and Gootenberg, J. S. (2017). Genome-scale CRISPR-Cas9 Knockout and Transcriptional Activation Screening. Nat. Protoc. 12, 828–863. doi:10.1038/nprot.2017.016
Kaeberlein, M. (2013). mTOR Inhibition: From Aging to Autism and beyond. Scientifica 2013, 1–17. doi:10.1155/2013/849186
Kageyama, R., Ohtsuka, T., Hatakeyama, J., and Ohsawa, R. (2005). Roles of bHLH Genes in Neural Stem Cell Differentiation. Exp. Cell Res. 306, 343–348. doi:10.1016/j.yexcr.2005.03.015
Kang, H. T., Park, J. T., Choi, K., Choi, H. J. C., Jung, C. W., Kim, G. R., et al. (2017). Chemical Screening Identifies ROCK as a Target for Recovering Mitochondrial Function in Hutchinson-Gilford Progeria Syndrome. Aging Cell 16, 541–550. doi:10.1111/acel.12584
Kashani, M. S., Tavirani, M. R., Talaei, S. A., and Salami, M. (2011). Aqueous Extract of Lavender (Lavandula Angustifolia) Improves the Spatial Performance of a Rat Model of Alzheimer's Disease. Neurosci. Bull. 27, 99–106. doi:10.1007/s12264-011-1149-7
Kashima, R., and Hata, A. (2018). The Role of TGF-β Superfamily Signaling in Neurological Disorders. Acta Biochim. Biophys. Sin. (Shanghai) 50, 106–120. doi:10.1093/abbs/gmx124
Kim, J., Efe, J. A., Zhu, S., Talantova, M., Yuan, X., Wang, S., et al. (2011). Direct Reprogramming of Mouse Fibroblasts to Neural Progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843. doi:10.1073/pnas.1103113108
Kimura, T., Horikoshi, Y., Kuriyagawa, C., and Niiyama, Y. (2021). Rho/ROCK Pathway and Noncoding RNAs: Implications in Ischemic Stroke and Spinal Cord Injury. Ijms 22, 11573. doi:10.3390/ijms222111573
Koch, J. C., Tatenhorst, L., Roser, A.-E., Saal, K.-A., Tönges, L., and Lingor, P. (2018). ROCK Inhibition in Models of Neurodegeneration and its Potential for Clinical Translation. Pharmacol. Ther. 189, 1–21. doi:10.1016/j.pharmthera.2018.03.008
Koga, Y., Tsurumaki, H., Aoki-Saito, H., Sato, M., Yatomi, M., Takehara, K., et al. (2019). Roles of Cyclic AMP Response Element Binding Activation in the ERK1/2 and P38 MAPK Signalling Pathway in Central Nervous System, Cardiovascular System, Osteoclast Differentiation and Mucin and Cytokine Production. Ijms 20, 1346. doi:10.3390/ijms20061346
Kouzarides, T. (2007). Chromatin Modifications and Their Function. Cell 128, 693–705. doi:10.1016/j.cell.2007.02.005
Kretsovali, A., Hadjimichael, C., and Charmpilas, N. (2012). Histone Deacetylase Inhibitors in Cell Pluripotency, Differentiation, and Reprogramming. Stem Cells Int. 2012, 1–10. doi:10.1155/2012/184154
Krol, J., Loedige, I., and Filipowicz, W. (2010). The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat. Rev. Genet. 11, 597–610. doi:10.1038/nrg2843
Ladewig, J., Koch, P., Brüstle, O., and Waddington, L. (2013). Leveling Waddington: the Emergence of Direct Programming and the Loss of Cell Fate Hierarchies. Nat. Rev. Mol. Cell Biol. 14, 225–236. doi:10.1038/nrm3543
Ladewig, J., Mertens, J., Kesavan, J., Doerr, J., Poppe, D., Glaue, F., et al. (2012). Small Molecules Enable Highly Efficient Neuronal Conversion of Human Fibroblasts. Nat. Methods 9, 575–578. doi:10.1038/nmeth.1972
Lamas, N. J., Johnson-Kerner, B., Roybon, L., Kim, Y. A., Garcia-Diaz, A., Wichterle, H., et al. (2014). Neurotrophic Requirements of Human Motor Neurons Defined Using Amplified and Purified Stem Cell-Derived Cultures. Plos One 9, e110324. doi:10.1371/journal.pone.0110324
Lee, C., Robinson, M., and Willerth, S. M. (2018). Direct Reprogramming of Glioblastoma Cells into Neurons Using Small Molecules. ACS Chem. Neurosci. 9, 3175–3185. doi:10.1021/acschemneuro.8b00365
Lee, J.-H., Mitchell, R. R., McNicol, J. D., Shapovalova, Z., Laronde, S., Tanasijevic, B., et al. (2015). Single Transcription Factor Conversion of Human Blood Fate to NPCs with CNS and PNS Developmental Capacity. Cell Rep. 11, 1367–1376. doi:10.1016/j.celrep.2015.04.056
Lesbats, P., Engelman, A. N., and Cherepanov, P. (2016). Retroviral DNA Integration. Chem. Rev. 116, 12730–12757. doi:10.1021/acs.chemrev.6b00125
Leung, T., Manser, E., Tan, L., and Lim, L. (1995). A Novel Serine/threonine Kinase Binding the Ras-Related RhoA GTPase Which Translocates the Kinase to Peripheral Membranes. J. Biol. Chem. 270, 29051–29054. doi:10.1074/jbc.270.49.29051
Li, B., Carey, M., and Workman, J. L. (2007). The Role of Chromatin during Transcription. Cell 128, 707–719. doi:10.1016/j.cell.2007.01.015
Li, H., and Chen, G. (2016). In Vivo Reprogramming for CNS Repair: Regenerating Neurons from Endogenous Glial Cells. Neuron 91, 728–738. doi:10.1016/j.neuron.2016.08.004
Li, R., Liang, J., Ni, S., Zhou, T., Qing, X., Li, H., et al. (2010). A Mesenchymal-To-Epithelial Transition Initiates and Is Required for the Nuclear Reprogramming of Mouse Fibroblasts. Cell Stem Cell 7, 51–63. doi:10.1016/j.stem.2010.04.014
Li, S., Mattar, P., Zinyk, D., Singh, K., Chaturvedi, C.-P., Kovach, C., et al. (2012). GSK3 Temporally Regulates Neurogenin 2 Proneural Activity in the Neocortex. J. Neurosci. 32, 7791–7805. doi:10.1523/jneurosci.1309-12.2012
Li, S., Zhang, H., Wang, A., Liu, Y., Liu, H., Yue, F., et al. (2019). Differentiation of Adult Human Retinal Pigment Epithelial Cells into Dopaminergic-like Cells In Vitro and in the Recipient Monkey Brain. Mol. Med. 25, 9. doi:10.1186/s10020-019-0076-3
Li, T., Xu, R., Xia, H., Hu, X., Wang, S., Li, Y., et al. (2020). ASK1 Phosphorylation Regulates Astrocytic Reactive Gliosis In Vitro and In Vivo. Neurosci. Lett. 716, 134675. doi:10.1016/j.neulet.2019.134675
Li, W., Zhou, H., Abujarour, R., Zhu, S., Young Joo, J., Lin, T., et al. (2009). Generation of Human-Induced Pluripotent Stem Cells in the Absence of Exogenous Sox2. Stem Cells 27, 2992–3000. doi:10.1002/stem.240
Li, X., Zuo, X., Jing, J., Ma, Y., Wang, J., Liu, D., et al. (2015). Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 17, 195–203. doi:10.1016/j.stem.2015.06.003
Li, Z., Düllmann, J., Schiedlmeier, B., Schmidt, M., von Kalle, C., Meyer, J., et al. (2002). Murine Leukemia Induced by Retroviral Gene Marking. Science 296, 497. doi:10.1126/science.1068893
Liang, G., Lin, J. C. Y., Wei, V., Yoo, C., Cheng, J. C., Nguyen, C. T., et al. (2004). Distinct Localization of Histone H3 Acetylation and H3-K4 Methylation to the Transcription Start Sites in the Human Genome. Proc. Natl. Acad. Sci. U.S.A. 101, 7357–7362. doi:10.1073/pnas.0401866101
Liang, X.-G., Tan, C., Wang, C.-K., Tao, R.-R., Huang, Y.-J., Ma, K.-F., et al. (2018). Myt1l Induced Direct Reprogramming of Pericytes into Cholinergic Neurons. Cns Neurosci. Ther. 24, 801–809. doi:10.1111/cns.12821
Lie, D.-C., Colamarino, S. A., and Song, H.-J. (2005). Wnt Signalling Regulates Adult Hippocampal Neurogenesis. Nature 437, 1370–1375. doi:10.1038/nature04108
Lin, T., Ambasudhan, R., Yuan, X., Li, W., Hilcove, S., Abujarour, R., et al. (2009). A Chemical Platform for Improved Induction of Human iPSCs. Nat. Methods 6, 805–808. doi:10.1038/nmeth.1393
Lindsell, C. E., Boulter, J., and DiSibio, G. (1996). Expression Patterns ofJagged, Delta1, Notch1, Notch2,andNotch3Genes Identify Ligand-Receptor Pairs that May Function in Neural Development. Mol. Cell. Neurosci. 8, 14–27. doi:10.1006/mcne.1996.0040
Lipton, J. O., and Sahin, M. (2014). The Neurology of mTOR. Neuron 84, 275–291. doi:10.1016/j.neuron.2014.09.034
Liu, D., Rychkov, G., Al-Hawwas, M., Manaph, N. P. A., Zhou, F., Bobrovskaya, L., et al. (2020). Conversion of Human Urine-Derived Cells into Neuron-like Cells by Small Molecules. Mol. Biol. Rep. 47, 2713–2722. doi:10.1007/s11033-020-05370-1
Liu, M.-L., Zang, T., and Zhang, C.-L. (2016). Direct Lineage Reprogramming Reveals Disease-specific Phenotypes of Motor Neurons from Human ALS Patients. Cell Rep. 14, 115–128. doi:10.1016/j.celrep.2015.12.018
Liu, M.-L., Zang, T., Zou, Y., Chang, J. C., Gibson, J. R., Huber, K. M., et al. (2013). Small Molecules Enable Neurogenin 2 to Efficiently Convert Human Fibroblasts into Cholinergic Neurons. Nat. Commun. 4, 2183. doi:10.1038/ncomms3183
Lu, M.-H., Ji, W.-L., Chen, H., Sun, Y.-Y., Zhao, X.-Y., Wang, F., et al. (2021). Intranasal Transplantation of Human Neural Stem Cells Ameliorates Alzheimer's Disease-like Pathology in a Mouse Model. Front. Aging Neurosci. 13, 650103. doi:10.3389/fnagi.2021.650103
Lunt, S. Y., and Vander Heiden, M. G. (2011). Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464. doi:10.1146/annurev-cellbio-092910-154237
Ma, N.-X., Yin, J.-C., and Chen, G. (2019). Transcriptome Analysis of Small Molecule-Mediated Astrocyte-To-Neuron Reprogramming. Front. Cell Dev. Biol. 7, 82. doi:10.3389/fcell.2019.00082
Ma, Y., Xie, H., Du, X., Wang, L., Jin, X., Zhang, Q., et al. (2021). In Vivo chemical Reprogramming of Astrocytes into Neurons. Cell Discov. 7, 12. doi:10.1038/s41421-021-00243-8
MacDonald, B. T., Tamai, K., and He, X. (2009). Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 17, 9–26. doi:10.1016/j.devcel.2009.06.016
Madhu, V., Dighe, A. S., Cui, Q., and Deal, D. N. (2016). Dual Inhibition of Activin/Nodal/TGF-Βand BMP Signaling Pathways by SB431542 and Dorsomorphin Induces Neuronal Differentiation of Human Adipose Derived Stem Cells. Stem Cells Int. 2016, 1–13. doi:10.1155/2016/1035374
Magistretti, P. J., and Allaman, I. (2015). A Cellular Perspective on Brain Energy Metabolism and Functional Imaging. Neuron 86, 883–901. doi:10.1016/j.neuron.2015.03.035
Magnusson, J. P., Göritz, C., Tatarishvili, J., Dias, D. O., Smith, E. M. K., Lindvall, O., et al. (2014). A Latent Neurogenic Program in Astrocytes Regulated by Notch Signaling in the Mouse. Science 346, 237–241. doi:10.1126/science.346.6206.237
Man, J. H. K., Groenink, L., and Caiazzo, M. (2018). Cell Reprogramming Approaches in Gene- and Cell-Based Therapies for Parkinson's Disease. J. Control. Release 286, 114–124. doi:10.1016/j.jconrel.2018.07.017
Maryanovich, M., and Gross, A. (2013). A ROS Rheostat for Cell Fate Regulation. Trends Cell Biol. 23, 129–134. doi:10.1016/j.tcb.2012.09.007
Masai, I., Yamaguchi, M., Tonou-Fujimori, N., Komori, A., and Okamoto, H. (2005). The Hedgehog-PKA Pathway Regulates Two Distinct Steps of the Differentiation of Retinal Ganglion Cells: the Cell-Cycle Exit of Retinoblasts and Their Neuronal Maturation. Development 132, 1539–1553. doi:10.1242/dev.01714
Massagué, J., Blain, S. W., and Lo, R. S. (2000). TGFβ Signaling in Growth Control, Cancer, and Heritable Disorders. Cell 103, 295–309. doi:10.1016/s0092-8674(00)00121-5
Masserdotti, G., Gascón, S., and Götz, M. (2016). Direct Neuronal Reprogramming: Learning from and for Development. Development 143, 2494–2510. doi:10.1242/dev.092163
Matsui, T., Amano, M., Yamamoto, T., Chihara, K., Nakafuku, M., Ito, M., et al. (1996). Rho-associated Kinase, a Novel Serine/threonine Kinase, as a Putative Target for Small GTP Binding Protein Rho. EMBO J. 15, 2208–2216. doi:10.1002/j.1460-2075.1996.tb00574.x
McMurran, C. E., Kodali, S., and Young, A. (2018). Clinical Implications of Myelin Regeneration in the Central Nervous System. Expert Rev. Neurother. 18, 111–123. doi:10.1080/14737175.2018.1421458
Medvedev, S. P., Shevchenko, A. I., and Zakian, S. M. (2010). Induced Pluripotent Stem Cells: Problems and Advantages when Applying Them in Regenerative Medicine. Acta Naturae 2, 18–27. doi:10.32607/20758251-2010-2-2-18-27
Meng, F., Chen, S., Miao, Q., Zhou, K., Lao, Q., Zhang, X., et al. (2012). Induction of Fibroblasts to Neurons through Adenoviral Gene Delivery. Cell Res. 22, 436–440. doi:10.1038/cr.2011.185
Mirakhori, F., Zeynali, B., Rassouli, H., Shahbazi, E., Hashemizadeh, S., Kiani, S., et al. (2015). Induction of Neural Progenitor-like Cells from Human Fibroblasts via a Genetic Material-free Approach. Plos One 10, e0135479. doi:10.1371/journal.pone.0135479
Modlich, U., Bohne, J., Schmidt, M., von Kalle, C., Knöss, S., Schambach, A., et al. (2006). Cell-culture Assays Reveal the Importance of Retroviral Vector Design for Insertional Genotoxicity. Blood 108, 2545–2553. doi:10.1182/blood-2005-08-024976
Mollinari, C., and Merlo, D. (2021). Direct Reprogramming of Somatic Cells to Neurons: Pros and Cons of Chemical Approach. Neurochem. Res. 46, 1330–1336. doi:10.1007/s11064-021-03282-5
Mollinari, C., Zhao, J., and Lupacchini, L. (2018). Transdifferentiation: a New Promise for Neurodegenerative Diseases. Cell Death Dis. 9, 830. doi:10.1038/s41419-018-0891-4
Morabito, G., Giannelli, S. G., Ordazzo, G., Bido, S., Castoldi, V., Indrigo, M., et al. (2017). AAV-PHP.B-Mediated Global-Scale Expression in the Mouse Nervous System Enables GBA1 Gene Therapy for Wide Protection from Synucleinopathy. Mol. Ther. 25, 2727–2742. doi:10.1016/j.ymthe.2017.08.004
Morrison, D. K. (2012). MAP Kinase Pathways. Cold Spring Harb. Perspect. Biol. 4, a011254. doi:10.1101/cshperspect.a011254
Nabavi, S. M., Ahmed, T., and Nawaz, M. (2019). Targeting STATs in Neuroinflammation: The Road Less Traveled!. Pharmacol. Res. 141, 73–84. doi:10.1016/j.phrs.2018.12.004
Nakagawa, O., Fujisawa, K., Ishizaki, T., Saito, Y., Nakao, K., and Narumiya, S. (1996). ROCK-I and ROCK-II, Two Isoforms of Rho-Associated Coiled-Coil Forming Protein Serine/threonine Kinase in Mice. Febs Lett. 392, 189–193. doi:10.1016/0014-5793(96)00811-3
Naldini, L., Blömer, U., and Gallay, P. (1996). In Vivo gene Delivery and Stable Transduction of Nondividing Cells by a Lentiviral Vector. Science 272, 263–267. doi:10.1126/science.272.5259.263
Nebreda, A. R., and Porras, A. (2000). p38 MAP Kinases: beyond the Stress Response. Trends Biochem. Sci. 25, 257–260. doi:10.1016/s0968-0004(00)01595-4
Nemoto, A., Kobayashi, R., Yoshimatsu, S., Sato, Y., Kondo, T., Yoo, A. S., et al. (2020). Direct Neuronal Reprogramming of Common Marmoset Fibroblasts by ASCL1, microRNA-9/9*, and microRNA-124 Overexpression. Cells 10, 6. doi:10.3390/cells10010006
O' Neill, C. (2013). PI3-kinase/Akt/mTOR Signaling: Impaired On/off Switches in Aging, Cognitive Decline and Alzheimer's Disease. Exp. Gerontol. 48, 647–653. doi:10.1016/j.exger.2013.02.025
O'Shea, J. J., Schwartz, D. M., Villarino, A. V., Gadina, M., McInnes, I. B., and Laurence, A. (2015). The JAK-STAT Pathway: Impact on Human Disease and Therapeutic Intervention. Annu. Rev. Med. 66, 311–328. doi:10.1146/annurev-med-051113-024537
Oh, J., Kim, Y., Che, L., Kim, J. B., Chang, G. E., Cheong, E., et al. (2017). Regulation of cAMP and GSK3 Signaling Pathways Contributes to the Neuronal Conversion of Glioma. Plos One 12, e0178881. doi:10.1371/journal.pone.0178881
Ono, K., and Han, J. (2000). The P38 Signal Transduction Pathway Activation and Function. Cell. Signal. 12, 1–13. doi:10.1016/s0898-6568(99)00071-6
Oproescu, A.-M., Han, S., and Schuurmans, C. (2021). New Insights into the Intricacies of Proneural Gene Regulation in the Embryonic and Adult Cerebral Cortex. Front. Mol. Neurosci. 14, 642016. doi:10.3389/fnmol.2021.642016
Pan, Y., Bai, C. B., Joyner, A. L., and Wang, B. (2006). Sonic Hedgehog Signaling Regulates Gli2 Transcriptional Activity by Suppressing its Processing and Degradation. Mol. Cell. Biol.; Res. Support 26, 3365–3377. doi:10.1128/mcb.26.9.3365-3377.2006
Park, J., Lee, N., Lee, J., Choe, E. K., Kim, M. K., Lee, J., et al. (2017). Small Molecule-Based Lineage Switch of Human Adipose-Derived Stem Cells into Neural Stem Cells and Functional GABAergic Neurons. Sci. Rep. 7, 10166. doi:10.1038/s41598-017-10394-y
Patel, S. S., Tomar, S., Sharma, D., Mahindroo, N., and Udayabanu, M. (2017). Targeting Sonic Hedgehog Signaling in Neurological Disorders. Neurosci. Biobehav. Rev. 74, 76–97. doi:10.1016/j.neubiorev.2017.01.008
Phiel, C. J., Zhang, F., and Huang, E. Y. (2001). Histone Deacetylase Is a Direct Target of Valproic Acid, a Potent Anticonvulsant, Mood Stabilizer, and Teratogen. J. Biol. Chem. 276, 36734–36741. doi:10.1074/jbc.M101287200
Politano, S. F., Salemme, R. R., Ashley, J., López‐Rivera, J. A., Bakula, T. A., Puhalla, K. A., et al. (2019). Tao Negatively Regulates BMP Signaling during Neuromuscular Junction Development in Drosophila. Dev. Neurobiol. 79, 335–349. doi:10.1002/dneu.22681
Prapainop, K., Witter, D. P., and Wentworth, P. (2012). A Chemical Approach for Cell-specific Targeting of Nanomaterials: Small-Molecule-Initiated Misfolding of Nanoparticle Corona Proteins. J. Am. Chem. Soc. 134, 4100–4103. doi:10.1021/ja300537u
Pu, J.-L., Chen, X.-Z., Dai, S.-B., Shen, T., and Zheng, T.-T. (2019). Plasmid-based Generation of Neural Cells from Human Fibroblasts Using Non-integrating Episomal Vectors. Neural Regen. Res. 14, 501–505. doi:10.4103/1673-5374.245476
Puls, B., Ding, Y., Zhang, F., Pan, M., Lei, Z., Pei, Z., et al. (2020). Regeneration of Functional Neurons after Spinal Cord Injury via In Situ NeuroD1-Mediated Astrocyte-To-Neuron Conversion. Front. Cell Dev. Biol. 8, 591883. doi:10.3389/fcell.2020.591883
Qi, Y., Zhang, X.-J., Renier, N., Wu, Z., Atkin, T., Sun, Z., et al. (2017). Combined Small-Molecule Inhibition Accelerates the Derivation of Functional Cortical Neurons from Human Pluripotent Stem Cells. Nat. Biotechnol. 35, 154–163. doi:10.1038/nbt.3777
Qin, H., Zhao, A.-D., Sun, M.-L., Ma, K., and Fu, X.-B. (2020). Direct Conversion of Human Fibroblasts into Dopaminergic Neuron-like Cells Using Small Molecules and Protein Factors. Mil. Med. Res. 7, 52. doi:10.1186/s40779-020-00284-2
Qin, H., Zhao, A., and Fu, X. (2017). Small Molecules for Reprogramming and Transdifferentiation. Cell. Mol. Life Sci. 74, 3553–3575. doi:10.1007/s00018-017-2586-x
Qin, H., Zhao, A., Ma, K., and Fu, X. (2018). Chemical Conversion of Human and Mouse Fibroblasts into Motor Neurons. Sci. China Life Sci. 61, 1151–1167. doi:10.1007/s11427-018-9359-8
Qin, L., Dai, X., and Yin, Y. (2016). Valproic Acid Exposure Sequentially Activates Wnt and mTOR Pathways in Rats. Mol. Cell. Neurosci. 75, 27–35. doi:10.1016/j.mcn.2016.06.004
Raingeaud, J., Gupta, S., and Rogers, J. S. (1995). Pro-inflammatory Cytokines and Environmental Stress Cause P38 Mitogen-Activated Protein Kinase Activation by Dual Phosphorylation on Tyrosine and Threonine. J. Biol. Chem. 270, 7420–7426. doi:10.1074/jbc.270.13.7420
Rao, Y., Du, S., Yang, B., Wang, Y., Li, Y., Li, R., et al. (2021). NeuroD1 Induces Microglial Apoptosis and Cannot Induce Microglia-To-Neuron Cross-Lineage Reprogramming. Neuron 109, 4094–4108. doi:10.1016/j.neuron.2021.11.008
Rawal, N., Parish, C., Castelo-Branco, G., and Arenas, E. (2007). Inhibition of JNK Increases Survival of Transplanted Dopamine Neurons in Parkinsonian Rats. Cell Death Differ. 14, 381–383. doi:10.1038/sj.cdd.4402010
Restan Perez, M., Sharma, R., Masri, N. Z., and Willerth, S. M. (2021). 3D Bioprinting Mesenchymal Stem Cell-Derived Neural Tissues Using a Fibrin-Based Bioink. Biomolecules 11, 1250. doi:10.3390/biom11081250
Rivetti di Val Cervo, P., Romanov, R. A., and Spigolon, G. (2017). Induction of Functional Dopamine Neurons from Human Astrocytes In Vitro and Mouse Astrocytes in a Parkinson's Disease Model. Nat. Biotechnol. 35, 444–452. doi:10.1038/nbt.3835
Rodriguez-Martinez, G., and Velasco, I. (2012). Activin and TGF-β Effects on Brain Development and Neural Stem Cells. Cnsnddt 11, 844–855. doi:10.2174/1871527311201070844
Roese-Koerner, B., Stappert, L., and Brüstle, O. (2017). Notch/Hes Signaling and miR-9 Engage in Complex Feedback Interactions Controlling Neural Progenitor Cell Proliferation and Differentiation. Neurogenesis 4, e1313647. doi:10.1080/23262133.2017.1313647
Roudnicky, F., Zhang, J. D., Kim, B. K., Pandya, N. J., Lan, Y., Sach-Peltason, L., et al. (2020). Inducers of the Endothelial Cell Barrier Identified through Chemogenomic Screening in Genome-Edited hPSC-Endothelial Cells. Proc. Natl. Acad. Sci. U.S.A. 117, 19854–19865. doi:10.1073/pnas.1911532117
Rubio, A., Luoni, M., Giannelli, S. G., Radice, I., Iannielli, A., Cancellieri, C., et al. (2016). Rapid and Efficient CRISPR/Cas9 Gene Inactivation in Human Neurons during Human Pluripotent Stem Cell Differentiation and Direct Reprogramming. Sci. Rep. 6, 37540. doi:10.1038/srep37540
Russo, E., Citraro, R., and Constanti, A. (2012). The mTOR Signaling Pathway in the Brain: Focus on Epilepsy and Epileptogenesis. Mol. Neurobiol. 46, 662–681. doi:10.1007/s12035-012-8314-5
Sabatini, D. M., Barrow, R. K., and Blackshaw, S. (1999). Interaction of RAFT1 with Gephyrin Required for Rapamycin-Sensitive Signaling. Science 284, 1161–1164. doi:10.1126/science.284.5417.1161
Salikhova, D., Bukharova, T., Cherkashova, E., Namestnikova, D., Leonov, G., Nikitina, M., et al. (2021). Therapeutic Effects of hiPSC-Derived Glial and Neuronal Progenitor Cells-Conditioned Medium in Experimental Ischemic Stroke in Rats. Ijms 22, 4694. doi:10.3390/ijms22094694
Samavarchi-Tehrani, P., Golipour, A., David, L., Sung, H.-k., Beyer, T. A., Datti, A., et al. (2010). Functional Genomics Reveals a BMP-Driven Mesenchymal-To-Epithelial Transition in the Initiation of Somatic Cell Reprogramming. Cell Stem Cell 7, 64–77. doi:10.1016/j.stem.2010.04.015
Sato, M., Takizawa, H., Nakamura, A., Turner, B. J., Shabanpoor, F., and Aoki, Y. (2019). Application of Urine-Derived Stem Cells to Cellular Modeling in Neuromuscular and Neurodegenerative Diseases. Front. Mol. Neurosci. 12, 297. doi:10.3389/fnmol.2019.00297
Saura, C. A., Choi, S.-Y., and Beglopoulos, V. (2004). Loss of Presenilin Function Causes Impairments of Memory and Synaptic Plasticity Followed by Age-dependent Neurodegeneration. Neuron 42, 23–36. doi:10.1016/s0896-6273(04)00182-5
Shao, L., and Wu, W.-S. (2010). Gene-delivery Systems for iPS Cell Generation. Expert Opin. Biol. Ther. 10, 231–242. doi:10.1517/14712590903455989
Shi, Y., Desponts, C., Do, J. T., Hahm, H. S., Schöler, H. R., and Ding, S. (2008). Induction of Pluripotent Stem Cells from Mouse Embryonic Fibroblasts by Oct4 and Klf4 with Small-Molecule Compounds. Cell Stem Cell 3, 568–574. doi:10.1016/j.stem.2008.10.004
Shi, Y., and Massagué, J. (2003). Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell 113, 685–700. doi:10.1016/s0092-8674(03)00432-x
Shyh-Chang, N., Daley, G. Q., and Cantley, L. C. (2013). Stem Cell Metabolism in Tissue Development and Aging. Development 140, 2535–2547. doi:10.1242/dev.091777
Silva, J., Barrandon, O., Nichols, J., Kawaguchi, J., Theunissen, T. W., and Smith, A. (2008). Promotion of Reprogramming to Ground State Pluripotency by Signal Inhibition. Plos Biol. 6, e253. doi:10.1371/journal.pbio.0060253
Sirko, S., Behrendt, G., Johansson, P. A., Tripathi, P., Costa, M. R., Bek, S., et al. (2013). Reactive Glia in the Injured Brain Acquire Stem Cell Properties in Response to Sonic Hedgehog. Cell Stem Cell 12, 426–439. doi:10.1016/j.stem.2013.01.019
Sivandzade, F., and Cucullo, L. (2021). Regenerative Stem Cell Therapy for Neurodegenerative Diseases: An Overview. Ijms 22, 2153. doi:10.3390/ijms22042153
Smith, D. K., Yang, J., and Liu, M.-L. (2016). Small Molecules Modulate Chromatin Accessibility to Promote NEUROG2-Mediated Fibroblast-To-Neuron Reprogramming. Stem Cell Rep. 7, 955–969. doi:10.1016/j.stemcr.2016.09.013
Srivastava, D., and DeWitt, N. (2016). In Vivo Cellular Reprogramming: The Next Generation. Cell 166, 1386–1396. doi:10.1016/j.cell.2016.08.055
Stark, G. R., and Darnell, J. E. (2012). The JAK-STAT Pathway at Twenty. Immunity 36, 503–514. doi:10.1016/j.immuni.2012.03.013
Sternberg, N., and Hamilton, D. (1981). Bacteriophage P1 Site-specific Recombination. J. Mol. Biol. 150, 467–486. doi:10.1016/0022-2836(81)90375-2
Stricker, S. H., and Gotz, M. (2021). Epigenetic Regulation of Neural Lineage Elaboration: Implications for Therapeutic Reprogramming. Neurobiol. Dis., 148. doi:10.1016/j.nbd.2020.105174
Struhl, K. (1998). Histone Acetylation and Transcriptional Regulatory Mechanisms. Genes & Dev. 12, 599–606. doi:10.1101/gad.12.5.599
Sun, Y., Yang, T., and Xu, Z. (2007). The JNK Pathway and Neuronal Migration. J. Genet. Genomics 34, 957–965. doi:10.1016/S1673-8527(07)60108-8
Sutherland, E. W., and Rall, T. W. (1958). Fractionation and Characterization of a Cyclic Adenine Ribonucleotide Formed by Tissue Particles. J. Biol. Chem. 232, 1077–1091. doi:10.1016/s0021-9258(19)77423-7
Suzuki, Y. J., and Shults, N. V. (2019). Antioxidant Regulation of Cell Reprogramming. Antioxidants 8, 323. doi:10.3390/antiox8080323
Tanabe, K., Ang, C. E., and Chanda, S. (2018). Transdifferentiation of Human Adult Peripheral Blood T Cells into Neurons. Proc. Natl. Acad. Sci. U.S.A. 115, 6470–6475. doi:10.1073/pnas.1720273115
Tang, B. (2017). The Potential of Targeting Brain Pathology with Ascl1/Mash1. Cells 6, 26. doi:10.3390/cells6030026
Tedeschi, A., Dupraz, S., Laskowski, C. J., Xue, J., Ulas, T., Beyer, M., et al. (2016). The Calcium Channel Subunit Alpha2delta2 Suppresses Axon Regeneration in the Adult CNS. Neuron 92, 419–434. doi:10.1016/j.neuron.2016.09.026
Tessier, S. N., Ingelson-Filpula, W. A., and Storey, K. B. (2021). Epigenetic Regulation by DNA Methyltransferases during Torpor in the Thirteen-Lined Ground Squirrel Ictidomys Tridecemlineatus. Mol. Cell. Biochem. 476, 3975–3985. doi:10.1007/s11010-021-04214-1
Toledo, E. M., and Inestrosa, N. C. (2010). Activation of Wnt Signaling by Lithium and Rosiglitazone Reduced Spatial Memory Impairment and Neurodegeneration in Brains of an APPswe/PSEN1ΔE9 Mouse Model of Alzheimer's Disease. Mol. Psychiatry 15, 272–285. doi:10.1038/mp.2009.72
Tournier, C., Hess, P., and Yang, D. D. (2000). Requirement of JNK for Stress- Induced Activation of the Cytochrome C-Mediated Death Pathway. Science 288, 870–874. doi:10.1126/science.288.5467.870
Tsacopoulos, M., and Magistretti, P. (1996). Metabolic Coupling between Glia and Neurons. J. Neurosci. 16, 877–885. doi:10.1523/jneurosci.16-03-00877.1996
Tu, W.-z., Fu, Y.-b., and Xie, X. (2019). RepSox, a Small Molecule Inhibitor of the TGFβ Receptor, Induces Brown Adipogenesis and Browning of White Adipocytes. Acta Pharmacol. Sin. 40, 1523–1531. doi:10.1038/s41401-019-0264-2
Tukachinsky, H., Petrov, K., Watanabe, M., and Salic, A. (2016). Mechanism of Inhibition of the Tumor Suppressor Patched by Sonic Hedgehog. Proc. Natl. Acad. Sci. U.S.A. 113, E5866–E5875. doi:10.1073/pnas.1606719113
Urbán, N., and Guillemot, F. o. (2014). Neurogenesis in the Embryonic and Adult Brain: Same Regulators, Different Roles. Front. Cell. Neurosci. 8, 396. doi:10.3389/fncel.2014.00396
Valvezan, A. J., and Klein, P. S. (2012). GSK-3 and Wnt Signaling in Neurogenesis and Bipolar Disorder. Front. Mol. Neurosci. 5, 1. doi:10.3389/fnmol.2012.00001
Vandana, J. J., Lacko, L. A., and Chen, S. (2021). Phenotypic Technologies in Stem Cell Biology. Cell Chem. Biol. 28, 257–270. doi:10.1016/j.chembiol.2021.02.001
Vasan, L., Park, E., David, L. A., Fleming, T., and Schuurmans, C. (2021). Direct Neuronal Reprogramming: Bridging the Gap between Basic Science and Clinical Application. Front. Cell Dev. Biol. 9, 681087. doi:10.3389/fcell.2021.681087
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Südhof, T. C., and Wernig, M. (2010). Direct Conversion of Fibroblasts to Functional Neurons by Defined Factors. Nature 463, 1035–1041. doi:10.1038/nature08797
Vignoles, R., Lentini, C., and d’Orange, M. (2019). Direct Lineage Reprogramming for Brain Repair: Breakthroughs and Challenges. Trends Mol. Med. 25, 897–914. doi:10.1016/j.molmed.2019.06.006
Villarino, A. V., Kanno, Y., and O'Shea, J. J. (2017). Mechanisms and Consequences of Jak-STAT Signaling in the Immune System. Nat. Immunol. 18, 374–384. doi:10.1038/ni.3691
Vuilleumier, R., Lian, T., Flibotte, S., Khan, Z. N., Fuchs, A., Pyrowolakis, G., et al. (2019). Retrograde BMP Signaling Activates Neuronal Gene Expression through Widespread Deployment of a Conserved BMP-Responsivecis-Regulatoryactivation Element. Nucleic Acids Res. 47, 679–699. doi:10.1093/nar/gky1135
Wan, X. Y., Xu, L. Y., Li, B., Sun, Q. H., Ji, Q. L., Huang, D. D., et al. (2018). Chemical Conversion of Human Lung Fibroblasts into Neuronal Cells. Int. J. Mol. Med. 41, 1463–1468. doi:10.3892/ijmm.2018.3375
Wang, H., Yang, Y., Liu, J., and Qian, L. (2021). Direct Cell Reprogramming: Approaches, Mechanisms and Progress. Nat. Rev. Mol. Cell Biol. 22, 410–424. doi:10.1038/s41580-021-00335-z
Wang, L.-L., Serrano, C., Zhong, X., Ma, S., Zou, Y., and Zhang, C.-L. (2021). Revisiting Astrocyte to Neuron Conversion with Lineage Tracing In Vivo. Cell 184, 5465–5481. doi:10.1016/j.cell.2021.09.005
Watanabe, K., Ueno, M., Kamiya, D., Nishiyama, A., Matsumura, M., Wataya, T., et al. (2007). A ROCK Inhibitor Permits Survival of Dissociated Human Embryonic Stem Cells. Nat. Biotechnol. 25, 681–686. doi:10.1038/nbt1310
Wen, L., and Tang, F. (2016). Single-cell Sequencing in Stem Cell Biology. Genome Biol. 17, 71. doi:10.1186/s13059-016-0941-0
Wendt, M. K., Balanis, N., Carlin, C. R., and Schiemann, W. P. (2014). STAT3 and Epithelial-Mesenchymal Transitions in Carcinomas. JAK-STAT 3, e28975. doi:10.4161/jkst.28975
Xiao, Y.-T., Xiang, L.-X., and Shao, J.-Z. (2007). Bone Morphogenetic Protein. Biochem. Biophysical Res. Commun. 362, 550–553. doi:10.1016/j.bbrc.2007.08.045
Xie, X., Fu, Y., and Liu, J. (2017). Chemical Reprogramming and Transdifferentiation. Curr. Opin. Genet. Dev. 46, 104–113. doi:10.1016/j.gde.2017.07.003
Xin, P., Xu, X., Deng, C., Liu, S., Wang, Y., Zhou, X., et al. (2020). The Role of JAK/STAT Signaling Pathway and its Inhibitors in Diseases. Int. Immunopharmacol. 80, 106210. doi:10.1016/j.intimp.2020.106210
Xu, G., Wu, F., Gu, X., Zhang, J., You, K., Chen, Y., et al. (2019). Direct Conversion of Human Urine Cells to Neurons by Small Molecules. Sci. Rep. 9, 16707. doi:10.1038/s41598-019-53007-6
Xu, J., Du, Y., and Deng, H. (2015). Direct Lineage Reprogramming: Strategies, Mechanisms, and Applications. Cell Stem Cell 16, 119–134. doi:10.1016/j.stem.2015.01.013
Xu, Y., Shi, Y., and Ding, S. (2008). A Chemical Approach to Stem-Cell Biology and Regenerative Medicine. Nature 453, 338–344. doi:10.1038/nature07042
Xu, Z., Su, S., Zhou, S., Yang, W., Deng, X., Sun, Y., et al. (2020). How to Reprogram Human Fibroblasts to Neurons. Cell Biosci. 10, 116. doi:10.1186/s13578-020-00476-2
Xue, Y., Ouyang, K., Huang, J., Zhou, Y., Ouyang, H., Li, H., et al. (2013). Direct Conversion of Fibroblasts to Neurons by Reprogramming PTB-Regulated microRNA Circuits. Cell 152, 82–96. doi:10.1016/j.cell.2012.11.045
Yang, D.-W., Moon, J.-S., Ko, H.-M., Shin, Y.-K., Fukumoto, S., Kim, S.-H., et al. (2020). Direct Reprogramming of Fibroblasts into Diverse Lineage Cells by DNA Demethylation Followed by Differentiating Cultures. Korean J. Physiol. Pharmacol. 24, 463–472. doi:10.4196/kjpp.2020.24.6.463
Yang, J., Cao, H., Guo, S., Zhu, H., Tao, H., Zhang, L., et al. (2020). Small Molecular Compounds Efficiently Convert Human Fibroblasts Directly into Neurons. Mol. Med. Rep. 22, 4763–4771. doi:10.3892/mmr.2020.11559
Yang, L. (2018). Neuronal cAMP/PKA Signaling and Energy Homeostasis. Adv. Exp. Med. Biol. 1090, 31–48. doi:10.1007/978-981-13-1286-1_3
Yang, N., Ng, Y. H., Pang, Z. P., Südhof, T. C., and Wernig, M. (2011). Induced Neuronal Cells: How to Make and Define a Neuron. Cell Stem Cell 9, 517–525. doi:10.1016/j.stem.2011.11.015
Yang, T., Xing, L., Yu, W., Cai, Y., Cui, S., and Chen, G. (2020). Astrocytic Reprogramming Combined with Rehabilitation Strategy Improves Recovery from Spinal Cord Injury. FASEB J. 34, 15504–15515. doi:10.1096/fj.202001657RR
Yang, Y., Chen, R., Wu, X., Zhao, Y., Fan, Y., Xiao, Z., et al. (2019). Rapid and Efficient Conversion of Human Fibroblasts into Functional Neurons by Small Molecules. Stem Cell Rep. 13, 862–876. doi:10.1016/j.stemcr.2019.09.007
Ye, J., Ge, J., Zhang, X., Cheng, L., Zhang, Z., He, S., et al. (2016). Pluripotent Stem Cells Induced from Mouse Neural Stem Cells and Small Intestinal Epithelial Cells by Small Molecule Compounds. Cell Res. 26, 34–45. doi:10.1038/cr.2015.142
Yi, B., Ding, T., Jiang, S., Gong, T., Chopra, H., Sha, O., et al. (2021). Conversion of Stem Cells from Apical Papilla into Endothelial Cells by Small Molecules and Growth Factors. Stem Cell Res. Ther. 12, 266. doi:10.1186/s13287-021-02350-5
Yin, J.-C., Zhang, L., Ma, N.-X., Wang, Y., Lee, G., Hou, X.-Y., et al. (2019). Chemical Conversion of Human Fetal Astrocytes into Neurons through Modulation of Multiple Signaling Pathways. Stem Cell Rep. 12, 488–501. doi:10.1016/j.stemcr.2019.01.003
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., et al. (2011). MicroRNA-mediated Conversion of Human Fibroblasts to Neurons. Nature 476, 228–231. doi:10.1038/nature10323
Yuan, Z.-D., Zhu, W.-N., Liu, K.-Z., Huang, Z.-P., and Han, Y.-C. (2020). Small Molecule Epigenetic Modulators in Pure Chemical Cell Fate Conversion. Stem Cells Int. 2020, 1–12. doi:10.1155/2020/8890917
Zarubin, T., and Han, J. (2005). Activation and Signaling of the P38 MAP Kinase Pathway. Cell Res. 15, 11–18. doi:10.1038/sj.cr.7290257
Zeng, J., Li, Y., Ma, Z., and Hu, M. (2021). Advances in Small Molecules in Cellular Reprogramming: Effects, Structures, and Mechanisms. Cscr 16, 115–132. doi:10.2174/1574888X15666200621172042
Zhang, F., Zhang, L., Qi, Y., and Xu, H. (2016). Mitochondrial cAMP Signaling. Cell. Mol. Life Sci. 73, 4577–4590. doi:10.1007/s00018-016-2282-2
Zhang, K., Yang, C., Chang, L., Sakamoto, A., Suzuki, T., Fujita, Y., et al. (2020). Essential Role of Microglial Transforming Growth Factor-Β1 in Antidepressant Actions of (R)-ketamine and the Novel Antidepressant TGF-Β1. Transl. Psychiatry 10, 32. doi:10.1038/s41398-020-0733-x
Zhang, L., Yang, X., Yang, S., and Zhang, J. (2011). The Wnt/β-Catenin Signaling Pathway in the Adult Neurogenesis. Eur. J. Neurosci. 33, 1–8. doi:10.1111/j.1460-9568.2010.7483.x
Zhang, L., Yin, J.-C., Yeh, H., Ma, N.-X., Lee, G., Chen, X. A., et al. (2015). Small Molecules Efficiently Reprogram Human Astroglial Cells into Functional Neurons. Cell Stem Cell 17, 735–747. doi:10.1016/j.stem.2015.09.012
Zhang, M., Lin, Y.-H., Sun, Y. J., Zhu, S., Zheng, J., Liu, K., et al. (2016). Pharmacological Reprogramming of Fibroblasts into Neural Stem Cells by Signaling-Directed Transcriptional Activation. Cell Stem Cell 18, 653–667. doi:10.1016/j.stem.2016.03.020
Zhang, S., Chung, W.-c., Wu, G., Egan, S. E., and Xu, K. (2014). Tumor-suppressive Activity of Lunatic Fringe in Prostate through Differential Modulation of Notch Receptor Activation. Neoplasia 16, 158–167. doi:10.1593/neo.131870
Zhang, Y., Zhou, L., and Miller, C. A. (1998). A Splicing Variant of a Death Domain Protein that Is Regulated by a Mitogen-Activated Kinase Is a Substrate for C-Jun N-Terminal Kinase in the Human Central Nervous System. Proc. Natl. Acad. Sci. U.S.A. 95, 2586–2591. doi:10.1073/pnas.95.5.2586
Zhao, A.-D., Qin, H., Sun, M.-L., Ma, K., and Fu, X.-B. (2020). Efficient and Rapid Conversion of Human Astrocytes and ALS Mouse Model Spinal Cord Astrocytes into Motor Neuron-like Cells by Defined Small Molecules. Mil. Med. Res. 7, 42. doi:10.1186/s40779-020-00271-7
Zheng, X., Boyer, L., and Jin, M. (2016). Metabolic Reprogramming during Neuronal Differentiation from Aerobic Glycolysis to Neuronal Oxidative Phosphorylation. Elife 5. doi:10.7554/eLife.13374
Zheng, Y., Huang, Z., Xu, J., Hou, K., Yu, Y., Lv, S., et al. (2021). MiR-124 and Small Molecules Synergistically Regulate the Generation of Neuronal Cells from Rat Cortical Reactive Astrocytes. Mol. Neurobiol. 58, 2447–2464. doi:10.1007/s12035-021-02345-6
Zhong, J., and Zou, H. (2014). BMP Signaling in Axon Regeneration. Curr. Opin. Neurobiol. 27, 127–134. doi:10.1016/j.conb.2014.03.009
Zhou, C., Gu, H., Fan, R., Wang, B., and Lou, J. (2015). MicroRNA 302/367 Cluster Effectively Facilitates Direct Reprogramming from Human Fibroblasts into Functional Neurons. Stem Cells Dev. 24, 2746–2755. doi:10.1089/scd.2015.0123
Nomenclature
Abbreviations
AAV-FLEX-EGFP flexed Enhanced Green Fluorescent Protein-adeno-associated virus
AD Alzheimer’s Disease
ALDH1L1 Aldehyde dehydrogenase 1 family member L1
ALK Activin-receptor-like kinase
ALS amyotrophic lateral sclerosis
ATRA all-trans retinoic acid
BBB blood-brain barrier
BDNF brain-derived neurotrophic factor
bFGF fibroblast growth factor-basic
BMP bone morphogenetic protein
cAMP cyclic adenosine monophosphate
CAYTFVB CHIR99021, A8301, Y27632, TTNPB, Forskolin, Valproic acid (VPA), NaB
CFLSSVY CHIR99021, Forskolin, LDN193189, SB431542, SP600125, VPA, and Y27632
CiNs Chemically-induced neurons
CNS Central Nervous System
CREB cyclic AMP-responsive element-binding protein
CRISPR/Cas9 Clustered regularly interspaced short palindromic repeat/Cas9
CRISPR Clustered Regularly Interspaced Short Palindromic Repeats
DCX doublecortin
DEGs differentially expressed genes
DFICBY DBcAMP, Forskolin, ISX9, CHIR99021, I-BET151, Y-27632
DNA deoxyribonucleic acid
DNMTs DNA methyltransferases
EMT Epithelial-to-Mesenchymal transition
ERK extracellular signal-regulated kinases
FAD familial Alzheimer’s disease
FGF8 fibroblast growth factor 8
FICSB Forskolin, ISX9, CHIR99021, SB431542, and I-BET151
FoxD3 Forkhead box D3
FRSCGYIB Forskolin, RepSox, SP600125, CHIR99021, Go6983, Y27632, IXS9, I BET151
GAPs GTPase-activating proteins
GEFs guanine nucleotide-exchange factors
GFAP glial fibrillary acidic protein
GFP green fluorescent protein
GO Gene Ontology
GSK3 glycogen synthase kinase-3
H3K27 histone H3 at lysine 27
H3K9 histone H3 at lysine 9
HAT histone acetyltransferases
hCiNs human chemical-induced neurons
HDACs histone deacetylases
hGFAP human GFAP
HIF-1α Hypoxia-inducible factor 1-alpha
hiMNs human-induced motor neurons
iNs Induced neurons
iNSCs Induced multipotent neural stem cells
iPSCs Induced pluripotent stem cells
JAG1/2 jagged 1/2
JAK-STAT Janus family tyrosine kinase–signal transducer and activator of transcription
JNK Jun N-terminal Kinase
KFYPR kenpaullone, forskolin, Y-27632, purmorphamine, RA
LRP5/6 receptor-related protein 5/6
MAP2 microtubule association protein-2
MAPKs mitogen-activated protein kinase
MET Mesenchymal-to-Epithelial transition
MSCs mesenchymal stromal cells
mTOR Mammalian Target of Rapamycin
NeuN neuronal nuclei
NF-κB nuclear factor kappa B
NICD Notch intracellular domain
NPCs neural progenitor cells
OXPHOS oxidative phosphorylation
PD Parkinson’s Disease
PTCH1 patched 1
RA retinoic acid
RBPJ recombining binding protein suppressor of hairless
RNA-seq RNA-sequencing
ROCK Rho-associated protein kinase
ROS reactive oxygen species
RT-qPCR real-time quantitative polymerase chain reaction
SCI spinal cord injury
SH2 Src homology 2
SHH Sonic Hedgehog
SLCD SB431542, LDN193189, CHIR99021, DAPT
SMO smoothened
sPSCs spontaneous postsynaptic currents
TCF/LEF1 T-cell factor/lymphoid enhancer factor
TFs Transcription factors
TGF-β transforming growth factor-beta
TSS transcription start site
TUJ1 β-tubulin III
VCRF VPA, CHIR99021, Repsox, Forskolin
VCRFSGY VPA, CHIR99021, Repsox, Forskolin, SP600125, GO6983, Y27632
VCRFSGYIBRTQ VPA, CHIR99021, Repsox, Forskolin, SP600625, GO6983, Y27632, ISX9, I-BET151, RA, Vit C, QVD-OPH
Wnt Wingless/integrated
Keywords: small molecule, direct neuronal reprogramming, transdifferentiation, regeneration medicine, neuron, neurological disease
Citation: Wang J, Chen S, Pan C, Li G and Tang Z (2022) Application of Small Molecules in the Central Nervous System Direct Neuronal Reprogramming. Front. Bioeng. Biotechnol. 10:799152. doi: 10.3389/fbioe.2022.799152
Received: 21 October 2021; Accepted: 09 June 2022;
Published: 07 July 2022.
Edited by:
Antonella Motta, University of Trento, ItalyReviewed by:
Ozgul Gok, Acıbadem University, TurkeyHuihua Yuan, Nantong University, China
Juergen Hescheler, University of Cologne, Germany
Pinghui Zhou, Bengbu Medical College, China
Copyright © 2022 Wang, Chen, Pan, Li and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaigai Li, bGdnaHVzdGVyQDE2My5jb20=; Zhouping Tang, ZGRqdHpwQDE2My5jb20=
 Jingyi Wang
Jingyi Wang Shiling Chen
Shiling Chen Chao Pan
Chao Pan Zhouping Tang
Zhouping Tang