- 1The Second Hospital of Hebei Medical University, Shijiazhuang, China
- 2Department of Oncology, The Second Hospital of Hebei Medical University, Shijiazhuang, China
- 3Department of Thoracic Surgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China
- 4Institute of Precision Medicine and Pathology, Jinan University, Guangzhou, China
Objective: LINC00662 is oncogenic in some human cancers, but no much was revealed concerning to its specific action in tumor angiogenesis. Given that, our study investigated the role of LINC00662 from esophageal squamous cell carcinoma (ESCC) cells-derived extracellular vehicles (EVs) in angiogenesis through microRNA (miR)-195-5p/vascular endothelial growth factor A (VEGFA) axis.
Methods: Clinical tissue samples were collected from patients with ESCC, in which LINC00662, miR-195-5p and VEGFA expression was analyzed. ESCC cells were transfected, from which EVs were isolated. Human umbilical vein endothelial cells (HUVECs) were co-cultured with the pretreated EVs. After that, viability, colony formation ability, invasion, migration and tube formation ability of HUVECs were observed. Tumor xenograft in nude mice was performed to detect the effect of LINC00662, miR-195-5p or EV specific inhibitor GW4869 on tumor development.
Results: LINC00662 and VEGFA were upregulated while miR-195-5p was downregulated in the cancer tissue of patients with ESCC. EVs derived from ESCC cells promoted viability, colony formation ability, invasion and tube formation ability of HUVECs. Downregulation of LINC00662 or upregulation of miR-195-5p reversed the promotion of EVs derived from ESCC cells on the viability, colony formation ability, invasion and tube formation ability of HUVECs in vitro and in vivo. VEGFA overexpression reversed EVs carrying restored miR-195-5p induced effects on HUVECs in vitro.
Conclusion: In summary, elevated LINC00662 transferred by ESCC cells-derived EVs induces angiogenesis through downregulating miR-195-5p and upregulating VEGFA.
Introduction
Esophageal squamous cell carcinoma (ESCC) is the main subtype of esophageal cancer (EC) which originates from the stratified squamous epithelium of the esophagus (Reichenbach et al., 2019). Because of repeated inflammation and mucosal damage of the esophagus, it is most often diagnosed by tissue biopsy during upper gastrointestinal endoscopy (Batra et al., 2019). ESCC mostly happens in late adulthood, and patients often experience symptoms such as dysphagia and weight loss (Lam, 2020). For advanced ESCC, cytotoxic agents (docetaxel, cisplatin and 5-FU), molecular-targeting therapy and immunotherapy are systematically applied to palliate symptoms and improve survival (Hirano and Kato, 2019). In tumor progression, tumor cells secrete high levels of pro-angiogenic factors, which contribute to the formation of abnormal vascular networks, leading to poor tumor perfusion (Viallard and Larrivée, 2017). Considering that, fighting against tumor angiogenesis may offer alternative therapeutic targets.
Extracellular vesicles (EVs) is a collective term involving in many subtypes of cell-released, membranous structures, named as exosomes, ectosomes, oncosomes, microvesicles, microparticles, apoptotic bodies, as well as many other names (Théry et al., 2018). EV are cell membrane-derived vesicles, and cancer cells-derived EVs exert multiple roles in malignant progression, including metastasis and angiogenesis (Knox et al., 2020). Hypoxia-conditioned EVs from ESCC cells have been reported to induce tube formation of human umbilical vein endothelial cells (HUVECs) (Mao et al., 2019). As effective carriers, EVs can transfer nucleic acids including miRNA and long noncoding RNA into specific target cells, regulating cell function and tumor microenvironment (Lin et al., 2019). More and more studies have confirmed the pro-angiogenesis effect of cancer-derived EVs carrying tumor-promoting cargos (Lang et al., 2017; Zeng et al., 2018; He et al., 2019). LINC00662 refers to an oncogene in many cancers, which acts notoriously in the fields of tumor metastasis (Cheng B et al., 2020) and radioresistance (Chen et al., 2020). In ESCC, a report has described specifically the pro-metastatic property of LINC00662 (Zhang et al., 2020), but little was discovered in relation to LINC00662-mediated angiogenesis. miR-195-5p is an anti-angiogenic modulator in human cancers (Cai et al., 2018; Dai et al., 2019). In the setting of ESCC, miR-195-5p has been identified as a novel biomarker for cancer diagnosis, classification and prognosis (Li C. Y et al., 2019), but miR-195-5p-dependent angiogenesis remains unknown. Tumor angiogenesis mainly relies on the response driven by vascular endothelial growth factor A (VEGFA) (Claesson-Welsh and Welsh, 2013). VEGFA is commonly amplified in esophageal adenocarcinomas (Cancer Genome Atlas Research, Network et al., 2017) and regulation of VEGFA by miRNAs serves a therapeutic option for EC (Kong et al., 2016; Shen W et al., 2020). For reasons of treatments, our research mainly discussed the effect of LINC00662 delivered by ESCC cells-derived EVs on angiogenesis through modification of miR-195-5p and VEGFA.
Methods and Materials
Tissue Collection
A total of 84 ESCC patients (44–79 years old, mean age of 62.02 ± 9.49 years) were recruited from The Second Hospital of Hebei Medical University. Among them, 42 cases were in stage I, 28 cases in stage II, and 14 cases in stage III. All patients underwent tumor resection before chemotherapy. ESCC tissues and adjacent normal tissues were collected and preserved at −80°C. All patients were pathologically diagnosed as ESCC, and no radiotherapy or chemotherapy before surgery was performed. Patients who died from diseases unrelated to ESCC during the follow-up were excluded (He et al., 2020).
Cell Culture and Transfection
Human normal esophageal epithelial cells (HEEC) and ESCC cell lines EC9706, Eca109, TE-13, TE-1 and TTN were provided by China Center for Type Culture Collection (Wuhan, China). Cells (3 × 105 cells/well) with 80% confluence were subjected to transfection using Lipofectamine 2000 (Invitrogen, CA, United States). Overexpressed (oe)-VEGFA, small interfering RNA (si)-LINC00662, oe-LINC00662, miR-195-5p mimic and their corresponding negative control (NC) were all from GenePharma (Shanghai, China) (Li Z et al., 2019).
EVs Collection and Identification
ESCC cells were placed in Roswell Park Memorial Institute (RPMI) 1640 medium containing EVs-free 10% fetal bovine serum (FBS) and cultured for 3 d. Then, the cell supernatant was collected and centrifuged to remove cell debris. The EVs were extracted using Hieff™ Quick exosome isolation kit (41201ES50, YEASEN, Shanghai, China). The supernatant and exosome separation reagent were added into the Eppendorf (EP) tube with a proportion of 2:1 overnight, and then centrifuged at 100,00 g for 1–2 h and the precipitate was resuspended in phosphate buffered saline (PBS). The EV suspension (30 μl) together with an equal volume of radioimmunoprecipitation assay (RIPA) lysis buffer was continuously dissolved in microwave (2 times, 10 s/time). Then, the EVs were purified at 4°C at 12,000 × g for 2 min, and the supernatant was retained and stored at −80°C. Protein quantification of EV was performed by bicinchoninic acid (BCA) kit (Beyotime, Nantong, China).
EVs identification: Transmission electron microscope (TEM; JEM-1010, JEOL, Tokyo, Japan) was adopted to observe EV morphology, tunable resistive pulse sensing (TRPS) to examine concentration and diameter of EVs, and Western blot analysis to test EV markers (CD81 and CD9) and non-marker protein (Calnexin and GM130) (Li Z et al., 2019).
EVs Uptake
EVs uptake was monitored using PKH26 fluorescent labeling kit (Sigma-Aldrich, CA, United States). EV sample was diluted in diluent C, and then PKH26 dye was added and kept at room temperature. After 4 min incubation avoiding light exposure, the reaction was neutralized by 5% bovine serum albumin. Then, EVs, resuspended in PBS were centrifuged at 100,000 g for 70 min, and incubated with HUVECs for 4 h. Subsequently, HUVECs (ATCC, VA, United States) were fixed with 4% paraformaldehyde, labeled with 4′,6-diamidino-2-phenylindole (0.5 μg/ml; Invitrogen) and imaged under a confocal microscope (Carl Zeiss, Oberkochen, Germany) (Zhang et al., 2021).
When HUVECs reached 50–60% confluence, they were incubated with dextran 498 (Dx498, 100 μg/ml) for 2 h to label cell membrane lysosomes, washed with PBS and cultured with complete culture medium for 18 h. Finally, HUVECs were incubated with PKH26-labeled EVs for colocalization analysis.
Co-Culture of HUVECs and EVs
HUVECs were allocated into groups: PBS group (HUVECs without any treatment), EV group (HUVECs co-cultured with EV from ESCC cells), si-NC-EV group (HUVECs co-cultured with EVs from si-NC-modified ESCC cells), si-LINC00662-EV group (HUVECs co-cultured with EVs from si-LINC00662-modified ESCC cells), mimic-NC-EV group (HUVECs co-cultured with EVs from miR-195-5p mimic NC-modified ESCC cells), miR-195-5p mimic-EV group (HUVECs co-cultured with EVs from miR-195-5p mimic-modified ESCC cells), si-NC group (HUVECs transfected with si-NC), si-LINC00662 group (HUVECs transfected with si-LINC00662), oe-NC group (HUVECs transfected with oe-NC), oe-LINC0066 group (HUVECs transfected with oe-LINC0066), miR-195-5p mimic-EV + oe-NC group (HUVECs co-cultured with EVs from miR-195-5p mimic-modified ESCC cells and further transfected with oe-NC), miR-195-5p mimic-EV + oe-VEGFA (HUVECs co-cultured with EVs from miR-195-5p mimic-modified ESCC cells and further transfected with oe-VEGFA).
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-Diphenyltetrazolium Bromide (MTT) Assay
HUVECs were added with 5 mg/ml MTT solution (20 μl, Solarbio, Beijing, China), and cell viability was measured after 12, 24, and 36 h. Using a microplate spectrophotometer (BioTek Instruments, VT, United States), the absorbance at 570 nm was measured (You et al., 2021).
Colony Formation Test
HUVECs were cultured in RPMI 1640 medium supplemented with 10% FBS, during which the medium was changed every 48 h. After 14 d, visible colonies were fixed with ethanol, stained with 0.1% crystal violet and manually counted.
Angiogenesis Test
Matrigel (BD Biosciences) was spread over 24-well plates at 100 ml/well and polymerized. HUVECs (resuspended in FBS-free medium) were transferred to the plates at 1 × 105 cells/well and cultured for 6 h. Angiogenesis quantification was judged by the number of branch points of the formed tube in at least five fields of view (Zhou et al., 2019).
Transwell Assay
HUVECs were resuspended in serum-free medium, and the resuspension (1 × 105 cells/mL, 200 μl) was added to the top side of Matrigel-coated Transwell chamber (Matrigel was not used to examine cell migration). RPMI 1640 medium (600 μl) containing 20% FBS was dispersed in the basolateral chamber. After 48 h of culture, cells were fixed with 4% paraformaldehyde, stained with crystal violet and observed under an optical microscope in five fields of view (Li Z et al., 2019).
Dual Luciferase Reporter Gene Assay
LINC00662-Wt, LINC00662-Mut, VEGFA-Wt and VEGFA-Mut were cloned and merged with psiCHECK-2 vector (Promega, WI, United States). Employing Lipofectamine 2,000, the vectors were co-transfected with mimic-NC or miR-195-5p mimic into cells and detected by the dual luciferase reporter gene detection system (Promega) (Liu and Meng, 2020).
RNA-Pull Down Assay
Cells were transfected with 50 nM biotin-labeled Wt-bio-miR-195-5p and Mut-bio-miR-195-5p for 48 h, and subjected to reaction in a specific lysis buffer (Ambion, Texas, United States) for 10 min. The lysate was incubated with M-280 streptavidin magnetic beads (Sigma) pre-coated with RNase-free BSA and yeast tRNA (TRNABAK-RO, Sigma). The beads were incubated at 4°C for 3 h and eluted, and the bound RNA was purified by Trizol, and the LINC00662 enrichment was detected by reverse transcription quantitative polymerase chain reaction (RT-qPCR).
RIP Assay
The binding of LINC00662/miR-195-5p to AGO2 protein was analyzed by Magna RIP kit (Millipore, United States). Cells were lysed with an equal volume of RIPA Lysis Buffer (P0013B, Beyotime) in an ice bath for 5 min and centrifuged at 14,000 rpm, 4°C for 10 min. The magnetic beads (50 µl) were resuspended in 100 µl RIP Wash Buffer, and added 5 µg antibody AGO2 (1:50, Abcam) or IgG (1:100, Abcam). The magnetic bead-antibody complex was resuspended in 900 µL RIP Wash Buffer and incubated with 100 μl cell lysate overnight at 4°C. The resulting complex was digested with proteinase K and RNA was extracted for PCR detection.
RT-qPCR
Total RNA in cells and tissues was extracted by Trizol (Takara, Dalian, China) and expression of miR-195-5p was tested by qRT-PCR with the All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia) and All-in-One miRNA qPCR Primer (GeneCopoeia). Expression of LINC00662 and VEGFA was determined by qRT-PCR RevertAid First Strand cDNA Synthesis Kit (Fermentas) and SYBR Premix Ex Taq (TaKaRa). Primer sequences (Supplementary Table S1) were synthesized by Genepharma. Target expression was analyzed using ABI 7500 (ABI, CA, United States), and standardized by U6 or glyceraldehyde phosphate dehydrogenase. Determination of gene expression was dependent on 2−ΔΔCt method (Li Z et al., 2019).
Western Blot Assay
Total protein was separated with RIPA lysis buffer (Beyotime) and quantified by BCA kit (Pierce, MA, United States). After separation by 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (Beyotime), the protein sample was electroblotted onto nitrocellulose membrane (HTS112 M), covered with 5% skim milk and kept overnight with primary antibodies VEGFA (1:1,000), CD81 (1 μg/ml), CD9 (1:1,000), Calnexin (1 μg/ml), GM130 (1 μg/ml) and GAPDH (1:1,000, all from Abcam). Next, horseradish peroxidase-conjugated secondary antibodies goat anti-mouse IgG (1:5,000) and goat anti-rabbit IgG (1:5,000, both from Abcam) were incubated with the membrane, after which signals were detected using SuperSig-nal-West Pico chemiluminescent substrate (Thermo Fisher Scientific, MA, United States). Data analysis was carried out with Image Lab™ 3.0 (Bio-Rad, CA, United States) (Sui et al., 2021).
Tumor Xenografts in Nude Mice
Eca109 cells (2 × 106) were injected subcutaneously into the hind limb of BALB/c nude mice (6 weeks old, athymic) for the establishment of the ESCC xenograft model. When the nude mice generated the tumors (100 mm3), 10 μg EVs from ESCC cells that had been transfected with si-NC, si-LINC00662, mimic-NC, miR-195-5p mimic, PBS, and exosome release inhibitor (GW4869, 1 mg/kg) were subsequently injected into the center of tumor sites every 2 days. Nude mice were euthanized 4 weeks later, and the subcutaneous tumors were obtained to calculate tumor volume (maximum diameter × vertical diameter2/2).
Statistical Analysis
GraphPad Prism 6.0 (GraphPad Software Inc.) and SPSS 20.0 (IBM, NY, United States) were used for data analysis. Values were expressed as mean ± standard deviation and compared by t-test (two groups) or analysis of variance (multiple groups). p < 0.05 was statistically significant.
Results
High LINC00662 and VEGFA and Low miR-195-5p in Cancer Tissues of Patients With ESCC
LINC00662, miR-195-5p, and VEGFA levels in tissue specimens of 84 patients with ESCC were determined by RT-qPCR, as the results reflecting that LINC00662 and VEGFA were upregulated and miR-195-5p was downregulated in ESCC tissues (Figures 1A–C).
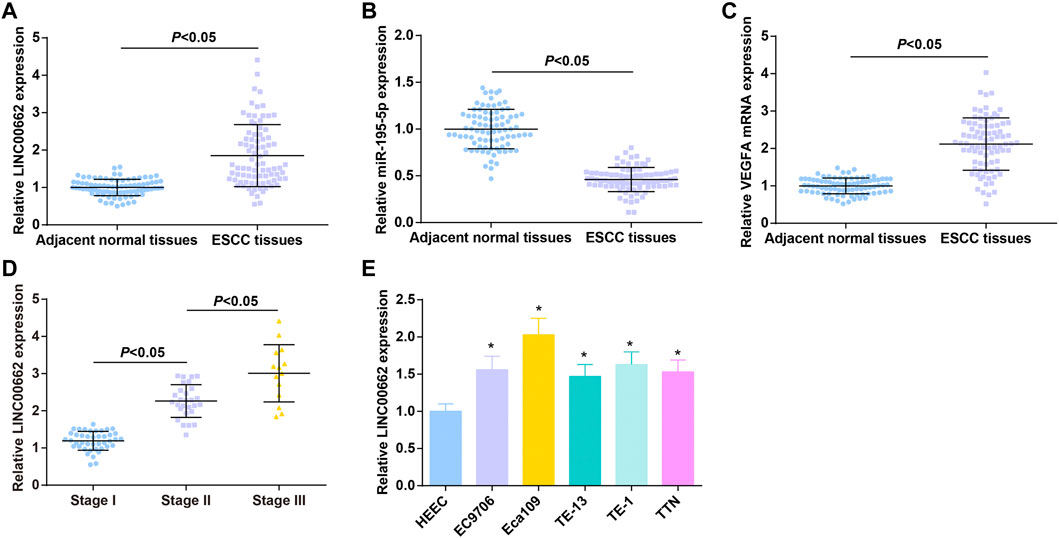
FIGURE 1. High LINC00662 and VEGFA and low miR-195-5p in cancer tissues of patients with ESCC. (A–C). RT-qPCR detection of LINC00662, miR-195-5p, VEGFA expression in ESCC tissues and adjacent normal tissues of patients; (D) RT-qPCR detection of LINC00662 expression in ESCC patients at different TNM stages; (E) RT-qPCR detection of LINC00662 expression in HEEC and ESCC cell lines. * p < 0.05 vs. HEEC cells.
ESCC patients were grouped according to TNM staging, and RT-qPCR analysis found higher LINC00662 expression in patients in the late stage than in the early stage (Figure 1D). LINC00662 expression was compared in HEEC and ESCC cell lines (EC9706, Eca109, TE13, TE1 and TTN) by RT-qPCR, and the outcomes indicated that LINC00662 was abundantly expressed in ESCC cell lines (Figure 1E). Because Eca109 had the highest LINC00662 level, it was selected for subsequent experiments.
Repressive Role of LINC00662 Downregulation in Angiogenesis in ESCC, and Upregulation of LINC00662 has the Opposite Effect
To clarify the effect of LINC00662 on Eca109 angiogenesis in vitro, LINC00662 was overexpressed or specifically interfered. RT-qPCR test results showed that oe-LINC00662 effectively promoted LINC00662 expression, while si-LINC00662 down-regulated LINC00662 expression in HUVECs (Figure 2A). As reflected by MTT assay, colony formation assay, Transwell assay and angiogenesis test, downregulated LINC00662 depressed proliferation, invasion, migration and angiogenesis but up-regulated LINC00662 promoted these phenotypes of HUVECs (Figures 2B–F). Collectively, LINC00662 downregulation inhibits tumor angiogenesis.
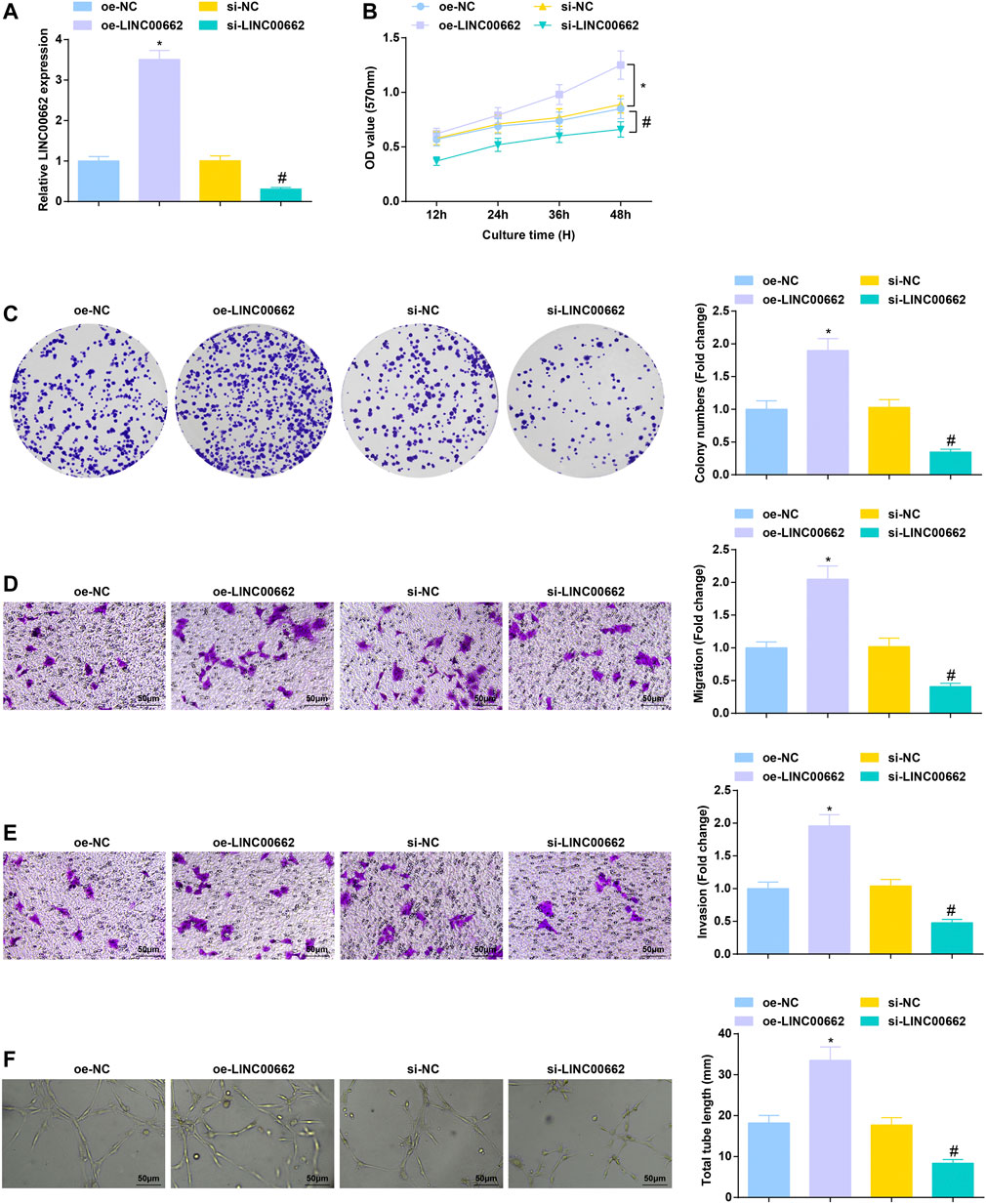
FIGURE 2. Repressive role of LINC00662 downregulation in angiogenesis. (A) RT-qPCR detection of LINC00662 expression in HUVECs after transfection with silenced LINC00662; (B,C) Viability and colony formation ability of HUVECs after transfection with silenced LINC00662; (D,E) Migration and invasion of HUVECs after transfection with silenced LINC00662; (F) Angiogenesis of HUVECs after transfection with silenced LINC00662. * p < 0.05 vs. the oe-NC group; # p < 0.05 vs. the si-NC group.
EVs Collection and Identification
A white precipitate was separated from the culture medium of Eca109 cells by differential centrifugation. Under the TEM, the white precipitate was round or elliptical vesicles with a double-layer membrane structure and a diameter of 40–100 nm (Figure 3A); TRPS showed that the mode of particle diameter was 90 nm and the average diameter was 130 nm (Figure 3B); Western blot detected that CD81 and CD9 were both positively expressed whilst calnexin and GM130 were not expressed in these vesicles (Figure 3C), confirming that EVs were extracted successfully.
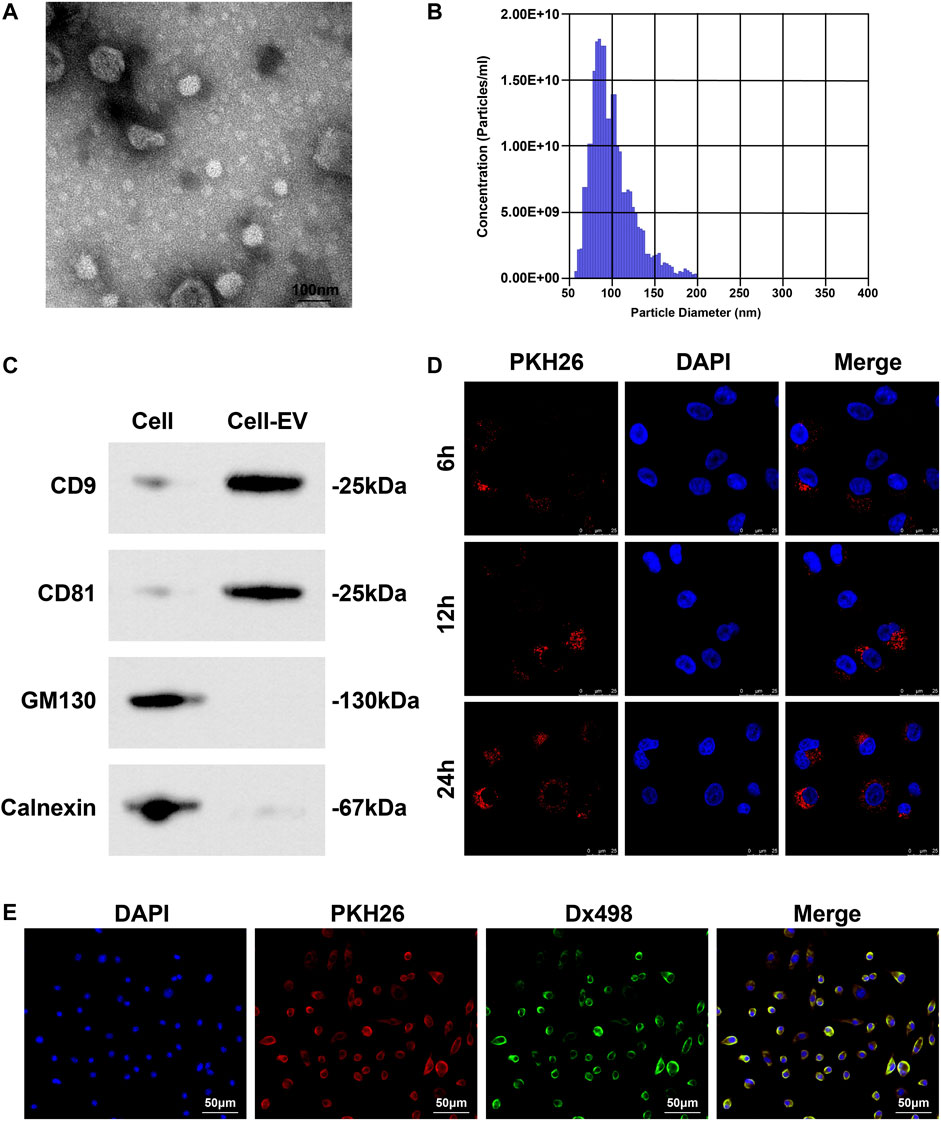
FIGURE 3. EVs collection and identification. (A) Morphology and structure of EVs under TEM; (B) Concentration and diameter of EVs examined by TRPS; (C) EV markers (CD81, CD9, Calnexin and GM130) analyzed by Western blot; (D) Distribution of PKH26-labled EVs in HUVECs; (E) Co-localization of lysosome and EVs in HUVECs.
PKH26-labeled EVs were co-cultured with HUVECs. Observed under a fluorescence microscope, EVs could enter HUVECs, and over time, the more EVs taken up by HUVECs (Figure 3D). Fluorescence colocalization experiment showed that HUVECs incubated with Dx498 were co-cultured with PKH26-labeled EVs, and EVs and lysosomes were colocalized (Figure 3E).
Anti-Angiogenesis Effect of EVs-Mediated Delivery of Inhibited LINC00662
After co-culture with Eca109 cells-derived EVs, RT-qPCR detection found that LINC00662 expression was increased in HUVECs, while LINC00662 expression was decreased after co-culture with EVs extracted from Eca109 cells transfected with si-LINC00662 (Figures 4A,B). Subsequently, analysis of cell functions revealed that Eca109 cells-derived EVs augmented proliferation, invasion, migration and angiogenesis of HUVECs, while downregulated LINC00662 reversed the effects of Eca109 cells-derived EVs (Figures 4C–G). In conclusion, Eca109 cells-derived EVs transfer of silenced LINC00662 blocks angiogenesis.
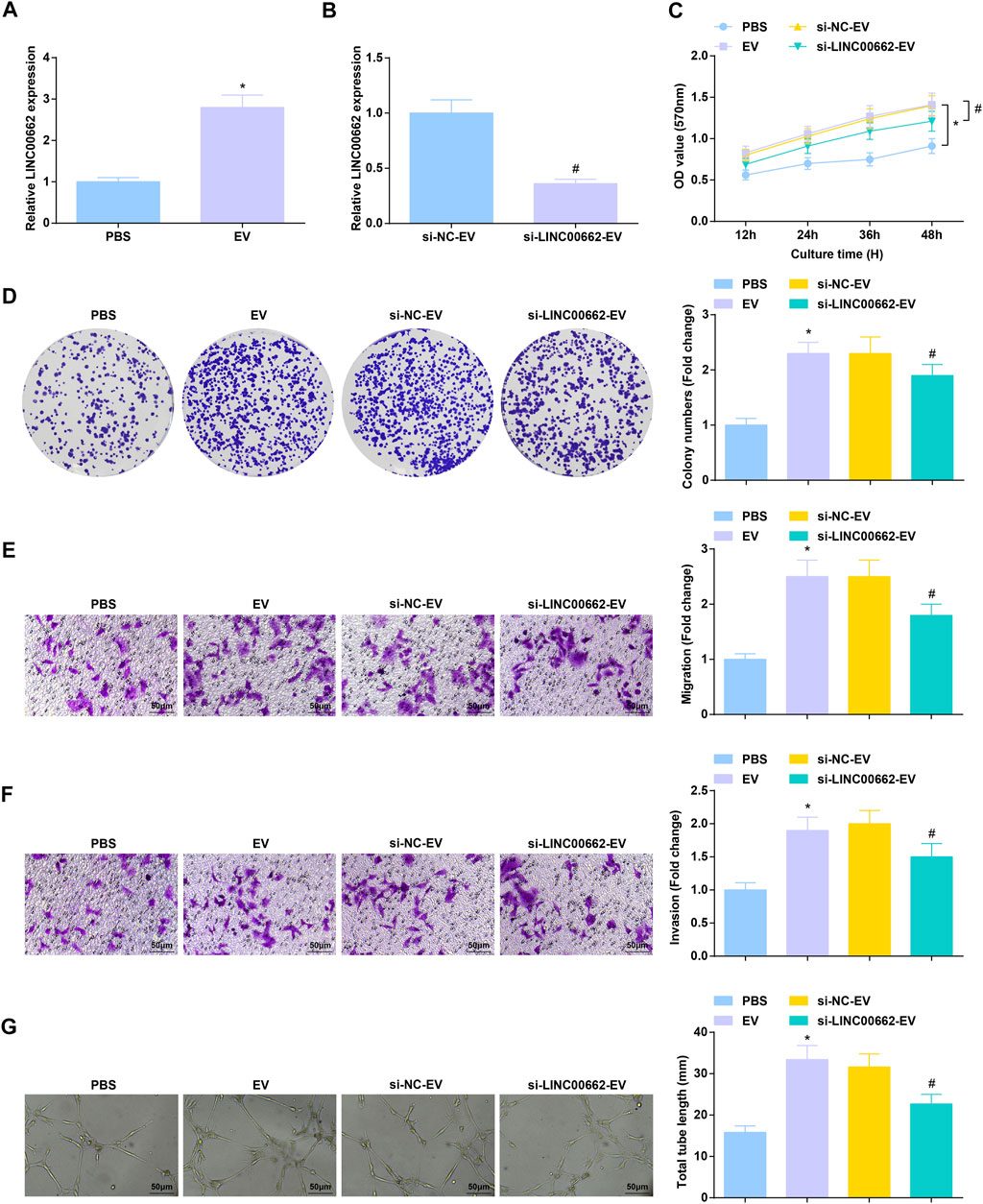
FIGURE 4. Anti-angiogenesis effect of EVs transfer of inhibited LINC00662. (A,B) RT-qPCR detection of LINC00662 expression in HUVECs after co-culture with EVs; (C,D) Viability and colony formation ability of HUVECs after co-culture with EVs; (E,F) Migration and invasion of HUVECs after co-culture with EVs; (G) Angiogenesis of HUVECs after co-culture with EVs. * p < 0.05 vs. the PBS group; #p < 0.05 vs. the si-NC-EV group.
Anti-Angiogenic Role of EVs Carrying Upregulated miR-195-5p
RT-qPCR test measured that co-culture with EVa from miR-195-5p mimic-treated Eca109 cells augmented miR-195-5p expression in HUVECs (Figure 5A). As a result of miR-195-5p upregulation, Eca109 cells-derived EVs-induced aggressive behaviors and angiogenesis of HUVECs were all constrained (Figures 5B–F). In short, EVa transfer of upregulated miR-195-5p exerts anti-angiogenic effect.
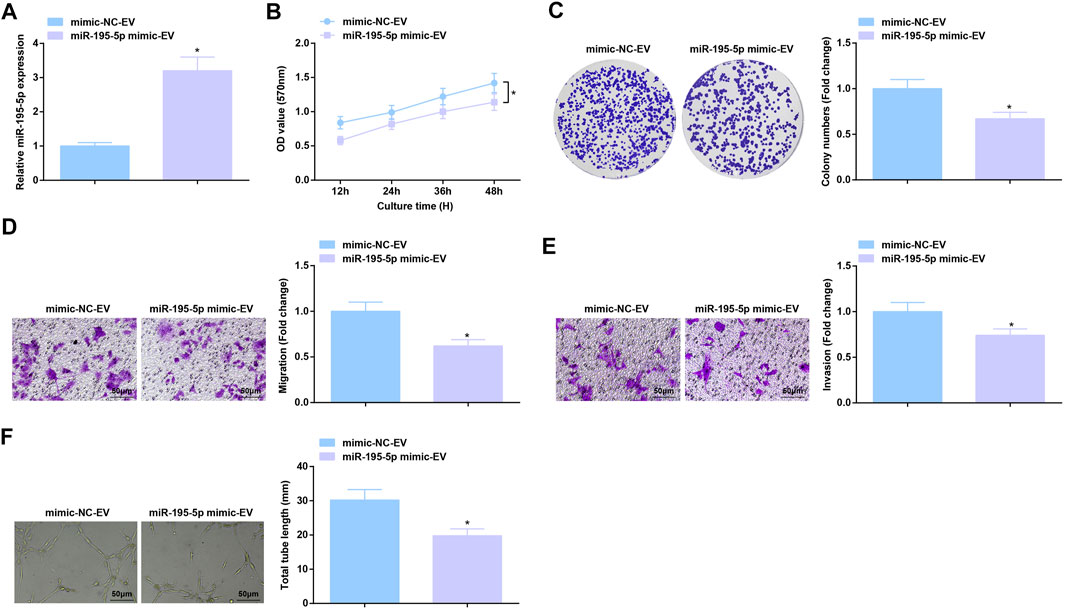
FIGURE 5. Anti-angiogenic role of EVs expressing upregulated miR-195-5p. (A) RT-qPCR detection of miR-195-5p expression in HUVECs after co-culture with EVs; (B,C) Viability and colony formation ability of HUVECs after co-culture with EVs; (D,E) Migration and invasion of HUVECs after co-culture with EVs; (F) Angiogenesis of HUVECs after co-culture with EVs. * p < 0.05 vs. the mimic-NC-EV group.
Binding Relation Between LINC00662 and miR-195-5p; Targeting Relation Between miR-195-5p and VEGFA
On the bioinformatics website, a binding relationship was predicted between LINC00662 and miR-195-5p (Figure 6A), as well as miR-195-5p and VEGFA mRNA (Figure 6B). Using dual luciferase reporter assay, it was estimated that miR-195-5p mimic reduced the luciferase activity of LINC00662-Wt and that of VEGFA-Wt (Figures 6C,D). RNA-pull down assay showed that Mut-bio-miR-195-5p had no significant effect on LINC00662 enrichment and Wt-bio-miR-195-5p could increase the amount of LINC00662 enrichment (Figure 6E). RIP experiment suggested an elevation in Ago2-bound LINC00662, indicating that LINC00662 could directly bind with miR-195-5p (Figure 6F).
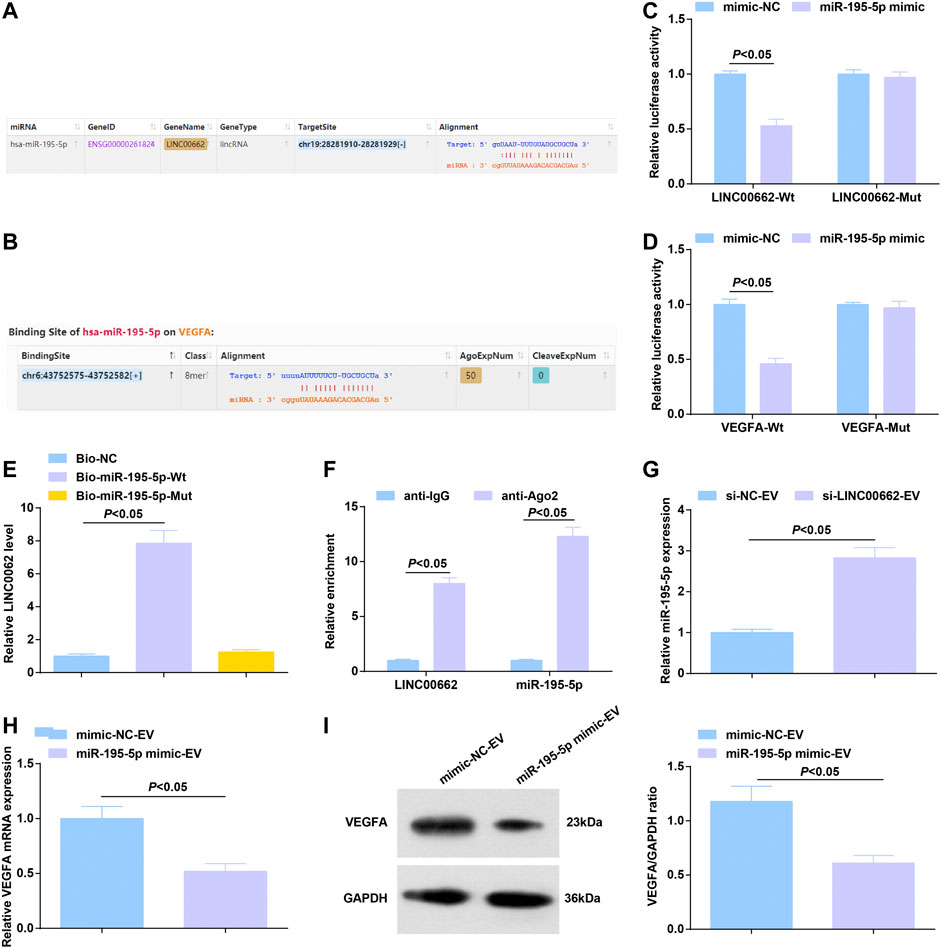
FIGURE 6. Binding relation between LINC00662 and miR-195-5p; targeting relation between miR-195-5p and VEGFA. (A,B) Binding sites of LINC00662 and miR-195-5p, miR-195-5p and VEGFA; (C,D) Dual luciferase report results; (E) RNA pull-down test detected the enrichment of LINC00662; (F) RIP detected the binding of LINC00662 and miR-195-5p to AGO2; (G) RT-qPCR detection of miR-195–5p expression in HUVECs after co-culture with EVs; (H,I) RT-qPCR and Western blot analysis of VEGFA mRNA and protein expression in HUVECs after co-culture with EVs.
Alternations of gene expression were observed: EVs delivery of silenced LINC00662 elevated miR-195-5p expression in HUVECs (Figure 6G) while that of upregulated miR-195-5p lowered VEGFA levels (Figures 6H,I).
Reversal of miR-195-5p-Mediated Effects on HUVECs by Overexpressed VEGFA
miR-195-5p/VEGFA axis-induced function for HUVECs was further evaluated. After co-culture with EVs carrying upregulated miR-195-5p, HUVECs were transfected with oe-VEGFA, which successfully elevated VEGFA expression (Figures 7A,B). Overexpression of VEGFA reversed the effects of EVs carrying upregulated miR-195-5p on proliferation, invasion, migration and angiogenesis of HUVECs (Figures 7C–G).
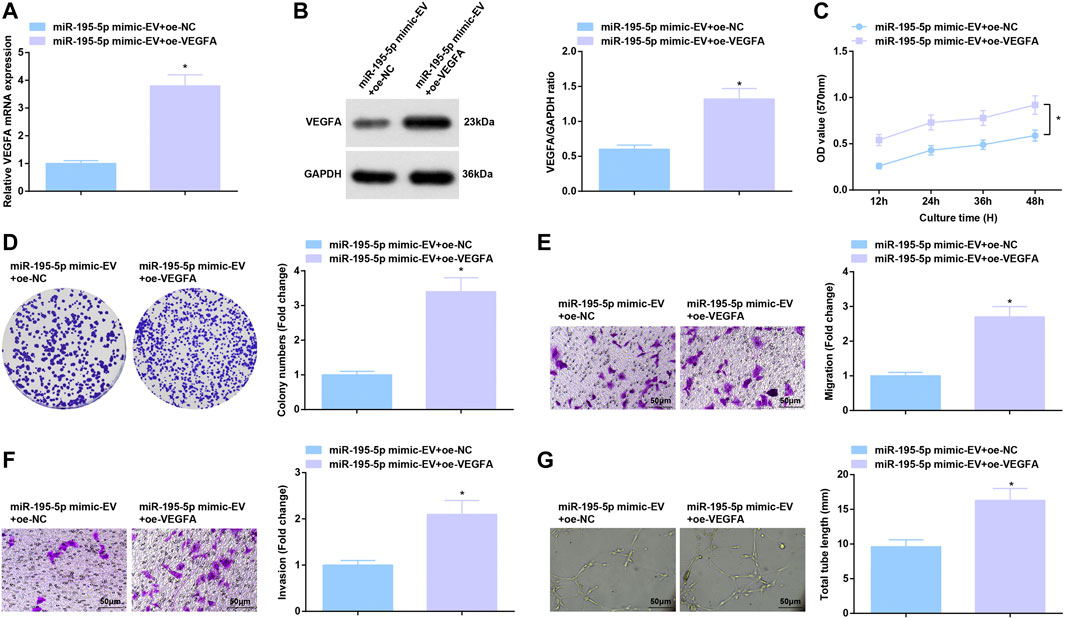
FIGURE 7. Reversal of miR-195-5p-mediated effects on HUVECs by overexpressed VEGFA. (A,B) RT-qPCR and Western blot analysis of VEGFA expression in HUVECs after co-culture with EVs and transfection with overexpressed VEGFA; (C,D) Viability and colony formation ability of HUVECs after co-culture with EVs and transfection with overexpressed VEGFA; (E,F) Migration and invasion of HUVECs after co-culture with EVs and transfection with overexpressed VEGFA; (G) Angiogenesis of HUVECs after co-culture with EVs and transfection with overexpressed VEGFA. * p < 0.05 vs. the miR-195-5p mimic-EV + oe-NC group.
Tumor Suppression Effect of EVs Carrying Downregulated LINC00662 or Upregulated miR-195-5p in Mice
To further determine the angiogenic role of EVs in vivo, an ESCC tumor-bearing nude mice model was established and treated with EVs. In mice xenograft model, after treatment with EVs delivering depleted LINC00662 or restored miR-195-5p, tumors grew slowly, accompanied by smaller tumor volume and lighter tumor weight (Figures 8A–C). Furthermore, the effect of EV-specific inhibitor GW4869 on the growth of nude mice was observed, reflecting that GW4869 treatment had a better efficacy to repress tumor growth (Figures 8D–F). Simply, LINC00662 suppression or miR-195-5p overexpression inhibits the growth of tumor.
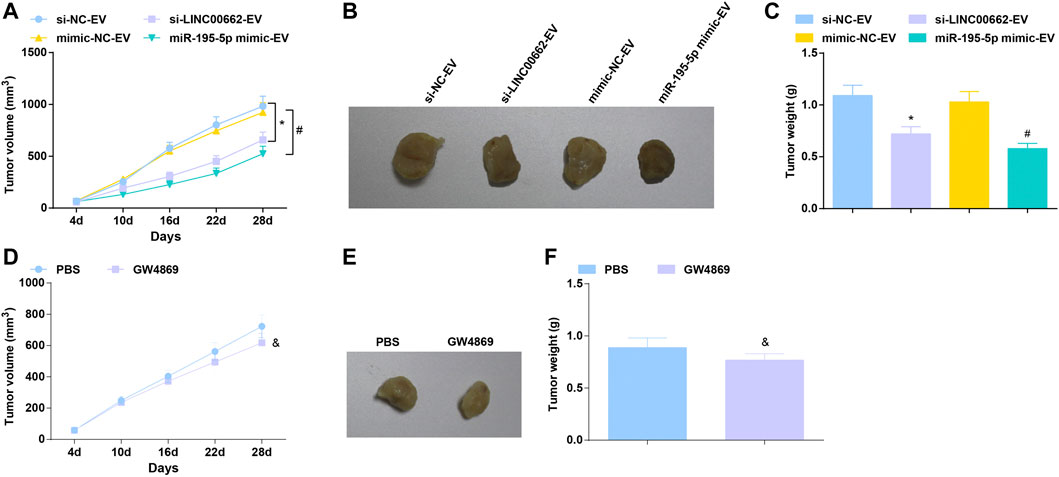
FIGURE 8. Tumor suppression effect of EVs expressing downregulated LINC00662 or upregulated miR-195-5p. (A,D) Tumor volume changes in nude mice; (B,E). Representative tumors in nude mice; (C,F) Tumor weight in nude mice. * p < 0.05 vs. the si-NC-EV; #p < 0.05 vs. the mimic-NC-EV group; & p < 0.05 vs. the PBS group.
Discussion
In the tumor microenvironment in EC, induction of angiogenesis is pro-tumorigenic, and its management is critical to the development of anti-angiogenic therapy (Barzi and Lenz, 2012; Lin et al., 2016). In the present article, it was clearly described that silenced LINC00662 transfer by ESCC cells-derived EVs attenuates angiogenesis by miR-195-5p-dependent VEGFA downregulation (Figure 9).
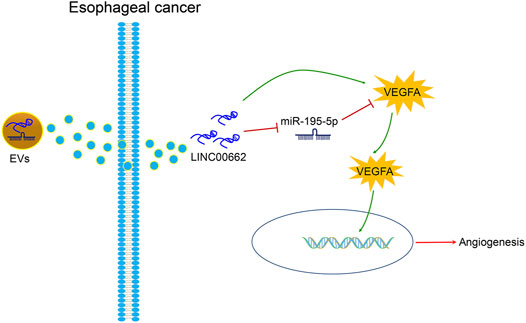
FIGURE 9. ESCC cells-derived EVs transferal of up-regulation of LINC00662 to promote angiogenesis in esophageal cancer by downregulating miR-195-5p and upregulating VEGFA.
To begin with, LINC00662 expression was found to be heightened in cancer tissues of patients with ESCC and was associated with advanced TNM stage. Consistently, LINC00662 high expression has been examined in cancer tissues of patients with colorectal cancer, indicating a positive correlation with tumor T stage (Wang et al., 2019). Elevation of LINC00662 has also been measured in patients with ESCC, which is negatively correlated with patients’ clinical outcome (Zhang et al., 2020). For HUVECs, knocking down LINC00662 made relives of proliferation, invasion, migration and tube formation abilities, confirming that LINC00662 silencing possesses anti-angiogenic effect. Bo Cheng et al. have displayed that LINC00662 overexpression increases proliferation, invasion and migration of colon cancer cells in vitro and tumor growth in vivo (Cheng B et al., 2020). In addition, a report by Long Cheng et al. has implicated the inhibitory role of LINC00662 knockdown as to proliferation, migration and invasion of breast cancer cells (Cheng L et al., 2020). At present, very few analysis has evaluated the complete mechanism of LINC00662-mediated angiogenesis.
Afterwards, ESCC cells-derived EVs were found to augment angiogenesis, but inhibited LINC00662, effectively transported by EVs could attenuate angiogenesis in vitro and tumor formation in vivo. In the light of an article composed by Yu Mao et al., ESCC cells-secreted EVs under hypoxia stimulate angiogenesis by inducing HUVECs to proliferate, invade, migrate and form tubes in vitro (Mao et al., 2019). In addition to EVs from ESCC cells, EVs from other cancer cells present pivotal actions in angiogenesis. As exampled by a late paper, EVs secreted by hypoxic papillary thyroid cancer cells enhance endothelial tube formation (Wu et al., 2019). On the other hand, EVs extracted from gastric cancer cells could be internalized by vascular endothelial cells, so as to activate angiogenesis and tumor growth (Yang et al., 2018). Nils Ludwig et al. have experimentally observed that after internalization of EVs from head and neck squamous cell carcinoma cells by HUVECs, proliferation, migration and tube formation of HUVECs are all promoted (Ludwig et al., 2018). Remarkably, hypoxia-induced EVs from lung cancer cells cause formidable force in accelerating proliferation and tube-forming abilities of HUVECs (Mo et al., 2020).
Subsequently, miR-195-5p, as a downstream actor of LINC00662, attracted our attention. Gene expression measurement found lowly expressed miR-195-5p in ESCC tissues, showing consistency with a former article (Shen S et al., 2020). Next, experimental assays proved that EVs delivery of restored miR-195-5p reduced angiogenesis in vitro and tumor growth in vivo. Indeed, much efforts have been paid to disclose the anti-angiogenic function of miR-195-5p in cancers, including squamous cell lung cancer cells (Liu et al., 2019), ovarian cancer (Dai et al., 2019) and prostate cancer (Cai et al., 2018).
It is of interest that miR-195-5p could directly inhibit VEGFA (Maroof et al., 2019), and the finding was echoed with our research. Importantly, data analysis further proved that VEGFA overexpression reversed miR-195-5p-dependent inhibition of angiogenesis. Based on the observation of Ruoqin W et al., the conditioned medium of CRC cells enhances migration, invasion, and tube formation of HUVECs, which could be reversed by VEGFA silencing (Wang et al., 2020). Meanwhile, Lijun L et al. have elaborated that in the treatment of non-small cell lung cancer with anlotinib, suppressed VEGFA signal facilitates to restrain the activities of HUVECs (Liang et al., 2019). Accordingly, it has been established that VEGFA overexpression can eliminate the anti-angiogenic effect induced by miR-130 b in prostate cancer (Mu et al., 2020).
Overall, our study analysis obtained that ESCC cells-derived EVs transferal of downregulated LINC00662 decreases miR-195-5p-targeted VEGFA and relives angiogenesis induction. This research helps the development of potential molecule-targeted therapy for angiogenesis in tumor progression.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
All patients signed an informed consent form. All animal experiments adhered to the protocol of Animal Protection and Use Committee in The Second Hospital of Hebei Medical University.
Author Contributions
SL finished study design, FL, RN, and HZ| finished experimental studies, SG, ZD, and HZ finished data analysis, FZ, ZD, and SL finished manuscript editing. All authors read and approved the final manuscript.
Funding
This study was supported by The Natural Science Fundation of Hebei Province (Grant No. H2018206249).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.772514/full#supplementary-material
References
Barzi, A., and Lenz, H.-J. (2012). Angiogenesis-related Agents in Esophageal Cancer. Expert Opin. Biol. Ther. 12 (10), 1335–1345. doi:10.1517/14712598.2012.707180
Batra, R., Malhotra, G. K., Singh, S., and Are, C. (2019). Managing Squamous Cell Esophageal Cancer. Surg. Clin. North America 99 (3), 529–541. doi:10.1016/j.suc.2019.02.006
Cai, C., He, H., Duan, X., Wu, W., Mai, Z., Zhang, T., et al. (2018). miR-195 Inhibits Cell Proliferation and Angiogenesis in Human Prostate Cancer by Downregulating PRR11 Expression. Oncol. Rep. 39 (4), 1658–1670. doi:10.3892/or.2018.6240
Cancer Genome Atlas Research, NetworkAnalysis Working Group: Asan UniversityB.C.Cancer AgencyBrigham and Women’s HospitalBroad InstituteBrown Universityet al. (2017). Integrated Genomic Characterization of Oesophageal Carcinoma. Nature 541 (7636), 169–175. doi:10.1038/nature20805
Chen, Y., Bao, C., Zhang, X., Lin, X., and Fu, Y. (2020). Knockdown of LINC00662 Represses AK4 and Attenuates Radioresistance of Oral Squamous Cell Carcinoma. Cancer Cel Int 20, 244. doi:10.1186/s12935-020-01286-9
Cheng, B., Rong, A., Zhou, Q., and Li, W. (2020). LncRNA LINC00662 Promotes colon Cancer Tumor Growth and Metastasis by Competitively Binding with miR-340-5p to Regulate CLDN8/IL22 Co-expression and Activating ERK Signaling Pathway. J. Exp. Clin. Cancer Res. 39 (1), 5. doi:10.1186/s13046-019-1510-7
Cheng, L., Xing, Z., Zhang, P., and Xu, W. (2020). Long Non-coding RNA LINC00662 Promotes Proliferation and Migration of Breast Cancer Cells via Regulating the miR-497-5p/EglN2 axis. Acta Biochim. Pol. 67 (2), 229–237. doi:10.18388/abp.2020_5203
Claesson-Welsh, L., and Welsh, M. (2013). VEGFA and Tumour Angiogenesis. J. Intern. Med. 273 (2), 114–127. doi:10.1111/joim.12019
Dai, J., Wei, R., Zhang, P., and Kong, B. (2019). Overexpression of microRNA-195-5p Reduces Cisplatin Resistance and Angiogenesis in Ovarian Cancer by Inhibiting the PSAT1-dependent GSK3β/β-Catenin Signaling Pathway. J. Transl Med. 17 (1), 190. doi:10.1186/s12967-019-1932-1
He, L., Zhu, W., Chen, Q., Yuan, Y., Wang, Y., Wang, J., et al. (2019). Ovarian Cancer Cell-Secreted Exosomal miR-205 Promotes Metastasis by Inducing Angiogenesis. Theranostics 9 (26), 8206–8220. doi:10.7150/thno.37455
He, Z., Li, W., Zheng, T., Liu, D., and Zhao, S. (2020). Human Umbilical Cord Mesenchymal Stem Cells-Derived Exosomes Deliver microRNA-375 to Downregulate ENAH and Thus Retard Esophageal Squamous Cell Carcinoma Progression. J. Exp. Clin. Cancer Res. 39 (1), 140. doi:10.1186/s13046-020-01631-w
Hirano, H., and Kato, K. (2019). Systemic Treatment of Advanced Esophageal Squamous Cell Carcinoma: Chemotherapy, Molecular-Targeting Therapy and Immunotherapy. Jpn. J. Clin. Oncol. 49 (5), 412–420. doi:10.1093/jjco/hyz034
Knox, M. C., Ni, J., Bece, A., Bucci, J., Chin, Y., Graham, P. H., et al. (2020). A Clinician's Guide to Cancer-Derived Exosomes: Immune Interactions and Therapeutic Implications. Front. Immunol. 11, 1612. doi:10.3389/fimmu.2020.01612
Kong, R., Ma, Y., Feng, J., Li, S., Zhang, W., Jiang, J., et al. (2016). The Crucial Role of miR-126 on Suppressing Progression of Esophageal Cancer by Targeting VEGF-A. Cell Mol Biol Lett 21, 3. doi:10.1186/s11658-016-0004-2
Lam, A. K. (2020). Introduction: Esophageal Squamous Cell Carcinoma-Current Status and Future Advances. Methods Mol. Biol. 2129, 1–6. doi:10.1007/978-1-0716-0377-2_1
Lang, H.-L., Hu, G.-W., Zhang, B., Kuang, W., Chen, Y., Wu, L., et al. (2017). Glioma Cells Enhance Angiogenesis and Inhibit Endothelial Cell Apoptosis through the Release of Exosomes that Contain Long Non-coding RNA CCAT2. Oncol. Rep. 38 (2), 785–798. doi:10.3892/or.2017.5742
Li, C.-Y., Zhang, W.-W., Xiang, J.-L., Wang, X.-H., Li, J., and Wang, J.-L. (2019). Identification of microRNAs as Novel Biomarkers for Esophageal Squamous Cell Carcinomaa: A Study Based on the Cancer Genome Atlas (Tcga) and Bioinformatics. Chin. Med. J. (Engl) 132 (18), 2213–2222. doi:10.1097/cm9.0000000000000427
Li, Z., Qin, X., Bian, W., Li, Y., Shan, B., Yao, Z., et al. (2019). Exosomal lncRNA ZFAS1 Regulates Esophageal Squamous Cell Carcinoma Cell Proliferation, Invasion, Migration and Apoptosis via microRNA-124/STAT3 axis. J. Exp. Clin. Cancer Res. 38 (1), 477. doi:10.1186/s13046-019-1473-8
Liang, L., Hui, K., Hu, C., Wen, Y., Yang, S., Zhu, P., et al. (2019). Autophagy Inhibition Potentiates the Anti-angiogenic Property of Multikinase Inhibitor Anlotinib through JAK2/STAT3/VEGFA Signaling in Non-small Cell Lung Cancer Cells. J. Exp. Clin. Cancer Res. 38 (1), 71. doi:10.1186/s13046-019-1093-3
Lin, E. W., Karakasheva, T. A., Hicks, P. D., Bass, A. J., and Rustgi, A. K. (2016). The Tumor Microenvironment in Esophageal Cancer. Oncogene 35 (41), 5337–5349. doi:10.1038/onc.2016.34
Lin, M., Zhou, C., He, S., Yu, H., Guo, T., Ye, J., et al. (2019). The Research Advances of Exosomes in Esophageal Cancer. Biomarkers Med. 13 (8), 685–695. doi:10.2217/bmm-2018-0314
Liu, H., Chen, Y., Li, Y., Li, C., Qin, T., Bai, M., et al. (2019). miR-195 S-uppresses M-etastasis and A-ngiogenesis of S-quamous C-ell L-ung C-ancer by I-nhibiting the E-xpression of VEGF. Mol. Med. Rep. 20 (3), 2625–2632. doi:10.3892/mmr.2019.10496
Liu, S., and Meng, X. (2020). LINC00662 Long Non-coding RNA Knockdown Attenuates the Proliferation, Migration, and Invasion of Osteosarcoma Cells by Regulating the microRNA-15a-5p/Notch2 Axis. Ott 13, 7517–7530. doi:10.2147/ott.s256464
Ludwig, N., Yerneni, S. S., Razzo, B. M., and Whiteside, T. L. (2018). Exosomes from HNSCC Promote Angiogenesis through Reprogramming of Endothelial Cells. Mol. Cancer Res. 16 (11), 1798–1808. doi:10.1158/1541-7786.mcr-18-0358
Mao, Y., Wang, Y., Dong, L., Zhang, Y., Zhang, Y., Wang, C., et al. (2019). Hypoxic Exosomes Facilitate Angiogenesis and Metastasis in Esophageal Squamous Cell Carcinoma through Altering the Phenotype and Transcriptome of Endothelial Cells. J. Exp. Clin. Cancer Res. 38 (1), 389. doi:10.1186/s13046-019-1384-8
Maroof, H., Irani, S., Arianna, A., Vider, J., Gopalan, V., and Lam, A. K.-y. (2019). Interactions of Vascular Endothelial Growth Factor and P53 with miR-195 in Thyroid Carcinoma: Possible Therapeutic Targets in Aggressive Thyroid Cancers. Ccdt 19 (7), 561–570. doi:10.2174/1568009618666180628154727
Mo, F., Xu, Y., Zhang, J., Zhu, L., Wang, C., Chu, X., et al. (2020). Effects of Hypoxia and Radiation-Induced Exosomes on Migration of Lung Cancer Cells and Angiogenesis of Umbilical Vein Endothelial Cells. Radiat. Res. 194 (1), 71–80. doi:10.1667/rr15555.1
Mu, H. Q., He, Y. H., Wang, S. B., Yang, S., Wang, Y. J., Nan, C. J., et al. (2020). MiR-130b/TNF-α/NF-κB/VEGFA Loop Inhibits Prostate Cancer Angiogenesis. Clin. Transl Oncol. 22 (1), 111–121. doi:10.1007/s12094-019-02217-5
Reichenbach, Z. W., Murray, M. G., Saxena, R., Farkas, D., Karassik, E. G., Klochkova, A., et al. (2019). Clinical and Translational Advances in Esophageal Squamous Cell Carcinoma. Adv. Cancer Res. 144, 95–135. doi:10.1016/bs.acr.2019.05.004
Shen S, S., Li, K., Liu, Y., Liu, X., Liu, B., Ba, Y., et al. (2020). Silencing lncRNA AGAP2-AS1 Upregulates miR-195-5p to Repress Migration and Invasion of EC Cells via the Decrease of FOSL1 Expression. Mol. Ther. - Nucleic Acids 20, 331–344. doi:10.1016/j.omtn.2019.12.036
Shen W, W., Yu, L., Cong, A., Yang, S., Wang, P., Han, G., et al. (2020). Silencing lncRNA AFAP1-AS1 Inhibits the Progression of Esophageal Squamous Cell Carcinoma Cells via Regulating the miR-498/VEGFA Axis. Cmar 12, 6397–6409. doi:10.2147/cmar.s254302
Sui, S., Sun, L., Zhang, W., Li, J., Han, J., Zheng, J., et al. (2021). LncRNA MEG8 Attenuates Cerebral Ischemia after Ischemic Stroke through Targeting miR-130a-5p/VEGFA Signaling. Cell Mol Neurobiol 41 (6), 1311–1324. doi:10.1007/s10571-020-00904-4
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): a Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell Vesicles 7 (1), 1535750. doi:10.1080/20013078.2018.1535750
Viallard, C., and Larrivée, B. (2017). Tumor Angiogenesis and Vascular Normalization: Alternative Therapeutic Targets. Angiogenesis 20 (4), 409–426. doi:10.1007/s10456-017-9562-9
Wang, H., Yu, M., Hu, W., Chen, X., Luo, Y., Lin, X., et al. (2019). Linc00662 Promotes Tumorigenesis and Progression by Regulating miR-497-5p/AVL9 Axis in Colorectal Cancer. Front. Genet. 10, 1385. doi:10.3389/fgene.2019.01385
Wang, R., Ma, Y., Zhan, S., Zhang, G., Cao, L., Zhang, X., et al. (2020). B7-H3 Promotes Colorectal Cancer Angiogenesis through Activating the NF-Κb Pathway to Induce VEGFA Expression. Cell Death Dis 11 (1), 55. doi:10.1038/s41419-020-2252-3
Wu, F., Li, F., Lin, X., Xu, F., Cui, R.-R., Zhong, J.-Y., et al. (2019). Exosomes Increased Angiogenesis in Papillary Thyroid Cancer Microenvironment. Endocr. Relat. Cancer 26 (5), 525–538. doi:10.1530/erc-19-0008
Yang, H., Zhang, H., Ge, S., Ning, T., Bai, M., Li, J., et al. (2018). Exosome-Derived miR-130a Activates Angiogenesis in Gastric Cancer by Targeting C-MYB in Vascular Endothelial Cells. Mol. Ther. 26 (10), 2466–2475. doi:10.1016/j.ymthe.2018.07.023
You, L.-N., Tai, Q.-W., Xu, L., Hao, Y., Guo, W.-J., Zhang, Q., et al. (2021). Exosomal LINC00161 Promotes Angiogenesis and Metastasis via Regulating miR-590-3p/ROCK axis in Hepatocellular Carcinoma. Cancer Gene Ther. 28 (6), 719–736. doi:10.1038/s41417-020-00269-2
Zeng, Z., Li, Y., Pan, Y., Lan, X., Song, F., Sun, J., et al. (2018). Cancer-derived Exosomal miR-25-3p Promotes Pre-metastatic Niche Formation by Inducing Vascular Permeability and Angiogenesis. Nat. Commun. 9 (1), 5395. doi:10.1038/s41467-018-07810-w
Zhang, L., Ouyang, P., He, G., Wang, X., Song, D., Yang, Y., et al. (2021). Exosomes from microRNA‐126 Overexpressing Mesenchymal Stem Cells Promote Angiogenesis by Targeting the PIK3R2‐mediated PI3K/Akt Signalling Pathway. J. Cel Mol Med 25 (4), 2148–2162. doi:10.1111/jcmm.16192
Zhang, Z., Liang, X., Ren, L., Zhang, S., Li, S., Wan, T., et al. (2020). LINC00662 Promotes Cell Viability and Metastasis in Esophageal Squamous Cell Carcinoma by Sponging miR ‐340‐5p and Upregulating HOXB2. Thorac. Cancer 11 (8), 2306–2315. doi:10.1111/1759-7714.13551
Keywords: esophageal squamous cell carcinoma, extracellular vehicles, LINC00662, microRNA-195-5p, vascular endothelial growth factor a
Citation: Li F, Niu R, Gao S, Zhao F, Dong Z, Zhang H and Li S (2022) Pro-Angiogenesis Role of LINC00662 From Esophageal Squamous Cell Carcinoma Cells-Derived Extracellular Vehicles. Front. Bioeng. Biotechnol. 10:772514. doi: 10.3389/fbioe.2022.772514
Received: 08 September 2021; Accepted: 14 February 2022;
Published: 01 April 2022.
Edited by:
Eder Lilia Romero, National University of Quilmes, ArgentinaReviewed by:
Honglin Jin, Huazhong University of Science and Technology, ChinaMarie Morille, Institut de chimie moléculaire et des matériaux Charles Gerhardt Montpellier (ICGM), France
Copyright © 2022 Li, Niu, Gao, Zhao, Dong, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shujun Li, TGlzaHVqdW4xOTk5QG91dGxvb2suY29t
†These authors have contributed equally to this work and share first authorship
 Feng Li1†
Feng Li1† Shujun Li
Shujun Li