- 1Jiangsu Co Innovation Center of Efficient Processing and Utilization of Forest Resources, College of Chemical Engineering, Nanjing Forestry University, Nanjing, China
- 2Jiangsu Provincial Key Lab for Chemistry and Utilization of Agro Forest Biomass, Jiangsu Key Lab of Biomass Based Green Fuels and Chemicals, Nanjing, China
- 3International Innovation Center for Forest Chemicals and Materials, Nanjing Forestry University, Nanjing, China
Microbial cell surface display technology provides a powerful platform for engineering proteins/peptides with enhanced properties. Compared to the classical intracellular and extracellular expression (secretion) systems, this technology avoids enzyme purification, substrate transport processes, and is an effective solution to enzyme instability. Saccharomyces cerevisiae is well suited to cell surface display as a common cell factory for the production of various fuels and chemicals, with the advantages of large cell size, being a Generally Regarded As Safe (GRAS) organism, and post-translational processing of secreted proteins. In this review, we describe various strategies for constructing modified S. cerevisiae using cell surface display technology and outline various applications of this technology in industrial processes, such as biofuels and chemical products, environmental pollution treatment, and immunization processes. The approaches for enhancing the efficiency of cell surface display are also discussed.
1 Background
Cell surface display technology is an attractive method for immobilizing functional proteins/peptides and imparting specific functions to microbial cells. In the cell surface display system, functional proteins/peptides are fused with anchoring protein genes and expressed on the cell surface or plasma membrane under the guidance of signal peptides. The cell-surface display technology has unique advantages over intracellular expression and secretion: 1) Proteins displayed on the cell surface can maintain their functions and properties more stably under harsh temperature or pH conditions than free proteins. 2) The proteins immobilized on the cell surface can be recycled by filtration or centrifugation, avoiding the complicated process of lysing cells or purifying proteins from the cultural liquid. 3) The multi-enzyme cell surface co-display system can shorten the substrate transfer distance, and the co-display of enzymes with synergistic effects can trigger synergistic proximity effects and improve catalytic efficiency. 4) Engineered yeast with functional proteins displayed on the surface can act on large molecular masses of substrates (e.g., cellulose, hemicellulose) that cannot enter the cell, and the resulting monomers (e.g., glucose) can be directly utilized by the cell to produce valuable products (e.g., bioethanol). This process contributes to the integrated bioprocessing process that combines hydrolysis, saccharification and fermentation. Cell surface display systems have many potential applications, including biocatalysts, high-throughput library screening, biological sorbents, oral vaccines, etc. (Teymennet-Ramírez et al., 2022).
Cell surface display systems have been successfully applied to various microbial cells. Among them, yeast cells are one of the most suitable host cells for cell surface display, with relatively large cell size, rigid cell walls, and allowing post-translational processing (including folding and glycosylation, etc.) of expressed heterologous eukaryotic proteins (Liu et al., 2016a). In yeast, cell surface display was first developed in S. cerevisiae with remarkable results. Subsequently, surface display systems using full-length or anchored structural domains of S. cerevisiae cell wall proteins were applied to other yeasts, including Pichia pastoris (Zhao N. et al., 2017; Yang et al., 2017; Ce et al., 2022) and Yarrowia lipolytica (An et al., 2016). S. cerevisiae is easy to manipulate genetically and has Generally Regarded As Safe (GRAS) status due to its widespread use in the food industry (Cha et al., 2022). In recent years, various applications of yeast surface display systems have been reported by many researchers, and there are also related review articles outlining strategies for improving yeast surface display systems (Teymennet-Ramírez et al., 2022). However, few have provided detailed overviews for one yeast alone. This paper focuses on the progress of research using S. cerevisiae as a cell surface display object and not only introduces strategies to improve cell surface display but also discusses in more detail the wide range of applications of surface-engineered S. cerevisiae and provides insights into the challenges of future commercialization.
2 Cell surface display systems in S. cerevisiae
Cell surface display is a powerful tool for endowing novel functions on S. cerevisiae cells by displaying functional proteins/peptides on the cell surface. The cell surface display system has been enriched by the research on cell wall structure of S. cerevisiae. Its inner layer consists mainly of β-linked glucans, which are cross-linked with chitin to maintain cell wall strength, and the outer layer consists mainly of mannoproteins, which are covalently linked to the inner layer, and heterologous proteins can be anchored to the mannoprotein layer to determine cell surface properties (Ye et al., 2021). In the cell surface display platform, different enzymes can be located on the same cell surface and the proximity could improve their synergism. This platform simplifies protein purification in some biocatalytic processes and promotes the construction and reuse of whole-cell catalysts.
2.1 Direct cell surface display
In cell surface display, the target protein fused with specific vector sequence is introduced into the yeast cell and then the fusion protein is expressed. The signal peptide guides the fusion protein to extracellular secretion, the anchor protein contained in the fusion protein can bind to the cell wall structure of yeast to immobilize the target protein on the surface of the yeast cell (Kondo & Ueda, 2004; Pepper et al., 2008). According to the different fusion modes of anchor proteins, cell surface display systems can be divided into glycosylphosphatidylinositol (Xing et al.) anchor system, FS/FL (truncated forms of Flo1p (Matsumoto et al., 2002), amino acids (FS) 1–1,099 and amino acids 1–1,417) anchor system and Pir protein anchor system (Figure 1). GPI anchor proteins such as Cwp1p, Cwp2p, Tip1p, Flo1p, Sed1p, YCR89w, and Tir1 link to β-1, 6-glucan of the cell wall by the C-terminal GPI-anchor structures, and the target gene is usually fused to the N- terminus (Van der Vaart et al., 1997). Particularly, the a-agglutinin, composed of Aga1 and Aga2, covalently binds to the cell wall by the C-terminal GPI-anchor structure of Aga1, the target protein gene fuses with the C-terminal or N-terminal of Aga2 and Aga2 is connected with Aga1 through a disulfide bond. The FL/FS gene encoding the Flo1p flocculation functional domain is located near the N- terminus of Flo1p and can adhere to mannans in the cell wall by non-covalent interaction, with the target protein generally fused at its C- terminus. Some proteins with active sites near the C-terminus have inhibitory activity when fused with GPI-anchored proteins, and this inhibition can be mitigated using the FL/FS display system (Matsumoto et al., 2002). In addition, the Pir protein display system is suitable for C-terminal fusion, N-terminal fusion and insertion fusion. The protein differs from the first two anchoring modes in that it can form an ester linkage with β-1, 3-glucose in the cell wall via N-terminal repeat sequences, or it can attach to specific components of the cell wall via disulfide bonds using C-terminal cysteine residues (Yang et al., 2014).
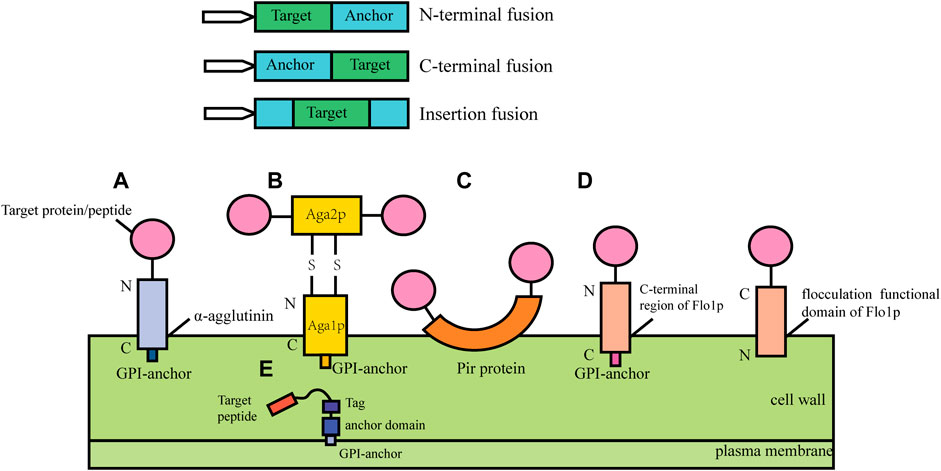
FIGURE 1. Cell surface display systems in S. cerevisia. (A) α-agglutinin-based display system; (B) a-agglutinin-based display system; (C) Pir-based display system; (D) Flo1p-based display system; (E) Membrane display system.
2.2 Scaffold-mediated cell surface display
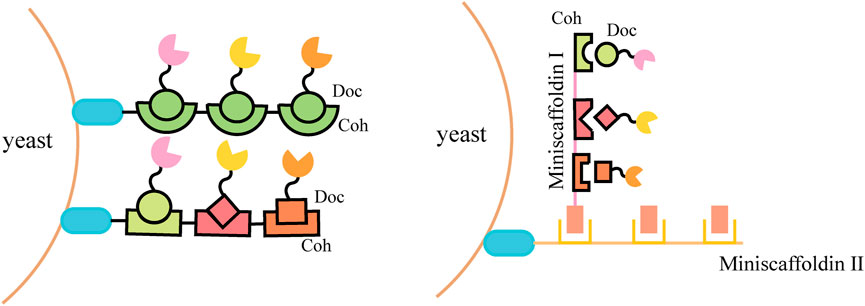
Natural cellulosomes are often found in anaerobic bacteria (Felix and Ljungdahl, 1993) (such as Clostridium thermocellum, C. cellulolyticum, C. cellulovorans, Ruminococcus flavefaciens, etc.) and the complex composed of scaffoldins, anchoring domains, cohesins, fiber binding domains, and catalytic units, which provides a novel scaffold-based strategy for cell surface display. This cell-surface display system usually triggers enzyme-enzyme proximity synergistic effects and enzyme-substrate-cell complex synergistic effects. A display method was described to allow the production of all cellulosomal components (miniscaffoldin CipA3 and enzymes) in vivo, avoiding the labor-intensive protein purification step (Wen et al., 2010). The complex composition of natural cellulosomes and the considerable molecular weight of scaffolding proteins create a metabolic burden for cell surface display. When multi-enzyme presentation is required, its specificity is not high and the efficiency of self-assembly is reduced. Therefore, the researchers selected cohesin domains and cellulose binding domain (CBD) from different strains to form artificial scaffolds, and selected the anchor proteins from the yeast cell wall as anchor elements to display the scaffolds on the yeast cell surface (Fierobe et al., 2005; Mingardon et al., 2007; Wieczorek and Martin, 2010). Target proteins fused with corresponding dockerins can bind to the scaffold through the interaction between the cohesin and dockerin (Figure 2). Cellulosomal components can be produced within the same cell and assembled on the cell surface, or they can be produced by cells of different strains and then co-incubated to assemble on the cell surface of the displayed scaffold. For example, cells displaying scaffoldins on the surface can be incubated directly with Escherichia coli cell lysates containing cellulases to form the cellulosome complex for ethanol production (Tsai S.-L. et al., 2009).
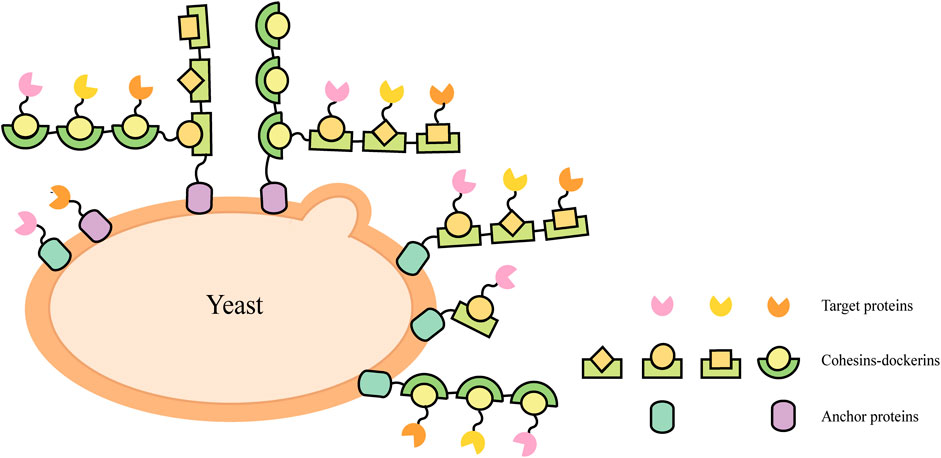
FIGURE 2. Schematic illustrations of direct and indirect cell-surface display strategies in S. cerevisiae.
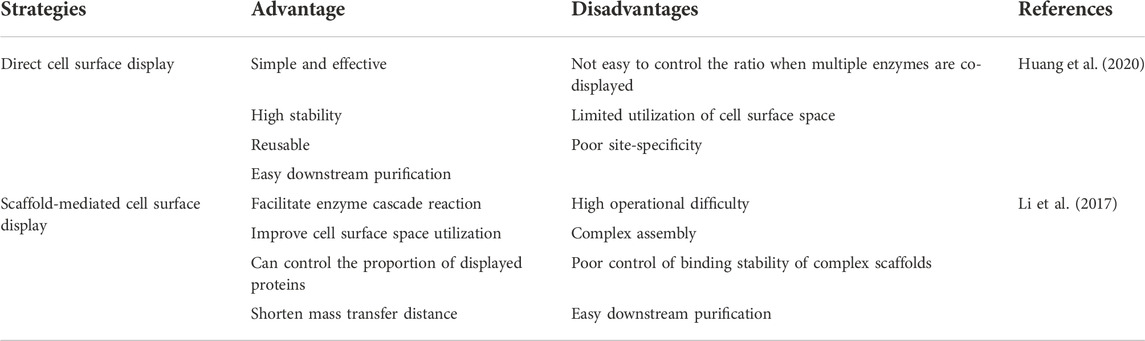
Direct cell surface display is a simple and effective strategy. The enzymes immobilized on the cell surface are stable, and the recycling of the whole cells shortens the purification process of the protein, but it cannot effectively use the cell surface space. When multiple enzymes with synergistic effects are co-displayed, the ratio of various enzymes cannot be well controlled. The scaffold-mediated surface display system can make up for the deficiency of the direct display system, shorten the transfer distance of the substrate and enhance the proximity effect between enzymes. However, the design is more complicated and the assembly stability on the cell surface cannot be guaranteed (Table 1). Therefore, a more appropriate design is needed to construct cell surface display engineering yeasts to make it more practical.
2.3 Improvement of cell-surface display efficiency
The improvement of cell surface display efficiency depends on the effective expression and secretion of target proteins and the activity of the displayed target proteins. To this end, many attempts have been made, such as selecting appropriate anchor proteins and expression vectors, constructing highly expressed gene cassettes, strengthening protein folding and post-translational modification, so as to improve the production and activity of proteins. However, further efforts are needed to achieve universal commercialization.
2.3.1 The important role and localization effect of anchoring domains
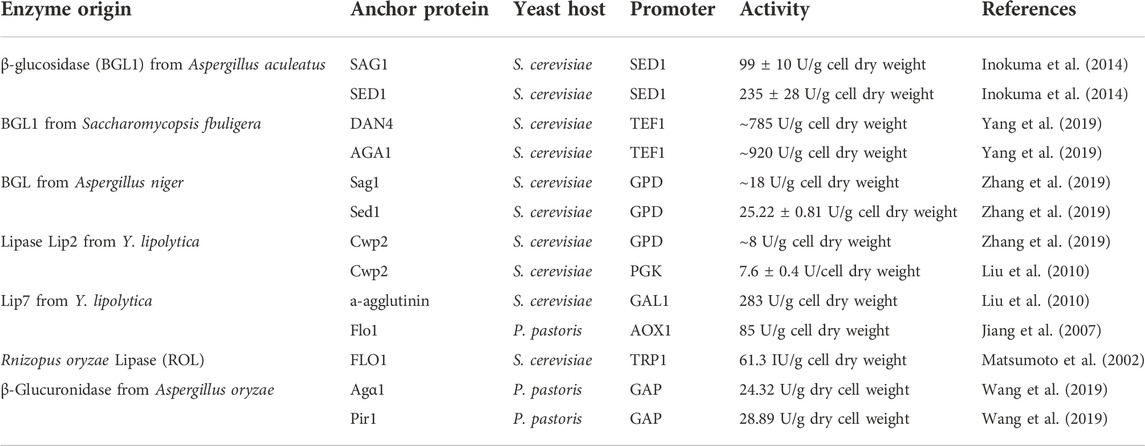
The display efficiency of enzymes on the cell surface is related to the anchoring proteins attached (Table 2). Attempts have been made to improve cell surface display efficiency by screening for contrasting new anchoring proteins. Phienluphon et al. (2019) selected 37 GPI-type anchor proteins from different sources by computational prediction, fused with yEGFP, pre-screened by comparing the fluorescence intensity, and further fused several vigorous fluorescence anchor proteins with β-glucosidase to determine the enzymatic activity. Finally, 6_Kl from Kluyveromyces lactis was found to have higher transcript levels and superior target protein display efficiency. By comparing the display efficiency and enzyme activity of several display systems, Yang et al. (2019) adapted the a-agglutinin anchoring system to develop a new surface display system by using α-galactosidase as the target enzyme and fusing it directly to Aga1p. The enzyme was also fused to conventional a-agglutinin and to six other anchoring proteins selected. A comparison of enzyme activity revealed that Aga1p, Dan4p, and Sed1p had high display efficiency and were promising anchoring systems for immobilizing recombinant proteins on the yeast surface. The display efficiency of the anchor protein domain varies with the molecular weight of the displayed protein. The localization effect of structural anchoring domains may also affect the display efficiency of target proteins on the cell surface. Inokuma et al. (2020) fused the anchored domains (Sed1p and Sag1p) with heterologous proteins and expressed them in S. cerevisiae. It was observed by fluorescence and immunoelectron microscopy that the two domains had different anchorings on the cell surface, with sed1p-anchoring domain mainly localized on the outer surface of the cell wall, while sag1p-anchoring domain was mainly localized inside the cell wall. The cell wall space can be effectively utilized by properly controlling the anchoring position, making the cell surface display technology more useful in various fields.
2.3.2 Improvement of protein secretion
Protein secretion is an essential factor in improving the efficiency of cell surface display. The main strategies to improve protein secretion include increasing protein expression levels and signal peptide engineering. The strength of the promoters often determines the expression level of heterologous proteins in S. cerevisiae. Different types of promoters differ in their strength. In addition, the strength of promoters can be influenced by other factors such as growth conditions (Sun et al., 2012a) (e.g., glucose concentration), introns (Hoshida et al., 2017) and other factors. Synthetic promoters were constructed using promoter engineering to further improve protein expression levels. The upstream activating sequences (UASs) of different promoters were combined with core promoters to construct a synthetic promoter library. Using this approach, galactose-inducible promoters stronger than PGAL1 were successfully constructed by fusing UASGAL1 to the core promoters of TDH3 and TEF1 (Deng et al., 2021). The selection of efficient signal peptides helps to enhance protein secretion and increase the number of proteins displayed on the cell surface. A signal peptide from S. cerevisiae SED1 was shown to be an efficiently secreted A. aculeatus β-glucosidase (BGL1). The extracellular activity of BGL1 was 1.3-fold and 1.9-fold higher than that of the Rhizopus oryzae glucoamylase (GLUASP) and S. cerevisiae α-mating pheromone (MFα1SP), respectively (Inokuma et al., 2016). The design mutations of the a-factor preproleader of the MFα1SP have the opportunity to obtain enhanced signal peptides for the secretion of a variety of fungal enzymes (Aza et al., 2021). In addition, some researchers proposed that the fusion of the target protein with a secretion-enhancing peptide cassette and rational optimization of the original peptide could enhance the secretion of the heterologous proteins (Cho et al., 2022). The expression of the enzyme can be further improved by optimizing the combination of promoter and signal peptide. Different promoters and signal peptides constituting nine gene cassettes were selected for fusion expression with Kluyveromyces marxianus inulinase gene. The inulinase activity of S. cerevisiae carrying the PGK1 promoter and MFα1 signal sequence was the highest (Hong et al., 2015). Terminators affect gene expression by terminating transcription and controlling the half-life of mRNA. Synthetic terminators are short in sequence, easy to synthesize, and more functional. A panel of short (35–70 bp) synthetic terminators was reported to increase fluorescent protein output by 3.7-fold and transcript levels by 4.4-fold compared to the commonly used CYC1 terminator (Curran et al., 2015). The synthetic terminators might function better in S. cerevisiae compared to a native promoter.
Other strategies to improve protein secretion include: 1) Overexpression of related genes involved in protein transport or cell surface display (Wentz & Shusta, 2007; Hou et al., 2012; Van Zyl et al., 2016; Bao et al., 2017; Li et al., 2022) and combinatorial regulation of gene expression (Yang et al., 2022). 2) Point mutations and gene deletions (Matsuoka et al., 2014). Point mutation target genes identified from UV-mutagenized strains were analyzed to be involved in multiple intracellular biological processes. Detecting the effects of point mutations and gene deletions on the secretion of α-amylase could help to balance these biological processes and thus improve protein secretion (Wang et al., 2022). 3) Optimization of secretion pathways (Tang et al., 2017; Huang et al., 2018; Besada-Lombana & Da Silva, 2019) and stress modulation (Lamour et al., 2019). 4) Co-overexpression of chaperones (Bae et al., 2016; Bae et al., 2022). 5) The choice of integration method. Auxotrophic integration, cocktail δ-integration (Yamada et al., 2010), CRISPR-δ-integration (Sasaki et al., 2019), and other integration methods (Zhang et al., 2021; Zheng et al., 2022) have been developed successively to increase the copy number of integrated genes, thereby enhancing protein secretory overexpression.
2.3.3 Reduction of steric hindrance and optimization of space utilization on the cell surface of S. cerevisiae
The activity of the target protein can be limited by the competition with other GPI proteins for the limited binding sites on the cell wall when displayed on the cell surface (Van der Vaart et al., 1997). For example, the β-Glucosidase activity of the SED1 disruptant was 22% higher than that of the Sed1-undisrupted strain (Bamba et al., 2018), the disruption of CWP2 and YGP1 could increase BGL activity by 63% and 24%, respectively, compared with the original strain (Arnthong et al., 2022), and CCW12 and CCW14 co-knockout resulted in increased cell wall thickness, which may increase the number of heterologous proteins displayed on the cell surface (Inokuma et al., 2021).
The addition of a suitable length of linker peptide between the anchor and target proteins seems to help to separate the active part of the protein from the cell wall, creating space for substrate access and improving the activity of the display enzyme (Washida et al., 2001; Ullah et al., 2017; Yang et al., 2019).
To further improve the space utilization and display efficiency of the cell surface, the introduction of scaffoldin is a potential strategy. Scaffoldin could be enhanced by increasing cohesion number and utilizing double-layered scaffoldins (Figure 3). The ex vivo assembly of a functional tetravalent designer cellulosome on the yeast cell surface was developed and exhibited a 4.2-fold enhancement in the hydrolysis of phosphoric acid swollen cellulose (PASC) compared with free enzymes (Tsai et al., 2013). Combining a two-scaffoldin-based cellulosome with an intracellular cellodextrin pathway in S. cerevisiae enabled the co-utilization of cellulose-mixed sugars (Fan et al., 2016; Li et al., 2017). A synthetic scaffoldin ScafAGA3 was constructed using the repeated N-terminus of Aga1p. In addition, the scaffoldin ScafCipA3 from C. thermocellum fused with Aga2p was attached to the scaffoldin ScafAGA3 via disulfide bonds, and the secreted cellulases assembled to ScafCipA3, allowing the formation of a complex cellulosome with two scaffoldins. The newly designed cellulosomes enabled yeast to ferment cellulose directly to ethanol (1.52 g/L) by regulating the ratio of scaffoldins and cellulases (Tang et al., 2018).
3 Applications of cell-surface display engineered S. cerevisiae
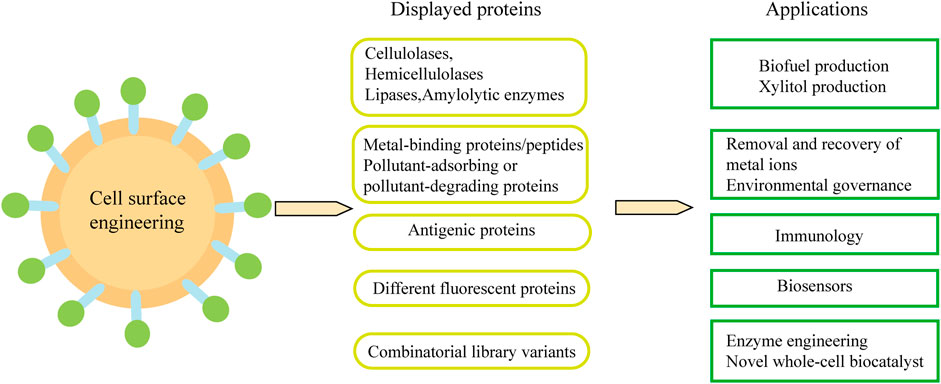
Cell-surface display technology can be used to produce biofuels and various chemicals from cheap sources rich in sugars, triglycerides and waste protein. In addition, it is also widely used in biological adsorbents and oral vaccines. Combined with metabolic engineering and synthetic biology methods, engineered yeast cells with desired functions can be constructed (Figure 4).
3.1 Applications in bioconversion of agricultural and forestry waste
Agricultural and forestry wastes are produced in large quantities worldwide, resulting in a certain amount of resource waste and environmental problems. From the perspective of the circular economy, the central task is to refine agricultural and forestry wastes to produce valuable energy and chemicals (Song et al., 2020). A variety of enzymes including cellulase, hemicellulases, lipase, amylase, cyclodextrin glucan invertase, amylase and other enzymes have been successfully expressed on the surface of yeast cells with biological activity. The successfully constructed engineered strains can not only effectively degrade agroforestry waste rich in lignocellulose and starch, but also lay the foundation for the simultaneous saccharification and fermentation (SSF)/consolidation bioprocessing (CBP) process.
3.1.1 Bioconversion of cellulose catalyzed by engineered S. cerevisiae
Cellulose, composed of glucose connected by β-1, 4-glycosidic bonds, is the most abundant macromolecular polysaccharide in nature. At least three cellulases are required to hydrolyze cellulose into glucose: endoglucanase (EG), cellobiohydrolase (CBH) and β-glucosidase (BGL). EG randomly hydrolyzes the β-1,4-glycosidic bonds by acting on amorphous regions of cellulose, and shortens the long-chain cellulose molecules to produce a larger number of shorter cellulose chains with reducing and non-reducing ends; CBH cuts off cellobiose units from the reducing/non-reducing ends of cellulose, and acts mainly on the crystalline region of cellulose (Hu et al., 2015); BGL hydrolyzes cellobiose into glucose. These three enzymes have been successfully displayed on the cell surface of S. cerevisiae to hydrolyze cellulose to glucose (Table 3). Placing EGII from Trichoderma reesei and CBHI from Talaromyces emersonii in the same space (on the cell surface or in the medium) facilitates the fermentation of amorphous cellulose-based ethanol (Liu et al., 2015). Engineering S. cerevisia demonstrated that these two enzymes, along with two others (CBH2 from Chrysosporium lucknowense and BGL from Aspergillus aculeatus) acted on amorphous cellulose and crystalline cellulose, exhibiting higher ethanol yields because CBH2 reduced the bumpy surface of the cellulose and facilitated the movement of CBH1 by enhanced synergistic hydrolysis (Liu et al., 2016b). Many attempts have been made to improve the fermentation process and introduce new lignocellulose-degrading enzymes (e.g., hydrolytic polysaccharide monooxygenases (LPMOs) and cellobiose dehydrogenases (CDHs)) to further enhance the degradation and product yield of cellulose (Quinlan et al., 2011; Horn et al., 2012; Matano et al., 2012; Nakatani et al., 2013; Bae et al., 2015; Cunha et al., 2021).
The development of multiple scaffoldins (e.g., CipA, ZZ-Coh-Coh, and ScafAGA3) can help to control the ratio of cell surface display proteins and improve ethanol production more effectively (Dong et al., 2020; Qi et al., 2021). A pentafunctional minicellulosome composed of LPMOs, CDHs, CBH, EG and BGL was generated and grown with phosphoric acid swollen cellulose as the sole carbon source (Liang et al., 2014). The delicate balance between the oxidative activity and classical hydrolyase is of importance for the degradation of cellulosic materials (Cannella and Jørgensen, 2014). Expression of all cellulosome components in a single strain may cause heavy metabolic burden and blockage of potential secretion mechanism, thus reducing enzyme activity (Wen et al., 2010). Therefore, displaying anchoring scaffoldins and secreting catalytic and non-catalytic units should be considered to be divided into two independent steps, which can decrease the metabolic burden of the yeast host (Tsai S. L. et al., 2009; Goyal et al., 2011). Researchers achieved an enhancement in the yield by increasing the complexity of the scaffold, that is, by constructing the cellulosome construction and its attachment by two individual miniscaffoldins. The engineered S. cerevisae EBY100 could directly convert avicel to bioethanol (1,412 mg/L) (Fan et al., 2012). Others constructed novel cellulolytic yeast consortiums for cellulosic ethanol production (yield up to 1.87 g/L) (Tsai et al., 2010; Kim et al., 2013). The major drawbacks of hydrolyzing cellulose into glucose before cellular uptake are: 1) Inefficient cellulose hydrolysis caused by glucose inhibition on extracellular cellulases. 2) Glucose-induced carbon catabolite repression affects the co-utilization of other sugars. Reconstitution of the cellodextrin transport system in S. cerevisiae has been reported (Galazka Jonathan et al., 2010; Choi et al., 2022; Ylinen et al., 2022). This system promotes the efficient utilization of cellulose and mitigates the inhibitory effect of glucose (Fan et al., 2016; Li et al., 2017).
3.1.2 Bioconversion of hemicellulose catalyzed by engineered S. cerevisiae
Xylan, the main component of hemicellulose is the second-most polysaccharide in lignocellulosic materials. It is more easily degraded into monomers than cellulose through pretreatment processes, making it an attractive source of sugar for bioethanol fermentation. With the gradual maturity of cell surface display engineering and the heterologous expression of xylose metabolizing enzymes, a one-step process for xylan assimilation can be realized by engineered S. cerevisiae. Xylose-utilizing S. cerevisiae can be constructed by introducing the genes encoding enzymes related to the xylose assimilation pathway (Katahira et al., 2006). A series of multifunctional minihemicellulosomes were constructed for the hydrolysis of arabinoxylans. The D-xylose utilization pathway consisting of xylose reductase (XR), xylitol dehydrogenase (XDH), and D-xylose kinase (XK) from Scheffersomyces stipitis was integrated into the genome of S. cerevisiae L2612, combined with the display of bifunctional minihemicellulosome. Engineering S. cerevisiae could convert birchwood xylan directly to ethanol (Sun et al., 2012b). A strategy was proposed to efficiently degrade xylan for ethanol production by controlling the mixed cultures of xylose-utilizing engineered yeast that displaying different hemicellulases (Tabañag et al., 2018). Industrial S. cerevisiae strains is considered to be a powerful host for displaying hemicellulose hydrolase on the cell surface and optimizing xylose assimilation because of its good heat resistance and high resistance to inhibitors. The use of industrial strains as gene recombination hosts may be more beneficial for the commercial production of ethanol. By introducing xylan-degrading enzymes and expressed xylose-assimilating enzymes into the recombinant yeast, hemicellulose substrates can be directly degraded to produce a variety of high-value-added products other than ethanol (Cunha et al., 2020).
Xylitol, a high-value-added pentose alcohol, can be produced by one-step reduction of xylose. Industrial production of xylitol from purified D-xylose requires an expensive catalytic hydrogenation process. A recombinant xylose-utilizing S. cerevisiae was constructed to directly degrade hemicellulose in rice straw hydrolysate to produce xylitol. The cell surface display technology combined with the incorporation of membrane separation technology further improved xylitol production, with a twofold increase in xylitol production by the multi-enzyme co-display engineered S. cerevisiae from the membrane separated hydrolysate (compared to the hydrolysate without membrane separation) (Guirimand et al., 2016). An engineered S. cerevisiae expressing cytosolic xylose reductase with β-D-glucosidase, xylosidase and xylanase co-displayed on the cell surface was constructed to produce xylitol from woody Kraft pulp (KP) (Guirimand et al., 2019). These results set the stage for large-scale xylitol production from lignocellulose.
3.1.3 Bioconversion of starch catalyzed by engineered S. cerevisiae
Starch, a polymer of α-D-glucose, is generally more easily degraded than cellulose and found in large quantities in many agricultural and industrial wastes. Using surface display engineering S. cerevisiae to degrade starch raw materials can not only produce biofuel but also improve the process quality of the food industry. Alpha-amylase and glucoamylase codisplayed S. cerevisiae was constructed to produce ethanol directly from corn starch with a yield of 86.5% of the theoretical value (Yamada et al., 2011). The activity of α-amylase was dependent on the anchor protein, and the activity of the strains based on flocculin system was 40 times higher than that based on agglutinin (Shigechi et al., 2002). Most of the insoluble starch is partially non-degradable (Shigechi et al., 2004), for which some attempts such as increasing enzyme activity to improve starch utilization efficiency have been reported. A diploid yeast strain displaying structurally optimized α-amylase and glucoamylase was successfully constructed. The modified yeast was used to ferment 100 g/L of raw starch with an average ethanol yield of up to 1.61 g/L/h over 23 successive cycles, yielding 76.6% of the theoretical value (Yamakawa et al., 2012). Cyclodextrin glucan transferase (CGTase) was demonstrated on S. cerevisiae cell surface to effectively hydrolyze starch to produce glucose and maltose for yeast fermentation and to enhance the bread-baking process (Shim et al., 2007).
3.1.4 Bioconversion of waste oil catalyzed by engineered S. cerevisiae
Lipases catalyze a variety of reactions and are widely used in industry, so the cell surface display of lipases is a potential way to construct whole cell catalysts. Biodiesel is a clean and renewable fuel that can be produced from waste rich in triglycerides. The production costs can be reduced by using engineered yeast cells that display lipases on the cell surface as recyclable biocatalysts. Through the flocculation function of Flo1p, the lipase ROL can be displayed on the cell surface of S. cerevisae, and the yield of synthesized methylesters from triglyceride and methanol reached 78.3% after 72 h of reaction (Matsumoto et al., 2002). Later Candida antarctica lipase B (CALB) and Yarrowia lipolytica lipases were also displayed on S. cerevisae by α-agglutinin- or Flo1p-display systems (Kato et al., 2007; Liu et al., 2010). Pichia pastoris can use methanol as the sole carbon source, and its promoter AOX1 is regulated by methanol and corresponding integrated vectors. The transformants have strong genetic stability and few native proteins are expressed extracellularly. P. pastoris lacks α-1, 3-mannosyltransferase, so lipase activity is not affected by excessive glycosylation. Compared with S. cerevisiae, P. pastoris displaying lipases on the cell surface is more suitable for the high-density culture and generally produce enzyme in higher yield. With the ability to utilize a variety of hydrophobic substrates as carbon sources, high resistance to adversity, and tolerance to high salinity and low temperatures, Y. lipolytica is an attractive host for cell surface display (Yuzbasheva et al., 2015; Qiao et al., 2018; Yang et al., 2020).
3.2 Applications in environmental control
Heavy metals are frequently detected in industrial wastewater, which are usually not biodegradable and can only be detoxified by means of absorption, enrichment and removal from wastewater (Fu and Wang, 2011). The recovery of rare metals can save resources and reduce environmental pollution. The cell wall of S. cerevisiae is mainly composed of mannoproteins, chitin, β-1, 6-glucan and β-1, 3- glucan (Kollár et al., 1995). These biomolecules consist of carboxylate, phosphate, hydroxyl group, amine, sulfhydryl group and other functional groups, which provide a range of different metal binding sites. Different metal-binding peptides combined with anchor systems can be displayed on the cell surface of S. cerevisiae. These engineered strains could recover metals from waste solutions and become useful tools for bioreremediation and bioadsorption of environmental pollutants (Mashangoane and Chirwa, 2022). Four types of Solanum nigrum metallothionein (SMT) were displayed on S. cerevisiae cell surface by using an α-agglutinin-based display system to adsorb ultra-trace cadmium effectively (Qinguo et al., 2016). Similarly, by displaying MerR on the cell surface, the adsorption capacity of S. cerevisiae to Hg2+ was much higher than that of the original and the control strains, while the engineered yeast strain also exhibited higher tolerance to Hg2+ (Wei et al., 2018). In addition to heavy metals, there are many emerging environmental pollutants such as parabens with disruptive effects and ethyl carbamate (EC), a possible human carcinogen. The removal of parabens could be mediated by Fusarium solani pisi cutinase (FsC) expressed on the cell surface of S. cerevisiae (Zhu and Wei, 2019). The EC degradation could be improved by displaying urethanease (UreA) from Micrococcus species on the cell surface of S. cerevisiae (Han et al., 2022).
3.3 Applications in immunology
In view of the safety of S. cerevisiae, it is an excellent vector for vaccine production. An efficient antibody response can be obtained by displaying antigenic proteins on the yeast cell surface and then feeding the vaccine to animals. The antibody response can be enhanced by increasing the number of antigenic proteins displayed on the cell surface. Using the single-chain variable fragment (scFv) of the anti-infectious hematopoietic necrosis virus isolate Sn1203 antibody as a model protein, the a-agglutinin system was used to display E. coli-derived and yeast-derived ScFV on the cell surface of S. cerevisiae, which effectively increased the number of proteins displayed on the cell surface and enhanced the possibility of the engineered yeast as an oral vaccine (Zhao J.-Z. et al., 2017). VP28 is an envelope protein of White Spot Syndrome Virus (WSSV), which induces a high immune response in shrimp. Probiotics combined with VP28 anchored yeast cell extract could be used as an oral vaccine in shrimp aquaculture (Le Linh et al., 2021). VP24 is another important envelope protein of WSSV. When applied as an oral vaccine, the protective effect of S. cerevisiae with VP28 and VP24 fusion protein on cell surface against WSSV attack was more significant than that of with VP28 only, and the survival rate was up to 100% (Lei et al., 2021a). Vaccines made from recombinant viral proteins displayed on the surface of S. cerevisiae cells can be more easily recognized by the host mucosal immune system through oral administration, thus triggering protective immunity. This method is not only easy to operate and safe, but also has a short production cycle, which can be used to treat poultry in a short time. EBY100/PYD1-HA was developed as an oral vaccine against avian influenza A (H5N1) infected chickens (Lei et al., 2021b). Timely vaccination is the most effective solution to the outbreak epidemics. An engineered S. cerevisiae displaying the receptor binding domains (RBDs) of the spike protein of the initial strain of SARS-CoV-2 and its variants was constructed as a vaccine candidate against the coronavirus (COVID-19) (Xing et al., 2022). In view of the clear genetic background and easy genetic modification of S. cerevisiae, the vaccines constructed based on S. cerevisiae cell surface display are expected to achieve large-scale production to prevent various infectious diseases.
3.4 Applications in other areas
Cell surface engineering yeast can also be used in biosensors, protein library screening and other fields (Han et al., 2018; Zhao et al., 2021). Cell surface display of fluorescent proteins exerts less metabolic stress on cells than intracellular expression. Displaying a visible reporter on the cell surface may help characterize cells at the single-cell level. Under the control of different promoters, different fluorescent protein variants were displayed on the surface of S. cerevisiae, and the engineered yeast could be used as a reporting system to monitor environmental changes and the production of foreign proteins. The GFP cell surface display gene cassette controlled by GAPDH promoter and the BFP cell surface display gene cassette controlled by UPR-ICL promoter were both integrated into the genome of S. cerevisiae. The GAPDH promoter acted when the glucose concentration was high in the early stage of culture, and GFP was displayed on the cell surface emitting green fluorescence. When the glucose concentration was low, the UPR-ICL promoter acted and BFP was co-displayed on the cell surface emitting blue fluorescence. The fluorescence intensity and color on the cell surface changed with the intracellular and extracellular glucose concentrations, so the surface display system controlled by the promoters could be used to monitor the glucose concentration inside and outside the cell (Shibasaki et al., 2001). Furthermore, a system was constructed to monitor the production of foreign proteins in yeast by the same promoter controlling the production of foreign proteins and the surface display of reporter gene EGFP. This detection system is expected to be a powerful tool in the field of biological processes (Shibasaki et al., 2003).
Yeast display library technology is a powerful tool for isolation and identification of proteins with unique or modified properties or selection of specific substances (including functional antibodies, enzymes, active protein sites, and chemical probes, etc.), and further identification in combination with fluorescence-activated cell sorting (FACS) (Jeong et al., 2019; Bacon et al., 2020; Banach et al., 2022; Fiebig et al., 2022; Lerma Romero et al., 2022). S. cerevisiae surface display system could be designed using inefficient ribosomal skipping, and the protein of interest could be simultaneously displayed on the cell surface and secreted out of the cell (Cruz-Teran et al., 2017). A Multiple Navigation of Antibody Structures (MINAS) method that combines CRISPR/Cas9-based traceable editing and fluorescence-activated cell sorting (FACS) of yeast-display libraries has been successfully designed to act on any region of the antibody, introducing hundreds of thousands of mutations and mapping the effect of these mutations on the desired phenotype (Oh et al., 2020). The optimization of the yeast surface display system is helpful in improving the application of the system in screening (Kajiwara et al., 2020).
In addition, cell surface display engineered S. cerevisiae encapsulated by biological materials can be used for protein purification. Taking advantage of the high affinity between E. coli vegetal E7 deoxyribonuclease (CL7) and its inhibitor immunoprotein 7 (IM7), calcium alginate beads encapsulated with S. cerevisiae cells displaying IM7 could be used as column filler materials to purify proteins fused with CL7 tags. Calcium alginate beads encapsulated with yeast cells displaying cellulosomes could act as a whole-cell catalyst to degrade cellulose substrates, which could be reused at least six times with very low activity reduction (Yin et al., 2022). This strategy of encapsulating cell surface displaying engineered bacteria will play an important role in pharmaceutical and bioengineering fields.
4 Current challenges and future prospects
Yeast cell surface display technology has developed rapidly in the past few decades, and different display systems have been designed. There are some bottlenecks in the development process of widely used S. cerevisiae cell surface display engineering: 1) Inefficient production of heterologous proteins leads to the lack of activity of engineered yeast strains; 2) with the increases in recycling times, the activity of engineered yeast may decrease more; 3) more attempts are needed in terms of improving the surface space utilization of yeast cells; 4) most of the current studies are mainly at the laboratory level, and further investigation are needed on the suitability of the constructed engineered strains for industrial mass production; 5) molecular and physiological knowledge on the tolerance of engineered yeast to many inhibitory compounds is limited.
To solve the above bottlenecks, future research can focus on the following aspects: 1) more novel anchoring proteins should be developed to extend the cell surface displayable sites and new gene editing systems combined with cell surface display should be designed to increase the copy number of target genes and simplify the process of plasmid transformation; 2) more biomaterials for encapsulating engineered yeasts should be developed to increase the times of reuses, maintain the activity of display proteins, and further expand the application areas. In addition, the system design to induce automatic flocculation of S. cerevisiae is also helpful to promote the yeast recovery; 3) constructing S. cerevisiae cell surface display systems by rational and systematic design, including the regulation of the location and ratio of multiple target proteins displayed on the cell surface and the way multifunctional scaffold proteins assembled on the cell surface. Exploring effective and versatile linkers is essential to improve the efficiency of cell surface display; 4) the combination of cell surface display technology, metabolic engineering and synthetic biology engineering is helpful to improve the ability of engineered yeast to utilize mixed substrates, reduce the production of by-products, and construct multifunctional “super yeast”; 5) to improve the tolerance of engineered strains to inhibitors or toxic substances in the fermentation process by means of strain mutagenesis, directed evolution, genetic engineering. The capability of engineered S. cerevisiae should be evaluated in a simulated industrial production environment.
5 Conclusion
In this paper, recent advances of cell-surface display engineering and the strategies to improve the display efficiency of S. cerevisiae are reviewed. In spite of the obvious requirement for increasing the amount of proteins that displayed on the cell surface and the efficient expression and secretion of target proteins, it is expected that more products using cell surface display will be commercialized in the future.
Author contributions
CZ: Drafted the manuscript. HC and YIZ: Illustration. YUZ and XL: Data curation. FW: Reviewing and Editing.
Funding
This work was financially supported by National Key Research & Development Program of China (No. 2019YFB1504002) and Topnotch Academic Programs Project (TAPP) Education Institutions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, J., Zhang, L., Li, L., Liu, D., Cheng, H., Wang, H., et al. (2016). An alternative approach to synthesizing galactooligosaccharides by cell-surface display of β-galactosidase on Yarrowia lipolytica. J. Agric. Food Chem. 64 (19), 3819–3827. doi:10.1021/acs.jafc.5b06138
Arnthong, J., Ponjarat, J., Bussadee, P., Deenarn, P., Prommana, P., Phienluphon, A., et al. (2022). Enhanced surface display efficiency of β-glucosidase in Saccharomyces cerevisiae by disruption of cell wall protein-encoding genes YGP1 and CWP2. Biochem. Eng. J. 179, 108305. doi:10.1016/j.bej.2021.108305
Aza, P., Molpeceres, G., de Salas, F., and Camarero, S. (2021). Design of an improved universal signal peptide based on the α-factor mating secretion signal for enzyme production in yeast. Cell. Mol. Life Sci. 78 (7), 3691–3707. doi:10.1007/s00018-021-03793-y
Bacon, K., Blain, A., Burroughs, M., McArthur, N., Rao, B. M., and Menegatti, S. (2020). Isolation of chemically cyclized peptide binders using yeast surface display. ACS Comb. Sci. 22 (10), 519–532. doi:10.1021/acscombsci.0c00076
Bae, J.-H., Sung, B. H., Seo, J.-W., Kim, C. H., and Sohn, J.-H. (2016). A novel fusion partner for enhanced secretion of recombinant proteins in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 100 (24), 10453–10461. doi:10.1007/s00253-016-7722-2
Bae, J.-H., Yun, S.-H., Kim, M.-J., Kim, H.-J., Sung, B. H., Kim, S. I., et al. (2022). Secretome-based screening of fusion partners and their application in recombinant protein secretion in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 106 (2), 663–673. doi:10.1007/s00253-021-11750-9
Bae, J., Kuroda, K., and Ueda, M. (2015). Proximity effect among cellulose-degrading enzymes displayed on the Saccharomyces cerevisiae cell surface. Appl. Environ. Microbiol. 81 (1), 59–66. doi:10.1128/AEM.02864-14
Bamba, T., Inokuma, K., Hasunuma, T., and Kondo, A. (2018). Enhanced cell-surface display of a heterologous protein using SED1 anchoring system in SED1-disrupted Saccharomyces cerevisiae strain. J. Biosci. Bioeng. 125 (3), 306–310. doi:10.1016/j.jbiosc.2017.09.013
Banach, B. B., Tripathi, P., Da Silva Pereira, L., Gorman, J., Nguyen, T. D., Dillon, M., et al. (2022). Highly protective antimalarial antibodies via precision library generation and yeast display screening. J. Exp. Med. 219 (8), e20220323. doi:10.1084/jem.20220323
Bao, J., Huang, M., Petranovic, D., and Nielsen, J. (2017). Moderate expression of SEC16 increases protein secretion by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 83 (14), e03400–e03416. doi:10.1128/AEM.03400-16
Besada-Lombana, P. B., and Da Silva, N. A. (2019). Engineering the early secretory pathway for increased protein secretion in Saccharomyces cerevisiae. Metab. Eng. 55, 142–151. doi:10.1016/j.ymben.2019.06.010
Cannella, D., and Jørgensen, H. (2014). Do new cellulolytic enzyme preparations affect the industrial strategies for high solids lignocellulosic ethanol production? Biotechnol. Bioeng. 111 (1), 59–68. doi:10.1002/bit.25098
Ce, D., Jie, Q., Xinping, W., Wenli, S., Lixia, C., Shuntang, L., et al. (2022). Engineering Pichia pastoris with surface-display minicellulosomes for carboxymethyl cellulose hydrolysis and ethonol production. Biotechnol. Biofuels Bioprod. doi:10.21203/rs.2.23317/v2
Cha, Y., Li, W., Wu, T., You, X., Chen, H., Zhu, C., et al. (2022). Probing the synergistic ratio of P450/CPR to improve (+)-Nootkatone production in Saccharomyces cerevisiae. J. Agric. Food Chem. 70 (3), 815–825. doi:10.1021/acs.jafc.1c07035
Cho, J. S., Oh, H. J., Jang, Y. E., Kim, H. J., Kim, A., Song, J.-A., et al. (2022). Synthetic pro-peptide design to enhance the secretion of heterologous proteins by Saccharomyces cerevisiae. MicrobiologyOpen 11 (3), e1300. doi:10.1002/mbo3.1300
Choi, H.-J., Jin, Y.-S., and Lee, W.-H. (2022). Effects of engineered Saccharomyces cerevisiae fermenting cellobiose through low-energy-consuming phosphorolytic pathway in simultaneous saccharification and fermentation. J. Microbiol. Biotechnol. 32 (1), 117–125. doi:10.4014/jmb.2111.11047
Cruz-Teran, C. A., Tiruthani, K., Mischler, A., and Rao, B. M. (2017). Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae. ACS Synth. Biol. 6 (11), 2096–2107. doi:10.1021/acssynbio.7b00144
Cunha, J. T., Gomes, D. G., Romaní, A., Inokuma, K., Hasunuma, T., Kondo, A., et al. (2021). Cell surface engineering of Saccharomyces cerevisiae for simultaneous valorization of corn cob and cheese whey via ethanol production. Energy Convers. Manag. 243, 114359. doi:10.1016/j.enconman.2021.114359
Cunha, J. T., Romaní, A., Inokuma, K., Johansson, B., Hasunuma, T., Kondo, A., et al. (2020). Consolidated bioprocessing of corn cob-derived hemicellulose: Engineered industrial Saccharomyces cerevisiae as efficient whole cell biocatalysts. Biotechnol. Biofuels 13 (1), 138. doi:10.1186/s13068-020-01780-2
Curran, K. A., Morse, N. J., Markham, K. A., Wagman, A. M., Gupta, A., and Alper, H. S. (2015). Short synthetic terminators for improved heterologous gene expression in yeast. ACS Synth. Biol. 4 (7), 824–832. doi:10.1021/sb5003357
Deng, J., Wu, Y., Zheng, Z., Chen, N., Luo, X., Tang, H., et al. (2021). A synthetic promoter system for well-controlled protein expression with different carbon sources in Saccharomyces cerevisiae. Microb. Cell. Fact. 20 (1), 202. doi:10.1186/s12934-021-01691-3
Dong, C., Qiao, J., Wang, X., Sun, W., Chen, L., Li, S., et al. (2020). Engineering Pichia pastoris with surface-display minicellulosomes for carboxymethyl cellulose hydrolysis and ethanol production. Biotechnol. Biofuels 13 (1), 108. doi:10.1186/s13068-020-01749-1
Fan, L.-H., Zhang, Z.-J., Mei, S., Lu, Y.-Y., Li, M., Wang, Z.-Y., et al. (2016). Engineering yeast with bifunctional minicellulosome and cellodextrin pathway for co-utilization of cellulose-mixed sugars. Biotechnol. Biofuels 9 (1), 137. doi:10.1186/s13068-016-0554-6
Fan, L.-H., Zhang, Z.-J., Yu, X.-Y., Xue, Y.-X., Wang, M.-M., and Tan, T.-W. (2013). In vitro assembly of minicellulosomes with two scaffoldins on the yeast cell surface for cellulose saccharification and bioethanol production. Process Biochem. 48 (3), 430–437. doi:10.1016/j.procbio.2013.01.012
Fan, L. H., Zhang, Z. J., Yu, X. Y., Xue, Y. X., and Tan, T. W. (2012). Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc. Natl. Acad. Sci. U. S. A. 109 (33), 13260–13265. doi:10.1073/pnas.1209856109
Felix, C. R., and Ljungdahl, L. G. (1993). The cellulosome: The exocellular organelle of Clostridium. Annu. Rev. Microbiol. 47 (1), 791–819. doi:10.1146/annurev.mi.47.100193.004043
Fiebig, D., Bogen, J. P., Carrara, S. C., Deweid, L., Zielonka, S., Grzeschik, J., et al. (2022). Streamlining the transition from yeast surface display of antibody fragment immune libraries to the production as IgG format in mammalian cells. Front. Bioeng. Biotechnol. 10, 794389. doi:10.3389/fbioe.2022.794389
Fierobe, H.-P., Mingardon, F., Mechaly, A., Bélaïch, A., Rincon, M. T., Pagès, S., et al. (2005). Action of Designer Cellulosomes on Homogeneous versus Complex Substrates: Controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280 (16), 16325–16334. doi:10.1074/jbc.M414449200
Fu, F., and Wang, Q. (2011). Removal of heavy metal ions from wastewaters: A review. J. Environ. Manag. 92 (3), 407–418. doi:10.1016/j.jenvman.2010.11.011
Galazka Jonathan, M., Tian, C., Beeson William, T., Martinez, B., Glass, N. L., and Cate Jamie, H. D. (2010). Cellodextrin transport in yeast for improved biofuel production. Science 330 (6000), 84–86. doi:10.1126/science.1192838
Goyal, G., Tsai, S. L., Madan, B., DaSilva, N. A., and Chen, W. (2011). Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microb. Cell. Fact. 10, 89. doi:10.1186/1475-2859-10-89
Guirimand, G., Inokuma, K., Bamba, T., Matsuda, M., Morita, K., Sasaki, K., et al. (2019). Cell-surface display technology and metabolic engineering of Saccharomyces cerevisiae for enhancing xylitol production from woody biomass. Green Chem. 21 (7), 1795–1808. doi:10.1039/C8GC03864C
Guirimand, G., Sasaki, K., Inokuma, K., Bamba, T., Hasunuma, T., and Kondo, A. (2016). Cell surface engineering of Saccharomyces cerevisiae combined with membrane separation technology for xylitol production from rice straw hydrolysate. Appl. Microbiol. Biotechnol. 100 (8), 3477–3487. doi:10.1007/s00253-015-7179-8
Han, K., Lee, H., Kang, T.-G., Lee, J., and Kim, S.-K. (2022). Direct and efficient elimination of ethyl carbamate by engineered Saccharomyces cerevisiae displaying urethanase. Food control. 142, 109236. doi:10.1016/j.foodcont.2022.109236
Han, L., Zhao, Y., Cui, S., and Liang, B. (2018). Redesigning of microbial cell surface and its application to whole-cell biocatalysis and biosensors. Appl. Biochem. Biotechnol. 185 (2), 396–418. doi:10.1007/s12010-017-2662-6
Hong, S.-J., Kim, H. J., Kim, J.-W., Lee, D.-H., and Seo, J.-H. (2015). Optimizing promoters and secretory signal sequences for producing ethanol from inulin by recombinant Saccharomyces cerevisiae carrying Kluyveromyces marxianus inulinase. Bioprocess Biosyst. Eng. 38 (2), 263–272. doi:10.1007/s00449-014-1265-7
Horn, S. J., Vaaje-Kolstad, G., Westereng, B., and Eijsink, V. (2012). Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5 (1), 45. doi:10.1186/1754-6834-5-45
Hoshida, H., Kondo, M., Kobayashi, T., Yarimizu, T., and Akada, R. (2017). 5´-UTR introns enhance protein expression in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 101 (1), 241–251. doi:10.1007/s00253-016-7891-z
Hou, J., Tyo, K., Liu, Z., Petranovic, D., and Nielsen, J. (2012). Engineering of vesicle trafficking improves heterologous protein secretion in Saccharomyces cerevisiae. Metab. Eng. 14 (2), 120–127. doi:10.1016/j.ymben.2012.01.002
Hu, J., Gourlay, K., Arantes, V., Van Dyk, J. S., Pribowo, A., and Saddler, J. N. (2015). The accessible cellulose surface influences cellulase synergism during the hydrolysis of lignocellulosic substrates. ChemSusChem 8 (5), 901–907. doi:10.1002/cssc.201403335
Huang, M., Wang, G., Qin, J., Petranovic, D., and Nielsen, J. (2018). Engineering the protein secretory pathway of Saccharomyces cerevisiae enables improved protein production. Proc. Natl. Acad. Sci. U. S. A. 115 (47), E11025–E11032. doi:10.1073/pnas.1809921115
Huang, X., Bai, S., Liu, Z., Hasunuma, T., Kondo, A., and Ho, S.-H. (2020). Fermentation of pigment-extracted microalgal residue using yeast cell-surface display: Direct high-density ethanol production with competitive life cycle impacts. Green Chem. 22 (1), 153–162. doi:10.1039/C9GC02634G
Inokuma, K., Bamba, T., Ishii, J., Ito, Y., Hasunuma, T., and Kondo, A. (2016). Enhanced cell-surface display and secretory production of cellulolytic enzymes with Saccharomyces cerevisiae Sed1 signal peptide. Biotechnol. Bioeng. 113 (11), 2358–2366. doi:10.1002/bit.26008
Inokuma, K., Hasunuma, T., and Kondo, A. (2014). Efficient yeast cell-surface display of exo- and endo-cellulase using the SED1 anchoring region and its original promoter. Biotechnol. Biofuels 7 (1), 8. doi:10.1186/1754-6834-7-8
Inokuma, K., Kitada, Y., Bamba, T., Kobayashi, Y., Yukawa, T., den Haan, R., et al. (2021). Improving the functionality of surface-engineered yeast cells by altering the cell wall morphology of the host strain. Appl. Microbiol. Biotechnol. 105 (14), 5895–5904. doi:10.1007/s00253-021-11440-6
Inokuma, K., Kurono, H., den Haan, R., van Zyl, W. H., Hasunuma, T., and Kondo, A. (2020). Novel strategy for anchorage position control of GPI-attached proteins in the yeast cell wall using different GPI-anchoring domains. Metab. Eng. 57, 110–117. doi:10.1016/j.ymben.2019.11.004
Jeong, M.-Y., Rutter, J., and Chou, D. H.-C. (2019). Display of single-chain insulin-like peptides on a yeast surface. Biochemistry 58 (3), 182–188. doi:10.1021/acs.biochem.8b01094
Jiang, Z.-B., Song, H.-T., Gupta, N., Ma, L.-X., and Wu, Z.-B. (2007). Cell surface display of functionally active lipases from Yarrowia lipolytica in Pichia pastoris. Protein Expr. Purif. 56 (1), 35–39. doi:10.1016/j.pep.2007.07.003
Kajiwara, K., Aoki, W., and Ueda, M. (2020). Evaluation of the yeast surface display system for screening of functional nanobodies. Amb. Expr. 10 (1), 51. doi:10.1186/s13568-020-00983-y
Katahira, S., Mizuike, A., Fukuda, H., and Kondo, A. (2006). Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl. Microbiol. Biotechnol. 72 (6), 1136–1143. doi:10.1007/s00253-006-0402-x
Kato, M., Fuchimoto, J., Tanino, T., Kondo, A., Fukuda, H., and Ueda, M. (2007). Preparation of a whole-cell biocatalyst of mutated Candida Antarctica lipase B (mCALB) by a yeast molecular display system and its practical properties. Appl. Microbiol. Biotechnol. 75 (3), 549–555. doi:10.1007/s00253-006-0835-2
Kim, S., Baek, S.-H., Lee, K., and Hahn, J.-S. (2013). Cellulosic ethanol production using a yeast consortium displaying a minicellulosome and β-glucosidase. Microb. Cell. Fact. 12 (1), 14. doi:10.1186/1475-2859-12-14
Kollár, R., Petráková, E., Ashwell, G., Robbins, P. W., and Cabib, E. (1995). Architecture of the yeast cell wall: The linkage between chitin and β(1 →3)-glucan (∗). J. Biol. Chem. 270 (3), 1170–1178. doi:10.1074/jbc.270.3.1170
Kondo, A., and Ueda, M. (2004). Yeast cell-surface display-applications of molecular display. Appl. Microbiol. Biotechnol. 64 (1), 28–40. doi:10.1007/s00253-003-1492-3
Lamour, J., Wan, C., Zhang, M., Zhao, X., and Den Haan, R. (2019). Overexpression of endogenous stress-tolerance related genes in Saccharomyces cerevisiae improved strain robustness and production of heterologous cellobiohydrolase. FEMS Yeast Res. 19 (4), foz035. doi:10.1093/femsyr/foz035
Le Linh, H., Thu, N. P. A., Dung, T. T. X., Van Hau, N., Nghia, N. H., and Thao, D. T. P. (2021). Yeast cell surface displaying VP28 antigen and its potential application for shrimp farming. Appl. Microbiol. Biotechnol. 105 (16), 6345–6354. doi:10.1007/s00253-021-11493-7
Lei, H., Li, S., Lu, X., and Ren, Y. (2021a). Oral administration of Saccharomyces cerevisiae displaying VP28-VP24 confers protection against white spot syndrome virus in shrimp. Virus Res. 302, 198467. doi:10.1016/j.virusres.2021.198467
Lei, H., Lu, X., Li, S., and Ren, Y. (2021b). High immune efficacy against different avian influenza H5N1 viruses due to oral administration of a Saccharomyces cerevisiae-based vaccine in chickens. Sci. Rep. 11 (1), 8977. doi:10.1038/s41598-021-88413-2
Lerma Romero, J. A., Meyners, C., Christmann, A., Reinbold, L. M., Charalampidou, A., Hausch, F., et al. (2022). Binding pocket stabilization by high-throughput screening of yeast display libraries. Front. Mol. Biosci. 9, 1023131. doi:10.3389/fmolb.2022.1023131
Li, J., Zeng, Y., Wang, W.-B., Wan, Q.-Q., Liu, C.-G., den Haan, R., et al. (2022). Increasing extracellular cellulase activity of the recombinant Saccharomyces cerevisiae by engineering cell wall-related proteins for improved consolidated processing of carbon neutral lignocellulosic biomass. Bioresour. Technol. 365, 128132. doi:10.1016/j.biortech.2022.128132
Li, Y.-J., Lu, Y.-Y., Zhang, Z.-J., Mei, S., Tan, T.-W., and Fan, L.-H. (2017). Co-Fermentation of cellulose and sucrose/xylose by engineered yeasts for bioethanol production. Energy fuels. 31 (4), 4061–4067. doi:10.1021/acs.energyfuels.7b00032
Liang, Y., Si, T., Ang Ee, L., Zhao, H., and Kelly, R. M. (2014). Engineered pentafunctional minicellulosome for simultaneous saccharification and ethanol fermentation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 80 (21), 6677–6684. doi:10.1128/AEM.02070-14
Liu, W.-S., Pan, X.-X., Jia, B., Zhao, H.-Y., Xu, L., Liu, Y., et al. (2010). Surface display of active lipases Lip7 and Lip8 from Yarrowia lipolytica on Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 88 (4), 885–891. doi:10.1007/s00253-010-2782-1
Liu, Z., Ho, S.-H., Hasunuma, T., Chang, J.-S., Ren, N.-Q., and Kondo, A. (2016a). Recent advances in yeast cell-surface display technologies for waste biorefineries. Bioresour. Technol. 215, 324–333. doi:10.1016/j.biortech.2016.03.132
Liu, Z., Ho, S.-H., Sasaki, K., den Haan, R., Inokuma, K., Ogino, C., et al. (2016b). Engineering of a novel cellulose-adherent cellulolytic Saccharomyces cerevisiae for cellulosic biofuel production. Sci. Rep. 6 (1), 24550. doi:10.1038/srep24550
Liu, Z., Inokuma, K., Ho, S.-H., den Haan, R., van Zyl, W. H., Hasunuma, T., et al. (2017). Improvement of ethanol production from crystalline cellulose via optimizing cellulase ratios in cellulolytic Saccharomyces cerevisiae. Biotechnol. Bioeng. 114 (6), 1201–1207. doi:10.1002/bit.26252
Liu, Z., Inokuma, K., Ho, S.-H., Haan, R. d., Hasunuma, T., van Zyl, W. H., et al. (2015). Combined cell-surface display- and secretion-based strategies for production of cellulosic ethanol with Saccharomyces cerevisiae. Biotechnol. Biofuels 8 (1), 162. doi:10.1186/s13068-015-0344-6
Mashangoane, B. F., and Chirwa, E. N. (2022). Cell surface display of palladium binding peptide on Saccharomyces cerevisiae EBY100 cells using the a-agglutinin anchor system developed for the biosorption of Pd (II). Miner. Eng. 176, 107325. doi:10.1016/j.mineng.2021.107325
Matano, Y., Hasunuma, T., and Kondo, A. (2012). Display of cellulases on the cell surface of Saccharomyces cerevisiae for high yield ethanol production from high-solid lignocellulosic biomass. Bioresour. Technol. 108, 128–133. doi:10.1016/j.biortech.2011.12.144
Matsumoto, T., Fukuda, H., Ueda, M., Tanaka, A., and Kondo, A. (2002). Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Appl. Environ. Microbiol. 68 (9), 4517–4522. doi:10.1128/AEM.68.9.4517-4522.2002
Matsuoka, H., Hashimoto, K., Saijo, A., Takada, Y., Kondo, A., Ueda, M., et al. (2014). Cell wall structure suitable for surface display of proteins in Saccharomyces cerevisiae. Yeast 31 (2), 67–76. doi:10.1002/yea.2995
Mingardon, F., Chanal, A., López-Contreras Ana, M., Dray, C., Bayer Edward, A., and Fierobe, H.-P. (2007). Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl. Environ. Microbiol. 73 (12), 3822–3832. doi:10.1128/AEM.00398-07
Nakatani, Y., Yamada, R., Ogino, C., and Kondo, A. (2013). Synergetic effect of yeast cell-surface expression of cellulase and expansin-like protein on direct ethanol production from cellulose. Microb. Cell. Fact. 12 (1), 66. doi:10.1186/1475-2859-12-66
Oh, E. J., Liu, R., Liang, L., Freed, E. F., Eckert, C. A., and Gill, R. T. (2020). Multiplex evolution of antibody fragments utilizing a yeast surface display platform. ACS Synth. Biol. 9 (8), 2197–2202. doi:10.1021/acssynbio.0c00159
Pepper, L. R., Cho, Y. K., Boder, E. T., and Shusta, E. V. (2008). A decade of yeast surface display technology: Where are we now? Comb. Chem. high throughput Screen. 11 (2), 127–134. doi:10.2174/138620708783744516
Phienluphon, A., Mhuantong, W., Boonyapakron, K., Deenarn, P., Champreda, V., Wichadakul, D., et al. (2019). Identification and evaluation of novel anchoring proteins for cell surface display on Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 103 (7), 3085–3097. doi:10.1007/s00253-019-09667-5
Qi, K., Chen, C., Yan, F., Feng, Y., Bayer, E. A., Kosugi, A., et al. (2021). Coordinated β-glucosidase activity with the cellulosome is effective for enhanced lignocellulose saccharification. Bioresour. Technol. 337, 125441. doi:10.1016/j.biortech.2021.125441
Qiao, Y., Yang, K., Zhou, Q., Xu, Z., Yan, Y., Xu, L., et al. (2018). Engineering Yarrowia lipolytica for sustainable production of fatty acid methyl esters using in situ self-cycled glycerol as a carbon source. ACS Sustain. Chem. Eng. 6 (6), 7645–7651. doi:10.1021/acssuschemeng.8b00492
Qinguo, W., Honghai, Z., Dongge, G., and Ma, S. (2016). Cell surface display of four types of Solanum nigrum metallothionein on Saccharomyces cerevisiae for biosorption of cadmium. J. Microbiol. Biotechnol. 26 (5), 846–853. doi:10.4014/jmb.1512.12041
Quinlan, R. J., Sweeney, M. D., Lo Leggio, L., Otten, H., Poulsen, J. C., Johansen, K. S., et al. (2011). Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U. S. A. 108 (37), 15079–15084. doi:10.1073/pnas.1105776108
Sasaki, Y., Mitsui, R., Yamada, R., and Ogino, H. (2019). Secretory overexpression of the endoglucanase by Saccharomyces cerevisiae via CRISPR-δ-integration and multiple promoter shuffling. Enzyme Microb. Technol. 121, 17–22. doi:10.1016/j.enzmictec.2018.10.014
Shibasaki, S., Tanaka, A., and Ueda, M. (2003). Development of combinatorial bioengineering using yeast cell surface display—Order-made design of cell and protein for bio-monitoring. Biosens. Bioelectron. 19 (2), 123–130. doi:10.1016/S0956-5663(03)00169-6
Shibasaki, S., Ueda, M., Ye, K., Shimizu, K., Kamasawa, N., Osumi, M., et al. (2001). Creation of cell surface-engineered yeast that display different fluorescent proteins in response to the glucose concentration. Appl. Microbiol. Biotechnol. 57 (4), 528–533. doi:10.1007/s002530100767
Shigechi, H., Fujita, Y., Koh, J., Ueda, M., Fukuda, H., and Kondo, A. (2004). Energy-saving direct ethanol production from low-temperature-cooked corn starch using a cell-surface engineered yeast strain co-displaying glucoamylase and α-amylase. Biochem. Eng. J. 18 (2), 149–153. doi:10.1016/j.bej.2003.08.003
Shigechi, H., Uyama, K., Fujita, Y., Matsumoto, T., Ueda, M., Tanaka, A., et al. (2002). Efficient ethanol production from starch through development of novel flocculent yeast strains displaying glucoamylase and co-displaying or secreting α-amylase. J. Mol. Catal. B Enzym. 17 (3), 179–187. doi:10.1016/S1381-1177(02)00026-7
Shim, J.-H., Seo, N.-S., Roh, S.-A., Kim, J.-W., Cha, H., and Park, K.-H. (2007). Improved bread-baking process using Saccharomyces cerevisiae displayed with engineered cyclodextrin glucanotransferase. J. Agric. Food Chem. 55 (12), 4735–4740. doi:10.1021/jf070217d
Song, C., Zhang, C., Zhang, S., Lin, H., Kim, Y., Ramakrishnan, M., et al. (2020). Thermochemical liquefaction of agricultural and forestry wastes into biofuels and chemicals from circular economy perspectives. Sci. Total Environ. 749, 141972. doi:10.1016/j.scitotenv.2020.141972
Sun, J., Shao, Z., Zhao, H., Nair, N., Wen, F., Xu, J.-H., et al. (2012a). Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109 (8), 2082–2092. doi:10.1002/bit.24481
Sun, J., Wen, F., Si, T., Xu, J.-H., and Zhao, H. (2012b). Direct conversion of xylan to ethanol by recombinant Saccharomyces cerevisiae strains displaying an engineered minihemicellulosome. Appl. Environ. Microbiol. 78 (11), 3837–3845. doi:10.1128/AEM.07679-11
Tabañag, I. D. F., Chu, I. M., Wei, Y.-H., and Tsai, S.-L. (2018). Ethanol production from hemicellulose by a consortium of different genetically-modified sacharomyces cerevisiae. J. Taiwan Inst. Chem. Eng. 89, 15–25. doi:10.1016/j.jtice.2018.04.029
Tang, H., Song, M., He, Y., Wang, J., Wang, S., Shen, Y., et al. (2017). Engineering vesicle trafficking improves the extracellular activity and surface display efficiency of cellulases in Saccharomyces cerevisiae. Biotechnol. Biofuels 10 (1), 53. doi:10.1186/s13068-017-0738-8
Tang, H., Wang, J., Wang, S., Shen, Y., Petranovic, D., Hou, J., et al. (2018). Efficient yeast surface-display of novel complex synthetic cellulosomes. Microb. Cell. Fact. 17 (1), 122. doi:10.1186/s12934-018-0971-2
Teymennet-Ramírez, K. V., Martínez-Morales, F., and Trejo-Hernández, M. R. (2022). Yeast surface display system: Strategies for improvement and biotechnological applications. Front. Bioeng. Biotechnol. 9, 794742. doi:10.3389/fbioe.2021.794742
Tsai, S.-L., DaSilva, N. A., and Chen, W. (2013). Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth. Biol. 2 (1), 14–21. doi:10.1021/sb300047u
Tsai, S.-L., Oh, J., Singh, S., Chen, R., and Chen, W. (2009a). Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 75 (19), 6087–6093. doi:10.1128/AEM.01538-09
Tsai, S. L., Goyal, G., and Chen, W. (2010). Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 76 (22), 7514–7520. doi:10.1128/AEM.01777-10
Tsai, S. L., Oh, J., Singh, S., Chen, R., and Chen, W. (2009b). Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 75 (19), 6087–6093. doi:10.1128/AEM.01538-09
Ullah, J., Chen, H., Vastermark, A., Jia, J., Wu, B., Ni, Z., et al. (2017). Impact of orientation and flexibility of peptide linkers on T. maritima lipase Tm1350 displayed on Bacillus subtilis spores surface using CotB as fusion partner. World J. Microbiol. Biotechnol. 33 (9), 166. doi:10.1007/s11274-017-2327-1
Van der Vaart, J. M., te Biesebeke, R., Chapman, J. W., Toschka, H. Y., Klis, F. M., and Verrips, C. T. (1997). Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl. Environ. Microbiol. 63 (2), 615–620. doi:10.1128/aem.63.2.615-620.1997
Van Zyl, J. H. D., Den Haan, R., and Van Zyl, W. H. (2016). Overexpression of native Saccharomyces cerevisiae ER-to-Golgi SNARE genes increased heterologous cellulase secretion. Appl. Microbiol. Biotechnol. 100 (1), 505–518. doi:10.1007/s00253-015-7022-2
Wang, X., Feng, X., Lv, B., Zhou, A., Hou, Y., and Li, C. (2019). Enhanced yeast surface display of β-glucuronidase using dual anchor motifs for high-temperature glycyrrhizin hydrolysis. AIChE J. 65 (9), e16629. doi:10.1002/aic.16629
Wang, Y., Li, X., Chen, X., and Siewers, V. (2022). CRISPR/Cas9-mediated point mutations improve α-amylase secretion in Saccharomyces cerevisiae. FEMS Yeast Res. 22 (1), foac033. doi:10.1093/femsyr/foac033
Washida, M., Takahashi, S., Ueda, M., and Tanaka, A. (2001). Spacer-mediated display of active lipase on the yeast cell surface. Appl. Microbiol. Biotechnol. 56 (5), 681–686. doi:10.1007/s002530100718
Wei, Q., Yan, J., Chen, Y., Zhang, L., Wu, X., Shang, S., et al. (2018). Cell surface display of MerR on Saccharomyces cerevisiae for biosorption of mercury. Mol. Biotechnol. 60 (1), 12–20. doi:10.1007/s12033-017-0039-2
Wen, F., Sun, J., and Zhao, H. (2010). Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 76 (4), 1251–1260. doi:10.1128/AEM.01687-09
Wentz, A. E., and Shusta, E. V. (2007). A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl. Environ. Microbiol. 73 (4), 1189–1198. doi:10.1128/AEM.02427-06
Wieczorek, A. S., and Martin, V. J. J. (2010). Engineering the cell surface display of cohesins for assembly of cellulosome-inspired enzyme complexes on Lactococcus lactis. Microb. Cell. Fact. 9 (1), 69. doi:10.1186/1475-2859-9-69
Xing, H., Zhu, L., Wang, P., Zhao, G., Zhou, Z., Yang, Y., et al. (2022). Display of receptor-binding domain of SARS-CoV-2 Spike protein variants on the Saccharomyces cerevisiae cell surface. Front. Immunol. 13. doi:10.3389/fimmu.2022.935573
Yamada, R., Taniguchi, N., Tanaka, T., Ogino, C., Fukuda, H., and Kondo, A. (2010). Cocktail δ-integration: A novel method to construct cellulolytic enzyme expression ratio-optimized yeast strains. Microb. Cell. Fact. 9 (1), 32. doi:10.1186/1475-2859-9-32
Yamada, R., Yamakawa, S.-i., Tanaka, T., Ogino, C., Fukuda, H., and Kondo, A. (2011). Direct and efficient ethanol production from high-yielding rice using a Saccharomyces cerevisiae strain that express amylases. Enzyme Microb. Technol. 48 (4), 393–396. doi:10.1016/j.enzmictec.2011.01.002
Yamakawa, S.-i., Yamada, R., Tanaka, T., Ogino, C., and Kondo, A. (2012). Repeated fermentation from raw starch using Saccharomyces cerevisiae displaying both glucoamylase and α-amylase. Enzyme Microb. Technol. 50 (6), 343–347. doi:10.1016/j.enzmictec.2012.03.005
Yang, J., Huang, K., Xu, X., Miao, Y., Lin, Y., and Han, S. (2020). Cell surface display of thermomyces lanuginosus lipase in Pichia pastoris. Front. Bioeng. Biotechnol. 8, 544058. doi:10.3389/fbioe.2020.544058
Yang, N., Yu, Z., Jia, D., Xie, Z., Zhang, K., Xia, Z., et al. (2014). The contribution of Pir protein family to yeast cell surface display. Appl. Microbiol. Biotechnol. 98 (7), 2897–2905. doi:10.1007/s00253-014-5538-5
Yang, S., Lv, X., Wang, X., Wang, J., Wang, R., and Wang, T. (2017). Cell-surface displayed expression of trehalose synthase from Pseudomonas putida ATCC 47054 in Pichia pastoris using Pir1p as an anchor protein. Front. Microbiol. 8, 2583. doi:10.3389/fmicb.2017.02583
Yang, S., Shen, J., Deng, J., Li, H., Zhao, J., Tang, H., et al. (2022). Engineering cell polarization improves protein production in Saccharomyces cerevisiae. Microorganisms 10, 2005. doi:10.3390/microorganisms10102005
Yang, X., Tang, H., Song, M., Shen, Y., Hou, J., and Bao, X. (2019). Development of novel surface display platforms for anchoring heterologous proteins in Saccharomyces cerevisiae. Microb. Cell. Fact. 18 (1), 85. doi:10.1186/s12934-019-1133-x
Ye, M., Ye, Y., Du, Z., and Chen, G. (2021). Cell-surface engineering of yeasts for whole-cell biocatalysts. Bioprocess Biosyst. Eng. 44 (6), 1003–1019. doi:10.1007/s00449-020-02484-5
Yin, W., Wang, X., Liao, Y., Ma, L., Qiao, J., Liu, H., et al. (2022). Encapsulating IM7-displaying yeast cells in calcium alginate beads for one-step protein purification and multienzyme biocatalysis. Front. Bioeng. Biotechnol. 10, 849542. doi:10.3389/fbioe.2022.849542
Ylinen, A., de Ruijter, J. C., Jouhten, P., and Penttilä, M. (2022). PHB production from cellobiose with Saccharomyces cerevisiae. Microb. Cell. Fact. 21 (1), 124. doi:10.1186/s12934-022-01845-x
Yuzbasheva, E. Y., Yuzbashev, T. V., Perkovskaya, N. I., Mostova, E. B., Vybornaya, T. V., Sukhozhenko, A. V., et al. (2015). Cell surface display of Yarrowia lipolytica lipase Lip2p using the cell wall protein YlPir1p, its characterization, and application as a whole-cell biocatalyst. Appl. Biochem. Biotechnol. 175 (8), 3888–3900. doi:10.1007/s12010-015-1557-7
Zhang, Y., Min, Z., Qin, Y., Ye, D.-Q., Song, Y.-Y., and Liu, Y.-L. (2019). Efficient display of Aspergillus Niger β-glucosidase on Saccharomyces cerevisiae cell wall for aroma enhancement in wine. J. Agric. Food Chem. 67 (18), 5169–5176. doi:10.1021/acs.jafc.9b00863
Zhang, Z.-X., Wang, Y.-Z., Xu, Y.-S., Sun, X.-M., and Huang, H. (2021). Developing GDi-CRISPR system for multi-copy integration in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 193 (7), 2379–2388. doi:10.1007/s12010-021-03532-w
Zhao, J.-Z., Xu, L.-M., Liu, M., Cao, Y.-S., LaPatra, S. E., Yin, J.-S., et al. (2017a). An efficient and simple method to increase the level of displayed protein on the yeast cell surface. J. Microbiol. Methods 135, 41–47. doi:10.1016/j.mimet.2017.02.002
Zhao, N., Xu, Y., Wang, K., and Zheng, S. (2017b). Synthesis of isomalto-oligosaccharides by Pichia pastoris displaying the Aspergillus Niger α-glucosidase. J. Agric. Food Chem. 65 (43), 9468–9474. doi:10.1021/acs.jafc.7b04140
Zhao, S., Guo, D., Zhu, Q., Dou, W., and Guan, W. (2021). Display of microbial glucose dehydrogenase and cholesterol oxidase on the yeast cell surface for the detection of blood biochemical parameters. Biosensors 11 (1), 13. doi:10.3390/bios11010013
Zheng, H., Wang, K., Xu, X., Pan, J., Sun, X., Hou, J., et al. (2022). Highly efficient rDNA-mediated multicopy integration based on the dynamic balance of rDNA in Saccharomyces cerevisiae. Microb. Biotechnol. 15 (5), 1511–1524. doi:10.1111/1751-7915.14010
Keywords: Saccharomyces cerevisiae, cell-surface display technology, scaffoldin, bioconversion, cell factory
Citation: Zhang C, Chen H, Zhu Y, Zhang Y, Li X and Wang F (2022) Saccharomyces cerevisiae cell surface display technology: Strategies for improvement and applications. Front. Bioeng. Biotechnol. 10:1056804. doi: 10.3389/fbioe.2022.1056804
Received: 29 September 2022; Accepted: 25 November 2022;
Published: 07 December 2022.
Edited by:
Benyamin Khoshnevisan, University of Southern Denmark, DenmarkReviewed by:
Mingfeng Cao, Xiamen University, ChinaC. French, University of Edinburgh, United Kingdom
Copyright © 2022 Zhang, Chen, Zhu, Zhang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Wang, aGd3ZkBuamZ1LmVkdS5jbg==
 Chenmeng Zhang
Chenmeng Zhang Hongyu Chen1,2,3
Hongyu Chen1,2,3 Yu Zhang
Yu Zhang Fei Wang
Fei Wang