- State key Laboratory of Wheat and Maize Crop Science/Henan International Laboratory for Green Pest Control/College of Plant Protection, Henan Agricultural University, Zhengzhou, China
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a key enzyme in glycolysis, is commonly used as an internal reference gene in humans, mice, and insects. However, the function of GAPDH in insect development, especially in metamorphosis, has not been reported. In the present study, Helicoverpa armigera and Spodoptera frugiperda ovarian cell lines (Sf9 cells) were used as materials to study the function and molecular mechanism of GAPDH in larval metamorphosis. The results showed that HaGAPDH was more closely related to GAPDH of S. frugiperda and Spodoptera litura. The transcript peaks of HaGAPDH in sixth instar larvae were 6L-3 (epidermal and midgut) and 6L-1 (fat body) days, and 20E and methoprene significantly upregulated the transcripts of HaGAPDH of larvae in qRT-PCR. HaGAPDH–GFP–His was specifically localized in mitochondria in Sf9 cells. Knockdown of HaGAPDH by RNA interference (RNAi) in sixth instar larvae resulted in weight loss, increased mortality, and decreases in the pupation rate and emergence rates. HaGAPDH is directly bound to soluble trehalase (HaTreh1) physically and under 20E treatment in yeast two-hybrid, coimmunoprecipitation, and colocalization experiments. In addition, knockdown of HaGAPDH increased the Treh1 activity, which in turn decreased the trehalose content but increased the glucose content in larvae. Therefore, these data demonstrated that GAPDH controlled the glucose content within the normal range to ensure glucose metabolism and metamorphosis by directly binding with HaTreh1.
1 Introduction
The best-known function of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is serving as the housekeeping gene. GAPDH is widely used as an internal reference in real-time PCR and Western blot (Chapman and Waldenstrom, 2015; Nie et al., 2017). In fact, GAPDH catalyzes glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate (1,3-BPG), the first reaction that consumes some energy in glucose after several steps without energy production in glycolysis (Yang et al., 2018). Therefore, the multifunctional enzyme GAPDH acts as the key enzyme in glycolysis and the center of glucose metabolism (Kornberg et al., 2018).
In humans, GAPDH has been shown to be involved in a variety of pathologies, such as diabetes (Yego and Mohr 2010) and several types of cancer (Zhou et al., 2008; Colell et al., 2009). For instance, Müller cells are a crucial component of the retinal tissue and maintain the blood–retinal barrier, and diabetes usually caused the loss of Müller cells. In the diabetic retina of rats, nuclear accumulation of GAPDH caused the loss of Müller cells (Kusner et al., 2004). GAPDH is also considered a new potential therapeutic target for cardiac disease states characterized by oxidative stress. Knockdown of GAPDH by siRNA in the H9C2 cardiomyoblasts decreased oxidant stress and cell apoptosis, enhanced autophagy in rotenone-induced H9C2 cells, increased antioxidant pathways, and preserved cell energy (Liang et al., 2015). Colony-stimulating factor 1 (CSF1), a macrophage cytokine, is strongly correlated with poor prognosis in ovarian cancer patients. Also, serum CSF-1 was used as a sensitive tumor marker, its elevated levels heralding disease recurrence or progression. In the human ovarian surface epithelial cell line, GAPDH stabilized the transcript and enhanced CSF1 protein levels by binding to CSF1 mRNA. The half-lives of CSF-1 mRNA were decreased by 50% in the presence of GAPDH siRNA (Zhou et al., 2008).
Research studies on GAPDH in insects are less than those of humans and mice. In insects, GAPDH is almost used as the reference gene. GAPDH was the development stage reference gene in Rhopalosiphum padi (Li et al., 2021), the tissue reference gene in Lymantria dispar (Yin et al., 2020), the reference gene in Apolygus lucorum for different sexes and entomopathogen infection studies (Luo et al., 2020), and the development profile study reference gene in Tuta absoluta (Yan et al., 2021). In Helicoverpa armigera, GAPDH was used as the reference gene for nuclear polyhedrosis viral infection (Zhang et al., 2015). In insects, GAPDH has other functions in addition to the reference gene. In Laodelphax striatellus, GAPDH is involved in resistance to autophagy induced by rice black-streaked dwarf virus (RBSDV) infection. RBSDV interacts with LsGAPDH in vivo and in vitro. Knockdown of LsGAPDH significantly reduces RBSDV infection-induced autophagy. RBSDV promotes the phosphorylation of AMPK and then leads to the phosphorylation and translocation of GAPDH from the cytoplasm to the nucleus, and phosphorylated GAPDH activates autophagy to inhibit RBSDV infection (Wang et al., 2022). Bisphenol S (BPS) is the industrial alternative to the endocrine disruptor bisphenol A (BPA).
Glycolysis starts with glucose (Zhong et al., 2018). In insects, trehalose is the blood sugar (Tatun N et al., 2008). The hydrolysis of trehalose to glucose, which is catalyzed by trehalase (Treh), is necessary for insect life activities. As the only known enzyme that irreversibly decomposes trehalose into glucose, Treh has two forms including soluble trehalase (soluble trehalase, Treh1) and membrane-bound trehalase (membrane-bound trehalase, Treh2), which are mainly classified by the transmembrane structure at the C-terminus (Shukla et al., 2015). Treh1 is crucial for insects. In the larvae midgut of Omphisa fuscidentalis, the Treh1 activity accounts for most of the total Treh activity, and 20-hydroxyecdysone (20E) increases Treh1 transcripts and the enzymatic activity, thus ensuring the normal development of the larvae (Tatun et al., 2008). In Spodoptera exigua, dsTreh1-injected larvae had a 50% mortality rate and molting failure (Chen et al., 2010). Knockdown of NlTreh1 in N. lugens caused larval abnormities in molting and wings (Tang et al., 2017). In Tribolium castaneum, RNAi of Treh1a resulted in a mortality rate of up to 30% (Tang et al., 2016). In Leptinotarsa decemlineata, RNAi of Treh1a induced a mortality rate of up to 80% (Shi et al., 2016). In H. armigera, knockdown of Treh1 in the larvae led to weight loss and mortality increased. 20E enhanced the direct binding of HaTreh1 to HaATPs-α and controlled the production of ATP, thus affecting the growth and development of larvae (Chang et al., 2022).
In our previous study, HaGAPDH was identified as a HaTreh1-binding protein by a yeast two-hybrid (Y2H) screening library experiment (Chang et al., 2022). Considering the key roles of HaTreh1 in molting and metamorphosis of H. armigera, HaGAPDH in glycolysis, and HaTreh1 in glucose production, we proposed the following questions, what is the relationship between these two enzymes HaTreh1 and HaGAPDH? In addition to serving as an internal reference gene in H. armigera larvae, does GAPDH have other functions, and is it involved in larvae metamorphosis similar to HaTreh1? To answer the aforementioned scientific questions, quantitative real-time PCR (qRT-PCR) was performed to study the expression profile of HaGAPDH, RNA interference was used to explore the function of HaGAPDH in larval metamorphosis, and Y2H and coimmunoprecipitation (Co-IP) experiments were performed to prove the interaction between HaGAPDH and HaTreh1. We explored the function and mechanism of HaGAPDH in larval metamorphosis and provided experimental evidence and theoretical basis for targeting GAPDH for pest control in the future.
2 Materials and methods
2.1 Materials
2.1.1 Insects
Helicoverpa armigera eggs were purchased from the Baiyun Industrial Company (Baiyun, Jiyuan, China) and reared in a laboratory using an artificial diet at 26°C, 75% humidity, and a 14L: 10D photoperiod (Zhao et al., 2005).
2.1.2 Cell culture
The Spodoptera frugiperda ovarian cell line (Sf9 cells) was cultured at 28°C with the Sf-900 II serum-free medium, which contains 10% fetal bovine serum (Gibco) and 0.5% penicillin–streptomycin liquid (HyClone).
2.1.3 Compound
Ecdysterone was purchased from the Solarbio Company (Solarbio, Beijing, China) and dissolved with DMSO. Methoprene was purchased from the MedChemExpress Company (MCE, Princeton, United States) and dissolved with DMSO.
3. Methods
3.1 Bioinformatic analysis
The open reading frame (ORF) of HaGAPDH (LOC110377691) was amplified by specific primers (Table 1) and sequenced by the Tsingke Biotechnology Company (Tsingke, Beijing, China). The phylogenetic tree was constructed by the neighbor-joining (NJ) method with MEGA 11.0 software.
3.2 Quantitative real-time PCR (qRT-PCR)
The TRIzol method and HiScript III RT SuperMix for qPCR (+gDNA wiper) (R323–01, Vazyme) were used to get the total RNA and cDNA, respectively. Then, qRT-PCR was performed by using the ChamQ Universal SYBR qPCR Master Mix (Q711-02-03) (Vazyme) and a real-time qPCR instrument (Eppendorf). There were 10 μl 2 × ChamQ Universal SYBR qPCR Master Mix, 0.4 μl forward primer, 0.4 μl reverse primer, 1 μl cDNA template, and 8.2 μl ddH2O in the qRT-PCR system. The reaction program was as follows: 95°C (5 min), followed by 40 cycles of 95°C (15 s) and 60°C (20 s). Two reference genes Ha18s (AB620126.1, product size 230 bp, PCR efficiency: 0.97) and Ha-β-Actin (EU527017.1, product size 144 bp, PCR efficiency: 0.979) were selected to normalize the expression of HaGAPDH. The PCR efficiency of HaGAPDH primers is 1.07 (product size 204 bp). Then, three larvae were collected as a single sample, and three biological replicates and three technical replicates were performed for each experiment. The Student’s t-test or Tukey test was used to compare the significant differences.
3.3 Hormone treatment
The sixth instar larvae about 2 h after molting were selected, and there were six larvae in each group. The control group was injected with 5 μl DMSO (diluted to 1:10,000 with 1 × PBS) per larvae, and the experimental group was injected with 5 μl 20E (1.2 μg) or 5 μl methoprene (0.5 μg) per larvae. Then, the epidermis and fat body were collected at 0, 1, 3, and 6 h after hormone injection. The experiments were performed in three biological replicates.
3.4 Subcellular localization
3.4.1 Subcellular localization of HaGAPDH
The ORF (without stop codon) sequence of HaGAPDH was ligated into the GFP-pIEx vector to construct HaGAPDH-GFP-pIEx using specific primers (Table 1). Then, the HaGAPDH-GFP-pIEx plasmid was transfected into the Sf9 cells using the FuGENE HD Transfection Reagent (4 μl of the Transfection Reagent per 1 μg plasmid) (Promega, E2311) referred to the previous description (Blochlinger and Diggelmann, 1984). The Sf9 cells transfected with a GFP-pIEx plasmid were used as control. After 24 h, the LSM710 laser confocal microscope (Zeiss) was used to observe the green fluorescence and take pictures.
3.4.2 The mitochondrial localization
The Sf9 cells that transfected with the HaGAPDH-GFP-pIEx plasmid for 24 h were incubated with Mito-Tracker Red CMXRos (C1049B, Beyotime, 50 nM) at 37°C for 15 min. Then, fluorescence images were photographed using an LSM710 laser confocal microscope (Zeiss).
3.4.3 Colocalization
The HaGAPDH-GFP-pIEx and HaTreh1-RFP-pIEx plasmids were co-transfected into Sf9 cells. About 48 h later, the green and red fluorescence were observed and photographed using an LSM710 laser confocal microscope (Zeiss).
3.5 RNA interference
The DNA templates for dsHaGAPDH (507 bp) and dsGFP (MN623123, 420 bp, the control) synthesis were amplified by PCR using specific primers containing the T7 promoter core sequence (Table 1). Subsequently, dsRNA was synthesized using the MEGAscript RNAi kit (Thermo Fisher Scientific), and the quality of dsRNA was detected using a BioPhotometer (Eppendorf) and by agarose electrophoresis. The sixth instar larvae about 2 h after molting were selected, and there were 30 larvae in each group. The control group was injected with 5 μg dsGFP per larvae, and the experimental group was injected with 5 μg dsHaGAPDH. After 24 h, the abovementioned injection was repeated one time. The larval weights were recorded at 0, 24, 48, 72, 96, and 144 h after dsRNA injection, and the mortality rate, pupation rate, and emergence rate were also recorded. These experiments were performed in three biological replicates. The Student’s t-test was used to compare the significant differences.
3.6 Yeast two-hybrid
The HaGAPDH-pGADT7 and HaTreh1-pGBKT7 plasmids were constructed using specific primers (Table 1). Then, HaGAPDH-pGADT7 and HaTreh1-pGBKT7 were co-transformed into the AH109 yeast competent cells, according to the methods described previously (Wang et al., 2020), and then, the yeast was spread on the SD-TL, SD-THL, SD-THLA, and SD-TLHA + x-α- gal plates. The aforementioned plates were cultured at 37°C for 3 days. The AH109 cells that co-transformed pGBKT7-P53 and pGADT7-LargeT were the positive control, and the AH109 cells that co-transformed pGBKT7-laminC and pGADT7-LargeT were the negative control.
3.7 Coimmunoprecipitation
The HaGAPDH-GFP-pIEx and HaTreh1-RFP-pIEx plasmids were co-transfected into Sf9 cells (4 μg each plasmid for 7.2 × 106 cells). After 24 h, the cells were treated with 20E (1 μM) for 3 h. Then, the cells were collected on ice using the radio immunoprecipitation assay (RIPA) lysis buffer. A part of the solution was taken out as the input, and the remaining solution was used to purify GFP-tagged proteins by using the GFP-tag IP/Co-IP Kit (Bio LinkedIn). Finally, the protein samples were subjected to Western blot experiments.
3.8 Endogenous substance content measurement
The sixth instar larvae about 2 h after molting were selected, and there were six larvae in each group. The control group was injected with 5 μg dsGFP per head, and the experimental group was injected with 5 μg dsHaGAPDH, and then, the midguts were collected at 24 h after dsRNA injection. Then, the soluble trehalase activity, trehalose content, and glucose content were measured according to the instructions of the trehalase activity assay kit (A150-1-1, Nanjing Jiancheng), trehalose content assay kit (G0552W, Grace), and glucose content assay kit (G0504W, Grace), respectively. Three biological replicates were performed, and each biological replicate included three technical replicates. The Student’s t-test was used to compare the significant differences.
4 Results
4.1 Bioinformatic analysis of the HaGAPDH sequence
The length of HaGAPDH ORF is 525 bp and encodes 174 amino acid residues (Figure 1A). Phylogenetic analysis of HaGAPDH sequences in 30 species of insects, including Lepidoptera, Hemiptera, Diptera, and Coleoptera, showed that the HaGAPDH sequence of H. armigera was more closely related to those of Spodoptera frugiperda and Spodoptera litura (Figure 1B).
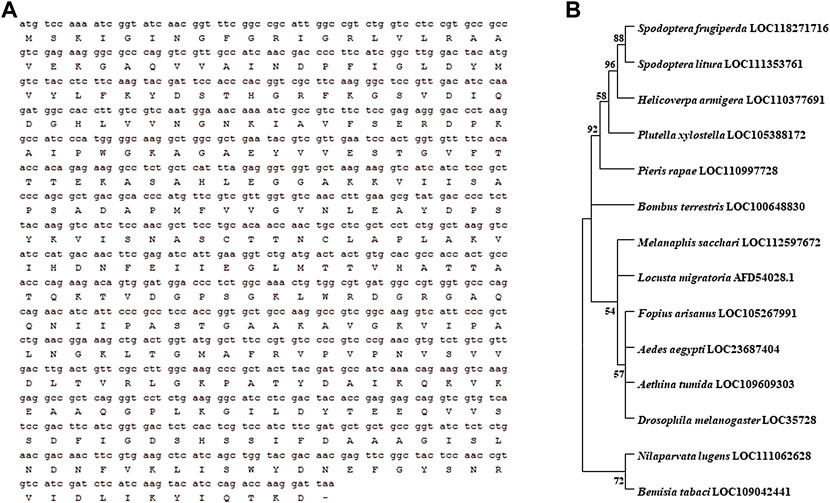
FIGURE 1. Bioinformatic analysis of the HaGAPDH sequence. (A) ORF sequence and the amino acid sequence of HaGAPDH. (B) Phylogenetic tree analysis of GAPDH. The tree was constructed by MEGA 11.0 software.
4.2 The expression profile of HaGAPDH
To study the function of HaGAPDH in larval development, its expression pattern was analyzed first. The results of qRT-PCR showed that HaGAPDH was expressed in all three tissues, namely, the epidermis, midgut, and fat body (Figures 2A–C). Also, during the period from the molting stage of the fifth instar (5L-M) to the fifth day of the sixth instar (6L-5), the transcript peaks of HaGAPDH were 6L-3 days in the epidermis (Figure 2A), 6L-1 and 6L-3 days in the midgut (Figure 2B), and 6L-1 day in the fat body (Figure 2C), respectively. Furthermore, the hormone treatment experimental data showed that both 20E and methoprene upregulated the transcript levels of HaGAPDH in the epidermis and fat body (Figures 2D–G).
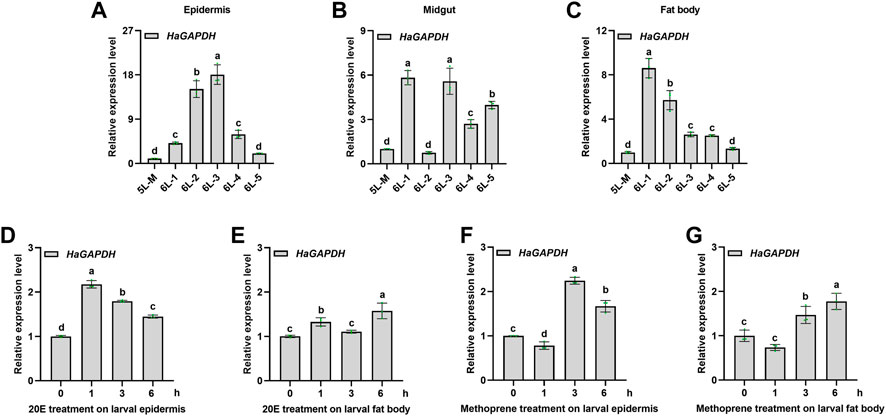
FIGURE 2. Expression profile of HaGAPDH. (A–C) Transcript analyses of HaGAPDH in the epidermis (A), midgut (B), and fat body (C) from 5L-M to 6L-5 days by qRT-PCR. 5L-M: the molting stage of the fifth instar larvae, 6L-1: the first day of the sixth instar larvae, (D) and (E) the transcript changes of HaGAPDH in the epidermis (D) and fat body (E) under the 20E treatment. Larvae were treated with 1.2 μg 20E (5 μl) or DMSO (5 μl) for 1, 3, and 6 h (F) and (G) the transcript changes of HaGAPDH in the epidermis (F) and fat body (G) under methoprene treatment. Larvae were treated with 0.5 μg methoprene (5 μl) or DMSO (5 μl) for 1, 3, and 6 h. Ha18s and β-actin were used as the reference genes. The error bars indicated the mean ± s.d. of three independent biological experiments and three technical repetitions. Different letters indicate significant differences at the p < 0.05 level using the Tukey test.
4.3 HaGAPDH localized in mitochondria
To explore the organelle in which HaGAPDH works, the subcellular localization of HaGAPDH in the Sf9 cells was studied. Compared to the GFP control that localized both in the cytoplasm and nucleus, the green fluorescence of the HaGAPDH-GFP protein was distributed in the cytoplasm (Figure 3A). The Western blot also proved the successful expression of GFP and HaGAPDH-GFP proteins in Sf9 cells (Figure 3B). Further mitochondrial staining experiments proved that the green fluorescence of the HaGAPDH-GFP protein specifically overlapped with the red fluorescence of mitochondria, which suggested the mitochondrial distribution of HaGAPDH-GFP (Figure 3C).
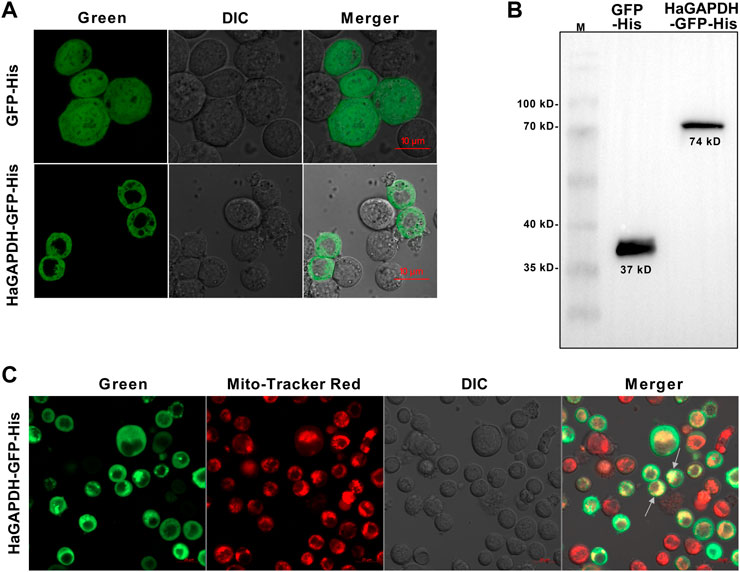
FIGURE 3. Mitochondrial localization of HaGAPDH. (A) Subcellular localization of HaGAPDH-GFP-His in Sf9 cells using an LSM710 confocal microscope. GFP-His was the control. The merger was the overlap of the green and bright fluorescence. (B) Expression of GFP-His (37 kD) and HaGAPDH-GFP-His (74 kD) in Sf9 cells by Western blot. M: protein marker. (C) HaGAPDH–GFP–His merged with mitochondria. The pictures were taken by using an LSM710 microscope. HaGAPDH–GFP–His overexpression cells were dyed with Mito-Tracker (50 nM) for 15 min. The merger was the overlap of the green fluorescence, Mito-Tracker red, and bright.
4.4 Knockdown of HaGAPDH caused the larval development defect
RNAi was employed to specifically analyze the function of HaGAPDH in larval development. Compared with the dsGFP control, after the knockdown of HaGAPDH, the growth and development of dsHaGAPDH-injected larvae were abnormal (Figures 4A,B). In the dsHaGAPDH-injected group, the larval body weights were markedly reduced at 48, 72, 96, and 144 h after dsRNA injection (Figure 4C), and the final mortality rate was significantly increased (Figure 4D). In addition, the pupation rate and emergence rate of the experimental group were both significantly decreased (Figures 4E,F). The aforementioned data suggested the necessary role of HaGAPDH in larval development and metamorphosis.
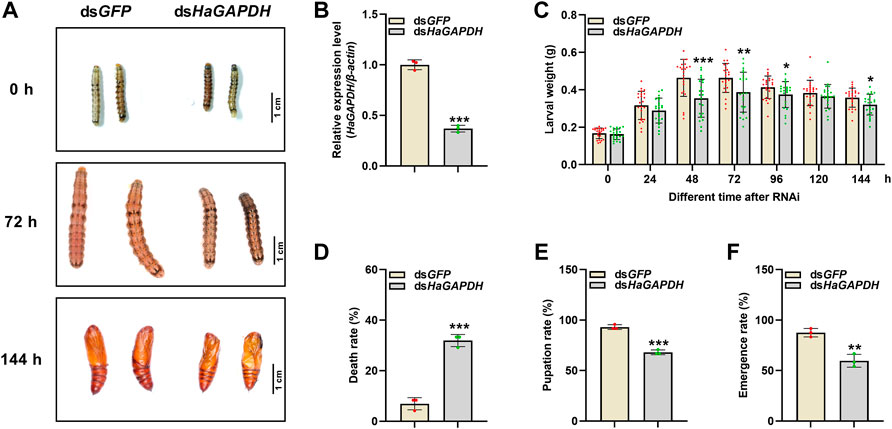
FIGURE 4. Effects on the sixth instar larvae after knockdown of HaGAPDH (A) and (B) the knockdown phenotypes (A) and efficiency (B) of HaGAPDH on the sixth instar larvae. The larvae were injected with 5 μg dsHaGAPDH or dsGFP (the control); then, photographs were taken at different timepoints, and midguts were collected for qRT-PCR at 24 h. β-actin was used as the reference gene. Error bars indicated the mean ± s.d. of three independent biological experiments and three technical repetitions. ***p < 0.001; Student’s t-test. (C) Larval weight changes after being injected with dsRNA for 24, 48, 72, 96, 120, and 144 h. There were 30 larvae in each group, and three independent biological experiments were performed. Each dot represented a repetition, and the error bars indicated mean ± SE. *p < 0.05, **p < 0.01, and ***p < 0.001; Student’s t-test. (D–F) Statistical analyses of the death rate (D), pupation rate (E), and emergence rate (F) after dsRNA injection. There were 30 larvae in each group. The error bars indicated the mean ± s.d. of three independent biological experiments. **p < 0.01 and ***p < 0.001; Student’s t-test.
4.5 HaGAPDH directly bound with HaTreh1
To further analyze the functional mechanism of HaGAPDH, a series of experiments were performed to identify the binding protein of HaGAPDH. HaGAPDH was proven as the HaTreh1 binding protein in the previous Y2H library screening. Therefore, the Y2H point-to-point experiment was performed. Also, the results showed that the growth of yeast co-transfected with pGADT7-HaGAPDH and pGBKT7-HaTreh1 was consistent with that of the positive control pGADT7-LargeT + pGBKT7-P53 and was opposite to that of the negative control pGADT7-LargeT + pGBKT7-laminC (Figure 5A), indicating the direct binding of HaGAPDH and HaTreh1. The Co-IP experiment data proved the direct binding of HaGAPDH and HaTreh1 both in the control and under the 20E treatment (Figure 5B). The fluorescence photographs showed that the red fluorescence of HaTreh1-RFP overlapped the green fluorescence of HaGAPDH-GFP, thus showing yellow fluorescence (Figure 5C), which suggested the colocalization of HaGAPDH and HaTreh1. Accordingly, we inferred that HaGAPDH is directly bound with HaTreh1.
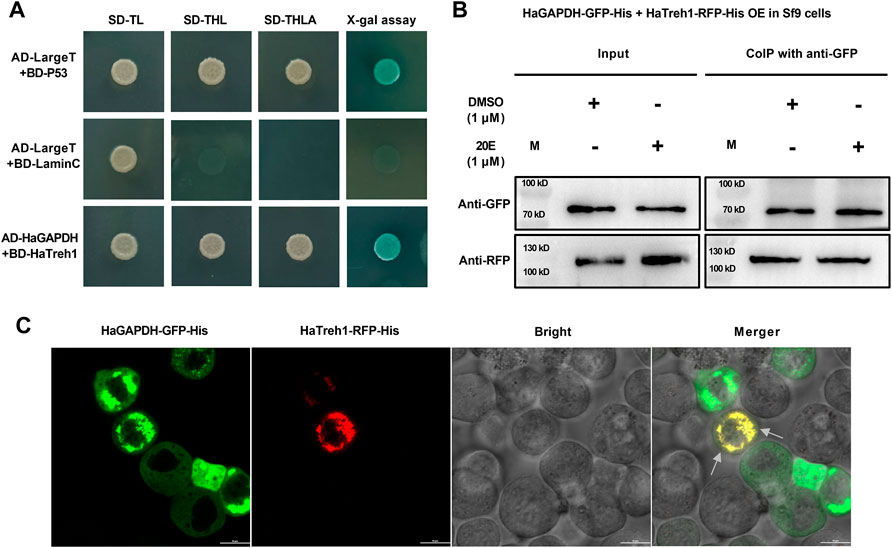
FIGURE 5. HaGAPDH bound to HaTreh1. (A) Y2H demonstrated the direct binding of BD-HaTreh1 and AD-HaGAPDH. AD-LargeT + BD-P53 was the positive control, and AD-LargeT + BD-laminC was the negative control. (B) Co-IP experiment proved the interaction between HaTreh1–RFP–His (103 kD) and HaGAPDH–GFP–His (74 kD) under the 20E treatment in Sf9 cells. M: protein marker. (C) Colocalization of HaTreh1–RFP–His and HaGAPDH–GFP–His in Sf9 cells by using the LSM710 microscope. The merger was the overlap of red and green fluorescence. The arrow indicates the overlapped yellow color.
4.6 Knockdown of HaGAPDH changed the Treh1 activity and glucose content
Considering the binding of HaGAPDH and HaTreh1, the corresponding endogenous sugar contents in larvae were measured. After the knockdown of HaGAPDH by RNAi, the enzymatic activity of HaTreh1 in the midgut was significantly increased by contrast with the dsGFP control (Figure 6A), and the trehalose content decreased but glucose increased correspondingly (Figures 6B,C).
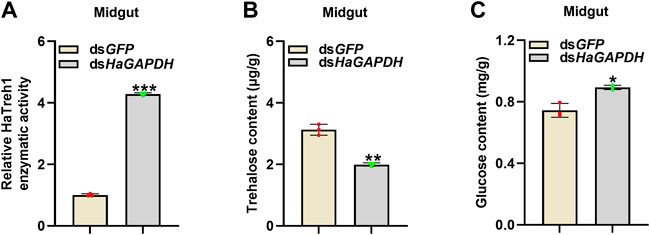
FIGURE 6. HaGAPDH controlled the Treh1 activity and sugar contents. (A–C) Treh1 activity (A), trehalose content (B), and glucose content (C) in the midguts after being injected with dsGFP (5 μg) or dsHaGAPDH (5 μg) for 24 h. Error bars indicated the mean ± s.d. of three independent biological experiments and three technical repetitions. *p < 0.05, **p < 0.01, and ***p < 0.001; Student’s t-test.
5 Discussion
Glucose is the main source of energy for insect life. Therefore, GAPDH, a key enzyme in glycolysis, plays a crucial role in insects. In the present study, we proved that HaGAPDH plays an important role in maintaining the growth and promoting normal metamorphosis of H. armigera larvae. The transcript level of HaGAPDH was upregulated by 20E, and the knockdown of HaGAPDH by RNAi resulted in decreased larval weight, increased mortality, reduced pupation rate, and emergence rate. GAPDH controlled the Treh1 activity and glucose content of larvae by directly binding to HaTreh1. Our results revealed the critical function and molecular mechanism of GAPDH in larval metamorphosis.
5.1 Glyceraldehyde-3-phosphate dehydrogenase is crucial for larval development and metamorphosis
GAPDH has been considered the housekeeping gene in various species including humans, mice, and insects (Chapman and Waldenstrom, 2015; Nie et al., 2017). However, studies in humans have showed that the mRNA level of GAPDH in different tissues is different. The mRNA expression of GAPDH in the skeletal muscle was the highest in 72 human tissues, and GAPDH had a high expression trend in the high-energy-requirement tissues (Barber et al., 2005). In the present study, our results also proved that the transcript level peaks of HaGAPDH were attained in 6L-3 days in the larval epidermis and midgut, the period with high energy consumption (Figure 2). However, the transcripts of HaGAPDH in the fat body declined from 6L-1 and were the lowest from 6L-3 to 6L-5 days, with the pre-pupation period and huge energy demand (Figure 2). Since the fat body is the tissue that biosynthesizes trehalose and stores glycogen, the HaGAPDH expression pattern is reasonable for glycogen storage in the fat body. The transcript peaks of HaGAPDH in the epidermis and midgut appeared at 6L-3 (Figure 2) when the 20E titers increased in H. armigera larvae (Kang et al., 2019). The 20E treatment upregulated the transcripts of HaGAPDH (Figure 2), which is consistent with its expression peak at 6L-3 (Figure 2). It also suggested the potential role of HaGAPDH in larval metamorphosis. 20E is a steroid hormone (Dubrovsky, 2005), and our results are consistent with the fact that the steroid hormone estradiol increased GAPDH transcripts in the endometrium of ovariectomized ewes (Ing and Zhang, 2004). In addition, we also found that methoprene, an important hormone regulating larval growth and metamorphosis (Dubrovsky, 2005), also upregulated the transcripts of HaGAPDH (Figure 2). This dynamically regulated expression pattern of GAPDH is similar to the studies in other species (Jacob et al., 2013. In human ovarian cell lines HOSE6-3 and HOSE17-1, the GAPDH transcript is highly dynamic (Chao et al., 1990; Graven et al., 1994; Polati et al., 2012; Jacob et al., 2013). Northern blot results in monolayers of human WI-38/va13 cells, several cancer cells, and monkey cos-1 cells revealed that GAPDH transcripts were upregulated by the calcium ionophore A23187 (Chao et al., 1990). The GAPDH mRNA levels were increased under hypoxia treatment in ovine endothelial cells (Graven et al., 1994). In mouse bone marrow macrophages, GAPDH expression was upregulated by ferric ammonium citrate (Polati et al., 2012).
More importantly, further RNAi results proved the key role of HaGAPDH in larval development and metamorphosis (Figure 4). The larval phenotypes in the dsHaGAPDH-injected larvae were similar to those of important genes in the 20E pathway, such as the ecdysone receptor (EcR). Decreased EcR transcripts by RNAi in Apolygus lucorum, Leptinotarsa decemlineata, Nilaparvata lugens, and Tribolium castaneum led to molting defects and larval death (Tan and Palli, 2008; Wu et al., 2012; Tan et al., 2015; Xu et al., 2020). The transcript peaks, the upregulated transcript level by 20E, the larval metamorphosis defects after RNAi, and the previously mentioned results all fully demonstrated that HaGAPDH indeed plays an important role in larval metamorphosis. Our study is the first report on the function of GAPDH in insect metamorphosis.
5.2 Glyceraldehyde-3-phosphate dehydrogenase controls the glucose content in larvae by directly binding with HaTreh1
In the present study, our data proved that HaGAPDH is the mitochondria protein (Figure 3). This is the first evidence on the mitochondrial localization of GAPDH in insects. There are lots of reports about the sublocalization of GAPDH in human and mouse cells. For instance, the non-tetrameric forms of GAPDH localized in the nucleus of HeLa cells (Arutyunova et al., 2003). In HEK293 cells, GAPDH accumulated in the nucleus during apoptosis (Hara et al., 2005). In rat insulinoma INS-1 cells, GAPDH primarily showed cytoplasm localization (Park et al., 2009). The mitochondria-localized HaGAPDH was similar to those of PC12 cells and mice. Rotenone induces PC12 cell apoptosis and the translocation to mitochondria of GAPDH (Huang et al., 2009). GAPDH targets the mitochondria in the mouse heart (Kohr et al., 2014).
For the first time, HaGAPDH was proven as the HaTreh1 binding protein, and this binding also exists under 20E treatment (Figure 3). HaTreh1, a key enzyme for trehalose hydrolysis, directly binds to HaATPs-α which controls ATP production (Chang et al., 2022). HaGAPDH is another protein that directly binds to HaTreh1 in the present study. In a previous study, HaTreh1 has been reported to be working in the mitochondria (Chang et al., 2022). Also, the combination between HaGAPDH and HaTreh1 was consistent with the mitochondrial localization of HaGAPDH. There have been many studies on the binding protein of GAPDH. In HEK293 cells, GAPDH bounds to Sirtuin 1 (SIRT1), a nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylase (Chang et al., 2022). GAPDH was demonstrated as tubulin-binding protein (Tisdale et al., 2009; Landino et al., 2014). Similarly, GAPDH is also bound to actin (Waingeh et al., 2004). In mouse cell lines, GAPDH interacts with lactoferrin (Chauhan et al., 2015).
Interestingly, the knockdown of HaGAPDH increased the HaTreh1 activity and glucose content but decreased the trehalose content in larvae (Figure 6). Accordingly, we inferred that the activity of HaTreh1 was inhibited after interaction with HaGAPDH. The aforementioned data were reasonable for the phenotypes of dsHaGAPDH-injected larvae (Figure 4). After knockdown of the key enzyme in glycolysis HaGAPDH, the interaction between GAPDH and Treh1 led to an increase in the enzymatic activity of Treh1, thus hydrolyzing more trehalose into glucose; however, the dsHaGAPDH-injected larvae cannot use these sugar sources; finally, the imbalance of glucose metabolism led to the death and pupation failure of larvae. From this, we inferred that during the normal growth of larvae, GAPDH ensures the glucose content is at a normal level by interacting with Treh1 and controlling its enzymatic activity. This regulation model can prevent larvae from being unable to complete the process of sugar decomposition and energy production for too much glucose and ensure the development of larvae.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization: WZ and SA. Data curation: BZ and WZ. Formal analysis: BZ. Funding acquisition: WZ. Investigation: BZ, ZG, and YC. Methodology: JW. Writing—original draft: WZ. Writing—review and editing: WZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the Key Scientific Research Projects of Universities in Henan Province (22A210005), the Key R&D and technology promotion projects in Henan Province (222102110063), and the Henan Province Young Talent Support Project (2022HYTP030). National Natural Science Foundation of China (31970472), Henan Postgraduate Quality Course (HNYJS2020KC16).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arutyunova, E. I., Danshina, P. V., Domnina, L. V., Pleten, A. P., and Muronetz, V. I. (2003). Oxidation of glyceraldehyde-3-phosphate dehydrogenase enhances its binding to nucleic acids. Biochem. Biophys. Res. Commun. 307, 547–552. doi:10.1016/s0006-291x(03)01222-1
Barber, R. D., Harmer, D. W., Coleman, R. A., and Clark, B. J. (2005). GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics 21, 389–395. doi:10.1152/physiolgenomics.00025.2005
Blochlinger, K., and Diggelmann, H. (1984). Hygromycin B phosphotransferase as a selectable marker for DNA transfer experiments with higher eucaryotic cells. Mol. Cell. Biol. 4, 2929–2931. doi:10.1128/mcb.4.12.2929-2931.1984
Chang, Y., Zhang, B., Du, M., Geng, Z., Wei, J., Guan, R., et al. (2022). The vital hormone 20-hydroxyecdysone controls ATP production by upregulating binding of trehalase 1 with ATP synthase subunit α in Helicoverpa armigera. J. Biol. Chem. 298, 101565. doi:10.1016/j.jbc.2022.101565
Chao, C. C., Yam, W. C., and Lin-Chao, S. (1990). Coordinated induction of two unrelated glucose-regulated protein genes by a calcium ionophore: Human BiP/GRP78 and GAPDH. Biochem. Biophys. Res. Commun. 171, 431–438. doi:10.1016/0006-291x(90)91411-k
Chapman, J. R., and Waldenström, J. (2015). With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS One 10, e0141853. doi:10.1371/journal.pone.0141853
Chauhan, A. S., Rawat, P., Malhotra, H., Sheokand, N., Kumar, M., Patidar, A., et al. (2015). Secreted multifunctional Glyceraldehyde-3-phosphate dehydrogenase sequesters lactoferrin and iron into cells via a non-canonical pathway. Sci. Rep. 5, 18465. doi:10.1038/srep18465
Chen, J., Tang, B., Chen, H., Yao, Q., Huang, X., Chen, J., et al. (2010). Different functions of the insect soluble and membrane-bound trehalase genes in chitin biosynthesis revealed by RNA interference. PLoS One 5, e10133. doi:10.1371/journal.pone.0010133
Colell, A., Green, D. R., and Ricci, J. E. (2009). Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 16 (12), 1573–1581. doi:10.1038/cdd.2009.137
Dubrovsky, E. B. (2005). Hormonal cross talk in insect development. Trends Endocrinol. Metab. 16 (1), 6–11. doi:10.1016/j.tem.2004.11.003
Graven, K. K., Troxler, R. F., Kornfeld, H., Panchenko, M. V., and Farber, H. W. (1994). Regulation of endothelial cell glyceraldehyde-3-phosphate dehydrogenase expression by hypoxia. J. Biol. Chem. 269 (39), 24446–24453. doi:10.1016/s0021-9258(19)51104-8
Hara, M. R., Agrawal, N., Kim, S. F., Cascio, M. B., Fujimuro, M., Ozeki, Y., et al. (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674. doi:10.1038/ncb1268
Huang, J., Hao, L., Xiong, N., Cao, X., Liang, Z., Sun, S., et al. (2009). Involvement of glyceraldehyde-3-phosphate dehydrogenase in rotenone-induced cell apoptosis: Relevance to protein misfolding and aggregation. Brain Res. 1279, 1–8. doi:10.1016/j.brainres.2009.05.011
Ing, N. H., and Zhang, Y. (2004). Cell-specific expression of estrogen-responsive genes in the uteri of cyclic, early pregnant and ovariectomized ewes. Theriogenology 62, 403–414. doi:10.1016/j.theriogenology.2003.10.017
Jacob, F., Guertler, R., Naim, S., Nixdorf, S., Fedier, A., Hacker, N. F., et al. (2013). Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS One 8, e59180. doi:10.1371/journal.pone.0059180
Kang, X. L., Zhang, J. Y., Wang, D., Zhao, Y. M., Han, X. L., Wang, J. X., et al. (2019). The steroid hormone 20-hydroxyecdysone binds to dopamine receptor to repress lepidopteran insect feeding and promote pupation. PLoS Genet. 15, e1008331. doi:10.1371/journal.pgen.1008331
Kohr, M. J., Murphy, E., and Steenbergen, C. (2014). Glyceraldehyde-3-phosphate dehydrogenase acts as a mitochondrial trans-S-nitrosylase in the heart. PLoS One 9, e111448. doi:10.1371/journal.pone.0111448
Kornberg, M. D., Bhargava, P., Kim, P. M., Putluri, V., Snowman, A. M., Putluri, N., et al. (2018). Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453. doi:10.1126/science.aan4665
Kusner, L. L., Sarthy, V. P., Mohr, S., Domb, A. J., and Frucht-Pery, J. (2004). Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase: A role in high glucose-induced apoptosis in retinal müller cells. Invest. Ophthalmol. Vis. Sci. 45 (5), 2543–1561. doi:10.1167/iovs.03-1294
Landino, L. M., Hagedorn, T. D., and Kennett, K. L. (2014). Evidence for thiol/disulfide exchange reactions between tubulin and glyceraldehyde-3-phosphate dehydrogenase. Cytoskelet. Hob. 71, 707–718. doi:10.1002/cm.21204
Li, M., Li, X., Wang, C., Li, Q., Zhu, S., Zhang, Y., et al. (2021). Selection and validation of reference genes for qRT-PCR analysis of Rhopalosiphum padi (Hemiptera: Aphididae). Front. Physiol. 12, 663338. doi:10.3389/fphys.2021.663338
Liang, S., Figtree, G., Aiqun, M., and Ping, Z. (2015). GAPDH-knockdown reduce rotenone-induced H9C2 cells death via autophagy and anti-oxidative stress pathway. Toxicol. Lett. 234, 162–171. doi:10.1016/j.toxlet.2015.02.017
Luo, J., Wang, A., Cheng, Y., Rong, H., Guo, L., Peng, Y., et al. (2020). Selection and validation of suitable reference genes for RT-qPCR analysis in Apolygus lucorum (Hemiptera: Miridae). J. Econ. Entomol. 113, 451–460. doi:10.1093/jee/toz301
Nie, X., Li, C., Hu, S., Xue, F., Kang, Y. J., and Zhang, W. (2017). An appropriate loading control for Western blot analysis in animal models of myocardial ischemic infarction. Biochem. Biophys. Rep. 12, 108–113. doi:10.1016/j.bbrep.2017.09.001
Park, J., Han, D., Kim, K., Kang, Y., and Kim, Y. (2009). O-GlcNAcylation disrupts glyceraldehyde-3-phosphate dehydrogenase homo-tetramer formation and mediates its nuclear translocation. Biochimica Biophysica Acta - Proteins Proteomics 1794, 254–262. doi:10.1016/j.bbapap.2008.10.003
Polati, R., Castagna, A., Bossi, A. M., Alberio, T., De Domenico, I., Kaplan, J., et al. (2012). Murine macrophages response to iron. J. Proteomics 76, 10–27. doi:10.1016/j.jprot.2012.07.018
Shi, J. F., Xu, Q. Y., Sun, Q. K., Meng, Q. W., Mu, L. L., Guo, W. C., et al. (2016). Physiological roles of trehalose in Leptinotarsa larvae revealed by RNA interference of trehalose-6-phosphate synthase and trehalase genes. Insect biochem. Mol. Biol. 77, 52–68. doi:10.1016/j.ibmb.2016.07.012
Shukla, E., Thorat, L. J., Nath, B. B., and Gaikwad, S. M. (2015). Insect trehalase: Physiological significance and potential applications. Glycobiology 25, 357–367. doi:10.1093/glycob/cwu125
Tan, A., and Palli, S. R. (2008). Edysone receptor isoforms play distinct roles in controlling molting and metamorphosis in the red flour beetle, Tribolium castaneum. Mol. Cell Endocrinol. 291, 42–49. doi:10.1016/j.mce.2008.05.006
Tan, Y. A., Xiao, L. B., Zhao, J., Sun, Y., and Bai, L. X. (2015). Molecular and functional characterization of the ecdysone receptor isoform-A from the cotton mirid bug, Apolygus lucorum (Meyer-Dür). Gene 574, 88–94. doi:10.1016/j.gene.2015.07.085
Tang, B., Wei, P., Zhao, L., Shi, Z., Shen, Q., Yang, M., et al. (2016). Knockdown of five trehalase genes using RNA interference regulates the gene expression of the chitin biosynthesis pathway in Tribolium castaneum. BMC Biotechnol. 16, 67. doi:10.1186/s12896-016-0297-2
Tang, B., Yang, M., Shen, Q., Xu, Y., Wang, H., and Wang, S. (2017). Suppressing the activity of trehalase with validamycin disrupts the trehalose and chitin biosynthesis pathways in the rice Brown planthopper, Nilaparvata lugens. Pestic. Biochem. Physiol. 137, 81–90. doi:10.1016/j.pestbp.2016.10.003
Tatun, N., Singtripop, T., and Sakurai, S. (2008). Dual control of midgut trehalase activity by 20-hydroxyecdysone and an inhibitory factor in the bamboo borer Omphisa fuscidentalis Hampson. J. Insect Physiol. 54, 351–357. doi:10.1016/j.jinsphys.2007.10.006
Tisdale, E. J., Azizi, F., and Artalejo, C. R. (2009). Rab2 utilizes glyceraldehyde-3-phosphate dehydrogenase and protein kinase cι to associate with microtubules and to recruit dynein. J. Biol. Chem. 284, 5876–5884. doi:10.1074/jbc.M807756200
Waingeh, V. F., Lowe, S. L., and Thomasson, K. A. (2004). Brownian dynamics of interactions between glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mutants and F-actin. Biopolymers 73, 533–541. doi:10.1002/bip.10560
Wang, K., Li, M. Q., Chang, Y. P., Zhang, B., Zhao, Q. Z., and Zhao, W. L. (2020). The basic helix-loop-helix transcription factor OsBLR1 regulates leaf angle in rice via brassinosteroid signalling. Plant Mol. Biol. 102, 589–602. doi:10.1007/s11103-020-00965-5
Wang, Q., Lu, L., Zeng, M., Wang, D., Zhang, T. Z., Xie, Y., et al. (2022). Rice black-streaked dwarf virus P10 promotes phosphorylation of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) to induce autophagy in Laodelphax striatellus. Autophagy 18, 745–764. doi:10.1080/15548627.2021.1954773
Wu, W. J., Wang, Y., Huang, H. J., Bao, Y. Y., and Zhang, C. X. (2012). Ecdysone receptor controls wing morphogenesis and melanization during rice planthopper metamorphosis. J. Insect Physiol. 58 (3), 420–426. doi:10.1016/j.jinsphys.2012.01.012
Xu, Q. Y., Deng, P., Zhang, Q., Li, A., Fu, K. Y., Guo, W. C., et al. (2020). Ecdysone receptor isoforms play distinct roles in larval-pupal-adult transition in Leptinotarsa decemlineata. Insect Sci. 27, 487–499. doi:10.1111/1744-7917.12662
Yan, X., Zhang, Y., Xu, K., Wang, Y., and Yang, W. (2021). Selection and validation of reference genes for gene expression analysis in Tuta absoluta meyrick (Lepidoptera: Gelechiidae). Insects 12, 589. doi:10.3390/insects12070589
Yang, J. S., Hsu, J. W., Park, S. Y., Li, J., Oldham, W. M., Beznoussenko, G. V., et al. (2018). GAPDH inhibits intracellular pathways during starvation for cellular energy homeostasis. Nature 561, 263–267. doi:10.1038/s41586-018-0475-6
Yego, E. C., and Mohr, S. (2010). siah-1 Protein is necessary for high glucose-induced glyceraldehyde-3-phosphate dehydrogenase nuclear accumulation and cell death in Muller cells. J. Biol. Chem. 285, 3181–3190. doi:10.1074/jbc.M109.083907
Yin, J., Sun, L., Zhang, Q., and Cao, C. (2020). Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 749, 144712. doi:10.1016/j.gene.2020.144712
Zhang, S., An, S., Li, Z., Wu, F., Yang, Q., Liu, Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555, 393–402. doi:10.1016/j.gene.2014.11.038
Zhao, X. F., An, X. M., Wang, J. X., Dong, D. J., Du, X. J., Sueda, S., et al. (2005). Expression of the Helicoverpa cathepsin B-like proteinase during embryonic development. Arch. Insect Biochem. Physiol. 58, 39–46. doi:10.1002/arch.20030
Zhong, X. Y., Yuan, X. M., Xu, Y. Y., Yin, M., Yan, W. W., Zou, S. W., et al. (2018). CARM1 methylates GAPDH to regulate glucose metabolism and is suppressed in liver cancer. Cell Rep. 24, 3207–3223. doi:10.1016/j.celrep.2018.08.066
Zhou, Y., Yi, X., Stoffer, J. B., Bonafe, N., Gilmore-Hebert, M., McAlpine, J., et al. (2008). The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol. Cancer Res. 6, 1375–1384. doi:10.1158/1541-7786.MCR-07-2170
Keywords: GAPDH, 20E, metamorphosis, Treh1, glucose content
Citation: Zhao W, Zhang B, Geng Z, Chang Y, Wei J and An S (2022) The uncommon function and mechanism of the common enzyme glyceraldehyde-3-phosphate dehydrogenase in the metamorphosis of Helicoverpa armigera. Front. Bioeng. Biotechnol. 10:1042867. doi: 10.3389/fbioe.2022.1042867
Received: 13 September 2022; Accepted: 04 October 2022;
Published: 18 October 2022.
Edited by:
Haichao Li, Center for Excellence in Molecular Plant Sciences (CAS), ChinaReviewed by:
Jin-Xing Wang, Shandong University, ChinaSongdou Zhang, China Agricultural University, China
Copyright © 2022 Zhao, Zhang, Geng, Chang, Wei and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiheng An, YW5zaGloZW5nQGFsaXl1bi5jb20=
†These authors have contributed equally to this work
‡ORCID: Jizhen Wei, https://orcid.org/0000-0001-6487-4725
 Wenli Zhao
Wenli Zhao Bo Zhang†
Bo Zhang† Yanpeng Chang
Yanpeng Chang Shiheng An
Shiheng An