- 1College of Plant Protection, Hunan Agricultural University, Changsha, China
- 2State Key Laboratory for Quality and Safety of Agro-products, Institute of Plant Virology, Ningbo University, Ningbo, China
Rhizosphere-associated microbes have important implications for plant health, but knowledge of the association between the pathological conditions of soil-borne virus-infected wheat and soil microbial communities, especially changes in fungal communities, remains limited. We investigated the succession of fungal communities from bulk soil to wheat rhizosphere soil in both infected and healthy plants using amplicon sequencing methods, and assessed their potential role in plant health. The results showed that the diversity of fungi in wheat rhizosphere and bulk soils significantly differed post wheat yellow mosaic virus disease onset. The structure differences in fungal community at the two wheat health states or two compartment niches were evident, soil physicochemical properties (i.e., NH4+) contribute to differences in fungal community structure and alpha diversity. Comparison analysis showed Mortierellomycetes and Dothideomycetes as dominant communities in healthy wheat soils at class level. The genus Pyronemataceae and Solicoccozyma were significantly are significantly enriched in rhizosphere soil of diseased plant, the genus Cystofilobasidium, Cladosporium, Mortierella, and Stephanonectria are significantly enriched in bulk soil of healthy plant. Co-occurrence network analysis showed that the fungi in healthy wheat soil has higher mutual benefit and connectivity compared with diseased wheat. The results of this study demonstrated that the occurrence of wheat yellow mosaic virus diseases altered both fungal community diversity and composition, and that NH4+ is the most important soil physicochemical factor influencing fungal diversity and community composition.
Highlights
• The soil physicochemical properties of diseased wheat and healthy wheat differed
• The composition of soil fungal community of healthy and diseased wheats differed.
• The co-occurrence networks of soil fungal communities significantly decreased in the soil of diseased wheat.
Introduction
The diversity, composition, function and ecological relationship of soil microbial community are closely correlated with the occurrence of soil-borne diseases (T. Kühne, 2009; H. Yang et al., 2017). Previous studies have illustrated that the greater community diversity, the greater its resistance to invasion (C.S. Elton, 1958; C. Roscher et al., 2009; R.W. Heckman et al., 2017; C. Shen et al., 2021). The more significant the diversity of the community, the more stable the ecosystem (T.W. Schoener, 1974). The reduction in soil microbial diversity is soil-borne plant diseases (J.D. Van Elsas et al., 2002). These finding indicate the important role of soil microbial diversity in combating plant diseases. Soil physicochemical properties significantly influence soil microbial diversity. For instance, the contents of organic carbon and total nitrogen in wheat rhizosphere soil were positively correlated with wheat yellow mosaic (WYM) (H. Zhang et al., 2022). Increasing organic input can improve soil microbial diversity and stability (Y. Wang et al., 2017). When continuously diluted soil suspensions were inoculated into sterilized soil, the phylogenetic diversity of inoculated soil was significantly reduced, the modularization of functional gene co-occurrence network was simplified, and the stability of soil microbial community was reduced (W. Xun et al., 2021). In the process of plant resistance to pathogen infection, plants themselves also have the recruitment behavior of beneficial microbial communities, which may be active. The enriched microbial communities can enhance soil resistance by interacting with other microbial communities (C. Wu et al., 2021a; C. Wu et al., 2021b). Therefore, we believe that microbial diversity plays a positive role in soil quality and soil inhibition (P. Garbeva et al., 2004).
Fungi comprises different species: different kinds of fungi have different roles in plant growth and development. Some species of fungi are pathogenic to plant, while others are beneficial (J.M. Raaijmakers et al., 2009), and play a key role in nutrient cycling, pest control, and plant community succession in terrestrial ecosystems (Y.-R. Liu et al., 2021). Pathogenic fungi such as Fusarium pseudograminearum can infect many crops, including wheat, leading to Fusarium crown rot (A. Blum et al., 2019) and Fusarium graminearum can cause Fusarium head blight (FHB) of wheat (Goswami and Kistler, 2004); In contrast, beneficial fungi such as Trichoderma have strong biological control effects on plants (M. Ghorbanpour et al., 2018). Arbuscular mycorrhizal fungi serve as protective agents against pathogens and virulence stresses for plants (P. Jeffries et al., 2003). In addition, many fungi secrete proteins that inhibit plant viruses. A fungal protease called AsES can induce plant resistance to viruses (M.D.P. Caro et al., 2022). Crude methanolic extracts of Neosartorya fischeri and Penicillium oxalicum have the activity of inhibiting Tobacco mosaic virus (TMV) (S. Shen et al., 2009). Soil fungal communities affected by plant diseases may also be affected. Studies have reported that viral infection of plants may regulate plants and their defense response pathways, thereby affecting other interactions with plant pathogenic fungi and beneficial fungi (Schoelz and Stewart, 2018). The number of Bacillus subtilis, Bacillus licheniformis and Bacillus velezensis colonizing the rhizosphere was higher in healthy cotton rhizosphere than in cotton rhizosphere infected with Tobacco streak virus (TSV) (V. S et al., 2018).
WYM is a soil-borne wheat disease mainly caused by Chinese wheat yellow mosaic virus (CWMV) in China (Yang et al., 2001). It is a new virus within the genus Furovirus and transmitted by an obligate root-infected fungus-like organism, Polymyxa graminis (J. Yang et al., 2022). Cases of this virus have been reported in North America, Europe and East Asia and causes different levels of damage to wheat grain yield (T. Kühne, 2009). In severe cases, CWMV causes chlorotic streaking, dwarf symptoms and even death of wheat plants. In wheat producing areas such as Shandong and Henan provinces of China (B.J. Sun et al., 2013). CWMV remains one of the main pathogenic viruses causing mixed infection with Wheat yellow mosaic virus (WYMV), thereby reducing wheat yield by an average of 20–30%, but in severe cases, results in no harvest (J.P. Yang et al., 2002). Although wheat yellow mosaic virus disease has caused significant damage to wheat yield, there is currently a lack of effective control methods. The direct and effective control method is the breeding and screening of resistant varieties (J.P. Yang et al., 2002). Effective transgenic varieties have been cultivated in recent years (W. Fu et al., 2016), however, there remain concerns regarding genetic modification affecting the normal wheat metabolism and increase undesirable metabolites in wheat (Kok and Kuiper, 2003).
Due to the important role of soil fungi in plant virus diseases, we attempted to explore and describe the effects of wheat yellow mosaic virus disease on soil fungal community, to provide evidence for the biological control of wheat yellow mosaic virus disease. We propose the following hypotheses: 1) the occurrence of wheat yellow mosaic virus disease can affect the composition of soil fungal community, soil physicochemical properties, and the associations between fungal communities; 2) the enrichment variations of the fungal communities in soil were correlated with the occurrence of wheat yellow mosaic virus disease. To test these hypotheses, this study 1) investigated the differences in fungal community composition, soil physicochemical properties, and fungal community between healthy and diseased wheat soils; 2) assessed, via co-occurrence network the differences in fungal enrichment communities between healthy and diseased wheat soils.
Materials and methods
Sampling sites and growing conditions
Soil samples were collected in Banquan town (35°11′N, 118°64′E), Junan, Shandong, China which has a temperate monsoon climate with an annual average temperature of 25.5°C (http://www.junan.gov.cn/xq/jngk/dlhj.htm). The climate is humid, and the field soil is aquic brown soil. The sampling plots were sown in winter wheat/summer maize rotation system for nine consecutive years (2012–2021), and the wheat variety was Lin Mai 4. Before sowing, the soil was turned over and plowed deeply so that the overall structure and physiochemical properties of the soil were similar, and the cultivation management was consistent with the local cultivation management.
Sample collection
Wheat yellow mosaic virus disease occurred in wheat returning-green stage. The health status of wheat was assessed and classified before sampling. Healthy plant samples were wheat plants with no symptom, while diseased plant samples were wheat plants with yellowing, shrinkage of leaves, or stunted whole plants. The rhizosphere soil and bulk soil of the diseased and healthy wheat were collected evenly on the diagonal of each plot, and the sampling sites were about 3 m apart. Rhizosphere soil refers to soil particles attached to the root system and can be obtained by strong shaking of the root system Collected soil samples were sealed in valve bag and labeled (A. Lombini et al., 2003). The bulk soil was collected from the soil between ridges where there was no wheat root distribution, sealed, and labeled with self-sealing bags. All samples were stored in low-temperature iceboxes and transported to the laboratory. In a follow-up test, all grouped soil samples were screened (<2 mm) and mixed evenly. Rhizosphere soil and bulk soil were divided into two parts one for DNA extraction and sequencing, and the other for dry rhizosphere soil to determine its chemical properties. The contents of ammonium nitrogen (NH4+) and nitrate nitrogen (NO3−) in fresh soil were determined. The sample name: BSD, the bulk soil of the diseased plant; BSH, the bulk soil of the healthy plant; RSD, the rhizosphere soil of disease plants; RSH, the rhizosphere soil of healthy plants.
Soil physiochemical properties analysis
The soil samples should be treated with 1% HNO3 solution and digested with HF to determine the contents of total potassium (TK), total sulfur (S), total copper (Cu), total calcium (Ca), total magnesium (Mg), total iron (Fe), total zinc (Zn), total boron (B) and molybdenum (Mo) (C. Wu et al., 2021b; H. Zhang et al., 2021) Atomic absorption spectrometer (Novaa350, Jena, Germany) was then used to determine the content of TK, and the contents of the other elements were measured using an inductively coupled plasma spectrometer (ICP-OS-5110, Agilent, America). Soil available phosphorus (AP) and soil total phosphorus (TP) contents were extracted with NaHCO3 and measured using a UV-Vis spectrophotometer (UV-2600, Shimadzu, Japan) (C. Wu et al., 2021b). The soil organic carbon (SOC) content was calculated using the K2Cr2O7 volumetric method. The soil total nitrogen (TN) was extracted with sulfuric acid and then measured using a Vario Max Element Analyzer (Vario Max, Elementar, Germany) (Z. Zhang et al., 2017). After extraction with 1 M KCl solution, NH4+ and NO3− were measured using a continuous flow analysis system (AA3, Seal Analytical, Germany) (Y. Shi et al., 2018).
DNA extraction, PCR assays, high-throughput sequencing
A 0.5 g of rhizosphere soil and bulk soil from five duplicated wheat samples were used for DNA extraction. Rhizosphere soil DNA and bulk soil DNA were extracted using the DNeasy Power Soil Kit and detailed steps were referred to the built-in instructions. The concentration of extracted DNA was detected by Nanodrop RND-2000 (NanoDrop Technologies, Wilmington, DE, United States).
The internal transcribed spacer 1 (ITS1) region of fungi were amplified with the primer pair ITS5-1737F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2-2043R (5′-GCTGCGTTCTTCATCGATGC-3′). PCR reaction consisted of 12.5 μL 2 × Premix Taq™ (TaKaRa. Bio Inc. Shiga, Japan), 1 μL of each primer (10 μm), 2 μL of DNA extract (5–20 ng), and 9 μL of ddH2O to a final volume of 25 µL. PCR amplification was sequenced as follows: desaturated at 95°C for 5 min; followed by 35 cycles at 94°C, 50 s at 58°C and 30 s at 68°C and a final extension step at 68°C for 10 min. MiSeq sequences of purified amplicons were high-throughput sequenced by Novogene Biopharm Technology Co. Ltd (Tianjin, China) using Illumina® MiSeq sequencer (Illumina, San Diego, CA, United States).
The DNA sequencing data were uploaded to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under accession number SRR 6008834.
Bioinformatics analysis
Fast Length Adjustment of Short reads was used to merge the DNA sequences obtained by sequencing (Magoč and Salzberg, 2011) and the merged sequences were processed with QIIME2 (E. Bolyen et al., 2019). According to the method of Callahan et al., the obtained original sequences were demultiplexed and quality filtered with Q2-demux, then denoised with DADA2 (B.J. Callahan et al., 2016). According to the methods of Nakamura et al. ‘s and Price et al., amplicon sequence variants (ASVs) were compared with MAFFT and inducted phylogenetic with FastTree 2 (M.N. Price et al., 2009; T. Nakamura et al., 2018). Fungal identifications were compared in the UNITE database (R.H. Nilsson et al., 2019). Before the community analysis, we deleted the OTU sequence less than two reads (E.M. Bach et al., 2018), and standardized the read depth (19,279 reads per sample) to ensure the reliability of the data.
Statistical analysis
Duncan’s test in IBM SPSS Statistics 19 was used to analyze the differences in soil physicochemical properties among the treatments (M.-Y. Zheng et al., 2021) after one-way variance analysis (ANOVA) to detected significance. Based on ASVs data, microbial diversity analysis was performed on the sampled plots. Diversity indices including Shannon diversity index, Richness index, Simpson index and ACE index were calculated using the “vegan” R package (https://cran.r-project.org/). Principal co-ordinates analysis (PCoA) was carried out based on the Bray–Curtis distance matrices to assess beta diversity of fungal communities, analysis of similarities (ANOSIM) tests were used for testing significant differences. The Mantel test was used to evaluate the relationship between soil fungal communities and soil physicochemical properties (S. Sunagawa et al., 2015). In order to determine the most important predictors of disease index, a randomForest model was built on R platform (4.0.2) using the “randomForest” R package (Liaw and Wiener, 2001). The importance of the model and each predictor was assessed using the “A3” and “rfPermute” R packages, respectively (M. Delgado-Baquerizo et al., 2020).
STAMP software was used to compare soil fungal community composition under different plant states, The differences were tested using Welch’s t-test (p < 0.05) (Parks et al., 2014). To maintain stability (Spearman’s R > 0.8 or R < -0.8) and statistically significant (p < 0.01) correlation, the “Psych” R package was used to conduct species-level symbiotic network analysis of fungal communities on the R platform. To avoid possible bias, more than 60% of the genera present in the sample were reserved for network analysis (C. Wu et al., 2021b). The completed Co-occurrence network relationship was visualized graphically in Gephi. The obtained analysis results were visualized by Gephi0.9.2 software (P. Du et al., 2021). We determined the within-module connectivity (Zi) and among-module connectivity (Pi) of the nodes. The nodes are classified using Zi and Pi according to their topological role in the network. Node topologies are classified as peripherals (Zi < 2.5 and Pi < 0.62), connector (Pi > 0.62), network hub (Zi > 2.5 and Pi > 0.62), and module hubs (Zi > 2.5) (Y. Deng et al., 2012), the “ggplot2” R package (ZiPi plots) was used for drawing (H. Wickham, 2011).
Results
Soil physicochemical properties
In healthy plants soil, the contents of pH, NH4+, TN, S, Ca, Mg, and Fe in rhizosphere soil were significantly higher than that of bulk soil, and the contents of NO3−, AP, SOC, TP, TK, Cu, Zn, B, and Mo had no significant difference. In diseased plants soil, the contents of NO3-, TP, Cu, and Zn in rhizosphere soil was significantly lower than that in bulk soil, and the contents of NH4+, TN, and S were significantly higher than that of bulk soil. Notably, we found that the contents of SOC, TN, S, Cu, and Zn in diseased plants were significantly higher in both rhizosphere and bulk soils than in healthy plants. In the bulk soil, the contents of NO3−, NH4+, AP, SOC, TN, TP, Cu, Ca, Fe, Zn, and pH in diseased soil were significantly higher than those of healthy soil (p < 0.05, Duncan’s test), while the contents of TK, Mg, B and Mo showed no observably differences. In rhizosphere soil, the contents of AP, SOC, TN, S, Cu and Zn in diseased soil was significantly higher than those in healthy soil (p < 0.05, Duncan’s test, Table 1), while the content of NO3−, NH4+, Ca, Mg, Fe in the healthy soil was significantly higher than that in those diseased soil (p < 0.05, Duncan’s test, Table 1), and there were no significant differences in the contents of TP, TK, B, Bo and pH. The contents of NO3−, TP, Cu, and Zn in the bulk soil of diseased plants were signally higher than those of the other three types of soil (p < 0.05, Duncan’s test, Table 1). The contents of NH4+, Ca, Mg, and Fe in the rhizosphere soil of healthy plants were significantly higher than those of the other three types of soil (p < 0.05, Duncan’s test, Table 1).
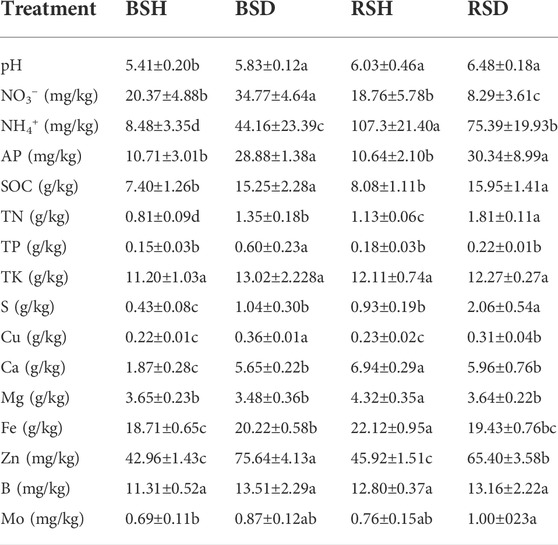
TABLE 1. Physicochemical properties of the bulk soil of healthy plants, the bulk soil of the diseased plants, the rhizosphere soil of the diseased plants and the rhizosphere soil of healthy plants. BSH, the bulk soil of healthy plants. BSD, the bulk soil of the diseased plants. RSH, the rhizosphere soil of healthy plants. RSD, different letters show significant difference among treatments. Values are means ± SEs (Duncan test, p < 0.05, n = 5). The letters in the table represent different significances (a, b, c). AP = available phosphorus, SOC = soil organic carbon, TN = total nitrogen, TP = total phosphorus, TK = total potassium, S = total sulfur, Cu = total copper, Ca = total calcium, Mg=total magnesium, Fe = total iron, Zn = total zinc, B = total boron, Mo = total molybdenum.
Analysis of fungal diversity
We investigated the alpha diversity of four different soil fungal communities. The results showed that the differences of the richness index of the four soil fungal communities were the same as the differences of Shannon index (Figure 1). The richness index and Shannon index of fungal communities in BSH treatment were significantly higher than those in the other three soils (p < 0.05, Tukey test), followed by BSD treatment, but there was no significant difference in richness index and Shannon index between RSD and RSH treatments. The results also showed that the alpha diversity of soil fungal community in diseased wheat significantly decreased compared with that in bulk soil, while the alpha diversity of rhizosphere soil fungal community did not significantly change. We performed principal co-ordinate analysis (PCoA) and found differences in fungal community structure among different soil compartments (Figure 2A). Correlation analysis showed NH4+ was the most important physicochemical factors affecting fungal diversity and community composition (Figure 2B).
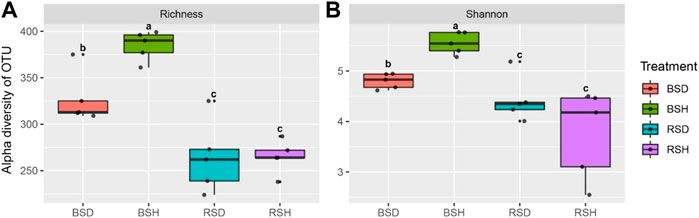
FIGURE 1. Difference analysis of alpha and beta diversity of fungi among the bulk soil of the diseased plant, the bulk soil of healthy plants, the rhizosphere soil of disease plants and the rhizosphere soil of healthy plants. (A) Richness index of BSD, BSH, RSD, and RSH; (B) Shannon index of BSD, BSH, RSD, and RSH; BSD, the bulk soil of the diseased plant; BSH, the bulk soil of healthy plants; RSD, the rhizosphere soil of disease plants; RSH, the rhizosphere soil of healthy plants. Significance satisfies p < 0.05 based on Tukey test.
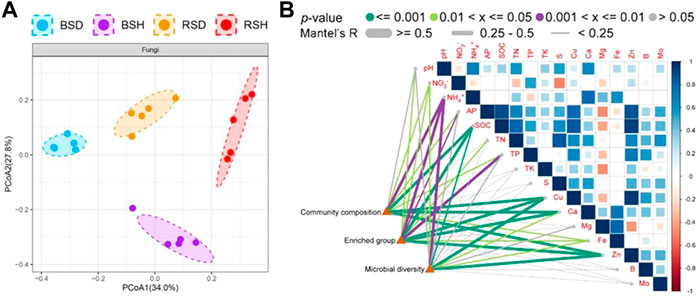
FIGURE 2. Difference analysis of beta diversity of fungi among the bulk soil of the diseased plant, the bulk soil of healthy plants, the rhizosphere soil of disease plants and the rhizosphere soil of healthy plants and pairwise correlation analysis between environmental factors and community composition, microbial diversity and enriched group (A) Principal co-ordinates analysis (PCoA) indicated the beta diversity of fungi community. Significance satisfies p < 0.05 based on Tukey test. BSD, the bulk soil of the diseased plant; BSH, the bulk soil of healthy plants; RSD, the rhizosphere soil of disease plants; RSH, the rhizosphere soil of healthy plants. (B) Different colors represent different ranges of p-values and different degrees of significance, Significance satisfies p < 0.05 based on Tukey test. Different line thicknesses represent different R values obtained through Mantel detection, and represent different correlation degrees. Microbial diversity was analyzed at the level of species, Enriched group was analyzed at the level of genus, and community composition was analyzed at the level of family.
Fugal community composition
To further explore the differences in soil fungal community structure between healthy and diseased plants, we analyzed the fungal community composition. The results showed the proportion of Mortierellomycetes and Dothideomycetes in the healthy treatment group was higher than that in the disease treatment group (Figure 3; Supplementary Table S1). Tremellomycetes (21.43%), Sordariomycetes (18.71%) and Pezizomycetes (13.65%) were the dominant in BSD treatment; Sordariomycetes (30.38%), Tremellomycetes (17.22%) and Mortierellomycetes (11.29%) were the dominant in BSH. In addition, the proportion of Mortierellomycetes and Sordariomycetes in BSH treatment was higher than that in BSD treatment (Figure 3; Supplementary Table S1). In the rhizosphere soil, Mortierellomycetes (27.44%), Tremellomycetes (24.95%) and Sordariomycetes (23.54%) were the dominant classes in RSD treatment; Mortierellomycetes (47.39%), Tremellomycetes (21.91%) and Sordariomycetes (7.48%) were the dominant classes in RSH treatment (Supplementary Table S1). Comparative analysis revealed that Fusarium, Pyronemataceae and Solicoccozyma were significantly enriched in RSD treatment (Supplementary Figure S1A) and P. Pseudaleur was significantly enriched in the BSD treatment (Supplementary Figure S1B). Cystofilobasidium, Cladosporium, Mortierella, Stephanonectria were significantly enriched in BSH treatment (Supplementary Figure S1B). In conclusion, fungal community structure and composition in rhizosphere and bulk soils of wheat yellow mosaic virus disease were changed compared with those of healthy plants.
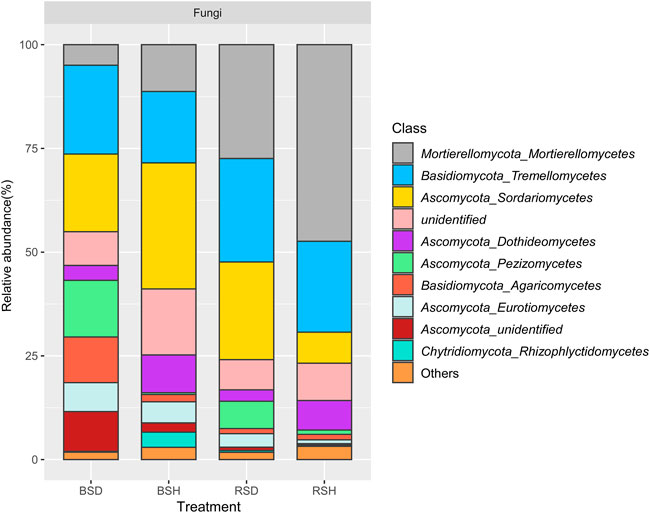
FIGURE 3. Relative abundance of 10 dominant classes in fungi among the bulk soil of the diseased plant, the bulk soil of healthy plants, the rhizosphere soil of disease plants and the rhizosphere soil of healthy plants. Family accounting for less than 1% of total reads were classified as “others”. BSD, the bulk soil of the diseased plant; BSH, the bulk soil of healthy plants; RSD, the rhizosphere soil of disease plants; RSH, the rhizosphere soil of healthy plants Significance satisfies p < 0.05 based on Tukey test.
Network association in different treatments
To better understand the interrelationships among different fungal communities in the four treatments, we generated fungal networks to describe the symbiotic patterns among fungal communities at the genus level (Figures 4, 5). In these bulk soils, the network nodes were mainly divided into Sordariomycetes, Dothideomycetes, Agaricomycetes, Leotiomycetes, Eurotiomycetes, Tremellomycetes, Microbotryomycetes, Mortierellomycetes, Pezizomycetes, Chytridiomycetes (Figure 4). The proportions of Sordariomycetes (31.75%), Dothideomycetes (14.85%), and Agaricomycetes (10.14%) (Figure 4A) were the three classes with the largest proportions in healthy plant soils were different from that of Sordariomycetes (33.95%), and Agaricomycetes (13.58%) (Figure 4B) in the soil of diseased plants. The analysis of network topological properties revealed that the soil fungal community network of BSH treatment was simpler than that of BSD treatment, and the constructed BSH fungal network contained 148 nodes, 608 links, with an average connectivity of 0.66, and a modularity of 2.43 while the BSD network had 164 nodes, 777 links, with an average connectivity of 0.77, and modularity of 0.80 (Table 2). In these rhizosphere soils, the RSH network nodes were mainly divided into Sordariomycetes, Agaricomycetes, Dothideomycetes, Eurotiomycetes, Leotiomycetes, Tremellomycetes, Microbotryomycetes, Pezizomycetes, Mortierellomycetes, Chytridiomycetes, while the RSD network nodes were mainly divided into Agaricomycetes, Saccharomycetes, Sordariomycetes, Dothideomycetes, Eurotiomycetes, Leotiomycetes, Tremellomycetes, Microbotryomycetes, Mortierellomycetes, Pezizomycetes (Figure 5). The top four classes of the RSH treatment included four keystone taxa, Sordariomycetes (29.53%), Agaricomycetes (15.44%), Dothideomycetes (14.09%), Eurotiomycetes (9.4%) (Figure 5A), while the top four classes of the RSD treatment included four keystone taxa, Agaricomycetes (16.54%), Saccharomycetes (16.54%), Sordariomycetes (12.78%), Dothideomycetes (12.03%) (Figure 5B). However, when we compared the co-occurrence networks of RSH and RSD treatments, we found that the network nodes were decreased from 149 in RSH treatment to 133 in RSD treatment (Table 2), the network edges were decreased from 678 in RSH treatment to 521 in RSH treatment (Table 2), but the network positive edges were increased from 82.57% in RSH treatment to 85.91% in RSD treatment (Table 2). These network topology parameters indicated that the bulk soil fungal community network became larger, more nodes and more complex after the occurrence of disease. Although the number of nodes and edges of the rhizosphere soil fungal community decreased after the occurrence of the disease, the mutually beneficial relationship among the communities within the network was enhanced.
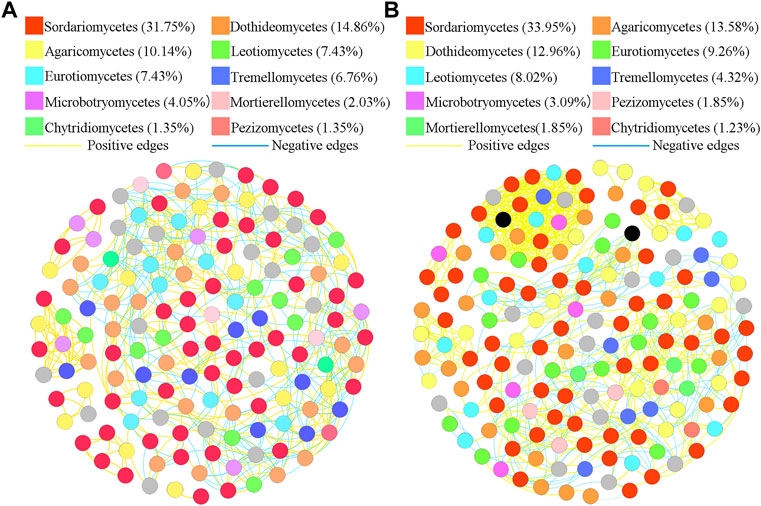
FIGURE 4. Schematic representation of symbiotic network patterns of soil fungi under different treatments at the genus level. These dots represented different genera of fungi that were clustered at class level. Different colors of the squares represent different dominant fungal genera that were clustered (only list the top ten fungal classes). Yellow lines between any connected two nodes indicates a positive relationship and blue lines indicate a negative relationship (A) Schematic diagram of fungal community network in healthy bulk soils. (B) Schematic diagram of fungal community network in diseased bulk soils.
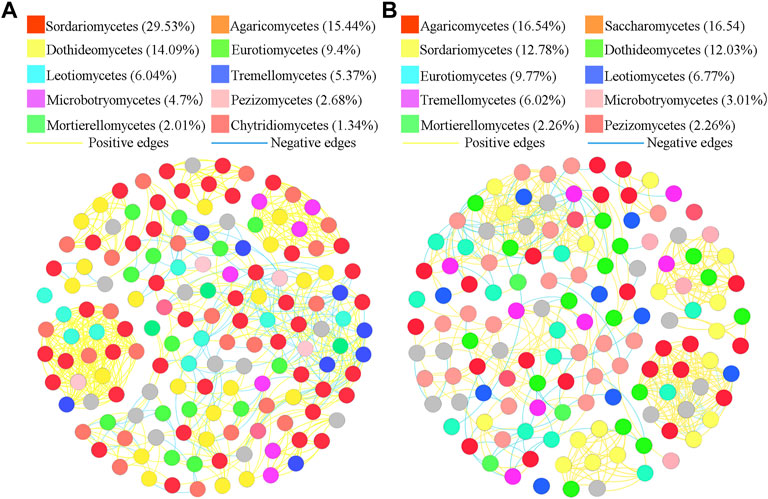
FIGURE 5. Schematic representation of symbiotic network patterns of soil fungi under different treatments at the genus level. These dots represented different genera of fungi that were clustered at class level. Different colors of the squares represent different dominant fungal genera that were clustered (only list the top ten fungal classes). Yellow lines between any connected two nodes indicates a positive relationship and blue lines indicate a negative relationship (A) Schematic diagram of fungal community network in healthy rhizosphere soils. (B) Schematic diagram of fungal community network in diseased rhizosphere soils.

TABLE 2. Microbial co-occurrence network indices. BSD, the bulk soil of the diseased plants. RSH, the rhizosphere soil of healthy plants. RSD, different letters show significant difference among treatments.
To evaluate the potential topological role of groups in soil fungal symbiotic networks, nodes were divided into four types (peripherals, connectors, modular hubs and network hubs) according to their within-module connectivity (Zi) and among-module connectivity (Pi) (Figure 6) values. The results show that the genus Pseudopithomyces belonged to connectors in BSH treatment (Figure 6A), the family Sympoventuriaceae belonged to Module hubs in BSD treatment (Figure 6B), the genus Exophiala belonged to Module hubs in RSH treatment (Figure 6C), and all nodes were classified as peripheral nodes in RSD treatment (Figure 6D). They were all highly connected to other important nodes.
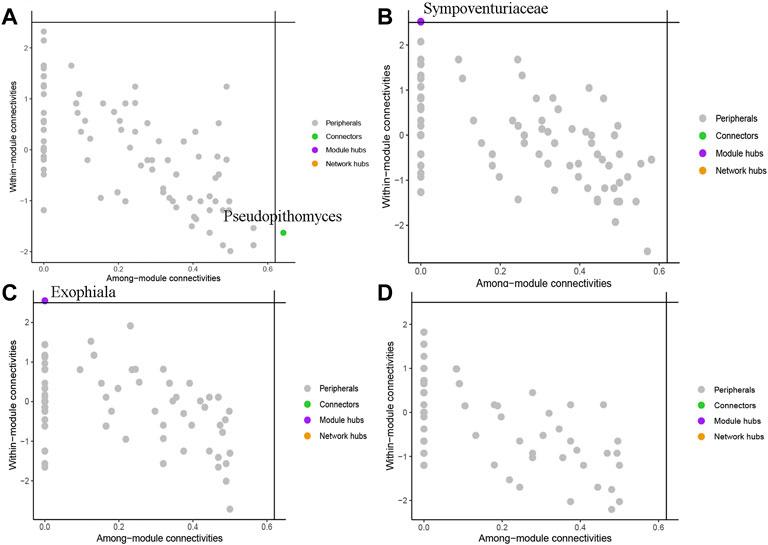
FIGURE 6. Soil fungi under different treatments were divided into network nodes by within-module connectivity (Zi) among-module connectivity (Pi) (A) the bulk soil of healthy plants (BSH) (B) the bulk soil of the diseased plant (BSD) (C) the rhizosphere soil of healthy plants (RSH) (D) the rhizosphere soil of disease plants (RSD).
Discussion
Previous reports have suggested NH4+ as one of the main factors causing the difference of fungal community among different land use types and plant species in the eastern mountainous area of Liaoning province (Q. Zhang et al., 2021), Arbuscular mycorrhizal fungi were positively correlated with NH4+ in degraded alpine grassland soil system of Qinghai-Tibet Plateau (J. Li et al., 2020b). NH4+ was also a major factor affecting the diversity and composition of fungal communities in our study (Figure 2B). This conclusion was consistent with findings of Smithwick et al. and Chen et al. that ammonium nitrogen can significantly affect soil microbial community (E.A.H. Smithwick et al., 2005; W. Chen et al., 2017). In our study, the proportion of NH4+ in rhizosphere soil was higher than that in bulk soil, which was consistent with previous (Table 1) (M. Koranda et al., 2011; Y. Cui et al., 2021; X. Zhu et al., 2021). This may be attributed to the fact that plant roots stimulate the production and fixation of NH4+, but limit the production of NO3− to meet the needs of plants for NH4+ (X. Zhu et al., 2021). In this study, the alpha diversity in bulk soil was significantly higher than that in rhizosphere soil, and NH4+ was closely related to the diversity and community composition of fungal communities, which may be due to the different responses of fungal communities in soils with different treatments to NH4+, resulting in the growth differences among fungal communities (Y.-C. Bai et al., 2020).
Through analysis of relative abundance, our results showed that Ascomycota and Basidiomycota were the two most common fungal phyla in the four soils (Figure 3), which is consistent with the results of previous studies (X.-g. Li et al., 2014b; J. Jiang et al., 2019). We observed changes in community composition between treatments at the class level. Notably, compared with healthy wheat, both in rhizosphere soil and bulk soil, the proportion of Mortierellomycetes and Dothideomycetes in healthy wheat was higher than that in diseased wheat (Supplementary Table S1). These two classes of fungi exist in a variety of soils with resistance to pathogen infection (X. Liu et al., 2021; T. Xu et al., 2021). Many genera of the class Mortierellomycetes belong to plant growth-promoting fungi (PGPF), which play a positive role in plant resistance to pathogen invasion (Ozimek and Hanaka, 2021). The class Dothideomycetes are the largest class of fungi, many of which are important for plant resistance to disease and stress (D. Pem et al., 2021). Cystofilobasidium, Cladosporium, Mortierella and Stephanonectria attracted our attention for their significant enrichment in BSH treatment. These genera have been reported to produce positive responses to plant growth or soil resistance by themselves in the soil. For instance, the genus Cystofilobasidium usually exists as the dominant community in compost and humus (J. Li et al., 2020a). The genus Cladosporium is a widely distributed endophytic fungus that protects plants from biological and abiotic stresses. These species secrete beneficial secondary metabolites that improve the ability of plants to adapt to new habitats and maintain plant health and performance (Achard and Genschik, 2009; Răut et al., 2021). The genus Mortierella helps crops and mycorrhizal fungi capture phosphorus (P) as well as decompose plant litter and degrade aromatics (L. Ellegaard-Jensen et al., 2013; Tamayo-Velez and Osorio, 2016). When investigating fungal enrichment communities in different soils, we found that the genera Fusarium was significantly enriched in RSD treatment, this phenomenon has been found in other plants as well (M. Guan et al., 2018; H. Liu et al., 2019). We determined that the genus Fusarium was significantly enriched in RSD treatment as a common pathogenic fungus (Steinkellner and Langer, 2004). The enrichment of Fusarium may lead to the occurrence of wheat root diseases and damage of wheat root surface (M. Winter et al., 2019), which may be more conducive to the parasitism of P. graminis on wheat roots (K. Hansen et al., 2013; Mestre and Fontenla, 2021). In conclusion, the occurrence of Wheat yellow mosaic virus disease affects the composition of soil fungal community, but there are many reasons for this change, such as soil physicochemical interaction and soil nutrient imbalance (C.H. Kong et al., 2008; C. Li et al., 2010), may also be due to the promoting or inhibiting effect of plant rhizosphere exudates on the growth of different fungal communities (X.-g. Li et al., 2014a), more detailed reasons need to be explored.
In recent years, network co-occurrence analysis has provided a reliable method to study the symbiotic mode and mutual relationship of soil microbial communities in complex environments (Faust and Raes, 2012). Previous reports have confirmed that the interaction and connectivity between communities affect the stability and persistence of communities, and the network structure of these interactions is correlated with the community invasion resistance (Thébault and Fontaine, 2010; Xu et al., 2020; Ji et al., 2021). We found that the soil fungal community network of BSH treatment was simpler than that of BDH treatment, it may be that the contents of NO3− and NH4+ in the rhizosphere soil is significantly higher than healthy wheat soil (Table 1), thus resulting in a loss of some fungal community richness (L.C. Cline et al., 2018; S. Jiang et al., 2018). In rhizosphere soil, RSH treatment had more edges and nodes than the RSD treatment, suggesting that the soil fungal network of healthy plants was more larger. Complex fungal networks in the soil lead to better community stability and provide better anti-interference (W. Yang et al., 2019; L. Xu et al., 2020) Simultaneously, a higher positive correlation edge in soil fungal communities of healthy plants may mean more cooperation (J. Zhang et al., 2021) and a greater connected network provides more functional redundancy (Mougi and Kondoh, 2012). In addition, more complex networks can cope with diverse and complex environmental changes or inhibit infection of plants by soil-transmitted pathogens (H. Yang et al., 2017; J. Tao et al., 2018). Highly diverse microbial communities have a better ability to resist pathogen infection, which makes the soil itself more resistant (C.A. Mallon et al., 2015). The mutualistic relationship between fungal communities in diseased wheat was broken, and the complexity and stability of the community were reduced, which may provide a more favorable environment for pathogen infection (N.L. Nwokolo et al., 2021).
Due to the importance of dominant groups, biomarker groups and network hub groups in ecology and evolutionary biology, many studies of microorganisms are aimed at finding and identifying them in networks (S. Banerjee et al., 2018; C. Xiong et al., 2021). We found two key genera and one family (Pseudopithomyces, Exophial, and Sympoventuriaceae) in different treatments. There are many species in the family Pseudopithomyces, including both pathogenic fungi and fungi that inhibit pathogens (A. Perelló et al., 2017; B.N.S. Ningsih et al., 2021). The family Sympoventuriaceae is a family of saprophytic and pathogenic fungi (M. Shen et al., 2020). Most species of Exophiala can cause plant diseases and human diseases (E.J. Otis et al., 1985; De Hoog et al., 2005). This study demonstrates the critical role of these fungal communities in the fungal community network after the onset of wheat yellow mosaic virus disease. Our results suggest that these fungal communities play an important role in the fungal community network after the occurrence of wheat yellow mosaic virus disease, so as to better understand the changes of fungal communities after the occurrence of wheat yellow mosaic virus disease and the role of these changes in the plant defense response.
In recent years, biological control through biological control agents has been recognized as a promising method to reduce the incidence of many field crop diseases (S. Munir et al., 2022). For example, Neorhizobium galegae, Lysobacter dokdonensis, and Ensifer adhaerens can prey on bacteria and can be used to control Tobacco bacterial wilt (W. Ahmed et al., 2022); P. camelliae-sinensis infection can be used to prevent and treat tea gray blight disease (Q. Wang et al., 2021). Our studies identified dominant fungal communities in healthy and diseased plant soils respectively, and we suggest that core communities in healthy plant soils help promote WYM resistance. These identified fungal communities can be developed into biological control agents for WYM control in the future. This will provide a theoretical basis for fungal microbial resistance to WYM to improve wheat yield and a reference community for biological control.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/?term=. Accession Number: 6008834.
Author contributions
JC and LD provided the idea and experimental design for this study; QL, CH, and LC carried out and completed the study with the assistance of CW, HZ, LW, TZ, HH, SL, JL, TG, and JY, All authors participate in the finalization of the article and are allowed to submit the modified article.
Funding
This study was supported by the earmarked fund for CARS (CARS-03) and sponsored by K.C. Wong Magna Funding in Ningbo University.
Acknowledgments
Authors are extremely thankful to the earmarked fund for CARS (CARS-03) and sponsored by K.C. Wong Magna Funding in Ningbo University. Authors also acknowledge the research platform provided by Ningbo University and Hunan Agricultural University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.1033991/full#supplementary-material
References
Achard, P., and Genschik, P. (2009). Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 60 (4), 1085–1092. doi:10.1093/jxb/ern301
Ahmed, W., Dai, Z., Liu, Q., Munir, S., Yang, J., Karunarathna, S. C., et al. (2022). Microbial cross-talk: Dissecting the core microbiota associated with flue-cured tobacco (Nicotiana tabacum) plants under healthy and diseased state. Front. Microbiol. 13, 845310. doi:10.3389/fmicb.2022.845310
Bach, E. M., Williams, R. J., Hargreaves, S. K., Yang, F., and Hofmockel, K. S. (2018). Greatest soil microbial diversity found in micro-habitats. Soil Biol. Biochem. 118, 217–226. doi:10.1016/j.soilbio.2017.12.018
Bai, Y.-C., Chang, Y.-Y., Hussain, M., Lu, B., Zhang, J.-P., Song, X.-B., et al. (2020). Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 8 (5), 694. doi:10.3390/microorganisms8050694
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16 (9), 567–576. doi:10.1038/s41579-018-0024-1
Blum, A., Benfield, A. H., Sørensen, J. L., Nielsen, M. R., Bachleitner, S., Studt, L., et al. (2019). Regulation of a novel Fusarium cytokinin in Fusarium pseudograminearum. Fungal Biol. 123 (3), 255–266. doi:10.1016/j.funbio.2018.12.009
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi:10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 581–583. doi:10.1038/nmeth.3869
Caro, M. D. P., Venturuzzi, A. L., Moschen, S., Salazar, S. M., Díaz-Ricci, J. C., and Asurmendi, S. (2022). A fungal protease named AsES triggers antiviral immune responses and effectively restricts virus infection in arabidopsis and Nicotiana benthamiana plants. Ann. Bot. 129, 593–606. doi:10.1093/aob/mcac013
Chen, W., Gu, Y., Xu, H., Liu, Z., Lu, C., and Lin, C. (2017). Variation of microbial communities and functional genes during the biofilm formation in raw water distribution systems and associated effects on the transformation of nitrogen pollutants. Environ. Sci. Pollut. Res. 24 (18), 15347–15359. doi:10.1007/s11356-017-9125-z
Cline, L. C., Huggins, J. A., Hobbie, S. E., and Kennedy, P. G. (2018). Organic nitrogen addition suppresses fungal richness and alters community composition in temperate forest soils. Soil Biol. Biochem. 125, 222–230. doi:10.1016/j.soilbio.2018.07.008
Cui, Y., Bing, H., Fang, L., Jiang, M., Shen, G., Yu, J., et al. (2021). Extracellular enzyme stoichiometry reveals the carbon and phosphorus limitations of microbial metabolisms in the rhizosphere and bulk soils in alpine ecosystems. Plant Soil 458 (1), 7–20. doi:10.1007/s11104-019-04159-x
de Hoog, G. S., Matos, T., Sudhadham, M., Luijsterburg, K. F., and Haase, G. (2005). Intestinal prevalence of the neurotropic black yeast Exophiala (Wangiella) dermatitidis in healthy and impaired individuals. Mycoses 48 (2), 142–145. doi:10.1111/j.1439-0507.2004.01083.x
Delgado-Baquerizo, M., Reich, P. B., Trivedi, C., Eldridge, D. J., Abades, S., Alfaro, F. D., et al. (2020). Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat. Ecol. Evol. 4 (2), 210–220. doi:10.1038/s41559-019-1084-y
Deng, Y., Jiang, Y.-H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinforma. 13 (1), 113. doi:10.1186/1471-2105-13-113
Du, P., He, H., Wu, X., Xu, J., Dong, F., Liu, X., et al. (2021). Mesosulfuron-methyl influenced biodegradability potential and N transformation of soil. J. Hazard. Mater. 416, 125770. doi:10.1016/j.jhazmat.2021.125770
Ellegaard-Jensen, L., Aamand, J., Kragelund, B. B., Johnsen, A. H., and Rosendahl, S. (2013). Strains of the soil fungus Mortierella show different degradation potentials for the phenylurea herbicide diuron. Biodegradation 24 (6), 765–774. doi:10.1007/s10532-013-9624-7
Elton, C. S. (1958). The ecology of invasion by animals and plants. Chicago: The University of Chicago Press.
Faust, K., and Raes, J. (2012). Microbial interactions: From networks to models. Nat. Rev. Microbiol. 10 (8), 538–550. doi:10.1038/nrmicro2832
Fu, W., Du, Z., He, Y., Zheng, W., Han, C., Liu, B., et al. (2016). Metabolic profiling of virus-infected transgenic wheat with resistance to wheat yellow mosaic virus. Physiological Mol. Plant Pathology 96, 60–68. doi:10.1016/j.pmpp.2016.08.001
Garbeva, P., van Veen, J. A., and van Elsas, J. D. (2004). Microbial diversity in soil: Selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42 (1), 243–270. doi:10.1146/annurev.phyto.42.012604.135455
Ghorbanpour, M., Omidvari, M., Abbaszadeh-Dahaji, P., Omidvar, R., and Kariman, K. (2018). Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 117, 147–157. doi:10.1016/j.biocontrol.2017.11.006
Goswami, R. S., and Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5 (6), 515–525. doi:10.1111/j.1364-3703.2004.00252.x
Guan, M., Pan, X.-C., Wang, S., Wei, X.-L., Zhang, C.-B., Wang, J., et al. (2018). Comparison of fungal communities among ten macrophyte rhizospheres. Fungal Biol. 122 (9), 867–874. doi:10.1016/j.funbio.2018.05.001
Hansen, K., Perry, B. A., Dranginis, A. W., and Pfister, D. H. (2013). A phylogeny of the highly diverse cup-fungus family Pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traits. Mol. Phylogenetics Evol. 67 (2), 311–335. doi:10.1016/j.ympev.2013.01.014
Heckman, R. W., Halliday, F. W., Wilfahrt, P. A., and Mitchell, C. E. (2017). Effects of native diversity, soil nutrients, and natural enemies on exotic invasion in experimental plant communities. Ecology 98 (5), 1409–1418. doi:10.1002/ecy.1796
Jeffries, P., Gianinazzi, S., Perotto, S., Turnau, K., and Barea, J.-M. (2003). The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol. Fertil. Soils 37 (1), 1–16. doi:10.1007/s00374-002-0546-5
Ji, L., Tian, L., Nasir, F., Chang, J., Chang, C., Zhang, J., et al. (2021). Impacts of replanting American ginseng on fungal assembly and abundance in response to disease outbreaks. Arch. Microbiol. 203 (5), 2157–2170. doi:10.1007/s00203-021-02196-8
Jiang, J., Yu, M., Hou, R., Li, L., Ren, X., Jiao, C., et al. (2019). Changes in the soil microbial community are associated with the occurrence of Panax quinquefolius L. root rot diseases. Plant Soil 438 (1), 143–156. doi:10.1007/s11104-018-03928-4
Jiang, S., Liu, Y., Luo, J., Qin, M., Johnson, N. C., Öpik, M., et al. (2018). Dynamics of arbuscular mycorrhizal fungal community structure and functioning along a nitrogen enrichment gradient in an alpine meadow ecosystem. New Phytol. 220 (4), 1222–1235. doi:10.1111/nph.15112
Kok, E. J., and Kuiper, H. A. (2003). Comparative safety assessment for biotech crops. Trends Biotechnol. 21(10), 439–444. doi:10.1016/j.tibtech.2003.08.003
Kong, C. H., Wang, P., Zhao, H., Xu, X. H., and Zhu, Y. D. (2008). Impact of allelochemical exuded from allelopathic rice on soil microbial community. Soil Biol. Biochem. 40 (7), 1862–1869. doi:10.1016/j.soilbio.2008.03.009
Koranda, M., Schnecker, J., Kaiser, C., Fuchslueger, L., Kitzler, B., Stange, C. F., et al. (2011). Microbial processes and community composition in the rhizosphere of European beech – the influence of plant C exudates. Soil Biol. Biochem. 43 (3), 551–558. doi:10.1016/j.soilbio.2010.11.022
Kühne, T. (2009). Soil-borne viruses affecting cereals—known for long but still a threat. Virus Res. 141 (2), 174–183. doi:10.1016/j.virusres.2008.05.019
Li, C., Li, X., Kong, W., Wu, Y., and Wang, J. (2010). Effect of monoculture soybean on soil microbial community in the Northeast China. Plant Soil 330 (1), 423–433. doi:10.1007/s11104-009-0216-6
Li, J., Bao, H., Xing, W., Yang, J., Liu, R., Wang, X., et al. (2020a). Succession of fungal dynamics and their influence on physicochemical parameters during pig manure composting employing with pine leaf biochar. Bioresour. Technol. 297, 122377. doi:10.1016/j.biortech.2019.122377
Li, J., Shao, X., Huang, D., Shang, J., Liu, K., Zhang, Q., et al. (2020b). The addition of organic carbon and nitrogen accelerates the restoration of soil system of degraded alpine grassland in Qinghai-Tibet Plateau. Ecol. Eng. 158, 106084. doi:10.1016/j.ecoleng.2020.106084
Li, X.-g., Ding, C.-f., Hua, K., Zhang, T.-l., Zhang, Y.-n., Zhao, L., et al. (2014a). Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159. doi:10.1016/j.soilbio.2014.07.019
Li, X.-g., Ding, C.-f., Zhang, T.-l., and Wang, X.-x. (2014b). Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 72, 11–18. doi:10.1016/j.soilbio.2014.01.019
Liu, H., Pan, F., Han, X., Song, F., Zhang, Z., Yan, J., et al. (2019). Response of soil fungal community structure to long-term continuous soybean cropping. Front. Microbiol. 9 (3316), 3316. doi:10.3389/fmicb.2018.03316
Liu, X., Hannula, S. E., Li, X., Hundscheid, M. P. J., klein Gunnewiek, P. J. A., Clocchiatti, A., et al. (2021). Decomposing cover crops modify root-associated microbiome composition and disease tolerance of cash crop seedlings. Soil Biol. Biochem. 160, 108343. doi:10.1016/j.soilbio.2021.108343
Liu, Y.-R., Eldridge, D. J., Zeng, X.-M., Wang, J., Singh, B. K., and Delgado-Baquerizo, M. (2021). Global diversity and ecological drivers of lichenised soil fungi. New Phytol. 231 (3), 1210–1219. doi:10.1111/nph.17433
Lombini, A., Llugany, M., Poschenrieder, C., Dinelli, E., and Barceló, J. (2003). Influence of the Ca/Mg ratio on Cu resistance in three Silene armeria ecotypes adapted to calcareous soil or to different, Ni- or Cu-enriched, serpentine sites. J. Plant Physiology 160 (12), 1451–1456. doi:10.1078/0176-1617-01002
Magoč, T., and Salzberg, S. L. (2011). Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. doi:10.1093/bioinformatics/btr507
Mallon, C. A., Elsas, J. D. v., and Salles, J. F. (2015). Microbial invasions: The process, patterns, and mechanisms. Trends Microbiol. 23 (11), 719–729. doi:10.1016/j.tim.2015.07.013
Mestre, M. C., and Fontenla, S. (2021). Yeast communities associated with ectomycorrhizal fungi in different Nothofagus forests of northwestern Patagonia. Symbiosis 84 (2), 179–193. doi:10.1007/s13199-021-00782-4
Mougi, A., and Kondoh, M. (2012). Diversity of interaction types and ecological community stability. Science 337 (6092), 349–351. doi:10.1126/science.1220529
Munir, S., Ahmed, A., Li, Y., He, P., Singh, B. K., He, P., et al. (2022). The hidden treasures of citrus: Finding huanglongbing cure where it was lost. Crit. Rev. Biotechnol. 42 (4), 634–649. doi:10.1080/07388551.2021.1942780
Nakamura, T., Yamada, K. D., Tomii, K., and Katoh, K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34 (14), 2490–2492. doi:10.1093/bioinformatics/bty121
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47 (D1), D259–D264. doi:10.1093/nar/gky1022
Ningsih, B. N. S., Rukachaisirikul, V., Pansrinun, S., Phongpaichit, S., Preedanon, S., and Sakayaroj, J. (2021). New aromatic polyketides from the marine-derived fungus Pseudopithomyces maydicus PSU-AMF350 and their antimicrobial activity. Nat. Prod. Res. 36, 4982–4989. doi:10.1080/14786419.2021.1915309
Nwokolo, N. L., Enebe, M. C., Chigor, C. B., Chigor, V. N., and Dada, O. A. (2021). The contributions of biotic lines of defence to improving plant disease suppression in soils: A review. Rhizosphere 19, 100372. doi:10.1016/j.rhisph.2021.100372
Otis, E. J., Wolke, R. E., and Blazer, V. S. (1985). Infection of Exophiala salmonis in atlantic salmon (Salmo salar L.). J. Wildl. Dis. 21 (1), 61–64. doi:10.7589/0090-3558-21.1.61
Ozimek, E., and Hanaka, A. (2021). Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture 11 (1), 7. doi:10.3390/agriculture11010007
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). Stamp: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30 (21), 3123–3124. doi:10.1093/bioinformatics/btu494
Pem, D., Jeewon, R., Chethana, K. W. T., Hongsanan, S., Doilom, M., Suwannarach, N., et al. (2021). Species concepts of Dothideomycetes: Classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Divers. 109 (1), 283–319. doi:10.1007/s13225-021-00485-7
Perelló, A., Aulicino, M., Stenglein, S. A., Labuda, R., and Moreno, M. V. (2017). Pseudopithomyces chartarum associated with wheat seeds in Argentina, pathogenicity and evaluation of toxigenic ability. Eur. J. Plant Pathol. 148 (2), 491–496. doi:10.1007/s10658-016-1093-5
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26 (7), 1641–1650. doi:10.1093/molbev/msp077
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321 (1), 341–361. doi:10.1007/s11104-008-9568-6
Răut, I., Călin, M., Capră, L., Gurban, A.-M., Doni, M., Radu, N., et al. (2021). Cladosporium sp. isolate as fungal plant growth promoting agent. Agronomy 11 (2), 392. doi:10.3390/agronomy11020392
Roscher, C., Beßler, H., Oelmann, Y., Engels, C., Wilcke, W., and Schulze, E.-D. (2009). Resources, recruitment limitation and invader species identity determine pattern of spontaneous invasion in experimental grasslands. J. Ecol. 97 (1), 32–47. doi:10.1111/j.1365-2745.2008.01451.x
Schoelz, J. E., and Stewart, L. R. (2018). The role of viruses in the phytobiome. Annu. Rev. Virol. 5 (1), 93–111. doi:10.1146/annurev-virology-092917-043421
Schoener, T. W. (1974). Stability and complexity in model ecosystms. Evolution 28 (3), 510–511. doi:10.1111/j.1558-5646.1974.tb00784.x
Shen, C., Wang, J., He, J.-Z., Yu, F.-H., Ge, Y., and Cann, I. (2021). Plant diversity enhances soil fungal diversity and microbial resistance to plant invasion. Appl. Environ. Microbiol. 87 (11), e00251-21–e00221. doi:10.1128/AEM.00251-21
Shen, M., Zhang, J. Q., Zhao, L. L., Groenewald, J. Z., Crous, P. W., and Zhang, Y. (2020). Stud. Mycol. 96, 185–308. doi:10.1016/j.simyco.2020.03.001
Shen, S., Li, W., Ouyang, M., Wu, Z., Lin, Q., and Xie, L. (2009). Identification of two marine fungi and evaluation of their antivirus and antitumor activities. Wei Sheng Wu Xue Bao 49 (9), 1240–1246. doi:10.1016/S1003-6326(09)60084-4
Shi, Y., Li, Y., Xiang, X., Sun, R., Yang, T., He, D., et al. (2018). Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 6 (1), 27. doi:10.1186/s40168-018-0409-4
Smithwick, E. A. H., Turner, M. G., Metzger, K. L., and Balser, T. C. (2005). Variation in NH4+ mineralization and microbial communities with stand age in lodgepole pine (Pinus contorta) forests, Yellowstone National Park (USA). Soil Biol. Biochem. 37 (8), 1546–1559. doi:10.1016/j.soilbio.2005.01.016
Steinkellner, S., and Langer, I. (2004). Impact of tillage on the incidence of Fusarium spp. in soil. Plant Soil 267 (1), 13–22. doi:10.1007/s11104-005-2574-z
Sun, B. J., Sun, L. Y., Tugume, A. K., Adams, M. J., Yang, J., Xie, L. H., et al. (2013). Selection pressure and founder effects constrain genetic variation in differentiated populations of soilborne bymovirus wheat yellow mosaic virus (potyviridae) in China. Phytopathology® 103 (9), 949–959. doi:10.1094/PHYTO-01-13-0013-R
Sunagawa, S., Coelho Luis, P., Chaffron, S., Kultima Jens, R., Labadie, K., Salazar, G., et al. (2015). Ocean plankton. Structure and function of the global ocean microbiome. Science 348 (6237), 1261359. doi:10.1126/science.1261359
Tamayo-Velez, A., and Osorio, N. W. (2016). Co-inoculation with an arbuscular mycorrhizal fungus and a phosphate-solubilizing fungus promotes the plant growth and phosphate uptake of avocado plantlets in a nursery. Botany 95, 539–545. doi:10.1139/cjb-2016-0224
Tao, J., Meng, D., Qin, C., Liu, X., Liang, Y., Xiao, Y., et al. (2018). Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl. Microbiol. Biotechnol. 102 (8), 3805–3818. doi:10.1007/s00253-018-8837-4
Thébault, E., and Fontaine, C. (2010). Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329 (5993), 853–856. doi:10.1126/science.1188321
van Elsas, J. D., Garbeva, P., and Salles, J. (2002). Effects of agronomical measures on the microbial diversity of soils as related to the suppression of soil-borne plant pathogens. Biodegradation 13 (1), 29–40. doi:10.1023/A:1016393915414
Vinodkumar, S., Nakkeeran, S., Renukadevi, P., and Mohankumar, S. (2018). Diversity and antiviral potential of rhizospheric and endophytic Bacillus species and phyto-antiviral principles against tobacco streak virus in cotton. Agric. Ecosyst. Environ. 267, 42–51. doi:10.1016/j.agee.2018.08.008
Wang, Q., Yang, R., Peng, W., Yang, Y., Ma, X., Zhang, W., et al. (2021). Tea plants with gray blight have altered root exudates that recruit a beneficial rhizosphere microbiome to prime immunity against aboveground pathogen infection. Front. Microbiol. 12, 774438. doi:10.3389/fmicb.2021.774438
Wang, Y., Li, C., Tu, C., Hoyt, G. D., DeForest, J. L., and Hu, S. (2017). Long-term no-tillage and organic input management enhanced the diversity and stability of soil microbial community. Sci. Total Environ. 609, 341–347. doi:10.1016/j.scitotenv.2017.07.053
Winter, M., Samuels, P. L., Dong, Y., and Dill-Macky, R. (2019). Trichothecene production is detrimental to early root colonization by Fusarium culmorum and F. graminearum in fusarium crown and root rot of wheat. Plant Pathol. 68 (1), 185–195. doi:10.1111/ppa.12929
Wu, C., Wang, F., Ge, A., Zhang, H., Chen, G., Deng, Y., et al. (2021a). Enrichment of microbial taxa after the onset of wheat yellow mosaic disease. Agric. Ecosyst. Environ. 322, 107651. doi:10.1016/j.agee.2021.107651
Wu, C., Wang, F., Zhang, H., Chen, G., Deng, Y., Chen, J., et al. (2021b). Enrichment of beneficial rhizosphere microbes in Chinese wheat yellow mosaic virus-resistant cultivars. Appl. Microbiol. Biotechnol. 105 (24), 9371–9383. doi:10.1007/s00253-021-11666-4
Xiong, C., Zhu, Y.-G., Wang, J.-T., Singh, B., Han, L.-L., Shen, J.-P., et al. (2021). Host selection shapes crop microbiome assembly and network complexity. New Phytol. 229 (2), 1091–1104. doi:10.1111/nph.16890
Xu, L., Nicolaisen, M., Larsen, J., Zeng, R., and Dai, F. (2020). Pathogen infection and host-resistance interactively affect root-associated fungal communities in watermelon. Front. Microbiol. 11, 605622. doi:10.3389/fmicb.2020.605622
Xu, T., Jiang, W., Qin, D., Liu, T., Zhang, J., Chen, W., et al. (2021). Characterization of the microbial communities in wheat tissues and rhizosphere soil caused by dwarf bunt of wheat. Sci. Rep. 11 (1), 5773. doi:10.1038/s41598-021-85281-8
Xun, W., Liu, Y., Li, W., Ren, Y., Xiong, W., Xu, Z., et al. (2021). Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 9 (1), 35. doi:10.1186/s40168-020-00985-9
Yang, J., Chen, J., Chen, J., Jiang, H., Zhao, Q., and Adams, M. (2001). Sequence of a second isolate of Chinese wheat mosaic Furovirus. J. Phytopathol. 149 (3-4), 135–140. doi:10.1046/j.1439-0434.2001.00598.x
Yang, H., Li, J., Xiao, Y., Gu, Y., Liu, H., Liang, Y., et al. (2017). An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front. Microbiol. 8, 2179. doi:10.3389/fmicb.2017.02179
Yang, J., Liu, P., Zhong, K. L., Ge, T. D., Chen, L., Hu, H. C., et al. (2022). Advances in understanding the soil-borne viruses of wheat: From the laboratory bench to strategies for disease control in the field. Phytopathol. Res. 4 (1), 27. doi:10.1186/s42483-022-00132-2
Yang, J. P., Chen, J. P., Cheng, Y., Chen, J., Jiang, H. M., Liu, Q., et al. (2002). Responses of some American, European and Japanese wheat cultivars to soil-borne wheat viruses in ChinaCNKI:SUN:ZGNX. Agric. Sci. China 1 (010), 1141–1150.
Yang, W., Jing, X., Guan, Y., Zhai, C., Wang, T., Shi, D., et al. (2019). Response of fungal communities and Co-occurrence network patterns to compost amendment in black soil of northeast China. Front. Microbiol. 10, 1562. doi:10.3389/fmicb.2019.01562
Zhang, H., Wu, C., Wang, F., Wang, H., Chen, G., Cheng, Y., et al. (2021). Wheat yellow mosaic enhances bacterial deterministic processes in a plant-soil system. Sci. Total Environ. 812, 151430. doi:10.1016/j.scitotenv.2021.151430
Zhang, H., Wu, C., Wang, F., Wang, H., Chen, G., Cheng, Y., et al. (2022). Wheat yellow mosaic enhances bacterial deterministic processes in a plant-soil system. Sci. Total Environ. 812, 151430. doi:10.1016/j.scitotenv.2021.151430
Zhang, J., Liu, S., Liu, C., Wang, H., Luan, J., Liu, X., et al. (2021). Soil bacterial and fungal richness and network exhibit different responses to long-term throughfall reduction in a warm-temperate oak forest. Forests 12 (2), 165. doi:10.3390/f12020165
Zhang, Q., Deng, J., Zhu, W., Wu, S., Bai, X., Zhou, Y., et al. (2021). Land-use types combined with plant species alter soil fungal community and functional guilds in the eastern mountainous region of liaoning province, China. Pol. J. Environ. Stud. 30 (1), 477–495. doi:10.15244/pjoes/121047
Zhang, Z., Yuan, Y., Zhao, W., He, H., Li, D., He, W., et al. (2017). Seasonal variations in the soil amino acid pool and flux following the conversion of a natural forest to a pine plantation on the eastern Tibetan Plateau, China. Soil Biol. Biochem. 105, 1–11. doi:10.1016/j.soilbio.2016.11.002
Zheng, M.-Y., Chen, C.-C., Lin, H.-H., Tseng, C.-H., and Hsu, C.-H. (2021). Research on the impact of popular tourism program involvement on rural tourism image, familiarity, motivation and willingness. Sustainability 13 (9), 4906. doi:10.3390/su13094906
Keywords: fungal community, bulk soil, rhizosphere soil, co-occurrence network, plant health
Citation: Lu Q, Hu C, Cai L, Wu C, Zhang H, Wei L, Zhang T, Hu H, Liu S, Lei J, Ge T, Dai L, Yang J and Chen J (2022) Changes in soil fungal communities after onset of wheat yellow mosaic virus disease. Front. Bioeng. Biotechnol. 10:1033991. doi: 10.3389/fbioe.2022.1033991
Received: 01 September 2022; Accepted: 30 September 2022;
Published: 17 October 2022.
Edited by:
Mengcen Wang, Zhejiang University, ChinaReviewed by:
Feng Chen, Henan Agricultural University, ChinaTong Zhou, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2022 Lu, Hu, Cai, Wu, Zhang, Wei, Zhang, Hu, Liu, Lei, Ge, Dai, Yang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Yang, bmF0aGVyMjAwOEAxNjMuY29t; Jianping Chen, Y2hlbmppYW5waW5nQG5idS5lZHUuY24=
†These authors have contributed equally to this work
 Qisen Lu
Qisen Lu Cailin Hu1†
Cailin Hu1† Liang Wei
Liang Wei Tida Ge
Tida Ge Jian Yang
Jian Yang