
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 06 January 2023
Sec. Industrial Biotechnology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.1028151
This article is part of the Research Topic Methods In Industrial Biotechnology & Bioprocess Engineering – Microalgae As A Source of Valuable Compounds View all 5 articles
The unicellular halotolerant cyanobacterium Aphanothece halophytica is known as a potential hydrogen (H2) producer. This study aimed to investigate the enhancement of H2 production under nutrient deprivation. The results showed that nitrogen and potassium deprivation induced dark fermentative H2 production by A. halophytica, while no differences in H2 production were found under sulfur and phosphorus deprivation. In addition, deprivation of nitrogen and potassium resulted in the highest H2 production in A. halophytica due to the stimulation of hydrogenase activity. The effect of adaptation time under nitrogen and potassium deprivation on H2 production was investigated. The results showed that the highest H2 accumulation of 1,261.96 ± 96.99 µmol H2 g dry wt−1 and maximum hydrogenase activity of 179.39 ± 8.18 µmol H2 g dry wt−1 min−1 were obtained from A. halophytica cells adapted in the nitrogen- and potassium-deprived BG11 medium supplemented with Turk Island salt solution (BG110-K) for 48 h. An increase in hydrogenase activity was attributed to the decreased O2 concentration in the system, due to a reduction of photosynthetic O2 evolution rate and a promotion of dark respiration rate. Moreover, nitrogen and potassium deprivation stimulated glycogen accumulation and decreased specific activity of pyruvate kinase. Transcriptional analysis of genes involved in H2 metabolism using RNA-seq confirmed the above results. Several genes involved in glycogen biosynthesis (glgA, glgB, and glgP) were upregulated under both nitrogen and potassium deprivation, but genes regulating enzymes in the glycolytic pathway were downregulated, especially pyk encoding pyruvate kinase. Interestingly, genes involved in the oxidative pentose phosphate pathway (OPP) were upregulated. Thus, OPP became the favored pathway for glycogen catabolism and the generation of reduced nicotinamide adenine dinucleotide phosphate (NADPH), which resulted in an increase in H2 production under dark anaerobic condition in both nitrogen- and potassium-deprived cells.
Huge amounts of energy are used by humans in households, in industry, and in agriculture. Fossil fuel is the main energy source worldwide, but it is non-renewable and limited in supply. Due to annual increases in energy consumption, there is a high demand for fossil fuel. As a result, greenhouse gases produced from the combustion of fossil fuels have increased. This has led to environmental problems, including global warming. In addition, the amount of fossil fuel will not be sufficient for human energy consumption in the future. Hydrogen (H2) is an alternative energy carrier that can be used instead of the limited fossil fuel resources since its combustion releases a high heating value of 141.6 MJ kg−1 (Perry 1963). This combustion of H2 is an environmentally friendly process that does not produce greenhouse gases as by-products (Gupta and Parkhey 2017). H2 can be produced by various types of microorganisms, such as purple bacteria, green sulfur bacteria, green algae, and cyanobacteria.
Cyanobacteria or blue-green algae are prokaryotic and photoautotrophic microorganisms that have oxygenic photosynthetic activity (Bothe et al., 2010). Cyanobacteria can convert abundant raw materials for photosynthesis such as water, sunlight, and CO2 into carbohydrate and O2. Cyanobacteria can produce H2 through many processes, depending on the cyanobacterial type and metabolism. Three enzymes are involved in H2 metabolism in cyanobacteria, namely, bidirectional hydrogenase, uptake hydrogenase, and nitrogenase (Khetkorn et al., 2013). Despite the fact that cyanobacteria are capable of photobiological H2 production, all enzymes involved are sensitive to O2. To sustain and increase the yield of H2 production and prevent enzyme inactivity from O2, a two-stage H2 production scheme was proposed (Ananyev et al., 2012). In the first stage, cyanobacteria are grown in complete media to accumulate biomass. Then, cells enter the second stage by incubating under stress conditions and produce H2 when anaerobic conditions are established.
The deliberate depletion of nutrients in media is a technique used to induce H2 production and is an effective way to maintain sustainable H2 production in cyanobacteria (Srirangan et al., 2011). Under nitrogen deprivation, A. halophytica shows high H2 production in dark anaerobic conditions (Taikhao et al., 2013; Taikhao et al., 2015). Moreover, nitrogen deprivation also induces H2 evolution in Oscillatoria brevis, Calothrix membranacea (Lambert and Smith 1977), and Anabaena siamensis TISIR 8012 (Khetkorn et al., 2010; Taikhao and Phunpruch 2017). Under nitrogen deprivation, the accumulation of glycogen is carried out via photoautotrophic processes. The accumulated glycogen is degraded into glucose-6-phosphate with the generation of reduced nicotinamide adenine dinucleotide phosphate (NADPH), the electron donor of [NiFe]-H2ase, and hydrogen is generated thereafter under dark anaerobic conditions (Ananyev et al., 2008). Previous research showed that under nitrogen deprivation, the bidirectional H2ase activity of A. halophytica was obviously induced during dark anaerobic incubation and the expression of narB encoding ferredoxin-nitrate reductase was downregulated (Phunpruch et al., 2016). Consequently, reduced nicotinamide adenine dinucleotide (NADH) was enhanced and subsequently H2 was increasingly generated (Phunpruch et al., 2016). In addition, sulfur deprivation enhanced H2 production in Gloeocapsa alpicola (Antal and Lindblad 2005) and Microcystis aeruginosa (Rashid et al., 2009). Potassium deficiency increased H2 production by Synechocystis sp. PCC 6803 (Ueda et al., 2016).
The unicellular halotolerant cyanobacterium Aphanothece halophytica, originally isolated from Solar Lake (Israel), has the ability to grow at high salinity at concentrations of NaCl up to 3 M (Takabe et al., 1988). Since it is an obligately halophilic strain that is unable to grow under NaCl-deprived conditions, it is a model microorganism for studying the halotolerance mechanism in cyanobacteria. A. halophytica is well able to produce dark fermentative H2 under nitrogen depletion in enriched medium (Taikhao et al., 2013). It has advantages of growing and producing H2 when cells were incubated in natural seawater without any supplementation of NaNO3 (Taikhao et al., 2015). The bidirectional [NiFe]-H2ase was identified in A. halophytica (Phunpruch et al., 2016). Moreover, H2 production by A. halophytica was enhanced by cell immobilization using alginate and agar (Pansook et al., 2016; Pansook et al., 2019a). In addition, A. halophytica treated with photosystem II inhibitors clearly increased H2 production (Pansook et al., 2019b). Recently, simazine, an herbicide of the triazine class, was shown to significantly enhance H2 production by A. halophytica under dark anaerobic condition (Pansook et al., 2022). In addition, sodium sulfide was an effective reducing agent for enhancing H2 production by A. halophytica due to the reduced O2 concentration in the system, thus increasing hydrogenase activity (Chinchusak et al., 2022).
In this study, we focused on H2 production by A. halophytica under nutrient-deprived conditions and investigated H2 metabolism under these conditions. Photosynthesis and carbon and nitrogen assimilation pathways in N- and K-deprived cells were studied by RNA-seq based transcriptome analysis. H2ase activity, glycogen content, and pyruvate kinase activity were also determined. The patterns of the transcriptome and experimental evidence were mutually supportive.
Aphanothece halophytica was grown photoautotrophically in a 250-ml Erlenmeyer flask containing 100 ml of BG11 medium (pH 7.4) (Rippka et al., 1979) supplemented with Turk Island salt solution (Garlick et al., 1977). The BG11 medium consisted of 17.6 mM NaNO3 as a nitrogen source. The Turk Island salt solution contained high concentrations of minerals, including 0.5 M NaCl, 49 mM MgCl2•6H2O, 30 mM MgSO4•7H2O, and 8.9 mM KCl as main components. Cells were initially adjusted to a density of OD730 at approximately 0.1 and subsequently shaken at 120 rpm on a rotary shaker at 30°C under a white-light fluorescence intensity of 30 µmol photons m−2 s−1 with a light (18 h/day): dark (6 h/day) cycle for 7 days.
H2 production by A. halophytica was measured as part of its two-stage cultivation. Firstly, A. halophytica was grown in BG11 with Turk Island salt solution for 7 days to accumulate biomass. The seven-day-old cells were harvested by centrifugation at 8,000 × g at 4 C for 10 min before resuspension in various types of single nutrient-depleted media: nitrogen-deprived BG11 supplemented with Turk Island salt solution (BG110), phosphorus-deprived BG11 supplemented with Turk Island salt solution (BG11-P), potassium-deprived BG11 supplemented with Turk Island salt solution (BG11-K), and sulfur-deprived BG11 supplemented with Turk Island salt solution (BG11-S). To remove nitrogen in BG110, NaNO3 was omitted from BG11. In BG11-P, NaH2PO4 was omitted from BG11. To remove potassium from BG11-K, KCl was omitted from Turk Island salt solution. In addition, K2HPO4 was also omitted from BG11 and replaced by the addition of Na2HPO4. In BG11-S, ZnSO4•7H2O and CuSO4•5H2O were omitted from BG11 and replaced by ZnCl2 and CuCl2, respectively. In addition, MgSO4•7H2O was omitted from both BG11 and Turk Island salt solution and replaced by the addition of MgCl2. Thereafter, the cell suspensions were incubated for 24 h before H2 was measured, under anaerobic conditions.
Aphanothece halophytica adapted in each single nutrient-deprived media for 24 h was harvested by centrifugation and resuspended in a fresh medium. Five ml of cell suspension was transferred to a 10-ml glass vial. The vial was sealed with a rubber stopper and the cyanobacterial cells were made to enter anaerobic conditions by purging with argon gas for 10 min. Each cell suspension was then incubated at 30°C under darkness for 2 h before H2 measurement was undertaken. H2 measurement was performed using a Gas Chromatograph (Hewlett-Packard HP5890A, Japan) with a molecular sieve 5 °A 60/80 mesh packed column and a thermal conductivity detector using argon as the carrier gas (Baebprasert et al., 2010). H2 production was expressed as µmol H2 evolved per milligram dry cell weight. All experiments were done in triplicate.
Bidirectional hydrogenase activity was determined by measuring H2 evolution in the presence of dithionite-reduced methyl viologen. The reaction mixture comprised 1 ml of cell suspension and 1 ml of 25 mM sodium phosphate buffer (pH 7.6) containing 10 mM methyl viologen dichloride hydrate (Roche, Germany) and 40 mM sodium dithionite (Sigma, Germany). The mixture was incubated under argon atmospheric conditions at 30°C under darkness for 30 min. H2 from headspace was subsequently analyzed by a gas chromatograph.
For each dry cell weight determination, 5 ml of cyanobacterial cell suspension was filtered through a glass microfiber filter GF/C (Whatman, United Kingdom). Sterile distilled water was used to wash the cells collected on the microfiber filter. Then, the filter containing cells was dried in an oven at 70°C until weight was constant. The dry cell weight was calculated. For glycogen content determination, glycogen extraction and hydrolysis were performed following the previous studied procedure (Ernst et al., 1984). A. halophytica cells that had been adapted in various media were harvested by centrifugation at 8,000 × g at 4°C for 10 min and resuspended in fresh medium that contained chlorophyll a at a concentration of 300 µg chl a mL−1. Fifty µL of each cell suspension was added to 200 µL of 30% (w/v) KOH. The mixture was then incubated at 100 °C in a water bath for 90 min. Cells were lysed by ultrasonication at 20% pulse for 5 min on ice. To precipitate glycogen, 600 µL of absolute ethanol was added into the extract. Each sample was kept on ice for 1 h. Glycogen was collected by centrifugation at 12,000 × g for 5 min at 4 C. Then, each glycogen pellet was washed twice by absolute ethanol and subsequently dried at 60°C for 10 min. Glycogen was resuspended in 300 µL of 100 mM acetate buffer (pH 4.75). For each sample, glycogen was digested into glucose by adding amyloglucosidase from Aspergillus niger (Sigma, United States) and α-amylase from Aspergillus oryzae (Sigma, Switzerland) at final concentrations of 4 and 8 units, respectively. The reaction was incubated at 25°C for 1 h. Insoluble membrane fragments were removed by centrifugation at 12,000 × g at 4°C for 5 min. Total sugar in each supernatant was determined by phenol-sulfuric acid assay (Dubois et al., 1956). Standard glycogen from bovine liver (Sigma, United States) (0–100 µg) was used. The glycogen content was calculated using a standard calibration curve. Each experiment was performed in triplicate.
Pyruvate kinase activity was determined indirectly by measuring the oxidation of NADH in the reaction catalyzed by lactate hydrogenase. The activity of pyruvate kinase was determined by a protocol previously described (Malcovati and Valentini, 1982). To prepare cell-free extract of A. halophytica, 100 ml of A. halophytica cell culture was harvested by centrifugation at 8,000 × g at 4°C for 10 min and subsequently resuspended in 1 ml of 10 mM (2-[4-(2-hydroxyethyl) piperazine-1-yl] ethanesulfonic acid (HEPES) buffer (pH 7.5). Cells were lysed using 20% pulse of ultrasonication on ice for 5 min. The cell-free extract was obtained after centrifugation at 8,000 × g at 4°C for 10 min. To assay pyruvate kinase activity, 50 µL of cell-free extract was added into 950 µL of reaction mixture containing 10 mM HEPES buffer, 10 mM MgCl2, 50 mM KCl, 20 mM ADP (Adenosine 5′-diphosphate sodium salt) (Sigma, United States), 10 mM PEP (Phosphoenolpyruvic acid monopotassium salt) (Sigma, Switzerland), 5 mM NADH (β-nicotinamide adenine dinucleotide, reduced disodium salt hydrate) (Sigma, United States), and 0.5 U of L-lactate dehydrogenase (Roche, Germany). The reaction mixture without PEP was used as a negative control. The decrease of NADH was measured spectrophotometrically at a wavelength of 340 nm at 25°C. One unit of pyruvate kinase activity is defined as the amount of enzyme that catalyzes the production of 1.0 µmol pyruvate from the substrate PEP in 1 min. The specific activity of pyruvate kinase was measured as activity per total protein concentration. Protein concentration was determined by Bradford assay (Bradford 1976).
Seven-day old A. halophytica cells adapted in BG110, BG11-K, and BG110K for 2 days were harvested by centrifugation at 12,000 × g at 4 C for 10 min. The total RNA of each sample was extracted using QIAzol Lysis Reagent (Qiagen, United States). Then, total RNA was qualified and quantified using a NanoDrop 2000/2000c spectrophotometer (Thermo Fisher Scientific, United States). Ribosomal RNA (rRNA) was removed by using a Ribo-Zero rRNA removal Kit (Epicentre, United States) following the manufacturer’s instructions. The construction of the next generation sequencing library was conducted by Genewiz (China) according to the standard protocols. The different indices of libraries were multiplexed and loaded on an Illumina HiSeq/Novaseq instrument according to the instructions of the manufacturer (Illumina, United States). RNA sequencing was performed using the 2 × 150 paired end (PE) configuration. Image analysis and base calling were conducted using the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument. All RNA sequencing and alignment processes were conducted by Genewiz (China).
All experiments were performed in triplicate. All data are shown as the mean ± standard deviation. One-way analysis of variance (ANOVA) was used for analysis to the 95% confidence level with IBM SPSS version 19 software.
The result showed that A. halophytica cells incubated in BG110 and BG11-K showed clearly higher H2 production than those in BG11, BG11-P, and BG11-S (Figure 1). The highest H2 accumulation of 332.82 ± 34.02 µmol H2 g dry cell wt−1 was shown by cells incubated in BG11-K for 24 h. It was 33-fold higher than that of cells cultivated in normal BG11 medium (10.01 ± 5.92 µmol H2 g dry cell wt−1) (Figure 1). In addition, cells incubated in BG110 accumulated maximum H2 of 263.75 ± 31.58 µmol H2 g dry cell wt−1, or 26-fold higher than that of cells incubated in normal BG11 medium (Figure 1). This study demonstrated the activation of H2 production by A. halophytica under nitrogen and potassium deprivation.
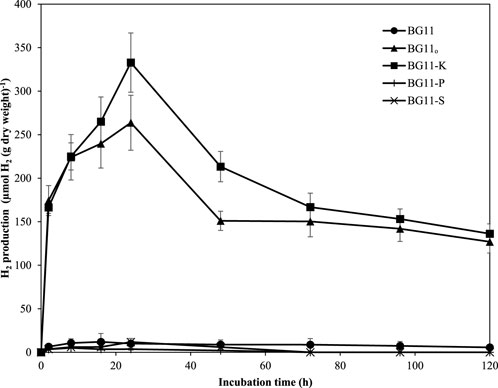
FIGURE 1. H2 production (µmol H2 g dry wt−1) by A. halophytica incubated for various times in BG11 (•), BG110 (▲), BG11-K (■), BG11-P (+), and BG11-S (*) medium under dark anaerobic condition. Data are means ± SD (n = 3).
In this study, the effects of various concentrations of nitrogen and potassium on H2 production and H2ase activity by A. halophytica with focus on potassium and nitrogen deprivation was investigated. The results showed that during potassium deprivation, a decrease in nitrogen concentration resulted in higher H2ase activity and H2 production by A. halophytica (Table 1). Similarly, during nitrogen deprivation, lower potassium concentration resulted in significantly higher H2 production and H2ase activity by A. halophytica (Table 1). The highest H2ase activity with 120.05 ± 8.98 µmol H2 g dry wt−1 min−1 and maximum H2 accumulation with 507.51 ± 13.78 µmol H2 g dry wt−1 was shown in cells incubated under a combined K- and N-deprived conditions (Table 1). This H2ase activity and H2 production was approximately 40 and 50-fold higher than those under normal conditions (incubated in BG11), respectively. The deprivation of both nitrogen and potassium together promoted H2ase activity, and thus resulted in higher H2 production.
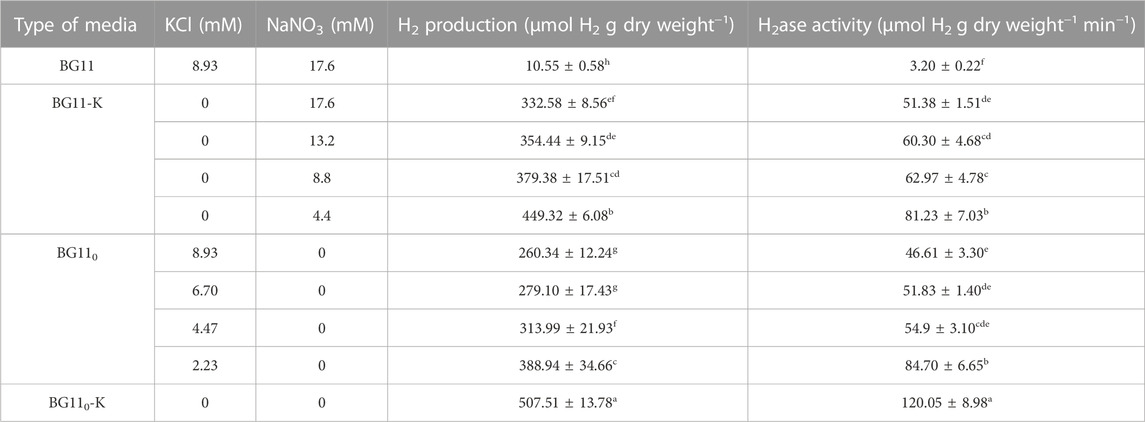
TABLE 1. H2 production and H2ase activity by A. halophytica cells incubated in various types of media: BG11 supplemented with Turk Island salt solution (BG11), potassium-deprived BG11 supplemented with Turk Island salt solution (BG11-K) containing various concentrations of NaNO3, nitrogen-deprived BG11 supplemented with Turk Island salt solution (BG110) containing various concentrations of KCl, and potassium- and nitrogen-deprived BG11 supplemented with Turk Island salt solution (BG110-K) for 24 h. Data are means ± SD (n = 3). Different letters in columns indicate a significant difference, and the same letter indicates no significant difference according to Duncan’s multiple range test at p < 0.05.
In this study, A. halophytica cells were adapted in BG11, BG110, BG11-K, and BG110-K media under photoautotrophic conditions with light exposure (18 h/day) for 1, 2, 3, 4, and 5 days. The hydrogenase activity, H2 production, and O2 production of adapted cells were measured after incubation under dark anaerobic condition for 24 h. The result showed that under nitrogen and/or potassium deprivation, A. halophytica cells gave higher H2 production and hydrogenase activity than cells under normal condition (Figures 2A, B). On the other hand, O2 production was highest in cells adapted in normal BG11 medium (Figure 2C). The combined deprivation of nitrogen and potassium promoted higher H2ase activity and H2 production in A. halophytica than did single nutrient deprivation. A. halophytica incubated under nitrogen and potassium deprivation showed maximum H2 production rate of 40.87 ± 1.07 µmol H2 g dry wt−1 h−1. The highest H2 production of 1,261.96 ± 96.99 µmol H2 g dry wt−1 was obtained in cells adapted in BG110-K for 2 days (Figure 2A). The lowest O2 evolution rate of 8.81 ± 2.93 nmol O2 g dry wt−1 h−1 was obtained in cells adapted in BG110-K after 3 days (Figure 2C). It was found that H2 production was related to H2ase activity (Figures 2A, B). H2ase activity increased dramatically at the first 48 h of adaptation incubation and the highest H2ase activity with 179.39 ± 8.18 µmol H2 g dry wt−1 min−1 was found in cells adapted in BG110-K for 2 days (Figure 2B).
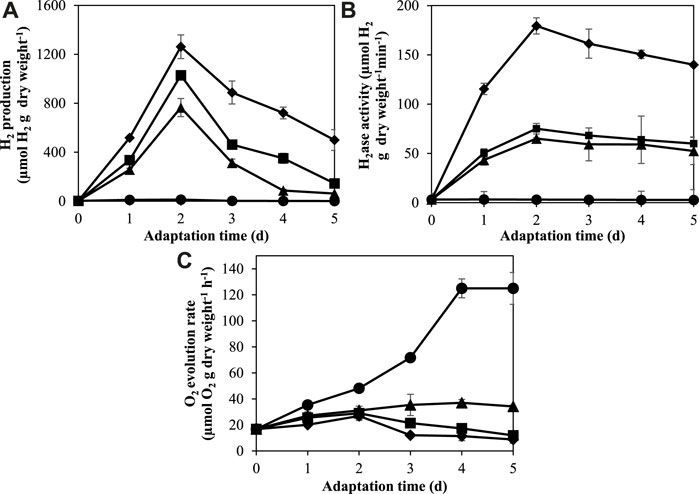
FIGURE 2. Effect of adaptation time on H2 production (A), H2ase activity (B), and O2 evolution rate (C) by A. halophytica cells. A. halophytica was adapted in various kinds of media for various times, BG11 (•), BG110 (▲), BG11-K (■), and BG110-K (⬧) for 1–5 days before harvesting and resuspending in a fresh medium. H2 production was measured in incubated cells under dark anaerobic condition for 24 h. Data are means ± SD (n = 3).
The glycogen content of A. halophytica cells adapted in BG11, BG110, BG11-K, and BG110-K media for 1, 2, 3, 4, and 5 days was measured. The results revealed that under nitrogen and/or potassium deprivation, A. halophytica clearly produced and accumulated higher intracellular glycogen than under normal conditions. The glycogen content of cells adapted in BG110, BG11-K, and BG110-K clearly increased during the first 3 days of adaptation, whereas in the case of BG11 adapted cells, it was almost constant over the period of adaptation (Figure 3A). The highest glycogen content at 65.53 ± 0.78% of cell dry wt was obtained in cells adapted in BG110-K for 3 days (Figure 3A). It was approximately three times higher than glycogen content of cells adapted in BG11 (24.66 ± 1.97% of cell dry wt) (Figure 3A). In the case of only single-nutrient deprivation, glycogen accumulated under potassium deprivation was higher than that under nitrogen deprivation. Under K-deprivation, cells accumulated the maximum glycogen content of 56.13 ± 0.66% of cell dry wt whereas under N-deprivation, cells accumulated the maximum glycogen content at 51.95 ± 2.11% of cell dry wt after 3 days of adaptation.
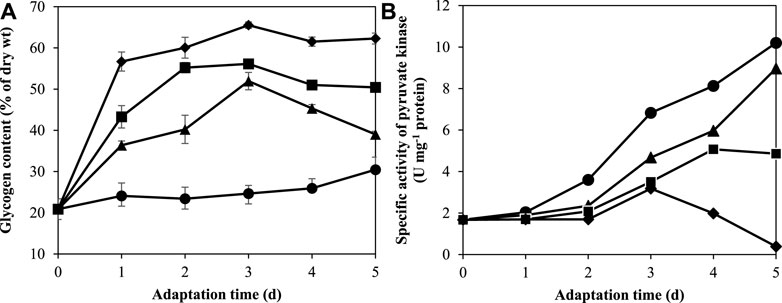
FIGURE 3. Effect of adaptation time on glycogen content (A) and specific activity of pyruvate kinase (B) by A. halophytica cells. A. halophytica was adapted in various kinds of media for various times, BG11 (•), BG110 (▲), BG11-K (■), and BG110-K (⬧) for 1–5 days before harvesting and resuspending in a fresh medium. H2 production was measured in incubated cells under dark anaerobic condition for 24 h. Data are means ± SD (n = 3).
Pyruvate kinase (PK) is known to be activated by K+ (Oria-Hernández et al., 2005). It has an important role in regulating the glycolytic pathway in carbohydrate metabolism. The measurement of pyruvate kinase (PK) activity was performed in A. halophytica cells adapted in BG11, BG110, BG11-K, and BG110-K for 1, 2, 3, 4, and 5 days. The results showed that specific activity of PK was highest with 10.20 ± 0.60 U mg−1 protein in A. halophytica adapted in BG11 for 5 days (Figure 3B). The deprivation of either nitrogen or potassium reduced specific activity of PK significantly compared with normal conditions (Figure 3B). Similarly, combined deprivation of both nitrogen and potassium had significant effects on PK activity. It was especially notable that after 5 days of adaptation time, cells clearly showed very low PK specific activity at 0.39 ± 0.01 U mg−1 protein (Figure 3B).
Since O2 is a strong inhibitor of H2ase, the presence of O2 in systems plays an important role in H2 production. In cyanobacteria, O2 is generated via oxygenic photosynthesis and the produced O2 is consumed in cellular respiration via the electron transport chain. To investigate how O2 evolution decreased under nitrogen and potassium deprivation, the photosynthetic O2 evolution rate and respiration rate of A. halophytica cells adapted in BG11, BG110, BG11-K, and BG110-K for 2 days was measured. The results showed that the highest photosynthetic O2 evolution rate of 16.94 ± 0.64 nmol O2 mg dry wt−1 min−1 was found in cells adapted in BG11. Under single or combined nitrogen and potassium deprivation, its photosynthetic O2 evolution rate decreased (Table 2). The combined deprivation of both nitrogen and potassium resulted in the lowest photosynthetic O2 evolution rate of 2.78 ± 0.09 nmol O2 mg dry wt−1 min−1 (Table 2). On the contrary, O2 consumption by respiration increased under nitrogen and/or potassium deprivation (Table 2). The ratio of photosynthetic O2 evolution rate and respiratory rate was calculated and evaluated. This ratio implies the quantity of O2 in a vial container. The highest ratio between photosynthetic O2 evolution rate and respiratory rate of 22.49 ± 0.81 was found in cells adapted in BG11 for 24 h whereas the lowest ratio of 2.57 ± 0.04 was shown in cells adapted in BG110-K for 24 h (Table 2).
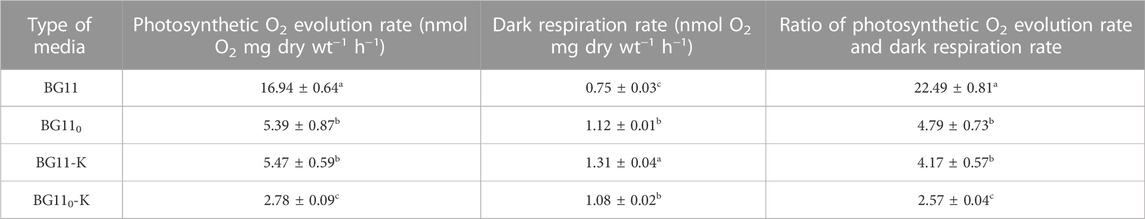
TABLE 2. Photosynthetic O2 evolution rate, dark respiration rate, and ratio of photosynthetic O2 evolution rate and respiratory rate in A. halophytica cells incubated in BG11, BG110, BG11-K, and BG110-K under dark anaerobic condition for 24 h before measuring. Data are means ± SD (n = 3). Different letters in columns indicate a significant difference, and the same letter indicates no significant difference according to Duncan’s multiple range test at p < 0.05.
Gene expression based on the RNA-seq of A. halophytica under nitrogen and potassium deprivation was compared with that under normal conditions. The results are shown in Figure 4. Transcripts of genes involved in photosynthetic, carbon, and nitrogen assimilation pathways were analyzed. In this study, log2FoldChange (log2FC) was used to indicate the differences between transcript expression in the experimental and control groups. The results showed that genes encoding allophycocyanin and phycobilisome under N and K deprivation were expressed as log2FC
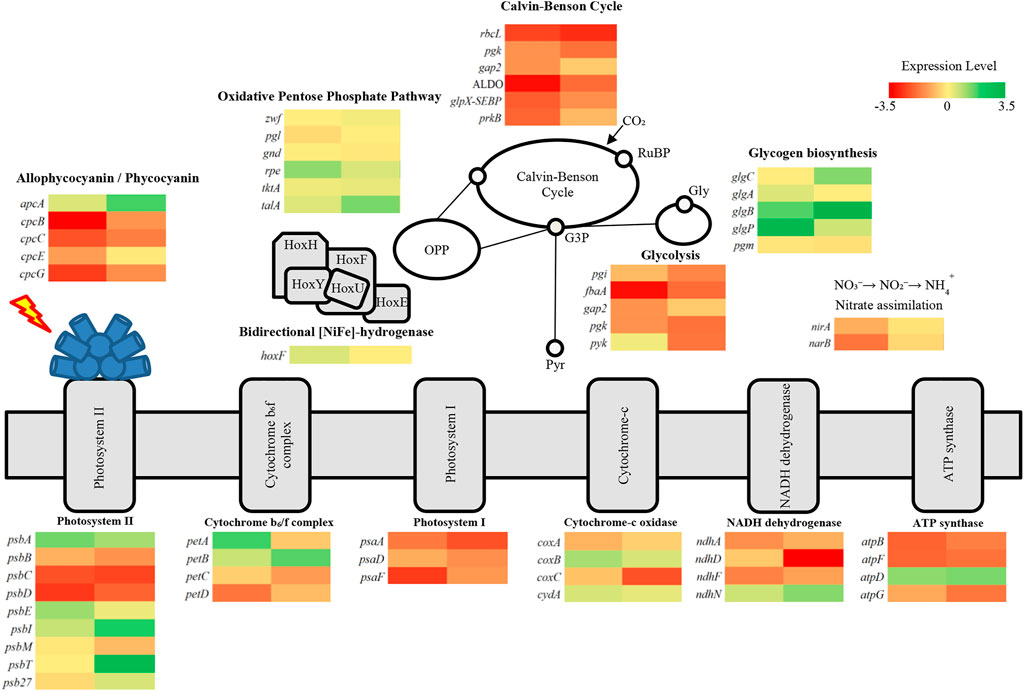
FIGURE 4. Transcriptional level of genes involved in photosynthetic pathway, carbon assimilation such as Calvin-Benson cycle, glycolysis, oxidative pentose phosphate pathway, and nitrate assimilation in A. halophytica. The first column shows differences of gene expression between N-deprived adapted cells and normal cells and the second column shows differences of gene expression between K-deprived adapted cells and normal cells. Expression levels are shown as heat maps with gene names, demonstrating different gene expression in many metabolic pathways involved in H2 production. The metabolic pathways are photosynthesis, oxidative pentose phosphate, glycolytic pathway, glycogen biosynthesis, and nitrate assimilation. Abbreviations: oxidative pentose phosphate pathway, OPP; glucose-3-phosphate, G3P; glycogen, gly; pyruvate, pyr; [NiFe]-bidirectional H2ase enzyme subunits, HoxEFUYH.
Under N and K deprivation, genes involved in the Calvin-Basham-Benson (CBB) cycle were downregulated (Figure 4). Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit encoded by rbcL was significantly down-regulated by log2FC of −2.46 and −1.83 under N and K deprivation, respectively. All genes in this pathway were differently expressed by log2FC
Cyanobacteria are oxygenic microorganisms that can produce H2 under dark anaerobic conditions, especially when they are incubated in nutrient-deprived media (Ananyev et al., 2008; Baebprasert et al., 2010; Min and Sherman 2010). In a previous study, the unicellular halotolerant cyanobacterium A. halophytica showed the ability to produce H2 under nitrogen starvation (Taikhao et al., 2013; Taikhao et al., 2015; Phunpruch et al., 2016). Previously, H2 production was shown to be catalyzed by bidirectional [NiFe]-H2ase (Phunpruch et al., 2016). The results in this study demonstrated that, apart from nitrogen starvation, potassium deprivation could also significantly enhance H2 production by A. halophytica under dark anaerobic conditions compared with normal conditions (Figure 1). Potassium and nitrogen deprivation could induce glycogen catabolism with subsequent generation of increased electrons and hydrogenase activity was simultaneously induced after entering anaerobic conditions. After 24 h of incubation, H2 production was decreased, as a result of the decreased number of electrons and the lower hydrogenase activity. Under potassium and nitrogen deprivation, biomass was not much changed due to the low level of nutrient concentration for cell division. However, A. halophytica incubated in seawater with the supplementation of 378 mmol C L−1 glucose, 0.25 M NaCl, and 0.4 µM Fe3+ at 35°C, pH six gave the highest H2 production at day 8 of dark incubation under anoxic condition, and the high yield of H2 was sustained at least up to 14 days (Taikhao et al., 2015).
Under potassium deprivations, a limitation of the NaNO3 concentration available in BG11 medium induced both H2ase activity and H2 production (Table 1). Similarly, under nitrogen deprivations, a limited KCl concentration in BG11 medium induced both H2ase activity and H2 production (Table 1). In a previous study, it was shown that the presence of potassium reduced H2ase activity in Synechocystis 6,803, under dark anaerobic conditions, which might have been due to competitive utilization of limited NADPH for H2 and organic acid production (Ueda et al., 2016). Moreover, it has been suggested that potassium deprivation decreased PSII activity, lowering the production of O2, which is an inhibitor of H2ase enzyme. Furthermore, potassium deprivation increased the degradation of starch in green alga Scenedesmus obliquus (Papazi et al., 2014) and in Tetraspora sp. CU2551 (Pewnual et al., 2022). With nitrogen deprivation, H2ase activity increased in Anabaena siamensis TISTR8012, resulting in enhanced H2 production (Taikhao and Phunpruch 2017). On the other hand, phosphorus and sulfur deprivation did not promote H2 production by A. halophytica. In addition, potassium, sulfur, and phosphate deprivation was observed to likely have important effects on nucleic acids, protein synthesis, and other metabolic pathways in cells, such as photosynthesis, cellular respiration, and H2 metabolism (Warichanan and Phunpruch 2019). Moreover, a combination of nitrogen and sulfur depletion enhanced H2 production in Synechocystis sp. strain PCC 6803 (Baebprasert et al., 2010) and in Arthrospira sp. PCC 8005 (Raksajit et al., 2012). Table 3 shows the comparison of maximum H2 production rate by A. halophytica under various conditions. It was shown that deprivation of potassium together with nitrogen promoted higher H2 production by A. halophytica than only nitrogen deprivation. However, H2 production rate by free cells of A. halophytica in this study was lower than that of agar-immobilized cells (Pansook et al., 2019b) and free cells incubated in BG110 treated with 25 µM simazine (Pansook et al., 2022) and 50 mM sodium sulfide (Chinchusak et al., 2022).
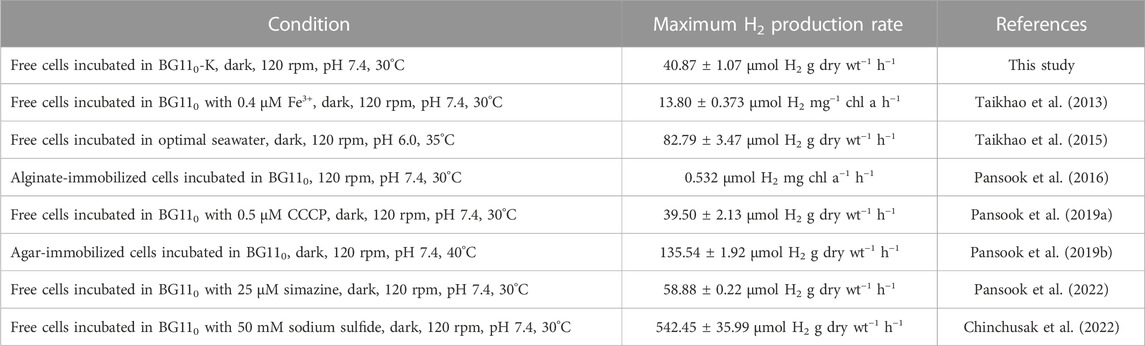
TABLE 3. Maximum dark fermentative H2 production rate by A. halophytica incubated under various conditions.
H2ase, including the [NiFe]-H2ase enzyme in cyanobacteria, is usually sensitive to the presence of O2 (McIntosh et al., 2011). The fundamental pathways involved in O2 generation and consumption in cyanobacteria are photosynthesis and dark respiration, respectively. Our result showed that a combined deprivation of both nitrogen and potassium promoted H2 production by reducing O2 concentration in the vial (Figure 2). A decrease in O2 concentration induced H2ase activity, resulting in a higher H2 production (Figure 2). The balance of O2 concentration in a system therefore plays a decisive role in H2 production. In cyanobacteria, O2 in the system is normally generated by photosynthetic activity of PSII. However, a measurement of H2 production in this study was performed under darkness when photosynthetic activity should have been less than under light conditions. The deprivation of either nitrogen or potassium reduced the photosynthetic O2 evolution rate by 3-fold, whereas combined deprivation of both nitrogen and potassium reduced the photosynthetic O2 evolution rate by 6-fold (Table 2). A decrease in photosynthetic O2 evolution rate caused the reduction of O2 in the system. This in turn induced H2ase activity, resulting in higher H2 production. In the case of nitrogen deprivation, the result was in agreement with a previous study showing the reduction of photosynthetic activities in Synechococcus elongatus PCC 7942 under nitrogen starvation (Choi et al., 2016). Potassium deprivation decreased photosynthetic pigments and activity in Synechocystis sp. strain PCC 6803 (Nanatani et al., 2015) and in Anabaena torulosa (Alahari and Apte 1998).
In addition, the produced O2 can be consumed by dark intracellular respiration. An increased respiration rate for A. halophytica was observed in cells deprived of nitrogen or potassium or both (Table 2). As a result, a decreased level of O2 in the vial containing cells incubated under nitrogen and potassium deprivation was obtained. The ratio of photosynthetic O2 evolution rate and dark respiration rate was used to monitor the level of O2 concentration in the system (Table 2). A higher ratio indicated higher activity of photosynthetic O2 evolution activity over respiration activity, suggesting the presence of high O2 concentration in the vial, and thus lower H2ase activity and lower H2 production. A low ratio of photosynthetic O2 evolution rate to dark respiration rate was found in nitrogen- and potassium-free adapted cells (Table 2). This probably involved a combined effect of nitrogen and potassium deprivation suppressing related genes. In terms of photosynthetic O2 evolution: respiratory rate ratio (Table 2), the results implied that the lower the ratio, the lower the O2 concentration. Cells incubated in deprived media showed lower photosynthetic O2 evolution rate and dark respiration rate than did normal cells. Several genes encoding cytochrome c oxidase and NADH dehydrogenase were upregulated in cells under both nitrogen and potassium starvation (Figure 4). The results suggested that A. halophytica increased dark respiration under nitrogen and potassium starvation. However, no significant differences of expression of genes encoding respiratory electron transport system were reported in Synechococcus elongatus PCC 7942 under nitrogen deprivation (Choi et al., 2016).
H2 production by A. halophytica was established by the two-stage regime: cyanobacterial growth followed by adaptation in nutrient deprivation condition to induce an increase of H2 production. Firstly, cells were grown in rich BG11 medium for 7 days to accumulate biomass. Then, the cells were made to enter the adaptation period, or the second stage, by incubation in nutrient-deprived media to accumulate glycogen under illumination and aerobic conditions. Subsequently, cells were transferred into a glass vial and H2 production was induced under dark anaerobic condition. The adaptation period during nutrient deficiency was very crucial since cells incubated in deprived media needed to accumulate a high content of glycogen, or other chemical compounds, in order to provide electrons for H2ase to produce H2. Individually and combined nitrogen and potassium deprivation were chosen for this study. The result showed that H2ase activity and H2 production was highest in cells adapted in nitrogen- and potassium-deprived media for 48 h (Figures 2A, B), suggesting a high accumulation of glycogen in adapted cells. Glycogen in the adapted cells was extracted and determined. Glycogen was seen to have accumulated more in cells incubated in nitrogen- and potassium-deprived media compared to that of cells incubated in BG11 (Figure 3B). The highest glycogen was found in combined nitrogen- and potassium-free cells during the adaptation period (Figure 3B). This result was confirmed with transcriptional analysis. Transcriptional analysis by RNA-seq showed that the glgA and glgB genes encoding the starch synthase and 1,4-alpha-glucan branching enzymes, respectively, were upregulated under both nitrogen and potassium deprivation (Figure 4). The results suggested that a lack of nitrogen and/or potassium causes an increase in glycogen content, giving rise to higher H2 production (Taikhao et al., 2015).
Under dark anaerobic condition, cyanobacterial H2 can be produced via [NiFe] H2ase reduced by NAD(P)H (Gutekunst et al., 2014) and reduced ferredoxin (Meuer et al., 1999; Gutekunst et al., 2014). The main sources of reductants for H2 generation under dark anaerobic condition are NADH and reduced ferredoxin from glycolysis and NADPH from catabolism of accumulated glycogen, provided by oxidative pentose phosphate (OPP) pathway (Kumaraswamy et al., 2013). In this study, the specific activity of PK in glycolytic pathway decreased in A. halophytica under K deprivation (Figure 3A). The specific activity of PK in K- and NK- free adapted cells was less than that in abundant and N-free adapted cells throughout the period of adaptation time. PK is known to be induced by K+ and it is less activated when K is absent in a system (Oria-Hernández et al., 2005). Therefore, this associated enzyme regulation might have controlled the flux of glycogen catabolism in A. halophytica. Furthermore, GAPDH-2 has a major role in the CBB cycle, using NADPH as a preferred electron donor to reduce 1,3-bis-phosphoglycerate to GAP (Koksharova et al., 1998). Under potassium deprivation, transcriptional analysis revealed that pyk and gap2 encoding PK and GAPDH-2, respectively, were downregulated (Figure 4). Consequently, in potassium-free adapted cells, it could be suggested that upper-glycolytic metabolites could be in excess, especially glucose-6-phophate (G6P). G6P is a broken-down molecule of glycogen, and the branching point between glycolysis and OPP pathway. For a reduced activity of both PK and GADPH-2, it could be the case that G6P preferentially enters the OPP pathway, generating more NADPH and using less NADPH in CBB and glycolysis. Therefore, OPP pathway may well be the favored pathway for H2 production in potassium-deprived adapted cells of A. halophytica under dark anaerobic conditions. In a previous study, the pykF knocking out Escherichia coli mutant provided higher activity of enzymes in OPP, but the activities of glycolytic enzymes decreased compared with those in wild-type cells (Siddiquee et al., 2004). Moreover, low activity of pyruvate kinase was shown to promote respiration in yeast (Grüning et al., 2011). Prior to this study, H2 production was induced in nitrogen-free cells because the expression of genes involved in glycogen catabolism had increased (Osanai et al., 2006). Moreover, several genes in the OPP pathway were extremely upregulated in Synechocystis sp. PCC 6803 incubated in nitrogen-free medium (Osanai et al., 2006). This was probably due to G6P being degraded through the OPP pathway. It seems that glycogen catabolism might preferentially take place via the OPP pathway, producing more NAD(P)H compared to that via the glycolytic pathway in cells incubated in both nitrogen- and potassium-deprived conditions. Therefore, the described effects of both nitrogen and potassium deprivation on H2 production may have been synergistic, and accelerated H2 production in A. halophytica under dark anaerobic conditions.
The schematic H2 production by A. halophytica during deprivation of individual nitrogen and potassium, and combined nitrogen and potassium, under dark anaerobic conditions is shown in Figure 5. The reductants involved in H2 metabolism by cyanobacteria, NADH, and NADPH are generated mainly by the glycolytic and OPP pathways (Kumaraswamy et al., 2013). In the former, one molecule of G6P can provide two molecules of NADH whereas seven molecules of reduced pyridine nucleotides can be produced from the same molecule of G6P (6 NADPH per glucose and one NADH per glucose) via the OPP pathway (Kumaraswamy et al., 2013). One molecule of NAD(P)H can be oxidized by H2ase to provide one molecule of H2. Therefore, the reduced pyridine nucleotides from the OPP pathway can produce more H2 compared with that from the glycolytic pathway (seven molecules vs. two molecules of H2). In plentiful conditions or in the case of the BG11 (Figure 5B (+NK)), cells were more likely to generate organic acid compounds than potassium-deprived adapted cells were. This corresponded with a previous study of Synechocystis sp. PCC 6803 that suggested that, under dark anaerobic conditions, where NADPH was limited, there was a competing demand to consume NADPH for H2 and organic acid production. In the presence of potassium, cells preferred to utilize NADPH for production of organic acids (Ueda et al., 2016). Therefore, NAD(P)H is normally used for organic acid production and the nitrate assimilation pathway rather than H2 production. In nitrogen-deprived cells, it may be that glycogen catabolism occurs via both the OPP and glycolytic pathway, as shown in Synechocystis sp. PCC 6803 (Osanai et al., 2006). In potassium-free cells, PK activity (Figure 3) and the gene expression of pyk encoding PK was lower than that found in normal cells (Figure 4). Consequently, glycogen was mainly degraded via the OPP pathway and provided more NADPH for H2 production.
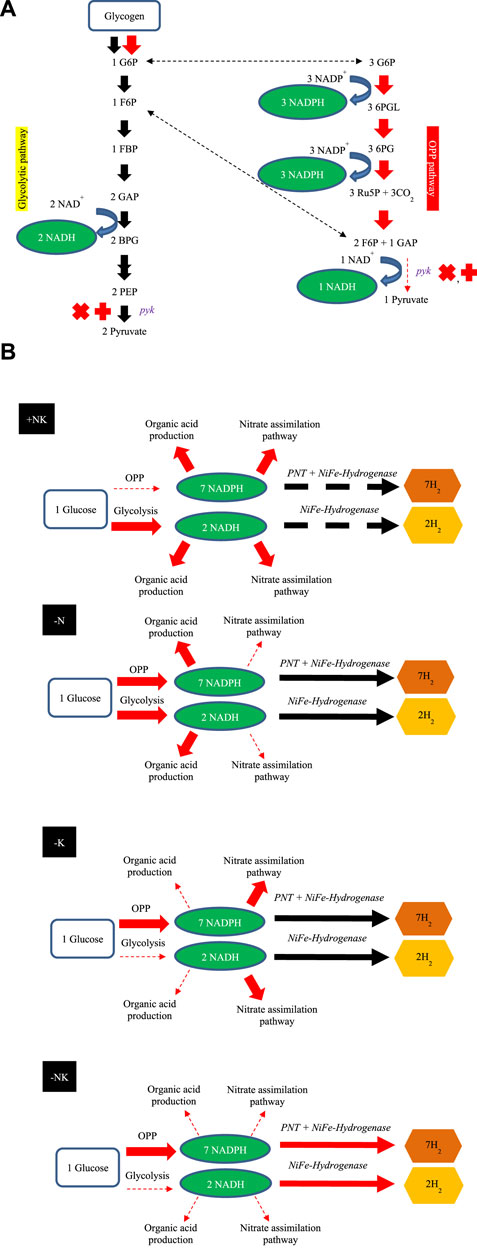
FIGURE 5. Proposed schematic mechanism of H2 production by A. halophytica based on the RNA-seq after adaptation in nitrogen and/or potassium deprivation for 2 days. Glycolytic and OPP pathways are involved in NAD(P)H generation via glycogen catabolism (A). Proposed H2 metabolism and involved pathways of A. halophytica incubated in BG11 (+NK), BG110 (-N), BG11-K (-K), and BG110-K (-NK) under dark anaerobic conditions (B). Arrows show electron transport direction in each pathway.
Under dark anaerobic conditions, the maximum H2 production of 1,261.96 ± 96.99 µmol H2 g dry wt−1 and the maximum hydrogenase activity of 179.39 ± 8.18 µmol H2 g dry wt−1 min−1 was found in A. halophytica cells incubated in the nitrogen- and potassium-deprived BG11 medium supplemented with Turk Island salt solution for 48 h. The increased hydrogenase activity was due to a reduction of O2 in the system, resulting from the lower photosynthetic O2 evolution and higher dark respiration. Under nitrogen and potassium deprivation, A. halophytica cells promoted the production and accumulation of glycogen and reduced pyruvate kinase activity. Transcriptional analysis by RNA-seq helped to understand the effect of nitrogen and potassium deprivation on H2 production by A. halophytica. Several genes involved in glycogen biosynthesis (glgA, glgB, and glgP) were upregulated in both nitrogen- and potassium-deprived cells. However, pyk and most genes that regulated enzymes in the glycolytic pathway were down-regulated in both nitrogen- and potassium-deprived cells. Interestingly, most genes that regulated enzymes in the oxidative pentose phosphate pathway (OPP) were upregulated. Accordingly, the OPP was suggested as a promising pathway for enhancing H2 production under dark anaerobic conditions in both nitrogen- and potassium-deprived A. halophytica cells. This study indicated that combined nitrogen and potassium deprivation in media is a promising strategy to promote sustainable H2 production by the halotolerant cyanobacterium A. halophytica.
The original contributions presented in the study are included in the article/SupplementaryMaterial; further inquiries can be directed to the corresponding author.
SP contributed to conception and design of the study. SP received the research grant. NC performed the experiments and statistical analysis. NC wrote the first draft of the manuscript. SP and NC wrote sections of the manuscript. All authors contributed to manuscript revision, and read and approved the submitted version.
This work was financially supported by King Mongkut’s Institute of Technology Ladkrabang (2563-02-05-33). NC would like to thank School of Science, King Mongkut’s Institute of Technology Ladkrabang for a Ph.D. scholarship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alahari, A., and Apte, S. K. (1998). Pleiotropic effects of potassium deficiency in a heterocystous, nitrogen-fixing cyanobacterium, Anabaena torulosa. Microbiology 144 (6), 1557–1563. doi:10.1099/00221287-144-6-1557
Ananyev, G., Carrieri, D., and Dismukes, G. C. (2008). Optimization of metabolic capacity and flux through environmental cues to maximize hydrogen production by the cyanobacterium “Arthrospira (Spirulina) maxima”. Appl. Environ. Microbiol. 74 (19), 6102–6113. doi:10.1128/AEM.01078-08
Ananyev, G. M., Skizim, N. J., and Dismukes, G. C. (2012). Enhancing biological hydrogen production from cyanobacteria by removal of excreted products. J. Biotechnol. 162 (1), 97–104. doi:10.1016/j.jbiotec.2012.03.026
Antal, T. K., and Lindblad, P. (2005). Production of H2 by sulphur-deprived cells of the unicellular cyanobacteria Gloeocapsa alpicola and Synechocystis sp. PCC 6803 during dark incubation with membrane or at various extracellular pH. J. Appl. Microbiol. 98 (1), 114–120. doi:10.1111/j.1365-2672.2004.02431.x
Baebprasert, W., Lindblad, P., and Incharoensakdi, A. (2010). Response of H2 production and Hox-hydrogenase activity to external factors in the unicellular cyanobacterium Synechocystis sp. Strain PCC 6803. Int. J. Hydrog. Energy 35 (13), 6611–6616. doi:10.1016/j.ijhydene.2010.04.047
Bothe, H., Schmitz, O., Yates, M. G., and Newton, W. E. (2010). Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol. Mol. Biol. Rev. 74 (4), 529–551. doi:10.1128/mmbr.00033-10
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 (1), 248–254. doi:10.1016/0003-2697(76)90527-3
Chinchusak, N., Incharoensakdi, A., and Phunpruch, S. (2022). Enhancement of dark fermentative hydrogen production in nitrogen-deprived halotolerant unicellular cyanobacterium Aphanothece halophytica by treatment with reducing agents. Biomass Bioenergy 167, 106624. doi:10.1016/j.biombioe.2022.106624
Choi, S. Y., Park, B., Choi, I. G., Sim, S. J., Lee, S. M., Um, Y., et al. (2016). Transcriptome landscape of Synechococcus elongatus PCC 7942 for nitrogen starvation responses using RNA-seq. Sci. Rep. 6 (1), 30584. doi:10.1038/srep30584
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. T., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28 (3), 350–356. doi:10.1021/ac60111a017
Ernst, A., Kirschenlohr, H., Diez, J., and Böger, P. (1984). Glycogen content and nitrogenase activity in Anabaena variabilis. Arch. Microbiol. 140 (2-3), 120–125. doi:10.1007/bf00454913
Garlick, S., Oren, A., and Padan, E. (1977). Occurrence of facultative anoxygenic photosynthesis among filamentous and unicellular cyanobacteria. J. Bacteriol. 129 (2), 623–629. doi:10.1128/jb.129.2.623-629.1977
Grüning, N. M., Rinnerthaler, M., Bluemlein, K., Mülleder, M., Wamelink, M. M., Lehrach, H., et al. (2011). Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 14 (3), 415–427. doi:10.1016/j.cmet.2011.06.017
Gupta, P., and Parkhey, P. (2017). Electrohydrogenesis: Energy efficient and economical technology for biohydrogen production. Adv. Biofeedstocks Biofuels Prod. Technol. Biofuels 2, 201–233. doi:10.1002/9781119117551.ch8
Gutekunst, K., Chen, X., Schreiber, K., Kaspar, U., Makam, S., and Appel, J. (2014). The bidirectional NiFe-hydrogenase in Synechocystis sp. PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. J. Biol. Chem. 289 (4), 1930–1937. doi:10.1074/jbc.m113.526376
Khetkorn, W., Lindblad, P., and Incharoensakdi, A. (2010). Enhanced biohydrogen production by the N2-fixing cyanobacterium Anabaena siamensis strain TISTR 8012. Int. J. Hydrog. Energy 35 (23), 12767–12776. doi:10.1016/j.ijhydene.2010.08.135
Khetkorn, W., Khanna, N., Incharoensakdi, A., and Lindblad, P. (2013). Metabolic and genetic engineering of cyanobacteria for enhanced hydrogen production. Biofuels 4 (5), 535–561. doi:10.4155/bfs.13.41
Koksharova, O., Schubert, M., Shestakov, S., and Cerff, R. (1998). Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 36 (1), 183–194. doi:10.1023/a:1005925732743
Kumaraswamy, G. K., Guerra, T., Qian, X., Zhang, S., Bryant, D. A., and Dismukes, G. C. (2013). Reprogramming the glycolytic pathway for increased hydrogen production in cyanobacteria: Metabolic engineering of NAD+-dependent GAPDH. Energy & Environ. Sci. 6 (12), 3722–3731. doi:10.1039/c3ee42206b
Lambert, G. R., and Smith, G. D. (1977). Hydrogen formation by marine blue-green algae. FEBS Lett. 83 (1), 159–162. doi:10.1016/0014-5793(77)80664-9
Malcovati, M., and Valentini, G. (1982). AMP- and Fructose-1, 6, -biphosphate-activated pyruvate kinases from Escherichia coli. Methods Enzym. 90, 170–179. doi:10.1016/s0076-6879(82)90123-9
McIntosh, C. L., Germer, F., Schulz, R., Appel, J., and Jones, A. K. (2011). The [NiFe]-hydrogenase of the cyanobacterium Synechocystis sp. PCC 6803 works bidirectionally with a bias to H2 production. J. Am. Chem. Soc. 133 (29), 11308–11319. doi:10.1021/ja203376y
Meuer, J., Bartoschek, S., Koch, J., Künkel, A., and Hedderich, R. (1999). Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265 (1), 325–335. doi:10.1046/j.1432-1327.1999.00738.x
Min, H., and Sherman, L. A. (2010). Hydrogen production by the unicellular, diazotrophic cyanobacterium Cyanothece sp. strain ATCC 51142 under conditions of continuous light. Appl. Environ. Microbiol. 76 (13), 4293–4301. doi:10.1128/aem.00146-10
Nanatani, K., Shijuku, T., Takano, Y., Zulkifli, L., Yamazaki, T., Tominaga, A., et al. (2015). Comparative analysis of kdp and ktr mutants reveals distinct roles of the potassium transporters in the model cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 197 (4), 676–687. doi:10.1128/jb.02276-14
Oria-Hernández, J., Cabrera, N., Pérez-Montfort, R., and Ramírez-Silva, L. (2005). Pyruvate kinase revisited the activating effect of K+. J. Biol. Chem. 280 (45), 37924–37929. doi:10.1074/jbc.m508490200
Osanai, T., Imamura, S., Asayama, M., Shirai, M., Suzuki, I., Murata, N., et al. (2006). Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res. 13 (5), 185–195. doi:10.1093/dnares/dsl010
Pansook, S., Incharoensakdi, A., and Phunpruch, S. (2016). Hydrogen production by immobilized cells of unicellular halotolerant cyanobacterium Aphanothece halophytica in alginate beads. Asia Pac. J. Sci. Technol. 21 (2), 248–255. doi:10.14456/kkurj.2016.34
Pansook, S., Incharoensakdi, A., and Phunpruch, S. (2019a). Effects of the photosystem II inhibitors CCCP and DCMU on hydrogen production by the unicellular halotolerant cyanobacterium Aphanothece halophytica. Sci. World J. 2019, 1030236. doi:10.1155/2019/1030236
Pansook, S., Incharoensakdi, A., and Phunpruch, S. (2019b). Enhanced dark fermentative H2 production by agar-immobilized cyanobacterium Aphanothece halophytica. J. Appl. Phycol. 31 (5), 2869–2879. doi:10.1007/s10811-019-01822-9
Pansook, S., Incharoensakdi, A., and Phunpruch, S. (2022). Simazine enhances dark fermentative H2 production by unicellular halotolerant cyanobacterium Aphanothece halophytica. Front. Bioeng. Biotechnol. 10, 904101. doi:10.3389/fbioe.2022.904101
Papazi, A., Gjindali, A. I., Kastanaki, E., Assimakopoulos, K., Stamatakis, K., and Kotzabasis, K. (2014). Potassium deficiency, a “smart” cellular switch for sustained high yield hydrogen production by the green alga Scenedesmus obliquus. Int. J. Hydrog. Energy 39 (34), 19452–19464. doi:10.1016/j.ijhydene.2014.09.096
Pewnual, T., Jampapetch, N., Saladtook, S., Raksajit, W., Klinsalee, R., and Maneeruttanarungroj, C. (2022). Response of green alga Tetraspora sp. CU2551 under potassium deprivation: A new promising strategy for hydrogen production. J. Appl. Phycol. 34 (2), 811–819. doi:10.1007/s10811-021-02672-0
Phunpruch, S., Taikhao, S., and Incharoensakdi, A. (2016). Identification of bidirectional hydrogenase genes and their co-transcription in unicellular halotolerant cyanobacterium Aphanothece halophytica. J. Appl. Phycol. 28 (2), 967–978. doi:10.1007/s10811-015-0664-8
Raksajit, W., Satchasataporn, K., Lehto, K., Mäenpää, P., and Incharoensakdi, A. (2012). Enhancement of hydrogen production by the filamentous non-heterocystous cyanobacterium. Arthrospira sp. PCC 8005. Int. J. Hydrog. Energy 37 (24), 18791–18797. doi:10.1016/j.ijhydene.2012.10.011
Rashid, N., Song, W., Park, J., Jin, H. F., and Lee, K. (2009). Characteristics of hydrogen production by immobilized cyanobacterium Microcystis aeruginosa through cycles of photosynthesis and anaerobic incubation. J. Ind. Eng. Chem. 15 (4), 498–503. doi:10.1016/j.jiec.2008.12.013
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111 (1), 1–61. doi:10.1099/00221287-111-1-1
Siddiquee, K. A. Z., Arauzo-Bravo, M. J., and Shimizu, K. (2004). Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol. Lett. 235 (1), 25–33. doi:10.1111/j.1574-6968.2004.tb09563.x
Srirangan, K., Pyne, M. E., and Chou, C. P. (2011). Biochemical and genetic engineering strategies to enhance hydrogen production in photosynthetic algae and cyanobacteria. Bioresour. Technol. 102 (18), 8589–8604. doi:10.1016/j.biortech.2011.03.087
Taikhao, S., Junyapoon, S., Incharoensakdi, A., and Phunpruch, S. (2013). Factors affecting biohydrogen production by unicellular halotolerant cyanobacterium Aphanothece halophytica. J. Appl. Phycol. 25 (2), 575–585. doi:10.1007/s10811-012-9892-3
Taikhao, S., Incharoensakdi, A., and Phunpruch, S. (2015). Dark fermentative hydrogen production by the unicellular halotolerant cyanobacterium Aphanothece halophytica grown in seawater. J. Appl. Phycol. 27 (1), 187–196. doi:10.1007/s10811-014-0292-8
Taikhao, S., and Phunpruch, S. (2017). Effect of metal cofactors of key enzymes on biohydrogen production by nitrogen fixing cyanobacterium Anabaena siamensis TISIR 8012. Energy Procedia 138, 360–365. doi:10.1016/j.egypro.2017.10.166
Takabe, T., Incharoensakdi, A., Arakawa, K., and Yokota, S. (1988). CO2 fixation rate and RuBisCO content increase in the halotolerant cyanobacterium, Aphanothece halophytica, grown in high salinities. Plant Physiol. 88 (4), 1120–1124. doi:10.1104/pp.88.4.1120
Ueda, S., Kawamura, Y., Iijima, H., Nakajima, M., Shirai, T., Okamoto, M., et al. (2016). Anionic metabolite biosynthesis enhanced by potassium under dark, anaerobic conditions in cyanobacteria. Sci. Rep. 6, 32354. doi:10.1038/srep32354
Keywords: Aphanothece halophytica, hydrogen production, gene expression, potassium deprivation, nitrogen deprivation
Citation: Chinchusak N, Incharoensakdi A and Phunpruch S (2023) Dark fermentative hydrogen production and transcriptional analysis of genes involved in the unicellular halotolerant cyanobacterium Aphanothece halophytica under nitrogen and potassium deprivation. Front. Bioeng. Biotechnol. 10:1028151. doi: 10.3389/fbioe.2022.1028151
Received: 25 August 2022; Accepted: 19 December 2022;
Published: 06 January 2023.
Edited by:
Suyun Xu, University of Shanghai for Science and Technology, ChinaReviewed by:
Amit Srivastava, University of Jyväskylä, FinlandCopyright © 2023 Chinchusak, Incharoensakdi and Phunpruch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saranya Phunpruch, c2FyYW55YS5waEBrbWl0bC5hYy50aA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.