- 1Department of Clinical Laboratory, The First People’s Hospital of Guiyang, Guiyang, China
- 2School of Public Health, The Key Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education, Guizhou Medical University, Guiyang, China
- 3Laboratory of Bacterial Infectious Disease of Experimental Center, Guizhou Provincial Centre for Disease Control and Prevention, Guiyang, China
Candida albicans is an opportunistic pathogenic yeast that predominantly causes invasive candidiasis. Conventional methods for detecting Candida species are costly, take 3–5 days, and require skilled technicians. Rapid pathogen identification is important in managing invasive candidiasis infection. Here, a novel molecular diagnostic assay termed loop-mediated isothermal amplification combined with nanoparticles-based lateral flow biosensor (LAMP-LFB) was developed for C. albicans rapid detection. A set of six primers was designed based on the C. albicans species-specific internal transcribed spacer 2 (ITS2) gene. The C. albicans-LAMP results were visually reported by LFB within 2 min. Various fungal strains representing Candida species, as well as several Gram-negative and Gram-positive bacterial species, were used to determine the analytical sensitivity and specificity of the assay. The optimal LAMP conditions were 64 °C for 40 min, with a sensitivity of 1 fg of genomic DNA template from C. albicans pure cultures. No cross-reactions were obtained with non-albicans strains. Thus, the analytical specificity of the LAMP-LFB assay was 100%. The entire procedure could be completed within 85 min, including specimen processing (40 min), isothermal reaction (40 min), and result reporting (within 2 min). In 330 clinical samples (including 30 whole blood, 100 middle segment urine, and 200 sputum samples), all C. albicans-positive (62/330) samples were identified by LAMP-LFB assay, and the diagnostic accuracy was 100% when compared to the traditional clinical cultural-based methods. Thus, this assay can be used as a diagnostic tool for the rapid, accurate, sensitive, low-cost and specific detection of C. albicans strains, especially in resource-limited settings.
Introduction
The invasive fungal disease is a neglected disease that threatens public health, is difficult to diagnose with long treatment time, costly, and poor treatment efficacy (Fisher et al., 2020; Rodrigues and Nosanchuket al., 2020). Epidemiological statistics depict that invasive fungi cause more than two million infections yearly, and the number of deaths is equivalent to tuberculosis or malaria (Schmiedel and Zimmerli, 2016). Invasive candidiasis is the most common invasive fungal disease with high morbidity and mortality. Even when the patients are treated with antifungal therapy, the mortality rate is as high as 45%, and approximately 50% of cases are attributed to C. albicans (Zilberberg et al., 2008; Dadar et al., 2018; Quindós et al., 2018). C. albicans is a symbiotic yeast on the human mucosal surface. Simultaneously, C. albicans is an opportunistic pathogen responsible for invasive candidiasis such as pyelonephritis, endocarditis, or candidemia (Sherry et al., 2014). According to several recent studies, biofilm formation strengthens C. albicans drug resistance and the immune system (Ben-Ami 2018). Additionally, the emergence of drug-resistant strains such as Candida auris is partially attributable to empirical treatment without an early and clear diagnosis (Adams et al., 2018; Eyre et al., 2018). Thus, patients lose the optimum time for treatment leading to increased mortality, and drug-resistant strains become prevalent (Fortún et al., 2012). Therefore, rapid and accurate C. albicans detection is critical in the prevention, control, and timely treatment of invasive candidiasis, and it can also prevent the emergence of drug-resistant strains.
Unfortunately, current methods for detecting C. albicans infections are inadequate to meet clinical requirements. The traditional culture method is the gold standard for diagnosing fungal infections, but it is time-consuming and insensitive (Pincus et al., 2007). Moreover, C. albicans detection is easily confused with Candida dubliniensis in some colorimetric methods; C. albicans is not always detected accurately and rapidly. Therefore, it is important to establish a rapid and effective method for C. albicans detection in grass-roots hospitals. Rapid detection receives considerable attention due to the high morbidity and mortality of invasive candidiasis and the increasing prevalence of drug-resistant candidiasis. In recent years, many methods that are more sensitive and faster than traditional culture have emerged for the identification of C. albicans have been developed, such as immunofluorescence (Gunasekera et al., 2015) and PCR-based assay (i.e., multiplex PCR and real-time PCR) (Kasai et al., 2006) and mass spectrometry (Zehm et al., 2012). These methods are unsuitable for popularization, especially in resource-poor areas, because they are complex and require expensive equipment and professionals.
In recent years, various isothermal amplification methods, including loop-mediated isothermal amplification (LAMP), multiple cross displacement amplification (MCDA), and recombinase polymerase amplification (RPA), have been designed for nucleic acid analysis to overcome the disadvantages of traditional culture and PCR-based methods and have been reported to detection of C. albicans (Zhao et al., 2019; Fallahi et al., 2020; Wang F. et al., 2021). Among them, loop-mediated isothermal amplification (LAMP) is a nucleic acid amplification technique under isothermal conditions that is simpler, economical, sensitive, and rapidly applied in many fields, especially in molecular medical diagnostics (Notomi et al., 2000; Dolka et al., 2019). For example, the LAMP method has been applied successfully to detect Staphylococcus aureus, C. auris, and the COVID-19 virus (Wang et al., 2017; Yamamoto et al., 2018; Zhu et al., 2020). Conventionally, LAMP amplification products can be tested using various methods, including agarose gel electrophoresis and turbidimetry changes (Hill et al., 2008; Zhao et al., 2020). However, these methods been restricted due to the need for expensive special equipment (real-time turbidimeter or fluorescence apparatus), expensive regents (calcein), and additional analysis procedure (agarose gel electrophoresis) to indicate assay’s results. Therefore, an economical, rapid, simple, accurate, and suitable method for C. albicans detection is needed urgently.
In the past few years, nanomaterial-based biosensors have been widely used for the bioassay detection of a broad variety of targets in specimens from cancer patients (Patel et al., 2022a; Azzouz et al., 2022), cardiovascular disease (Azzouz et al., 2021), metabolites in sweat (Wang et al., 2022), proteins (Patel et al., 2022b), bacteria (Li et al., 2019) and fungi (Wang Y. et al., 2021). In the diagnosis of fungi and bacterial infections, biosensors are extremely accurate and quick (Li et al., 2019; Wang Y. et al., 2021). In the current study, we aim to combine classic Loop-mediated isothermal amplification (LAMP) with the gold nanoparticle-based lateral flow biosensors (LFB) to detect C. albicans, which is a new method to detect specific gene fragments of microbial with convenient and visual (Li et al., 2019). Moreover, we evaluated the ability to detect clinical samples with C. albicans-LAMP-LFB, to establish a fast, economical, accurate, and convenient tool for C. albicans detection in clinical applications.
Materials and methods
Reagents and instruments
Fungal and Bacterial genomic DNA extraction kits (Baitaike DNA mini kits, China) were obtained from Baitaike. Co., Ltd (Beijing, China). DNA Isothermal amplification kits, colorimetric indicator (Visual detection reagent, VDR), and biotin-14-dCTP were supplied by Tian-Jin HuiDeXin Biotech. Co., Ltd (Tianjin, China). The LFB materials, including the backing card, sample pad, conjugate pad, absorbent pad, and nitrocellulose membrane (NC), were obtained from Jie-Yi Biotech. Co., Ltd (Shanghai, China). Rabbit anti-fluorescein antibody (Anti-FITC) and biotinylated bovine serum albumin (biotin-BSA) were acquired from Abcam. Co., Ltd (Shanghai, China). Dye (Crimson red) streptavidin-coated polymer nanoparticles (129 nm, 10 mg ml−1; 100 mm borate, pH 8.5, with 0.1% BSA, 0.05% Tween 20 and 10 mm Ethylene Diamine Tetraacetic Acid, EDTA) were purchased from Bangs Laboratories, Inc (Indiana, United States).
LAMP assay primers design and synthesis
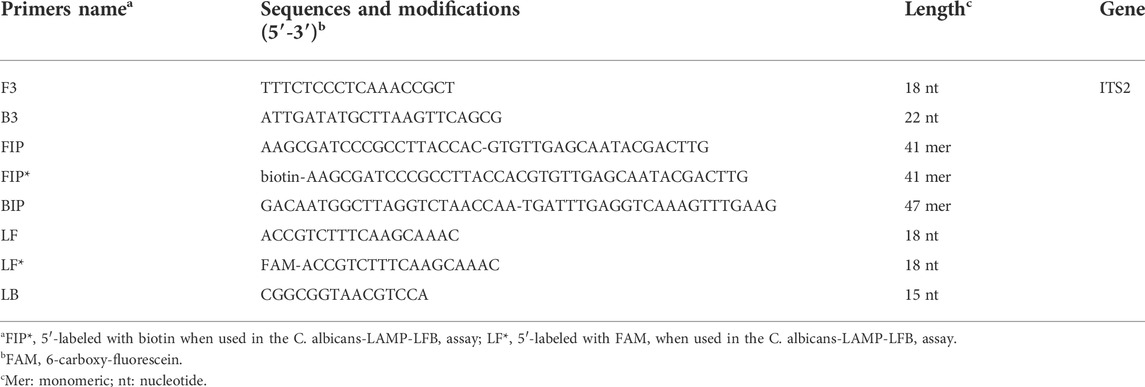
A set of six primes including two inner primers (FIP and BIP), two outer primers (F3 and B3), and two loop primers (LF and LB), were designed based on the reaction mechanism of LAMP to target the sequence of internal transcribed spacer 2 (ITS2) gene (Genbank accession no. AF455531.1) of C. albicans for the LAMP assay. All primers were designed using Primer Explorer V5 (http://primer-explorer.jp/e/; Eiken Chemical Co., Ltd, Tokyo, Japan) and validated using the basic local alignment search tool (BLAST). Figure 1 displays primer positions, while Table 1 depicts the primer sequences and modifications. In this study, all the primers were synthesized by Kunming-Tsingke Biotechnology Co., Ltd (Kunming, China) with HPLC purification grade.
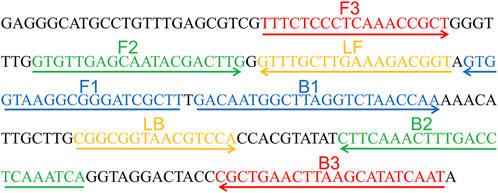
FIGURE 1. Sequence and location of ITS2 gene used to design loop-mediated isothermal amplification primers. The nucleotide sequences of the sense strand of ITS2 are listed. Right arrows and left arrows indicate sense and complementary sequences that are used.
Fungal and bacterial strains genomic DNA preparation
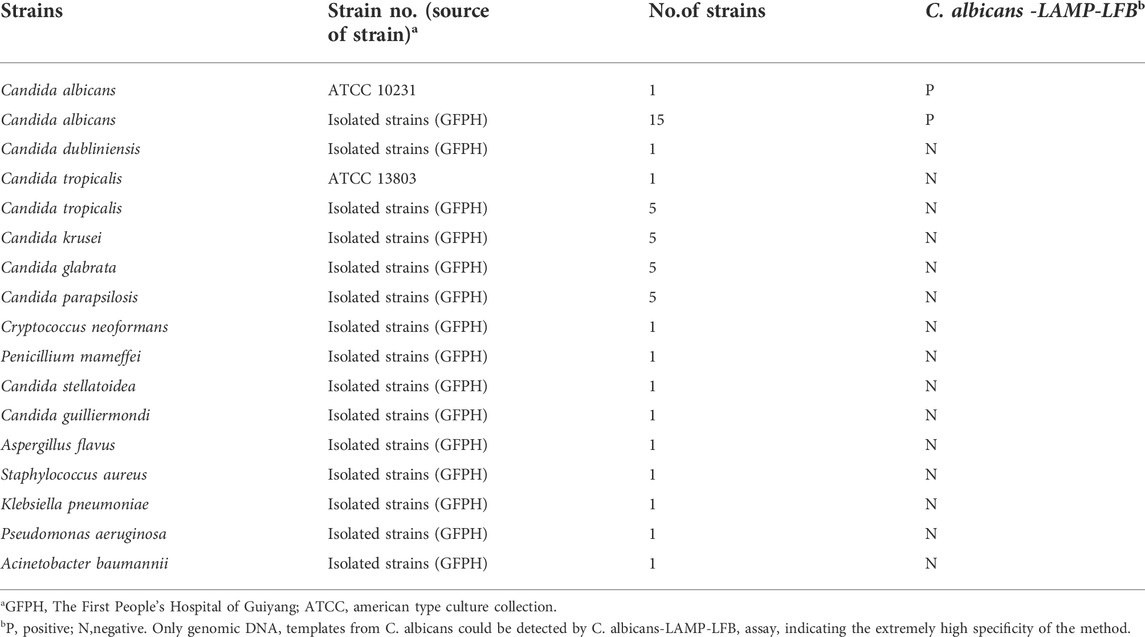
The 43 fungal and four bacterial strains were employed in the current study (Table 2), including the C. albicans reference strain (ATCC10231), 15 isolated C. albicans, and 27 non-C. albicans fungal strains from clinical samples. All genomic DNA templates were extracted using DNA extraction kits following the manufacturer’s instructions, and the concentration and purity were measured using a Nanodrop 2000 (Thermo Fisher Scientific) at A260/280. The extracted DNA templates were stored at −20°C until further analysis. The genomic DNA of the C. albicans reference strain (ATCC10231) was serially diluted to concentrations ranging from 10 ng/μL to 100 ag/μL (10 ng/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL, 1 fg/μL, and 100 ag/μL), to optimize the reaction temperature, reaction time, specificity and sensitivity.
Gold nanoparticle-based lateral flow biosensor preparation
LFB was constructed and used for reporting LAMP results according to the description of a previous publication in our study (Li et al., 2019). Briefly, LFB included an absorbent pad, an immersion pad, an NC membrane, a conjugate pad, and a backing pad. Dye streptavidin-coated polymer nanoparticles (SA-PNPs) were impregnated on the conjugate pad. Then, anti-FITC and biotin-BSA were immobilized at the test line (TL) and control line (CL), respectively. According to our design, the LFB was manufactured timely by Tian-Jin HuiDeXin Biotech. Co., Ltd (Tianjin, China). The LFB was stored in a dry place away from light at a storage temperature of 2°C–25°C, valid for 18 months.
The standard C. albicans-LAMP assay
C. albicans-LAMP reactions were conducted per the manufacturer’s instructions for the DNA Isothermal amplification kits. Each reaction mixture contained 12.5 μl of supplied buffer (2×), 0.4 μm of each outer primer, F3 and B3, 0.8 μm of each loop primer, LF* and LB, 1.6 μm of each inner primer, FIP* and BIP, 0.4 mm biotin-14-dCTP, 1 μl (8 U) 2.0 Bst DNA polymerase, and one μ1 DNA template and 1 μl VDR, and distilled water (DW) to a total volume of 25.0 µl. The reaction mixture was incubated for 40 min at 64°C. Moreover, genomic DNA from non-C. albicans strains, including Candida tropicalis and Klebsiella pneumoniae, were used as a negative control (NC), and blank control mixtures contained 1 μl of distilled water (DW). The C. albicans-LAMP products were determined and verified using two distinct techniques, including VDR and LFB methods. The color of the amplified products effectively changed from colorless to light green in the VDR assay. However, the negative and blank controls remained colorless. The strategy for visualizing LAMP products with LFB was as previously described.
Reaction temperature optimization for C. albicans-LAMP assay
In this study, the optimal temperature of the C. albicans-LAMP assay was confirmed by setting the different reaction temperatures (61–68°C, with 1°C intervals) and using C. albicans reference strain (ATCC10231) template DNA (10 pg/μl). Amplification mixtures containing 1 μl of C. tropicalis and K. pneumoniae template were used as NC, and 1 μl of DW was used as a blank control. The assay was performed according to the standard LAMP assay and was monitored using the Loopamp Realtime Turbidimeter LA-500 (Eiken Chemical Co., Ltd., Japan). Moreover, the threshold value (turbidity) was 0.1, and turbidity of >0.1 was considered positive amplification (Li et al., 2019).
Sensitivity of the C. albicans-LAMP-LFB assays
The genomic DNA templates from C. albicans (ATCC10231) pure culture were serially diluted 10-fold (10 ng, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg, and 100 ag per microliter) to confirm the limit of detection (LoD) and to test the analytical sensitivity of LAMP-LFB assay. The LAMP-LFB reactions were carried out under the standard conditions described above, and the results were examined using a VDR and LFB. The LoD of the LAMP-LFB assay was verified as the last dilution of each positive test. Each dilution was tested at least three times.
Reaction time optimization for C. albicans-LAMP assay
Four amplification reaction times (30, 40, 50, and 60 min, with 10 min intervals) were evaluated to optimize the amplification reaction time of C. albicans-LAMP. The C. albicans-LAMP reactions were performed under standard conditions, and the results were reported using LFB. Each amplification time was repeated three times.
Specificity of the C. albicans-LAMP-LFB assays
Genomic DNA (at least 10 ng/μl) was detected from 16 C. albicans strains, 27 non-C. albicans fungal strains and four bacterial strains according to the optimal amplification temperature and time to evaluate the specificity of C. albicans-LAMP-LFB assay (Table 2). All LAMP results were verified using LFB. Each sample was analyzed thrice independently.
Application of C. albicans-lamp-LFB assay in clinical samples
A total of 330 clinical samples (including 30 whole blood, 100 middle segment urine, and 200 sputum samples) were collected from the First People’s Hospital of Guiyang suspected of having Candida infection to assess the applicability of the C. albicans-LAMP-LFB assay. All clinical samples were detected for C. albicans using traditional clinical cultural-based and C. albicans-LAMP-LFB methods. The routine diagnosis of Candida species in the laboratory was carried out using diagnostic methods such as CHROMagar Candida and biochemical identification. The C. albicans-LAMP-LFB detection was performed as described above.
Results
Detection and confirmation of C. albicans-LAMP products
The LAMP amplification reaction was incubated for 40 min at a constant temperature of 64°C to confirm the availability of C. albicans-LAMP primers. Then, the C. albicans-LAMP products were examined using the VDR and LFB methods. The C. albicans-LAMP amplified products appeared bright green under visible light, while the negative samples and blank control reactions remained colorless (Figure 2A). C. albicans-LAMP amplification was further confirmed by the appearance of two crimson red bands (CL and TL) in LFB, while the negatives samples and blank control only appeared as a crimson red line (CL) in LFB (Figure 2B). Therefore, the results suggested that the LAMP primer set targeting the ITS2 gene was valid for establishing the LAMP-LFB assay for C. albicans detection.
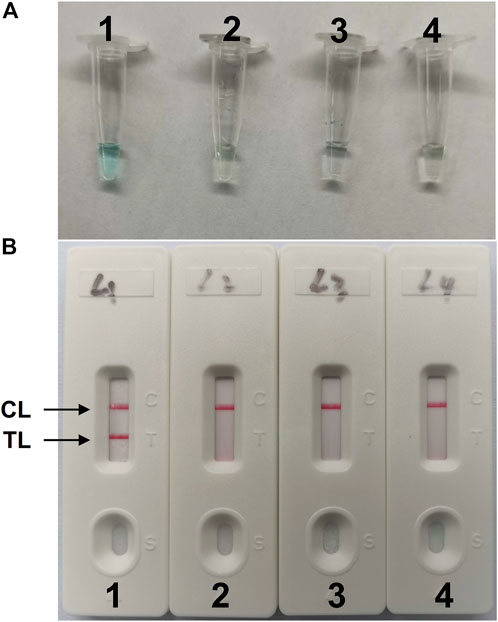
FIGURE 2. Detection and validation of (C) albicans-LAMP products (A), Color change of C. albicans-LAMP tubes; (B), biosensor applied for visual detection of C. albicans-LAMP products. Tube A1 (biosensor B1), positive amplification; tube A2 (biosensor B2), negative amplification (Candida tropicalis), tube A3 (biosensor B3), negative amplification (Klebsiella pneumoniae), tube A4 (biosensor B4), negative control (DW); TL, test line; CL, control line.
Optimal temperature of C. albicans-LAMP assay
The optimization test was performed to obtain the optimal reaction temperature for the C. albicans-LAMP experiment by setting a series of amplification temperatures (ranging from 61 to 68°C, with 1°C interval). The results were monitored using real-time turbidity measurements. The kinetics graphs were produced from the eight temperatures, with the faster reactions obtained for C. albicans-LAMP reactions at approach temperature of 62–65°C (Figure 3). Hence, we selected 64°C as the optimal amplification temperature for the C. albicans-LAMP-LFB assay.
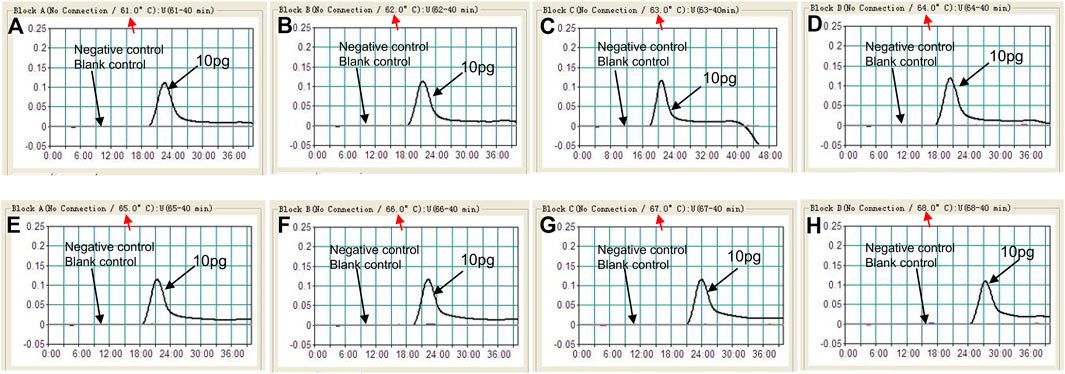
FIGURE 3. Optimization of amplification temperature for the C. albicans-LAMP primer set. The LAMP reactions for detection of C. albicans were monitored through realtime turbidity (LA-500), and the corresponding curves of concentrations of templates were marked in the figures. Eight kinetic graphs (A–H) were generated at various temperatures (61–68°C, 1°C intervals) with target pathogens DNA at the level of 10 pg per tube (The threshold value was 0.1 and the turbidity of >0.1 was considered to be positive). The graphs from B (62°C) to E (65°C) showed robust reaction. The optimal amplification temperature was 64°C.
Sensitivity of C. albicans-lamp-LFB detection
In our study, repeated detection of serial dilutions of C. albicans genomic DNA confirmed the sensitivity of the C. albicans-LAMP-LFB method. The C. albicans-LAMP-LFB assay LoD of template DNA was 1 fg/μl. Visual inspection of LAMP products with VDR reagents (Figure 4A) revealed CL and TL lines (red) on LFB, indicating positive amplification of the ITS2 gene (Figure 4B). The biosensors of the blank controls displayed only the CL line.
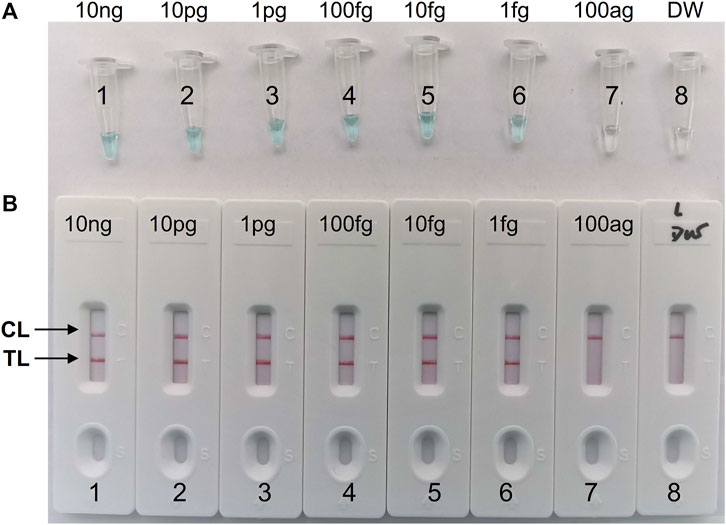
FIGURE 4. Sensitivity analysis of the C. albicans-LAMP-LFB assay with serial dilutions of C. albicans genomic DNA concentrations. Two detection methods involving a colorimetric indictor (VDR) (A) and LFB (B) were used to analyze the LAMP products. The genomic DNA was serially diluted (10 ng, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg, and 100 ag per microliter) and subjected to standard LAMP. Tubes A1-7 (Biosensors B1-7), C. albicans strain (ATCC 10231) genomic templates (10 ng-100 ag); Tube A8 (Biosensor B8), blank control (DW), respectively. The LoD of C. albicans-LAMP was one fg per reaction.
Optimal time of C. albicans -lamp-LFB assay
Four reaction time intervals (30, 40, 50, and 60 min) were tested at 64°C to obtain an optimal reaction time for C. albicans-LAMP-LFB. The C. albicans-LAMP products were identified through VDR and LFB methods. The results confirmed that the C. albicans (ATCC 10231) genomic DNA template was detected at the LoD level (1 fg) when the C. albicans-LAMP reaction lasted for 40 and 50 min at 64°C (Figure 5). There is no significant difference between the 40 min and 50 min lines. Therefore, a response time of 40 min was chosen for the LAMP-LFB test of C. albicans. Incubating C. albicans-LAMP-LFB at 64°C for 40 min was the optimal time. Thus, the whole diagnostic procedure of the C. albicans-LAMP-LFB assay, including Genomic DNA extraction (40 min), LAMP reaction (40 min) and result indicating (<2 min), can be completed within 85 min (Figure 6).
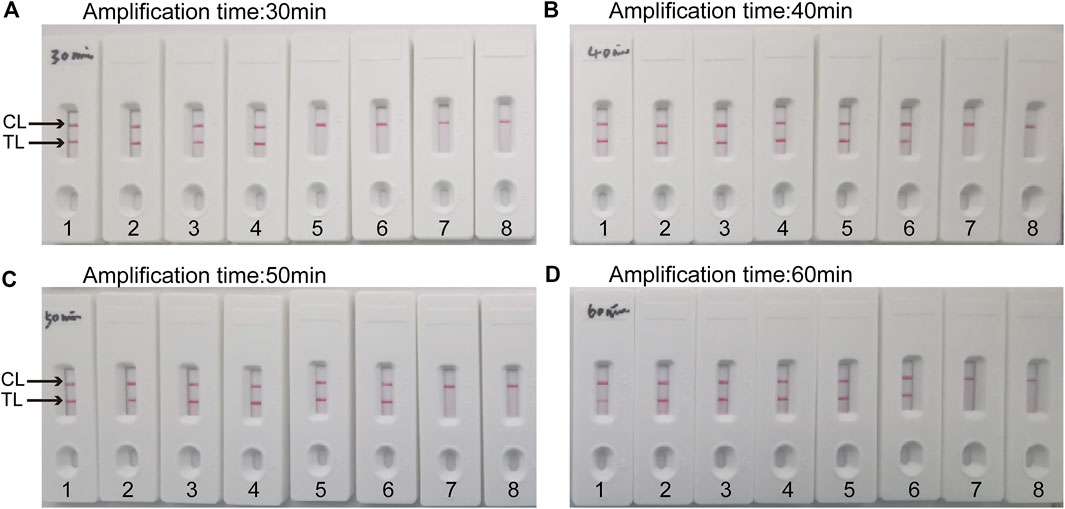
FIGURE 5. Optimal duration of time required for C. albicans-LAMP-LFB assay Four amplification times (A), 30 min; (B), 40 min; (C), 50 min; (D), 60 min) were evaluated at 64 °C. Biosensors 1, 2, 3, 4, 5, 6, 7, and eight represent genomic DNA (C.albicans (ATCC 10231)) levels of 10 ng, 10 pg, 1 pg, 100 fg, 10 fg, one fg and 100 ag target template per reaction and blank control (DW), respectively. The optimal time was showed when the amplification lasted for 40 min (B).
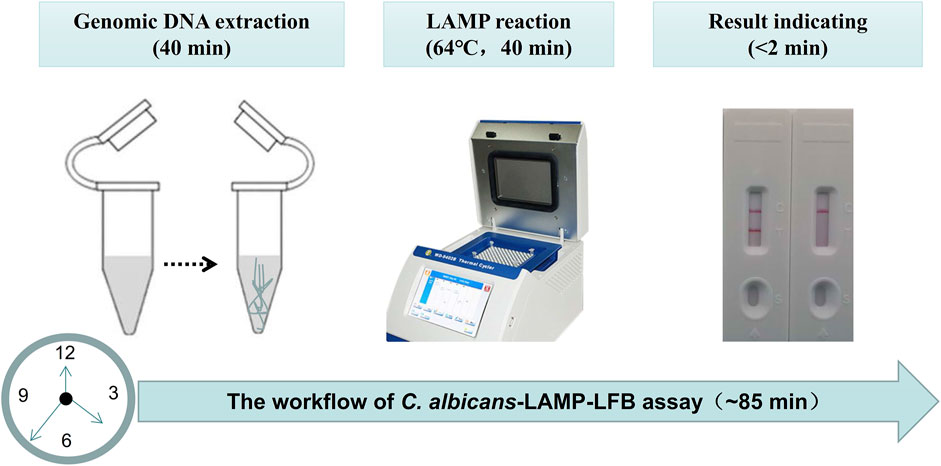
FIGURE 6. The workflow of C. albicans-LAMP-LFB assay. Three steps, including Genomic DNA extraction (40 min), LAMP reaction (40 min) and result reporting (<2 min), are required for the C. albicans-LAMP-LFB assay, and the total procedure can be completed within 85 min.
Specificity of C. albicans-lamp-LFB detection
The specificity of the C. albicans-LAMP-LFB assay was evaluated using DNA from 47 strains, including 1 C. albicans reference strain (ATCC10231), 15 C. albicans isolated strains, 27 non-C. albicans fungal strains and four bacterial strains (Table 2). Figure 7 illustrates that the C. albicans-LAMP-LFB assay detected all C. albicans strains while not detecting non-C. albicans fungal or bacterial strains. Hence, the C. albicans-LAMP-LFB assay can accurately differentiate C. albicans from other microbes.
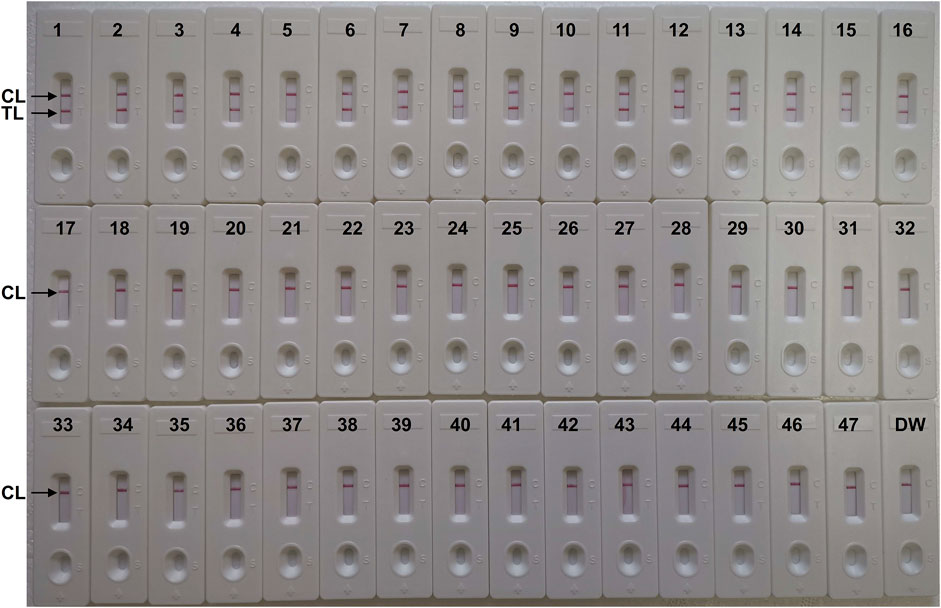
FIGURE 7. The specificity of C. albicans-LAMP-LFB assay for different strains. The C. albicans-LAMP-LFB assay was evaluated with different strains genomic DNA as templates. Two crimson lines (TL and CL) were appeared on the LFB, indicating positive results for the C. albicans isolates. Only a visible red line (CL) at the detection zone of LFB, reporting the negative results for non-C. albicans strains and blank control (DW). Biosensor1, Positive control (C. albicans ATCC 10231), Biosensors 2-16, isolated C. albicans strains; Biosensor 17, Candida dubliniensis; Biosensor18, Candida tropicalis (ATCC 13803); Biosensors19-23, isolated Candida tropicalis strains; Biosensors 24-28, isolated Candida krusei strains; Biosensors 29-33, isolated Candida glabrata strains; Biosensors 34-38, isolated Candida parapsilosis strains; Biosensor 39, Cryptococcus neoformans; Biosensor 40, Penicillium mameffei; Biosensor 41, Candida stellatoidea; Biosensor 42, Candida guilliermondi; Biosensor 43, Aspergillus flavus; Biosensor 44, Staphylococcus aureus; Biosensor 45, Klebsiella pneumoniae; Biosensor 46, Pseudomonas aeruginosa; Biosensor 47, Acinetobacter baumannii; Biosensor 48, blank control (DW).
Application of the LAMP-LFB assay in clinical samples for C. albicans detection
To verify the practical application of C. albicans-LAMP-LFB as a valuable tool for target pathogen detection. Traditional culture method identified C. albicans in 62 (18.8%) of the 330 samples (including two of the 30 whole blood, 18 of the 100 middle segment urine, and 42 of the 200 sputum samples), while the remaining 268 samples contained other bacteria or were microbe-free. The C. albicans-LAMP-LFB assay results were consistent with the traditional cultivation detection results. Table 3 suggests that the C. albicans-LAMP-LFB assay established in the current study could be used as an advanced tool to detect C. albicans in clinical samples.
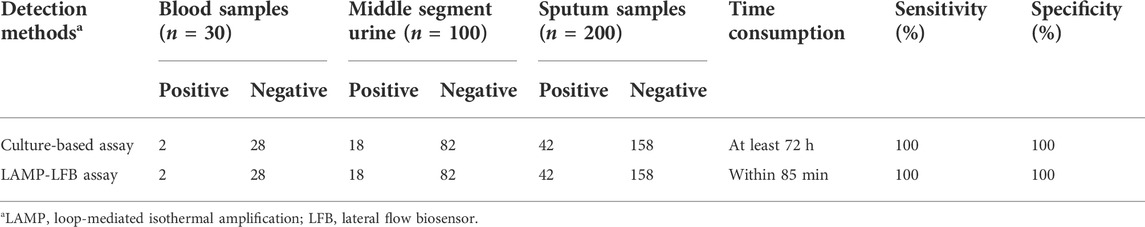
TABLE 3. Comparison of conventional culture and C. albicans-LAMP-LFB methods to identify C. albicans in clinical samples.
Discussion
C. albicans is a common but important opportunistic pathogen that frequently causes superficial and deep fungal infections and is primarily responsible for invasive candidiasis such as pyelonephritis, endocarditis, and candidemia (Gow and Yadav, 2017). It is also considered the fourth leading cause of hospital bloodstream infection, with unacceptably high mortality rates (Dadar et al., 2018). Currently, invasive candidiasis is difficult to diagnose and has a poor prognosis, posing a huge burden on public health worldwide (Casadevall, 2019). However, antifungal drugs can be employed to treat fungal infections; early and accurate diagnosis results in effective treatment outcomes. Moreover, C. albicans has increased drug resistance by forming biofilms, making antifungal treatments more intractable (Ben-Ami, 2018). Therefore, developing a rapid and accurate diagnostic method for C. albicans is a high priority to improve patient quality of life, reduce patient mortality and drug resistance. Rapid pathogen identification is important in controlling and managing invasive candidiasis infection (McCarty and Pappas, 2015). However, current methods for diagnosing C. albicans infection are time-consuming, cumbersome, and uneconomical. Hence, an accurate, rapid, simple, and economical method for detecting C. albicans is urgently needed. This report combined classic LAMP with LFB to detect C. albicans. LAMP primer sets, including two inner primers (FIP and BIP), two outer primers (F3 and B3), and two loop primers (LF and LB), were designed per LAMP rules to recognize the sequence of the ITS2 gene of C. albicans with a high degree of specificity (Notomi et al., 2000). The entire procedure could be completed within 85 min, including specimen processing (40 min), isothermal reaction (40 min), and result reporting (within 2 min). Importantly, we could visually evaluate the C. albicans-LAMP results based on the bands on the LFB within 2 min; even non-experts can correctly interpret the results.
LAMP is more convenient and cost-effective than traditional methods such as direct microscopy, but it is not sensitive enough to distinguish all Candida species. Thus, the diagnosis can be ambiguous, delaying precise treatment. Although the CHROMagar Candida method is simple, it is time-consuming (maybe spend 3 days or more). Moreover, it is impossible to distinguish all Candida species, especially due to the color similarity of the colonies, such as Candida dubliniensis (Ells et al., 2011). Furthermore, compared to other molecular diagnostic methods that are more accurate and time efficient than traditional culture methods, real-time PCR, for example, requires professionals to operate special instruments to change the temperature to complete the experiment, making it cumbersome and uneconomical, limiting its application, especially in areas where resources are scarce (Asadzadeh et al., 2018). In this reaction system, the optimal LAMP conditions were 64°C for 40 min; a very simple and inexpensive device, such as a water bath or a heater, is sufficient to keep the reaction temperature at 64°C for 40 min. It reveals that LAMP is time-saving, economical, and easy to operate. We also compared LAMP to other isothermal amplification assays, for example, Multiple cross displacement amplification (MCDA), for C. albicans detection (Zhao et al., 2019). In Zhao’s report, the LoD to detect C. albicans was 200 fg, while in our study, the sensitivity was one fg of genomic DNA template from C. albicans pure cultures. The results demonstrated that LAMP was more sensitive than MCDA in our study. In addition, the LoD of the C. albicans-LAMP-LFB method developed in this report was 1 fg/μl of C. albicans genomic DNA, which was 10 times more sensitive that of the conventional LAMP added SYBR Green I amplification indicator to the reaction mixture in the experiment (Fallahi et al., 2020). Moreover, we demonstrated the assay specificity of the C. albicans-LAMP method using DNA from 47 strains, including one C. albicans reference strain (ATCC10231), 15 C. albicans isolated strains, 27 non-C. albicans fungal strains and four bacterial strains. No cross-reactions to non-albicans strains were obtained, and the analytical specificity of the LAMP-LFB assay is 100%; therefore, the LAMP-LFB assay could be employed to detect C. albican with high specificity.
The LAMP assay has been used to detect various pathogens, including C. albicans (Wang et al., 2017; Yamamoto et al., 2018; Fallahi et al., 2020; Zhu et al., 2020). Unfortunately, traditional LAMP assay results were tedious to interpret. It requires special materials and instruments, such as agarose gel electrophoresis, real-time turbidity equipment, and an SYBR green I color indicator (Hill et al., 2018; Zhao et al., 2020). Worse, it requires a professional to operate. Hence, the factors mentioned above limited their point-of-care testing. This study used VDR and LFB methods to detect LAMP products, avoiding the shortcomings of traditional LAMP assays. The color of the amplified products effectively changed from colorless to light green in the VDR assay, while the negative and blank controls remained colorless. Moreover, CL and TL appeared on the LFB, revealing positive LAMP results for the ITS2 gene. However, only the CL line was observed on the biosensor for the blank controls. These two methods are visual and very simple; non-specialists can interpret the results accurately. Although the VDR assay can detect LAMP products, the color is fuzzy when the product concentration is low, which may lead to interpretation errors (Chen et al., 2021). So, the biosensor LFB is considered a suitable method for monitoring C. albicans-LAMP products due to its high sensitivity, simplicity of operation, and cost savings. The total cost of one test, including fungi genomic DNA extraction (approximately $1 USD), C. albicans-LAMP reaction (approximately $3.5 USD) and LFB reporting (approximately $2 USD), is estimated to be $6.5 USD, which is cheaper than conventional PCR-based methods. Combined with the elimination of labor costs because of the requirements for trained personnel in a certified laboratory, the C. albicans-LAMP-LFB assay becomes more cost-effective.
We evaluated the clinical application of C. albicans-LAMP-LFB using 330 clinical samples (including 30 whole blood, 100 middle segment urine, and 200 sputum samples); the LAMP-LFB assay identified all C. albicans-positive (62/330) samples. The diagnostic accuracy was 100% compared to the traditional clinical cultural-based methods. The results suggested that this assay can be used as a diagnostic tool for the rapid, accurate, sensitive, and specific detection of C. albicans strains. It is especially useful in implementing point-of-care testing, which is more conducive to developing resource-poor areas. Our study has some limitations LAMP-LFB assay is a qualitative test without quantitative detection. Hence, it is difficult to determine whether it is a pathogen or a colonizing bacterium. Therefore, in the future research, this method should focus on the detection of blood, middle segment urine, bronchoalveolar lavage fluid and aseptic body fluid, and try to avoid the detection of sputum samples, which is conducive to the correct use of antifungal drugs. In addition, LAMP is an isothermal amplification technique that requires multiple primer pairs; contamination can easily occur, leading to incorrect results. Therefore, we will address the above deficiencies in subsequent studies.
Conclusion
In conclusion, a visual, rapid, simple, sensitive, and cost-saving C. albicans-LAMP-LFB assay based on the ITS2 gene was successfully devised for identifying C. albicans agents in the current study. The C. albicans-LAMP-LFB assay showed high sensitivity and high specificity, which could successfully detect C. albicans isolates, and had the LoD of 1 fg genomic DNA template per tube. Besides, the clinical effectiveness of this assay was successfully upheld by clinical samples, including whole blood, middle segment urine, and sputum samples. Hence, our results indicate that C. albicans-LAMP-LFB strategy is an effective tool for C. albicans rapid detection in clinical samples, especially in resource-limited settings.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The study was approved by the Human Ethics Committee of the First People’s Hospital of Guiyang (Approval No. 2020-S025) and complied with the Declaration of Helsinki. Before our team obtained the samples/isolates and conducted the study, any personal identifiers of the suspected C. albicans-infected patients had been removed by the monitoring stations. So, the patient informed consent was waived by the Human Ethics Committee of the First People’s Hospital of Guiyang.
Author contributions
YW, XZ, and SL conceived and designed the experiments. YW, XZ, YZ, and JL performed the experiments. YW, JL, XZ, and HL analyzed the data. YW, XZ, and SL wrote the paper.
Funding
This study was supported by the Qian Ke He Zhi Cheng [2021] Yi Ban 440 from the Science and Technology Department of Guizhou Province. Zhu Ke He Tong (2020)-10-6, Zhu Ke He Tong (2021)43-25, and (2019) Zhu Wei Jian Ke Ji He Tong Zi Di 001 from the Science and Technology Department of Guiyang city of Guizhou Province.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, E., Quinn, M., Tsay, S., Poirot, E., Chaturvedi, S., Southwick, K., et al. (2018). Candida auris investigation workgroup Candida auris in healthcare facilities, New York, USA, 2013-2017. Emerg. Infect. Dis. 24 (10), 1816–1824. doi:10.3201/eid2410.180649
Asadzadeh, M., Ahmad, S., Al-Sweih, N., and Khan, Z. (2018). Rapid and accurate identification of Candida albicans and Candida dubliniensis by real-time PCR and melting curve analysis. Med. Princ. Pract. 27 (6), 543–548. doi:10.1159/000493426
Azzouz, A., Hejji, L., Kim, K. H., Kukkar, D., Souhail, B., Bhardwaj, N., et al. (2022). Advances in surface plasmon resonance-based biosensor technologies for cancer biomarker detection. Biosens. Bioelectron. X. 197, 113767. doi:10.1016/j.bios.2021.113767
Azzouz, A., Hejji, L., Sonne, C., Kim, K. H., Kumar, V., Bhardwaj, N., et al. (2021). Nanomaterial-based aptasensors as an efficient substitute for cardiovascular disease diagnosis: Future of smart biosensors. Biosens. Bioelectron. X. 193, 113767. doi:10.1016/j.bios.2021.113767
Ben-Ami, R. (2018). Treatment of invasive candidiasis: A narrative review. J. Fungi (Basel). 4, 97. doi:10.3390/jof4030097
Casadevall, A. (2019). Global catastrophic threats from the fungal kingdom: Fungal catastrophic threats. Curr. Top. Microbiol. Immunol. 424, 21–32. doi:10.1007/82_2019_161
Chen, X., Wang, S., Tan, Y., Huang, J., Yang, X., and Li, S. (2021). Nanoparticle-based lateral flow biosensors integrated with loop-mediated isothermal amplification for the rapid and visual diagnosis of hepatitis B virus in clinical application. Front. Bioeng. Biotechnol. 9, 731415. doi:10.3389/fbioe.2021.731415
Dadar, M., Tiwari, R., Karthik, K., Chakraborty, S., Shahali, Y., and Dhama, K. (2018). Candida albicans - biology, molecular characterization, pathogenicity, and advances in diagnosis and control - an update. Microb. Pathog. 117, 128–138. doi:10.1016/j.micpath.2018.02.028
Dolka, B., Cisek, A. A., and Szeleszczuk, P. (2019). The application of the loop-mediated isothermal amplification (LAMP) method for diagnosing Enterococcus hirae-associated endocarditis outbreaks in chickens. BMC Microbiol. 19 (1), 48. doi:10.1186/s12866-019-1420-z
Ells, R., Kock, J. L., and Pohl, C. H. (2011). Candida albicans or Candida dubliniensis. Mycoses 54 (1), 1–16. doi:10.1111/j.1439-0507.2009.01759.x
Eyre, D. W., Sheppard, A. E., Madder, H., Moir, I., Moroney, R., Quan, T. P., et al. (2018). A Candida auris outbreak and its control in an intensive care setting. N. Engl. J. Med. Overseas. Ed. 379 (14), 1322–1331. doi:10.1056/NEJMoa1714373
Fallahi, S., Babaei, M., Rostami, A., Rostami, A., Mirahmadi, H., Arab-Mazar, Z., et al. (2020). Diagnosis of Candida albicans: Conventional diagnostic methods compared to the loop-mediated isothermal amplification (lamp) assay. Arch. Microbiol. 202 (17), 275–282. doi:10.1007/s00203-019-01736-7
Fisher, M. C., Gurr, S. J., Cuomo, C. A., Blehert, D. S., Jin, H., Stukenbrock, E. H., et al. (2020). Threats posed by the fungal kingdom to humans, wildlife, and agriculture. mBio 11 (3), e00449-20. doi:10.1128/mBio.00449-20
Fortún, J., Martín-Dávila, P., Gómez-García, d., Pedrosa, E., Pintado, V., Cobo, J., et al. (2012). Emerging trends in candidemia: A higher incidence but a similar outcome. J. Infect. 65 (1), 64–70. doi:10.1016/j.jinf.2012.02.011
Gow, N. A. R., and Yadav, B. (2017). Microbe profile: Candida albicans: A shape-changing, opportunistic pathogenic fungus of humans. Microbiol. Read. 163 (8), 1145–1147. doi:10.1099/mic.0.000499
Gunasekera, M., Narine, M., Ashton, M., and Esfandiari, J. (2015). Development of a dual path platform (DPP®) immunoassay for rapid detection of Candida albicans in human whole blood and serum. J. Immunol. Methods 424, 7–13. doi:10.1016/j.jim.2015.04.014
Hill, J., Beriwal, S., Chandra, I., Paul, V. K., Kapil, A., Singh, T., et al. (2008). Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J. Clin. Microbiol. 46 (8), 2800–2804. doi:10.1128/JCM.00152-08
Kasai, M., Francesconi, A., Petraitiene, R., Petraitis, V., Kelaher, A. M., Kim, H. S., et al. (2006). Use of quantitative real-time PCR to study the kinetics of extracellular DNA released from Candida albicans, with implications for diagnosis of invasive candidiasis. J. Clin. Microbiol. 44 (1), 143–150. doi:10.1128/JCM.44.1.143-150.2006
Li, S., Liu, Y., Wang, Y., Chen, H., Liu, C., and Wang, Y. (2019). Lateral flow biosensor combined with loop-mediated isothermal amplification for simple, rapid, sensitive, and reliable detection of Brucella spp. Infect. Drug Resist. 30 (12), 2343–2353. doi:10.2147/IDR.S211644
McCarty, T. P., and Pappas, P. G. (2015). Invasive candidiasis. Infect. Dis. Clin. North Am. 30, 103–124. doi:10.1016/j.idc.2015.10.013
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28 (12), E63–E63. doi:10.1093/nar/28.12.e63
Patel, S. K., Surve, J., and Parmar, J. (2022a). Detection of cancer with graphene metasurface-based highly efficient sensors. Diam. Relat. Mat. 129, 109367. doi:10.1016/j.diamond.2022.109367
Patel, S. K., Surve, J., Parmar, J., Natesan, A., and Katkar, V. (2022b). Graphene-based metasurface refractive index biosensor for hemoglobin detection: Machine learning assisted optimization. IEEE Trans. Nanobioscience, 1. doi:10.1109/TNB.2022.3201237
Pincus, D. H., Orenga, S., and Chatellier, S. (2007). Yeast identification-past, present, and future methods. Med. Mycol. 45 (2), 97–121. doi:10.1080/13693780601059936
Quindós, G., Marcos-Arias, C., San-Millán, R., Mateo, E., and Eraso, E. (2018). The continuous changes in the aetiology and epidemiology of invasive candidiasis: From familiar Candida albicans to multiresistant Candida auris. Int. Microbiol. 21 (3), 107–119. doi:10.1007/s10123-018-0014-1
Rodrigues, M. L., and Nosanchuk, J. D. (2020). Fungal diseases as neglected pathogens: A wake-up call to public health officials. PLoS Negl. Trop. Dis. 14 (2), e0007964. doi:10.1371/journal.pntd.0007964
Schmiedel, Y., and Zimmerli, S. (2016). Common invasive fungal diseases: An overview of invasive candidiasis, aspergillosis, cryptococcosis, and pneumocystis pneumonia. Swiss Med. Wkly. 146, w14281. doi:10.4414/smw.2016.14281
Sherry, L., Rajendran, R., Lappin, D. F., Borghi, E., Perdoni, F., Falleni, M., et al. (2014). Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC Microbiol. 14, 182. doi:10.1186/1471-2180-14-182
Wang, F., Ge, D. B., Wang, L., Li, N., Chen, H. M., Zhang, Z. X., et al. (2021a). Rapid and sensitive recombinase polymerase amplification combined with lateral flow strips for detecting Candida albicans. Anal. Biochem. 15 (633), 114428. doi:10.1016/j.ab.2021.114428
Wang, M., Yang, Y., Min, J., Song, Y., Tu, J. B., Mukasa, D., et al. (2022). A wearable electrochemical biosensor for the monitoring of metabolites and nutrients. Nat. Biomed. Eng. doi:10.1038/s41551-022-00916-z
Wang, Y., Li, H., Wang, Y., Zhang, L., Xu, J., and Ye, C. (2017). Loop-mediated isothermal amplification label-based gold nanoparticles lateral flow biosensor for detection of Enterococcus faecalis and Staphylococcus aureus. Front. Microbiol. 8, 192. doi:10.3389/fmicb.2017.00192
Wang, Y., Zhao, X., Cheng, J., Tang, X., Chen, X., Yu, H. L., et al. (2021b). Development and application of a multiple cross displacement amplification combined with nanoparticle-based lateral flow biosensor assay to detect Candida tropicalis. Front. Microbiol. 12, 681488. doi:10.3389/fmicb.2021.681488
Yamamoto, M., Alshahni, M. M., Tamura, T., Satoh, K., Iguchi, S., Kikuchi, K., et al. (2018). Rapid detection of Candida auris based on loop-mediated isothermal amplification (LAMP). J. Clin. Microbiol. 56 (9), e00591-18. doi:10.1128/JCM.00591-18
Zehm, S., Schweinitz, S., Würzner, R., Colvin, H. P., and Rieder, J. (2012). Detection of Candida albicans by mass spectrometric fingerprinting. Curr. Microbiol. 64 (3), 271–275. doi:10.1007/s00284-011-0064-5
Zhao, F., Niu, L., Yan, L., Nong, J., Wang, C., Wang, J., et al. (2019). Establishment and application of multiple cross displacement amplification coupled with lateral flow biosensor (MCDA-LFB) for visual and rapid detection of Candida albicans in clinical samples. Front. Cell. Infect. Microbiol. 9, 102. doi:10.3389/fcimb.2019.00102
Zhao, V. X. T., Wong, T. I., Zheng, X. T., Tan, Y. N., and Zhou, X. (2020). Colorimetric biosensors for point-of-care virus detections. Mat. Sci. Energy Technol. 3, 237–249. doi:10.1016/j.mset.2019.10.002
Zhu, X., Wang, X., Han, L., Chen, T., Wang, L., Li, H., et al. (2020). Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. X. 166, 112437. doi:10.1016/j.bios.2020.112437
Keywords: Candida albicans, loop-mediated isothermal amplification, lateral flow biosensor, LAMP-LFB, limit of detection
Citation: Wang Y, Zhao X, Zhou Y, Lu J, Yu H and Li S (2022) Establishment and application of loop-mediated isothermal amplification coupled with nanoparticle-based lateral flow biosensor (LAMP-LFB) for visual and rapid diagnosis of Candida albicans in clinical samples. Front. Bioeng. Biotechnol. 10:1025083. doi: 10.3389/fbioe.2022.1025083
Received: 22 August 2022; Accepted: 24 October 2022;
Published: 07 November 2022.
Edited by:
Kawsar Ahmed, Mawlana Bhashani Science and Technology University, BangladeshReviewed by:
Angel León-Buitimea, Universidad Autonoma de Nuevo Leon, MexicoPedro Fernández-Soto, University of Salamanca, Spain
Md Bellal Hossain, The University of Sydney, Australia
Shobhitkumar Kiritkumar Patel, Marwadi University, India
Copyright © 2022 Wang, Zhao, Zhou, Lu, Yu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honglan Yu, MTA0OTI1MTQ2M0BxcS5jb20=; Shijun Li, emp1bWVkanVuQDE2My5jb20=
†These authors have contributed equally to this work
 Yu Wang
Yu Wang Xue Zhao1†
Xue Zhao1† Yuhong Zhou
Yuhong Zhou Shijun Li
Shijun Li