- 1Department of Orthopaedics, The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, China
- 2Institute of Orthopedics of Jiangxi Province, Nanchang University, Nanchang, Jiangxi, China
- 3Institute of Minimally Invasive Orthopedics, Nanchang University, Nanchang, Jiangxi, China
- 4Jiangxi Key Laboratory of Intervertebral Disc Disease, Nanchang University, Nanchang, Jiangxi, China
- 5Department of Paediatrics, The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, China
Idiopathic short stature (ISS) is the most common clinical cause of the short stature with an unclear aetiology and a lack of effective treatment. Circular RNAs have been shown to play a significant regulatory role through various signal transduction pathways in a variety of diseases in recent years. However, the role of circular RNAs on ISS is not yet well-understood and requires a special attention. The differentially expressed circular RNAs were screened by microarray chip analysis, and RT-qPCR was used to verify the expression of hsa_circ_0008870 in ISS patients. Subsequently, in vitro and in vivo experiments were conducted to determine the biological functions of hsa_circ_0008870 in ISS. The authors first confirmed that hsa_ circ_0008870 was downregulated in ISS children. Meanwhile, we also observed that the downregulated hsa_circ _0008870 significantly inhibited chondrocyte proliferation and endochondral ossification in vivo and in vitro. Mechanistically, hsa_circ_0008870 regulates MAPK1 expression by sponge miR-185-3p. This mechanism of action was further verified through rescue experiments. Finally, the authors revealed that the silencing of hsa_circ_0008870 induces low expression of MAPK1 by impairing the sponge action of miR-185-3p, thereby inhibiting chondrocyte proliferation, hypertrophy, and endochondral ossification, which results in a short stature phenotype. In addition to these, we also observed an interesting phenomenon that upregulated of miR-185-3p can in turn inhibit the expression of hsa_circ_0008870 in chondrocytes. This suggests that hsa_circ_0008870 could potentially serve as a therapeutic target for the treatment of ISS.
1 Introduction
Short stature consistently remains the most common reason for referral to a pediatric endocrinologist (Collett-Solberg et al., 2019). Idiopathic short stature (ISS) accounts for 60%–80% of cases with short stature in children (Cohen et al., 2008). ISS is characterized with a height below the two standard deviations or the third percentile of the normal reference levels for the same ethnic group, gender and age, without chronic systemic, endocrine, nutritional, skeletal diseases or chromosomal abnormalities (Lee et al., 2020). To date, no breakthrough has been made in the field of treatment to improve height in ISS children due to the uncertain etiology.
Recombinant human growth hormone (rhGH) was first introduced in 1985 to increase height in short children (Bryant et al., 2007; Ross et al., 2015). Since 2003, rhGH has been used for the treatment of ISS (Grimberg et al., 2016; Wu et al., 2020). However, as all known, the children with ISS are not deficient in growth hormone (Lee, 2006). Therefore, the efficacy of the rhGH to treat ISS is controversial so far (Pedicelli et al., 2009). Although Paltoglou et al. reported that ISS children who received rhGH treatment increased significantly in height after 1 year, and Barton et al. reported that increasing the dose of rhGH could further improve the height growth of patients (Barton et al., 1995; Paltoglou et al., 2020), Kamp et al. study found that epiphyseal closure was advanced in ISS children, the growing period was greatly shortened and no improvement in height at the termination of growth after the treatment of rhGH (Kamp et al., 2002). In addition, several studies have reported many potential risks to children and complications during rhGH therapy, including cancer, scoliosis, slipped capital femoral epiphysis, idiopathic intracranial hypertension, carbohydrate metabolism disorders, and hypertension, suggesting that rhGH therapy may require stricter consideration and regulation (Vyas and Menon, 2021). Worse yet, rhGH treatment for patients with ISS proved extremely expensive, expected to spend $52,000 per inch of height gain (Lee et al., 2006). The main reason for the difficulty of diagnosis and poor treatment of ISS disease is that the pathogenesis of ISS is still unclear. Therefore, elucidating the molecular mechanisms of ISS initiation and progression is of great significance for improving the diagnostic and therapeutic efficiency of ISS.
As a special type of endogenous non-coding RNAs, circular RNAs (circRNAs) are a research hotspot in RNA biology (Zhao et al., 2019). Unlike linear RNA, circRNAs have no 5′caps and 3′polyadenylic acid tails (Chen and Yang, 2015). With their closed-loop structure, circRNAs can avoid degradation by RNase enzymes. Hence, its expression is more stable (Greene et al., 2017; Zhao et al., 2019). Circular RNAs have been shown to play a significant regulatory role through various signal transduction pathways in a variety of diseases in recent years (Qu et al., 2015). For instance, CircACTN4 was demonstrated to be upregulated in ICC (intrahepatic cholangiocarcinoma) patients, acting as a molecular sponge of miR-424-5p and interacting with YBX1, to transcriptionally activate FZD7, thereby promoting proliferation and metastasis of ICC (Chen et al., 2022). CircFndc3b enhanced the expression and signal of vascular endothelial growth factor-A by interacting with the RNA binding protein fused in sarcoma to promote cardiac function and remodeling after myocardial infarction (Garikipati et al., 2019). Additionally, circSERPINE2, acting as the sponge for miR-1271, suppressed human chondrocytes apoptosis and promoted the synthesis of extracellular matrix, thus inhibiting its apoptosis and delaying the progress of osteoarthritis (Shen et al., 2019). Related studies showed that circRNAs have been tightly associated with the occurrence and development of heart vascular, digestive system diseases, malignant tumours, osteoarthritis and neurological diseases (Qu et al., 2017). To determine the role of circRNAs in the molecular mechanisms of ISS, researchers identified differential expression of circRNAs in the peripheral blood of patients with ISS versus healthy controls. According to the outcomes of the circRNAs microarray, 145 differentially expressed circRNAs were screened (83 upregulated and 62 downregulated) (Liu et al., 2020; Liu et al., 2021). In subsequent studies, it was verified that circANAPC2 and hsa_circularRNA_0079201 can serve as potential targets for treatment of ISS (Liu et al., 2020; Liu et al., 2021).
To further evaluated the role of other circRNAs in pathogenesis of ISS, hsa_circ_0008870 was detected to be downregulated in ISS, and its downregulation was subsequently confirmed using RT-qPCR. Then, it was observed that the hsa_circ_0008870 downregulation suppressed the expression of MAPK1 via upregulating miR-185-3p, and finally inhibited chondrocyte proliferation, chondrocyte hypertrophic differentiation and endochondral ossification in vitro and in vivo. Interestingly, the authors observed that the upregulation of miR-185-3p can further repress the hsa_circ_0008870 expression via a positive feedback loop.
2 Materials and methods
2.1 Patient information and specimen acquisition
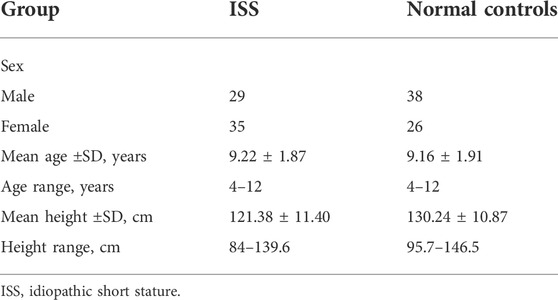
64 idiopathic short stature (ISS) patients comprised the experimental group. These patients received treatment in the Second Affiliated Hospital of Nanchang University (Nanchang, China) from August 2018 to August 2020.64 healthy children comprising the normal group, received physical examination during the same period. The conditions for patient enrollment in this study were consistent with our previous studies (Liu et al., 2021).
The ISS patients group comprised of 29 males and 35 females at the estimated average age of 9.22 ± 1.87 years with the average height of 121.38 ± 11.40 cm. The normal group comprised of 38 males and 26 females at the average age of 9.16 ± 1.91 years. The estimated average height for the control group was 130.24 ± 10.87 cm (Table 1). All blood samples were frozen immediately in liquid nitrogen after collection to prevent the degradation of biomolecules and contamination in samples. All samples were stored in a refrigerator at -80 °C until use. Blood samples from four ISS patients and four normal children were used for circRNAs microarray chip analysis, while all 64 pairs of blood samples were used for quantitative Real-Time PCR (RT-qPCR) to quantify the circRNAs expression.
2.2 Cell culture
Human chondrocyte cell line C28/I2 was sourced from Procell Life Science & Technology Co., Ltd. In Wuhan, China. The complete medium consisted of 89% Dulbecco’s modified Eagle medium (DMEM/F12; Gibco, United States), supplemented with 10% fetal bovine serum (FBS; Gibco, United States) and 1% penicillin-streptomycin (Invitrogen). Cells were incubated in a sterile incubator at 5% CO2 saturation and 37°C constant temperature.
2.3 Cell transfection
Small interfering RNAs (siRNAs) for circ-0008870 (designated as si-circ-0008870) and miR-185-3p mimics, miR-185-3p inhibitor and their negative control (NC) were obtained from RiboBio (Guangzhou, China). According to the instructions of the manufacturer, transient transfection was conducted employing Lipofectamine 3,000 (Invitrogen).
The lentiviral of sh-circ-0008870, circ-0008870-overexpressing and corresponding NC were constructed by the GeneChem (Shanghai, China). When chondrocytes confluence reaches about 30%–40%, then transfection was performed. Stable transfected cells were sifted with puromycin.
2.4 RT-qPCR
The chondrocytes were lysed following the instructions of the Trizol reagent RNA extraction kit (Invitrogen, United States) and total RNA was extracted. The RNA was reverse transcribed using the PrimeScript RT Master Mix Kit (Takara). Quantitative PCR was executed utilizing a real-time PCR kit (Takara) and the specific primers. For primers, see Supplementary Table S1. Each experiment was conducted in triplicates. The endogenous controls were GAPDH and U6 small nuclear RNAs. The relative expression level of RNA was analyzed by the 2−ΔΔCt method.
2.5 Western blotting
Cells were collected by cell scraping. RIPA lysis buffer rich in protease inhibitors (Beyotime, China) was applied to extracted the proteins. BCA protein quantification kit (Beyotime, China) was applied to detect the proteins concentrations following the instructions of the manufacturer. Proteins were separated by electrophoresis using SDS-PAGE (Beijing Biosynthetic Biotechnology, China) and transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad, United States). The PVDF membrane was put into 5% skimmed milk powder blocking solution (Solarbio, China), and sealed for 2 h at room temperature. The sealed PVDF membrane was incubated with the anti-OCN (1:1000; Abcam, ab133612), OPN (1:1000; Abcam, ab214050), RUNX2 (1:2000; Abcam, ab76956), Collagen X (1:1000; Abcam, ab182563), MAPK1 (1:5000; Abcam, ab265600), GAPDH (1:2500; Abcam, ab9485), MEF2A (1:1000; Abcam, ab109420) overnight at 4°C.According to the source of the primary antibody, the secondary antibody was diluted with TBST solution and the PVDF membrane was put into the corresponding secondary antibody. An ultra-sensitive ECL chemiluminescence kit was applied to expose the membrane. The chemiluminescence of the protein bands was quantified by ImageJ software.
2.6 Flow cytometry
The cell cycle detection was performed using the KeyGEN Cell Cycle Kit (China). Briefly, samples were centrifugated to collect the cells, which were rinsed with PBS. The collected cells were fixed with ice-cold 70% ethanol for 4 h at 4°C. RNase A and propidium iodide (PI) were mixed and kept at RT without exposure to sunlight for 30 min. Flow cytometry was conducted to detect DNA, the red fluorescence was recorded at the excitation wavelength of 488 nm. Flow cytometry (Becton Dickinson, United States) was used to analyze the cell distribution ratios in each cell cycle stage.
2.7 Cell proliferation
Cell proliferation was detected using the Edu Cell Proliferation Assay Kit (RiboBio, China) with the protocol of manufacturer. In 12-well plates, chondrocytes were seeded and incubated for 24 h at 37°C. The cells transfected with siRNA or lentivirus were cultured in the medium containing 50 µM Edu for 2 h. After fixing the cells, they were stained with Apollo and were observed under laser scanning confocal microscope (Nikon Instrument, Japan). Data quantitation was done by counting the Edu positive (red) and Hoechst 33342 positive cells (blue). The proliferation rate was calculated by the ratio between the Edu positive vs Hoechst 33342 positive cells.
2.8 Luciferase reporter assay
Luciferase reporter gene vectors (circ-0008870-wt and MAPK1-wt; circ-0008870-Mut and MAPK1-Mut) (GenePharma) were constructed. Lipofectamine 3,000 (Invitrogen, United States) was used for co-transfection of the cells with circ-0008870-wt, circ-0008870-mut, MAPK1-wt, MAPK1-mut, miR-185-3p mimics or negative control (NC). After culturing for 48 h and the luciferase activity was detected using the Dual-Luciferase Assay System (Promega, United States).
2.9 In situ hybridization
The has-cir-008870 was labelled with Cy3 whereas miR-185-3p was with FAM. DAPI (4′,6-diamino-2-phenylindole) was also used for staining. All staining procedures were done following the instructions in the fluorescence in situ hybridization kit (FISH) kit (GenePharma, China). Imaging was conducted by laser scanning confocal microscope (Nikon Instrument, Japan).
2.10 RNase R treatment assay
Chondrocytes were collected and Trizol reagent was added to lyse and extract RNA. RNase R was added to the mixture, which was incubated for 30 min at 37°C. Additionally, the expression of hsa_circ_0008870 and the corresponding linear RNAs were detected by RT-qPCR.
2.11 Alkaline phosphatase (ALP) and calcium staining
ALP staining solution (azo-coupling method) kit (Solarbio, China) was used for ALP staining. Chondrocytes were seeded in 12-well plates and mixed with ALP fixative solution for 3 min at RT. Added the ALP incubation solution (AS-BI: FBB = 1:1) dropwise and incubated the mixture in the dark for 20 min. Chondrocytes were counterstained with nuclear fast red staining solution for 5 min. Finally, viewing cells with a microscope (Nikon, Japan).
Chondrocytes were fixed in 4% paraformaldehyde and exposed to ultraviolet light for 30 min after Von Kossa silver solution staining. Then cells were mixed with Hypo solution for 2 min and hematoxylin staining solution for 25 min, Finally, eosin staining solution was added for 5 min. All staining procedures followed the instructions of the manufacturer (Solarbio, China).
2.12 Animal experiments in vivo
15 Sprague Dawley (SD) rats (3 weeks old) were divided into two groups and were injected with LV-sh-circ-0008870 and LV-NC lentiviral expression vectors, which were diluted with saline from the tail vein. All rats were fed for 6 weeks. During this period, the body length and weight were measured every 2 weeks. The Animal Ethics Committee of Nanchang University (Nanchang, China) gave the formal ethical approval for animal experiments.
Bone and cartilage specimens from the knee joints were collected and stored in 4% formalin. The samples were decalcified for 2months using 10% EDTA and then embedded in paraffin. The thickness of the sagittal section was 5 μm. Subsequently, slices are used for safranin O, immunohistochemistry (IHC) analysis and calcein staining. The specific methods refer to previous studies (Liu et al., 2022). RT-PCR was used to detect the expression levels of hsa_circ_0008870 and miR-185-3p. The expression levels of OCN, OPN, RUNX2, and collagen type X were detected by RT-qPCR and Western blot.
2.13 Statistical analysis
Experiments were conducted in triplicates and data are analyzed by GraphPad Prism 9.3 software. The experimental data are expressed as mean ± standard deviation (SD). Differences between groups were analyzed using student’s t test or one-way ANOVA. p values <0.05 indicated the statistically significant differences.
3 Results
3.1 Hsa_circ_0008870 is downregulated in ISS patients
The blood samples of four ISS and four normal children were used for circRNA microarray analysis. Downregulated circRNA expression profile was mapped by hierarchical clustering. The microarray result showed that 62 circRNAs were downregulated in ISS patients (Figure 1A). Because of the hsa_circ_0008870 expression downregulated significantly (p < 0.05, |logFC|-value > 2.0), hsa_circ_0008870 was selected as the candidate for further analyses in 42 ISS children and 42 normal children (Figure 1B). The expression levels of hsa_circ_0008870 were verified by RT-qPCR after RNase R was used to treat total RNA from ISS and normal groups (Figures 1C,D). These results revealed that hsa_circ_0008870 in ISS presented up-regulated expression compared with normal children instead of linear RNAs.
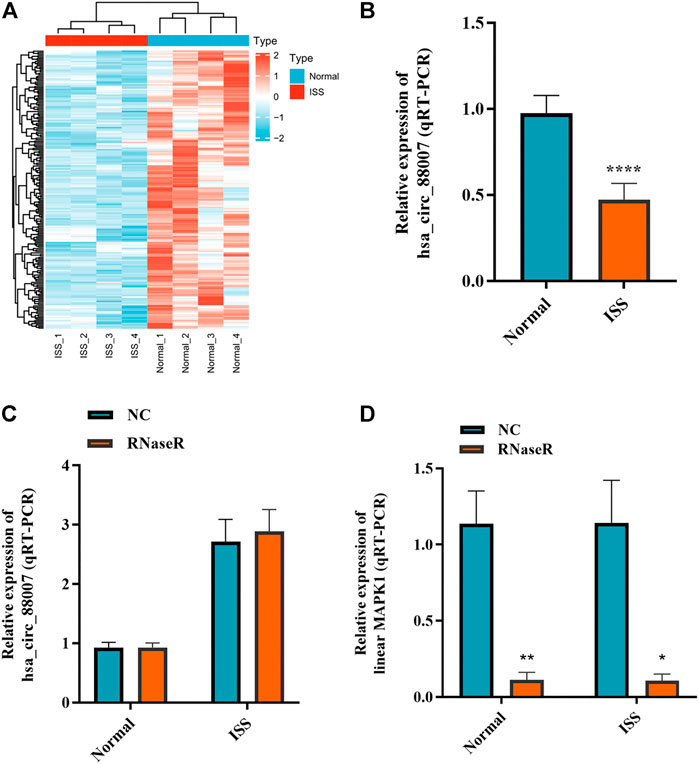
FIGURE 1. The expression of hsa_circ_0008870 in peripheral blood of ISS is downregulated. (A) Heatmap changes of downregulated circRNAs between the two groups. (B) Downregulated hsa_circ_0008870 in ISS patients were validated by RT-qPCR. (C) The expression of hsa_circ_0008870 was detected between the RNase R group and the normal control group. (D) The expression of liner MAPK1 was detected between the RNase R group and the normal control group. p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.3.1.22.
3.2 Knockdown of hsa_circ_0008870 suppresses chondrocyte proliferation and endochondral ossification
To figure out the function of hsa_circ_0008870 downregulation in ISS, the expression of hsa_circ_0008870 was silenced in human chondrocytes (Figure 2A). The Edu assays showed that the chondrocyte proliferation was markedly inhibited in the group of hsa_circ_0008870 downregulation compared with control group (Figure 2B). Meanwhile, flow cytometry analysis demonstrated that compared with the control group, a larger proportion of chondrocytes in the hsa_circ_0008870 down-regulated group were arrested in the G0/G1 phase (Figure 2C).
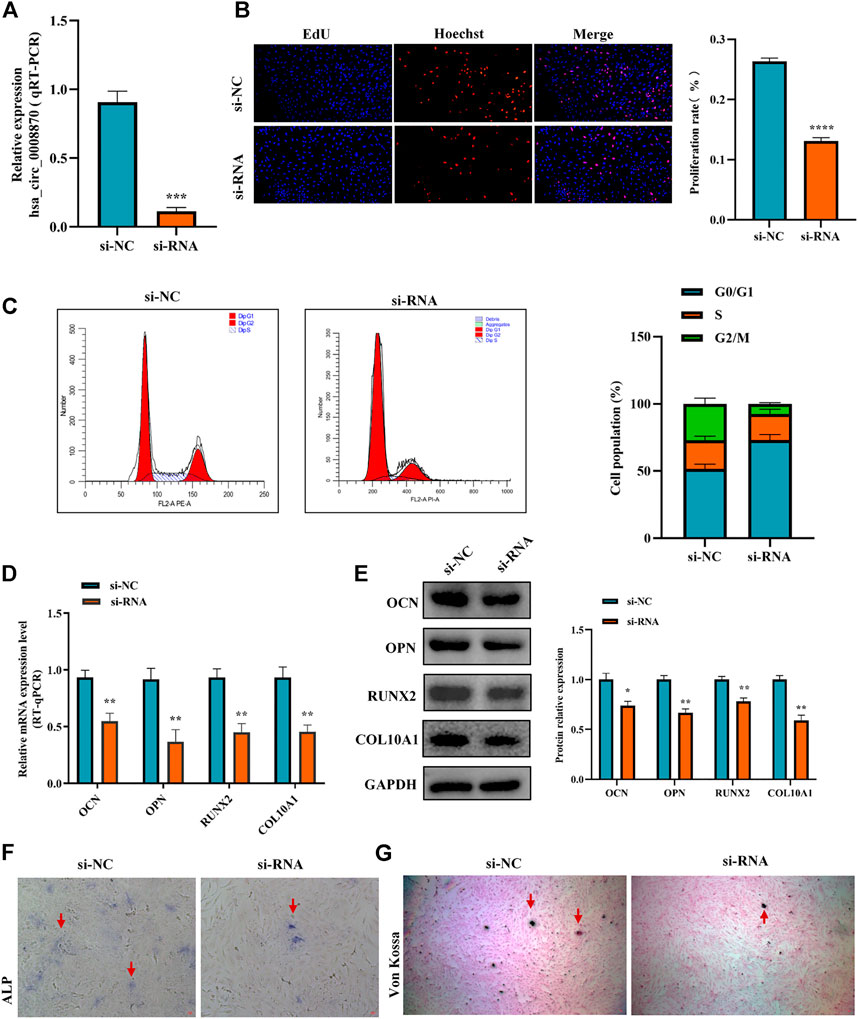
FIGURE 2. Silencing hsa_circ_0008870 inhibits chondrocyte proliferation, hypertrophy and endochondral ossification. (A) The expression of hsa_circ_0008870 in chondrocytes transfected with siRNA was detected by RT-qPCR. (B) Edu assays was used to detect changes in cell proliferation after knockdown of hsa_circ_0008870. (C) Flow cytometry assays indicate changes of cell cycle by knockdown of hsa_circ_0008870. (D) Following knockdown of hsa_circ_0008870, the mRNA expressions of OCN, OPN, RUNX2 and collagen type X were detected by RT-qPCR. (E) Following knockdown of hsa_circ_0008870, the protein expression levels of OCN, OPN, RUNX2 and collagen type X were detected by Western blot. (F) The activity changes of ALP were observed following knockdown of hsa_circ_0008870. (G) Von Kossa staining demonstrated changes in mineralization after knockdown of hsa_circ_0008870. P > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.3.1.23.
To clarify the effect of hsa_circ_0008870 downregulation on the hypertrophy, mineralization, and endochondral osteogenesis in chondrocytes, the expression of OCN, OPN, RUNX2 and collagen type X genes were determined using RT-qPCR and Western blot (Figures 2D,E). These results displayed that the expression of OCN, OPN, RUNX2, and collagen type X genes were significantly suppressed in the group of hsa_circ_0008870 downregulation. Furthermore, the ALP active nodules and mineralized nodules were decreased in the group of hsa_circ_0008870 downregulation using ALP activity analysis and Von Kossa staining (Figures 2F,G). These experimental outcomes indicated that silencing of hsa_circ_0008870 suppressed chondrocyte proliferation and endochondral ossification.
3.3 Hsa_circ_0008870 suppresses chondrocytes proliferation and endochondral ossification by modulating miR-185-3p
To further elucidate the molecular mechanism of hsa_circ_0008870 in ISS, possible interactions between hsa_circ_0008870 and downstream miRNAs were predicted using Circinteractome (http://circinteractome.irp.nia.nih.gov) and manually, and to build the hsa_circ_0008870 targeted miRNAs-mRNA interaction network (Figure 3A). Subsequently, knockdown of hsa_circ_0008870 and RT-qPCR were used to validate the expression of all predicted miRNAs. As shown in Figure 3B, miR-185-3p and miR-616-3p were upregulated after hsa_circ_0008870 downregulation. The miR-185-3p was chosen as the candidate miRNA of hsa_circ_0008870 for further research due to the differential expression was the highest with a 2.41-fold (Figure 3B). The potential binding sites between this miR-185-3p and hsa_circ_0008870 are illustrated in Figure 3C. Afterwards, the luciferase results displayed that, compared with the control group, the fluorescence intensity of the hsa_circ_0008870-wt group decreased significantly, while the fluorescence intensity of the hsa_circ_0008870-mut group recovered (Figures 3C,D). This indicated that hsa_circ_0008870 could target and bind to miR-185-3p. Moreover, in situ hybridization proved the co-localization of hsa_circ_0008870 and miR-185-3p in human chondrocytes and mouse femoral growth plates (Figure 3E).
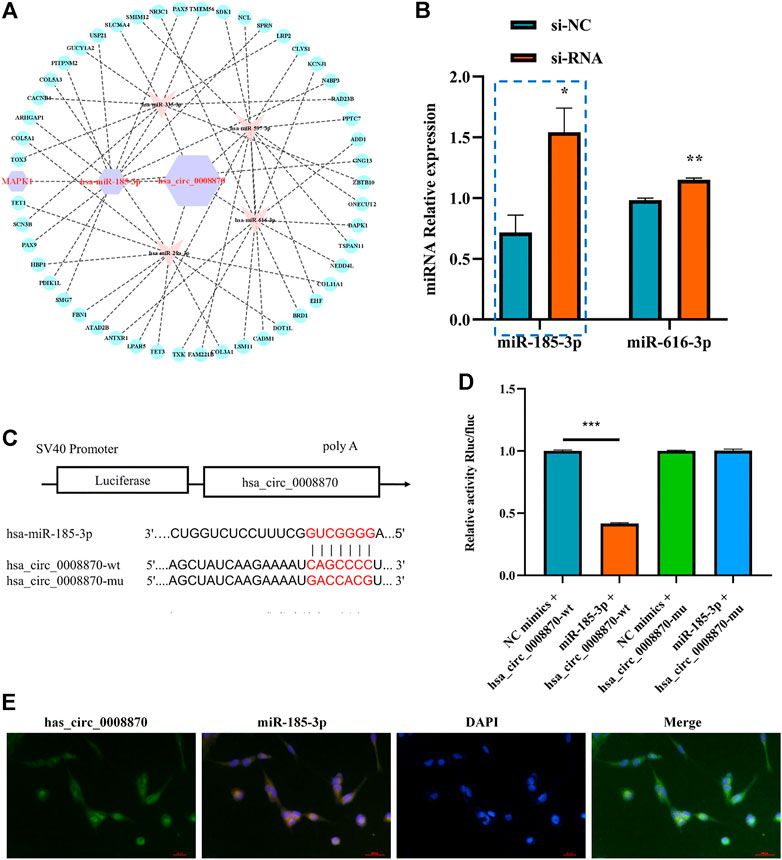
FIGURE 3. Hsa_circ_0008870 acted as an miR-185-3p sponge. (A) hsa_circ_0008870 targeted miRNAs-mRNAs network prediction. (B) The expression of predicted miRNAs in chondrocytes following downregulation of hsa_circ_0008870 expression. (C) The potential target binding sites between hsa_circ_0008870 and miR-185-3p. (D) Luciferase activity was repressed in human chondrocyte cotransfected with WT hsa_circ_0008870 and miR-185-3p mimics, while it was restored in cells cotransfected with Mut hsa_circ_0008870 and miR-185-3p mimics. (E) Fluorescence in situ hybridization proved the co-localization of hsa_circ_0008870 and miR-185-3p in human chondrocytes and mouse femoral growth plates. P > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.3.1.24.
The authors confirmed the function of miR-185-3p in chondrocytes. Firstly, we overexpressed miR-185-3p in chondrocytes (Figure 4A). Subsequent experiments revealed that overexpression of miR-185-3p obviously inhibited the proliferation level of chondrocytes (Figures 4B,C), decreased the expression levels of OCN, OPN, RUNX2 and collagen type X genes (Figure 4D), and reduced ALP activity and mineralized nodules (Figures 4E,F).
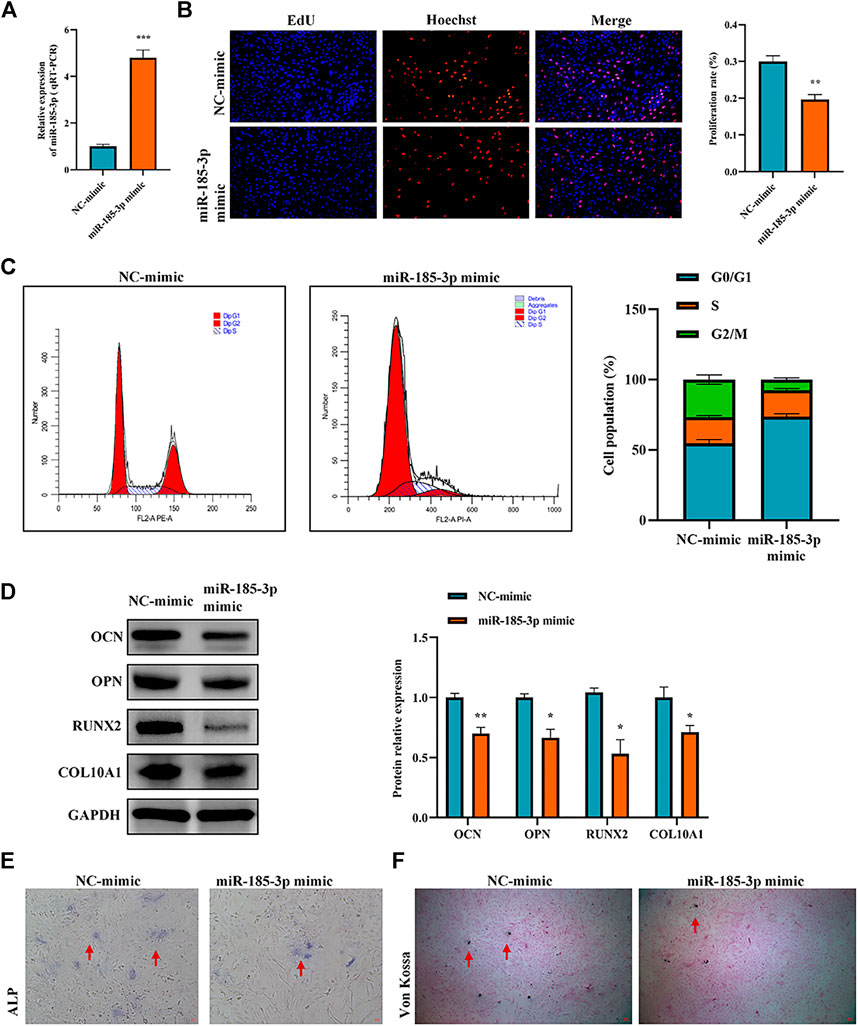
FIGURE 4. Upregulation of miR-185-3p suppresses chondrocytes growth and proliferation and cartilage osteogenesis. (A) The RNA levels of miR-185-3p in human chondrocytes transfected with miR-185-3p mimics were measured by RT-qPCR. (B) The effect of overexpression of miR-185-3p on chondrocyte proliferation was detected by Edu assays. (C) The effect of overexpression of miR-185-3p on chondrocyte cell cycle was detected by flow cytometry. (D) The protein expressions of OCN, OPN, RUNX2 and collagen type X were measured in miR-185-3p overexpressed chondrocytes. (E) ALP activity was measured in miR-185-3p overexpressed chondrocytes. (F) Von Kossa staining demonstrated changes in mineralization in miR-185-3p overexpressed chondrocytes. P > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.3.1.25.
To clarify the interaction between hsa_circ_0008870 and miR-185-3p, we transfected miR-185-3p inhibitor in chondrocytes and assessed the transfection efficiency of inhibitor by RT-qPCR (Figure 5A). The results of rescue experiments indicated that silencing miR-185-3p reversed the inhibition of knockout hsa_circ_0008870 on chondrocyte proliferation compared with the negative control group (Figures 5B,C). Western blot results revealed that although knockdown of hsa_circ_0008870 downregulated the expression levels of OCN, OPN, RUNX2 and collagen type X, silencing miR-185-3p restored their expression levels (Figure 5D). Furthermore, ALP activity and mineralized nodules were restored in si-circ-0008870+ miR-185-3p inhibitor group (Figures 5E,F). To summarize, hsa_circ_0008870 suppresses chondrocytes proliferation and cartilage osteogenesis by targeted upregulation of miR-185-3p.
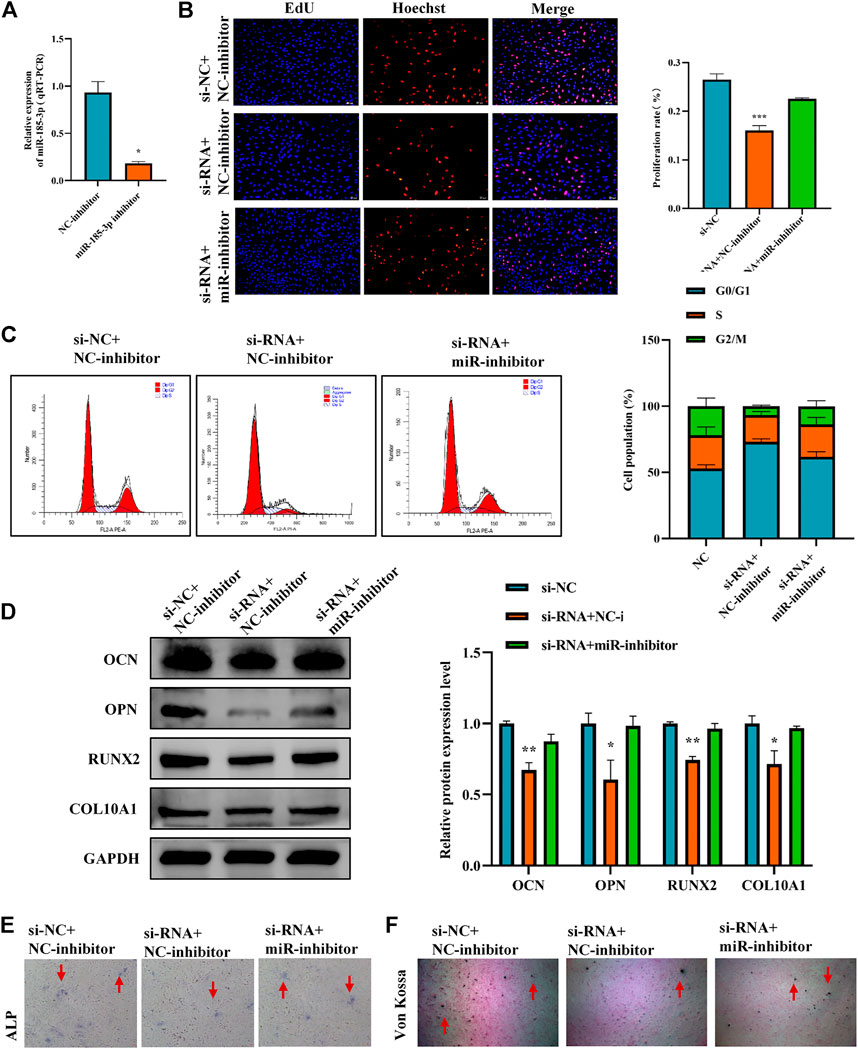
FIGURE 5. Hsa_circ_0008870 suppresses chondrocytes growth and proliferation and cartilage osteogenesis by targeted upregulation of miR-185-3p. (A) The RNA levels of miR-185-3p in human chondrocytes transfected with miR-185-3p inhibitor were measured by RT-qPCR. (B) Proliferation of chondrocytes downregulated by hsa_circ_0008870 was detected by Edu assays, with or without miR-185-3p inhibitor. (C) The effect of hsa_circ_0008870 downregulation on chondrocyte cell cycle was detected by flow cytometry, with or without miR-185-3p inhibitor. (D) The protein expressions of OCN, OPN, RUNX2 and collagen type X were measured in hsa_circ_0008870 knockdown chondrocytes, with or without miR-185-3p inhibitor. (E) ALP activity was measured in hsa_circ_0008870 knockdown chondrocytes, with or without miR-185-3p inhibitor. (F) Von Kossa staining demonstrated changes in mineralization in hsa_circ_0008870 knockdown chondrocytes, with or without miR-185-3p inhibitor. P > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.3.1.26.
3.4 The silencing of hsa_circ_0008870 suppresses chondrocytes proliferation, hypertrophy, as well as endochondral osteogenesis via miR-185-3p/MAPK1 axis
To determine the downstream gene of miR-185-3p, the differential expression of mRNAs was screened by high-throughput sequencing analysis after silencing of the hsa_circ_0008870 in human chondrocytes (Figures 6A,B). The result show that 853 differentially expressed mRNAs were recognized, 433 of them were downregulated whereas the rest 420 were upregulated (p < 0.05, |logFC|-value > 2.0). The top 30 KEGG pathways and GO enrichment analyses were summarized in Figures 6C,D. In addition, we predicted potential downstream target genes of miR-185-3p using two online databases (TargetScan and miRTarBase). According to the obtained Venn diagram (Figure 6E), there are three genes in common. According to the high-throughput sequencing analysis results, the differential expression fold of MAPK1 ranks among the top ten, and studies have confirmed that MAPK1 can maintain the metabolism and homeostasis of chondrocytes, thus playing a protective role (Feng et al., 2010). Therefore, MAPK1 was selected as the target gene of miR-185-3p. The binding sites of miR-185-3p and MAPK1 are shown in Figures 6F,G. The expression levels of MAPK1 was significantly downregulated by upregulating the expression of miR-185-3p (Figures 7A,B.
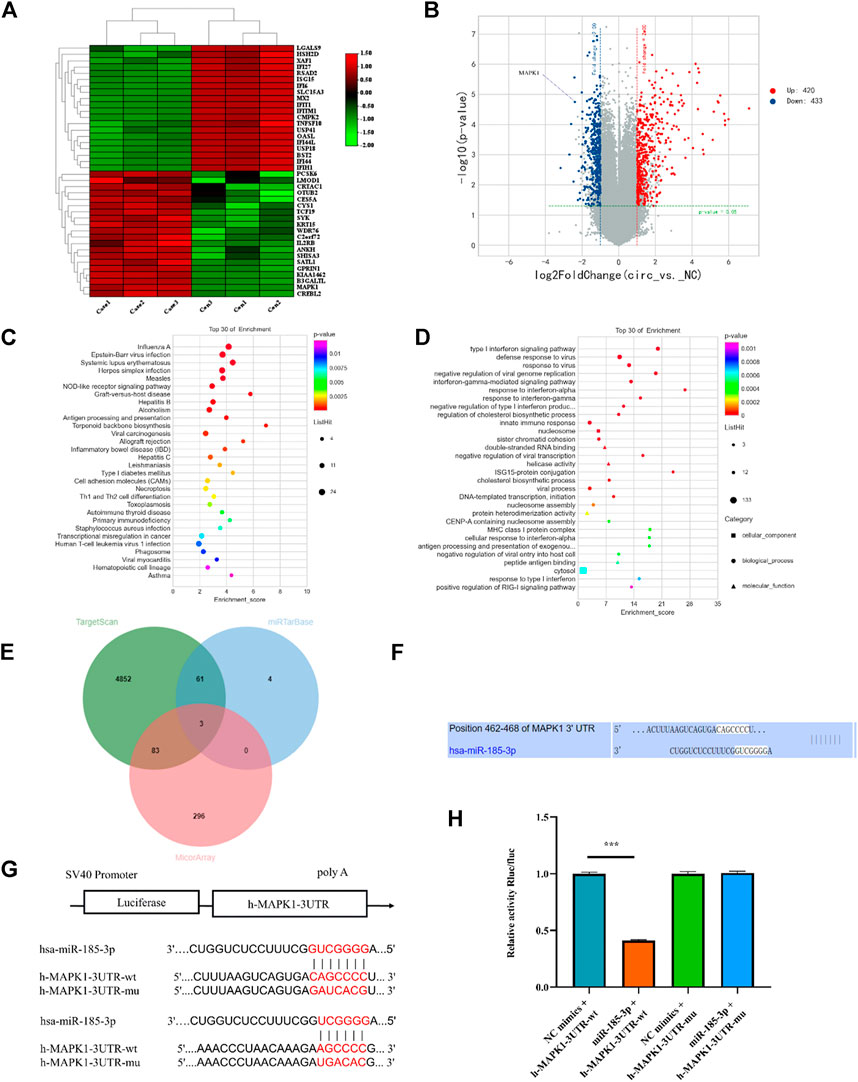
FIGURE 6. mRNA high-throughput sequencing and bioinformatics analyses following silencing hsa_circ_0008870, and luciferase activity between miR-185-3p and MAPK1. (A) The heatmap of differently expressed mRNAs in hsa_circ_0008870 knockdown chondrocytes (n = 3) and normal control chondrocytes (n = 3). (B) Volcano plot of the significantly upregulated and downregulated mRNAs. (C,D) The top 30 KEGG pathway and GO enrichment are exhibited. (E) Venn diagram demonstrating the intersection of downregulated mRNAs and predicted target mRNAs. (F) The potential target binding sites between miR-185-3p and MAPK1. (G,H) Luciferase activity was repressed in human chondrocyte cotransfected with WT MAPK1 and miR-185-3p mimics, while it was restored in cells cotransfected with Mut MAPK1 and miR-185-3p mimics. ***, p < 0.001.3.1.27.
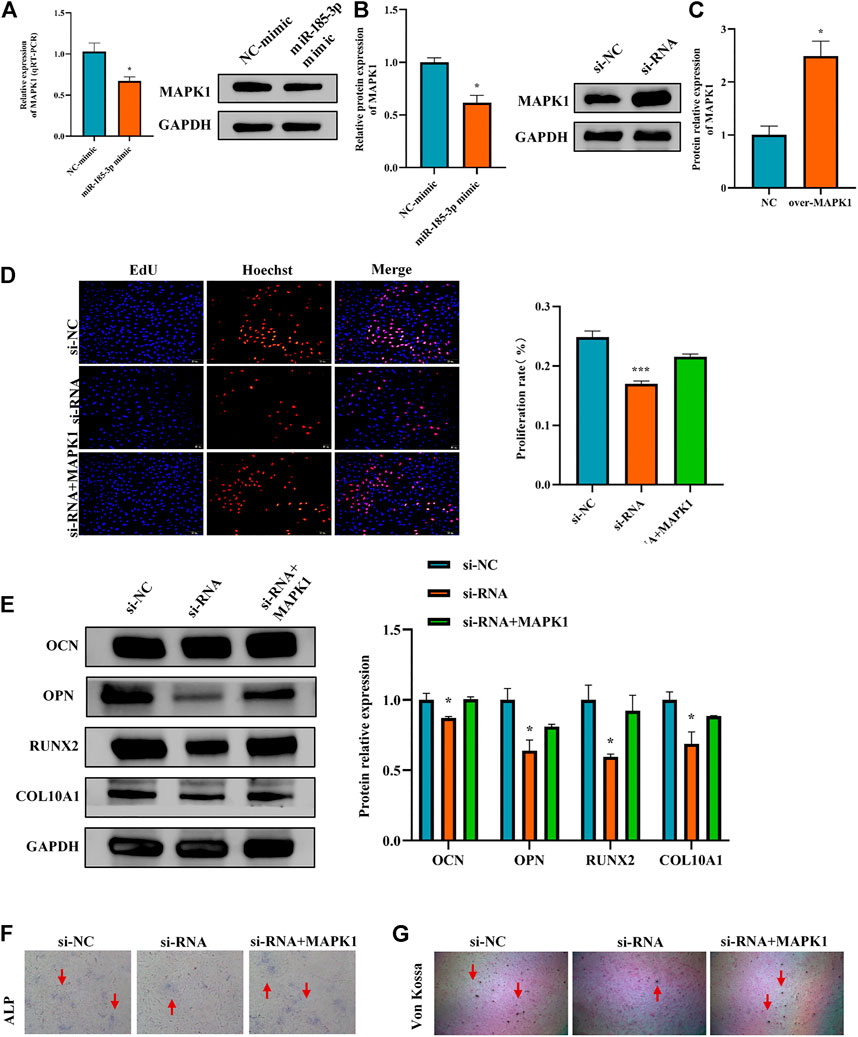
FIGURE 7. Hsa_circ_0008870 suppressed chondrocyte proliferation, hypertrophy and endochondral ossification by regulating miR-185-3p/MAPK1. (A,B) The mRNA and protein expression of MAPK1 was assessed with miR-185-3p mimics. (C) The protein expression of MAPK1 expression levels was assessed following overexpressing MAPK1. (D) The effect of MAPK1 overexpression on hsa_circ_0008870 knockdown-mediated cell proliferation was measured by Edu assays. (E) The protein expressions of OCN, OPN, RUNX2 and collagen type X were analyzed by Western blot. (F,G) The effect of MAPK1 overexpression on hsa_circ_0008870 knockdown-mediated ALP activity and Von Kossa staining. *, p < 0.05 and ***, p < 0.001.3.1.28.
The Luciferase reporter assay indicated that the relative luciferase activity of miR-185-3p + MAPK1 3′UTR-wt was significantly inhibited, while the relative luciferase activity of miR-185-3p + MAPK1 3′UTR-mut group had no significant change (Figures 6G,H). Previous studies had shown that MAPK1 not only promoted the proliferation, growth and differentiation of chondrocytes but also stimulated the formation of osteoblasts in the process of endochondral osteogenesis. To further evaluate whether hsa_circ_0008870 regulated the progress of ISS by regulating the expression levels of MAPK1, we carried rescue experiments. The transfection of MAPK1 protein in chondrocytes significantly promoted the expression levels of MAPK1, confirming the successful transfection of MAPK1 protein (Figure 7C). Transfected human chondrocytes with both si-circ-0008870 and MAPK1 protein showed that the si-circ-0008870 + MAPK1 protein group reversed the inhibition of hsa_circ_0008870 knockdown on chondrocyte proliferation (Figure 7D). Function experiments manifested that the inhibitory effect of hsa_circ_0008870 knockdown on the proliferation and hypertrophy of chondrocytes was reversed by adding MAPK1 protein. Consistently, the inhibition of hsa_circ_0008870 knockdown on the expression levels of OCN, OPN, RUNX2, and collagen type X proteins was suppressed by the overexpression of MAPK1 (Figure 7E). This was indicated by the increase of the number of mineralized nodules and ALP activity in the silenced hsa_circ_0008870 after the overexpression of MAPK1 (Figures 7F,G). The in vitro experimental results showed that hsa_circ_0008870 knockdown inhibited chondrocytes proliferation, hypertrophy, as well as endochondral osteogenesis via miR-185-3p/MAPK1 axis.
3.5 Upregulation of hsa_circ_0008870 enhanced chondrocytes hypertrophy and endochondral osteogenesis by regulating miR-185-3p/MAPK1
To further reveal the effect of hsa_circ_0008870 in chondrocytes proliferation, hypertrophy, as well as endochondral osteogenesis, we first constructed hsa_circ_0008870 overexpressing lentivirus (Figure 8A) and the hsa_circ_0008870 was successfully transfected and overexpressed in human chondrocytes (Figures 8B,C). Subsequent results revealed that miR-185-3p was downregulated (Figure 8D). Although there was no significant difference in the proliferation of chondrocyte between the hsa_circ_0008870 over-expression and the control groups (Figures 8E,F) via Edu assays and flow cytometry analysis, the expression of OCN, OPN, RUNX2, collagen type X and MAPK1 were significantly upregulated in hsa_circ_0008870 over-expression group (Figures 8G,H). In addition, the ALP active nodules and mineralized nodules were promoted (Figures 8I,J). These results indicated that the upregulated hsa_circ_0008870 promoted chondrocytes hypertrophy and endochondral osteogenesis.
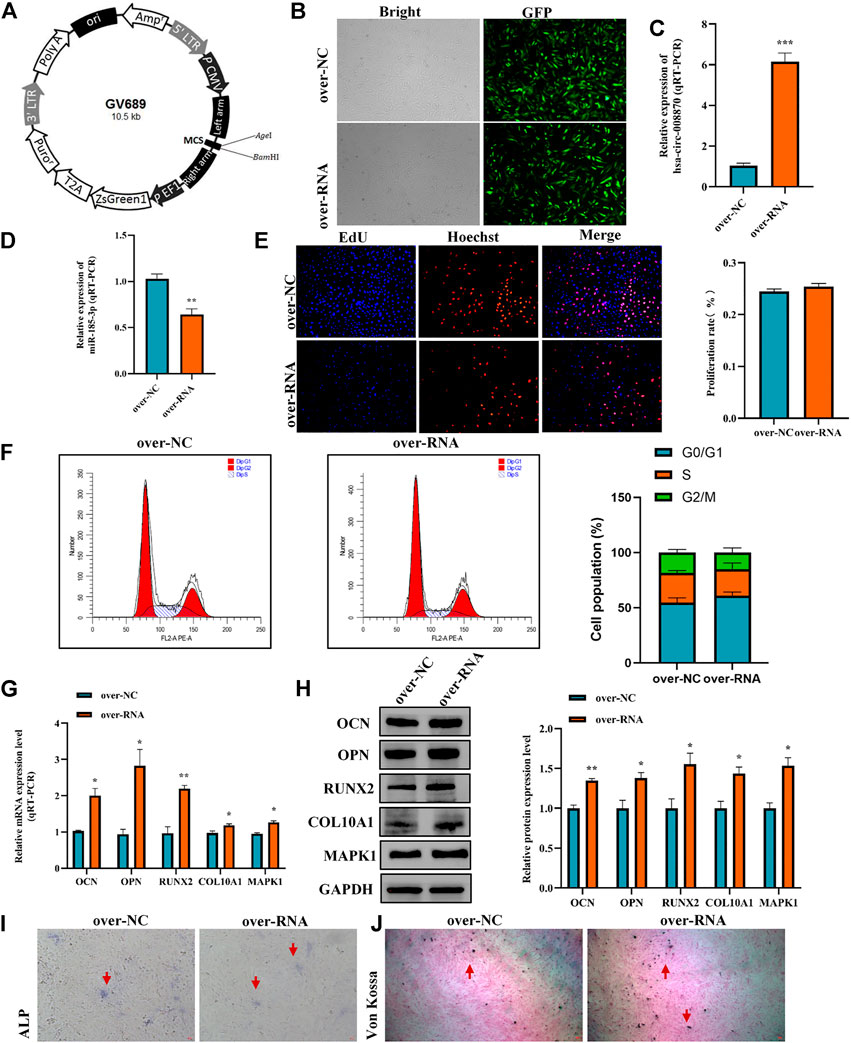
FIGURE 8. Overexpression of hsa_circ_0008870 promoted endochondral osteogenesis, hypertrophy and the growth and proliferation of chondrocytes by regulating miR-185-3p/MAPK1. (A) Hsa_circ_0008870 overexpressing lentivirus. (B) Efficiency of lentiviral transduction were detected by fluorescence of green fluorescent protein. (C,D) The RNA levels of hsa_circ_0008870 and miR-185-3p in human chondrocytes transfected with over_circ_0008870 lentivirus were measured by RT-qPCR. (E) The effect of overexpression of hsa_circ_0008870 on cell proliferation was measured by Edu assays. (F) The effect of overexpression of hsa_circ_0008870 on cell cycle was detected by flow cytometry assays. (G,H) The expressions of OCN, OPN, RUNX2, collagen type X and MAPK1 were analyzed by RT-qPCR and Western blot. (I,J) The effect of hsa_circ_0008870 overexpression on ALP activity and Von Kossa staining. *p < 0.05, **p < 0.01 and ***p < 0.001.3.1.29.
3.6 The miR-185-3p upregulation can inhibit the expression of hsa_circ_0008870 in chondrocytes via a positive feedback loop
It is an interesting phenomenon that we observed that not only hsa_circ_0008870 can regulate the expression of miR-185-3p via the sponge mechanism, but also the upregulated miR-185-3p could inhibit the expression of hsa_circ_0008870 (Figure 9A). For example, as shown Figure 5, the silencing of hsa_circ_0008870 inhibited chondrocytes proliferation and endochondral osteogenesis via upregulating miR-185-3p. Subsequently, it was observed that the expression of MAPK1 and hsa_circ_0008870 were downregulated when miR-185-3p was upregulated in human chondrocytes. Hence, we hypothesized that there might be a positive feedback loop underlying the co-regulation between hsa_circ_0008870 and miR-185-3p in ISS. Bioinformatics analysis demonstrated that the MEF2A is the transcription factor of hsa_circ_0008870 (Figure 9B). Meanwhile, several studies have suggested that MAPK1 can downregulate the expression of MEF2. And there are four mammalian MEF2 genes, MEF2A, -B, -C, and -D (McKinsey et al., 2002). Therefore, we speculate that upregulated miR-185-3p suppresses the expression of MAPK1, which induces the downregulation of MEF2A expression. As a result, hsa_circ_0008870 expression was inhibited (Figure 9C). Though our results observed that phosphorylated active of MEF2A was inhibited after upregulation of miR-185-3p, no statistical significance was shown (Figure 9D). Unfortunately, our experiments do not reveal the regulation mechanism of positive feedback loop between miR-185-3p and hsa_circ_0008870.
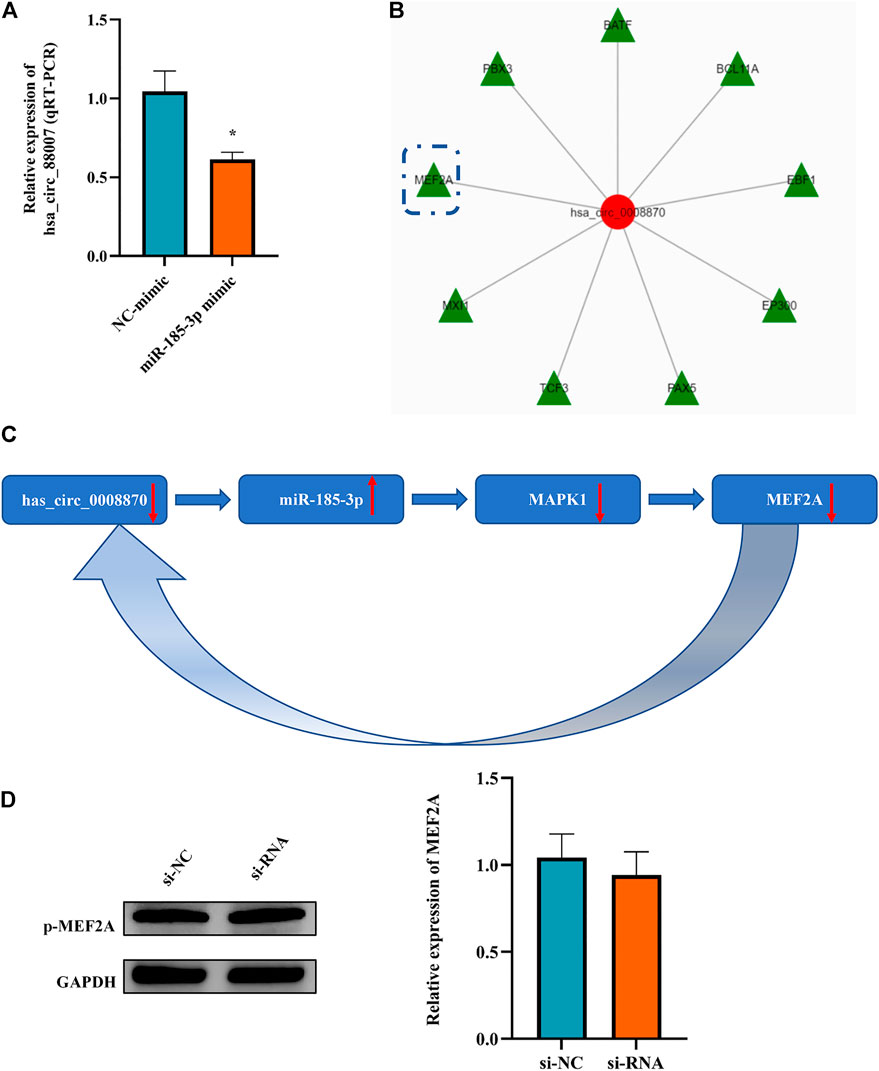
FIGURE 9. The miR-185-3p upregulation can inhibit the expression of hsa_circ_0008870 in chondrocytes via a positive feedback loop. (A) The expression of hsa_circ_0008870 was assessed with miR-185-3p mimics. (B) Transcription factors of hsa_circ_0008870 were analyzed by bioinformatics. (C) The summary of the positive feedback regulation of hsa_circ_0008870/miR-185-3p/MAPK1/MEF2A loop. (D) The protein expressions of phosphorylated MEF2A after knockdown of hsa_circ_0008870 were detected by Western blot. *p < 0.05.3.1.30.
3.7 In vivo experiment further verified that hsa_circ_0008870 inhibited proliferation, hypertrophy and bone formation of growth plate via miR-185-3p/MAPK1 axis
To clarify the effect of the hsa_circ_0008870 knockdown on the rat’s stature, a lentivirus carrying si-circ-0008870 was constructed and used to downregulate the expression of hsa_circ_0008870 in wild-type rats via tail vein (Figure 10A). Compared with the rats in the negative control group, the body size, tibia and femur length of the hsa_circ_0008870 knockdown rats were reduced (Figure 10B). RT-qPCR results manifested that hsa_circ_0008870 was upregulated in rat growth plate cartilage, and miR-185-3p was downregulated after downregulation of hsa_circ_0008870 expression (Figures 11A, 1B). Calcein staining revealed that the rate of new bone formation was lower in the rats of hsa_circ_0008870 downregulation than that in the control rats (Figure 11C). Safranin O staining demonstrated that the height of the femoral growth plate in the rats of hsa_circ_0008870 downregulation was significantly shorter compared to control rats (Figure 11D).
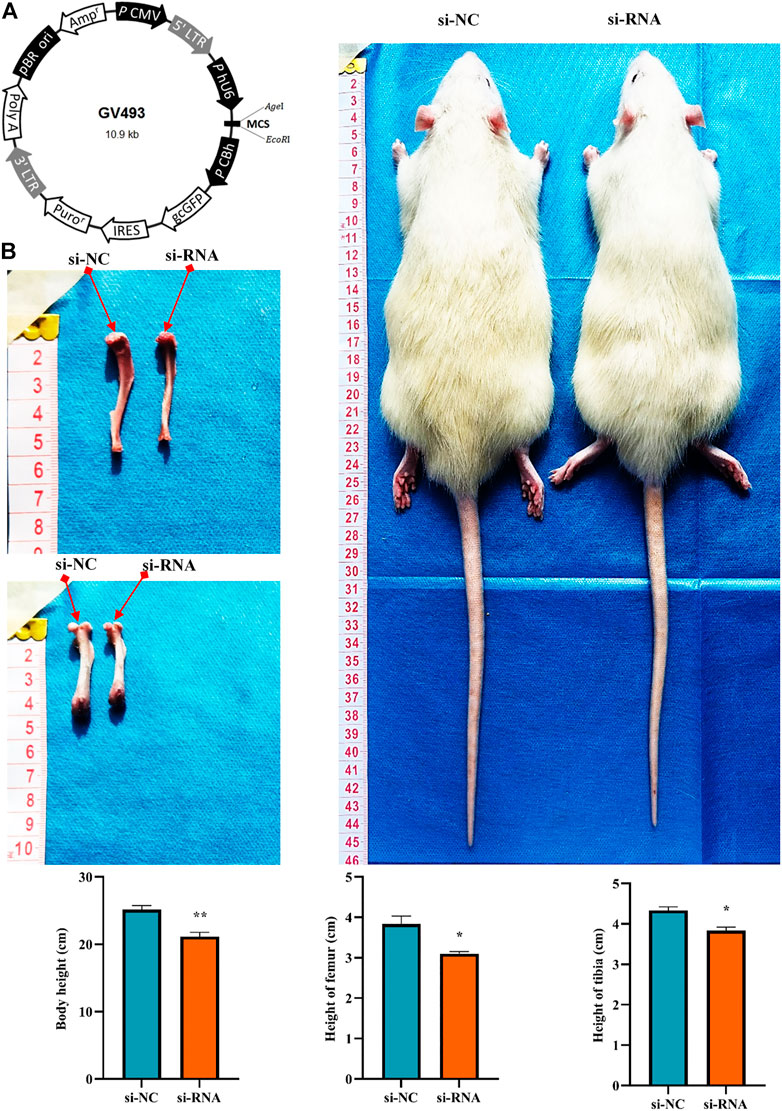
FIGURE 10. Knockdown of hsa_circ_0008870 resulted in smaller stature in rats. (A) Hsa_circ_0008870 knockdown lentivirus. (B) Compared with the rats in the negative control group, changes in body size, tibia and femur length of hsa_circ_0008870 knockdown rats. *p < 0.05 and **p < 0.01.3.1.31.
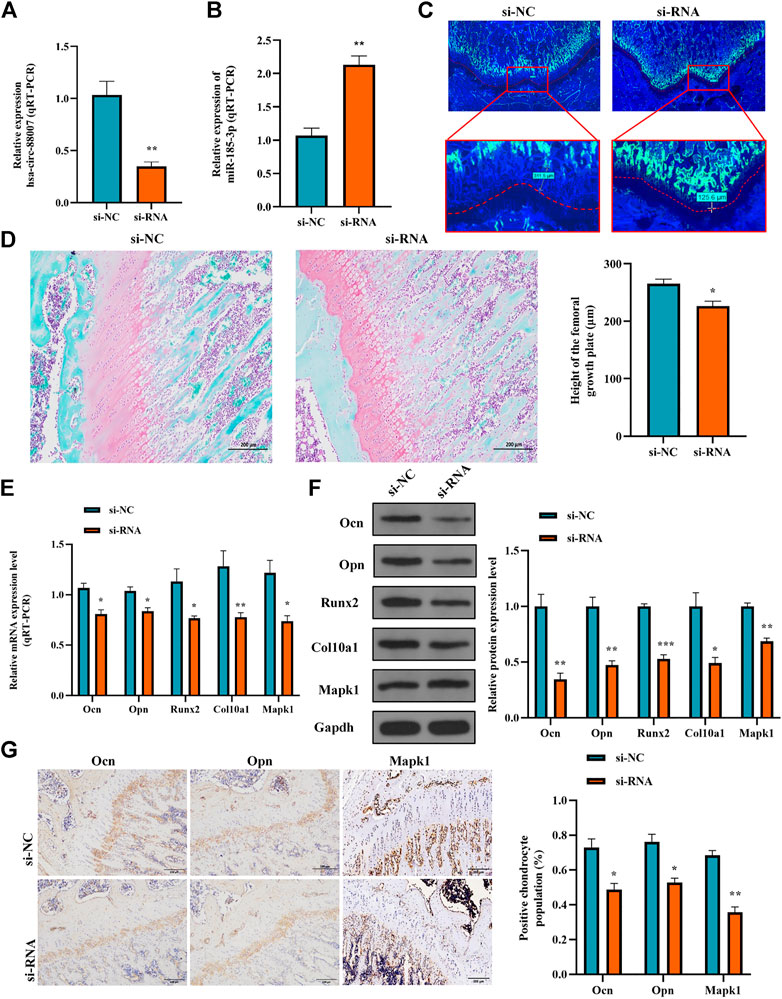
FIGURE 11. Hsa_circ_0008870/miR-185-3p/MAPK1 axis inhibits endochondral osteogenesis as well as the hypertrophy, proliferation and growth of chondrocytes in vivo. (A,B) The RNA levels of hsa_circ_0008870 and miR-185-3p in rats injected with si-circ-0008870 or normal control lentivirus were measured by RT-qPCR. (C) Analysis of the new bone formation rate in the rats by calcein labeling. (D) Assessment of the height of the femoral Growth plate in rats by Safranin O staining. (E,F) The mRNA and protein expressions of Ocn, Opn, Runx2, collagen type X and Mapk1 were detected by RT-qPCR. (G) Expression levels of Ocn, Opn and Mapk1 in rat growth plates were analyzed by immunohistochemical staining. *p < 0.05, **p < 0.01 and ***p < 0.001.
Afterwards, the mRNA and protein expression levels of Ocn, Opn, Runx2, the collagen type-X and Mapk1 in rat growth plates were determined by RT-qPCR and Western blot (Figures 11E, 1F). Compared to control rats, the expression of Ocn, Opn, Runx2 collagen type X and Mapk1 genes were significantly downregulated in the rats of hsa_circ_0008870 downregulation. The immunohistochemical analysis indicated that the expression of Ocn, Opn and Mapk1 were also markedly downregulated in the rats of hsa_circ_0008870 downregulation (Figure 11G). To sum up the above, these results demonstrate that knock down the expression of hsa_circ_0008870 damages the proliferation and bone formation of growth plate in rats. As a result, the rats showed a short stature phenotype.
4 Discussion
Circular RNAs are found in stable forms in human peripheral blood, according to recent studies, and the expression of circRNAs in blood may contain disease-related information (Memczak et al., 2015; Zhang et al., 2020; Yang et al., 2022). For example, Yang et al. discovered that the patients with ischemic stroke had significantly reduced expression of circUSP36 in their peripheral blood, and it alleviated the injury of ischemic stroke injury via miR-139-3p/SMAD3 axis (Yang et al., 2022). circPPM1F is evidently upregulated in peripheral blood of children with type 1 diabetes mellitus, and overexpression of circPPM1F can promote islet injury by enhancing the activation of M1 macrophages (Zhang et al., 2020). In our study, authors first confirmed that hsa_circ_0008870 was downregulated in ISS patients. However, to date, the biological function of hsa_circ_0008870 has not been addressed in the literature published.
In order to determine whether hsa_circ_0008870 is the cause or result of ISS, the authors down regulated the expression of hsa_circ_0008870 in human chondrocytes. These experiments outcomes demonstrated that hsa_circ_0008870 knockdown inhibited chondrocytes proliferation, hypertrophy, and endochondral ossification, whereas overexpression of hsa_circ_0008870 had the opposite results. This suggests that hsa_circ_0008870 may play a key role on ISS pathogenesis. Subsequent in situ hybridization experiment showed hsa_circ_0008870 was localized in the cytoplasm, which indicates that hsa_circ_0008870 could function as miRNA sponges. The targeted miRNAs of hsa_circ_0008870 were predicted by bioinformatics analysis and validity was further assessed by RT-qPCR after hsa_circ _0008870 silencing. MiR-185-3p presented the greatest differential expression in chondrocytes after hsa_circ_0008870 silencing. Thus, it was selected as a candidate target microRNA for further study in the present study. Previous studies observed that overexpression of miR-185-3p improved the renal function of diabetic nephropathy mice by inhibiting and reducing the expression of AGER (Xue et al., 2020). Researchers found that miR-185-3p overexpression enhanced the chemical sensitivity of drug-resistant colorectal cancer (CRC) cells through inhibiting AQP5, offering potential therapeutic targets for CRC cells that are resistant to 5-FU (Zhou et al., 2020). Li et al. demonstrated that downregulated miR-185-3p regulated WNT2B expression in vitro inhibiting the proliferation and transfer of nasopharyngeal carcinoma cells (Li C et al., 2021). However, few study explained the role of miR-185-3p on chondrocytes and growth plates.
To further identify the biological function of miR-185-3p on chondrocytes, the authors have applied fluorescence in situ hybridization. These outcomes showed that hsa_circ_0008870 and miR-185-3p expression co-localized in human chondrocytes and femur growth plates of SD rats. Upregulation of miR-185-3p suppressed the proliferation and hypertrophy, and endochondral osteogenesis in chondrocytes. Subsequent experiments observed that these phenotypes caused by has_circ_0008870 downregulated were rescued via silencing miR-185-3p in chondrocytes. The dual-luciferase reporter gene experiment further confirmed that hsa_circ_0008870 negatively regulated miR-185-3p expression. The results suggested that hsa_circ_0008870 mediated the progression of ISS via targeting miR-185-3p.
To verify the targeted genes of miR-185-3p, the differentially expressed mRNA in human chondrocytes was screened by high-throughput sequencing analysis after silencing of hsa_circ_0008870. According to these results, the top six mRNAs were screened. Moreover, the targeted mRNA of miR-185-3p was selected by bioinformatic analysis. Among all overlap mRNAs between the top six mRNAs and bioinformatic analysis, MAPK1 has attracted attention as it can promote chondrocyte proliferation and alleviate chondrocytes damage. For example, Li et al. reported that the inflammation damage of chondrocytes induced by IL-1 was improved by MAPK1 upregulation (Li Q et al., 2021). Zhao et al. indicated that overexpression of MAPK1 reduced the inhibitory effect of miR-320c on articular chondrocyte proliferation and attenuated the promoting effect of miR-320c on apoptosis (Zhao et al., 2021). Wang et al. observed that the positive feedback regulation between ERK2 (MAPK1) and USP15 played a critical role in the regulation of TGF-β/SMAD2 signaling and the maintaining of cartilage phenotype (Wang et al., 2021).
In the present study, the authors observed that miR-185-3p overexpression significantly suppressed the expression of MAPK1. Meanwhile, luciferase reporter assay showed that miR-185-3p can target MAPK1. To further confirm the biological role of MAPK1 in hsa_circ_0008870/miR-185-3p, the rescue experiments were carried out. As predicted, the over-regulation of MAPK1 reversed the inhibition of the proliferation and hypertrophy of chondrocytes and endochondral osteogenesis caused by the downregulation of hsa_circ_0008870. Meanwhile, we also found that hsa_circ_ 0008870 overexpression can enhance chondrocyte hypertrophy and endochondral ossification. These outcomes indicated that hsa_circ_0008870 can regulate the proliferation and hypertrophy of chondrocytes and endochondral osteogenesis via the miR-185-3p/MAPK1 axis. Subsequently, the effect of the hsa_circ_0008870/miR-185-3p/MAPK1 axis in the progress of ISS was further confirmed in vivo.
In the present study, the authors also observed an interesting phenomenon that upregulated of miR-185-3p can in turn inhibit hsa_circ_0008870 expression in chondrocytes. The upstream transcription factors of hsa_circ_0008870 was screened via UCSC analysis. The outcomes demonstrated that MEF2A might be a transcription factor binding to hsa_circ_0008870 promoter sequences. There are four mammalian MEF2 genes, MEF2A, -B, -C and -D (McKinsey et al., 2002). Rampalli et al. reported P38 MAPK kinase regulates transcription by phosphorylation of myocyte enhancer factor-2 (MEF2) protein to recruit methylase to MEF2 target gene for epigenetic modification of its target gene (Rampalli et al., 2007). Previous studies reported that one of the most typical targets of MAPK1 was the transcription factor of MEF2 family (Yang et al., 1998; Fukuhara et al., 2000; Kato et al., 2000). MAPK1 increased the phosphorylation of MEF2 promoting the activation of MEF2 (Olson, 2004). To further analyze whether MAPK1 influences MEF2A gene expression by its phosphorylation in our study, the expression of phosphorylated MEF2A was detected after silencing hsa_circ_0008870. These results revealed that phosphorylated active of MEF2A was inhibited, while there was no statistical significance. The current study still has several limitations. To date, the underlying mechanism of positive feedback loop between hsa_circ_0008870 and miR-185-3p is still uncertain and requires further attention in future study. Additionally, though no significant difference in sex distribution was observed between the ISS and normal children using Chi square test (p = 0.175), our samples present a discordant sex distribution. Multi center research may be beneficial to further confirm and refute our findings.
In conclusions, our study revealed a novel role of hsa_circ_0008870 in the progression of ISS through miR-185-3p/MAPK1 axis. This suggests the hsa_circ_0008870 might be a potential biomarker and new therapeutic target for ISS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Clinical Ethics Committee of the Second Affiliated Hospital of Nanchang University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. The animal study was reviewed and approved by Animal Care and Use Committee at the Second Affiliated Hospital of Nanchang University.
Author contributions
JJ and XL conceived of the study and designed the experiments jointly. ZW, JY, MY, ZD and JL carried out the experiments and analyzed the experimental results. ZW, JY, XC and JJ wrote the manuscript. All authors approved the final manuscript.
Funding
This study was supported by the National Nature Science Foundation of China (grant no. 82060169 and 81960392), the project of Science and Technology Department of Jiangxi Province (grant no. 20212ACB216010) and Jiangxi Provincial Department of Education (grant nos. GJJ200120).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.1022830/full#supplementary-material
References
Barton, J. S., Gardineri, H. M., Cullen, S., Hindmarsh, P. C., Brook, C. G., and Preece, M. A. (1995). The growth and cardiovascular effects of high dose growth hormone therapy in idiopathic short stature. Clin. Endocrinol. (Oxf). 42 (6), 619–626. doi:10.1111/j.1365-2265.1995.tb02689.x
Bryant, J., Baxter, L., Cave, C. B., and Milne, R. (2007). Recombinant growth hormone for idiopathic short stature in children and adolescents. Cochrane Database Syst. Rev. (3), Cd004440. doi:10.1002/14651858.CD004440
Chen, L. L., and Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12 (4), 381–388. doi:10.1080/15476286.2015.1020271
Chen, Q., Wang, H., Li, Z., Li, F., Liang, L., Zou, Y., et al. (2022). Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J. Hepatology 76 (1), 135–147. doi:10.1016/j.jhep.2021.08.027
Cohen, P., Rogol, A. D., Deal, C. L., Saenger, P., Reiter, E. O., Ross, J. L., et al. (2008). Consensus statement on the diagnosis and treatment of children with idiopathic short stature: a summary of the growth hormone research society, the lawson wilkins pediatric endocrine society, and the European society for paediatric endocrinology workshop. J. Clin. Endocrinol. Metab. 93 (11), 4210–4217. doi:10.1210/jc.2008-0509
Collett-Solberg, P. F., Jorge, A. A. L., Boguszewski, M. C. S., Miller, B. S., Choong, C. S. Y., Cohen, P., et al. (2019). Growth hormone therapy in children; research and practice - a review. Growth Horm. IGF Res. 44, 20–32. doi:10.1016/j.ghir.2018.12.004
Feng, J. Q., Guo, F. J., Jiang, B. C., Zhang, Y., Frenkel, S., Wang, D. W., et al. (2010). Granulin epithelin precursor: a bone morphogenic protein 2-inducible growth factor that activates erk1/2 signaling and JunB transcription factor in chondrogenesis. FASEB J. 24 (6), 1879–1892. doi:10.1096/fj.09-144659
Fukuhara, S., Marinissen, M. J., Chiariello, M., and Gutkind, J. S. (2000). Signaling from G Protein-coupled receptors to ERK5/big MAPK 1 involves gαq and gα12/13 families of heterotrimeric G proteins. J. Biol. Chem. 275 (28), 21730–21736. doi:10.1074/jbc.m002410200
Garikipati, V. N. S., Verma, S. K., Cheng, Z., Liang, D., Truongcao, M. M., Cimini, M., et al. (2019). Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 10 (1), 4317. doi:10.1038/s41467-019-11777-7
Greene, J., Baird, A. M., Brady, L., Lim, M., Gray, S. G., McDermott, R., et al. (2017). Circular RNAs: Biogenesis, function and role in human diseases. Front. Mol. Biosci. 4, 38. doi:10.3389/fmolb.2017.00038
Grimberg, A., DiVall, S. A., Polychronakos, C., Allen, D. B., Cohen, L. E., Quintos, J. B., et al. (2016). Guidelines for growth hormone and insulin-like growth factor-I treatment in children and adolescents: Growth hormone deficiency, idiopathic short stature, and primary insulin-like growth factor-I deficiency. Horm. Res. Paediatr. 86 (6), 361–397. doi:10.1159/000452150
Kamp, G. A., Waelkens, J. J., de Muinck Keizer-Schrama, S. M., Delemarre-Van de Waal, H. A., Verhoeven-Wind, L., Zwinderman, A. H., et al. (2002). High dose growth hormone treatment induces acceleration of skeletal maturation and an earlier onset of puberty in children with idiopathic short stature. Arch. Dis. Child. 87 (3), 215–220. doi:10.1136/adc.87.3.215
Kato, Y., Zhao, M., Morikawa, A., Sugiyama, T., Chakravortty, D., Koide, N., et al. (2000). Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275 (24), 18534–18540. doi:10.1074/jbc.m001573200
Lee, J. M., Davis, M. M., Clark, S. J., Hofer, T. P., and Kemper, A. R. (2006). Estimated cost-effectiveness of growth hormone therapy for idiopathic short stature. Arch. Pediatr. Adolesc. Med. 160 (3), 263–269. doi:10.1001/archpedi.160.3.263
Lee, B., Kwon, C. Y., and Jang, S. (2020). Comparative effectiveness of east asian traditional medicine for treatment of idiopathic short stature in children: A protocol for systematic review and network meta-analysis. Med. Baltim. 99 (43), e22856. doi:10.1097/md.0000000000022856
Lee, M. M. (2006). Idiopathic short stature. N. Engl. J. Med. Overseas. Ed. 354 (24), 2576–2582. doi:10.1056/nejmcp060828
Li, C., Li, W., Cao, S., Xu, J., Qian, Y., Pan, X., et al. (2021). Circ_0058106 promotes proliferation, metastasis and EMT process by regulating Wnt2b/β-catenin/c-Myc pathway through miR-185-3p in hypopharyngeal squamous cell carcinoma. Cell Death Dis. 12 (11), 1063. doi:10.1038/s41419-021-04346-8
Li, Q., Wu, M., Fang, G., Li, K., Cui, W., Li, L., et al. (2021). MicroRNA-186-5p downregulation inhibits osteoarthritis development by targeting MAPK1. Mol. Med. Rep. 23 (4), 253. doi:10.3892/mmr.2021.11892
Liu, X., Yan, C., Deng, X., and Jia, J. (2020). Hsa_circularRNA_0079201 suppresses chondrocyte proliferation and endochondral ossification by regulating the microRNA-140-3p/SMAD2 signaling pathway in idiopathic short stature. Int. J. Mol. Med. 46 (6), 1993–2006. doi:10.3892/ijmm.2020.4737
Liu, X., Du, Z., Yi, X., Sheng, T., Yuan, J., and Jia, J. (2021). Circular RNA circANAPC2 mediates the impairment of endochondral ossification by miR-874-3p/SMAD3 signalling pathway in idiopathic short stature. J. Cell. Mol. Med. 25 (7), 3408–3426. doi:10.1111/jcmm.16419
Liu, X., Zhang, J., Yuan, J., Ding, R., Liu, T., and Jia, J. (2022). LCN2 is a new diagnostic biomarker and potential therapeutic target in idiopathic short stature. J. Cell. Mol. Med. 26 (12), 3568–3581. doi:10.1111/jcmm.17408
McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002). MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends biochem. Sci. 27 (1), 40–47. doi:10.1016/s0968-0004(01)02031-x
Memczak, S., Papavasileiou, P., Peters, O., and Rajewsky, N. (2015). Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10 (10), e0141214. doi:10.1371/journal.pone.0141214
Olson, E. N. (2004). Undermining the endothelium by ablation of MAPK-MEF2 signaling. J. Clin. Invest. 113 (8), 1110–1112. doi:10.1172/jci200421497
Paltoglou, G., Dimitropoulos, I., Kourlaba, G., and Charmandari, E. (2020). The effect of treatment with recombinant human growth hormone (rhGH) on linear growth and adult height in children with idiopathic short stature (ISS): a systematic review and meta-analysis. J. Pediatr. Endocrinol. Metab. 33 (12), 1577–1588. doi:10.1515/jpem-2020-0287
Pedicelli, S., Peschiaroli, E., Violi, E., and Cianfarani, S. (2009). Controversies in the definition and treatment of idiopathic short stature (ISS). J. Clin. Res. Pediatr. Endocrinol. 1 (3), 105–115. doi:10.4008/jcrpe.v1i3.53
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: A new star of noncoding RNAs. Cancer Lett. 365 (2), 141–148. doi:10.1016/j.canlet.2015.06.003
Qu, S., Zhong, Y., Shang, R., Zhang, X., Song, W., Kjems, J., et al. (2017). The emerging landscape of circular RNA in life processes. RNA Biol. 14 (8), 992–999. doi:10.1080/15476286.2016.1220473
Rampalli, S., Li, L., Mak, E., Ge, K., Brand, M., Tapscott, S. J., et al. (2007). p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat. Struct. Mol. Biol. 14 (12), 1150–1156. doi:10.1038/nsmb1316
Ross, J. L., Lee, P. A., Gut, R., and Germak, J. (2015). Attaining genetic height potential: Analysis of height outcomes from the ANSWER Program in children treated with growth hormone over 5 years. Growth Horm. IGF Res. 25 (6), 286–293. doi:10.1016/j.ghir.2015.08.006
Shen, S., Wu, Y., Chen, J., Xie, Z., Huang, K., Wang, G., et al. (2019). CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 78 (6), 826–836. doi:10.1136/annrheumdis-2018-214786
Vyas, V., and Menon, R. K. (2021). Management of short stature: Use of growth hormone in GH-deficient and non-GH-deficient conditions. Indian J. Pediatr. 88 (12), 1203–1208. doi:10.1007/s12098-021-03892-5
Wang, W., Zhu, Y., Sun, Z., Jin, C., and Wang, X. (2021). Positive feedback regulation between USP15 and ERK2 inhibits osteoarthritis progression through TGF-β/SMAD2 signaling. Arthritis Res. Ther. 23 (1), 84. doi:10.1186/s13075-021-02456-4
Wu, D., Chen, R. M., Chen, S. K., Liu, G. L., Chen, L. Q., Yang, Y., et al. (2020). Final adult height of children with idiopathic short stature: a multicenter study on GH therapy alone started during peri-puberty. BMC Pediatr. 20 (1), 138. doi:10.1186/s12887-020-02034-8
Xue, X. X., Lei, H. Q., Zhao, L., Wang, X. Y., Wang, Z., Xie, L. Y., et al. (2020). MiR-185-3p downregulates advanced glycosylation end product receptor expression and improves renal function in diabetic nephropathy mice. Eur. Rev. Med. Pharmacol. Sci. 24 (9), 5018–5027. doi:10.26355/eurrev_202005_21193
Yang, C. C., Ornatsky, O. I., McDermott, J. C., Cruz, T. F., and Prody, C. A. (1998). Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res. 26 (20), 4771–4777. doi:10.1093/nar/26.20.4771
Yang, J., He, W., Gu, L., Long, J., Zhu, L., Zhang, R., et al. (2022). CircUSP36 attenuates ischemic stroke injury through the miR-139-3p/SMAD3/Bcl2 signal axis. Clin. Sci. (Lond). 136 (12), 953–971. doi:10.1042/cs20220157
Zhang, C., Han, X., Yang, L., Fu, J., Sun, C., Huang, S., et al. (2020). Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus. Theranostics 10 (24), 10908–10924. doi:10.7150/thno.48264
Zhao, W., Dong, M., Pan, J., Wang, Y., Zhou, J., Ma, J., et al. (2019). Circular RNAs: A novel target among non-coding RNAs with potential roles in malignant tumors (review). Mol. Med. Rep. 20 (4), 3463–3474. doi:10.3892/mmr.2019.10637
Zhao, L., Zhou, R., Wang, Q., Cheng, Y., Gao, M., and Huang, C. (2021). MicroRNA-320c inhibits articular chondrocytes proliferation and induces apoptosis by targeting mitogen-activated protein kinase 1 (MAPK1). Int. J. Rheum. Dis. 24 (3), 402–410. doi:10.1111/1756-185x.14053
Keywords: circular RNA, growth plate, idiopathic short stature, miR-185-3p, MAPK1
Citation: Wu Z, Yuan J, Li J, Du Z, Yin M, Cheng X, Liu X and Jia J (2022) Hsa_circ_0008870 suppresses bone formation of growth plate through inhibition of miR-185-3p/ MAPK1 axis in idiopathic short stature. Front. Bioeng. Biotechnol. 10:1022830. doi: 10.3389/fbioe.2022.1022830
Received: 19 August 2022; Accepted: 26 September 2022;
Published: 11 October 2022.
Edited by:
Yiqiang Hu, Huazhong University of Science and Technology, ChinaReviewed by:
Bo Huang, Xinqiao Hospital, ChinaLianyong Li, Shengjing Hospital of China Medical University, China
Copyright © 2022 Wu, Yuan, Li, Du, Yin, Cheng, Liu and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xijuan Liu, bGl1eGlqdWFuOUAxNjMuY29t; Jingyu Jia, amlheGludG9uZzlAMTYzLmNvbQ==
 Zhiwen Wu
Zhiwen Wu Jinghong Yuan
Jinghong Yuan Jiantian Li1,4
Jiantian Li1,4 Zhi Du
Zhi Du Jingyu Jia
Jingyu Jia