
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 14 October 2022
Sec. Bioprocess Engineering
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.1017190
This article is part of the Research Topic Biotransformation of Carbohydrates View all 5 articles
A correction has been applied to this article in:
Corrigendum: Engineered biosynthesis of plant polyketides by type III polyketide synthases in microorganisms
Plant specialized metabolites occupy unique therapeutic niches in human medicine. A large family of plant specialized metabolites, namely plant polyketides, exhibit diverse and remarkable pharmaceutical properties and thereby great biomanufacturing potential. A growing body of studies has focused on plant polyketide synthesis using plant type III polyketide synthases (PKSs), such as flavonoids, stilbenes, benzalacetones, curcuminoids, chromones, acridones, xanthones, and pyrones. Microbial expression of plant type III PKSs and related biosynthetic pathways in workhorse microorganisms, such as Saccharomyces cerevisiae, Escherichia coli, and Yarrowia lipolytica, have led to the complete biosynthesis of multiple plant polyketides, such as flavonoids and stilbenes, from simple carbohydrates using different metabolic engineering approaches. Additionally, advanced biosynthesis techniques led to the biosynthesis of novel and complex plant polyketides synthesized by diversified type III PKSs. This review will summarize efforts in the past 10 years in type III PKS-catalyzed natural product biosynthesis in microorganisms, especially the complete biosynthesis strategies and achievements.
Plant natural products (PNPs, also called specialized metabolites) occupy unique structural space and are of great significance in human medicine, agriculture, and the cosmetic industry. Representative examples include the anticancer agent Taxol (Kingston, 1994), antimalarial drug artemisinin (Klayman, 1985), and analgesic opiates (Pastemak Gavril et al., 1980). Sourcing the valuable PNPs from their native producers relies on plant growth and is usually susceptible to environmental changes. The rapid development of synthetic biology has enabled microbial biomanufacturing as a powerful alternative approach due to microorganisms’ shorter cultivation time and less reliance on climate. Microorganisms are more feasible hosts to engineer than plants due to their simple genetic background and easy genetic operation. Various PNPs have been synthesized in engineered microorganisms in the past decade, including medicinal alkaloids (Galanie et al., 2015; Li et al., 2018b; Srinivasan and Smolke, 2020), terpenes (Belcher et al., 2020), and flavonoids (Shah et al., 2019). This review will focus on the microbial biomanufacturing of another PNP family, namely polyketides, which are valuable PNPs with medicinal, nutraceutical, or agricultural potential.
Plant polyketides are a large family of PNPs extensively existing in the plant kingdom. The committed reaction in plant polyketide synthesis is a condensation reaction catalyzed by type III polyketide synthases (PKSs) (shown in Figure 1), which condenses a coenzyme A (CoA)-linked starter molecule with several dicarboxyl-CoA extender units (mainly malonyl-CoA) to generate the polyketide scaffold (Dibyendu, 2015). Type III PKSs are mostly found in plants, while type I and type II PKSs are mainly found in bacteria or fungi (Hertweck, 2009). Different types of PKSs vary in both structure and mechanism. Shen (2003) has discussed the differences and provided a detailed diagram of the catalytic mechanisms: Generally, type I PKSs are larger modular and multifunctional enzymes, and each of modules has a different, non-iteratively acting activity that catalyzes one cycle of polyketide chain elongation. Type II PKSs are dissociable complexes and carry out iteratively acting activities. In contrast, type III PKSs are homodimeric proteins that have active sites comprising a Cys, His, Asn catalytic triad in each monomer and carry out iteratively acting condensing activities as well (Morita et al., 2019). The conserved structure of these PKSs allows them to accept various CoA starters and different numbers malonyl-CoAs (Bisht et al., 2021). The substrate preferences of different type III PKSs, along with the downstream tailoring reactions catalyzed by various enzymes (e.g., methyltransferases, oxidoreductases, prenyltransferases, and glycosyltransferases), leads to the large diversity of plant polyketides (Noel et al., 2005). The syntheses of these plant polyketides by type III PKSs also include different catalytic mechanisms, such as C6-C1 Claisen Cyclization and C2-C7 Aldol Cyclization. Morita et al. (2019) discussed the diversity in mechanisms of type III PKSs and gave a hint on how structural subtleties lead to molecular diversities. In this review, we focus on the microbial biosynthesis of plant polyketides from simple sugars by de novo pathway reconstruction and engineering, with a particular emphasis on the heterologous expression and engineering of type III PKSs.
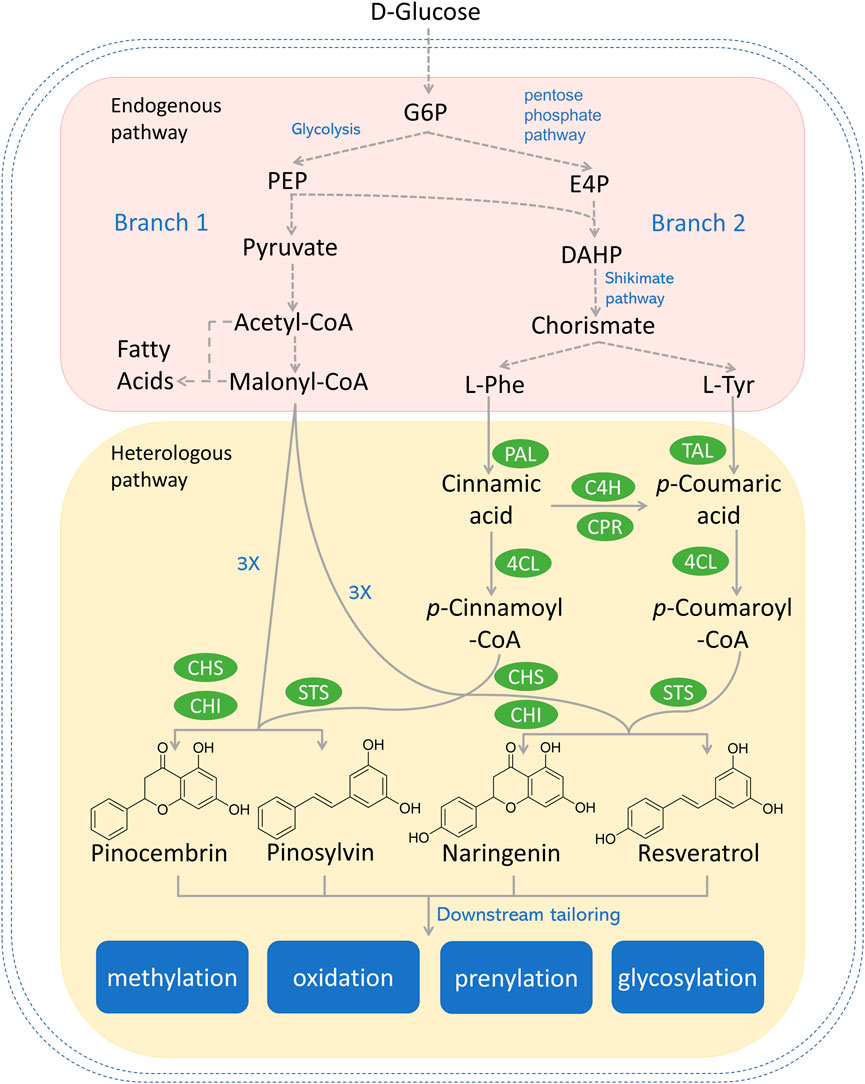
FIGURE 1. Reconstructed biosynthetic pathway for most explored type III PKS-derived polyketides (e.g., pinocembrin, pinosylvin, naringenin, and resveratrol) in microbial hosts. Dotted arrows refer to multiple steps. Genes and enzymes in green circle are heterologous genes from plants or bacterium. G6P, glucose-6-phosphate; PEP, phosphoenolpyruvate; E4P, erythrose-4-phosphate; DAHP, 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate; L-Phe, L-phenylalanine; L-Tyr, L-tyrosine; PAL, phenylalanine ammonia lyase; TAL, tyrosine ammonia lyase; C4H, cinnamic acid hydroxylase; CPR, P450 reductase; 4CL, 4-coumaroyl-coA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; STS, stilbene synthase.
There are two upstream branch pathways in plants, which lead to the polyketide precursors in the committed step catalyzed by the type III PKS. Similar pathways have been reconstructed in various microorganisms. The graphical summary of the reconstructed type III PKS-centered biosynthetic pathway in microorganisms is shown in Figure 1. Branch 1 starts with D-glucose, which is converted to acetyl-CoA through a series of enzymatic activities. While most acetyl-CoA will enter the tricarboxylic acid (TCA) cycle, a small portion of acetyl-CoA will be converted to malonyl-CoA by the action of acetyl-CoA carboxylase (ACC) (Krivoruchko et al., 2015). Branch 2 starts with aromatic amino acids (AAA, i.e., L-phenylalanine and L-tyrosine), which are deaminated to cinnamic acid or p-coumaric acid by the action of monofunctional phenylalanine ammonia lyase (PAL) or bifunctional phenylalanine/tyrosine ammonia lyase (PTAL) in plants (Barros and Dixon, 2020). Alternatively, in reconstructed pathways, L-tyrosine can be deaminated to p-coumaric acid catalyzed by tyrosine ammonia lyase (TAL) from bacteria (Jendresen et al., 2015). 4-coumarate-CoA ligase (4CL) conjugates hydroxycinnamic acids to CoAs to form the corresponding CoA esters (e.g., cinnamoyl-CoA and coumaroyl-CoA) (Schneider et al., 2003). Diverse type III PKSs [e.g., chalcone synthase (CHS); stilbene synthase (STS), curcuminoid synthase (CUS)] subsequently condense the starter CoA ester with multiple malonyl-CoAs to form diversified polyketides such as naringenin, pinocembrin, and resveratrol.
Many microbial hosts have been developed for the heterologous production of plant natural polyketides derived from type III PKSs. Escherichia coli and Saccharomyces cerevisiae are the two most developed hosts due to their unique advantages in biomanufacturing, including fast growth speed and massive genetic manipulation tools that enable rapid pathway construction and optimization (Lian et al., 2018; Pontrelli et al., 2018). As the model bacterial host, E. coli can produce a large amount of aromatic amino acids, which can be leveraged for optimal polyketide production (Lütke-Eversloh and Stephanopoulos, 2005; Lütke-Eversloh and Stephanopoulos, 2007). S. cerevisiae is a model eukaryotic host particularly suitable for various PNPs’ production. It has similar post-translational modifications as plants to enable functional expression of plant-derived enzymes; the membrane-bound organelles such as endoplasmic reticulum (ER) in yeast provide a similar intracellular environment as in plants for the expression of plant-derived cytochrome P450 enzymes, which are necessary for downstream tailoring of polyketide scaffolds (Huang et al., 2008; Krivoruchko and Nielsen, 2015). Both E. coli and S. cerevisiae can be engineered to produce naringenin, pinocembrin, and resveratrol from feeding substrates supplemented in the culture medium (e.g., L-phenylalanine and L-tyrosine, cinnamic acid, p-coumaric acid, and caffeic acid). Recently, the oleaginous yeast Yarrowia lipolytica has emerged as another eukaryotic host to produce plant polyketides due to the high flux through acetyl-CoA precursors, but the genetic operations are still limited by the lack of genetic engineering tools (Abdel-Mawgoud et al., 2018; Markham et al., 2018). More genetic modification strategies are expected in future research. A few non-conventional microorganisms have also been developed to produce plant polyketides, including Streptomyces venezuelae and Corynebacterium glutamicum. S. venezuelae can express large biosynthetic gene clusters and is the common microbial host for the production of complex polyketide production catalyzed by type I and II PKSs (Liu et al., 2018). However, the production of type III PKS-derived polyketides is quite limited. C. glutamicum is widely used in amino acid production and has a strong tolerance to aromatic compounds (Becker and Wittmann, 2012; Eberhardt et al., 2014). Engineered C. glutamicum can produce naringenin and resveratrol (Kallscheuer et al., 2016; Braga et al., 2018), but endogenous aromatic catabolic pathways must be deleted to inhibit the degradation of aromatic compounds. The key features of each host are summarized in Table 1.
One outstanding challenge in plant polyketide biosynthesis is the low production due to the lack of upstream precursors. Feeding expensive upstream precursors is not economical or compatible for larger-scale biomanufacturing. Therefore, engineering the complete plant polyketides biosynthesis pathway from glucose in microbial hosts is important and commercially favorable. The predominant strategy is to increase the availability of both the CoA starter and malonyl-CoA extender. Other strategies include co-culture, flux balance, and gene selection.
Engineering the shikimate pathway in microbial hosts can increase AAA supply and polyketide production. The shikimate pathway widely exists in plants, algae, fungi, and bacteria. It includes seven enzymatic reactions that combine phosphoenolpyruvate (PEP) and D-erythrose 4-phosphate (E4P) to make chorismate. Chorismate is then converted to AAA, such as L-tyrosine and L-phenylalanine (Rodriguez et al., 2014).
Successful methods that overproduce AAA in E. coli are widely explored (Rodriguez et al., 2014; Jiang and Zhang, 2016). Popular AAA pathway engineering strategies are summarized in Figure 2, including 1) Deleting tyrR, which is a global repressor of AAA biosynthetic pathway and negatively regulates the expression of genes aroF, aroG, aroH, and aroL using AAA as cofactors (Hong et al., 2020; Yuan et al., 2020); 2) Improving PEP and E4P production by overexpressing the phosphoenolpyruvate synthase (ppsA) and transketolase (tktA) genes (Zhang et al., 2017); 3) Alleviating feedback inhibition from L-tyrosine by using 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate (DAHP) synthase feedback inhibition resistant variant (aroGfbr) and chorismate mutase-prephenate dehydrogenase feedback inhibition resistant variant (tyrAfbr) (Zhang et al., 2017; Hong et al., 2020; Yuan et al., 2020). Combining several strategies, the E. coli strain was able to produce abundant p-coumaric acids, which paves the way for polyketide production, especially in co-culture strategies.
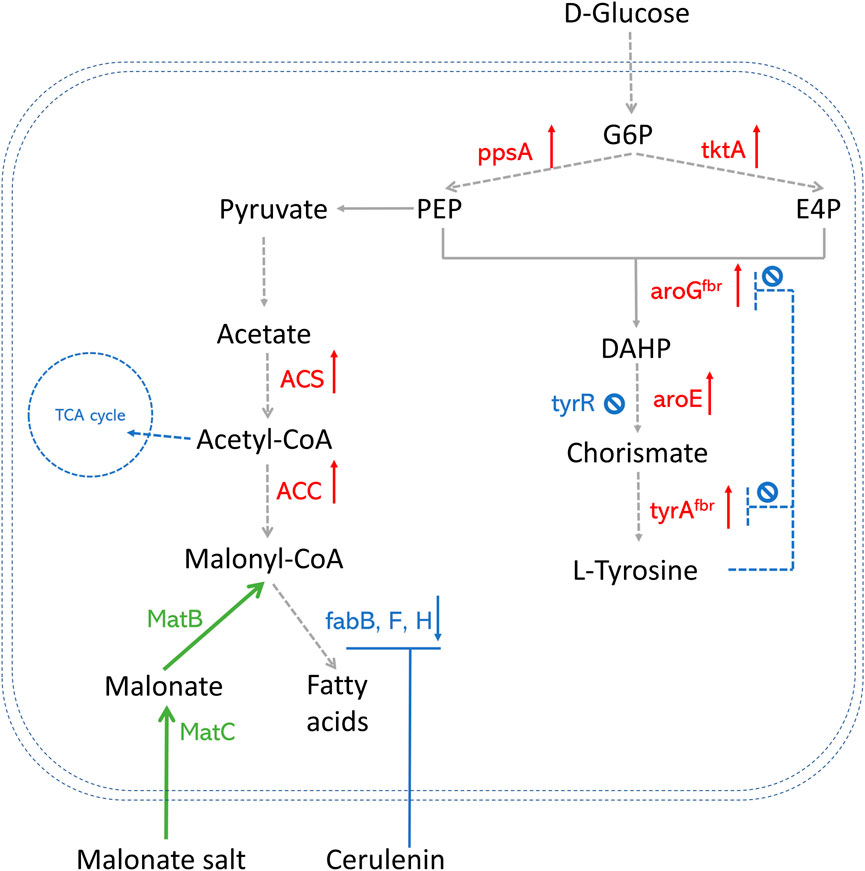
FIGURE 2. An overview of precursor enhancement engineering in E. coli. Genes and enzymes in red are overexpressed. Genes and enzymes in blue are deleted or downregulated. Genes and enzymes in green are heterologous expressed. Dotted arrows refer to multiple steps. G6P, glucose-6-phosphate; PEP, phosphoenolpyruvate; E4P, erythrose-4-phosphate; DAHP, 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate; fabH, gene that encodes 3-oxoacyl carrier protein synthase III; fabB/fabF, genes that encode the beta-ketoacyl-acp synthase I/II protein; MatB, malonylCoA synthetase; MatC, malonate carrier protein; ACS, acetyl-CoA synthase; ACC, acetyl-CoA carboxylase; TCA cycle, tricarboxylic acid cycle; ppsA, phosphoenolpyruvate synthase; tktA, transketolase; tyrAfbr, chorismate mutase-prephenate dehydrogenase feedback inhibition resistant variant; aroGfbr, DAHP synthase feedback inhibition resistant variant; TyrR, a DNA binding transcriptional regulatory protein.
In S. cerevisiae, significant efforts have been made to enhance the production of AAA-derived products, as shown in Figure 3. One of the most successful strategies is overexpressing the feedback-insensitive enzyme mutants (i.e., 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase ARO4K229L and chorismate mutase ARO7G141S) (Li et al., 2015; Sáez-Sáez et al., 2020). In doing so, the titer of resveratrol in an engineered yeast strain was enhanced by 78% in batch cultures. Downregulating competing branch pathways is also a promising approach. For example, pyruvate decarboxylase (PDC5) and phenylpyruvate decarboxylase (ARO10), which convert hydroxyphenylpyruvate to fusel alcohol or acid, were deleted to direct more metabolic flux to the L-tyrosine production (Li et al., 2021). Combining both strategies, Li et al. (2021) got strains that produced (2S)-naringenin and p-coumaric acid with titers of 43.4 mg/L and 177.9 mg/L. In contrast, the highest titer was only 5.4 mg/L in the strains that only expressed naringenin pathway genes. A new strategy is to increase E4P availability by introducing a phosphoketalose-based pathway into yeast (Liu et al., 2019). Liu et al. (2019) constructed an S. cerevisiae strain to produce high levels of p-coumaric acid by increasing the E4P availability. They further replaced the promoters of several important genes at key nodes to optimize carbon flux distribution between glycolysis and the AAA biosynthesis pathway. The maximum p-coumaric acid titer yielded by the strain was 12.5 g/L. This p-coumaric overproducing strain was then used as a background strain to de novo biosynthesize bioactive isoflavonoid daidzein with the expression of a CHS (Liu et al., 2021).
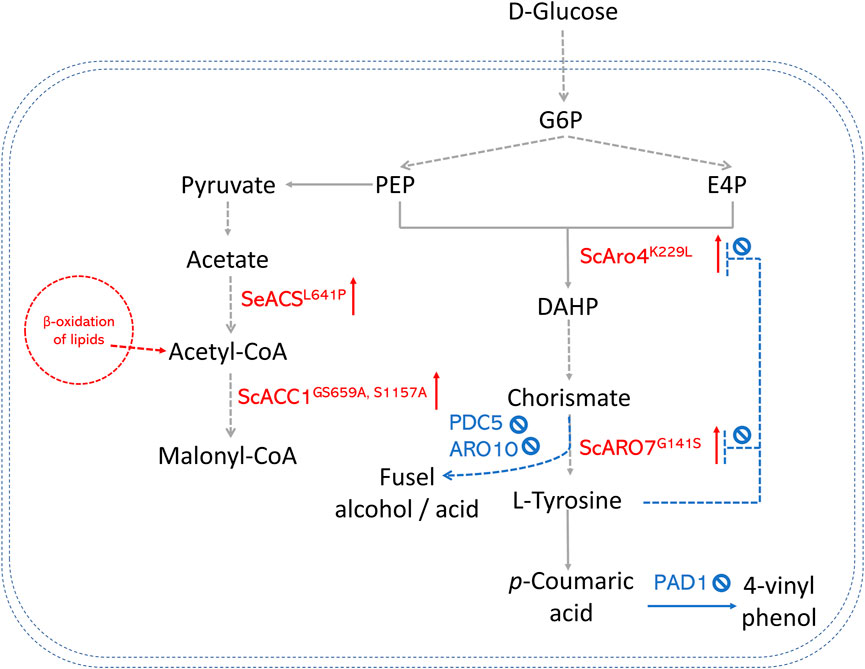
FIGURE 3. An overview of precursor enhancement engineering in S. cerevisiae. Genes and enzymes in red are overexpressed. Genes and enzymes in blue are deleted. Dotted arrows refer to multiple steps. G6P, glucose-6-phosphate; PEP, phosphoenolpyruvate; E4P, erythrose-4-phosphate; DAHP, 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate; ARO4K229L, DAHP synthase feedback inhibition resistant variants; ARO7G141S, chorismate mutase feedback inhibition resistant variants; PAD1, phenyl acrylic acid decarboxylase; PDC5, pyruvate decarboxylase; ARO10, phenylpyruvate decarboxylase; ACS, acetyl-CoA synthase; ACC, acetyl-CoA carboxylase.
The availability of malonyl-CoA in many microbial hosts is limited and considered the rate-limiting step towards diverse polyketides. Multiple strategies have been developed to overcome this bottleneck, summarized in Figures 2, 3. In polyketide-producing microorganisms, malonyl-CoA is usually synthesized from acetyl-CoA catalyzed by acetyl-CoA carboxylases (ACC). Native ACC enzyme in E. coli is feedback-inhibited by acyl carrier proteins (ACPs) from the fatty acid biosynthetic pathway (Davis and Cronan, 2001). A widely applied strategy to increase ACC activity is heterologously expressing the ACC enzyme. Zha et al. (2009) enhanced malonyl-CoA titer by 3-fold after introducing C. glutamicum ACC into E. coli. Wu et al. (2021) found novel ACC families from Salmonella enterica that exhibited the highest activities among all the candidates tested by performing a comprehensive phylogenomic and experimental analysis and obtained 1,073.8 mg/L of naringenin production in E. coli. Phosphorylation of ACC in S. cerevisiae by the sucrose non-fermenting protein 1 (Snf1p) decrease ACC’s activity (Shirra et al., 2001). To address this inhibition, mutant ScAcc1pS659A, S1157A was overexpressed to increase ACC activity (Shi et al., 2014). Li et al. (2015) overexpressed this mutated ScAcc1pS659A, S1157A in a resveratrol-producing strain and improved resveratrol production from 4.85 mg/L to 6.39 mg/L.
Rewiring the central carbon flux towards malonyl-CoA and malonyl-CoA-derived polyketides is also widely explored (Zha et al., 2009; Lian et al., 2014; Cardenas and da Silva, 2016; Zhang et al., 2019). In E. coli, optimization of the glycolytic pathway has increased acetyl-CoA availability and led to 474 mg/L naringenin production (Xu et al., 2011). Deleting the competing acetate and ethanol pathways, inhibiting fatty acid synthesis that consumes malonyl-CoA, and acetyl-CoA synthase (ACS) overexpression resulted in a 15-fold improvement of malonyl-CoA availability and phloroglucinol production of 1,280 mg/L (Zha et al., 2009). Heterologous expression of an ACS mutant from Salmonella enterica (SeACSL641P) with increased activity in S. cerevisiae can boost acetyl-CoA availability from acetate (Chen et al., 2013). β-oxidation of fatty acids in yeast peroxisomes, the main pathway for fatty acid degradation, can also be optimized to increase malonyl-CoA supply (Zhang et al., 2021).
Alternatively, in E. coli, the intracellular malonyl-CoA level can be elevated by the heterologous expression of the malonate assimilation pathway, including gene matB (malonyl-CoA synthetase) and matC (malonate carrier protein) from Rhizobium trifolii and supplied with malonate at the same time (Wu et al., 2014; Zhou et al., 2019). Moreover, unfavored malonyl-CoA consumption by fatty acid synthesis can be reduced by using cerulenin, a well-characterized chemical inhibitor for fatty acid elongation (Santos et al., 2011; Kallscheuer et al., 2016). The clustered regularly interspaced short palindromic repeats interference (CRISPRi) system was also proposed to identify the candidate genes that can efficiently channel carbon flux toward malonyl-CoA (Wu et al., 2017b; Wu et al., 2021).
Co-culture of different strains (E. coli and S. cerevisiae, or different E. coli strains) can fully utilize the innate metabolic flux towards AAA and malonyl-CoA and/or reduce the metabolic burden of each strain by dividing extensively long and complicated pathways into individual modules. Various PNPs have been produced by microbial co-culture, including naringenin and resveratrol. E. coli-E. coli co-culture systems have enabled the heterologous production of naringenin (Ganesan et al., 2017) and resveratrol (Hong et al., 2020), in which the upstream strain produced p-coumaric acid and the downstream strain produced naringenin or resveratrol (55.7 mg/L). An E. coli-S. cerevisiae co-culture system produced naringenin from xylose (Zhang et al., 2017). E. coli can utilize xylose as the carbon source and excrete acetate, which is then utilized by S. cerevisiae as the carbon source (Song et al., 2014) to produce naringenin, leading to 21.16 ± 0.41 mg/L naringenin production. Another E. coli-S. cerevisiae co-culture system produced resveratrol from glucose (Yuan et al., 2020), in which an E. coli strain produced p-coumaric acid, and a downstream S. cerevisiae strain produced malonyl-CoA and resveratrol (36 mg/L). In addition to the common polyketides naringenin and resveratrol, some uncommon polyketides and their derivatives have been successfully produced from glucose or feeding substrates (e.g., p-coumaric acid or malonate) in microbial hosts through co-culture strategies, including curcuminoids (Fang et al., 2018), anthocyanins (Jones et al., 2017), apigetrin (Thuan et al., 2018), sakuranetin (Wang et al., 2020), and other flavonoids (Du et al., 2020). As the optimal fermentation conditions of each strain in the co-culture (e.g., pH, oxygen, and temperature) are usually different, future improvements in controlling the culture condition dynamically will be helpful to address current challenges in manufacturing. In addition, dynamic tuning of the ratio of different strains during co-culture will be another promising approach to optimize the strategy.
There are other bottlenecks hindering the production of plant polyketides. Many studies have found that the conversion of AAAs to polyketides is the limiting step (Santos et al., 2011; Lv et al., 2019). Both enzyme variants and gene expression levels of the essential genes were engineered to address this challenge. For example, the 4CL gene from Medicago truncatula and CHS gene from Vitis vinifera led to the highest naringenin production among various combinations (Mark et al., 2019). TALs and CHSs from different organisms can also lead to optimized flavonoid production (Santos et al., 2011; Tong et al., 2022).
Optimizing gene expression levels can also increase polyketide production by increasing gene copy number (enhancing naringenin production in S. cerevisiae) (Li et al., 2021), promoter strength, and RBS engineering (enhanced shikimic acid production in C. glutamicum) (Zhang et al., 2015), and by tuning gene transcriptional level (enhanced curcumin production) (Kang et al., 2018). Multi-module optimization strategy has been developed to optimize the production of naringenin (Wu et al., 2014) and pinocembrin (Wu et al., 2013): The de novo pathway was divided into different modules at the key nodes; Modular expression levels were combinatorially tuned by modifying plasmid gene copy numbers and promoter strengths. Zhou et al. (2019) constructed a library of constitutive promoters in E. coli randomly cloned upstream of pathway genes and screened the library to identify a balanced strain with the highest titer. Biosensor-based strategy combined with metabolic circuits was established to dynamically regulate the de novo naringenin pathway (Zhou et al., 2021). In this system, the dynamic regulation network contains three layers: the naringenin biosynthesis pathway, p-coumaric acid-responsive regulation pathway, and real-time control of intracellular supply of malonyl-CoA. By doing so, 523.7 ± 51.8 mg/L of naringenin was produced. To address the challenge in possible genetic degeneration during long-term cultivation or scale-up, a promising strategy that combines metabolic addiction and negative autoregulation was recently applied in naringenin production in Y. lipolytica (Lv et al., 2020). The negative autoregulation using CRISPRi and inducible promoters lowered the competition of malonyl-CoA between the naringenin pathway and the fatty acid pathway; naringenin-responsive transcription factor FdeR coupled naringenin level with leucine synthesis and cell fitness for improved naringenin production. The naringenin-producing population was enriched and 90.9% of naringenin titer still persisted after 324 generations.
In this section, we will discuss about recent efforts on the complete synthesis of diverse plant polyketides using different type III PKSs. We will also discuss about a variety of rare polyketides, the complete biosynthetic pathway of which remain yet to be constructed. All these achievements and their strategies are summarized in Table 2 and Figure 4.
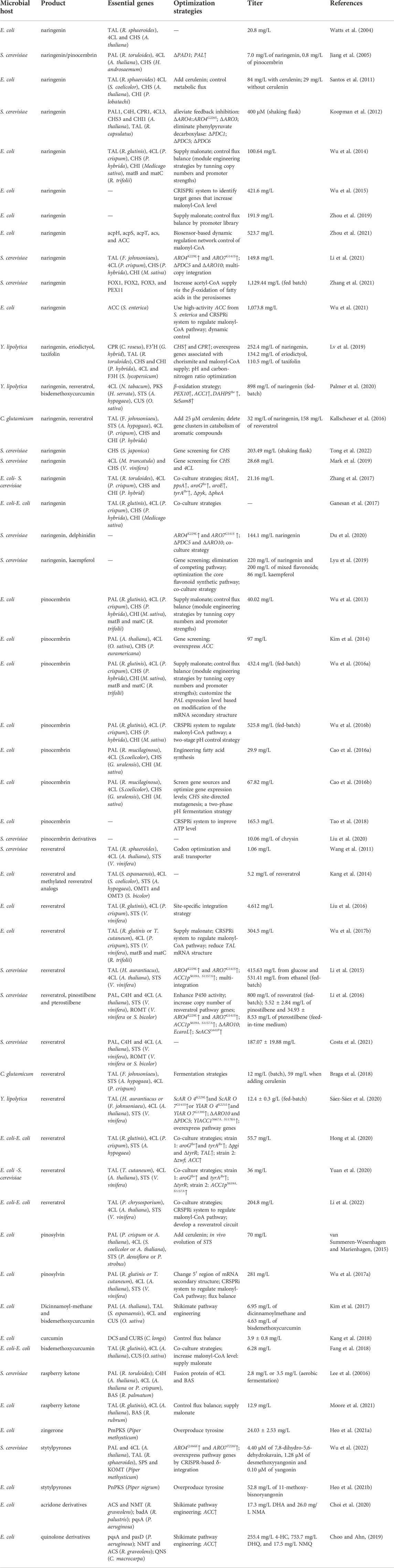
TABLE 2. Summary of achievements and metabolic engineering strategies for de novo production of type III-PKS-derived polyketides.
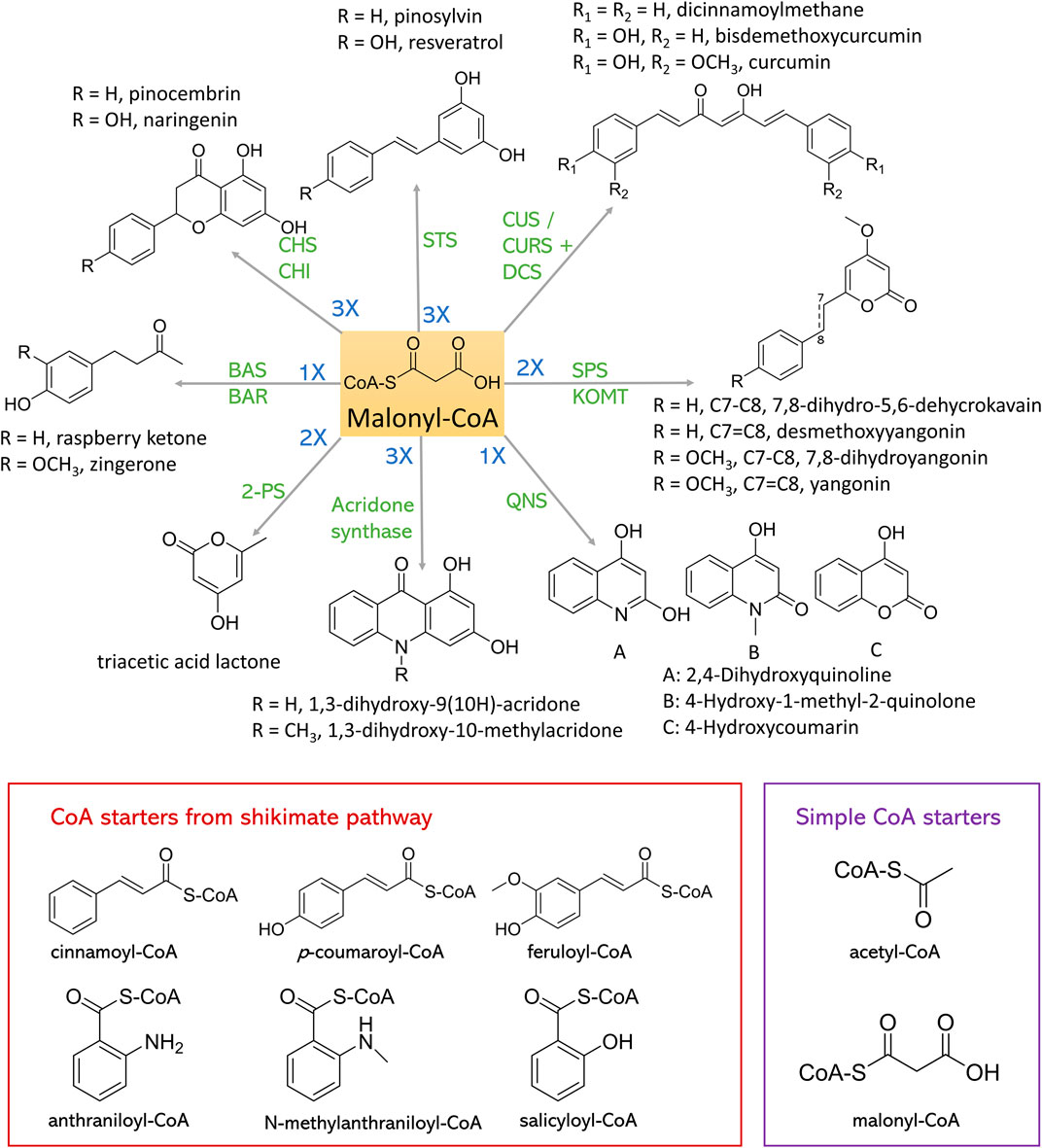
FIGURE 4. Synthesis of plant polyketides from malonyl-CoA and CoA starters. CHS, chalcone synthase; CHI, chalcone isomerase; STS, stilbene synthase; CUS, curcuminoid synthase; CURS, curcumin synthase; DCS, diketide-CoA synthase; SPS, styrylpyrone synthase; KOMT, kava O-methyltransferase; QNS, quinolone synthase; 2-PS, 2-pyrone synthase; BAS, benzalacetone synthase; BAR, benzalacetone reductase.
Chalcone Synthase (CHS) is one of the most explored type III PKSs, essential in flavonoid chalcone synthesis. Various chalcones are important intermediates for flavonoid synthesis in plants. The CHS is ubiquitous in almost any plant species (Rammohan et al., 2020). CHS uses C6-C1 Claisen cyclization to produce naringenin chalcone, pinocembrin chalcone, and eriodictyol chalcone from p-coumaroyl-CoA, cinnamoyl-CoA, and caffeoyl-CoA, respectively, with three extender malonyl-CoAs. These flavonoid chalcones can further convert to corresponding flavonoids via spontaneous reaction or catalyzed by chalcone isomerase (CHI). CHS could also produce isoliquiritigenin with chalcone reductase (CHR) in a concerted manner. Given the antiviral, antioxidant, antitumor, and anti-inflammatory effects of plant flavonoids and their high safety profile (Dao et al., 2011), these CHS-derived flavonoids have very high medical or nutritional value. Therefore, the demand for flavonoid biosynthesis in microorganisms is increasing rapidly. Numerous achievements have been accomplished in recent years, paving the way for industrial biomanufacturing.
The first CHS gene sequence was reported by Reimold et al. (1983) in 1983 from cultured parsley cells (Petroselinum Hortense). Later, more than 650 CHS-like gene sequences were identified in various plants (Bisht et al., 2021). The first CHS structure was elucidated from alfalfa CHS in 1999, revealing its specific homodimeric structure (Ferrer et al., 1999). CHSs combined with 4CLs and CHIs are widely used in chalcone biosynthesis. To achieve de novo naringenin chalcone and naringenin biosynthesis in microbial hosts, TAL is introduced to convert L-tyrosine to p-coumaric acid, which is the precursor of naringenin chalcone. Naringenin was firstly biosynthesized from simple carbon source glucose in E. coli (Watts et al., 2004) and S. cerevisiae (Jiang et al., 2005) decades ago, though with a quite low titer. In the following years, numerous strategies were applied to increase the production, such as increasing essential precursors malonyl-CoA (Wu et al., 2014; Wu et al., 2015; Zhou et al., 2019; Wu et al., 2021; Zhang et al., 2021) and L-tyrosine or L-phylalanine (Koopman et al., 2012; Li et al., 2021), screening and overexpressing essential genes (Lv et al., 2019; Mark et al., 2019; Tong et al., 2022), and co-culture strategies (Ganesan et al., 2017; Zhang et al., 2017). Some major achievements are summarized in Table 2. Till now, the highest naringenin production from glucose reported is 1,129.44 mg/L in yeast fed-batch fermentation (Zhang et al., 2021). All these efforts combined to build a solid and stable naringenin-producing platform. Naringenin biosynthesis pathway in microbial hosts especially E. coli and S. cerevisiae, have been widely used to develop novel synthetic biology tools, such as malonyl-CoA biosensor development (Yang et al., 2018), evolution-guided optimization (Raman et al., 2014). Strategies developed in naringenin de novo biosynthesis laid a solid foundation for other valuable plant natural polyketides biosynthesis in microbial hosts. Pinocembrin, another plant polyketide derived from CHS, can serve as a good example. PAL was introduced in pinocembrin-producing strains to convert L-phenylalanine to cinnamic acid. Similar strategies, such as ACC overexpression (Kim et al., 2014) and flux balance control (Wu et al., 2013; Wu et al., 2016a), were applied to enhance pinocembrin titers. Knowing the pH impact on different stages of fermentation, a two-stage pH strategy was explored (Cao et al., 2016a; Wu et al., 2016b). Tao et al., 2018 used the CRISPRi system to identify genes regulating ATP levels and increased pinocembrin titer to 165.3 mg/L.
The de novo naringenin and pinocembrin pathways were further engineered to produce numerous medically important derivatives from glucose. The bioactive isoflavonoids genistein and daidzein are downstream products of naringenin and liquiritigenin catalyzed by isoflavone synthase (IFS), which can serve as good examples. Liu et al. (2019) used a p-coumaric acid overproducing yeast strain to produce isoflavonoids (Liu et al., 2021). Chalcone reductase (CHR), IFS, and 2-hydroxyisoflavanone dehydratase (HID) were co-expressed with the naringenin biosynthesis pathway. By enhancing the activity of cytochrome P450, which are monooxygenases (i.e., cinnamic acid hydroxylase C4H and IFS), and improving metabolic flux, up to 85.4 mg/L daidzein was produced. Kaempferol, which has positive effects on cancer cell regulation (Chen and Chen, 2013), is a downstream product of naringenin via the catalysis of flavanone 3-hydroxylase (F3H) and flavonol synthase (FLS) enzymes. Lyu et al. (2019) optimized kaempferol production to 86 mg/L by gene screening, core flavonoid synthetic pathway optimization, and relocalization into mitochondria of F3H and FLS. Combined with co-culture strategies, 220 mg/L of naringenin and 200 mg/L of mixed flavonoids were obtained. Other examples are delphinidin (Du et al., 2020), ponciretin and sakuranetin (Kim et al., 2013), and anthocyanin (Levisson et al., 2018), all of which are naringenin derivatives in plants. Though de novo biosynthesis has not been reported yet, some derivative compounds can be synthesized in microbial hosts by adding precursors. For example, prenylated naringenin can be synthesized in yeast by using prenyltransferases (PTs) from bacteria and adding L-phenylalanine and Dimethylallyl pyrophosphate (DMAPP) (Isogai et al., 2021). With the developed de novo metabolic engineering strategies, the complete biosynthesis of these compounds can be realized in the future.
Stilbene is another most studied plant natural polyketide, which is biosynthesized by a type III PKS stilbene synthase (STS). STS shares 75%–90% sequence similarity with CHS but only exists in a few species, such as Vitaceae, Gnetaceae, Dipterocarpaceae, Pinaceae, Poaceae, Fabaceae, Leguminosae, and Cyperaceae families (Austin et al., 2004). The catalytic mechanisms of STS and CHS are also different. STS enables iterative decarboxylative condensation of three molecules of malonyl-CoA with one molecule of p-coumaroyl-CoA or cinnamoyl-CoA following a C2-C7 aldol cyclization to produce resveratrol and pinosylvin respectively.
Resveratrol is medically valuable to many diseases, such as diabetes, cardiovascular diseases, and neurological disorders (Bhatt et al., 2012; Tomé-Carneiro et al., 2012; Turner et al., 2015) and is also an important additive in foods and cosmetics. De novo biosynthesis of resveratrol in microbial hosts has been explored for over a decade with numerous achievements, listed in Table 2. Complete resveratrol biosynthesis was achieved in S. cerevisiae for supplementing extra resveratrol in white wine, in which codon-optimized TAL mutant and araE transporter were used to improve resveratrol production (1.9 mg/L when supplied with tyrosine, and 1.06 mg/L without supplementation) (Wang et al., 2011). An E. coli strain was developed to produce resveratrol, and methylated resveratrol that has better bioavailability from glucose (Kang et al., 2014), resulting in a production of 5.2 mg/L resveratrol and four distinct methylated resveratrol analogues were produced by the action of a series O-methyltransferase genes from sorghum. A site-specific integration strategy led to 4.61 mg/L resveratrol from glucose in E. coli (Liu et al., 2016). Li et al. (2015) used a stepwise engineering strategy to produce resveratrol, which led to 415.65 mg/L or 531.41 mg/L resveratrol from glucose or ethanol, respectively. The same group also constructed another yeast strain to produce resveratrol from the phenylalanine pathway rather than the tyrosine pathway (Li et al., 2016). After enhancing P450 activity, overexpressing pathway genes, and increasing the precursor supply, a final production of 800 mg/L was achieved in fed-batch fermentation from glucose. In 2021, Costa et al. (2021) used a mixed medium with 2% glucose and 5% ethanol to produce resveratrol and achieved a maximum 187.07 ± 19.88 mg/L production. Multiple similar strategies used in naringenin production were also applied in resveratrol production in microbial hosts to obtain higher production (Wu et al., 2017b; Hong et al., 2020; Sáez-Sáez et al., 2020; Yuan et al., 2020; Li et al., 2022).
Pinosylvin has important medical functions in treating various cancers and cardiovascular, inflammatory diseases (Jeong et al., 2013; Park et al., 2013). van Summeren-Wesenhagen and Marienhagen (2015) increased pinosylvin production by adding cerulenin to increase intracellular malonyl-CoA level and in vivo evolving stilbene synthase from Pinus strobus for higher activity. 70 mg/L pinosylvin from glucose was achieved. Muti-module optimization strategy combined with malonyl-CoA availability enhancement strategy, 281 mg/L pinosylvin was obtained from glucose (Wu et al., 2017a). Xu et al. used pinosylvin biosynthesis pathway to explore the correlation between the acylation level of proteins and the titer of malonyl-CoA-derived metabolites and found oversupply of malonyl-CoA leads to malonylation of important enzymes involved in biosynthetic pathway, thus inhibiting pinosylvin productivity (Xu et al., 2018).
Curcuminoids are highly produced in the spice turmeric and possess many bioactivities, including anti-inflammatory (Aggarwal and Sung, 2009), anticancer (Wilken et al., 2011), antioxidant (Jayaprakasha et al., 2006) activities. Curcuminoids synthesized in Curcuma longa require the actions of two type III PKSs, namely diketide-CoA synthase (DCS) and curcumin synthase (CURS) to form the C6-C7-C6 core structure (Katsuyama et al., 2009). DCS condenses one p-coumaroyl-CoA or feruloyl-CoA with one malonyl-CoA to synthesize p-coumaroyl-diketide-CoA and feruloyl-diketide-CoA, which will be further condensed with another p-coumaroyl-CoA or feruloyl-CoA to form bisdemethoxycurcumin or curcumin by the action of CURS, respectively. Meanwhile, curcuminoids synthesized in Oryza sativa require only one type III PKS, named curcuminoid synthase (CUS). By employing similar three-step reaction, bisdemethoxycurcumin can be synthesized from two molecules of p-coumaroyl-CoA and one molecule of malonyl-CoA (Katsuyama et al., 2009). When using cinnamoyl-CoA or feruloyl-CoA as the substrate, dicinnamoylmethane or curcumin can be produced, respectively.
Co-expression of ClDCS with ClCURS or OsCUS in microbial hosts have produced a variety of curcuminoids. The highest curcumin production (563.4 mg/L) was achieved in E. coli when ferulic acid was fed as the substrate (Rodrigues et al., 2020). However, as a more complex polyketide, the titer of de novo production is much lower. Engineered E. coli strains with TAL, PAL, 4CL, and OsCUS and optimized shikimate pathway can synthesize two curcuminoids, dicinnamoylmethane and bisdemethoxycurcumin, from glucose (Kim et al., 2017), but the productions are only 4.63 mg/L (bisdemethoxycurcumin) and 6.95 mg/L (dicinnamoylmethane). Multiplex automatic genome engineering (MAGE) was used to construct a library of 5′- UTR sequences for curcumin pathway optimization, which improved curcurmin production by 38.2-fold compared to the parental strain to 3.9 ± 0.8 mg/L (Kang et al., 2018). E. coli-E. coli co-culture system produced bisdemethoxycurcumin from glucose (Fang et al., 2018), in which an E. coli strain with 4CL and CUS produced bisdemethoxycurcumin from p-coumaric acid, and the other strain harboring TAL produces p-coumaric acid from glucose. Malonyl-CoA level was also improved by introducing MatB and MatC to transform supplemented malonate directly into malonyl-CoA. Finally, 6.28 mg/L bisdemethoxycurcumin was produced from glucose when malonate was supplied (Fang et al., 2018).
BAS shares 70% amino acid sequence identity with CHS. It condenses one p-coumaroyl-CoA with one malonyl-CoA to produce C6-C4 diketide benzalacetone, which can lead to valuable flavoring agent such as raspberry ketone used in cosmetics, food industry, and pharmaceutics (Wang et al., 2012; Bredsdorff et al., 2015; Kim et al., 2016). Till now, BAS have been cloned from Rheum palmatum, Wachendorfia thyrsiflora, Rubus idaeus, and Polygonum cuspidatum (Shimokawa et al., 2012).
Plant-derived and chemical synthesized raspberry ketone are expensive. Therefore, synthesizing this compound via synthetic biology tools in microbial hosts is appealing. A lot of efforts have been made in producing raspberry ketone from p-coumaric acid or tyrosine (Wang et al., 2019; Milke et al., 2020; Chang et al., 2021; Yang et al., 2021). De novo production has been recently achieved by the co-expression of four heterologous genes (PAL/TAL, C4H, 4CL, and BAS) in an S. cerevisiae strain (Lee et al., 2016). Fusion protein of 4CL and BAS further increased the titer by five-fold to 7.5 mg/L from 3 mM p-coumaric acid, or 2.8 mg/L de novo. A similar five-step raspberry ketone-producing pathway was reconstructed in E. coli (Moore et al., 2021), and led to 12.9 mg/L raspberry ketone production by fine-tuning the expression level of each pathway gene using a promoter library.
Zingerone is another phenylbutanones and the key component responsible for the pungency of ginger. Zingerone has various medical importance, including anti-inflammatory, antioxidant, antihyperlipidemic, anticancer, and antibacterial activities (Ahmad et al., 2015). The biosynthetic pathway of zingerone is similar with that of raspberry ketone, while zingerone starts with ferulic acid as the precursor and raspberry ketone biosynthesis starts with p-coumaric acid. Recently, a type III PKS gene (PmPKS) from Piper methysticum (kava) which exhibits feruloyl-CoA-preferred BAS activity has been identified (Heo et al., 2021a). Consequently, an E. coli strain harboring 4CL, PmPKS and benzalacetone reductase (BAR) produced zingerone from ferulic acid, which led to a de novo zingerone pathway assembled in an L-tyrosine-overproducing E. coli, that produced 24.03 ± 2.53 mg/L zingerone.
Plant stytylpyrones are mainly found in angiosperm plants, including Piperaceae, Lauraceae, Annonaceae, Ranuculaceae and Zingiberaceae (Beckert et al., 1997). They usually serve as defense role when attacked by bacterium or fungi, and exhibit anxiolytic, analgesic, or neuroprotective effects (Tzeng and Lee, 2015). Plant styrylpyrone scaffold is formed by C5-C1 Claisen cyclization condensation of one hydroxycinnamoyl-CoA with two malonyl-CoA catalyzed by SPS (Pluskal et al., 2019).
Kavalactones are methylated stytylpyrones derived from kava, which have well-established anxiolytic and analgesic properties and interact with the human central nervous system through mechanisms distinct from those of conventional psychiatric drugs (Chua et al., 2016). In 2019, Pluskal et al. (2019) characterized two CHS-like genes encode functional SPSs from P. methysticum, namely PmSPS1 and PmSPS2. Complete biosynthesis of plant styrylpyrones was achieved in yeast (Wu et al., 2022). Firstly, a styrylpyrone-producing strain containing PmSPS1 and kava O-methyltransferase 1 (PmKOMT1) enable diverse plant styrylpyrones production from fed substrates (e.g., p-coumaric acid and cinnamic acid). Then the rate-limiting steps were identified and optimized by enzyme overexpression via yeast genomic integration. To further enhance the production, a CRISPR-based δ-integration method was used to overexpress the codon-optimized yPmSPS and yPmKOMT1 in yeast. Finally, the strains produced various plant stytylpyrones de novo, with the titers of 7,8-dihydro-5,6-dehydrokavain, desmethoxyyangonin and yangonin at 4.40, 1.28, and 0.10 μM, respectively. In another example, an SPS-like enzyme, PnPKS, was recently found from the leaves of black pepper (Piper nigrum), catalyzing two-chain elongation with feruloyl CoA-linked starter substrates to form a styrylpyrone 11-methoxy-bisnoryangonin (Heo et al., 2021b). After characterizing the SPS function with feruloyl-CoA of PnPKS, an artificial pathway that can produce 11-methoxy-bisnoryangonin was introduced into an engineered L-tyrosine overproducing E. coli strain, resulting in complete production with a titer up to 52.8 mg/L. These studies laid the foundation for industrial scale styrylpyrone biosynthesis and the complete biosynthesis of more complicated plant styrylpyrones from simple carbon source.
Both acridone synthase and QNS are from Rutaceae plant family, the products of which lead to anthranilic acid-derived alkaloids including acridone alkaloids and quinolone alkaloids. These PNPs exhibit numerous pharmaceutical potentials including anticancer, antimicrobial, antimalarial, antipsoriatic activities (Whitlon et al., 1978; Foley and Tilley, 1998; Michael, 2003; Goodell et al., 2006; Putic et al., 2010). Acridone synthase and QNS mainly use N-methylanthraniloyl-CoA instead of p-coumaroyl-CoA as starter substrate. Despite the structural similarities, acridone synthase and QNS catalyze different reactions. Acridone synthase performs the C6-C1 Claisen cyclization of one N-methylanthraniloyl-CoA and three malonyl-CoA units to produce 1,3-dihydroxy-N-methylacridone, a common intermediate which can form more complex acridones such as rutacridone. QNS catalyzes the decarboxylation condensation of starter one N-methylanthraniloyl-CoA and one malonyl-CoA which spontaneously cyclize to 4-hydroxy-1-methyl-2(1H)-quinolone. In 2020, Choi et al. (2020) constructed E. coli strains to produce 1,3-dihydroxy-9(10H)-acridone (DHA) and 1,3-dihydroxy-10-methylacridone (NMA) de novo. The pathway contains acridone synthase and N-methyltransferase (NMT) from Ruta graveolens, benzoate CoA ligase (badA) from Rhodopseudomonas palustris and anthranilate coenzyme A ligase (pqsA) from Pseudomonas. aeruginosa. Overexpression of shikimate pathway gene and an ACC from Photorhabdus luminescens optimized the production to 17.3 mg/L DHA and 26.0 mg/L NMA. Similar strategies were used to construct E. coli strains that can produce 4-hydroxycoumarin (4-HC), 2,4-dihydroxyquinoline (DHQ), and 4-hydroxy-1-methyl-2(1H)-quinolone (NMQ). Type III PKS acridone synthase from R. graveolens, QNS from Citrus macrocarpa and the P. aeruginosa quinolone signal synthase pqsD were used in this study. With the use of strategies that increased the availability of chorismate, anthranilate, and malonyl-CoA, 255.4 mg/L 4-HC, 753.7 mg/L DHQ, and 17.5 mg/L NMQ were synthesized.
There are a variety of type III PKSs-derived polyketides, such as triacetic acid lactone, phloroglucinol, flaviolin, and aleosones, the starters of all of which are from acetyl-CoA or malonyl-CoA instead of shikimate pathway. In Figure 4 these starters are named simple CoA starters. Triacetic acid lactone is from the condensation of one acetyl-CoA starter and two malonyl-CoA extender units by the action of 2-pyrone synthase (2-PS). 2-PS from Gerbera hybrida (Gh2PS1) was identified and has been imported into S. cerevisiae (Cardenas and da Silva, 2014; Saunders et al., 2015), and Y. lipolytica (Markham et al., 2018; Yu et al., 2018) hosts to produce triacetic acid lactone. Numerous achievements that can biosynthesize triacetic acid lactone from glucose in microbial hosts have been accomplished. Till now, the highest production was achieved in Y. lipolytica (Markham et al., 2018). Bypassing pyruvate pathway and the overexpression of a β-oxidation-related enzyme PEX10 enhanced triacetic acid lactone production to 35.9 ± 3.9 g/L in bioreactor.
Type III PKSs are one of the most studied enzymes. This review has discussed the specific strategies and achievements of de novo plant polyketides biosynthesis in different microbial hosts, of which the biosynthetic pathways are almost fully elucidated. Other universal strategies to optimize the production of high value PNPs have been covered in a series of recent reviews (Li et al., 2018a; Cravens et al., 2019), including reconstructing biosynthetic pathways that are not fully elucidated, engineering cytochrome P450, or enhancing efficiency of multi-enzyme pathways.
The abilities to reconstruct and tune characterized plant polyketide biosynthetic pathways efficiently are important to the complete biosynthesis of valuable plant polyketides. High throughput and automation methods have the potential to transform current pace in increasing valuable plant polyketide synthesis in microbial hosts by generating and coupling modular genetic parts and modules in larger scale and faster speed, which have been highlighted by the development of multiplex automatic genome engineering (MAGE) (Kang et al., 2018) and various automation platforms (Radivojević et al., 2020). CRISPR/Cas is another powerful tool that can enable pathway construction in short time by assembling multiple genetic parts/genes simultaneously. For example, Gong et al. (2022) has developed a CRISPR/Cas mediated method customized for complicated plant pathway reconstruction in yeast, which has succeeded in reconstructing a 12-gene, de novo isoflavone pathway in one pot. Apel et al. (2017) developed a cloning-free toolkit for integration by employing CRISPR/Cas9 that can be applied to PNP synthesis too. These new genome editing methods are expected to advance the microbial biosynthesis of plant polyketides and other PNPs.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
National Science Foundation Grant No. DBI-2019674 and the National Institutes of Health—National Institute on Deafness and Other Communication Disorders award R21DC019206.
We thank Yinan Wu for the valuable feedback in the preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Mawgoud, A. M., Markham, K. A., Palmer, C. M., Liu, N., Stephanopoulos, G., and Alper, H. S. (2018). Metabolic engineering in the host Yarrowia lipolytica. Metab. Eng. 50, 192–208. doi:10.1016/j.ymben.2018.07.016
Aggarwal, B. B., and Sung, B. (2009). Pharmacological basis for the role of curcumin in chronic diseases: an age-old spice with modern targets. Trends Pharmacol. Sci. 30, 85–94. doi:10.1016/j.tips.2008.11.002
Ahmad, B., Rehman, M. U., Amin, I., Arif, A., Rasool, S., Bhat, S. A., et al. (2015). A review on pharmacological properties of zingerone (4-(4-hydroxy-3-methoxyphenyl)-2-butanone). Sci. World J. 2015, 1–6. doi:10.1155/2015/816364
Apel, A. R., D’Espaux, L., Wehrs, M., Sachs, D., Li, R. A., Tong, G. J., et al. (2017). A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 45, 496–508. doi:10.1093/nar/gkw1023
Austin, M., Bowman, M., Ferrer, J., Schroder, J., and Noel, J. (2004). An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem. Biol. 11, 1179–1194. doi:10.1016/j.chembiol.2004.05.024
Barros, J., and Dixon, R. A. (2020). Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci. 25, 66–79. doi:10.1016/j.tplants.2019.09.011
Becker, J., and Wittmann, C. (2012). Systems and synthetic metabolic engineering for amino acid production - the heartbeat of industrial strain development. Curr. Opin. Biotechnol. 23, 718–726. doi:10.1016/j.copbio.2011.12.025
Beckert, C., Horn, C., Schnitzler, J.-P., Lehning, A., Hellert, W., and Veit, M. (1997). Styrylpyrone biosynthesis in Equisetum arvense. Phytochemistry 44, 275–283. doi:10.1016/S0031-9422(96)00543-2
Belcher, M. S., Mahinthakumar, J., and Keasling, J. D. (2020). New frontiers: harnessing pivotal advances in microbial engineering for the biosynthesis of plant-derived terpenoids. Curr. Opin. Biotechnol. 65, 88–93. doi:10.1016/j.copbio.2020.02.001
Bhatt, J. K., Thomas, S., and Nanjan, M. J. (2012). Resveratrol supplementation improves glycemic control in type 2 diabetes mellitus. Nutr. Res. 32, 537–541. doi:10.1016/j.nutres.2012.06.003
Bisht, R., Bhattacharyya, A., Shrivastava, A., and Saxena, P. (2021). An overview of the medicinally important plant type III PKS derived polyketides. Front. Plant Sci. 12, 746908. doi:10.3389/fpls.2021.746908
Braga, A., Oliveira, J., Silva, R., Ferreira, P., Rocha, I., Kallscheuer, N., et al. (2018). Impact of the cultivation strategy on resveratrol production from glucose in engineered Corynebacterium glutamicum. J. Biotechnol. 265, 70–75. doi:10.1016/j.jbiotec.2017.11.006
Bredsdorff, L., Wedebye, E. B., Nikolov, N. G., Hallas-Møller, T., and Pilegaard, K. (2015). Raspberry ketone in food supplements - high intake, few toxicity data - a cause for safety concern? Regul. Toxicol. Pharmacol. 73, 196–200. doi:10.1016/j.yrtph.2015.06.022
Cao, W., Ma, W., Wang, X., Zhang, B., Cao, X., Chen, K., et al. (2016a). Enhanced pinocembrin production in Escherichia coli by regulating cinnamic acid metabolism. Sci. Rep. 6, 32640. doi:10.1038/srep32640
Cao, W., Ma, W., Zhang, B., Wang, X., Chen, K., Li, Y., et al. (2016b). Improved pinocembrin production in Escherichia coli by engineering fatty acid synthesis. J. Ind. Microbiol. Biotechnol. 43, 557–566. doi:10.1007/s10295-015-1725-3
Cardenas, J., and da Silva, N. A. (2014). Metabolic engineering of Saccharomyces cerevisiae for the production of triacetic acid lactone. Metab. Eng. 25, 194–203. doi:10.1016/j.ymben.2014.07.008
Cardenas, J., and da Silva, N. A. (2016). Engineering cofactor and transport mechanisms in Saccharomyces cerevisiae for enhanced acetyl-CoA and polyketide biosynthesis. Metab. Eng. 36, 80–89. doi:10.1016/j.ymben.2016.02.009
Chang, C., Liu, B., Bao, Y., Tao, Y., and Liu, W. (2021). Efficient bioconversion of raspberry ketone in Escherichia coli using fatty acids feedstocks. Microb. Cell Fact. 20, 68. doi:10.1186/s12934-021-01551-0
Chen, A. Y., and Chen, Y. C. (2013). A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 138, 2099–2107. doi:10.1016/j.foodchem.2012.11.139
Chen, Y., Daviet, L., Schalk, M., Siewers, V., and Nielsen, J. (2013). Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab. Eng. 15, 48–54. doi:10.1016/j.ymben.2012.11.002
Choi, G. S., Choo, H. J., Kim, B. G., and Ahn, J. H. (2020). Synthesis of acridone derivatives via heterologous expression of a plant type III polyketide synthase in Escherichia coli. Microb. Cell Fact. 19, 73. doi:10.1186/s12934-020-01331-2
Choo, H. J., and Ahn, J. H. (2019). Synthesis of three bioactive aromatic compounds by introducing polyketide synthase genes into engineered Escherichia coli. J. Agric. Food Chem. 67, 8581–8589. doi:10.1021/acs.jafc.9b03439
Chua, H. C., Christensen, E. T. H., Hoestgaard-Jensen, K., Hartiadi, L. Y., Ramzan, I., Jensen, A. A., et al. (2016). Kavain, the major constituent of the anxiolytic kava extract, potentiates gabaa receptors: Functional characteristics and molecular mechanism. PLoS ONE 11, e0157700. doi:10.1371/journal.pone.0157700
Costa, C. E., Møller-Hansen, I., Romaní, A., Teixeira, J. A., Borodina, I., and Domingues, L. (2021). Resveratrol production from hydrothermally pretreated Eucalyptus wood using recombinant industrial Saccharomyces cerevisiae strains. ACS Synth. Biol. 10, 1895–1903. doi:10.1021/acssynbio.1c00120
Cravens, A., Payne, J., and Smolke, C. D. (2019). Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 10, 2142. doi:10.1038/s41467-019-09848-w
Dao, T. T. H., Linthorst, H. J. M., and Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. doi:10.1007/s11101-011-9211-7
Davis, M. S., and Cronan, J. (2001). Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J. Bacteriol. 183, 1499–1503. doi:10.1128/JB.183.4.1499-1503.2001
Dibyendu, D. M. (2015). A brief review on plant type III polyketide synthases, an important group of enzyme of secondary metabolism. Available at: www.isca.me.
Du, Y., Yang, B., Yi, Z., Hu, L., and Li, M. (2020). Engineering Saccharomyces cerevisiae coculture platform for the production of flavonoids. J. Agric. Food Chem. 68, 2146–2154. doi:10.1021/acs.jafc.9b07916
Eberhardt, D., Jensen, J. V. K., and Wendisch, V. F. (2014). L-citrulline production by metabolically engineered Corynebacterium glutamicum from glucose and alternative carbon sources. Amb. Express 4, 85. doi:10.1186/s13568-014-0085-0
Fang, Z., Jones, J. A., Zhou, J., and Koffas, M. A. G. (2018). Engineering Escherichia coli Co-cultures for production of curcuminoids from glucose. Biotechnol. J. 13, 1700576. doi:10.1002/biot.201700576
Ferrer, J.-L., Jez, J. M., Bowman, M. E., Dixon, R. A., and Noel, J. P. (1999). Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6, 775–784. doi:10.1038/11553
Foley, M., and Tilley, L. (1998). Quinoline antimalarials: Mechanisms of action and resistance and prospects for new agents. Pharmacol. Ther. 79, 55–87. doi:10.1016/S0163-7258(98)00012-6
Galanie, S., Thodey, K., Trenchard, I. J., Interrante, M. F., Smolke, C. D., Lindsey, E. O., et al. (2015). Slip pulse and resonance of the Kathmandu basin during the 2015 Gorkha earthquake, Nepal. Science 349, 1091–1095. doi:10.1126/science.aac6383
Ganesan, V., Li, Z., Wang, X., and Zhang, H. (2017). Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synthetic Syst. Biotechnol. 2, 236–242. doi:10.1016/j.synbio.2017.08.003
Gong, F. L., Han, J., and Li, S. (2022). MULTI-SCULPT: Multiplex integration via selective, CRISPR-mediated, ultralong pathway transformation in yeast for plant natural product synthesis. ACS Synth. Biol. 11, 2484–2495. doi:10.1021/acssynbio.2c00135
Goodell, J. R., Madhok, A. A., Hiasa, H., and Ferguson, D. M. (2006). Synthesis and evaluation of acridine- and acridone-based anti-herpes agents with topoisomerase activity. Bioorg. Med. Chem. 14, 5467–5480. doi:10.1016/j.bmc.2006.04.044
Heo, K. T., Lee, B., Jang, J. H., Ahn, J. O., and Hong, Y. S. (2021a). Construction of an artificial biosynthetic pathway for the styrylpyrone compound 11-methoxy-bisnoryangonin produced in engineered Escherichia coli. Front. Microbiol. 12, 714335. doi:10.3389/fmicb.2021.714335
Heo, K. T., Park, K. W., Won, J., Lee, B., Jang, J. H., Ahn, J. O., et al. (2021b). Construction of an artificial biosynthetic pathway for zingerone production in Escherichia coli using benzalacetone synthase from piper methysticum. J. Agric. Food Chem. 69, 14620–14629. doi:10.1021/acs.jafc.1c05534
Hertweck, C. (2009). The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 48, 4688–4716. doi:10.1002/anie.200806121
Hong, J., Im, D. K., and Oh, M. K. (2020). Investigating E. coli coculture for resveratrol production with 13C metabolic flux analysis. J. Agric. Food Chem. 68, 3466–3473. doi:10.1021/acs.jafc.9b07628
Huang, B., Guo, J., Yi, B., Yu, X., Sun, L., and Chen, W. (2008). Heterologous production of secondary metabolites as pharmaceuticals in Saccharomyces cerevisiae. Biotechnol. Lett. 30, 1121–1137. doi:10.1007/s10529-008-9663-z
Isogai, S., Okahashi, N., Asama, R., Nakamura, T., Hasunuma, T., Matsuda, F., et al. (2021). Synthetic production of prenylated naringenins in yeast using promiscuous microbial prenyltransferases. Metab. Eng. Commun. 12, e00169. doi:10.1016/j.mec.2021.e00169
Jayaprakasha, G. K., Jaganmohan Rao, L., and Sakariah, K. K. (2006). Antioxidant activities of curcumin, demethoxycurcumin and bisdemethoxycurcumin. Food Chem. 98, 720–724. doi:10.1016/j.foodchem.2005.06.037
Jendresen, C. B., Stahlhut, S. G., Li, M., Gaspar, P., Siedler, S., Förster, J., et al. (2015). Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 81, 4458–4476. doi:10.1128/AEM.00405-15
Jeong, E., Lee, H. R., Pyee, J., and Park, H. (2013). Pinosylvin induces cell survival, migration and anti-adhesiveness of endothelial cells via nitric oxide production. Phytother. Res. 27, 610–617. doi:10.1002/ptr.4770
Jiang, M., and Zhang, H. (2016). Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli. Curr. Opin. Biotechnol. 42, 1–6. doi:10.1016/j.copbio.2016.01.016
Jiang, H., Wood, K. v., and Morgan, J. A. (2005). Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 2962–2969. doi:10.1128/AEM.71.6.2962-2969.2005
Jones, J. A., Vernacchio, V. R., Collins, S. M., Shirke, A. N., Xiu, Y., Englaender, J. A., et al. (2017). Complete biosynthesis of anthocyanins using E. coli polycultures. mBio 8, e00621. doi:10.1128/mBio.00621-17
Kallscheuer, N., Vogt, M., Stenzel, A., Gätgens, J., Bott, M., and Marienhagen, J. (2016). Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 38, 47–55. doi:10.1016/j.ymben.2016.06.003
Kang, S. Y., Lee, J. K., Choi, O., Kim, C. Y., Jang, J. H., Hwang, B. Y., et al. (2014). Biosynthesis of methylated resveratrol analogs through the construction of an artificial biosynthetic pathway in E. coli. BMC Biotechnol. 14, 67. doi:10.1186/1472-6750-14-67
Kang, S. Y., Heo, K. T., and Hong, Y. S. (2018). Optimization of artificial curcumin biosynthesis in E. coli by randomized 5′-UTR sequences to control the multienzyme pathway. ACS Synth. Biol. 7, 2054–2062. doi:10.1021/acssynbio.8b00198
Katsuyama, Y., Kita, T., Funa, N., and Horinouchi, S. (2009). Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J. Biol. Chem. 284, 11160–11170. doi:10.1074/jbc.M900070200
Kim, M. J., Kim, B. G., and Ahn, J. H. (2013). Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli. Appl. Microbiol. Biotechnol. 97, 7195–7204. doi:10.1007/s00253-013-5020-9
Kim, B. G., Lee, H., and Ahn, J. H. (2014). Biosynthesis of pinocembrin from glucose using engineered Escherichia coli. J. Microbiol. Biotechnol. 24, 1536–1541. doi:10.4014/jmb.1406.06011
Kim, M., Baek, H. S., Lee, M., Park, H., Shin, S. S., Choi, D. W., et al. (2016). Rhododenol and raspberry ketone impair the normal proliferation of melanocytes through reactive oxygen species-dependent activation of GADD45. Toxicol. Vitro 32, 339–346. doi:10.1016/j.tiv.2016.02.003
Kim, E. J., Cha, M. N., Kim, B. G., and Ahn, J. H. (2017). Production of curcuminoids in engineered Escherichia coli. J. Microbiol. Biotechnol. 27, 975–982. doi:10.4014/jmb.1701.01030
Kingston, D. G. I. (1994). Taxol: the chemistry and structure-activity relationships of a novel anticancer agent. Trends Biotechnol. 12, 222–227. doi:10.1016/0167-7799(94)90120-1
Klayman, D. L. (1985). Qinghaosu (artemisinin): An antimalarial drug from China. Science 228, 1049–1055. doi:10.1126/science.3887571
Koopman, F., Beekwilder, J., Crimi, B., van Houwelingen, A., Hall, R. D., Bosch, D., et al. (2012). De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Available at: http://www.microbialcellfactories.com/content/11/1/155.
Krivoruchko, A., and Nielsen, J. (2015). Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 35, 7–15. doi:10.1016/j.copbio.2014.12.004
Krivoruchko, A., Zhang, Y., Siewers, V., Chen, Y., and Nielsen, J. (2015). Microbial acetyl-CoA metabolism and metabolic engineering. Metab. Eng. 28, 28–42. doi:10.1016/j.ymben.2014.11.009
Lee, D., Lloyd, N. D. R., Pretorius, I. S., and Borneman, A. R. (2016). Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion. Microb. Cell Fact. 15, 49. doi:10.1186/s12934-016-0446-2
Levisson, M., Patinios, C., Hein, S., de Groot, P. A., Daran, J. M., Hall, R. D., et al. (2018). Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb. Cell Fact. 17, 103. doi:10.1186/s12934-018-0951-6
Li, M., Kildegaard, K. R., Chen, Y., Rodriguez, A., Borodina, I., and Nielsen, J. (2015). De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab. Eng. 32, 1–11. doi:10.1016/j.ymben.2015.08.007
Li, M., Schneider, K., Kristensen, M., Borodina, I., and Nielsen, J. (2016). Engineering yeast for high-level production of stilbenoid antioxidants. Sci. Rep. 6, 36827. doi:10.1038/srep36827
Li, S., Li, Y., and Smolke, C. D. (2018a). Strategies for microbial synthesis of high-value phytochemicals. Nat. Chem. 10, 395–404. doi:10.1038/s41557-018-0013-z
Li, S., Li, Y., Thodey, K., Trenchard, I., Cravens, A., and Smolke, C. D. (2018b). Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U. S. A. 115, E3922–E3931. doi:10.1073/pnas.1721469115
Li, H., Gao, S., Zhang, S., Zeng, W., and Zhou, J. (2021). Effects of metabolic pathway gene copy numbers on the biosynthesis of (2S)-naringenin in Saccharomyces cerevisiae. J. Biotechnol. 325, 119–127. doi:10.1016/j.jbiotec.2020.11.009
Li, J., Qiu, Z., and Zhao, G. R. (2022). Modular engineering of E. coli coculture for efficient production of resveratrol from glucose and arabinose mixture. Synthetic Syst. Biotechnol. 7, 718–729. doi:10.1016/j.synbio.2022.03.001
Lian, J., Si, T., Nair, N. U., and Zhao, H. (2014). “Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains,” in Food, Pharmaceutical and Bioengineering Division 2014 - Core Programming Area at the 2014 AIChE Annual Meeting, USA, 16-21 Nov 2014, 750–760. (American Institute of Chemical Engineers). doi:10.1016/j.ymben.2014.05.010
Lian, J., Mishra, S., and Zhao, H. (2018). Recent advances in metabolic engineering of Saccharomyces cerevisiae: New tools and their applications. Metab. Eng. 50, 85–108. doi:10.1016/j.ymben.2018.04.011
Liu, X., Lin, J., Hu, H., Zhou, B., and Zhu, B. (2016). De novo biosynthesis of resveratrol by site-specific integration of heterologous genes in Escherichia coli. FEMS Microbiol. Lett. 363, fnw061. doi:10.1093/femsle/fnw061
Liu, R., Deng, Z., and Liu, T. (2018). Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab. Eng. 50, 74–84. doi:10.1016/j.ymben.2018.05.015
Liu, Q., Yu, T., Li, X., Chen, Y., Campbell, K., Nielsen, J., et al. (2019). Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10, 4976. doi:10.1038/s41467-019-12961-5
Liu, X., Cheng, J., Zhu, X., Zhang, G., Yang, S., Guo, X., et al. (2020). De novo biosynthesis of multiple pinocembrin derivatives in Saccharomyces cerevisiae. ACS Synth. Biol. 9, 3042–3051. doi:10.1021/acssynbio.0c00289
Liu, Q., Liu, Y., Li, G., Savolainen, O., Chen, Y., and Nielsen, J. (2021). De novo biosynthesis of bioactive isoflavonoids by engineered yeast cell factories. Nat. Commun. 12, 6085. doi:10.1038/s41467-021-26361-1
Lütke-Eversloh, T., and Stephanopoulos, G. (2005). Feedback inhibition of chorismate mutase/prephenate dehydrogenase (TyrA) of Escherichia coli: Generation and characterization of tyrosine-insensitive mutants. Appl. Environ. Microbiol. 71, 7224–7228. doi:10.1128/AEM.71.11.7224-7228.2005
Lütke-Eversloh, T., and Stephanopoulos, G. (2007). L-Tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75, 103–110. doi:10.1007/s00253-006-0792-9
Lv, Y., Marsafari, M., Koffas, M., Zhou, J., and Xu, P. (2019). Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth. Biol. 8, 2514–2523. doi:10.1021/acssynbio.9b00193
Lv, Y., Gu, Y., Xu, J., Zhou, J., and Xu, P. (2020). Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab. Eng. 61, 79–88. doi:10.1016/j.ymben.2020.05.005
Lyu, X., Zhao, G., Ng, K. R., Mark, R., and Chen, W. N. (2019). Metabolic engineering of Saccharomyces cerevisiae for de Novo production of kaempferol. J. Agric. Food Chem. 67, 5596–5606. doi:10.1021/acs.jafc.9b01329
Mark, R., Lyu, X., Ng, K. R., and Chen, W. N. (2019). Gene source screening as a tool for naringenin production in engineered Saccharomyces cerevisiae. ACS Omega 4, 12872–12879. doi:10.1021/acsomega.9b00364
Markham, K. A., Palmer, C. M., Chwatko, M., Wagner, J. M., Murray, C., Vazquez, S., et al. (2018). Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation. Proc. Natl. Acad. Sci. U. S. A. 115, 2096–2101. doi:10.1073/pnas.1721203115
Michael, J. P. (2003). Quinoline, quinazoline and acridone alkaloids. Nat. Prod. Rep. 20, 476–493. doi:10.1039/b208140g
Milke, L., Mutz, M., and Marienhagen, J. (2020). Synthesis of the character impact compound raspberry ketone and additional flavoring phenylbutanoids of biotechnological interest with Corynebacterium glutamicum. Microb. Cell Fact. 19, 92. doi:10.1186/s12934-020-01351-y
Moore, S. J., Hleba, Y. B., Bischoff, S., Bell, D., Polizzi, K. M., and Freemont, P. S. (2021). Refactoring of a synthetic raspberry ketone pathway with EcoFlex. Microb. Cell Fact. 20, 116. doi:10.1186/s12934-021-01604-4
Morita, H., Wong, C. P., and Abe, I. (2019). How structural subtleties lead to molecular diversity for the type III polyketide synthases. J. Biol. Chem. 294, 15121–15136. doi:10.1074/jbc.REV119.006129
Noel, J. P., Austin, M. B., and Bomati, E. K. (2005). Structure-function relationships in plant phenylpropanoid biosynthesis. Curr. Opin. Plant Biol. 8, 249–253. doi:10.1016/j.pbi.2005.03.013
Palmer, C. M., Miller, K. K., Nguyen, A., and Alper, H. S. (2020). Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a β-oxidation mediated strategy. Metab. Eng. 57, 174–181. doi:10.1016/j.ymben.2019.11.006
Park, E. J., Chung, H. J., Park, H. J., Kim, G. D., Ahn, Y. H., and Lee, S. K. (2013). Suppression of Src/ERK and GSK-3/β-catenin signaling by pinosylvin inhibits the growth of human colorectal cancer cells. Food Chem. Toxicol. 55, 424–433. doi:10.1016/j.fct.2013.01.007
Pastemak Gavril, W., Childers Stephen, R., and Snyder Solomon, H. (1980). Opiate analgesia: Evidence for mediation by a subpopulation of opiate receptors. Science 208, 514–516. doi:10.1126/science.6245448
Pluskal, T., Torrens-Spence, M. P., Fallon, T. R., de Abreu, A., Shi, C. H., and Weng, J. K. (2019). The biosynthetic origin of psychoactive kavalactones in kava. Nat. Plants 5, 867–878. doi:10.1038/s41477-019-0474-0
Pontrelli, S., Chiu, T. Y., Lan, E. I., Chen, F. Y. H., Chang, P., and Liao, J. C. (2018). Escherichia coli as a host for metabolic engineering. Metab. Eng. 50, 16–46. doi:10.1016/j.ymben.2018.04.008
Putic, A., Stecher, L., Prinz, H., and Müller, K. (2010). Structure-activity relationship studies of acridones as potentialantipsoriatic agents.1.Synthesis and antiproliferative activity of simple N-unsubstituted 10H-acridin-9-ones against human keratinocyte growth. Eur. J. Med. Chem. 45, 3299–3310. doi:10.1016/j.ejmech.2010.04.013
Radivojević, T., Costello, Z., Workman, K., and Garcia Martin, H. (2020). A machine learning Automated Recommendation Tool for synthetic biology. Nat. Commun. 11 (1), 4879. doi:10.1038/s41467-020-18008-4
Raman, S., Rogers, J. K., Taylor, N. D., and Church, G. M. (2014). Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. U. S. A. 111, 17803–17808. doi:10.1073/pnas.1409523111
Rammohan, A., Reddy, J. S., Sravya, G., Rao, C. N., and Zyryanov, G. v. (2020). Chalcone synthesis, properties and medicinal applications: a review. Environ. Chem. Lett. 18, 433–458. doi:10.1007/s10311-019-00959-w
Reimold, U., Kröger, M., Kreuzaler, F., and Hahlbrock, K. (1983). Coding and 3′ non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. EMBO J. 2, 1801–1805. doi:10.1002/j.1460-2075.1983.tb01661.x
Rodrigues, J. L., Gomes, D., and Rodrigues, L. R. (2020). A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front. Bioeng. Biotechnol. 8, 59. doi:10.3389/fbioe.2020.00059
Rodriguez, A., Martínez, J. A., Flores, N., Escalante, A., Gosset, G., and Bolivar, F. (2014). Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Fact. 13, 126. doi:10.1186/s12934-014-0126-z
Sáez-Sáez, J., Wang, G., Marella, E. R., Sudarsan, S., Cernuda Pastor, M., and Borodina, I. (2020). Engineering the oleaginous yeast Yarrowia lipolytica for high-level resveratrol production. Metab. Eng. 62, 51–61. doi:10.1016/j.ymben.2020.08.009
Santos, C. N. S., Koffas, M., and Stephanopoulos, G. (2011). Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab. Eng. 13, 392–400. doi:10.1016/j.ymben.2011.02.002
Saunders, L. P., Bowman, M. J., Mertens, J. A., da Silva, N. A., and Hector, R. E. (2015). Triacetic acid lactone production in industrial Saccharomyces yeast strains. J. Ind. Microbiol. Biotechnol. 42, 711–721. doi:10.1007/s10295-015-1596-7
Schneider, K., Hö vel, K., Witzel, K., rn Hamberger, B., Schomburg, D., Kombrink, E., et al. (2003). The substrate specificity-determining amino acid code of 4-coumarate: CoA ligase. Proc. Natl. Acad. Sci. 100, 8601. doi:10.1073/pnas.1430550100
Shah, F. L. A., Ramzi, A. B., Baharum, S. N., Noor, N. M., Goh, H. H., Leow, T. C., et al. (2019). Recent advancement of engineering microbial hosts for the biotechnological production of flavonoids. Mol. Biol. Rep. 46, 6647–6659. doi:10.1007/s11033-019-05066-1
Shen, B. (2003). Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 7, 285–295. doi:10.1016/S1367-5931(03)00020-6
Shi, S., Chen, Y., Siewers, V., and Nielsen, J. (2014). Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. mBio 5, e01130–e01114. doi:10.1128/mBio.01130-14
Shimokawa, Y., Morita, H., and Abe, I. (2012). Benzalacetone synthase. Front. Plant Sci. 3, 57. doi:10.3389/fpls.2012.00057
Shirra, M. K., Patton-Vogt, J., Ulrich, A., Liuta-Tehlivets, O., Kohlwein, S. D., Henry, S. A., et al. (2001). Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 5710–5722. doi:10.1128/mcb.21.17.5710-5722.2001
Song, H., Ding, M. Z., Jia, X. Q., Ma, Q., and Yuan, Y. J. (2014). Synthetic microbial consortia: From systematic analysis to construction and applications. Chem. Soc. Rev. 43, 6954–6981. doi:10.1039/c4cs00114a
Srinivasan, P., and Smolke, C. D. (2020). Biosynthesis of medicinal tropane alkaloids in yeast. Nature 585, 614–619. doi:10.1038/s41586-020-2650-9
Tao, S., Qian, Y., Wang, X., Cao, W., Ma, W., Chen, K., et al. (2018). Regulation of ATP levels in Escherichia coli using CRISPR interference for enhanced pinocembrin production. Microb. Cell Fact. 17, 147. doi:10.1186/s12934-018-0995-7
Thuan, N. H., Chaudhary, A. K., van Cuong, D., and Cuong, N. X. (2018). Engineering co-culture system for production of apigetrin in Escherichia coli. J. Ind. Microbiol. Biotechnol. 45, 175–185. doi:10.1007/s10295-018-2012-x
Tomé-Carneiro, J., Gonzálvez, M., Larrosa, M., Yáñez-Gascón, M. J., García-Almagro, F. J., Ruiz-Ros, J. A., et al. (2012). One-year consumption of a grape nutraceutical containing resveratrol improves the inflammatory and fibrinolytic status of patients in primary prevention of cardiovascular disease. Am. J. Cardiol. 110, 356–363. doi:10.1016/j.amjcard.2012.03.030
Tong, Y., Lv, Y., Yu, S., Lyu, Y., Zhang, L., and Zhou, J. (2022). Improving (2S)-naringenin production by exploring native precursor pathways and screening higher-active chalcone synthases from plants rich in flavonoids. Enzyme Microb. Technol. 156, 109991. doi:10.1016/j.enzmictec.2022.109991
Turner, R. S., Thomas, R. G., Craft, S., van Dyck, C. H., Mintzer, J., Reynolds, B. A., et al. (2015). A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology 85, 1383–1391. doi:10.1212/WNL.0000000000002035
Tzeng, Y. M., and Lee, M. J. (2015). Neuroprotective properties of kavalactones. Neural Regen. Res. 10, 875–877. doi:10.4103/1673-5374.158335
van Summeren-Wesenhagen, P. V., and Marienhagen, J. (2015). Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin. Appl. Environ. Microbiol. 81, 840–849. doi:10.1128/AEM.02966-14
Wang, Y., Halls, C., Zhang, J., Matsuno, M., Zhang, Y., and Yu, O. (2011). Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab. Eng. 13, 455–463. doi:10.1016/j.ymben.2011.04.005
Wang, L., Meng, X., and Zhang, F. (2012). Raspberry ketone protects rats fed high-fat diets against nonalcoholic steatohepatitis. J. Med. Food 15, 495–503. doi:10.1089/jmf.2011.1717
Wang, C., Zheng, P., and Chen, P. (2019). Construction of synthetic pathways for raspberry ketone production in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 103, 3715–3725. doi:10.1007/s00253-019-09748-5
Wang, X., Li, Z., Policarpio, L., Koffas, M. A. G., and Zhang, H. (2020). De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering. Appl. Microbiol. Biotechnol. 104, 4849–4861. doi:10.1007/s00253-020-10576-1
Watts, K. T., Lee, P. C., and Schmidt-Dannert, C. (2004). Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. ChemBioChem 5, 500–507. doi:10.1002/cbic.200300783
Whitlon, D. S., Sadowski, J. A., and Suttie, J. W. (1978) Mechanism of Coumarin Action: Significance of Vitamin K Epoxide Reductase Inhibition1-V itamin K functions in the postribosomal modification of liver microsomal protein precursors to form biologically active prothrombin (factor II) and the other vitamin K dependent plasma clotting proteins, factors VII, IX and X (Suttie &. Available at: https://pubs.acs.org/sharingguidelines.
Wilken, R., Veena, M. S., Wang, M. B., and Srivatsan, E. S. (2011). Curcumin: A review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma. Mol. Cancer 10, 12. doi:10.1186/1476-4598-10-12
Wu, J., Du, G., Zhou, J., and Chen, J. (2013). Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab. Eng. 16, 48–55. doi:10.1016/j.ymben.2012.11.009
Wu, J., Zhou, T., Du, G., Zhou, J., and Chen, J. (2014). Modular optimization of heterologous pathways for de Novo synthesis of (2S)-Naringenin in Escherichia coli. PLoS ONE 9, e101492. doi:10.1371/journal.pone.0101492
Wu, J., Du, G., Chen, J., and Zhou, J. (2015). Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 5, 13477. doi:10.1038/srep13477
Wu, J., Zhang, X., Dong, M., and Zhou, J. (2016a). Stepwise modular pathway engineering of Escherichia coli for efficient one-step production of (2S)-pinocembrin. J. Biotechnol. 231, 183–192. doi:10.1016/j.jbiotec.2016.06.007
Wu, J., Zhang, X., Zhou, J., and Dong, M. (2016b). Efficient biosynthesis of (2S)-pinocembrin from D-glucose by integrating engineering central metabolic pathways with a pH-shift control strategy. Bioresour. Technol. 218, 999–1007. doi:10.1016/j.biortech.2016.07.066
Wu, J., Zhang, X., Zhu, Y., Tan, Q., He, J., and Dong, M. (2017a). Rational modular design of metabolic network for efficient production of plant polyphenol pinosylvin. Sci. Rep. 7, 1459. doi:10.1038/s41598-017-01700-9
Wu, J., Zhou, P., Zhang, X., and Dong, M. (2017b). Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 44, 1083–1095. doi:10.1007/s10295-017-1937-9
Wu, J., Zhou, L., Duan, X., Peng, H., Liu, S., Zhuang, Q., et al. (2021). Applied evolution: Dual dynamic regulations-based approaches in engineering intracellular malonyl-CoA availability. Metab. Eng. 67, 403–416. doi:10.1016/j.ymben.2021.08.004
Wu, Y., Chen, M. N., and Li, S. (2022). De novo biosynthesis of diverse plant-derived styrylpyrones in Saccharomyces cerevisiae. Metab. Eng. Commun. 14, e00195. doi:10.1016/j.mec.2022.e00195
Xu, P., Ranganathan, S., Fowler, Z. L., Maranas, C. D., and Koffas, M. A. G. (2011). Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab. Eng. 13, 578–587. doi:10.1016/j.ymben.2011.06.008
Xu, J. Y., Xu, Y., Chu, X., Tan, M., and Ye, B. C. (2018). Protein acylation affects the artificial biosynthetic pathway for pinosylvin production in engineered E. coli. ACS Chem. Biol. 13, 1200–1208. doi:10.1021/acschembio.7b01068
Yang, D., Kim, W. J., Yoo, S. M., Choi, J. H., Ha, S. H., Lee, M. H., et al. (2018). Repurposing type III polyketide synthase as a malonyl- CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. U. S. A. 115, 9835–9844. doi:10.1073/pnas.1808567115
Yang, B., Zheng, P., Wu, D., and Chen, P. (2021). Efficient biosynthesis of raspberry ketone by engineered Escherichia coli coexpressing zingerone synthase and glucose dehydrogenase. J. Agric. Food Chem. 69, 2549–2556. doi:10.1021/acs.jafc.0c07697
Yu, J., Landberg, J., Shavarebi, F., Bilanchone, V., Okerlund, A., Wanninayake, U., et al. (2018). Bioengineering triacetic acid lactone production in Yarrowia lipolytica for pogostone synthesis. Biotechnol. Bioeng. 115, 2383–2388. doi:10.1002/bit.26733
Yuan, S. F., Yi, X., Johnston, T. G., and Alper, H. S. (2020). De novo resveratrol production through modular engineering of an Escherichia coli-Saccharomyces cerevisiae co-culture. Microb. Cell Fact. 19, 143. doi:10.1186/s12934-020-01401-5
Zha, W., Rubin-Pitel, S. B., Shao, Z., and Zhao, H. (2009). Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab. Eng. 11, 192–198. doi:10.1016/j.ymben.2009.01.005
Zhang, B., Zhou, N., Liu, Y. M., Liu, C., Lou, C. B., Jiang, C. Y., et al. (2015). Ribosome binding site libraries and pathway modules for shikimic acid synthesis with Corynebacterium glutamicum. Microb. Cell Fact. 14, 71. doi:10.1186/s12934-015-0254-0
Zhang, W., Liu, H., Li, X., Liu, D., Dong, X. T., Li, F. F., et al. (2017). Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae. Eng. Life Sci. 17, 1021–1029. doi:10.1002/elsc.201700039
Zhang, S., Yang, W., Chen, H., Liu, B., Lin, B., and Tao, Y. (2019). Metabolic engineering for efficient supply of acetyl-CoA from different carbon sources in Escherichia coli. Microb. Cell Fact. 18, 130. doi:10.1186/s12934-019-1177-y
Zhang, Q., Yu, S., Lyu, Y., Zeng, W., and Zhou, J. (2021). Systematically engineered fatty acid catabolite pathway for the production of (2 S)-naringenin in Saccharomyces cerevisiae. ACS Synth. Biol. 10, 1166–1175. doi:10.1021/acssynbio.1c00002
Zhou, S., Lyu, Y., Li, H., Koffas, M. A. G., and Zhou, J. (2019). Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy. Biotechnol. Bioeng. 116, 1392–1404. doi:10.1002/bit.26941
Zhou, S., Yuan, S. F., Nair, P. H., Alper, H. S., Deng, Y., and Zhou, J. (2021). Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metab. Eng. 67, 41–52. doi:10.1016/j.ymben.2021.05.007
Keywords: type III polyketide synthases, plant polyketides, complete biosynthesis, microorganisms, biosynthesis strategies, biosynthesis achievements
Citation: Liu C and Li S (2022) Engineered biosynthesis of plant polyketides by type III polyketide synthases in microorganisms. Front. Bioeng. Biotechnol. 10:1017190. doi: 10.3389/fbioe.2022.1017190
Received: 11 August 2022; Accepted: 04 October 2022;
Published: 14 October 2022.
Edited by:
Ye-Wang Zhang, Jiangsu University, ChinaReviewed by:
Jian Zha, Shaanxi University of Science and Technology, ChinaCopyright © 2022 Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sijin Li, c2lqaW4ubGlAY29ybmVsbC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.