- 1School of Animal Science and Technology, Guangxi University, Nanning, Guangxi, China
- 2State Key Laboratory of Non-Food Biomass and Enzyme Technology, Guangxi Key Laboratory of Marine Natural Products and Combinatorial Biosynthesis Chemistry, Guangxi Academy of Sciences, Nanning, Guangxi, China
- 3Guangxi Key Laboratory for Polysaccharide Materials and Modifications, School of Marine Sciences and Biotechnology, Guangxi University for Nationalities, Nanning, China
- 4Nanning Dabeinong Feed Technology Co., Ltd., Nanning, Guangxi, China
- 5Guangxi Huaren Medical Technolgoy Group, Nanning, Guangxi, China
N-acetylglucosamine (GlcNAc) is widely used in nutritional supplement and is generally produced from chitin using chitinases. While most GlcNAc is produced from colloidal chitin, it is essential that chitinases be acidic enzymes. Herein, we characterized an acidic, highly salinity tolerance and thermostable chitinase AfChiJ, identified from the marine fungus Aspergillus fumigatus df673. Using AlphaFold2 structural prediction, a truncated Δ30AfChiJ was heterologously expressed in E. coli and successfully purified. It was also found that it is active in colloidal chitin, with an optimal temperature of 45°C, an optimal pH of 4.0, and an optimal salt concentration of 3% NaCl. Below 45°C, it was sound over a wide pH range of 2.0–6.0 and maintained high activity (≥97.96%) in 1–7% NaCl. A notable increase in chitinase activity was observed of Δ30AfChiJ by the addition of Mg2+, Ba2+, urea, and chloroform. AfChiJ first decomposed colloidal chitin to generate mainly N-acetyl chitobioase, which was successively converted to its monomer GlcNAc. This indicated that AfChiJ is a bifunctional enzyme, composed of chitobiosidase and β-N-acetylglucosaminidase. Our result suggested that AfChiJ likely has the potential to convert chitin-containing biomass into high-value added GlcNAc.
Introduction
Chitin is homopolymer consisting of β-1, 4-N-acetyl-D-glucosamine (GlcNAc) units in a linear form (Chen et al., 2021). It exists in the skeletal organic matrices of coraline algae (Rahman and Halfar, 2014) and in the extracellular matrix of various invertebrates, including sponges, molluscs, nematodes, arthropods, and fungi (Lambris, 2021). Chitin is also a white, hard, inelastic polysaccharide and is a substantial contributor to pollution in coastal zones (Kumar, 2000). It is commonly thought that chitin is the second most plentiful and renewable polysaccharide on earth, after cellulose (Duo-Chuan, 2006; Yang et al., 2009). At least 10 GT chitin is synthesized and decay each year in the ecosystem (Jolles et al.). Approximately 1010–1012 kg chitin are yielded yearly by living organisms in seas and oceans (Dhillon et al., 2013), but decomposing chitin under natural environmental conditions is extremely tricky. However, this massive quantity of chitin would supply gigantic amounts of raw material. However, the currently huge amounts of chitin-containing waste produced by seafood processing industries is burned, landfilled, discarded at sea, or left to decay (Xu et al., 2013). This may cause environmental stress and is a waste of raw chitinous materials.
The conversion of chitin into derivatives, i.e., chito-oligosaccharides (COS), N-acetylglucosamine, and chitosan, exhibits a tremendously wide range of biological properties and outstanding potential applications, such as in the medicine and cosmetics (Jimtaisong and Saewan, 2014; Kumar et al., 2015), agriculture and aquaculture (Razdan and Pettersson, 1994; Wang and Chen, 2005), and materials science and membranes (Wang et al., 2016; Chen et al., 2018). GlcNAc, the terminal hydrolysis of chitin, is a crucial functional amino sugar complex that could serve as healthcare, medicine, and skin care products with a large requirement. Consequently, it may be possible to convert inexpensive chitin into high-value GlcNAc.
The enzymatic hydrolyzation of chitin-containing debris has been suggested as a cost-effective and sustainable way of producing GlcNAc (Liu et al., 2020). Chitinase (EC 3.2.1.14) is a type of glycosyl hydrolase enzyme that cleaves the bond between the C1 or C4 of two successive N-acetyl-β-D-glucosamine molecules in chitin (Xia et al., 2001; Hou et al., 2020). Chitinases are classified into two major groups, namely, endochitinase (EC 3.2.1.14) and exochitinase (EC 3.2.1.29). Endochitinases randomly hydrolyze chitin at internal sites to generate multimers of GlcNAc (Saima et al., 2013). Exochitinases contain two subcategories, called chitobiosidases (EC 3.2.1.29) and β-N-acetylglucosaminidases (EC 3.2.1.52). Chitobiosidases can progressively liberate (GlcNAc)2, beginning at the nonreducing end of the chitin. β-N-acetylglucosaminidases can crack the oligomeric products of endochitinases and chitobiosidases, further converting them to monomer GlcNAc (Wang and Wu, 2020). The thorough hydrolyzation of chitin for GlcNAc production requires the combining action of endochitinase and β-N-acetylglucosaminidases or chitobiosidases and β-N-acetylglucosaminidases (Du et al., 2019). The assembly of different chitinases of SmChiA, SmChiB, and SmChiC from the microorganism Serratia marcescens and a β-N-acetylglucosaminidases of OfHex1 from the insect Ostrinia furnacalis have been used to yield GlcNAc from the mycelial product in the processing waste of the fermentation industry (Zhu et al., 2016). However, the isolation of various chitinases from the cytoplasm of E. coli for industrial application is costly and difficult. For this reason, an enzyme that combines the activity of endochitinase and NAGase or chitobiosidases and NAGase would be much more suitable for industrial application.
Chitin has a highly organized crystalline structure, consisting of powerful intermolecular hydrogen bonding and intramolecular hydrogen bonding networks, such that this hydrogen bonding contributes to the strength and rigidity of the chitin chain (Cohen, 2001; Khan et al., 2017). Chitin has three polymorphic isomers viz. α-chitin, β-chitin, and γ-chitin. Among these, α-chitin is the most common form in nature, and α-chitin decays more slowly than the other two kinds, as it has higher recalcitrance, which decreases the accessibility of the chitin chains. Chitin is insoluble in water and most inorganic and organic solvents, except for concentrated acid (Hou et al., 2019). Colloidal chitin is usually produced by processing chitin with concentrated acids to destroy the crystalline structure and enhance the approach of chitin to chitinase and lytic polysaccharide monooxygenases. Therefore, colloidal chitin is commonly applied as the substrate for chitinase to prepare COSs and GlcNAc (Ren et al., 2022). Meanwhile, resources of ocean-derived chitin contain about 3% NaCl. Thus, the stability of chitinases at certain concentrations of salt, high temperature, and low pH can magnify their commercial utilization relative to their salt-free environment and their mesophilic and neutrally stable counterparts using the raw materials of colloidal chitin collected from ocean debris.
Marine chitinases possess higher pH and salinity tolerance than chitinases from terrestrial organisms (Han et al., 2009). These chitinases exist in the marine bacteria Pseudoalteromonas flavipulchra DSM 14401 (Ren et al., 2022) and the Paenibacillus strain UMBR 0002 (Liu et al., 2020). Researchers have identified chitinases produced by more than 30 marine bacteria (Xinyue et al., 2021), but few studies have investigated chitinase in marine fungus. Further, many thermostable chitinases have been discovered from fungi (Krolicka et al., 2018). The genomes of the fungal Aspergillus fumigatus are well known to be rich in chitinase genes. This species contains 17 chitinases at least, including 5 class III chitinases. Class III chitinases are normally acidic and may play a nutritional role during autolysis (Alcazar-Fuoli et al., 2011). Nevertheless, the majority of previous reports on A. fumigatus have focused on terrestrial organisms (Xia et al., 2009; Wang et al., 2016). Herein, we adopted A. fumigatus df673 as a research strain, stored in our lab, previously obtained from the tidal flat sediments of mangrove in Beibu Gulf, Guangxi Province, China. To date, little research has reported on the chitinases of oceanic A. fumigatus. The analysis of the marine A. fumigatus genome is hoped to extract thermostable chitinases with acid-resistance, highly salinity tolerance, and high enzyme activity adapted to industrial applications. Herein, we successfully cloned and expressed the chitinases AfChiJ, Δ19AfChiJ, and Δ30AfChiJ with an E. coli BL21 (DE3) expression system. The N-truncated chitinase of Δ30AfChiJ was successfully purified to homogeneity and biochemically characterized. AfChiJ is acid-resistant, has a high salinity tolerance, and is a thermostable exochitinase, a GH18 chitinase belong to the fungal/plant class chitinase (class III) family of filamentous fungi (Alcazar-Fuoli et al., 2011). AfChiJ degrades colloidal chitin to generate mainly N-acetyl chitobioase at first, which is further hydrolyzed to GlcNAc. The results suggest that the marine chitinase AfChiJ is an exochitinase that contains the properties of both chitobiosidases and β-N-acetylglucosaminidase. It is concluded that AfChiJ is a promising and sustainable option for GlcNAc preparation using chitin-containing biomass resources.
Materials and methods
Materials
We obtained QuickCut NcoI, QuickCut NdeI, and QuickCut XhoI restriction endonucleases and a recombinant DNase I (RNase-free) and PrimeScript II 1st Strand cDNA Synthesis Kit from TaKaRa (Dalian, China). The FastPure Gel DNA Extraction Mini Kit, DNA polymerases and 2 × ClonExpress Mix were all purchased from Vazyme Biotech CO., Ltd. (Nanjing, China). StarPrep Plasmid Miniprep Kit, Direct-load Color Prestained Protein Marker (15–150 kDa), Direct-load Star Marker D2000, and Star Marker D5000 were purchased from GenStar (Beijing, China). Host cell strains, E. coli DH 5α, E. coli BL21 (DE3), and the pET-22b (+) were stored the lab in Guangxi Academy of Sciences. Chitin, chitosan, cellulose microcrystalline, sodium carboxymethylcellulose (CMC-Na), and N-acetyl-D-glucosamine (GlcNAc) were purchased from BBI Co. Ltd. (Shanghai, China). Chitin oligosaccharides with degrees of polymerization (DP) between 1 and 6 were purchased from Tokyo Chemical Industry Co. Ltd. (Tokyo, Japan). Ni NTA Beads 6FF was purchased from Smart-Lifesciences Biotech (Changzhou, China). Redprussiate, isopropyl-beta-D-thiogalactoside (IPTG), aniline, and diphenylamine were purchased from 9dingchem (Shanghai, China). All other reagents were of analytical grade or the guaranteed grade available unless otherwise mentioned.
Strains and culture conditions
Aspergillus fumigatus df673 had previously been isolated by our lab from tidal flat sediments of mangrove in Beibu Gulf, Guangxi Province, China. It was stored in the Guangxi Beibu Gulf Marine Research Center and was cultured with potato dextrose broth medium at 28°C. E. coli strains were grown in LB culture medium (10 g tryptone, 5 g yeast extract, and 10 g sodium chloride per liter) at 37°C (1.5% agar powder should also be added to the solid medium).
Chitinous substrates
α-Chitin was obtained from crab shells (BBI; Shanghai, China) and then ball-milled by the inverter planetary ball mill XQM-2L (Kexi, Nanjing, China). The chitin powder was ground at 300 rpm for total of 9 h with 3 h grinding cycles interrupted 1 h pauses, producing an ultimate average particle size of 0.15 mm (Mekasha et al., 2020).
β-Chitin was acquired from a squid pen with an average particle size of 0.3 mm. The squid pen was collected from Nanning seafood wholesale trading market (Lecun Road 4, Nanning, Guangxi Province) and stored at −20°C until being handled in the lab. β-Chitin was prepared according to (Lavall et al., 2007): the squid pens were repeatedly washed with water, dried at 50°C, and then ground to particulates of less than 0.3 mm by the inverter planetary ball mill XQM-2L. In all, 1 g powder was dissolved in 10 ml hydrochloric acid (0.55 M) at 25°C for 2 h, and the treatments were repeated. Then, 1 g product was dissolved in 20 ml sodium hydroxide (0.3 M) at 80°C for 1 h, in two sequential treatments (Tolaimate et al., 2000). Finally, the product was washed to neutral with double distilled water, dried at 80°C, and stored at −40°C for following use.
Colloidal chitin was prepared as previously described by Wu H et al. (2022). Briefly, 20 g chitin (Aladdin, Shanghai, China) was stirred in 200 ml concentrated phosphoric acid (4 M) at room temperature for 2 h and then placed at 4°C for 24 h. Subsequently, 2 L cooled double distilled water and gelatinous chitin were mixed and placed for 24 h at 4°C. The colloidal chitin was separated by centrifugation at 6,000 rpm for 15 min and washed to neutral with double distilled water. Finally, the colloidal chitin was dissolved in 1 L phosphate-citrate buffer (pH = 7) to an ultimate concentration of 2%.
Enzyme assay
The activities of the purified recombinant Δ30AfChiJ and crude proteins toward the substrate were assayed according to the Schales method with some subtle modifications (Imoto and Yagishita, 1971). To detect the chitinase activity, the reaction containing 100 µL enzyme solution, 400 µL substrate (at a ultimate concentration of 2%) dissolved in phosphate buffer was incubated for 30 min at 45°C. The reaction was stopped by the addition of 1,000 µL Schales buffer and heating for 5 min in boiling water. The optical density (OD) of the supernatant was assayed at 420 nm using the Infinite M200 pro microplate reader (TECAN, Switzerland). The chitinase activity was determined by colorimetry and defined as the amount of enzyme that produces 1 µmol GlcNAc from the substrate per min.
Isolation of total RNA from A. fumigatus df673 and reverse transcribed to cDNA
A. fumigatus df673 was cultured in conical flask with potato dextrose broth (PDB) medium in a constant temperature incubator of 200 rpm for 3 days at 28°C. Total RNA was isolated from A. fumigatus df673 with Trizol solution (Invitrogen). The degradation of DNA was performed in the isolated total RNA samples (20 µg) using Recombinant DNase I (RNase-free) (Takara, Japan). The total RNA was reverse transcribed to cDNA using a PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Japan). The kit instructions were followed for all steps. cDNA was stored at −20°C (Reddy and Sarma, 1995).
Bioinformatic analyses
The analysis and searches of sequenced data were carried out by NCBI. Signal peptides of the chitinases were predicted by SignalP 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) (Almagro Armenteros et al., 2019). The domain of the chitinases were predicted on the PROSITE website (https://prosite.expasy.org) (Hulo et al., 2006). The generated sequences from the current study and reference sequences from Cazy (http://www.cazy.org) were used to create a phylogenetic tree using the neighbor-joining method (NJM) with 2000 replications for each bootstrap value, using the MEGA 11 software version (Tamura et al., 2021). Multi-sequence alignment results were visualized using the ENDscript 2.0 (https://endscript.ibcp.fr/ESPript/ENDscript/). The three-dimensional structure of AfChiJ was predicted by AlphaFold2 (Cramer, 2021). Ligand molecule (GlcNAc)4 was drawn using ChemDraw software. Molecular docking was performed by AutoDock 4.0 (Morris et al., 2009) and flexible docking methods. PyMOL (https://pymol.org/) (Rigsby and Parker, 2016) was carried out visualization of the modeled structure and the analysis of detailed interactions.
Cloning of the chitinase genes and design of the recombinant plasmid
The chitinase (AfChiJ, Δ19AfChiJ, Δ30AfChiJ) genes were amplified using the primer sets listed in Table 1. The nucleotide sequence of AfChiJ gene was uploaded in GenBank with the accession number: ON932820. cDNA of A. fumigatus df673 was used as the PCR template. PCR products were cleaned with PCR clean-up kit according to manufacturer’s description. The purified AfChiJ fragment was ligated into pET-22b (+) that was digested with Nde I and Xho I by ClonExpress II One Step Cloning Kit (Vazyme, China), generating the recombinant plasmid named pET-22b (+)-AfChiJ. Other expression plasmids pET-22b (+)-Δ19AfChiJ and pET-22b (+)-Δ30AfChiJ were constructed by the same strategy. The plasmids pET-22b (+)-AfChiJ, pET-22b (+)-Δ19AfChiJ, and pET-22b (+)-Δ30AfChiJ were respectively transformed into expression host BL21 (DE3).
Expression and purification of chitinase
The recombinant plasmids pET-22b (+)-AfChiJ, pET-22b (+)-Δ19AfChiJ, and pET-22b (+)-Δ30AfChiJ were respectively transformed into the expression host BL21 (DE3) to express chitinase. Cells were grown at 37°C to OD600 about 0.6–1.0, then induced by adding IPTG to ultimate concentration of 0.1 mM, and the cells were cultured for 8 h at 25°C. The cell pellet was separated by centrifugation (Avanti J-26S XP, Beckman Coulter, United States) at 7,000 rpm for 10 min and resuspended in NaH2PO4-NaCl buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0), then lysed on ice using an ultrasonic cell disrupter (Sonics VCX-750, SONICS, United States), and the cell fragments was precipitated by centrifugation at 4°C for 30 min (12,000 rpm). The supernatant was loaded onto a column containing Ni-NTA Beads 6FF (Smart-Lifesciences, Changzhou, China) for His-tag affinity purification. The column was washed with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 25 mM imidazole, pH 8.0) to remove contaminating proteins. Chitinase was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). The protein that was finally obtained was detected using 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The elution buffer was concentrated and desalted with a 10 kDa centrifuge ultrafiltration tube (Millipore, Massachusetts, United States) with 100 mM phosphate-citrate buffer (pH 7.0). The protein concentration of Δ30AfChiJ was measured using the NanoDrop 2000 (Thermo Fisher Scientific, United States).
Biochemical characterization of Δ30AfChiJ
The optimal temperature for Δ30AfChiJ activity was measured between 25–80°C in 100 mM phosphate-citrate buffer at pH 7.0 for 30 min. The thermal stability of Δ30AfChiJ was studied at different temperatures from 25°C to 70°C substrate-free for 60 or 90 min, then the residual enzyme activity was detected according to the standard assay condition. The optimum pH of the Δ30AfChiJ activity was determined using phosphate-citrate buffer (pH 2–8) and glycine-NaOH buffer (pH 8–10). Chitinase was incubated in above buffer at pH 1.0–11.0 at 30°C for 3 h to determine the remaining activity.
The influences of metal ions and chemical reagents (Ca2+, Co2+, Al3+, NH4+, Zn2+, Cu2+, K+, Ni2+, Na+, Mn2+, Li+, Mg2+, Ba2+, Tris, ethylenediaminetetraacetic acid (EDTA), sodium dodecyl sulfate (SDS), and urea) on Δ30AfChiJ activities were measured in the presence of 10 and 50 mM; at the same time, the effects of organic solvents (methanol, ethanol, acetonitrile, ethyl acetate, chloroform, dimethylsulfoxide (DMSO), tween-20, tween-80, glycerin, and isopropyl alcohol) were measured at ultimate concentrations of 10 and 20% (v/v). Each compound was added to the standard enzyme reaction system and assayed under optimal conditions.
For the halotolerance assay, the purified chitinase of Δ30AfChiJ was incubated with different ultimate concentrations of NaCl (1, 2, 3, 4, 5, 7, 9, or 11%) at 4°C for 3 h, and the residual Δ30AfChiJ activity toward colloidal chitin was detected under optimum temperature and pH.
Substrate specificity
The zymolyte specificity assay of the purified Δ30AfChiJ was studied at 45°C in 100 mM phosphate-citrate buffer (pH 4) with zymolyte including α-chitin powder, β-chitin powder, colloidal chitin, cellulose microcrystalline, sodium carboxymethylcellulose (CMC-Na), and chitosan. Each compound was appended to the standard enzyme reaction system at an ultimate concentration of 2% (w/v) and assayed under optimal conditions with the highest activity, defined as 100%.
Product assay
Reaction products from colloidal chitin hydrolyzed by Δ30AfChiJ were inspected by silica gel thin-layer chromatography (GF254, Qingdao, China). The reaction mixture was put into effect with the basis of the way (Morimoto et al., 2001) with some subtle changes. The reaction containing 400 µL colloidal chitin (2%) and 100 µL Δ30AfChiJ dissolved 100 mM phosphate-citrate buffer (pH 4) were incubated at 45°C for 1 h, 2 h, 3 h, 4 h, 6 h, and 8 h. The reaction mixturesysis was heated for 5 min in boiling water and centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was detected by TLC, which was spread out with a solvent of n-butanol, acetic acid, and aqueous (2:1:1). The products could be visualized with the method of spraying a diphenylamine-aniline-phosphate reagent (a blend of 1 ml aniline, 1 g diphenylamine, 5 ml concentrated phosphoric acid, and 50 ml acetone) evenly on the plate and heating the plate at 80°C for 20 min.
The main products of colloidal chitin degraded by Δ30AfChiJ were determined by high performance liquid chromatography (HPLC, Thermo Fisher Scientific, United States). The reaction mixturesysis, including 400 µL colloidal chitin (2%) and 100 µL Δ30AfChiJ dissolved in 100 mM phosphate-citrate buffer (pH 4) was incubated at 45°C for 1 h, 3 h, and 6 h. The reaction was stopped by adding 500 µL acetonitrile (75%) and then centrifuged at 12,000 rpm for 10 min at 4°C. After centrifugation, 10 µL supernatant was pushed into the HPLC system with a differential refractive index detector and a Luna NH2 column (250 mm × 4.6 mm, Phenomenex), at the temperature of 45°C. The mobile phase was acetonitrile: water (75:25, v/v), and its flow rate was 0.6 ml/min.
Detect the activity of β-N-acetylglucosaminidases
β-N-acetylglucosaminidase activity of Δ30AfChiJ was measured by breaking 4-methylumbelliferyl-GlcNAc (4MU-GlcNAc) zymolyte. The activity was calculated with a fluorometric assay based on detecting of chromophores of the released 4MU from chitinase-specific zymolyte (Guthrie et al., 2005). To determine the β-N-acetylglucosaminidases activity, a reaction mixture containing 5 µL enzyme solution and 95 µL 5 mM 4MU-GlcNAc dissolved in Mcllvaine buffer (pH 7.0) was incubated for 10 min at 45°C. The reaction mixture was stopped by the adding of 100 µL 1M glycine/NaOH buffer of pH 10.6 for readings and were taken immediately by use of a Fluorescence Spectrophotometer (HITACHI F-7000, Japan), the fluorescence was monitored (emission 460 nm, excitation 360 nm). β-N-acetylglucosaminidases activity was measured by 4MU released from 4MU-GlcNAc. The activity was counted by a standard curve of fluorescence units and 4MU (from 1 to 14 µM). One unit of β-N-acetylglucosaminidases activity was specified as the amount of enzyme which produces 1 µmol of 4MU from 4MU-GlcNAc per min (Mccret and Gooday, 1992).
Results and discussion
Bioinformatic analyses and construction of three constructs
A target gene band was produced by PCR from marine A. fumigatus df673, designated as AfChiJ. The size of the PCR production was 1,002 bp (No. ON932820), which encoded 334 amino acids containing GH18 domains. The first 19 amino acids of AfChiJ were determined as the signal peptide using the SignalP 5.0 analysis. The pH of AfChiJ was 4.6, and the theoretical molecular mass was 35.4 kDa, calculated using the ExPASy Tool. The tertiary structure of AfChiJ was predicted using AlphaFold2. The simulated structure indicated that AfChiJ consisted of a (β/α)8 TIM barrel fold, accompanied by an additional α/β structural domain, formed of four antiparallel β-strands, inset in the loop connecting helix α2 and barrel chain β2 (Figure 1A). The 3 disulfide bridges in AfChiJ were annotated in yellow, and the three-dimensional structure was stabilized by three disulfide bridges: C54–C107, C86–C97, and C205–C234 (Figure 1B). The homologue proteins previously reported were mostly expressed in Pichia pastoris (Landim et al., 2017). In this paper, we first try to use E. coli expression system with the high expression level of the target gene and short culture cycle to express AfChiJ. Meanwhile, we choose a plasmid of pET-22b (+) with a signal peptide of pelB to express this enzyme because signal peptides can bring proteins into the periplasmic space of E. coli, which is conducive to the correct formation of protein disulfide bonds and improves the stability of protein. At the same time, the N-terminal of AfChiJ contains a flexible region of 30 amino acids, as shown in tv-red color (Figure 1B). When E. coli is used to produce a eukaryotic protein, it is desirable to design the different constructs to produce as large an amount of the soluble and active protein as possible. With the aim of generating AfChiJ constructs with increased stability and expression, we selected the N-terminal flexible region and signal peptide in AfChiJ to test the influence of deleting them on protein expression and purification. Herein, we constructed three constructs in the schematic structure domains of chitinase (Figure 2A): a full length AfChiJ with native signal peptide (1-334 AAs), N-terminal truncation 19 amino acids of Δ19AfChiJ (20-334 AAs), and N-terminal truncation 30 amino acids of Δ30AfChiJ (31-334 AAs). Δ19AfChiJ and Δ30AfChiJ used the signal peptide of pelB.
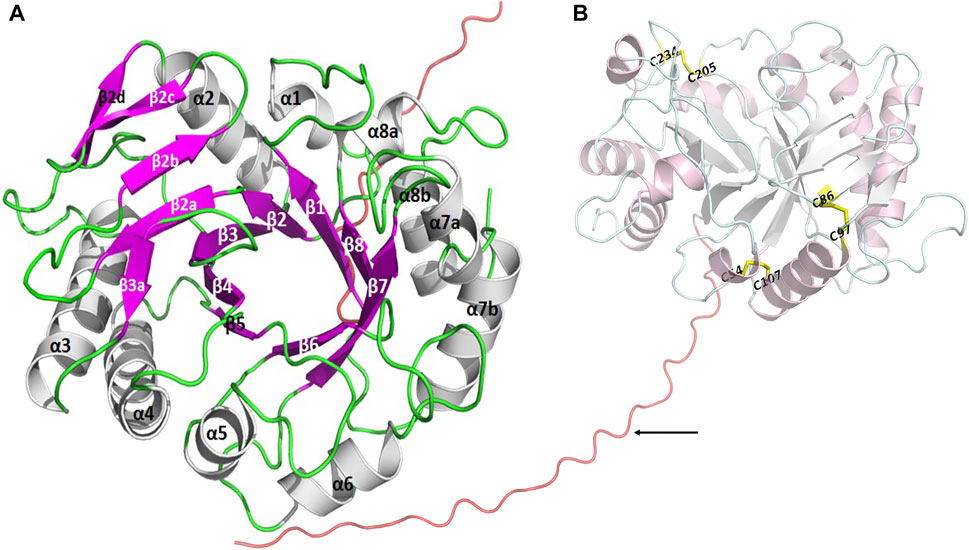
FIGURE 1. The tertiary structure of AfChiJ was predicted by AlphaFold2. (A) AfChiJ consisted of a (β/α)8 TIM barrel fold with an additional α/β structural domain, the additional α/β structural is composed of four antiparallel β-strands, inserted in the loop connecting helix α2 and barrel chain β2. (B) The N-terminal of AfChiJ contained a flexible region of 30 amino acids in tv-red color, with the 30 amino acids being identified by black arrows. The three disulfide bridges C54–C107, C86–C97, and C205–C234 are shown in yellow.
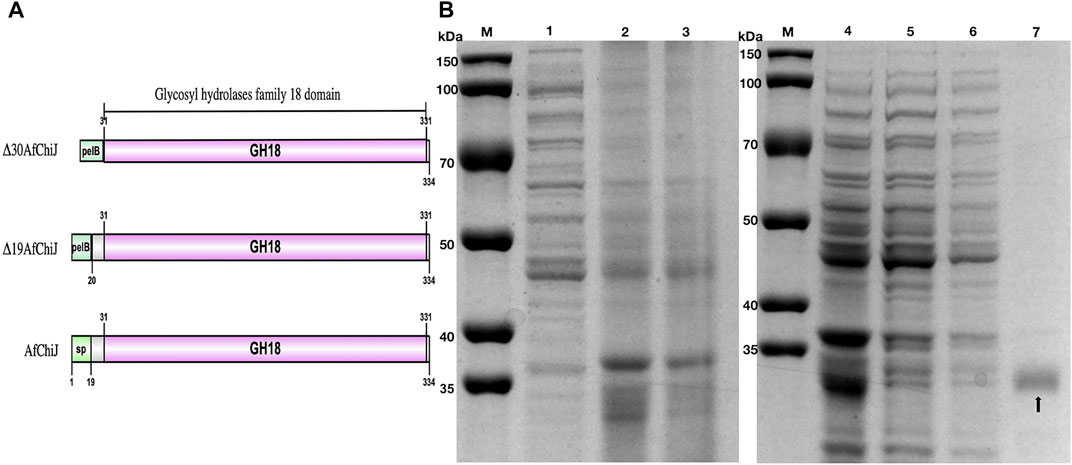
FIGURE 2. Schematic structure domains and SDS-PAGE analysis of chitinase Δ30AfChiJ. (A) Schematic structure domains of AfChiJ and its N-terminal truncation variants, Δ19AfChiJ and Δ30AfChiJ use signal peptide of pelB from pET-22b (+). (B) SDS-PAGE analysis of the purified chitinase Δ30AfChiJ. M, protein molecular weight markers; 1, crude extract of E. coli BL21 (DE3)/pET-22b; 2, crude extract of E. coli BL21 (DE3)/pET-22b-Δ30AfChiJ; 3, supernatant of E. coli BL21 (DE3)/pET-22b-Δ30AfChiJ; 4, supernatant of E. coli BL21 (DE3)/pET-22b-Δ30AfChiJ; 5, unbound protein through Ni-NTA Beads 6FF column; 6, protein eluted with buffer containing 25 mM imidazole; 7, protein eluted with buffer containing 250 mM imidazole. The gel was stained with Coomassie Brilliant Blue R-250.
Expression and purification of chitinases
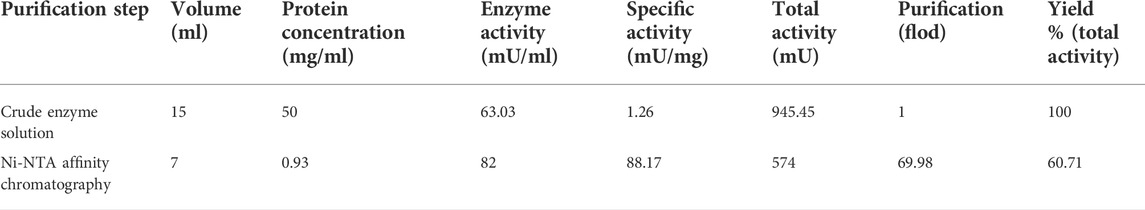
Because the recombinant AfChiJ mainly formed an insoluble protein, we calculated that the yield (total activity %) rate of AfChiJ was only 8.51% after passing through the Ni-NTA affinity chromatography column. Nevertheless, a large amount of protein was precipitated when Δ19AfChiJ was purified by passing through the Ni-NTA affinity chromatography column in 4°C. It was declared that Δ19AfChiJ was extremely unstable in the process of purification. Finally, we failed to obtain purified Δ19AfChiJ.
The yield rate of Δ30AfChiJ reached 60.71%, and the purification was 69.98-flod (Table 2). Thus, we purified a large amount of Δ30AfChiJ recombinant protein and proceeded to the next step of the research (Figure 2B). The 30 aa at the N-terminal of the chitinase of AfChiJ neither formed a secondary structure nor a tertiary one, based on the AlphaFold2 structure prediction (Figure 1B). In this study, the N-terminal truncation of Δ30AfChiJ can be successfully obtained from large amounts of solubly expressed protein. In a similar case, Fuciños et al. (2011) removed 16 or 26 aa from the N-terminal of esterase, which contributed to slight influences for substrate specificity. The results showed that the N-terminal fragment is pivotal for preserving the thermophilic property and stability, as well as the activity of the enzyme. Meanwhile, the ChiW protein was successfully overexpressed in E. coli by the method of N-terminally truncated (Itoh et al., 2014).
Effect of temperature and pH on Δ30AfChiJ activity and stability
The optimum temperature of Δ30AfChiJ activity was detected to be approximately 45°C by colloidal chitin as a zymolyte (Figure 3A). Δ30AfChiJ remains much more than 68% of the Δ30AfChiJ activity between 30 and 50°C. The activity of Δ30AfChiJ decreased markedly above 50°C. The optimum temperature was lower than that for other chitinase reported from A. fumigatus YJ-407 (60°C), but it was higher than that of chitinase from A. griseoaurantiacus KX010988 (40°C) (Xia et al., 2001; Shehata et al., 2018). The thermostability of Δ30AfChiJ was determined through preincubation for 60 or 90 min with different temperatures, from 25°C to 70°C, in the lack of substrate. The relative activity of Δ30AfChiJ decreased slowly below 45°C, with a relative activity of 67% at 45°C for 60 min (48% at 45°C for 90 min). The activity of Δ30AfChiJ dramatically decreased to about 24% at 50°C for 90 min, while more than 73% of the relative activity was maintained at 40°C for 60 min (Figure 3B). Therefore, Δ30AfChiJ has higher activity and stability than most thermostability chitinases. So far, the thermostability of chitinases have had advantages for some utilizations where high temperature was required or could not be eliminated, for instance, the production of chitin oligomers and monomers could be used for aquaculture or material science. Therefore, Δ30AfChiJ may demonstrate better utilization potential for such manufacture.
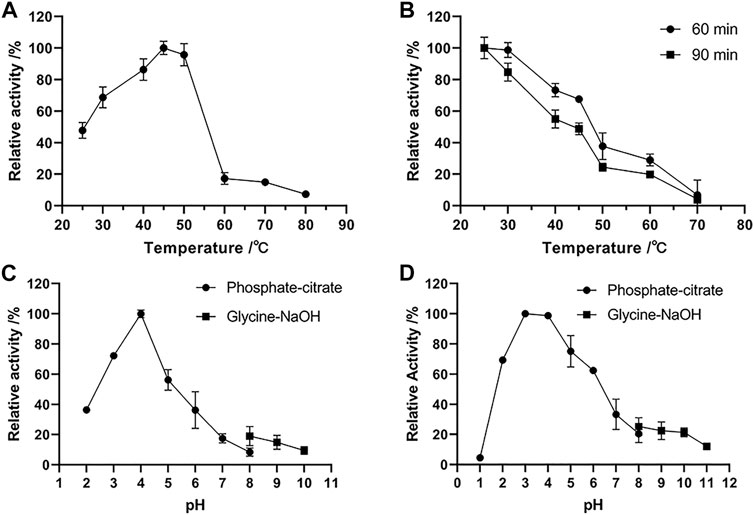
FIGURE 3. Effects of temperature and pH on the activity and stability of chitinase Δ30AfChiJ. (A) The optimum temperature of Δ30AfChiJ. The activity of Δ30AfChiJ was measured at pH 7.0 with colloidal chitin as the substrate. The highest activity of Δ30AfChiJ at 45°C was defined as 100%. (B) Thermostability of Δ30AfChiJ. The thermostability was measured at pH 4.0. The highest enzyme activity after preincubation was defined as 100%. (C) The optimum pH of Δ30AfChiJ. The activity of Δ30AfChiJ was measured at 45°C with colloidal chitin as the substrate. The activity of Δ30AfChiJ at pH 4.0 was defined as 100%. (D) pH stability of Δ30AfChiJ. Stability of Δ30AfChiJ after incubation at different pH at 30°C for 3 h without substrates. The activity of the enzyme after incubation at pH 3.0 was defined as 100%. Each data point represents the mean of three independent experiments, standard errors are shown (mean ± SD).
The optimal pH for Δ30AfChiJ activity was shown in Figure 3C. The activity of Δ30AfChiJ was maximal at pH 4, and it maintained high activity at pH 2.0 to 6.0, with relative activities of 36.42 and 36.24% at pH 2.0 and 6.0, respectively. There were have been a few reports on acid chitinases from A. fumigatus, and the majority of them exhibited optimal pH around 5.0. For instance, the optimum pH values of chitinase from A. fumigatus YJ-407, A. fumigatus CS-01, and Acremonium sp. YS2-2 were reported to be at 5.0, 5.0, and 6.0, respectively (Xia et al., 2001, 2009; Chung et al., 2019). Only the chitinase produced by Streptomyces and AMCase isolated from mouse stomach maintained the highest activity at pH 2.0 (Karthik et al., 2015; Du et al., 2021). The pH stability of Δ30AfChiJ was shown in Figure 3D by pre-incubating substrate-free for 3 h at 30°C, using pH values from 1 to 11. The residual activity of Δ30AfChiJ was the highest at pH 3.0 and was stabilization within the scope of pH 2–6 (relative activity >62%). It indicated that Δ30AfChiJ is alkaline sensitive, while can be work well at acidic conditions. The majority of chitinases from marine fungus appeared medium high stability within the scope of pH 4–7 (Xia et al., 2009; Han et al., 2016; Wu Y et al., 2022). The Δ30AfChiJ in this research was equivalent to the acidic chitinase from A. fumigatus CS-01, displaying stability at pH 2.0–6.0 for 3 h at 30°C, which was comparable to the chitinase from Pyrus ussuriensis (pH 2.5–6.0) (Xia et al., 2009; Han et al., 2016). On the other side, converting 20 g chitin to colloidal chitin requires approximately 10–12 L water to reach a neutral pH, while only 4–6 L is required to reach a pH of 4.0. Thus, it was saved half of the water consumption using an acidic chitinase of Δ30AfChiJ. In summary, its high stability at low pH conditions and its lower water consumption requirements for processing indicate that Δ30AfChiJ is a potential candidate for commercial and industrial applications (Ueda and Kurosawa, 2015).
Effect of metal ions and chemical reagents on Δ30AfChiJ activity
The influences of metal ions and chemical reagents on the activity of purified Δ30AfChiJ are shown in Figure 4. The Δ30AfChiJ activity was inhibited within 10 mM Cu2+ or Ni2+, moreover, the activity dropped sharply within 50 mM concentration, even 50 mM Ni2+ completely no activity at all. It has been suggested that Cu2+ catalyzes the amino acids of cysteine to form intramolecular disulfide bonds or sulfenic-acid (Vieille and Zeikus, 2001) and to hinder chitinase activities from different microorganisms such as Streptomyces sp. and Trichoderma harzianum GIM 3.442 (Pradeep et al., 2014; Deng et al., 2019), this phenomenon may indicate the potential role of cysteines involved in the enzyme catalysis. At the same time, the enzymatic activity was mightily inhibited within both concentrations of Co2+, Mn2+, and SDS (relative activity <60%). Moreover, the activity of Δ30AfChiJ was barely seen in the presence of Co2+ and Mn2+. This is because these heavy metal ions can deactivate proteins by disrupting the tertiary structure of enzymes (Liu et al., 2020). This is similar to the chitinase MDcht2 from Musca domestica, while enzymatic activities were thoroughly inhibited by Mn2+ and Fe2+ (Peipei et al., 2021). In this study, we also detected that the activity of Δ30AfChiJ increased by adding Mg2+and Ba2+ (10 and 50 mM), which increased the enzymatic activity about 1.2-fold relative to control. Studies have found that metal ions serve as cofactors for enzyme activities, and after acting as ion or salt bridges between two adjacent amino acid residues, maintain rigid confirmation of enzymes (Kamran et al., 2015), this is close to ChiEn1 from Coprinopsis cinerea, where enzymatic activity is moderately increased by Mg2+and Ba2+ at 1 mM concentration (Niu et al., 2017). The effect of 10 and 50 mM urea on Δ30AfChiJ is compatible with chitinase obtained from Paenibacillus UMBR 0002 (Liu et al., 2020). It is widely known that strong concentrations of urea denatures proteins, but we achieved the opposite result at 10 and 50 mM of urea. In addition, EDTA was a metal ion chelating agent, and the influence on enzyme activity demonstrated that Δ30AfChiJ was greatly affected by the enzyme activity of metal ions. The effect of EDTA on Δ30AfChiJ is similar to that on chitinase originated from Qinghai-Tibetan Plateau (Dai et al., 2021). Generally, these results provide a basis for which metals ions or chemical reagents to add for the commercial and industrial applications of Δ30AfChiJ. The roles of the metal ions are related to substrate activation or electrostatic stabilization and played a vital role in biocatalysts by stabilizing the complexes of substrate-enzyme (Andreini et al., 2008).
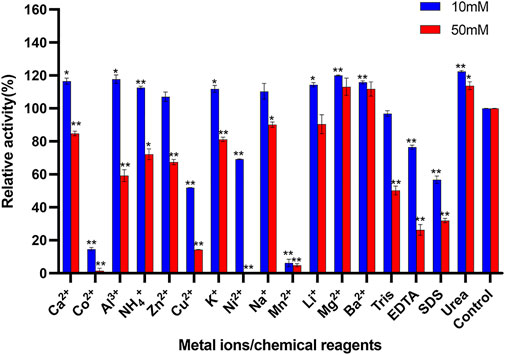
FIGURE 4. Effects of metal ions and chemical reagents on activity of Δ30AfChiJ. Untreated enzyme was used as the control, and the activity of this preparation defined as 100%. Experiments were carried out in triplicate, and standard errors are shown (mean ± SD). Asterisks indicate statistically significant differences from the control (Student’s t-test. *, p < 0.05; **, p < 0.01). Abbreviations: EDTA, ethylenediaminetetraacetic acid; SDS, sodium dodecyl sulfate.
Effects of solvents on Δ30AfChiJ activity
The effects of different organic compounds on the stability of Δ30AfChiJ are displayed in Figure 5. In the presence of chloroform, there is a notedly increased (10 and 20%). The influence of chloroform on Δ30AfChiJ is equivalent to the chitinase obtained from Melghiribacillus Nari2A (Mohamed et al., 2019). This indicates that the chitinase aqueous solution is mixed with chloroform, and the water molecules between the enzyme molecules are squeezed out by chloroform. Therefore, chloroform lowers the activation energy of the enzymatic reaction and promotes the progress of the enzyme reaction. On the other hand, for most organic solvents tested in his paper, including methanol, ethanol, acetonitrile, ethyl acetate, tween-20, tween-80, glycerin, and isopropyl alcohol, when using an ultimate concentration of 10%, the activity of Δ30AfChiJ decreased, while the 20% organic solvents notedly inhibited the activity. Moreover, ethyl acetate and tween-20 strongly inhibited Δ30AfChiJ activity (relative activity <50%) at both concentrations. In addition, Δ30AfChiJ activity was not significantly affected by 10% DMSO, while relative activity moderately decreased (<70%) in the presence of 20% DMSO. In summary, the influences of different organic compounds in this paper on Δ30AfChiJ activity approximated those of chitinases from Aeromonas hydrophila SBK1 (Halder et al., 2016). The property of Δ30AfChiJ that was stable at an ultimate 10% concentration of most organic solvents suggested that the enzyme could be used in practical industrial applications (Mohamed et al., 2019). Thus, Δ30AfChiJ might be a suitable candidate for commercial applications.
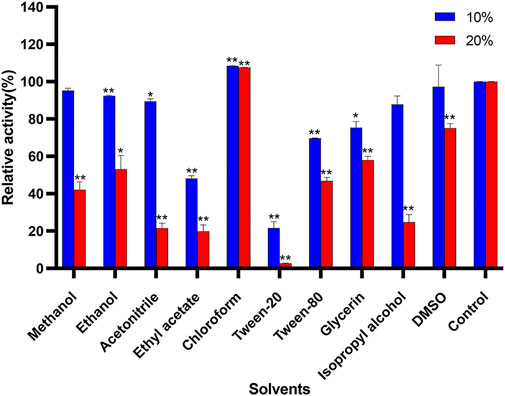
FIGURE 5. Effects of solvents on activity of Δ30AfChiJ. Untreated enzyme was used as the control, and the activity of this preparation defined as 100%. Each data represents the mean of three independent experiments, standard errors are shown (mean ± SD). Asterisks indicate statistically significant difference, compared with the control (Student’s t-test. *, p < 0.05; **, p < 0.01). Abbreviation: DMSO, dimethylsulfoxide.
Influence of NaCl concentration on Δ30AfChiJ stability
The Δ30AfChiJ showed highest relative activity of 157.71% at an optimal NaCl concentration of 3%. When it was incubated at 4°C after 3 h, the remained high activity (≥97.96%) was detected in the 1–7% concentration of NaCl. However, Δ30AfChiJ activity decreased under the 11% NaCl, and its relative activity reached <72.19% (Figure 6). The concentration of NaCl in the tidal flat sediments of mangrove is approximately 3%; therefore, Δ30AfChiJ could be used to hydrolyze chitin waste from oceanic and coastal environments (Liu et al., 2020).
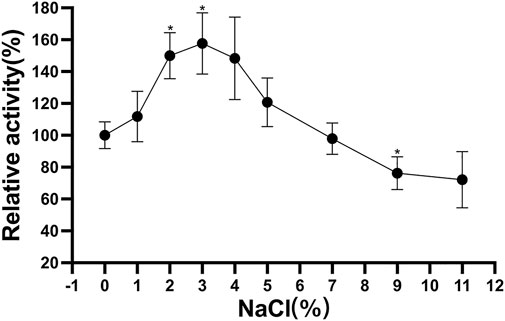
FIGURE 6. Effects of NaCl concentration on stability of Δ30AfChiJ. Stability of Δ30AfChiJ after incubation at various NaCl concentration at 30°C for 3 h. The residual activity of Δ30AfChiJ was measured at 45°C and pH 4.0 with colloidal chitin as the substrate. The highest enzyme activity after preincubation at no NaCl concentration was defined as 100%. Each data represents the mean of three independent experiments, standard errors are shown (mean ± SD). Asterisks indicate statistically significant differences from the control (Student’s t-test. *, p < 0.05; **, p < 0.01).
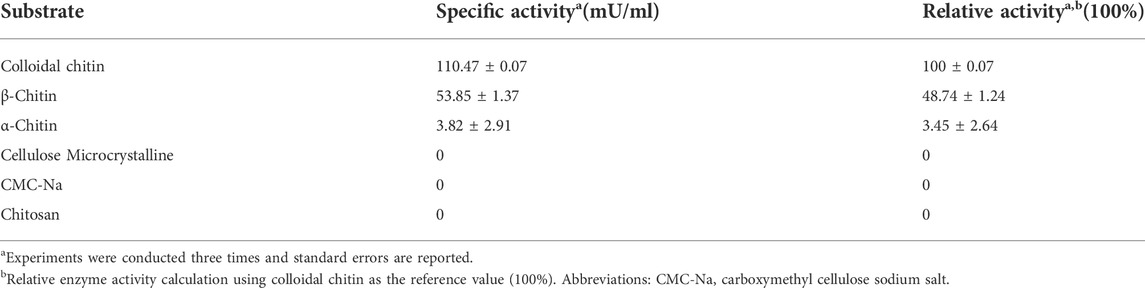
Substrate specificity of Δ30AfChiJ
The specific activities of Δ30AfChiJ (approximately 1 mg/ml) against different zymolyte are listed in Table 3. The specific activity to colloidal chitin was highest (110.47 mU/mL). The activity was 53.85 mU/mL when using β-chitin as a substrate. The relative activity was 48.74 ± 1.24% compared with colloidal chitin (100%). The activity against α-chitin was weak (3.82 mU/mL). No activity was detected toward cellulose microcrystalline, sodium carboxymethylcellulose (CMC-Na) and chitosan. Our results suggested that Δ30AfChiJ preferred colloidal chitin to β-chitin and α-chitin. At the same time, colloidal chitin was the most suitable substrate for Δ30AfChiJ. This suggested that the chitinase more easily accessed the treated colloidal chitin. For α-chitin, low Δ30AfChiJ activity (3.8%) was detected, which likely entailed that α-chitin was too difficult to dissolve in the phosphate buffer. Chitinases belonging to GH family 18 possessed a substrate-assisted catalytic mechanism (van Aalten, 2021). The glycosidic bond protonated by the acid was a conservative glutamate, and the nucleophile is an oxygen of the N-acetyl group on the -1 sugar, producing an intermediate of oxazolinium ion. Due to this mechanism, a GlcNAc residue in the -1 subsite was obligatory for catalytic cleavage to appear (Hartl et al., 2012). This, it is reasonable to be unable to detect any activity for the chitosan, even if chitosan was dissolved in the optimum pH 4.0 of Δ30AfChiJ. For cellulose microcrystalline and CMC-Na, the activity of Δ30AfChiJ cannot be measured in our study, which was similar to chitinase from T. harzianum GIM 3.442 (Deng et al., 2019). Δ30AfChiJ is a typical chitinase, and it can efficiently degrade colloidal chitin.
TLC and HPLC analysis of the hydrolysis product
In this study, we determined a mixed product including (GlcNAc)2 and GlcNAc during the degradation of colloidal chitin. As shown in Figure 7, (GlcNAc)2 and GlcNAc can be observed at 2 h by TLC assay, and the amount of product was gradually accumulated with the increase of reaction time. However, we could not detect any products at 1 h, and we presumed that the amount of reaction products at 1 h was below the minimum detection limit of the TLC method. Therefore, the colloidal chitin hydrolysate by Δ30AfChiJ at 1, 3, and 6 h was detected using HPLC. The results appeared that the products of (GlcNAc)2 and GlcNAc in the reaction of Δ30AfChiJ and colloidal chitin for 1 h were low. At the same time, the peak heights of (GlcNAc)2 and GlcNAc were nearly the same (Figure 8). This shows that the yields of (GlcNAc)2 and GlcNAc were basically comparable in the early stage of the enzyme-substrate reaction. The amount of GlcNAc increased obviously after 3 h, while the amount of (GlcNAc)2 increased slightly, which suggested that Δ30AfChiJ could hydrolyze colloidal chitin to obtain GlcNAc as the major product and (GlcNAc)2 as a minor product. Meanwhile, the Δ30AfChiJ retains stability and activity for 6 h, which could be an appropriate reaction time to obtain GlcNAc.
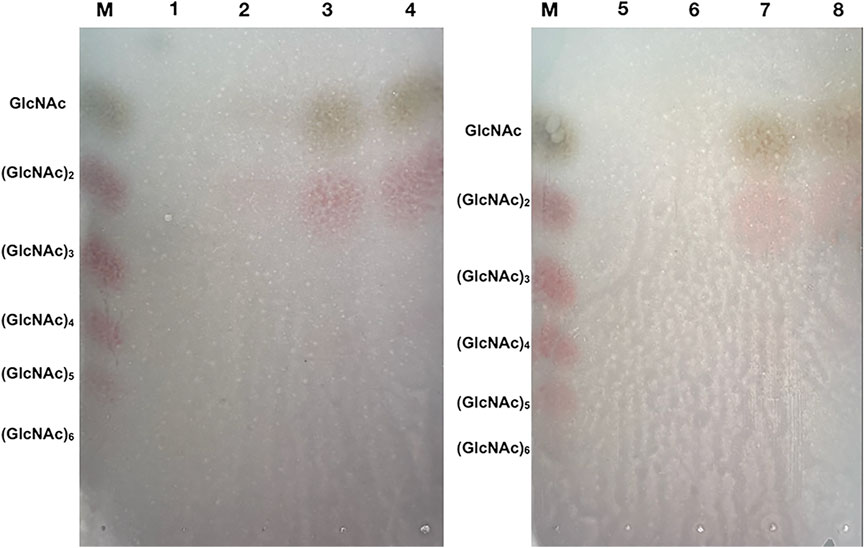
FIGURE 7. TLC analysis of the degradation products of Δ30AfChiJ. M, Standards of chitin oligosaccharides with degrees of polymerization between 1 and 6; 1 and 5, The degradation products of inactivation Δ30AfChiJ was used to be control; 2–4, The degradation products of Δ30AfChiJ with colloidal chitin for 2, 4, and 8 h; 6–8, The degradation products of Δ30AfChiJ with colloidal chitin for 1, 3, and 6 h. Developing solvent: n-butanol, acetic acid and aqueous (2:1:1). Color reagents: 1 g diphenylamine, 1 ml aniline, 5 ml 85% phosphoric acid, and 50 ml acetone.
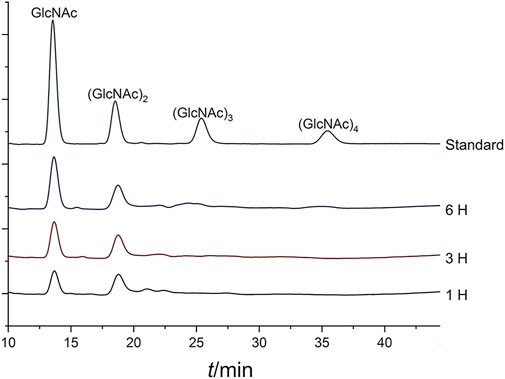
FIGURE 8. HPLC analysis of the degradation products of Δ30AfChiJ. Colloidal chitin was degraded by Δ30AfChiJ at 45°C and pH 4.0 for different lengths of time (1, 3, and 6 h). A refractive index detector and a Phenomenex Luna NH2 column (250 mm × 4.6 mm) were used to analyze 10 microliters supernatant of the reaction mixtures. The mobile phase was acetonitrile: water (75:25, v/v) with a flow rate of 0.6 ml/min, the column temperature was 45°C.
Chitinases can be divided into two major groups: endochitinase and exochitinase. Endochitinase hydrolyzes chitin randomly at internal sites, generating low molecular mass oligosaccharide, such as the forms (GlcNAc)2, (GlcNAc)3, and (GlcNAc)4 forms. Exochitinase can be classified into two subcategories: chitobiosidases progressively liberate (GlcNAc)2 beginning at the chain end of chitin, and β-N-acetylglucosaminidase cleave the (GlcNAc)2 to generate monomers of GlcNAc (Sahai and Manocha, 1993; Dahiya et al., 2006). The chitinase Δ30AfChiJ from A. fumigatus df673 may be an exochitinases, which represents both chitobiosidases and β-N-acetylglucosaminidase activity. In our study, GlcNAc and (GlcNAc)2 were detected to be hydrolysis products from colloidal chitin in the whole phase, and (GlcNAc)2 was detected, suggesting that the chitinase could cleave preferentially terminal glycosidic bonds, thereby demonstrating a chitobiosidases activity. Interestingly, (GlcNAc)2 was further hydrolyzed to GlcNAc, it was suggested that Δ30AfChiJ had β-N-acetylglucosaminidase activity. The hydrolysis mechanism and processing of Δ30AfChiJ is close to the chitinase of the Baozhu pear from Pyrus ussuriensis Maxim and PbChi74 from Paenibacillus barengoltzii (Fu et al., 2014; Han et al., 2016). These cleave colloidal chitin to obtain GlcNAc as the major product and possess two chitinase activities (exochitinase and β-N-acetylglucosaminidase activities). The majority of the 18 other GH family exochitinases exhibit a proceeding mode of action to produce (GlcNAc)2 as the main end product, while they cannot further degrade (GlcNAc)2 to GlcNAc (Horn et al., 2006). Meanwhile, the chitinase CmChi1 from meiyuanensis SYBC-H1 has shown the ability of endochitinase and chitobiosidase with β-N-acetylglucosaminidase activities (Zhang et al., 2018). In general, the biofacturing of GlcNAc is a valuable “green” method, and the special properties of Δ30AfChiJ may make chitinase a suitable candidate for the converting chitin into GlcNAc. Therefore, the hydrolysis activity of Δ30AfChiJ facilitates the commercial production of GlcNAc from chitin. Nevertheless, the catalytic mechanism of Δ30AfChiJ hydrolyzed chitin requires further research.
The β-N-acetylglucosaminidases activity assay
The yield of GlcNAc increased obviously with colloidal chitin hydrolysis by Δ30AfChiJ for 6 h (Figure 8), so we deduced Δ30AfChiJ may have β-N-acetylglucosaminidases activity. 4MU-GlcNAc was chosen to mimic (GlcNAc)2. The β-N-acetylglucosaminidase activity of Δ30AfChiJ was measured by cleavage of 4MU-GlcNAc substrates and released 4MU fluorescent monomers. The fluorescence value was 264.5, obtained by a Fluorescence Spectrophotometer monitored (excitation 360 nm, emission 460 nm). The β-N-acetylglucosaminidase activity of Δ30AfChiJ was 4.46 mU/mL with the standard curve calculation, and the specific activity of Δ30AfChiJ was 5.57 mU/mg, all monitored under the specified assay conditions. This result suggests that Δ30AfChiJ has β-N-acetylglucosaminidase activity; it is first demonstrated that β-N-acetylglucosaminidase activity existed in the Aspergillus family chitinases. There are many Aspergilli family chitinases with exochitinase activity, but little β-N-acetylglucosaminidase activity has been verified (Kuranda and Robbins, 1987; Rush et al., 2010).
Alignment and molecular docking simulation
Following the phylogenetic tree analysis of AfChiJ (Figure 9A), AfChiJ was compared to 11 sequences with different properties which were characterized in Uniport. Among of these, an endochitinase from A. fumigatus ChiA1 (Uniport id Q873Y0) has a homology (36.9%) with AfChiJ, another chitinase with exochitinase and β-N-acetylglucosaminidase properties from the Baozhu pear (Uniport id A0A5N5F715) has a homology (29.24%) with respect to AfChiJ. Due to the lack of sufficiently characterized chitinases from filamentous fungi, it is difficult to determine the endo, exo, or β-N-acetylglucosaminidase properties of AfChiJ based the homologous sequence alignment. Thus, we infer that it is a GH18 chitinase that belongs to the fungal/plant class chitinase (class III) family. Comparing to available structures of GH18 family member enzymes indicates that AfChiJ is most similar to A. fumigatus ChiA1 (PDB id 2XTK) and S. cerevisiae CTS1 (PDB id 2UY4), with an RMSD (root mean square derivation) of 1.6 Å (286 Cα atoms) and 1.8 Å (278 Cα atoms), respectively. In the meantime, some different conformations occur in the connecting loops, most of which harbor insertions/deletions. Herein, we use the ConSurf server to identify the functional regions of AfChiJ (Glaser et al., 2003). It is also demonstrated that the flexible region of the AfChiJ (colored turquoise) exist in the connecting loops (Figure 9B). However, these flexible regions endow AfChiJ with substrate specificity and catalytic properties.
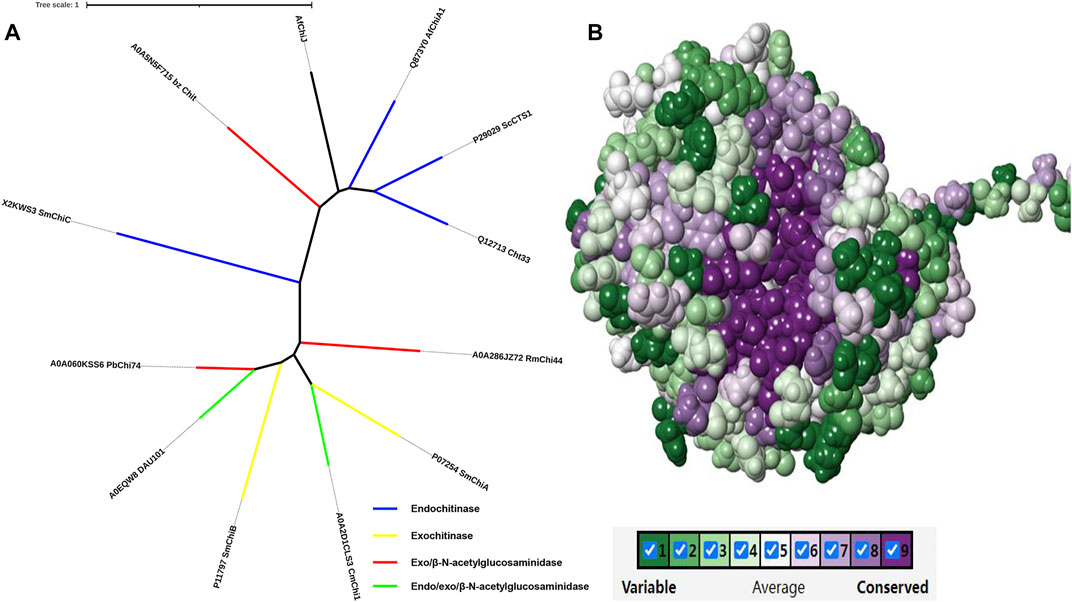
FIGURE 9. Phylogenetic tree analysis and evolutionary analysis of AfChiJ. (A) The phylogenetic tree analysis of AfChiJ. The endochitinases are shown in blue, exochitinases are shown in yellow, exo and β-N-acetylglucosaminidases are shown in red, endo/exo and β-N-acetylglucosaminidases are shown in green. Tree scale: 1 substitution per nucleotide position. (B) The evolutionary conservation is determined based on the structure and multiple sequence alignment. The amino acid residues are colored ingredient representing the conservation grades from the most variable (turquoise) to the most conserved (maroon) obtained with ConSurf program.
For the alignment of the amino acid sequence of the 11 homology proteins (Figure 10), analysis revealed that AfChiJ contain the catalytic signature motif of GH 18, DxDxE (residues 144–148), and a chitin-binding motif, SxGG (residues 104–107). The glutamate 144 residue is considered a proton donor, and the aspartate 148 residue play a prime role in supporting the acetamido group in it nucleophilic attack and supporting the forming a positive oxazolinium ion charge (Bokma et al., 2022). Substrates with the active sites groove were mimicked by docking (GlcNAc)4, using the model of AfChiJ simulated by AlphaFold2 (Figure 11). It was found that the active pocket formed a shallow and open groove. The catalytic motif DxDxE (residues 166–170) is located between β4-α4. It was found that the (GlcNAc)4 binding pocket of AfChiJ contained four glycosyl binding sites using molecular docking, and that the predicted catalytic residue of AfChiJ, Glu 170 is space-based near to the glycosidic bond among subsite −1 and subsite +1.
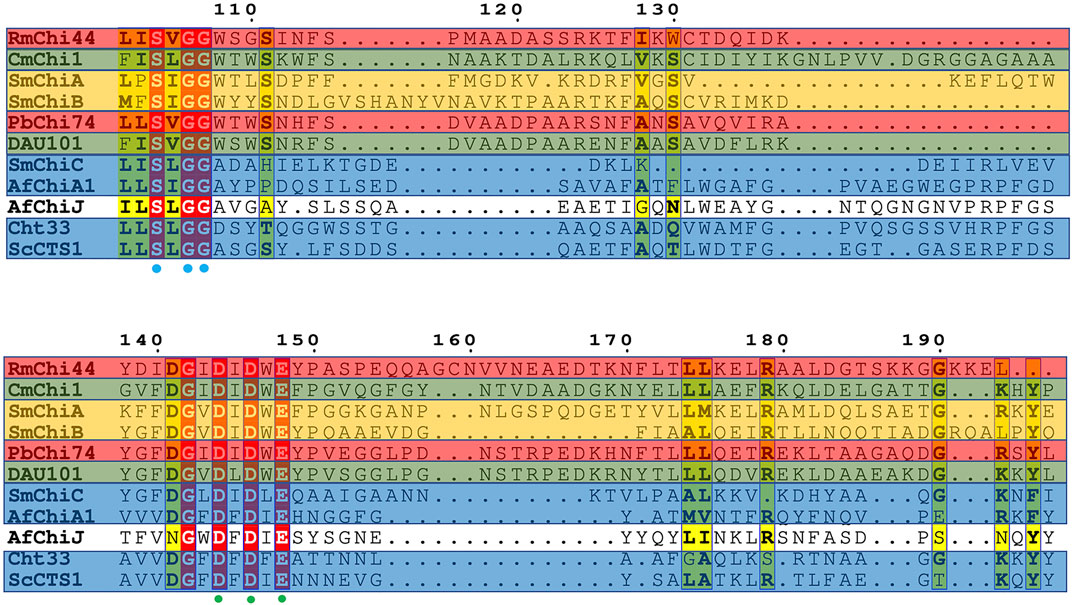
FIGURE 10. Alignment of amine acid sequence homology. Structure-based sequence alignment of 11 homology proteins. Residue numbers are given shaded by sequence similarity between the 11 enzymes shown (red = identical, yellow = chemically similar residues). Residues DXDXE are highlighted by green filled circles. Residues SXGG are highlighted by blue filled circles. Endochitinases are shown in blue background, exochitinases are shown against a yellow background, exo and β-N-acetylglucosaminidases are shown against a red background, endo/exo and β-N-acetylglucosaminidases are shown against a green background.
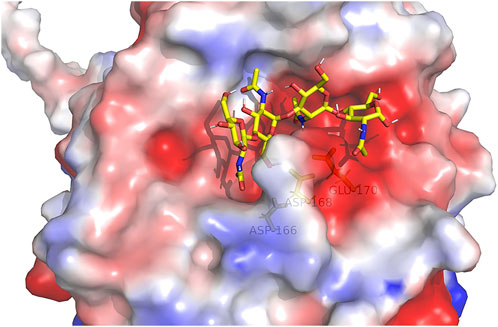
FIGURE 11. (GlcNAc)4 by molecularly docking with the AfChiJ. Substrates with the active sites channel were simulated by docking (GlcNAc)4 with the AlphaFold2 model of AfChiJ. Molecular docking revealed that the (GlcNAc)4 binding cavity of AfChiJ contains four glycosyl binding sites, and that the predicted catalytic residue of AfChiJ, Glu 170, is spatially close to the glycosidic bond between −1 and +1.
Conclusion
In summary, we successfully cloned and expressed chitinases AfChiJ, Δ19AfChiJ, and Δ30AfChiJ using E. coli BL21 (DE3). It is worth mentioning that Δ30AfChiJ was successfully purified. The molecular weight of enzyme Δ30AfChiJ was about 35 kDa and the hydrolytic activity of colloidal chitin was best at pH 4 and 45°C. TLC and HPLC analysis manifested that GlcNAc was the main hydrolysates of colloidal chitin. Furthermore, β-N-acetylglucosaminidase activity of Δ30AfChiJ was measured by the cleavage of 4MU-GlcNAc substrates, releasing 4MU fluorescent monomers. Finally, it was verified that Δ30AfChiJ represents both chitobiosidases and β-N-acetylglucosaminidase activities. A chitobiosidases showing β-N-acetylglucosaminidase activities from Aspergillus species was firstly reported. In addition, better enzymatic activity and stability were inspected over a wide scope of pH and temperature. The existence of Ca2+, Mg2+, and chloroform notably increased the activity of chitinase. The hydrolytic properties and great environmental adaptions made the Δ30AfChiJ a potential candidate for green industrial transformation chitin debris into monomer of chitin. Generally, Δ30AfChiJ might be applied for chitinous biomass high-value added utilization in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Writing-original draft, BH; writing-review and editing, LP and LY; project administration FJ and LP; resources, DY and HC; software, LP and LY; supervision, LP, HC, and FJ; methodology, BH and CL; conceptualization, LP; investigation, LY and MJ. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
The authors are grateful for the financial support from the Guangxi Innovation-driven Development Major Science and Technology Innovation Base Construction Project (Grant No. 2022-36-Z06, awarded to LP), the Basic research fund of Guangxi Academy of Sciences (Grant No. 2020YBJ701, awarded to LY), and the National Natural Science Foundation of China (Grant No. 31860245, 31960203, awarded to LP and DY).
Conflict of interest
CL was employed by the company Nanning Dabeinong Feed Technology Co, Ltd., and FJ was employed by the company Guangxi Huaren Medical Technolgoy Group. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alcazar-Fuoli, L., Clavaud, C., Lamarre, C., Aimanianda, V., Seidl-Seiboth, V., Mellado, E., et al. (2011). Functional analysis of the fungal/plant class chitinase family in Aspergillus fumigatus. Fungal Genet. Biol. 48, 418–429. doi:10.1016/j.fgb.2010.12.007
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi:10.1038/s41587-019-0036-z
Andreini, C., Bertini, I., Cavallaro, G., Holliday, G. L., and Thornton, J. M. (2008). Metal ions in biological catalysis: From enzyme databases to general principles. J. Biol. Inorg. Chem. 13, 1205–1218. doi:10.1007/s00775-008-0404-5
Bokma, E., tte Rozeboom, H. J., Sibbald, M., Dijkstra, B. W., and Beintema, J. J. (2022). Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis. Eur. J. Biochem. 269, 893–901. doi:10.1046/j.0014-2956.2001.02721.x
Chen, C., Wang, Y., Yang, Y., Pan, M., Ye, T., and Li, D. (2018). High strength gelatin-based nanocomposites reinforced by surface-deacetylated chitin nanofiber networks. Carbohydr. Polym. 195, 387–392. doi:10.1016/j.carbpol.2018.04.095
Chen, Y., Tao, Z., Zhang, M., Sun, L., Liu, G., and Cao, M. (2021). Identification of a chitinase from the hepatopancreas of Chinese black sleeper (Bostrychus sinensis). Acta Oceanol. Sin. 40, 50–60. doi:10.1007/s13131-021-1781-7
Chung, D., Baek, K., Bae, S. S., and Jung, J. (2019). Identification and characterization of a marine-derived chitinolytic fungus, Acremonium sp. YS2-2. J. Microbiol. 57, 372–380. doi:10.1007/s12275-019-8469-0
Cohen, E. (2001). Chitin synthesis and inhibition: A revisit. Pest Manag. Sci. 57, 946–950. doi:10.1002/ps.363
Cramer, P. (2021). AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 28, 704–705. doi:10.1038/s41594-021-00650-1
Dahiya, N., Tewari, R., and Hoondal, G. S. (2006). Biotechnological aspects of chitinolytic enzymes: A review. Appl. Microbiol. Biotechnol. 71, 773–782. doi:10.1007/s00253-005-0183-7
Dai, Y., Yang, F., Liu, X., and Wang, H. (2021). The discovery and characterization of a novel chitinase with dual catalytic domains from a Qinghai-Tibetan Plateau wetland soil metagenome. Int. J. Biol. Macromol. 188, 482–490. doi:10.1016/j.ijbiomac.2021.07.153
Deng, J. J., Shi, D., Mao, H., Li, Z., Liang, S., Ke, Y., et al. (2019). Heterologous expression and characterization of an antifungal chitinase (Chit46) from Trichoderma harzianum GIM 3.442 and its application in colloidal chitin conversion. Int. J. Biol. Macromol. 134, 113–121. doi:10.1016/j.ijbiomac.2019.04.177
Dhillon, G. S., Kaur, S., Brar, S. K., and Verma, M. (2013). Green synthesis approach: Extraction of chitosan from fungus mycelia. Crit. Rev. Biotechnol. 33, 379–403. doi:10.3109/07388551.2012.717217
Du, C., Jiang, S., Jiang, S., Zhou, Y., and Zhang, G. (2019). A Bacillus pumilus originated β-N-acetylglucosaminidase for chitin combinatory hydrolysis and exploration of its thermostable mechanism. Int. J. Biol. Macromol. 132, 1282–1289. doi:10.1016/j.ijbiomac.2019.04.054
Du, C., Zhao, X., Song, W., He, N., Jiang, S., Zhou, Y., et al. (2021). Combined strategies to improve the expression of acidic mammalian chitinase in Pichia pastoris for the production of N, N’-diacetylchitobiose. Biochem. Eng. J. 167, 107907. doi:10.1016/j.bej.2020.107907
Duo-Chuan, L. (2006). Review of fungal chitinases. Mycopathologia 161, 345–360. doi:10.1007/s11046-006-0024-y
Fu, X., Yan, Q., Yang, S., Yang, X., Guo, Y., and Jiang, Z. (2014). An acidic, thermostable exochitinase with β-N-acetylglucosaminidase activity from Paenibacillus barengoltzii converting chitin to N-acetyl glucosamine. Biotechnol. Biofuels 7, 174. doi:10.1186/s13068-014-0174-y
Fuciños, P., Atanes, E., López-López, O., Esperanza Cerdán, M., Isabel González-Siso, M., Pastrana, L., et al. (2011). Production and characterization of two N-terminal truncated esterases from Thermus thermophilus HB27 in a mesophilic yeast: Effect of N-terminus in thermal activity and stability. Protein Expr. Purif. 78, 120–130. doi:10.1016/j.pep.2011.04.002
Glaser, F., Pupko, T., Paz, I., Bell, R. E., Bechor-Shental, D., Martz, E., et al. (2003). ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164. doi:10.1093/bioinformatics/19.1.163
Guthrie, J. L., Khalif, S., and Castle, A. J. (2005). An improved method for detection and quantification of chitinase activities. Can. J. Microbiol. 51, 491–495. doi:10.1139/w05-020
Halder, S. K., Jana, A., Paul, T., Das, A., Ghosh, K., Pati, B. R., et al. (2016). Purification and biochemical characterization of chitinase of Aeromonas hydrophila SBK1 biosynthesized using crustacean shell. Biocatal. Agric. Biotechnol. 5, 211–218. doi:10.1016/j.bcab.2015.11.003
Han, P., Yang, C., Liang, X., and Li, L. (2016). Identification and characterization of a novel chitinase with antifungal activity from “Baozhu” pear (Pyrus ussuriensis Maxim.). Food Chem. 196, 808–814. doi:10.1016/j.foodchem.2015.10.006
Han, Y., Yang, B., Zhang, F., Miao, X., and Li, Z. (2009). Characterization of antifungal chitinase from marine Streptomyces sp. DA11 associated with South China sea sponge Craniella australiensis. Mar. Biotechnol. (NY). 11, 132–140. doi:10.1007/s10126-008-9126-5
Hartl, L., Zach, S., and Seidl-Seiboth, V. (2012). Fungal chitinases: Diversity, mechanistic properties and biotechnological potential. Appl. Microbiol. Biotechnol. 93, 533–543. doi:10.1007/s00253-011-3723-3
Horn, S. J., Sørbotten, A., Synstad, B., Sikorski, P., Sørlie, M., Vårum, K. M., et al. (2006). Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 273, 491–503. doi:10.1111/j.1742-4658.2005.05079.x
Hou, F., Ma, X., Fan, L., Wang, D., Ding, T., Ye, X., et al. (2020). Enhancement of chitin suspension hydrolysis by a combination of ultrasound and chitinase. Carbohydr. Polym. 231, 115669. doi:10.1016/j.carbpol.2019.115669
Hou, F., Ma, X., Fan, L., Wang, D., Wang, W., Ding, T., et al. (2019). Activation and conformational changes of chitinase induced by ultrasound. Food Chem. 285, 355–362. doi:10.1016/j.foodchem.2019.01.180
Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., de Castro, E., Langendijk-Genevaux, P. S., et al. (2006). The PROSITE database. Nucleic Acids Res. 34, D227–D230. doi:10.1093/nar/gkj063
Imoto, T., and Yagishita, K. (1971). A simple activity measurement of lysozyme. Agric. Biol. Chem. 35, 1154–1156.
Itoh, T., Hibi, T., Sugimoto, I., Suzuki, F., Fujii, Y., Taketo, A., et al. (2014). Crystallization and preliminary X-ray analysis of the catalytic domains of Paenibacillus sp. strain FPU-7 cell-surface-expressed chitinase ChiW. Acta Crystallogr. F. Struct. Biol. Commun. 70, 350–353. doi:10.1107/S2053230X14002325
Jimtaisong, A., and Saewan, N. (2014). Utilization of carboxymethyl chitosan in cosmetics. Int. J. Cosmet. Sci. 36, 12–21. doi:10.1111/ics.12102
Kamran, A., Ur Rehman, H., Ul Qader, S. A., Baloch, A. H., and Kamal, M. (2015). Purification and characterization of thiol dependent, oxidation-stable serine alkaline protease from thermophilic Bacillus sp. J. Genet. Eng. Biotechnol. 13, 59–64. doi:10.1016/j.jgeb.2015.01.002
Karthik, N., Binod, P., and Pandey, A. (2015). Purification and characterisation of an acidic and antifungal chitinase produced by a Streptomyces sp. Bioresour. Technol. 188, 195–201. doi:10.1016/j.biortech.2015.03.006
Khan, F. I., Rahman, S., Queen, A., Ahamad, S., Ali, S., Kim, J., et al. (2017). Implications of molecular diversity of chitin and its derivatives. Appl. Microbiol. Biotechnol. 101, 3513–3536. doi:10.1007/s00253-017-8229-1
Krolicka, M., Hinz, S. W. A., Koetsier, M. J., Joosten, R., Eggink, G., van den Broek, L. A. M., et al. (2018). Chitinase Chi1 from myceliophthora thermophila C1, a thermostable enzyme for chitin and chitosan depolymerization. J. Agric. Food Chem. 66, 1658–1669. doi:10.1021/acs.jafc.7b04032
Kumar, M. N. V. R. (2000). A review of chitin and chitosan applications. Available at: www.elsevier.com/locate/react.
Kumar, R. A., Sivashanmugam, A., Deepthi, S., Iseki, S., Chennazhi, K. P., Nair, S. v., et al. (2015). Injectable chitin-poly(#-caprolactone)/nano hydroxyapatite composite microgels prepared by simple regeneration technique for bone tissue engineering. Available at: http://pubs.acs.org.
Kuranda, M. J., and Robbins, P. W. (1987). Cloning and heterologous expression of glycosidase genes from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 84, 2585–2589. doi:10.1073/pnas.84.9.2585
Landim, P. G. C., Correia, T. O., Silva, F. D. A., Nepomuceno, D. R., Costa, H. P. S., Pereira, H. M., et al. (2017). Production in Pichia pastoris, antifungal activity and crystal structure of a class I chitinase from cowpea (Vigna unguiculata): Insights into sugar binding mode and hydrolytic action. Biochimie 135, 89–103. doi:10.1016/j.biochi.2017.01.014
Lavall, R. L., Assis, O. B. G., and Campana-Filho, S. P. (2007). β-Chitin from the pens of Loligo sp.: Extraction and characterization. Bioresour. Technol. 98, 2465–2472. doi:10.1016/j.biortech.2006.09.002
Liu, C., Shen, N., Wu, J., Jiang, M., Shi, S., Wang, J., et al. (2020). Cloning, expression and characterization of a chitinase from Paenibacillus chitinolyticus strain UMBR 0002. PeerJ 8, e8964. doi:10.7717/peerj.8964
Mccret, K. J., and Gooday, G. W. (1992). A rapid and sensitive microassay for determination of chitinolytic activity. J. Microbiol. Methods 14, 229–237. doi:10.1016/0167-7012(92)90055-9
Mekasha, S., Tuveng, T. R., Askarian, F., Choudhary, S., Schmidt-Dannert, C., Niebisch, A., et al. (2020). A trimodular bacterial enzyme combining hydrolytic activity with oxidative glycosidic bond cleavage efficiently degrades chitin. J. Biol. Chem. 295, 9134–9146. doi:10.1074/jbc.ra120.013040
Mohamed, S., Bouacem, K., Mechri, S., Addou, N. A., Laribi-Habchi, H., Fardeau, M. L., et al. (2019). Purification and biochemical characterization of a novel acido-halotolerant and thermostable endochitinase from Melghiribacillus thermohalophilus strain Nari2AT. Carbohydr. Res. 473, 46–56. doi:10.1016/j.carres.2018.12.017
Morimoto, K., Karita, S., Kimura, T., Sakka, K., and Ohmiya’, K. (2001). Characterization of Clostridium parapukjkum chitinase A from a recombinant Escherichia coli. J. Biosci. Bioeng. 92, 466–468. doi:10.1016/S1389-1723(01)80297-8
Morris, G. M., Ruth, H., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi:10.1002/jcc.21256
Muzzarelli, Riccardo A. A. (1999). Native, industrial and fossil chitins. Chitin Chitinases 87, 1–6. doi:10.1007/978-3-0348-8757-1_1
Niu, X., Zhou, J. S., Wang, Y. X., Liu, C. C., Liu, Z. H., and Yuan, S. (2017). Heterologous expression and characterization of a novel chitinase (ChiEn1) from Coprinopsis cinerea and its synergism in the degradation of chitin. J. Agric. Food Chem. 65, 6943–6956. doi:10.1021/acs.jafc.7b02278
Peipei, S., Xinyu, Z., Huiling, M., Yan, L., Longbing, Y., and Guo, G. 2021 Enzyme property analysis of its translated product and prokaryotic expres-sion of MDcht2 gene encoding chitinase of Musca domestica.
Pradeep, G. C., Choi, Y. H., Choi, Y. S., Suh, S. E., Seong, J. H., Cho, S. S., et al. (2014). An extremely alkaline novel chitinase from Streptomyces sp. CS495. Process Biochem. 49, 223–229. doi:10.1016/j.procbio.2013.12.002
Rahman, M. A., and Halfar, J. (2014). First evidence of chitin in calcified coralline algae: New insights into the calcification process of Clathromorphum compactum. Sci. Rep. 4, 6162. doi:10.1038/srep06162
Razdan, A., and Pettersson, D. (1994). Effect of chitin and chitosan on nutrient digestibility and plasma lipid concentrations in broiler chickens. Br. J. Nutr. 72, 277–288. doi:10.1079/bjn19940029
Reddy, P., Sarma, P. U., and Muralidhar, K. (1995). A rapid method for the isolation of genomic DNA from Aspergillus fumigatus. Prep. Biochem. 25, 171–181. doi:10.1080/10826069508010119
Ren, X. B., Dang, Y. R., Liu, S. S., Huang, K. X., Qin, Q. L., Chen, X. L., et al. (2022). Identification and characterization of three chitinases with potential in Direct conversion of crystalline chitin into N, N′-diacetylchitobiose. Mar. Drugs 20, 165. doi:10.3390/md20030165
Rigsby, R. E., and Parker, A. B. (2016). Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 44, 433–437. doi:10.1002/bmb.20966
Rush, C. L., Schüttelkopf, A. W., Hurtado-Guerrero, R., Blair, D. E., Ibrahim, A. F. M., Desvergnes, S., et al. (2010). Natural product-guided discovery of a fungal Chitinase inhibitor. Chem. Biol. 17, 1275–1281. doi:10.1016/j.chembiol.2010.07.018
Sahai, A. S., and Manocha, M. S. (1993). Chitinases of fungi and plants: Their in morphogenesis and host-parasite involvement interaction. FEMS Microbiol. Rev. 11, 317–338. doi:10.1111/j.1574-6976.1993.tb00004.x
Saima, , Kuddus, M., Roohi, , and Ahmad, I. Z. (2013). Isolation of novel chitinolytic bacteria and production optimization of extracellular chitinase. J. Genet. Eng. Biotechnol. 11, 39–46. doi:10.1016/j.jgeb.2013.03.001
Shehata, A. N., Abd El Aty, A. A., Darwish, D. A., Abdel Wahab, W. A., and Mostafa, F. A. (2018). Purification, physicochemical and thermodynamic studies of antifungal chitinase with production of bioactive chitosan-oligosaccharide from newly isolated Aspergillus griseoaurantiacus KX010988. Int. J. Biol. Macromol. 107, 990–999. doi:10.1016/j.ijbiomac.2017.09.071
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi:10.1093/molbev/msab120
Tolaimate, A., Desbrières, J., Rhazi, M., Alagui, A., Vincendon, M., and Vottero, P. (2000). On the influence of deacetylation process on the physicochemical characteristics of chitosan from squid chitin. Polymer 41, 2463–2469. doi:10.1016/S0032-3861(99)00400-0
Ueda, J., and Kurosawa, N. (2015). Characterization of an extracellular thermophilic chitinase from Paenibacillus thermoaerophilus strain TC22-2b isolated from compost. World J. Microbiol. Biotechnol. 31, 135–143. doi:10.1007/s11274-014-1754-5
van Aalten, D. M. F., Komander, D., Synstad, B., Gåseidnes, S., Peter, M. G., and Eijsink, V. G. H. (2021). Structural insights into the catalytic mechanism of a family 18 exo-chitinase. PNAS 98, 8979–8984. doi:10.1073/pnas.151103798
Vieille, C., and Zeikus, G. J. (2001). Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43. doi:10.1128/mmbr.65.1.1-43.2001
Wang, S. H., and Chen, J. C. (2005). The protective effect of chitin and chitosan against Vibrio alginolyticus in white shrimp Litopenaeus vannamei. Fish. Shellfish Immunol. 19, 191–204. doi:10.1016/j.fsi.2004.11.003
Wang, Y., Li, J., and Li, B. (2016). Nature-inspired one-step green procedure for enhancing the antibacterial and antioxidant behavior of a chitin film: Controlled interfacial assembly of tannic acid onto a chitin film. J. Agric. Food Chem. 64, 5736–5741. doi:10.1021/acs.jafc.6b01859
Wang, Y. T., and Wu, P. L. (2020). Gene cloning, characterization, and molecular simulations of a novel recombinant chitinase from chitinibacter tainanensis CT01 appropriate for chitin enzymatic hydrolysis. Polym. (Basel) 12, 1648. doi:10.3390/POLYM12081648
Wu, H., Liu, J., Yang, J., Hu, X., and Zhang, H. (2022). Synergistic enzymatic degradation of colloidal chitin and its mechanism. Shipin Kexue/Food Sci. 43, 74–80. doi:10.7506/spkx1002-6630-20210205-106
Wu, Y. L., Wang, S., Yang, D. F., Yang, L. Y., Wang, Q. Y., Yu, J., et al. (2022). The discovery, enzymatic characterization and functional analysis of a newly isolated chitinase from marine-derived fungus Aspergillus fumigatus df347. Mar. Drugs 20 (8), 520. doi:10.3390/md20080520
Xia, G., Jin, C., Zhou, J., Yang, S., Zhang, S., and Jin, C. (2001). A novel chitinase having a unique mode of action from Aspergillus fumigatus YJ-407. Available at: http://www.im.ac.cn/imcas/keylab.html.
Xia, J.-L., Xiong, J., Tao, X. U., Zhang, C.-G., Zhang, R.-Y., Zhang, Q., et al. (2009). Purification and characterization of extracellular chitinase from a novel strain Aspergillus fumigatus CS-01. J. Cent. South Univ. Technol. 16, 552–557. doi:10.1007/s11771−009−0092−5
Xinyue, Z., Yueqi, Z., Fengbiao, W., Jisheng, W., Haodong, N., and Zhipeng, W. (2021). Research progress of microbial chitinase from marine bacteria. Sci. Technol. Food Industry 42, 3832021–3832389. doi:10.13386/j.issn1002-0306.2020070096
Xu, Y., Bajaj, M., Schneider, R., Grage, S. L., Ulrich, A. S., Winter, J., et al. (2013). Transformation of the matrix structure of shrimp shells during bacterial deproteination and demineralization. Available at: http://www.microbialcellfactories.com/content/12/1/90.
Yang, C. Y., Ho, Y. C., Pang, J. C., Huang, S. S., and Tschen, J. S. M. (2009). Cloning and expression of an antifungal chitinase gene of a novel Bacillus subtilis isolate from Taiwan potato field. Bioresour. Technol. 100, 1454–1458. doi:10.1016/j.biortech.2008.07.039
Zhang, A., He, Y., Wei, G., Zhou, J., Dong, W., Chen, K., et al. (2018). Molecular characterization of a novel chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and its use in N-acetyl-d-glucosamine production. Biotechnol. Biofuels 11, 179. doi:10.1186/s13068-018-1169-x
Keywords: chitin, acidic chitinase, N-acetylglucosamine, AlphaFold2, N-terminal truncation, Aspergillus fumigatus, β-N-acetylglucosaminidase
Citation: He B, Yang L, Yang D, Jiang M, Ling C, Chen H, Ji F and Pan L (2022) Biochemical purification and characterization of a truncated acidic, thermostable chitinase from marine fungus for N-acetylglucosamine production. Front. Bioeng. Biotechnol. 10:1013313. doi: 10.3389/fbioe.2022.1013313
Received: 06 August 2022; Accepted: 22 August 2022;
Published: 04 October 2022.
Edited by:
Wei Zeng, Guilin Medical University, ChinaReviewed by:
Mingbo Qu, Dalian University of Technology, ChinaXiaoming Zhao, Shanxi University, China
Copyright © 2022 He, Yang, Yang, Jiang, Ling, Chen, Ji and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailan Chen, aGxjaGVuMzE5QDE2My5jb20=; Feng Ji, MzI5ODQyMjQxQHFxLmNvbQ==; Lixia Pan, cGFubGl4aWFAZ3hhcy5jbg==
†These authors have contributed equally to this work
 Bin He
Bin He Liyan Yang
Liyan Yang Dengfeng Yang
Dengfeng Yang Minguo Jiang3
Minguo Jiang3 Hailan Chen
Hailan Chen Lixia Pan
Lixia Pan