
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol., 06 September 2022
Sec. Biomaterials
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.1002967
This article is part of the Research TopicInsights In Biomaterials 2022 / 2023 - Novel Developments, Current Challenges, and Future PerspectivesView all 13 articles
Cells sense their environment through the cell membrane receptors. Interaction with extracellular ligands induces receptor clustering at the nanoscale, assembly of the signaling complexes in the cytosol and activation of downstream signaling pathways, regulating cell response. Nanoclusters of receptors can be further organized hierarchically in the cell membrane at the meso- and micro-levels to exert different biological functions. To study and guide cell response, cell culture substrates have been engineered with features that can interact with the cells at different scales, eliciting controlled cell responses. In particular, nanoscale features of 1–100 nm in size allow direct interaction between the material and single cell receptors and their nanoclusters. Since the first “contact guidance” experiments on parallel microstructures, many other studies followed with increasing feature resolution and biological complexity. Here we present an overview of the advances in the field summarizing the biological scenario, substrate fabrication techniques and applications, highlighting the most recent developments.
The compartmentalization of cellular functions is a ubiquitous strategy to increase efficiency, providing spatio-temporally discrete domains for dynamic processes to take place simultaneously, in close vicinity, and without interfering with each other. The plasma membrane is generally accepted as being compartmentalized (Garcia-Parajo et al., 2014; Nicolson, 2014). This characteristic emerges from the temporary limitation of lateral diffusion, promoting confinement and allowing lipids and proteins to be organized in specific locations of variable size and composition (Kusumi et al., 1993; Jacobson et al., 2019). Restrictions in lateral diffusion of membrane components have been mainly attributed to their association to the underlying cytoskeleton as described by the “membrane skeleton fence model”, in which fences or corrals are defined by transmembrane proteins acting as posts linked to the cytoskeletal structures (Kusumi and Sako, 1996; Kusumi et al., 2012). Also, the presence of extracellular lattices (Lajoie et al., 2009), or specific lipid domains or “lipid rafts” (van Meer et al., 1987) can create regions of restricted lateral diffusion in the plasma membrane. This compartmentalization is hierarchically organized from small nanoclusters of dynamic small protein oligomers (of 3–10 nm diameter) and lipid rafts (2–20 nm) to actin-cytoskeletal fence domains (40–300 nm) up to bigger domains of several micrometers, thereby allowing the multiscale regulation of the membrane protein function (Kusumi et al., 2011) (Figure 1A).
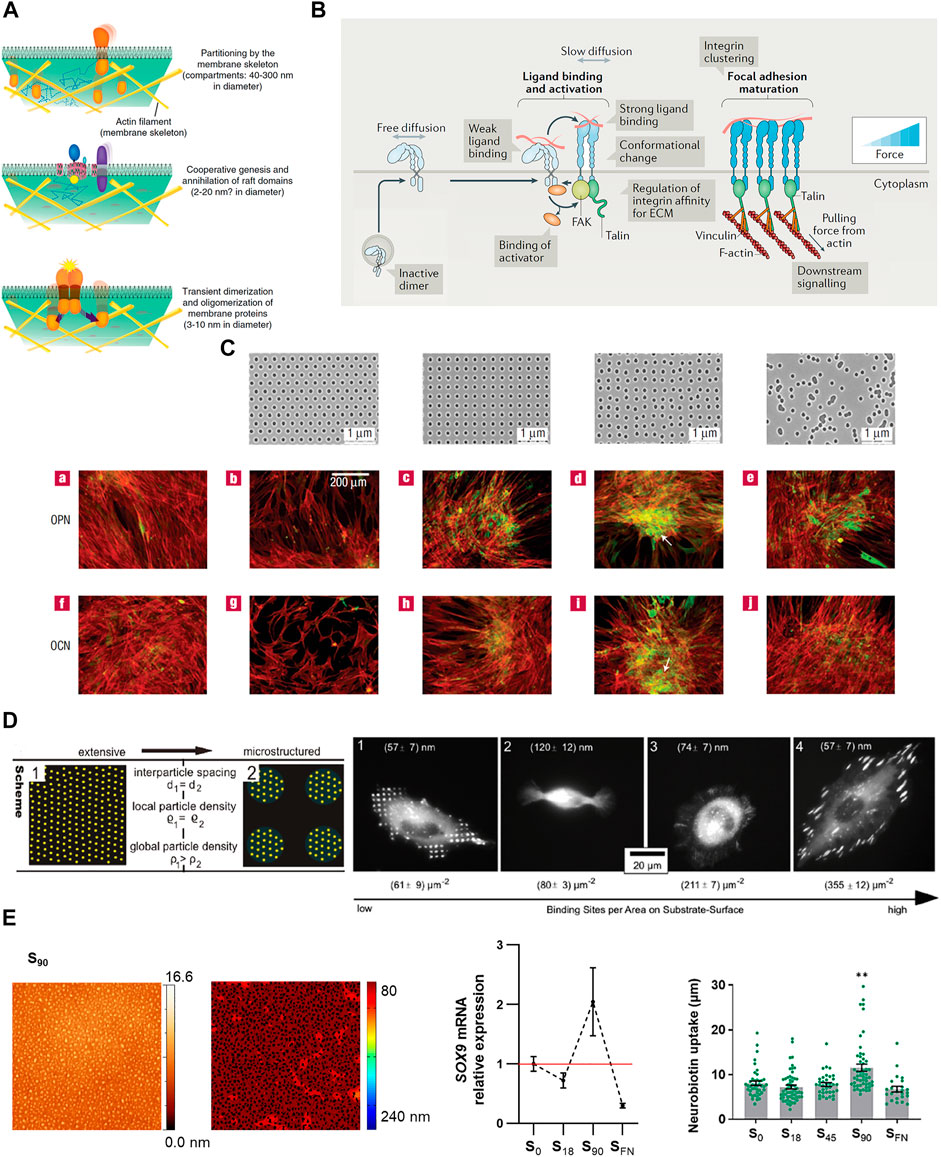
FIGURE 1. (A) Hierarchical organization of cell membrane compartments. Reprinted with permission from Kusumi et al., 2011. Copyright 2011 Elsevier. (B) Receptor nanoclustering: Ligand-mediated integrin clustering initiates the recruitment of adaptor proteins at FAs, leading to cytoskeleton engagement, force transmission and downstream signaling activation. Adapted with permission from Kechagia et al., 2019. Copyright 2019 Springer Nature. (C) Osteoprogenitor differentiation (osteopontin (OCP) and osteocalcin (OCN) expression, and bone nodule formation (white arrows)) on nanotopographies with different levels of disorder, fabricated by EBL. Reprinted with permission from Dalby et al., 2007b. Copyright 2007 Springer Nature. (D) BCML combined with photolithography were used to create micro- and nanopatterned surfaces of the cell adhesive ligand cyclic-(RGDfK). The development of stable FAs, number, size and adhesion strength is more influenced by local than global ligand density. Adapted with permission from Deeg et al., 2011. Copyright 2011 American Chemical Society. (E) The nanopatterning of RGD functionalized dendrimers revealed a threshold nanopattern configuration to induce cell response promoting chondrogenic differentiation and enhancing GJIC. Adapted from Casanellas et al., 2020 and Reprinted with permission Casanellas et al., 2022. Copyright 2022 Future Medicine Ltd.
The cellular microenvironment is also organized at the nanoscale, as seen for collagen, the main structural protein in the extracellular matrix (ECM). Collagen I is assembled by three peptide chains of collagen that conform a helical structure of around 1.5 nm diameter and 300 nm length, which then organize into microfibrils with a cross-section of around 3 × 5 nm (Jiang et al., 2004). The ECM protein fibronectin forms bundles of fibrils, in which the average span of a fibronectin molecule (a dimer of two polynucleotides) in each fibronectin fibril is ∼92 nm (Früh et al., 2015). This favors ligand interaction with the receptors at the cell membrane in a particular configuration that is confined to the nanometer scale.
Therefore, nanotopography represents an effective physical approach for studies on cell behavior mediated by cell-cell environment interactions. Nanotopographies (1–100 nm) lie in the same scale range as many ECM proteins, allowing the direct interaction of the material with single cell receptors and their nanoclusters. Nanoscale surface topography affects cellular and tissue responses, including adhesion, migration, growth, morphogenesis, and differentiation (Martínez et al., 2009; Luo et al., 2022).
Specific protein-protein and protein-lipid interactions promote oligomerization, aiding the formation of signaling complexes at the cell membrane. Glycosylphosphatidyl-anchored proteins (GPI-APs) are a class of soluble proteins attached to the external side of the plasma membrane. They form small clusters of up to four molecules (<5 nm) stabilized in sphingolipid- and cholesterol-dependent domains or rafts. These lipid rafts act as sorting platforms for the GPI-APs selective delivery to the apical membrane in polarized epithelial cells, where they exert specialized functions (Zurzolo and Simons, 2016). Besides lipid-linked proteins, many transmembrane proteins also cluster to exert their functions.
The clustering of transmembrane receptors is common among different types of immune cells (Dustin and Groves, 2012; Li and Yu, 2021). T cell receptors (TCRs) on resting T cells can be found as monomers and as cholesterol- and sphingomyelin-stabilized nanoclusters (<10%) containing 2–30 TCRs each (Molnár et al., 2012). Upon activation of TCRs, they assemble into larger clusters of ten up to hundreds of receptors, which recruit kinases and adaptor proteins including Lck, ZAP-70, Lat, and SLP76. These microclusters initiate and sustain TCR signaling at the immunological synapse. Moreover, TCR microclusters associate and are transported by cortical F-actin flows over micrometer distances along the synapse (Dustin and Groves, 2012; Yi et al., 2019; Balagopalan et al., 2021). The enrichment of oligomeric TCRs has been reported to increase the sensitivity of memory T cells compared to naïve T cells (Kumar et al., 2011). A similar hierarchical organization has been described for integrin receptors. Integrins are transmembrane receptors that mediate cell-cell and cell-extracellular matrix (ECM) adhesion (Kechagia et al., 2019). However, integrin binding alone is insufficient to elicit full adhesion. Instead, upon ligand binding, integrin receptors arrange into nanoclusters that build tension through the recruitment of adaptor proteins such as paxillin, vinculin, talin, FAK or SRC, cytoskeletal engagement into focal adhesions (FAs) and downstream signaling activation (Hu et al., 2015; Kechagia et al., 2019) (Figure 1B). Remarkably, within FAs, active and inactive β1 integrins segregate into different nanoclusters, thus suggesting integrin activity is not only regulated at the monomeric level but is subjected to collective or coordinate regulation at the level of the nanoclusters (Spiess et al., 2018). As integrins, Eph tyrosine kinase receptors cluster upon the interaction with their ligand, ephrin, which is presented on the surface of neighboring cell membranes. During development, Eph receptors act as positional cues in tissue patterning by regulating cell adhesion and repulsion. Ephrin ligands presented as concentration gradients guide axonal patterning in retinotectal mapping and stem cell migration in the developing intestines (Klein, 2012). Activation of Eph receptors occurs immediately after ligand interaction, inducing receptor polymerization. Maximum receptor activation is reached on clusters of five to eight receptors, after which oligomers cannot grow further by recruiting more monomers and instead, they grow through the condensation of oligomers into larger complexes that dampen the signaling. These polymerization–condensation dynamics provide a framework for the mechanism by which cells properly respond to variable concentrations and gradients of the ephrin ligand (Ojosnegros et al., 2017).
From the first contact guidance experiments (Curtis and Varde, 1964), microfabrication techniques first developed for the electronics industry came into use to produce micro- and nanopatterned surfaces for cell studies, with high feature resolution and increased biological complexity. Fabrication methods to produce controlled nanotopographies on cell culture substrates mainly rely on the use of lithographic techniques, which in general require specialized equipment and skilled personnel, thus limiting their widespread application. The later introduction of the directed self-assembly techniques to produce nanostructured surfaces greatly facilitated the implementation of nanotopography production in general wet labs. Here we summarize the main fabrication techniques used to generate nanotopographical substrates, and their characteristics.
Photolithography or optical lithography is a patterning technique in which a light-sensitive chemical (photoresist) coated on the substrate is selectively exposed to light through a mask. The photoresist either collapses or hardens in the regions exposed to light and the pattern emerges on the substrate by dissolving the softer parts of the coating, which can subsequently be transferred to the substrate material. The wavelength of the light used determines the minimum feature size that can be impressed on the photoresist: The use of incoherent, vacuum ultraviolet (VUV) radiation of 172 nm allowed the production of nanoscale features with a minimum lateral feature size of 350 nm (Mironov et al., 2020). To overcome the resolution limitations, surface plasmon polaritons (SPPs), able to surpass the diffraction-limits, have been used for the fabrication of nanopatterns with a half-pitch resolution of less than 15 nm (Luo and Ishihara, 2004; Dong et al., 2014).
In electron beam lithography or e-beam lithography (EBL), a focused beam of electrons is applied (direct-writing) on an electron sensitive coating on a substrate (Lercel et al., 1994). This is a maskless lithography technique in which custom nanopatterns can be transferred to a substrate with up to 3–5 nm resolution (Ermis et al., 2018). Like photolithography, the coating is degraded or crosslinked upon exposure and after a development process, patterns are revealed. While conventional lithography mostly relies on flat wafer-base processing, EBL can be applied on curved surfaces (Lee et al., 2019). However, compared to photolithography, only small areas can be patterned at a time and the equipment manipulation is tough, which makes the whole process significantly slower. Ion beam lithography (IBL) or focused ion beam lithography (FIBL) uses a narrow scanning ion beam source (typically of gallium ions) instead of a focused beam of electrons to pattern a resist. Compared to EBL, IBL offers higher resolution due to ions have much heavier mass than electrons and more momentum, thus leading to smaller wavelengths and reducing diffraction, but also minimizing the back scattering and radiation towards sensitive resists (Joshi-Imre and Bauerdick, 2014; Li et al., 2021).
SPL approaches are a set of maskless nanolithographic techniques based on the ability of scanning probe microscopy to create variable surface patterns either by adsorption, nanoshaving and/or nanografting (Rosa and Liang, 2009). They include dip-pen nanolithography (DPN), fluidic force microscopy (FluidFM), and polymer pen lithography (PPL), among others. DPN was pioneered by the group of Prof. Mirkin (Piner et al., 1999; Salaita et al., 2007), where an AFM tip is used to create patterns of 15–100 nm by direct writing on the substrate. A molecular ink is transferred from the atomic force microscope (AFM) tip to the substrate by the spontaneous formation of a water meniscus, which is facilitated by the ambient conditions. DPN can work in sequential or parallel modes (multiplexed DPN), where parallel DPN tip arrays are scanned on the substrate simultaneously, thus significantly improving the throughput limitations of the technique (Ma et al., 2018). FluidFM introduces microfluidic channels (300 nm-8 µm) into the AFM probes allowing to dispense volumes of fluid that can be below the femtoliter range. The patterns are created when the nanopipette contacts the surface and the ink is released from the probe with a short pressure pulse (few hPa) (Zambelli et al., 2018). Alternatively, a cantilever-free scanning probe molecular printing technology referred as polymer pen nanolithography (PPL) was introduced to overcome the throughput issues and the use of complicated pen arrays (Huo et al., 2008). Since the SPL techniques work under mild conditions, they allow patterning sensitive compounds such as DNA, proteins, lipids, viruses and even polymers for 3D additive manufacturing (Liu et al., 2022).
Nanoimprint lithography (NIL) is a simple and low-cost lithography technique in which a pattern is transferred by mechanical deformation of a polymer resist from a previously nanostructured mold (created by photolithography or EBL). The transfer of the nanopattern can be conducted in several ways: by thermocompression using high temperatures to soften the polymer resist while pressing it with the stamp, also known as hot embossing lithography, or by using UV light to cross-link and harden a soft polymer resist during the imprint (UV-NIL). UV-NIL requires the substrate and/or stamp to be transparent to UV wavelengths (Modaresialam et al., 2021).
First introduced by Bain and Whitesides in 1989 (Bain and Whitesides, 1989), soft lithography refers to a number of techniques that use elastomeric stamps (typically of polydimethylsiloxane (PDMS)) previously cast on a master, to transfer micro- or nanopatterns to a substrate. It includes replica molding, microcontact printing, micromolding in microcapillaries, microtransfer molding, and solvent-assisted micromolding. Soft lithography is a low-cost technique that does not require stringent control over the environment (such as a clean-room facilities), thus being accessible to general wet labs (Qin et al., 2010).
Supramolecular chemistry can be used as a bottom-up approach to achieve nanopatterned surfaces based on the self-organization of molecular scale architectures, allowing precision on the nanofeature position. Compared to the self-assembly of small molecules, polymers offer higher stability and durability due to their mechanical and physical properties. Self-assembly of block copolymers (BCPs) has attracted considerable attention in nanoscience due to its ability to self-assemble both in bulk and in solution into different types of nanostructures through the repulsion of their immiscible blocks (Mai and Eisenberg, 2012). BCP micelle nanolithography (BCML) has been extensively used to generate ordered and disordered nanopatterns of gold nanoclusters on surfaces with well-controlled interparticle distances (Glass et al., 2003). Dendrimers, presenting a highly branched and easily tunable size and chemical structure, have been used to create nanopatterns with a liquid-like order on low charged surfaces (Lagunas et al., 2014), and DNA and peptides have been used to build nanostructures presenting multiple epitopes with nanoscale spatial control (Stephanopoulos et al., 2015; Wang et al., 2021).
The first visible phenomena of nanostructures-cell interaction are the changes in cell adhesion, spreading and morphology, which provide cues to predict cellular functions. Studies have explored the influence of different nanotopographies on the adhesive/spreading behavior of various cell types, some of them summarized in Table 1. Nanotopography has been vastly employed to control cell differentiation with especial emphasis on enhancing tissue integration in bone implants (Chen et al., 2018). Due to the easy manufacturing, first attempts were conducted by using the surface roughness strategy. However, the lack of control in the produced structures, and poor reproducibility, prompted the use of lithographic techniques to fabricate nanostructured biocompatible materials that promote osteointegration (Figure 1C and Table 1).
In many cases, the assembly of membrane receptors into fully functional microcomplexes requires of both ligand occupancy and receptor clustering. Spatz and coworkers used BCML to create ordered gold nanopatterns coated with the integrin receptor ligand arginine-glycine-aspartic acid (RGD), present in many ECM proteins. The gold nanodots, of less than 8 nm in diameter, allowed the binding of one integrin per dot and were positioned at different interdot spacings on a non-fouling substrate. Authors observed that a ligand spacing of more than 73 nm impairs integrin clustering, cell adhesion and spreading, and dramatically reduces the formation of FAs and actin stress fibers (Arnold et al., 2004). Since this seminal work, BCML has been used in a number of cell studies, showing the prevalence of local over global ligand density (Deeg et al., 2011) (Figure 1D and Table 1), and that integrin clustering influences many aspects of cell behavior, including cell differentiation (Table 1). More recently, Roca-Cusachs and coworkers used BCML to create cell adhesive nanopatterns on substrates of different rigidity, and they found that the optimal ligand spacing for cell adhesion increases as substrate stiffness decreases (Oria et al., 2017) (Table1).
The multivalent interactions between ligand and receptors, in which the simultaneous binding of multiple ligands on receptor complexes takes place (Kiessling et al., 2006), have been extensively used to study receptor clustering and the downstream signaling in cells. Self-assembled diblock copolymers of polystyrene-blockpoly(methyl methacrylate) (PS-b-PMMA) were used to produce nanopatterned substrates able to establish multivalent interactions between surface-bound ephrinB1 ligands and membrane EphB2 receptors. The preclustering of ephrinB1 ligands in the nanopatterns resulted in a more efficient and faster receptor oligomerization kinetics compared to the traditional cross-linked ligand presentation (Hortigüela et al., 2018). Also, dendrimer nanopatterning of RGD-functionalized dendrimers was used to study the effects of the local RGD ligand density on the adhesion, differentiation, and gap junction intercellular communication (GJIC) of mesenchymal stem cells (Lagunas et al., 2017; Casanellas et al., 2020; Casanellas et al., 2022) (Figure 1E).
Nanoscale cell-environmental interactions regulate cell behavior. Nanotopography produced by lithographic techniques and/or by the self-assembly of molecular scale architectures effectively mimics those interactions, helping to direct particular cell responses and providing information about the underlying mechanisms. We expect that further advances in the field by including stimuli-responsive materials, combined with super-resolution microscopies, will bring more detailed information on the molecular mechanisms that direct cell function, unveiling traits that are normally hidden by the ensemble average in bulk experiments. This will provide an otherwise unavailable insight on the cell interactions at the nanoscale so that they can be used to systematically drive cell responses by fabricating the appropriate nanotopographical substrates, with potential applications in translational medicine.
AL: defined the focus of the review, writing, reviewing, and editing. IC and JS: writing, reviewing, and editing.
This work was supported by the Biomedical Research Networking Center (CIBER), Spain. CIBER is an initiative funded by the VI National R&D&i Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions, and the Instituto de Salud Carlos III, with the support of the European Regional Development Fund (ERDF). This work was funded by the CERCA Program and by the Commission for Universities and Research of the Department of Innovation, Universities, and Enterprise of the Generalitat de Catalunya (2017 SGR 1079). IC was supported by the Spanish Ministry of Science through the Subsidies for Predoctoral Contracts for the Training of Doctors open call, co-funded by the European Social Fund, grant number: BES-2016-076682.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allan, C., Ker, A., Smith, C. -A., Tsimbouri, P. M., Borsoi, J., O’Neill, S., et al. (2018). Osteoblast response to disordered nanotopography. J. Tissue Eng. 9, 204173141878409–7. doi:10.1177/2041731418784098
Altrock, E., Muth, C. A., Klein, G., Spatz, J. P., and Lee-Thedieck, C. (2012). The significance of integrin ligand nanopatterning on lipid raft clustering in hematopoietic stem cells. Biomaterials 33 (11), 3107–3118. doi:10.1016/j.biomaterials.2012.01.002
Bain, C. D., and Whitesides, G. M. (1989). Modeling organic surfaces with self-assembled monolayers. Angew. Chem. Int. Ed. Engl. 28 (4), 522–528. doi:10.1002/ange.19891010446
Balagopalan, L., Raychaudhuri, K., and Samelson, L. E. (2021). Microclusters as T Cell signaling hubs: Structure, kinetics, and regulation. Front. Cell Dev. Biol. 8, 608530. doi:10.3389/fcell.2020.608530
Casanellas, I., Lagunas, A., Vida, Y., Pérez-Inestrosa, E., Andrades, J. A., Becerra, J., et al. (2020). The janus role of adhesion in chondrogenesis. Int. J. Mol. Sci. 21 (15), 5269. doi:10.3390/ijms21155269
Casanellas, I., Lagunas, A., Vida, Y., Pérez-Inestrosa, E., Rodríguez-Pereira, C., Magalhaes, J., et al. (2022). Nanoscale ligand density modulates gap junction intercellular communication of cell condensates during chondrogenesis. Nanomedicine (Lond.) 17 (11), 775–791. doi:10.2217/nnm-2021–0399
Chen, S., Guo, Y., Liu, R., Wu, S., Fang, J., Huang, B., et al. (2018). Tuning surface properties of bone biomaterials to manipulate osteoblastic cell adhesion and the signaling pathways for the enhancement of early osseointegration. Colloids Surfaces B Biointerfaces 164, 58–69. doi:10.1016/j.colsurfb.2018.01.022
Curran, J. M., Stokes, R., Irvine, E., Graham, D., Amro, N. A., Sanedrin, R. G., et al. (2010). Introducing dip pen nanolithography as a tool for controlling stem cell behaviour: Unlocking the potential of the next generation of smart materials in regenerative medicine. Lab. Chip 10 (13), 1662–1670. doi:10.1039/C004149A
Curtis, A., and Varde, M. (1964). Control of cell behavior: Topological factors. J. Natl. Cancer Inst. 33 (1), 15–26. doi:10.1093/jnci/33.1.15
Dalby, M. J., Biggs, M. J. P., Gadegaard, N., Kalna, G., Wilkinson, C. D. W., and Curtis, A. S. G. (2007a). Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J. Cell. Biochem. 100 (2), 326–38. doi:10.1002/jcb.21058
Dalby, M. J., Gadegaard, N., Tare, R., Andar, A., Riehle, M. O., Herzyk, P., et al. (2007b). The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mat. 6, 997–1003. doi:10.1038/nmat2013
Deeg, J. A., Louban, I., Aydin, D., Selhuber-Unkel, C., Kessler, H., and Spatz, J. P. (2011). Impact of local versus global ligand density on cellular adhesion. Nano Lett. 11 (4), 1469–1476. doi:10.1021/nl104079r
Dong, J., Liu, J., Kang, G., Xie, J., and Wang, Y. (2014). Pushing the resolution of photolithography down to 15 nm by surface plasmon interference. Sci. Rep. 4, 5618. doi:10.1038/srep05618
Dustin, M. L., and Groves, J. T. (2012). Receptor signaling clusters in the immune synapse. Annu. Rev. Biophys. 41, 543–556. doi:10.1146/annurev-biophys-042910-155238
Ermis, M., Antmen, E., and Hasirci, V. (2018). Micro and nanofabrication methods to control cell-substrate interactions and cell behavior: A review from the tissue engineering perspective. Bioact. Mat. 3 (3), 355–369. doi:10.1016/j.bioactmat.2018.05.005
Früh, S. M., Schoen, I., Ries, J., and Vogel, V. (2015). Molecular architecture of native fibronectin fibrils. Nat. Commun. 6, 7275. doi:10.1038/ncomms8275
Garcia-Parajo, M. F., Cambi, A., Torreno-Pina, J. A., Thompson, N., and Jacobson, K. (2014). Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci. 127 (23), 4995–5005. doi:10.1242/jcs.146340
Glass, R., Möller, M., and Spatz, J. P. (2003). Block copolymer micelle nanolithography. Nanotechnology 14 (10), 1153–1160. doi:10.1088/0957-4484/14/10/314
Hortiguela, V., Larranaga, E., Cutrale, F., Seriola, A., García-Díaz, M., Lagunas, A., et al. (2018). Nanopatterns of surface-bound EphrinB1 produce multivalent Ligand−Receptor interactions that tune EphB2 receptor clustering. Nano Lett. 18 (1), 629–637. doi:10.1021/acs.nanolett.7b04904
Hu, S., Tee, Y. -H., Kabla, A., Zaidel-Bar, R., Bershadsky, A., and Hersen, P. (2015). Structured illumination microscopy reveals focal adhesions are composed of linear subunits. Cytoskeleton 72, 235–245. doi:10.1002/cm.21223
Huang, J., Gräter, S. V., Corbellini, F., Rinck, S., Bock, E., Kemkemer, R., et al. (2009). Impact of order and disorder in RGD nanopatterns on cell adhesion. Nano Lett. 9 (3), 1111–1116. doi:10.1021/nl803548b
Huo, F., Zheng, Z., Zheng, G., Giam, L. R., Zhang, H., and Mirkin, C. A. (2008). Polymer pen lithography. Science 321 (5896), 1658–1660. doi:10.1126/science.1162193
Jacobson, K., Liu, P., and Lagerholm, B. C. (2019). The lateral organization and mobility of plasma membrane components. Cell 177 (4), 806–819. doi:10.1016/j.cell.2019.04.018
Jiang, F., Hörber, H., Howard, J., and Müller, D. J. (2004). Assembly of collagen into microribbons: Effects of pH and electrolytes. J. Struct. Biol. X. 148 (3), 268–278. doi:10.1016/j.jsb.2004.07.001
Joshi-Imre, A., and Bauerdick, S. (2014). Direct-write ion beam lithography. J. Nanotechnol. 2014, 170415. doi:10.1155/2014/17041
Kechagia, J. Z., Ivaska, J., and Roca- Cusachs, P. (2019). Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20 (8), 457–473. doi:10.1038/s41580-019-0134-2
Kiessling, L. L., Gestwicki, J. E., and Strong, L. E. (2006). Synthetic multivalent ligands as probes of signal transduction. Angew. Chem. Int. Ed. 45 (15), 2348–2368. doi:10.1002/anie.200502794
Klein, R. (2012). Eph/ephrin signalling during development. Development 139, 4105–4109. doi:10.1242/dev.074997
Kumar, R., Ferez, M., Swamy, M., Arechaga, I., Rejas, M. T., Valpuesta, J. M., et al. (2011). Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity 35 (3), 375–387. doi:10.1016/j.immuni.2011.08.010
Kusumi, A., Fujiwara, T. K., Chadda, R., Xie, M., Tsunoyama, T. A., Kalay, Z., et al. (2012). Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of singer and nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 28, 215–250. doi:10.1146/annurev-cellbio-100809-151736
Kusumi, A., and Sako, Y. (1996). Cell surface organization by the membrane skeleton. Curr. Opin. Cell Biol. 8 (4), 566–574. doi:10.1016/s0955-0674(96)80036-6
Kusumi, A., Sako, Y., and Yamamoto, M. (1993). Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65 (5), 2021–2040. doi:10.1016/S0006-3495(93)81253-0
Kusumi, A., Suzuki, K. G., Kasai, R. S., Ritchie, K., and Fujiwara, T. K. (2011). Hierarchical mesoscale domain organization of the plasma membrane. Trends biochem. Sci. 36 (11), 604–615. doi:10.1016/j.tibs.2011.08.001
Lagunas, A., Castaño, A. G., Artés, J. M., Vida, Y., Collado, D., Pérez-Inestrosa, E., et al. (2014). Large-scale dendrimer-based uneven nanopatterns for the study of local arginine–glycine–aspartic acid (RGD) density effects on cell adhesion. Nano Res. 7 (3), 399–409. doi:10.1007/s12274-014-0406-2
Lagunas, A., Tsintzou, I., Vida, Y., Collado, D., Pérez-Inestrosa, E., Rodríguez Pereira, C., et al. (2017). Tailoring RGD local surface density at the nanoscale toward adult stem cell chondrogenic commitment. Nano Res. 10 (6), 1959–1971. doi:10.1007/s12274-016-1382-5
Lajoie, P., Goetz, J. G., Dennis, J. W., and Nabi, I. R. (2009). Lattices, rafts, and scaffolds: Domain regulation of receptor signaling at the plasma membrane. J. Cell Biol. 185 (3), 381–385. doi:10.1083/jcb.200811059
Lee, C. R., Ok, J. G., and Jeong, M. Y. (2019). Nanopatterning on the cylindrical surface using an E-beam pre-mapping algorithm. J. Micromech. Microeng. 29, 015004. doi:10.1088/1361-6439/aaea48
Lercel, M. J., Redinbo, G. F., Pardo, F. D., Rooks, M., Tiberio, R. C., Simpson, P., et al. (1994). Electron beam lithography with monolayers of alkylthiols and alkylsiloxanes. J. Vac. Sci. Technol. B 12, 3663–3667. doi:10.1116/1.587635
Li, M., and Yu, Y. (2021). Innate immune receptor clustering and its role in immune regulation. J. Cell Sci. 134 (4), jcs249318. doi:10.1242/jcs.249318
Li, P., Chen, S., Dai, H., Yang, Z., Chen, Z., Wang, Y., et al. (2021). Recent advances in focused ion beam nanofabrication for nanostructures and devices: Fundamentals and applications. Nanoscale 13, 1529–1565. doi:10.1039/d0nr07539f
Li, Z., Cao, B., Wang, X., Ye, K., Li, S., and Ding, J. (2015). Effects of RGD nanospacing on chondrogenic differentiation of mesenchymal stem cells. J. Mat. Chem. B 3, 5197–5209. doi:10.1039/C5TB00455A
Liu, G., Rong, M., Hu, H., Chen, L., Xie, Z., and Zheng, Z. (2022). 3D dip-pen nanolithography. Adv. Mat. Technol. 7, 2101493. doi:10.1002/admt.202101493
Luo, J., Walker, M., Xiao, Y., Donnelly, H., Dalby, M. J., and Salmeron-Sanchez, M. (2022). The influence of nanotopography on cell behaviour through interactions with the extracellular matrix – a review. Bioact. Mat. 15, 145–159. doi:10.1016/j.bioactmat.2021.11.024
Luo, X., and Ishihara, T. (2004). Subwavelength photolithography based on surface-plasmon polariton resonance. Opt. Express 12 (14), 3055–3065. doi:10.1364/OPEX.12.003055
Ma, H., Jiang, Z., Xie, X., Huang, L., and Huang, W. (2018). Multiplexed biomolecular arrays generated via parallel dip-pen nanolithography. ACS Appl. Mat. Interfaces 10 (30), 25121–25126. doi:10.1021/acsami.8b07369
Mai, Y., Eisenberg, A., Lin, J., Zhang, L., Cai, C., Yao, Y., et al. (2012). Self-assembly of rod-coil block copolymers on a substrate into micrometer-scale ordered stripe nanopatterns. Polym. Chem. 11 (47), 7487–7496. doi:10.1039/D0PY01404D
Martínez, E., Lagunas, A., Mills, C. A., Rodríguez-Seguí, S., Estévez, M., Oberhansl, S., et al. (2009). Stem cell differentiation by functionalized micro- and nanostructured surfaces. Nanomedicine (Lond) 4 (1), 65–82. doi:10.2217/17435889.4.1.65
Medda, R., Helth, A., Herre, P., Pohl, D., Rellinghaus, B., Perschmann, N., et al. (2014). Investigation of early cell-surface interactions of human mesenchymal stem cells on nanopatterned β-type titanium-niobium alloy surfaces. Interface Focus 4, 20130046. doi:10.1098/rsfs.2013.0046
Mironov, A. E., Kim, J., Huang, Y., Steinforth, A. W., Sievers, D. J., and Eden, J. G. (2020). Photolithography in the vacuum ultraviolet (172 nm) with sub-400 nm resolution: Photoablative patterning of nanostructures and optical components in bulk polymers and thin films on semiconductors. Nanoscale 12 (32), 16796–16804. doi:10.1039/d0nr04142d
Modaresialam, M., Chehadi, Z., Bottein, T., Abbarchi, M., and Grosso, D. (2021). Nanoimprint lithography processing of inorganic-based materials. Chem. Mat. 33 (14), 5464–5482. doi:10.1021/acs.chemmater.1c00693
Molnár, E., Swamy, M., Holzer, M., Beck-García, K., Worch, R., Thiele, C., et al. (2012). Cholesterol and sphingomyelin drive ligand-independent T-cell antigen receptor nanoclustering. J. Biol. Chem. 287 (51), 42664–42674. doi:10.1074/jbc.M112.386045
Nicolson, G. L. (2014). The fluid—mosaic model of membrane structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochimica Biophysica Acta - Biomembr. 1838 (6), 1451–1466. doi:10.1016/j.bbamem.2013.10.019
Ojosnegros, S., Cutrale, F., Rodrígueza, D., Otterstrom, J. J., Chiu, C. L., Hortigüela, V., et al. (2017). Eph-ephrin signaling modulated by polymerization and condensation of receptors. Proc. Natl. Acad. Sci. U. S. A. 114 (50), 13188–13193. doi:10.1073/pnas.1713564114
Oria, R., Wiegand, T., Escribano, J., Elosegui-Artola, A., Uriarte, J. J., Moreno-Pulido, C., et al. (2017). Force loading explains spatial sensing of ligands by cells. Nature 552, 219–224. doi:10.1038/nature24662
Piner, R. D., Zhu, J., Xu, F., Hong, S., and Mirkin, C. A. (1999). Dip-Pen" nanolithography. Science 283 (5402), 661–663. doi:10.1126/science.283.5402.661
Qin, D., Xia, Y., and Whitesides, G. M. (2010). Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 5, 491–502. doi:10.1038/nprot.2009.234
Rosa, L. G., and Liang, J. (2009). Atomic force microscope nanolithography: Dip-pen, nanoshaving, nanografting, tapping mode, electrochemical and thermal nanolithography. J. Phys. Condens. Matter 21 (48), 483001. doi:10.1088/0953-8984/21/48/483001
Salaita, K., Wang, Y., and Mirkin, C. A. (2007). Applications of dip-pen nanolithography. Nat. Nanotechnol. 2 (3), 145–55. doi:10.1038/nnano.2007.39
Spiess, M., Hernandez-Varas, P., Oddone, A., Olofsson, H., Blom, H., Waithe, D., et al. (2018). Active and inactive β1 integrins segregate into distinct nanoclusters in focal adhesions. J. Cell Biol. 217 (6), 1929–1940. doi:10.1083/jcb.201707075
Stephanopoulos, N., Freeman, R., North, H. A., Sur, S., Jeong, S. J., Tantakitti, F., et al. (2015). Bioactive DNA-peptide nanotubes enhance the differentiation of neural stem cells into neurons. Nano Lett. 15 (1), 603–609. doi:10.1021/nl504079q
van Meer, G., Stelzer, E. H., Wijnaendts-van-Resandt, R. W., and Simons, K. (1987). Sorting of sphingolipids in epithelial (Madin-Darby canine kidney) cells. J. Cell Biol. 105 (4), 1623–1635. doi:10.1083/jcb.105.4.1623
Wang, K., Park, S. H., Zhu, J., Kim, J. K., Zhang, L., and Yi, G. -R. (2021). Self-assembled colloidal nanopatterns toward unnatural optical meta-materials. Adv. Funct. Mat. 31, 2008246. doi:10.1002/adfm.202008246
Wang, X., Li, S., Yan, C., Liu, P., and Ding, J. (2015). Fabrication of RGD micro/nanopattern and corresponding study of stem cell differentiation. Nano Lett. 15 (3), 1457–1467. doi:10.1021/nl5049862
Yi, J., Balagopalan, L., Nguyen, T., McIntire, K. M., and Samelson, L. E. (2019). TCR microclusters form spatially segregated domains and sequentially assemble in calcium-dependent kinetic steps. Nat. Commun. 10, 277. doi:10.1038/s41467-018-08064-2
Yim, E. K., Darling, E. M., Kulangara, K., Guilak, F., and Leong, K. W. (2010). Nanotopography induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials 31 (6), 1299–1306. doi:10.1016/j.biomaterials.2009.10.037
Yim, E. K., Reano, R. M., Pang, S. W., Yee, A. F., Chen, C. S., and Leong, K. W. (2005). Nanopattern induced changes in morphology and motility of smooth muscle cells. Biomaterials 26 (26), 5405–5413. doi:10.1016/j.biomaterials.2005.01.058
Zambelli, T., Aebersold, M. J., Behr, P., Han, H., Hirt, L., Martinez, V., et al. (2018). “FluidFM: Development of the instrument as well as its applications for 2D and 3D lithography”. in open-space microfluidics: Concepts, implementations, applications. Editors E. Delamarche, and G. V. Kaigala (Weinheim, Germany: Wiley VCH), 295–323.
Keywords: receptor nanoclustering, nanofabrication, nanotopography, nanopatterning, cell response
Citation: Casanellas I, Samitier J and Lagunas A (2022) Recent advances in engineering nanotopographic substrates for cell studies. Front. Bioeng. Biotechnol. 10:1002967. doi: 10.3389/fbioe.2022.1002967
Received: 25 July 2022; Accepted: 16 August 2022;
Published: 06 September 2022.
Edited by:
Nihal Engin Vrana, Sparta Medical, FranceReviewed by:
Pietro Parisse, National Research Council (CNR), ItalyCopyright © 2022 Casanellas, Samitier and Lagunas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Lagunas, YWxhZ3VuYXNAaWJlY2JhcmNlbG9uYS5ldQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.