- 1Department of Cell & Developmental Biology, University College London, London, United Kingdom
- 2Department of Genetics, Evolution & Environment, University College London, London, United Kingdom
The human microbiota is implicated in many disease states, including neurological disorders, cancer, and inflammatory diseases. This potentially huge impact on human health has prompted the development of microbiome engineering methods, which attempt to adapt the composition and function of the human host-microbiota system for a therapeutic purpose. One promising method is the use of engineered microorganisms that have been modified to perform a therapeutic function. The majority of these products have only been demonstrated in laboratory models; however, in recent years more concepts have reached the translational stage. This has led to an increase in the number of clinical trials, which are designed to assess the safety and efficacy of these treatments in humans. Within this review, we highlight the progress of some of these microbiome engineering clinical studies, with a focus on engineered live biotherapeutic products.
1 Introduction
The human microbiota refers to all microorganisms and microbial communities that colonise the human body. The largest of these communities is found within the digestive tract. However, there are many other communities that play a role in human health, including the skin (Byrd et al., 2018), vaginal (Chen et al., 2021), and oral microbiota (Lamont et al., 2018). The microbiota potentially plays an integral role in human disease, and dysbiosis in these communities has been implicated in numerous conditions. To date, the microbiota has been linked to neurological diseases (via the gut-brain axis) (Morais et al., 2021; Liu et al., 2022), inflammatory diseases (such as ulcerative colitis) (Caruso et al., 2020), and cancer (Helmink et al., 2019), amongst others (Pascal et al., 2018; Gurung et al., 2020; Hao et al., 2021). It is this profound effect that the microbiota is thought to impart on the human host that has driven interest in microbiome engineering methods, which aim to modify the human host-microbiota system for a therapeutic purpose; particularly for diseases where current treatment is inadequate or non-existent (Brennan, 2022).
This review focuses on studies that use recombinant or engineered live biotherapeutic products (eLBPs); in particular, highlighting clinical trials that attempt to demonstrate their safety and efficacy in humans. eLBPs are microorganisms that have been genetically modified to perform a specific diagnostic or therapeutic function. Previous reviews have summarised the progress that has been made in developing these methods, but the majority focus on preclinical research, with only a few referring to clinical trials (Riglar and Silver, 2018; Charbonneau et al., 2020; Cubillos-Ruiz et al., 2021; McNerney et al., 2021). In this review we firstly introduce the concept of eLBPs and how they differ from other microbiome engineering techniques, before discussing relevant clinical trials that are using eLBPs.
2 Engineered live biotherapeutic products
Although the human microbiota is an incredibly complex community, there are many strategies that can be used to try and influence its composition and/or function (Lawson et al., 2019; Madhusoodanan, 2020). Some examples of previously reported strategies are given in Figure 1. One common method uses naturally occurring probiotic bacteria, which are thought to confer a health benefit to the host; although often their effects on the host and native microbiota are not fully understood and evidence of their ability to effectively colonise the gut remains sparse (Liu et al., 2018; Zmora et al., 2018; Wieërs et al., 2020). It should be noted that this short residency time can also occur with eLBPs depending on the host chassis chosen, as shown by an engineered E. coli Nissle 1917 strain that had a mean residence time of 48 h in human patients (Kurtz et al., 2019). Although this may be desirable in certain cases as it allows for more reproducible and predictable pharmacological properties (Adolfsen et al., 2021), repeated dosing may be required in order to provide prolonged residency of the therapeutic strain (Charbonneau et al., 2020). In addition, clinical evidence implies that the effectiveness of probiotics can vary, whereby some patients may benefit from treatment whilst others do not (Reid et al., 2010). Over 1,000 clinical studies have involved probiotics (Dronkers et al., 2020), exploring conditions such as Parkinson’s disease (NCT04389762) (Lu et al., 2021), COVID-19 infection (NCT04399252) (Tang et al., 2021), and atopic dermatitis (NCT02585986) (Climent et al., 2021). Additionally, it is possible to use multiple species as part of natural or synthetic consortia. Faecal microbiota transplants (FMTs) are a prominent example, where a “healthy” individual’s faecal matter is used to try and replace a patient’s dysbiotic microbiota (Gupta et al., 2016). Currently, there are many clinical trials exploring FMTs and the FDA permits their use under “enforced discretion” for Clostridioides difficile infection (CDI) that does not respond to standard therapy (Grigoryan et al., 2020). Other perturbation strategies include synbiotics (Panigrahi et al., 2017), phage (Schooley et al., 2017), diets and personalised nutrition (e.g. NCT01892956) (Zeevi et al., 2015). A non-exhaustive list of clinical trials investigating various microbiome engineering techniques is given in SI table 1.
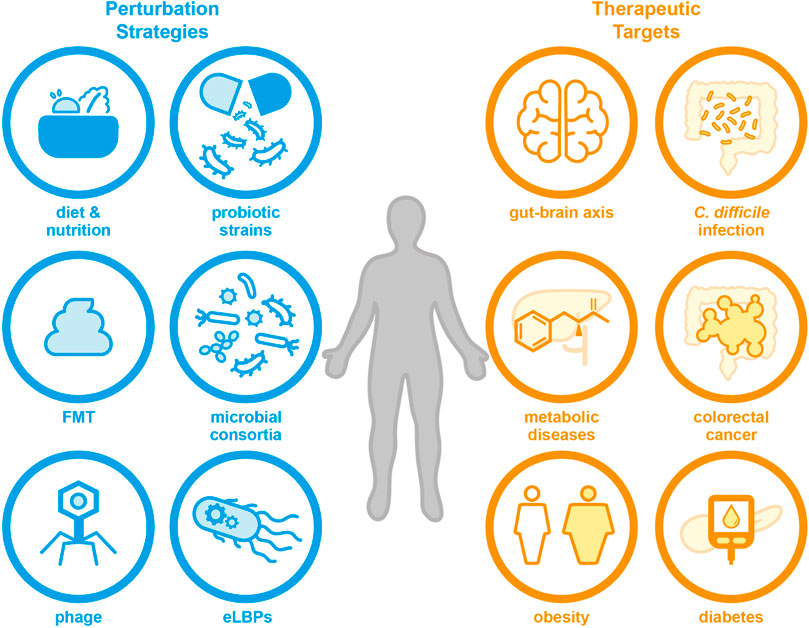
FIGURE 1. There are many techniques that can be used to engineer the human microbiota towards a therapeutic purpose, including probiotic strains, defined microbial consortia, personalised diet and nutrition, or eLBPs (which are the focus of this review). These methods may be useful for the treatment of a range of human diseases in future, for example metabolic diseases, diabetes and neurological conditions.
Engineered methods include eLBPs (Ozdemir et al., 2018; Tan et al., 2020). A simple illustration of this strategy is engineering a bacterial chassis to express a therapeutic molecule. In a ground-breaking study, Steidler et al. (2000) demonstrated this principle by engineering Lactococcus lactis to produce interleukin-10 for the treatment of murine colitis (Steidler et al., 2000). In a more recent example, Cubillos-Ruiz et al. (2022) engineered Lactococcus lactis to produce a heterodimeric β-lactamase, showing that the strain was able to reduce ampicillin-induced dysbiosis in a mouse model (Cubillos-Ruiz et al., 2022). Using eLBPs offers several advantages over the use of pre-, pro- or synbiotics (as defined in Figure 2A), as genetic engineering can confer functions that are not expressed by the native microbiota (Cubillos-Ruiz et al., 2021). Other advantages include the ability to choose a defined strain chassis, reducing the possibility of introducing pathogenic species and the secretion of non-native molecules (Charbonneau et al., 2020; Yadav and Chauhan, 2022). Furthermore, additional genetic elements (e.g. auxotrophies and inducible promoters) allow for greater control and flexibility of eLBPs in the host and environment (Brennan, 2022). For example, Harimoto et al. (2022) developed a programmable surface capsular polysaccharide (CAP) system in Escherichia coli Nissle 1917; through inducible expression the CAP system controlled bacterial encapsulation and subsequently, showed a ten-fold increase in the maximum tolerated dose and anti-tumour efficacy of the strain in a mouse model of cancer (Harimoto et al., 2022). However, with prolonged use of eLBPs there are concerns regarding loss of function, mutation, and biocontainmnent of both the whole organism and genetic material (Ozdemir et al., 2018; McNerney et al., 2021). In addition, outside of E. coli and Lactobacillus, there are relatively few tools available for engineering other relevant species (Charbonneau et al., 2020; Chen et al., 2021). To date, the vast majority of these methods have only been tested in vitro or animal models. Examples of these studies are provided in SI table 2. In recent years, an increasing number of eLBPs have entered clinical trials resulting in more data on the safety and efficacy of eLBPs in humans. Through the clinicaltrials.gov database and literature review, we identified 65 clinical trials involving eLBPs, although others may exist. Of these studies, 46.2% were reported as complete, 21.5% as terminated and the remainder as recruiting, active or unknown (Figure 2B). Within this review, we primarily focus on two major areas of research: 1) cancer therapeutics, and 2) metabolic diseases.
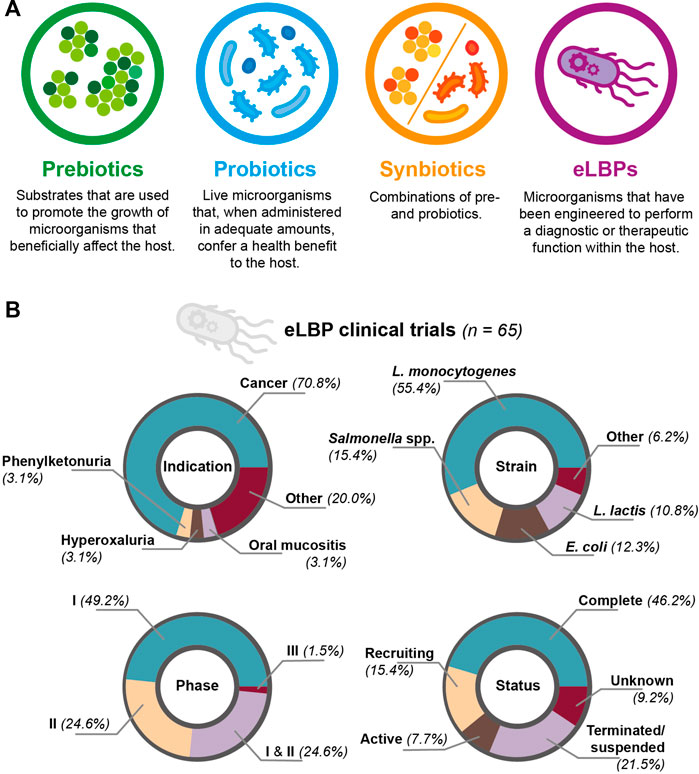
FIGURE 2. (A) Comparison of prebiotics, probiotics, synbiotics and eLBPs. All offer promising routes for modifying the human microbiotia (B) Summary of clinical trials involving eLBPs (genetically-modified microorganisms used for a therapeutic purpose), discovered during this literature review.
3 Cancer therapeutics
Treating cancer with bacteria is not a new concept. One of the earliest reported cases of immunotherapy dates back to the 19th century when mixtures of heat-killed bacteria, known as “Coley’s toxin”, were used to treat inoperable tumours (Coley, 1891; Bickels et al., 2002). Although not without criticism, as little was known about the underlying principles by which it worked, many patients were declared disease free after treatment (McCarthy, 2006). In addition, many species of bacteria have an inherent ability to preferentially colonise within the tumour microenvironment (Duong et al., 2019). It has also been shown that different tumour types have distinct microbiota compositions (Nejman et al., 2020) and that the gut microbiota is able to modulate the effects of chemotherapeutic drugs (Heshiki et al., 2020). These factors have led to interest in harnessing bacteria as vaccine products or as vehicles for the production of localised therapeutics directly within the tumour (Chowdhury et al., 2019). Therefore, it is of little surprise that cancer therapy is one of the most common targets for eLBP clinical trials (Figure 2B).
Currently, most cancer-focussed eLBPs in clinical trials can be grouped into two categories based on the chosen chassis: trials using Listeria monocytogenes (36 trials) or a Salmonella subspecies (10 trials). Attenuated strains of L. monocytogenes have been produced that display reduced virulence. For example, the double-knockout ΔactA/ΔinlB strain, where the genes for tropism and cell-to-cell transmission have been deleted; or the ΔactA/ΔplcB strain (Johnson et al., 2011; Flickinger et al., 2018). These attenuated strains are commonly engineered to act as vaccines for specific forms of cancer, via secretion of a target antigen (Morrow et al., 2019). A prominent example is ADXS11-001 (also known as AXAL, or ADXS-HPV), an eLBP developed by Advaxis for the treatment of HPV-associated cancers (Galicia-Carmona et al., 2021). This prfA-defective strain secretes a truncated fragment of listeriolysin O (LLO), an immunological pore-forming protein, fused to HPV-16 E7 (Miles et al., 2017). The ADXS11-001 strain has been involved in several clinical trials for the treatment of cervical cancer (e.g. NCT02164461, and NCT01266460). In addition, ADXS11-001 has been evaluated as a treatment for anal/rectal (NCT02399813), head and neck cancers (NCT02002182). Currently, the AIM2CERV double-blind, placebo-controlled randomised study investigating ADXS11-001 for high risk cervical cancer, is the only phase III clinical trial for a L. monocytogenes based eLBP (NCT02853604) (Flickinger et al., 2018). However, not all ADXS11-001 trials have been successful, as a phase I trial (NCT01598792) for oropharyngeal cancer was terminated early after a patient suffered from dose-limited toxicity (Sacco et al., 2016).
Advaxis also developed the ADXS31-142 and ADXS31-164 eLBPs. ADXS31-142, intended for the treatment of prostate cancer, was engineered to secrete a fusion of LLO and prostate specific antigen (a serine protease that displays elevated levels during prostate cancer progression) (Hannan et al., 2012). Whereas ADXS31-164 was engineered to secrete an LLO-HER2/neu chimeric protein (Shahabi et al., 2011). As with ADXS11-001, both of these products have now gone through phase I/II clinical trials (NCT02325557 and NCT02386501, respectively).
Salmonella subspecies are also commonly used, as they can penetrate and preferentially grow within tumour tissues (Pangilinan and Lee, 2019). The VXM01 strain, developed by Vaximm using a Salmonella typhi chassis, was engineered to produce vascular endothelial growth factor-2 (Wick et al., 2018). This strain was initially trialed with 45 patients suffering from advanced/stage IV pancreatic cancer (NCT01486329) (Niethammer et al., 2012). A further phase I clinical trial (NCT02718443) tested an oral administration of VXM01 on 14 patients with glioblastoma. VXM01 was found to produce a positive increase in the CD8/Treg ratio of post-vaccine tumour tissues (Wick et al., 2018). Following these promising results, a phase I/II clinical trial (NCT03750071) is now recruiting 30 patients with recurrent glioblastoma. This study will explore the efficacy of VXM01 in combination with Avelumab (a checkpoint inhibitor), following standard treatment. Further trials are exploring the use of engineered Salmonella for the treatment of liver cancer (NCT01099631), myeloma (NCT03762291), and neuroblastoma (NCT04049864).
Alongside the L. monocytogenes and Salmonella based studies, E. coli Nissle 1917 has been used as a chassis for cancer-targeting eLBPs. Synlogic’s SYNB1891 strain was engineered to target the stimulator of interferon genes (STING) agonists pathway to trigger anti-tumour immunity, via expression of cyclic di-AMP (CDA) (Leventhal et al., 2020). Alongside CDA production, a biocontainment system was added to the SYNB1891 strain. Leventhal et al. introduced ΔdapA (4-hydroxy-tetrahydropicolinate synthase) and ΔthyA (thymidylate synthesase) knockouts, showing that this dual-auxotroph was able to prevent in vivo proliferation of SYNB1891 in various tumour types (Leventhal et al., 2020). SYNB1891 was able to stimulate anti-tumour immunity in tumour-bearing mice and triggered the STING pathway in human antigen-presenting cells in vitro. SYNB1891 has subsequently entered a phase I clinical trial (NCT04167137). This trial is recruiting patients with advanced/metastatic solid tumours and lymphoma, with the aim to test SYNB1891 alone or in combination with Atezolizumab (an immunotherapy drug). These studies highlight the vast potential eLBPs hold for the treatment of a variety of cancer types.
4 Metabolic diseases
Synthetic biology engineering techniques allow for the creation of eLBPs that can produce non-native molecules, such as human proteins (Charbonneau et al., 2020). This allows for eLBPs that can correct conditions caused by errors of human metabolism. There are several promising eLBPs for treating metabolic diseases in clinical development. For example, the SYNB1618 E. coli Nissle 1917 strain developed by Synlogic, for the treatment of phenylketonuria (PKU). PKU is a rare disease caused by genetic mutations of the phenylalanine hydroxylase enzyme; which results in elevated levels of phenylalanine in the blood. If left untreated, PKU can cause severe neurological complications. Traditionally, PKU is managed through a phenylalanine-restricted diet, which can have a negative impact on a patient’s quality of life (Rocha and MacDonald, 2016). The SYNB1618 strain was engineered to consume phenylalanine within the digestive tract, via action of the phenylalanine ammonia lyase (PAL) and l-amino acid deaminase (LAAD) enzymes. Following positive results in mouse and non-human primate models of PKU (Isabella et al., 2018), SYNB1618 was evaluated in a Phase I/IIa clinical study (NCT03516487). This trial recruited both healthy volunteers and PKU patients; showing that SYNB1618 was safe and well-tolerated up to a maximum dose of 2 × 1011 colony-forming units (Puurunen et al., 2021). As discussed by the authors, the successful completion of this clinical trial provided proof of the potential to use eLBPs in the treatment of rare metabolic conditions (Puurunen et al., 2021). In addition, data from this study has been used to develop in vitro gut-on-chip (Nelson et al., 2021) and predictive pharmacology models (Charbonneau et al., 2021); showing that results from clinical trials can feed back into the preceding laboratory design process, in order to help address some of the limitations present in animal models (Brennan, 2022). Following the successful trial of SYNB1618, Synlogic developed the SYNB1934 strain (Adolfsen et al., 2021). SYNB1934 was created through optimisation of PAL enzyme activity, displaying a two-fold increase in vivo PAL activity relative to SYNB1618. A phase I clinical trial (NCT04984525) successfully showed that SYNB1934 was also safe and well-tolerated in healthy adult volunteers. Subsequently, a new clinical trial (NCT04534842) is recruiting patients, in order to perform a head-to-head comparison of the SYNB1618 and SYNB1934 eLBPs.
Another E. coli Nissle 1917 based eLBP, referred to as SYNB8802, has been designed to combat enteric hyperoxaluria (EH). EH is caused by excessive absorption of dietary oxalate, which can cause kidney failure. This can be due to genetic defects, or via gastrointestinal conditions that increase oxalate uptake (Witting et al., 2021). Currently, there are no approved pharmaceutical treatments for this condition (Lubkowicz et al., 2022). SYNB8802 was modified to express genes from the oxalate degradation pathway of Oxalobacter formigenes, alongside an oxalyl-CoA synthetase gene. These modifications conferred the ability for SYNB8802 to degrade oxalate in vitro (Lubkowicz et al., 2022). As with SYN1618, SYNB8802 was tested in mouse and non-human primate models; demonstrating that SYNB8802 could reduce the excreted levels of urinary oxalate (Lubkowicz et al., 2022). A phase I clinical trial (NCT04629170) is now recruiting healthy adults and EH patients, to evaluate the safety and efficacy of SYNB8802 in humans. Novome Biotechnologies have also developed an engineered Bacteroides strain for the treatment of EH, named NOV-001; which is currently recruiting for a phase I/II clinical trial (NCT04909723).
The Synlogic SYNB1020 strain was developed as a treatment option for hyperammonia; a condition caused by liver damage (i.e. cirrhosis), or defects in ammonia-detoxifying enzymes. SYNB1020, another E. coli Nissle 1917 strain, was engineered to convert NH3 to arginine in anaerobic conditions; with the strain promoting improved survival in a mouse model of hyperammonia (Kurtz et al., 2019). The strain completed phase I clinical trials (NCT03179878). However, SYNB1020 failed a subsequent phase Ib/IIa clinical trial (NCT03447730), which was terminated due to a reported lack of efficacy.
5 Other targets
Inflammatory bowel disease (IBD) is a chronic, inflammatory disease of the digestive tract, with two major subtypes: Crohn’s disease and ulcerative colitis (UC). The full mechanisms behind IBD are not yet understood, however there is a general consensus that the microbiota plays an integral role in the development and progression of IBD (Lee and Chang, 2021). Traditionally, IBD therapies attempt to target cytokines of the inflammatory cascade or systemic immunomodulation. However, these treatments are not always effective and may cause adverse side effects (Eindor-Abarbanel et al., 2021). As such, there are several clinical trials exploring the use of probiotics for the treatment of IBD (e.g. NCT04969679, NCT04842149). Additionally, ActoGenix have developed two eLBPs to treat these conditions that have entered clinical trials. The first, AG011, was an engineered L. lactis strain that secreted hIL-10 and was shown to be safe and well-tolerated in an UC patient cohort (NCT00729872). AG014, based on the same L. lactis platform as AG011, was engineered to produce an anti-TNFα antibody fragment of certolizumab (Vandenbroucke et al., 2010). AG014 was evaluated as part of a phase I clinical trial, but has not been developed further (Crowe et al., 2018).
Another eLBP, AG013, was created to treat oral mucositis (OM) in patients with head and neck cancer undergoing chemoradiation therapy. This L. lactis strain was engineered to secrete human Trefoil Factor 1. Initially, AG013 was shown to be effective in a hamster based model of radiation-induced OM (Caluwaerts et al., 2010). Following a successful phase I trial (NCT00938080), which demonstrated the safety of AG013 in patients receiving induction chemotherapy for the treatment of head and neck cancer, AG013 entered phase II clinical trials (NCT03234465). However, in a statement issued by Oragenics, Inc. this study was terminated as there was no statistically significant difference in the duration of OM between AG013 and the placebo.
6 Outlook
Currently, many eLBPs in trials are based on the constitutive expression of specific molecules. However, modern engineering tools allow for the development of more complex systems. For example biosensors that sense and respond to environmental cues in a dynamic manner (Riglar et al., 2017; Hicks et al., 2020; Rutter et al., 2021), engineered strains that display targeted or prolonged colonisation in a specific environment (Fedorec et al., 2021; Chien et al., 2022), and living materials that are embedded with engineered microbes (Rodrigo-Navarro et al., 2021; Omer et al., 2022). Recent examples include biosensors for acetoacetate, oxygen lactacte, pH and inflammation (Riglar et al., 2017; Rutter et al., 2021; Chien et al., 2022); ingestible micro-bio-electronic devices that use heme-sensitive bacteria to report on gastrointestinal health (Mimee et al., 2018); and engineered native strains that were used for prolonged transgene delivery in a mouse model (Russell et al., 2022). Furthermore, several methods for mitigating the issue of biocontainment for eLBPs have been developed. One method that has already been used in clinical trials are auxotrophies that attempt to limit the growth of engineered strains outside of the desired environment, for example the Synlogic ΔdapA auxotroph for the essential cell wall component diaminopimelate (Adolfsen et al., 2021). Other biocontainment strategies include ‘deadman’ and ‘passcode’ kill switches, which can be re-programmed for various environmental cues. These switches can be used to block the transcription of essential genes in the absence of a specific input or to trigger self-killing of engineered strains via the production of a toxin, preventing undesired cell growth (Chan et al., 2016). We expect clinical trials evaluating these more complex systems to arise; although it will be essential to ensure these systems behave in a predictable and robust manner. Furthermore, it should be noted that an integral feature of eLBPs is that a single strain can be engineered to target more than one condition simultaneously (Brennan, 2022). As stated by Puurunen et al. (2021) this is particularly promising for the treatment of metabolic diseases and differentiates eLBPs from traditional methods, such as enzyme replacement therapy (Puurunen et al., 2021). Therefore, it is possible that more advanced eLBPs, which are able to produce multiple therapeutic effectors from a single strain, will enter clinical trials.
As evidenced by the number of complete, and ongoing, clinical trials, the development of eLBPs is an exciting application of microbiome engineering research. Despite this, it is important to highlight the issues that can occur when eLBPs enter clinical trials. As shown by the number of terminated and unsuccessful clinical trials, a major issue is that positive in vitro and animal model data do not always translate into safe and efficacious treatment in humans. Of the 14 terminated eLBP clinical trials identified during this literature review 50% were terminated due to a reported lack of efficacy or clinical activity in humans. Although undoubtedly more needs to be done to alleviate these problems, considerable research is going into the development of novel model systems that can be used to characterise eLBPs more comprehensively before they reach clinical testing. These include novel animal models (Hwang et al., 2017; Rutter et al., 2019), innovative organ-on-chip systems that try to mimic human tissues (Bein et al., 2018; Nelson et al., 2021) and mechanistic models that attempt to predict the in vivo activity of eLBPs (Charbonneau et al., 2021). It is hoped that these new technologies will benefit the patients that need these therapies most by helping to translate some of the many developed eLBP strains into clinical treatments.
Finally, as discussed in detail by Charbonneau et al. (2020), it is important to consider the wider regulatory, environmental and societal impacts these new treatment modalities may have if they are to be approved for human use (Charbonneau et al., 2020). For example, manufacturing facilities will be needed that are able to produce standardised, high-quality eLBPs at large scales, while complying with current good manufacturing practices (cGMP) (Brennan, 2022). In addition, physicians, patients and the general public will need to be educated on the benefits and associated risks of these products if they are to become widely accepted (Charbonneau et al., 2020). Although it is evident that challenges remain, eLBPs hold vast potential to address unmet needs in the treatment of human disease. It will be interesting to watch how society reacts to these technologies as they continue to approach the clinic over the coming years.
Author contributions
JR and LD wrote the manuscript. All authors edited the manuscript and approved the submitted version.
Funding
JR was supported by an EPSRC grant (EP/W004674/1). KO was supported through an EPSRC studentship (2505625). CB and LD were supported through the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant no. 770835). CB was also supported by the Wellcome Trust (grant no. 209409/Z/17/Z).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.1000873/full#supplementary-material
References
Adolfsen, K. J., Callihan, I., Monahan, C. E., Greisen, P., Spoonamore, J., Momin, M., et al. (2021). Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat. Commun. 12, 6215. doi:10.1038/s41467-021-26524-0 | |
Bein, A., Shin, W., Jalili-Firoozinezhad, S., Park, M. H., Sontheimer-Phelps, A., Tovaglieri, A., et al. (2018). Microfluidic organ-on-a-chip models of human intestine. Cell. Mol. Gastroenterology Hepatology 5, 659–668. doi:10.1016/j.jcmgh.2017.12.010 | |
Bickels, J., Kollender, Y., Merinsky, O., and Meller, I. (2002). Coley’s toxin: Historical perspective. Isr. Med. Assoc. J. 4, 471–472. |
Brennan, A. M. (2022). Development of synthetic biotics as treatment for human diseases. Synth. Biol. 7, ysac001. doi:10.1093/synbio/ysac001 |
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi:10.1038/nrmicro.2017.157 | |
Caluwaerts, S., Vandenbroucke, K., Steidler, L., Neirynck, S., Vanhoenacker, P., Corveleyn, S., et al. (2010). Ag013, a mouth rinse formulation of lactococcus lactis secreting human trefoil factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncol. 46, 564–570. doi:10.1016/j.oraloncology.2010.04.008 | |
Caruso, R., Lo, B. C., and Núñez, G. (2020). Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 20, 411–426. doi:10.1038/s41577-019-0268-7 | |
Chan, C. T. Y., Lee, J. W., Cameron, D. E., Bashor, C. J., and Collins, J. J. (2016). ’deadman’ and ’passcode’ microbial kill switches for bacterial containment. Nat. Chem. Biol. 12, 82–86. doi:10.1038/nchembio.1979 | |
Charbonneau, M. R., Denney, W. S., Horvath, N. G., Cantarella, P., Castillo, M. J., Puurunen, M. K., et al. (2021). Development of a mechanistic model to predict synthetic biotic activity in healthy volunteers and patients with phenylketonuria. Commun. Biol. 4, 898. doi:10.1038/s42003-021-02183-1 | |
Charbonneau, M. R., Isabella, V. M., Li, N., and Kurtz, C. B. (2020). Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 11, 1738. doi:10.1038/s41467-020-15508-1 | |
Chen, J., Chen, X., and Ho, C. L. (2021). Recent development of probiotic bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 9, 770248. doi:10.3389/fbioe.2021.770248 | |
Chien, T., Harimoto, T., Kepecs, B., Gray, K., Coker, C., Hou, N., et al. (2022). Enhancing the tropism of bacteria via genetically programmed biosensors. Nat. Biomed. Eng. 6, 94–104. doi:10.1038/s41551-021-00772-3 | |
Chowdhury, S., Castro, S., Coker, C., Hinchliffe, T. E., Arpaia, N., and Danino, T. (2019). Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 25, 1057–1063. doi:10.1038/s41591-019-0498-z | |
Climent, E., Martinez-Blanch, J. F., Llobregat, L., Ruzafa-Costas, B., Ángel Carrión-Gutiérrez, M., Ramírez-Boscá, A., et al. (2021). Changes in gut microbiota correlates with response to treatment with probiotics in patients with atopic dermatitis. a post hoc analysis of a clinical trial. Microorganisms 9, 854. doi:10.3390/microorganisms9040854 | |
Coley, W. B. (1891). Ii. contribution to the knowledge of sarcoma. Ann. Surg. 14, 199–220. doi:10.1097/00000658-189112000-00015 | |
Crowe, J. S., Roberts, K. J., Carlton, T. M., Maggiore, L., Cubitt, M. F., Clare, S., et al. (2018). Preclinical development of a novel, orally-administered anti-tumour necrosis factor domain antibody for the treatment of inflammatory bowel disease. Sci. Rep. 8, 4941. doi:10.1038/s41598-018-23277-7 | |
Cubillos-Ruiz, A., Alcantar, M. A., Donghia, N. M., Cárdenas, P., Avila-Pacheco, J., and Collins, J. J. (2022). An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nat. Biomed. Eng. 6, 910–921. doi:10.1038/s41551-022-00871-9 | |
Cubillos-Ruiz, A., Guo, T., Sokolovska, A., Miller, P. F., Collins, J. J., Lu, T. K., et al. (2021). Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 20, 941–960. doi:10.1038/s41573-021-00285-3 | |
Dronkers, T. M., Ouwehand, A. C., and Rijkers, G. T. (2020). Global analysis of clinical trials with probiotics. Heliyon 6, e04467. doi:10.1016/j.heliyon.2020.e04467 | |
Duong, M. T.-Q., Qin, Y., You, S.-H., and Min, J.-J. (2019). Bacteria-cancer interactions: Bacteria-based cancer therapy. Exp. Mol. Med. 51, 1–15. doi:10.1038/s12276-019-0297-0 | |
Eindor-Abarbanel, A., Healey, G. R., and Jacobson, K. (2021). Therapeutic advances in gut microbiome modulation in patients with inflammatory bowel disease from pediatrics to adulthood. Int. J. Mol. Sci. 22, 12506. doi:10.3390/ijms222212506 | |
Fedorec, A. J. H., Karkaria, B. D., Sulu, M., and Barnes, C. P. (2021). Single strain control of microbial consortia. Nat. Commun. 12, 1977. doi:10.1038/s41467-021-22240-x | |
Flickinger, J., Rodeck, U., and Snook, A. (2018). Listeria monocytogenes as a vector for cancer immunotherapy: Current understanding and progress. Vaccines 6, 48. doi:10.3390/vaccines6030048 | |
Galicia-Carmona, T., Arango-Bravo, E., a Serrano-Olvera, J., de La Torre, C. F., Cruz-Esquivel, I., Villalobos-Valencia, R., et al. (2021). Adxs11-001 lm-llo as specific immunotherapy in cervical cancer. Hum. Vaccines Immunother. 17, 2617–2625. doi:10.1080/21645515.2021.1893036 |
Grigoryan, Z., Shen, M. J., Twardus, S. W., Beuttler, M. M., Chen, L. A., and Bateman-House, A. (2020). Fecal microbiota transplantation: Uses, questions, and ethics. Med. Microecology 6, 100027. doi:10.1016/j.medmic.2020.100027 | |
Gupta, S., Allen-Vercoe, E., and Petrof, E. O. (2016). Fecal microbiota transplantation: In perspective. Ther. Adv. Gastroenterol. 9, 229–239. doi:10.1177/1756283X15607414 | |
Gurung, M., Li, Z., You, H., Rodrigues, R., Jump, D. B., Morgun, A., et al. (2020). Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 51, 102590. doi:10.1016/j.ebiom.2019.11.051 | |
Hannan, R., Zhang, H., Wallecha, A., Singh, R., Liu, L., Cohen, P., et al. (2012). Combined immunotherapy with listeria monocytogenes-based psa vaccine and radiation therapy leads to a therapeutic response in a murine model of prostate cancer. Cancer Immunol. Immunother. 61, 2227–2238. doi:10.1007/s00262-012-1257-x | |
Hao, X., Shang, X., Liu, J., Chi, R., Zhang, J., and Xu, T. (2021). The gut microbiota in osteoarthritis: Where do we stand and what can we do? Arthritis Res. Ther. 23, 42. doi:10.1186/s13075-021-02427-9 | |
Harimoto, T., Hahn, J., Chen, Y.-Y., Im, J., Zhang, J., Hou, N., et al. (2022). A programmable encapsulation system improves delivery of therapeutic bacteria in mice. Nat. Biotechnol. 40, 1259–1269. doi:10.1038/s41587-022-01244-y | |
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V., and Wargo, J. A. (2019). The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388. doi:10.1038/s41591-019-0377-7 | |
Heshiki, Y., Vazquez-Uribe, R., Li, J., Ni, Y., Quainoo, S., Imamovic, L., et al. (2020). Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 8, 28. doi:10.1186/s40168-020-00811-2 | |
Hicks, M., Bachmann, T. T., and Wang, B. (2020). Synthetic biology enables programmable cell-based biosensors. ChemPhysChem 21, 132–144. doi:10.1002/cphc.201900739 | |
Hwang, I. Y., Koh, E., Wong, A., March, J. C., Bentley, W. E., Lee, Y. S., et al. (2017). Engineered probiotic escherichia coli can eliminate and prevent pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 8, 15028. doi:10.1038/ncomms15028 | |
Isabella, V. M., Ha, B. N., Castillo, M. J., Lubkowicz, D. J., Rowe, S. E., Millet, Y. A., et al. (2018). Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864. doi:10.1038/nbt.4222 | |
Johnson, P. V., Blair, B. M., Zeller, S., Kotton, C. N., and Hohmann, E. L. (2011). Attenuated listeria monocytogenes vaccine vectors expressing influenza a nucleoprotein: Preclinical evaluation and oral inoculation of volunteers. Microbiol. Immunol. 55, 304–317. doi:10.1111/j.1348-0421.2011.00322.x | |
Kurtz, C. B., Millet, Y. A., Puurunen, M. K., Perreault, M., Charbonneau, M. R., Isabella, V. M., et al. (2019). An engineered e. coli nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 11, eaau7975. doi:10.1126/scitranslmed.aau7975 | |
Lamont, R. J., Koo, H., and Hajishengallis, G. (2018). The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759. doi:10.1038/s41579-018-0089-x | |
Lawson, C. E., Harcombe, W. R., Hatzenpichler, R., Lindemann, S. R., Löffler, F. E., O’Malley, M. A., et al. (2019). Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. doi:10.1038/s41579-019-0255-9 | |
Lee, M., and Chang, E. B. (2021). Inflammatory bowel diseases (ibd) and the microbiome—Searching the crime scene for clues. Gastroenterology 160, 524–537. doi:10.1053/j.gastro.2020.09.056 | |
Leventhal, D. S., Sokolovska, A., Li, N., Plescia, C., Kolodziej, S. A., Gallant, C. W., et al. (2020). Immunotherapy with engineered bacteria by targeting the sting pathway for anti-tumor immunity. Nat. Commun. 11, 2739. doi:10.1038/s41467-020-16602-0 | |
Liu, L., Huh, J. R., and Shah, K. (2022). Microbiota and the gut-brain-axis: Implications for new therapeutic design in the cns. eBioMedicine 77, 103908. doi:10.1016/j.ebiom.2022.103908 | |
Liu, Y., Tran, D. Q., and Rhoads, J. M. (2018). Probiotics in disease prevention and treatment. J. Clin. Pharmacol. 58, S164–S179. doi:10.1002/jcph.1121 | |
Lu, C.-S., Chang, H.-C., Weng, Y.-H., Chen, C.-C., Kuo, Y.-S., and Tsai, Y.-C. (2021). The add-on effect of lactobacillus plantarum ps128 in patients with Parkinson’s disease: A pilot study. Front. Nutr. 8, 650053. doi:10.3389/fnut.2021.650053 | |
Lubkowicz, D., Horvath, N. G., James, M. J., Cantarella, P., Renaud, L., Bergeron, C. G., et al. (2022). An engineered bacterial therapeutic lowers urinary oxalate in preclinical models and in silico simulations of enteric hyperoxaluria. Mol. Syst. Biol. 18, e10539. doi:10.15252/msb.202110539 | |
Madhusoodanan, J. (2020). Editing the microbiome. Proc. Natl. Acad. Sci. U. S. A. 117, 3345–3348. doi:10.1073/pnas.2000108117 | |
McCarthy, E. F. (2006). The toxins of william b. coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop. J. 26, 154–158. |
McNerney, M. P., Doiron, K. E., Ng, T. L., Chang, T. Z., and Silver, P. A. (2021). Theranostic cells: Emerging clinical applications of synthetic biology. Nat. Rev. Genet. 22, 730–746. doi:10.1038/s41576-021-00383-3 | |
Miles, B. A., Monk, B. J., and Safran, H. P. (2017). Mechanistic insights into adxs11-001 human papillomavirus-associated cancer immunotherapy. Gynecol. Oncol. Res. Pract. 4, 9. doi:10.1186/s40661-017-0046-9 | |
Mimee, M., Nadeau, P., Hayward, A., Carim, S., Flanagan, S., Jerger, L., et al. (2018). An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918. doi:10.1126/science.aas9315 | |
Morais, L. H., Schreiber, H. L., and Mazmanian, S. K. (2021). The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 19, 241–255. doi:10.1038/s41579-020-00460-0 | |
Morrow, Z. T., Powers, Z. M., and Sauer, J.-D. (2019). Listeria monocytogenes cancer vaccines: Bridging innate and adaptive immunity. Curr. Clin. Microbiol. Rep. 6, 213–224. doi:10.1007/s40588-019-00133-4 | |
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 368, 973–980. doi:10.1126/science.aay9189 | |
Nelson, M. T., Charbonneau, M. R., Coia, H. G., Castillo, M. J., Holt, C., Greenwood, E. S., et al. (2021). Characterization of an engineered live bacterial therapeutic for the treatment of phenylketonuria in a human gut-on-a-chip. Nat. Commun. 12, 2805. doi:10.1038/s41467-021-23072-5 | |
Niethammer, A. G., Lubenau, H., Mikus, G., Knebel, P., Hohmann, N., Leowardi, C., et al. (2012). Double-blind, placebo-controlled first in human study to investigate an oral vaccine aimed to elicit an immune reaction against the vegf-receptor 2 in patients with stage iv and locally advanced pancreatic cancer. BMC Cancer 12, 361. doi:10.1186/1471-2407-12-361 | |
Omer, R., Mohsin, M. Z., Mohsin, A., Mushtaq, B. S., Huang, X., Guo, M., et al. (2022). Engineered bacteria-based living materials for biotherapeutic applications. Front. Bioeng. Biotechnol. 10, 870675. doi:10.3389/fbioe.2022.870675 | |
Ozdemir, T., Fedorec, A. J., Danino, T., and Barnes, C. P. (2018). Synthetic biology and engineered live biotherapeutics: Toward increasing system complexity. Cell. Syst. 7, 5–16. doi:10.1016/j.cels.2018.06.008 | |
Pangilinan, C. R., and Lee, C.-H. (2019). Salmonella-based targeted cancer therapy: Updates on a promising and innovative tumor immunotherapeutic strategy. Biomedicines 7, 36. doi:10.3390/biomedicines7020036 | |
Panigrahi, P., Parida, S., Nanda, N. C., Satpathy, R., Pradhan, L., Chandel, D. S., et al. (2017). A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412. doi:10.1038/nature23480 | |
Pascal, M., Perez-Gordo, M., Caballero, T., Escribese, M. M., Longo, M. N. L., Luengo, O., et al. (2018). Microbiome and allergic diseases. Front. Immunol. 9, 1584. doi:10.3389/fimmu.2018.01584 | |
Puurunen, M. K., Vockley, J., Searle, S. L., Sacharow, S. J., Phillips, J. A., Denney, W. S., et al. (2021). Safety and pharmacodynamics of an engineered e. coli nissle for the treatment of phenylketonuria: A first-in-human phase 1/2a study. Nat. Metab. 3, 1125–1132. doi:10.1038/s42255-021-00430-7 | |
Reid, G., Gaudier, E., Guarner, F., Huffnagle, G. B., Macklaim, J. M., Munoz, A. M., et al. (2010). Responders and non-responders to probiotic interventions. Gut Microbes 1, 200–204. doi:10.4161/gmic.1.3.12013 | |
Riglar, D. T., Giessen, T. W., Baym, M., Kerns, S. J., Niederhuber, M. J., Bronson, R. T., et al. (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35, 653–658. doi:10.1038/nbt.3879 | |
Riglar, D. T., and Silver, P. A. (2018). Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 16, 214–225. doi:10.1038/nrmicro.2017.172 | |
Rocha, J. C., and MacDonald, A. (2016). Dietary intervention in the management of phenylketonuria: Current perspectives. Pediatr. Health Med. Ther. 7, 155–163. doi:10.2147/PHMT.S49329 | |
Rodrigo-Navarro, A., Sankaran, S., Dalby, M. J., del Campo, A., and Salmeron-Sanchez, M. (2021). Engineered living biomaterials. Nat. Rev. Mat. 6, 1175–1190. doi:10.1038/s41578-021-00350-8 |
Russell, B. J., Brown, S. D., Siguenza, N., Mai, I., Saran, A. R., Lingaraju, A., et al. (2022). Intestinal transgene delivery with native e. coli chassis allows persistent physiological changes. Cell. 185, 3263–3277.e15. e15. doi:10.1016/j.cell.2022.06.050 | |
Rutter, J. W., Ozdemir, T., Galimov, E. R., Quintaneiro, L. M., Rosa, L., Thomas, G. M., et al. (2019). Detecting changes in the caenorhabditis elegans intestinal environment using an engineered bacterial biosensor. ACS Synth. Biol. 8, 2620–2628. doi:10.1021/acssynbio.9b00166 | |
Rutter, J. W., Dekker, L., Fedorec, A. J. H., Gonzales, D. T., Wen, K. Y., Tanner, L. E. S., et al. (2021). Engineered acetoacetate-inducible whole-cell biosensors based on the atosc two-component system. Biotechnol. Bioeng. 118, 4278–4289. doi:10.1002/bit.27897 | |
Sacco, J. J., Evans, M., Harrington, K. J., Man, S., Powell, N., Shaw, R. J., et al. (2016). Systemic listeriosis following vaccination with the attenuated listeria monocytogenes therapeutic vaccine, adxs11-001. Hum. Vaccines Immunother. 12, 1085–1086. doi:10.1080/21645515.2015.1121338 | |
Schooley, R. T., Biswas, B., Gill, J. J., Hernandez-Morales, A., Lancaster, J., Lessor, L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954–17. doi:10.1128/AAC.00954-17 | |
Shahabi, V., Seavey, M. M., Maciag, P. C., Rivera, S., and Wallecha, A. (2011). Development of a live and highly attenuated listeria monocytogenes-based vaccine for the treatment of her2/neu-overexpressing cancers in human. Cancer Gene Ther. 18, 53–62. doi:10.1038/cgt.2010.48 | |
Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., et al. (2000). Treatment of murine colitis by lactococcus lactis secreting interleukin-10. Science 289, 1352–1355. doi:10.1126/science.289.5483.1352 | |
Tan, Y., Shen, J., Si, T., Ho, C. L., Li, Y., and Dai, L. (2020). Engineered live biotherapeutics: Progress and challenges. Biotechnol. J. 15, 2000155. doi:10.1002/biot.202000155 | |
Tang, H., Bohannon, L., Lew, M., Jensen, D., Jung, S.-H., Zhao, A., et al. (2021). Randomised, double-blind, placebo-controlled trial of probiotics to eliminate Covid-19 transmission in exposed household contacts (protect-ehc): A clinical trial protocol. BMJ Open 11, e047069. doi:10.1136/bmjopen-2020-047069 | |
Vandenbroucke, K., de Haard, H., Beirnaert, E., Dreier, T., Lauwereys, M., Huyck, L., et al. (2010). Orally administered l. lactis secreting an anti-tnf nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol. 3, 49–56. doi:10.1038/mi.2009.116 | |
Wick, W., Wick, A., Sahm, F., Riehl, D., von Deimling, A., Bendszus, M., et al. (2018). Vxm01 phase i study in patients with progressive glioblastoma: Final results. J. Clin. Oncol. 36, 2017. doi:10.1200/JCO.2018.36.15_suppl.2017 | |
Wieërs, G., Belkhir, L., Enaud, R., Leclercq, S., de Foy, J.-M. P., Dequenne, I., et al. (2020). How probiotics affect the microbiota. Front. Cell. Infect. Microbiol. 9, 454. doi:10.3389/fcimb.2019.00454 | |
Witting, C., Langman, C. B., Assimos, D., Baum, M. A., Kausz, A., Milliner, D., et al. (2021). Pathophysiology and treatment of enteric hyperoxaluria. Clin. J. Am. Soc. Nephrol. 16, 487–495. doi:10.2215/CJN.08000520 | |
Yadav, M., and Chauhan, N. S. (2022). Microbiome therapeutics: Exploring the present scenario and challenges. Gastroenterol. Rep. 10, goab046. doi:10.1093/gastro/goab046 |
Zeevi, D., Korem, T., Zmora, N., Israeli, D., Rothschild, D., Weinberger, A., et al. (2015). Personalized nutrition by prediction of glycemic responses. Cell. 163, 1079–1094. doi:10.1016/j.cell.2015.11.001 | |
Keywords: microbiota, microbiome engineering, human health, clinical trials, engineered bacteria
Citation: Rutter JW, Dekker L, Owen KA and Barnes CP (2022) Microbiome engineering: engineered live biotherapeutic products for treating human disease. Front. Bioeng. Biotechnol. 10:1000873. doi: 10.3389/fbioe.2022.1000873
Received: 22 July 2022; Accepted: 30 August 2022;
Published: 16 September 2022.
Edited by:
Qing Sun, Texas A&M University, United StatesReviewed by:
Jing Wui Yeoh, National University of Singapore, SingaporeCopyright © 2022 Rutter, Dekker , Owen and Barnes . This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jack W. Rutter, amFjay5ydXR0ZXIuMTZAdWNsLmFjLnVr
 Jack W. Rutter
Jack W. Rutter Linda Dekker
Linda Dekker  Kimberley A. Owen 1
Kimberley A. Owen 1 Chris P. Barnes
Chris P. Barnes