- 1Institute of Food Processing, Heilongjiang Academy of Agricultural Sciences, Harbin, China
- 2Heilongjiang Province Key Laboratory of Food Processing, Harbin, China
- 3Soybean Institute, Heilongjiang Academy of Agricultural Sciences, Harbin, China
- 4Biotechnology Research Institute, Heilongjiang Academy of Agricultural Sciences, Harbin, China
Tannases are a family of esterases that catalyze the hydrolysis of ester and depside bonds present in hydrolyzable tannins to release gallic acid. Here, a novel tannase from Lachnospiraceae bacterium (TanALb) was characterized. The recombinant TanALb exhibited maximal activity at pH 7.0 and 50°C, and it maintained more than 70% relative activity from 30°C to 55°C. The activity of TanALb was enhanced by Mg2+ and Ca2+, and was dramatically reduced by Cu2+ and Mn2+. TanALb is capable of degrading esters of phenolic acids with long-chain alcohols, such as lauryl gallate as well as tannic acid. The Km value and catalytic efficiency (kcat /Km) of TanALb toward five substrates showed that tannic acid (TA) was the favorite substrate. Homology modeling and structural analysis indicated that TanALb contains an insertion loop (residues 341–450). Based on the moleculer docking and molecular dynamics (MD) simulation, this loop was observed as a flap-like lid to interact with bulk substrates such as tannic acid. TanALb is a novel bacterial tannase, and the characteristics of this enzyme make it potentially interesting for industrial use.
Introduction
Tannins are water-soluble polyphenolic compounds that constitute the fourth most abundant class of plant biomass components, richly secreted by defensive systems in wood, fruits, roots, and seeds (White, 1957; Smeriglio et al., 2016; Maisetta et al., 2019). Tannins and their derivatives are the key compounds of the astringent taste of tea, fruits, and vegetables, and determine the quality of their production such as muddy and astringency problem of juice and beer (Charlton et al., 2002). In addition, tannins inhibit the growth of microbes, exert antinutritional effect on animals and are even toxic to the human body in that they possess the capacity to bind and precipitate proteins and to bind iron, which could lead to many biological process disorders (Mcdonald et al., 1996; Baxter et al., 1997; Chung et al., 1998; Aguilar et al., 2007; Smeriglio et al., 2016). During millions of years of biological evolution, various microorganisms have adapted to utilize tannins as carbon and/or energy sources for growth and development using enzymes such as tannin acyl hydrolases, generally known as tannases (Lekha and Lonsane, 1997; Aguilar et al., 2007). Tannases (EC 3.1.1.20) catalyze the hydrolysis of ester and depside bonds present in gallotannins, complex tannins, and gallic acid esters, resulting in the release of glucose and gallic acid (Govindarajan et al., 2016). Tannase have broad practical applications in the food, feed, beverage, pharmaceutical, agricultural, and the leather industries. Compared with chemical hydrolysis methods catalyzed by HCl, nucleophiles and precious metals, the bio-hydrolysis of tannins has unique advantages due to its low environmental impact (Aharwar and Parihar, 2018). Thus, there is an increasing demand for tannases with desirable biochemical properties in industry.
Most tannases that are biochemically studied and characterized in detail come from fungi (Aboubakr et al., 2013), but not much is known about tannase of bacteria or yeasts living under extreme conditions, which are considered to be the potential of tannases with unexpected characteristics (Dhiman et al., 2018; Tomás-Cortázar, 2018). The studied tannases in bacteria are almost from the rumen, gut, oral cavity microbiota, or soils (Noguchi et al., 2007; Wu et al., 2014; Chaitanyakumar and Anbalagan, 2016; Tomás-Cortázar et al., 2018). Recently, it is reported that plant tannase was first identified from leaves of tea, strawberry, and crops, which hydrolyze tannin compounds, glycosides, and ph enolic glycosides to produce toxic small molecule phenolic compounds, so as to protect plants from attack by microorganisms and herbivore (Dai et al., 2020).
Tannase includes extracellular tannases encoded by the tanA gene and intracellular tannases encoded by tanB gene, with molecular size of 66 and 50 kDa, respectively (Tomás-Cortázar et al., 2018). Exceptionally, TanBSS encoded by a tanB gene from Streptomyces sviceus was identified as an extracellular tannase (Wu et al., 2014). The optimum temperature and pH value for biochemical characteristics of bacterial tannases are 30°C–50°C and 3.0–7.0, respectively (Lekshmi et al., 2021). Up to now, two kinds of bacterial tannase belonging to probiotics (TanALp, Lactobacillus plantarum) and the oral bacteria (TanBFnn, Fusobacterium nucleatum) have been structurally characterized by combining crystal structure, molecular dynamics, and mutation analysis, which confirmed the functional role of the flap domain and flap-like domain in the substrate recognition and specificity of tannase (Ren et al., 2013; Mancheño et al., 2020).
Here, a putative bacterial tannase from a species of Lachnospiraceae bacterium was characterized in detail. Recombinant TanALb was cloned, overexpressed, and purified in order to fully analyze the biochemical characteristics. To understand the biochemical characteristics of TanALb, three-dimensional structure of the enzyme was modeled. Assisted by molecular docking and MD simulation, the structure-oriented substrate specificity analysis was carried out. This work adds a new bacterial tannase with comprehensive biochemical and structural characterization to provide a platform for the manipulation of tannins in food and beverage products, or for transformation into biologically active products to benefit human health on the basis of specific requirements.
Materials and Methods
Cloning and Expression of Tannase From Lachnospiraceae Bacterium
The coding sequence of TanALb (GenBank accession No. MBQ6323131.1) was codon optimized for E. coli and synthesized by Genewiz (Beijing, China). The purified PCR products were subcloned into the pET-28a (+) vector (Novagen, Madison, WI, USA) between the Nde I and EcoR I restriction sites, resulting in the addition of a 6×His tag at the N-terminus. The primer sequences are TanALb_F (GGAATTCCATATGTCACAGTCAACAGCTACACGC) and TanALb_R (GGAATTCAAGGCCAGCAGCTTTAAGGCG). E. coli BL21 (DE3) was transformed with the resulting expression vector pET-28a-TanALb and grown in Luria–Bertani (LB) broth medium with 50 µg/ml kanamycin overnight at 37°C. On the next day, a fresh culture in the same medium was inoculated, and when it reached 0.6–0.8, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, to induce the expression of pET-28a-TanALb at 16°C overnight. The cells were harvested by centrifugation for 15 min at 5,000 × g and 4°C, and washed with 0.9% NaCl solution.
Purification of the Recombinant Tannase From Lachnospiraceae bacterium
The cells expressing recombinant TanALb were resuspended in lysis buffer [20 mM Tris-HCl, 20 mM imidazole, 500 mM NaCl, 1 mM dithiothreitol (DTT), pH 8.0] and ultrasonicated at 1.5-s pulse and 3-s output for 15 min. Then, the crude lysate was centrifuged at 40,000 × g and 4°C for 30 min, and the supernatant was bound to a 1 ml of Ni-NTA Superflow column (Qiagen, Hilden, Germany). After washing with 10 ml of wash buffer (20 mM Tris-HCl, 0.5 M NaCl, 1 mM DTT, 50 mM imidazole, pH 8.0), the TanALb protein was eluted with 10 ml of elution buffer (20 mM Tris-HCl, 0.3 M NaCl, 1 mM DTT, 0.5 M imidazole, pH 8.0). For further purification, the target protein was buffer exchanged into 20 mM Tris–HCl pH 8.0, 1 mM DTT, loaded onto a 5-ml MonoQ ion exchange column (GE Healthcare, Piscataway, NJ, USA), and eluted using a linear gradient using from 0 to 1 M NaCl. Then, gel filtration chromatography was used to determine the apparent mass of the TanALb protein using a Superdex 200 increase column (GE Healthcare, Piscataway, NJ, USA). The running buffer was 20 mM Tris-HCl (pH 8.0, containing 150 mM NaCl and 1 mM DTT), and the flow rate was 0.5 ml/min. Finally, the purified protein was stored at 4°C until further analysis. The molecular mass and purity of the target protein were assessed by SDS-PAGE.
Enzyme Activity Assay
The enzyme activity of TanALb was determined using the rhodamine assay published by Inoue and Hagerman (Jiménez et al., 2014). TanALb (100 μg) in 700 μl of 50 mM phosphate buffer, pH 6.5, was incubated with 40 μl of 25 mM methyl gallate (final substrate concentration 1 mM), for 5 min at 37°C. After incubation, 150 μl of 0.667% rhodamine in 100% methanol was added to the mixture. After incubation at 30°C for 5 min, 100 μl of 0.5 M KOH was added and incubated for an additional 5–10 min, after which the absorbance was measured at 520 nm using a spectrophotometer. A standard curve was prepared using gallic acid solutions ranging from 0.125 to 1 mM. One unit of TanALb activity was defined as the amount of enzyme that releases 1 μM gallic acid per minute under the described assay conditions. The concentration of TanALb was determined using a BCA Protein Assay Kit (Solarbio, China).
To determine the substrate specificity of TanALb, 1 mM methyl gallate (MG), ethyl gallate (EG), propyl gallate (PG), lauryl gallate (LG) and tannic acid (TA) were used as the substrate, and enzyme activity was measured at 50°C and pH 7.0. The kinetic parameters of the purified enzymes were determined by assaying the activity on various substrate concentrations. MG, EG, PG, LG (0.2–5 mM) and tannic acid (0.01–0.05 mM) were incubated with appropriate amount of enzyme to calculate the kinetics. The amount of gallic acid which was formed by the catalysis of tannase was calibrated using the absorbance at 520 nm. Kinetic parameters were obtained according to the Line-weaver and Burk method. Each measurement was conducted three times. All calculations were performed using the Origin 9.0 software.
Effects of Temperature, pH, and Additives on the Activity and Stability of the Enzyme
The optimal temperature of TanALb was determined by measuring the relative activities in the range of 20°C–70°C in 20 mM phosphate buffer saline at pH 7.0. To evaluate the thermal stability of TanALb, the residual activity was measured after pre-incubation in 20 mM phosphate buffer saline buffer (pH 7.0) at 20°C, 30°C, 40°C, 50°C, 60°C, and 70°C, respectively. Samples were taken every 30 min for 5 h.
Different buffers were used to assess the optimal pH of TanALb at 50°C, with methyl gallate as the substrate. The pH value was varied from 2.5 to 10 using 20 mM glycine-HCl buffer (pH 2.5–5.5), 20 mM 4-morpholineethanesulfonic acid (MES) (pH 5.5–6.5), 20 mM phosphate buffer saline (pH 6.5–8.0), 20 mM Tris-HCl (pH 8.0–9.5), and 20 mM glycine-NaOH (pH 9.5–10.0). In order to determine the pH stability of TanALb, the enzyme was incubated in the indicated buffers (pH 2.5–10.0) at 50°C for 30 min, and the residual activity was measured under the standard conditions.
The effects of various metal ions on the purified recombinant TanALb were investigated by measuring the relative enzyme activity in the presence of 1 mM K+, Ca2+, Co2+, Mn2+, Mg2+, Zn2+, and Cu2+, respectively. The enzyme activity was measured at pH 7.0 and 50°C. Additionally, the enzyme was incubated in 20 mM phosphate saline buffer pH 7.0 with 1 mM ethylene diamine tetraacetic acid (EDTA), sodium dodecyl sulfate (SDS), Tween-80, dimethyl sulfoxide (DMSO), and TritonX-100 at 25°C for 1.0 h to investigate the effect of a chelating agent. The activity without adding any additives was defined as 100%.
Homology Modeling of Tannase From Lachnospiraceae bacterium
Modeller 9.9 software (http://salilab.org/modeller/) was used to generate a homology model of TanALb (Gao et al., 2012). The crystal structure of a homologous protein from Fusobacterium nucleatum subsp. nucleatum (PDB ID: 6YQ4), which has 42.9% sequence identity with the target protein TanALb, was chosen as template. A sequence alignment between TanBFnn and TanALb was generated automatically using the align2d command. Subsequently, homology modeling was executed using the auto-model command. The best model was chosen based on the MODELLER objective function values and the DOPE assessment scores. Quality of the predicted 3D structural model was assessed by the PROCHECK (https://servicesn. mbi.ucla.edu/PROCHECK). The constructed model structure was visualized and analyzed using PyMOL software (http://www.pymol.org).
Molecular Docking
MOE package (Molecular Operating Environment, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2018) was used for molecular docking of TanALb with five ligands and predicting the binding affinity, respectively. The 2D structure of ligands were downloaded from the PubChem web site (http://pubchem.ncbi.nlm.nih.gov) and converted to 3D in MOE through energy minimization. Then the protonation state of target and the orientation of the hydrogens were optimized by LigX, at the PH of 7 and temperature of 300 K. Prior to docking, the force field of AMBER10:EHT and the implicit solvation model of Reaction Field (R-field) were selected (Case et al., 2008). The position of the native ligand (SPD) in the X-ray structure of 6YQ4 which is selected as the homology template of TanALb was defined as the binding site. The docking workflow followed the “induced fit” protocol, in which the side chains of the receptor pocket were allowed to move according to ligand conformations, with a constraint on their positions. The weight used for tethering side chain atoms to their original positions was 10. All docked poses of molecules were ranked by London dG scoring first, and then a force field refinement was carried out on the top 30 poses followed by a rescoring of GBVI/WSA dG.
Molecular Dynamics (MD) Simulation
For the complex structure of TanALb and tannic acid, whose structure was neutralized by adding sodium/chlorine counter ions and solvated in a cuboid box of TIP3P water molecules with solvent layers 12 Å between the box edges and solute surface.
All MD simulations were performed using AMBER20 (Case et al., 2021). The AMBER FF19SB and gaff2 force fields were applied, and the SHAKE algorithm was used to restrict all covalent bonds involving hydrogen atoms with a time step of 2 fs. The Particle mesh Ewald (PME) method was employed to treat long-range electrostatic interactions. For the solvated system, two steps of minimization were performed before the heating step. The first 4,000 cycles of minimization were performed with all heavy atoms restrained with 50 kcal/(mol·Å2), whereas solvent molecules and hydrogen atoms were free to move. Then non-restrained minimization was carried out involving 2,000 cycles of steepest descent minimization and 2,000 cycles of conjugated gradient minimization. Afterward, the whole system was first heated from 0 to 300 K in 50 ps using Langevin dynamics at a constant volume and then equilibrated for 400 ps at a constant pressure of 1 atm. A weak constraint of 10 kcal/ (mol·Å2) was used to restrain all the heavy atoms during the heating steps. Periodic boundary dynamics simulations were carried out for the whole system with an NPT (constant composition, pressure, and temperature) ensemble at a constant pressure of 1 atm and 300 K in the production step. In production phase, a 10-ns simulation was carried out. The trajectories were further analyzed using Cpptraj (Roe and Cheatham., 2013). The binding free energy of complexes was calculated using the MM-GBSA method.
Results and discussion
Multiple Sequence Alignment and Expression of Recombinant Tannase From Lachnospiraceae Bacterium
A search in the NCBI database for homologues tannase sequences yielded the putative TanALb protein from a species belonging to the family Lachnospiraceae (GenBank Accession No. MBQ6323131.1). A multiple sequence alignment was performed based on the protein sequence of TanALb with characterized tannase from other organisms (Supplementary Figure S1). The sequence alignment results revealed that TanALb showed 42.9% sequence similarity with TanBFnn from Fusobacterium nucleatum subsp. nucleatum (GenBank Accession No. ALQ42581.1) (Mancheño et al., 2020) and 40.4% with TanASl from Staphylococcus lugdunensis (GenBank Accession No. BAF03594.1) (Rusniok et al., 2010). However, it showed only 24% sequence similarity with TanBLp from Lactobacillus plantarum (GenBank Accession No. AB379685.1) (Matoba et al., 2013). TanALb exhibited the conserved catalytic triad (Ser205, His614, Asp582) typical for the tannase family (Supplementary Figure S1). Furthermore, TanALb also contained the conserved motif Gly203-X-Ser205-X-Gly207, which is typical of serine hydrolases (Ren et al., 2013). A phylogenetic analysis of TanALb and previously identified tannases indicated that TanALb is more similar with TanASl than the other investigated enzymes (Figure 1). Multiple sequence alignment and phylogenetic analysis, therefore, both indicated that TanALb may have tannase activity.
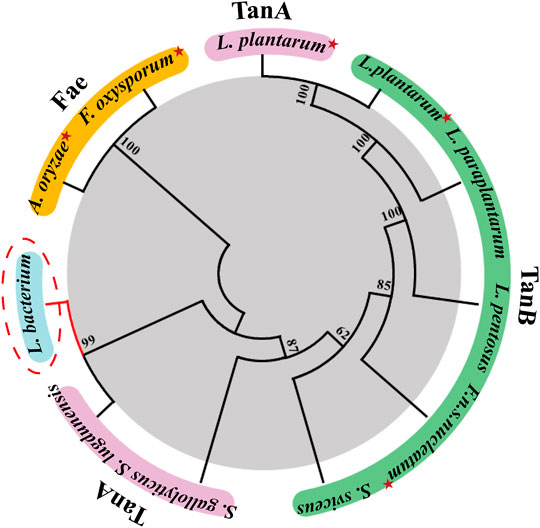
FIGURE 1. Phylogenetic tree of tannase from Lachnosperaceae bacterium (TanALb) and homologous enzymes. Tannases with similar amino acid sequences to TanALb were searched in the UniProt and NCBI databases, and the phylogenetic tree was constructed using MEGA 7.0. Enzymes with solved crystal structures are indicated with red star.
TanALb was purified using Ni-NTA affinity chromatography, ion exchange (Figure 2A) and size-exclusion chromatography (Figure 2B) of which the purification multiple was probably 6.9. The purified TanALb was analyzed via SDS-PAGE and showed a single protein band with an apparent molecular weight of approximately 68 kDa (Figure 2C), which was in agreement with the predicted molecular weight. Furthermore, size-exclusion chromatography of purified TanALb was performed to determine its native state in solution. A single peak was observed between the peaks of the standard protein markers actin from rabbit muscle (43 kDa) and conalbumin (75 kDa), which indicated that TanALb was a monomer in solution (Figure 2B).
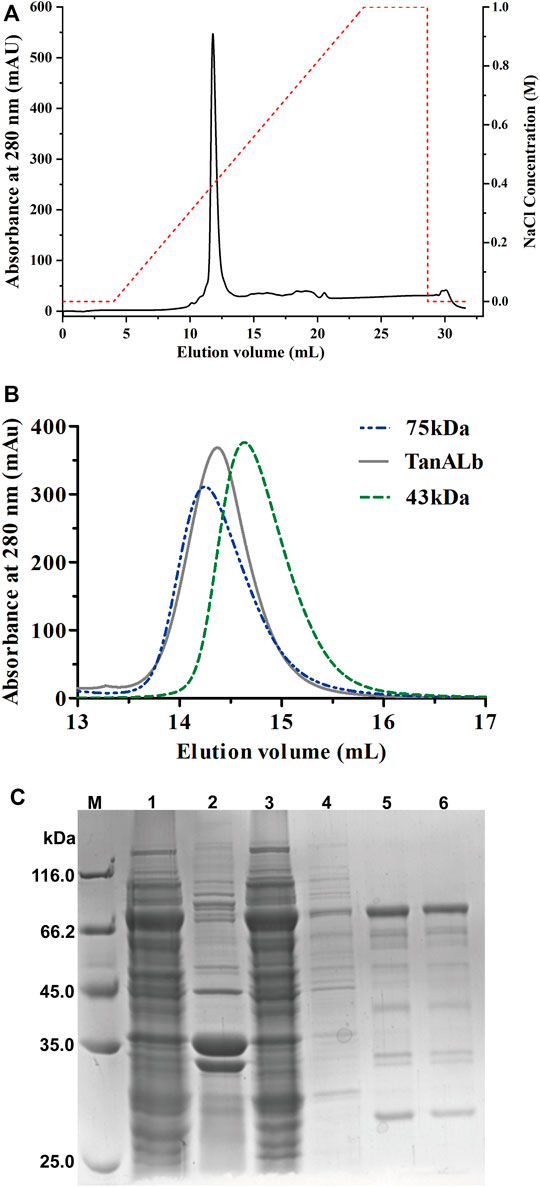
FIGURE 2. Purification of recombinant TanALb protein. (A) The purification of TanALb by ion exchange chromatography. (B) The purification of TanALb by size-exclusion chromatography. (C) SDS-PAGE analysis of the expression and purification of His6-TanALb. Lane M: protein molecular weight markers; Lanes 1–2: crude extract; Lane 3: flowthrough; Lane 4: unbound proteins; Lane 5: the sample before elution; Lane 6: fractions eluted from his affinity resin.
Biochemical Characterization of Tannase From Lachnospiraceae Bacterium
Since tannic acid is almost exclusively formed from poly-galloyl glucose derivatives, it was used as substrate to assess if TanALb can catalyze the hydrolysis of natural tannin. The gallic acid resulting from the degradation was identified using the rhodamine method, which indicated that tannic acid was hydrolyzed by TanALb.
The biochemical properties of TanALb were characterized using methyl gallate (MG) as the substrate. The temperature–activity profile revealed that the purified TanALb displayed the highest activity at 50°C, which was determined by incubating the TanALb in 20 mM phosphate buffer saline at pH 7.0. TanALb maintained more than 70% relative activity in a rather wide temperature range from 30°C to 55°C. However, the activity of TanALb decreased dramatically at temperatures over 60°C. (Figure 3A). Furthermore, the thermostability of the purified TanALb was also measured by pre-incubation at different temperatures in the range of 20°C–60°C. After pre-incubation of the enzyme between 20°C and 40°C, it maintained more than 70% residual activity, which indicated that TanALb was stable at moderate temperatures. However, incubation at temperature 60°C induced a dramatic decrease in TanALb activity within 30 min, and the enzyme was completely inactivated after 2 h (Figure 3B). Thus, TanALb was highly active in a wider range of temperatures, from 20°C to 50°C, exhibiting much higher thermostability than the previously reported tannases from L. plantarum (Curiel et al., 2009) and B. subtilis (Jana et al., 2013).
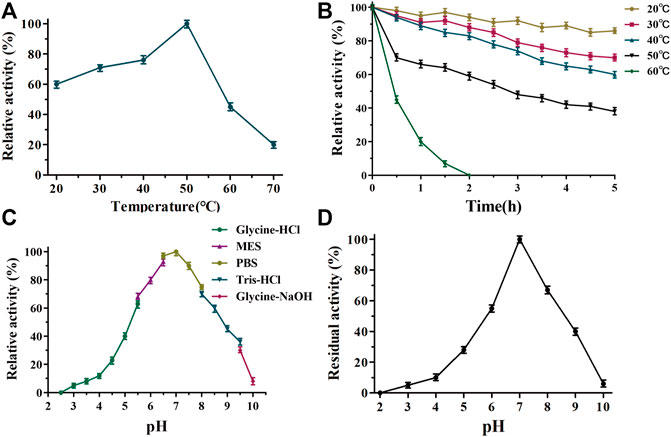
FIGURE 3. Determination of the optimal temperature (A), thermostability (B), pH optimum (C), and pH stability (D) of TanALb.
TanALb exhibited measurable activity toward methyl gallate at pH 2.5–10, with optimal activity in 20 mM phosphate buffer at pH 7.0. Furthermore, it maintained more than 80% of the maximal activity at pH 5.5–7.5 (Figure 3C). TanALb was also incubated for 2 h in different buffers to investigate its pH stability. Over 80% of the initial enzyme activity remained after incubation for 2 h at pH 6.5–7.5 (Figure 3D), while the residual activity decreased dramatically at pH values from 2–6.5 and 6.5–10.0.
The enzymatic activity of TanALb in the presence of various additives was determined by pre-incubating the enzyme at 50°C for 1.0 h, and the effects on the activity of TanALb are shown in Table 1. When Mg2+ and Ca2+ ions were added, the relative activity of TanALb was enhanced by 130% and 122%, respectively. Zn2+ and Co2+ had a weak influence on the activity of TanALb, while Cu2+ and Mn2+ had a strong inhibitory effect. Additionally, the enzyme activity was increased by surfactants, such as SDS, Tween-80, DSMO, and TritonX-100, while decreased by the metal chelating agent EDTA.
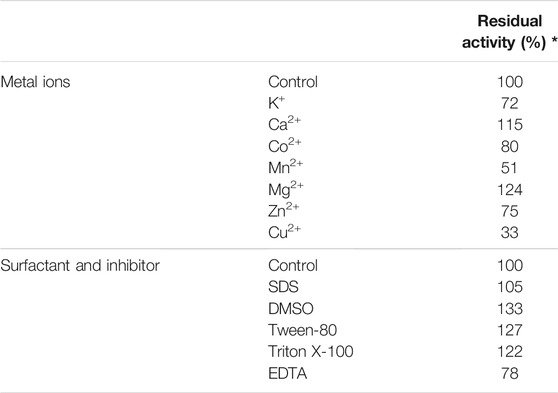
TABLE 1. Effects of metal ions, inhibitors, and surfactants on the activity of tannase from Lachnosperaceae bacterium (TanALb).
Kinetic Analysis on Substrate Preference of Tannase From Lachnospiraceae bacterium
Tannase catalyzes the hydrolysis of the galloyl ester bond, liberating gallic acid. Accordingly, tannase activity can be determined by measuring the concentration of gallic acid formed by the enzyme. In order to investigate the substrate specificity of TanALb different esters of gallic acid, methyl gallate (MG), ethyl gallate (EG), propyl gallate (PG), lauryl gallate (LG), and tannic acid (TA) were transformed into gallic acid by TanALb under the optimal catalytic conditions. The Km and kcat value were always used to compare the substrate specificity of enzymes. Km values of five kinds of substrates showed that tannic acid (TA) had the highest affinity, laurate gallate (LG) had lower activity than tannic acid, and methyl gallate (MG) had the lowest affinity for the TanALb (Table 2). The activity assay also showed that the catalytic efficiency (kcat/Km) of TanALb toward tannic acid (TA) was 5.3- to 12.5-fold larger than that toward the other four substrates. Analysis of catalytic efficiency implied that TanALb was conducive to using tannic acid (TA) as substrate. On the basis of these results, we can conclude that TanALb has high substrate affinity and catalytic efficiency for depside bonds. In addition, TanALb is capable of degrading not only esters of phenolic acid with short chain aliphatic alcohols but also esters of phenolic acids with long chain alcohols such as lauryl gallate, similar to TanBFnp and TanBFnn (Tomás-Cortázar et al., 2018; Mancheño et al., 2020).
Molecular Structure Simulation and Structural Analysis of Tannase From Lachnospiraceae bacterium
The Ramachandran plot (Supplementary Figure S2) for TanALb showed that 98% residues were in allowed regions, indicating that the 3D structure of the model was reasonable. The structure of TanALb was composed of nine β-strands and six α-helices. The β-strands formed a twisted β-sheet, in which β2 runs antiparallel to the other β-strands, similar to the crystal structure of TanBFnn. Furthermore, the structure of TanALb exhibited an almost parallel six-strand β-sheet surrounded by five α helices, two of which were located on the concave side of the sheet (α2 and α15) and three on the convex side (α3, α4, and α14), to form a central cavity (Figure 4A). In agreement with the sequence alignment, the structural characterization, therefore, indicated that TanALb belongs to the α/β-hydrolase superfamily (Remington et al., 1992). It is worth noting that the TanALb contained an insertion of a 109 residue stretch (residues 341–450) (Figure 4B), which is the largest structural deviation and may assume the role of the flap covering the active site of the enzyme (Mancheño et al., 2020).
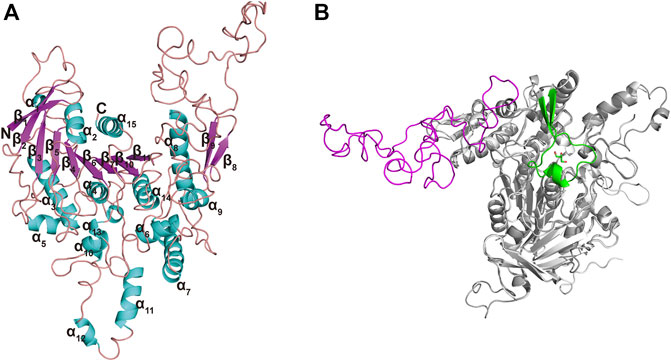
FIGURE 4. Ribbon representation of the TanALb homology model. (A) The modeled 3D-structure of TanALb based on the crystal structure of TanBFnn from Fusobacterium nucleatum subsp. Polymorphum (PDB code, 6YQ4). (B) Superimposition of the structures of TanALb (gray, ribbon model) and TanBFnn (white, ribbon model) reveals significant differences between both proteins predominantly found in the flap region. The flap lid in TanBFnn is shown in green and the hypothetic one in TanALb is shown in magenta.
Molecular Docking and Molecular Dynamics Simulation
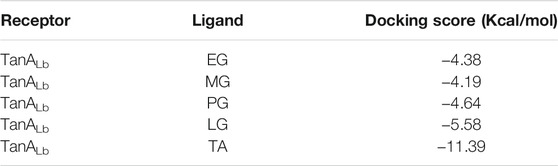
In order to further understand the mechanism of action of TanALb, molecular docking and MD simulation were carried out. In the overall docking conformation of TanALb, the substrates have formed a suitable steric complementarity with the binding site of TanALb. The docking scores (Table 3), the more negative, the better binding with protein, were consistent with the kinetic analysis. The contact residues during docking of ligands on the predicted structure of TanALb are presented in Figure 5 and Supplementary Figures S3–S5. Concretely, hydrogen bond interactions and H-π stacking were formed between TanALb and ethyl gallate. The oxygen atom of ethyl gallate, regarded as hydrogen bond donor, forms a hydrogen bond with the oxygen atom of backbone of Gly485 and the oxygen atom of side chain of Asn483. The benzene ring of ethyl gallate, forms a CH-π stacking with the alpha carbon atom of Lys482 (Figure 5A). The other three phenolic acid esters had similar binding modes (Supplementary Figures S3–S5). For the most suitable and largest substrate, the oxygen atom of the propyl group in tannic acid, regarded as hydrogen bond donor, forms hydrogen bonds with Gly485, Glu492, Gly493, Thr498, and Asn506. In addition, the two oxygen atoms of the phenolic hydroxyl group in tannic acid form two hydrogen bonds with the oxygen atom of the side chain of Asn483. VDW interactions were also formed between ethyl gallate and the surrounding residues (Figure 5B). These docking simulations may provide insights into the substrate specificity of TanALb, although the exact enzyme structures were not obtained (Wang et al., 2020).
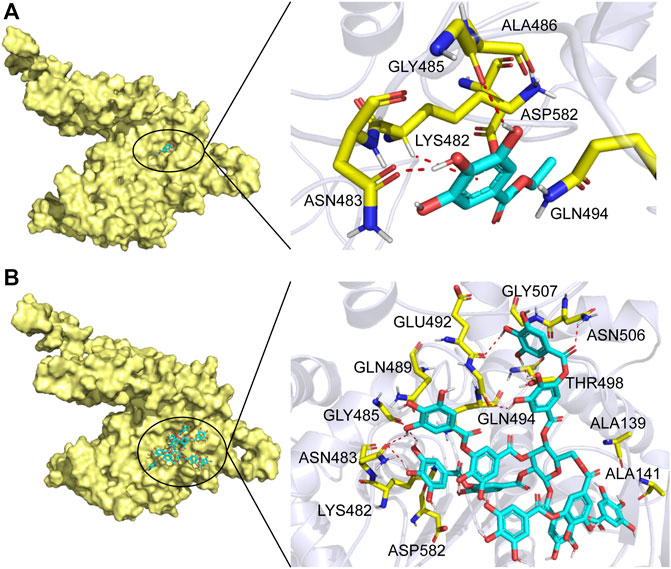
FIGURE 5. Substrate binding mode to TanALb. (A) The binding model of ethyl gallate on molecular surface of TanALb. (B) The binding model of tannic acid on molecular surface of TanALb. Substrates are colored cyan, and the molecular surface of TanALb is colored pale yellow. The residues involved in binding are shown in yellow sticks.
MD simulation was carried out to explore the role of the insertion part (residue 341–450). The results showed that the root means square deviation (RMSD) of heavy atoms of TanALb was less than 10.0 Å, that of tannic acid was less than 5.0 Å, and the system achieved equilibrium within 150 ns (Supplementary Figure S6). The final stable 3D structure binding model of the complex of TanALb and tannic acid is shown in Figure 6 and the binding energy of key residues are listed in Supplementary Tables S1, S2. During the MD simulation, tannic acid tended to approach the target loop structure (residue 341–450), and the other loop (residue 389–395) moved toward tannic acid to form a stable interaction (Figure 7). The main conformational change was the loop from amino acid 389 to 395, which swung like a pair of hands holding tannic acid. The remaining parts, which mainly interact with tannic acid, were the loop structures of 431–435 and 446–448, which were relatively stable here (Figure 8). It is supposed that the loop (residue 341–450) is a flap-like domain working for binding bulk substrates such as tannic acid, like the flexible structure in esterases (Truongvan et al., 2016).
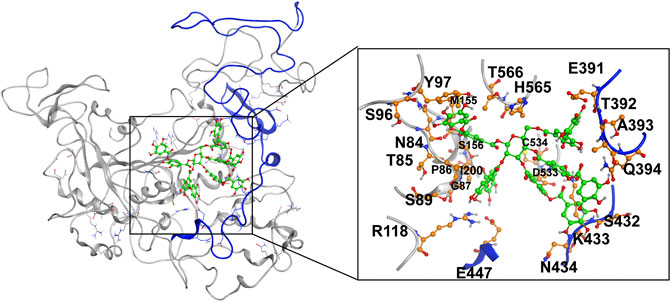
FIGURE 6. The final stable structure model after 500-ns molecular dynamics (MD) simulation. The sequence from 341 to 450 is highlighted with blue color. The atoms in tannic acid in binding area are depicted as green, and the residues in TanALb involved in binding are depicted as orange.
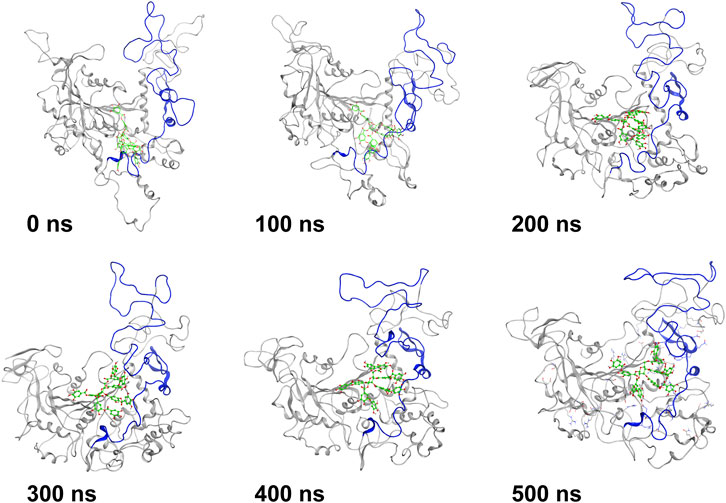
FIGURE 7. The structures at different simulation times of the complex. Residues 341–450 are shown as blue ribbons, and tannic acid is shown as green atoms. Other residues are shown as gray ribbons.
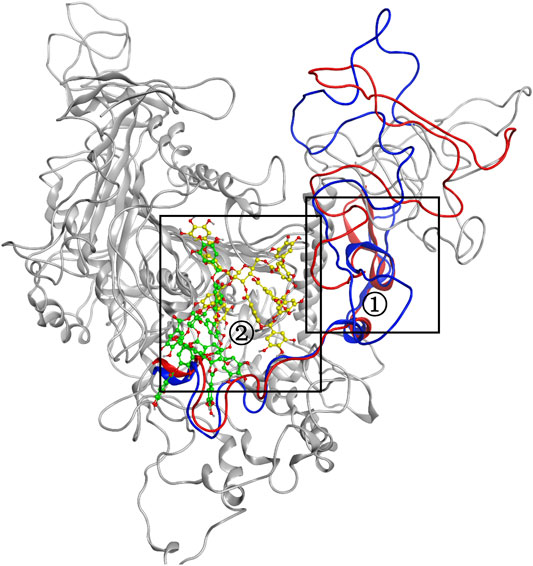
FIGURE 8. The conformational change for the complex of TanALb and tannic acid. Loops 341–450 of 0 -s simulation structure are shown in blue, and the ligand is shown in green. Meanwhile, loops 341–450 of 500-ns simulation structure are shown in red, and the ligand is shown in yellow.
Conclusion
A novel bacterial tannase from a species of Lachnospiraceae has been cloned and overexpressed in E. coli BL21 (DE3), purified, and characterized. It exhibited maximal activity at pH 7.0 and 50°C, and retained more than 70% relative activity from 30°C to 55°C. The enzyme retained more than 80% of the initial activity after incubation for 2 h in the pH range 6.5–7.5. TanALb is stable in the presence of surfactants such as SDS, DSMO, and TritonX-100. TanALb has broad substrate specificity, and investigating the detailed kinetic characteristics of TanALb advances our knowledge about this bacterial tannase at the molecular level. MD simulation revealed an assumed flap-like lid, which was beneficial to the interactions with the substrates. This may be helpful to guide the application of tannase and provide the theoretical basis for the modification of tannase. As a novel bacterial tannase, TanALb has demonstrated superior properties that may help improve the industrial biodegradation or bioconversion of tannins. Moreover, TanALb is from Lachnospiraceae bacterium in ruminant gastrointestinal tract, and the research probably helps to increase our understanding of the relationship between tannases and gut microbiota and health.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
LG contributed to conception and design of the study. Experiments were conceived and supervised by LG and performed by KW, YG, JL, JW, and SY. Data were analyzed by LG and JW. The manuscript was initially drafted by LG and then revised by CR and SL. NJ, CR, and JW coordinated the project. YZ contributed to project administration. BL and SL took charge of the conceptualization and funding acquisition. All authors read and approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31600642), the Agricultural Science and Technology Innovation Project of Heilongjiang Academy of Agricultural Sciences (HNK2019CX19), and the Excellent Young Scholars Fund of Heilongjiang Academy of Agricultural Sciences (2020JCQN004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Haibo Liu from the Chinese Academy of Medical Sciences for providing the aforementioned modeling software and Wecomput Technology for providing computation consulting.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.806788/full#supplementary-material
References
Aboubakr, H. A., El-Sahn, M. A., and El-Banna, A. A. (2013). Some Factors Affecting Tannase Production by Aspergillus niger Van Tieghem. Braz. J. Microbiol. 44 (2), 559–567. doi:10.1590/s1517-83822013000200036
Aguilar, C. N., Rodríguez, R., Gutiérrez-Sánchez, G., Augur, C., Favela-Torres, E., Prado-Barragan, L. A., et al. (2007). Microbial Tannases: Advances and Perspectives. Appl. Microbiol. Biotechnol. 76 (1), 47–59. doi:10.1007/s00253-007-1000-2
Aharwar, A., and Parihar, D. K. (2018). Tannases: Production, Properties, Applications. Biocatal. Agric. Biotechnol. 15, 322–334. doi:10.1016/j.bcab.2018.07.005
Baxter, N. J., Lilley, T. H., Haslam, E., and Williamson, M. P. (1997). Multiple Interactions between Polyphenols and a Salivary Proline-Rich Protein Repeat Result in Complexation and Precipitation. Biochemistry 36 (18), 5566–5577. doi:10.1021/bi9700328
Case, D. A., Aktulga, H. M., Belfon, K., Ben-Shalom, I. Y., Brozell, S. R., Cerutti, D. S., et al. (2021). Amber 2021. California, USA: University of California Press.
Case, D. A., Darden, T. A., Cheatham, T. E., Simmerling, C. L., Wang, J., Duke, R. E., et al. (2008). Amber 10. San Francisco CA: University of California.
Chaitanyakumar, A., and Anbalagan, M. (2016). Expression, Purification and Immobilization of Tannase from Staphylococcus Lugdunensis MTCC 3614. AMB Expr. 6 (1), 89. doi:10.1186/s13568-016-0261-5
Charlton, A. J., Baxter, N. J., Khan, M. L., Moir, A. J. G., Haslam, E., Davies, A. P., et al. (2002). Polyphenol/Peptide Binding and Precipitation. J. Agric. Food Chem. 50 (6), 1593–1601. doi:10.1021/jf010897z
Chung, K.-T., Wong, T. Y., Wei, C.-I., Huang, Y.-W., and Lin, Y. (1998). Tannins and Human Health: A Review. Crit. Rev. Food Sci. Nutr. 38 (6), 421–464. doi:10.1080/10408699891274273
Curiel, J. A., Rodríguez, H., Acebrón, I., Mancheño, J. M., De Las Rivas, B., and Muñoz, R. (2009). Production and Physicochemical Properties of Recombinant Lactobacillus Plantarum Tannase. J. Agric. Food Chem. 57 (14), 6224–6230. doi:10.1021/jf901045s
Dai, X., Liu, Y., Zhuang, J., Yao, S., Liu, L., Jiang, X., et al. (2020). Discovery and Characterization of Tannase Genes in Plants: Roles in Hydrolysis of Tannins. New Phytol. 226 (4), 1104–1116. doi:10.1111/nph.16425
Dhiman, S., Mukherjee, G., and Singh, A. K. (2018). Recent Trends and Advancements in Microbial Tannase-Catalyzed Biotransformation of Tannins: A Review. Int. Microbiol. 21 (4), 175–195. doi:10.1007/s10123-018-0027-9
Gao, S., Wang, J., Wu, M., Zhang, H., Yin, X., and Li, J. (2012). Engineering Hyperthermostability into a Mesophilic Family 11 Xylanase fromAspergillus Oryzaeby In Silico Design of N-Terminus Substitution. Biotechnol. Bioeng. 110 (4), 1028–1038. doi:10.1002/bit.24768
Govindarajan, R., Revathi, S., Rameshkumar, N., Krishnan, M., and Kayalvizhi, N. (2016). Microbial Tannase: Current Perspectives and Biotechnological Advances. Biocatal. Agric. Biotechnol. 6, 168–175. doi:10.1016/j.bcab.2016.03.011
Jana, A., Maity, C., Halder, S. K., Das, A., Pati, B. R., Mondal, K. C., et al. (2013). Structural Characterization of Thermostable, Solvent Tolerant, Cytosafe Tannase from Bacillus Subtilis PAB2. Biochem. Eng. J. 77, 161–170. doi:10.1016/j.bej.2013.06.002
Jiménez, N., Esteban-Torres, M., Mancheño, J. M., Rivas, B. D., and Muñoz, R. (2014). Tannin Degradation by a Novel Tannase Enzyme Present in Some Lactobacillus Plantarum Strains. Appl. Environ. Microbiol. 80 (10), 2991–2997. doi:10.1128/aem.00324-14
Lekha, P., and Lonsane, B. (1997). Production and Application of Tannin Acyl Hydrolase: State of the Art. Adv. Appl. Microbiol. 44, 215–260. doi:10.1016/s0065-2164(08)70463-5
Lekshmi, R., Nisha, S. A., Vasan, P. T., and Kaleeswaran, B. (2021). A Comprehensive Review on Tannase: Microbes Associated Production of Tannase Exploiting Tannin Rich Agro-Industrial Wastes with Special Reference to its Potential Environmental and Industrial Applications. Environ. Res. 201, 111625. doi:10.1016/j.envres.2021.111625
Maisetta, G., Batoni, G., Caboni, P., Esin, S., Rinaldi, A. C., and Zucca, P. (2019). Tannin Profile, Antioxidant Properties, and Antimicrobial Activity of Extracts from Two Mediterranean Species of Parasitic Plant Cytinus. BMC Complement. Altern. Med. 19 (1), 82. doi:10.1186/s12906-019-2487-7
Mancheño, J. M., Atondo, E., Tomás‐Cortázar, J., Lavín, J. L., Plaza‐Vinuesa, L., Martín‐Ruiz, I., et al. (2020). A Structurally Unique Fusobacterium Nucleatum Tannase Provides Detoxicant Activity against Gallotannins and Pathogen Resistance. Microb. Biotechnol., 1–20. [Epub ahead of print]. doi:10.1111/1751-7915.13732
Matoba, Y., Tanaka, N., Noda, M., Higashikawa, F., Kumagai, T., and Sugiyama, M. (2013). Crystallographic and Mutational Analyses of Tannase from Lactobacillus Plantarum. Proteins 81 (11), 2052–2058. doi:10.1002/prot.24355
Mcdonald, M., Mila, I., and Scalbert, A. (1996). Precipitation of Metal Ions by Plant Polyphenols:Optimal Conditions and Origin of Precipitation. J. Agric. Food Chem. 44 (2), 599–606. doi:10.1021/jf950459q
Noguchi, N., Ohashi, T., Shiratori, T., Narui, K., Hagiwara, T., Ko, M., et al. (2007). Association of Tannase-Producing Staphylococcus Lugdunensis with colon Cancer and Characterization of a Novel Tannase Gene. J. Gastroenterol. 42 (5), 346–351. doi:10.1007/s00535-007-2012-5
Remington, S. J., Franken, S. M., Sussman, J., Frolow, F., Ollis, D., Verschueren, K., et al. (1992). The Alpha/beta Hydrolase Fold. Prot. Eng. 5, 197–211. doi:10.1093/protein/5.3.197
Ren, B., Wu, M., Wang, Q., Peng, X., Wen, H., Mckinstry, W., et al. (2013). Crystal Structure of Tannase from from Lactobacillus Plantarum. J. Mol. Biol. 425, 2737–2751. doi:10.1016/j.jmb.2013.04.032
Roe, D. R., and Cheatham, T. E. (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem.Theory Comput. 9 (7), 3084–3095. doi:10.1021/ct400341p
Rusniok, C., Couvé, E., Cunha, V. D., Gana, R. E., Zidane, N., Bouchier, C., et al. (2010). Genome Sequence ofStreptococcus Gallolyticus: Insights into its Adaptation to the Bovine Rumen and its Ability to Cause Endocarditis. J. Bacteriol. 192 (8), 2266–2276. doi:10.1128/jb.01659-09
Smeriglio, A., Barreca, D., Bellocco, E., and Trombetta, D. (2016). Proanthocyanidins and Hydrolysable Tannins: Occurrence, Dietary Intake and Pharmacological Effects. Br. J. Pharmacol. 174 (11), 1244–1262. doi:10.1111/bph.13630
Tomás-Cortázar, J., Plaza-Vinuesa, L., Rivas, B. D., Lavín, J. L., Barriales, D., Abecia, L., et al. (2018). Identification of a Highly Active Tannase Enzyme from the Oral Pathogen Fusobacterium Nucleatum Subsp. Polymorphum. Microb. Cell. Fact. 17 (1). doi:10.1186/s12934-018-0880-4
Truongvan, N., Jang, S., and Lee, C. (2016). Flexibility and Stability Trade-Off in Active Site of Cold-Adapted Pseudomonas Mandelii Esterase EstK. Biochemistry 55 (25), 3542–3549. doi:10.1021/acs.biochem.6b00177
Wang, F., Zhu, M., Song, Z., Li, C., Wang, Y., Zhu, Z., et al. (2020). Reshaping the Binding Pocket of Lysine Hydroxylase for Enhanced Activity. ACS Catal. 10 (23), 13946–13956. doi:10.1021/acscatal.0c03841
White, T. (1957). Tannins—their Occurrence and Significance. J. Sci. Food Agric. 8 (7), 377–385. doi:10.1002/jsfa.2740080702
Keywords: bacterial tannase, biochemical characterization, kinetic analysis, homology modeling, structural analysis
Citation: Guan L, Wang K, Gao Y, Li J, Yan S, Ji N, Ren C, Wang J, Zhou Y, Li B and Lu S (2021) Biochemical and Structural Characterization of a Novel Bacterial Tannase From Lachnospiraceae bacterium in Ruminant Gastrointestinal Tract. Front. Bioeng. Biotechnol. 9:806788. doi: 10.3389/fbioe.2021.806788
Received: 01 November 2021; Accepted: 17 November 2021;
Published: 15 December 2021.
Edited by:
Weidong Liu, Tianjin Institute of Industrial Biotechnology (CAS), ChinaReviewed by:
Fei Wang, Jiangxi Agricultural University, ChinaFenghua Wang, Tianjin University of Science and Technology, China
Copyright © 2021 Guan, Wang, Gao, Li, Yan, Ji, Ren, Wang, Zhou, Li and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijun Guan, cXFhaXBwaGhAc2luYS5jb20=; Shuwen Lu, c2h1d2VubEBzaW5hLmNvbQ==
 Lijun Guan
Lijun Guan Kunlun Wang1,2
Kunlun Wang1,2