- Experiment Center, Captital Institute of Pediatrics, Beijing, China
Bordetella pertussis is the most frequent causative agent for pertussis, which is a highly contagious disease. Here, we developed a method based on loop-mediated isothermal amplification (LAMP) and nanoparticle-based lateral flow biosensor (LFB) for the timely diagnosis of B. pertussis infections. A set of six primers was designed for LAMP reactions, and the LAMP results were rapidly and visually indicated using LFB. The recommended condition for the B. pertussis LAMP reactions is 40 min at 66°C. Our results confirmed that the LAMP-LFB assay could specifically detect B. pertussis and did not cross-react with non-B. pertussis isolates. The sensitivity of the B. pertussis LAMP-LFB assay was 50 fg per reaction. In particular, 108 nasopharyngeal swab (NPS) samples were collected to evaluate the B. pertussis LAMP-LFB assay, and the results were compared with those of the quantitative PCR (qPCR) method. The positive rates of B. pertussis LAMP-LFB and qPCR were 40.7% and 38.8%, respectively, and the agreement between the LAMP-LFB and qPCR results was 98%, with a kappa value of 0.96. The whole process of LAMP-LFB can be completed within 1 h, which is much shorter than that of qPCR, including about 15 min of rapid DNA extraction, 40 min of LAMP reaction, and within 2 min of the LFB test. Collectively, the B. pertussis LAMP-LFB assay developed in this report offers a new option for the rapid, reliable, and simple diagnosis of B. pertussis infections.
Introduction
Bordetella pertussis mainly causes pertussis, a highly infectious, even fatal illness in children. In the past few years, the resurgence of pertussis has become a global public health issue in spite of high vaccination rates (Wood and McIntyre, 2008; Cherry, 2013; Mooi et al., 2014; Yeung et al., 2017; Del Valle-Mendoza et al., 2021; Pandolfi et al., 2021). In China, B. pertussis infections are becoming more and more prevalent even with over 99% vaccination coverage in children during the last 20 years (Liu et al., 2018; Fu et al., 2019; Zhang et al., 2019; Kang et al., 2022). Consequently, a rapid and reliable laboratory diagnosis of B. pertussis is particularly important (Cherry et al., 2005; de Greeff et al., 2010; Tao et al., 2019; Wu et al., 2019; Macina and Evans, 2021).
The current approaches to the diagnosis of pertussis include direct fluorescent antibody (DFA) assay, culture-based approaches, serodiagnosis, and PCR assays (van der Zee et al., 2015). DFA is a simple fluorescent antibody examination done through microscopic observation directed to the pathogen, but lacks both specificity and sensitivity (Chia et al., 2004; van der Zee et al., 2015). Culture is the gold standard diagnostic test, but with very low sensitivity. Meanwhile, the process of culture is laborious and time-consuming, which do not help with timely treatment, especially for infants too young to be vaccinated. Serodiagnosis is another technique earlier used for confirmation of the clinical diagnosis of pertussis, but it suffers persistent problems, including cross-reactivity with other bacteria, not only with Bordetella species, and the interference of previous vaccination or previous infections (Chia et al., 2004; Mertens et al., 2007; Bock et al., 2012). At present, PCR-based assays [e.g., conventional PCR, real-time PCR (RT-PCR), and quantitative PCR (qPCR)] have been established for the detection of B. pertussis (Roorda et al., 2011; Tatti et al., 2011; Abu Raya et al., 2012; Gao et al., 2014; Pittet et al., 2014). In particular, RT-PCR and qPCR use labeled probes to release a reporter or high-resolution melt (HRM) analysis to the amplicon, thus allowing the real-time monitoring of the amplification results. However, RT-PCR and qPCR examination is rarely available in primary medical institutions or in underdeveloped areas due to the high requirements of equipment and skilled professionals for a PCR laboratory.
Loop-mediated isothermal amplification (LAMP) is a newly developed amplification technique amplifying DNA at an isothermal condition, which can be satisfied merely by a water bath or a heater. By using six primers directing the different regions of the target sequence, this method showed high specificity, sensitivity, and efficiency (Notomi et al., 2000; Kamachi et al., 2006; Fujino et al., 2015; Notomi et al., 2015). In this report, we employed LAMP to amplify the target sequence of the pertussis toxin (PT) promoter, ptxA (pertussis toxin subunit 1), assumed to be specific for B. pertussis (Grimprel et al., 1993; Nygren et al., 2000). The LAMP products were judged using nanoparticle-based lateral flow biosensor (LFB), a method for the detection of nucleic acid and protein molecules (Huang et al., 2021; Huang et al., 2020; Huang et al., 2019), which can visually, rapidly and objectively indicate the results without the need for any extra instrument. The B. pertussis LAMP-LFB assay was further evaluated by applying it to clinical nasopharyngeal swab (NPS) samples.
Materials and Methods
Reagents and Instruments
The DNA isothermal amplification kit, visual detection reagent (VDR), and the nanoparticle-based LFB were obtained from Huidexin Biotech Co., Ltd. (Tianjin, China). The primers and labeled primers used in this study were synthesized by AoKe Biotech (Beijing) Co., Ltd. (Beijing, China). The B. pertussis isolate and qPCR kits were purchased from Beijing Transgen Biotech Co., Ltd. (Beijing, China) and Shanghai ZJ Bio-Tech Co., Ltd. (Shanghai, China). Real-time turbidimeter LA-320C was purchased from Eiken Chemical Co., Ltd. (Tokyo, Japan).
Primer Design
A set of six primers, including two outer primers (F3 and B3), two inner primers (FIP and BIP), and two loop primers (LF* and LB), was designed based on the specific pertussis toxin (PT) promoter gene of Bordetella pertussis (genome positions 159549–159755; GeneBank: BX640422) using Primer Premier 5.0. The sequences, locations, and modifications of the primers used in this report are shown in Figure 1 and Table 1.
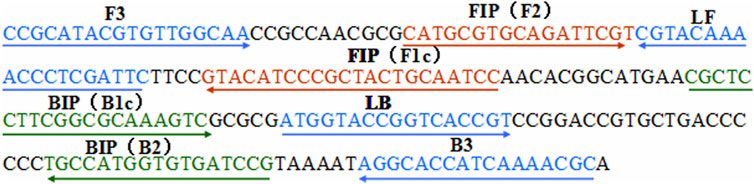
FIGURE 1. Locations of the primer sequences used in this study for targeting the pertussis toxin (PT) promoter region of Bordetella pertussis. Right and left arrows show the sense and complementary sequences, respectively. The colored text indicates the positions of the primers, including two outer primers (F3 and B3), two inner primers (FIP and BIP), and two loop primers (LF* and LB).
LAMP Reaction
LAMP reactions were performed as a one-step reaction in a 25-μl mixture containing 12.5 μl reaction buffer, 0.1 μmol L−1 each of the displacement primers (F3 and B3), 0.4 μmol L−1 each of the inner primers (FIP and BIP), 0.2 μmol L−1 each of the loop primers (LF* and LB), 1.0 μl Bst DNA polymerase (8 U), 0.5 μl biotin-14-dCTP (Huidexin Biotech Co., Ltd., Tianjin, China), 1.0 μl VDR, and 1.0 μl template for pure culture (5 µl for clinical sample).
Lateral Flow Biosensor (LFB) Test
LFB was constructed according to the previous report (Li et al., 2020). Briefly, LFB contained a sample pad, a conjugate pad, a nitrocellulose(NC)membrane (#Whatma99; Jie-Yi biotech Co., Ltd, Shanghai, China) and a absorbent pad (Huidexin Biotech Co., Ltd, Tianjin, China). On the conjugated pad, the detector reagents (dye streptavidin-coated gold nanoparticles (streptavidin-GNPs)) were laminated. As for the control line (CL) and test line (TL), Biotin-BSA and anti-FAM were immobilized on the NC membrane, respectively. The finally assembled biosensors were packaged in plastic box and conserved with silica gel desiccant at room temperature. For indicating the LAMP results, a 5 µl aliquot of LAMP reaction products was added to the sample pad, followed with 100 µl running buffer (10 mM PBS, PH 7.4 with 1% Tween 20), The results was indicated within 2 min, two red lines at TL and CL represent positive and one red line at CL means negative.
Optimal Temperature for the B. pertussis LAMP Assay
The amplification temperatures were optimized from 60°C to 67°C with 1°C intervals for the optimal temperature of the LAMP reaction. The DNA template of B. pertussis was used as a positive control and distilled water (DW) was used as the blank control. The LAMP reactions were monitored using real-time turbidity measurements.
Specificity of the B. pertussis LAMP-LFB Assay
To evaluate the specificity of the B. pertussis LAMP-LFB assay, the DNA templates from B. pertussis and non-B. pertussis strains (Table 2) were tested at least twice with the assay.
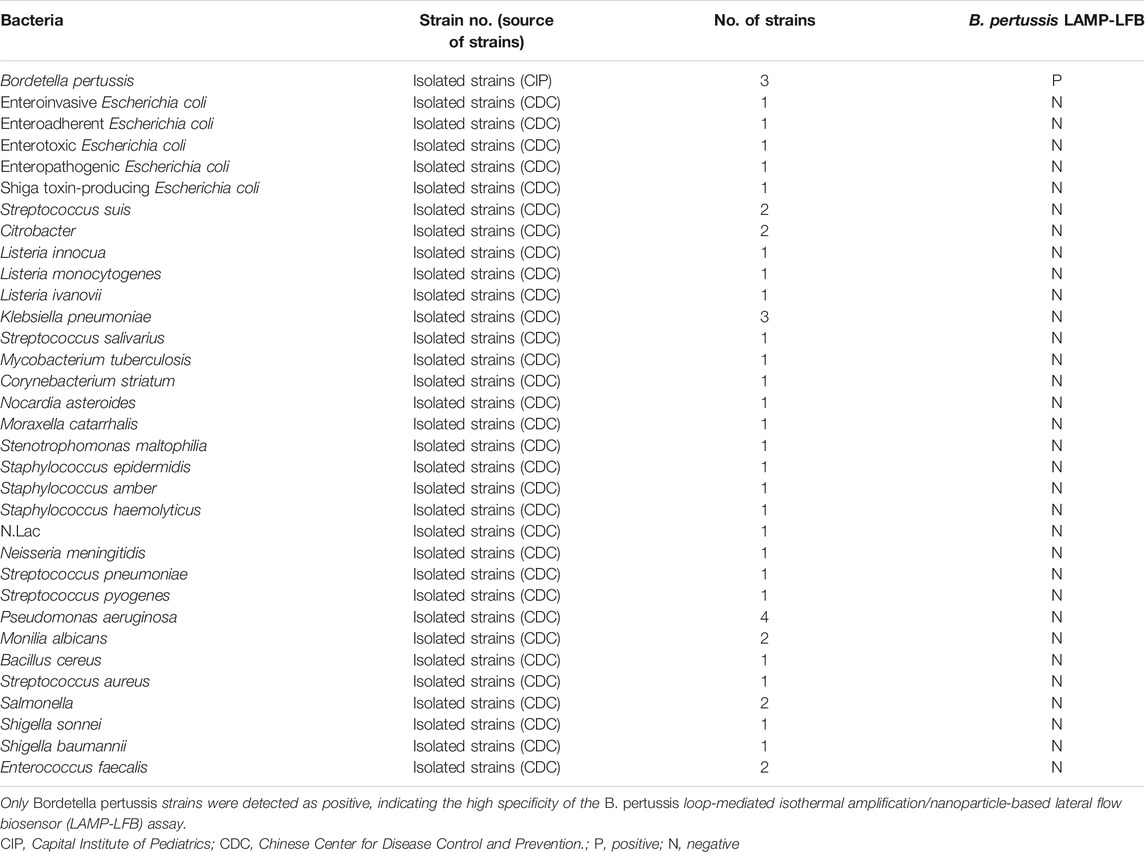
TABLE 2. Bacterial strains used to determine the specificity of loop-mediated isothermal amplification (LAMP)
Sensitivity of the B. pertussis LAMP-LFB Assay
To verify the limit of detection (LoD), the DNA templates of B. pertussis were serially diluted (5 ng ml−1; 500, 50, and 5 pg ml−1; and 500, 50, and 5 fg ml−1) for the LAMP assay, and 1 μl of each serial dilution or DW was added to the reaction mixtures. The LoD of the B. pertussis LAMP assay was determined using real-time turbidity measurement, VDR, and the LFB test. All tests were repeated at least twice.
Optimal Amplification Time for the B. pertussis LAMP Assay
Serially diluted templates were applied to obtain the optimal amplification time. LAMP reactions were conducted at 66°C with reaction times ranging from 10 to 40 min, with 10-min intervals. Each reaction time was verified twice.
Application of the B. pertussis LAMP-LFB Assay in Clinical Specimens
A total of 108 NPS samples collected from patients suspected of pertussis in the clinics of the Children’s Hospital affiliated with the Capital Institute of Pediatrics from January 1, 2019 to December 30, 2020 were retrospectively used. All samples were obtained with informed consent signed by the guardians of the participants. Nucleic extractions from these samples were firstly used for clinical and laboratory diagnosis. A volume of 5 μl DNA template was collected from the remaining samples for the B. pertussis LAMP-LFB assay. The results of the B. pertussis LAMP-LFB assay were compared with those of the qPCR assay for identical samples. All procedures were reviewed and approved by the Ethics Committee of the Capital Institute of Pediatrics.
Statistical Analysis
A comparison between two methods, qPCR and LAMP-LFB assay, was analyzed using the χ2 test with SPSS software (version 11.5). A p < 0.05 was considered statistically significant.
Results
Confirming the Feasibility of the B. pertussis LAMP Reaction
The feasibility of the B. pertussis LAMP primer set (Figure 1 and Table. 1) was confirmed using DNA templates extracted from B. pertussis strains. The LAMP reaction was conducted at 64°C for ∼60°min. The results showed that the templates were effectively amplified, and no amplifications were observed for DW (blank control) (Figure 2). Thus, the primer set designed in our report was used as the candidate to establish the B. pertussis LAMP-LFB assay.
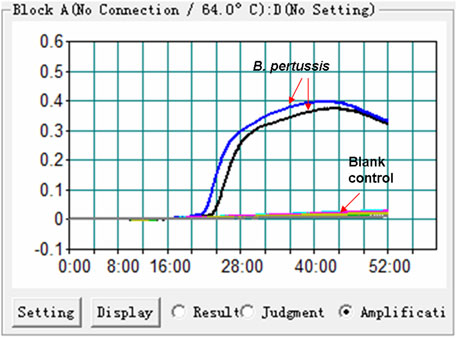
FIGURE 2. Effectiveness of the primer set for the Bordetella pertussis loop-mediated isothermal amplification (LAMP) reaction. The DNA templates extracted from B. pertussis strains were effectively amplified with LAMP reaction at 64°C, while there was no reaction for the blank controls (distilled water, DW).
Optimal Temperature for the B. pertussis LAMP Reaction
We used eight different temperatures ranging from 60°C to 67°C at 1°C intervals for 40°min to conduct the B. pertussis LAMP reaction for the optimal temperature. As shown in Figure 3, faster amplification was observed at 66°C, which was subsequently used for the B. pertussis LAMP-LFB reaction as the optimal temperature in this report.
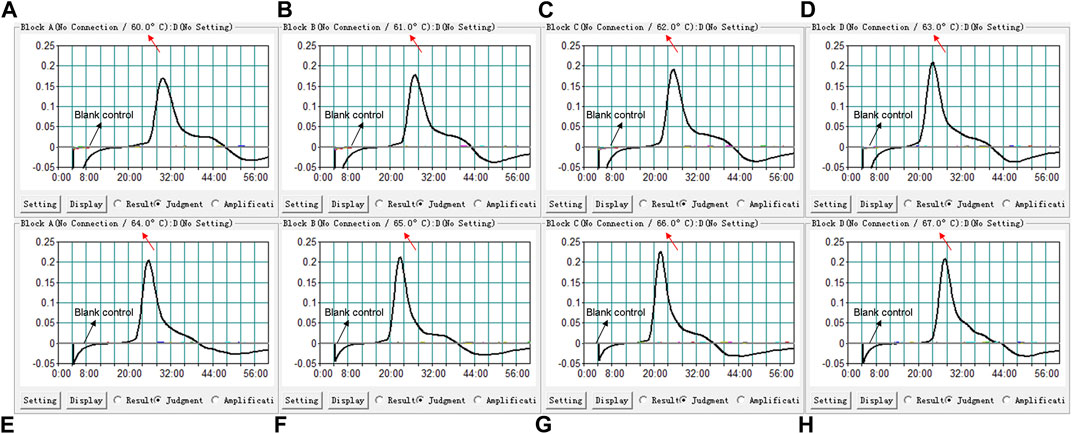
FIGURE 3. Temperature optimization for the loop-mediated isothermal amplification (LAMP) assay. LAMP reactions detecting Bordetella pertussis were conducted using real-time turbidimetry and kinetic curves (A–H) at different temperatures ranging from 60°C to 67°C were acquired, showing that 66°C was optimal for the B. pertussis LAMP reaction.
Sensitivity of the LAMP-LFB Assay for the Detection of B. pertussis
The DNA templates of B. pertussis were serially diluted to examine the LoD of the B. pertussis LAMP-LFB assay. The results were indicated by LFB and further confirmed by turbidity and VDR. As shown in Figure 4, the LoD of the B. pertussis LAMP-LFB assay was as low as 50 fg (∼12 copies) per reaction.

FIGURE 4. Sensitivity confirmation of the Bordetella pertussis loop-mediated isothermal amplification (LAMP) assay. The sensitivity of the assay was analyzed using 10-fold serial dilutions from 5 ng to 5 fg per reaction. The LAMP reactions with various levels of DNA templates were recorded with turbidity curves, and the products were indicated by a visual detection reagent (VDR) and a nanoparticle-based lateral flow biosensor (LFB). Turbidity curves/tubes/strips 1–7 represent different concentrations of DNA: 5 ng; 500, 50, and 5 pg; and 500, 50, and 5 fg per reaction. Turbidity curve/tube/strip 8 represents the blank control. TL, test line; CL, control line.
Optimal Time for the B. pertussis LAMP Reaction
We examined a total of four reaction times, 10–40 min with 10-min intervals, for the optimal amplification time of the B. pertussis LAMP assay. As shown in Figure 5, at 40 min, the amplicon of the diluted template at the LoD level was successfully detected by LFB, in which two red lines appeared respectively at the location of the test line (TL) and the control line (CL) on the strips. Therefore, 40 min was subsequently used as the optimal time for the B. pertussis LAMP assay. Hence, the whole procedure, which included rapid DNA extraction (15 min), LAMP reaction (40 min), and LFB indication (2 min), takes approximately 60 min, which is only half of that of qPCR.
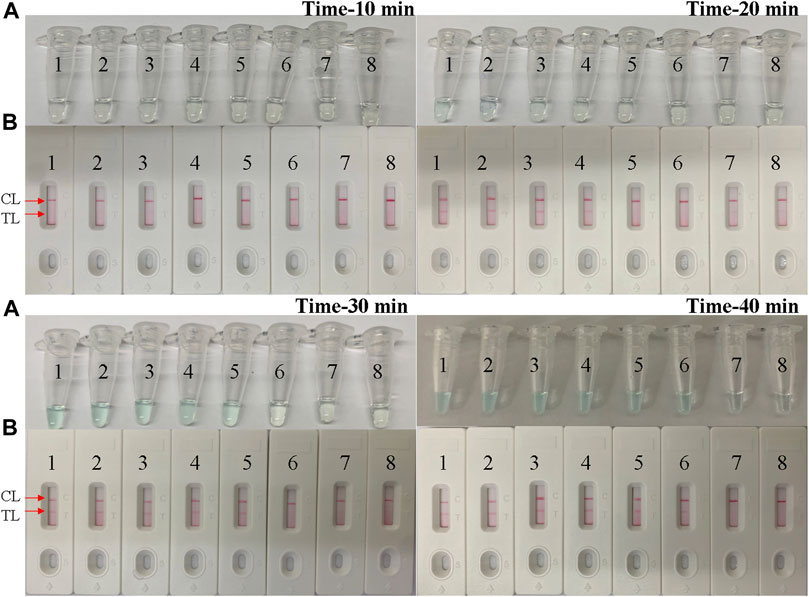
FIGURE 5. Optimal time for the Bordetella pertussis loop-mediated isothermal amplification (LAMP) assay. Tenfold serial dilutions of the B. pertussis templates were used for the optimization of time. The LAMP reactions were conducted at different times from 10 to 40 min, with 10-min intervals. The products were indicated by the visual detection reagent (A) and a nanoparticle-based lateral flow biosensor (LFB) test (B). Tubes/strips 1–7 represent serial dilutions of DNA: 5 ng; 500, 50, and 5 pg; and 500, 50, and 5 fg per reaction. Tube/strip 8 represents the blank control. TL, test line; CL, control line.
Specificity of the B. pertussis LAMP-LFB Assay
The specificity of the B. pertussis LAMP-LFB assay was examined using B. pertussis and non-B. pertussis strains (Table 2). As in the results shown in Figure 6, only CL lines appeared on the LFB strips of the non-B. pertussis strains and blank controls, while two red lines appeared at the CL and TL locations on the strips of the B. pertussis strains, suggesting the specificity of the primers in that only DNA isolates from B. pertussis strains could be amplified.
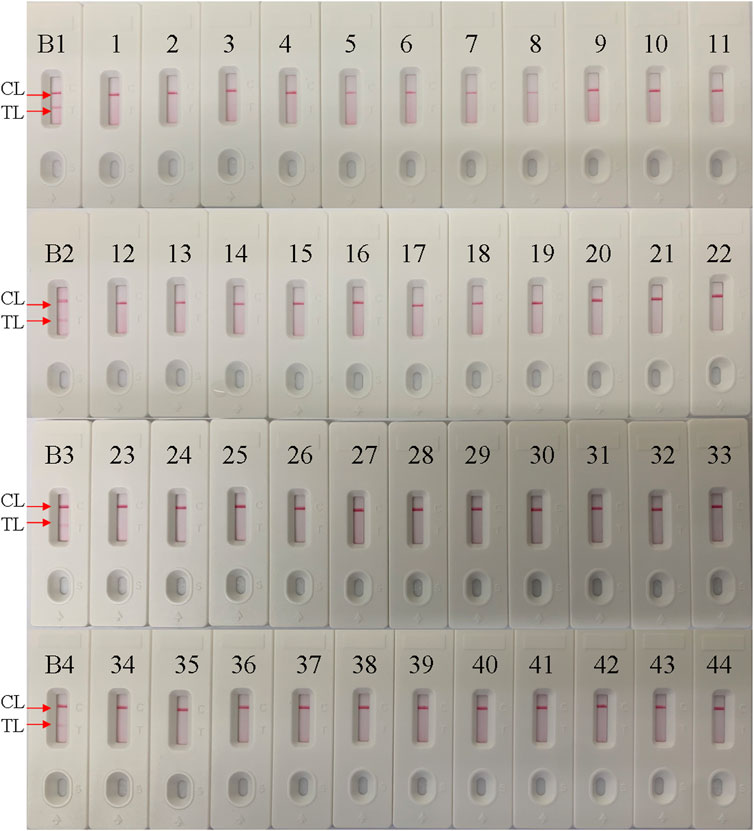
FIGURE 6. Specificity conformation for the Bordetella pertussis loop-mediated isothermal amplification (LAMP) assay. A lateral flow biosensor was applied for the LAMP products. Strips B1–B4 represent DNA isolation of the B. pertussis-positive clinical samples; strips 1–44 represent the other bacterial strains, shown in Table 2. TL, test line; CL, control line.
Application of the B. pertussis LAMP-LFB Assay in Clinical Specimens
In order to confirm its clinical application value, the optimized B. pertussis LAMP-LFB assay was used to detect 108 NPS samples, which were also detected using qPCR. The results (Figure 7) showed that 44 samples (40.7%) tested positive with the LAMP-LFB assay, while 42 samples (38.8%) tested positive with the qPCR. The agreement in the results between the qPCR and the LAMP-LFB assay was 98%, with a kappa value of 0.96.
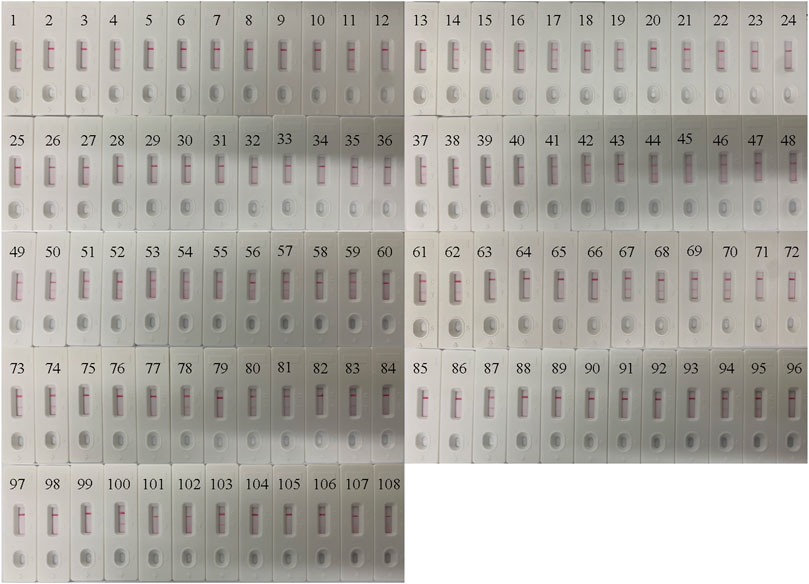
FIGURE 7. Application of the Bordetella pertussis loop-mediated isothermal amplification/nanoparticle-based lateral flow biosensor (LAMP-LFB) assay in clinical specimens. A total of 108 nasopharyngeal swab (NPS) samples were detected by the LAMP-LFB assay, with the results showing 44 samples testing positive.
Discussion
As a previous major cause of infant death, the morbidity and mortality of B. pertussis infections have significantly declined, benefitting from general vaccinations in childhood since 1950s. However, in the last 20 years, global resurgence was found in several highly vaccinated populations (Mooi et al., 2014; Yeung et al., 2017; Liu et al., 2018; Fu et al., 2019; Tao et al., 2019; Wu et al., 2019; Zhang et al., 2019; Kang et al., 2022). An estimation of the infection frequency derived from seroprevalence studies among adolescents and adults revealed a high circulation rate (1%–9% annually) in vaccinated populations (Wood and McIntyre, 2008; Cherry, 2013). Thus, the early detection of B. pertussis enables not only timely treatment, especially for infants much more fragile to the infection, but also the prevention of transmission and unnecessary diagnostic procedure, especially during an outbreak.
In this report, a simple LAMP-LFB assay for the detection of B. pertussis was designed and validated by its application in clinical samples. The assay takes less time, ∼60 min, with 15 min for rapid DNA extraction, 40 min for LAMP reaction, and 2 min for LFB detection, which was more rapid than that of traditional molecule-based diagnosis (e.g., PCR-based assay). Moreover, the significant advantage of the LAMP reaction is the isothermal condition, so the assay can be easily carried out under any experiment conditions with just a thermostatic water bath or a heater. The LFB test can subjectively indicate the results of the amplicons within 2 min.
As a molecular technique, the efficiency of the LAMP reaction is mostly decided by the primers and its targeting sequence. In previous studies, the sequence of the PT promoter has been considered as the very specific region for the diagnosis of B. pertussis. Comparison with previously used popular targeting sequences, such as insert sequence (IS) 1002 or IS481, demonstrated that primers targeting the PT gene showed marked reliability, selectivity, and sensitivity (van der Zee et al., 2015; Kilgore et al., 2016; Wu et al., 2019). We designed six primers targeting the PT sequence for the LAMP assay. The efficiency was conformed, as shown in Figure 3G, with the amplification peaking at 20–24 min under the condition of 66°C by turbidimetry. The specificity was confirmed by using the genomic templates of other bacterial strains, shown in Table 2. The high specificity (100%) indicated that the LAMP-LFB assay we investigated was reliable for the detection of B. pertussis.
In addition to the specificity, sensitivity was confirmed by using serial dilutions of genomic DNA. As shown in Figure 4, the lowest detection level of the LAMP-LFB assay was 50 fg of the DNA templates isolated from pure culture of B. pertussis. Since the LF primers were labeled with Fam at the 5’ end and biotin-14-dCTP was used in the reaction system, the amplicons positive in LAMP were simultaneously labeled with Fam and biotin, which can be detected by LFB. Thus, the positive LAMP amplicons displayed two red lines (CL and TL), while the negative reactions and the blank control displayed only the CL line when the reaction products were tested using LFB. The use of biotin-14-dCTP in the LAMP reaction instead of a biotin-labeled primer such as FIP absolutely avoided the interference of primer dimers containing Fam-labeled LF and biotin-labeled FIP.
For evaluation of the feasibility of the LAMP-LFB assay in the clinical diagnosis of B. pertussis infections, we compared the test with qPCR, the established method for B. pertussis diagnosis. Of the 108 clinical samples tested, 42 (38.8%) were positive by qPCR and 44 (40.7%) were positive by the LAMP-LFB assay. According to manual in the kit, the LoD of the qPCR assay used in this report is 250 copies, which is equal to about 1 pg DNA template, while the B. pertussis LAMP-LFB assay we conducted displayed better sensitivity with an LoD of 50 fg DNA template. For this reason, the LAMP-LFB assay yielded a higher positive rate than that of the qPCR assay in the clinical samples. Besides the lower LoD of the qPCR kit, the presence of some inhibitors specific to qPCR may have also contributed to the lower positive rates of detections. Therefore the application of the B. pertussis LAMP-LFB assay was verified to be sensitive and specific for the clinical diagnosis of B. pertussis infections. Moreover, the lower cost of the LAMP-LFB assay could also benefit its extensive application prospects in resource-limited laboratories.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Capital Institute of Pediatrics (ethical approval no. SHERLL2021031). The patients/participants legal guardian/next of kin provided written informed consent to participate in this study.
Author Contributions
YW and XC designed this study and revised the manuscript. CS, FX, and JF performed the experiments. CS analyzed the data and drafted the manuscript. CS, FX, JF, XH, NJ, and ZX contributed to the reagents and materials. YW conducted the software. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by National Key Research and Development Program of China (Grant Nos. 2021YFC2301101 (YW), 2021YFC2301102 (YW)) and the Research Foundation of Capital Institute of Pediatrics (Grant No. PY-2019-07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Rong Mi (Neonates Department, Children’s Hospital Affiliated to Capital Institute of Pediatrics) and Prof. Linqing Zhao (Virus Laboratory of Capital Institute of Pediatrics) for their kind help.
References
Abu Raya, B., Bamberger, E., Gershtein, R., Peterman, M., and Srugo, I. (2012). The Laboratory Diagnosis of Bordetella Pertussis Infection: a Comparison of Semi-nested PCR and Real-Time PCR with Culture. Eur. J. Clin. Microbiol. Infect. Dis. 31, 619–622. doi:10.1007/s10096-011-1327-6
Bock, J. M., Burtis, C. C., Poetker, D. M., Blumin, J. H., and Frank, M. O. (2012). Serum Immunoglobulin G Analysis to Establish a Delayed Diagnosis of Chronic Cough Due to Bordetella Pertussis. Otolaryngol. Head Neck Surg. 146 (1), 63–67. doi:10.1177/0194599811425145
Cherry, J. D. (2013). Pertussis: Challenges Today and for the Future. Plos Pathog. 9 (7), e1003418. doi:10.1371/journal.ppat.1003418
Cherry, J. D., Grimprel, E., Guiso, N., Heininger, U., and Mertsola, J. (2005). Defining Pertussis Epidemiology. Pediatr. Infect. Dis. J. 24 (Suppl. 5), S25–S34. doi:10.1097/01.inf.0000160926.89577.3b
Chia, J. H., Su, L. H., Lin, P. Y., Chiu, C. H., Kuo, A. J., Sun, C. F., et al. (2004). Comparison of Multiplex Polymerase Chain Reaction, Culture, and Serology for the Diagnosis of Bordetella Pertussis Infection. Chang Gung Med. J. 27, 408–415.
de Greeff, S. C., de Melker, H. E., van Gageldonk, P. G. M., Schellekens, J. F. P., van der Klis, F. R. M., Mollema, L., et al. (2010). Seroprevalence of Pertussis in the Netherlands: Evidence for Increased Circulation of Bordetella Pertussis. PLoS One 5, e14183. doi:10.1371/journal.pone.0014183
Del Valle-Mendoza, J., Del Valle-Vargas, C., Aquino-Ortega, R., Del Valle, L. J., Cieza-Mora, E., Silva-Caso, W., et al. (2021). Clinical Characteristics and Molecular Detection of Bordetella Pertussis in Hospitalized Children with a Clinical Diagnosis of Whooping Cough in Peru. Iran J. Microbiol. 13 (1), 23–30. doi:10.18502/ijm.v13i1.5488
Fu, P., Wang, C., Tian, H., Kang, Z., and Zeng, M. (2019). Bordetella Pertussis Infection in Infants and Young Children in Shanghai, China, 2016-2017: Clinical Features, Genotype Variations of Antigenic Genes and Macrolides Resistance. Pediatr. Infect. Dis. J. 38 (4), 370–376. doi:10.1097/INF.0000000000002160
Fujino, M., Suzuki, E., Watanabe, M., and Nakayama, T. (2015). Loop-Mediated Isothermal Amplification (LAMP) Aids the Clinical Diagnosis of Pertussis. Jpn. J. Infect. Dis. 68 (6), 532–533. doi:10.7883/yoken.JJID.2015.125
Gao, F., Mahoney, J. C., Daly, E. R., Lamothe, W., Tullo, D., and Bean, C. (2014). Evaluation of a Multitarget Real-Time PCR Assay for Detection of Bordetella Species during a Pertussis Outbreak in New Hampshire in 2011. J. Clin. Microbiol. 52, 302–306. doi:10.1128/JCM.01656-13
Grimprel, E., Bégué, P., Anjak, I., Betsou, F., and Guiso, N. (1993). Comparison of Polymerase Chain Reaction, Culture, and Western Immunoblot Serology for Diagnosis of Bordetella Pertussis Infection. J. Clin. Microbiol. 31 (10), 2745–2750. doi:10.1128/jcm.31.10.2745-2750.1993
Huang, Y., Cheng, Z., Han, R., Gao, X., Qian, L., Wen, Y., et al. (2020). Target-Induced Molecular-Switch On Triple-Helix DNA-Functionalized Carbon Nanotubes For Simultaneous Visual Detection Of Nucleic Acids And Proteins. Chem. Commun. (Camb) 56 (88), 13657–13660. doi:10.1039/d0cc05986b
Huang, Y., Xu, T., Luo, Y., Liu, C., Gao, X., Cheng, Z., et al. (2021). Ultra-Trace Protein Detection by Integrating Lateral Flow Biosensor with Ultrasound Enrichment. Anal. Chem. 93 (5), 2996–3001. doi:10.1021/acs.analchem.0c05032
Huang, Y., Xu, T., Wang, W., Wen, Y., Li, K., Qian, L., et al. (2019). Lateral Flow Biosensors Based On The Use Of Micro- And Nanomaterials: A Review On Recent Developments. Mikrochim Acta. 187 (1), 70. doi:10.1007/s00604-019-3822-x
Kang, L., Cui, X., Fu, J., Wang, W., Li, L., Li, T., et al. (2022). Clinical Characteristics of 967 Children with Pertussis: a Single-center Analysis over an 8-year Period in Beijing, China. Eur. J. Clin. Microbiol. Infect. Dis. 41 (1), 9–20. doi:10.1007/s10096-021-04336-w
Kamachi, K., Toyoizumi-Ajisaka, H., Toda, K., Soeung, S. C., Sarath, S., Nareth, Y., et al. (2006). Development and Evaluation of a Loop-Mediated Isothermal Amplification Method for Rapid Diagnosis of Bordetella Pertussis Infection. J. Clin. Microbiol. 44 (5), 1899–1902. doi:10.1128/JCM.44.5.1899-1902.2006
Kilgore, P. E., Salim, A. M., Zervos, M. J., and Schmitt, H-J. (2016). Pertussis: Microbiology, Disease, Treatment, and Prevention. Clin. Microbiol. Rev. 29 (3), 449–486. doi:10.1128/CMR.00083-15
Li, S., Jiang, W., Huang, J., Liu, Y., Ren, L., Zhuang, L., et al. (2020). Highly Sensitive And Specific Diagnosis Of COVID-19 By Reverse Transcription Multiple Cross-Displacement Amplification-Labelled Nanoparticles Biosensor. Eur. Respir. J. 56 (6), 2002060. doi:10.1183/13993003.02060-2020
Liu, X., Wang, Z., Zhang, J., Li, F., Luan, Y., Li, H., et al. (2018). Pertussis Outbreak in a Primary School in China. Pediatr. Infect. Dis. J. 37 (6), e145–e148. doi:10.1097/INF.0000000000001814
Macina, D., and Evans, K. E. (2021). Bordetella Pertussis in School-Age Children, Adolescents and Adults: A Systematic Review of Epidemiology and Mortality in Europe. Infect. Dis. Ther. 10, 2071–2118. doi:10.1007/s40121-021-00520-9
Mertens, P. L., Stals, F. S., Steyerberg, E. W., and Richardus, J. H. (2007). Sensitivity and Specificity of Single IgA and IgG Antibody Concentrations for Early Diagnosis of Pertussis in Adults: an Evaluation for Outbreak Management in Public Health Practice. BMC Infect. Dis. 7, 53. doi:10.1186/1471-2334-7-53
Mooi, F. R., Van Der Maas, N. A. T., and De Melker, H. E. (2014). Pertussis Resurgence: Waning Immunity and Pathogen Adaptation - Two Sides of the Same coin. Epidemiol. Infect. 142, 685–694. doi:10.1017/S0950268813000071
Notomi, T., Mori, Y., Tomita, N., and Kanda, H. (2015). Loop-mediated Isothermal Amplification (LAMP): Principle, Features, and Future Prospects. J. Microbiol. 53, 1–5. doi:10.1007/s12275-015-4656-9
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated Isothermal Amplification of DNA. Nucleic Acids Res. 28, 63e. doi:10.1093/nar/28.12.e63
Nygren, M., Reizenstein, E., Ronaghi, M., and Lundeberg, J. (2000). Polymorphism in the Pertussis Toxin Promoter Region Affecting the DNA-Based Diagnosis of Bordetella Infection. J. Clin. Microbiol. 38 (1), 55–60. doi:10.1128/JCM.38.1.55-60.2000
Pandolfi, E., Gesualdo, F., Rizzo, C., Russo, L., Campagna, I., Carloni, E., et al. (2021). The Impact of Pertussis in Infants: Insights from a Hospital-Based Enhanced Surveillance System, Lazio Region, Italy, 2016 to 2019. Euro Surveill. 26 (24), 2000562. doi:10.2807/1560-7917.ES.2021.26.24.2000562
Pittet, L. F., Emonet, S., François, P., Bonetti, E.-J., Schrenzel, J., Hug, M., et al. (2014). Diagnosis of Whooping Cough in Switzerland: Differentiating Bordetella Pertussis from Bordetella Holmesii by Polymerase Chain Reaction. PLoS One 9 (2), e88936. doi:10.1371/journal.pone.0088936
Roorda, L., Buitenwerf, J., Ossewaarde, J. M., and van der Zee, A. (2011). A Real-Time PCR Assay with Improved Specificity for Detection and Discrimination of All Clinically Relevant Bordetella Species by the Presence and Distribution of Three Insertion Sequence Elements. BMC Res. Notes 4, 11. doi:10.1186/1756-0500-4-11
Tao, Y., Tang, M., Luo, L., Xiang, L., Xia, Y., Li, B., et al. (2019). Identification of Etiologic Agents and Clinical Characteristics for Patients Suspected of Having Pertussis in a Large Children's Hospital in China. Ann. Transl. Med. 7 (18), 443. doi:10.21037/atm.2019.08.85
Tatti, K. M., Sparks, K. N., Boney, K. O., and Tondella, M. L. (2011). Novel Multitarget Real-Time PCR Assay for Rapid Detection of Bordetella Species in Clinical Specimens. J. Clin. Microbiol. 49, 4059–4066. doi:10.1128/JCM.00601-11
van der Zee, A., Schellekens, J. F. P., and Mooi, F. R. (2015). Laboratory Diagnosis of Pertussis. Clin. Microbiol. Rev. 28, 1005–1026. doi:10.1128/cmr.00031-15
Wood, N., and McIntyre, P. (2008). Pertussis: Review of Epidemiology, Diagnosis, Management and Prevention. Paediatric Respir. Rev. 9 (3), 201–212. doi:10.1016/j.prrv.2008.05.010
Wu, D.-X., Chen, Q., Yao, K.-H., Li, L., Shi, W., Ke, J.-W., et al. (2019). Pertussis Detection in Children with Cough of Any Duration. BMC Pediatr. 19 (1), 236. doi:10.1186/s12887-019-1615-3
Yeung, K. H. T., Duclos, P., Nelson, E. A. S., Hutubessy, R. C. W., and Hutubessy Rcw Lancet Infect Dis, (2017). An Update of the Global burden of Pertussis in Children Younger Than 5 years: a Modelling Study. Lancet Infect. Dis. 17 (9), 974–980. doi:10.1016/s1473-3099(17)30390-0
Keywords: Bordetella pertussis, LAMP, lateral flow biosensor, rapid diagnosis, qPCR
Citation: Sun C, Xiao F, Fu J, Huang X, Jia N, Xu Z, Wang Y and Cui X (2022) Loop-Mediated Isothermal Amplification Coupled With Nanoparticle-Based Lateral Biosensor for Rapid, Sensitive, and Specific Detection of Bordetella pertussis. Front. Bioeng. Biotechnol. 9:797957. doi: 10.3389/fbioe.2021.797957
Received: 19 October 2021; Accepted: 23 December 2021;
Published: 08 February 2022.
Edited by:
Tailin Xu, Shenzhen University, ChinaReviewed by:
Junjie Yue, Institute of Biotechnology (CAAS), ChinaYan Huang, University of Science and Technology Beijing, China
Copyright © 2022 Sun, Xiao, Fu, Huang, Jia, Xu, Wang and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodai Cui, eGRjdWk2MUAxNjMuY29t; Yi Wang, d2lsZHdvbGYwMTAxQDE2My5jb20=
 Chunrong Sun
Chunrong Sun Yi Wang
Yi Wang Xiaodai Cui
Xiaodai Cui