- 1School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan, South Korea
- 2Department of Energy Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan, South Korea
Methylorubrum extorquens AM1 is an efficient platform strain possessing biotechnological potential in formate- and methanol-based single carbon (C1) bioeconomy. Constitutive expression or costly chemical-inducible expression systems are not always desirable. Here, several glucose-, xylose-, and levulinic acid (LA)-inducible promoter systems were assessed for the induction of green fluorescent protein (GFP) as a reporter protein. Among them, the LA-inducible gene expression system (HpdR/PhpdH) showed a strong expression of GFP (51-fold) compared to the control. The system was induced even at a low concentration of LA (0.1 mM). The fluorescence intensity increased with increasing concentrations of LA up to 20 mM. The system was tunable and tightly controlled with meager basal expression. The maximum GFP yield obtained using the system was 42 mg/g biomass, representing 10% of the total protein content. The efficiency of the proposed system was nearly equivalent (90%–100%) to that of the widely used strong promoters such as PmxaF and PL/O4. The HpdR/PhpdH system worked equally efficiently in five different strains of M. extorquens. LA is a low-cost, renewable, and sustainable platform chemical that can be used to generate a wide range of products. Hence, the reported system in potent strains of M. extorquens is highly beneficial in the C1-biorefinery industry to produce value-added products and bulk chemicals.
Introduction
Microbial utilization of single-carbon (C1) compounds as a feedstock to produce fuels, chemicals, and other value-added products is an ideal strategy to combat the depletion of fossil fuels and environmental pollution (Humphreys and Minton, 2018). C1 gases such as CO and CO2 are abundant and low-cost waste products generated by various industries such as petroleum refining, thermal power plants, steel mills, and landfills (Humphreys and Minton, 2018; Hwang et al., 2020). Although several studies have reported the utilization of CO as an energy and/or carbon source for the production of fuels and chemicals (Rittmann et al., 2015; Kim et al., 2016), the direct conversion of CO-containing gaseous waste into value-added products is challenging owing to its high toxicity to most living organisms. At the same time, reduced C1 compounds such as formate and methanol can be synthesized through the electrochemical reduction of CO2 or the hydration of synthesis gas (a mixture of CO and H2) at a relatively low cost (Agarwal et al., 2011; Ganesh, 2014; Hwang et al., 2020). These compounds are considered sustainable and eco-friendly microbial feedstocks, as well as alternatives to sugar-based raw materials for industrial fermentation (Belkhelfa et al., 2019; Yoon et al., 2021).
Among the methylotrophic microorganisms, the pink-pigmented facultative methylotrophic alpha-proteobacterium Methylorubrum extorquens AM1 is widely used as a catalyst; it can use both C1 (formate and methanol) and multi-carbon compounds such as succinate (C4), pyruvate (C3), and acetate (C2) as the sole carbon and energy sources (Peyraud et al., 2012; Schneider et al., 2012; Ochsner et al., 2015; Green and Ardley, 2018). Furthermore, the availability of the complete genomic sequence (Vuilleumier et al., 2009), metabolic models (Peyraud et al., 2011), and the basic set of genetic tools for overexpression, knockout, and transposon mutagenesis (Marx and Lidstrom, 2001; Ochsner et al., 2015) makes the strain an attractive host for biotechnological applications. Strain AM1 employs a number of interlocked metabolic processes for carbon assimilation, such as C1 pathways, the serine cycle, the TCA cycle, the ethylmalonyl-CoA (EMC) pathway, the poly-3-hydroxybutyrate (PHB) cycle, and gluconeogenesis (Anthony, 2011; Peyraud et al., 2011). These processes are connected by overlapping metabolites and enzymatic reactions (Peyraud et al., 2011). When cultivated on media containing formate or methanol, the metabolic flux largely goes through the tetrahydrofolate, serine, and EMC pathways, generating a stable supply of acetyl-CoA, which could serve as a precursor for the production of value-added chemicals in engineered M. extorquens AM1 strains (Peyraud et al., 2011; Zhang et al., 2019; Hwang et al., 2020). This strain has been engineered to produce a variety of value-added chemicals or biofuels such as PHB (Hwang et al., 2020; Yoon et al., 2021), polyhydroxyalkanoate terpolymers (Orita et al., 2014), 1-butanol (Hu and Lidstrom, 2014), 3-hydroxypropionic acid (3-HP) (Yang et al., 2017), mevalonic acid (Zhu et al., 2016), butadiene precursor (Yang et al., 2018), terpenoids (Sonntag et al., 2015a), mesaconic acids (Sonntag et al., 2015b), crotonic acids (Schada Von Borzyskowski et al., 2018), and itaconic acids (Lim et al., 2019) from formate or methanol.
The overexpression of genes in M. extorquens AM1 is a challenging task, as the commonly used promoters (such as λPL and λPR) are inactive, and the Plac promoter and its derivatives confer weak or leaky expression in M. extorquens (Marx and Lidstrom, 2001; Chubiz et al., 2013). These data indicate a lack of dynamic control of gene expression in M. extorquens. The strong native PmxaF promoter, which drives the expression of methanol dehydrogenase in M. extorquens, has almost exclusively been used for constitutive gene expression. A set of constitutive promoters (PmxaF, PcoxB, PfumC, and Ptuf) of different strengths has been reported for the expression of genes in M. extorquens AM1 (Schada Von Borzyskowski et al., 2015). In general, uncontrolled high-level expression or constitutive expression systems are not always desirable because they are detrimental to cells due to uncontrolled production of target protein(s) and structural instability (Lee and Keasling, 2005; Chaves et al., 2020). Moreover, cumate- or anhydrotetracycline-inducible hybrid promoters (PR/cmtO and PR/tetO) with low basal expression and high expression upon induction have been developed (Chubiz et al., 2013). Recently, Carrillo et al. (2019) introduced a set of isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoters that were strong, tight, and controllable, even exceeding the promoter elements available thus far. However, IPTG is not suitable on an industrial scale because of its toxicity and cost (Makrides, 1996; Browning et al., 2017). Thus, the use of inexpensive renewable substrates as inducers could be a feasible strategy for the large-scale production of biochemicals (Sathesh-Prabu et al., 2021).
Therefore, to expand the biotechnological potential of M. extorquens AM1 in formate- and methanol-based C1 bioeconomy, we aimed to develop an efficient tunable gene expression system with low-cost renewable substrates such as glucose, xylose, and levulinic acid (LA). These substrates can be used as starting materials to produce a wide range of chemicals (Cha et al., 2020; Hayes and Becer, 2020; Sathesh-Prabu et al., 2020). In the present study, we characterized the LA-inducible expression system for the induction of the expression of green fluorescent protein (GFP) as a quantitative reporter of promoter activity.
To our knowledge, this is the first report describing the characterization of an LA-inducible and tunable promoter system for synthetic biological applications in M. extorquens AM1.
Materials and Methods
Microbial Strains, Plasmids, and Culture Conditions
The wild-type M. extorquens AM1 strain was used to analyze the efficiency of the inducible promoter systems. The Escherichia coli strain DH10B was used for cloning. All plasmids and strains used in this study are listed in Supplementary Table S1. M. extorquens AM1 was cultivated in minimal salt medium (MSM) at 30°C under aerobic conditions, with agitation in an orbital incubator shaker at 200 rpm. The MSM contained 1.62 g NH4Cl, 0.2 g MgSO4, 2.21 g K2HPO4, 1.25 g NaH2PO4.2H2O, 15 mg Na2EDTA.2H2O, 4.5 mg ZnSO4.7H2O, 0.3 mg CoCl2.6H2O, 1 mg MnCl2.4H2O, 1 mg H3BO3, 2.5 mg CaCl2, 0.4 mg of Na2MoO4.2H2O, 3 mg FeSO47H2O, 0.3 mg CuSO4.5H2O, and 4 g succinate per liter. E. coli was cultured in a lysogeny broth (5 g yeast extract, 10 g peptone, and 10 g NaCl per liter) at 37°C, with agitation in a shaker incubator at 200 rpm. Tetracycline (10 μg/ml) was added to the growth medium to cultivate all the recombinant strains. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). LA was neutralized with 10 N NaOH and sterilized prior to use.
Construction of Inducible Expression Systems
Restriction enzymes, the Q5 high-fidelity DNA polymerase, and the Gibson assembly cloning kit were purchased from New England Biolabs (Ipswich, MA, USA) and used for cloning and plasmid construction. Five different promoter systems for the M. extorquens AM1 strain, including glucose-inducible HexR/Pzwf1, LA-inducible LvaR/PlvaA, 3HP- and LA-inducible HpdR/PhpdH and MmsR/PmmsA, and xylose-inducible XutR/PxutA, were evaluated. In our previous study (Sathesh-Prabu et al., 2021), we evaluated the efficiency of these inducible expression systems in Pseudomonas putida KT2440 by constructing promoter candidates in the pPROBE_eGFP+ plasmid (pBBR1-ori, KmR: a broad-host-range expression vector) (Kim et al., 2019), which served as a template for PCR amplification in the present study. To construct the inducible expression systems in M. extorquens AM1, the fragments containing the coding sequence of the transcription regulator of interest, their respective promoter sequences, and eGFP+ were generated through PCR using the respective plasmids as templates. The fragments were then cloned into the XbaI/BamHI double-digested expression vector pCM110_PmxaF_Fdh1, a derivative of the pCM110 plasmid (Marx and Lidstrom, 2001), via the Gibson assembly to generate the following plasmids: pMHRZ_eGFP+, pMLRL_eGFP+, pMHRH_eGFP+, pMMRM_eGFP+, and pMXRX_eGFP+, harboring the HexR/Pzwf1, LvaR/PlvaA, HpdR/PhpdH, MmsR/PmmsA, and XutR/PxutA systems, respectively. In addition, to compare the efficiency of the constructed systems, LacI/PL/O4 (Carrillo et al., 2019), the strongest inducible promoter system reported to date in M. extorquens AM1, was similarly constructed in the pCM110 plasmid, thereby generating pMPLO4_eGFP+. The LacI/PL/O4 promoter was adapted from the pCM110_PL/O4 plasmid. The oligonucleotides used for the PCR amplification of each promoter system are listed in Supplementary Table S2.
Construction of Recombinant Strains
The constructed plasmids were introduced into the electrocompetent cells of M. extorquens AM1. The electrocompetent cells of M. extorquens AM1 were prepared as previously described (Toyama et al., 1998). The constructed plasmids were transformed into electrocompetent cells via electroporation (0.1-cm gap cuvette at a voltage of 1.8 kV) using a MicroPulser electroporator (Bio-Rad, Hercules, CA, USA). Subsequently, ice-cold sterile nutrient broth (3 g beef extract and 5 g peptone per liter) was added to the cuvette. The cell suspension was transferred into a test tube and incubated at 30°C and 200 rpm for 3 h prior to being spread onto MSM with 1.5% agar, 0.4% succinate, and tetracycline (10 μg/ml).
Promoter Expression Assay
The GFP fluorescence intensity of the recombinant strains was estimated using their respective inducers. For the primary evaluation of all the inducible expression systems constructed in this study, the recombinant strains were first cultured in MSM until the late exponential phase and then subcultured [initial optical density at 600 nm (OD600) set to 0.1] in 5 ml of fresh MSM medium. After the strains were incubated for at least 12 h (OD600 ≈ 0.5–0.7), they were induced with 10 mM solutions of their respective inducers (glucose, xylose, LA, or 3-HP) or 1 mM IPTG solution. Thereafter, 180 µl of culture was collected at 24 h after induction and inoculated into a clear bottom Corning 96-well plate to measure the GFP fluorescence intensity (a gain of 30 with an excitation wavelength at 485 nm and emission wavelength at 535 nm) of the constructed systems in a microplate fluorescence reader (Infinite F200 PRO; Tecan, Grődig, Austria). Subsequently, the culture was diluted appropriately with phosphate-buffered saline (pH 7.2), and flow cytometric analysis of GFP fluorescence was performed using fluorescence-activated cell sorting (FACSCalibur Flow Cytometer; BD Biosciences, San Jose, CA, USA). Approximately 2 × 105 cells were analyzed per sample. The fluorescence intensity was normalized based on the OD600 of the culture.
Optimization of LA Induction
To determine the optimal time for the induction of the HpdR/PhpdH system, following the preparation of the culture as described above, LA (10 mM) was added at 6-h intervals from the initial inoculation (OD600 ≈ 0.1) for 24 h. Samples were collected every 6 h to measure the GFP fluorescence intensity for 48 h. To analyze the degradation of the added LA, residual concentrations of LA in the supernatant free of cells from the culture media of recombinant strains at 0, 24, and 48 h after LA addition (10 mM) were measured using high-performance liquid chromatography (HPLC) (Shimadzu HPLC station equipped with a refractive index detector and an SIL-20A auto-sampler, all from Shimadzu, Kyoto, Japan). The samples were eluted through a 4.6-mm 150-mm, 5-μm Zorbax SB-Aq column (Agilent, Santa Clara, CA, USA) at 40°C using 25 mM ammonium formate (pH 2.0) as the mobile phase, at a flow rate of 1 ml/min. The cell density was monitored with the OD600 using a Biochrom Libra S22 spectrophotometer (Biochrom, Cambridge, UK). An OD600 of 1.0 was equivalent to 0.278 g/L dry cell weight (Skovran et al., 2010).
To determine the optimal LA dosage, different concentrations (0.1, 0.25, 0.5, 1, 2, 4, 8, 10, 20, 40, 50, and 100 mM) of LA were added as a single dose at 0 h. Samples were collected at 12 and 24 h after induction to measure the GFP fluorescence intensity.
Qualitative and Quantitative Measurement of eGFP+
Following the optimization of time and concentration of LA for the induction of the HpdR/PhpdH system, the expression of eGFP+ was induced with 10 mM of LA, and the cells were harvested after 24 h of induction. The cell pellet was resuspended in ice-cold cell lysis buffer (50 mM MOPS, pH 8.0, 10 mM imidazole), and cell lysates were prepared by ultrasonication (35% amplitude, 10 s ON/OFF, total sonication ON time = 6 min) using a programmable ultrasonic processor (VCX 130; Sonics, Newton, CT, USA). Protein concentration was determined with the Bradford assay reagent using bovine serum albumin (BSA) as a standard. The samples were then diluted in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, and 5 µg per well was loaded onto a 15% polyacrylamide gel. The protein bands were visualized by staining with Coomassie Brilliant Blue R-250. The concentration of eGFP+ in the sample was calculated based on a linear relationship between known concentrations of purified GFP (Abcam, Cambridge, MA, USA) and the fluorescence units, according to the manufacturer’s protocol.
Results
Evaluation of the Inducible Expression Systems
The constructed plasmids pMHRZ_eGFP+, pMLRL_eGFP+, pMHRH_eGFP+, pMMRM_eGFP+, pMXRX_eGFP+, and pMPLO4_eGFP+ were individually transformed into M. extorquens AM1 to generate the MRHZ01, MLRL01, MHRH01, MMRM01, MXRX01, and MPLO4 strains, respectively. The inducibility of the constructed expression systems was analyzed by measuring the GFP fluorescence of the strains cultivated in MSM supplemented with glucose (MRHZ01), LA or 3-HP (MLRL01, MHRH01, and MMRM01), xylose (MXRX01), or IPTG (MPLO4). The results showed that the HpdR/PhpdH and MmsR/PmmsA systems significantly (p < 0.05) increased the fluorescence intensity when induced by 3-HP or LA and 3-HP, respectively, compared to that in the control (Figure 1A). The GFP expression level of HpdR/PhpdH induced by LA was similar to that of HpdR/PhpdH induced by 3-HP (p > 0.05), a known inducer of the HpdR/PhpdH system (Zhou et al., 2015; Hanko et al., 2017; Sathesh-Prabu et al., 2021). In contrast, the MmsR/PmmsA system was not induced by LA (Figure 1A).
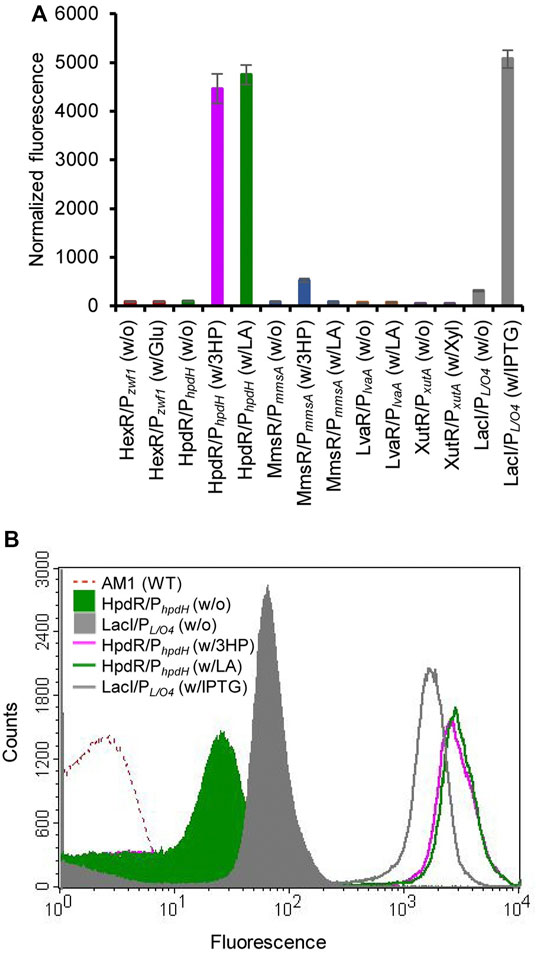
FIGURE 1. Evaluation of various inducible expression systems in Methylorubrum extorquens AM1. (A) Normalized green fluorescent protein (GFP) fluorescence intensity of the various inducible expression systems (HexR/Pzwf1, LvaR/PlvaA, HpdR/PhpdH, MmsR/PmmsA, XutR/PxutA, and LacI/PL/O4) induced by their respective inducers. System name (w/o) and System name (w/x) indicate the absence and presence of the respective inducers, respectively. The systems were induced with 10 mM solutions of the respective inducers (glucose, xylose, LA, or 3-HP) or 1 mM IPTG. The HpdR/PhpdH system showed the maximum expression upon induction either by LA or 3-HP. (B) Flow cytometric analysis of the HpdR/PhpdH system (strain MHRH01) and the LacI/PL/O4 system (strain MPLO4) induced by 10 mM LA and 1 mM IPTG, respectively. Data represent the mean ± SD of the three biological replicates. Glu, glucose; LA, levulinic acid; 3HP, 3-hydroxypropionic acid; Xyl, xylose; IPTG, isopropyl β-d-1-thiogalactopyranoside.
Other systems (HexR/Pzwf1, LvaR/PlvaA, and XutR/PxutA) did not show any comparable fluorescence intensity upon induction when compared to that of the control condition. Flow cytometric analysis of the inducible promoter systems (HexR/Pzwf1, LvaR/PlvaA, MmsR/PmmsA, and XutR/PxutA) is shown in Supplementary Figure S1. In the present study, we compared the efficiency of the constructed expression systems with IPTG-inducible LacI/PL/O4, demonstrating a maximum strength of 157% relative to the conventional PmxaF promoter. HpdR/PhpdH (strain MHRH01) induced with LA (10 mM) achieved 94% efficiency of the LacI/PL/O4 system induced with IPTG (1 mM). In contrast, MmsR/PmmsA (strain MMRM01) showed 10% of the efficiency exhibited by LacI/PL/O4. As the efficiency of the HpdR/PhpdH (strain MHRH01) system was similar to that of the strongest IPTG-inducible expression (LacI/PL/O4) system (p > 0.05), the HpdR/PhpdH system was selected for further study. Notably, without the inducer, the HpdR/PhpdH system was tightly controlled (no leaky expression) (Figure 1B and Supplementary Figure S2). The HpdR/PhpdH system was at least threefold less leaky than the uninduced LacI/PL/O4. Furthermore, the HpdR/PhpdH system was characterized using LA as the inducer.
Optimization of HpdR/PhpdH Induction
The induction time for LA was also evaluated. Figure 2A shows the GFP fluorescence intensity of strain MHRH01 (HpdR/PhpdH) induced by LA (10 mM) at 0, 6, 12, 18, and 24 h of cultivation. When analyzing the fluorescence intensity of the strains at 24 h of induction, the addition of LA at different time points did not show any significant difference (p > 0.05) in the expression of eGFP+ (Figure 2B). This property allows the addition of LA during the early or exponential growth phase, thereby avoiding the requirement of constant monitoring of cell growth to ensure that the inducer, such as IPTG, is added at the optimal cell density (Hu et al., 2015; Briand et al., 2016). The addition time of IPTG was critical for IPTG-inducible expression systems. IPTG induction during the early exponential growth phase was more efficient than that in the middle exponential growth phase or late exponential growth phase (Briand et al., 2016). Our system reported here eases the addition of an inducer to the culture medium.
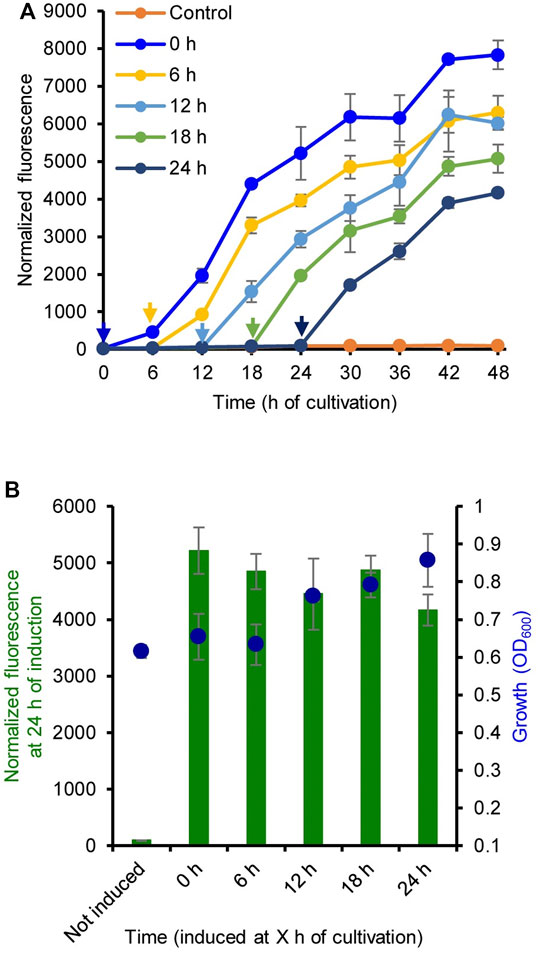
FIGURE 2. Optimization of the induction time of levulinic acid (LA) addition. (A) Normalized green fluorescent protein (GFP) fluorescence intensity of the strain MHRH01 induced by 10 mM LA. LA was added at different time points during cultivation. The arrows indicate the time of LA addition. (B) Normalized fluorescence intensity of the strain MHRH01 measured at 24 h after induction. Data represent the mean ± SD of three biological replicates.
To optimize the concentration of LA, the induction of the HpdR/PhpdH system was analyzed by estimating the fluorescence intensity of the strain MHRH01 with different concentrations of LA (0.1–100 mM). The fluorescence intensity of strain MHRH01 increased with increasing concentrations of LA (Figure 3A) up to 20 mM LA, demonstrating the dose-dependent expression of the reporter protein. The system was induced even at a low concentration of LA (0.1 mM), showing a fourfold stronger fluorescence intensity than that observed in the control. However, the intensity decreased beyond 20 mM LA. Approximately 10%, 28%, and 68% decreased intensities were observed at 40, 50, and 100 mM LA, respectively, compared to the fluorescence intensity observed at 20 mM LA (Figure 3A). The decreased efficiency was attributed to the reduced cell growth of the strain when cultivated in the presence of more than 20 mM LA (Figure 3B). A maximum 1.5-fold reduction in cell growth was observed with 50 mM LA. Flow cytometric analysis of HpdR/PhpdH in MHRH01 induced by different concentrations of LA showed tight control and homogeneity (cell populations with similar expression levels) of the expression system up to 20 mM of LA (Figure 3C).
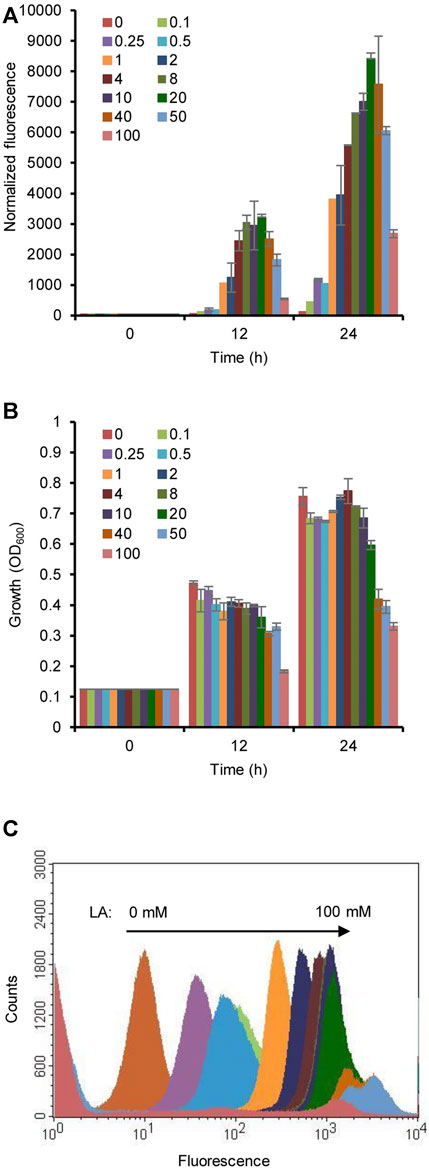
FIGURE 3. Optimization of the concentration of levulinic acid (LA). (A) Normalized green fluorescent protein (GFP) fluorescence intensity of the strain MHRH01 induced by different concentrations of LA. LA was added at 0 h of cultivation. The intensity increased with increasing concentrations of LA up to 20 mM of LA. (B) Measurement of the cell growth of strain MHRH01 induced by different concentrations of LA. Beyond 20 mM LA, growth retardation was observed. (C) Flow cytometric analysis of strain MHRH01 induced by different concentrations of LA. The samples were collected after 24 h of induction for analysis. MHRH01 showed tight control and homogeneity (cell populations with similar expression levels) of the HpdR/PhpdH expression system up to 20 mM of LA.
The residual concentration of LA was measured at 0, 24, and 48 h of cultivation. LA (10 mM) was added at 0 h of cultivation. As shown in Figure 4A, LA was not consumed by the strain MHRH01 even after sufficient cell growth and showed OD600 values of 0.125 and 1.94 at 0 and 48 h, respectively.
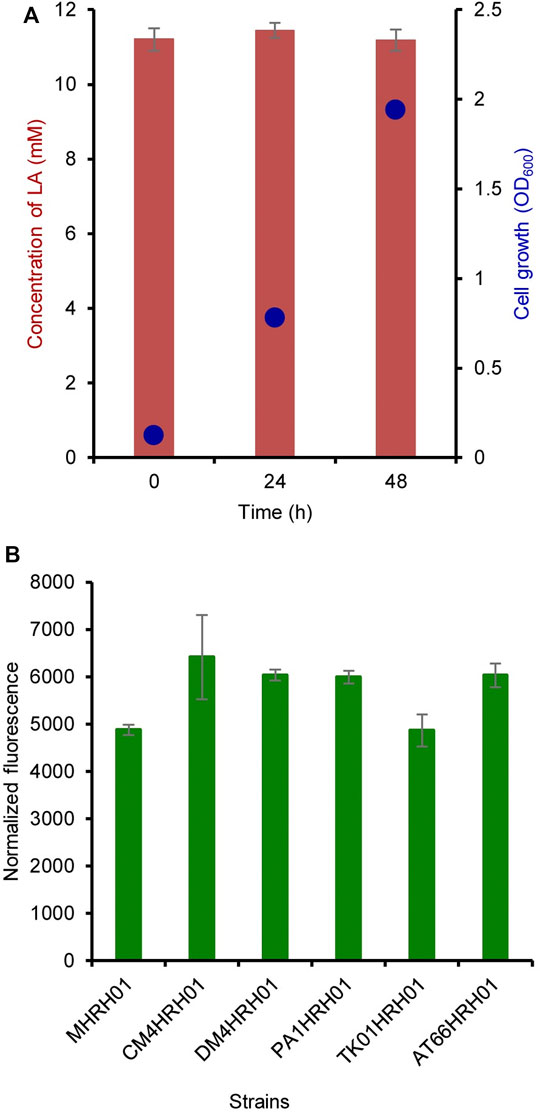
FIGURE 4. Non-degradation of levulinic acid (LA) and strain independence of the HpdR/PhpdH system. (A) Residual concentration of LA measured at 0, 24, and 48 h after its addition. The concentration of LA did not reduce over the cultivation period. (B) Strain independence of the HpdR/PhpdH system. The normalized green fluorescent protein (GFP) fluorescence intensities of the various Methylorubrum strains (CM4, DM4, PA1, TK0001, and ATCC55366) harboring pMHRH-eGFP+ were measured. The system showed similar levels of expression in all the strains induced with 10 mM LA. Data represent the mean ± SD of three biological replicates.
The plasmid pMHRH_eGFP+ was transformed into other Methylorubrum strains, such as CM4 (KCTC 32005), DM4 (DSM 6343), PA1 (DSM 23939), TK0001 (DSM1337), and ATCC55366, to evaluate the strain independence of the HpdR/PhpdH system. The system showed a similar level of expression in all strains upon induction with 10 mM LA (Figure 4B). The strain independence of the constructed expression system would be highly useful for a wide range of applications in different strains.
Quantitation of GFP
The efficiency of the constructed system for producing GFP was analyzed both qualitatively and quantitatively. The SDS-PAGE results shown in Figure 5A reveal a strong expression of eGFP+ (27 kDa) by the strain MHRH01 induced by 10 mM LA. The strain did not produce eGFP+ (no protein bands were visualized) in the absence of LA. The normalized GFP fluorescence intensities of strains MHRH01 and MPLO4 are shown in Figure 5B. In addition, the concentration of GFP produced by strain MHRH01 was measured. The strains MHRH01 and MPLO4 efficiently produced eGFP+, with yields of 42 and 46 mg/g of biomass (dry cell weight), representing approximately 10% and 11% of the total protein content, respectively (Figure 5C). The efficiency of the LA-inducible system was almost equal (90%) to that of the IPTG-inducible system (p > 0.05).
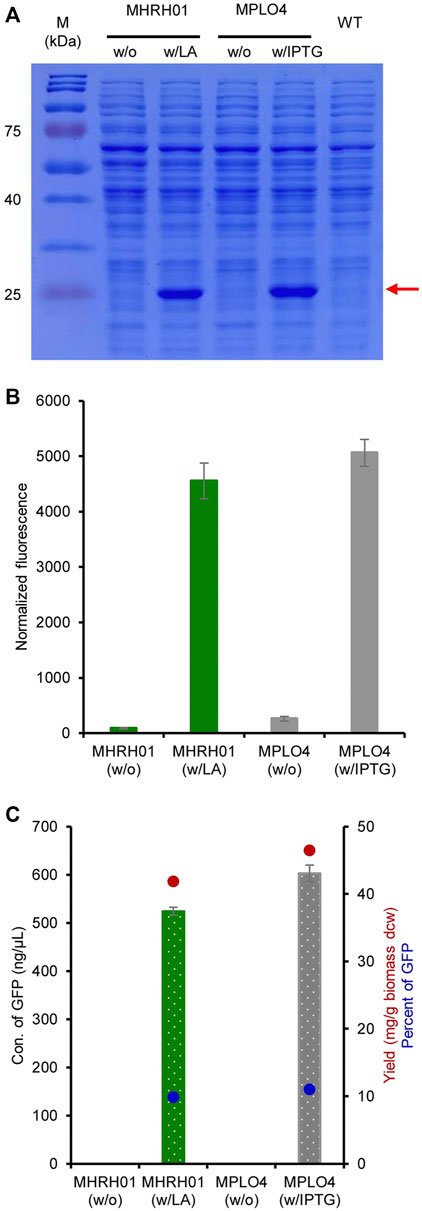
FIGURE 5. Efficiency of the HpdR/PhpdH system in the production of green fluorescent protein (GFP). (A) SDS-PAGE analysis of GFP. Strains MHRH01 and MPLO4 showed a strong expression of eGFP+ (27 kDa), indicated by the red arrow, upon induction with 10 mM LA and 1 mM IPTG, respectively. (B) Maximum normalized fluorescence intensity of strains MHRH01 and MPLO4 induced with 10 mM LA and 1 mM IPTG, respectively. (C) Quantitation of GFP. Strains MHRH01 and MPLO4 efficiently produced eGFP+ with yields of 42 and 46 mg/g of biomass (dry cell weight), representing around 10% and 11% of the total protein content, respectively. Data represent the mean ± SD of three biological replicates.
Discussion
Methylorubrum extorquens AM1 has been widely used in methylotrophic studies and has attracted great interest from the scientific community because of its versatile metabolism of C1 compounds. In the present study, we developed an LA-inducible expression system for use in M. extorquens AM1. The efficiency of the system was evaluated by inducing the expression of the fluorescent reporter gene (egfp+) as a quantitative reporter of relative promoter activity (Born and Pfeifer, 2019; Sathesh-Prabu et al., 2021). The strength of the GFP signal directly correlates with the promoter activity. GFP has been widely used as a model protein to study heterologous protein expression in different kinds of microorganisms, including M. extorquens AM1 (Bélanger et al., 2004; Choi et al., 2006), because of its unique characteristics, such as no requirement of additional substrates and/or cofactors and enzymes, no cell lysis, sensitivity, and the short analysis time (Lissemore et al., 2000; Born and Pfeifer, 2019).
In our previous study, we reported inducible and tunable gene expression systems for use in P. putida KT2440 (Sathesh-Prabu et al., 2021) and showed that the efficiency rates of the glucose-inducible HexR/PzwfA, LA-inducible LvaR/PlvaA, 3-HP- and LA-inducible HpdR/PhpdH, and xylose-inducible XutR/PxutA were comparable to that of conventional chemical-inducible systems. In contrast, in the present study, HexR/PzwfA, LvaR/PlvaA, and XutR/PxutA were non-functional in M. extorquens AM1. This result may be attributed to the lack of transporters for glucose and xylose. Notably, LA, the native inducer of PlvaA, failed to induce the LvaR/PlvaA system, but induced the HpdR/PhpdH system. This finding suggests that the transcriptional regulator LvaR might be unexpressed or non-functional or that the LA-bound LvaR may not be able to form an active form of LvaR–DNA complex or recruit RNA polymerase for transcription. Moreover, the orthogonality of PhpdH and PmmsA of P. putida KT2440 has been demonstrated in E. coli and Cupriavidus necator (Hanko et al., 2017). In the present study, we showed the functional eGFP+ expression of the HpdR/PhpdH and MmsR/PmmsA systems in M. extorquens AM1. The HpdR/PhpdH and MmsR/PmmsA systems are involved in the catabolism of 3-HP in Pseudomonas species (Zhou et al., 2015; Hanko et al., 2017). Interestingly, the HpdR/PhpdH system was found to be activated by two different inducers (3-HP and LA) in M. extorquens AM1. This result is in accordance with that of a recent study conducted on P. putida KT2440 (Sathesh-Prabu et al., 2021). The HpdR/PhpdH system was tightly controlled, with a basal expression of 2%. The minimum requirement for the ligand to be functional and induce the HpdR/PhpdH system is one carboxyl group, as present in 3-HP, propionate, butyrate, d-3-hydroxybutyrate, and l-3-hydroxybutyrate (Hanko et al., 2017). LA also contains one carboxyl group. The presence of a hydroxyl group at the β-position enhances regulator activity. To our knowledge, this is the first description of 3-HP- and LA-inducible systems in M. extorquens AM1.
The IPTG-inducible system is currently the most efficient method for the overexpression of recombinant proteins in many hosts of biotechnological interest, such as E. coli (Balzer et al., 2013; Briand et al., 2016), P. putida KT2440 (Calero et al., 2016), and M. extorquens AM1 (Carrillo et al., 2019). However, the extremely high level of transcription in the IPTG-inducible system resulted in a correspondingly high production of insoluble and inactive proteins and a high level of background expression (Balzer et al., 2013). In addition, it is not cost effective and is incompatible with scaling up on an industrial level. In contrast, the reported system was efficiently induced by the low-cost renewable LA, tightly controlled, and achieved a high level of expression relative to the IPTG-inducible system with a lower level of basal expression (without inducer).
Since the added LA was not degraded by strain MHRH01, a stable gene expression level can be envisaged. However, the addition of small doses of IPTG at multiple time points during cultivation is necessary to maintain an optimal concentration of IPTG for the efficient production of recombinant proteins (Hu et al., 2015). Furthermore, LA, when used as a substrate for biochemical production, can be efficiently utilized for both induction and production with a higher yield of the desired product. LA (a five-carbon compound) is a sustainable platform molecule and has been identified as one of the “top 12” promising sugar-based building blocks by the United States Department of Energy (Werpy and Petersen, 2004). LA can be obtained from renewable cellulosic biomass through acid-catalyzed dehydration and hydrolysis (Hayes and Becer, 2020). As LA contains both ketone and carboxylic acid functional groups, a wide range of valuable chemicals and fuels can be produced from it (Pileidis and Titirici, 2016). Moreover, low-cost LA is considered to be a promising organic intermediate for the synthesis of several chemicals such as polymers, plasticizers, fuels, resins, fragrances, pharmaceuticals, anti-freeze agents, and solvents (Bozell et al., 2000; Rackemann and Doherty, 2011; Pileidis and Titirici, 2016; Sathesh-Prabu and Lee, 2019; Cha et al., 2020; Hayes and Becer, 2020).
The system constructed here efficiently produced the reporter protein with a specific GFP production of 42 mg/g biomass, representing 10% of the total cell protein. The efficiency of our system was similar to that of the widely used promoter PmxaF in M. extorquens. The strong natural promoter PmxaF in M. extorquens produced 41–48 mg/g biomass of GFP at the shake-flask level, representing 8%–10% of the total cell protein. In addition, a maximum of 80 mg/g biomass of GFP (≈16% of the total cell protein) was obtained during the fed-batch cultivation of M. extorquens in a 20-L fermenter with a constant methanol concentration of 1.4 g/L for 50 h of cultivation (Bélanger et al., 2004). Thus, the LA-inducible system achieved an almost absolute PmxaF efficiency. Other inducible promoter systems (PR/cmtO and PR/tetO) were reported to have a maximum of 33% PmxaF in M. extorquens AM1 (Chubiz et al., 2013). Moreover, the constructed LA-inducible system achieved 90%–94% efficiency of that of PL/O4, the recently developed IPTG-inducible system that is considered to be stronger than PmxaF (Carrillo et al., 2019).
In summary, a renewable and eco-friendly LA-inducible and tunable gene expression system, HpdR/PhpdH, was evaluated for use in M. extorquens AM1. The system showed almost equal efficiency to the widely used strong promoters in M. extorquens AM1. The advantages of this system include the regulatable and tight control of the system, low cost, ease of LA induction, non-degradation of LA, and strain independence. Hence, the reported system is highly useful for expanding the biotechnological applicability of the potent strains of M. extorquens in C1 biorefinery.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
CS-P designed the experiments, performed all experiments, obtained and processed the data, and wrote the original draft of the manuscript. YR constructed pCM110_PL/O4 and analyzed the data. SL conceived and designed the study, validated the data, and revised the manuscript. All authors read, reviewed, and approved the contents of the manuscript.
Funding
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (MSIT) (NRF 2020R1A4A1018332). This research was also supported by the C1 Gas Refinery Program through the NRF, funded by the MSIT (2017K1A4A30154371).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.797020/full#supplementary-material
REFERENCES
Agarwal, A. S., Zhai, Y., Hill, D., and Sridhar, N. (2011). The Electrochemical Reduction of Carbon Dioxide to Formate/Formic Acid: Engineering and Economic Feasibility. ChemSusChem 4, 1301–1310. doi:10.1002/CSSC.201100220
Anthony, C. (2011). How Half a Century of Research Was Required to Understand Bacterial Growth on C1 and C2 Compounds; the Story of the Serine Cycle and the Ethylmalonyl-CoA Pathway. Sci. Prog. 94, 109–137. doi:10.3184/003685011X13044430633960
Balzer, S., Kucharova, V., Megerle, J., Lale, R., Brautaset, T., and Valla, S. (2013). A Comparative Analysis of the Properties of Regulated Promoter Systems Commonly Used for Recombinant Gene Expression in Escherichia coli. Microb. Cell Fact. 12, 26. doi:10.1186/1475-2859-12-26
Bélanger, L., Figueira, M. M., Bourque, D., Morel, L., Béland, M., Laramée, L., et al. (2004). Production of Heterologous Protein by Methylobacterium extorquens in High Cell Density Fermentation. FEMS Microbiol. Lett. 231, 197–204. doi:10.1016/S0378-1097(03)00956-X
Belkhelfa, S., Roche, D., Dubois, I., Berger, A., Delmas, V. A., Cattolico, L., et al. (2019). Continuous Culture Adaptation of Methylobacterium extorquens AM1 and TK 0001 to Very High Methanol Concentrations. Front. Microbiol. 10, 1313. doi:10.3389/fmicb.2019.01313
Born, J., and Pfeifer, F. (2019). Improved GFP Variants to Study Gene Expression in Haloarchaea. Front. Microbiol. 10, 1200. doi:10.3389/fmicb.2019.01200
Bozell, J. J., Moens, L., Elliott, D. C., Wang, Y., Neuenscwander, G. G., Fitzpatrick, S. W., et al. (2000). Production of Levulinic Acid and Use as a Platform Chemical for Derived Products. Resour. Conservation Recycling 28, 227–239. doi:10.1016/S0921-3449(99)00047-6
Briand, L., Marcion, G., Kriznik, A., Heydel, J. M., Artur, Y., Garrido, C., et al. (2016). A Self-Inducible Heterologous Protein Expression System in Escherichia coli. Sci. Rep. 6, 33037. doi:10.1038/srep33037
Browning, D. F., Richards, K. L., Peswani, A. R., Roobol, J., Busby, S. J. W., and Robinson, C. (2017). Escherichia coli "TatExpress" Strains Super-secrete Human Growth Hormone into the Bacterial Periplasm by the Tat Pathway. Biotechnol. Bioeng. 114, 2828–2836. doi:10.1002/BIT.26434
Calero, P., Jensen, S. I., and Nielsen, A. T. (2016). Broad-Host-Range ProUSER Vectors Enable Fast Characterization of Inducible Promoters and Optimization of P-Coumaric Acid Production in Pseudomonas putida KT2440. ACS Synth. Biol. 5, 741–753. doi:10.1021/acssynbio.6b00081
Carrillo, M., Wagner, M., Petit, F., Dransfeld, A., Becker, A., and Erb, T. J. (2019). Design and Control of Extrachromosomal Elements in Methylorubrum extorquens AM1. ACS Synth. Biol. 8, 2451–2456. doi:10.1021/acssynbio.9b00220
Cha, D., Ha, H. S., and Lee, S. K. (2020). Metabolic Engineering of Pseudomonas putida for the Production of Various Types of Short-Chain-Length Polyhydroxyalkanoates from Levulinic Acid. Bioresour. Technology 309, 123332. doi:10.1016/j.biortech.2020.123332
Chaves, J. E., Wilton, R., Gao, Y., Munoz, N. M., Burnet, M. C., Schmitz, Z., et al. (2020). Evaluation of Chromosomal Insertion Loci in the Pseudomonas putida KT2440 Genome for Predictable Biosystems Design. Metab. Eng. Commun. 11, e00139. doi:10.1016/J.MEC.2020.E00139
Choi, Y. J., Bourque, D., Morel, L., Groleau, D., and Míguez, C. B. (2006). Multicopy Integration and Expression of Heterologous Genes in Methylobacterium extorquens ATCC 55366. Appl. Environ. Microbiol. 72, 753–759. doi:10.1128/AEM.72.1.753-759.2006
Chubiz, L. M., Purswani, J., Carroll, S. M., and Marx, C. J. (2013). A Novel Pair of Inducible Expression Vectors for Use in Methylobacterium extorquens. BMC Res. Notes 6, 183. doi:10.1186/1756-0500-6-183
Ganesh, I. (2014). Conversion of Carbon Dioxide into Methanol - a Potential Liquid Fuel: Fundamental Challenges and Opportunities (A Review). Renew. Sustainable Energ. Rev. 31, 221–257. doi:10.1016/J.RSER.2013.11.045
Green, P. N., and Ardley, J. K. (2018). Review of the Genus Methylobacterium and Closely Related Organisms: a Proposal that Some Methylobacterium Species Be Reclassified into a New Genus, Methylorubrum Gen. Nov. Int. J. Syst. Evol. Microbiol. 68, 2727–2748. doi:10.1099/IJSEM.0.002856
Hanko, E. K. R., Minton, N. P., and Malys, N. (2017). Characterisation of a 3-Hydroxypropionic Acid-Inducible System from Pseudomonas putida for Orthogonal Gene Expression Control in Escherichia coli and Cupriavidus Necator. Sci. Rep. 7, 1724. doi:10.1038/s41598-017-01850-w
Hayes, G. C., and Becer, C. R. (2020). Levulinic Acid: a Sustainable Platform Chemical for Novel Polymer Architectures. Polym. Chem. 11, 4068–4077. doi:10.1039/d0py00705f
Hu, B., and Lidstrom, M. E. (2014). Metabolic Engineering of Methylobacterium extorquens AM1 for 1-Butanol Production. Biotechnol. Biofuels 7, 156. doi:10.1186/s13068-014-0156-0
Hu, J.-H., Wang, F., and Liu, C.-Z. (2015). Development of an Efficient Process Intensification Strategy for Enhancing pfu DNA Polymerase Production in Recombinant Escherichia coli. Bioproc. Biosyst. Eng. 38, 651–659. doi:10.1007/S00449-014-1304-4
Humphreys, C. M., and Minton, N. P. (2018). Advances in Metabolic Engineering in the Microbial Production of Fuels and Chemicals from C1 Gas. Curr. Opin. Biotechnol. 50, 174–181. doi:10.1016/j.copbio.2017.12.023
Hwang, H. W., Yoon, J., Min, K., Kim, M.-S., Kim, S.-J., Cho, D. H., et al. (2020). Two-Stage Bioconversion of Carbon Monoxide to Biopolymers via Formate as an Intermediate. Chem. Eng. J. 389, 124394. doi:10.1016/j.cej.2020.124394
Kim, S., Kim, H., Qiao, T., Cha, C., Lee, S. K., Lee, K., et al. (2019). Fluorescence Enhancement from Nitro-Compound-Sensitive Bacteria within Spherical Hydrogel Scaffolds. ACS Appl. Mater. Inter. 11, 14354–14361. doi:10.1021/acsami.9b02262
Kim, T. W., Bae, S. S., Lee, J. W., Lee, S.-M., Lee, J.-H., Lee, H. S., et al. (2016). A Biological Process Effective for the Conversion of CO-Containing Industrial Waste Gas to Acetate. Bioresour. Technology 211, 792–796. doi:10.1016/j.biortech.2016.04.038
Lee, S. K., and Keasling, J. D. (2005). A Propionate-Inducible Expression System for Enteric Bacteria. Appl. Environ. Microbiol. 71, 6856–6862. doi:10.1128/AEM.71.11.6856-6862.2005
Lim, C. K., Villada, J. C., Chalifour, A., Duran, M. F., Lu, H., and Lee, P. K. H. (2019). Designing and Engineering Methylorubrum extorquens AM1 for Itaconic Acid Production. Front. Microbiol. 10, 1027. doi:10.3389/fmicb.2019.01027
Lissemore, J. L., Jankowski, J. T., Thomas, C. B., Mascotti, D. P., and deHaseth, P. L. (2000). Green Fluorescent Protein as a Quantitative Reporter of Relative Promoter Activity in E. coli. Biotechniques 28, 82–89. doi:10.2144/00281st02
Makrides, S. C. (1996). Strategies for Achieving High-Level Expression of Genes in Escherichia coli. Microbiol. Rev. 60, 512–538. doi:10.1128/mr.60.3.512-538.1996
Marx, C. J., and Lidstrom, M. E. (2001). Development of Improved Versatile Broad-Host-Range Vectors for Use in Methylotrophs and Other Gram-Negative Bacteria. Microbiology 147, 2065–2075. doi:10.1099/00221287-147-8-2065
Ochsner, A. M., Sonntag, F., Buchhaupt, M., Schrader, J., and Vorholt, J. A. (2015). Methylobacterium extorquens: Methylotrophy and Biotechnological Applications. Appl. Microbiol. Biotechnol. 99, 517–534. doi:10.1007/s00253-014-6240-3
Orita, I., Nishikawa, K., Nakamura, S., and Fukui, T. (2014). Biosynthesis of Polyhydroxyalkanoate Copolymers from Methanol by Methylobacterium extorquens AM1 and the Engineered Strains under Cobalt-Deficient Conditions. Appl. Microbiol. Biotechnol. 98, 3715–3725. doi:10.1007/S00253-013-5490-9
Peyraud, R., Kiefer, P., Christen, P., Portais, J.-C., and Vorholt, J. A. (2012). Co-Consumption of Methanol and Succinate by Methylobacterium extorquens AM1. PLoS One 7, e48271. doi:10.1371/journal.pone.0048271
Peyraud, R., Schneider, K., Kiefer, P., Massou, S., Vorholt, J. A., and Portais, J.-C. (2011). Genome-scale Reconstruction and System Level Investigation of the Metabolic Network of Methylobacterium extorquens AM1. BMC Syst. Biol. 5, 189. doi:10.1186/1752-0509-5-189
Pileidis, F. D., and Titirici, M.-M. (2016). Levulinic Acid Biorefineries: New Challenges for Efficient Utilization of Biomass. ChemSusChem 9, 562–582. doi:10.1002/cssc.201501405
Rackemann, D. W., and Doherty, W. O. (2011). The Conversion of Lignocellulosics to Levulinic Acid. Biofuels, Bioprod. Bioref. 5, 198–214. doi:10.1002/bbb.267
Rittmann, S. K.-M. R., Lee, H. S., Lim, J. K., Kim, T. W., Lee, J.-H., and Kang, S. G. (2015). One-carbon Substrate-Based Biohydrogen Production: Microbes, Mechanism, and Productivity. Biotechnol. Adv. 33, 165–177. doi:10.1016/J.biotechadv.2014.11.004
Sathesh-Prabu, C., Kim, D., and Lee, S. K. (2020). Metabolic Engineering of Escherichia coli for 2,3-butanediol Production from Cellulosic Biomass by Using Glucose-Inducible Gene Expression System. Bioresour. Technology 309, 123361. doi:10.1016/j.biortech.2020.123361
Sathesh-Prabu, C., and Lee, S. K. (2019). Engineering the lva Operon and Optimization of Culture Conditions for Enhanced Production of 4-Hydroxyvalerate from Levulinic Acid in Pseudomonas putida KT2440. J. Agric. Food Chem. 67, 2540–2546. doi:10.1021/acs.jafc.8b06884
Sathesh-Prabu, C., Tiwari, R., Kim, D., and Lee, S. K. (2021). Inducible and Tunable Gene Expression Systems for Pseudomonas putida KT2440. Sci. Rep. 11, 18079. doi:10.1038/s41598-021-97550-7
Schada Von Borzyskowski, L., Remus-Emsermann, M., Weishaupt, R., Vorholt, J. A., and Erb, T. J. (2015). A Set of Versatile Brick Vectors and Promoters for the Assembly, Expression, and Integration of Synthetic Operons in Methylobacterium extorquens AM1 and Other Alphaproteobacteria. ACS Synth. Biol. 4, 430–443. doi:10.1021/sb500221v
Schada Von Borzyskowski, L., Sonntag, F., Pöschel, L., Vorholt, J. A., Schrader, J., Erb, T. J., et al. (2018). Replacing the Ethylmalonyl-CoA Pathway with the Glyoxylate Shunt Provides Metabolic Flexibility in the Central Carbon Metabolism of Methylobacterium extorquens AM1. ACS Synth. Biol. 7, 86–97. doi:10.1021/acssynbio.7b00229
Schneider, K., Peyraud, R., Kiefer, P., Christen, P., Delmotte, N., Massou, S., et al. (2012). The Ethylmalonyl-CoA Pathway Is Used in Place of the Glyoxylate Cycle by Methylobacterium extorquens AM1 during Growth on Acetate. J. Biol. Chem. 287, 757–766. doi:10.1074/JBC.M111.305219
Skovran, E., Crowther, G. J., Guo, X., Yang, S., and Lidstrom, M. E. (2010). A Systems Biology Approach Uncovers Cellular Strategies Used by Methylobacterium extorquens AM1 during the Switch from Multi- to Single-Carbon Growth. PLoS One 5, e14091. doi:10.1371/journal.pone.0014091
Sonntag, F., Kroner, C., Lubuta, P., Peyraud, R., Horst, A., Buchhaupt, M., et al. (2015a). Engineering Methylobacterium extorquens AM1 for De Novo Synthesis of the Sesquiterpenoid α-humulene from Methanol. Metab. Eng. 32, 82–94. doi:10.1016/J.YMBEN.2015.09.004
Sonntag, F., Müller, J. E. N., Kiefer, P., Vorholt, J. A., Schrader, J., and Buchhaupt, M. (2015b). High-level Production of Ethylmalonyl-CoA Pathway-Derived Dicarboxylic Acids by Methylobacterium extorquens under Cobalt-Deficient Conditions and by Polyhydroxybutyrate Negative Strains. Appl. Microbiol. Biotechnol. 99, 3407–3419. doi:10.1007/S00253-015-6418-3
Toyama, H., Anthony, C., and Lidstrom, M. E. (1998). Construction of Insertion and Deletion mxa Mutants of Methylobacterium extorquens AM1 by Electroporation. FEMS Microbiol. Lett. 166, 1–7. doi:10.1016/S0378-1097(98)00282-110.1111/j.1574-6968.1998.tb13175.x
Vuilleumier, S., Chistoserdova, L., Lee, M.-C., Bringel, F., Lajus, A., Zhou, Y., et al. (2009). Methylobacterium Genome Sequences: a Reference Blueprint to Investigate Microbial Metabolism of C1 Compounds from Natural and Industrial Sources. PLoS One 4, e5584. doi:10.1371/journal.pone.0005584
Werpy, T., and Petersen, G. (2004). Top Value Added Chemicals from Biomass: Volume I -- Results of Screening for Potential Candidates from Sugars and Synthesis Gas. USA_National Renewable Energy Laboratory, 76. doi:10.2172/15008859
Yang, J., Zhang, C.-T., Yuan, X.-J., Zhang, M., Mo, X.-H., Tan, L.-L., et al. (2018). Metabolic Engineering of Methylobacterium extorquens AM1 for the Production of Butadiene Precursor. Microb. Cell Fact. 17, 194. doi:10.1186/s12934-018-1042-4
Yang, Y.-M., Chen, W.-J., Yang, J., Zhou, Y.-M., Hu, B., Zhang, M., et al. (2017). Production of 3-Hydroxypropionic Acid in Engineered Methylobacterium extorquens AM1 and its Reassimilation through a Reductive Route. Microb. Cell Fact. 16, 179. doi:10.1186/s12934-017-0798-2
Yoon, J., Chang, W., Oh, S.-H., Choi, S.-H., Yang, Y.-H., and Oh, M.-K. (2021). Metabolic Engineering of Methylorubrum extorquens AM1 for Poly (3-Hydroxybutyrate-Co-3-Hydroxyvalerate) Production Using Formate. Int. J. Biol. Macromolecules 177, 284–293. doi:10.1016/j.ijbiomac.2021.02.092
Zhang, M., Yuan, X.-j., Zhang, C., Zhu, L.-p., Mo, X.-h., Chen, W.-j., et al. (2019). Bioconversion of Methanol into Value-Added Chemicals in Native and Synthetic Methylotrophs. Curr. Issues Mol. Biol. 33, 225–236. doi:10.21775/CIMB.033.225
Zhou, S., Ainala, S. K., Seol, E., Nguyen, T. T., and Park, S. (2015). Inducible Gene Expression System by 3-Hydroxypropionic Acid. Biotechnol. Biofuels 8, 169. doi:10.1186/s13068-015-0353-5
Zhu, W.-L., Cui, J.-Y., Cui, L.-Y., Liang, W.-F., Yang, S., Zhang, C., et al. (2016). Bioconversion of Methanol to Value-Added Mevalonate by Engineered Methylobacterium extorquens AM1 Containing an Optimized Mevalonate Pathway. Appl. Microbiol. Biotechnol. 100, 2171–2182. doi:10.1007/s00253-015-7078-z
Keywords: Methylorubrum extorquens AM1, levulinic acid, promoter, inducible expression, C1-biorefinery
Citation: Sathesh-Prabu C, Ryu YS and Lee SK (2021) Levulinic Acid-Inducible and Tunable Gene Expression System for Methylorubrum extorquens. Front. Bioeng. Biotechnol. 9:797020. doi: 10.3389/fbioe.2021.797020
Received: 18 October 2021; Accepted: 17 November 2021;
Published: 15 December 2021.
Edited by:
Dong-Yup Lee, Sungkyunkwan University, South KoreaReviewed by:
Dae-Hee Lee, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaYanning Zheng, Institute of Microbiology (CAS), China
Copyright © 2021 Sathesh-Prabu, Ryu and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung Kuk Lee, c2tsZWVAdW5pc3QuYWMua3I=
 Chandran Sathesh-Prabu1
Chandran Sathesh-Prabu1 Sung Kuk Lee
Sung Kuk Lee