
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol. , 10 January 2022
Sec. Industrial Biotechnology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.794742
This article is part of the Research Topic Designing Carrier-free Immobilized Enzymes for Biocatalysis View all 9 articles
Yeast surface display (YSD) is a “whole-cell” platform used for the heterologous expression of proteins immobilized on the yeast’s cell surface. YSD combines the advantages eukaryotic systems offer such as post-translational modifications, correct folding and glycosylation of proteins, with ease of cell culturing and genetic manipulation, and allows of protein immobilization and recovery. Additionally, proteins displayed on the surface of yeast cells may show enhanced stability against changes in temperature, pH, organic solvents, and proteases. This platform has been used to study protein-protein interactions, antibody design and protein engineering. Other applications for YSD include library screening, whole-proteome studies, bioremediation, vaccine and antibiotics development, production of biosensors, ethanol production and biocatalysis. YSD is a promising technology that is not yet optimized for biotechnological applications. This mini review is focused on recent strategies to improve the efficiency and selection of displayed proteins. YSD is presented as a cutting-edge technology for the vectorial expression of proteins and peptides. Finally, recent biotechnological applications are summarized. The different approaches described herein could allow for a better strategy cascade for increasing protein/peptide interaction and production.
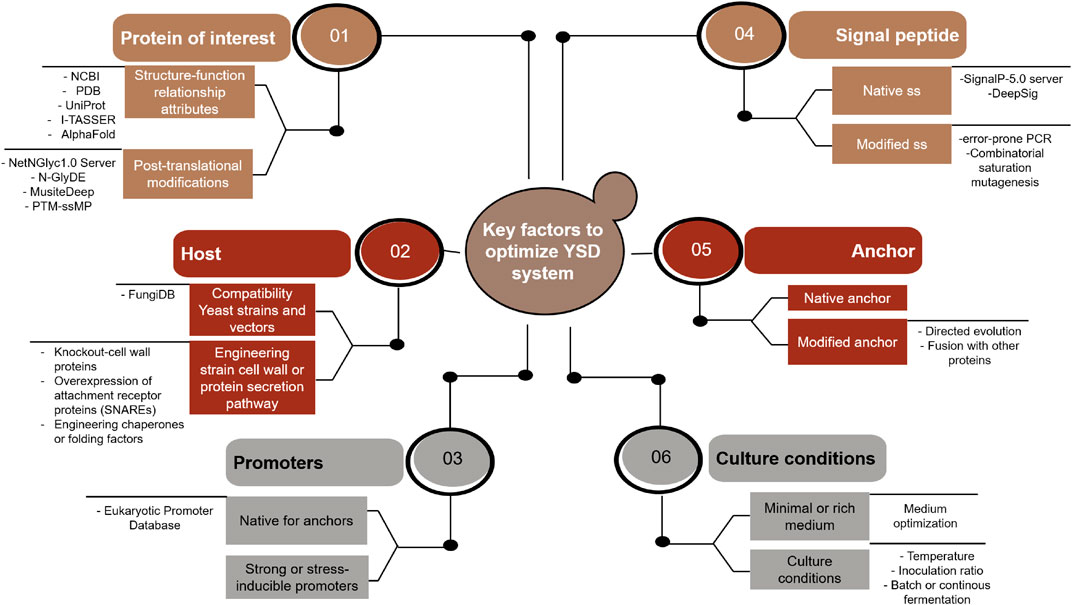
GRAPHICAL ABSTRACT. Different strategies and tools involved in the improvement of the YSD system performance. Abbreviatures found in the figure: NCBI (National Center for Biotechnology Information); PDB (Protein Data Bank); SS (signal sequence); CWPs (cell-wall proteins); SNARES (soluble N-ethylmaleimide-sensitive factor attachment receptor proteins); and CSM (combinatorial saturation mutagenesis).
Cell-surface display allows the expression of target peptides or proteins on the cell surface of bacteria, yeast, insect, or mammalian cells through the connection of a protein of interest (POI) fused to an anchor protein. In yeast, typically, this comprises a cell wall protein (CWP) linked to glycosylphosphatidylinositol (GPI) (Tanaka and Kondo, 2015; Lim et al., 2017). This platform combines fine-tuned gene expression and protein immobilization, which simplifies the purification process and allows the reuse and recovery of biocatalysts (Yuzbasheva et al., 2011; Liu et al., 2014; Yang et al., 2019).
The yeast cell wall landscape consists of an internal layer, mainly composed of polysaccharides, and a 100–200 nm thick fibrillar outer layer framework of 50% mannoproteins, 30–45% β-1,3 glucans, 5–10% β-1,6 glucans and 1.5–6% chitin (Klis et al., 2006; Inokuma et al., 2014; Ananphongmanee et al., 2015). The cell surface offers a topological environment with unique properties not found in other cell compartments. The high content of polysaccharides allows multiple interactions with the proteins embedded in the cell wall that can have a positive impact on the properties of the POI although negative effects have also been reported depending on the yeast strain used for display (Crowe et al., 1988; Allison et al., 2000; Li et al., 2010).
Yeast surface display has been extensively used due to several advantages over other, similar systems, which includes: 1) various yeast strains (Saccharomyces cerevisiae, Pichia pastoris and Yarrowia lipolytica) have a “generally recognized as safe” (GRAS) status by the United States Food and Drug Administration (FDA), 2) yeast cells are able to perform eukaryotic post-translational modifications, 3) the ease of cell culture and genetic modification allows the proper folding and secretion of large and complex protein scaffolds, and 4) compatibility with flow cytometric analysis (Angelini et al., 2015; Lim et al., 2017; Raeeszadeh-Sarmazdeh et al., 2019).
When soluble proteins or peptides are linked to the cell surface, they gain biotechnological advantages not met by the soluble state, e.g. ease of recovery, control of the spatial protein orientation, concerted protein interactions with co-displayed proteins to mimic supramolecular complexes and easy cell sorting formats. Another important advantage of this system is the possibility of coupling yeast metabolism with the function of the protein on the yeast cell surface. This can be useful for the study of transport systems, signal transduction proteins, capsule influences and biofuel production (Wen et al., 2010).
It has been reported that biochemical and catalytic properties can be improved by immobilizing a protein on the yeast cell surface (Shiraga et al., 2005; Li et al., 2014; Moura et al., 2015). Since the first YSD system was developed by Boder and Wittrup (1997), this platform has been employed for the directed evolution of antibodies, peptides and proteins. Nevertheless, promising results in biotechnological applications have been achieved by engineering YSD in different ways that will be discussed herein.
The directed evolution of proteins, antibodies and enzymes to increase their biochemical or catalytic properties has been discussed in excellent reviews (Traxlmayr and Obinger, 2012; Könning and Kolmar, 2018; Linciano et al., 2019). The strategies to improve the displayed protein expression and secretion by engineering the YSD directly are summarized in this review. Table 1 compares these strategies and their relevance on protein expression levels and/or activity.
The regulatory and structural elements that control YSD can be organized in synthetic expression plasmids or integrated into the yeast genome. Usually, plasmid vectors are the first choice to assess the function of regulatory and structural DNA sequences. The genetic construct can be integrated into the chromosome to gain a more stable genetic background, generally not offered by episomal constructs.
Synthetic yeast plasmids are extrachromosomal genetic elements used for the controlled heterologous protein expression, designed to drive gene expression under the control of regulatory sequences, i.e., promoters, terminators, transcription factors, among others. Additionally, plasmid copy number can affect the level of gene expression (Redden et al., 2015). YSD plasmid protein expression depends on promoter strength (Inokuma et al., 2016). Both constitutive and inducible promoters have been used for displaying proteins. The most common promoters used are the galactose-promoter (GAL1/GAL10) for expression in S. cerevisiae (Schröter et al., 2018; Zhao et al., 2020b), GAP and AOX1 promoters for expression in Pichia pastoris (Yang et al., 2017; Li et al., 2019), and TEF1 and hp4d promoters for expression in Y. lipolytica (Yuzbasheva et al., 2011; Moon et al., 2013). Recently, GAPDH, GPD or stress-induced (SED1) promoters have been used for protein expression in S. cerevisiae (Inokuma et al., 2014; Zhang et al., 2019; Nguyen et al., 2020), proving to be appropriate alternatives to the galactose-induced promoter. One drawback is that the promoter strength may vary in different yeast genetic backgrounds (Inokuma et al., 2014; Andreu and del Olmo, 2017). The recent development of software, based on synthetic and omics approaches, predicts in silico and in vivo changes in the level of expression can give substantial information to rationally modify conditions to optimize the selection of a particular promoter (Fiore et al., 2015).
Generally, an anchor contains 2 main parts: 1) a signal peptide sequence (SS), involved in protein transport through the protein secretion pathway, and 2) an anchor to which the POI is fused (Tanaka and Kondo, 2015). Modification of SS has an important impact over the improvement in production levels of the displayed POI. Generally, native signal peptides are used, i.e., a-agglutinin, SED1p, Pir1p and Flo1p have been shown to give good protein expression levels using their own signal peptides (Khasa et al., 2011; Andreu and del Olmo, 2017). Likewise, SS from other genes have been evaluated, for example, the SS of Rhizopus oryzae glucoamylase or the SS of Aspergillus niger α-amylase (Moon et al., 2013; Inokuma et al., 2014). Recent studies on directed evolution of signal peptides have shown that changes in the hydrophobic core of the SS significantly impacts protein secretion (Mateljak et al., 2017; Barrero et al., 2018; Aza et al., 2021). It is worth mentioning that SS modifications can affect the protein secretion pathway, as discussed later.
Selection of the anchor protein is crucial for display effectiveness and is dependent on the specific application and properties of the POI (Van der Vaart, 1997; Tanaka et al., 2012; Tanaka and Kondo, 2015; Yang et al., 2019). The most common anchor are the GPI-dependent CWPs (cell wall proteins), which provide a covalent bond between the target protein and the cell wall β-1,6 glucans. On the other hand, Pir-CWP contributes to the covalent linkage of fusion proteins both to cell wall β-1,3 glucans and to structural proteins via disulfide bonds (Duquesne et al., 2014). The Aga1-Aga2 anchor, initially developed by Boder and Wittrup (1997), has been used in the expression of several proteins (Blazic et al., 2013; Bertrand et al., 2016). Particularly in P. pastoris, SED1p and Pir anchors have shown the highest display efficiencies (Duquesne et al., 2014; Li et al., 2019; Dong et al., 2020).
Figure 1 depicts common and modified anchors mentioned in this review. Some anchors, such as Aga1-Aga2, allow the immobilization of proteins through their N-terminus or C-terminus. Optimal orientation of the POI has an impact on ligand binding affinity (Wang et al., 2005; Valldorf et al., 2021). Native or modified anchors have been successfully evaluated. A novel modified anchor (Li et al., 2019), which consists of the incorporation of the lm7 protein and CL7 protein between the SED1 anchor and the POI efficiently displays the green fluorescent protein (GFP) and a human arginase. Dong et al. (2020) also used the same assembly, SED1/Im7/CL7, to construct a minicellulosome assembly by endoglucanase, exoglucanase and β-glucosidase, which demonstrates the flexibility of this modified anchor for single or multiple enzyme display. Additionally, the search for novel GPI anchors has been performed in P. pastoris and H. polymorpha to find anchors with potential uses for YSD in other yeast strains (Zhang et al., 2013; Cheon et al., 2014).
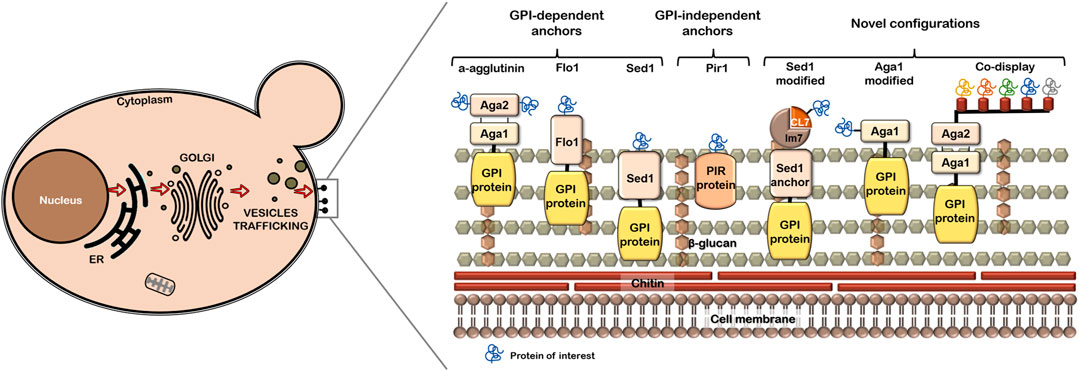
FIGURE 1. Scheme of cell wall proteins used as anchors in YSD system. Common anchors and the modified ones are shown in a representation of yeast cell wall.
The anchor length is another important factor to be considered, since the POI fused to short anchors may show steric hindrance of the active site, affecting ligand-receptor interactions (Sato et al., 2002). It is common to find small sequences between the anchor protein and the POI, called linkers or spacers. Linkers are O-glycosylated regions, which gives protection from protease degradation. The main function of linkers is the reduction of the effects of GPI fusion on the activity of the POI. A Ser/Thr-rich sequence and (G₄S)₃ linkers are the most widely used (Washida et al., 2001). It has been reported that the presence of linkers improves the display efficiencies and the activity of POI by preserving the conformation of the active site and the accessibility of substrates to the POI displayed at the outer yeast cell surface (Breinig and Schmitt, 2002; Sato et al., 2002; Liu et al., 2010; Yang et al., 2019). The anchor length enhancement and the addition of linkers between the anchor and the POI demonstrated a positive effect on accessibility of substrates to displayed proteins (Sato et al., 2002). Yang et al. (2019) reconstructed the a-agglutinin (Aga1-Aga2) by fusing the POI to the N-terminus of Aga1p and incorporating a flexible linker, containing 17 amino acids composed of Ser and Gly repeat sequence, between the POI and the anchor. This modification showed higher display efficiency and could be more suitable for larger proteins or protein complexes.
A strategy widely evaluated is the co-display, which enhances the expression of the target protein or a complex of multiple proteins (Sun et al., 2012). Cellulases can be anchored to yeasts used in fermentations, allowing a formally two-step process to be carried out as a one-step process for a consolidated bioprocessing development (Wen et al., 2010). In a displayed multi-enzyme complex, the distance between the enzymes and the number and nature of cohesion domains must be considered (Tsai et al., 2013). Enzyme-substrate interactions can be negatively affected if distance is not optimum, the opposite effect is detected when a favorable distance is established. Smith et al. (2019) highlighted that the enzyme density is a pivotal parameter to enhance cellulose hydrolytic performance when a multi-enzyme assembly is designed. Tsai et al. (2013) observed a 2-fold increase in ethanol production when cells displayed a tetravalent cellulosome instead of a divalent cellulosome. Novel protein complexes not found in nature can be designed on the surface of yeast or other cell systems by exploiting the extensive proteomic databases available today, e.g., Proteomics DB (https://www.proteomicsdb.org) (Tunyasuvunakool et al., 2021).
The genomic and metabolic background of the applied yeast strain is an important trait that influences YSD of native or heterologous proteins. Synthetic biology approaches combined with genomic techniques (e.g., CRISPR/Cas system) have allowed the development of yeast strains with fine-tuned heterologous protein expression. Novel yeast strains for YSD are designed by modifying their native features, such as their cell wall composition or their protein secretion pathway. The latter includes the engineering of vesicle trafficking and the development of a platform that secrete simultaneously soluble protein and surface-displayed protein to ease their characterization (Zhang et al., 2013; Cruz-Terán et al., 2017; Tang et al., 2017). S. cerevisiae strains, mainly EBY100 (ATCC MYA-4941) and BY4741 (Brachmann et al., 1998) have been successfully used as hosts for YSD, MATa and the ura3 gene disruption (used as an auxotrophic marker) being the main features of these strains. In addition, to obtain an optimized system other yeast strains have been evaluated, e.g., S. cerevisiae (AWY100, AWY101, AWY102), Pichia pastoris, Yarrowia lipolytica, Hansenula polymorpha and Saccharomyces boulardii (Cheon et al., 2014; Dong et al., 2020; Wang et al., 2020; Patent CN103031329B, 2014; Wentz and Shusta, 2007).
Some authors propose modifications of cell wall protein composition to obtain a suitable environment for display. However, one genetic defect can cause pleiotropic changes in the cell wall structure-function relationship (Matsuoka et al., 2014). Inokuma et al. (2021) increased the cell wall thickness of S. cerevisiae by performing the knockout of proteins CCW12 and CCW14, which allowed an increased amount of displayed β-glucosidase as compared to the parental strain.
For the secretion of heterologous proteins is the protein secretory pathway, which includes protein translocation, protein folding, post-translational modifications, protein sorting and trafficking. The secretory pathway can be influenced solely by the nature of SS (Tang et al., 2015). Additionally, the number of proteins that reach cell surface can be controlled by changes in the number of molecules that travel through the secretion pathway. Foreign proteins can be diverted to a vacuolar compartment for destruction due to misfolding (Fitzgerald and Glick, 2014). Efforts related to engineering chaperones, folding factors and translocation components have been performed exhibiting improvements in the secretion of heterologous proteins (Tang et al., 2015; Duan et al., 2019). Tang et al. (2017) improved the surface display efficiency of cellulases by over-expressing the components involved in vesicle trafficking, such as Snc2p, Exo70p and Sso1p. On the other hand, a system based on inefficient ribosomal skipping allows the simultaneous expression of soluble and cell surface displayed proteins to simplify the screening of combinatorial protein libraries and protein characterization (Cruz-Terán et al., 2017).
As previously mentioned, improvement in protein display efficiency is crucial for the implementation of YSD. However, a straightforward detection methodology for displayed proteins is also required for a good performance of combinatorial library screening. For example, GFP has shown high performance of yeast display library high-throughput screening of glucose oxidase (Kovačević et al., 2019). Uchański et al. (2019) developed a YSD platform for the screening of nanobody libraries. Their research group fused a nanobody to the N-terminal of Aga2p to avoid steric hindrance and nanobody detection was performed by using a fluorophore which was attached to an orthogonal acyl carrier protein tag by a one-step reaction catalyzed by a Sfp synthase. In addition, it has been reported that the visualization of endoglucanase displayed on the yeast surface can be performed by using atomic force microscopy (Takenaka et al., 2017).
Protein properties such as stability and activity can be improved by displaying them at yeast cell surface (Shiraga et al., 2005; Li et al., 2014). It is possible to improve the biochemical and catalytic properties of the POI by combining protein engineering with YSD (Antipov et al., 2008; Chen et al., 2011) or to increase the production of value-added chemicals by metabolic engineering (Takayama et al., 2018). Bacon et al. (2019) fused YSD with nanotechnology for the screening of combinatorial libraries. The group co-expressed the target protein with a protein showing affinity to iron oxide for separation of cells by magnetism. A combination of YSD with SELEX technology has aided in the characterization of endonucleases (Jacoby et al., 2017).
YSD can be used in a wide range of applications, such as the engineering proteins or peptides (Smith et al., 2015; Deweid et al., 2018; Raeeszadeh-Sarmazdeh et al., 2019), for whole-cell biocatalysis (Zheng et al., 2019; Wang et al., 2020), production of vaccines (Lei et al., 2016), antibody and nanobody production (McMahon et al., 2018; Sun et al., 2019), biofuel production (Yang et al., 2017; Dong et al., 2020), biofuel cells (Fan et al., 2020) and whole-proteome studies (Bidlingmaier and Liu, 2007; Procko, 2020). A description of some recent advances in YSD biotechnology applications is presented in this section.
Recent reviews on antibody engineering include novel strategies using approaches such as yeast mating and yeast endoplasmic reticulum sequestration screening (YESS) (Könning and Kolmar, 2018; Valldorf et al., 2021). The following applications focus mainly on enzyme biotechnology.
Recent advances in human health care include a vaccine against candidiasis (Shibasaki et al., 2013; Ueda, 2016) and a platform to detect SARS-CoV-2 (Maneira et al., 2021). Animal vaccines have been developed which include vaccines against Toxoplasma gondii (Wang et al., 2018), a vaccine against hemorrhagic disease of grass carp (Luo et al., 2015) and an anti-tick vaccine (Trentelman et al., 2021). Other relevant applications are the development of antibiotics (Chun et al., 2020), biosensors to detect blood biochemical parameters (Zhao et al., 2020b) and the expression of hydrophobins (Andreu et al., 2021).
In the food industry YSD can be used to immobilize enzymes that synthesize relevant compounds to produce sweeteners such as isomaltulose and fructooligosaccharides (Zhang et al., 2016; Zheng et al., 2019) and beneficial fats, e.g., omega 3-fatty acids (Singh et al., 2020). Additionally, whole-cell biocatalysts have been used to improve the sensory properties of beverages which include beer and wine (Cejnar et al., 2017; Zhang et al., 2019). In agri-food applications YSD can be used to develop biocontrol agents, e.g., expressing flagellin to increase the resistance of tomatoes towards Botrytis cinerea infection (Zhao et al., 2020a).
Related to bioethanol production, YSD can exploit the metabolic ability of yeast to ferment sugars to ethanol and the activity of hydrolytic enzymes immobilized on the yeast surface to achieve the simultaneous cellulose saccharification and ethanol fermentation for a consolidated bioprocessing (Fan et al., 2012; Tsai et al., 2013; Ishii et al., 2016; Chen, 2017; Khatun et al., 2017; Yang et al., 2017; Bamba et al., 2018; Tang et al., 2018; Anandharaj et al., 2020; Dong et al., 2020). Yuzbasheva et al. (2015) performed biodiesel production using the lipase Lip2 displayed by Y. lipolytica. The aforementioned examples couple the cell-surface exposure of hydrolytic enzymes with the yeast robustness to efficiently ferment sugars. Also, the yeast GRAS attributes allow a wide use in this and other fields.
Immobilized enzymes by YSD present a wide range of applications, converting YSD to a powerful alternative to conventional immobilization. It allows the advancement of engineered microorganisms, with special functions not found in nature, for biotechnological applications. Since developed the first YSD, different strategies have been followed to improve the production and detection of the POI in this platform. Particularly, it was demonstrated that anchors have a decisive impact on YSD of fully functional proteins. Additionally, the compatibility of the YSD system with metabolic or protein engineering offers vast opportunities for the introduction of new platforms fed by omics databases and predictive software coupled to in vivo or in vitro systems. It is still a long way to obtaining an optimal YSD platform, but the existing strategies are the foundations for the development of new bioengineering strategies.
KT-R proposed and developed the conceptualization of the topic. KT-R and FM-M wrote the paper draft and constructed the figures. MT-H edited and revised the manuscript. All co-authors read the manuscript. All authors contributed to the article and approved the final version.
This work was supported by the National Council of Science and Technology (CONACyT) under Grant CONACyT CB 2017-2018 A1-S-34559 and a doctoral fellowship CVU number 558865.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge the National Council of Science and Technology (CONACyT) under Grant CONACyT CB 2017-2018 A1-S-34559. KT-R. acknowledges the doctoral fellowship of CONACyT (CVU number 558865). We are grateful to Brandt Bertrand (ICF-UNAM) and Daniel Morales Guzmán (CEIB-UAEM) for their critical comments on the manuscript. English grammar was kindly reviewed by Lorraine Yomano (UFL, United States) and Barbara Selisko (AMU, France).
Allison, S. D., Manning, M. C., Randolph, T. W., Middleton, K., Davis, A., and Carpenter, J. F. (2000). Optimization of Storage Stability of Lyophilized Actin Using Combinations of Disaccharides and Dextran. J. Pharm. Sci. 89 (2), 199–214. doi:10.1002/(sici)1520-6017(200002)89:2<199:aid-jps7>3.3.co;2-2
Anandharaj, M., Lin, Y.-J., Rani, R. P., Nadendla, E. K., Ho, M.-C., Huang, C.-C., et al. (2020). Constructing a Yeast to Express the Largest Cellulosome Complex on the Cell Surface. Proc. Natl. Acad. Sci. USA 117 (5), 2385–2394. doi:10.1073/pnas.1916529117
Ananphongmanee, V., Srisala, J., Sritunyalucksana, K., and Boonchird, C. (2015). Yeast Surface Display of Two Proteins Previously Shown to Be Protective against white Spot Syndrome Virus (WSSV) in Shrimp. PLoS One 10 (6), e0128764. doi:10.1371/journal.pone.0128764
Andreu, C., and del Olmo, M. L. (2017). Development of a New Yeast Surface Display System Based on Spi1 as an Anchor Protein. Appl. Microbiol. Biotechnol. 101 (1), 287–299. doi:10.1007/s00253-016-7905-x
Andreu, C., Gómez-Peinado, J., Winandy, L., Fischer, R., and del Olmo, M. l. (2021). Surface Display of HFBI and DewA Hydrophobins on Saccharomyces cerevisiae Modifies Tolerance to Several Adverse Conditions and Biocatalytic Performance. Appl. Microbiol. Biotechnol. 105 (4), 1505–1518. doi:10.1007/s00253-021-11090-8
Angelini, A., Chen, T. F., de Picciotto, S., Yang, N. J., Tzeng, A., Santos, M. S., et al. (2015). Protein Engineering and Selection Using Yeast Surface Display. Methods Mol. Biol. 1319, 3–36. doi:10.1007/978-1-4939-2748-7_1
Antipov, E., Cho, A. E., Wittrup, K. D., and Klibanov, A. M. (2008). Highly L and D Enantioselective Variants of Horseradish Peroxidase Discovered by an Ultrahigh-Throughput Selection Method. Pnas 105 (46), 17694–17699. doi:10.1073/pnas.0809851105
Aza, P., Molpeceres, G., de Salas, F., and Camarero, S. (2021). Design of an Improved Universal Signal Peptide Based on the α-factor Mating Secretion Signal for Enzyme Production in Yeast. Cell. Mol. Life Sci. 78 (7), 3691–3707. doi:10.1007/s00018-021-03793-y
Bacon, K., Burroughs, M., Blain, A., Menegatti, S., and Rao, B. M. (2019). Screening Yeast Display Libraries against Magnetized Yeast Cell Targets Enables Efficient Isolation of Membrane Protein Binders. ACS Comb. Sci. 21 (12), 817–832. doi:10.1021/acscombsci.9b00147
Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., et al. (1998). Designer Deletion Strains Derived fromSaccharomyces Cerevisiae S288C: A Useful Set of Strains and Plasmids for PCR-Mediated Gene Disruption and Other Applications. Yeast 14, 115–132. doi:10.1002/(sici)1097-0061(19980130)14:2<115:aid-yea204>3.0.co;2-2
Bamba, T., Inokuma, K., Hasunuma, T., and Kondo, A. (2018). Enhanced Cell-Surface Display of a Heterologous Protein Using SED1 Anchoring System in SED1-Disrupted Saccharomyces cerevisiae Strain. J. Biosci. Bioeng. 125 (3), 306–310. doi:10.1016/j.jbiosc.2017.09.013
Barrero, J. J., Casler, J. C., Valero, F., Ferrer, P., and Glick, B. S. (2018). An Improved Secretion Signal Enhances the Secretion of Model Proteins from Pichia pastoris. Microb. Cell Fact 17 (1), 161. doi:10.1186/s12934-018-1009-5
Bertrand, B., Trejo-Hernández, M. R., Morales-Guzmán, D., Caspeta, L., Suárez Rodríguez, R., and Martínez-Morales, F. (2016). Functional Expression, Production, and Biochemical Characterization of a Laccase Using Yeast Surface Display Technology. Fungal Biol. 120 (12), 1609–1622. doi:10.1016/j.funbio.2016.08.009
Bidlingmaier, S., and Liu, B. (2007). Interrogating Yeast Surface-Displayed Human Proteome to Identify Small Molecule-Binding Proteins. Mol. Cell Proteomics 6 (11), 2012–2020. doi:10.1074/mcp.M700223-MCP200
Blazic, M., Kovacevic, G., Prodanovic, O., Ostafe, R., Gavrovic-Jankulovic, M., Fischer, R., et al. (2013). Yeast Surface Display for the Expression, Purification and Characterization of Wild-type and B11 Mutant Glucose Oxidases. Protein Expr. Purif. 89 (2), 175–180. doi:10.1016/j.pep.2013.03.014
Boder, E. T., and Wittrup, K. D. (1997). Yeast Surface Display for Screening Combinatorial Polypeptide Libraries. Nat. Biotechnol. 15 (6), 553–557. doi:10.1038/nbt0697-553
Cejnar, R., Hložková, K., Jelínek, L., Kotrba, P., and Dostálek, P. (2017). Development of Engineered Yeast for Biosorption of Beer Haze-Active Polyphenols. Appl. Microbiol. Biotechnol. 101 (4), 1477–1485. doi:10.1007/s00253-016-7923-8
Chen, I., Dorr, B. M., and Liu, D. R. (2011). A General Strategy for the Evolution of Bond-Forming Enzymes Using Yeast Display. Proc. Natl. Acad. Sci. 108 (28), 11399–11404. doi:10.1073/pnas.1101046108
Chen, X. (2017). Yeast Cell Surface Display: An Efficient Strategy for Improvement of Bioethanol Fermentation Performance. Bioengineered 8 (2), 115–119. doi:10.1080/21655979.2016.1212135
Cheon, S. A., Jung, J., Choo, J. H., Oh, D.-B., and Kang, H. A. (2014). Characterization of Putative Glycosylphosphatidylinositol-Anchoring Motifs for Surface Display in the Methylotrophic Yeast Hansenula Polymorpha. Biotechnol. Lett. 36, 2085–2094. doi:10.1007/s10529-014-1582-6
Chun, J., Bai, J., and Ryu, S. (2020). Yeast Surface Display System for Facilitated Production and Application of Phage Endolysin. ACS Synth. Biol. 9 (3), 508–516. doi:10.1021/acssynbio.9b00360
Crowe, J. H., Crowe, L. M., Carpenter, J. F., Rudolph, A. S., Wistrom, C. A., Spargo, B. J., et al. (1988). Interactions of Sugars with Membranes. Biochim. Biophys. Acta (Bba) - Rev. Biomembranes 947 (2), 367–384. doi:10.1016/0304-4157(88)90015-9
Cruz-Terán, C. A., Tiruthani, K., Mischler, A., and Rao, B. M. (2017). Inefficient Ribosomal Skipping Enables Simultaneous Secretion and Display of Proteins in Saccharomyces cerevisiae. ACS Synth. Biol. 6 (11), 2096–2107. doi:10.1021/acssynbio.7b00144
Deweid, L., Neureiter, L., Englert, S., Schneider, H., Deweid, J., Yanakieva, D., et al. (2018). Directed Evolution of a Bond‐Forming Enzyme: Ultrahigh‐Throughput Screening of Microbial Transglutaminase Using Yeast Surface Display. Chem. Eur. J. 24 (57), 15195–15200. doi:10.1002/chem.201803485
Dong, C., Qiao, J., Wang, X., Sun, W., Chen, L., Li, S., et al. (2020). Engineering Pichia pastoris with Surface-Display Minicellulosomes for Carboxymethyl Cellulose Hydrolysis and Ethanol Production. Biotechnol. Biofuels 13, 108. doi:10.1186/s13068-020-01749-1
Duan, G., Ding, L., Wei, D., Zhou, H., Chu, J., Zhang, S., et al. (2019). Screening Endogenous Signal Peptides and Protein Folding Factors to Promote the Secretory Expression of Heterologous Proteins in Pichia pastoris. J. Biotechnol. 306, 193–202. doi:10.1016/j.jbiotec.2019.06.297
Duquesne, S., Bozonnet, S., Bordes, F., Dumon, C., Nicaud, J.-M., and Marty, A. (2014). Construction of a Highly Active Xylanase Displaying Oleaginous Yeast: Comparison of Anchoring Systems. PLoS One 9 (4), e95128. doi:10.1371/journal.pone.0095128
Fan, L.-H., Zhang, Z.-J., Yu, X.-Y., Xue, Y.-X., and Tan, T.-W. (2012). Self-surface Assembly of Cellulosomes with Two Miniscaffoldins on Saccharomyces cerevisiae for Cellulosic Ethanol Production. Proc. Natl. Acad. Sci. 109 (33), 13260–13265. doi:10.1073/pnas.1209856109
Fan, S., Liang, B., Xiao, X., Bai, L., Tang, X., Lojou, E., et al. (2020). Controllable Display of Sequential Enzymes on Yeast Surface with Enhanced Biocatalytic Activity toward Efficient Enzymatic Biofuel Cells. J. Am. Chem. Soc. 142 (6), 3222–3230. doi:10.1021/jacs.9b13289
Fiore, G., Perrino, G., di Bernardo, M., and di Bernardo, D. (2015). In Vivo real-time Control of Gene Expression: a Comparative Analysis of Feedback Control Strategies in Yeast. ACS Synth. Biol. 5 (2), 154–162. doi:10.1021/acssynbio.5b00135
Fitzgerald, I., and Glick, B. S. (2014). Secretion of a Foreign Protein from Budding Yeasts Is Enhanced by Cotranslational Translocation and by Suppression of Vacuolar Targeting. Microb. Cell Fact 13 (1), 125. doi:10.1186/s12934-014-0125-0
Hossain, A. S., Teparić, R., and Mrša, V. (2019). Comparison of Two Models of Surface Display of Xylose Reductase in the Saccharomyces cerevisiae Cell wall. Enzyme Microb. Tech. 123, 8–14. doi:10.1016/j.enzmictec.2019.01.005
Inokuma, K., Bamba, T., Ishii, J., Ito, Y., Hasunuma, T., and Kondo, A. (2016). Enhanced Cell-Surface Display and Secretory Production of Cellulolytic Enzymes withSaccharomyces cerevisiaeSed1 Signal Peptide. Biotechnol. Bioeng. 113 (11), 2358–2366. doi:10.1002/bit.26008
Inokuma, K., Hasunuma, T., and Kondo, A. (2014). Efficient Yeast Cell-Surface Display of Exo- and Endo-Cellulase Using the SED1 Anchoring Region and its Original Promoter. Biotechnol. Biofuels 7 (1), 8. doi:10.1186/1754-6834-7-8
Inokuma, K., Kitada, Y., Bamba, T., Kobayashi, Y., Yukawa, T., den Haan, R., et al. (2021). Improving the Functionality of Surface-Engineered Yeast Cells by Altering the Cell wall Morphology of the Host Strain. Appl. Microbiol. Biotechnol. 105 (14-15), 5895–5904. doi:10.1007/s00253-021-11440-6
Ishii, J., Okazaki, F., Djohan, A. C., Hara, K. Y., Asai-Nakashima, N., Teramura, H., et al. (2016). From Mannan to Bioethanol: Cell Surface Co-display of β-mannanase and β-mannosidase on Yeast Saccharomyces cerevisiae. Biotechnol. Biofuels 9 (1), 188. doi:10.1186/s13068-016-0600-4
Jacoby, K., Lambert, A. R., and Scharenberg, A. M. (2017). Characterization of Homing Endonuclease Binding and Cleavage Specificities Using Yeast Surface Display SELEX (YSD-SELEX). Nucleic Acids Res. 45 (3), gkw864. doi:10.1093/nar/gkw864
Kajiwara, K., Aoki, W., and Ueda, M. (2020). Evaluation of the Yeast Surface Display System for Screening of Functional Nanobodies. AMB Expr. 10 (1), 51. doi:10.1186/s13568-020-00983-y
Katsurada, K., Tominaga, M., Kaishima, M., Kato, H., Matsuno, T., Ogino, C., et al. (2021). Constitutive Cell Surface Expression of ZZ Domain for the Easy Preparation of Yeast-Based Immunosorbents. J. Gen. Appl. Microbiol. doi:10.2323/jgam.2021.03.004
Khatun, M. M., Liu, C.-G., Zhao, X.-Q., Yuan, W.-J., and Bai, F.-W. (2017). Consolidated Ethanol Production from Jerusalem Artichoke Tubers at Elevated Temperature by Saccharomyces cerevisiae Engineered with Inulinase Expression through Cell Surface Display. J. Ind. Microbiol. Biotechnol. 44 (2), 295–301. doi:10.1007/s10295-016-1881-0
Klis, F. M., Boorsma, A., and De Groot, P. W. J. (2006). Cell wall Construction inSaccharomyces Cerevisiae. Yeast 23 (3), 185–202. doi:10.1002/yea.1349
Könning, D., and Kolmar, H. (2018). Beyond Antibody Engineering: Directed Evolution of Alternative Binding Scaffolds and Enzymes Using Yeast Surface Display. Microb. Cell Fact 17 (1), 32. doi:10.1186/s12934-018-0881-3
Kovačević, G., Ostafe, R., Balaž, A. M., Fischer, R., and Prodanović, R. (2019). Development of GFP-Based High-Throughput Screening System for Directed Evolution of Glucose Oxidase. J. Biosci. Bioeng. 127 (1), 30–37. doi:10.1016/j.jbiosc.2018.07.002
Lei, H., Jin, S., Karlsson, E., Schultz-Cherry, S., and Ye, K. (2016). Yeast Surface-Displayed H5N1 Avian Influenza Vaccines. J. Immunol. Res. 2016, 1–12. doi:10.1155/2016/4131324
Li, J., Jiang, Z., Wu, H., Liang, Y., Zhang, Y., and Liu, J. (2010). Enzyme-polysaccharide Interaction and its Influence on Enzyme Activity and Stability. Carbohydr. Polym. 82 (1), 160–166. doi:10.1016/j.carbpol.2010.04.045
Li, S., Qiao, J., Lin, S., Liu, Y., and Ma, L. (2019). A Highly Efficient Indirect P. Pastoris Surface Display Method Based on the CL7/Im7 Ultra-high-affinity System. Molecules 24 (8), 1483. doi:10.3390/molecules24081483
Li, X., Jin, X., Lu, X., Chu, F., Shen, J., Ma, Y., et al. (2014). Construction and Characterization of a Thermostable Whole-Cell Chitinolytic Enzyme Using Yeast Surface Display. World J. Microbiol. Biotechnol. 30 (10), 2577–2585. doi:10.1007/s11274-014-1681-5
Lim, S., Glasgow, J. E., Filsinger Interrante, M., Storm, E. M., and Cochran, J. R. (2017). Dual Display of Proteins on the Yeast Cell Surface Simplifies Quantification of Binding Interactions and Enzymatic Bioconjugation Reactions. Biotechnol. J. 12 (5), 1600696. doi:10.1002/biot.201600696
Linciano, S., Pluda, S., Bacchin, A., and Angelini, A. (2019). Molecular Evolution of Peptides by Yeast Surface Display Technology. Med. Chem. Commun. 10 (9), 1569–1580. doi:10.1039/c9md00252a
Liu, D., Cheng, H., and Deng, Z. (2019). One Step Production of Isomalto-Oligosaccharides by Engineered Yarrowia Lipolytica Yeast Co-displayed β-amylase and α-transglucosidase. Sheng Wu Gong Cheng Xue Bao 35 (1), 121–132. doi:10.13345/j.cjb.180077
Liu, W., Zhao, H., Jia, B., Xu, L., and Yan, Y. (2010). Surface Display of Active Lipase in Saccharomyces cerevisiae Using Cwp2 as an Anchor Protein. Biotechnol. Lett. 32 (2), 255–260. doi:10.1007/s10529-009-0138-7
Liu, Y., Zhang, R., Lian, Z., Wang, S., and Wright, A. T. (2014). Yeast Cell Surface Display for Lipase Whole Cell Catalyst and its Applications. J. Mol. Catal. B: Enzymatic 106, 17–25. doi:10.1016/j.molcatb.2014.04.011
Luo, S., Yan, L., Zhang, X., Yuan, L., Fang, Q., Zhang, Y.-A., et al. (2015). Yeast Surface Display of Capsid Protein VP7 of Grass Carp Reovirus: Fundamental Investigation for the Development of Vaccine against Hemorrhagic Disease. J. Microbiol. Biotechnol. 25 (12), 2135–2145. doi:10.4014/jmb.1505.05041
Maneira, C., Bermejo, P. M., Pereira, G. A. G., and de Mello, F. d. S. B. (2021). Exploring G Protein-Coupled Receptors and Yeast Surface Display Strategies for Viral Detection in baker's Yeast: SARS-CoV-2 as a Case Study. FEMS Yeast Res. 21 (1), foab004. doi:10.1093/femsyr/foab004
Mateljak, I., Tron, T., and Alcalde, M. (2017). Evolved α‐factor Prepro‐leaders for Directed Laccase Evolution in Saccharomyces cerevisiae. Microb. Biotechnol. 10 (6), 1830–1836. doi:10.1111/1751-7915.12838
Matsuoka, H., Hashimoto, K., Saijo, A., Takada, Y., Kondo, A., Ueda, M., et al. (2014). Cell wall Structure Suitable for Surface Display of Proteins inSaccharomyces Cerevisiae. Yeast 31 (2), 67–76. doi:10.1002/yea.2995
McMahon, C., Baier, A. S., Pascolutti, R., Wegrecki, M., Zheng, S., Ong, J. X., et al. (2018). Yeast Surface Display Platform for Rapid Discovery of Conformationally Selective Nanobodies. Nat. Struct. Mol. Biol. 25 (3), 289–296. doi:10.1038/s41594-018-0028610.1038/s41594-018-0028-6
Moon, H. Y., Van, T. L., Cheon, S. A., Choo, J., Kim, J.-Y., and Kang, H. A. (2013). Cell-surface Expression of Aspergillus Saitoi-Derived Functional α-1,2-mannosidase on Yarrowia Lipolytica for Glycan Remodeling. J. Microbiol. 51 (4), 506–514. doi:10.1007/s12275-013-3344-x
Moura, M. V. H., da Silva, G. P., Machado, A. C. d. O., Torres, F. A. G., Freire, D. M. G., and Almeida, R. V. (2015). Displaying Lipase B from Candida antarctica in Pichia pastoris Using the Yeast Surface Display Approach: Prospection of a New Anchor and Characterization of the Whole Cell Biocatalyst. PLoS One 10 (10), e0141454. doi:10.1371/journal.pone.0141454
Nguyen, T. P. A., Nguyen, T. T. M., Nguyen, N. H., Nguyen, T. N., and Dang, T. T. P. (2020). Application of Yeast Surface Display System in Expression of Recombinant Pediocin PA-1 in Saccharomyces cerevisiae. Folia Microbiol. 65 (6), 955–961. doi:10.1007/s12223-020-00804-6
N., S., T., M., M., U., A., T., H., F., and A., K. (2002). Long Anchor Using Flo1 Protein Enhances Reactivity of Cell Surface-Displayed Glucoamylase to Polymer Substrates. Appl. Microbiol. Biotechnol. 60 (4), 469–474. doi:10.1007/s00253-002-1121-6
Pal Khasa, Y., Conrad, S., Sengul, M., Plautz, S., Meagher, M. M., and Inan, M. (2011). Isolation of Pichia pastoris PIR Genes and Their Utilization for Cell Surface Display and Recombinant Protein Secretion. Yeast 28 (3), 213–226. doi:10.1002/yea.1832
Procko, E. (2020). Deep Mutagenesis in the Study of COVID-19: a Technical Overview for the Proteomics Community. Expert Rev. Proteomics 17 (9), 633–638. doi:10.1080/14789450.2020.1833721
Raeeszadeh-Sarmazdeh, M., Greene, K. A., Sankaran, B., Downey, G. P., Radisky, D. C., and Radisky, E. S. (2019). Directed Evolution of the Metalloproteinase Inhibitor TIMP-1 Reveals that its N- and C-Terminal Domains Cooperate in Matrix Metalloproteinase Recognition. J. Biol. Chem. 294 (24), 9476–9488. doi:10.1074/jbc.RA119.008321
Rakestraw, J. A., Sazinsky, S. L., Piatesi, A., Antipov, E., and Wittrup, K. D. (2009). Directed Evolution of a Secretory Leader for the Improved Expression of Heterologous Proteins and Full-Length Antibodies inSaccharomyces Cerevisiae. Biotechnol. Bioeng. 103 (6), 1192–1201. doi:10.1002/bit.22338
Redden, H., Morse, N., and Alper, H. S. (2014). The Synthetic Biology Toolbox for Tuning Gene Expression in Yeast. FEMS Yeast Res. 15, 1–10. doi:10.1111/1567-1364.12188
Schmitt, F. B. M., and Schmitt, M. J. (2002). Spacer-elongated Cell wall Fusion Proteins Improve Cell Surface Expression in the Yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 58 (5), 637–644. doi:10.1007/s00253-002-0939-2
Schröter, C., Krah, S., Beck, J., Könning, D., Grzeschik, J., Valldorf, B., et al. (2018). Isolation of pH-Sensitive Antibody Fragments by Fluorescence-Activated Cell Sorting and Yeast Surface Display. Methods Mol. Biol. 1685, 311–331. doi:10.1007/978-1-4939-7366-8_19
Shibasaki, S., Aoki, W., Nomura, T., Miyoshi, A., Tafuku, S., Sewaki, T., et al. (2013). An Oral Vaccine against Candidiasis Generated by a Yeast Molecular Display System. Pathog. Dis. 69 (3), 262–268. doi:10.1111/2049-632X.12068
Shiraga, S., Kawakami, M., Ishiguro, M., and Ueda, M. (2005). Enhanced Reactivity of Rhizopus Oryzae Lipase Displayed on Yeast Cell Surfaces in Organic Solvents: Potential as a Whole-Cell Biocatalyst in Organic Solvents. Appl. Environ. Microbiol. 71 (8), 4335–4338. doi:10.1128/aem.71.8.4335-4338.2005
Singh, N., Gupta, A., Mathur, A. S., Barrow, C., and Puri, M. (2020). Integrated Consolidated Bioprocessing for Simultaneous Production of Omega-3 Fatty Acids and Bioethanol. Biomass and Bioenergy 137, 105555. doi:10.1016/j.biombioe.2020.105555
Smith, M. R., Gao, H., Prabhu, P., Bugada, L. F., Roth, C., Mutukuri, D., et al. (2019). Elucidating Structure-Performance Relationships in Whole-Cell Cooperative Enzyme Catalysis. Nat. Catal. 2 (9), 809–819. doi:10.1038/s41929-019-0321-8
Smith, M. R., Khera, E., and Wen, F. (2015). Engineering Novel and Improved Biocatalysts by Cell Surface Display. Ind. Eng. Chem. Res. 54 (16), 4021–4032. doi:10.1021/ie504071f
Sun, W., Yang, Z., Lin, H., Liu, M., Zhao, C., Hou, X., et al. (2019). Improvement in Affinity and Thermostability of a Fully Human Antibody against interleukin-17A by Yeast-Display Technology and CDR Grafting. Acta Pharmaceutica Sinica B 9 (5), 960–972. doi:10.1016/j.apsb.2019.02.007
Sun, Y.-F., Lin, Y., Zhang, J.-H., Zheng, S.-P., Ye, Y.-R., Liang, X.-X., et al. (2012). Double Candida antarctica Lipase B Co-display on Pichia pastoris Cell Surface Based on a Self-Processing Foot-And-Mouth Disease Virus 2A Peptide. Appl. Microbiol. Biotechnol. 96 (6), 1539–1550. doi:10.1007/s00253-012-4264-0
Takayama, S., Ozaki, A., Konishi, R., Otomo, C., Kishida, M., Hirata, Y., et al. (2018). Enhancing 3-hydroxypropionic Acid Production in Combination with Sugar Supply Engineering by Cell Surface-Display and Metabolic Engineering of Schizosaccharomyces pombe. Microb. Cell Fact 17 (1), 176. doi:10.1186/s12934-018-1025-5
Takenaka, M., Kobayashi, T., Inokuma, K., Hasunuma, T., Maruyama, T., Ogino, C., et al. (2017). Mapping of Endoglucanases Displayed on Yeast Cell Surface Using Atomic Force Microscopy. Colloids Surf. B: Biointerfaces. 151, 134–142. doi:10.1016/j.colsurfb.2016.12.014
Tanaka, T., and Kondo, A. (2014). Cell-surface Display of Enzymes by the yeastSaccharomyces Cerevisiaefor Synthetic Biology. FEMS Yeast Res. 15 (1), 1–9. doi:10.1111/1567-1364.12212
Tanaka, T., Yamada, R., Ogino, C., and Kondo, A. (2012). Recent Developments in Yeast Cell Surface Display toward Extended Applications in Biotechnology. Appl. Microbiol. Biotechnol. 95 (3), 577–591. doi:10.1007/s00253-012-4175-0
Tang, H., Bao, X., Shen, Y., Song, M., Wang, S., Wang, C., et al. (2015). Engineering Protein Folding and Translocation Improves Heterologous Protein Secretion inSaccharomyces Cerevisiae. Biotechnol. Bioeng. 112 (9), 1872–1882. doi:10.1002/bit.25596
Tang, H., Song, M., He, Y., Wang, J., Wang, S., Shen, Y., et al. (2017). Engineering Vesicle Trafficking Improves the Extracellular Activity and Surface Display Efficiency of Cellulases in Saccharomyces cerevisiae. Biotechnol. Biofuels 10, 53. doi:10.1186/s13068-017-0738-8
Tang, H., Wang, J., Wang, S., Shen, Y., Petranovic, D., Hou, J., et al. (2018). Efficient Yeast Surface-Display of Novel Complex Synthetic Cellulosomes. Microb. Cell Fact 17 (1), 122. doi:10.1186/s12934-018-0971-2
Traxlmayr, M. W., and Obinger, C. (2012). Directed Evolution of Proteins for Increased Stability and Expression Using Yeast Display. Arch. Biochem. Biophys. 526 (2), 174–180. doi:10.1016/j.abb.2012.04.022
Trentelman, J. J. A., Tomás-Cortázar, J., Knorr, S., Barriales, D., Hajdusek, O., Sima, R., et al. (2021). Probing an Ixodes Ricinus Salivary Gland Yeast Surface Display with Tick-Exposed Human Sera to Identify Novel Candidates for an Anti-tick Vaccine. Sci. Rep. 11 (1), 15745. doi:10.1038/s41598-021-92538-9
Tsai, S.-L., DaSilva, N. A., and Chen, W. (2013). Functional Display of Complex Cellulosomes on the Yeast Surface via Adaptive Assembly. ACS Synth. Biol. 2 (1), 14–21. doi:10.1021/sb300047u
Tunyasuvunakool, K., Adler, J., Wu, Z., Green, T., Zielinski, M., Žídek, A., et al. (2021). Highly Accurate Protein Structure Prediction for the Human Proteome. Nature 596, 590–596. doi:10.1038/s41586-021-03828-1
Uchański, T., Zögg, T., Yin, J., Yuan, D., Wohlkönig, A., Fischer, B., et al. (2019). An Improved Yeast Surface Display Platform for the Screening of Nanobody Immune Libraries. Sci. Rep. 9 (1), 382. doi:10.1038/s41598-018-37212-3
Ueda, M. (2016). Establishment of Cell Surface Engineering and its Development. Biosci. Biotech. Biochem. 80 (7), 1243–1253. doi:10.1080/09168451.2016.1153953
Valldorf, B., Hinz, S. C., Russo, G., Pekar, L., Mohr, L., Klemm, J., et al. (2021). Antibody Display Technologies: Selecting the Cream of the Crop. Biol.Chem. doi:10.1515/hsz-2020-0377
Van der Vaart, J. M., te Biesebeke, R., Chapman, J. W., Toschka, H. Y., Klis, F. M., Verrips, C. T., et al. (2021). Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl. Environ. Microbiol 63, 615–620. doi:10.1128/AEM.63.2.615-620.1997
Wang, J.-m., Wang, C.-m., Men, X., Yue, T.-q., Madzak, C., Xiang, X.-h., et al. (2020). Construction of Arming Yarrowia Lipolytica Surface-Displaying Soybean Seed Coat Peroxidase for Use as Whole-Cell Biocatalyst. Enzyme Microb. Tech. 135, 109498. doi:10.1016/j.enzmictec.2019.109498
Wang, L.-j., Xiao, T., Xu, C., Li, J., Liu, G.-z., Yin, K., et al. (2018). Protective Immune Response against Toxoplasma Gondii Elicited by a Novel Yeast-Based Vaccine with Microneme Protein 16. Vaccine 36 (27), 3943–3948. doi:10.1016/j.vaccine.2018.05.072
Wang, Z., Mathias, A., Stavrou, S., and Neville, D. M. (2005). A New Yeast Display Vector Permitting Free scFv Amino Termini Can Augment Ligand Binding Affinities. Protein Eng. Des. Sel 18 (7), 337–343. doi:10.1093/protein/gzi036
Washida, M., Takahashi, S., Ueda, M., and Tanaka, A. (2001). Spacer-mediated Display of Active Lipase on the Yeast Cell Surface. Appl. Microbiol. Biotechnol. 56 (5-6), 681–686. doi:10.1007/s002530100718
Wen, F., Sun, J., and Zhao, H. (2010). Yeast Surface Display of Trifunctional Minicellulosomes for Simultaneous Saccharification and Fermentation of Cellulose to Ethanol. Appl. Environ. Microbiol. 76 (4), 1251–1260. doi:10.1128/AEM.01687-09
Wentz, A. E., and Shusta, E. V. (2007). A Novel High-Throughput Screen Reveals Yeast Genes that Increase Secretion of Heterologous Proteins. Appl. Environ. Microbiol. 73 (4), 1189–1198. doi:10.1128/AEM.02427-06
Yang, S., Lv, X., Wang, X., Wang, J., Wang, R., and Wang, T. (2017). Cell-surface Displayed Expression of Trehalose Synthase from Pseudomonas Putida ATCC 47054 in Pichia pastoris Using Pir1p as an Anchor Protein. Front. Microbiol. 8, 2583. doi:10.3389/fmicb.2017.02583
Yang, X., Tang, H., Song, M., Shen, Y., Hou, J., and Bao, X. (2019). Development of Novel Surface Display Platforms for Anchoring Heterologous Proteins in Saccharomyces cerevisiae. Microb. Cell Fact 18 (1), 85. doi:10.1186/s12934-019-1133-x
Yuzbasheva, E. Y., Yuzbashev, T. V., Laptev, I. A., Konstantinova, T. K., and Sineoky, S. P. (2011). Efficient Cell Surface Display of Lip2 Lipase Using C-Domains of Glycosylphosphatidylinositol-Anchored Cell wall Proteins of Yarrowia Lipolytica. Appl. Microbiol. Biotechnol. 91 (3), 645–654. doi:10.1007/s00253-011-3265-8
Yuzbasheva, E. Y., Yuzbashev, T. V., Perkovskaya, N. I., Mostova, E. B., Vybornaya, T. V., Sukhozhenko, A. V., et al. (2015). Cell Surface Display of Yarrowia Lipolytica Lipase Lip2p Using the Cell Wall Protein YlPir1p, its Characterization, and Application as a Whole-Cell Biocatalyst. Appl. Biochem. Biotechnol. 175 (8), 3888–3900. doi:10.1007/s12010-015-1557-7
Zhang, L., An, J., Li, L., Wang, H., Liu, D., Li, N., et al. (2016). Highly Efficient Fructooligosaccharides Production by an Erythritol-Producing Yeast Yarrowia Lipolytica Displaying Fructosyltransferase. J. Agric. Food Chem. 64 (19), 3828–3837. doi:10.1021/acs.jafc.6b00115
Zhang, L., Liang, S., Zhou, X., Jin, Z., Jiang, F., Han, S., et al. (2013). Screening for Glycosylphosphatidylinositol-Modified Cell wall Proteins in Pichia pastoris and Their Recombinant Expression on the Cell Surface. Appl. Environ. Microbiol. 79 (18), 5519–5526. doi:10.1128/AEM.00824-13
Zhang, Y., Min, Z., Qin, Y., Ye, D.-Q., Song, Y.-Y., and Liu, Y.-L. (2019). Efficient Display of Aspergillus niger β-Glucosidase on Saccharomyces cerevisiae Cell Wall for Aroma Enhancement in Wine. J. Agric. Food Chem. 67 (18), 5169–5176. doi:10.1021/acs.jafc.9b00863
Zhao, S., Guo, D., Zhu, Q., Dou, W., and Guan, W. (2020b). Display of Microbial Glucose Dehydrogenase and Cholesterol Oxidase on the Yeast Cell Surface for the Detection of Blood Biochemical Parameters. Biosensors 11 (1), 13. doi:10.3390/bios11010013
Zhao, S., Guo, Y., Wang, Q., Luo, H., He, C., and An, B. (2020a). Expression of Flagellin at Yeast Surface Increases Biocontrol Efficiency of Yeast Cells against Postharvest Disease of Tomato Caused by Botrytis Cinerea. Postharvest Biol. Tech. 162, 111112. doi:10.1016/j.postharvbio.2019.111112
Keywords: cell surface display, yeast, anchor, microbial engineering, whole cell biocatalyst
Citation: Teymennet-Ramírez KV, Martínez-Morales F and Trejo-Hernández MR (2022) Yeast Surface Display System: Strategies for Improvement and Biotechnological Applications. Front. Bioeng. Biotechnol. 9:794742. doi: 10.3389/fbioe.2021.794742
Received: 14 October 2021; Accepted: 20 December 2021;
Published: 10 January 2022.
Edited by:
José Cleiton Sousa dos Santos, University of International Integration of Afro-Brazilian Lusophony, BrazilReviewed by:
Roberta Bussons Rodrigues Valério, Federal University of Ceara, BrazilCopyright © 2022 Teymennet-Ramírez, Martínez-Morales and Trejo-Hernández. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: María R. Trejo-Hernández, bXRyZWpvQHVhZW0ubXg=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.