
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 30 November 2021
Sec. Synthetic Biology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.787881
This article is part of the Research Topic Cell-Free Synthetic Biology, Volume II View all 16 articles
Circular RNA (circRNA) is a unique type of noncoding RNA molecule. Compared with traditional linear RNA, circRNA is a covalently closed circle produced by a process called backsplicing. CircRNA is abundant in many cells and has rich functions in cells, such as acting as miRNA sponge, protein sponge, protein scaffold, and mRNA regulator. With the continuous development of circRNA study, circRNA has also played an important role in medical applications, including circRNA vaccines and gene therapy. In this review, we illustrate the synthesis of circRNAs in vitro. We focus on biological ligation methods, such as enzymatic ligation from the bacteriophage T4 and ribozyme method. In addition, we summarize the current challenges in the design, synthesis, application, and production of circRNAs, and propose possible solutions in the future. CircRNA is expected to play an essential role in basic research and medical applications.
A newly described RNA, called circular RNA (circRNA) (Sanger et al., 1976; Arnberg et al., 1980; Kos et al., 1986; Flores et al., 2011; Salzman et al., 2012; Jeck et al., 2013; Memczak et al., 2013; Guo et al., 2014; Jeck and Sharpless, 2014; Wang et al., 2014), has got much attention because of the development of high-throughput RNA-sequencing technology in recent years. Compared with traditional linear RNA, circRNA is a 3′-5′ covalently closed ring (Holdt et al., 2018) and does not need 5′-cap or 3′-poly(A) tails to keep it stable (Petkovic and Müller, 2015). These circRNAs have been found in a wide range of cells (Sanger et al., 1976; Arnberg et al., 1980; Kos et al., 1986; Salzman et al., 2012; Memczak et al., 2013; Wang et al., 2014), and most of them are noncoding RNAs (ncRNAs) (Santer et al., 2019). According to the components of circRNAs, circRNAs are classified as exonic (ecircRNA), exon-intron (EIcircRNA), or intronic (ciRNA) (He et al., 2021; Tao et al., 2021). Among them, ecircRNAs are the major circRNAs and are mainly produced by a process called backsplicing in vivo (Schindewolf et al., 1996; Salzman et al., 2012; Jeck et al., 2013; Memczak et al., 2013; Guo et al., 2014; Barrett et al., 2015; Starke et al., 2015; Szabo et al., 2015). EcircRNAs are mainly located in the cytoplasm and have various functions. The most well-known function of circRNAs is the microRNA (miRNA) sponge (Li et al., 2020; He et al., 2021), such as circRNA CDR1as and Sry (Hansen et al., 2013a; Memczak et al., 2013). Besides, circRNAs are also proved to serve as protein sponge (Ashwal-Fluss et al., 2014) and scaffold for protein complexes (Du et al., 2017a; Du et al., 2020). In addition, circRNAs also play a role in regulating the translation and stability of mRNA levels (Chen et al., 2020b; Huang et al., 2020) and the activity of proteins (Jeck et al., 2013; Ashwal-Fluss et al., 2014; Abdelmohsen et al., 2017; Du et al., 2017b). As biomarkers, circRNAs are also proved to be relative to the age-dependent neural accumulation in drosophila (Westholm et al., 2014). Moreover, circRNAs can also be involved in cancer or other diseases (Hansen et al., 2013b; Li et al., 2015; Qu et al., 2015).
With the development of RNA vaccines, the improvement of the stability of RNAs has been a great challenge. Fortunately, circRNAs show great potential ahead of this challenge. The structural advantage of circRNAs brings it higher stability, especially against the degradation of exonucleases (Wesselhoeft et al., 2018). Endogenously produced circRNAs are 2–5 times more stable than linear RNAs (Enuka et al., 2016; Holdt et al., 2018). Therefore, the possibility that circRNAs serve as vaccines has been the main concern for researchers. Although circRNAs lack the essential elements for cap-dependent translation, the discovery of internal ribosome entry sites (IRES) makes it possible for circRNAs to act as translation templates (Legnini et al., 2017; Pamudurti et al., 2017; Wesselhoeft et al., 2018). N6-methyladenosine (m6A) is also proved to promote the extensive translation of circRNAs (Yang et al., 2017). Based on these studies, circRNA vaccines are likely to become the dominant form of the next generation of vaccines.
In this review, we introduced the synthesis of circRNAs in vitro and focused on the challenges and opportunities of circRNA biosynthesis. Since circRNAs have unique structures and special functions different from linear RNAs, further study of circRNAs will help understand biogenesis and expand the application of circRNAs in biological therapy and vaccine research.
The concept of circRNAs was first proposed by Sanger et al., in 1976 (Sanger et al., 1976) (Figure 1A). They studied four different highly purified viroids and found that viroids were covalently closed circRNA molecules. For the next 20 years, circRNAs were found in several kinds of life, such as yeast (Arnberg et al., 1980) and hepatitis delta virus (HDV) (Kos et al., 1986). CircRNAs were first discovered in mammals in 1991. Nigro et al. identified several abnormally spliced transcripts, in which exons from a candidate tumor suppressor gene (DCC) were scrambled during the splicing process in vivo (Nigro et al., 1991). After that, more circRNAs were found in mammalian cells (Cocquerelle et al., 1992; Capel et al., 1993; Zaphiropoulos, 1996; Zaphiropoulos, 1997; Surono et al., 1999). However, since the expression level of these circRNAs observed was so low, researchers believed that these circRNAs were the products of aberrant RNA splicing (Nigro et al., 1991; Cocquerelle et al., 1993). Therefore, researchers only knew the existence of circRNAs, but had no further understanding of theirs functions or impacts, and circRNAs did not receive much attention.
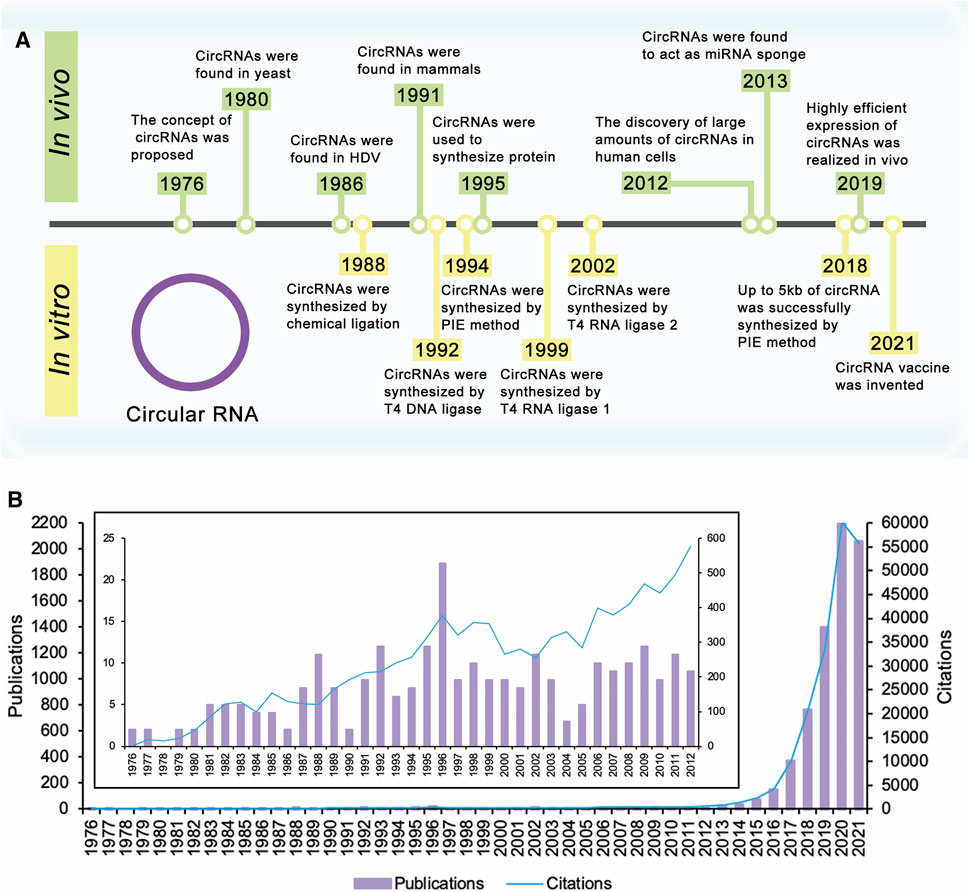
FIGURE 1. Timeline of circular RNA. (A) The concept of circRNAs was proposed in 1976. Extensive research on circRNAs began in 2012 with the discovery of large amounts of circRNAs in human cells. These researches have improved yields of circRNAs, realized the synthesis of large circRNA molecules, and expanded the applications of circRNAs. (B) Times cited and publications over time. There were a few publications about circRNAs before 2012. Since the discovery of large amounts of circRNAs in human cells in 2012, the number of publications and citations has increased year by year and is still rising.
Extensive research on circRNAs began in 2012 with the discovery of large amounts of circRNAs in human cells (Figure 1B). With the development of high-throughput sequencing technology and computational analysis, Salzman et al. found that a substantial fraction of the spliced transcripts from hundreds of genes were circRNAs by deep sequencing of RNA from a variety of normal and malignant human cells (Salzman et al., 2012). This result proved that the circRNA was not a mistake of aberrant RNA splicing, but a general feature of the gene expression program in human cells. The discovery revived great interest in circRNA research, and relative research was growing exponentially. In 2013, the function of circRNAs as the miRNA sponge was reported (Hansen et al., 2013a; Memczak et al., 2013). Besides, circRNAs were proved to be more stable than associated linear mRNAs in vivo (Jeck et al., 2013). The confirmation of the abundance, function, and stability of circRNAs laid the foundation of circRNAs research. Since then, more functions of circRNAs have been explored by researchers (Figure 2A).
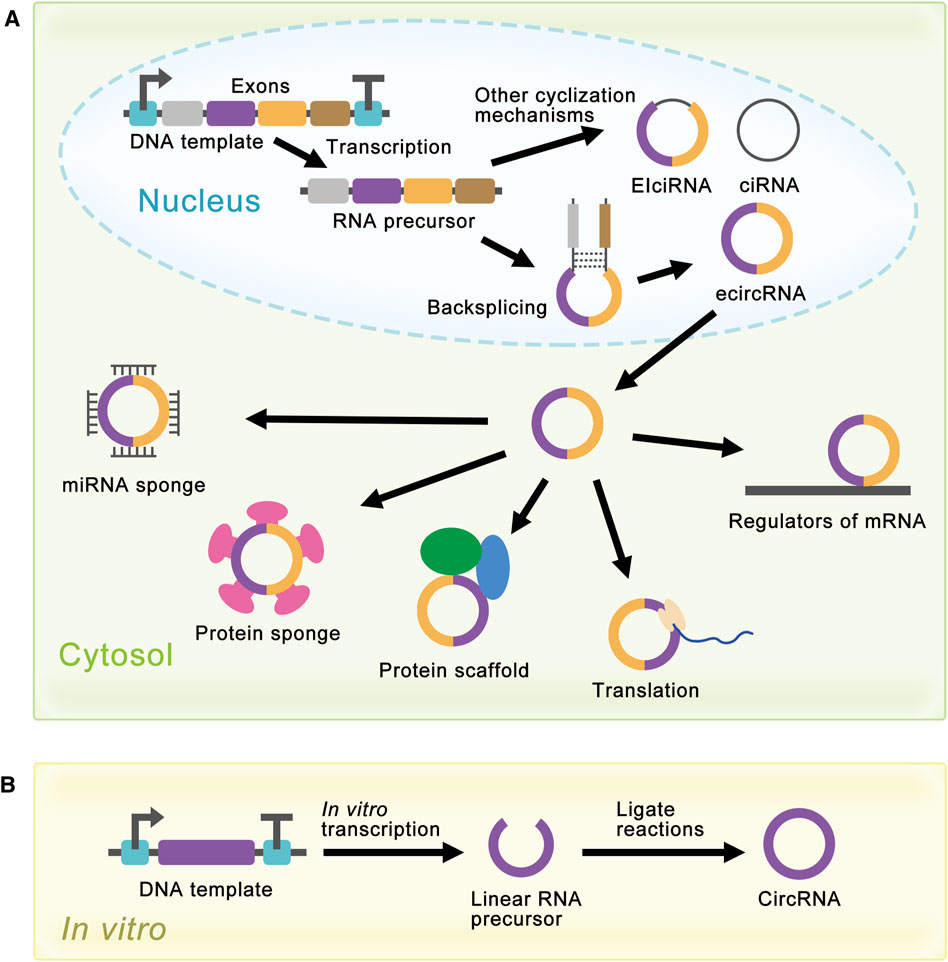
FIGURE 2. Schematic diagram of circRNA synthesis in vivo and in vitro. (A) EcircRNAs are the major circRNAs and are mainly produced by a process called backsplicing in vivo. EIciRNAs and ciRNAs are produced by other cyclization reactions and located in the nucleus. EcircRNAs are mainly located in the cytoplasm and have various functions. In the cytosol, circRNAs can act as miRNA sponges, protein sponges, protein scaffolds, translation templates, and regulators of mRNA. (B) Linear RNA precursor is produced from in vitro transcription, and researchers have developed several methods for linear RNA precursor ligation to synthesize circRNA in vitro.
Although natural circRNAs were ncRNAs and were considered incapable of translation, research had proved that manufactured circRNAs with IRES could be translated in vivo and in vitro (Chen and Sarnow, 1995; Perriman and Ares, 1998; Wang and Wang, 2015; Barrett and Salzman, 2016; Szabo and Salzman, 2016), which opened the way for the synthesis of proteins with circRNAs. More and more research was devoted to artificially controlling the expression level of circRNAs to realize the enrichment of circRNAs functions (Liang and Wilusz, 2014; Zhang et al., 2014; Kramer et al., 2015; Zhang et al., 2016; Garikipati et al., 2019). For the past few years, researchers have achieved highly efficient expression of circRNAs in cells using autocatalytic transcripts (Litke and Jaffrey, 2019) or viroid scaffolds (Daròs, 2021). At the same time, researchers had also developed several methods for the synthesis of circRNAs in vitro, such as chemical method (Sokolova et al., 1988), enzymatic method (Moore, 1999), and ribozyme method (Puttaraju and Been, 1992). These methods produced circRNAs by ligating the ends of linear RNA precursor, which would be covered later (Figure 2B).
At present, the main method of circRNA synthesis in vitro is ligating the ends of linear RNA precursor to produce a covalently closed circle. Linear RNA can be produced by chemical synthesis (Müller and Appel, 2017; Obi and Chen, 2021) or enzymatic strategy (Obi and Chen, 2021). The advantage of chemical synthesis is that the 5′ monophosphate can be directly introduced during the synthesis process for future cyclization. However, limited by the high cost of purification and low yield, chemical synthesis can only produce RNA of less than 50 to 70 nucleotides in length. Therefore, enzymatic strategy is the primary linear RNA synthesis method at present. Enzymatic strategy is usually realized through an in vitro transcription (IVT) reaction (Beckert and Masquida, 2011), which includes DNA template, reaction buffer, and phage RNA polymerase. The phage RNA polymerase usually derives from the T7, SP6, or T3 bacteriophages, and T7 RNA polymerase (Rio, 2013) is the most common phage RNA polymerase. IVT reaction allows for longer RNA synthesis at a lower cost. However, the run-off nature of phage polymerases may result in incomplete RNA. Some studies have improved transcription quality and reduced side reactions by mutating wild-type phage RNA polymerase.
Researchers have developed several methods for linear RNA precursor ligation in vitro. These ligation methods include chemical ligation, enzymatic ligation, and ribozyme method. Chemical ligation is realized by using cyanogen bromide (BrCN) or 1-ethyl-3-(3′-dimethylaminopropyl) carbodiimide to link DNA-RNA hybrids (Sokolova et al., 1988). However, this method suffers from low ligating efficiency (Dolinnaya et al., 1991) and biosafety concerns. In addition, chemical ligation forms 2′, 5′-phosphodiester bonds instead of the natural 3′, 5′-phosphodiester bonds (Table 1). Therefore, chemical ligation is not a common ligation method. Researchers are more interested in the biosynthesis of circRNAs, which includes enzymatic ligation and ribozyme method.
Enzymatic ligations are realized by catalytic reactions of several enzymes from the bacteriophage T4, including T4 DNA ligase (T4 Dnl), T4 RNA ligase 1 (T4 Rnl 1), and T4 RNA ligase 2 (T4 Rnl 2). It is worth noting that linear RNA precursor needs a 3′-OH on the acceptor substrate and a 5′ monophosphate on the donor substrate for enzymatic ligation (Moore, 1999). If the linear RNA precursor is produced by chemical synthesis, a 5′ monophosphate can be incorporated during the synthesis or added after the synthesis using ATP and T4 polynucleotide kinase. However, if the linear RNA precursor is synthesized by IVT reaction, it usually starts with 5′-pppG. Therefore, the 5′-terminus has to be dephosphorylated prior using calf intestinal (CIP) enzyme or other phosphatases (Petkovic and Müller, 2018). Then, 5′ monophosphate can be added using ATP and T4 polynucleotide kinase. In addition, the addition of GMP to the IVT reaction mixture is also proved to be useful for phosphorylating the transcript at its 5′ end (Abe et al., 2018). However, this method must ensure that the first base in the transcript is G, so there are some limitations to this method.
T4 Dnl ligation reaction includes inactivated kinase reaction and hybridized reaction. T4 Dnl can help ligate double-stranded duplexes, such as DNA/RNA hybrids (Moore, 1999) (Figure 3A). Therefore, this method needs a complementary DNA (cDNA) template or bridge to achieve RNA ligation. Generally, the cDNA bridge needs at least 10 nucleotides on either side of the ligation junction to guarantee high-quality ligation. The advantage of this method is that the accuracy of the linkage sites is greatly improved due to the addition of cDNA bridge. However, the ends of linear RNA precursor should be free of significant RNA secondary structure, and there should not be a high percentage of Us in the duplex region. Besides, T4 Dnl is less efficient in DNA/RNA hybrids linkage than double-stranded DNA (dsDNA) linkage. Due to these characteristics, only a few studies use this method for RNA ligation (Chen et al., 2017).
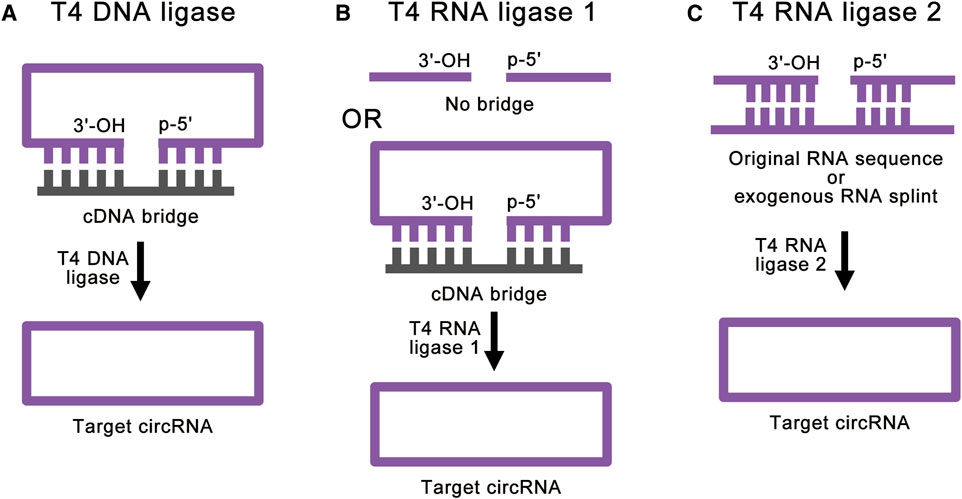
FIGURE 3. Strategies for enzymatic ligations. (A) T4 DNA ligase can help ligate double-stranded duplexes, such as DNA/RNA hybrids. With the help of cDNA bridge, T4 DNA ligase can achieve accurate RNA ligation. (B) T4 RNA ligase 1 catalyzes the nucleophilic attack of the 3′-OH terminus onto the activated 5′-terminus to form a covalent 5′, 3′-phosphodiester bond. The cDNA bridge can prevent the linear RNA precursor from folding into an unsuitable structure. (C) T4 RNA ligase 2 is suitable for linear RNA precursor with the ligation junction in a double-stranded region. With the help of RNA splint, T4 RNA ligase 2 can also realize the ligation of ends of ssRNA.
T4 Rnl 1 is a more common ligase for RNA ligation. T4 Rnl 1 catalyzes the nucleophilic attack of the 3′-OH terminus onto the activated 5′-terminus to form a covalent 5′, 3′-phosphodiester bond (Petkovic and Müller, 2018) and produces circRNAs (Figure 3B). Some studies have used cDNA bridge to prevent the linear RNA precursor from folding into an unsuitable structure (Wang and Ruffner, 1998). It is worth noting that T4 Rnl 1 has different preferences for the nucleotides of the 5′-terminus and 3′-terminus: A > G ≥ C > U for the 3′-terminal nucleotide acceptor, and pC > pU > pA > pG for the 5′-terminal nucleotide donor (England and Uhlenbeck, 1978; McLaughlin et al., 1982; Petkovic and Müller, 2015; Müller and Appel, 2017). In this way, as little as 6 to 8 nucleotides of circRNAs can be synthesized (Kaufmann et al., 1974; Petkovic and Müller, 2015). This method can achieve high-efficiency single-stranded RNA (ssRNA) linkage. However, the RNA ligation efficiency reduces with large RNA molecules (Costello et al., 2020). Similar to the T4 Dnl ligation reaction, significant RNA secondary structure at the ends of linear RNA precursor can greatly reduce the ligation efficiency of T4 Rnl 1. Besides, intermolecular end joining (oligomerization) is also a serious side reaction, which cannot be completely suppressed (Petkovic and Müller, 2018). This side reaction will increase with the increase of the concentration of linear RNA precursor, which limits the amount of RNA cyclization.
T4 Rnl 2 can also be used for RNA ligation (Ho and Shuman, 2002; Nandakumar and Shuman, 2004; Yin et al., 2004). Similar to T4 Rnl 1, T4 Rnl 2 also catalyzes the nucleophilic attack of the 3′-OH terminus onto the activated 5′-terminus to form a covalent 5′, 3′-phosphodiester bond. However, T4 Rnl 2 is much more active at joining nicks in double-stranded RNA (dsRNA) than at ligating the ends of ssRNA (Nandakumar et al., 2004; Bullard and Bowater, 2006) (Figure 3C). Based on this feature, when the linear RNA precursor folds into a secondary structure with the ligation junction in a double-stranded region, the efficiency of T4 Rnl 2 is much higher than that of T4 Rnl 1 (Petkovic and Müller, 2018). Besides, with the help of RNA splint, T4 Rnl 2 can also realize the ligation of ends of ssRNA. However, RNA splints cannot be used to ligate short RNA precursors, because the splint-precursor complex is sterically unstable when the length of linear RNA precursor is shorter than 30 nucleotides. Same as T4 Rnl 1, T4 Rnl 2 also suffers from low efficiency for large RNA molecules and side reactions. To overcome these challenges, there are studies using software to simulate the secondary structure of the target circRNA and hypothetically cut at one site so that a few intramolecular base pairs are formed at the terminal (Chen et al., 2020a). This method can achieve efficient RNA cyclization at high concentrations with T4 Rnl 2, but it is still affected by different RNA sequences.
In a word, T4 Dnl and T4 Rnl 1 are suitable for RNA ligations without complex secondary structures. T4 Rnl 2 is more suitable for linear RNA precursor with the ligation junction in a double-stranded region. Therefore, different T4 ligases need to be selected according to the secondary structure of the linear RNA precursor. However, all these enzymatic ligation methods cannot realize large RNA molecules ligation, and cannot totally avoid intermolecular end-joining side reactions. These problems still need to be solved (Table 1).
Modified group I intron self-splicing system is the most common ribozyme method, which is also called permuted introns and exons (PIE) method (Puttaraju and Been, 1992; Ford and Ares, 1994; Umekage and Kikuchi, 2009; Wesselhoeft et al., 2018; Rausch et al., 2021). PIE method requires only the addition of GTP and Mg2+ as cofactors and shows great potential for protein synthesis. This method realized RNA ligation through a regular group I intron self-splicing reaction, including two transesterifications at defined splice sites (Figure 4A). PIE method can be used for RNA cyclization in vitro and in vivo (Meganck et al., 2021), which broadens its application. Compared to chemical ligation and enzymatic ligation, PIE method could be applied for the cyclization of larger linear RNA precursor, and the reaction condition and purification method of PIE method is simpler. A recent study has realized accurate RNA ligation by designing custom-tailored PIE transcription templates from which synthetic circRNAs of almost any sequence may be efficiently synthesized without exogenous exon sequences (Rausch et al., 2021). Based on these advantages, PIE method is currently the most studied and most widely used RNA ligation method. However, PIE method still has its disadvantages. Due to the complexity of RNA secondary structure, different RNA sequences will lead to a great difference in the final circRNA field, which limits the application of PIE method. Due to the introduction of exogenous exon sequences, the final circRNA sequence will be different from the original linear RNA precursor sequence, which may negatively affect the validation of some circRNA functions.
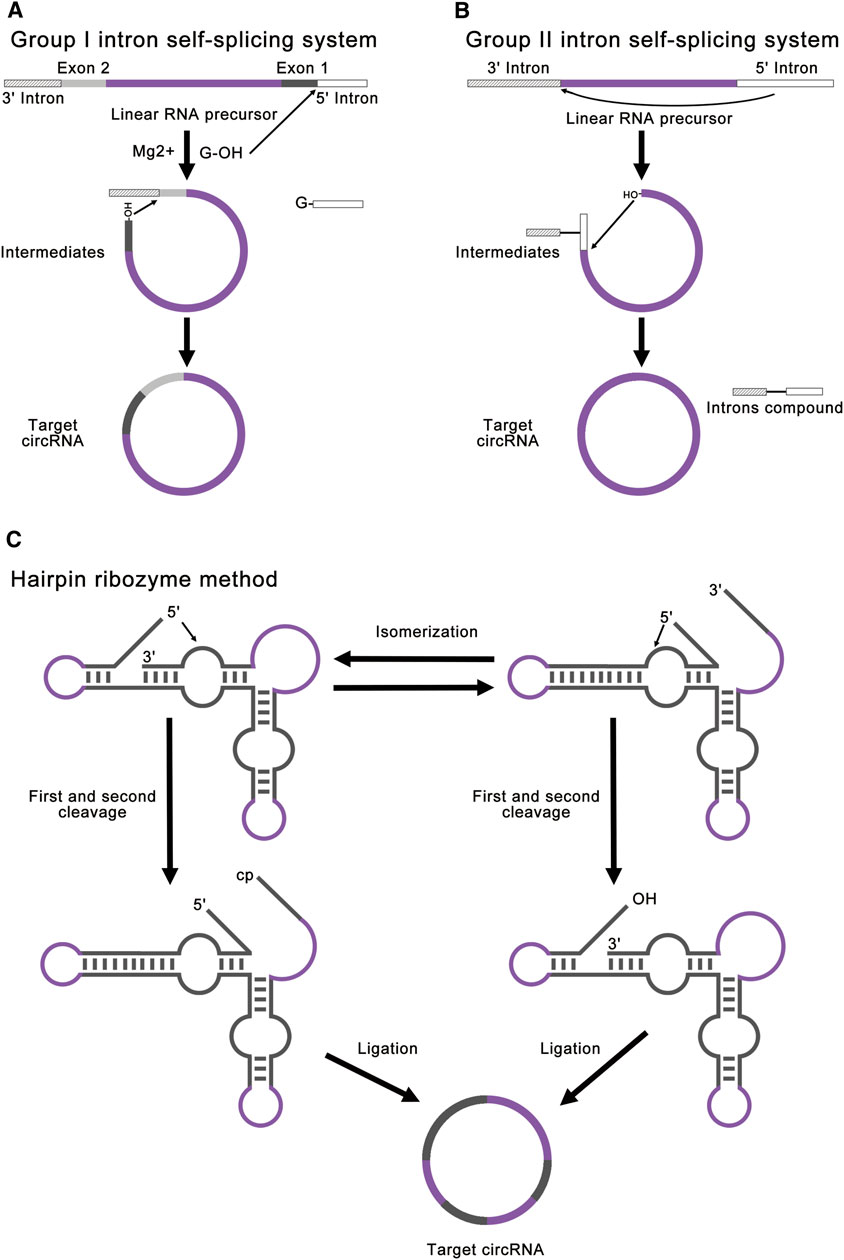
FIGURE 4. Strategies for ribozyme methods. (A) Group I intron self-splicing system requires only the addition of GTP and Mg2+ as cofactors and shows great potential for protein synthesis. This method realized RNA ligation through a normal group I intron self-splicing reaction, including two transesterifications at defined splice sites. The final circRNA will contain exogenous exon sequences. (B) Group II intron self-splicing system involves the joining of the 5′ splice site at the end of an exon to the 3′ splice site at the beginning of the same exon. All exon sequences are dispensable for group II intron catalyzed inverse splicing. This method can enable more accurate linear RNA precursor ligation. (C) Hairpin ribozyme method can produce circRNA through the rolling circle reaction and the self-splicing reaction. The linear RNA precursor with HPR will fold into two alternative cleavage-active conformations to remove the 3′-end and the 5′-end. As a result, the intermediate will contain a 5′-OH and a 2′, 3′-cyclic phosphate to produce target circRNA.
Group II introns can also be used for circRNA synthesis, which involves an inverse splicing reaction (Jarrell, 1993; Mikheeva et al., 1997). This splicing reaction involves the joining of the 5′ splice site at the end of an exon to the 3′ splice site at the beginning of the same exon (Figure 4B). Compared to the group I introns, all exon sequences are dispensable for group II intron-catalyzed inverse splicing. Therefore, this method can enable more accurate linear RNA precursor ligation. However, this method forms 2′, 5′-phosphodiester bonds at the ligation site instead of the natural 3′, 5′-phosphodiester bonds, and the mechanism remains unclear in vitro (Petkovic and Müller, 2015; Müller and Appel, 2017; Obi and Chen, 2021). At present, there are few studies concentrating on group II introns.
Hairpin ribozyme (HPR) can produce circRNAs through rolling circle reaction and the self-splicing reaction from circular single-strand DNA template (Diegelman and Kool, 1998; Kazakov et al., 2006; Dallas et al., 2008; Petkovic and Müller, 2013). The linear RNA precursor with HPR will fold into two alternative cleavage-active conformations to remove the 3′-end and the 5′-end. As a result, the intermediate will contain a 5′-OH and a 2′, 3′-cyclic phosphate to produce the target circRNA (Figure 4C). In this method, small circRNAs can be produced from long repeating RNAs transcribed by RNA polymerase through a rolling circle mechanism in vitro (Diegelman and Kool, 1998). This method is mainly used for efficient production of small circRNAs (Hieronymus and Müller, 2019). Freezing stimulation, ionic conditions, and additional cofactors are proved to affect the activity of HPR (Nesbitt et al., 1999; Kazakov et al., 2006; Strohbach et al., 2006; Dallas et al., 2008). The disadvantage of this method is that the circRNA is not stable due to the dynamic equilibrium of HPR-catalyzed cleavage and ligation (Müller and Appel, 2017). Besides, the circRNA contains HPR sequences, which may negatively affect the function of some circRNAs.
In a word, PIE method using group I introns is suitable for most circRNAs production at present, which is still limited by the secondary structure of linear RNA precursor. Group II introns method can enable more accurate linear RNA precursor ligation, but the mechanism remains unclear in vitro. HPR method can efficiently produce small circRNAs but suffers from unstable product and exogenous HPR sequences. Among these ribozyme methods, PIE method shows the greatest potential for circRNAs production. At present, PIE method has been widely used in basic research and industrial production (Table 1).
Although there are many ways to synthesize circRNAs in vitro, there are still many challenges and opportunities. CircRNA is still a hot area of research, and there are many difficulties to be solved (Table 2).
Although PIE method has realized accurate linear RNA precursor ligation (Rausch et al., 2021), the structure of the linear RNA precursor still has a marked effect on circRNA synthesis yield. To overcome this problem, embedding unnatural nucleotides into linear RNA precursor may help change its secondary structure and improve the efficiency of ligation. Besides, RNA-binding proteins (RBPs) may also help the cyclization of RNA.
In terms of enzyme design, although existing natural enzymes from the bacteriophage T4 are capable of RNA cyclization, there is still much room for improvement. It is necessary to improve the cyclization efficiency of the ligases by mutation or rational design, especially for large RNA molecules. The critical point of mutation and rational design is to improve the binding efficiency of ligase to substrate RNA and the selectivity of cyclic products.
Reducing side reactions during circRNAs synthesis is also a big challenge. At present, the main side reaction is the intermolecular end-joining reaction, which exists in most circRNA synthesis methods and cannot be completely avoided. Therefore, it is necessary to optimize the cyclization reaction conditions and control the linear RNA precursor concentration to reduce the occurrence of such side reactions. In addition, immobilizing the ligase with hydrogels or other materials may realize the compartmentalization of the reaction and the cyclization of a single RNA substrate.
With the growing application of circRNAs, the synthesis of circRNAs with chemically modified or unnatural nucleotides is also difficult to be solved. Since circRNA does not have a terminal structure, the modification of circRNA is more difficult than linear RNA. One solution is to use unnatural nucleotides for the synthesis of linear RNA precursors, which can embed unnatural nucleotides into the final circRNA to improve the stability of circRNA and diversify its functions. In addition, the incorporation of chemically modified groups or unnatural nucleotides during the ligation reaction is also a possible solution.
Although circRNAs have been proved to hold the potential to become a novel vaccine (Qu et al., 2021), the commercial potential of circRNAs has yet to be fully exploited. Because of the unique structure of circRNAs, circRNAs can be used in the synthesis of some specific proteins, such as protein materials with lots of repeated sequences. Besides, circRNA can also be used as an expression template in cell-free expression systems. At the same time, the function of circRNA as ncRNA is also important. CircRNAs can not only be used as biomarkers of some diseases but also be used for gene therapy (Holdt et al., 2018). Since the function and mechanism of circRNAs in cells have not been fully studied, applications for circRNAs are still being developed. Further efforts are still needed.
Faced with the growing demand for circRNAs, large quantities of circRNAs need to be produced. Generally, increasing the yield of circRNAs requires increasing the concentration of the ligation reaction components, such as linear RNA precursors. However, an increase in the concentration of linear RNA precursors will lead to an increase in side reactions. These side reactions will decrease the yield of target circRNAs and increase the difficulty of product purification. Therefore, further optimization of current reaction components and conditions may be required based on scaled-up production systems. This problem may also be solved by new-type reactors, such as microreactors. At the same time, the cost of current circRNAs production is still a concern for researchers. The main reason is the cost of raw materials, such as expensive nucleotides. One potential solution is to use cells to synthesize nucleotides or directly synthesize linear RNA precursors. However, how to efficiently purify these raw materials produced by cells still needs further research.
In a word, circRNA is still a hot area of current research and holds great potentials. With the continuous efforts in this field, circRNAs will play an essential role in basic research and medical applications, and become an important part of human health in the near future.
XC and YL contributed to conception and design of the study. XC wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by Beijing Natural Science Foundation (2192023), National Natural Science Foundation of China (21878173), and National Key R&D Program of China (2018YFA0901700).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelmohsen, K., Panda, A. C., Munk, R., Grammatikakis, I., Dudekula, D. B., De, S., et al. (2017). Identification of HuR Target Circular RNAs Uncovers Suppression of PABPN1 Translation by CircPABPN1. RNA Biol. 14, 361–369. doi:10.1080/15476286.2017.1279788
Abe, N., Kodama, A., and Abe, H. (2018). Preparation of Circular RNA In Vitro. Methods Mol. Biol. 1724, 181–192. doi:10.1007/978-1-4939-7562-4_15
Arnberg, A. C., Van Ommen, G.-J. B., Grivell, L. A., Van Bruggen, E. F. J., and Borst, P. (1980). Some Yeast Mitochondrial RNAs Are Circular. Cell 19, 313–319. doi:10.1016/0092-8674(80)90505-X
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). CircRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cel 56, 55–66. doi:10.1016/j.molcel.2014.08.019
Barrett, S. P., and Salzman, J. (2016). Circular RNAs: Analysis, Expression and Potential Functions. Dev 143, 1838–1847. doi:10.1242/dev.128074
Barrett, S. P., Wang, P. L., and Salzman, J. (2015). Circular RNA Biogenesis Can Proceed through an Exon-Containing Lariat Precursor. Elife 4, 1–18. doi:10.7554/eLife.07540
Beckert, B., and Masquida, B. (2011). Synthesis of RNA by In Vitro Transcription. Methods Mol. Biol. 703, 29–41. doi:10.1007/978-1-59745-248-9_3
Bullard, D. R., and Bowater, R. P. (2006). Direct Comparison of Nick-Joining Activity of the Nucleic Acid Ligases from Bacteriophage T4. Biochem. J. 398, 135–144. doi:10.1042/BJ20060313
Capel, B., Swain, A., Nicolis, S., Hacker, A., Walter, M., Koopman, P., et al. (1993). Circular Transcripts of the Testis-Determining Gene Sry in Adult Mouse Testis. Cell 73, 1019–1030. doi:10.1016/0092-8674(93)90279-Y
Chen, C.-y., and Sarnow, P. (1995). Initiation of Protein Synthesis by the Eukaryotic Translational Apparatus on Circular RNAs. Science 268, 415–417. doi:10.1126/science.7536344
Chen, Y. G., Kim, M. V., Chen, X., Batista, P. J., Aoyama, S., Wilusz, J. E., et al. (2017). Sensing Self and Foreign Circular RNAs by Intron Identity. Mol. Cel 67, 228–238. doi:10.1016/j.molcel.2017.05.022
Chen, H., Cheng, K., Liu, X., An, R., Komiyama, M., and Liang, X. (2020a). Preferential Production of RNA Rings by T4 RNA Ligase 2 without Any Splint through Rational Design of Precursor Strand. Nucleic Acids Res. 48, e54. doi:10.1093/nar/gkaa181
Chen, L., Kong, R., Wu, C., Wang, S., Liu, Z., Liu, S., et al. (2020b). Circ‐MALAT1 Functions as Both an mRNA Translation Brake and a microRNA Sponge to Promote Self‐Renewal of Hepatocellular Cancer Stem Cells. Adv. Sci. 7, 1900949. doi:10.1002/advs.201900949
Cocquerelle, C., Daubersies, P., Majérus, M. A., Kerckaert, J. P., and Bailleul, B. (1992). Splicing with Inverted Order of Exons Occurs Proximal to Large Introns. EMBO J. 11, 1095–1098. doi:10.1002/j.1460-2075.1992.tb05148.x
Cocquerelle, C., Mascrez, B., Hétuin, D., and Bailleul, B. (1993). Mis-splicing Yields Circular RNA Molecules. FASEB J. 7, 155–160. doi:10.1096/fasebj.7.1.7678559
Costello, A., Lao, N. T., Barron, N., and Clynes, M. (2020). Reinventing the Wheel: Synthetic Circular RNAs for Mammalian Cell Engineering. Trends Biotechnol. 38, 217–230. doi:10.1016/j.tibtech.2019.07.008
Dallas, A., Balatskaya, S. V., Kuo, T.-C., Ilves, H., Vlassov, A. V., Kaspar, R. L., et al. (2008). Hairpin Ribozyme-Antisense RNA Constructs Can Act as Molecular Lassos. Nucleic Acids Res. 36, 6752–6766. doi:10.1093/nar/gkn637
Daròs, J.-A. (2021). Production of Circular Recombinant RNA in Escherichia coli Using Viroid Scaffolds. Methods Mol. Biol. 99, 99–107. doi:10.1007/978-1-0716-1499-0_8
Diegelman, A., and Kool, E. T. (1998). Generation of Circular RNAs and Trans-cleaving Catalytic RNAs by Rolling Transcription of Circular DNA Oligonucleotides Encoding Hairpin Ribozymes. Nucleic Acids Res. 26, 3235–3241. doi:10.1093/nar/26.13.3235
Dolinnaya, N. G., Sokolova, N. I., Ashirbekova, D. T., and Shabarova, Z. A. (1991). The Use of BrCN for Assembling Modified DNA Duplexes and DNA-RNA Hybrids; Comparison with Water-Soluble Carbodiimide. Nucl. Acids Res. 19, 3067–3072. doi:10.1093/nar/19.11.3067
Du, W. W., Fang, L., Yang, W., Wu, N., Awan, F. M., Yang, Z., et al. (2017a). Induction of Tumor Apoptosis through a Circular RNA Enhancing Foxo3 Activity. Cell Death Differ. 24, 357–370. doi:10.1038/cdd.2016.133
Du, W. W., Yang, W., Chen, Y., Wu, Z.-K., Foster, F. S., Yang, Z., et al. (2017b). Foxo3 Circular RNA Promotes Cardiac Senescence by Modulating Multiple Factors Associated with Stress and Senescence Responses. Eur. Heart J. 38, ehw001–1412. doi:10.1093/eurheartj/ehw001
Du, W. W., Yang, W., Li, X., Fang, L., Wu, N., Li, F., et al. (2020). The Circular RNA circSKA3 Binds Integrin β1 to Induce Invadopodium Formation Enhancing Breast Cancer Invasion. Mol. Ther. 28, 1287–1298. doi:10.1016/j.ymthe.2020.03.002
England, T. E., and Uhlenbeck, O. C. (1978). Enzymic Oligoribonucleotide Synthesis with T4 RNA Ligase. Biochemistry 17, 2069–2076. doi:10.1021/bi00604a008
Enuka, Y., Lauriola, M., Feldman, M. E., Sas-Chen, A., Ulitsky, I., and Yarden, Y. (2016). Circular RNAs Are Long-Lived and Display Only Minimal Early Alterations in Response to a Growth Factor. Nucleic Acids Res. 44, 1370–1383. doi:10.1093/nar/gkv1367
Flores, R., Grubb, D., Elleuch, A., Nohales, M.-Á., Delgado, S., and Gago, S. (2011). Rolling-circle Replication of Viroids, Viroid-like Satellite RNAs and Hepatitis delta Virus: Variations on a Theme. RNA Biol. 8, 200–206. doi:10.4161/rna.8.2.14238
Ford, E., and Ares, M. (1994). Synthesis of Circular RNA in Bacteria and Yeast Using RNA Cyclase Ribozymes Derived from a Group I Intron of Phage T4. Proc. Natl. Acad. Sci. 91, 3117–3121. doi:10.1073/pnas.91.8.3117
Garikipati, V. N. S., Verma, S. K., Cheng, Z., Liang, D., Truongcao, M. M., Cimini, M., et al. (2019). Circular RNA CircFndc3b Modulates Cardiac Repair after Myocardial Infarction via FUS/VEGF-A axis. Nat. Commun. 10. doi:10.1038/s41467-019-11777-7
Guo, J. U., Agarwal, V., Guo, H., and Bartel, D. P. (2014). Expanded Identification and Characterization of Mammalian Circular RNAs. Genome Biol. 15, 409. doi:10.1186/s13059-014-0409-z
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013a). Natural RNA Circles Function as Efficient microRNA Sponges. Nature 495, 384–388. doi:10.1038/nature11993
Hansen, T. B., Kjems, J., and Damgaard, C. K. (2013b). Circular RNA and miR-7 in Cancer. Cancer Res. 73, 5609–5612. doi:10.1158/0008-5472.CAN-13-1568
He, A. T., Liu, J., Li, F., and Yang, B. B. (2021). Targeting Circular RNAs as a Therapeutic Approach: Current Strategies and Challenges. Sig. Transduct. Target. Ther. 6, 1–14. doi:10.1038/s41392-021-00569-5
Hieronymus, R., and Müller, S. (2019). Engineering of Hairpin Ribozyme Variants for RNA Recombination and Splicing. Ann. N.Y. Acad. Sci. 1447, 135–143. doi:10.1111/nyas.14052
Ho, C. K., and Shuman, S. (2002). Bacteriophage T4 RNA Ligase 2 (gp24.1) Exemplifies a Family of RNA Ligases Found in All Phylogenetic Domains. Proc. Natl. Acad. Sci. 99, 12709–12714. doi:10.1073/pnas.192184699
Holdt, L. M., Kohlmaier, A., and Teupser, D. (2018). Circular RNAs as Therapeutic Agents and Targets. Front. Physiol. 9, 1262. doi:10.3389/fphys.2018.01262
Huang, Q., Guo, H., Wang, S., Ma, Y., Chen, H., Li, H., et al. (2020). A Novel Circular RNA, circXPO1, Promotes Lung Adenocarcinoma Progression by Interacting with IGF2BP1. Cell Death Dis. 11, 1031. doi:10.1038/s41419-020-03237-8
Jarrell, K. A. (1993). Inverse Splicing of a Group II Intron. Proc. Natl. Acad. Sci. 90, 8624–8627. doi:10.1073/pnas.90.18.8624
Jeck, W. R., and Sharpless, N. E. (2014). Detecting and Characterizing Circular RNAs. Nat. Biotechnol. 32, 453–461. doi:10.1038/nbt.2890
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 19, 141–157. doi:10.1261/rna.035667.112
Kaufmann, G., Klein, T., and Littauer, U. Z. (1974). T4 RNA Ligase: Substrate Chain Length Requirements. FEBS Lett. 46, 271–275. doi:10.1016/0014-5793(74)80385-6
Kazakov, S. A., Balatskaya, S. V., and Johnston, B. H. (2006). Ligation of the Hairpin Ribozyme in Cis Induced by Freezing and Dehydration. RNA 12, 446–456. doi:10.1261/rna.2123506
Kos, A., Dijkema, R., Arnberg, A. C., Van Der Meide, P. H., and Schellekens, H. (1986). The Hepatitis delta (δ) Virus Possesses a Circular RNA. Nature 323, 558–560. doi:10.1038/323558a0
Kramer, M. C., Liang, D., Tatomer, D. C., Gold, B., March, Z. M., Cherry, S., et al. (2015). Combinatorial Control ofDrosophilacircular RNA Expression by Intronic Repeats, hnRNPs, and SR Proteins. Genes Dev. 29, 2168–2182. doi:10.1101/gad.270421.115
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cel 66, 22–37. doi:10.1016/j.molcel.2017.02.017
Li, F., Zhang, L., Li, W., Deng, J., Zheng, J., An, M., et al. (2015). Circular RNA ITCH Has Inhibitory Effect on ESCC by Suppressing the Wnt/β-Catenin Pathway. Oncotarget 6, 6001–6013. doi:10.18632/oncotarget.3469
Li, J., Mohammed‐Elsabagh, M., Paczkowski, F., and Li, Y. (2020). Circular Nucleic Acids: Discovery, Functions and Applications. ChemBioChem 21, 1547–1566. doi:10.1002/cbic.202000003
Liang, D., and Wilusz, J. E. (2014). Short Intronic Repeat Sequences Facilitate Circular RNA Production. Genes Dev. 28, 2233–2247. doi:10.1101/gad.251926.114
Litke, J. L., and Jaffrey, S. R. (2019). Highly Efficient Expression of Circular RNA Aptamers in Cells Using Autocatalytic Transcripts. Nat. Biotechnol. 37, 667–675. doi:10.1038/s41587-019-0090-6
McLaughlin, L. W., Romaniuk, E., Romaniuk, P. J., and Neilson, T. (1982). The Effect of Acceptor Oligoribonucleotide Sequence on the T4 RNA Ligase Reaction. Eur. J. Biochem. 125, 639–643. doi:10.1111/j.1432-1033.1982.tb06730.x
Meganck, R. M., Liu, J., Hale, A. E., Simon, K. E., Fanous, M. M., Vincent, H. A., et al. (2021). Engineering Highly Efficient Backsplicing and Translation of Synthetic circRNAs. Mol. Ther. Nucleic Acids 23, 821–834. doi:10.1016/j.omtn.2021.01.003
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 495, 333–338. doi:10.1038/nature11928
Mikheeva, S., Hakim-Zargar, M., Carlson, D., and Jarrell, K. (1997). Use of an Engineered Ribozyme to Produce a Circular Human Exon. Nucleic Acids Res. 25, 5085–5094. doi:10.1093/nar/25.24.5085
Moore, M. J. (1999). Joining RNA Molecules with T4 DNA Ligase. Methods Mol. Biol. 118, 11–19. doi:10.1385/1-59259-676-2:11
Müller, S., and Appel, B. (2017). In Vitro circularization of RNA. RNA Biol. 14, 1018–1027. doi:10.1080/15476286.2016.1239009
Nandakumar, J., and Shuman, S. (2004). How an RNA Ligase Discriminates RNA versus DNA Damage. Mol. Cel 16, 211–221. doi:10.1016/j.molcel.2004.09.022
Nandakumar, J., Ho, C. K., Lima, C. D., and Shuman, S. (2004). RNA Substrate Specificity and Structure-Guided Mutational Analysis of Bacteriophage T4 RNA Ligase 2. J. Biol. Chem. 279, 31337–31347. doi:10.1074/jbc.M402394200
Nesbitt, S. M., Erlacher, H. A., and Fedor, M. J. (1999). The Internal Equilibrium of the Hairpin Ribozyme: Temperature, Ion and pH Effects. J. Mol. Biol. 286, 1009–1024. doi:10.1006/jmbi.1999.2543
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled Exons. Cell 64, 607–613. doi:10.1016/0092-8674(91)90244-S
Obi, P., and Chen, Y. G. (2021). The Design and Synthesis of Circular RNAs. Methods S1046-2023, 00065–00067. doi:10.1016/j.ymeth.2021.02.020
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol. Cel 66, 9–21. doi:10.1016/j.molcel.2017.02.021
Perriman, R., and Ares, M. (1998). Circular mRNA Can Direct Translation of Extremely Long Repeating-Sequence Proteins In Vivo. RNA 4, 1047–1054. doi:10.1017/S135583829898061X
Petkovic, S., and Müller, S. (2013). RNA Self-Processing: Formation of Cyclic Species and Concatemers from a Small Engineered RNA. FEBS Lett. 587, 2435–2440. doi:10.1016/j.febslet.2013.06.013
Petkovic, S., and Müller, S. (2015). RNA Circularization Strategies In Vivo and In Vitro. Nucleic Acids Res. 43, 2454–2465. doi:10.1093/nar/gkv045
Petkovic, S., and Müller, S. (2018). Synthesis and Engineering of Circular RNAs. Methods Mol. Biol. 1724, 167–180. doi:10.1007/978-1-4939-7562-4_14
Puttaraju, M., and Been, M. (1992). Group I Permuted Intron-Exon (PIE) Sequences Self-Splice to Produce Circular Exons. Nucl. Acids Res. 20, 5357–5364. doi:10.1093/nar/20.20.5357
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: A New star of Noncoding RNAs. Cancer Lett. 365, 141–148. doi:10.1016/j.canlet.2015.06.003
Qu, L., Yi, Z., Shen, Y., Xu, Y., Wu, Z., Tang, H., et al. (2021). Circular RNA Vaccines against SARS-CoV-2 and Emerging Variants. bioRxiv. doi:10.1101/2021.03.16.435594
Rausch, J. W., Heinz, W. F., Payea, M. J., Sherpa, C., Gorospe, M., and Le Grice, S. F. J. (2021). Characterizing and Circumventing Sequence Restrictions for Synthesis of Circular RNA In Vitro. Nucleic Acids Res. 49, E35. doi:10.1093/nar/gkaa1256
Rio, D. C. (2013). Expression and Purification of Active Recombinant T7 RNA Polymerase from E. coli. Cold Spring Harb. Protoc. 2013, pdb.prot078527. doi:10.1101/pdb.prot078527
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS One 7, e30733. doi:10.1371/journal.pone.0030733
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. 73, 3852–3856. doi:10.1073/pnas.73.11.3852
Santer, L., Bär, C., and Thum, T. (2019). Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Mol. Ther. 27, 1350–1363. doi:10.1016/j.ymthe.2019.07.001
Schindewolf, C., Braun, S., and Domdey, H. (1996). In Vitro generation of a Circular Exon from a Linear Pre-mRNA Transcript. Nucleic Acids Res. 24, 1260–1266. doi:10.1093/nar/24.7.1260
Sokolova, N. I., Ashirbekova, D. T., Dolinnaya, N. G., and Shabarova, Z. A. (1988). Chemical Reactions within DNA Duplexes Cyanogen Bromide as an Effective Oligodeoxyribonucleotide Coupling Agent. FEBS Lett. 232, 153–155. doi:10.1016/0014-5793(88)80406-X
Starke, S., Jost, I., Rossbach, O., Schneider, T., Schreiner, S., Hung, L.-H., et al. (2015). Exon Circularization Requires Canonical Splice Signals. Cel Rep. 10, 103–111. doi:10.1016/j.celrep.2014.12.002
Strohbach, D., Novak, N., and Müller, S. (2006). Redox-active Riboswitching: Allosteric Regulation of Ribozyme Activity by Ligand-Shape Control. Angew. Chem. Int. Ed. 45, 2127–2129. doi:10.1002/anie.200503820
Surono, A., Takeshima, Y., Wibawa, T., Ikezawa, M., Nonaka, I., and Matsuo, M. (1999). Circular Dystrophin RNAs Consisting of Exons that Were Skipped by Alternative Splicing. Hum. Mol. Genet. 8, 493–500. doi:10.1093/hmg/8.3.493
Szabo, L., and Salzman, J. (2016). Detecting Circular RNAs: Bioinformatic and Experimental Challenges. Nat. Rev. Genet. 17, 679–692. doi:10.1038/nrg.2016.114
Szabo, L., Morey, R., Palpant, N. J., Wang, P. L., Afari, N., Jiang, C., et al. (2015). Statistically Based Splicing Detection Reveals Neural Enrichment and Tissue-specific Induction of Circular RNA during Human Fetal Development. Genome Biol. 16, 126. doi:10.1186/s13059-015-0690-5
Tao, M., Zheng, M., Xu, Y., Ma, S., Zhang, W., and Ju, S. (2021). CircRNAs and Their Regulatory Roles in Cancers. Mol. Med. 2794, 94. doi:10.1186/s10020-021-00359-3
Umekage, S., and Kikuchi, Y. (2009). In Vitro and In Vivo Production and Purification of Circular RNA Aptamer. J. Biotechnol. 139, 265–272. doi:10.1016/j.jbiotec.2008.12.012
Wang, L., and Ruffner, D. E. (1998). Oligoribonucleotide Circularization by 'template-Mediated' Ligation with T4 RNA Ligase: Synthesis of Circular Hammerhead Ribozymes. Nucleic Acids Res. 26, 2502–2504. doi:10.1093/nar/26.10.2502
Wang, Y., and Wang, Z. (2015). Efficient Backsplicing Produces Translatable Circular mRNAs. RNA 21, 172–179. doi:10.1261/rna.048272.114
Wang, P. L., Bao, Y., Yee, M.-C., Barrett, S. P., Hogan, G. J., Olsen, M. N., et al. (2014). Circular RNA Is Expressed across the Eukaryotic Tree of Life. PLoS One 9, e90859. doi:10.1371/journal.pone.0090859
Wesselhoeft, R. A., Kowalski, P. S., and Anderson, D. G. (2018). Engineering Circular RNA for Potent and Stable Translation in Eukaryotic Cells. Nat. Commun. 9, 2629. doi:10.1038/s41467-018-05096-6
Westholm, J. O., Miura, P., Olson, S., Shenker, S., Joseph, B., Sanfilippo, P., et al. (2014). Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-dependent Neural Accumulation. Cel Rep. 9, 1966–1980. doi:10.1016/j.celrep.2014.10.062
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res 27, 626–641. doi:10.1038/cr.2017.31
Yin, S., Kiong Ho, C., Miller, E. S., and Shuman, S. (2004). Characterization of Bacteriophage KVP40 and T4 RNA Ligase 2. Virology 319, 141–151. doi:10.1016/j.virol.2003.10.037
Zaphiropoulos, P. G. (1996). Circular RNAs from Transcripts of the Rat Cytochrome P450 2C24 Gene: Correlation with Exon Skipping. Proc. Natl. Acad. Sci. 93, 6536–6541. doi:10.1073/pnas.93.13.6536
Zaphiropoulos, P. G. (1997). Exon Skipping and Circular RNA Formation in Transcripts of the Human Cytochrome P-450 2C18 Gene in Epidermis and of the Rat Androgen Binding Protein Gene in Testis. Mol. Cel. Biol. 17, 2985–2993. doi:10.1128/mcb.17.6.2985
Zhang, X.-O., Wang, H.-B., Zhang, Y., Lu, X., Chen, L.-L., and Yang, L. (2014). Complementary Sequence-Mediated Exon Circularization. Cell 159, 134–147. doi:10.1016/j.cell.2014.09.001
Keywords: circular RNA, RNA synthesis, in vitro transcription, enzymatic ligation, ligase, permuted intron-exon
Citation: Chen X and Lu Y (2021) Circular RNA: Biosynthesis in vitro. Front. Bioeng. Biotechnol. 9:787881. doi: 10.3389/fbioe.2021.787881
Received: 01 October 2021; Accepted: 16 November 2021;
Published: 30 November 2021.
Edited by:
Dawei Zhang, Tianjin Institute of Industrial Biotechnology (CAS), ChinaReviewed by:
Marcos De La Peña, Polytechnic University of Valencia, SpainCopyright © 2021 Chen and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Lu, eXVhbmx1QHRzaW5naHVhLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.