- 1School of Life Sciences, Anhui University, Hefei, China
- 2Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China
- 3Department of Chemistry, School of Science, Tianjin University, Tianjin, China
- 4Department of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 5National Technology Innovation Center of Synthetic Biology, Tianjin, China
Chiral amino alcohols are prevalent synthons in pharmaceuticals and synthetic bioactive compounds. The efficient synthesis of chiral amino alcohols using ammonia as the sole amino donor under mild conditions is highly desired and challenging in organic chemistry and biotechnology. Our previous work explored a panel of engineered amine dehydrogenases (AmDHs) derived from amino acid dehydrogenase (AADH), enabling the one-step synthesis of chiral amino alcohols via the asymmetric reductive amination of α-hydroxy ketones. Although the AmDH-directed asymmetric reduction is in a high stereoselective manner, the activity is yet fully excavated. Herein, an engineered AmDH derived from a leucine dehydrogenase from Sporosarcina psychrophila (SpAmDH) was recruited as the starting enzyme, and the combinatorial active-site saturation test/iterative saturation mutagenesis (CAST/ISM) strategy was applied to improve the activity. After three rounds of mutagenesis in an iterative fashion, the best variant wh84 was obtained and proved to be effective in the asymmetric reductive amination of 1-hydroxy-2-butanone with 4-fold improvements in kcat/Km and total turnover number (TTN) values compared to those of the starting enzyme, while maintaining high enantioselectivity (ee >99%) and thermostability (T5015 >53°C). In preparative-scale reaction, the conversion of 100 and 200 mM 1-hydroxy-2-butanone catalyzed by wh84 was up to 91–99%. Insights into the source of an enhanced activity were gained by the computational analysis. Our work expands the catalytic repertoire and toolbox of AmDHs.
Introduction
As essential structural moieties, chiral α-amino alcohols are widely applied to produce synthetic and natural bioactive molecules (Ager et al., 1996; Erlanson et al., 2011). For instance, many pharmaceuticals consist of α-amino alcohols that serve as chiral building blocks (Supplementary Scheme S1). Direct asymmetric reductive amination of ketones with free ammonia to produce chiral amino alcohols is a highly aspirational transformation (Abrahamson et al., 2013; Pushpanath et al., 2017). In the traditional chemical synthesis, it mainly relies on stoichiometric amounts of chemical reducing agents or organometallic catalysts, which are limited by low stereoselectivities, the formation of the alcohol as side product, and the requirement of extreme reaction conditions (Larrow et al., 1996; Breuer et al., 2004; Ma et al., 2010; Nugent and El-Shazly, 2010; Karjalainen and Koskinen, 2012; Xie et al., 2020; Hollmann et al., 2021). Alternatively, enzymes as catalysts are increasingly explored as essential tools in asymmetric reductive aminations (Reetz, 2011; Abrahamson et al., 2013; Schrittwieser et al., 2015; Chen and Arnold, 2020; Winkler et al., 2021). In recent years, a number of enzymes have been identified that are capable of catalyzing the asymmetric reductive amination of ketones (Wu et al., 2021), including lipases (Francalanci et al., 1987), acylases (Wang et al., 2016), transaminases (Wu et al., 2017), and imine reductases (Matzel et al., 2017). Moreover, native amine dehydrogenases (AmDHs) have been identified that they can directly utilize ammonia as a sole amino source in reductive amination but with insufficient enantioselectivity (Itoh et al., 2000; Mayol et al., 2019).
Apart from the native AmDHs, the Bommarius group at Georgia Tech has engineered two natural amino acid dehydrogenases (AADHs), including a leucine dehydrogenase from Bacillus stearothermophilus (Abrahamson et al., 2012) and a phenylalanine dehydrogenase from Bacillus badius (Abrahamson et al., 2013). After altering two determinant residues of carboxylate recognition, the natural AADHs were transformed to AmDHs, thereby eliminating the activity toward ketone acids while affording new activity toward ketones. Taking this advantage, more natural AADHs from diverse organisms have been explored and engineered to AmDHs based on the introduction of two-point mutations, which were then harnessed in the asymmetric production of chiral amines (Au et al., 2014; Chen et al., 2015; Ye et al., 2015; Franklin et al., 2020; Liu et al., 2020). In addition, the engineered AmDHs derived from AADHs were also utilized in the preparation of chiral amino alcohols, which are usually with very high enantioselectivity (>99% ee), making these enzymes of potential value in biocatalysis (Chen et al., 2019). Our previous work has characterized and engineered five novel AmDHs from natural AADHs by genome mining, these newly identified AmDHs provided reductive amination of a broad range of prochiral α- and β-hydroxy ketones in a high stereoselective manner (Wang et al., 2020). As an example, the engineered AmDH derived from the leucine dehydrogenase from Sporosarcina psychrophila (SpAmDH), enabled the reduction of 1-hydroxybutan-2-one (1a) to (S)-2-aminobutan-1-ol ((S)-1b, Scheme 1) with >99% selectivity, while the conversion is modest (∼60%) at a substrate concentration of 50 mM (Wang et al., 2020).
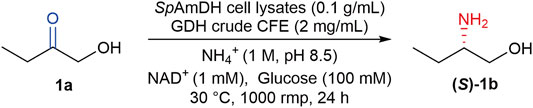
SCHEME 1. SpAmDH-catalyzed asymmetric reductive amination of 1a to (S)-1b by using ammonia as the sole amino donor, and glucose dehydrogenase (GDH) cell-free extract (CFE) for NADH cofactor regeneration.
In this work, we sought to optimize the activity of SpAmDH in the biosynthesis of (S)-1b, which is an important intermediate desired in the preparation of antitubercular drugs ethambutol (Pablos-Méndez et al., 1998; Supplementary Scheme S1). When aiming at the improvement of activity and/or selectivity, the combinatorial active-site saturation test (CAST) combined with iterative saturation mutagenesis (ISM) has been emerged as a powerful means in protein engineering (Reetz et al., 2005; Yanai et al., 2013; Qu et al., 2020; Li D. et al., 2021; Li J. et al., 2021; Qu et al., 2021; Zheng et al., 2021). Taking the advantage of CAST/ISM, three robust variants of SpAmDH with improved activity and high stereoselectivity were obtained, and their potential as biocatalysts in the preparative scale reactions was explored. Computational docking simulations were also performed to rationalize the elevated activity of SpAmDH variants.
Materials and Methods
Chemicals and Reagents
Hydroxy ketones, chiral amino alcohols, and other chemical reagents were purchased from Bidepharm (Shanghai, China), Energy Chemical (Shanghai, China), Kaiwei Chemical (Shanghai, China), Acmec (Shanghai, China), Arkpharm (Chicago, United States), Accela (Shanghai, China), Aladdin (Shanghai, China), Macklin (Shanghai, China), Heowns (Tianjin, China), and CINC (Shanghai, China). PrimeSTAR DNA polymerase and restriction enzyme Dpn I were ordered from TAKARA and NEB, respectively. The primer synthesis and sequencing were carried out by GENEWIZ. A plasmid preparation kit was purchased from TIANGEN Biotech. All other chemical reagents can be obtained through commercialization, unless otherwise noted.
Site-Directed Mutagenesis
Mutagenesis was constructed by using the overlap PCR and megaprimer approach (Tyagi et al., 2004) with high-fidelity master mix polymerase. Reaction mixtures (50 μl) typically contained ddH2O (22 μl), 2× high-fidelity master mix polymerase (25 μl), template DNA (1 μl, 50 ng), forward primer (1 μl, 0.2 μM), and reverse primer (1 μl, 0.2 μM). The PCR conditions for short fragment were as follows: 98°C, 2 min (98°C, 10 s; 55°C, 15 s; 72°C, 30 s) 30 cycles; 72°C, 3 min. For mega-PCR (Tyagi et al., 2004), 1st PCR product (2 μl, 800 ng) was used as primer, and the PCR conditions as below: 98°C, 2 min (98°C, 10 s; 60°C, 15 s; 72°C, 3.5 min) 30 cycles, 72°C, 5 min. The PCR products were treated with restriction endonuclease Dpn I for 3 h and then electroporated into E. coli BL21 (DE3). After culturing for 12 h, the colonies on the plate were washed with ddH2O, and the plasmids were extracted and sequenced. The primers used for constructing single-site saturation mutagenesis and combinatory saturation libraries were listed in Supplementary Tables S1, S2, respectively.
Screening of Saturation Mutagenesis Libraries
Clones from the plate were transferred to 96-well deep-well culture plates containing 300 μl of LB medium (50 μg/ml kanamycin), shaking at 37°C and 800 rpm for 10 h. Then 120 μl of the aforementioned culture broth was transferred to a 96-well glycerol plate, 60 μl of glycerol (60%, v/v) was added, and stored at −80°C. At the same time, 800 μl of the TB medium was added to the 96-well deep-well culture plates, IPTG (0.2 mM) and kanamycin (50 μg/ml) were added, and the culture was shaken at 30°C, 800 rpm for 12 h for protein expression. The cells were collected by centrifugation at 4,000 rpm, 4°C for 10 min. The cells were washed and resuspended with 400 μl of potassium phosphate buffer (PBK, 50 mM, pH 7.4). DNase I (6 U/ml) and lysozyme (1 mg/ml) were added to the culture and were shaken at 30°C for 1 h to lysis cells. After low-temperature centrifugation (4°C, 4,000 rpm, for 30 min), the supernatant was used for enzyme activity determination. The reductive amination reaction was performed in NH4Cl/NH3·H2O buffer (1 M, pH 8.5) containing 200 μl supernatant, 10 mM substrate 1a, 1 mM NAD+, 100 mM glucose, and 2 mg/ml GDH at 30°C, 800 rpm for 24 h. After that, 1 μl of 1M para-methoxy-2-amino benzamidoxime (PMA) was added to 99 μl of the reaction solution, and fluorescence was measured using an excitation wavelength of 380 nm and emission wavelength of 520 nm for substrate consumption (Mei et al., 2020).
Active Assay
For single point saturation mutation library rescreening, the best clones from 96-well glycerol plates were cultivated, expressed in a shake flask and used for biotransformation. Reductive amination reactions were performed in a reaction mixture (0.5 ml) containing 1 M NH4Cl/NH3·H2O buffer (pH 8.5), 1 mM NAD+, 100 mM glucose, 2 mg/ml GDH cell-free extract (CFE), 6 U/ml DNase I, 20 mM 1a, and 20 mg/ml mutant CFE in 2 ml Eppendorf tubes at 30°C, 1,000 rpm for 24 h in a thermostatic metal bath. For the combinatorial saturation mutation library screening, using the same procedure as before, except for that, the concentration of 1a and the wet cell of mutant were 40 mM and 0.1 g/ml, respectively. After the reaction was over, the aforementioned reaction solution was boiled for 5 min and then centrifuged at 12,000 rpm for 10 min to remove the precipitate. The supernatant was measured by HPLC to detect the conversion of 1a. The conversions of hydroxy ketones substrates (1a–8a) to chiral amino alcohol products (1b-8b) were measured with Marfey’s reagent (1-fluoro-2, 4-dinitrophenyl-5-L-alanine amide) for pre-column derivatization. The reaction mixture was mixed with 100 μl sample, 30 μl of Marfey’s reagent (14 mM), 80 μl NaHCO3 (1 M), and 200 μl DMSO at 80°C, 1,000 rpm for 10 min. Finally, 10 μl HCl (4 M) was added to stop the reaction. Detection conditions were given as follows: Zorbax SB-C18 column (4.6 × 150 mm, 5 μm), detection wavelength: 340 nm, temperature: 25°C, flow rate: 1 ml/min, loading volume: 10 μl, mobile phase buffer A: ddH2O (0.1% trifluoroacetic acid), buffer B: methanol (0.1% trifluoroacetic acid), gradient program: 40% B, hold for 6 min, increase B to 60% in 9 min, hold for 3 min, decrease B to 40% in 2 min, and hold for 5 min. More details are listed in Supplementary Table S3.
Protein Expression and Purification
E. coli BL21(DE3) glycerol bacteria containing SpAmDH gene were cultivated in 5 ml LB liquid medium (50 μg/ml kanamycin) for 10 h. The previous culture was transferred to the TB medium (100 ml) (50 μg/ml kanamycin) and was cultured at 37°C, 220 rpm. The culture was induced by the addition of IPTG (0.1 mM) when OD600 reached 0.8 and was then allowed to grow for an additional 12 h at 20°C. The cells expressing AmDHs were harvested, sonicated, and centrifuged (4°C, 12,000 rpm) for 60 min to remove the precipitate. The supernatant with soluble His-tagged protein was filtered using a 0.45-µm filter membrane and was verified by SDS-PAGE analysis (Supplementary Figure S1). The column (HisTrap FF, 5 ml) was washed with A buffer (50 mM PBK containing 300 mM NaCl and 20 mM imidazole, pH 8.0) before and after the supernatant was loaded. The proteins were eluted with B buffer containing a high concentration of salt (50 mM PBK containing 300 mM NaCl and 500 mM imidazole, pH 8.0). The eluates were ultrafiltered (4°C, 3,500 rpm) with an ultrafiltration tube (10,000 Da) to concentrate and replace the buffer (25 mM PBK, 100 mM NaCl, 5% glycerol, pH 8.0). The protein concentration was confirmed by measuring the absorbance at 280 nm using a Nano-300 micro-spectrophotometer.
Total Turnover Numbers Assay for Conversion of Substrate 1a to 1b Using SpAmDH Variants
The asymmetric reductive amination reactions were performed with 0.32–0.64 mg/ml purified enzyme, 1 M NH4Cl/NH3·H2O buffer (pH 8.5), 1 mM NAD+, 100 mM glucose, 2 mg/ml GDH CFE, and 40 mM 1a. The reaction mixture was proceeded at 30°C, 1,000 rpm for 24 h. The product was then detected by HPLC. TTN was defined as the molar number of the product yield divided by the catalyst concentration (Qu et al., 2019).
Determination of Kinetic Parameters and Thermostability
The kinetic parameters were obtained by measuring the initial velocities of NADH consumption (the initial rate of change in absorbance at 340 nm) in the enzymatic reaction and fitting the curve according to the Michaelis–Menten equation (Supplementary Figure S2). The activity assay was performed in a mixture containing 0.2 mM NADH, 1–30 mM 1-hydroxy-2-butanone, 1 M NH4Cl/NH3·H2O (pH 8.5), and the purified enzyme (1 mg/ml). The reaction was initiated by the addition of the enzyme and was monitored for 5 min at 30°C. The activity was determined by measuring NADH oxidation from a decrease in the absorbance at 340 nm (ε = 6,220 M−1 cm−1) and then Origin was used to perform the nonlinear fitting of the Michaelis–Menten equation. One unit (1 U) of activity is defined as the amount of enzyme required to consume 1 μM NADH in 1 minute. For thermostability, 1 mg/ml pure enzyme solution was incubated at different temperatures (30–70°C) for 15 min, followed by measuring the residual activity in 1 M NH4Cl/NH3·H2O (pH 8.5) containing 0.2 mM NADH, 10 mM substrate 1a at 50°C for 2 min. All experiments were conducted in triplicate.
Preparative-Scale Reactions Using SpAmDH Mutants
The SpAmDH mutants wh18 and wh84 were inoculated in the 5 ml LB medium (50 μg/ml kanamycin) for 10 h (37°C, 220 rpm). The aforementioned culture was inoculated into the 1,000 ml TB medium (50 μg/ml kanamycin) and cultured at 37°C, 220 rpm until the OD600 reached 0.8–0.9. IPTG was then added to a final concentration of 0.1 mM, and the culture was allowed to grow for additional 16 h at 20°C, 220 rpm. The cells were pelleted by centrifugation for 20 min (4°C, 4,000 rpm) and washed once with phosphate buffer (50 mM, pH 7.4). Subsequently, 0.1 g/ml wet cells, 1 M NH4Cl/NH3·H2O buffer (pH 8.5), 1 mM NAD+, 100 mM glucose, 2 mg/ml GDH CFE, 6 U/ml DNase I, 1 mg/ml lysozyme, and 1a (100, 200 mM) were mixed for 10 ml in a Erlenmeyer flask. The reaction was performed at 30°C, 220 rpm for 24 h in a shaker with triple replica. Then 200 μl samples were taken at 0, 1, 2, 3, 4, 8, 12, 18, and 24 h and then prepared and analyzed by HPLC. The 100 mM scaled-up reaction of 1a catalyzed by wh84 was terminated by adding 5% H2SO4 to pH <2 and then centrifuged at an rpm of 4,000 for 20 min at 4°C, to collect the supernatant. The product ((S)-2a) was purified via an ion exchange method with Dowex® 50WX8 ion exchange resin (Chen et al., 2015). The column was prepared by washing with 100 ml ddH2O and 50 ml 5% w/v H2SO4. Then, the acidified reaction supernatant was loaded into the column at a low flow rate, washed with ddH2O until pH ∼7.0, eluted with 9% w/v NH4OH (100 ml), and dried via rotary evaporation to harvest the final product (S)-2a. 1H NMR (400 MHz, D2O) δ 3.61 (dd, J = 11.8, 4.1 Hz, 1H), 3.51–3.33 (m 1H), 3.06–2.82 (m, 1H), 1.57–1.33 (m, 2H), and 0.98–0.81 (m, 3H).
Model Generation and Substrate Docking
The structure of the mutant wh84 (K68S/N261L/I111F/V294C/E114V) was generated by PyMol program (http://www.pymol.org) using the crystallographic structure of SpLeuDH (PDB ID: 3VPX, Zhao et al., 2012) as a template. The initial structure of wh84 was relaxed/repacked before docking using Rosetta relax program (Conway et al., 2014). The generated pose with the lowest Rosetta_total_score was selected as input file for docking. The cofactor NADH, substrate 1a, and NH4+ were prepared in Schrödinger Maestro software (Schrödinger, 2015). Thereafter, NADH and 1a were stepwise docked into the active site of wh84 by using Rosetta docking program (Combs et al., 2013). Thereafter, 1a was docked to the protein–NADH complex using the same procedure as NADH, only by replacing the input files. After that, NH4+ was docked to the protein–NADH-1a complex by using AutoDock Vina (Trott and Olson, 2010). For docking NH4+, a total of 16 poses were generated by Vina, and only the pose with proper interactions with D115 and 1a was kept.
Results and Discussion
Knowledge-Based and Structural Guided Engineering of SpLeuDH
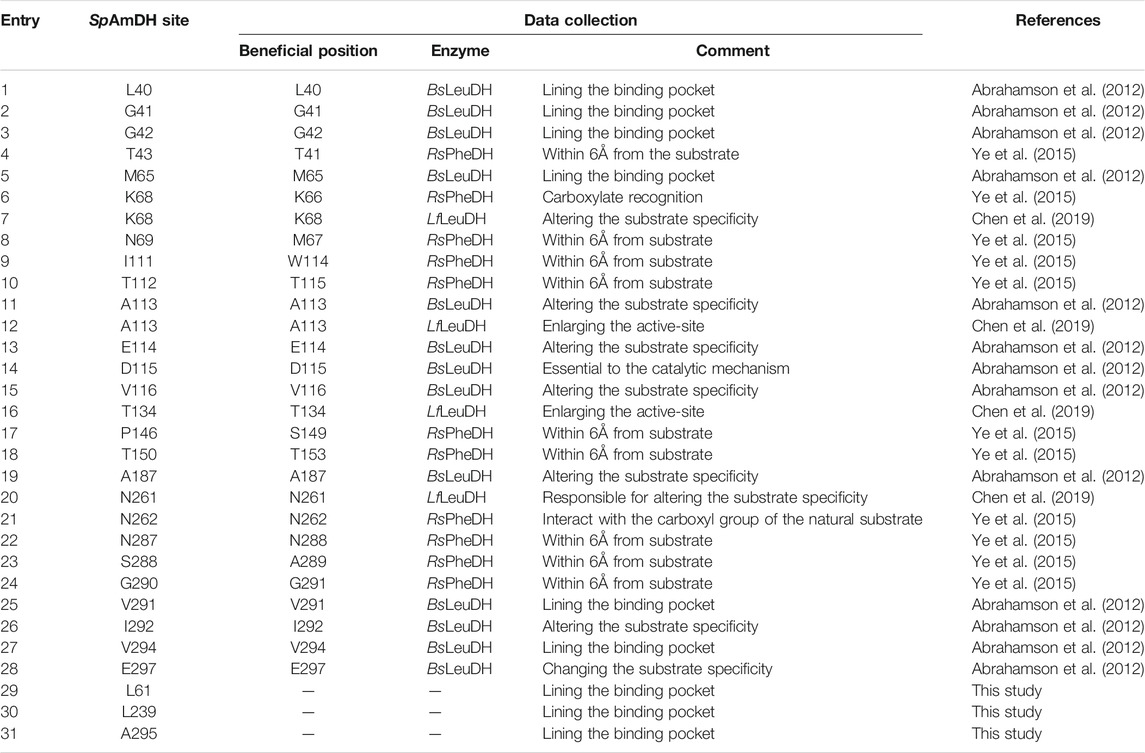
Wild-type (WT) leucine dehydrogenase from Sporosarcina psychrophila (SpLeuDH, Accession No.: WP_067209859) has been successfully engineered to an amine dehydrogenase (SpAmDH) by implanting the double mutations K68S/N261L in our recent work (Wang et al., 2020). Based on the crystallographic structure of SpLeuDH (PDB ID: 3VPX, Zhao et al., 2012), this SpAmDH variant (K68S/N261L, dubbed wh18) showed excellent stereoselectivity (>99% ee) in the transformation of 1a to 1b, while the catalytic efficiency was modest when it was applied in large-scale production (Wang et al., 2020). Thus, the CAST/ISM strategy was employed with the aim of enhancing the activity of SpAmDH. To identify the hot spot positions that may manipulate the activity, the beneficial mutations reported in other engineering studies toward AmDHs were collected (Table 1). We anticipated that targeting these residues at the equivalent positions in SpAmDH may give rise to more proficient variants. Moreover, three additional positions L61, L239, and A295 were also selected as they are situated at the substrate binding pocket (Table 1; Figure 1).
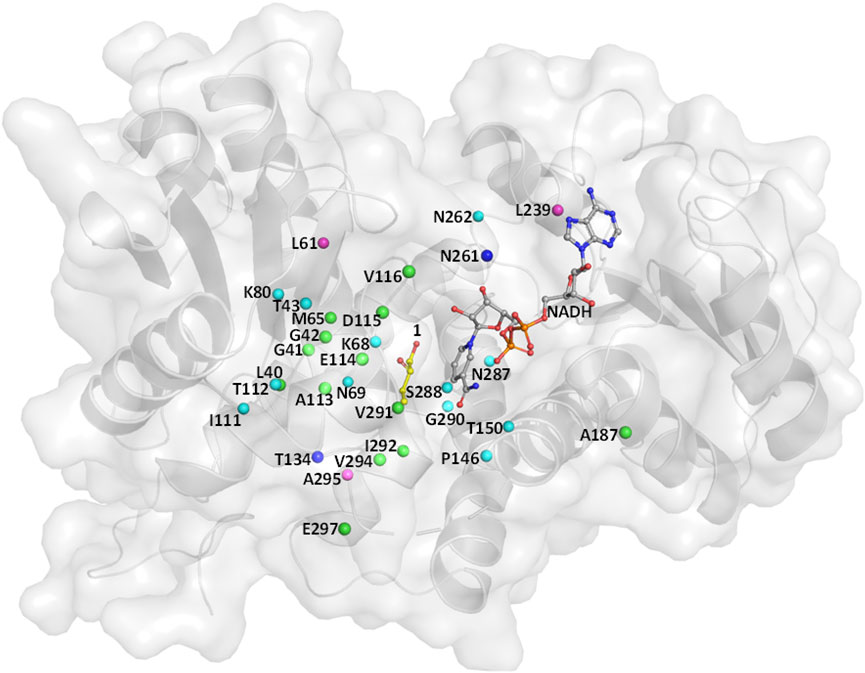
FIGURE 1. Residues lining the substrate-binding pocket of SpLeuDH (PDB ID: 3VPX). The sites marked in green, cyan, and blue are obtained from the three studies Abrahamson et al. (2012); Ye et al. (2015); Chen et al. (2019), respectively. The sites marked in magenta are found in this study.
The 27 positions were then performed site-directed saturation mutagenesis using wh18 as the template, each site was substituted by the other 19 amino acids with NNK codon degeneracy (32 codons). To satisfy the 95% library coverage, 96 clones were screened for each position by using para-methoxy-2-amino benzamidoxime (PMA) as colorimetric probe (Mei et al., 2020). After the initial assaying, the improved variants (hits) were then picked up for further evaluation using the reaction conditions shown in Scheme 1. As a result, several active variants showing pronounced degrees of conversion for substrate 1a were obtained (Figure 2). Five of them including wh27, wh43, wh50, wh53, and wh59 showed more than 10% increment on the formation of (S)-1b compared to the starting enzyme wh18 (Figure 2).
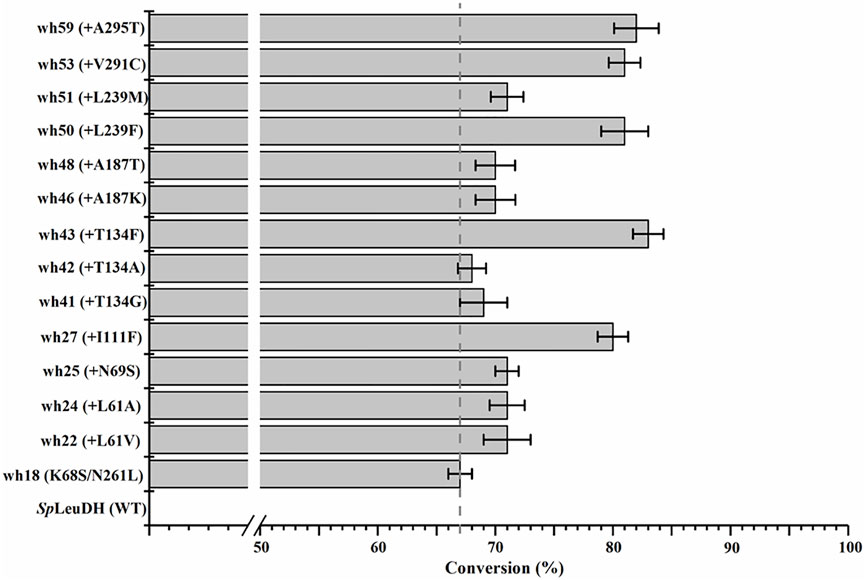
FIGURE 2. Improved variants obtained by screening the single point saturation mutagenesis libraries on the 27 positions. Reductive amination reactions were performed in a reaction mixture (0.5 ml) containing 1 M NH4Cl/NH3·H2O buffer (pH 8.5), 1 mM NAD+, 100 mM glucose, 2 mg/ml GDH cell-free extract (CFE), 6 U/ml DNase I, 20 mM 1a, and 20 mg/ml mutant CFE at 30°C, 1,000 rpm for 24 h. The conversion was detected by HPLC at least three times.
Combinatorial Engineering of SpAmDH Using Reduced Amino Acid Alphabets
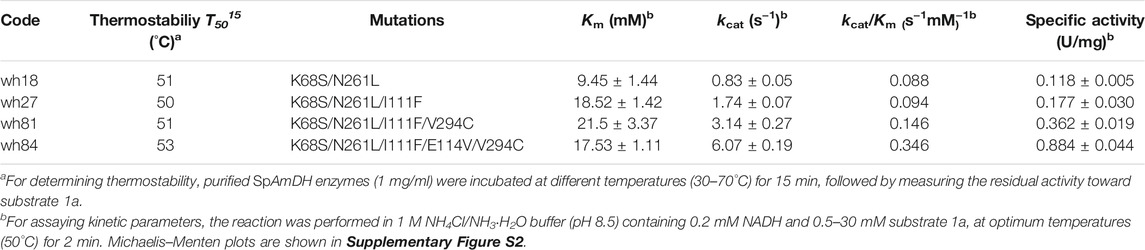
After purification, the five variants were then assayed for the specific activity, three of which including wh27, wh50, and wh53 displayed a higher specific activity than that of wh18 (Table 2). In order to investigate the potential epistatic interactions operating between the individual point mutations shown in Table 2, ten additional double mutants (wh61–wh70, Supplementary Table S4) were constructed and tested using the whole-cell lysate system. As a result, the variant wh43 showed the highest conversion of 85%, while none of the ten combinatorial mutants can exceed the conversion of 80%. It suggests that no additive effects among the five single mutations I111F, T134F, L239F, V291C, and A295T were in pairs, and the triple to quintuple mutants were therefore not constructed further.
In parallel, we noted that the three mutants with improved activity, shown in Table 2, are all composed of the substitutions of phenylalanine and cysteine (e.g., I111F, L239F, and V291C), indicating that hydrophobic interaction and steric hindrance may play key roles in tailoring the activity. Therefore, phenylalanine and cysteine were chosen as building blocks for the further saturation mutagenesis to determine the interactions among the positions in adjacent to residues 111, 239, and 291. As such, two focused libraries A and B were constructed using F and C as reduced amino acid alphabets based on the double-code saturation mutagenesis (DCSM) concept (Sun et al., 2016): library A using wh53 as template with residues M65, S68, N69, and S288 involved, while library B applying wh27 as template with residues L40, A113, T134, and V294 arrested. As a result, two improved mutants wh76 (K68S/N261L/V291C/S68C/N69C) and wh81 (K68S/N261L/I111F/V294C) were obtained from libraries A and B, respectively. The variants wh76 and wh81 gave a conversion of 87 and 90% in the transformation of 1a, respectively, with >99% ee.
Third Round of Mutagenesis Toward SpAmDH, Characterization, and Preparative-Scale Reduction
The variant wh81 was further used as template for the third round of mutagenesis. In this scenario, an important single mutation E114V was considered because it has been reported that this residue can function on the ammonia activation, and thereby affecting the enzyme activity (Abrahamson et al., 2012; Chen et al., 2018; Patil et al., 2018; Lee et al., 2021). To our delight, the resultant quintuple variant wh84 (K68S/N261L/I111F/V294C/E114V) showed 99% conversion and >99% ee toward 1a. Taken together, after three rounds of mutagenesis, the conversion of 1a directed by the SpAmDH mutants was successfully elevated from 46 to 99% (Supplementary Figure S3; Figure 3A). Thereafter, the best mutants in each round of mutagenesis were then purified and characterized by total turnover numbers (TTN), thermostability, and enzyme kinetics. Likewise, wh27, wh81, and wh84 increased the TTN stepwise for substrate 1a (Figure 3B). In particular, wh84 increased the TTN up to 32108, which is 3.2-fold in contrast to that of wh18.
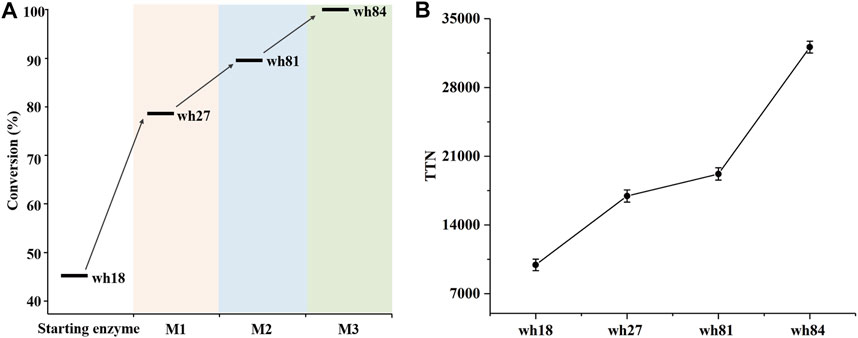
FIGURE 3. Conversion (A) and TTN (B) analysis of the key variants using 40 mM substrate 1a. M1∼M3 depicted the three rounds of mutagenesis.
To evaluate the enzyme robustness of the engineered variants, thermostability was assessed by measuring T5015, the temperature at which 50% of the enzyme activity is lost following a heat treatment for 15 min. Intriguingly, all of the three variants constructed based on wh18 showed comparable T5015 values, reflecting no trade-off between the thermostability and the improved activity (Table 3). Kinetic studies showed that wh84 has the highest catalytic efficiency (kcat/Km) (0.346 s−1mM−1) among all the mutants, a more than 3.9-fold increase to the starting template wh18.
Preparative-scale amination reactions were performed using whole-cell lysates as catalysts for the selected variants in 10 ml of reaction volume, with 100 mM substrate 1a. The mutant wh84 achieved an excellent conversion (>99%), with the high enantioselectivity of >99% ee within 18 h, while wh18 used as a control only showed ca. 80% conversion (Figure 4A). (S)-2b was subsequently isolated as pure form (164 mg, 62% yield) in the reaction system catalyzed by wh84. To further examine the catalytic potential of wh84, the concentration of 1a was raised up to 200 mM. Interestingly, wh84 enabled 91% conversion (Figure 4B).
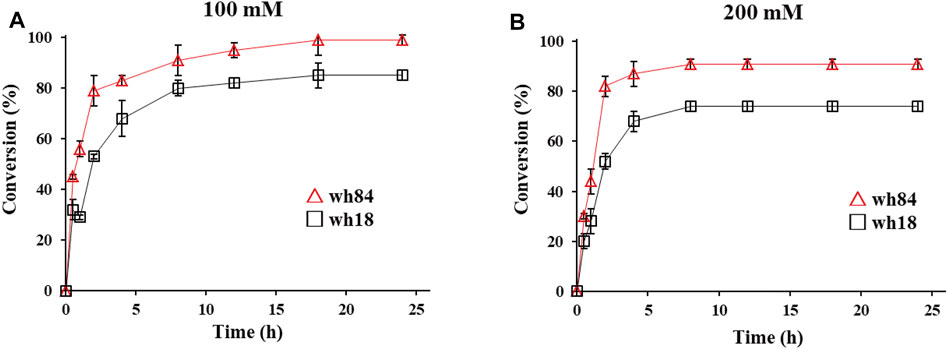
FIGURE 4. Preparative-scale of the asymmetric amination reactions of substrate 1a 100 mM (A) and 200 mM (B) catalyzed by wh18 and wh84 (□: wh18, △: wh84). Reaction system: 0.1 g/ml wet cell, 1 M NH4Cl/NH3·H2O buffer (pH 8.5), 1 mM NAD+, 100 mM glucose, 2 mg/ml GDH CFE, 6 U/mL DNase I, 1 mg/ml lysozyme, and 1a (100 and 200 mM) were mixed in Erlenmeyer flask. The reaction was performed at 30°C, 220 rpm for 24 h. The samples taken at 0, 1, 2, 3, 4, 8, 12, 18, and 24 h were prepared and analyzed by HPLC.
Gaining Insight on the Improved Activity of SpAmDH
In order to shed light on the improved activity of the SpAmDH variant wh84, computational docking analyses were performed to gain insights into the relationship between reshaping of the active site and the effect on activity. The homology structure of wh84 was directly constructed based on the X-ray structure of SpLeuDH (PDB ID: 3VPX, Zhao et al., 2012) by introducing the corresponding mutations in PyMol program (http://www.pymol.org). After relaxing the initial structure of wh84, cofactor NADH, substrate 1a, and ammonium ion (NH4+) were docked into the active site one after another to generate the protein–NADH-1a–NH4+ quaternary complex. It is of interest to note that the catalytic sites K80 and D115 form hydrogen bond interactions with the substrate carbonyl group and the ammonium ion, while the substitutions I111F, E114V, and V294C contribute to hydrophobic interactions with the substrate carbon chain (Figure 5). These strengthened interactions may benefit the substrate recognition and orientation, thereby promoting the activity and maintaining the stereoselectivity. Moreover, on the basis of the proposed mechanism of the engineered AmDHs and their parent AADHs (Sharma et al., 2017), this preferred binding orientation of 1a with respect to NADH determines the Re face of the C=O bond undergoes nucleophilic attack and profits the (S)-configure product, which is consistent with the experimental results.
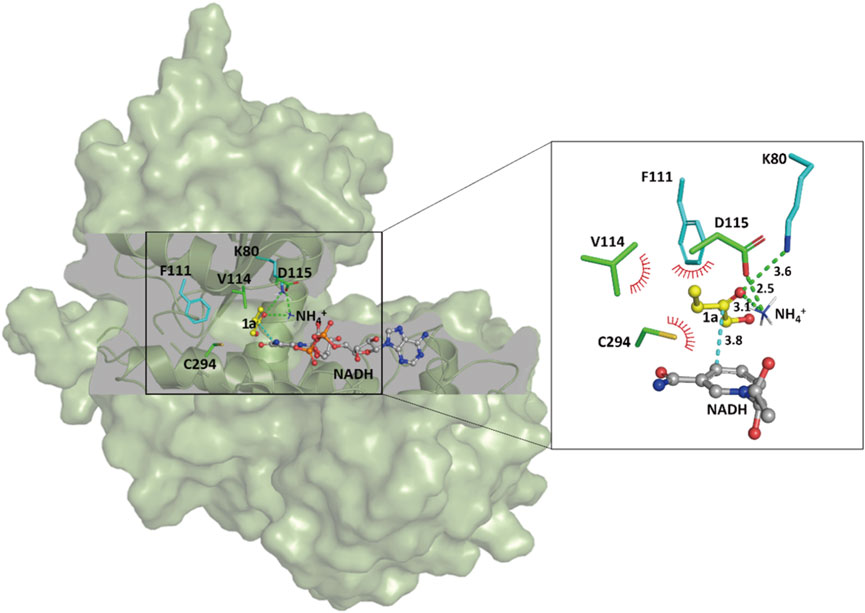
FIGURE 5. Docking analysis of substrate 1a in the catalytic pocket of the SpAmDH variant wh84. Green lines and red spikes represent the hydrogen bonds and the penitential hydrophobic interactions, respectively. The cyan line indicates the nucleophilic attack. Distances are shown in angstroms.
Substrate Scope Analysis Toward Other Prochiral Hydroxy ketones
With the aim to explore the catalytic potential of the engineered variants toward other prochiral hydroxy ketones, seven structurally different substrates were assayed using wh18, wh27, wh81, and wh84 as catalysts (Figure 6). For the four aliphatic compounds 2a–4a and cyclic ketone 8a, the variants showed high stereoselectivity (>99%), and interestingly, the conversions were elevated from wh18 to wh84 in all the four cases (Supplementary Figures S4–S6, S10). For example, the conversion of 3a was improved from 43% (wh18) to 97% (wh84). However, in the scenario of the aromatic substrates, the four variants displayed relatively lower activity in the asymmetric reduction of 5a–7a (Figure 6), while the stereoselectivities were still inherently maintained (>99%, Supplementary Figures S7–S9).
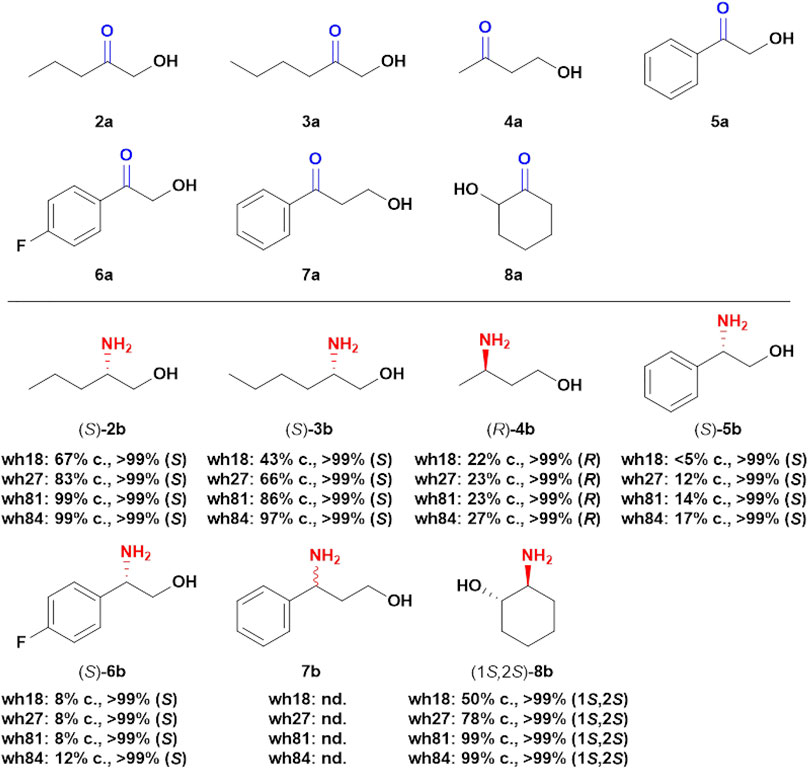
FIGURE 6. Asymmetric transformations of hydroxy ketones. Reaction system: 0.1 g/ml wet cell, 1 M NH4Cl/NH3·H2O buffer (pH 8.5), 1 mM NAD+, 100 mM glucose, 2 mg/ml GDH CFE, 6 U/ml DNase I, 1 mg/ml lysozyme, and 2a-8a (5 mM) were mixed in 2 ml Eppendorf tubes. The reaction was performed at 30°C, 1,000 rpm for 24 h in a thermostatic metal bath. c, conversion; nd, not detectable.
Conclusion
Reductive amination of carbonyl compounds employing AmDHs is an attractive route for the biosynthesis of chiral amino alcohols. It provides several advantages, including lower costs (the amino donor (NH4+) is inexpensive, and water is the main byproduct), elimination of heavy metals, and high stereoselectivity. As such, AmDHs have been reported in the selective synthesis of chiral compounds used as pharmaceutical intermediates. However, the limited catalytic efficiency can be a major obstacle to its industrial application. This work reports the engineering of SpAmDH with the aim to improve the activity. After three rounds of CAST/ISM-guided mutagenesis, mutant wh84 was obtained with the best performance toward substrate 1a, resulting in an excellent TTN (32108) and a kcat/Km value of 0.346 s−1 mM−1, amounting to 3.2-fold and 3.9-fold improvements relative to the starting enzyme, respectively, while maintaining the high enantioselectivity (>99% ee). In the 100 mM preparative reaction, the conversion of 1a catalyzed by wh84 was up to 99% with a 62% yield, which is comparable with the recent work that employed an engineered AmDH from Lysinibacillus fusiformis as catalyst (Chen et al., 2019).
Overall, this work paves the way toward engineering AmDHs with increased activity and also expands the biocatalytic toolbox for asymmetric reductive aminations, and should prove useful insights for further development of other AmDHs as catalysts in the biosynthesis of enantiopure amino alcohols.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding authors.
Author Contributions
YX, ZS, and GQ designed and planned the study. FT, ZQ, HW, YJ, HM, and JL performed the experiments. GQ wrote the manuscript. All authors were involved in the discussions, planning the experiments, and editing manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China (No. 2019YFA0906400), the National Natural Science Foundation of China (No. 31870779), and the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-CXRC-009, TSBICIP-KJGG-003). GQ also thanks financial supports from the Youth Innovation Promotion Association (2021175), CAS.
Conflict of Interest
This work has been included in patent applications by the Tianjin Institute of Industrial Biotechnology.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.778584/full#supplementary-material
References
Abrahamson, M. J., Vázquez-Figueroa, E., Woodall, N. B., Moore, J. C., and Bommarius, A. S. (2012). Development of an Amine Dehydrogenase for Synthesis of Chiral Amines. Angew. Chem. Int. Ed. 51, 3969–3972. doi:10.1002/anie.201107813
Abrahamson, M. J., Wong, J. W., and Bommarius, A. S. (2013). The Evolution of an Amine Dehydrogenase Biocatalyst for the Asymmetric Production of Chiral Amines. Adv. Synth. Catal. 355, 1780–1786. doi:10.1002/adsc.201201030
Ager, D. J., Prakash, I., and Schaad, D. R. (1996). 1,2-Amino Alcohols and Their Heterocyclic Derivatives as Chiral Auxiliaries in Asymmetric Synthesis. Chem. Rev. 96, 835–876. doi:10.1021/cr9500038
Au, S. K., Bommarius, B. R., and Bommarius, A. S. (2014). Biphasic Reaction System Allows for Conversion of Hydrophobic Substrates by Amine Dehydrogenases. ACS Catal. 4, 4021–4026. doi:10.1021/cs4012167
Breuer, M., Ditrich, K., Habicher, T., Hauer, B., Kesseler, M., Stürmer, R., et al. (2004). Industrial Methods for the Production of Optically Active Intermediates. Angew. Chem. Int. Ed. 43, 788–824. doi:10.1002/anie.200300599
Chen, F.-F., Cosgrove, S. C., Birmingham, W. R., Mangas-Sanchez, J., Citoler, J., Thompson, M. P., et al. (2019). Enantioselective Synthesis of Chiral Vicinal Amino Alcohols Using Amine Dehydrogenases. ACS Catal. 9, 11813–11818. doi:10.1021/acscatal.9b03889
Chen, F.-F., Liu, Y.-Y., Zheng, G.-W., and Xu, J.-H. (2015). Asymmetric Amination of Secondary Alcohols by Using a Redox-Neutral Two-Enzyme Cascade. ChemCatChem 7, 3838–3841. doi:10.1002/cctc.201500785
Chen, F.-F., Zheng, G.-W., Liu, L., Li, H., Chen, Q., Li, F.-L., et al. (2018). Reshaping the Active Pocket of Amine Dehydrogenases for Asymmetric Synthesis of Bulky Aliphatic Amines. ACS Catal. 8, 2622–2628. doi:10.1021/acscatal.7b04135
Chen, K., and Arnold, F. H. (2020). Engineering New Catalytic Activities in Enzymes. Nat. Catal. 3, 203–213. doi:10.1038/s41929-019-0385-5
Combs, S. A., Deluca, S. L., Deluca, S. H., Lemmon, G. H., Nannemann, D. P., Nguyen, E. D., et al. (2013). Small-molecule Ligand Docking into Comparative Models with Rosetta. Nat. Protoc. 8, 1277–1298. doi:10.1038/nprot.2013.074
Conway, P., Tyka, M. D., DiMaio, F., Konerding, D. E., and Baker, D. (2014). Relaxation of Backbone Bond Geometry Improves Protein Energy Landscape Modeling. Protein Sci. 23, 47–55. doi:10.1002/pro.2389
Erlanson, D. A., Arndt, J. W., Cancilla, M. T., Cao, K., Elling, R. A., English, N., et al. (2011). Discovery of a Potent and Highly Selective PDK1 Inhibitor via Fragment-Based Drug Discovery. Bioorg. Med. Chem. Lett. 21, 3078–3083. doi:10.1016/j.bmcl.2011.03.032
Francalanci, F., Cesti, P., Cabri, W., Bianchi, D., Martinengo, T., and Foa, M. (1987). Lipase-Catalyzed Resolution of Chiral 2-amino 1-alcohols. J. Org. Chem. 52, 5079–5082. doi:10.1021/jo00232a002
Franklin, R. D., Mount, C. J., Bommarius, B. R., and Bommarius, A. S. (2020). Separate Sets of Mutations Enhance Activity and Substrate Scope of Amine Dehydrogenase. ChemCatChem 12, 2436–2439. doi:10.1002/cctc.201902364
Hollmann, F., Opperman, D. J., and Paul, C. E. (2021). Biocatalytic Reduction Reactions From a Chemist’s Perspective. Angew. Chem. Int. Ed. 60, 5644–5665. doi:10.1002/anie.20180034310.1002/anie.202001876
Itoh, N., Yachi, C., and Kudome, T. (2000). Determining a Novel NAD+-dependent Amine Dehydrogenase with a Broad Substrate Range from Streptomyces Virginiae IFO 12827: Purification and Characterization. J. Mol. Catal. B: Enzymatic 10, 281–290. doi:10.1016/S1381-1177(00)00111-9
Karjalainen, O. K., and Koskinen, A. M. P. (2012). Diastereoselective Synthesis of Vicinal Amino Alcohols. Org. Biomol. Chem. 10, 4311–4326. doi:10.1039/C2OB25357G
Larrow, J. F., Schaus, S. E., and Jacobsen, E. N. (1996). Kinetic Resolution of Terminal Epoxides via Highly Regioselective and Enantioselective Ring Opening with TMSN3. An Efficient, Catalytic Route to 1,2-Amino Alcohols. J. Am. Chem. Soc. 118, 7420–7421. doi:10.1021/ja961708+
Lee, S., Jeon, H., Giri, P., Lee, U.-J., Jung, H., Lim, S., et al. (2021). The Reductive Amination of Carbonyl Compounds Using Native Amine Dehydrogenase from Laribacter hongkongensis. Biotechnol. Bioproc. E. 26, 384–391. doi:10.1007/s12257-021-0113-2
Li, D., Chen, X., Chen, Z., Lin, X., Xu, J., and Wu, Q. (2021). Directed Evolution of Lipase A from Bacillus Subtilis for the Preparation of Enantiocomplementary Sec-Alcohols. Green. Synth. Catal. 2, 290–294. doi:10.1016/j.gresc.2021.07.003
Li, J., Qu, G., Shang, N., Chen, P., Men, Y., Liu, W., et al. (2021). Near-Perfect Control of the Regioselective Glucosylation Enabled by Rational Design of Glycosyltransferases. Green. Synth. Catal. 2, 45–53. doi:10.1016/j.gresc.2021.01.005
Liu, L., Wang, D.-H., Chen, F.-F., Zhang, Z.-J., Chen, Q., Xu, J.-H., et al. (2020). Development of an Engineered Thermostable Amine Dehydrogenase for the Synthesis of Structurally Diverse Chiral Amines. Catal. Sci. Technol. 10, 2353–2358. doi:10.1039/D0CY00071J
Ma, S. K., Gruber, J., Davis, C., Newman, L., Gray, D., Wang, A., et al. (2010). A Green-by-Design Biocatalytic Process for Atorvastatin Intermediate. Green. Chem. 12, 81–86. doi:10.1039/B919115C
Matzel, P., Gand, M., and Höhne, M. (2017). One-step Asymmetric Synthesis of (R)- and (S)-rasagiline by Reductive Amination Applying Imine Reductases. Green. Chem. 19, 385–389. doi:10.1039/c6gc03023h
Mayol, O., Bastard, K., Beloti, L., Frese, A., Turkenburg, J. P., Petit, J.-L., et al. (2019). A Family of Native Amine Dehydrogenases for the Asymmetric Reductive Amination of Ketones. Nat. Catal. 2, 324–333. doi:10.1038/s41929-019-0249-z
Mei, Z., Zhang, K., Qu, G., Li, J.-K., Liu, B., Ma, J.-A., et al. (2020). High-throughput Fluorescence Assay for Ketone Detection and its Applications in Enzyme Mining and Protein Engineering. ACS Omega 5, 13588–13594. doi:10.1021/acsomega.0c00245
Nugent, T. C., and El-Shazly, M. (2010). Chiral Amine Synthesis - Recent Developments and Trends for Enamide Reduction, Reductive Amination, and Imine Reduction. Adv. Synth. Catal. 352, 753–819. doi:10.1002/adsc.200900719
Pablos-Méndez, A., Raviglione, M. C., Laszlo, A., Binkin, N., Rieder, H. L., Bustreo, F., et al. (1998). Global Surveillance for Antituberculosis-Drug Resistance, 1994-1997. N. Engl. J. Med. 338, 1641–1649. doi:10.1056/nejm199806043382301
Patil, M. D., Grogan, G., Bommarius, A., and Yun, H. (2018). Oxidoreductase-Catalyzed Synthesis of Chiral Amines. ACS Catal. 8, 10985–11015. doi:10.1021/acscatal.8b02924
Pushpanath, A., Siirola, E., Bornadel, A., Woodlock, D., and Schell, U. (2017). Understanding and Overcoming the Limitations of Bacillus badius and Caldalkalibacillus thermarum Amine Dehydrogenases for Biocatalytic Reductive Amination. ACS Catal. 7, 3204–3209. doi:10.1021/acscatal.7b00516
Qu, G., Li, A., Acevedo‐Rocha, C. G., Sun, Z., and Reetz, M. T. (2020). The Crucial Role of Methodology Development in Directed Evolution of Selective Enzymes. Angew. Chem. Int. Ed. 59, 13204–13231. doi:10.1002/anie.201901491
Qu, G., Liu, B., Jiang, Y., Nie, Y., Yu, H., and Sun, Z. (2019). Laboratory Evolution of an Alcohol Dehydrogenase Towards Enantioselective Reduction of Difficult-to-Reduce Ketones. Bioresour. Bioproc. 6, 18. doi:10.1186/s40643-019-0253-9
Qu, G., Sun, Z., and Reetz, M. T. (2021). “Iterative Saturation Mutagenesis for Semi‐rational Enzyme Design,” in Protein Engineering: Tools and Applications. Editor H. Zhao (Weinheim: Wiley-VCH Press), 105–132. doi:10.1002/9783527815128.ch5
Reetz, M. T., Bocola, M., Carballeira, J. D., Zha, D., and Vogel, A. (2005). Expanding the Range of Substrate Acceptance of Enzymes: Combinatorial Active-Site Saturation Test. Angew. Chem. Int. Ed. 44, 4192–4196. doi:10.1002/anie.200500767
Reetz, M. T. (2011). Laboratory Evolution of Stereoselective Enzymes: a Prolific Source of Catalysts for Asymmetric Reactions. Angew. Chem. Int. Ed. 50, 138–174. doi:10.1002/anie.201000826
Schrittwieser, J. H., Velikogne, S., and Kroutil, W. (2015). Biocatalytic Imine Reduction and Reductive Amination of Ketones. Adv. Synth. Catal. 357, 1655–1685. doi:10.1002/adsc.201500213
Sharma, M., Mangas-Sanchez, J., Turner, N. J., and Grogan, G. (2017). NAD(P)H-dependent Dehydrogenases for the Asymmetric Reductive Amination of Ketones: Structure, Mechanism, Evolution and Application. Adv. Synth. Catal. 359, 2011–2025. doi:10.1002/adsc.201700356
Sun, Z., Lonsdale, R., Li, G., and Reetz, M. T. (2016). Comparing Different Strategies in Directed Evolution of Enzyme Stereoselectivity: Single- Versus Double-Code Saturation Mutagenesis. ChemBioChem 17, 1865–1872. doi:10.1002/cbic.201600296
Trott, O., and Olson, A. J. (2009). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Tyagi, R., Lai, R., and Duggleby, R. G. (2004). A New Approach to ‘megaprimer’ Polymerase Chain Reaction Mutagenesis Without an Intermediate Gel Purification Step. BMC Biotechnol. 4, 2. doi:10.1186/1472-6750-4-2
Wang, H., Qu, G., Li, J.-K., Ma, J.-A., Guo, J., Miao, Y., et al. (2020). Data Mining of Amine Dehydrogenases for the Synthesis of Enantiopure Amino Alcohols. Catal. Sci. Technol. 10, 5945–5952. doi:10.1039/D0CY01373K
Wang, J., Liu, N., Cheng, X., and Chen, L. (2016). Efficient Continuous Kinetic Resolution of Racemic 2-Aminobutanol Over Immobilized Penicillin G Acylase. Synth. Commun. 46, 956–962. doi:10.1080/00397911.2016.1181763
Winkler, C. K., Schrittwieser, J. H., and Kroutil, W. (2021). Power of Biocatalysis for Organic Synthesis. ACS Cent. Sci. 7, 55–71. doi:10.1021/acscentsci.0c01496
Wu, H.-L., Zhang, J.-D., Zhang, C.-F., Fan, X.-J., Chang, H.-H., and Wei, W.-L. (2017). Characterization of Four New Distinct ω-Transaminases from Pseudomonas Putida NBRC 14164 for Kinetic Resolution of Racemic Amines and Amino Alcohols. Appl. Biochem. Biotechnol. 181, 972–985. doi:10.1007/s12010-016-2263-9
Wu, S., Snajdrova, R., Moore, J. C., Baldenius, K., and Bornscheuer, U. T. (2021). Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 60, 88–119. doi:10.1002/anie.202006648
Xie, C., Song, J., Hua, M., Hu, Y., Huang, X., Wu, H., et al. (2020). Ambient-temperature Synthesis of Primary Amines via Reductive Amination of Carbonyl Compounds. ACS Catal. 10, 7763–7772. doi:10.1021/acscatal.0c01872
Yanai, N., Sindoro, M., Yan, J., and Granick, S. (2013). Electric Field-Induced Assembly of Monodisperse Polyhedral Metal-Organic Framework Crystals. J. Am. Chem. Soc. 135, 34–37. doi:10.1021/ja309361d
Ye, L. J., Toh, H. H., Yang, Y., Adams, J. P., Snajdrova, R., and Li, Z. (2015). Engineering of Amine Dehydrogenase for Asymmetric Reductive Amination of Ketone by Evolving Rhodococcus Phenylalanine Dehydrogenase. ACS Catal. 5, 1119–1122. doi:10.1021/cs501906r
Zhao, Y., Wakamatsu, T., Doi, K., Sakuraba, H., and Ohshima, T. (2012). A Psychrophilic Leucine Dehydrogenase from Sporosarcina psychrophila: Purification, Characterization, Gene Sequencing and Crystal Structure Analysis. J. Mol. Catal. B Enzym. 83, 65–72. doi:10.1016/j.molcatb.2012.06.018
Keywords: biosynthesis, protein engineering, directed evolution, amine dehydrogenase, chiral amino alcohol
Citation: Tong F, Qin Z, Wang H, Jiang Y, Li J, Ming H, Qu G, Xiao Y and Sun Z (2022) Biosynthesis of Chiral Amino Alcohols via an Engineered Amine Dehydrogenase in E. coli. Front. Bioeng. Biotechnol. 9:778584. doi: 10.3389/fbioe.2021.778584
Received: 17 September 2021; Accepted: 15 November 2021;
Published: 05 January 2022.
Edited by:
Hua Ling, National University of Singapore, SingaporeReviewed by:
Wen Shan Yew, National University of Singapore, SingaporeLei Shao, Shanghai University of Medicine and Health Sciences, China
Shuke Wu, Huazhong Agricultural University, China
Copyright © 2022 Tong, Qin, Wang, Jiang, Li, Ming, Qu, Xiao and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yazhong Xiao, eXp4aWFvQGFodS5lZHUuY24=; Zhoutong Sun, c3Vuemh0QHRpYi5jYXMuY24=
†These authors have contributed equally to this work
 Feifei Tong1,2†
Feifei Tong1,2† Ge Qu
Ge Qu Zhoutong Sun
Zhoutong Sun