- 1Laboratory of Biotechnology, Dalian Institute of Chemical Physics (CAS), Dalian, China
- 2Dalian Key Laboratory of Energy Biotechnology, Dalian Institute of Chemical Physics (CAS), Dalian, China
- 3State Key Laboratory of Catalysis, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, China
Conversion of lignocellulosic biomass into lipids and related chemicals has attracted much attention in the past two decades, and the oleaginous yeast Rhodosporidium toruloides has been widely used in this area. While R. toruloides species naturally have physiological advantages in terms of substrate utilization, lipid accumulation, and inhibitor resistance, reduced lipid production and cell growth are noticed when biomass hydrolysates are used as feedstocks. To improve the robustness of R. toruloides, here, we devised engineered strains by overexpressing genes responsible for phenolic compound degradation. Specifically, gene expression cassettes of the manganese peroxidase gene (MNP) and versatile peroxidase gene (VP) were constructed and integrated into the genome of R. toruloides NP11. A series of engineered strains were evaluated for lipid production in the presence of typical phenolic inhibitors. The results showed that R. toruloides strains with proper expression of MNP or VP indeed grew faster in the presence of vanillin and 5-hydroxymethylfurfural than the parental strain. When cultivated in concentrated mode biomass hydrolysates, the strain VP18 had improved performance as the cell mass and lipid content increased by 30% and 25%, respectively. This study provides more robust oleaginous yeast strains for microbial lipid production from lignocellulosic biomass, and similar efforts may be used to devise more advanced lipid producers.
Introduction
The bioconversion of lignocellulose into biofuels and other high value-added chemicals is of significant potential for its environmental protection and energy sustainability (Jin et al., 2015). Lignocellulose contains roughly 40% of cellulose, 25% of hemicellulose, and 20% of lignin (Kim et al., 2021). After pretreatment and hydrolysis, cellulose and hemicellulose are hydrolyzed into sugars, along with the formation of some inhibitory compounds, including acids (formic, acetic, and levulinic acids) and furnaldehydes (furfural and 5-hydroxymethylfurfural HMF) (Tramontina et al., 2020). Lignin is a phenolic heteropolymer composed of p-hydroxyphenyl, guaiacyl, and syringyl groups (Chen and Wan, 2017). During chemical or biological pretreatment, lignin is degraded into monomeric compounds, mainly as p-hydroxybenzaldehyde (PHB), vanillin, syringaldehyde, and their corresponding reduced or oxidized products (Zhu et al., 2017; Osorio–González et al., 2019). These lignin-derived phenols, associated with organic acids and furnaldehydes, usually have inhibitive and toxic effects on microorganisms, thus playing negative roles in biological processes (Ragauskas et al., 2014).
Rhodosporidium toruloides is a typical oleaginous yeast. It is considered an ideal platform for the bioconversion of lignocellulose into fine chemical products (Wen et al., 2020). This yeast can generate cell mass up to a high density of 130 g/L with a capacity to produce lipids over 70% of its dry cell weight (Li et al., 2007; Park et al., 2018). Besides being endowed with an endogenous mevalonate pathway for the synthesis of carotenoids and some other terpenes (Jiao et al., 2019; Zhuang et al., 2019; Geiselman et al., 2020), R. toruloides is also designed with the ability to utilize a number of biomass and carbon sources. As a robust organism, this yeast has demonstrated to utilize some forms of biomass-derived inhibitors (Hu and Zhao, 2009; Tramontina et al., 2020). However, study has shown that R. toruloides are strongly inhibited by 1 g/L of furfuran, PHB, or vanillin (Hu and Zhao, 2009; Zhao et al., 2012). This therefore suggests a weaker tolerance and the utilization ability of R. toruloides to phenolic aldehydes and furnaldehydes. Specially, in order to improve the product yield, the hydrolysate is usually concentrated to increase the sugar content, resulting in a corresponding rise of by-products (Mussatto, 2004; Agu et al., 2016). Thus, it is challenging for R. toruloides to improve robustness during fermentation.
Fungal lignin peroxidase (Lip), manganese peroxidase (Mnp), and versatile peroxidase (Vp), usually participating in lignin degradation, have shown relevance in the detoxification of lignocellulosic hydrolysates (Tramontina et al., 2020). Lip catalyzes H2O2-dependent oxidative depolymerization, resulting in demethylation and intramolecular rearrangements of lignin-derived phenols (Chandra et al., 2017). Mnp is also a known phenol-oxidizing enzyme that decomposes the substrate by forming a radical on the phenolic hydroxyl group. In addition, Mnp is also reported to degrade non-phenolic compounds by generating a peroxyl radical (Watanabe et al., 2000). A peroxyl radical can degrade bonds in a number of different structures, including a non-phenolic β-O-4–linked lignin, polyethylene, and phenanthrene. Due to their strong oxidizing properties, Mnp has recently been reported to degrade furfural and HMF (Yee et al., 2018). Vp is a hybrid peroxidase that has catalytic functions of both Lip and Mnp (Tramontina et al., 2020).
Here, we hypothesize that engineered R. toruloides could improve the robustness of R. toruloides. This strategy includes the overexpressing the manganese peroxidase gene (MNP) and the versatile peroxidase gene (VP) responsible for by-products degradation (Figure 1A). After high-throughput screening, fermentation was employed to investigate the lipid production and typical inhibitor utilization of the engineered strains. This study provides more robust oleaginous yeast strains for microbial lipid production from lignocellulosic biomass, and similar efforts may be used to devise more advanced lipid producers.
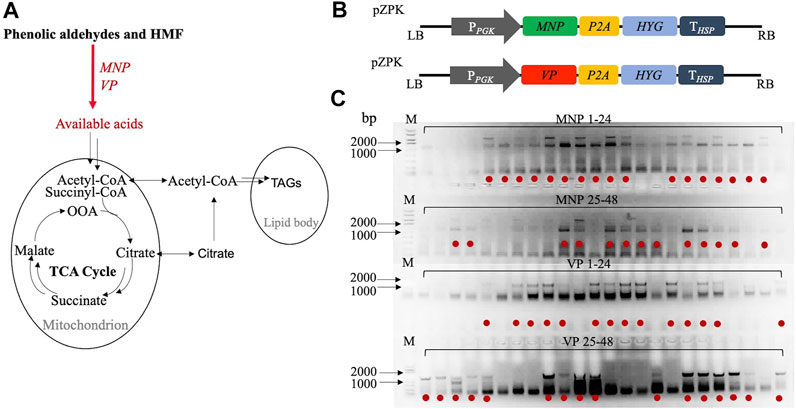
FIGURE 1. Integration of exogenous genes MNP and VP with R. toruloides chromosomes: (A) hypothetic metabolize pathway of phenolic aldehydes and HMF; (B) information of gene expression cassettes; (C) R. toruloides colony PCR results for MNP and VP genes verification. Red dots represent right genotypes chose for further screening.
Materials and Methods
Strains, Media, and Reagents
All strains used in this study are listed in Supplementary Table S1. Rhodosporidium toruloides CGMCC 2.1389 was purchased from the China General Microbiological Culture Collection Centre (Beijing, China), and Rhodosporidium toruloides NP11 was its haploid separated by our laboratory (Zhu et al., 2012). Escherichia coli DH10B and Agrobacterium tumefaciens AGL1 used for routine molecular manipulation were cultured in the Luria–Bertani medium (10.0 g/L NaCl, 10.0 g/L tryptone, and 5.0 g/L yeast extract) at 37°C and 30°C, respectively. Yeast cells were cultured in the yeast extract–peptone–dextrose (YPD) medium (20.0 g/L glucose, 10.0 g/L yeast extract, and 20.0 g/L peptone) at 30°C. Antibiotics were used at the following concentrations: kanamycin of 50 μg/ml and hygromycin of 50 μg/ml.
The media used in preliminary screening consist of yeast nitrogen base (YNB), with 2.5 g/L of PHB, vanillin, syringaldehyde or HMF as the sole carbon source, 1.0 g/L of KH2PO4. Mother selection media used for secondary screening were as follows: PHB (2.5 g/L), vanillin (2.5 g/L), syringaldehyde (2.5 g/L), and HMF (5.0 g/L) which served as the sole carbon source in the YNB medium. Each of these media, respectively, contains 1.0 g/L of KH2PO4.
The concentrated model hydrolysate was made up of by-products such as vanillin 16 g/L, PHB 10 g/L, syringaldehyde 15 g/L, vanillic acid 4 g/L, p-hydroxybenzoic acid 4 g/L, vanillyl alcohol 4 g/L, syringic acid 5 g/L, guaiacol 1 g/L, syringol 2 g/L, and HMF 5 g/L. These by-products were dissolved in ddH2O and filter-sterilized. The nitrogen-limited medium was formulated with glucose 70.0 g/L, yeast extract 0.8 g/L, (NH4)2SO4 0.1 g/L, MgSO4 1.5 g/L, and KH2SO4 1.0 g/L at a C/N ratio of 100 and then sterilized using an autoclave. As a fermentation substrate, a concentrated model hydrolysate was ten times diluted in the nitrogen-limited medium. The initial pH of the substrate was adjusted to 5.6 using 3 M of NaOH.
The aforementioned by-products and chemicals were purchased from Aladdin (Shanghai, China). Yeast extract, peptone, and YNB were from Oxoid (Basingstoke, Hampshire, United Kingdom). Enzymes used for PCR amplification were purchased from Takara (Dalian, China). DNA gel purification and plasmid extraction kits were supplied by Sangon Biotech (Shanghai, China). All other chemicals were purchased from Bonuo Biological and Chemical Reagent Co. (Dalian, China).
Plasmid Construction, Transformation, and Verification
To construct plasmids pZPK-MNP-HYG and pZPK-VP-HYG, MNP (GenBank: M60672.1) gene from Phanerochaete chrysosporium and VP (GenBank: AF175710.1) gene from Pleurotus eryngii were codon-optimized according to the codon preference of R. toruloides, synthesized by Synbio Technologies (Suzhou, China) and linked to the pUC57 plasmid. First, the primer pair P2A-HYG-F and HYG-THSP-R was used to amplify the fragment P2A-HYG-THSP from the plasmid pJX14 (Jiao et al., 2018). The fragments were then assembled into the plasmid pJX14 to obtain pZPK-PPGK-HYG-P2A-HYG-THSP by the RF cloning method (Van den Ent and Löwe, 2006). Primer pairs PPGK-MNP/VP-F and MNP/VP-P2A-R were used, respectively, to amplify PPGK-MNP-P2A and PPGK-VP-P2A fragments from pUC57 plasmids. The fragments were then assembled into the plasmid pZPK-PPGK-HYG-P2A-HYG-THSP by the RF cloning method. The genotype of the clone was verified by Sanger sequencing (Synbio Technologies, Suzhou, China). All plasmids and primers used in this study are listed in Supplementary Tables S1, S2, respectively.
The correct plasmids pZPK-MNP-HYG and pZPK-VP-HYG were transformed into AGL1 by electroporation, and strains were selected on LB plates supplemented with 50 μg/ml kanamycin. The transformation of the R. toruloides was done according to a published Agrobacterium tumefaciens–mediated transformation (ATMT) method (Lin et al., 2014). R. toruloides colony PCR analysis for transformant verification was assayed according to previous reports (Lin et al., 2012; Lin et al., 2014). Primer pairs used in this study were PPGK-MNP-F/HYG-R and PPGK-VP-F/HYG-R, as shown in Table S2.
Preliminary and Secondary Screening
Randomized selected transformants from selective plates were subjected to repeated sub-cultivation 5 times. A single colony of the transformant was added to 3 ml of YPD medium in a 24-well plate for 24-h culturing to prepare seeds.
For preliminary screening, 15 µL of the seed broth was added to 400 µL of selection medium in 96-well plates (Corning 3960, Corning, NY). For secondary screening, the seed broth was inoculated into the selection medium in 96-well plates to make an initial optical density (measured at a wavelength of 600 nm, OD600) of 0.2. Mother media were diluted into four concentration gradients to keep phenolic aldehydes to 2.5-, 1.2-, 0.6-, and 0.3 g/L, and HMF to 5.0-, 2.5-, 1.3- and 0.6 g/L, respectively. All plates were covered with a sterile breathable sealing film (Axygen, United States), vortexed in a Multitron (ZQZY-88BH, Shanghai Zhichu Instrument Co., Ltd., China) at 30°C and 800 rpm for 48 h. The samples were collected after 48 h to measure OD600. Inoculation and measurement processes were done by the fluid handing workstation (Biomek i7) combined with an automation workflow scheduling software (SAMI EX) and a microplate reader and controlled by Beckman software (Beckman Coulter Inc., United States) (Sobreira et al., 2020).
Fermentation and Lipid Production Procedures
Seed cultures were prepared by inoculating a single colony from selective plates into 50 ml YPD medium for 24 h; 5 ml of seed cultures were added to 45 ml nitrogen-limited medium in a 250-ml shake flask at 30°C and 200 rpm for 152 h. Fermentation experiments were performed in triplicate.
During fermentation in the nitrogen-limited substrate, glucose concentration and OD600 were quantified every 24 h by a glucose analyzer (SBA-50B; Shandong Academy of Sciences, Jinan, China) and a spectrophotometer (Evolution 220; Thermo, United States). At the fermentation endpoint, the lipids were extracted and detected, as described previously (Zhu et al., 2015). By-product degradation was analyzed by thin layer chromatography (TLC) with chloroform and ethyl acetate (3:1, v/v) as the elution solvent and was visualized under iodine vapor and UV 230 nm.
Results
Engineering R. toruloides and Verification
Plasmids were constructed using the RF cloning method (Figure 1B) and transformed into R. toruloides using the ATMT method, which resulted in the random chromosomal integration of transgenes. To further ensure the integration of exogenous genes, the colony PCR analysis was carried out (Figure 1C). The results clearly indicated the presence of MNP and VP genes for most transformants. For each genotype, thirty of the right transformants were chosen for the following experiment.
Screening of Engineered Strains
Thirty transformants of each genotype were selected for preliminary screening. The selection medium was prepared by the YNB substrate with four different kinds of inhibitors as the sole carbon source. These inhibitors, “phenolic aldehydes, PHB, vanillin, and syringaldehyde,” were selected to represent the typical lignin degradation monomers of hydroxyphenyl, guaiacyl, and syringyl groups. All the transformants were cultured for 48 h, and the initial and the terminal OD600 were detected using the Biomek i7 fluid handling workstation. The results showed that 2.5 g/L of PHB, vanillin, and syringaldehyde, except HMF, inhibited the cell growth of all strains (Supplementary Figures S1, S2). Thus, seven strains with the highest cell density under 2.5 g/L of HMF were selected as dominant strains.
For secondary screening, substrates within various inhibitors were set for four concentration gradients (Figure 2). Seven dominant strains and the control were cultured with an initial OD600 of 0.2 for 48 h and screened by detecting the highest OD600 under high concentrations. The strain MNP28 and VP28 showed an increase in cell growth under 1.3- and 0.6 g/L of vanillin compared with the control. The cell growth of strains MNP28 and VP9 had an increase under 2.5 g/L of HMF. These results suggest that some engineered strains could indeed utilize vanillin and HMF. All strains used in the study were found to grow under 1.3 g/L of PHB and 2.5 g/L of syringaldehyde, which indicated that the wild-type R. toruloides NP11 was naturally capable of using or tolerating certain concentrations of the two inhibitors. This finding is also consistent with our previous study (Hu and Zhao, 2009). In this study, concentrations of PHB and syringaldehyde concentrations were higher than those in real lignocellulose hydrolysates (Persson et al., 2002; Min et al., 2014). Thus, we did not explore the optimal concentration range of these two inhibitors. In addition, we selected the engineered strains (MNP28, VP9, and VP28) with superior performance for further test.
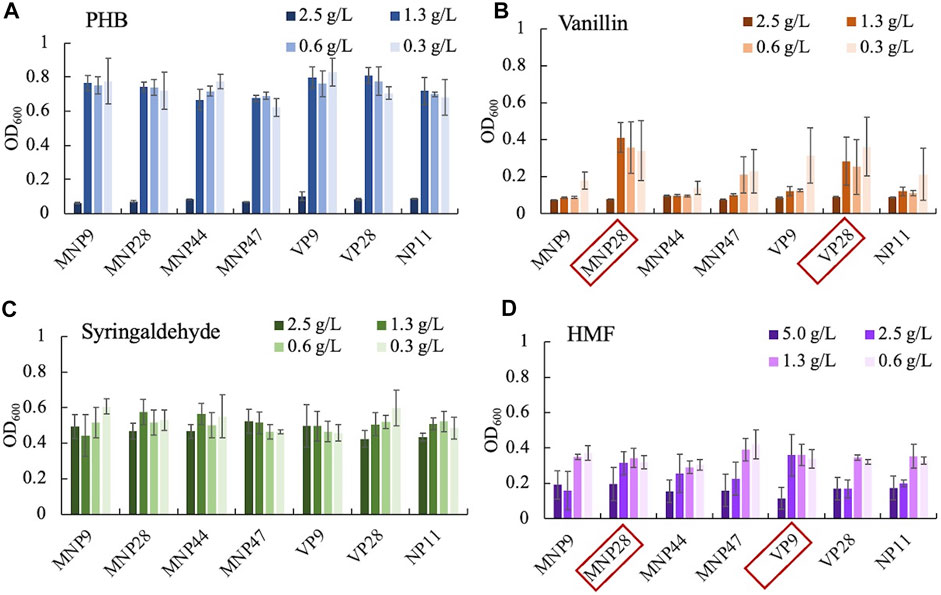
FIGURE 2. Growth of dominant strains and the control under gradient concentrations (g/L) of PHB-p-hydroxy benzaldehyde (A), vanillin (B), syringaldehyde (C), and HMF-hydroxymethyl furfural (D). Initial OD600 was controlled at 0.2 by computer programming, and final OD600 was reached in 48 h. Red boxes represent the best growing strains selected for further test. Experiments were performed in triplicate.
Fermentation Performance of Engineered Strains
To detect the fermentation performance and the by-product utilization of engineered strains in a concentrated hydrolysate, a corn stover hydrolysate with three times the concentration was simulated. The composition of the model hydrolysate was carried out according to the alkaline pretreatment method (Persson et al., 2002; Mcintosh and Vancov, 2011; Min et al., 2014). First, it was necessary to test whether simulated hydrolysates inhibited R. toruloides. Two wild-type strains R. toruloides NP11 and CGMCC 2.1389 were fermented by the concentrated hydrolysate (Supplementary Figure S3). Compared with the control (wild strains cultured under the nitrogen-limited medium), the concentrated model hydrolysate showed serious inhibition on R. toruloides strains. After 48 h of fermentation, cell densities of the NP11 and the CGMCC 2.1389 were slightly decreased and with almost no glucose consumed.
Furthermore, the concentrated model hydrolysate was used as the fermentation substrate. As shown in Figures 3A,B, cell growth was still restrained in the first 48 h, and glucose was hardly consumed. Then strains VP9 and VP28 released form the by-product inhibition and had a quick recovery of cell growth and glucose consumption. These results suggest that the VP gene in R. toruloides play a more important role than the MNP during by-product degradation. Compared with the control, the VP28 had a significant improvement in cell mass and lipid accumulation, which increased by 30 and 25%, respectively (Figures 3C,D). This indicates that the VP gene had a positive effect on the bioconversion of lignin-derived phenols and HMF. The MNP28 strain also had an increase of 32% on cell mass but remained unchanged in the lipid content, which might be caused by its long inhibitory stage.
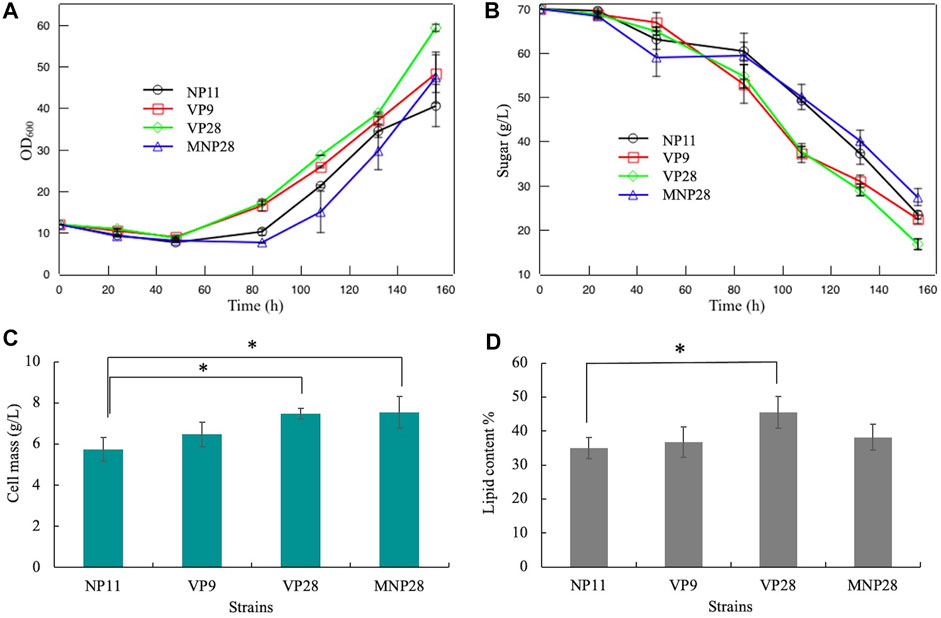
FIGURE 3. Fermentation performance of engineering strains and the control: (A) cell growth; (B) glucose consumption; (C) cell mass at the fermentation endpoint; (D) lipid content at the fermentation endpoint. Error bars represent the standard deviation of three independent experiments. Significantly different values were performed by the ANOVA and post hoc Dunnett test. * means p < 0.05.
The by-product conversion of engineered strains was then examined by TLC (Supplementary Figure S4). Syringol and guaiacol could not be colored by iodine vapor due to their low concentrations in the simulated hydrolysate. The TLC result gave a qualitative analysis about lignin-derived phenol bioconversion. Three phenolic aldehydes and HMF were completely conversed within 48 h in all fermentation supernatants. The VP9 and the VP28 strains exhibited a faster bioconversion rate than the MNP47 and the control. p-Hydroxybenzyl alcohol generated along with the PHB consumption indicates that the endogenous aldehyde reductase of R. toruloides catalyzed the reduction of the PHB aldehyde group (Hu et al., 2018). Compared with the control, three engineered strains rapidly convert p-hydroxybenzoic acid, vanillic acid, and syringic acid in 48 h without obvious accumulation (Supplementary Figure S5), thus suggesting an efficient decomposition of phenolic acids by Mnp and Vp enzymes.
Discussion
This study integrated two peroxidase genes (MNP and VP) into the oleaginous yeast R. toruloides. After high-throughput screening, the strains MNP28, VP9, and VP28 with superior performance were chosen for fermentation. There were only three strains obtained from sixty transformants; this might be due to the low expression level of Mnp and Vp in R. toruloides. Thus, high-throughput screening is required for fast identification of the engineered strains.
During the batch test, the seed broth was first inoculated into the simulated hydrolysate to reach an initial OD600 of 2. However, after fermentation for 96 h, the engineered and the control (NP11) strains were all inhibited by the high concentration of by-products (data were not shown). Due to the additive inhibition caused by inhibitor combinations (Hu and Zhao, 2009), such high concentration of by-products would greatly affect the cell growth in a short time. Moreover, within a certain range, high-density cells could form quorum sensing and improve stress resistance to the environment (Gao et al., 2016). Thus, efforts were made to increase the inoculum amount of fermentation and finally set an initial OD600 of 12.
This study also gives a hypothetical model for by-product utilization in the engineered R. toruloides (Figure 4). On one hand, benzene rings of phenolic aldehydes would be directly and oxidatively decomposed by Mnp (Supplementary Figure S6A) (Hofrichter, 2002). On the other hand, Lip is capable of demethoxylation or changing the methoxy group into the phenolic hydroxyl group (Supplementary Figure S6B) (Chandra et al., 2017; Wang et al., 2018). Consequently, the hybrid enzyme Vp, which combines the structural–functional properties of Lip and Mnp, could decompose phenolic aldehydes to organic acids. PHB needs to be carboxylated by PHBH first and participates in the latter peroxidation ways (Zhu et al., 2012). Otherwise, phenolic aldehydes and alcohols could also be reduced into their less toxic acids by alcohol dehydrogenases, aldehyde reductases, and aldehyde dehydrogenases (Supplementary Figure S6C) (Hu et al., 2018). The HMF might be easily degraded into the available acid by Mnp through its peroxidation effect (Supplementary Figure S6D). These organic acids are able to be further converted into the intermediates to enter the TCA cycle, or into precursor compounds for lipid synthesis. Therefore, the complex by-products of the lignocellulosic hydrolysate would be fast and globally decomposed by the engineered strains with the VP gene expression.
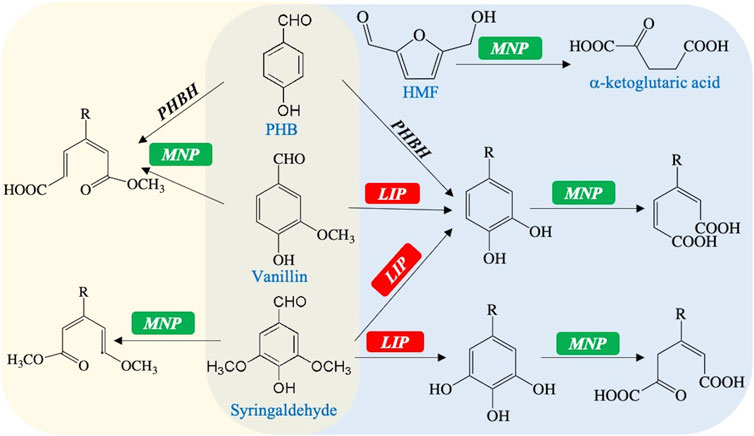
FIGURE 4. Hypothetical model for the degradation of typical by-products mediated by MNP and VP: PHBH, para-hydroxybenzoate hydroxylase (R. toruloides endogenous, gene symbol RHTO_07012); R represents the group of −CHO, −COOH, or −CH2OH.
In order to further improve the capability of engineered strains, fermentation conditions can be optimized by, for example, medium optimization, pH control, and continuous feeding. Adaptive laboratory evolution (ALE) could also be introduced to promote the robustness of engineered strains (Liu et al., 2021). Through these techniques, oxidases and/or peroxidases, which are major participants in the degradation of lignin-derived phenols such as laccase and dye-decolorizing peroxidases (Dyp), could also be expressed in R. toruloides.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author Contributions
ZZ contributed to conception and design of the study. LL organized the database and wrote the first draft of the manuscript. YC involved in the methodology and the software. ZZ and SZ acquired funding. YZ performed statistical analysis. QH and SW revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2019YFA0904902) and Dalian Institute of Chemical Physics, CAS (No. DICP I201947).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Chuks Kenneth Odoh for English improvement and Energy Biotechnology Platform of Dalian Institute of Chemical Physics for providing advanced facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.768934/full#supplementary-material
References
Agu, C. V., Ujor, V., Gopalan, V., and Ezeji, T. C. (2016). Use of Cupriavidus Basilensis-Aided Bioabatement to Enhance Fermentation of Acid-Pretreated Biomass Hydrolysates by Clostridium Beijerinckii. J. Ind. Microbiol. Biotechnol. 43 (9), 1215–1226. doi:10.1007/s10295-016-1798-7
Chandra, R., Kumar, V., and Yadav, S. (2017). “Extremophilic Ligninolytic Enzymes,” in Extremophilic Enzymatic Processing of Lignocellulosic Feedstocks to Bioenergy. Editors R.K. Sani, and R.N. Krishnaraj (Cham: Springer International Publishing), 115–154. doi:10.1007/978-3-319-54684-1_8
Chen, Z., and Wan, C. (2017). Biological Valorization Strategies for Converting Lignin Into Fuels and Chemicals. Renew. Sustainable Energ. Rev. 73, 610–621. doi:10.1016/j.rser.2017.01.166
Gao, M., Song, H., Zheng, H., Ren, Y., Li, S., Liu, X., et al. (2016). Culture of Low Density E. coli Cells in Alginate-Chitosan Microcapsules Facilitates Stress Resistance by Up-Regulating LuxS/AI-2 System. Carbohydr. Polym. 141, 160–165. doi:10.1016/j.carbpol.2016.01.011
Geiselman, G. M., Zhuang, X., Kirby, J., Tran-Gyamfi, M. B., Prahl, J.-P., Sundstrom, E. R., et al. (2020). Production of Ent-Kaurene From Lignocellulosic Hydrolysate in Rhodosporidium Toruloides. Microb. Cell Fact. 19 (1), 24. doi:10.1186/s12934-020-1293-8
Hofrichter, M. (2002). Review: Lignin Conversion by Manganese Peroxidase (MnP). Enzyme Microb. Technology. 30 (4), 454–466. doi:10.1016/S0141-0229(01)00528-2
Hu, C., Zhao, X., Zhao, J., Wu, S., and Zhao, Z. K. (2009). Effects of Biomass Hydrolysis By-Products on Oleaginous Yeast Rhodosporidium Toruloides. Bioresour. Technology. 100 (20), 4843–4847. doi:10.1016/j.biortech.2009.04.041
Hu, M., Wang, J., Gao, Q., and Bao, J. (2018). Converting Lignin Derived Phenolic Aldehydes Into Microbial Lipid by Trichosporon Cutaneum. J. Biotechnol. 281, 81–86. doi:10.1016/j.jbiotec.2018.06.341
Jiao, X., Zhang, Q., Zhang, S., Yang, X., Wang, Q., and Zhao, Z. K. (2018). Efficient Co-Expression of Multiple Enzymes From a Single Promoter Mediated by Virus 2A Sequence in the Oleaginous Yeast Rhodosporidium Toruloides. FEMS Yeast Res. 18, foy086. doi:10.1093/femsyr/foy086
Jiao, X., Zhang, Y., Liu, X., Zhang, Q., Zhang, S., and Zhao, Z. K. (2019). Developing a CRISPR/Cas9 System for Genome Editing in the Basidiomycetous Yeast Rhodosporidium Toruloides. Biotechnol. J. 14 (7), 1900036. doi:10.1002/biot.201900036
Jin, M., Slininger, P. J., Dien, B. S., Waghmode, S., Moser, B. R., Orjuela, A., et al. (2015). Microbial Lipid-Based Lignocellulosic Biorefinery: Feasibility and Challenges. Trends Biotechnol. 33 (1), 43–54. doi:10.1016/j.tibtech.2014.11.005
Kim, H., Lee, S., Lee, J., and Won, W. (2021). Simultaneous Production of 1,6-Hexanediol, Furfural, and High-Purity Lignin From White Birch: Process Integration and Techno-Economic Evaluation. Bioresour. Technology. 331, 125009. doi:10.1016/j.biortech.2021.125009
Li, Y., Zhao, Z., and Bai, F. (2007). High-Density Cultivation of Oleaginous Yeast Rhodosporidium Toruloides Y4 in Fed-Batch Culture. Enzyme Microb. Technology. 41 (3), 312–317. doi:10.1016/j.enzmictec.2007.02.008
Lin, X., Wang, Y., Zhang, S., Zhu, Z., Zhou, Y. J., Yang, F., et al. (2014). Functional Integration of Multiple Genes into the Genome of the Oleaginous Yeast Rhodosporidium Toruloides. FEMS Yeast Res. 14 (4), 547–555. doi:10.1111/1567-1364.12140
Lin, X., Yang, F., Zhou, Y., Zhu, Z., Jin, G., Zhang, S., et al. (2012). Highly-Efficient colony PCR Method for Red Yeasts and its Application to Identify Mutations Within Two Leucine Auxotroph Mutants. Yeast. 29 (11), 467–474. doi:10.1002/yea.2926
Liu, Z., Radi, M., Mohamed, E. T. T., Feist, A. M., Dragone, G., and Mussatto, S. I. (2021). Adaptive Laboratory Evolution of Rhodosporidium Toruloides to Inhibitors Derived From Lignocellulosic Biomass and Genetic Variations Behind Evolution. Bioresour. Technology. 333, 125171. doi:10.1016/j.biortech.2021.125171
Mcintosh, S., and Vancov, T. (2011). Optimisation of Dilute Alkaline Pretreatment for Enzymatic Saccharification of Wheat Straw. Biomass and Bioenergy. 35, 3094–3103. doi:10.1016/j.biombioe.2011.04.018
Min, D.-y., Chang, H.-m., Jameel, H., Lucia, L., Wang, Z.-g., and Jin, Y.-c. (2014). The Structure of Lignin of Corn stover and its Changes Induced by Mild Sodium Hydroxide Treatment. Bioresources. 9 (2), 2405–2414. doi:10.15376/biores.9.2.2405-2414
Mussatto, S. (2004). Alternatives for Detoxification of Diluted-Acid Lignocellulosic Hydrolyzates for Use in Fermentative Processes: a Review. Bioresour. Technology. 93 (1), 1–10. doi:10.1016/j.biortech.2003.10.005
Osorio-González, C. S., Hegde, K., Brar, S. K., Kermanshahipour, A., and Avalos‐Ramírez, A. (2019). Challenges in Lipid Production from Lignocellulosic Biomass Using Rhodosporidium sp.; A Look at the Role of Lignocellulosic Inhibitors. Biofuels, Bioprod. Bioref. 13 (3), 740–759. doi:10.1002/bbb.1954
Park, Y.-K., Nicaud, J.-M., and Ledesma-Amaro, R. (2018). The Engineering Potential of Rhodosporidium Toruloides as a Workhorse for Biotechnological Applications. Trends Biotechnol. 36 (3), 304–317. doi:10.1016/j.tibtech.2017.10.013
Persson, P., Andersson, J., Gorton, L., Larsson, S., Nilvebrant, N.-O., and Jönsson, L. J. (2002). Effect of Different Forms of Alkali Treatment on Specific Fermentation Inhibitors and on the Fermentability of Lignocellulose Hydrolysates for Production of Fuel Ethanol. J. Agric. Food Chem. 50, 5318–5325. doi:10.1021/jf025565o
Ragauskas, A. J., Beckham, G. T., Biddy, M. J., Chandra, R., Chen, F., Davis, M. F., et al. (2014). Lignin Valorization: Improving Lignin Processing in the Biorefinery. Science. 344, 1246843. doi:10.1126/science.1246843
Sobreira, T. J. P., Avramova, L., Szilagyi, B., Logsdon, D. L., Loren, B. P., Jaman, Z., et al. (2020). High-throughput Screening of Organic Reactions in Microdroplets Using Desorption Electrospray Ionization Mass Spectrometry (DESI-MS): Hardware and Software Implementation. Anal. Methods. 12, 3654–3669. doi:10.1039/d0ay00072h
Tramontina, R., Brenelli, L. B., Sodré, V., Franco Cairo, J. P., Travália, B. M., Egawa, V. Y., et al. (2020). Enzymatic Removal of Inhibitory Compounds from Lignocellulosic Hydrolysates for Biomass to Bioproducts Applications. World J. Microbiol. Biotechnol. 36 (11), 166. doi:10.1007/s11274-020-02942-y
Van den Ent, F., and Löwe, J. (2006). RF Cloning: A Restriction-free Method for Inserting Target Genes into Plasmids. J. Biochem. Biophysical Methods 67 (1), 67–74. doi:10.1016/j.jbbm.2005.12.008
Wang, J., Liang, J., and Gao, S. (2018). Biodegradation of Lignin Monomers Vanillic, P-Coumaric, and Syringic Acid by the Bacterial Strain, Sphingobacterium Sp. HY-H. Curr. Microbiol. 75 (9), 1156–1164. doi:10.1007/s00284-018-1504-2
Watanabe, T., Katayama, S., Enoki, M., Honda, Y., and Kuwahara, M. (2000). Formation of Acyl Radical in Lipid Peroxidation of Linoleic Acid by Manganese-Dependent Peroxidase From Ceriporiopsis Subvermispora and Bjerkandera Adusta. Eur. J. Biochem. 267, 4222–4231. doi:10.1046/j.1432-1033.2000.01469.x
Wen, Z., Zhang, S., Odoh, C. K., Jin, M., and Zhao, Z. K. (2020). Rhodosporidium Toruloides - A Potential Red Yeast Chassis for Lipids and beyond. FEMS Yeast Res. 20 (5), foaa038. doi:10.1093/femsyr/foaa038
Yee, K. L., Jansen, L. E., Lajoie, C. A., Penner, M. H., Morse, L., and Kelly, C. J. (2018). Furfural and 5-Hydroxymethyl-Furfural Degradation Using Recombinant Manganese Peroxidase. Enzyme Microb. Technology. 108, 59–65. doi:10.1016/j.enzmictec.2017.08.009
Zhao, X., Peng, F., Du, W., Liu, C., and Liu, D. (2012). Effects of Some Inhibitors on the Growth and Lipid Accumulation of Oleaginous Yeast Rhodosporidium Toruloides and Preparation of Biodiesel by Enzymatic Transesterification of the Lipid. Bioproc. Biosyst. Eng. 35 (6), 993–1004. doi:10.1007/s00449-012-0684-6
Zhu, D., Dai, L., Shao, X., Zhou, Q., Itti, L., Luo, Y., et al. (2017). Image Salient Object Detection With Refined Deep Features via Convolution Neural Network. J. Electron. Imaging. 26 (6), 1. doi:10.1117/1.JEI.26.6.063018
Zhu, Z., Ding, Y., Gong, Z., Yang, L., Zhang, S., Zhang, C., et al. (2015). Dynamics of the Lipid Droplet Proteome of the Oleaginous Yeast Rhodosporidium Toruloides. Eukaryot. Cell. 14 (3), 252–264. doi:10.1128/EC.00141-14
Zhu, Z., Zhang, S., Liu, H., Shen, H., Lin, X., Yang, F., et al. (2012). A Multi-Omic Map of the Lipid-Producing Yeast Rhodosporidium Toruloides. Nat. Commun. 3, 1112. doi:10.1038/ncomms2112
Keywords: Rhodosporidium toruloides, microbial lipids, lignocellulose, inhibitors, genetic engineering, screening
Citation: Lyu L, Chu Y, Zhang S, Zhang Y, Huang Q, Wang S and Zhao ZK (2021) Engineering the Oleaginous Yeast Rhodosporidium toruloides for Improved Resistance Against Inhibitors in Biomass Hydrolysates. Front. Bioeng. Biotechnol. 9:768934. doi: 10.3389/fbioe.2021.768934
Received: 01 September 2021; Accepted: 01 October 2021;
Published: 15 November 2021.
Edited by:
Rongming Liu, University of Colorado Boulder, United StatesReviewed by:
Qiuqiang Gao, Columbia University, United StatesDavid T. Stuart, University of Alberta, Canada
Copyright © 2021 Lyu, Chu, Zhang, Zhang, Huang, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liting Lyu, bGx0QGRpY3AuYWMuY24=; Zongbao K. Zhao, emhhb3piQGRpY3AuYWMuY24=
 Liting Lyu
Liting Lyu Yadong Chu
Yadong Chu Sufang Zhang1
Sufang Zhang1 Qitian Huang
Qitian Huang Zongbao K. Zhao
Zongbao K. Zhao