- 1Department of Microbiology, Gulbarga University, Gulbarga, India
- 2Department of Toxicology, Chaudhary Charan Singh University, Meerut, India
- 3Department of Life Science, School of Life Sciences, Central University of Karnataka, Kadaganchi, India
- 4Department of Psychology, School of Social and Behavioural Sciences, Central University of Karnataka, Kadaganchi, India
- 5Department of Biotechnology, University of Mysore, Mysore, India
- 6Sanorva Biotech Pvt. Ltd., Mysuru, India
A biosurfactant producing bacterium was identified as Pseudomonas aeruginosa DNM50 based on molecular characterization (NCBI accession no. MK351591). Structural characterization using MALDI-TOF revealed the presence of 12 different congeners of rhamnolipid such as Rha-C8-C8:1, Rha-C10-C8:1, Rha-C10-C10, Rha-C10-C12:1, Rha-C16:1, Rha-C16, Rha-C17:1, Rha-Rha-C10:1-C10:1, Rha-Rha-C10-C12, Rha-Rha-C10-C8, Rha-Rha-C10-C8:1, and Rha-Rha-C8-C8. The radical scavenging activity of rhamnolipid (DNM50RL) was determined by 2, 3-diphenyl-1-picrylhydrazyl (DPPH) assay which showed an IC50 value of 101.8 μg/ ml. The cytotoxic activity was investigated against MDA-MB-231 breast cancer cell line by MTT (4,5-dimethylthiazol-2-yl-2,5-diphenyl tetrazolium bromide) assay which showed a very low IC50 of 0.05 μg/ ml at 72 h of treatment. Further, its activity was confirmed by resazurin and trypan blue assay with IC50 values of 0.01 μg/ml and 0.64 μg/ ml at 72 h of treatment, respectively. Thus, the DNM50RL would play a vital role in the treatment of breast cancer targeting inhibition of p38MAPK.
Introduction
Biosurfactants are a group of amphipathic compounds that moderate the surface and interfacial tension of liquids (De almeida et al., 2016; Gudiña et al., 2016). They harbor saturated and unsaturated fatty acids as hydrophobic moieties along with polysaccharides, peptides, amino acids, and anions/cations as hydrophilic moieties. There are different groups of biosurfactants, out of which glycolipids are the most preferred due to their low molecular weight (Banat et al., 2014a). Also, they can be synthesized using hydrocarbons, agro-industrial waste, frying and olive oil wastes (Inès and Dhouha, 2015). Rhamnolipids (RLs) are the extensively studied glycolipid based biosurfactant due to its properties such as low critical micellar concentration (CMC: 10–200 mg/ L), surface tension reduction (up to 28–31 mN/ m), high emulsification index (60–70%), and high production in a short period of time (Gudiña et al., 2016).
Different microorganisms can produce different congeners or homologs of RLs by using a variety of sugars, hydrocarbons, and other oil/dairy product wastes (Shao et al., 2017; Pathania and Jana, 2020). Pseudomonas aeruginosa, a predominant microbial species, can produce various RL congeners with the most common ones as Rha-Rha-C10-C10, Rha-C10, Rha-C10-C10, and Rha-Rha-C10 (Hošková et al., 2015). RLs have gained importance in various industries such as food, healthcare, pharmaceuticals, and petrochemicals due to their wide range of properties along with enhanced oil recovery, biodegradation, and bioremediation. RLs are known to inhibit proliferation of cancer cells by inducing apoptosis. They regulate humoral and cellular immune response by acting as immunomodulators. They act as antimicrobial agents as they can lower the surface hydrophobicity, thus destroying the cytoplasmic membrane causing killing of bacterial cells. They are also involved in synthesis of nanoparticles. Thus, RLs have high biological applications because of its antimicrobial, anticancer, antioxidant, immunomodulatory property and their capability of nanoparticles synthesis (Banat et al., 2014b; Thakur et al., 2021).
Rhamnolipid is recovered from cell free culture broth by solvent extraction or acid precipitation method in partially purified form. Structural elucidation of any surface-active compound is of utmost importance and cannot be confirmed merely with TLC and FTIR as both of them carries some drawbacks and hence it should be accompanied by other methods such HPLC-MS/UPLC-MS and NMR spectroscopy (Twigg et al., 2020). MALDI-TOF is the emerging technology and can be used to characterize biosurfactants (Price et al., 2009). As RLs are composed of mono-RLs and di-RLs along with different congeners, MALDI-TOF has emerged as the screening strategy where new congeners can be determined at intimal stages (Sato et al., 2019).
Breast cancer is one of the most prominent malignant diseases in the world. Worldwide, there are 3,465,951 incidences and 1,121,413 deaths respectively, according to GLOBOCAN 2020. In 2020, 1,204,532 new cases and 436,417 deaths were recorded in India (Sung et al., 2021). Breast cancer affecting the female population continues to increase annually and there is no ideal treatment for the disease (Wu et al., 2017). Doxorubicin, etoposide, and mitoxantrone are some of the widely prescribed anticancer drugs. One of the major causes of high mortality in people with cancer is resistance developed to the classical chemotherapeutic agent, which is being considered to be a major hindrance, leading to major obstructions in cancer treatment (Housman et al., 2014; Wambang et al., 2016; Wang et al., 2019). Besides this, the limited solubility, stability, bio-distribution, and cytotoxic effect on normal cells significantly reduces the effectiveness of chemotherapeutic drugs. Thus, it is essential to create efficient alternative formulations, selectively targeting cancer cells without causing substantial harm to healthy cells (Haque et al., 2021). The life expectancy of cancer patients would be significantly influenced by the development of new chemotherapeutic/chemical substances/agents. RLs have recently been discovered as a promising antitumor agent interfering with the proliferation of cancer cells of different origins (Chen et al., 2017).
Rhamnolipids have shown a significant effect on human as well as animal cancer cells in the recent past. It is reported that di-RLs produced from Pseudomonas aeruginosa B189 inhibit the proliferation of a breast cancer cell line MCF-7 and an insect cell line C6/36 (Thanomsub et al., 2006). This anticancer property of RLs was further tested on various cell lines such as BV-173, SKW-3, HL-60, and JMSU-1 derived from tumors of different origin with IC50 values of 50–140 µM (Christova et al., 2013). RLs isolated from Pseudomonas aeruginosa MR01 inhibited HeLa cancer cells growth at the concentration of 5 μg/ ml (Loftabad et al., 2010). Burgos-Díaz et al. (2013) reported anticancer activity of glycolipid from Sphinobacterium detergens against colorectal cancer cell line. Trehalose lipid, a glycolipid from Mycobateria, was used as an antitumor agent (Fracchia et al., 2015).
Similarly, Rahimi et al. (2019) investigated the cytotoxic effect of RL-1 and RL-2 isolated from Pseudomonas aeruginosa MR01 against MCF-7, a breast cancer cell line. The results clearly indicate the antiproliferative effect of RLs, emphasizing induction of apoptosis. They reported increased expression of p53 gene in mRNA levels, indicating induction of cell cycle control by RLs in cancer cells. All these reports give insight into the use of RLs as a potential therapeutic antitumor agent.
TNBC, commonly referred to as triple negative breast cancer cell line, lacks expression of ER−a estrogen receptor, PR−a progesterone receptor, and HER 2-a human epidermal growth factor receptor 2 (Chavez et al., 2010). Poor long-term outcomes are associated with it, as compared to other breast cancer (Li et al., 2017). The MDA-MB-231 cell line also lacks expression of these receptors and thus is an aggressive and invasive breast cancer cell line (Cailleau et al., 1978). It is insensitive to anti-hormone-based therapies, including tamoxifen due, to the absence of ER expression (Osborne and Schiff, 2011). It accounts for approximately 15% of breast cancer diagnosed worldwide, which amounts to almost 200,000 cases each year. It is more commonly diagnosed in women younger than 40 years, compared with hormone receptor-positive breast cancer (Swain, 2008). These types of breast cancer have limited treatment options and thus it is commonly used for development of novel therapeutic approaches by exploring molecular basis of this type of breast cancer (Mohammed et al., 2020).
The involvement of p38MAPK signaling pathway, a group of stress-activated kinases in apoptotic cell death phenomenon, makes this pathway striking for cancer researchers (García-Cano et al., 2016). The inhibition of p38MAPK plays a remarkable role in cancer therapy. P38MAPK is a crucial player in response to chemotherapy, as apoptosis is the main mechanisms associated with it (Sanchez-Prieto et al., 2000; Cai et al., 2006). In few cases of colorectal cancer, experimental evidence supports the fact that p38MAPK inhibition by itself is a promising target in cancer therapy (Gupta et al., 2015; Igea and Nebreda 2015). In primary acute myeloid leukemia, the p38MAPK inhibition overcomes the resistance to compounds such as Birinpant (Lalaoui et al., 2016). Ongoing trials showed these inhibitors safety for ralimetinib and ARRY-614 (Garcia-Manero et al., 2015; Patnaik et al., 2016). The implication of active p38MAPK in cancer is not a genetic alteration in the MAPK, instead a pathological context of a tumor. Thus, it is advisable to search for P38MAPK inhibitor for cancer researchers (Pritchard and Hayward 2013). Hence, the use of p38MAPK can be considered as a potential target for cancer therapy. A new era of better prognosis and personalized treatment can be led by a deep understanding of the role of p38MAPK in cancer therapy.
The DNM50RL (a mixture of mono-RL and di-RL congeners) used in the present study was produced using Pseudomonas aeruginosa DNM50. MALDI-TOF was used to characterize RLs in view to identify its different congeners. Radical scavenging activity was assessed though DPPH assay. Cytotoxic effect of DNM50RL was assessed against MDA-MB-231 triple negative cell line through MTT, Resazurin, and Trypan blue assay. Since p38 plays a crucial role in anticancer therapy, inhibition of phosphorylated p38 was examined using Western blot analysis.
Materials and Methods
Chemicals Used
Chemicals and media used in the present study were obtained from Himedia (Mumbai, India), Sigma Aldrich Pvt. Ltd. (United States), and Merck and Co. Inc. (United States).
Characterization of Microbial Strain
Pseudomonas aeruginosa DNM50 used in the present study was previously isolated from oil contaminated soil sourced from railway tracks of Wadi, Kalaburagi, Karnataka, India, in A-DBT Research Laboratory, Gulbarga University, Kalaburagi. Seven different screening methods were used to confirm its efficiency for biosurfactant production, and the data was reported in our earlier study (Mishra et al., 2021).
Pseudomonas aeruginosa DNM50 was identified and characterized using 16 S rRNA sequencing. Spin column kit (Qiagen, Hilden, Germany) was used to extract chromosomal DNA while purification of bacterial 16 S rRNA (1500) was done by using exonuclease I-Shrimp Alkaline phosphatase (Exo-SAP) (Darby et al., 2005). For the amplification of 200 ng/ µL of 16 S rRNA genes, PCR technique was applied using universal primers, 27F (AGAGTTTGATC(C/A)TGGCTCAG) and 1492R (TACGG (C/T) TACCTTGTTACGACTT) (Clarridge 2004; Mulla et al., 2016; 2018). Amplicons of PCR amplified genes were sequenced by following the method of Sanger (Sanger et al., 2000) using ABI 3500xL genetic analyzer (Life Technologies, United States). Sequencing files (.ab1 format) were edited with CHROMASLITE (version1.5) and then analyzed with the Basic Local Alignment Search Tool (BLAST) against the closest culture sequence retrieved from the National Centre for Biotechnology Information (NCBI) database, which identifies regions of local similarity between sequences. The evolutionary history and evolutionary analysis were deduced by Neighbor-Joining method and MEGA 6, respectively (Altschul et al., 1990; Tamura et al., 2013).
Characterization of Rhamnolipids
The RLs produced by Pseudomonas aeruginosa DNM50 under submerged process was subjected for its chemical characterization. The production medium used was neem oil cake extract (19.5%), as a sole source of nutrition. The extract of neem oil cake (pH 7.5) was prepared (Amena et al., 2010), sterilized and inoculated with 10% of 24 h culture followed by incubation at 40°C with agitation of 200 rpm for 5 days. Growth stages of the culture, production levels of RL, and surface tension (SFT) reduction of the production medium were recorded for a period of 5 days. All experiments were carried out in triplicates. The mean values were plotted along with standard deviation (Hazra et al., 2010; Samykannu and Achary, 2017; Chopra et al., 2020).
Cell free broth was used for extraction of RLs as per the modified procedure described by Chandankere et al. (2013). After 3 days of fermentation, culture broth was centrifuged for 10 min at 10,000 rpm. The cell free broth was filtered through 0.45 µM filter and extracted three times with equal volume of chloroform. The organic phase was collected, and solvent evaporated using a rotavapour. The resulting brownish semisolid compound was dried in an oven at 70°C and subjected to TLC (compared with reference standard of RLs from AGAE technologies, USA). The extracted RL (DNM50RL) was further analyzed by HPLC (not reported here) and MALDI—TOF.
Thin Layer Chromatography
For thin layer chromatography, a sample of DNM50RL was dissolved in chloroform and 20 µL of the aliquot was applied to pre-coated silica gel (F 1500 LS. 254; Schleicher-Schull, Germany). A mixture of butanol:acetic acid:water in the ratio of 2:1:1 (v/v/v) was used as the mobile phase. Molisch reagent (α-naphthol in ethanol with 10% H2SO4), ninhydrin reagent, and iodine vapors were used for detection of carbohydrate, proteins, and lipids, respectively (Loftabad et al., 2010).
Matrix Assisted Laser Desorption/Ionization Time of Flight
To elucidate the structure of RLs, MALDI-TOF was performed using an Applied Biosystem Ultraflextreme, Bruker Daltonic Germany Mass Spectrometer in reflection mode (Maiqian et al., 2010).
Antioxidant Activity
DPPH Assay
The 2,2,-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging potential of DNM50RL was examined according to the method described by Li et al. (2014). A concentration range of 0.00625–5 mg/ml was used for both DNM50RL and ascorbic acid (standard). The absorbance at 517 nm determines the reduction of DPPH radical and the calculation for radical scavenging activity was done as follows,
DPPH radical scavenging % = [(A0-A1)/A0] x 100. where DPPH absorbance is denoted by A0 and absorbance of sample is denoted by A1.
The antioxidant property is measured in % radical scavenging activity and the results are presented as the average of three independent experiments.
The IC50 (effective concentration to scavenge DPPH radical) value of DNM50RL was determined by GraphPad prism software version 9.1.0.221. The dose response curve is plotted between DPPH radical scavenging activity and the concentration.
Cytotoxic Activity
Cell Cultures
MDA-MB-231, a tumorigenic, invasive, metastatic, and TNBC representative cell line was procured and authenticated from National Centre for Cell Sciences (NCCS), Pune, India. The cells were cultured in L-15 (Leibovitz’s 15) medium with 2 mM L-glutamine and 10% FBS (fetal bovine serum) and were maintained at 37°C.
MTT Assay
5 × 104 cells (MDA-MB-231) were cultured in 96-well plate (Nunc MicroWell™) in complete Dulbecco’s Modified Eagle Medium (DMEM) medium and incubated overnight at 37°C in 5% CO2. Cells were exposed to different concentrations (0.01, 0.05, 0.1, 0.5, 1.0, and 5.0 μg/ ml) of compound in triplicates with vehicle control (0.05% DMSO served as negative control) and positive control (Etoposide in the concentration range of 1—1000 µM). 24 h of post-treatment, 10 μL of 4,5-dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide (MTT) (5 mg/ ml) was added in each well and incubated for 4 h at 37°C. The resultant purple formazan crystals were solubilized in 150 μL of DMSO. The color development was recorded at a test wavelength of 570 nm and a reference wavelength of 630 nm (Varioskan™ Flash Multimode Reader, Thermo Scientific, Switzerland). The cytotoxicity in percentage was derived against control cells as 100% (Riss et al., 2016; Byrappa et al., 2017). The cytotoxicity measurements were depicted in the dose-response curve for a period of 24, 48, and 72 h. The IC50 value (half-maximal inhibition concentration) was determined using GraphPad Prism software version 9.1.0.221 using nonlinear regression (curve fit).
Resazurin Assay
5 × 104 MDA-MB-231 cells were grown in 96-well plate (Nunc MicroWell™) in complete L-15 medium and incubated overnight at 37°C. Cells were treated with increasing concentrations (0.01, 0.05, 0.1, 0.5, 1.0, and 5.0 μg/ml) of the compound in triplicates with vehicle control (0.05% DMSO served as negative control) and a positive control (Etoposide in the concentration range of 1—1000 µM). After 24 h of treatment, cells were incubated with 100 μL of resazurin sodium salt (0.01 g/2 ml sterile PBS—phosphate buffered saline) at 37°C for 4 h. The resultant change from blue to pink color indicates the presence of viable cells. The optical density of the colored solution was determined at a reference wavelength of 630 nm and test wavelength of 570 nm (Infinite Pro Tecan Multimode reader, Thermo Scientific, Finland). The percentage of cytotoxicity was calculated against control cells as 100% (Riss et al., 2016; Byrappa et al., 2017). The cytotoxicity measurements were analyzed in the dose-response curve for 24, 48, and 72 h. The IC50 value (half-maximal inhibition concentration) was demonstrated using GraphPad Prism software version 9.1.0.221 using nonlinear regression (curve fit).
Trypan Blue Assay
5×104 MDA-MB-231 cells were seeded in 6-well plate in complete L-15 medium and cells were treated with increasing concentrations (0.01, 0.05, 0.1, 0.5, 1.0, and 5.0 μg/ ml) of the compound with control. After 24 h of treatment, cells were stained with trypan blue (0.4% trypan blue in PBS—phosphate buffered saline, pH 7.2–7.3). A mixture of treated cells (10 µL), trypan blue (10 µL), and PBS (980 µL) were resuspended and 10 µL of the mixture was loaded from the edge of a coverslip placed on a hemocytometer. The number of viable cells/mL was enumerated by using the following formula.
Average of cells counted from four squares x 10,000 x dilution factor (i.e., 100)
= Average cells count x 106 cells.
Cell viability (%) = (Number of live cells ÷ Number of total cells) × 100.
Three independent readings were taken. All the measurements were depicted in the dose-response curve for 24, 48, and 72 h (Phillips, 1973). Etoposide in the concentration range of 1–1000 µM served as the positive control along with 0.05% DMSO as negative control. The IC50 value (half-maximal inhibition concentration) was determined using GraphPad Prism software version 9.1.0.221 using nonlinear regression (curve fit).
Western Blotting for Regulation of P38
MDA-MB-231 cells were revived and grown until 80% confluent, the cells were trypsinized (trypsin-EDTA 0.25%), split, and then seeded into sterile Petri dishes. After 80–90% confluency, the media was aspirated, and cells were starved overnight with basal media. MDA-MB 231 cells were treated with 2 mM sodium orthovanadate for 2 h, followed by 5 µg of compound for 24, 48, and 72 h. The cell lysate was prepared using chilled lysis RIPA buffer. Cell lysate of 24, 48, and 72 h was collected. After estimating protein (Bradford, 1976), the supernatant was loaded on gel (150 µg/well) and processed for Western blotting using a semidry blotter. The unwanted protein binding sites on the membrane were blocked by incubating membrane in 5% bovine serum albumin (BSA) for 2 h at room temperature with moderate agitation. Blot was washed three times with TBST buffer (tris-buffered saline, 0.1% tween 20) for 10 min each. Membrane was incubated overnight at 4°C with moderate agitation in primary antibody (phosphorylated p38, polyclonal rabbit antibody), diluted as 1:1000 ratio in blocking buffer. After washing the blot, the blot was incubated with secondary goat anti-rabbit HRP tagged antibody in 1:3000 ratio for 2 h at room temperature with gentle agitation. Following washing, the blot was developed using enhanced chemiluminescence (ECL). Blot was stripped and re-probed with total p38 antibody to show equal loading in all blots. β actin served as the loading control. Membrane was washed with TBST extensively and continued with blocking procedure. Quantification of protein bands were determined by ImageJ software (Towbin et al., 1979).
Statistical Analysis
All the results were expressed as the Mean ± SD values obtained in triplicate from three independent tests. SPSS version 22.0 was used to perform all statistical analysis. One-way ANOVA followed by Tukey’s HSD Post-Hoc test and Student’s t test were used to make multiple comparisons with control groups. *p < 0.05 represents statistically significant values.
Results and Discussion
Strain Characterization
The isolated genomic DNA of the potential isolate was amplified to obtain a PCR product, which was further purified. It was then subjected to 16SrRNA gene sequencing and identified by phylogenetic analysis. The consensus sequences available from National Centre for Biotechnology Information (NCBI) were used to perform Nucleotide Blast (Blast-n). Sequence alignment was done using CLUSTAL–W and a phylogenetic tree was constructed using MEGA 6 software as shown in Figure 1. The potential isolate DNM50 was identified based on phylogenetic analysis and designated as Pseudomonas aeruginosa DNM50 (99% similarity with Pseudomonas aeruginosa). The sequence was submitted to NCBI and the GenBank accession number MK351591 was obtained.
Characterization of Rhamnolipids
A linear increase in the growth of the culture was observed up to 60 h and was declined further. Maximum production of RLs (2.9 g/ L) was achieved at 72 h with no significant increase up to 120 h. A consistent decrease in SFT (from 62 mN/ m to 30 mN/ m) of the medium was recorded up to 120 h (Supplementary Figure S1).
Thin layer chromatogram (Figure 2A) of RLs produced by Pseudomonas aeruginosa DNM50 showed two characteristic spots corresponding to standard RLs on reaction with Molisch reagent with retardation factor (Rf) values of 0.84 and 0.68 confirming the presence of mono-RLs and di-RLs, respectively. Positive reaction with Molisch reagent and iodine vapors ratifies glycolipid nature of RLs. No spots were observed with ninhydrin reagent indicating absence of amino acids/proteins. Many researchers reported similar RLs mixture with varying composition (Haba et al., 2003; Silva et al., 2010; Abbasi et al., 2012).
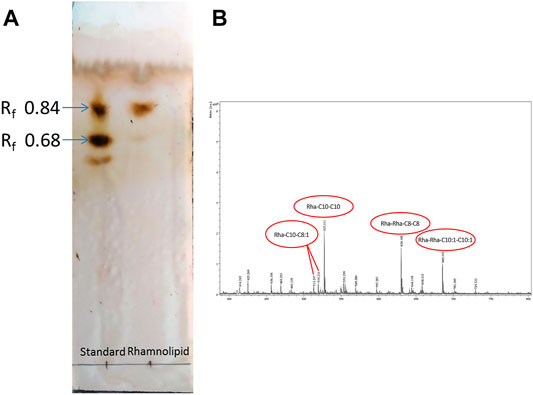
FIGURE 2. Chemical characterization of Rhamnolipid (DNM50RL) produced from Pseudomonas aeruginosa DNM50 by (A) thin-layer chromatography (TLC): TLC plate run in a solvent system of butanol:acetic acid:water (2:1:1), developed with Molisch reagent (α—naphthol in ethanol and H2SO4) showing typical brown color spots of Rhamnolipid. (B) MALDI TOF peaks.
MALDI-TOF (Figure 2B) confirmed the presence of mono-RL and di-RL congeners, specifically Rha-C8-C8:1, Rha-C10-C8:1, Rha-C10-C10, Rha-C10-C12:1, Rha-C16:1, Rha-C16, Rha-C17:1, Rha-Rha-C10:1-C10:1, Rha-Rha-C10-C12, Rha-Rha-C10-C8, Rha-Rha-C10-C8:1, and Rha-Rha-C8-C8 as shown in Table 1. The spectrum was characterized by molecular ions for mono-RLs (m/z 470-575) and di-RLs (m/z 640-730). Major ions at m/z 527 were attributed to [M + Na]+ adduct ions of the major mono-RL, Rha-C10-C10 and m/z 630 and 685 were attributed to [M + K]+ adduct ions of the major di-RLs, Rha-Rha-C8-C8, and Rha-Rha-C10:1-C10:1, respectively. These findings were in accordance with the work previously reported for analysis of RLs produced by different strains of Pseudomonas aeruginosa (Price et al., 2009; Sarachat et al., 2010; Abeer Mohammed et al., 2018).
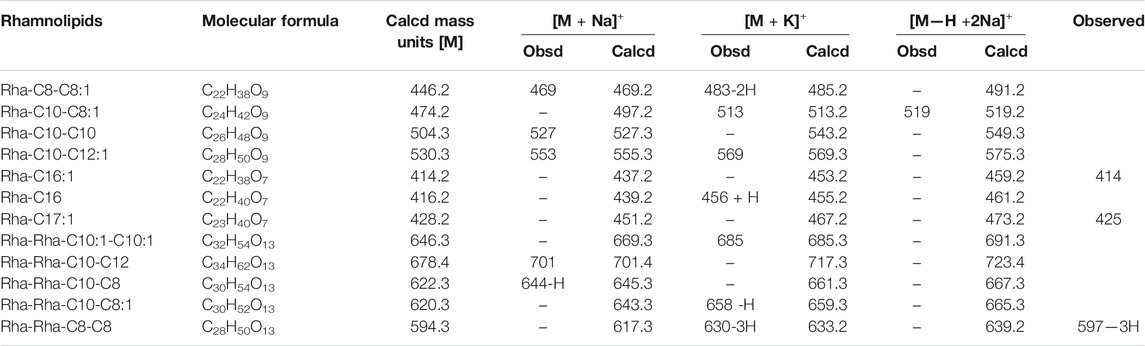
TABLE 1. Molecular ions observed in MALDI-TOF for Rhamnolipids produced by Pseudomonas aeruginosa DNM50.
Based on chromatographic and spectroscopic analyses, it can be inferred that the biosurfactant produced by Pseudomonas aeruginosa DNM50 is comprised of congeners of mono-RLs and di-RLs.
Radical Scavenging Attribute
DPPH assay was developed to examine the antioxidant activity, which relies on the measurement of scavenging ability of antioxidants to a stable free radical, DPPH. Electron of nitrogen atom from DPPH was reduced to corresponding hydrazine by a hydrogen atom from the antioxidant. Reduced form of DPPH shows a color change from deep violet to yellow, thus assessing antioxidant activity of the given compound (Kedare and Singh, 2011). The DPPH radical scavenging activity of DNM50RL and ascorbic acid was as shown in Figures 3A,B.
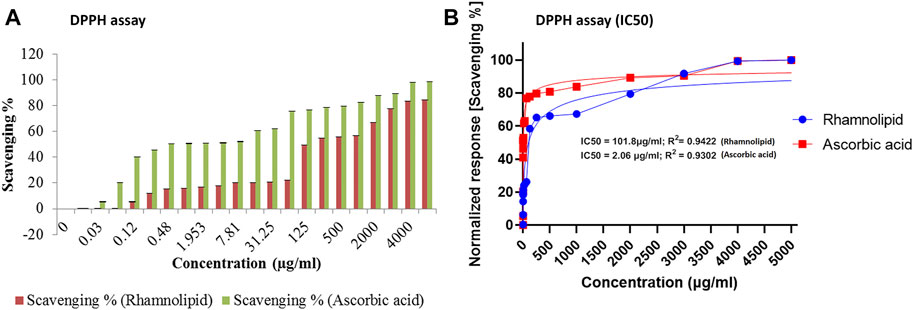
FIGURE 3. The DPPH radical scavenging activity of DNM50RL and ascorbic acid (A) Scavenging %. Values represented as mean ± SD of three independent experiments. Error bars represent SD (*p < 0.05). (B) Non-linear regression curve fit (DPPH assay). IC50 determined by nonlinear regression (curve fit [Inhibitor] vs. normalized response–Variable slope) using GraphPad prism software.
At 5 mg/ml concentration, DNM50RL and ascorbic acid showed a radical scavenging activity of 84.25 and 98.4%, respectively. This activity increases with an increase in concentration. The IC50 value was 101.8 μg/ml for DNM50RL and 2.064 μg/ml for ascorbic acid as determined from GraphPad Prism software. At 1 mg/ml, MCTG107b and MCTG214 (3b1) derived biosurfactant exhibited only 9.67 ± 3.27% and 15.46 ± 4.03% inhibition for DPPH assay (Voulgaridou et al., 2021).
DPPH radical scavenging activity of 69.1 and 73.5% were reported at 5.0 mg/ml of BS-VSG4 and BS-VS16 biosurfactants from Bacillus strains (Giri et al., 2019). An IC50 value of 4.15 mM for RLs from Pseudomonas aeruginosa was deduced (Abdollahi et al., 2020). Similarly, IC50 value of 357 μg/ml was observed for DCS1 lipopeptide biosurfactants produced from Bacillus methylotrophicus (Jemil et al., 2017). The DNM50RL has shown weak antioxidant activity than standard ascorbic acids but is better than the activity reported by other related studies. The radical scavenging activity for biosurfactants was due to the transfer of protons or electrons, thus neutralizing free radicals. This activity was enhanced by hydrocarbon fatty acid in RLs and was not affected by their variation in chain length (Tofani et al., 2010; Tabbene et al., 2012).
Cytotoxic Attribute
Cell viability assays such as dye exclusion, colorimetric, fluorometric, luminometric, and flowcytometric are being used by various researchers. However, measurement of cell viability/proliferation cannot be examined by only one method, since it is influenced by types and origins of cell lines and the mechanism of action of the cytotoxic agents explored. Thus, it is recommended to apply more than one assay for cell viability/proliferation (Kamiloglu et al., 2020).
MTT assay was performed to assess the antiproliferative effect of different concentrations of DNM50RL against MDA-MB-231 cell lines. There is a linear relationship between cell activity and absorbance measuring growth rate of cells, thus MTT is a quantitative and sensitive detection of cell proliferation (Mahajan et al., 2012). MTT can be reduced through the mitochondrial enzyme, which is possible only in the cells with active metabolism. Hence, measurement of cell proliferation is indicated directly by the number of metabolically active/viable cells. With an increase in the concentration of DNM50RL, there is an increase in inhibition of cell proliferation at 24 h. Similar dose-dependent inhibition was observed at 48 and 72 h, with the best results obtained when treatment was given for 72 h showing an IC50 value of 0.05 μg/ml (Figures 4A,B).
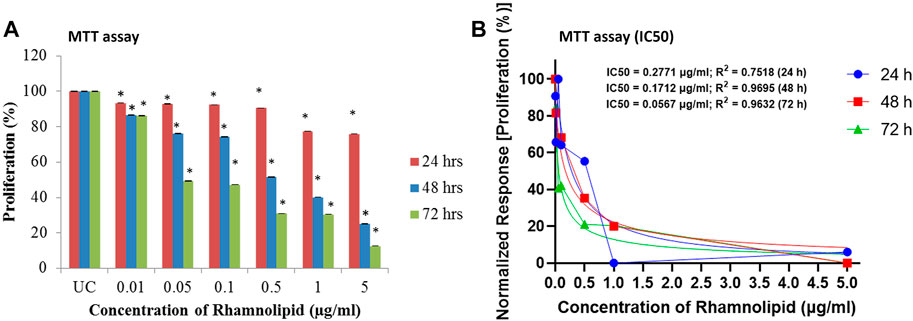
FIGURE 4. Antiproliferative effect of DNM50RL on MDA-MB 231 cell line by MTT assay after 24, 48, and 72 h of treatment. (A) Dose dependent inhibition. Values represented as mean ± SD of three independent experiments. Error bars represent SD (*p < 0.05). (B) IC50 determination by nonlinear regression [curve fit (Inhibitor) vs. normalized response–Variable slope] using GraphPad prism software.
In resazurin assay, viable cells with active metabolism cause reduction of resazurin to resofurin product, which is pink and fluorescent (Riss et al., 2016). The assay is more sensitive than the MTT assay (Shum et al., 2008), hence it is used to confirm further the antiproliferative activity of DNM50RL against MDA-MB-231 cell lines. A dose dependent trend was observed with IC50 value of 1.5 μg/ml at 48 h of treatment, though the best effect was observed at 72 h of treatment with IC50 of 0.01 μg/ml (Figures 5A,B).
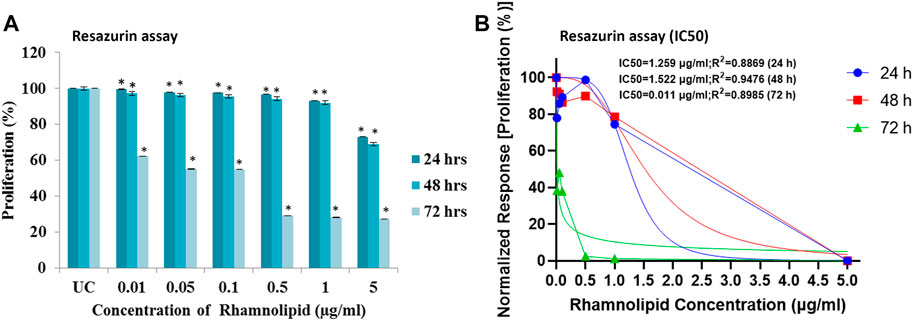
FIGURE 5. Antiproliferative effect of DNM50RL on human breast cancer cell lines MDA-MB- 231 cell line by resazurin assay after 24, 48, and 72 h of treatment. (A) Dose dependent inhibition. Values represented as mean ± SD of three independent experiments. Error bars represent SD (*p < 0.05). (B) IC50 determined by nonlinear regression [curve fit (Inhibitor) vs. normalized response–Variable slope] using GraphPad prism software.
Trypan blue dye exclusion assay was further used for evaluation of cytotoxicity activity of DNM50RL. This assay easily differentiates live and dead cells as the dead cells take up the dye and live cells exclude trypan blue, hence the name. It is because live cells will retain their membrane intact and dead cells will lose membrane integrity due to apoptosis/necrosis (Farghadani et al., 2017). Figure 6A shows the decrease in cell viability in 72 h by DNM50RL in a dose dependent manner confirming its role in cytotoxicity activity against MDA-MB-231 cell line (Figure 6B). When the treatment was given for 72 h, the IC50 value was 0.64 μg/ml. The IC50 value of DNM50RL was lower than the positive control, Etoposide (IC50 value of 20.65, 25.77, 51.10 µM by MTT, resazurin and Trypan blue assay at 72 h of treatment) (Supplementary Figure S2).
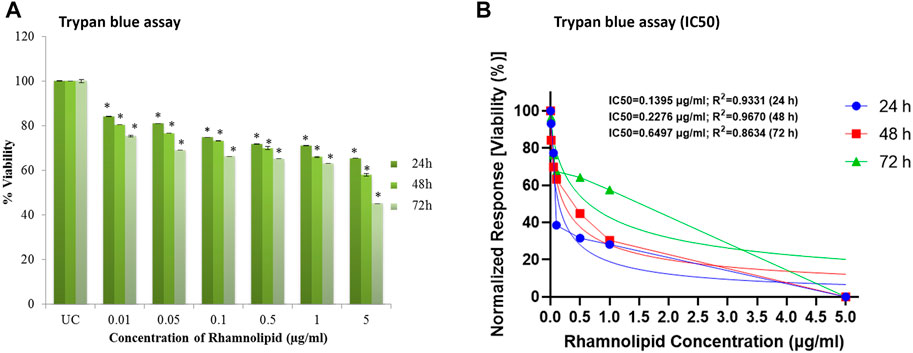
FIGURE 6. Cell viability efficacy of DNM50RL on human breast cancer cell lines MDA-MB-231 cell line by trypan blue assay after 24, 48, and 72 h of treatment. (A) Dose dependent inhibition. Values represented as mean ± SD of three independent experiments. Error bars represent SD (*p < 0.05). (B) IC50 determined by nonlinear regression [curve fit (Inhibitor) vs. normalized response–Variable slope] using GraphPad prism software.
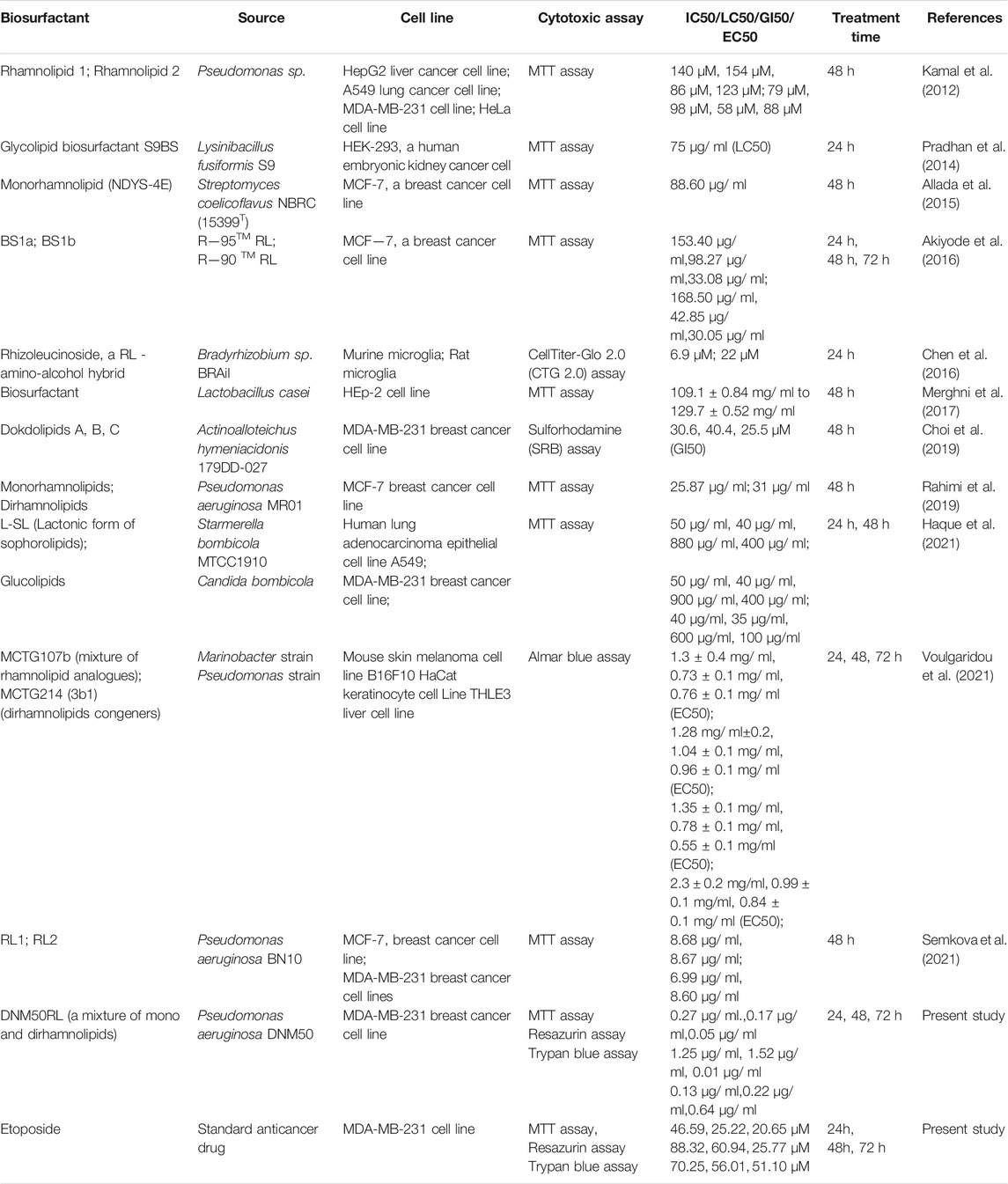
A number of researchers worked on cytotoxic effect of different glycolipid based biosurfactants against different cell lines (Table 2). However, there is limited data available on the cytotoxic effect of RLs against MDA-MB-231 cell line.
It can be inferred from the results of all cell proliferation/viability assays that DNM50RL is inhibiting proliferation of MDA-MB-231 cell line in a dose dependent and time dependent manner. Moreover, few reports showed that mono-RLs are more effective for inhibition of breast cancer cells and some emphasize di-RLs to be more effective (Thanomsub et al., 2006; Jiang et al., 2014; Rahimi et al., 2019). Due to such variations found in the biological potentials of purified mono-RLs and di-RLs from previous reports, the present study was aimed to reveal the biological attributes of natural RLs in their native form. This is because an appropriate proportion/mixture of mono-RLs and di-RLs may have much pronounced activity than the individual congeners. Many researchers reported a lack of cytotoxic activity of RLs against normal human cells, e.g., Vero cells, PBMN-peripheral blood mononuclear cells. This may be due to the presence of more negatively charged functional groups on cancer cells and its variation in fatty acid profiles with respect to normal cells, which facilitates greater endocytosis activity of cancer cells (Gudiña et al., 2013; Dwivedi et al., 2015). Reports from various studies on RLs against different cancerous and normal healthy cells show discrete results. This may be due to the nature of normal and cancerous cells studied, purity and types of RL variants, and the degree of surface tension reduction (Rahimi et al., 2019).
Downregulation of P38MAPK
Western blot analysis was done to compare the phosphorylated (p-p38MAPK) and total (t-p38MAPK) p38MAPK protein levels after treatment with DNM50RL (5 μg/ ml). This was done to determine whether p38MAPK is responsible for DNM50RL cytotoxicity in triple negative breast cancer cell line MDA-MB-231. As shown in Figures 7A,B, the protein levels of total p38 were higher in these cells until 72 h in control and treated cells (lower panel) while the phosphorylated p38 was downregulated (upper panel).
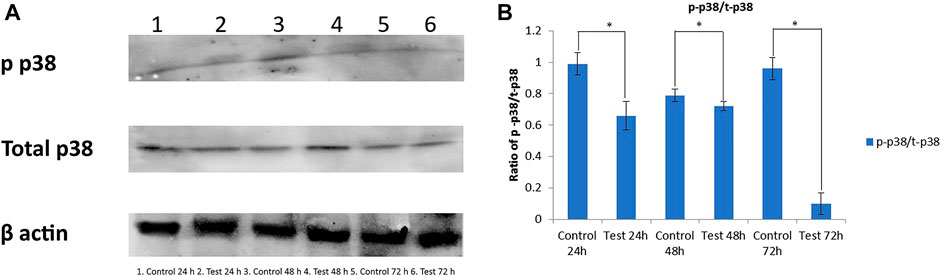
FIGURE 7. (A) Western blot for antibody phosphorylated p38 and total p38. β actin served as loading control. (B) Graph representing the ratio of phosphorylated p38 (p-p38)/total p38 (t-p38). The intensity related to expression of p-p38 and t-p38 was analyzed using ImageJ software. All the results are presented as the means ± SD of three independent measurements. Error bars represent standard deviation. (*) Differences were considered significant at p < 0.05 as compared with control groups.
Several studies showed that p38 mitogen activated protein kinases (MAPKs) play a critical role in cellular responses, proliferation, survival, cell cycle, and migration in cancer. As DNM50RL can downregulate p38 phosphorylation, it can be proposed as a novel and potent anticancer agent with its effectiveness at a very low concentration. RL a (Rha-Rha-C10-C10) isolated from Pseudomonas aeruginosa B189 inhibited the proliferation of MCF-7 cells, human breast cancer cell lines with a MIC of 6.25 μg/ml, whereas RL b (Rha-Rha-C10-C12) did not exhibit significant cytotoxic activity. At 50 μg/ml, Rha-Rha-C10-C10 or Rha-Rha-C10-C12 did not show cytotoxic effect against Vero cells, a normal cell line. Thus, this report demonstrates the toxic sensitivity of different congeners of these RLs to other cell lines used (Thanomsub et al., 2006).
Di-rhamnolipids (Rha-Rha-C10-C10 as a major component) produced by Pseudomonas aeruginosa M14808 showed antiproliferative efficiency against MCF-7 and H460, a human non-small lung cancer line with a MIC of 1 μg/ml and 5 μg/ml, whereas mono-RLs (Rha-C10-C10 as a major component) and crude extract did not show any antiproliferative activity (Zhao et al., 2013).
Similar sensitivity of RLs (mono-RLs and di-RLs) against two cancer cell lines, HepG2 (a human hepatocellular carcinoma cell line), Caco-2 (a human colon carcinoma cell line), and a normal cell HK-2 (a human proximal tubular epithelial cell line) were reported (Jiang et al., 2014). Mono-RLs presented higher cytotoxicity than di-RLs, which is due to large surface tension reduction by mono-RLs than di-RLs. They stated that similar sensitivity was observed for normal as well as cancerous cell lines with or without fetal bovine serum (FBS). Addition of FBS attenuates the cytotoxic effect of RLs. These observations revealed that RLs reduce surface tension of the culture medium, eliciting its cytotoxic effect, instead of causing any modification in molecular structure. Further, mainly decreased cytotoxic effect was observed in vivo as RLs get dissolved in simulated gastric juice. Thus, further research to illustrate high level of biosafety of RLs in vivo offers valuable suggestions for cancer researchers (Jiang et al., 2014).
An efficient cytotoxic potency of RLs (Rha-Rha-C10-C10 and Rha-Rha-C10) isolated from Pseudomonas sp. strain ICTB -745 was reported against different human cancer cell lines, e.g., lung adenocarcinoma, hepatocellular liver carcinoma, breast adenocarcinoma, and cervical cancer cell lines (Kamal et al., 2012). Mono-RLs (Rha-C10-C10, as a major component) from Pseudomonas aeruginosa BN10 had noticeable cytotoxic activity against human urinary bladder carcinoma cell line, human promyelocytic leukemia cell line, human pre-B leukemia cell line, and T-cell chronic lymphocytic leukemia cell line. The probable reason may be the structure of mono-RLs molecule, causing high penetration in tumor cell, leading to cell viability reduction. Mono-RLs induced cell death, are mediated by induction of apoptosis, inactivation of cellular oncogenes or activation/alteration of tumor suppressor process. The dose dependent apoptotic progression of mono-RLs may be due to efflux pump or the receptors present on cell membrane gets saturated, interfering with the permeation of active molecules within the cancerous cells (Christova et al., 2013). Bcl-2 and c-myc gene overexpression in human BV-173 pre-B leukemia cell line were observed with high doses of mono-RLs, resulting in high proliferative and anti-apoptotic effect, which is overcome by prolonged incubation period (≥72 h). Low cytotoxicity against Balb/c 3T3, a non-tumorigenic mouse fibroblast cell line was confirmed by Balb/c 3T3 neutral red uptake test (Christova et al., 2013).
RL-AgNPs from Pseudomonas aeruginosa JS-11, at concentration of 10 μg/ ml, showed a remarkable decrease in cell viability of MCF-7 breast cancer cell lines and no significant effect on PBMN—a normal human peripheral blood mononuclear cell was reported. It might be because the anionic metal nanoparticles cause greater attachment into plasma membrane of MCF-7 cells due to the presence of a negatively charged group on the cancer cell surface. This causes greater endocytic activity causing internalization of metal nanoparticles through vesicular transport pathway. Thus, the RL-AgNPs attachment to the cell surface leads to oxidative stress and cell membrane disruption, which assures it as a primary candidate for novel cancer therapy (Dwivedi et al., 2015).
Although many reports are available showing the anticancer activity of the RLs against various other cancer cell lines, there is limited studies available on MDA-MB-231 cell lines. Also, the mechanism of action of RLs at the molecular level is still insufficiently understood. A future study to elucidate the mechanism is needed to learn its biochemical and cellular interaction followed by preclinical studies and clinical trials at different stages (Abbasi et al., 2013; Abalos et al., 2017). The structure and activity relationship studies of pure or mixture of RLs would also offer advantages for researchers to elucidate their biological characterization and predict the biological function of various RL congeners in combination or in particular (Chen et al., 2017).
Conclusion
Lower toxicity and higher biodegradability attributes of biosurfactants are always advantageous over chemical surfactants for significant biotechnological applications. Isolated and identified strain of Pseudomonas aeruginosa DNM50 in the present study is novel due to the significant cytotoxic effect of its RL against triple negative breast cancer cell lines. Twelve different congeners were detected by MALDI-TOF, which contributes to the functional efficiency of RLs. DNM50RL seems to be a potential molecule in the future for cancer therapy due to its very low IC50 value against MDA-MB-231 cell line and its significant role in inhibition of p38MAPK. Therefore, the DNM50RL can be a prospective natural, therapeutic anticancer agent.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
NM, DA, and BS designed the experiments. NM, SD, and KR performed the experiments. NM, RK, and VP performed the data analysis and wrote the manuscript. DA and BS critically reviewed the whole manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by the Department of Science and Technology, Women Scientist Scheme-A (SR/WOS-A/LS-262/2017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
NM and DA thank BS, Department of Biotechnology, University of Mysuru, Manasgangothri, Mysuru for extending laboratory facilities. NM and DA thank NCIM, Pune for extending support for sequencing. The authors also wish to acknowledge Proteomics facility, Molecular Biophysics Unit (IISC Bangalore) for MALDI-TOF facility.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.761266/full#supplementary-material
Supplementary Figure 1 | Growth curve and production of rhamnolipids by Pseudomonas aeruginosa DNM50 in neem oil cake extract as the production medium.
Supplementary Figure 2 | Antiproliferative effect of Etoposide (Positive control) on MDA-MB 231 cell lines for 24, 48, and 72 h of treatment by (A) MTT assay showing dose dependent inhibition (B) IC50 representative graph for MTT assay (C) Resazurin assay showing dose dependent inhibition (D) IC50 representative graph for Resazurin assay (E) Trypan blue assay showing dose dependent inhibition (F) IC50 representative graph for Trypan blue assay. For dose dependent inhibition, values are represented as mean ± SD of three independent experiments. Error bars represent SD. Differences with control groups were significant at *p < 0.05. IC50 was determined by nonlinear (curve fit [Inhibitor] vs. normalized response-Variable slope) using GraphPad prism software.
References
Abalos, A., Pinazo, A., Infante, M. R., Casals, M., García, F., and Manresa, A. (2001). Physicochemical and Antimicrobial Properties of New Rhamnolipids Produced by Pseudomonas aeruginosa AT10 from Soybean Oil Refinery Wastes. Langmuir 17, 1367–1371. doi:10.1021/la0011735
Abbasi, H., Hamedi, M. M., Lotfabad, T. B., Zahiri, H. S., Sharafi, H., Masoomi, F., et al. (2012). Biosurfactant-producing Bacterium, Pseudomonas aeruginosa MA01 Isolated from Spoiled Apples: Physicochemical and Structural Characteristics of Isolated Biosurfactant. J. Biosci. Bioeng. 113, 211–219. doi:10.1016/j.jbiosc.2011.10.002
Abbasi, H., Noghabi, K. A., Hamedi, M. M., Zahiri, H. S., Moosavi-Movahedi, A. A., Amanlou, M., et al. (2013). Physicochemical Characterization of a Monorhamnolipid Secreted by Pseudomonas aeruginosa MA01 in Aqueous Media. An Experimental and Molecular Dynamics Study. Colloids Surf. B: Biointerfaces 101, 256–265. doi:10.1016/j.colsurfb.2012.06.035
Abdollahi, S., Tofighi, Z., Babaee, T., Shamsi, M., Rahimzadeh, G., Rezvanifar, H., et al. (2020). Evaluation of Anti-oxidant and Anti-biofilm Activities of Biogenic Surfactants Derived from Bacillus Amyloliquefaciens and Pseudomonas aeruginosa. Iran J. Pharm. Res. 19, 115–126. doi:10.22037/IJPR.2020.1101033
Abeer Mohammed, A. B., Tayel, A. A., and Elguindy, N. M. (2018). Production of New Rhamnolipids Rha C16-C16 by Burkholderia sp. Through Biodegradation of Diesel and Biodiesel. Beni-Suef Univ. J. Basic Appl. Sci. 7, 492–498. doi:10.1016/j.bjbas.2018.05.003
Akiyode, O., George, D., Getti, G., and Boateng, J. (2016). Systematic Comparison of the Functional Physico-Chemical Characteristics and Biocidal Activity of Microbial Derived Biosurfactants on Blood-Derived and Breast Cancer Cells. J. Colloid Interf. Sci. 479, 221–233. doi:10.1016/j.jcis.2016.06.051
Allada, L. T. K., Guntuku, G. S., Muthyala, M. K. K., Duddu, M. K., and Golla, N. S. (2015). Characterization of Bioactive Compound from Streptomyces Coelicoflavus NBRC (15399T) and its Anticancer Activity. Int. J. Chem. Pharm. Anal. 2, 1–13. https://www.ijcpa.in/articles/characterization-of-bioactive-compound-obtained-from-streptomyces-coelicoflavus-nbrc-15399t-and-its-anticancer-activity.pdf. https://www.ijcpa.in/abstract/characterization-of-bioactive-compound-obtained-from-streptomyces-coelicoflavus-nbrc-15399t-and-its-anticancer-activity-80732.html.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Amena, S., Vishalakshi, N., Prabhakar, M., Dayanand, A., and Lingappa, K. (2010). Production, Purification and Characterization of L-Asparaginase from Streptomyces Gulbargensis. Braz. J. Microbiol. 41, 173–178. doi:10.1590/S1517-83822010000100002510.1590/s1517-83822010000100025
Banat, I. M., De Rienzo, M. A. D., and Quinn, G. A. (2014a). Microbial Biofilms: Biosurfactants as Antibiofilm Agents. Appl. Microbiol. Biotechnol. 98, 9915–9929. doi:10.1007/s00253-014-6169-6
Banat, I. M., Satpute, S. K., Cameotra, S. S., Patil, R., and Nyayanit, N. V. (2014b). Cost Effective Technologies and Renewable Substrates for Biosurfactants Production. Front. Microbiol. 5, 697. doi:10.3389/fmicb.2014.00697
Bradford, M. M. (1976). A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 72, 248–254. doi:10.1006/abio.1976.999910.1016/0003-2697(76)90527-3
Burgos-Díaz, C., Martín-Venegas, R., Martínez, V., Storniolo, C. E., Teruel, J. A., Aranda, F. J., et al. (2013). In Vitro study of the Cytotoxicity and Antiproliferative Effects of Surfactants Produced by Sphingobacterium Detergens. Int. J. Pharm. 453, 433–440. doi:10.1021/acsomega.0c04933
Byrappa, S., Harsha Raj, M., Kungyal, T., Kudva, N., and Narayana, U. (2017). Synthesis and Biological Evaluation of Novel Isoxazolines Linked via Piperazine to 2- Benzoisothiazoles as Potent Apoptotic Agents. Eur. J. Med. Chem. 126, 218–224. doi:10.1016/j.ejmech.2016.09.094
Cai, B., Chang, S. H., Becker, E. B. E., Bonni, A., and Xia, Z. (2006). p38 MAP Kinase Mediates Apoptosis through Phosphorylation of BimEL at Ser-65. J. Biol. Chem. 281, 25215–25222. doi:10.1074/jbc.M512627200
Cailleau, R., Olivé, M., and Cruciger, Q. V. J. (1978). Long-term Human Breast Carcinoma Cell Lines of Metastatic Origin: Preliminary Characterization. In Vitro 14, 911–915. doi:10.1007/BF02616120
Chandankere, R., Yao, J., Choi, M. M. F., Masakorala, K., and Chan, Y. (2013). An Efficient Biosurfactant-Producing and Crude-Oil Emulsifying Bacterium Bacillus Methylotrophicus USTBa Isolated from Petroleum reservoir Bacillus methylotrophicus USTBa Isolated from Petroleum Reservoir. Biochem. Eng. J. 74, 46–53. doi:10.1016/j.bej.2013.02.018
Chavez, K. J., Garimella, S. V., and Lipkowitz, S. (2011). Triple Negative Breast Cancer Cell Lines: One Tool in the Search for Better Treatment of Triple Negative Breast Cancer. Breast. Dis. 32, 35–48. doi:10.3233/BD-2010-0307
Chen, J., Sun, J., Deering, R. W., DaSilva, N., Seeram, N. P., Wang, H., et al. (2016). Rhizoleucinoside, a Rhamnolipid-Amino Alcohol Hybrid from the Rhizobial Symbiont Bradyrhizobium sp. BTAi1. Org. Lett. 18, 1490–1493. doi:10.1021/acs.orglett.6b00461
Chen, J., Wu, Q., Hua, Y., Chen, J., Zhang, H., and Wang, H. (2017). Potential Applications of Biosurfactant Rhamnolipids in Agriculture and Biomedicine. Appl. Microbiol. Biotechnol. 101, 8309–8319. doi:10.1007/s00253-017-8554-4
Choi, B.-K., Lee, H.-S., Kang, J. S., and Shin, H. J. (2019). Dokdolipids A−C, Hydroxylated Rhamnolipids from the Marine-Derived Actinomycete Actinoalloteichus hymeniacidonis. Mar. Drugs 17, 237. doi:10.3390/md17040237
Chopra, A., Bobate, S., Rahi, P., Banpurkar, A., Mazumder, P. B., and Satpute, S. (2020). Pseudomonas aeruginosa RTE4: A tea Rhizobacterium with Potential for Plant Growth Promotion and Biosurfactant Production. Front. Bioeng. Biotechnol. 8, 861. doi:10.3389/fbioe.2020.00861
Christova, N., Tuleva, B., Kril, A., Georgieva, M., Konstantinov, S., Terziyski, I., et al. (2013). Chemical Structure and In Vitro Antitumor Activity of Rhamnolipids from Pseudomonas aeruginosa BN10. Appl. Biochem. Biotechnol. 170, 676–689. doi:10.1007/s12010-013-0225-z
Clarridge, J. E. (2004). Impact of 16S rRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Rev. 17, 840–862. doi:10.1128/CMR.17.4.840-862.2004
Darby, A. C., Chandler, S. M., Welburn, S. C., and Douglas, A. E. (2005). Aphid-symbiotic Bacteria Cultured in Insect Cell Lines. Appl. Environ. Microbiol. 71, 4833–4839. doi:10.1128/AEM.71.8.4833-4839.2005
De Almeida, D. G., Soares Da Silva, R. d. C. F., Luna, J. M., Rufino, R. D., Santos, V. A., Banat, I. M., et al. (2016). Biosurfactants: Promising Molecules for Petroleum Biotechnology Advances. Front. Microbiol. 7, 1718. doi:10.3389/fmicb.2016.01718
Dwivedi, S., Saquib, Q., Al-Khedhairy, A. A., Ahmad, J., Siddiqui, M. A., and Musarrat, J. (2015). Rhamnolipids Functionalized AgNPs-Induced Oxidative Stress and Modulation of Toxicity Pathway Genes in Cultured MCF-7 Cells. Colloids Surf. B: Biointerfaces 132, 290–298. doi:10.1016/j.colsurfb.2015.05.034
Farghadani, R., Rajarajeswaran, J., Mohd Hashim, N. B., Abdulla, M. A., and Muniandy, S. (2017). A Novel β-diiminato manganese III Complex as the Promising Anticancer Agent Induces G0/G1 Cell Cycle Arrest and Triggers Apoptosis via Mitochondrial-dependent Pathways in MCF-7 and MDA-MB-231 Human Breast Cancer Cells. RSC Adv. 7, 24387–24398. doi:10.1039/c7ra02478a
Fracchia, L., Banat, J. J., J. Banat, J., Cavallo, M., Ceresa, C., and M. Banat, I. (2015). Potential Therapeutic Applications of Microbial Surface-Active Compounds. AIMS Bioeng. 2, 144–162. doi:10.3934/bioeng.2015.3.144
García-Cano, J., Roche, O., Cimas, F. J., Pascual-Serra, R., Ortega-Muelas, M., Fernández-Aroca, D. M., et al. (2016). p38MAPK and Chemotherapy: We Always Need to Hear Both Sides of the story. Front. Cel Dev. Biol. 4, 1–8. doi:10.3389/fcell.2016.00069
Garcia-Manero, G., Khoury, H. J., Jabbour, E., Lancet, J., Winski, S. L., Cable, L., et al. (2015). A Phase I Study of Oral ARRY-614, a P38 MAPK/Tie2 Dual Inhibitor, in Patients with Low or Intermediate-1 Risk Myelodysplastic Syndromes. Clin. Cancer Res. 21, 985–994. doi:10.1158/1078-0432.CCR-14-1765
Giri, S. S., Ryu, E., Sukumaran, V., and Park, S. C. (2019). Antioxidant, Antibacterial, and Anti-adhesive Activities of Biosurfactants Isolated from Bacillus Strains. Microb. Pathogenesis 132, 66–72. doi:10.1016/j.micpath.2019.04.035
Gudiña, E. J., Rangarajan, V., Sen, R., and Rodrigues, L. R. (2013). Potential Therapeutic Applications of Biosurfactants. Trends Pharmacol. Sci. 34, 667–675. doi:10.1016/j.tips.2013.10.002
Gudiña, E. J., Rodrigues, A. I., de Freitas, V., Azevedo, Z., Teixeira, J. A., and Rodrigues, L. R. (2016). Valorization of Agro-Industrial Wastes towards the Production of Rhamnolipids. Bioresour. Tech. 212, 144–150. doi:10.1016/j.biortech.2016.04.027
Gupta, J., Igea, A., Papaioannou, M., Lopez-Casas, P. P., Llonch, E., Hidalgo, M., et al. (2015). Pharmacological Inhibition of P38 MAPK Reduces Tumor Growth in Patient-Derived Xenografts from colon Tumors. Oncotarget 6, 8539–8551. doi:10.18632/oncotarget.3816
Haba, E., Pinazo, A., Jauregui, O., Espuny, M. J., Infante, M. R., and Manresa, A. (2003). Physicochemical Characterization and Antimicrobial Properties of Rhamnolipids Produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 81, 316–322. doi:10.1002/bit.10474
Haque, F., Khan, M. S. A., and AlQurashi, N. (2021). ROS-mediated Necrosis by Glycolipid Biosurfactants on Lung, Breast, and Skin Melanoma Cells. Front. Oncol. 11, 622470. doi:10.3389/fonc.2021.622470
Hazra, C., Kundu, D., Ghosh, P., Joshi, S., Dandi, N., and Chaudhari, A. (2010). Screening and Identification of Pseudomonas aeruginosa AB4 for Improved Production, Characterization and Application of a Glycolipid Biosurfactant Using Low-Cost Agro-Based Raw Materials. J. Chem. Technol. Biotechnol. 86, 185–198. doi:10.1002/jctb.2480
Hošková, M., Ježdík, R., Schreiberová, O., Chudoba, J., Šír, M., Čejková, A., et al. (2015). Structural and Physiochemical Characterization of Rhamnolipids Produced by Acinetobacter Calcoaceticus, Enterobacter Asburiae and Pseudomonas aeruginosa in Single Strain and Mixed Cultures. J. Biotechnol. 193, 45–51. doi:10.1016/j.jbiotec.2014.11.014
Housman, G., Byler, S., Heerboth, S., Lapinska, K., Longacre, M., Snyder, N., et al. (2014). Drug Resistance in Cancer: An Overview. Cancers 6, 1769–1792. doi:10.3390/cancers6031769
Igea, A., and Nebreda, A. R. (2015). The Stress Kinase P38α as a Target for Cancer Therapy. Cancer Res. 75, 3997–4002. doi:10.1158/0008-5472.CAN-15-0173
Inès, M., and Dhouha, G. (2015). Glycolipid Biosurfactants: Potential Related Biomedical and Biotechnological Applications. Carbohydr. Res. 416, 59–69. doi:10.1016/j.carres.2015.07.016
Jemil, N., Ben Ayed, H., Manresa, A., Nasri, M., and Hmidet, N. (2017). Antioxidant Properties, Antimicrobial and Anti-adhesive Activities of DCS1 Lipopeptides from Bacillus Methylotrophicus DCS1. BMC Microbiol. 17, 144. doi:10.1186/s12866-017-1050-2
Jiang, L., Shen, C., Long, X., Zhang, G., and Meng, Q. (2014). Rhamnolipids Elicit the Same Cytotoxic Sensitivity between Cancer Cell and Normal Cell by Reducing Surface Tension of Culture Medium. Appl. Microbiol. Biotechnol. 98, 10187–10196. doi:10.1007/s00253-014-6065-0
Kamal, A., Shaik, A. B., Kumar, C. G., Mongolla, P., Rani, P. U., Krishna, K. V., et al. (2012). Metabolic Profiling and Biological Activities of Bioactive Compounds Produced by Pseudomonas Sp. Strain ICTB-745 Isolated from Ladakh, India. J. Microbiol. Biotechnol. 22, 69–79. doi:10.4014/jmb.1105.05008
Kamiloglu, S., Sari, G., Ozdal, T., and Capanoglu, E. (2020). Guidelines for Cell Viability Assays. Food Front. 1, 332–349. doi:10.1002/fft2.44
Kedare, S. B., and Singh, R. P. (2011). Genesis and Development of DPPH Method of Antioxidant Assay. J. Food Sci. Technol. 48, 412–422. doi:10.1007/s13197-011-0251-1
Lalaoui, N., Hänggi, K., Brumatti, G., Chau, D., Nguyen, N.-Y. N., Vasilikos, L., et al. (2016). Targeting P38 or MK2 Enhances the Anti-leukemic Activity of Smac-Mimetics. Cancer Cell 29, 145–158. doi:10.1016/j.ccell.2016.01.006
Li, H., Guo, W., Ma, X.-j., Li, J.-s., and Song, X. (2017). In Vitro and In Vivo Anticancer Activity of Sophorolipids to Human Cervical Cancer. Appl. Biochem. Biotechnol. 181, 1372–1387. doi:10.1007/s12010-016-2290-6
Li, S., Huang, R., Shah, N. P., Tao, X., Xiong, Y., and Wei, H. (2014). Antioxidant and Antibacterial Activities of Exopolysaccharides from Bifidobacterium Bifidum WBIN03 and Lactobacillus Plantarum R315. J. Dairy Sci. 97, 7334–7343. doi:10.3168/jds.2014-7912
Lotfabad, T. B., Abassi, H., Ahmadkhaniha, R., Roostaazad, R., Masoomi, F., Zahiri, H. S., et al. (2010). Structural Characterization of a Rhamnolipid-type Biosurfactant Produced by Pseudomonas aeruginosa MR01: Enhancement of Di-rhamnolipid Proportion Using Gamma Irradiation. Colloids Surf. B: Biointerfaces 81, 397–405. doi:10.1016/j.colsurfb.2010.06.026
Mahajan, S. D., Law, W.-C., Aalinkeel, R., Reynolds, J., Nair, B. B., Yong, K.-T., et al. (2012). Nanoparticle-mediated Targeted Delivery of Antiretrovirals to the Brain. Methods Enzymol. 509, 41–60. doi:10.1016/B978-0-12-391858-1.00003-4
Merghni, A., Dallel, I., Noumi, E., Kadmi, Y., Hentati, H., Tobji, S., et al. (2017). Antioxidant and Antiproliferative Potential of Biosurfactants Isolated from Lactobacillus Casei and Their Anti-biofilm Effect in Oral Staphylococcus aureus Strains. Microb. Pathogenesis 104, 84–89. doi:10.1016/j.micpath.2017.01.017
Mishra, N., Agsar, D., Seelam, S. D., Kattegauda, U. M., Shetty, P. R., and Pandey, V. (2021). Novel Application of Nerium Leaf and Image J Software in Drop Collapse Assay for Rapid Screening of Biosurfactant Producing Microorganism. Indian J. Exp. Biol. 59, 484–492. http://nopr.niscair.res.in/handle/123456789/57850.
Mohammed, F., Rashid-Doubell, F., Taha, S., Cassidy, S., and Fredericks, S. (2020). Effects of Curcumin Complexes on MDA MB 231 Breast Cancer Cell Proliferation. Int. J. Oncol. 57, 445–455. doi:10.3892/ijo.2020.5065
Mulla, S. I., Hu, A., Sun, Q., Li, J., Suanon, F., Ashfaq, M., et al. (2018). Biodegradation of Sulfamethoxazole in Bacteria from Three Different Origins. J. Environ. Manage. 206, 93–102. doi:10.1016/j.jenvman.201710.1016/j.jenvman.2017.10.029
Mulla, S. I., Wang, H., Sun, Q., Hu, A., and Yu, C.-P. (2016). Characterization of Triclosan Metabolism in Sphingomonas sp. Strain YL-Jm2c. Sci. Rep. 6, 21965. doi:10.1038/srep21965
Nie, M., Yin, X., Ren, C., Wang, Y., Xu, F., and Shen, Q. (2010). Novel Rhamnolipid Biosurfactants Produced by a Polycyclic Aromatic Hydrocarbon-Degrading Bacterium Pseudomonas aeruginosa Strain. Biotechnol. Adv. 28, 635–643. doi:10.1016/j.biotechadv.2010.05.013
Osborne, C. K., and Schiff, R. (2011). Mechanisms of Endocrine Resistance in Breast Cancer. Annu. Rev. Med. 62, 233–247. doi:10.1146/annurev-med-070909-182917
Pathania, A. S., and Jana, A. K. (2020). Improvement in Production of Rhamnolipids Using Fried Oil with Hydrophilic Co-substrate by Indigenous Pseudomonas aeruginosa NJ2 and Characterizations. Appl. Biochem. Biotechnol. 191, 1223–1246. doi:10.1007/s12010-019-03221-9
Patnaik, A., Haluska, P., Tolcher, A. W., Erlichman, C., Papadopoulos, K. P., Lensing, J. L., et al. (2016). A First-In-Human Phase I Study of the Oral P38 MAPK Inhibitor, Ralimetinib (LY2228820 Dimesylate), in Patients with Advanced Cancer. Clin. Cancer Res. 22, 1095–1102. doi:10.1158/1078-0432.CCR-15-1718
Phillips, H. J. (1973). “Dye Exclusion Tests for Cell Viability,” in Tissue Culture: Methods and Applications. Editors P. F. KruseJr, and M. K. PattersonJr (New York: Academic Press), 406–408. doi:10.1016/b978-0-12-427150-0.50101-7
Pradhan, A. K., Pradhan, N., Mohapatra, P., Kundu, C. N., Panda, P. K., and Mishra, B. K. (2014). Cytotoxic Effect of Microbial Biosurfactants against Human Embryonic Kidney Cancerous Cell: HEK-293 and Their Possible Role in Apoptosis. Appl. Biochem. Biotechnol. 174, 1850–1858. doi:10.1007/s12010-014-1168-8
Price, N. P. J., Ray, K. J., Vermillion, K., and Kuo, T.-M. (2009). MALDI-TOF Mass Spectrometry of Naturally Occurring Mixtures of Monorhamnolipids and Dirhamnolipids. Carbohydr. Res. 344, 204–209. doi:10.1016/j.carres.2008.10.013
Pritchard, A. L., and Hayward, N. K. (2013). Molecular Pathways: Mitogen-Activated Protein Kinase Pathway Mutations and Drug Resistance. Clin. Cancer Res. 19, 2301–2309. doi:10.1158/1078-0432.CCR-12-0383
Rahimi, K., Lotfabad, T. B., Jabeen, F., and Mohammad Ganji, S. (2019). Cytotoxic Effects of Mono- and Di-rhamnolipids from Pseudomonas aeruginosa MR01 on MCF-7 Human Breast Cancer Cells. Colloids Surf. B: Biointerfaces 181, 943–952. doi:10.1016/j.colsurfb.2019.06.058
Riss, T. L., Moravec, R. A., Niles, A. L., Duellman, S., Benink, H. A., and Worzella, T. J. (20162016). Cell Viability Assays. Assay Guidance Manual [Internet]. Maryland, United StatesEli Lilly & Company and the National Center for Advancing Translational Sciences. Available from: https://www.ncbi.nlm.nih.gov/books/NBK144065/.
Samykannu, M., and Achary, A. (2017). Utilization of Agro-Industry Residue for Rhamnolipid Production by P. aeruginosa AMB AS7 and its Application in Chromium Removal. Appl. Biochem. Biotechnol. 183, 70–90. doi:10.1007/s12010-017-2431-6
Sanchez-Prieto, R., Rojas, J. M., Taya, Y., and Gutkind, J. S. (2000). A Role for the P38 Mitogen-Acitvated Protein Kinase Pathway in the Transcriptional Activation of P53 on Genotoxic Stress by Chemotherapeutic Agents. Cancer Res. 60, 2464–2472.
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. United States of America 74 (12), 5463–5467. doi:10.1073/pnas.74.12.5463
Sarachat, T., Pornsunthorntawee, O., Chavadej, S., and Rujiravanit, R. (2010). Purification and Concentration of a Rhamnolipid Biosurfactant Produced by Pseudomonas aeruginosa SP4 Using Foam. Bioresour. Tech. 101, 324–330. doi:10.1016/j.biortech.2009.08.012
Sato, S., Fukuoka, T., Saika, A., Koshiyama, T., and Morita, T. (2019). A New Screening Approach for Glycolipid-type Biosurfactant Producers Using MALDI-TOF/MS. J. Oleo Sci. 68, 1287–1294. doi:10.5650/jos.ess19177
Semkova, S., Antov, G., Iliev, I., Tsoneva, I., Lefterov, P., Christova, N., et al. (2021). Rhamnolipid Biosurfactants-Possible Natural Anticancer Agents and Autophagy Inhibitors. Separations 8, 92. doi:10.3390/separations8070092
Shao, B., Liu, Z., Zhong, H., Zeng, G., Liu, G., Yu, M., et al. (2017). Effects of Rhamnolipids on Microorganism Characteristics and Applications in Composting: a Review. Microbiol. Res. 200, 33–44. doi:10.1016/j.micres.2017.04.005
Shum, D., Radu, C., Kim, E., Cajuste, M., Shao, Y., Seshan, V. E., et al. (2008). A High Density Assay Format for the Detection of Novel Cytotoxic Agents in Large Chemical Libraries. J. Enzyme Inhib. Med. Chem. 23 (6), 931–945. doi:10.1080/14756360701810082
Silva, S. N. R. L., Farias, C. B. B., Rufino, R. D., Luna, J. M., and Sarubbo, L. A. (2010). Glycerol as Substrate for the Production of Biosurfactant by Pseudomonas aeruginosa UCP0992. Colloids Surf. B: Biointerfaces 79, 174–183. doi:10.1016/j.colsurfb.2010.03.050
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Swain, S. (2008). Triple-Negative Breast Cancer: Metastatic Risk and Role of Platinum Agents 2008 ASCO Clinical Science Symposium. Alexandria, Virginia: American Society of Clinical Oncology.
Tabbene, O., Gharbi, D., Slimene, I. B., Elkahoui, S., Alfeddy, M. N., Cosette, P., et al. (2012). Antioxidative and DNA Protective Effects of Bacillomycin D-like Lipopeptides Produced by B38 Strain. Appl. Biochem. Biotechnol. 168, 2245–2256. doi:10.1111/j.1574-6968.2010.02199.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi:10.1093/molbev/mst197
Thakur, P., Saini, N. K., Thakur, V. K., Gupta, V. K., Saini, R. V., and Saini, A. K. (2021). Rhamnolipid the Glycolipid Biosurfactant: Emerging Trends and Promising Strategies in the Field of Biotechnology and Biomedicine. Microb. Cel Fact 20, 1–15. doi:10.1186/s12934-020-01497-9
Thanomsub, B., Pumeechockchai, W., Limtrakul, A., Arunrattiyakorn, P., Petchleelaha, W., Nitoda, T., et al. (2006). Chemical Structures and Biological Activities of Rhamnolipids Produced by Pseudomonas aeruginosa B189 Isolated from Milk Factory Waste. Bioresour. Tech. 97, 2457–2461. doi:10.1016/j.biortech.2005.10.029
Tofani, D., Balducci, V., Gasperi, T., Incerpi, S., and Gambacorta, A. (2010). Fatty Acid Hydroxytyrosyl Esters: Structure/antioxidant Activity Relationship by ABTS and in Cell-Culture DCF Assays. J. Agric. Food Chem. 58, 5292–5299. doi:10.1021/jf1000716
Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets: Procedure and Some Applications. Proc. Natl. Acad. Sci. 76, 4350–4354. doi:10.1073/pnas.76.9.4350
Twigg, M. S., Baccile, N., Banat, I. M., Déziel, E., Marchant, R., Roelants, S., et al. (2020). Microbial Biosurfactant Research: Time to Improve the Rigour in the Reporting of Synthesis, Functional Characterization and Process Development. Microb. Biotechnol. 14, 147–170. doi:10.1111/1751-7915.13704
Voulgaridou, G.-P., Mantso, T., Anestopoulos, I., Klavaris, A., Katzastra, C., Kiousi, D.-E., et al. (2021). Toxicity Profiling of Biosurfactants Produced by Novel marine Bacterial Strains. Int. J. Mol. Sci. 22, 2383. doi:10.3390/ijms22052383
Wambang, N., Schifano-Faux, N., Aillerie, A., Baldeyrou, B., Jacquet, C., Bal-Mahieu, C., et al. (2016). Synthesis and Biological Activity of Ferrocenyl Indeno[1,2-C]isoquinolines as Topoisomerase II Inhibitors. Bioorg. Med. Chem. 24, 651–660. doi:10.1016/j.bmc.2015.12.033
Wang, X., Zhang, H., and Chen, X. (2019). Drug Resistance and Combating Drug Resistance in Cancer. Cancer Drug. Resist. 2, 141–160. doi:10.20517/cdr.2019.10
Wu, D., Si, M., Xue, H.-Y., and Wong, H. L. (2017). Nanomedicine Applications in the Treatment of Breast Cancer: Current State of the Art. I. J. Nano. Vol. 12, 5879–5892. doi:10.2147/IJN.S123437
Zhao, J., Wu, Y., Alfred, A. T., Xin, X., and Yang, S. (2013). Chemical Structures and Biological Activities of Rhamnolipid Biosurfactants Produced by Pseudomonas aeruginosa M14808. J. Chem. Pharm. Res. 5, 177–182. https://www.jocpr.com/abstract/chemical-structures-and-biological-activities-of-rhamnolipid-biosurfactants-produced-by-pseudomonas-aeruginosa-m14808-2394.html.
Keywords: MDA-MB-231 cell lines, rhamnolipid, MTT, trypan blue, p38MAPK, TNBC, resazurin, cytotoxic
Citation: Mishra N, Rana K, Seelam SD, Kumar R, Pandey V, Salimath BP and Agsar D (2021) Characterization and Cytotoxicity of Pseudomonas Mediated Rhamnolipids Against Breast Cancer MDA-MB-231 Cell Line. Front. Bioeng. Biotechnol. 9:761266. doi: 10.3389/fbioe.2021.761266
Received: 19 August 2021; Accepted: 27 October 2021;
Published: 30 November 2021.
Edited by:
Surekha K. Satpute, Savitribai Phule Pune University, IndiaReviewed by:
Nurul Farahana Kamaludin, National University of Malaysia, MalaysiaDeepansh Sharma, Amity University Jaipur, India
Copyright © 2021 Mishra, Rana, Seelam, Kumar, Pandey, Salimath and Agsar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayanand Agsar, ZGF5YW5hbmRhZ3NhckBnbWFpbC5jb20=
 Neelam Mishra
Neelam Mishra Kavita Rana
Kavita Rana Siva Deepthi Seelam
Siva Deepthi Seelam Rakesh Kumar
Rakesh Kumar Vijyendra Pandey
Vijyendra Pandey Bharathi P. Salimath5,6
Bharathi P. Salimath5,6 Dayanand Agsar
Dayanand Agsar