- 1Institute of Biosciences and Bioresources, National Research Council of Italy, Naples, Italy
- 2Department of Applied Chemistry, Institute of Natural Science, Kyung Hee University, Yongin, South Korea
- 3Department of Biomedical Science and Technology, Kyung Hee Medical Science Research Institute, Kyung Hee University, Seoul, South Korea
- 4Department of Biomolecular Sciences, University of Urbino Carlo Bo, Urbino, Italy
- 5Allergy Data Laboratories (ADL), Latina, Italy
- 6Associated Centers for Molecular Allergology (CAAM), Rome, Italy
Background: Nanometer-sized membrane-surrounded vesicles from different parts of plants including fruits are gaining increasing attention due to their anti-inflammatory and anticancer effects demonstrated by in vitro and in vivo studies, and as nanovectors for molecular delivery of exogenous substances. These nanomaterials are very complex and contain a diverse arsenal of bioactive molecules, such as nucleic acids, proteins, and lipids. Our knowledge about the transport of allergens in vesicles isolated from plant food is limited today.
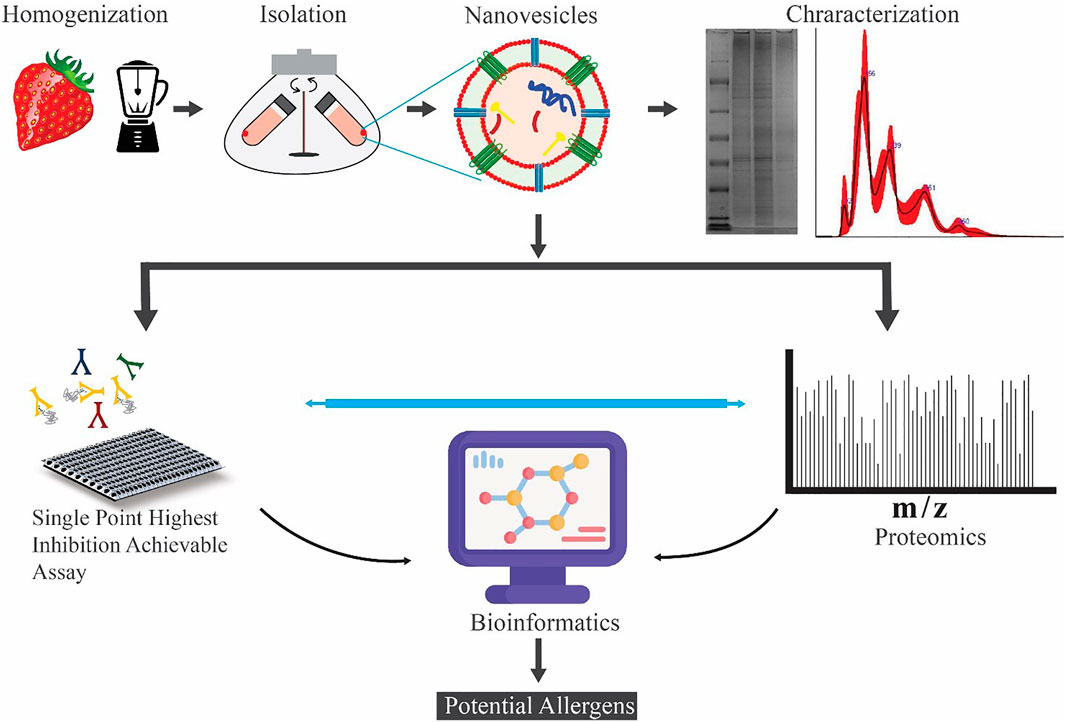
Methods: Here, to investigate the allergenicity of strawberry-derived microvesicles (MVs), nanovesicles (NVs), and subpopulations of NV, we have set up a multidisciplinary approach. The strategy combines proteomics-based protein identification, immunological investigations, bioinformatics, and data mining to gain biological insights useful to evaluate the presence of potential allergens and the immunoglobulin E (IgE) inhibitory activity of vesicle preparations.
Results: Immunological test showed that several proteins of strawberry-derived vesicles compete for IgE binding with allergens spotted on the FABER biochip. This includes the known strawberry allergens Fra a 1, Fra a 3, and Fra a 4, and also other IgE-binding proteins not yet described as allergens in this food, such as gibberellin-regulated proteins, 2S albumin, pectate lyase, and trypsin inhibitors. Proteomics identified homologous sequences of the three strawberry allergens and their isoforms in total protein extract (TPE) but only Fra a 1 and Fra a 4 in the vesicle samples. Label-free quantitative proteomic analysis revealed no significant enrichment of these proteins in strawberry vesicles with respect to TPE.
Conclusion: Immunological tests and bioinformatics analysis of proteomics data sets revealed that MVs and NVs isolated from strawberries can carry functional allergens their isoforms as well as proteins potentially allergenic based on their structural features. This should be considered when these new nanomaterials are used for human nutraceutical or biomedical applications.
Introduction
Extracellular vesicles (EVs) are nanosized lipid bilayer-surrounded vesicles that are released by cells. They are packed with a multimolecular cargo of proteins, lipids, nucleic acids, and small molecules, which makes them a valuable source of information on the cell of origin. EVs can be uptaken by local or distant cells with which they can interact. In this way, they contribute to cell–cell communication with important functional consequences. Membrane-bound vesicles that are similar to the mammalian EVs in size, shape, and density have been isolated and studied from various plant sources (Mu et al., 2014) (Zhang et al., 2011) (Perut et al., 2021) (Pocsfalvi et al., 2018) (Bokka et al., 2020). Recently, substantial attention has been given to plant-derived vesicles because of their frequently observed anti-inflammatory and anticancer activities in cellular and animal models (Mu et al., 2014) (Zhang et al., 2011) (Perut et al., 2021). Like EVs secreted by mammalian cells, vesicles in plants play an important role in intracellular and cell–cell communications, and according to recent hypothesis, they can take part in interspecies communication too (Mu et al., 2014). Recently, nanovesicles (NVs) were successfully isolated from strawberries using widely employed differential centrifugation (DC) and filtration-based methods (Perut et al., 2021). The NVs were shown to have the typical morphology and size distribution of mammalian EVs. They also found that the internalization of these vesicles by cells did not affect cell viability negatively and prevented oxidative stress in a dose-dependent manner.
Strawberries are berries cultivated and consumed worldwide because they represent a delicious and nutritious food. Taxonomically, they belong to the genus Fragaria, included in the Rosaceae family. Fragaria vesca, known as wild strawberry, is a species with small berries, whereas Fragaria ananassa has higher size berries, and it is widely present on the market. Strawberries are described as false fruits bearing on the surface the real fruits, namely, the achenes, where the development of seeds occurs. The NCBI database reports 32,886 protein sequences of F. vesca, while only 1,671 records are present for the less sequenced and annotated F. x ananassa (dated July 26, 2021).
Strawberry extracts were shown to be rich in polyphenols, and they exhibit important biological activities in vitro against different cancer cell lines and in vivo against invasive breast cancer cells (Amatori et al., 2016) (Perut et al., 2021). Strawberries can also trigger allergic reactions in some people sensitized to this food (Hyun and Kim, 2011; Muñoz et al., 2010). There are three known strawberry allergens (F. x ananassa) that have been registered by the World Health Organization and International Union of Immunological Societies (WHO/IUIS, www.allergen.org) (Pomes et al., 2018) with the names Fra 1, Fra a 3, and Fra a 4 (Zuidmeer et al., 2006). Fra a 1 is a 17-kDa protein homologous to the major birch pollen allergen Bet v 1, a member of the ubiquitous PR-10 family of plant pathogenesis-related proteins. Fra a 3 is a 9-kDa lipid transfer protein (LTP), belonging to the PR-14 protein family. Fra a 4 is a 13-kDa protein belonging to the profilin family, the members of which play a key role in cell physiology. Profilins are highly cross-reacting allergens. This property is linked to the high conservation of their structural features highlighted by the high amino acid sequence identity between homologous molecules (Offermann et al., 2016). Profilins and Bet v 1-like proteins are classified as class 2 food allergens that are heat-labile, easily degraded by the gastrointestinal proteases, and responsible for localized oral allergy symptoms (OAS) (Alessandri et al., 2020). In contrast, LTP belongs to class 1 food allergens, which are heat and protease stable. They are clinically relevant allergens because their ingestion, inhalation, and skin contact can cause symptoms ranging from mild to very severe, including food-dependent exercise-induced anaphylaxis and anaphylactic shock. Fra a 1, Fra a 3, and Fra a 4 belong to multigene families, and therefore, their various isoallergens/isoforms are also reported in protein databases. In particular, 12 Fra a 1, 7 Fra a 3, and 2 Fra a 4 isoallergens/isoforms are reported today in UniProt protein database (July 26, 2021). Interestingly, the protein homologous of Fra 1, Fra a 3, and Fra a 4 have also been identified in F. vesca (Hyun and Kim, 2011), but they are not available in public protein databases.
In this study, we investigate if strawberry-derived vesicles carry protein sequences having homologs to known allergens using a quantitative proteomics and bioinformatics approach. Label-free shotgun proteomics was used to identify and quantify proteins in the total protein extract, and three vesicle isolates, microvesicles (MVs), NVs, and the density gradient ultracentrifugation (DGUC) separated NVs. BLAST searches against the COMPARE allergen and the UniProt Fragaria x ananassa databases were used to point out already known and potential allergens among the identified proteins based on structural features. Parallel to the proteomics analysis, we used the Single Point Highest Inhibition Achievable assay (SPHIAa) method associated with the FABER test (Tuppo et al., 2019) (Tuppo et al., 2018; Giangrieco et al., 2019) to identify the presence of functional allergens. FABER is a nanotechnology-based multiplex in vitro serological test, which takes advantage of database and bioinformatics tools of the Allergome platform (http://www.allergome.org/). FABER test exploits a comprehensive panel of 244 allergens, including the most important allergy markers, in addition to exclusive allergens not available in other test systems. Finally, the results obtained from FABER and the quantitative proteomics were merged to evaluate the enrichment of the potential allergens in the different vesicle fractions with respect to the total proteins extracted from strawberries.
Materials and Methods
Fruit Material and Isolation of Nanovesicles by Differential Ultracentrifugation
Strawberries grown without any pre- and postharvest treatments were acquired from a local vendor in Naples, Italy. 250 g of fresh fruits were weighed and washed thrice with cold running tap water and once with Milli-Q water. The calyx was removed, berries were air dried and transferred to a blender containing 1:1 weight-to-volume ratio extraction buffer composed of 100 mM phosphate and 10 mM ethylenediamine tetraacetic acid (EDTA) (pH 8), and protease inhibitor cocktail [0.25 ml leupeptin (1 mg/ ml), 1.25 ml 100 mM phenylmethylsulfonyl fluoride (PMSF), and 0.8 ml 1 M sodium azide for every 250 g of strawberry]. The sample was homogenized three times at a maximum velocity for 10 s. Crude vesicles were isolated by differential centrifugation (DC) following the procedure described by Stanly et al. (2016). Briefly, sequential low-velocity centrifugations were performed at 400, 800, and 2,000 ×g using a swinging-out bucket rotor for 30 min for each step at 22°C. The supernatant was centrifuged at 15,000 ×g in a fixed-angle rotor for 30 min at 22°C to collect the pellet containing the microvesicles (MVs). The supernatant was centrifuged at 150,000 ×g for 60 min at 4°C using a Type 70 Ti Beckman rotor in a Beckman Coulter Optima L-90K ultracentrifuge. The pellet was resuspended in 28.5 ml of 50 mM Tris-HCl (pH 8.6) buffer and centrifuged at 150,000 ×g for 60 min at 4°C. The resulting Pellet (crude NVs) crude NVs were suspended in 50 mM Tris-HCl pH 8.6 and further separated by DGUC.
Density Gradient Ultracentrifugation Using Iodixanol
DGUC was performed according to Karimi et al. (2018) with some modifications. OptiPrepTM (60 w/v% iodixanol in distilled water; Axis-Shield, Oslo, Norway) was diluted to 50%, 30%, and 10%, (w/v), and 3 ml of each solution was layered in 13.5-ml polypropylene centrifugation tubes (Beckman Coulter, Pasadena CA, United States). Three milliliters of the NVs isolated by dUC was gently layered on the top and centrifuged at 110,000 ×g for 24 h at 4°C using an SW 41 Ti Beckman rotor in a Beckman Coulter Optima L-90K ultracentrifuge. Twelve vesicle fractions (layers) were collected, and the density of each fraction was determined. The standard curve was prepared using 2 μL of iodixanol solution with concentrations between 1% and 50%. Standards and DGUC fraction samples previously diluted 5,000-fold in distilled water were measured using Nanodrop 2000 (Thermo Fisher Scientific Inc., Waltham, MA, United States) at 244 nm wavelength according to the instruction of the manufacturer. The density of samples was calculated based on their iodixanol concentration.
Density Gradient Ultracentrifugation Using Sucrose/Deuterium Oxide Cushions
The sample was underlaid with a sucrose/D2O double cushion consisting of 6 ml of 1 M and 4 ml of 2 M sucrose prepared in 20 mM Tris pH 8.6/D2O in 38.5-ml thin-wall polypropylene tubes (Beckman Coulter, United States) and centrifuged at 110,000 ×g for 3 h at 4°C using an SW 28 Ti rotor (Beckman Coulter, United States). The fractions were collected by piercing the bottom of the tube, diluted in solubilization buffer, and centrifuged at 110,000 ×g for 1 h at 4°C using an SW 28 Ti rotor to remove sucrose. The resulting pellets were resuspended in 10–20 µl of solubilization buffers. Protein concentration was determined by the microbicinchoninic acid protein assay (Pierce/Thermo Scientific, Rockford, IL, United States) using a nanodrop 200 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States). Samples were stored at −80°C until used.
Nanoparticle Tracking Analysis
Particle number concentration was measured by nanoparticle tracking analysis (NTA) using a Nanosight NS300 (Malvern Panalytical, United States) instrument. The samples were diluted to obtain less than 100 particles per frame; then 5 × 60-s measurements were performed with a moderate flow and analyzed by the built-in software of the instrument (NanoSight NTA software 3.4 version 003).
Preparation of Total Protein Extract of Strawberries
Strawberries were frozen using liquid N2 and then transferred into a stainless-steel blender. The sample was homogenized using the blender at maximum velocity for 10 s three times. In between blending intervals, some liquid N2 was added to grind the fruit completely into a powder. Two grams of powdered fruit was weighed and transferred into a 50-ml test tube. Six milliliters of extraction buffer [2% SDS, 60 mM dithiothreitol (DTT), 20% glycerol, and 40 mM Tris-HCl, pH 8.5] was added per gram of powdered fruit and incubated at 90°C for 8 min. After incubation, the sample was centrifuged at 8,000 × g for 15 min at 4°C using a fixed angle rotor. The supernatant was collected in a 50-ml Falcon tube, and 25 ml of precipitation solution [(containing 10% w/v) trichloroacetic acid and 20 mM DTT prepared in ice-cold acetone) was added. The solution was kept at −20°C for 45 min and then centrifuged at 18,000 ×g for 10 min at 4°C. The supernatant was discarded, and the pellet was washed three times with a 7.5-ml washing solution containing 20 mM DTT prepared in ice-cold acetone. The pellet was suspended in 35 ml of washing solution, incubated at -20°C for 1 h, and centrifuged at 20,000 xg for 10 min at 4°C. The supernatant was discarded, and the pellet was air dried. The air-dried pellet was resuspended in 700 μl of rehydration buffer containing 7 M urea, 2 M thiourea, 30 mM Tris HCl, pH 8.5, and 43 mM DTT. The sample was kept at constant shaking for 1 h at room temperature and centrifuged at 12,000 × g for 10 min at room temperature. The resulting supernatant was used as the total protein extract (TPE).
Protein Quantification and Profiling
The protein concentrations were measured by a microbicinchoninic (BCA) acid assay kit (Thermo Scientific, Rockford, IL, United States) using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, United States). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used for protein separation. Samples (20 μg protein measured by the micro-BCA assay) were electrophoretically separated under reducing conditions on a precast Novex Bolt 4–12% Bis-Tris Plus gel using Bolt MOPS SDS running buffer (Life Technologies, Carlsbad, CA, United States) according to the instructions of the manufacturer and stained with colloidal Coomassie blue (Applichem GmbH, Darmstadt, Germany).
Lysis of the Vesicles and Enzymatic Digestion
For in-solution digestion proteomics analysis, samples (50 μg of protein measured by micro-BCA assay) were resuspended in 0.2% RapiGest SF (Waters Corp., Milford, MA, United States), and vesicles were lysed using five freeze and thaw cycles under sonication. Proteins were reduced using 5 mM DTT (Sigma-Aldrich, Saint Louis, MO, United States), alkylated using 15 mM iodoacetamide (Sigma-Aldrich, Saint Louis, MO, United States), and proteolytically digested using mass spectrometry-grade trypsin (Pierce, Thermo Fisher Sci Rockford, IL, United States). Samples were vacuum dried and solubilized in 5% acetonitrile and 0.5% formic acid (LC-MS grade, Thermo Sci. Rockford, IL, United States) before nano-HPLC-MS/MS analysis.
LC-ESI-MS/MS
The peptide samples were dissolved in 0.1% formic acid by gentle vortexing. Shotgun proteomics analysis was carried out using an Acquity UPLC system (Waters, Milford, MA, United States) directly coupled to a Q-Exactive Orbitrap Hybrid Mass Spectrometer (Thermo Fisher Scientific). For the proteome profiling analysis, the optimized gradient elution program of solvent A (Sol A: 0.1% formic acid) and solvent B (Sol B: 0.1% formic acid in acetonitrile) was set as follows: (T min/% of Sol B): 0/5, 4/5, 9/12, 50/30, 110/40, 112/80, 118/80, 120/5, 140/5. The dissolved peptides were separated using an EASY-spray column (50 cm × 75 µm i.d., C18, 2 μm, 100 Å, Thermo Fisher Scientific). MS acquisitions were performed in positive ion mode using an electrospray voltage of 2.4 kV. The operating conditions for full MS scan were set as follows: scan range, 400–2,000 m/z; resolution, 70,000; automated gain control target value (AGC target), 3.0 × 106; maximum ion injection time (IT), 100 ms. The operating conditions for MS/MS scan were set as follows: resolution, 17,500; AGC target, 5.0 × 105; minimum AGC target, 2.0 × 103; maximum IT, 50 ms; normalized HCD collision energy (NCE), 30; dynamic exclusion time, 30 s; isolation window, 2.0 m/z. Samples were analyzed in three technical replicates.
Protein Identification and Quantitation
The acquired raw LC-ESI-MS/MS data were processed using the SEQUEST HT search engine of the Proteome Discoverer v.2.4 program (Thermo Fisher Scientific). The tandem MS spectra were analyzed against the Fragaria vesca protein database (January 2020, 32,886 entries) in the NCBI database, which generated decoy spectra. The following SEQUEST search parameters were used: cleavage of arginine and lysine residues using trypsin, allows for two missed cleavages, carbamidomethylation of cysteines, variable modifications of oxidation of methionine and protein N-terminal acetylation. The threshold for identification of the target peptide spectrum was applied to a 1% false discovery rate (FDR) in protein level analysis. Tolerances were set at 30 ppm for precursor mass and 0.02 Da for fragment mass, respectively.
IgE Inhibition Experiments with the FABER® Testing System
FABER is a multiplex in vitro serological test allowing the detection of IgE antibodies specifically recognizing allergens immobilized on a biochip (Tuppo et al., 2018; Giangrieco et al., 2019). The FABER version used to perform this study (FABER 244–122–122) contains 122 purified allergens and 122 multiple protein allergenic extracts, coupled to chemically activated nanoparticles and then spotted on a biochip (Figure 3). The FABER biochip is used for an ELISA-like testing procedure that allows the detection of specific IgE to each of the 244 allergenic preparations. The final signal for each allergen is obtained with the use of an optical scanner and elaborated with specific software. The signals detected for each spot are interpolated with values obtained with an internal IgE standard reference curve present in each biochip and also used as a reference between chips. Signal intensities are then used to calculate the arbitrary units, FIU (FABER International Units), correlated with the level of IgE bound to each allergen spot. To obtain information on the content of possible allergens in the strawberry samples, the SPHIAa (Pasquariello et al., 2012; Tuppo et al., 2019) (Yakhlef et al., 2021) was performed on the FABER platform. Sera of allergic patients for which the IgE profile was already known was used as providers of IgE recognizing specific allergenic proteins spotted on the FABER biochip. The assay was performed by incubating 0.12 ml of the sera pool with 0.12 ml of a solution containing 0.1 mg of the NVs samples. The IgE-binding inhibition was evaluated by running the FABER test and recording the residual IgE binding on the allergens spotted on the biochip. Experiments were carried out in duplicates, and the mean values are reported. Reference values for lack of IgE-binding inhibition were obtained by running control samples where the allergen solution was substituted with buffer only. The inhibition values were calculated in real time by a specific procedure developed within the InterAll software (version 5.0, ADL s.r.l.) (Alessandri et al., 2017). Values equal to or higher than 30% inhibition were considered as reliable inhibitions and reported, whereas lower values were discarded.
Sera of Patients
Sera of patients that had proven to be sensitized to different plant foods and had been analyzed with the FABER test were selected. To carry out the SPHIAa assays (Figure 3), four sera of patients sensitized to different plant foods and containing IgE-recognizing relevant plant allergens, such as LTP, profilin, Bet v 1-like, GRP, and seed storage proteins, were selected in the InterAll databank (version 5.0, ADL S.r.l.) (Alessandri et al., 2017). A volume of 0.5 ml was collected from each one of the four sera and pooled to be used for IgE-binding inhibition experiments.
All patients gave their informed consent to the use of their clinical data for research purposes in an anonymous form. Because of the purely comparative nature of this study, along with the fact that all venous blood samplings were part of routine clinical practice and that a residual part of the routine sample was used for inhibition experiments, formal approval by the Ethical Committee was not necessary.
Bioinformatics
The OmicsBox 2.0.10 software package was used for in silico data analysis (Conesa and Gotz, 2008). Protein sequences were blasted against i) the NCBI public database with taxonomy filter green plants (Viridiplantae, 20,952,768 entries) and ii) the COMPARE allergen database (2348 entries) (Van Ree et al., 2021) (https://comparedatabase.org/) using BlastP service with a maximum number of blast hits 20 and expectation value of 1.0 × 10−3. Blast databases of F. x ananassa, NCBI green plants, and COMPARE were created by OmicsBox 2.0.10. The InterPro domain searches were performed using the public European Molecular Biology Lab-European Bioinformatics Institute to identify sequences against the signatures of CDD, HAMAP, HMMpfam, HMMPIR, Fprintscan, and BlastproDom Interpro. All sequences generated InterPro results. Annotated sequences were mapped against exclusively created GO-annotated proteins to obtain functional labels of GO associated and ID mapping of Uniprot. The FunRich program was used to prepare Venn diagrams (Pathan et al., 2015). Sequence homologs of the three known strawberry allergens (Fra a 1, Fra a 3, and Fra a 4) were identified by blasting the sequences of F. vesca (wild strawberry, Taxonomy ID: 57,918) in the UniProt database used for the protein identification against F. x ananassa (Taxonomy ID: 3747, 1671 entries on July 21, 2021).
Results
Isolation and Characterization of Microvesicles and Nanovesicles
MVs and crude NV samples from strawberries were isolated by DUC as low- (15,000 × g) and high-velocity (110,000 ×g) pellets according to the protocol (Supplementary Figure S1). NV samples were further separated by DGUC using iodixanol, a nonionic density gradient medium with relatively low osmolality and high density (Figure 1B), or sucrose/D2O cushions. The isolated vesicles were characterized regarding their size, density, zeta-potential, protein concentration, and bio-cargo composition according to the recent MISEV guidelines (Théry et al., 2018).
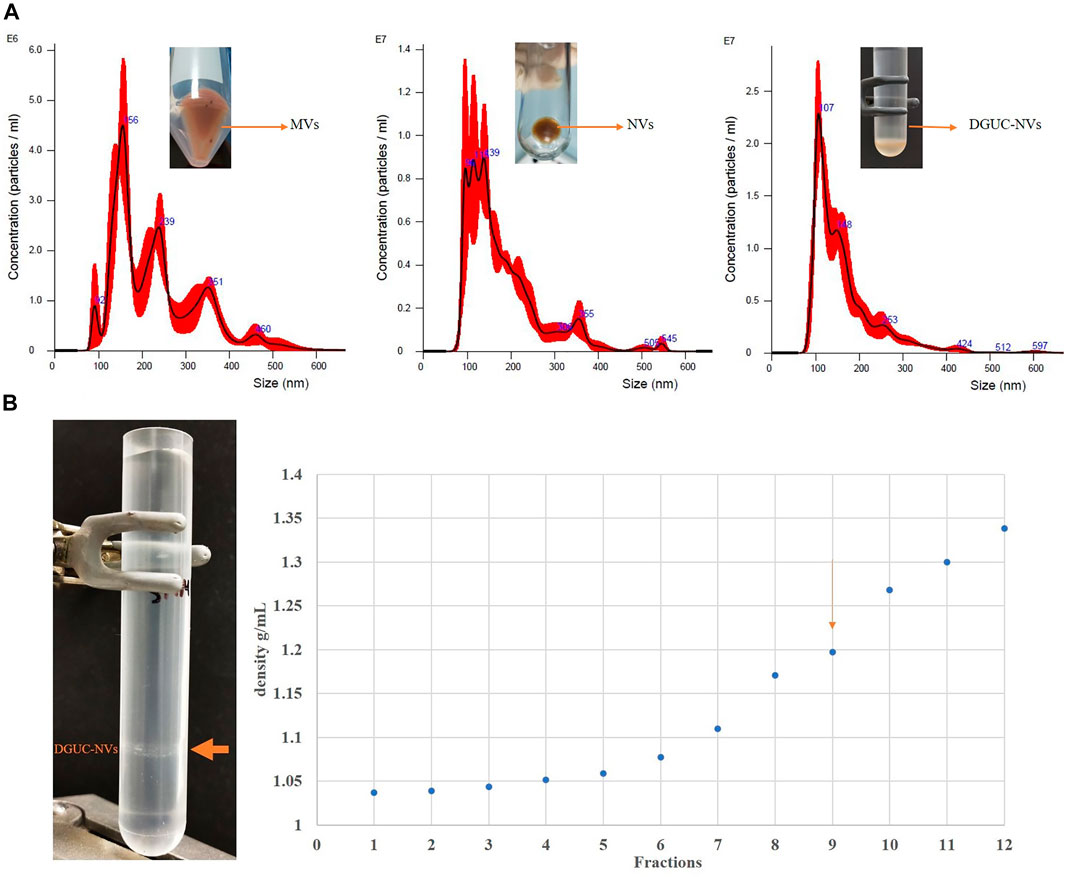
FIGURE 1. (A) Nanoparticle tracking analysis showing the size distribution of microvesicles (MVs), nanovesicles (NVs), and the sucrose density gradient purified nanovesicle fraction (DGUC-NVs). Particle numbers, mean diameter values, and standard deviations are reported in Table 1. (B) The pictures on the left show the sample of strawberry-derived NVs after iodixanol density gradient ultracentrifugation (DGUC). After DGUC, a single visible band could be observed (orange arrow). The densities measured in the 12 different fractions from top to bottom are shown in the chart on the right. The visible band was observed in fraction 9. The vesicles floating in fraction nine are similar in density to mammalian extracellular vesicles, which have a density of 1.13–1.19 g/ ml (Konoshenko et al., 2018).
Figure 1A and Table 1 show the size distribution of particles in MVs, NVs, and the sucrose DGUC separated NV fraction (DGUC-NVs) analyzed by NTA. The size distribution of vesicles in the density gradient purified fraction (DGUC-NVs) was more homogenous with abundant small-sized (<150 nm) vesicles compared with MVs, which had a larger size distribution. The small vesicles that are present in the MV fraction could have cosedimented during the 15,000 ×g centrifugation step. The measured z-potential showed a negative surface charge of −26 mV for DGUC-NVs. The protein amounts associated with the vesicles were determined by the micro-BCA assay. The isolation (Supplementary Figure S1) yielded 20 mg of proteins in the MVs, 38 mg in the NVs, and 5 mg in the DGUC-NVs from 250 g of strawberry. The calculated particle per µg of protein ratio for the NVs, MV samples, and DGUC-NVs fraction were 8.0E8, 2.2E9, and 3.0E9 particles/µg of protein, respectively. The increased particles to protein ratios indicate the enrichment of the vesicles in the samples.
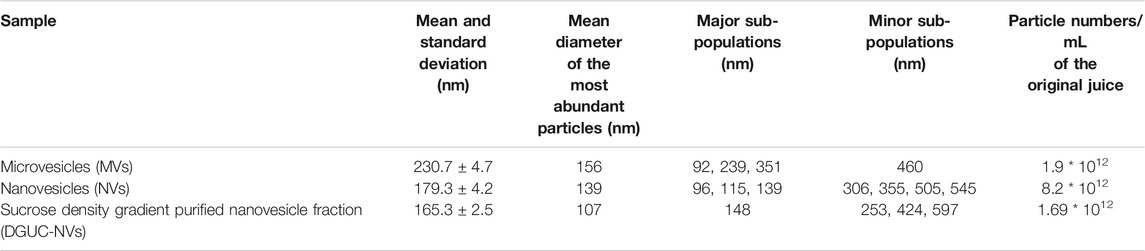
TABLE 1. The particle concentration and the size range of different vesicle populations isolated from strawberries.
Proteomic Analysis of Vesicles Isolated from Strawberry
Figure 2A shows the SDS-PAGE protein profile of the MVs, NVs, and DGUC-NVs samples isolated from strawberries. The gel image shows protein profiles with numerous bands that are evenly distributed throughout the gel. The three vesicle samples exhibit similar SDS-PAGE protein profiles different from that of TPE.
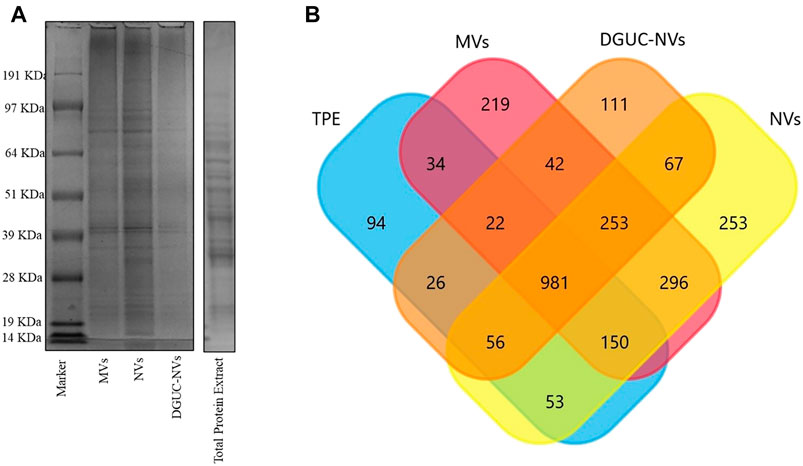
FIGURE 2. (A) SDS-PAGE protein profile of MVs, NVs, DGUC-NVs, and total protein extract (TPE). (B) Venn diagram showing the number of identified proteins and their overlappings in the TPE, MVs, crude NVs, and the sucrose gradient density DGUC-NV fraction.
Samples TPE, MVs, NVs, and DGUC-NVs were subjected to in-solution digestion-based label-free shotgun proteomic analysis. Supplementary Table S1 reports the proteins identified against the F. vesca NCBI database in each sample, and Figure 2B shows the numbers of identified proteins. Proteins (981) were commonly identified in all samples, while 219, 253, and 111 sequences were unique to MVs, NVs, and DGUC-NVs, respectively (Supplementary Table S1). Vesicle isolates express highly abundant heat shock proteins (HSP83, HSP70, and HSP90), villin-3-like protein, myosin-10, ATPases, and clathrin heavy chain-1 protein. However, it should be noted that these proteins were also abundant in the TPE. A label-free quantitative study was performed to highlight the differentially expressed proteins over the three vesicle isolates (MVs, NVs, and DGUC-NVs) with respect to TPE. Four hundred one commonly expressed proteins were quantified over the sample set (Supplementary Table S2). Volcano plots highlight the statistically significant differential expression according to the t-test (Supplementary Table S2). There are 75 MVs, 30 NVs, and 51 DGUC-NVs proteins that showed significantly higher expression levels compared with their expression in TPE (Supplementary Table S3). In particular, six proteins were enriched more than two times in all the vesicle samples (MVs, NVs, and DGUC-NVs), and these were tubulin beta chain, protein SSUH2 homolog, HSP90, pyruvate kinase (PK), malonyl-CoA decarboxylase, and a probable mediator of RNA polymerase II transcription. The function of SSUH2 homolog proteins in the plant is largely unknown, and its increased expression in vesicles would need further investigation. HSP90 and the glycolytic enzymes PK are frequently identified also in mammalian cell-derived EVs. Malonyl-CoA decarboxylase is involved in fatty acid biosynthesis and, its expression, secretion, or transportation in vesicles has not been reported, although it is known that some EVs carry proteins involved in lipid metabolism including different enzymes (Clement et al., 2020). ATPases, similar to other plant-derived vesicles, such as, for example, that from citrus juice fruit (Pocsfalvi et al., 2018) and tomato fruit (Bokka et al., 2020), were also significantly enriched in strawberry vesicles (Supplementary Table S3). The specificity of these six proteins enriched in strawberry vesicles could be further elucidated as general markers for fruit-derived vesicles.
Quantitative Analysis of Homologs of Strawberry Allergens
Homologs of so far reported strawberry allergens were identified in the proteomics data by performing a blast search against the F x ananassa UniProt database that contains several isoforms of Fra a 1, Fra a 3, and Fra a 4 allergens. The analysis resulted in the detection of the following homologs for each of the three allergens: Fra a 1 (major allergen Pru av 1-like, XP_004296887.1), Fra a 3 (three non-specific lipid transfer proteins, XP_004297800.1, XP_004300702.1, and XP_011466421.1), and Fra a 4 (profilin, XP_004287490.1). Supplementary Table S4 reports the expression levels measured in all samples and their fold change with respect to TPE. While all three known strawberry allergens were expressed in TPE, we could only identify Fra a 1 and Fra a 4 in the vesicle samples by MS-based proteomics analysis. Fold-changes of Fra a 1 and Fra a 4 homologs were measured less than two indicating no significant enrichment for these allergens in the vesicle samples with respect to TPE. Specifically, Fra a 1 was expressed at a lower level in MVs (fold change 0.2) and NVs (fold change 0.5) as in TPE, and the same level in DGUC-NVs. Fra a 4 expression was lower in MVs (fold change 0.4), similar in DGUC-NVs (fold change 0.9), and higher in NVs (fold change 1.5) than in TPE. Thus, quantitative proteomics indicate that while vesicles carry two out of the three strawberry allergens, they were less expressed (Fra a 1) or not significantly enriched (Fra a 4) within the different vesicle populations.
Identification of IgE-Binding Proteins in Strawberry Vesicle Isolates by the SPHIAa Assay
The presence in strawberry-derived vesicle isolates (MVs, NVs, and DGUC-NVs) of proteins recognized by IgE antibodies contained in the sera of allergic patients was investigated by in vitro immunological tests. In particular, the SPHIAa method was used to probe allergens included in the FABER system (Figure 3).
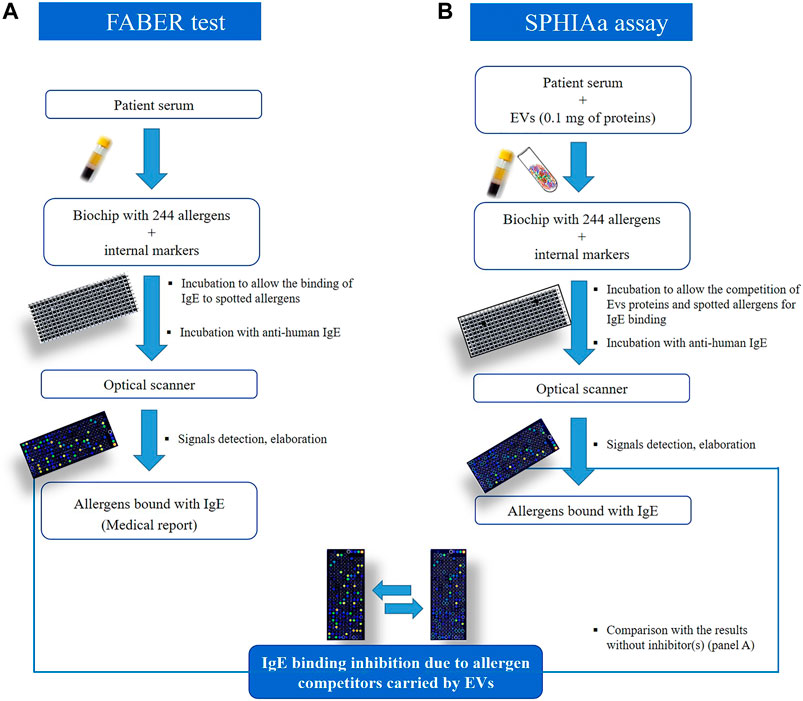
FIGURE 3. Schematic representation to compare the FABER test on (A), which is used for allergy diagnosis and research purposes, to the procedure used for the Single Point Highest Inhibition Achievable assay (SPHIAa) assay (B).
The isolated strawberry allergens (Fra a 1, Fra a 3, and Fra a 4) were not present on the FABER biochip, but several Bet v1, LTP, and profilin homologs from other sources were available. In the assay, the proteins transported by the vesicles compete with the allergens spotted on the FABER biochip for binding to specific IgE. Vesicle isolates were applied without lysis. Table 2 shows the allergens of relevant allergen families, contained in the FABER biochip, that were analyzed for IgE-binding inhibitions. In particular, we observed inhibition on seven protein allergen groups: Bet v 1-like proteins (Fra a 1), profilins (Fra a 4), seed storage proteins (2S-albumin/11S globulin or legumin-like protein), gibberellin-regulated proteins (GRPs), LTP (Fra a 3), trypsin inhibitors, cross-reactive carbohydrate determinant (CCD) markers and other CCD-bearing allergens. It can be observed that components of the same protein family are inhibited to a different extent (Figure 4) producing different inhibition profiles of MVs, NVs, and DGUC-NVs. Five allergens of the Bet v 1-like family (Bet v 1, Api g 1, Ara h 8, Cor a 1, and Mal d 1) were available in the test system for the SPHIAa assay. The celery Api g 1 was completely inhibited by DGUC-NVs proteins, and it was partially inhibited by NVs, whereas no inhibition at all was produced by the proteins of MVs. This trend is in line with the quantitative data measured by proteomics. The hazelnut Cor a 1 was partially inhibited at a similar extent by all the three vesicles fractions, whereas the peanut, apple, and birch pollen homologous allergens were not inhibited. Among the three analyzed profilins, the latex Hev b 8 is the only one partially inhibited by all the three vesicle fractions, whereas the birch pollen Bet v 2 and Mer a 1 from annual mercury are inhibited at about 80% by MVs only. Three seed storage proteins competed with vesicle proteins for IgE binding. The cashew 2S albumin, Ana o 3, displayed 100% IgE-binding inhibition with all the vesicle fractions, whereas the hazelnut 11S globulin, Cor a 9, was completely inhibited by NVs only. Conversely, the peanut 11S globulin, Ara h 3, was partially inhibited by all the vesicles studied.
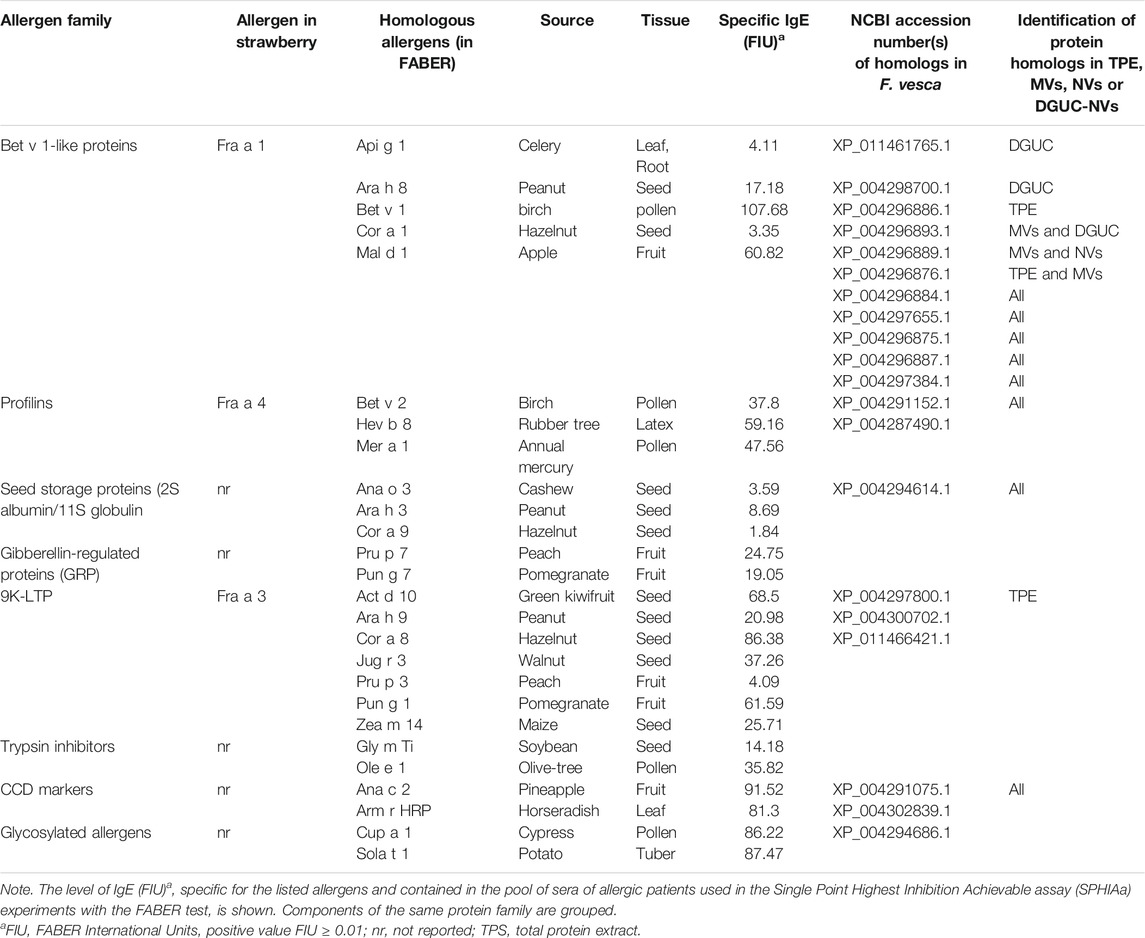
TABLE 2. Details of plant allergens analyzed for competition in the IgE binding with the proteins contained in the vesicles samples.
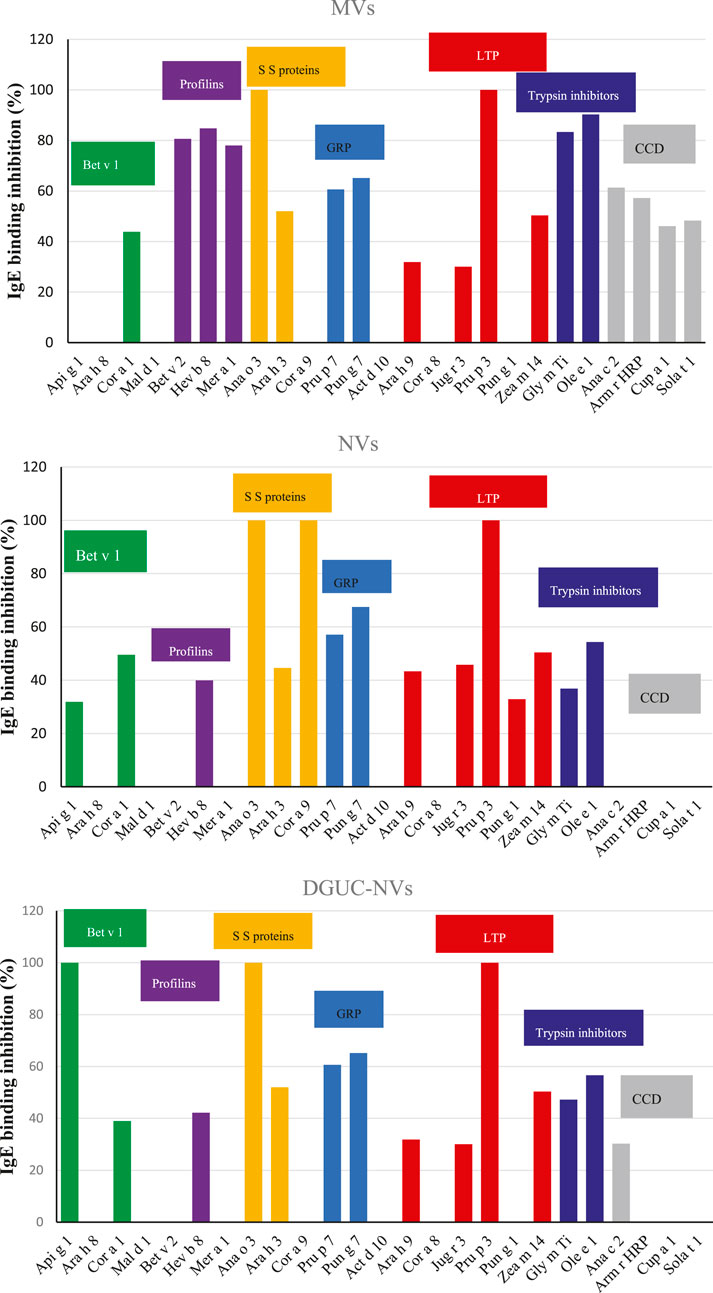
FIGURE 4. IgE-binding inhibition assays using the FABER testing system with the SPHIAa method and a pool of sera from allergic patients as the provider of specific IgE. The three panels show the percentage of inhibition on the indicated allergens using NVs, MVs, and DGUC-NVs as inhibitors. The allergens belonging to the same protein family have been grouped. The protein families are highlighted with different colors: Bet v 1-like allergens in green, profilins in purple, seed storage proteins in yellow, gibberellin-regulated proteins (GRP) in light blue, LTP in dark red, trypsin inhibitors in blue. In addition, cross-reactive carbohydrate determinant (CCD)-bearing proteins are highlighted in gray.
The two analyzed GRP, the peach Pru p 7 and the pomegranate Pun g 7, were partially inhibited by all the three vesicle samples with values around 60%. Six components of the LTP family were available for SPHIa assays. All the vesicle samples produced 100% IgE-binding inhibition on the peach LTP, Pru p 3, and a partial inhibition on the peanut Ara h 9, the walnut Jug r 3, and the maize Zea m 14. The hazelnut Cor a 8 was never inhibited, whereas the pomegranate Pun g 1 showed a value of about 34% with NVs.
The trypsin inhibitors, namely, the soybean Gly m Ti and the olive tree Ole e 1, were inhibited by all three vesicle isolates, but the highest values were produced by MVs. Only MVs caused inhibition on both the CCD markers, the pineapple Ana c 2, and the horseradish Arm r HRP Altmann, 2016. In addition, this isolate caused IgE-binding inhibition on two glycosylated allergens, Cup a 1 and Sola t 1, which are frequently recognized by CCD-specific IgE.
Identification of Sequence Homologs to known Allergens Enriched in Vesicle Samples
To find homologs to known allergens in strawberry vesicles, a BLAST sequence similarity searching of the quantified proteins (Supplementary Table S2) was performed against the COMPARE database. The analysis identified 56 homologs with 50% or higher sequence identity, and these, together with their expression values, are reported in Supplementary Table S5. Among these proteins, we can find that beta-tubulin (BTUB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), cysteine proteinase inhibitor (CPI, cystatin), and peptidyl-prolyl cis-trans isomerase 1 (PPIL1) were highly expressed in all vesicle samples (Supplementary Table S5 and Figure 5) compared with TPE. BTUB was reported as an allergen from dust mites, but a recent study reported on plant-specific tubulin as a respiratory allergen from pollen grains too (Ghosh et al., 2015). BTUB was especially highly enriched in the MV isolate (7.6 times higher than in TPE). GAPDH was about threefold enriched in all vesicle samples in comparison with the extract. GAPDH is frequently found to be expressed in EVs. A recent study identified GAPDH as one of the in vitro and in vivo IgE suppressors present in strawberry extract (Iwamoto et al., 2021). Our study is the first indication that GAPDH could be accumulated in, and perhaps transmitted by, membrane-enclosed vesicles. The sequence homolog of CPI 12 (XP_004307209.1) Act d 4 is a kiwifruit allergen that contributes to the clinical symptoms of kiwifruit allergy (Popovic et al., 2010). The homolog of Act d 4 in strawberry has not yet been reported. However, plant EVs have been reported to carry a complete set of defense molecules; thus, it was not surprising that CPI, an important molecule involved in plant development and defense, was found to accumulate in all strawberry vesicles and especially in the NVs. Bet v 7 from birch pollen and Cat r 1 from periwinkle pollen are considered pan-allergens due to frequently reported cross-reactivity. PPIL1 (XP_004289844.1) of strawberry shows structural similarity with Bet v 7 cyclophilin, and it was enriched in all vesicle samples though at a higher rate in DGUC-NVs.
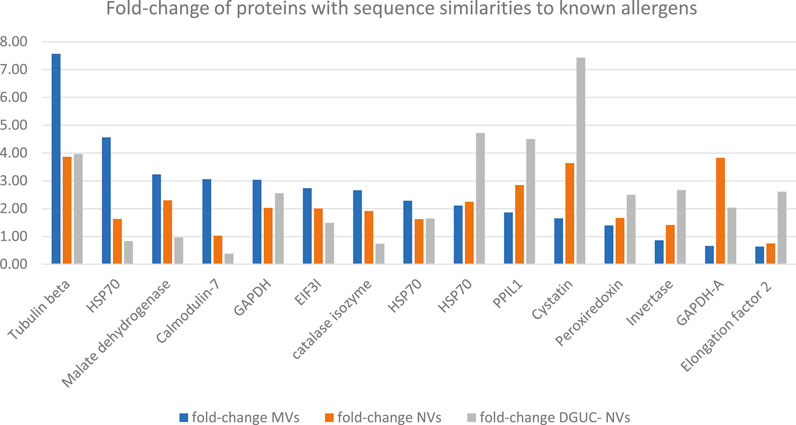
FIGURE 5. Fold-change of proteins with sequence similarities to known allergens in the COMPARE database and highly expressed in MVs, NVs, or sucrose gradient ultracentrifugation separated NVs fraction. Fold-change with respect to the total protein extract measured by quantitative shotgun proteomics (Supplementary Table S5).
To find the sequence homologs to the allergens identified by the SPHIAa in vitro immunological tests among the identified proteins (F. vesca), we performed a blast search of allergens inhibited in FABER and their isoforms published in the Allergome database against COMPARE database. This resulted in 24 sequences out of which we identified six structurally similar proteins in our proteomics dataset. This comprises the Fra a 4 (profilin), Fra a 1 (Bet v 1), CCD-bearing proteins, and seed storage proteins. Figure 6 shows the fold change of these proteins with respect to TPE measured by label-free quantitative proteomics. While different expression patterns of these proteins were found in the different vesicle populations (MVs, NVs, and DGUC-NVs), it is important to note that their fold changes were less than two in all cases (Figure 6). This indicates that these homologs to allergens measured by FABER are present, but they are not significantly enriched in the vesicle populations.
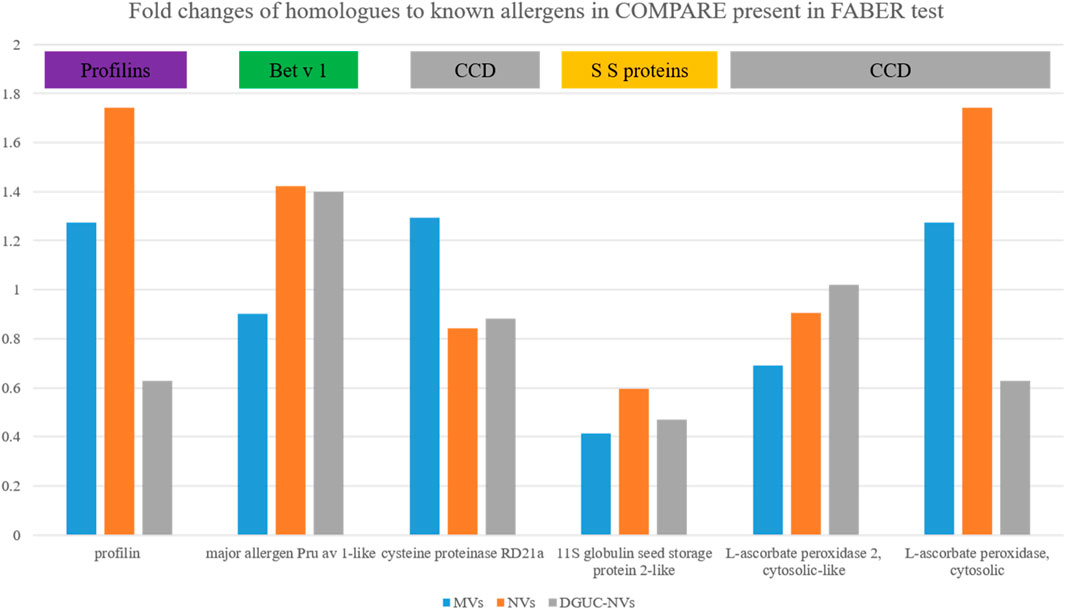
FIGURE 6. The fold changes measured by quantitative proteomics of the six proteins, homologs to allergens commonly identified in MVs, NVs, and DGUC-NVs (Supplementary Table S5).
Discussion
Here, we isolated three different vesicle populations from the false fruit of strawberries based on size (MVs and NVs) and density (DGUC-NVs). We obtained very high yields that could put a strawberry in an interesting position for the production of nanovesicles from this plant source. NVs were separated by the density gradient to obtain vesicle populations similar to mammalian EVs in density. NTA analysis showed the vesicle character of the samples and proved that MVs, although not considerable, were bigger than NVs. The SDS-PAGE and shotgun proteomic analyses revealed complex protein profiles of all three vesicle isolates studied comparable with that of the total protein extract of strawberries. This is in line with the observations reported in other fruit-derived vesicles, such as tomato (Bokka et al., 2020) and citrus (Pocsfalvi et al., 2018), and share characteristics with mammalian EVs isolated from different sources, such as biofluids and cell cultures. The proteomes of the different vesicle isolates showed overlapping protein contents: 1,234 proteins were commonly expressed (Figure 2B), although in different amounts in all vesicle samples (Supplementary Table S3). However, distinct features could also be recognized since 219, 252, and 111 proteins were exclusively identified in the MVs, NVs, and DGUC-NVs vesicle isolates (Figure 2B). Quantitative proteomics highlighted a set of proteins that were significantly overexpressed in the vesicles isolated from strawberries and which could be useful as markers for their enrichment. Among these, tubulin, HSPs, pyruvate kinase, and ATPases are often reported in both plant-derived vesicles and mammalian EVs.
Proteomics and bioinformatics analysis identified and quantified several proteins in TPE and vesicle isolates with high sequence similarities to known strawberry allergens and allergens from other sources reported in the COMPARE database. Immunological investigations revealed that vesicle isolates from strawberries were carrying proteins bearing functional IgE epitopes. Competition between the proteins of the vesicle isolates and components of several relevant allergen families, included in the FABER biochip, was detected by IgE-binding inhibition experiments. The results obtained showed that the vesicles carry all the three allergens so far described in strawberry, belonging to the protein families of Bet v 1-like protein, (Fra a 1), LTP (Fra a 3), and profilins (Fra a 4). Proteomics identified Fra a 3 in TPE, but not in of the vesicle preparations. This could be explained by its lower than the detectable of this allergen level in the vesicles. Importantly, the quantitative proteomic analysis did not show a significant fold enrichment of the three strawberry allergens in the vesicle samples with respect to TPE. Moreover there, we identified 14 proteins that were homologs of known allergens present in the COMPARE database.
In addition to the already reported allergenic proteins of strawberries, immunological and proteomics results suggest that additional allergenic proteins and homologs to known allergens contained in this food can be carried by vesicles. In particular, IgE-binding proteins, belonging to seed storage proteins, GRP, and trypsin inhibitor allergen families were detected as part of the vesicular cargo.
For instance, all the three vesicle fractions competed with IgE specific for the cashew 2S albumin immobilized on the FABER biochip giving 100% inhibition. In addition, inhibition on peanut and hazelnut 11S globulins was observed.
The immunological investigation also suggests that vesicles of strawberry carry homologous to allergenic GRP. All the vesicle isolates caused a similar partial inhibition (about 60%) IgE-binding inhibition on peach (Pru p 7) and pomegranate (Pun g 7) GRP. The first allergenic protein of the GRP family was discovered quite recently (Tuppo et al., 2013), but later, the number of components reported to cause allergic reactions has increased. Since severe symptoms, including anaphylaxis, have been associated with these proteins, GRP represents a clinically relevant allergen family (Inomata, 2020). The detection of proteins competing with Pru p 7 and Pun g 7 for IgE binding suggests that strawberries could be an additional source of allergenic GRP, and they are part of the protein cargo of the vesicles. Nevertheless, the sensitization revealed by IgE binding is not sufficient to understand the clinical relevance of strawberry GRP, and further studies are required.
In this study, as per our knowledge, the presence of functional allergens was shown for the first time in plant food-derived vesicles. These novel nanomaterials have promising characteristics as native carriers of endogenous substances against inflammation and cancer and they are under exploitation as nanovectors for the delivery of exogenous substances for human health. The allergens detected were not enriched in strawberry vesicles with respect to the fruit extract showing that vesicles are not specific vehicles of these allergens. Before the application of these nanomaterials in human health and nutrition, however, it is important to accumulate information on their possible allergenicity and, thus, carry out a similar analysis in other plant food derived vesicles.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CS participated in the study design and performed the experiments, interpreted the data, and drafted the manuscript. GA performed the experiments. HK performed the proteomics analysis. MG performed the sizing studies. IF performed the experiments and data analysis. MAC performed the immunological data analysis and wrote the manuscript. IG analyzed the data and wrote the manuscript. AM designed the immunological experiments and selected the patient sera. GP and KK designed the study, analyzed the data, revised the manuscript, and provided professional advice. All authors read and approved the final manuscript.
Funding
This study was supported by funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 801338, the Green and Circular Economy (GECE) project N.856, the year 2019, funded by the Italian Ministry of University and Research (MUR), and by the institutional funding at Gyeonggi-do Regional Research Center (GRRC) program of Gyeonggi Province GRRC-Kyung Hee 2018 (B03).
Conflict of Interest
AM as, an ADL stakeholder, develops, manufactures, and distributes the FABER test as a diagnostic service. IG as an employee of ADL contributed to the development, manufacture, and performance of the FABER test. MAC receives funding from ADL.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Chiara Rafaiani and Claudia Rafaiani for performing the immunological experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.760730/full#supplementary-material
References
Alessandri, C., Ferrara, R., Bernardi, M. L., Zennaro, D., Tuppo, L., Giangrieco, I., et al. (2020). Molecular Approach to a Patient's Tailored Diagnosis of the Oral Allergy Syndrome. Clin. Transl. Allergy 10, 22. doi:10.1186/s13601-020-00329-8
Alessandri, C., Ferrara, R., Bernardi, M. L., Zennaro, D., Tuppo, L., Giangrieco, I., et al. (2017). Diagnosing Allergic Sensitizations in the Third Millennium: Why Clinicians Should Know Allergen Molecule Structures. Clin. Transl. Allergy 7, 21. doi:10.1186/s13601-017-0158-7
Altmann, F. (2016). Coping with Cross-Reactive Carbohydrate Determinants in Allergy Diagnosis. Allergo J. Int. 25, 98–105. doi:10.1007/s40629-016-0115-3
Amatori, S., Mazzoni, L., Alvarez-Suarez, J. M., Giampieri, F., Gasparrini, M., Forbes-Hernandez, T. Y., et al. (2016). Polyphenol-rich Strawberry Extract (PRSE) Shows In Vitro and In Vivo Biological Activity against Invasive Breast Cancer Cells. Sci. Rep. 6, 30917. doi:10.1038/srep30917
Bokka, R., Ramos, A. P., Fiume, I., Manno, M., Raccosta, S., Turiák, L., et al. (2020). Biomanufacturing of Tomato-Derived Nanovesicles. Foods 9, 1852. doi:10.3390/foods9121852
Clement, E., Lazar, I., Attané, C., Carrié, L., Dauvillier, S., Ducoux‐Petit, M., et al. (2020). Adipocyte Extracellular Vesicles Carry Enzymes and Fatty Acids that Stimulate Mitochondrial Metabolism and Remodeling in Tumor Cells. EMBO J. 39, e102525. doi:10.15252/embj.2019102525
Conesa, A., and Götz, S. (2008). Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genomics 2008, 1–12. doi:10.1155/2008/619832
Ghosh, N., Sircar, G., Saha, B., Pandey, N., and Gupta Bhattacharya, S. (2015). Search for Allergens from the Pollen Proteome of Sunflower (Helianthus Annuus L.): A Major Sensitizer for Respiratory Allergy Patients. PLoS One 10, e0138992. doi:10.1371/journal.pone.0138992
Giangrieco, I., Ricciardi, T., Alessandri, C., Farina, L., Crescenzo, R., Tuppo, L., et al. (2019). ENEA, a Peach and Apricot IgE-Binding Protein Cross-Reacting with the Latex Major Allergen Hev B 5. Mol. Immunol. 112, 347–357. doi:10.1016/j.molimm.2019.05.007
Hyun, T. K., and Kim, J.-S. (2011). Genomic Identification of Putative Allergen Genes in woodland Strawberry (Fragaria Vesca) and Mandarin orange (Citrus Clementina). Plant Omi. J. 4, 428–434. Available at: https://www.pomics.com/kim_hyun_4_7_2011_428_434.pdf.
Inomata, N. (2020). Gibberellin-regulated Protein Allergy: Clinical Features and Cross-Reactivity. Allergol. Int. 69, 11–18. doi:10.1016/j.alit.2019.10.007
Iwamoto, A., Inoue, Y., Tachibana, H., and Kawahara, H. (2021). Immunomodulatory Effect of Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) in Allergic Conditions In Vitro and In Vivo. Cytotechnology 73, 333–342. doi:10.1007/s10616-020-00438-z
Karimi, N., Cvjetkovic, A., Jang, S. C., Crescitelli, R., Hosseinpour Feizi, M. A., Nieuwland, R., et al. (2018). Detailed Analysis of the Plasma Extracellular Vesicle Proteome after Separation from Lipoproteins. Cell. Mol. Life Sci. 75, 2873–2886. doi:10.1007/s00018-018-2773-4
Konoshenko, M. Y., Lekchnov, E. A., Vlassov, A. V., and Laktionov, P. P. (2018). Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 1–27. doi:10.1155/2018/8545347
Mu, J., Zhuang, X., Wang, Q., Jiang, H., Deng, Z. B., Wang, B., et al. (2014). Interspecies Communication between Plant and Mouse Gut Host Cells through Edible Plant Derived Exosome‐like Nanoparticles. Mol. Nutr. Food Res. 58, 1561–1573. doi:10.1002/mnfr.201300729
Offermann, L. R., Schlachter, C. R., Perdue, M. L., Majorek, K. A., He, J. Z., Booth, W. T., et al. (2016). Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art V 4, and Bet V 2. J. Biol. Chem. 291, 15447–15459. doi:10.1074/jbc.M116.733659
Pasquariello, M. S., Palazzo, P., Tuppo, L., Liso, M., Petriccione, M., Rega, P., et al. (2012). Analysis of the Potential Allergenicity of Traditional Apple Cultivars by Multiplex Biochip-Based Immunoassay. Food Chem. 135, 219–227. doi:10.1016/j.foodchem.2012.04.075
Pathan, M., Keerthikumar, S., Ang, C.-S., Gangoda, L., Quek, C. Y. J., Williamson, N. A., et al. (2015). FunRich: An Open Access Standalone Functional Enrichment and Interaction Network Analysis Tool. Proteomics 15, 2597–2601. doi:10.1002/pmic.201400515
Perut, F., Roncuzzi, L., Avnet, S., Massa, A., Zini, N., Sabbadini, S., et al. (2021). Strawberry-Derived Exosome-like Nanoparticles Prevent Oxidative Stress in Human Mesenchymal Stromal Cells. Biomolecules 11, 87. doi:10.3390/biom11010087
Pocsfalvi, G., Turiák, L., Ambrosone, A., del Gaudio, P., Puska, G., Fiume, I., et al. (2018). Protein Biocargo of Citrus Fruit-Derived Vesicles Reveals Heterogeneous Transport and Extracellular Vesicle Populations. J. Plant Physiol. 229, 111–121. doi:10.1016/j.jplph.2018.07.006
Popovic, M. M., Milovanovic, M., Burazer, L., Vuckovic, O., Hoffmann-Sommergruber, K., Knulst, A. C., et al. (2010). Cysteine Proteinase Inhibitor Act D 4 Is a Functional Allergen Contributing to the Clinical Symptoms of Kiwifruit Allergy. Mol. Nutr. Food Res. 54, 373–380. doi:10.1002/mnfr.200900035
Stanly, C., Fiume, I., Capasso, G., and Pocsfalvi, G. (2016). Isolation of Exosome-like Vesicles from Plants by Ultracentrifugation on Sucrose/deuterium Oxide (D2O) Density Cushions. Methods Mol. Biol. 1459, 259–269. doi:10.1007/978-1-4939-3804-9_18
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): a Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 7, 1. doi:10.1080/20013078.2018.1535750
Tuppo, L., Alessandri, C., Giangrieco, I., Ciancamerla, M., Rafaiani, C., Tamburrini, M., et al. (2019). Isolation of cypress Gibberellin-Regulated Protein: Analysis of its Structural Features and IgE Binding Competition with Homologous Allergens. Mol. Immunol. 114, 189–195. doi:10.1016/j.molimm.2019.07.023
Tuppo, L., Giangrieco, I., Alessandri, C., Ricciardi, T., Rafaiani, C., Ciancamerla, M., et al. (2018). Pomegranate Chitinase III: Identification of a New Allergen and Analysis of Sensitization Patterns to Chitinases. Mol. Immunol. 103, 89–95. doi:10.1016/j.molimm.2018.09.009
van Ree, R., Sapiter Ballerda, D., Berin, M. C., Beuf, L., Chang, A., Gadermaier, G., et al. (2021). The COMPARE Database: A Public Resource for Allergen Identification, Adapted for Continuous Improvement. Front. Allergy 2, 39. doi:10.3389/falgy.2021.700533
Yakhlef, M., Giangrieco, I., Ciardiello, M. A., Fiume, I., Mari, A., Souiki, L., et al. (2021). Potential Allergenicity of Medicago Sativa Investigated by a Combined IgE ‐binding Inhibition, Proteomics and In Silico Approach. J. Sci. Food Agric. 101, 1182–1192. doi:10.1002/jsfa.10730
Keywords: strawberry, nanovesicles, allergens, proteomics, sequence similarity
Citation: Stanly C, Kim H, Antonucci G, Fiume I, Guescini M, Kim KP, Ciardiello MA, Giangrieco I, Mari A and Pocsfalvi G (2021) Crosstalk Between the Immune System and Plant-Derived Nanovesicles: A Study of Allergen Transporting. Front. Bioeng. Biotechnol. 9:760730. doi: 10.3389/fbioe.2021.760730
Received: 18 August 2021; Accepted: 05 October 2021;
Published: 26 November 2021.
Edited by:
Wojciech Chrzanowski, The University of Sydney, AustraliaReviewed by:
Reiaz Ul Rehman, University of Kashmir, IndiaClaudia Tortiglione, Institute of Applied Sciences and Intelligent Systems (ISASI), Italy
Copyright © 2021 Stanly, Kim, Antonucci, Fiume, Guescini, Kim, Ciardiello, Giangrieco, Mari and Pocsfalvi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriella Pocsfalvi, Z2FicmllbGxhLnBvY3NmYWx2aUBpYmJyLmNuci5pdA==
†These authors have contributed equally to this work and share the first authorship
 Christopher Stanly
Christopher Stanly Hyoseon Kim
Hyoseon Kim Giuseppe Antonucci1
Giuseppe Antonucci1 Michele Guescini
Michele Guescini Kwang Pyo Kim
Kwang Pyo Kim Maria Antonietta Ciardiello
Maria Antonietta Ciardiello Ivana Giangrieco
Ivana Giangrieco Adriano Mari
Adriano Mari Gabriella Pocsfalvi
Gabriella Pocsfalvi