- 1National Engineering Laboratory for Cereal Fermentation Technology, Jiangnan University, Wuxi, China
- 2Engineering Research Center of Ministry of Education on Food Synthetic Biotechnology, Jiangnan University, Wuxi, China
- 3Science Center for Future Foods, Jiangnan University, Wuxi, China
- 4Jiangsu Provisional Research Center for Bioactive Product Processing Technology, Jiangnan University, Wuxi, China
- 5Jiangsu Province Engineering Research Center of Food Synthetic Biotechnology, Jiangnan University, Wuxi, China
Gluconobacter oxydans is important in the conversion of D-sorbitol into l-sorbose, which is an essential intermediate for industrial-scale production of vitamin C. In a previous study, the strain G. oxydans WSH-004 could directly produce 2-keto-l-gulonic acid (2-KLG). However, its D-sorbitol tolerance was poor compared with that of other common industrial G. oxydans strains, which grew well in the presence of more than 200 g/L of D-sorbitol. This study aimed to use the microbial microdroplet culture (MMC) system for the adaptive evolution of G. oxydans WSH-004 so as to improve its tolerance to high substrate concentration and high temperature. A series of adaptively evolved strains, G. oxydans MMC1-MMC10, were obtained within 90 days. The results showed that the best strain MMC10 grew in a 300 g/L of D-sorbitol medium at 40°C. The comparative genomic analysis revealed that genetic changes related to increased tolerance were mainly in protein translation genes. Compared with the traditional adaptive evolution method, the application of microdroplet-aided adaptive evolution could improve the efficiency in terms of reducing time and simplifying the procedure for strain evolution. This research indicated that the microdroplet-aided adaptive evolution was an effective tool for improving the phenotypes with undemonstrated genotypes in a short time.
Introduction
Gluconobacter strains belong to the group of acetic acid bacteria (Prust et al., 2005; Deppenmeier and Ehrenreich, 2009). They have a unique capacity to incompletely oxidize polyol substrates to produce a high titer of oxidation products, such as l-sorbose (vitamin C synthesis) (Azar and Alemzadeh, 2020) and 6-(N-hydroxyethyl)-amino-6-deoxy-α-l-sorbofuranose (a key intermediate for the synthesis of miglitol) (Liu et al., 2020). In the current industrial-scale production of vitamin C via the two-step fermentation method, G. oxydans is used to convert D-sorbitol into l-sorbose, which is the substrate for the second-step fermentation process (Pappenberger and Hohmann, 2014). However, l-sorbose fermentation is severely inhibited by D-sorbitol when the initial concentrations are more than 200 g/L in industry (Giridhar and Srivastava, 2002). Fed-batch fermentation is usually employed to alleviate substrate inhibition, and subsequently to improve l-sorbose productivity. The titer of the final product (l-sorbose) can be reduced and the fermentation time extended with this fermentation process (Zhou et al., 2019). Besides, the application of fed-batch fermentation can greatly increase the production cost and the risk of contamination. It is necessary to obtain a G. oxydans strain that can tolerate higher D-sorbitol concentration so as to release the substrate inhibition.
Improving the tolerance of industrial microorganisms to more harsh conditions, such as higher substrate concentration and higher temperature, is a long pursued goal for a more economic industrial fermentation process (Pereira et al., 2019; Wu et al., 2020). Enhanced tolerance to higher substrate/product concentration can not only further improve the final titer of the target product but also significantly simplify the fermentation process by avoiding complicated fed-batch or in situ product removing operations (Heerema et al., 2011; Gaber et al., 2021). Higher fermentation temperature can decrease the use of cooling water, thus decreasing the energy cost, and extend effective production time, especially in warmer areas (Wang et al., 2019). Because these two phenotypes play important roles in improving the economy of the fermentation process, researchers have conducted a number of studies that have revealed many important regulatory mechanisms. For example, altering the sterol composition could improve the thermotolerance of Saccharomyces cerevisiae (Caspeta et al., 2014), and strengthening the opposing potassium and proton electrochemical membrane gradients could facilitate the general resistance of S. cerevisiae to multiple alcohols (Lam et al., 2014). However, up to now, the general mechanisms of thermotolerance in microorganisms are still unclear. Adaptive evolution continues to be of interest to accelerate the breeding of robust microorganisms with improved high-temperature fermentation performance.
For directly improving the tolerance of microorganisms to adverse environmental stresses, adaptive evolution is an effective method to evolve microorganisms under specific environmental conditions, such as tolerance to high substrate/product concentration, high temperature, and low/high pH (d’oelsnitz and Ellington, 2018; Sandberg et al., 2019). Adaptive evolution has many strategies to obtain desired phenotype strains. For example, the classical laboratory evolution is to culture an organism under selective conditions (temperature and presence of inhibitors) so that mutant clones with beneficial mutations are enriched in the population (Rudolph et al., 2010; Ju et al., 2016; Zhu et al., 2018). However, it has inherent limitations because conventional adaptive evolution methods are typically labor-intensive, extremely time-consuming (from months to years generally), with low throughput; and are poorly parallelized. For example, Jin et al. (2019) improved the conversion rate of all nonglucose sugars to the corresponding sugar acids by several folds after 420 days of continuous culture. Recently, advances in miniaturization and parallelization of microbial cultivation systems have translated into advances in the capabilities of microbioreactors, which are now widely applied in studies of evolution (Jakiela et al., 2013; Wong et al., 2018). Compared with traditional microbial cultivation methods, microcultivation systems have many advantages, such as high throughput, less reagent and labor cost, and superior cultivation properties in mixing and parallelization (Jian et al., 2020; Zeng et al., 2020).
In this study, an efficient and reliable adaptive evolution approach was designed to improve the tolerance of G. oxydans WSH-004 to high D-sorbitol concentration and high temperature in a microbial microdroplet culture (MMC) system. Within 90 days of continuous adaptive evolution in MMC, ten adaptive evolutionary strains (MMC1–MMC10) were obtained. The optimum evolved strain MMC10 achieved the productivity of 3.13 g/(Lh) in a 5-L fermentor cultured in a medium containing 300 g/L of initial D-sorbitol, which was similar to that of the original strain G. oxydans WSH-004 cultured in a medium containing 80 g/L of initial D-sorbitol. Genome sequencing of evolved strains showed that most mutations were related to the translational process, which was rarely mentioned for improving tolerance to stresses in previous studies. The evolved strains and the genes identified could not only be used for more efficient production of l-sorbose and its derivatives but also provide clues for the rational engineering of microorganisms for enhanced tolerances to stresses.
Materials and Methods
Strains, Culture Media, and Reagents
Gluconobacter oxydans WSH-004, which used D-sorbitol to produce l-sorbose and 2-KLG, was screened in a previous study (Chen et al., 2019). The D-sorbitol medium (10 g/L of yeast extract and 80 g/L of D-sorbitol) was used for culturing the G. oxydans strains. In agar plates, 2% agar was added. Adaptive evolution of a D-sorbitol medium (10 g/L of yeast extract and 300 g/L of D-sorbitol) was used for adaptive evolution. Carrier oil (mineral oil with 12 g/L of Span 80; Sigma–Aldrich, Germany) and MMC chips for MMC were purchased from Wuxi Tmaxtree Biotechnology Co., Ltd. (Wuxi, China).
Adaptive Evolution of G. oxydans to High D-Sorbitol Concentration
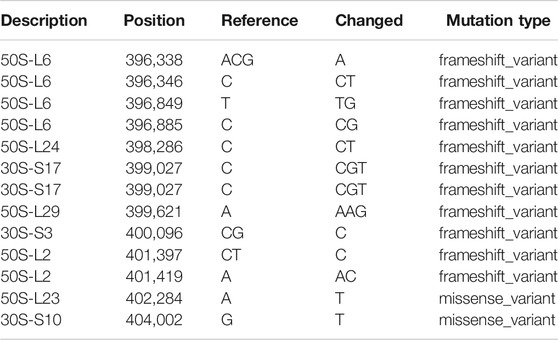
G. oxydans WSH-004 preserved in glycerol tubes was inoculated to solid D-sorbitol medium plates and cultured for 48 h at 30°C. A single colony was inoculated into 250-mL flasks containing 25 mL of D-sorbitol medium for 48 h at 30°C and 220 rpm. The seed was injected into a special sterilized seed bottle (Figure 1, bottle 3) for adaptive evolution in MMC (Jian et al., 2020). Meanwhile, the D-sorbitol medium was injected into a special sterilized medium bottle (Figure 1, bottle 2), while the adaptive evolution D-sorbitol medium was injected into another special sterilized medium bottle (Figure 1, bottle 1). The first step for adaptive evolution in MMC was the microdroplet formation (Figure 1A), in which pumps 2 and 3 were operated to form 2 μL microdroplets. The second step involved culturing the microdroplets in MMC (Figure 1B), in which pumps 1 and 2 were operated to move the microdroplets in special tubes that could pass through the air but not through water and cells. A key step for the adaptive evolution and continuous culture is the droplet segmentation fusion-generation (Figures 1C,D). After a period of adaptive evolution in MMC, the droplet with a higher OD600 value (Figure 1F) was collected and spread on the D-sorbitol medium plates with selective pressure (high D-sorbitol concentration or/and high-temperature conditions).
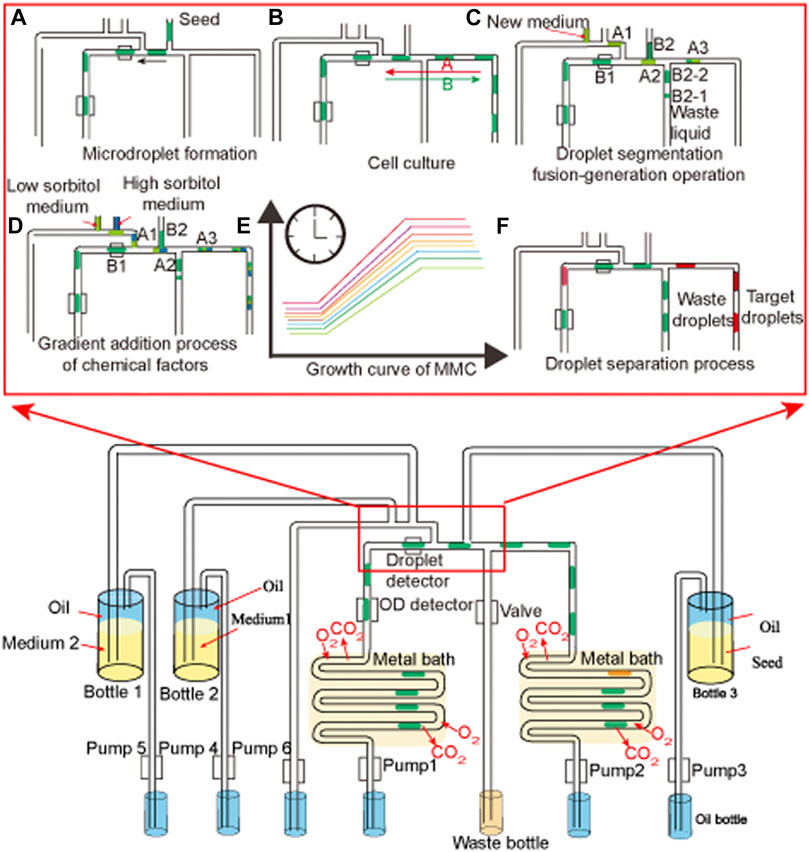
FIGURE 1. Diagram of the operation of the microdroplet culture system. (A) Microdroplet formation, (B) cell culture, (C) droplet segmentation fusion-generation operation, (D) gradient addition process of chemical factors, (E) growth curve of MMC, and (F) droplet separation process.
Adaptive Evolution of G. oxydans to High Temperature
G. oxydans MMC4 obtained in the last round of adaptive evolution to the high D-sorbitol concentration was used as the start strain. The single colony of G. oxydans MMC4 was inoculated into 250-ml flasks containing 25 ml of D-sorbitol medium for 48 h at 30°C and 220 rpm to get the seeds. All steps were the same (microdroplet formation, incubation, droplet segmentation fusion-generation, and sorting), except for the gradual increase in the metal bath temperature instead of increasing the D-sorbitol concentration.
Culture Conditions in Shake Flasks
For verifying the tolerance of evolved strains to different D-sorbitol concentrations and temperature, a single colony of the strain was inoculated into 250-ml flasks containing 25 ml of D-sorbitol medium for 48 h at 30°C and 220 rpm for seed culture. Then, 5 ml of the seed culture was transferred to 25 ml of the culture medium containing different D-sorbitol concentrations (50–400 g/L of D-sorbitol, 10 g/L of yeast extract, and 50 μg/ml of cefoxitin). The flasks were then cultured at 220 rpm at 30, 35, or 40°C.
Genome Sequencing and Data Analysis
A single colony of the evolved strains isolated from the evolved populations was cultivated for 36–48 h in 25 ml of D-sorbitol medium until the OD600 value reached 3. A genomic DNA extraction kit (Sangon Biotech, Shanghai) was used to extract genomic DNA of G. oxydans strains using the protocol recommended by the manufacturer. High-throughput genome sequencing was performed with the Illumina Hiseq using 2 × 150 paired-end reads targeting a genome. The average coverage for each sample was more than 300X. Genome sequencing was performed by Sangon Biotech. Referring to the GATK best practice–recommended design process, BWA was used to compare the effective data of the sample to the reference genome (Kumar et al., 2019). The SAM tool was used to compare the results for format conversion and sorting, and statistical comparison results (Li et al., 2009). The HaplotypeCaller of GATK was employed to analyze the genetic differences between the adaptive evolution strains and wild-type strain, merge and integrate the analysis of different samples, and obtain sample variation information (McKenna et al., 2010).
Culture Conditions in Bioreactors
Fermentation studies were performed in a 5-L bioreactor (T&J Bioengineering, Shanghai, China) with a working volume of 3 L. For batch fermentation, a single colony of the strain was inoculated into 500-ml flasks containing 50 ml of D-sorbitol medium for 48 h at 30°C and 220 rpm. Then, 300 ml of the seed culture was inoculated into a bioreactor with 2.7 L of the culture medium containing different D-sorbitol concentrations (300 g/L of D-sorbitol and 10 g/L of yeast extract, or 80 g/L of D-sorbitol and 10 g/L of yeast extract). The temperature was maintained at 30 or 37°C, the agitation speed was controlled at 400 rpm, and the aeration rate was 1 vvm.
Analysis Procedures
D-sorbitol and l-sorbose in the fermentation broth were evaluated by Shimadzu high-performance liquid chromatography equipped with an Aminex HPX-87H column (Bio-Rad, Hercules, CA, United States) at 40°C with a flow rate of 0.5 ml/min and 5 mmol/L of H2SO4 as the eluent. A refractive index detector was used for detection (Chen et al., 2019).
Results
Effects of D-Sorbitol and Temperature on the Cell Growth of G. oxydans WSH-004
G. oxydans WSH-004 was screened from the soil in a previous study (Chen et al., 2019). It directly converted D-sorbitol into 2-KLG without obvious accumulation of by-products during the fermentation process. Therefore, G. oxydans WSH-004 had the potential to be a one-step fermentation strain for the production of 2-KLG, which is the direct precursor of vitamin C. G. oxydans WSH-004 was grown with different D-sorbitol concentrations (Figure 2). The results showed that the OD600 value of G. oxydans WSH-004 in the stationary phase could reach 2.7 in 50 g/L of D-sorbitol medium, which was similar to that of most other commonly available G. oxydans strains. When the concentration of D-sorbitol reached 300 g/L, which is the commonly used concentration for the industrial-scale production of l-sorbose from D-sorbitol (Zhou et al., 2019), the growth of G. oxydans WSH-004 was significantly inhibited with a final OD600 value of 0.7. The results showed that the tolerance of G. oxydans WSH-004 to high D-sorbitol concentration was poor and should be significantly improved to fulfill the industrial-scale production of l-sorbose and 2-KLG.
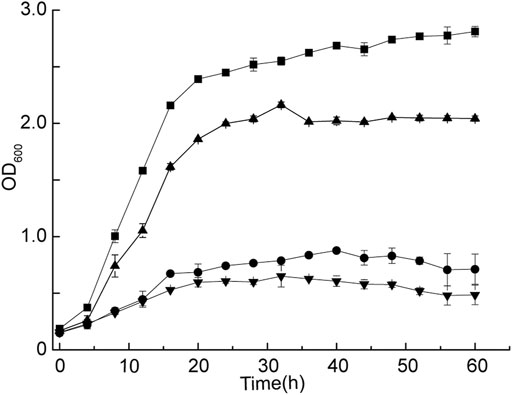
FIGURE 2. Growth curve of G. oxydans with different D-sorbitol concentrations. Square: Growth curve of G. oxydans at 30°C in 50 g/L of D-sorbitol medium. Circle: Growth curve of G. oxydans at 30°C in 300 g/L of D-sorbitol medium. Up-triangle: Growth curve of G. oxydans at 35°C in 50 g/L of D-sorbitol medium. Down-triangle: Growth curve of G. oxydans at 35°C in 300 g/L of D-sorbitol medium.
Adaptive Evolution of G. oxydans WSH-004 to High D-Sorbitol Concentration
The strain G. oxydans WSH-004 was continuously cultured in the MMC system with different D-sorbitol concentrations to improve the tolerance of the strain to higher D-sorbitol concentrations. The growth curve of G. oxydans WSH-004 was first checked in the MMC system. The result showed that the OD600 value reached the stationary phase after 10 h with a final OD600 value of about 1.2 and a typical growth curve in the MMC system (Figure 3A). The value was different from that in shaking flask fermentation because of the difference in both culture conditions and detection methods. The evolution of tolerance to high D-sorbitol concentration was carried out. The automatic passage time was set to 15 h. After every three passages, the D-sorbitol concentration was automatically increased according to the set program. During the growth of G. oxydans WSH-004, the OD600 value of every droplet was recorded by MMC (Figure 3B). When the D-sorbitol concentration of G. oxydans WSH-004 reached 300 g/L, the OD600 values of different drops were quite different. The droplets with a higher OD600 value were harvested, cultured in 300 g/L of D-sorbitol medium at 30°C, and then diluted and spread on D-sorbitol plates containing 300 g/L of D-sorbitol. Two single colonies of evolved strains, namely G. oxydans MMC1 and MMC2, that could grow well on 300 g/L of D-sorbitol plates, were picked and verified.
FIGURE 3. Adaptive evolutionary growth curve of G. oxydans. (A) Growth curve of G. oxydans WSH-004 in MMC. (B) Growth curve of G.oxydans in adaptive evolution. (C) Adaptive evolution of G. oxydans was carried out in a high-concentration D-sorbitol medium. The controlled passage time was 15 h, the culture temperature was 30°C, and the continuous passage of G. oxydans in MMC lasted for 15 days.
G. oxydans MMC2 was selected as the starting strain for the second round of adaptive evolution to high D-sorbitol concentration in MMC to stabilize the capacity of G. oxydans to tolerate high D-sorbitol concentrations. In the second round of adaptive evolution, no significant growth difference was found among different drops. All drops could be cultured stably in MMC (Figure 3C). After 14 days of continuous culture in MMC, the droplets with higher OD600 values were harvested, cultured in 300 g/L of D-sorbitol medium at 30°C, and then diluted and spread on D-sorbitol plates containing 300 g/L of D-sorbitol. Two single colonies of evolved strains, namely G. oxydans MMC3 and MMC4, that could grow well on 300 g/L of D-sorbitol plates, were picked and verified.
Adaptive Evolution of G. oxydans to a Higher Temperature
High-temperature fermentation has many advantages in industrial production. In the process of adaptive evolution to high-temperature tolerance, G. oxydans MMC4 was used as the starting strain for performing adaptive evolution. The D-sorbitol concentration and culture temperature were initially set at 300 g/L and 30°C, respectively. The result showed that the OD600 value could reach to about 0.7 in the MMC system with 300 g/L of D-sorbitol medium at 30°C. Then, the culture temperature was gradually raised from 30 to 40°C (Figure 4), and the D-sorbitol concentration was maintained at 300 g/L. When the incubation temperature was increased to 35°C, the maximum OD600 value reached to about 0.5 (Figure 4A). In the later adaptive evolution of temperature tolerance (the temperature was gradually increased from 35 to 39°C), the growth of G. oxydans was basically unaffected with a maximum OD600 value of about 0.5 (Figure 4A). After 26 days of adaptive evolution in MMC, the droplets with higher OD600 value were harvested and then cultured in 300 g/L of D-sorbitol concentration at 39°C. As strains isolated from all drops could not grow at less than 40°C, 39°C was the highest temperature that G. oxydans could withstand in this round (Figure 4A). After 27 days of continuous culture in MMC, the droplets with higher OD600 value were harvested, cultured in 300 g/L of D-sorbitol medium at 39°C, diluted and spread on D-sorbitol plates containing 300 g/L of D-sorbitol, and cultured at 39°C. Two single colonies of evolved strains, namely G. oxydans MMC5 and MMC6, that grew well on 300 g/L of D-sorbitol plates, were picked and verified.
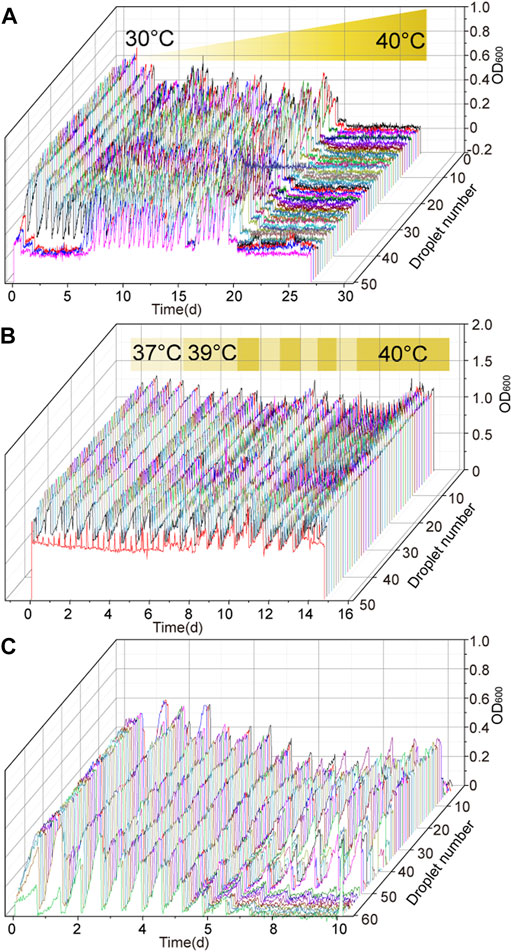
FIGURE 4. Temperature tolerance adaptive evolution of G. oxydans. (A) Adaptability evolution of G. oxydans to high temperature: the culture temperature and passage time were changed according to the growth of G. oxydans in MMC. (B) Adaptability evolution of G. oxydans to high temperature: the culture temperature was alternatively switched between 39 and 40°C. (C) Adaptability evolution of G. oxydans to high temperature: it was continuously cultured at 40°C.
Taking G. oxydans MMC6 as the starting strain, the adaptive evolution was further carried out. When the culture temperature increased to 39°C, the evolved strain grew normally (Figure 4B). Then, the evolution temperature was altered between 39 and 40°C (Figure 4B). Finally, when the strains grew stably at 40°C, the droplets with higher OD600 value were harvested, cultured in 300 g/L of D-sorbitol medium at 40°C, diluted and spread on D-sorbitol plates containing 300 g/L of D-sorbitol, and cultured at 40°C. Two single colonies of evolved strains, namely G. oxydans MMC7 and MMC8, that grew well on 300 g/L of D-sorbitol plates, were picked and verified.
Further adaptive evolution of G. oxydans MMC8 was conducted to stabilize the capacity of G. oxydans to tolerate both high D-sorbitol concentration and high temperature. The D-sorbitol concentration and culture temperature were initially set at 300 g/L and 37°C, respectively. Once the growth of cells reached the stationary phase, the adaptive evolution temperature was gradually increased from 37 to 40°C (Figure 4C). Finally, when the cells grew stably at 40°C, the droplets with higher OD600 value were harvested, cultured in 300 g/L of D-sorbitol medium at 40°C, diluted and spread on D-sorbitol plates containing 300 g/L of D-sorbitol, and cultured at 40°C. Two single colonies of evolved strains, namely G. oxydans MMC9 and MMC10, that grew well on 300 g/L of D-sorbitol plates, were picked and verified.
Verification of Adaptive Evolutionary Strains in Shake Flasks
Both wild-type and evolved strains were cultured under different D-sorbitol concentrations to verify the D-sorbitol tolerance of the evolved strains. In 50 g/L of D-sorbitol medium, the final OD600 values of both G. oxydans MMC2 and MMC4 reached to about 2.9, which was higher than that of G. oxydans WSH-004 (Figure 5A). G. oxydans MMC2 and MMC4 grew better in 100 g/L of D-sorbitol medium than in 50 g/L of the medium. The final OD600 values of G. oxydans MMC2 and MMC4 was about 3.2 (Figure 4B). In 100–300 g/L of D-sorbitol medium, G. oxydans MMC2 and MMC4 grew better than G. oxydans WSH-004 (Figures 5C–F). When the D-sorbitol concentration was increased to 350 g/L, G. oxydans WSH-004 could not grow, while G. oxydans MMC2 and MMC4 grew to a final OD600 value of about 1.2 (Figure 5G). When D-sorbitol concentration was increased to 400 g/L, none of the strains could grow (Figure 5H).
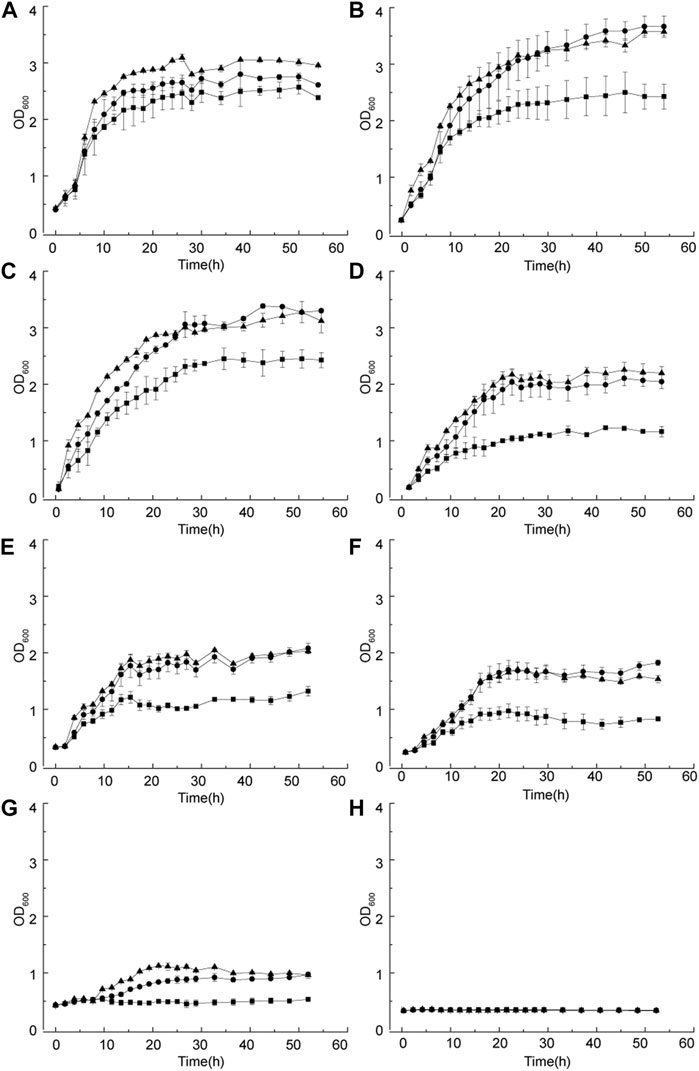
FIGURE 5. Growth curve of G. oxydans with different D-sorbitol concentrations. (A) 50 g/L, (B) 100 g/L, (C) 150 g/L, (D) 200 g/L, (E) 250 g/L, (F) 300 g/L, (G) 350 g/L, and (H) 400 g/L. Square: G. oxydans WSH-004; circle: G. oxydans MMC2; and up-triangle: G. oxydans MMC4.
G. oxydans MMC4-MMC10 grew better than the wild-type strain in the 50 g/L D-sorbitol media at 30 and 35°C, and the OD600 value of the evolved strains (about 3.5) was higher than that of the wild-type strains (about 2.0) (Figures 6A-D). When the temperature was 40°C, the growth of all strains was inhibited in 50 g/L of D-sorbitol medium (Figure 6E). However, 300 g/L of D-sorbitol could benefit cell growth at 40°C. The evolved strains grew much better at 40°C than in 50 g/L of D-sorbitol medium. The final OD600 value reached to about 0.75 (Figure 6F). Therefore, high D-sorbitol concentration could protect G. oxydans under high-temperature conditions.
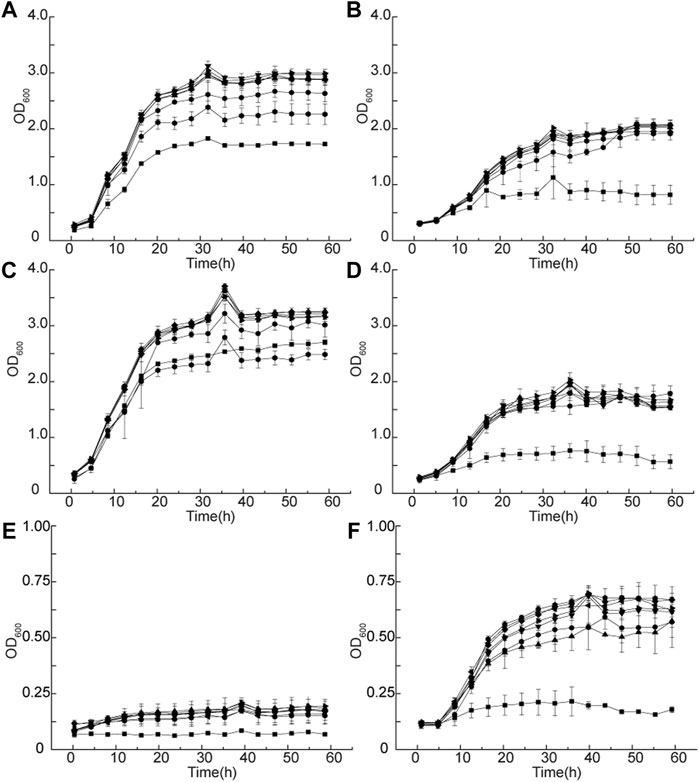
FIGURE 6. Growth curve of G. oxydans at different temperatures. Growth curves at (A) 50 g/L of D-sorbitol, 30°C; (B) 300 g/L of D-sorbitol, 30°C; (C) 50 g/L of D-sorbitol, 35°C; (D) 300 g/L of D-sorbitol, 35°C; (E) 50 g/L of D-sorbitol, 40°C; and (F) 300 g/L of D-sorbitol, 40°C. Square: G. oxydans WSH-004; circle: G. oxydans MMC4; up-triangle: G. oxydans MMC5; down-triangle: G. oxydans MMC6; rhombus: G. oxydans MMC7; left triangle: G. oxydans MMC8; right triangle: G. oxydans MMC9; and pentagon: G. oxydans MMC10.
Bioreactor Fermentation for l-Sorbose Synthesis
Batch fermentation of G. oxydans WSH-004 and evolved strain G. oxydans MMC10 was conducted in a 5-L bioreactor to verify the production capacity of l-sorbose. Both G. oxydans WSH-004 and G. oxydans MMC10 could completely convert 80 g/L of D-sorbitol into l-sorbose at 30°C. However, the required fermentation time was quite different. It took about 60 and 25 h for G. oxydans WSH-004 and G. oxydans MMC10, respectively, to completely convert the D-sorbitol to l-sorbose, with productivities of 1.33 g/(Lh) and 3.20 g/(Lh), respectively (Figures 7A,B). When the concentration of D-sorbitol was increased to 300 g/L, the growth of G. oxydans WSH-004 was completely inhibited, while G. oxydans MMC10 could still completely convert D-sorbitol into l-sorbose in 96 h (Figures 7C,D). The productivity of G. oxydans MMC10 with the 300 g/L of D-sorbitol fermentation medium was 3.13 g/(Lh), which was similar to that in the 80 g/L of D-sorbitol fermentation medium.
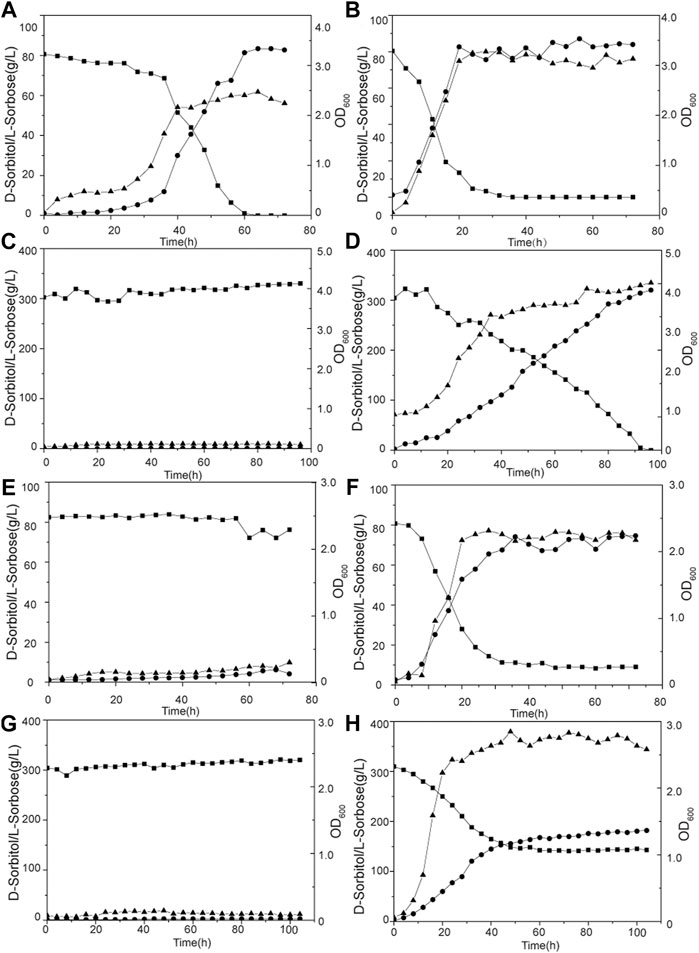
FIGURE 7. Production of l-sorbose by G. oxydans in a 5-L fermentor. (A) G. oxydans WSH-004 in 80 g/L of D-sorbitol at 30°C; (B) G. oxydans MMC10 in 80 g/L of D-sorbitol at 30°C; (C) G. oxydans WSH-004 in 300 g/L of D-sorbitol at 30°C; (D) G. oxydans MMC10 in 300 g/L of D-sorbitol at 30°C; (E) G. oxydans WSH-004 in 80 g/L of D-sorbitol at 37°C; (F) G. oxydans MMC10 in 80 g/L of D-sorbitol at 37°C; (G) G. oxydans WSH-004 in 300 g/L of D-sorbitol at 37°C; and (H) G. oxydans MMC10 in 300 g/L of D-sorbitol at 37°C. Square: D-sorbitol; circle: l-sorbose; and up-triangle: OD600.
Then, the fermentation temperature was raised to 37°C. G. oxydans MMC10 grew normally, while G. oxydans WSH-004 could not grow with 80 g/L or 300 g/L of D-sorbitol (Figures 7E,F). G. oxydans MMC10 could not completely convert D-sorbitol into l-sorbose at 37°C in 80 g/L of D-sorbitol medium. The final concentration of l-sorbose was 74 g/L at 35 h, with a productivity of 2.11 g/(Lh). Besides, G. oxydans MMC10 could not completely convert D-sorbitol into l-sorbose in 300 g/L of D-sorbitol medium at 37°C. However, the strain grew better than in the 80 g/L of D-sorbitol medium, and the OD600 value reached to 2.8. The final l-sorbose concentration was 160 g/L after 60 h, with a productivity of 2.67 g/(Lh) (Figures 7G,H). It was presumed that the enzyme activity and stability of D-sorbitol dehydrogenase were affected under high-temperature conditions.
Comparative Genomic Analysis of G. oxydans
Genome sequencing was performed on evolved strains to identify mutations that might contribute to the improved phenotypes in the evolved populations. Based on the comparative genomic analysis, 104 mutations were detected among the ten evolved strains comprising 53 point mutations and 51 insertions or deletions. The genetic variations have been listed in Supplementary Material 1 and Supplementary Material 2. Functional annotations of the mutated genes (Figure 8A) showed that the mutated genes were mainly related to translation (ribosomal small subunits S17 and S3, and the major ribosomal subunits L2, L3, L6, L24, and L29), amino acid and nucleotide metabolism, carbohydrate metabolism, and energy metabolism. In the same time, all evolved strains have the ribosome mutation-related genes and this gene was listed in Table 1. A phylogenetic tree of evolved strains and wild-type strains was built based on the SNP of each sample using the adjacency NJ algorithm (Figure 8B). According to the clusterProfiler for feature enrichment analysis, mutant genes were mainly involved in ribosomes, ribosomal protein complexes, and protein mutations that constituted ribosomes (Figure 8C).
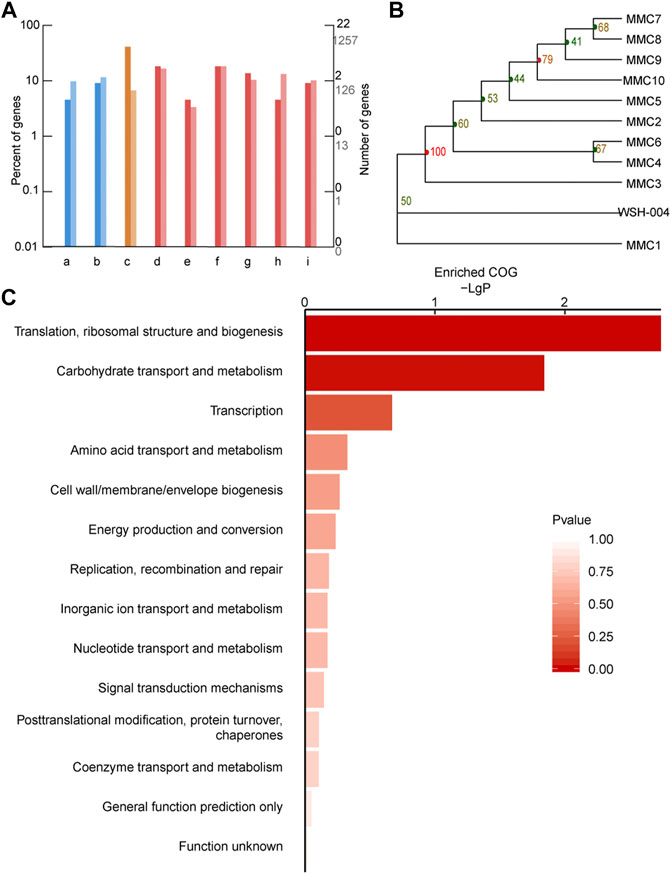
FIGURE 8. Comparative genomic analysis of G. oxydans strains. (A) GO annotation classification column map of variant genes. The horizontal axis is the functional classification, and the vertical axis is the number of genes in the classification (right) and the percentage of the total number of genes on the annotation (left). Different colors represent different categories. The dark color on the histogram and the axis represents the variant gene, and the light color represents all genes (background genes). (i) Membrane transport, (ii) signal transduction, (iii) translation, (iv) amino acid metabolism, (v) biosynthesis of other secondary metabolites, (vi) carbohydrate metabolism, (vii) energy metabolism, (viii) metabolism of cofactors and vitamins, and (i) nucleotide metabolism. (B) Phylogenetic tree of a series of adaptive evolution strains. G. oxydans phylogenetic tree based on the SNP of each sample. (C) Significant enrichment function bar chart. The vertical axis represents function annotation information, and the horizontal axis represents the degree of enrichment corresponding to the function, that is, the −log10 (p value) value. The higher the enrichment degree, the longer the column and the darker the color.
Discussion
In a previous study, a strain G. oxydans WSH-004 could directly produce 2-KLG from D-sorbitol by a high-throughput screening method based on 2-keto-gulonic acid dehydrogenase (Chen et al., 2019). In the present study, adaptive evolution of G. oxydans WSH-004 was conducted using an automated, high-throughput microbial cultivation MMC system (Jian et al., 2020) to improve its tolerance to high temperature and high D-sorbitol concentration. A series of evolved strains were obtained within 90 days. These strains achieved significantly enhanced growth and conversion rates and productivity with higher substrate concentration, and at higher temperature compared with G. oxydans WSH-004. Analysis of the genome of G. oxydans WSH-004 and the evolved strains showed that mutated genes were mainly related to translation, thus providing new research directions for the tolerance mechanism of G. oxydans. The obtained results suggested that the MMC system greatly improved the efficiency of the adaptive evolution of microorganisms and obviously reduced labor and cost involved in the adaptive evolution process.
The substrate/product resistance of microorganisms is vital for their applications in industrial fermentation processes (Mohamed et al., 2020; Wang et al., 2021). In recent years, many reports on substrate/product tolerance of microbial strains, such as Clostridium acetobutylicum and Escherichia coli (Alsaker et al., 2010; Zhang et al., 2015), have been published, but studies on the tolerance of G. oxydans are few. Mechanisms of substrate/product stress have been studied mostly in Gram-negative microorganisms. For example, Si et al. (2016) found a series of key genes related to organic solvent tolerance of Escherichia coli through gene chip technology, and Zhang et al. (2015) constructed a global transcription factor mutation library and finally screened two strains with high organic solvent tolerance. In the present study, D-sorbitol tolerant strains were obtained through adaptive evolution in MMC, and the genomes of D-sorbitol-tolerant strains were re-sequenced. The findings showed that the mutated genes were mainly related to translation. However, due to the difficulty in genetic modification, future studies need to verify these mutation sites for enhancing specific phenotypes rather than simply modifying the membrane components or other known processes.
High-temperature fermentation has great advantages in industrial production, especially for reducing cooling costs and risk of contaminations, and simultaneous saccharification and fermentation of pretreated lignocellulosic for biofuel production (Zhang et al., 2016; Vishnu Prasad et al., 2020). However, most of the strains produced in the industry have poor heat resistance. Fermentation process at 30°C could be a big challenge for cooling water supply in a majority of temperate, subtropical, and tropical regions. Previous studies have demonstrated the mechanism of heat resistance in bacteria, such as reduction of the level of intracellular reactive oxygen species (Xu et al., 2018; Qin et al., 2020), regulation of chaperones for maintaining protein-folding homeostasis (Kim et al., 2020), and mutation of F-type ATPase (Huang et al., 2018). Some of the thermotolerant Gluconobacter strains, which can withstand a temperature of 36–37°C, have been isolated for producing 5-keto-d-gluconic acid or coenzyme Q10 (Saichana et al., 2009; Moghadami et al., 2019). However, the specific heat resistance mechanism remains unclear. In this study, G. oxydans MMC10 could withstand 40°C in 300 g/L of D-sorbitol medium through adaptive evolution in MMC. The identified mutations related to thermotolerance could be useful for further rational engineering of thermotolerant bacteria to facilitate their applications in industrial fermentation processes.
Many strategies can be applied to improve the specific phenotypes of microorganisms for industrial applications, such as metabolic engineering (Tian et al., 2020), mutagenesis screening (Jiang et al., 2018), and adaptive evolution (Cubas-Cano et al., 2019), with their own merits and demerits. Compared with metabolic engineering, adaptive evolution is a simple and efficient way to improve microbial tolerance because identifying gene targets for tolerance remains an overwhelming challenge and microbial metabolic networks complexity and interconnectivity. However, conventional adaptive evolution for microorganisms is typically time-consuming and labor-intensive, for example, 420 days for improving the conversion rate of nonglucose sugars of G. oxydans (Jin et al., 2019), 150 days for improving the salinity resistance of Schizochytrium sp. (Sun et al., 2018), 200 days for co-utilization of glucose and xylose of Zymomonas mobilis (Sarkar et al., 2020), and 230 generations for enhanced native methanol assimilation in S. cerevisiae (Espinosa et al., 2020). Reports on enhanced heat resistance by adaptive evolution or other rational strategies are rare (Cox et al., 2010). In this study, tolerant G. oxydans strains could be obtained within 90 days because the MMC system provided an automated, high-throughput microbial cultivation approach for adaptive evolution. Meanwhile, the growth conditions could be easily evaluated by an online detection system. Besides, the automated process could greatly facilitate the performance of experimental capabilities, throughput, clarity, and repeatability.
In conclusion, G. oxydans strains with enhanced tolerance to higher D-sorbitol concentration and higher temperature were obtained in only 90 days using an established continuous MMC system. Compared with the wild-type strain, G. oxydans WSH-004 not only developed tolerance to higher substrate concentration and higher temperature but also showed significantly improved titer, yield, and productivity under different conditions. The results proved that the droplet-based adaptive evolution system was a powerful tool to obtain the target phenotypes in acceptable durations. Comparative genomic analysis of evolved strains showed that the mutations were mainly located on ribosomal subunits. Further systematic investigation should be performed to reveal the mechanism underlying the phenotypes because of the complexity in establishing the relationships between the genotype and the phenotype at a global translational level.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
LL and WZ designed the experiments. LL and WZ performed the experiments. LL, SY and JL wrote the manuscript. JZ conceived the project. All authors contributed to the manuscript, read and approved the final version.
Funding
This work was supported by grants from the National Key Research and Development Program of China (No. 2019YFA0904900), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (No. 32021005), and the National Science Fund for Excellent Young Scholars (No. 21822806).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YX declared a shared affiliation with the authors to the Handling Editor at the time of review.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.731247/full#supplementary-material
References
Alsaker, K. V., Paredes, C., and Papoutsakis, E. T. (2010). Metabolite Stress and Tolerance in the Production of Biofuels and Chemicals: Gene-Expression-Based Systems Analysis of Butanol, Butyrate, and Acetate Stresses in the anaerobe Clostridium Acetobutylicum. Biotechnol. Bioeng. 105, a–n. doi:10.1002/bit.22628
Azar, S. A. D., and Alemzadeh, I. (2020). L-sorbose Production by Gluconobacter Oxydans Using Submerged Fermentation in a Bench Scale Fermenter. Appl. Food Biotechnol. 7, 41–48. doi:10.22037/afb.v7i1.26582
Caspeta, L., Chen, Y., Ghiaci, P., Feizi, A., Buskov, S., Hallström, B. M., et al. (2014). Altered Sterol Composition Renders Yeast Thermotolerant. Science 346, 75–78. doi:10.1126/science.1258137
Chen, Y., Liu, L., Shan, X., Du, G., Zhou, J., and Chen, J. (2019). High-Throughput Screening of a 2-Keto-L-Gulonic Acid-Producing Gluconobacter Oxydans Strain Based on Related Dehydrogenases. Front. Bioeng. Biotechnol. 7, 9. doi:10.3389/fbioe.2019.00385
Cox, J., Schubert, A. M., Travisano, M., and Putonti, C. (2010). Adaptive Evolution and Inherent Tolerance to Extreme thermal Environments. BMC Evol. Biol. 10, 75. doi:10.1186/1471-2148-10-75
Cubas-Cano, E., González-Fernández, C., and Tomás-Pejó, E. (2019). Evolutionary Engineering of Lactobacillus Pentosus Improves Lactic Acid Productivity from Xylose-Rich media at Low pH. Bioresour. Technol. 288, 121540. doi:10.1016/j.biortech.2019.121540
Deppenmeier, U., and Ehrenreich, A. (2009). Physiology of Acetic Acid Bacteria in Light of the Genome Sequence of Gluconobacter Oxydans. J. Mol. Microbiol. Biotechnol. 16, 69–80. doi:10.1159/000142895
d’Oelsnitz, S., and Ellington, A. (2018). Continuous Directed Evolution for Strain and Protein Engineering. Curr. Opin. Biotechnol. 53, 158–163. doi:10.1016/j.copbio.2017.12.020
Espinosa, M. I., Gonzalez-Garcia, R. A., Valgepea, K., Plan, M. R., Scott, C., Pretorius, I. S., et al. (2020). Adaptive Laboratory Evolution of Native Methanol Assimilation in Saccharomyces cerevisiae. Nat. Commun. 11. doi:10.1038/s41467-020-19390-9
Gaber, M. A., Abdel-Rahman, M. A., Hassan, S. E.-D., and Azab, M. S. (2021). Efficient Biorefinery Process for Lactic Acid Production from Date Wastes with Alleviating Substrate Inhibition Effect Using Thermo-Alkaline Repeated Batch Fermentation. Biomass Conv. Bioref. 11, 1053–1066. doi:10.1007/s13399-020-01043-y
Giridhar, R. N., and Srivastava, A. K. (2002). Productivity Improvement in L-Sorbose Biosynthesis by Fedbatch Cultivation of Gluconobacter Oxydans. J. Biosci. Bioeng. 94, 34–38. doi:10.1263/jbb.94.34
Heerema, L., Wierckx, N., Roelands, M., Hanemaaijer, J. H., Goetheer, E., Verdoes, D., et al. (2011). In Situ phenol Removal from Fed-Batch Fermentations of Solvent Tolerant Pseudomonas Putida S12 by Pertraction. Biochem. Eng. J. 53, 245–252. doi:10.1016/j.bej.2010.11.002
Huang, C.-J., Lu, M.-Y., Chang, Y.-W., and Li, W.-H. (2018). Experimental Evolution of Yeast for High-Temperature Tolerance. Mol. Biol. Evol. 35, 1823–1839. doi:10.1093/molbev/msy077
Jakiela, S., Kaminski, T. S., Cybulski, O., Weibel, D. B., and Garstecki, P. (2013). Bacterial Growth and Adaptation in Microdroplet Chemostats. Angew. Chem. Int. Ed. 52, 8908–8911. doi:10.1002/anie.201301524
Jian, X., Guo, X., Wang, J., Tan, Z. L., Xing, X. h., Wang, L., et al. (2020). Microbial Microdroplet Culture System (MMC): An Integrated Platform for Automated, High‐throughput Microbial Cultivation and Adaptive Evolution. Biotechnol. Bioeng. 117, 1724–1737. doi:10.1002/bit.27327
Jiang, A.-L., Hu, W., Li, W.-J., Liu, L., Tian, X.-J., Liu, J., et al. (2018). Enhanced Production of L -lactic Acid by Lactobacillus thermophilus SRZ50 Mutant Generated by High-Linear Energy Transfer Heavy Ion Mutagenesis. Eng. Life Sci. 18, 626–634. doi:10.1002/elsc.201800052
Jin, C., Hou, W., Yao, R., Zhou, P., Zhang, H., and Bao, J. (2019). Adaptive Evolution of Gluconobacter Oxydans Accelerates the Conversion Rate of Non-glucose Sugars Derived from Lignocellulose Biomass. Bioresour. Technol. 289, 121623. doi:10.1016/j.biortech.2019.121623
Ju, S. Y., Kim, J. H., and Lee, P. C. (2016). Long-term Adaptive Evolution of Leuconostoc Mesenteroides for Enhancement of Lactic Acid Tolerance and Production. Biotechnol. Biofuels 9, 240. doi:10.1186/s13068-016-0662-3
Kim, S., Kim, Y., Suh, D. H., Lee, C. H., Yoo, S. M., Lee, S. Y., et al. (2020). Heat-responsive and Time-Resolved Transcriptome and Metabolome Analyses of Escherichia coli Uncover Thermo-Tolerant Mechanisms. Sci. Rep. 10. doi:10.1038/s41598-020-74606-8
Kumar, S., Agarwal, S., and Ranvijay, (2019). Fast and Memory Efficient Approach for Mapping NGS Reads to a Reference Genome. J. Bioinform. Comput. Biol. 17, 1950008. doi:10.1142/s0219720019500082
Lam, F. H., Ghaderi, A., Fink, G. R., and Stephanopoulos, G. (2014). Engineering Alcohol Tolerance in Yeast. Science 346, 71–75. doi:10.1126/science.1257859
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Liu, D., Ke, X., Hu, Z.-C., and Zheng, Y.-G. (2020). Combinational Expression of D-Sorbitol Dehydrogenase and Pyrroloquinoline Quinone Increases 6-(n-Hydroxyethyl)-Amino-6-Deoxy-α-L-Sorbofuranose Production by Gluconobacter oxydans through Cofactor Manipulation. Enzyme Microb. Technol. 141, 109670. doi:10.1016/j.enzmictec.2020.109670
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 20, 1297–1303. doi:10.1101/gr.107524.110
Moghadami, F., Fooladi, J., and Hosseini, R. (2019). Introducing a Thermotolerant Gluconobacter japonicus Strain, Potentially Useful for Coenzyme Q10 Production. Folia Microbiol. 64, 471–479. doi:10.1007/s12223-018-0666-4
Mohamed, E. T., Werner, A. Z., Salvachúa, D., Singer, C. A., Szostkiewicz, K., Rafael Jiménez-Díaz, M., et al. (2020). Adaptive Laboratory Evolution of Pseudomonas Putida KT2440 Improves P-Coumaric and Ferulic Acid Catabolism and Tolerance. Metab. Eng. Commun. 11, e00143. doi:10.1016/j.mec.2020.e00143
Pappenberger, G., and Hohmann, H.-P. (2013). Industrial Production of L-Ascorbic Acid (Vitamin C) and D-Isoascorbic Acid. Biotechnol. Food Feed Additives, 143–188. doi:10.1007/10_2013_243
Pereira, R., Wei, Y., Mohamed, E., Radi, M., Malina, C., Herrgård, M. J., et al. (2019). Adaptive Laboratory Evolution of Tolerance to Dicarboxylic Acids in Saccharomyces cerevisiae. Metab. Eng. 56, 130–141. doi:10.1016/j.ymben.2019.09.008
Prust, C., Hoffmeister, M., Liesegang, H., Wiezer, A., Fricke, W. F., Ehrenreich, A., et al. (2005). Complete Genome Sequence of the Acetic Acid Bacterium Gluconobacter Oxydans. Nat. Biotechnol. 23, 195–200. doi:10.1038/nbt1062
Qin, L., Dong, S., Yu, J., Ning, X., Xu, K., Zhang, S.-J., et al. (2020). Stress-driven Dynamic Regulation of Multiple Tolerance Genes Improves Robustness and Productive Capacity of Saccharomyces cerevisiae in Industrial Lignocellulose Fermentation. Metab. Eng. 61, 160–170. doi:10.1016/j.ymben.2020.06.003
Rudolph, B., Gebendorfer, K. M., Buchner, J., and Winter, J. (2010). Evolution of Escherichia coli for Growth at High Temperatures. J. Biol. Chem. 285, 19029–19034. doi:10.1074/jbc.M110.103374
Saichana, I., Moonmangmee, D., Adachi, O., Matsushita, K., and Toyama, H. (2009). Screening of Thermotolerant Gluconobacter Strains for Production of 5-Keto- D -Gluconic Acid and Disruption of Flavin Adenine Dinucleotide-Containing D -Gluconate Dehydrogenase. Appl. Environ. Microbiol. 75, 4240–4247. doi:10.1128/aem.00640-09
Sandberg, T. E., Salazar, M. J., Weng, L. L., Palsson, B. O., and Feist, A. M. (2019). The Emergence of Adaptive Laboratory Evolution as an Efficient Tool for Biological Discovery and Industrial Biotechnology. Metab. Eng. 56, 1–16. doi:10.1016/j.ymben.2019.08.004
Sarkar, P., Mukherjee, M., Goswami, G., and Das, D. (2020). Adaptive Laboratory Evolution Induced Novel Mutations in Zymomonas Mobilis ATCC ZW658: a Potential Platform for Co-utilization of Glucose and Xylose. J. Ind. Microbiol. Biotechnol. 47, 329–341. doi:10.1007/s10295-020-02270-y
Si, H.-M., Zhang, F., Wu, A.-N., Han, R.-Z., Xu, G.-C., and Ni, Y. (2016). DNA Microarray of Global Transcription Factor Mutant Reveals Membrane-Related Proteins Involved in N-Butanol Tolerance in Escherichia coli. Biotechnol. Biofuels 9, 14. doi:10.1186/s13068-016-0527-9
Sun, X.-M., Ren, L.-J., Bi, Z.-Q., Ji, X.-J., Zhao, Q.-Y., and Huang, H. (2018). Adaptive Evolution of Microalgae Schizochytrium Sp. Under High Salinity Stress to Alleviate Oxidative Damage and Improve Lipid Biosynthesis. Bioresour. Technol. 267, 438–444. doi:10.1016/j.biortech.2018.07.079
Tian, X., Liu, X., Zhang, Y., Chen, Y., Hang, H., Chu, J., et al. (2021). Metabolic Engineering Coupled with Adaptive Evolution Strategies for the Efficient Production of High-Quality L-Lactic Acid by Lactobacillus Paracasei. Bioresour. Technol. 323, 124549. doi:10.1016/j.biortech.2020.124549
Vishnu Prasad, J., Sahoo, T. K., Naveen, S., and Jayaraman, G. (2020). Evolutionary Engineering of Lactobacillus Bulgaricus Reduces Enzyme Usage and Enhances Conversion of Lignocellulosics to D-Lactic Acid by Simultaneous Saccharification and Fermentation. Biotechnol. Biofuels 13. doi:10.1186/s13068-020-01812-x
Wang, Z., Qi, Q., Lin, Y., Guo, Y., Liu, Y., and Wang, Q. (2019). QTL Analysis Reveals Genomic Variants Linked to High-Temperature Fermentation Performance in the Industrial Yeast. Biotechnol. Biofuels 12, 18. doi:10.1186/s13068-019-1398-7
Wang, Z., Zhou, L., Lu, M., Zhang, Y., Perveen, S., Zhou, H., et al. (2021). Adaptive Laboratory Evolution of Yarrowia Lipolytica Improves Ferulic Acid Tolerance. Appl. Microbiol. Biotechnol. 105, 1745–1758. doi:10.1007/s00253-021-11130-3
Wong, B. G., Mancuso, C. P., Kiriakov, S., Bashor, C. J., and Khalil, A. S. (2018). Precise, Automated Control of Conditions for High-Throughput Growth of Yeast and Bacteria with eVOLVER. Nat. Biotechnol. 36, 614–623. doi:10.1038/nbt.4151
Wu, T., Liu, J., Li, M., Zhang, G., Liu, L., Li, X., et al. (2020). Improvement of Sabinene Tolerance of Escherichia coli Using Adaptive Laboratory Evolution and Omics Technologies. Biotechnol. Biofuels 13, 15. doi:10.1186/s13068-020-01715-x
Xu, K., Gao, L., Hassan, J. U., Zhao, Z., Li, C., Huo, Y.-X., et al. (2018). Improving the Thermo-Tolerance of Yeast Base on the Antioxidant Defense System. Chem. Eng. Sci. 175, 335–342. doi:10.1016/j.ces.2017.10.016
Zeng, W., Guo, L., Xu, S., Chen, J., and Zhou, J. (2020). High-Throughput Screening Technology in Industrial Biotechnology. Trends Biotechnol. 38, 888–906. doi:10.1016/j.tibtech.2020.01.001
Zhang, B., Zhang, J., Wang, D., Han, R., Ding, R., Gao, X., et al. (2016). Simultaneous Fermentation of Glucose and Xylose at Elevated Temperatures Co-produces Ethanol and Xylitol through Overexpression of a Xylose-specific Transporter in Engineered Kluyveromyces Marxianus. Bioresour. Technol. 216, 227–237. doi:10.1016/j.biortech.2016.05.068
Zhang, F., Qian, X., Si, H., Xu, G., Han, R., and Ni, Y. (2015). Significantly Improved Solvent Tolerance of Escherichia coli by Global Transcription Machinery Engineering. Microb. Cel Fact. 14. doi:10.1186/s12934-015-0368-4
Zhou, X., Hua, X., Zhou, X., Xu, Y., and Zhang, W. (2019). Continuous Co-production of Biomass and Bio-Oxidized Metabolite (Sorbose) Using Gluconobacter Oxydans in a High-Oxygen Tension Bioreactor. Bioresour. Technol. 277, 221–224. doi:10.1016/j.biortech.2019.01.046
Keywords: adaptive evolution, evolutionary strategies, Gluconobacter oxydans, microbial microdroplet culture system, MMC
Citation: Liu L, Zeng W, Yu S, Li J and Zhou J (2021) Rapid Enabling of Gluconobacter oxydans Resistance to High D-Sorbitol Concentration and High Temperature by Microdroplet-Aided Adaptive Evolution. Front. Bioeng. Biotechnol. 9:731247. doi: 10.3389/fbioe.2021.731247
Received: 26 June 2021; Accepted: 10 August 2021;
Published: 03 September 2021.
Edited by:
Chong Li, Agricultural Genomics Institute at Shenzhen (CAAS), ChinaReviewed by:
Jian-Zhong Liu, Sun Yat-Sen University, ChinaChandrabose Selvaraj, Alagappa University, India
Yu Xia, Jiangnan University, China
Copyright © 2021 Liu, Zeng, Yu, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwen Zhou, emhvdWp3MTk4MkBqaWFuZ25hbi5lZHUuY24=
 Li Liu1,2,3,4,5
Li Liu1,2,3,4,5 Jingwen Zhou
Jingwen Zhou