- 1Nucleus of Multidisciplinary Research in Biology (Numpex-Bio), Federal University of Rio de Janeiro Xerém, Duque de Caxias, Brazil
- 2Laboratory of Tissue Bioengineering, National Institute of Metrology, Quality and Technology (Inmetro), Duque de Caxias, Brazil
- 3Post-graduation Program of Translational Biomedicine (Biotrans), Unigranrio, Duque de Caxias, Brazil
- 4Post-graduation Program in Biotechnology, National Institute of Metrology, Quality and Technology (Inmetro), Duque de Caxias, Brazil
Cancer is considered one of the most predominant diseases in the world and one of the principal causes of mortality per year. The cellular and molecular mechanisms involved in the development and establishment of solid tumors can be defined as tumorigenesis. Recent technological advances in the 3D cell culture field have enabled the recapitulation of tumorigenesis in vitro, including the complexity of stromal microenvironment. The establishment of these 3D solid tumor models has a crucial role in personalized medicine and drug discovery. Recently, spheroids and organoids are being largely explored as 3D solid tumor models for recreating tumorigenesis in vitro. In spheroids, the solid tumor can be recreated from cancer cells, cancer stem cells, stromal and immune cell lineages. Organoids must be derived from tumor biopsies, including cancer and cancer stem cells. Both models are considered as a suitable model for drug assessment and high-throughput screening. The main advantages of 3D bioprinting are its ability to engineer complex and controllable 3D tissue models in a higher resolution. Although 3D bioprinting represents a promising technology, main challenges need to be addressed to improve the results in cancer research. The aim of this review is to explore (1) the principal cell components and extracellular matrix composition of solid tumor microenvironment; (2) the recapitulation of tumorigenesis in vitro using spheroids and organoids as 3D culture models; and (3) the opportunities, challenges, and applications of 3D bioprinting in this area.
Introduction
Cancer remains one of the most predominant diseases in the world in the 21st century, affecting millions of patients per year (Roy and Saikia, 2016). Rather than responding appropriately to signals that maintain cell behavior, cancer cells grow and proliferate without control, invading normal tissues and organs, and eventually spreading throughout the organism (Chambers et al., 2002). The cellular and molecular mechanisms involved in the development and establishment of solid tumors is known as tumorigenesis. It is widely accepted that tumorigenesis is a multistep process, depending on a sequential accumulation of mutations of tissue cells (Ashkenazi et al., 2008). The tumor microenvironment is composed of non-cancerous cells with functions in all stages of tumorigenesis by both stimulating and/or facilitating abnormal cell proliferation (Arneth, 2019).
In recent years, literature has advanced in the better understanding of tumor microenvironment (DeBerardinis, 2020). The non-cancerous cell types include fibroblasts, endothelial cells, and immune cells (Casey et al., 2015; Jarosz-Biej et al., 2019). In addition, depending on the type of tumor, organ-specific interstitial cells are also present. According to previous descriptions, these cells are denominated as “tumor stroma” and, together with the extracellular matrix (ECM), oxygen levels and pH, constitute the tumor microenvironment (Briest et al., 2012; Hirata and Sahai, 2017). This complex interaction between tumor and non-tumor cells leads to an altered metabolism and ECM production. The better understanding of tumor microenvironment is a key challenge to address, contributing to the development of new drugs and treatments (Valkenburg et al., 2018).
In this context, 3D cell culture has gained space in literature due to its advantages compared with “classical” 2D cell culture. 3D cell culture can recreate a sort of tissue microenvironment, providing more accurate data about cell-to-cell interactions, cell-to-extracellular matrix interactions, tumorigenesis, drug discovery, gene expression, metabolic profiling, and protein profiling of the cells. 3D cell culture, such as spheroids and organoids, has the potential to provide alternative models to study tumor microenvironments (Nath and Devi, 2016; Jensen and Teng, 2020). In tumor biology, spheroids are represented by cancer cell lineages self-assembled in rounded shape and organoids by cells derived from tumor biopsies, including cancer stem cells, self-assembled in amorphous shape. Furthermore, cell culture platforms of tumor spheroids and organoids start to be adapted as a model for drug assessment and high-throughput screening (HTS) (Kondo et al., 2019; Heredia-Soto et al., 2020; Renner et al., 2020).
3D bioprinting is a promising emergent bottom–up technology to develop complex tissue models in vitro. 3D bioprinting is a form of additive manufacturing, where cells, biomaterials, and soluble factors can be assembled layer by layer (Mandrycky et al., 2016). From 3D bioprinting, it is possible to hierarchically organize tissues, as they are found in vivo, and faithfully recapitulate their morphology as well as functional aspects (Datta et al., 2018). Although 3D bioprinting represents a promising technology, main challenges still remain such as the speed of bioprinters and better bioinks for improving cell survival and function in cancer research. The main objective of this review is to explore the cellular and molecular composition of solid tumor microenvironment, the recapitulation of tumorigenesis and drug assessment using spheroids and organoids, and the opportunities and challenges of 3D bioprinting in this field.
The Tumor Microenvironment
Background
The tumor microenvironment is heterogeneous, composed mainly of tumor cells and endogenous stromal cells (non-cancerous) that are later recruited by the tumor itself. This microenvironment also contains extracellular components: ECM proteins, extracellular vesicles, cytokines, growth factors, and hormones nourished by a vascular network. The stromal cells are represented by endothelial cells, mesenchymal stem/stromal cells (MSCs), fibroblasts, and macrophages (Wu and Dai, 2017; O’Loghlen, 2018). During tumorigenesis, tumor cells interact greatly and evolve with this surrounding microenvironment, having profound effects on therapeutic efficacy (Bussard et al., 2016).
All tumor microenvironment components communicate continuously with each other mainly by (1) cell-to-cell interactions, (2) cell-to-extracellular matrix interactions, and (3) the network of cytokines, proteins, and chemokines that can favor the immune system or the tumor growth. Thus, any disruption in tumor microenvironment signaling will reflect changes of the balance between immune system and tumor (Hui and Chen, 2015; Merlano et al., 2019).
One of the most crucial factors for tumor microenvironment maintenance and progression to metastasis is the vascular network (Naumov et al., 2008; Quail and Joyce, 2013). Tumor vasculature is characterized as being disorganized and leaky, which is associated with altered endothelial cell adherents junction and tight junction formations, both critical to maintain vascular barrier functions. In addition, tumor cells induce programmed necrosis of endothelial cells, thus, increasing vascular leakiness and tumor cell extravasation and metastasis (Yang and Lin, 2017).
Cell Components of Tumor Microenvironment
In solid tumors, mesenchymal stem cells and fibroblasts, also named as cancer-associated fibroblasts (CAFs), are the main cellular components of the microenvironment. It is well known that in healthy tissues, fibroblasts support tissue repair and homeostasis; however, CAFs is a heterogeneous population that serves a different function compared with resident fibroblasts (Petrova et al., 2018; Ayan et al., 2020), as suggested by Sugimoto et al. (2006) and Kobayashi et al. (2019). The principal functions of CAFs in the tumor microenvironment are: (1) stimulate tumor cell proliferation by growth factor secretion, (2) modify cancer ECM, which will induce tumor progression and metastasis, and (3) modulate the inflammatory components that facilitate tumor initiation, progression, and metastasis (Servais and Erez, 2013; Raffaghello and Dazzi, 2015). Furthermore, CAFs support endothelial cells to start tumor angiogenesis. Endothelial cells offer nutritional support for tumor growth and development, showing a key role in tumor cell protection from the immune system (Arneth, 2019). Tumor endothelial cells are considered one of the main targets of anti-angiogenic therapy (Hida et al., 2013). A study published by Maishi et al. (2016) showed with two different tumor models that endothelial cells in the tumor microenvironment are able to promote tumor metastasis by direct interaction with tumor cells.
Myo-fibroblasts are specialized fibroblasts, a subpopulation of CAFs, which express the alpha-smooth muscle actin protein and are considered major players in the development of different fibrotic diseases, mainly due to their capacity to remodel the ECM (Yazdani et al., 2017; Ribatti and Tamma, 2019). In tumors, these activated fibroblasts can enhance tumorigenesis, angiogenesis, and metastasis by secreting growth factors and cytokines. Besides fibroblasts and endothelial cells, MSCs are present in the tumor microenvironment as well, interacting with tumor cells via the secretion of growth factors or cytokines, and by transferring mitochondria or microRNAs. Residing in tumors, MSCs form a fibrovascular network by differentiating into smooth muscle cells and vascular pericytes, contributing to vascular network extension (Guo and Deng, 2018). At the beginning of tumorigenesis, MSCs have been shown to drive tumor cells toward an invasive, premetastatic state. However, some studies showed that MSCs can also have an inhibitory effect on tumor growth by reducing cytotoxicity effects, pluripotency, and even by influencing macrophage polarization (Ridge et al., 2017).
Pericytes are multipotent perivascular cells with an established role in vasculature development. Studies have already shown that these cells present immune properties and might serve as a reservoir of MSCs to influence in the in vivo regeneration of diverse tissues. Pericytes located in the vessels play a significant role in the homeostasis of these vessels, and when recruited, they change their activation stage to MSCs in order to participate in injury events of the tissue (Meirelles et al., 2013). In addition, pericytes are capable of realizing tumor homing and are considered an important cell component of the tumor microenvironment (Ribeiro and Okamoto, 2015). In cancer, pericytes have been explored because of their capacity to stabilize blood vessel structure and permeability. Due to this, it was discovered that pericytes can affect tumor growth and metastasis positively or negatively. The effects of tumor growth are related to establishing a stable vascular network, which will ensure a proper delivery of nutrients to allow tumor cells maintenance and proliferation. However, these cells can prevent tumor cell dissemination by maintaining the permeability of blood vessels (Barrow and Colonna, 2019). Furthermore, many studies have shown that cancer vessels are characterized by abnormal pericyte population of cells and altered pericytes/endothelial cell interactions, which can effectively contribute to metastasis process and progression of cancers, especially perivascular ones such as glomus tumor, myopericytoma, and solitary fibrous tumor/hemangiopericytoma (Mravic et al., 2014; Chen et al., 2016).
Another cell type whose role is largely explored in tumor microenvironment is the adipocyte. Adipose tissue is composed of adipocytes and non-adipocyte cells, including MSCs from adipose tissue and macrophages. These cells release a variety of molecules that enable them to play a paracrine effect in pathological processes such as breast and ovarian cancer (Robado de Lope et al., 2018).
The macrophage is the most prominent immune cell type in the tumor microenvironment (Arneth, 2019). Macrophages have an active role from early carcinogenesis to tumor progression and metastasis, constituting up to 50% of a tumor mass depending on the type of tumor. Previous studies suggest that after infiltrating tumors, macrophages polarize to a M2 phenotype, take on the functions of tumor growth and angiogenesis, tissue remodeling, and suppression of antitumor immunity (Kim and Bae, 2016). Zhang A. et al. (2017) reported that CAFs promoted M2 polarization of macrophages in pancreatic ductal adenocarcinoma, which enhanced tumor cell growth, migration, and invasion.
Another immune population of cells present in the tumor microenvironment is the natural killer cells (NK). NK cells are large granular lymphocytes that control tumor growth by interaction with tumor cells or because they can affect the function of other innate and adaptative cell populations (Melaiu et al., 2020). Interestingly, NK cells show antitumor activity as they have the efficient and fast capacity to recognize and kill tumor cells. This function is mediated through cell-surface receptors, which examine tissue microenvironments for changes in surface and secretory phenotypes, and then alerts the immune system for the presence of infection or of a malignancy agent. Therefore, this function is largely explored for cancer immunotherapy treatments (Bi and Tian, 2017; Barrow and Colonna, 2019; Zhang et al., 2020). According to Fang et al. (2017), the main approaches used for cancer immunotherapy with NK cells are based on the use of cytokines, as IL-2 and isoforms, antibodies, and the adoptive transfer of ex vivo NK cells.
T cells also play important functions in the tumor microenvironment, where it is common to find inhibitory receptors. These can inhibit T cell metabolism and influence T cell signaling, both directly and through release of extracellular vesicles. When isolated from tumors, T cells generally show signs of exhaustion and present distinct metabolic features (Lim et al., 2020). Other immune cells that are present and modulate the tumor microenvironment are granulocytes, such as the mastocytes. Early mastocyte cell infiltration has been reported in human and animal tumors, especially in malignant melanoma, breast, and colorectal cancer (Liu et al., 2011; Komi and Redegeld, 2020). Mastocytes have different functions in the tumor microenvironment such as: (1) modulating tumor biology, by influencing in cell proliferation, survival, angiogenesis, and metastasis; and (2) establishing crosstalk with other tumor-infiltrating cells in the microenvironment (Aponte-López and Muñoz-Cruz, 2020).
Currently, different studies discuss the concept and functions of cancer stem cells (CSC) in tumor microenvironments. These cells, also called stem-like cells or tumor-initiating cells (TICs), were first described in 1994 and are a distinct subpopulation of tumor cells. Recently, this subpopulation of cells has been described as having a unique ability to initiate tumor growth and maintenance. In this context, CSC is considered an important target for cancer immunotherapies (Nassar and Blanpain, 2016; Codd et al., 2018). The quantity of CSC in the tumor microenvironment varies according to the tumor type. These cells can be responsible for preserving tumor heterogeneity by retaining self-renewal and differentiation properties. In addition, CSC also plays a role in innate resistance to cancer therapies, which in turn links to their persistence of the tumor in a specific tissue, which can lead to disease recurrence and metastatic spread (Albini et al., 2015). A study performed by Chen et al. (2014) demonstrated that CAFs enrich CSCs through de-differentiation process and reacquisition of stem cell-like properties in lung cancer. Briefly, the main results showed that CAFs develop a paracrine signaling that induce Nanog expression and promote stemness in cancer niche. What is interesting is that it is possible to discover new therapeutic targets to act in this paracrine signaling of CAFs to CSCs.
The Extracellular Matrix in Tumor Microenvironment
The ECM contains a diversity of proteins, which influence the cell phenotype of specific tissues due to their biochemical and biophysical properties. The principal ECM proteins secreted by cells in the tumor microenvironment are collagen, fibronectin, laminin, vitronectin, and tenascin (Cheng et al., 2020). It is well known that the ECM is highly dynamic because it is constantly being remodeled and degraded from embryogenesis until maturity. This remodeling is crucial for tissues homeostasis; however, dysregulation of ECM dynamics is common in the development of diseases as cancer (Bonnans et al., 2014; Walker et al., 2018).
In the tumor microenvironment, two main modifications are commonly observed in the ECM: stiffness (rigidity) and degradation. The increase in cross-linking between ECM proteins can cause stiffness (Najafi et al., 2019). The enhancement of tumor ECM stiffness is mainly induced by ECM deposition, remodeling by resident fibroblasts and by the transformed epithelium. In addition, the presence of chemokines and growth factors lead to an inflammation state. The inflammation state induces CAFs activation and their transdifferentiation into myofibroblasts, causing tissue desmoplasia. Then, myofibroblasts deposit ECM proteins, secrete growth factors, and apply contraction forces on the tumor ECM. In the end, newly deposited ECM proteins will generate larger and rigid fibers that turn the ECM rigid (Frantz et al., 2010). However, the disruption in the signaling between these ECM proteins will result in degradation, mainly caused by the activation of metalloproteinases (MMPs) (Najafi et al., 2019). The MMPs cleave collagen fibers of tumor ECM and reorganize them into tube-like structures to facilitate cell migration in the microenvironment (Malik et al., 2015).
The MMP genes were previously associated with increased risk and evolution of breast cancer. In the study developed by Slattery et al. (2013), the genetic variation of MMP1 (nine SNPs), MMP2 (eight SNPs), MMP3 (four SNPs), and MMP9 (three SNPs) together with breast cancer risk was evaluated in Hispanic and Non-Hispanic women. The results showed that MMPs have associations with breast cancer progression and prognosis. Overall, MMP-2 showed the strongest gene association with breast cancer development.
Regarding ECM modifications in breast cancer, another study, published by Boghaert et al. (2012) showed, with a 3D cell culture model, that the regions where the tumor cells invaded the breast tissue more was directly correlated with a higher mechanical stress of the host epithelial tissue. The use of a 3D cell culture model to recapitulate the breast tumor microenvironment can then aid in the better understanding of in vivo mechanisms.
One of the first studies published correlating abnormal ECM and the progression of cancer was performed by Neglia et al. (1991), which investigated the risk of cancer in patients with cystic fibrosis. The study was developed with North American and European patients with cystic fibrosis, and the results showed that, in fact, these patients had an increased risk to develop digestive tract cancers. In cancer, the abnormal ECM affects the progression of the disease by promoting changes in host cells normal functions. In addition, ECM anomalies are also capable of (1) deregulating the behavior of stromal cells, (2) promoting angiogenesis and inflammation associated with the tumor, (3) leading to the generation and maintenance of an established tumorigenic microenvironment, and (4) can also induce metastatic dissemination (Lu et al., 2012; Seager et al., 2017).
Not only cancer cells but also CAFs lead the modification and remodeling of the ECM during cancer progression. The biochemical cross talk between the cancer cells and CAFs, and the biomechanical changes of the ECM are major contributors to tumor cell migration and invasion, which will influence tumor progression to metastatic state. Additionally, growth factors, chemokines, and metabolic changes released from the ECM contribute to the maintenance and progression of the tumor microenvironment (Erdogan and Webb, 2017; Eble and Niland, 2019).
Due to the importance of ECM modification in the tumor microenvironment, studies are being conducted in order to develop therapeutic treatments to target the cancer ECM. Van der Steen et al. (2017) explored the functionalization of drug-loaded lyophilisomes (albumin-based biocapsules) loaded with doxorubicin and functionalized with antibodies, to act in the ECM, or stroma, of ovarian carcinomas, in order to evaluate its potential to eliminate cancer cells. The principal results showed that drug-loaded lyophilisomes were effective to induce cancer cell death and can be considered as a therapeutic agent to specifically target ECM components of the tumors. In addition, Zhang et al. (2018) explored the use of cyclopamine, a special inhibitor of the hedgehog-signaling pathway, which contributes to ECM formation of pancreatic ductal adenocarcinoma, to ameliorate solid stress and improve nanomedicine delivery to tumor site. The principal results showed that the drug was able to disrupt ECM in pancreatic ductal adenocarcinoma, reduced solid stress of the tumor together with an improvement of function of tumor vessels, which allowed a better perfusion in the tumor area.
Although the drugs discovered recently to target tumor ECM might effectively reduce the number of cancer cells and reduce solid stress, there are still many challenges that ECM components in tumor microenvironment can set that could interfere with therapeutic treatments. Briefly: (1) ECM proteins act as a physical barrier, which makes drug delivery more difficult, (2) ECM proteins can de-differentiate non-CSCs into CSCs, and this can make it harder for the elimination process of CSCs in the microenvironment, (3) the ability of ECM to modulate immune responses, and (4) complex nature of ECM, with its different molecules and isoforms (Nallanthighal et al., 2019). Therefore, the ECM in the tumor microenvironment has a considerable impact in cancer progression and further metastasis. Due to this, a better understanding of the interactions between cancer cells and ECM is needed and might only be addressed by 3D cell culture models, especially in order to have more faith in the results of drug screening to target cancer (Drost and Clevers, 2018).
3D Models Recapitulating the Tumorigenesis in vitro
Background
The tumorigenesis of cancer disease is heterogeneous in growth rate, invasiveness, drug sensibility, and individual patient derived characteristics (McGranahan and Swanton, 2017; Fan et al., 2019). Therefore, the in vitro and in vivo preclinical studies fail in emulating the microenvironment of the tumor to predict its sensibility or its resistance to drugs, or the metabolic and molecular pathways. This explains the low success rate of drug acceptance for oncologic drugs at 3.4% (Wong et al., 2019).
Immortalized cell lines are a valuable resource to investigate the physiological mechanisms and body–environmental interactions between healthy cells and cancerous cells due to their ease of growing and manipulating in vitro. Monolayer assays employing immortalized cancer cells are characterized by low cost, less complexity, and are readily employed in the HTS of drug trials and molecular biomarkers (Fan et al., 2019). However, because of the fast proliferation of the monolayers, it is likely that the culture might be affected by problems such as de-differentiation or abnormal gene expression profiles, which may influence the result of experiments as well as be contrasting to in vivo tests (Shah et al., 2018). Furthermore, monolayer assays glean so little about the gene expression, reorganization, and responses involved in the tumorigenesis, mainly due the absence of a tumor microenvironment (Gao and Chen, 2015).
To fill the gap between these insufficient or inappropriate models, 3D cultures arise as an urgent tool to improve the prediction system and mechanism of understanding tumorigenesis in humans. 3D cultures allow for systematic investigation into the several unidentified metabolic pathways and cascades (Sawant et al., 2016).
The classical scaffold-based approach in tissue engineering has focused on devising cells, bioactive factors, and scaffolds with biocompatible biomaterials to produce models able to maintain the tumor phenotype (Molina et al., 2020). In these models, it is possible to co-cultivate epithelial and stromal cells and observe the crosstalk of multiple cell types interacting, which regulate normal and neoplastic development (Sawant et al., 2016).
In contrast to scaffold-based methods, scaffold-free approaches emerge as 3D tumor models. The scaffold-free approaches are aggregates of cells, producing several common features that are similar to the solid tumor in vivo such as cellular heterogeneity, cell-cell signaling, hypoxia, membrane protein distribution, and gene expression patterns (Zhao et al., 2019).
Tumor Spheroids
The development of 3D models such as spheroids made it possible to engineer several cancer-like microenvironments in vitro. Many papers claim to have developed their protocols to build tumors such as glioblastomas, colorectal, breast, liver, lungs, among others (Kelm et al., 2003; Hirschhaeuser et al., 2010; Chimenti et al., 2017; Eilenberger et al., 2018; Froehlich et al., 2018; Oraiopoulou et al., 2019; Foglietta et al., 2020; Lee et al., 2020).
The breast cell line MCF-7 is an adenocarcinoma-luminal subtype one. The cell morphology is epithelium-like resulting in their ability to self-aggregate into a steady shape, which makes it easier to maintain their viability and to use it for implantation in mice for in vivo studies (Do Amaral et al., 2011; Comşa et al., 2015; Froehlich et al., 2018).
HEPG-2 is an epithelial-like hepatocellular carcinoma that, due to the liver cells’ role of the metabolism, is considered a valuable option to study cell genotoxicity (Luckert et al., 2017; Shah et al., 2018). 3D models using HEPG-2 can be used alone in drug screening or as a co-culture with other tumor cell lines (Lan et al., 2010; Jung et al., 2017).
Some aspects must be considered when working with spheroids. One of them is the quality of the 3D protocol, which is related to some variables such as the kind of support for the culture, the non-adherent medium used, the number of cells that are seeded, the spheroid formation technique, the temperature, and the amount of CO2 and O2 available (Mironov et al., 2009; Mehta et al., 2012; Däster et al., 2017). All these factors are highly changeable according to the tumor line chosen.
The role of hypoxia and the capacity of a tumor to induce neovascularization in its microenvironment using spheroid models have been debated since the early 1990s. It has been established that genetic changes can cause an “angiogenic switch” as the newly mutated cells acquire the ability to upregulate the production of angiogenic factors in comparison to healthy cells, especially in hypoxic niches (Shweiki et al., 1995; Catalano et al., 2013).
Studies using colorectal spheroids and the 5-Fluorouracil drug have indicated that hypoxia and necrosis induction is associated with tumor progression and cell resistance to chemotherapy treatments. The difference in the spheroid size is a variation that also shows its importance in determining whether the mentioned effects moderately or intensely impact the aggressiveness of the tumor (Karlsson et al., 2012; Däster et al., 2017). On the contrary, other studies developed with multicellular spheroids also have demonstrated that when hypoxia–reoxygenation is induced, the levels of vascular endothelial growth factor (VEGF) are downregulated by the tumor cells, as well as it activated DNA damage repair markers (Kondoh et al., 2013; Riffle et al., 2017).
Nevertheless, managing these elements and controlling the long-term viability of the spheroids is an arduous task due to their natural propensity of apoptosis, as a result of poor gaseous and nutrients diffusion (Zhang W. et al., 2016). One possible solution is to use microfluidic systems to allow continued flow of the molecules needed for the spheroids to keep metabolizing and proliferating (Moshksayan et al., 2018). Human lung adenocarcinoma A549 cells, for instance, can be seeded with human endothelial cells in a collagen-I–Matrigel microfluidic device containing a micro-pump to supply the system with oxygen and nutrients. It is a useful protocol for further respiratory system cancer studies (Lee et al., 2019).
Colorectal tumors are likely to be formed at elderly ages, especially over 50 years. It is also the third cause of death among men and women in the United States (Siegel et al., 2020). The communication promoted by cells in the spheroid allows studies to explore the interactions between drugs and the 3D model (Elliott and Yuan, 2011). Concerning this approach, it was shown through spheroid models that the anticancer drug KP1339 triggers an immune cell death in vitro, which matches arrays that showed preclinical activity in vivo (Wernitznig et al., 2019).
Coming up with a model that mimics the microenvironment of mammary tissue requires a complex mixture of several cell types and tissues, as well as functional ECM and long-term sustainable cell–cell and cell–ECM interactions. In this regard, adipose tissue might work well when co-cultured with mammary cell lines (Kim et al., 2004; Picollet-D’hahan et al., 2016). As a complex tissue, adipose is constituted of several populations of cells such as adipocytes, MSCs, endothelial progenitor cells, pre-adipocytes, lymphocytes, pericytes, and macrophages (Schäffler and Büchler, 2007; Hu and Polyak, 2008).
Studies with co-culture between MCF7 line and MSCs have shown that this mesenchymal population can improve tumor aggressiveness in vivo in comparison with MCF7 culture alone. Similar to immune cells, MSCs demonstrate tropism for spots consisting of damaged tissue including tumor microenvironmental sites, cooperating with migration and metastasis (Koellensperger et al., 2017; Chen et al., 2019).
Tumor Organoids
Different from spheroids, tumor organoids must be derived from human tumor biopsies (Drost and Clevers, 2018; Wang et al., 2020). The advantages and applications of tumor organoids are related to the tissue-specific mutagenic processes accumulating specific types of somatic mutations during malignant transformation in patients. Single stem cell-derived and long-term-cultured organoids were used to determine the genome-wide mutation patterns in distinct healthy stem cells (Wang et al., 2020).
The ability to grow organoids with high efficiency from healthy human adult stem cells has paved the way to grow tumor tissue patient-derived organoids (PDO). So far, long-term organoid cultures have been established from primary colon, esophagus, pancreas, stomach, liver, endometrium, and breast cancer tissues, as well as from metastatic colon, prostate, and breast cancer biopsy samples (Drost and Clevers, 2018).
Another 3D model is the cultivation and testing of the patient-derived tumor xenografts (PDTX) generated in animal models. PDTX is about the implantation of small pieces of tumors from human biopsies into highly immunodeficient mice. After tumor growth, the tumor is transferred into secondary recipient mice. PDTXs often maintain the structures of the original tumors at molecular, cellular, and tissue levels (Drost and Clevers, 2018). Thus, it is able to recapitulate the heterogeneity of the tumor and its native microenvironment; however, it is more incompatible to HTS due to its expensive, time consuming and complex procedure (Hidalgo et al., 2014). Besides PDTX, it is also possible to induce the tumor directly into animal models. However, animals present great phylogenetic distance to humans, have different metabolism, size, and lifespan, which all misdirect the drug development during human clinical trials (Wang, 2019).
The generation of cancer spheroids and organoids, like PDO are low cost, fast compared with PDTX, can be adapted to HTS and allow investigation of the alterations occurring during the initiation and progression of tumorigenesis (Fatehullah et al., 2016; Fan et al., 2019). This is one of the reasons why tumor organoids have been increasingly used as a faithful in vitro model system to study cancer metastasis (Fan et al., 2019).
Tumor organoids keep the main pathophysiological features required to identify the critical factors in the acquisition of cancer metastatic potential, which may elucidate mechanisms involved in the metastasis cascade (Fan et al., 2019). On the other hand, one of the intrinsic limitations is the lack of stroma, blood vessels, and immune cells in cultured organoids, especially the immune cells (Wang et al., 2020) due to their regulatory roles in epithelial cell growth and differentiation, invasion, and metastasis (Mueller and Fusenig, 2002; Sawant et al., 2016).
One very interesting strategy when studying tumor organoids is to associate healthy organoids with tumor ones in a fluidic platform called organ-on-a-chip aiming to study metastasis via the circulatory system. These devices mentioned before are microfabricated to emulate a precise microenvironment, controlled, with continuous flow perfusion culture, and high-throughput format (Fan et al., 2019). Jeon et al. (2015) studied 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation, while Xu et al. (2016) projected a four-organ chip to assess lung cancer metastasis. Huang et al. (2009) found out that the laminar flow properties of microfluidic devices have been leveraged to compartmentalize human mammary fibroblasts in an ECM gel side-by-side with another ECM gel containing breast ductal carcinoma in situ cells; this setup revealed that the fibroblasts had to be in contact with the tumor cells to induce the transition to the invasive phenotype (Huang et al., 2009; Benam et al., 2015).
High-Throughput Screening and 3D Models
Pre-clinical studies fail around 85% in the oncological drug trials, not demonstrating sufficient safety or efficacy (Gao et al., 2015). To overcome this issue, the approaches that enable high-throughput (thousands of cells per experiment) are best suited to efficiently sample the complex cellular diversity in organoids and to understand organoid-to-organoid variability (Brazovskaja et al., 2019).
High-throughput screening provides a practical method to investigate large numbers of pharmaceutical compounds in in vitro monolayers assays, being a universal assay in pharmaceutical and Biotech industries (Pereira and Williams, 2007). It has also spawned a billion-dollar industry that supports the increasing demands for speed, capacity, and cost-effective screening of vast libraries of compounds (Pereira and Williams, 2007). The accessibility of HTS data merged with the ToxCastTM/Tox21 databases allows for elucidative toxicological considerations seen below (Suh et al., 2018).
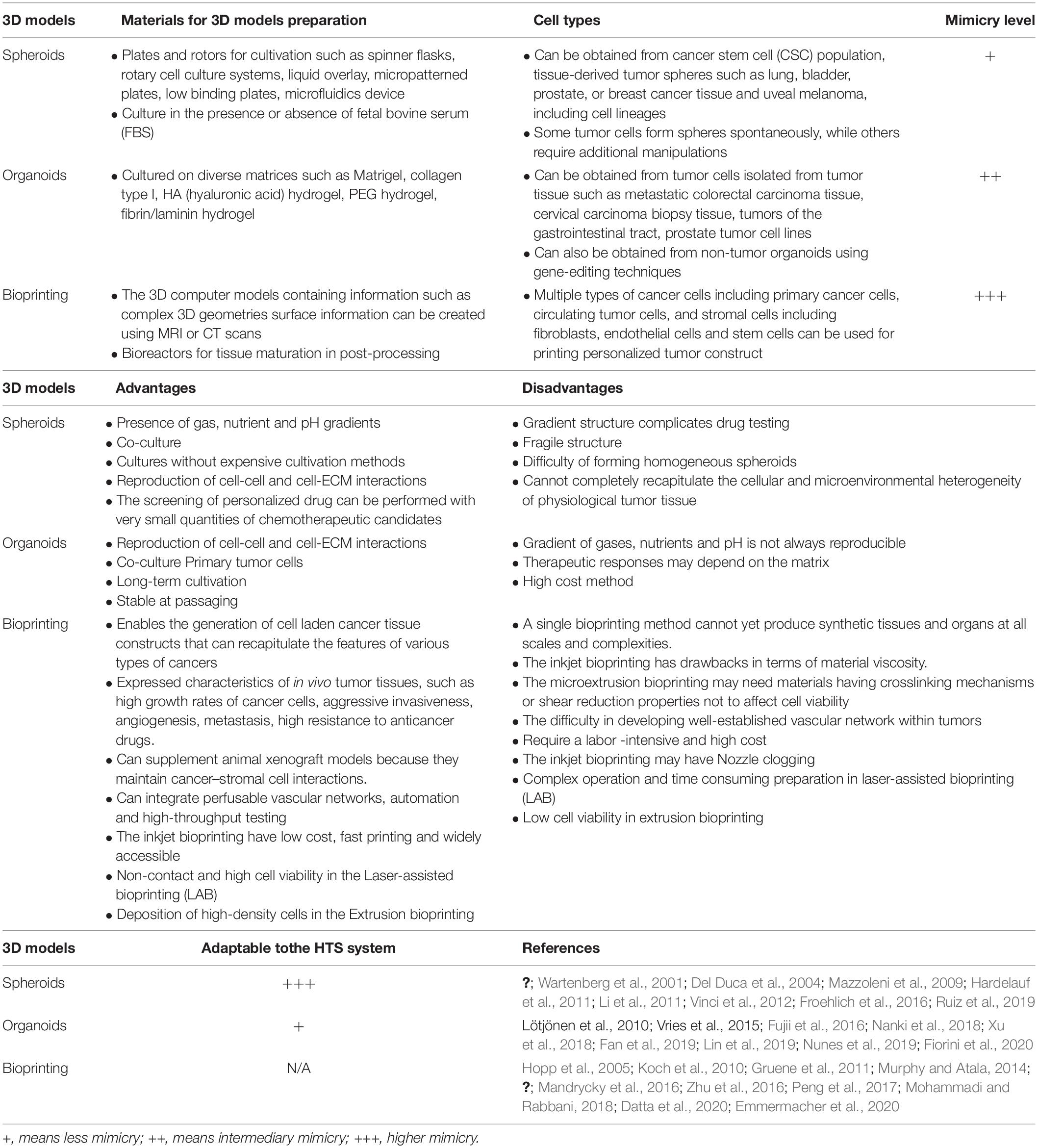
Based on the advantages of tissue engineering scaffold-free approaches in recapitulating the tumor microenvironment, mainly represented by spheroids and organoids, a paradigm shift in HTS placing them at the forefront of drug discovery (Li et al., 2016) together with the need to adapt the protocols for the HTS. Spheroids have been adapted for use with several HTS technologies. On the other hand, organoids represent a challenge, mainly due to the presence of hydrogels and their heterogeneity of shape (Figure 1). Furthermore, the most common read-out of HTS technologies is still based on imaging systems making spheroids and organoid depths and their associated light scattering a technical challenge (Li et al., 2016).
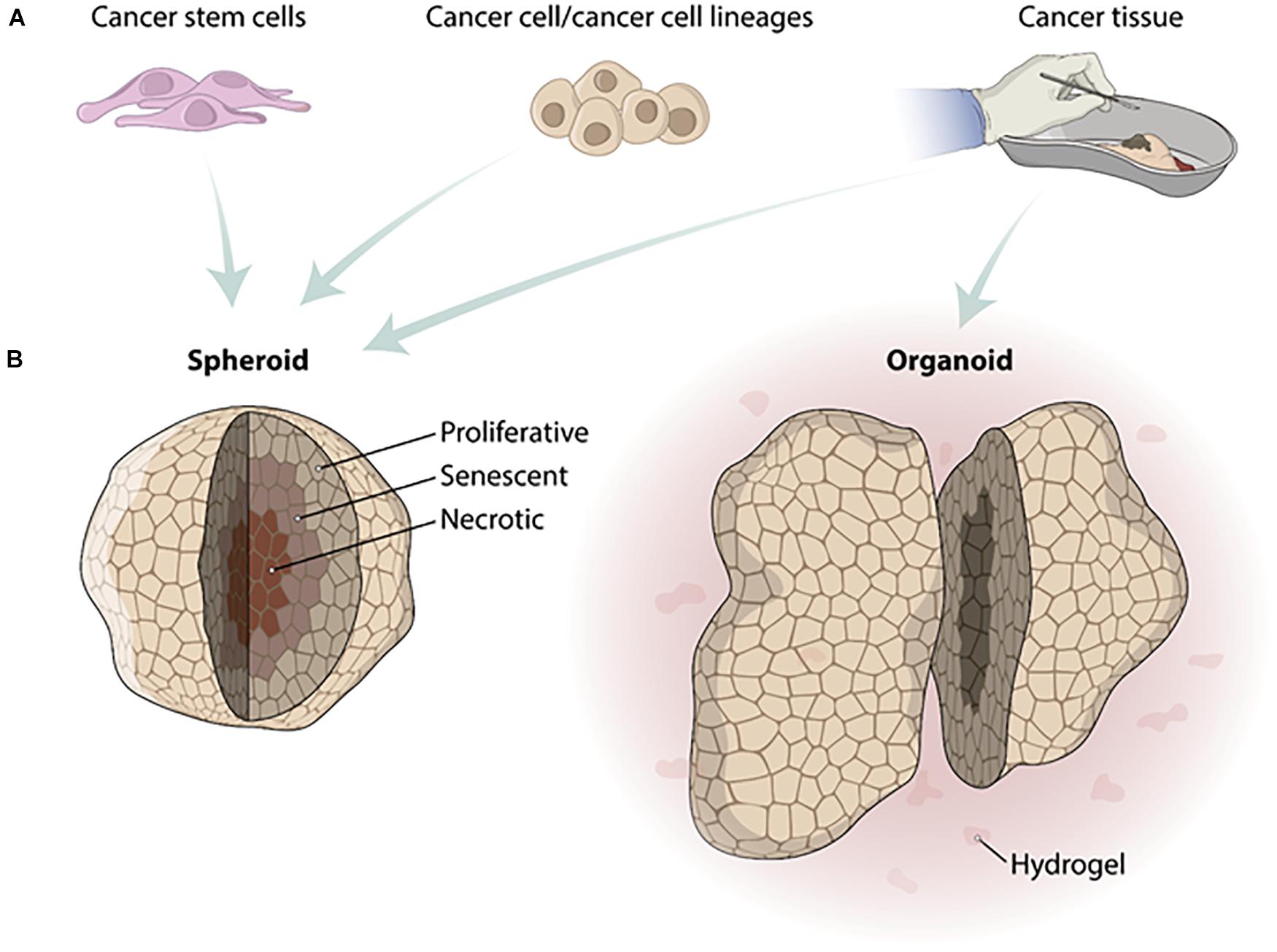
Figure 1. Differences in fabrication of tumor spheroids and organoids. (A) Cell types used to produce tumor spheroids and organoids. Spheroids can be fabricated from cancer stem cells, cancer cells/cancer cell lineages, or cancer tissue. Organoids must be fabricated from human cancer biopsies. (B) After the fabrication process, tumor spheroids show different zones because of the distinct gradient concentrations of O2 and CO2. The zones from spheroids inside out are necrotic, senescent, and proliferative. Organoids are usually produced in a hydrogel substrate and do not present a homogeneous size and shape.
So far, tumor spheroids, tumor organoids, and PDTXs are allowed for testing of multiple individual drugs prior to in vivo analysis (Beshiri et al., 2018). Gao et al. (2015) established ∼1,000 PDXs with a diverse set of driver mutations against 62 treatments across six indications. Mateo et al. (2015) showed the presence of a homologous recombination deficiency genotype in Metastatic castrate-resistant prostate cancer and predicted responsiveness to Olaparib, which is the first genomic biomarker-driven therapy on track for FDA approval. Another example is for human kidney organoids, where Czerniecki et al. (2018) produced automated organoids and assessed drug effects by HTS.
Liu et al. (2020) review that Kita et al. (2019) screened 2,098 compounds in bladder cancer organoid cell lines. They also discovered that Disulfiram, an anti-alcoholism drug, and cisplatin had a cooperative effect. Lampis et al. (2018), after screening 484 compounds in six cholangiocarcinoma’s organoid cell lines, presented that the sensitivity of HSP90 inhibitors was related to the mutation of MIR21 gene. Kondo and Inoue (2019) reported an advanced system for the HTS of 2,427 drugs using the cancer tissue-originated spheroid; those lines exhibited diverse sensitivities to the hit compounds, demonstrating the usefulness of this system for investigating highly heterogeneous disease.
There is now increasing evidence that the tumor microenvironment affects the efficacy of drugs on the cancer cells (Lal-Nag et al., 2017). Several complex ovarian cancer models have already been published, such as the 3D omental mesothelium model and models that include microfluidics, which demonstrates this (Watters et al., 2018). Currently, the mesothelium model is the only 3D organotypic microenvironment model of ovarian cancer that is used by multiple research groups (Kenny et al., 2007). The mesothelium model recapitulates the main physiological aspects of ovarian cancer cells in the mesothelium lining (Watters et al., 2018). Lal-Nag et al. (2017) proved that several classes of targets were more efficacious in cancer cells growing in the absence of the metastatic microenvironment, and other target classes were less efficacious in cancer cells in pre-formed spheres compared with forming spheroids cultures. These methods were adapted to HTS and to more than 100,000 small-molecule compounds that can potentially identify novel treatments (Watters et al., 2018). Hasan and group reported the use of bioprinting for in vitro ovarian cancer tissue modeling for research applicable to HTS. Human ovarian cancer was printed on MatrigelTM to form multicellular acini (Hasan et al., 2011). This approach allows for physiologically relevant cell fabrications and can also provide an alternative to animal testing (Matai et al., 2020).
Tumor Organoids and Personalized Medicine
As explained in the sections before, tumor organoids must be derived from human biopsies. This outstanding characteristic from tumor organoids has given rise to the creation of tumor biobanks highlighting the concept of personalized medicine to predict effective drugs before the start of the treatment. One crucial challenge to be addressed related to drug testing for cancer models is that the majority of drugs show intratumor heterogeneity, while others are uniformly toxic in all cases. Furthermore, as organoids can be produced from a patient’s own cells, the genetic analyses and drug screening results will be specific to the patient’s tumor (Tellez-Gabriel et al., 2018; Kondo et al., 2019). Some examples are described below.
An organoid biobank of breast cancer tissues from >100 patients was established (Sachs et al., 2018). These organoids represented genetic and histopathological features of breast cancer and maintained the expression of breast cancer biomarkers. This means organoid biobanks have predictive value for drug efficacy in the treatment of individual patients (Wang, 2019), allowing personalized cancer treatment.
Van de Wetering et al. (2015) established tumor organoid cultures from 20 consecutive colorectal carcinoma patients. The results showed that organoids were able to resemble the original tumor characteristics, and gene expression analysis indicated that the majority of consecutive colorectal carcinoma molecular subtypes were properly represented.
Sachs et al. (2018) described a protocol to produce a biobank of human mammary epithelial organoids. The organoids were able to recapitulate the diversity of the disease. Additionally, histological, hormonal, and gene expression analysis resembled the status of the original tumor. Furthermore, the organoids allowed proper drug screening tests when compared with in vivo xeno-transplantations.
Yan et al. (2017) developed a primary gastric cancer organoid biobank that comprises normal, dysplastic, cancer, and lymph node metastases from 34 patients. The results showed that organoids were able to closely mimic the morphology, transcriptome, and gene expression profiles when compared with in vivo original tumors. It was also seen that organoids were sensitive to unexpected drugs (recently approved or in clinical trials) after drug screening tests.
3D Bioprinting
Background
As discussed previously, 3D cell culture models as spheroids and organoids are capable of better mimicking the tumor microenvironment that is found in vivo by recapitulating cellular and molecular events. However, spheroids and organoids follow a non-guided spontaneous formation of tissues and organs by self-assembly mechanism. In this context, because of the ability to precisely guide and organize the position of different cell types and growth factors and also perfusable networks, 3D bioprinting has a potential to improve current models and guide recapitulation of the tumor microenvironment (Datta et al., 2020). The ability to engineer controllable cancer tissue models in high resolution can considerably accelerate cancer research and improve personalized medicine, improving the treatment and life expectancy of cancer patients in the future (Knowlton et al., 2015; Belgodere et al., 2018; Langer et al., 2019).
3D bioprinting is one of the most widely used technologies in tissue engineering and regenerative medicine to develop complex tissues and organs that mimic their native microenvironment (Murphy and Atala, 2014; Moroni et al., 2018). As 3D bioprinting is a process where bioinks, usually composed of hydrogels, and cells are turned into functional tissue-engineered constructs from digital models, it is constantly showing more advantages compared with classical scaffold-based tissue engineering. One of the principal aims of using 3D bioprinting techniques so far is to biofabricate vascular structures (Vijayavenkataraman et al., 2018). This technique integrates biomaterials, living cells, and automated controlled systems to create complex microstructures and precise control over the structures developed compared with other currently available methods (Mandrycky et al., 2016).
Usually, 3D bioprinting begins with a computer-assisted process in order to deposit biologically relevant biomaterials, growth factors, and living cells to generate a desired tissue or organ model. Basically, it is possible to divide the 3D bioprinting process in three: (1) pre-processing for acquiring the 3D computer-aided design (CAD) model of the tissue to be bioprinted, (2) automated deposition of cells, spheroids, biomaterials, or other biological component of interest, and (3) maturation of the tissue constructs (Zhang Y. S. et al., 2017; Datta et al., 2018).
The principal 3D bioprinting techniques are (1) inkjet, (2) extrusion-based, and (3) laser-assisted bioprinting (Huang et al., 2017). In inkjet bioprinting, it is possible to precisely control both the size of the desired tissue pattern, as well as the generated droplets. In this way, it is possible to determine the volume, size, and quantity of a sample to be bioprinted. In terms of precision, it is possible to control the number of the cells per droplet, which is an advantage when scaffolds are being used (Zhang and Zhang, 2015).
Extrusion-based bioprinting is the most used technique that uses the principles of a fluid-dispensing system with a robotic one for extruding materials, which can then be applied to different 3D bioprinting approaches. The fluid-dispensing system can be directed by pneumatic, mechanical, or solenoid forces. Through extrusion-based bioprinting, it is possible to precisely deposit cells, which can be encapsulated in a pre-established design of geometrical filaments and then bioprinted (Ozbolat and Hospodiuk, 2016). However, one of the biggest challenges of this technique is the resolution level that can be reached (Ning and Chen, 2017).
Laser-assisted bioprinting is based on the laser-induced forward transfer (LIFT) principle and is considered a “direct-write” method, which can precisely control the virtual deposition of cells, growth factors, and biomaterial containing droplets at a MHz range speed. Therefore, with this technique, it is possible to achieve high resolution (Devillard et al., 2014). However, the principal disadvantage is the use of the laser directly on the cells, which can damage cell viability (Derakhshanfar et al., 2018).
In order to authentically develop the desired tissue construct, the hydrogel choice is crucial, mainly because the hydrogel will provide the physical and biochemical properties to guide cell proliferation, differentiation, and the final maturation of the engineered construct. In this way, the hydrogel must contain similar properties of the desired tissue when in vivo (Gopinathan and Noh, 2018). Several hydrogel formulations have been developed, such as decellularized ECM, alginate, gelatin, hyaluronic acid, and polymers (such as methacrylated gelatin, polyethylene glycol and poly lactic acid) to serve as functional bioinks (Parak et al., 2019).
Scaffold-Free 3D Bioprinting
The use of bioinks is the foundation of bioprinting. This approach is based on cells and/or biomaterials with specific formulations for each type of cell (Hospodiuk et al., 2017). The ideal formulation of bioinks should meet each cell type’s biological requirements without toxicity to the cells (Gungor-Ozkerim et al., 2018). Their desired properties include printing, mechanical properties, biodegradation, and post-bioprinter maturation (Hong et al., 2018). These properties depend on different parameters such as solution viscosity, surface tension of the bioink, the ability to interconnect on its own, and the properties of the printer nozzle surface. The living cells encapsulated in the bioink grow and occupy the space to form predefined tissue structures (Huang et al., 2017; Gungor-Ozkerim et al., 2018).
However, an important limitation of this approach is that, although cells can be manipulated individually, they do not form mechanically stable assemblies in many cases unless intercellular adhesions are made very strong, possibly by chemical means, which is not ideal for mimicking the tissue microenvironment (Goulart et al., 2019). Additional structural cohesion needs to be produced by the cells, like their own secreted ECM. However, this is a long-term process and depends on the cell type and the ECM deposition quality (Ong et al., 2018; Heo et al., 2020).
The alternative approach of using cells with a pre-assembly of spheroids has been widely studied, as it improves the production capacity of its ECM, in addition to providing greater biomechanical cohesion in larger-scale constructs for bioprinting (Swaminathan et al., 2019). Also, spheroid-based methods are generally milder and, therefore, induce much less or no cell damage during bioprinting (De Moor et al., 2018). Another attractive feature of spheroid bioprinting is its efficiency, as the speed of bioprinting can be increased using large building blocks such as spheroids (Gutzweiler et al., 2017).
An alternative method, still considered to be scaffold-free, can provide temporary support to the spheroids and, thus, facilitate their fusion and maturation in tissue models using a set of microneedles (”Kenzan”). The Kenzan bioprinting method provides a high-resolution biofabrication process, facilitating the fusion of spheroids into larger tissue constructions in a needle matrix removed after spheroid fusion. This method is used in the Bio-3D Regenova Printer marketed by Cyfuse Biomedical (Moldovan, 2018; Murata et al., 2020).
Recently, bioinks were developed using formulations composed only of spheroids with several thousand cells. Studies have shown that spheroids can form tissue threads up to 8 cm in length with rapid spheroid fusion without using aggressive chemicals as crosslinkers or as support materials (Bakirci et al., 2017; Ji and Guvendiren, 2017; Osidak et al., 2019). The spheroid bioinks showed better results than the individual cells because they preserved the integrity of the ECM. The use of bioinks without structure, composed only of cells, has been attracting more and more attention as a bioprinting method for the 3D construction of complex tissues, through which the application of ball-beading constructions is widely addressed (Skardal et al., 2016).
Several spheroid bioprinting techniques have been reported in the literature. One of the first techniques widely explored was extrusion-based bioprinting, in which the spheroids were loaded into a syringe cylinder and extruded into a controlled distribution gel medium. However, the spheroid tips easily deform in the syringe and are subject to breakage during the extrusion process. Simultaneously, the support structures need to be printed in 3D to facilitate the aggregation of extruded ball tips (Mandrycky et al., 2016). A significant advance was made using the Kenzan method. However, the method has limitations inherent to the accuracy of the 3D bioprinting process (Moldovan et al., 2017). To overcome some of the greatest challenges of the current techniques, recent studies have shown that aspiration-assisted bioprinting allows accurate bioprinting of spheroids over sacrificial or functional gel substrates (Chimene et al., 2016). In a mold of sacrificial material, such as alginate or agarose, the material is discarded as the bioprinted tissue matures and subsequently deposits its components in the ECM (Ayan et al., 2020).
Although methods using sacrificial gel substrates do not present the common problems of inkjet and microextrusion (such as nozzle clogging), they still have their technical limitations (Vijayavenkataraman et al., 2018; Adhikari et al., 2021). One of these limitations is the time necessary to form and maturate spheroids prior to bioprinting. In addition, the development of specific bioinks compatible with the characteristics of most spheroid types is essential for the viability and correct maturation of each tissue (Li et al., 2016).
Large-scale production is also still a great challenge (Datta et al., 2020). Studies on standardization and automation of spheroid production are essential for building more complex and genuine-sized tissues in the future. Moreover, an important implication in biofabrication is training for the unique skills and techniques required of this technology’s users and operators. Finally, the bioprinting of 3D cell constructs originating from spheroids composed of various types of cells has been studied to increase the functionality of these 3D constructs (Sasmal et al., 2018; Swaminathan et al., 2019).
3D Bioprinting of Tumor Models
For recreating the tumor microenvironment, there is a need of tumor ECM reconstitution and the recreation of tumor vasculature (Liu et al., 2020). In Figure 2, the essential steps and bioinks to recreate the tumor microenvironment by 3D bioprinting are proposed. The ECM of the tumor microenvironment is composed of different proteins and stromal cells, but it is known that the composition of tumor ECM is tumor and patient specific. In addition, the biomechanical properties of tumor ECM can regulate tumor behavior and progression (Zhang Y. S. et al., 2016).
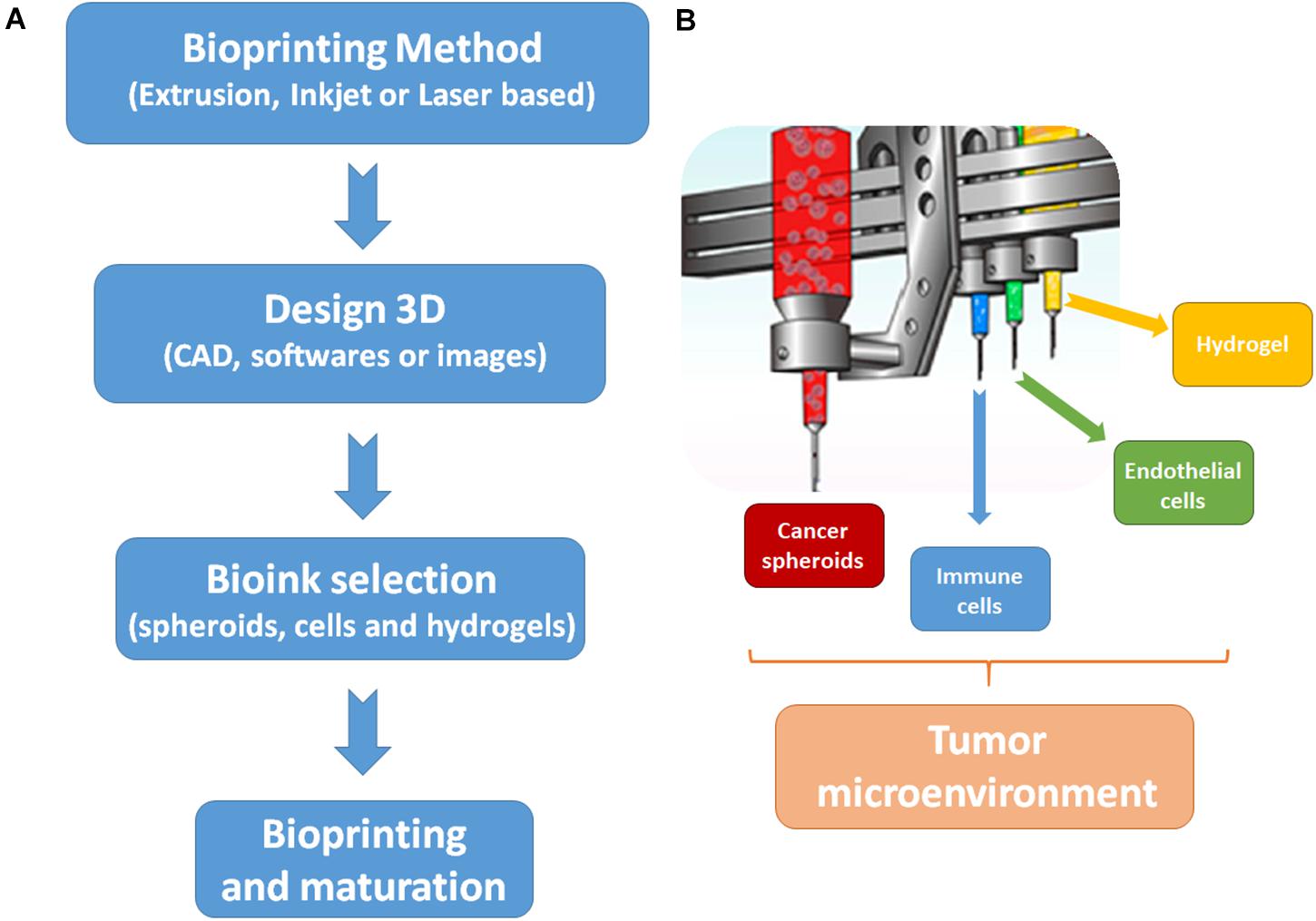
Figure 2. Steps and bioinks to biofabricate the tumor microenvironment by 3D bioprinting. (A) Steps to start the biofabrication process. First, it is necessary to choose a bioprinting method, which will complement the desired output. The majority of studies to develop cancer models are done with extrusion-base techniques. Then, the 3D design of the cancer model must be made by software or can be based on images. Next, it is necessary to choose the biological (cells or spheroids) and biomaterial (usually hydrogels) components of the bioinks. Finally, the bioprinting process can be started and tissue maturation can be carried out post printing. (B) Bioinks to replicate tumor microenvironments. In order to mimic the tumor microenvironment, the main bioinks are tumor spheroids, immune cells, endothelial cells, and a hydrogel to support cells proliferation and survival.
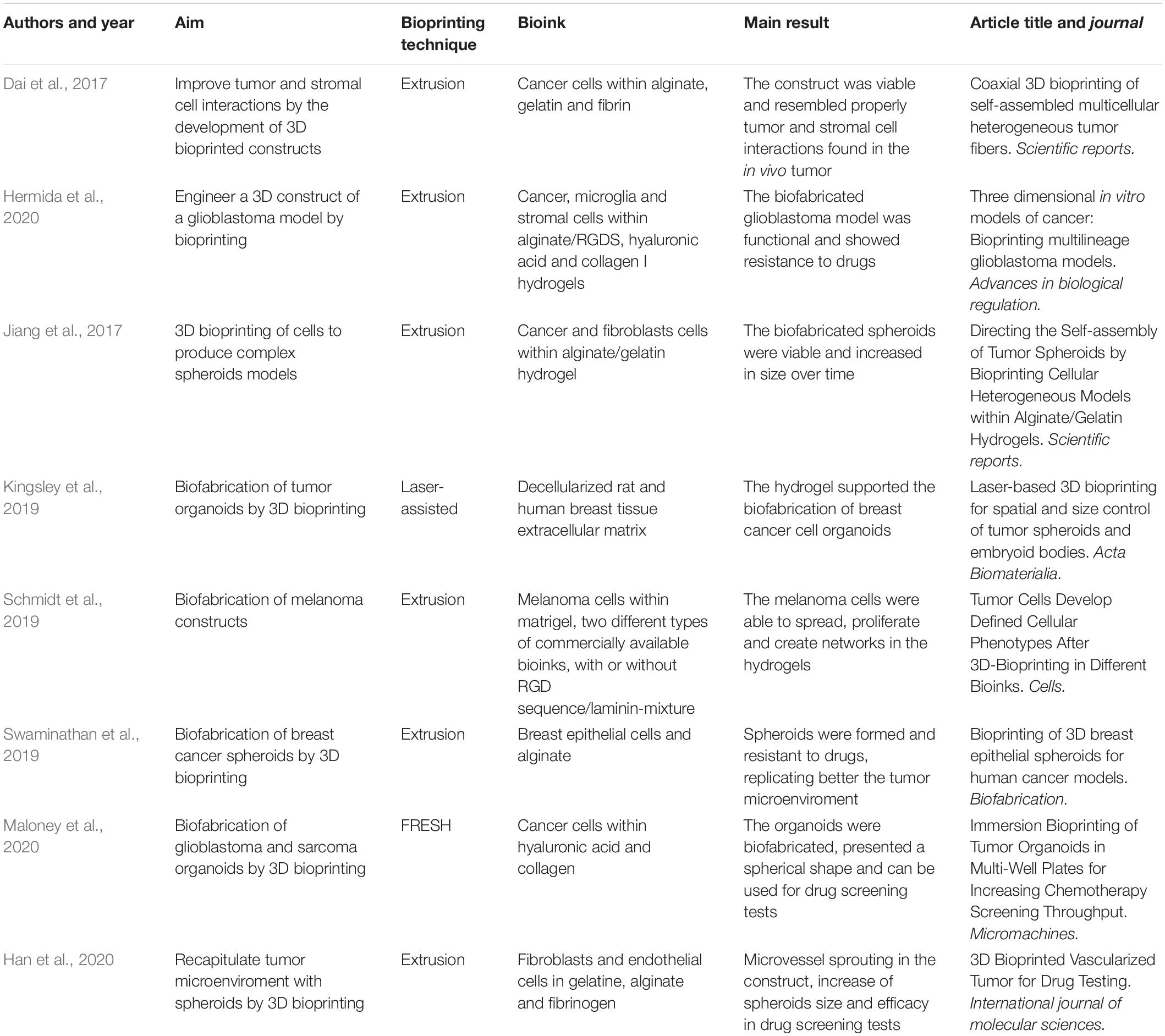
Recently, a considerable number of studies were performed to develop tumor models by 3D bioprinting. Table 1 reviews some of these studies. Dai et al. (2017) focused on replicating tumor microenvironments by improving tumor and stromal cell interactions in 3D bioprinted constructs. Their strategy relied on the self-assembly of multicellular heterogeneous brain tumor cell fibers by extrusion-based bioprinting. These fibers were part of the tumor ECM of the brain tumor. The morphological results showed that the construct was viable, proliferative, and presented tumor-stromal cell interactions. Hermida et al. (2020) used extrusion-based bioprinting to engineer glioblastoma models made of cancer, microglia, and stromal cells bioprinted within alginate modified with RGDS cell adhesion peptides, hyaluronic acid, and type I collagen. The glioblastoma cells presented more resistance to chemotherapeutic drugs in 3D engineered bioprinted constructs compared with monolayer cultures.
Despite the development described above for spheroid bioprinting strategies, several studies have shown the spontaneous formation of spheroids after 3D bioprinting, reaching the mimicry of specific cancer types. Jiang et al. (2017) developed a proof of concept study by bioprinting a cross-linked alginate/gelatin hydrogel composed of breast cancer lineage cells and fibroblasts. After 1 week in culture, breast cancer cells formed viable spheroids that increased in size over time and attracted migrating fibroblasts through a matrix region of the hydrogel, which infiltrated the breast cancer spheroids.
Using the technique of 3D bioprinting named “laser direct write,” Kingsley et al. (2019) used microbeads to allow the formation and growth of multicellular tumor spheroids with homogeneous size and shape. The decellularized rat and human breast tissue ECM was used as a bioink for organoid formation by 3D bioprinting (Mollica et al., 2019). The principal advantage in this strategy is that these ECM hydrogels keep the structural and signaling cues of the breast cancer environment, which can determine a cell’s fate. The results showed that the hydrogel supported the production of breast cancer cell organoids allowing their use to engineer more complex organoids models to pre-clinical assays.
Schmidt et al. (2019) compared the interaction of different bioprinted hydrogels with melanoma cells. In total, five hydrogels were tested: matrigel and two different types of commercially available bioinks, with or without RGD sequence/laminin mixture. In Matrigel, melanoma cells were able to spread, proliferate, and produce networks in the construct, while in gelatin methacrylate melanoma cells grow in clusters. As expected, the choice of the bioink is crucial for the behavior of cancer cells in engineered constructs.
Human breast epithelial cell lines can be bioprinted as a cell suspension or as formed spheroids in alginate-based bioinks. These cells only formed spheroids in Matrigel-based biolinks and pre-formed spheroids kept their morphology and viability after bioprinting. When spheroids were formed, breast cancer cells were more resistant to drug assessment, replicating the tumor microenvironment (Swaminathan et al., 2019).
Maloney et al. (2020) used an immersion printing technique approach to perform the 3D bioprinting of tissue organoids in 96-well plates. The results showed that the bioink allowed the maintenance of the organoid structure. In the study, the bioink was composed of hyaluronic acid and collagen and was printed in a support bath made of gelatin. This innovative strategy, named as “Freeform Reversible Embedding of Suspended Hydrogels” (FRESH) is being largely explored to bioprint soft tissues without a scaffold, because it allows the maintenance of the biological structure after the removal of the support bath. To the best of our knowledge, this is the only technique at the moment that can be used to bioprint organoids. More importantly, the authors proved with patient-derived glioblastoma and sarcoma organoids that it is possible to use the method for drug screening tests in vitro.
Han et al. (2020) used 3D bioprinting to recapitulate the tumor microenvironment using spheroids. The method consisted of the biofabrication of a blood vessel layer engineered by fibroblasts and endothelial cells in gelatin, alginate, and fibrinogen, followed by the seeding of multicellular tumor spheroids of glioblastoma cells onto this blood vessel layer. The main results showed the sprouting of blood vessels with an increase in spheroid size. In addition, drug testing was performed and the biofabricated construct was sensitive to the treatment, showing that it can be used for drug efficacy tests in vitro.
However, there is an important limitation of these models that use hydrogels for drug testing. HTS analysis based on luminescence/fluorescence cannot be applied to these models due to the presence of hydrogels which are high viscous biomaterials. Another issue related to hydrogels is the small volume used in some applications because it can impair HTS tests (Yu et al., 2018). Table 2 summarizes the main characteristics, advantages, and disadvantages of 3D models described in this review.
Perspectives
Some studies already used 3D bioprinting to develop successful tumor models; however, to the best of our knowledge, the use of spheroids as a printable bioink to biofabricate tumor models has not been largely explored yet. As spheroids are a 3D model with complex cell-to-cell and cell-to-extracellular matrix interactions, it would be advantageous to use them as the main component of the bioink associated with the stromal components and immune cells. Tumor organoids show the main advantage of being derived from human cancer biopsies; however, their 3D bioprinting is still in its infancy due to their shape heterogeneity, lack of reproducibility, and complexity.
Furthermore, patient-derived 3D bioprinted tumor models could be successfully used for in vitro drug screening of anticancer drugs in large scale. However, some challenges need to be addressed before this step, especially related to the hydrogel composition. Some studies are already exploring how to optimize the hydrogel to not impair HTS tests and analysis (Barata et al., 2016; Sarkar and Kumar, 2016; Lee et al., 2018). In addition, ongoing studies are focusing on the development of combined microfluidic/bioprinted constructs to minimize the cost and facilitate HTS of a large number of cancer drugs for a particular patient in order to improve personalized medicine approaches (Augustine et al., 2021).
Conclusion
3D bioprinting is a recent and innovative approach that offers the ability to create highly complex hierarchical 3D constructs with cells, biomaterials, and growth factors. Bioprinting methods have been developed and optimized in recent years in order to accurately replicate the morphology, functions, and physiology of a specific tissue and their in vivo microenvironment. As tumor microenvironments are complex in cell and extracellular matrix composition, 3D bioprinting holds great potential for applications in cancer research, in order to mimic more reliable tumor models and their vasculature (Knowlton et al., 2015; Albritton and Miller, 2017).
The use of 3D bioprinting can allow the positioning of tumor spheroids or organoids and the surrounding stromal and immune cells, commonly associated with this complex tumor microenvironment. The recapitulation of tumorigenesis will provide more reliable results to drug screening tests (Satpathy et al., 2018; Meng et al., 2019) and personalized medicine (Ma et al., 2018).
Author Contributions
GK and LB contributed to conception and design of the manuscript. GK, GM, RT, and LB wrote the first draft of the manuscript. GK, GM, RT, and ÚK wrote sections of the manuscript. BM designed the table and performed the formatting of the manuscript. LB performed the main corrections and revised the manuscript. All the authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This review was supported by the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, grant number: E26/202.682/2018 and by the Office of Naval Research (ONR), grant number: #N62909-21-1-2091.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank the Office of Naval Research (ONR) for the financial support. The authors wish to thank researcher Grace Brogan, Staff Scientist at DPS Engineering providing Lab Management services to the Johnson and Johnson 3D printing center (Trinity College Dublin, Dublin, Ireland), for reviewing the English of the manuscript.
References
Adhikari, J., Roy, A., Das, A., Ghosh, M., Thomas, S., Sinha, A., et al. (2021). Effects of processing parameters of 3D bioprinting on the cellular activity of bioinks. Macromol. Biosci. 21:2000179. doi: 10.1002/mabi.202000179
Albini, A., Bruno, A., Gallo, C., Pajardi, G., Noonan, D. M., and Dallaglio, K. (2015). Cancer stem cells and the tumor microenvironment: interplay in tumor heterogeneity. Connect. Tissue Res. 56, 414–425.
Albritton, J. L., and Miller, J. S. (2017). 3D bioprinting: improving in vitro models of metastasis with heterogeneous tumor microenvironments. Dis. Models Mech. 10, 3–14. doi: 10.1242/dmm.025049
Aponte-López, A., and Muñoz-Cruz, S. (2020). Mast cells in the tumor microenvironment. Adv. Exp. Medi. Biol. 1273, 159–173. doi: 10.1007/978-3-030-49270-0_9
Ashkenazi, R., Gentry, S. N., and Jackson, T. L. (2008). Pathways to tumorigenesis-modeling mutation acquisition in stem cells and their progeny. Neoplasia (New York, N.Y.) 10, 1170–1182. doi: 10.1593/neo.08572
Augustine, R., Kalva, S. N., Ahmad, R., Zahid, A. A., Hasan, S., Nayeem, A., et al. (2021). 3D Bioprinted cancer models: Revolutionizing personalized cancer therapy. Transl. Oncol. 14:101015. doi: 10.1016/j.tranon.2021.101015
Ayan, B., Celik, N., Zhang, Z., Zhou, K., Kim, M. H., Banerjee, D., et al. (2020). Aspiration-assisted freeform bioprinting of pre-fabricated tissue spheroids in a yield-stress gel. Commun. Phys. 3:183.
Bakirci, E., Toprakhisar, B., Zeybek, M. C., Ince, G. O., and Koc, B. (2017). Cell sheet based bioink for 3D bioprinting applications. Biofabrication 9:024105. doi: 10.1088/1758-5090/aa764f
Barata, D., van Blitterswijk, C., and Habibovic, P. (2016). High-throughput screening approaches and combinatorial development of biomaterials using microfluidics. Acta Biomater. 34, 1–20. doi: 10.1016/j.actbio.2015.09.009
Barrow, A. D., and Colonna, M. (2019). Exploiting NK cell surveillance pathways for cancer therapy. Cancers 11:55. doi: 10.3390/cancers11010055
Belgodere, J. A., King, C. T., Bursavich, J. B., Burow, M. E., Martin, E. C., and Jung, J. P. (2018). Engineering breast cancer microenvironments and 3D bioprinting. Front. Bioeng. Biotechnol. 6:66.
Benam, K. H., Dauth, S., Hassell, B., Herland, A., Jain, A., Jang, K. J., et al. (2015). Engineered in vitro disease models. Annu. Rev. Pathol. 10, 195–262.
Beshiri, M. L., Tice, C. M., Tran, C., Nguyen, H. M., Sowalsky, A. G., Agarwal, S., et al. (2018). A PDX/organoid biobank of advanced prostate cancers captures genomic and phenotypic heterogeneity for disease modeling and therapeutic screening. Clin. Cancer Res. 24, 4332–4345. doi: 10.1158/1078-0432.ccr-18-0409
Boghaert, E., Gleghorn, J. P., Lee, K., Gjorevski, N., Radisky, D. C., and Nelson, C. M. (2012). Host epithelial geometry regulates breast cancer cell invasiveness. Proc. Natl. Acad. Sci. U.S.A. 109, 19632–19637. doi: 10.1073/pnas.1118872109
Bonnans, C., Chou, J., and Werb, Z. (2014). Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801. doi: 10.1038/nrm3904
Brazovskaja, A., Treutlein, B., and Camp, J. G. (2019). High-throughput single-cell transcriptomics on organoids. Curr. Opin. Biotechnol. 55, 167–171. doi: 10.1016/j.copbio.2018.11.002
Briest, F., Berndt, A., Clement, J., Junker, K., Eggeling, F. V., Grimm, S., et al. (2012). Tumor-stroma interactions in tumorigenesis: lessons from stem cell biology. Front. Biosci. (Elite Edition) 4, 1871–1887. doi: 10.2741/509
Bussard, K. M., Mutkus, L., Stumpf, K., Gomez-Manzano, C., and Marini, F. C. (2016). Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 18:84.
Casey, S. C., Amedei, A., Aquilano, K., Azmi, A. S., Benencia, F., Bhakta, D., et al. (2015). Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin. Cancer Biol. 35 Suppl(Suppl), S199–S223.
Catalano, V., Turdo, A., Di Franco, S., Dieli, F., Todaro, M., and Stassi, G. (2013). Tumor and its microenvironment: a synergistic interplay. Semin. Cancer Biol. 23, 522–532. doi: 10.1016/j.semcancer.2013.08.007
Chambers, A. F., Groom, A. C., and MacDonald, I. C. (2002). Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2, 563–572. doi: 10.1038/nrc865
Chen, B., Wu, Y., Ao, Z., Cai, H., Nunez, A., Liu, Y., et al. (2019). High-throughput acoustofluidic fabrication of tumor spheroids. Lab on a Chip 19, 1755–1763. doi: 10.1039/C9LC00135B
Chen, W. J., Ho, C. C., Chang, Y. L., Chen, H. Y., Lin, C. A., Ling, T. Y., et al. (2014). Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nat. Commun. 5:3472.
Chen, Z., Xu, X. H., and Hu, J. (2016). Role of pericytes in angiogenesis: focus on cancer angiogenesis and anti-angiogenic therapy. Neoplasma 63, 173–182.
Cheng, Y. Q., Wang, S. B., Liu, J. H., Jin, L., Liu, Y., Li, C. Y., et al. (2020). Modifying the tumour microenvironment and reverting tumour cells: new strategies for treating malignant tumours. Cell Prolif. 53:e12865.
Chimene, D., Lennox, K. K., Kaunas, R. R., and Gaharwar, A. K. (2016). Advanced bioinks for 3D printing: a materials science perspective. Ann. Biomed. Eng. 44, 2090–2102. doi: 10.1007/s10439-016-1638-y
Chimenti, I., Pagano, F., Angelini, F., Siciliano, C., Mangino, G., Picchio, V., et al. (2017). Human lung spheroids as in vitro niches of lung progenitor cells with distinctive paracrine and plasticity properties. Stem Cells Transl. Med. 6, 767–777. doi: 10.5966/sctm.2015-0374
Codd, A. S., Kanaseki, T., Torigo, T., and Tabi, Z. (2018). Cancer stem cells as targets for immunotherapy. Immunology 153, 304–314. doi: 10.1111/imm.12866
Comşa, Ş, Cîmpean, A. M., and Raica, M. (2015). The story of MCF-7 breast cancer cell line: 40 years of experience in research.. Anticancer Res. 35, 3147–3154.
Czerniecki, S. M., Cruz, N. M., Harder, J. L., Menon, R., Annis, J., Otto, E. A., et al. (2018). High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940.e4.
Dai, X., Liu, L., Ouyang, J., Li, X., Zhang, X., Lan, Q., et al. (2017). Coaxial 3D bioprinting of self-assembled multicellular heterogeneous tumor fibers. Sci. Rep. 7:1457.
Däster, S., Amatruda, N., Calabrese, D., Ivanek, R., Turrini, E., Droeser, R. A., et al. (2017). Induction of hypoxia and necrosis in multicellular tumor spheroids is associated with resistance to chemotherapy treatment. Oncotarget 8, 1725–1736. doi: 10.18632/oncotarget.13857
Datta, P., Barui, A., Wu, Y., Ozbolat, V., Moncal, K. K., and Ozbolat, I. T. (2018). Essential steps in bioprinting: From pre-to post-bioprinting. Biotechnol. Adv. 36, 1481–1504. doi: 10.1016/j.biotechadv.2018.06.003
Datta, P., Dey, M., Ataie, Z., Unutmaz, D., and Ozbolat, I. T. (2020). 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis. Oncol. 4:18.
De Moor, L., Merovci, I., Baetens, S., Verstraeten, J., Kowalska, P., Krysko, D. V., et al. (2018). High-throughput fabrication of vascularized spheroids for bioprinting. Biofabrication 10:035009. doi: 10.1088/1758-5090/aac7e6
DeBerardinis, R. J. (2020). Tumor Microenvironment, Metabolism, and Immunotherapy. New Engl. J. Med. 382, 869–871.
Del Duca, D., Werbowetski, T., and Del Maestro, R. F. (2004). Spheroid preparation from hanging drops: characterization of a model of brain tumor invasion. J. Neuro Oncol. 67, 295–303. doi: 10.1023/b:neon.0000024220.07063.70
Derakhshanfar, S., Mbeleck, R., Xu, K., Zhang, X., Zhong, W., and Xing, M. (2018). 3D bioprinting for biomedical devices and tissue engineering: a review of recent trends and advances. Bioact. Mater. 3, 144–156. doi: 10.1016/j.bioactmat.2017.11.008
Devillard, R., Pagès, E., Correa, M. M., Kériquel, V., Rémy, M., Kalisky, J., et al. (2014). Cell patterning by laser-assisted bioprinting. Methods Cell Biol. 119, 159–174. doi: 10.1016/b978-0-12-416742-1.00009-3
Do Amaral, J. B., Rezende-Teixeira, P., Freitas, V. M., and Machado-Santelli, G. M. (2011). MCF-7 cells as a three-dimensional model for the study of human breast cancer. Tissue Eng. Part C Methods 17, 1097–1107. doi: 10.1089/ten.tec.2011.0260
Drost, J., and Clevers, H. (2018). Organoids in cancer research. Nat. Rev. Cancer 18, 407–418. doi: 10.1038/s41568-018-0007-6
Eble, J. A., and Niland, S. (2019). The extracellular matrix in tumor progression and metastasis. Clin. Exp. Metastasis 36, 171–198. doi: 10.1007/s10585-019-09966-1
Eilenberger, C., Kratz, S., Rothbauer, M., Ehmoser, E. K., Ertl, P., and Küpcü, S. (2018). Optimized alamarBlue assay protocol for drug dose-response determination of 3D tumor spheroids. MethodsX 5, 781–787. doi: 10.1016/j.mex.2018.07.011
Elliott, N. T., and Yuan, F. (2011). A review of three-dimensional in vitro tissue models for drug discovery and transport studies. J. Pharm. Sci. 100, 59–74. doi: 10.1002/jps.22257
Emmermacher, J., Spura, D., Cziommer, J., Kilian, D., Wollborn, T., Fritsching, U., et al. (2020). Engineering considerations on extrusion-based bioprinting: interactions of material behavior, mechanical forces and cells in the printing needle. Biofabrication 12:025022. doi: 10.1088/1758-5090/ab7553
Erdogan, B., and Webb, D. J. (2017). Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem. Soc. Trans. 45, 229–236. doi: 10.1042/bst20160387
Fan, H., Demirci, U., and Chen, P. (2019). Emerging organoid models: leaping forward in cancer research. J. Hematol. Oncol. 12:142.
Fang, F., Xiao, W., and Tian, Z. (2017). NK cell-based immunotherapy for cancer. Semin. Immunol. 31, 37–54.
Fatehullah, A., Tan, S. H., and Barker, N. (2016). Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254. doi: 10.1038/ncb3312
Fiorini, E., Veghini, L., and Corbo, V. (2020). Modeling cell communication in cancer with organoids: making the complex simple. Front. Cell and Dev. Biol. 8:166.
Foglietta, F., Canaparo, R., Muccioli, G., Terreno, E., and Serpe, L. (2020). Methodological aspects and pharmacological applications of three-dimensional cancer cell cultures and organoids. Life Sci. 254:117784. doi: 10.1016/j.lfs.2020.117784
Frantz, C., Stewart, K. M., and Weaver, V. M. (2010). The extracellular matrix at a glance. J. Cell Sci. 123(Pt 24), 4195–4200. doi: 10.1242/jcs.023820
Froehlich, K., Haeger, J. D., Heger, J., Pastuschek, J., Photini, S. M., Yan, Y., et al. (2018). Generation of multicellular breast cancer tumor spheroids: comparison of different protocols. J. Mammary Gland Biol. Neoplasia 21, 89–98. doi: 10.1007/s10911-016-9359-2
Fujii, M., Shimokawa, M., Date, S., Takano, A., Matano, M., Nanki, K., et al. (2016). A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 18, 827–838. doi: 10.1016/j.stem.2016.04.003
Gao, D., and Chen, Y. (2015). Organoid development in cancer genome discovery. Curr. Opin. Genet. Dev. 30, 42–48. doi: 10.1016/j.gde.2015.02.007
Gao, H., Korn, J. M., Ferretti, S., Monahan, J. E., Wang, Y., Singh, M., et al. (2015). High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325.
Goulart, E., de Caires-Junior, L. C., Telles-Silva, K. A., Araujo, B. H. S., Rocco, S. A., Sforca, M., et al. (2019). 3D bioprinting of liver spheroids derived from human induced pluripotent stem cells sustain liver function and viability in vitro. Biofabrication 12:015010. doi: 10.1088/1758-5090/ab4a30
Gruene, M., Deiwick, A., Koch, L., Schlie, S., Unger, C., Hofmann, N., et al. (2011). Laser printing of stem cells for biofabrication of scaffold-free autologous grafts. Tissue Eng. Part C Methods 17, 79–87. doi: 10.1089/ten.tec.2010.0359
Gungor-Ozkerim, P. S., Inci, I., Zhang, Y. S., Khademhosseini, A., and Dokmeci, M. R. (2018). Bioinks for 3D bioprinting: an overview. Biomater. Sci. 6, 915–946.
Guo, S., and Deng, C. X. (2018). Effect of stromal cells in tumor microenvironment on metastasis initiation. Int. J. Biol. Sci. 14:2083. doi: 10.7150/ijbs.25720
Gutzweiler, L., Kartmann, S., Troendle, K., Benning, L., Finkenzeller, G., Zengerle, R., et al. (2017). Large scale production and controlled deposition of single HUVEC spheroids for bioprinting applications. Biofabrication 9:025027. doi: 10.1088/1758-5090/aa7218
Han, S., Kim, S., Chen, Z., Shin, H. K., Lee, S. Y., Moon, H. E., et al. (2020). 3D bioprinted vascularized tumour for drug testing. Int. J. Mol. Sci. 21:2993. doi: 10.3390/ijms21082993
Hardelauf, H., Frimat, J. P., Stewart, J. D., Schormann, W., Chiang, Y. Y., Lampen, P., et al. (2011). Microarrays for the scalable production of metabolically relevant tumour spheroids: a tool for modulating chemosensitivity traits. Lab Chip 11, 419–428. doi: 10.1039/c0lc00089b
Hasan, M. R., Ho, S. H., Owen, D. A., and Tai, I. T. (2011). Inhibition of VEGF induces cellular senescence in colorectal cancer cells. Int. J. Cancer 129, 2115–2123. doi: 10.1002/ijc.26179
Heo, D. N., Ayan, B., Dey, M., Banerjee, D., Wee, H., Lewis, G. S., et al. (2020). Aspiration-assisted bioprinting of co-cultured osteogenic spheroids for bone tissue engineering. Biofabrication 13:015013. doi: 10.1088/1758-5090/abc1bf
Heredia-Soto, V., Redondo, A., Kreilinger, J., Martínez-Marín, V., Berjón, A., and Mendiola, M. (2020). 3D culture modelling: an emerging approach for translational cancer research in sarcomas. Curr. Med. Chem. 27, 4778–4788. doi: 10.2174/0929867326666191212162102
Hermida, M. A., Kumar, J. D., Schwarz, D., Laverty, K. G., Di Bartolo, A., Ardron, M., et al. (2020). Three dimensional in vitro models of cancer: bioprinting multilineage glioblastoma models. Adv. Biol. Regul. 75:100658. doi: 10.1016/j.jbior.2019.100658
Hida, K., Akiyama, K., Ohga, N., Maishi, N., and Hida, Y. (2013). Tumour endothelial cells acquire drug resistance in a tumour microenvironment. J. Biochem. 153, 243–249. doi: 10.1093/jb/mvs152
Hidalgo, M., Amant, F., Biankin, A. V., Budinská, E., Byrne, A. T., Caldas, C., et al. (2014). Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 4, 998–1013. doi: 10.1158/2159-8290.cd-14-0001
Hirata, E., and Sahai, E. (2017). Tumor microenvironment and differential responses to therapy. Cold Spring Harb. Perspect. Med. 7:a026781. doi: 10.1101/cshperspect.a026781
Hirschhaeuser, F., Menne, H., Dittfeld, C., West, J., Mueller-Klieser, W., and Kunz-Schughart, L. A. (2010). Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 148, 3–15. doi: 10.1016/j.jbiotec.2010.01.012
Hong, N., Yang, G. H., Lee, J., and Kim, G. (2018). 3D bioprinting and its in vivo applications. J. Biomed. Mater. Res. Part B Appl. Biomater. 106, 444–459. doi: 10.1002/jbm.b.33826
Hopp, B., Smausz, T., Kresz, N., Barna, N., Bor, Z., Kolozsvári, L., et al. (2005). Survival and proliferative ability of various living cell types after laser-induced forward transfer. Tissue Eng. 11, 1817–1823. doi: 10.1089/ten.2005.11.1817
Hospodiuk, M., Dey, M., Sosnoski, D., and Ozbolat, I. T. (2017). The bioink: a comprehensive review on bioprintable materials. Biotechnol. Adv. 35, 217–239. doi: 10.1016/j.biotechadv.2016.12.006
Hu, M., and Polyak, K. (2008). Molecular characterisation of the tumour microenvironment in breast cancer. Eur. J. Cancer (Oxford, England 1990) 44, 2760–2765. doi: 10.1016/j.ejca.2008.09.038
Huang, C. P., Lu, J., Seon, H., Lee, A. P., Flanagan, L. A., Kim, H. Y., et al. (2009). Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab Chip 9, 1740–1748. doi: 10.1039/b818401a
Huang, Y., Zhang, X. F., Gao, G., Yonezawa, T., and Cui, X. (2017). 3D bioprinting and the current applications in tissue engineering. Biotechnol. J. 12:1600734.
Hui, L., and Chen, Y. (2015). Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 368, 7–13. doi: 10.1016/j.canlet.2015.07.039
Jarosz-Biej, M., Smolarczyk, R., Cichoń, T., and Kułach, N. (2019). Tumor microenvironment as a “game changer” in cancer radiotherapy. Int. J. Mol. Sci. 20:3212. doi: 10.3390/ijms20133212
Jensen, C., and Teng, Y. (2020). Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 7:33.
Jeon, J. S., Bersini, S., Gilardi, M., Dubini, G., Charest, J. L., Moretti, M., et al. (2015). Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. U.S.A. 112, 214–219. doi: 10.1073/pnas.1417115112
Ji, S., and Guvendiren, M. (2017). Recent advances in bioink design for 3D bioprinting of tissues and organs. Front. Bioeng. Biotechnol. 5:23.
Jiang, T., Munguia-Lopez, J. G., Flores-Torres, S., Grant, J., Vijayakumar, S., Leon-Rodriguez, A., et al. (2017). Directing the self-assembly of tumour spheroids by bioprinting cellular heterogeneous models within alginate/gelatin hydrogels. Sci. Rep. 7:4575.
Jung, H. R., Kang, H. M., Ryu, J. W., Kim, D. S., Noh, K. H., Kim, E. S., et al. (2017). Cell spheroids with enhanced aggressiveness to mimic human liver cancer in vitro and in vivo. Sci. Rep. 7:10499.
Karlsson, H., Fryknäs, M., Larsson, R., and Nygren, P. (2012). Loss of cancer drug activity in colon cancer HCT-116 cells during spheroid formation in a new 3-D spheroid cell culture system. Exp. Cell Res. 318, 1577–1585. doi: 10.1016/j.yexcr.2012.03.026
Kelm, J. M., Timmins, N. E., Brown, C. J., Fussenegger, M., and Nielsen, L. K. (2003). Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 83, 173–180. doi: 10.1002/bit.10655
Kenny, H. A., Krausz, T., Yamada, S. D., and Lengyel, E. (2007). Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int. J. Cancer 121, 1463–1472. doi: 10.1002/ijc.22874
Kim, J. B., Stein, R., and O’Hare, M. J. (2004). Three-dimensional in vitro tissue culture models of breast cancer – a review. Breast Cancer Res. Treat. 85, 281–291. doi: 10.1023/b:brea.0000025418.88785.2b
Kim, J., and Bae, J. S. (2016). Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm. 2016:6058147.
Kingsley, D. M., Roberge, C. L., Rudkouskaya, A., Faulkner, D. E., Barroso, M., Intes, X., et al. (2019). Laser-based 3D bioprinting for spatial and size control of tumor spheroids and embryoid bodies. Acta Biomater. 95, 357–370. doi: 10.1016/j.actbio.2019.02.014
Kita, Y., Hamada, A., Saito, R., Teramoto, Y., Tanaka, R., Takano, K., et al. (2019). Systematic chemical screening identifies disulfiram as a repurposed drug that enhances sensitivity to cisplatin in bladder cancer: a summary of preclinical studies. Br. J. Cancer 121, 1027–1038. doi: 10.1038/s41416-019-0609-0
Knowlton, S., Onal, S., Yu, C. H., Zhao, J. J., and Tasoglu, S. (2015). Bioprinting for cancer research. Trends Biotechnol. 33, 504–513.
Kobayashi, H., Enomoto, A., Woods, S. L., Burt, A. D., Takahashi, M., and Worthley, D. L. (2019). Cancer-associated fibroblasts in gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 282–295. doi: 10.1038/s41575-019-0115-0
Koch, L., Kuhn, S., Sorg, H., Gruene, M., Schlie, S., Gaebel, R., et al. (2010). Laser printing of skin cells and human stem cells. Tissue Eng. Part C Methods 16, 847–854. doi: 10.1089/ten.tec.2009.0397
Koellensperger, E., Bonnert, L. C., Zoernig, I., Marmé, F., Sandmann, S., Germann, G., et al. (2017). The impact of human adipose tissue-derived stem cells on breast cancer cells: implications for cell-assisted lipotransfers in breast reconstruction. Stem Cell Res. Ther. 8:121.
Komi, D., and Redegeld, F. A. (2020). Role of mast cells in shaping the tumor microenvironment. Clin. Rev. Allergy Immunol. 58, 313–325. doi: 10.1007/s12016-019-08753-w
Kondo, J., and Inoue, M. (2019). Application of cancer organoid model for drug screening and personalized therapy. Cells 8:470. doi: 10.3390/cells8050470
Kondo, J., Ekawa, T., Endo, H., Yamazaki, K., Tanaka, N., Kukita, Y., et al. (2019). High-throughput screening in colorectal cancer tissue-originated spheroids. Cancer Sci. 110, 345–355. doi: 10.1111/cas.13843
Kondoh, M., Ohga, N., Akiyama, K., Hida, Y., Maishi, N., Towfik, A. M., et al. (2013). Hypoxia-induced reactive oxygen species cause chromosomal abnormalities in endothelial cells in the tumor microenvironment. PLoS One 8:e80349. doi: 10.1371/journal.pone.0080349
Lal-Nag, M., McGee, L., Guha, R., Lengyel, E., Kenny, H. A., and Ferrer, M. (2017). A high-throughput screening model of the tumor microenvironment for ovarian cancer cell growth. SLAS Discov. Adv. Life Sci. R D 22, 494–506. doi: 10.1177/2472555216687082
Lampis, A., Carotenuto, P., Vlachogiannis, G., Cascione, L., Hedayat, S., Burke, R., et al. (2018). MIR21 drives resistance to heat shock protein 90 inhibition in cholangiocarcinoma. Gastroenterology 154, 1066–1079.e5.
Lan, S. F., Safiejko-Mroczka, B., and Starly, B. (2010). Long-term cultivation of HepG2 liver cells encapsulated in alginate hydrogels: a study of cell viability, morphology and drug metabolism. Toxicol. Vitro 24, 1314–1323. doi: 10.1016/j.tiv.2010.02.015
Langer, E. M., Allen-Petersen, B. L., King, S. M., Kendsersky, N. D., Turnidge, M. A., Kuziel, G. M., et al. (2019). Modeling tumor phenotypes in vitro with three-dimensional bioprinting. Cell Rep. 26, 608–623.e6.
Lee, J. M., Yang, L., Kim, E. J., Ahrberg, C. D., Lee, K. B., and Chung, B. G. (2018). Generation of uniform-sized multicellular tumor spheroids using hydrogel microwells for advanced drug screening. Sci. Rep. 8:17145.
Lee, J., Lee, J. E., Kim, S., Kang, D., and Yoo, H. M. (2020). Evaluating cell death using cell-free supernatant of probiotics in three-dimensional spheroid cultures of colorectal cancer cells. J. Vis. Exp. JoVE. 160:10.3791/61285. doi: 10.3791/61285
Lee, S. W., Hong, S., Jung, B., Jeong, S. Y., Byeon, J. H., Jeong, G. S., et al. (2019). In vitro lung cancer multicellular tumor spheroid formation using a microfluidic device. Biotechnol. Bioeng. 116, 3041–3052. doi: 10.1002/bit.27114
Li, L., Zhou, Q., Voss, T. C., Quick, K. L., and LaBarbera, D. V. (2016). High-throughput imaging: focusing in on drug discovery in 3D. Methods (San Diego, Calif.) 96, 97–102. doi: 10.1016/j.ymeth.2015.11.013
Li, Q., Chen, C., Kapadia, A., Zhou, Q., Harper, M. K., Schaack, J., et al. (2011). 3D models of epithelial-mesenchymal transition in breast cancer metastasis: high-throughput screening assay development, validation, and pilot screen. J. Biomol. Screen. 16, 141–154. doi: 10.1177/1087057110392995
Lim, A. R., Rathmell, W. K., and Rathmell, J. C. (2020). The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. eLife 9:e55185.
Lin, M., Gao, M., Cavnar, M. J., and Kim, J. (2019). Utilizing gastric cancer organoids to assess tumor biology and personalize medicine. World J. Gastrointest. Oncol. 11, 509–517. doi: 10.4251/wjgo.v11.i7.509
Liu, C., Qin, T., Huang, Y., Li, Y., Chen, G., and Sun, C. (2020). Drug screening model meets cancer organoid technology. Transl. Oncol. 13:100840. doi: 10.1016/j.tranon.2020.100840
Liu, J., Zhang, Y., Zhao, J., Yang, Z., Li, D., Katirai, F., et al. (2011). Mast cell: insight into remodeling a tumor microenvironment. Cancer Metastasis Rev. 30, 177–184. doi: 10.1007/s10555-011-9276-1
Lu, P., Weaver, V. M., and Werb, Z. (2012). The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196, 395–406. doi: 10.1083/jcb.201102147
Luckert, C., Schulz, C., Lehmann, N., Thomas, M., Hofmann, U., Hammad, S., et al. (2017). Comparative analysis of 3D culture methods on human HepG2 cells. Arch. Toxicol. 91, 393–406. doi: 10.1007/s00204-016-1677-z
Ma, X., Liu, J., Zhu, W., Tang, M., Lawrence, N., Yu, C., et al. (2018). 3D bioprinting of functional tissue models for personalized drug screening and in vitro disease modeling. Adv. Drug Deliv. Rev. 132, 235–251. doi: 10.1016/j.addr.2018.06.011
Maishi, N., Ohba, Y., Akiyama, K., Ohga, N., Hamada, J., Nagao-Kitamoto, H., et al. (2016). Tumour endothelial cells in high metastatic tumours promote metastasis via epigenetic dysregulation of biglycan. Sci. Rep. 6:28039.
Malik, R., Lelkes, P. I., and Cukierman, E. (2015). Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 33, 230–236. doi: 10.1016/j.tibtech.2015.01.004
Maloney, E., Clark, C., Sivakumar, H., Yoo, K., Aleman, J., Rajan, S., et al. (2020). Immersion bioprinting of tumor organoids in multi-well plates for increasing chemotherapy screening throughput. Micromachines 11:208. doi: 10.3390/mi11020208
Mandrycky, C., Wang, Z., Kim, K., and Kim, D. H. (2016). 3D bioprinting for engineering complex tissues. Biotechnol. Adv. 34, 422–434. doi: 10.1016/j.biotechadv.2015.12.011
Matai, I., Kaur, G., Seyedsalehi, A., McClinton, A., and Laurencin, C. T. (2020). Progress in 3D bioprinting technology for tissue/organ regenerative engineering. Biomaterials 226:119536. doi: 10.1016/j.biomaterials.2019.119536
Mateo, J., Carreira, S., Sandhu, S., Miranda, S., Mossop, H., Perez-Lopez, R., et al. (2015). DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708.
Mazzoleni, G., Di Lorenzo, D., and Steimberg, N. (2009). Modelling tissues in 3D: the next future of pharmaco-toxicology and food research? Genes Nutr. 4, 13–22. doi: 10.1007/s12263-008-0107-0
McGranahan, N., and Swanton, C. (2017). Clonal heterogeneity and tumor evolution: past present, and the future. Cell 168, 613–628. doi: 10.1016/j.cell.2017.01.018
Mehta, G., Hsiao, A. Y., Ingram, M., Luker, G. D., and Takayama, S. (2012). Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Controll. Release 164, 192–204. doi: 10.1016/j.jconrel.2012.04.045
Meirelles, L. S., Caplan, A. I. N., and Nardi, B. N. (2013). “Pericytes as the source of mesenchymal stem cells,” in Resident Stem Cells and Regenerative Therapy, eds R. C. Goldenberg and A. C. C. Carvalho (Amsterdam: Elsevier), 233–250. doi: 10.1016/b978-0-12-416012-5.00012-8
Melaiu, O., Lucarini, V., Cifaldi, L., and Fruci, D. (2020). Influence of the tumor microenvironment on NK cell function in solid tumors. Front. Immunol. 10:3038.
Meng, F., Meyer, C. M., Joung, D., Vallera, D. A., McAlpine, M. C., and Panoskaltsis-Mortari, A. (2019). 3D Bioprinted In Vitro Metastatic Models via Reconstruction of Tumor Microenvironments. Adv. Mater. (Deerfield Beach, Fla.) 31:e1806899.
Merlano, M. C., Abbona, A., Denaro, N., and Garrone, O. (2019). Knowing the tumour microenvironment to optimise immunotherapy. Acta Otorhinolaryngol. Ital. 39:2. doi: 10.14639/0392-100x-2481
Mironov, V., Visconti, R. P., Kasyanov, V., Forgacs, G., Drake, C. J., and Markwald, R. R. (2009). Organ printing: tissue spheroids as building blocks. Biomaterials 30, 2164–2174. doi: 10.1016/j.biomaterials.2008.12.084
Mohammadi, Z., and Rabbani, M. (2018). Bacterial bioprinting on a flexible substrate for fabrication of a colorimetric temperature indicator by using a commercial inkjet printer. J. Med. Signal. Sens. 8, 170–174. doi: 10.4103/jmss.jmss_41_17
Moldovan, N. I. (2018). Progress in scaffold-free bioprinting for cardiovascular medicine. J. Cell. Mol. Med. 22, 2964–2969. doi: 10.1111/jcmm.13598
Moldovan, N. I., Hibino, N., and Nakayama, K. (2017). Principles of the Kenzan method for robotic cell spheroid-based three-dimensional bioprinting. Tissue Eng. Part B Rev. 23, 237–244. doi: 10.1089/ten.teb.2016.0322
Molina, E. R., Chim, L. K., Barrios, S., Ludwig, J. A., and Mikos, A. G. (2020). Modeling the tumor microenvironment and pathogenic signaling in bone sarcoma. Tissue Eng. Part B Rev. 26, 249–271. doi: 10.1089/ten.teb.2019.0302
Mollica, P. A., Booth-Creech, E. N., Reid, J. A., Zamponi, M., Sullivan, S. M., Palmer, X. L., et al. (2019). 3D bioprinted mammary organoids and tumoroids in human mammary derived ECM hydrogels. Acta Biomater. 95, 201–213. doi: 10.1016/j.actbio.2019.06.017
Moroni, L., Boland, T., Burdick, J. A., De Maria, C., Derby, B., Forgacs, G., et al. (2018). Biofabrication: a guide to technology and terminology. Trends Biotechnol. 36, 384–402. doi: 10.1016/j.tibtech.2017.10.015
Moshksayan, K., Kashaninejad, N., Warkiani, M. E., Lock, J. G., Moghadas, H., Firoozabadi, B., et al. (2018). Spheroids-on-a-chip: recent advances and design considerations in microfluidic platforms for spheroid formation and culture. Sens. Actuators B Chem. 263, 151–176. doi: 10.1016/j.snb.2018.01.223
Mravic, M., Asatrian, G., Soo, C., Lugassy, C., Barnhill, R. L., Dry, S. M., et al. (2014). From pericytes to perivascular tumours: correlation between pathology, stem cell biology, and tissue engineering. Int. Orthop. 38, 1819–1824. doi: 10.1007/s00264-014-2295-0
Mueller, M. M., and Fusenig, N. E. (2002). Tumor-stroma interactions directing phenotype and progression of epithelial skin tumor cells. Differentiation 70, 486–497. doi: 10.1046/j.1432-0436.2002.700903.x
Murata, D., Arai, K., and Nakayama, K. (2020). Scaffold-Free Bio-3D printing using spheroids as “bio-inks” for tissue (re-)construction and drug response tests. Adv. Healthc. Mater. 9:e1901831.
Murphy, S. V., and Atala, A. (2014). 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785. doi: 10.1038/nbt.2958
Najafi, M., Farhood, B., and Mortezaee, K. (2019). Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 120, 2782–2790. doi: 10.1002/jcb.27681
Nallanthighal, S., Heiserman, J. P., and Cheon, D. J. (2019). The role of the extracellular matrix in cancer stemness. Front. Cell Dev. Biol. 7:86.
Nanki, K., Toshimitsu, K., Takano, A., Fujii, M., Shimokawa, M., Ohta, Y., et al. (2018). Divergent routes toward wnt and r-spondin niche independency during human gastric carcinogenesis. Cell 174, 856–869.e17.
Nassar, D., and Blanpain, C. (2016). Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 11, 47–76. doi: 10.1146/annurev-pathol-012615-044438
Nath, S., and Devi, G. R. (2016). Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Therap. 163, 94–108. doi: 10.1016/j.pharmthera.2016.03.013
Naumov, G. N., Folkman, J., Straume, O., and Akslen, L. A. (2008). Tumor-vascular interactions and tumor dormancy. APMIS 116, 569–585. doi: 10.1111/j.1600-0463.2008.01213.x
Neglia, J. P., Wielinski, C. L., and Warwick, W. J. (1991). Cancer risk among patients with cystic fibrosis. J. Pediatr. 119, 764–766.
Ning, L., and Chen, X. (2017). A brief review of extrusion-based tissue scaffold bio-printing. Biotechnol. J. 12:1600671. doi: 10.1002/biot.201600671
Nunes, A. S., Barros, A. S., Costa, E. C., Moreira, A. F., and Correia, I. J. (2019). 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 116, 206–226. doi: 10.1002/bit.26845
O’Loghlen, A. (2018). Role for extracellular vesicles in the tumour microenvironment. Philos. Trans. R. Soc. B Biol. Sci. 373:20160488. doi: 10.1098/rstb.2016.0488
Ong, C. S., Yesantharao, P., Huang, C. Y., Mattson, G., Boktor, J., Fukunishi, T., et al. (2018). 3D bioprinting using stem cells. Pediatr. Res. 83, 223–231. doi: 10.1038/pr.2017.252
Oraiopoulou, M. E., Tampakaki, M., Tzamali, E., Tamiolakis, T., Makatounakis, V., Vakis, A. F., et al. (2019). A 3D tumor spheroid model for the T98G glioblastoma cell line phenotypic characterization. Tissue Cell 59, 39–43. doi: 10.1016/j.tice.2019.05.007
Orloff, N., Truong, C., Cira, N., Koo, S., Hamilton, A., Choi, S., et al. (2014). Integrated bioprinting and imaging for scalable. Royal Soc. Chemis. 4, 34721–34728. doi: 10.1039/C4RA05932H
Osidak, E. O., Karalkin, P. A., Osidak, M. S., Parfenov, V. A., Sivogrivov, D. E., Pereira, F. D., et al. (2019). Viscoll collagen solution as a novel bioink for direct 3D bioprinting. J. Mater. Sci. 30:31.
Ozbolat, I. T., and Hospodiuk, M. (2016). Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 76, 321–343. doi: 10.1016/j.biomaterials.2015.10.076
Parak, A., Pradeep, P., du Toit, L. C., Kumar, P., Choonara, Y. E., and Pillay, V. (2019). Functionalizing bioinks for 3D bioprinting applications. Drug Discov. Today 24, 198–205. doi: 10.1016/j.drudis.2018.09.012
Peng, W., Datta, P., Ayan, B., Ozbolat, V., Sosnoski, D., and Ozbolat, I. T. (2017). 3D bioprinting for drug discovery and development in pharmaceutics. Acta Biomat. 57, 26–46. doi: 10.1016/j.actbio.2017.05.025
Pereira, D. A., and Williams, J. A. (2007). Origin and evolution of high throughput screening. Br. J. Pharmacol. 152, 53–61. doi: 10.1038/sj.bjp.0707373
Petrova, V., Annicchiarico-Petruzzelli, M., Melino, G., and Amelio, I. (2018). The hypoxic tumour microenvironment. Oncogenesis 7:10.
Picollet-D’hahan, N., Dolega, M. E., Liguori, L., Marquette, C., Le Gac, S., Gidrol, X., et al. (2016). A 3D toolbox to enhance physiological relevance of human tissue models. Trends Biotechnol. 34, 757–769. doi: 10.1016/j.tibtech.2016.06.012
Quail, D. F., and Joyce, J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. doi: 10.1038/nm.3394
Raffaghello, L., and Dazzi, F. (2015). Classification and biology of tumour associated stromal cells. Immunol. Lett. 168, 175–182. doi: 10.1016/j.imlet.2015.06.016
Renner, H., Grabos, M., Becker, K. J., Kagermeier, T. E., Wu, J., Otto, M., et al. (2020). A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. eLife 9:e52904.
Ribatti, D., and Tamma, R. (2019). Giulio Gabbiani and the discovery of myofibroblasts. Inflamm. Res. 68, 241–245. doi: 10.1007/s00011-018-01211-x
Ribeiro, A. L., and Okamoto, O. K. (2015). Combined effects of pericytes in the tumor microenvironment. Stem Cells Int. 2015:868475.
Ridge, S. M., Sullivan, F. J., and Glynn, S. A. (2017). Mesenchymal stem cells: key players in cancer progression. Mol. Cancer 16:31.
Riffle, S., Pandey, R. N., Albert, M., and Hegde, R. S. (2017). Linking hypoxia, DNA damage and proliferation in multicellular tumor spheroids. BMC Cancer 17:338.
Robado de Lope, L., Alcíbar, O. L., Amor López, A., Hergueta-Redondo, M., and Peinado, H. (2018). Tumour-adipose tissue crosstalk: fuelling tumour metastasis by extracellular vesicles. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 373:20160485. doi: 10.1098/rstb.2016.0485
Roy, P. S., and Saikia, B. J. (2016). Cancer and cure: a critical analysis. Indian J. Cancer 53, 441–442.
Ruiz, M. C., Kljun, J., Turel, I., Di Virgilio, A. L., and León, I. E. (2019). Comparative antitumor studies of organoruthenium complexes with 8-hydroxyquinolines on 2D and 3D cell models of bone, lung and breast cancer. Metallomics 11, 666–675. doi: 10.1039/c8mt00369f
Sachs, N., de Ligt, J., Kopper, O., Gogola, E., Bounova, G., Weeber, F., et al. (2018). A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 856–869.e17.
Sarkar, J., and Kumar, A. (2016). Thermo-responsive polymer aided spheroid culture in cryogel based platform for high throughput drug screening. Analyst 141, 2553–2567. doi: 10.1039/c6an00356g
Sasmal, P., Datta, P., Wu, Y., and Ozbolat, I. T. (2018). 3D bioprinting for modelling vasculature. Microphysiol. Syst. 2:9.
Satpathy, A., Datta, P., Wu, Y., Ayan, B., Bayram, E., and Ozbolat, I. T. (2018). Developments with 3D bioprinting for novel drug discovery. Expert Opin Drug Discov. 13, 1115–1129. doi: 10.1080/17460441.2018.1542427
Sawant, S., Dongre, H., Singh, A. K., Joshi, S., Costea, D. E., Mahadik, S., et al. (2016). Establishment of 3D co-culture models from different stages of human tongue tumorigenesis: utility in understanding neoplastic progression. PLoS One 11:e0160615. doi: 10.1371/journal.pone.0160615
Schäffler, A., and Büchler, C. (2007). Concise review: adipose tissue-derived stromal cells–basic and clinical implications for novel cell-based therapies. Stem Cells (Dayton, Ohio) 25, 818–827. doi: 10.1634/stemcells.2006-0589
Schmidt, S. K., Schmid, R., Arkudas, A., Kengelbach-Weigand, A., and Bosserhoff, A. K. (2019). Tumor cells develop defined cellular phenotypes after 3d-bioprinting in different bioinks. Cells 8:1295. doi: 10.3390/cells8101295
Seager, R. J., Hajal, C., Spill, F., Kamm, R. D., and Zaman, M. H. (2017). Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg. Sci. Phys. Oncol. 3:034002. doi: 10.1088/2057-1739/aa7e86
Servais, C., and Erez, N. (2013). From sentinel cells to inflammatory culprits: cancer-associated fibroblasts in tumour-related inflammation. J. Pathol. 229, 198–207. doi: 10.1002/path.4103
Shah, U. K., Mallia, J. O., Singh, N., Chapman, K. E., Doak, S. H., and Jenkins, G. (2018). A three-dimensional in vitro HepG2 cells liver spheroid model for genotoxicity studies. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 825, 51–58. doi: 10.1016/j.mrgentox.2017.12.005
Shweiki, D., Neeman, M., Itin, A., and Keshet, E. (1995). Induction of vascular endothelial growth factor expression by hypoxia and by glucose deficiency in multicell spheroids: implications for tumor angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 92, 768–772. doi: 10.1073/pnas.92.3.768
Siegel, R. L., Miller, K. D., Goding Sauer, A., Fedewa, S. A., Butterly, L. F., Anderson, J. C., et al. (2020). Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164. doi: 10.1016/j.clcc.2020.07.001
Skardal, A., Devarasetty, M., Kang, H. W., Seol, Y. J., Forsythe, S. D., Bishop, C., et al. (2016). Bioprinting cellularized constructs using a tissue-specific hydrogel bioink. J. Vis. Exp. JoVE 110:e53606.
Slattery, M. L., John, E., Torres-Mejia, G., Stern, M., Lundgreen, A., Hines, L., et al. (2013). Matrix metalloproteinase genes are associated with breast cancer risk and survival: the breast cancer health disparities study. PLoS One 8:e63165. doi: 10.1371/journal.pone.0063165
Sugimoto, H., Mundel, T. M., Kieran, M. W., and Kalluri, R. (2006). Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 5, 1640–1646. doi: 10.4161/cbt.5.12.3354
Suh, M., Proctor, D., Chappell, G., Rager, J., Thompson, C., Borghoff, S., et al. (2018). A review of the genotoxic, mutagenic, and carcinogenic potentials of several lower acrylates. Toxicology 402-403, 50–67. doi: 10.1016/j.tox.2018.04.006
Sutherland, R. M. (1988). Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 240, 177–184. doi: 10.1126/science.2451290
Swaminathan, S., Hamid, Q., Sun, W., and Clyne, A. M. (2019). Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication 11:025003. doi: 10.1088/1758-5090/aafc49
Tellez-Gabriel, M., Cochonneau, D., Cadé, M., Jubellin, C., Heymann, M. F., and Heymann, D. (2018). Circulating tumor cell-derived pre-clinical models for personalized medicine. Cancers 11:19. doi: 10.3390/cancers11010019
Valkenburg, K. C., de Groot, A. E., and Pienta, K. J. (2018). Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 15, 366–381. doi: 10.1038/s41571-018-0007-1
Van der Steen, S. C., Raavé, R., Langerak, S., van Houdt, L., van Duijnhoven, S. M., van Lith, S. A., et al. (2017). Targeting the extracellular matrix of ovarian cancer using functionalized, drug loaded lyophilisomes. Eur. J. Pharm. Biopharm. 113, 229–239. doi: 10.1016/j.ejpb.2016.12.010
Van de Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi: 10.1016/j.cell.2015.03.053
Vijayavenkataraman, S., Yan, W. C., Lu, W. F., Wang, C. H., and Fuh, J. (2018). 3D bioprinting of tissues and organs for regenerative medicine. Adv. Drug Deliv. Rev. 132, 296–332. doi: 10.1016/j.addr.2018.07.004
Vinci, M., Gowan, S., Boxall, F., Patterson, L., Zimmermann, M., Court, W., et al. (2012). Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 10:29. doi: 10.1186/1741-7007-10-29
Walker, C., Mojares, E., and Del Río Hernández, A. (2018). Role of extracellular matrix in development and cancer progression. Int. J. Mol. Sci. 19:3028. doi: 10.3390/ijms19103028
Wang, X. (2019). Stem cells in tissues, organoids, and cancers. Cell. Mol. Life Sci. CMLS 76, 4043–4070. doi: 10.1007/s00018-019-03199-x
Wang, X., Li, X., Ding, J., Long, X., Zhang, H., Zhang, X., et al. (2020). 3D bioprinted glioma microenvironment for glioma vascularization. J. Biomed. Mater. Res. Part A 109, 915–925.
Wartenberg, M., Dönmez, F., Ling, F. C., Acker, H., Hescheler, J., and Sauer, H. (2001). Tumor-induced angiogenesis studied in confrontation cultures of multicellular tumor spheroids and embryoid bodies grown from pluripotent embryonic stem cells. FASEB J. 15, 995–1005. doi: 10.1096/fsb2fj000350com
Watters, K. M., Bajwa, P., and Kenny, H. A. (2018). Organotypic 3D models of the ovarian cancer tumor microenvironment. Cancers 10:265. doi: 10.3390/cancers10080265
Wernitznig, D., Kiakos, K., Del Favero, G., Harrer, N., Machat, H., Osswald, A., et al. (2019). First-in-class ruthenium anticancer drug (KP1339/IT-139) induces an immunogenic cell death signature in colorectal spheroids in vitro. Metallomics 11, 1044–1048. doi: 10.1039/c9mt00051h
Wong, C. H., Siah, K. W., and Lo, A. W. (2019). Estimation of clinical trial success rates and related parameters. Biostatistics (Oxford, England) 20, 273–286. doi: 10.1093/biostatistics/kxx069
Wu, T., and Dai, Y. (2017). Tumor microenvironment and therapeutic response. Cancer Lett. 387, 61–68. doi: 10.1016/j.canlet.2016.01.043
Xu, H., Lyu, X., Yi, M., Zhao, W., Song, Y., and Wu, K. (2018). Organoid technology and applications in cancer research. J. Hematol. Oncol. 11:116.
Xu, Z., Li, E., Guo, Z., Yu, R., Hao, H., Xu, Y., et al. (2016). Design and construction of a multi-organ microfluidic chip mimicking the in vivo microenvironment of lung cancer metastasis. ACS Appl. Mater. Interfaces 8, 25840–25847. doi: 10.1021/acsami.6b08746
Yan, H., Siu, H. C., Law, S., Ho, S. L., Yue, S., Tsui, W. Y., et al. (2017). Mechanisms that drive inflammatory tumor microenvironment, tumor heterogeneity, and metastatic progression. Semin. Cancer Biol. 47, 185–195. doi: 10.1016/j.semcancer.2017.08.001
Yang, L., and Lin, P. C. (2017). “Mechanisms that drive inflammatory tumor microenvironment, tumor heterogeneity, and metastatic progression,” in Seminars in Cancer Biology, Vol. 47(Cambridge, MA: Academic Press), 185–195.
Yazdani, S., Bansal, R., and Prakash, J. (2017). Drug targeting to myofibroblasts: Implications for fibrosis and cancer. Adv. Drug Deliv. Rev. 121, 101–116. doi: 10.1016/j.addr.2017.07.010
Yu, Y. J., Kim, Y. H., Na, K., Min, S. Y., Hwang, O. K., Park, D. K., et al. (2018). Hydrogel-incorporating unit in a well: 3D cell culture for high-throughput analysis. Lab Chip 18, 2604–2613. doi: 10.1039/c8lc00525g
Zhang, A., Qian, Y., Ye, Z., Chen, H., Xie, H., Zhou, L., et al. (2017). Cancer-associated fibroblasts promote M2 polarization of macrophages in pancreatic ductal adenocarcinoma. Cancer Med. 6, 463–470. doi: 10.1002/cam4.993
Zhang, B., Wang, H., Jiang, T., Jin, K., Luo, Z., Shi, W., et al. (2018). Cyclopamine treatment disrupts extracellular matrix and alleviates solid stress to improve nanomedicine delivery for pancreatic cancer. J. Drug Target. 26, 913–919. doi: 10.1080/1061186x.2018.1452243
Zhang, C., Hu, Y., and Shi, C. (2020). Targeting natural killer cells for tumor immunotherapy. Front. Immunol. 11:60.
Zhang, W., Li, C., Baguley, B. C., Zhou, F., Zhou, W., Shaw, J. P., et al. (2016). Optimization of the formation of embedded multicellular spheroids of MCF-7 cells: how to reliably produce a biomimetic 3D model. Anal. Biochem. 515, 47–54. doi: 10.1016/j.ab.2016.10.004
Zhang, X., and Zhang, Y. (2015). Tissue engineering applications of three-dimensional bioprinting. Cell Biochem. Biophys. 72, 777–782. doi: 10.1007/s12013-015-0531-x
Zhang, Y. S., Duchamp, M., Oklu, R., Ellisen, L. W., Langer, R., and Khademhosseini, A. (2016). Bioprinting the cancer microenvironment. ACS Biomater. Sci. Eng. 2, 1710–1721. doi: 10.1021/acsbiomaterials.6b00246
Zhang, Y. S., Yue, K., Aleman, J., Moghaddam, K. M., Bakht, S. M., Yang, J., et al. (2017). 3D Bioprinting for Tissue and Organ Fabrication. Ann. Biomed. Eng. 45, 148–163.
Zhao, L., Shi, M., Liu, Y., Zheng, X., Xiu, J., Liu, Y., et al. (2019). Systematic analysis of different cell spheroids with a microfluidic device using scanning electrochemical microscopy and gene expression profiling. Anal. Chem. 91, 4307–4311. doi: 10.1021/acs.analchem.9b00376
Keywords: tumor microenvironment, tumorigenesis, 3D cell culture, spheroids, organoids, drug assessment, high-throughput screening, 3D bioprinting
Citation: Kronemberger GS, Miranda GASC, Tavares RSN, Montenegro B, Kopke ÚA and Baptista LS (2021) Recapitulating Tumorigenesis in vitro: Opportunities and Challenges of 3D Bioprinting. Front. Bioeng. Biotechnol. 9:682498. doi: 10.3389/fbioe.2021.682498
Received: 18 March 2021; Accepted: 29 April 2021;
Published: 22 June 2021.
Edited by:
Ângela Sousa, University of Beira Interior, PortugalReviewed by:
Hilary Ann Kenny, University of Chicago, United StatesKevin Dzobo, University of Cape Town, South Africa
Copyright © 2021 Kronemberger, Miranda, Tavares, Montenegro, Kopke and Baptista. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leandra S. Baptista, bGVhbmRyYWJhcHRpc3RhQHhlcmVtLnVmcmouYnI=
 Gabriela S. Kronemberger1,2,3
Gabriela S. Kronemberger1,2,3 Bianca Montenegro
Bianca Montenegro Leandra S. Baptista
Leandra S. Baptista