
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 09 April 2021
Sec. Synthetic Biology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.673844
This article is part of the Research Topic Recent Advances in Application of Synthetic Biology for Production of Bioactive Compounds View all 11 articles
The acetic acid bacterium Gluconobacter oxydans is known for its unique incomplete oxidation and therefore widely applied in the industrial production of many compounds, e.g., 2-keto-L-gulonic acid (2-KLG), the direct precursor of vitamin C. However, few molecular tools are available for metabolically engineering G. oxydans, which greatly limit the strain development. Promoters are one of vital components to control and regulate gene expression at the transcriptional level for boosting production. In this study, the low activity of SDH was found to hamper the high yield of 2-KLG, and enhancing the expression of SDH was achieved by screening the suitable promoters based on RNA sequencing data. We obtained 97 promoters from G. oxydans’s genome, including two strong shuttle promoters and six strongest promoters. Among these promoters, P3022 and P0943 revealed strong activities in both Escherichia coli and G. oxydans, and the activity of the strongest promoter (P2703) was about threefold that of the other reported strong promoters of G. oxydans. These promoters were used to overexpress SDH in G. oxydans WSH-003. The titer of 2-KLG reached 3.7 g/L when SDH was under the control of strong promoters P2057 and P2703. This study obtained a series of gradient promoters, including two strong shuttle promoters, and expanded the toolbox of available promoters for the application in metabolic engineering of G. oxydans for high-value products.
Gluconobacter oxydans has been widely applied in the industrial production of L-sorbose from D-sorbitol (De Wulf et al., 2000), dihydroxyacetone from glycerol (De La Morena et al., 2020), 1-amino-L-sorbose from 1-amino-D-sorbitol (Schedel, 2000), and levan-type fructans from sucrose (Hövels et al., 2020). Furthermore, G. oxydans is also an excellent workhorse for the biosynthesis of 2-keto-D-gluconate (Li et al., 2016; Zhou et al., 2020), 5-keto-D-gluconate (Merfort et al., 2006), xylonic acid (Hahn et al., 2020; Shen et al., 2020), 5-keto-D-fructose (Battling et al., 2020; Hoffmann et al., 2020), and many other products (Deppenmeier et al., 2002; De Muynck et al., 2007). The wide applications of G. oxydans are mainly due to its unique dehydrogenases in the periplasm (Deppenmeier et al., 2002). Many protein engineering approaches have been used to improve the catalytic efficiency of these dehydrogenases, including enzyme immobilization (Kim et al., 2016), cofactor regeneration (Gao et al., 2019), ligand docking, and molecular dynamics simulations (Selvaraj et al., 2016). On the other hand, many metabolic engineering strategies of G. oxydans were based on the overexpression of related dehydrogenases or enzymes associated with the respiratory chains (Li et al., 2016; Yuan et al., 2016). However, the expression of key enzymes were often impeded by the accessible promoters.
Though the first genome of G. oxydans was reported in 2005 (Prust et al., 2005), only a few studies on the promoters of G. oxydans have been carried out. Generally, most studies directly selected some promoters from high expression level of genes. Nishikura-Imamura et al. (2014) cloned the putative promoter region of G. oxydans PQQ-dependent alcohol dehydrogenase (PadhAB) to overexpress 3-dehydroquinate dehydratase. Mientus et al. (2017) characterized promoters of six membrane-bound dehydrogenases of G. oxydans 621H, and used the constitutive promoter of the alcohol dehydrogenase and the glucose-repressed promoter of inositol dehydrogenase to construct a shuttle vector system. Though some progress were achieved in promoter discovery, it was hard to apply the promoters to other metabolic pathways because the promoters were relatively weak without systematic comparison. Saito et al., found some strong promoters of G. oxydans, such as PtufB, P0169, and P264 (Saito et al., 1997; Yuan et al., 2016; Blank and Schweiger, 2018). Moreover, Kallnik and Hu reported some promoters of different strengths in G. oxydans (Kallnik et al., 2010; Hu et al., 2015). Nevertheless, the available promoters are still insufficient, especially the strong promoters are highly needed to support engineering G. oxydans for their industrial application.
RNA sequencing (RNA-Seq) in the study of prokaryotic and eukaryotic organisms has become more accessible in the last decade (Poulsen and Vinther, 2018; Stark et al., 2019). It has become an excellent strategy to mine strong promoters in many microorganisms. Lee et al. (2015) screened a novel strong promoter PTN0510 from Thermococcus onnurineus by RNA-Seq and applied it to the production of H2. Liao et al. (2015) identified a strong promoter, Pr2, from the RNA-Seq data of Bacillus amyloliquefaciens and verified it by measuring beta-galactosidase activity. Several studies about the transcriptome analysis of G. oxydans has been reported to reveal the secretion pathways of PQQ (Wan et al., 2017) and the response to osmotic and oxidative stress of 2-keto-L-gulonic acid (2-KLG) (Fang et al., 2020). Kranz et al. (2018) provided deep insights into the transcriptional landscapes of G. oxydans including promoters and other regulatory elements. However, no further experimental studies were performed to characterize these promoters and regulatory elements. Thus, RNA-Seq of G. oxydans WSH-003 was first conducted in this study, followed with the characterization of promoters by using mCherry as a report to compare the strength of the screened promoters.
2-KLG is an important precursor of vitamin C in industry (Wang et al., 2018). However, there are only a few G. oxydans strains that can produce 2-KLG naturally (Hoshino et al., 1990; Saito et al., 1998; Chen et al., 2019), although many sequenced G. oxydans possess the entire set of 2-KLG biosynthesis genes (Wang et al., 2018). In our previous study, we identified the key SDH from a G. oxydans that was able to naturally produce 2-KLG, and successfully constructed a high-throughput screening platform for an FAD-dependent SDH (Shan et al., 2020). Different from SSDHs from Ketogulonigenium vulgare, SDH from G. oxydans showed higher substrate specificity to L-sorbose and did not require PQQ as a cofactor (Saito et al., 1997; Wang et al., 2018). In the present study, a group of gradient promoters was identified and applied in the biosynthesis of 2-KLG in the strain G. oxydans WSH-003. The titer of 2-KLG reached 3.7 g/L when used the strongest promoter (P2703) to overexpress SDH. The results implied the low expression level of SDH may be the main problem for 2-KLG production in many G. oxydans strains. In conclusion, this study obtained a series of gradient promoters, and these promoters revealed promising prospects in metabolic engineering of G. oxydans for high-value products.
Escherichia coli JM109 was used for plasmid construction. G. oxydans WSH-003 was used for PCR amplification of promoters and protein expression. G. oxydans ATCC 621H was used for PCR amplification of promoters. G. oxydans WSH-004 was screened in our previous research (Chen et al., 2019). G. oxydans WSH-003-Δgdh was used for 2-KLG production. The plasmids p2-5 and pBBR1MCS-5 were used to overexpress mCherry (Li et al., 2020) and sorbose dehydrogenase in G. oxydans, respectively. All strains and plasmids are listed in Table 1. The nucleotide sequences of p2-5 and pBBR1MCS-5 were listed in Supplementary Table 5.
The strain G. oxydans WSH-003 was cultured to mid-log phase in sorbitol medium (50 g/L sorbitol and 10 g/L yeast extract) at 30°C with shaking at 220 rpm. Then the cells were harvested and washed twice with PBS. The total RNA was extracted by RNeasy Mini Kit (Qiagen, Hilden, Germany), and ribosomal RNAs were removed by Ribo-ZeroTM rRNA Removal Kits (Epicentre, Wisconsin, United States). The RNA sequencing libraries were constructed by TruSeq RNA Sample Preparation Kit v2 (Illumina, California, United States) and sequenced on MiSeq (Illumina, California, United States) using MiSeq Reagent Kit v3 (Illumina, California, United States). RNA sequencing was performed by Shanghai Biotechnology Corporation (Shanghai Biotechnology Co., Shanghai, China). The abundance of transcripts was determined using bowtie2 (Langmead and Salzberg, 2012; Bolger et al., 2014) and cufflinks (Trapnell et al., 2010) by mapping the appropriate reads to the genome of G. oxydans WSH-003 (Gao et al., 2012).
All the promoters were worked with a native ribosomal binding site (RBS), because it was hard to find a proper RBS with guaranteed strength. Promoters PtufB and Pdnak and all the screened potential promoters were obtained by PCR amplification from the genomic DNA of G. oxydans WSH-003. Promoters P264 and Php0169 were obtained by PCR amplification from the genomic DNA of G. oxydans ATCC 621H. The gene sdh was PCR-amplified from the genomic DNA of G. oxydans WSH-004. The gene mCherry was kept in our laboratory and obtained by PCR amplification. The mCherry gene was first ligated into the vector p2-5 to form the skeleton plasmid p2-5-mCherry by a one-step cloning kit (Takara, Dalian, China). Then different promoters were individually inserted into the plasmid p2-5-mCherry by the one-step cloning kit (Takara, Dalian, China). The sdh gene and gradient promoters were ligated into the vector pBBR1MCS-5 in the same way. All promoters and genes were verified by Sanger sequencing (Sangon Biotech, Shanghai, China). All vectors were constructed and amplified in the strain E. coli JM109. The vectors p2-5 and pBBR1MCS-5 were transferred by electroporation into G. oxydans WSH-003 (Zhang et al., 2010), which were selected using kanamycin and gentamycin, respectively. All primers are listed in Supplementary Table 1.
Single colonies of G. oxydans WSH-003 were picked into 14 mL tubes containing 2 mL of sorbitol medium and cultured for 24 hours at 30°C with shaking at 220 rpm. Then 2% of these cultures were inoculated into a 250 mL flask containing 25 mL of sorbitol medium and cultured at 30°C with shaking at 220 rpm. The cell fluorescence and cell density (OD600) were measured every 4 hours on a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek Instruments, Winooski, VT, United States) with excitation and emission wavelengths of 580 and 610 nm, respectively. The relative activity of mCherry was defined as the ratio of relative fluorescence unit (RFUs) divided by the optical density (OD600). The strain G. oxydans WSH-003 harboring p2-5-mCherry without promoters was used as the control.
The fermentation medium was formed with sorbitol medium (50 g/L sorbitol and 10 g/L yeast extract) containing 20 g/L CaCO3. Single colonies of G. oxydans WSH-003 were picked into 250 mL flasks containing 25 mL of sorbitol medium and cultured for 24 hours at 30°C with shaking at 220 rpm. Then, 10% of these cultures were inoculated into 250 mL flasks containing 25 mL of fermentation medium and cultured at 30°C with shaking at 220 rpm.
The cell concentration was measured using a Microplate Reader (BioTek Instruments, Winooski, VT, United States). The concentrations of D-sorbitol, L-sorbose, and 2-KLG were detected by HPLC using an Aminex HPX-87H column (BioRad, Hercules, CA, United States) at 35°C with 5 mmol/L H2SO4 as the eluent at a flow rate of 0.5 mL/min (Chen et al., 2019).
The results were interpreted with mean values and its standard. The paired two-tailed Student t tests were performed to demonstrate statistically significant differences between data points. A p-value of ≤ 0.05 was thought to be statistically significant. For data illustration, bar charts with error bars were used.
The isolated mRNAs of G. oxydans WSH-003 were subjected to high-throughput Illumina paired-end sequencing to obtain a global view of the transcriptome after removing ribosomal RNAs. The RNA sequencing data was submitted to sequence read archive (SRA) with the accession number of PRJNA706889. A total of 19.84 million mapped reads with an average length of 100 bp were obtained; 19.31 million reads were uniquely mapped to the genome of G. oxydans WSH-003, which represented a 500-fold coverage of the genome. The transcriptome data were analyzed with the software bowtie2 (Langmead and Salzberg, 2012; Bolger et al., 2014) and cufflinks (Trapnell et al., 2010). Only 188 genes were transcribed with transcript per million (TPM) values higher than 1000, and nearly 95% of the predicted 3545 genes were transcribed with TPM values below 1000 (Supplementary Table 2).
Because we aimed to identify strong promoters in G. oxydans, the genes that exhibited strong transcriptional activity were studied. Genes were excluded from our analysis if they encoded tRNAs or lacked a RBS. Genes that belonged to a gene cluster were also excluded because the same promoter probably controlled their transcription. Gene clusters were defined as contiguous genes with similar functions and with spacers smaller than 50 nucleotides. A total of 97 potential promoters were obtained based on the TPM values in the transcriptome (Figure 1). The sequences around 500 bp upstream of the open reading frame (ORF) were chosen as the potential promoters because little information on the promoter elements of G. oxydans has been reported.

Figure 1. The strongly transcribed genes of G. oxydans WSH-003 obtained by RNA sequencing. The promoters were selected based on transcript per million (TPM) values of the transcriptome. The abscissa axis shows the TPM values. The ordinate axis shows the different genes in G. oxydans WSH-003.
All 97 potential promoters were transferred into G. oxydans WSH-003 to determine their strength by measuring mCherry expression. The fluorescence intensity was assayed every 4 hours. The highest value of relative mCherry activity was defined as the relative strength of the promoter. Most of the screened promoters showed remarkable intensity compared to the control (Figure 2). Among the promoters, the six strongest promoters were P2703, P2564, P0365, P2057, P0295, and P2038. Besides, it was found that most of the screened promoters had the highest strength at about 36 hours, when the strain was grown in stationary phase (Supplementary Figure 1).
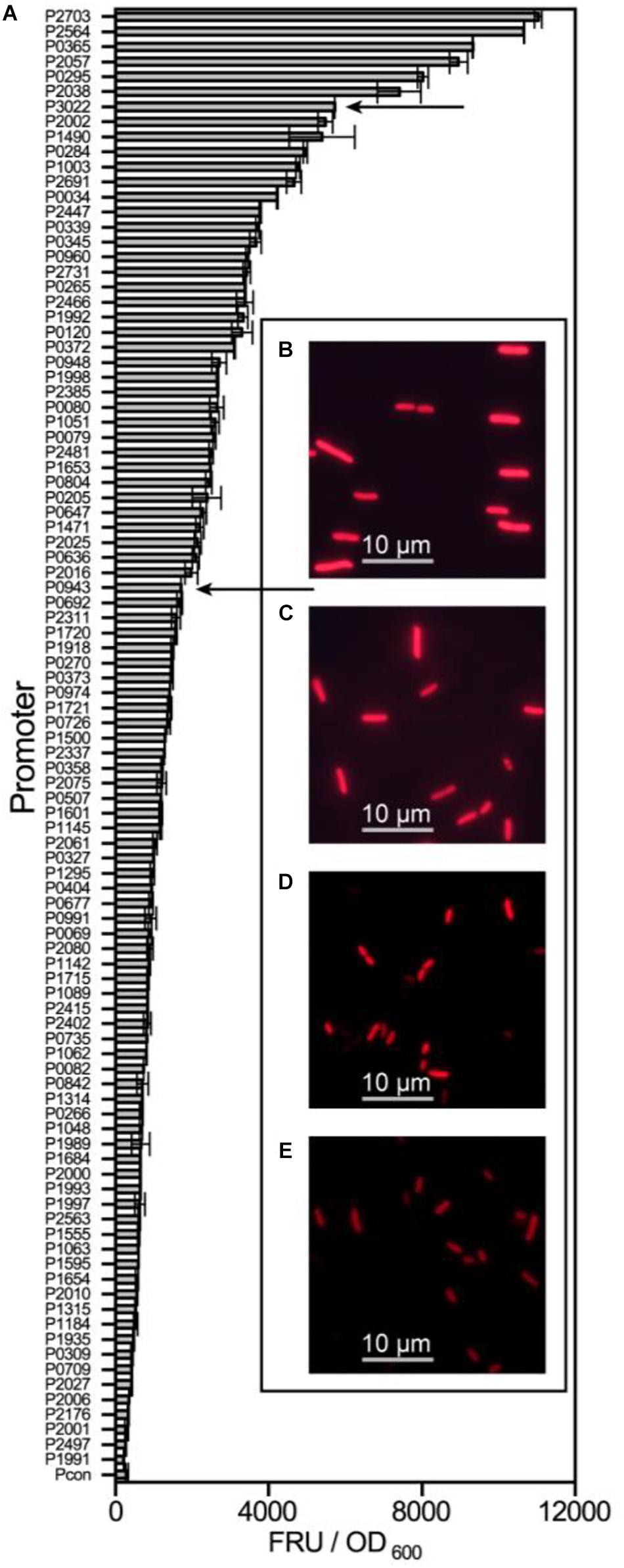
Figure 2. The strength determination of screened promoters of G. oxydans WSH-003. The strength of screened promoters and microscope pictures of two shuttle promoters. (A) The strength of screened promoters as measured by evaluating mCherry activity. The abscissa axis shows the relative activity of mCherry. The ordinate axis shows different promoters of G. oxydans WSH-003. Arrows indicate the two strong shuttle promoters P3022 and P0943. (B) E. coli JM109 expressing mCherry under the control of P3022. (C) E. coli JM109 expressing mCherry under the control of P0943. (D) G. oxydans WSH-003 expressing mCherry under the control of P3022. (E) G. oxydans WSH-003 expressing mCherry under the control of P0943.
As mentioned previously, a few strong G. oxydans promoters have been reported (Hu et al., 2015; Li et al., 2016; Blank and Schweiger, 2018). To verify the strength of our strongest screened promoters, we also obtained four reported strong promoters (PtufB, Pdnak, Php0169, and P264) from the genomes of WSH-003 and ATCC 621H. Compared with these four reported strong promoters, the promoter P2703 has the highest strength, which was about 2.8-fold higher than that of P264 and about 3.1-fold higher than that of Pdnak (Figure 3). The results showed that P2703 was the strongest promoter discovered in G. oxydans at present. Interestingly, two strong shuttle promoters (P3022 and P0943) in E. coli and G. oxydans were discovered in this study. The E. coli JM109 containing the above plasmids showed visible red fluorescence (Supplementary Figure 2). When detecting the strength of these promoters in G. oxydans WSH-003, strong red fluorescence could also be observed (Figure 2). The shuttle promoters were often applied to the construction of shuttle vectors to express resistance genes or used to build a broad host expression system. To the best of our knowledge, such strong shuttle promoters have rarely been reported in E. coli and G. oxydans, although many shuttle promoters have been reported in other strains (Yang et al., 2018; Khan et al., 2020).

Figure 3. The comparison of screened strongest promoter with reported strong promoters. The abscissa axis shows different promoters of G. oxydans. Pcon is the negative control. P0169 and P264 are reported strong promoters of G. oxydans ATCC 621H. PtufB and Pdnak are reported strong promoters of G. oxydans WSH-003. P2703 is the strongest promoter screened in this study. The ordinate axis shows the relative activity of mCherry. **P < 0.01 compared with the Pcon by two-tailed t test; ***P < 0.001 compared with the Pcon by two-tailed t test. +++P < 0.001 compared with the P2703 by two-tailed t test.
The structures of these screened promoters were also analyzed in this study. The promoter sequences are listed in Supplementary Table 3. Analysis of the above promoter region and transcription start site was performed by Softberry1 (Salamov and Solovyevand, 2011) and Neural Network Promoter Prediction2 (Reese, 2001). The analyzed results are listed in Supplementary Table 4 and shown in Figure 4 as mapped by the website3 (Schneider and Stephens, 1990; Crooks et al., 2004). As shown in Figure 4, a “TTGnnn” region, with a highly conserved “TTG,” near position −35 and a “TATAAT” region near position −10 were found in the screened promoters. High frequencies of “A” or “G” were also observed at transcription initiation sites. At last, a region enriched in “A” and “G” was discovered at about eight nucleotides before the initiation codon “ATG” or “GTG” (Figure 4C), and many “AGGAg” regions were observed when strong promoters were analyzed (Supplementary Table 4).
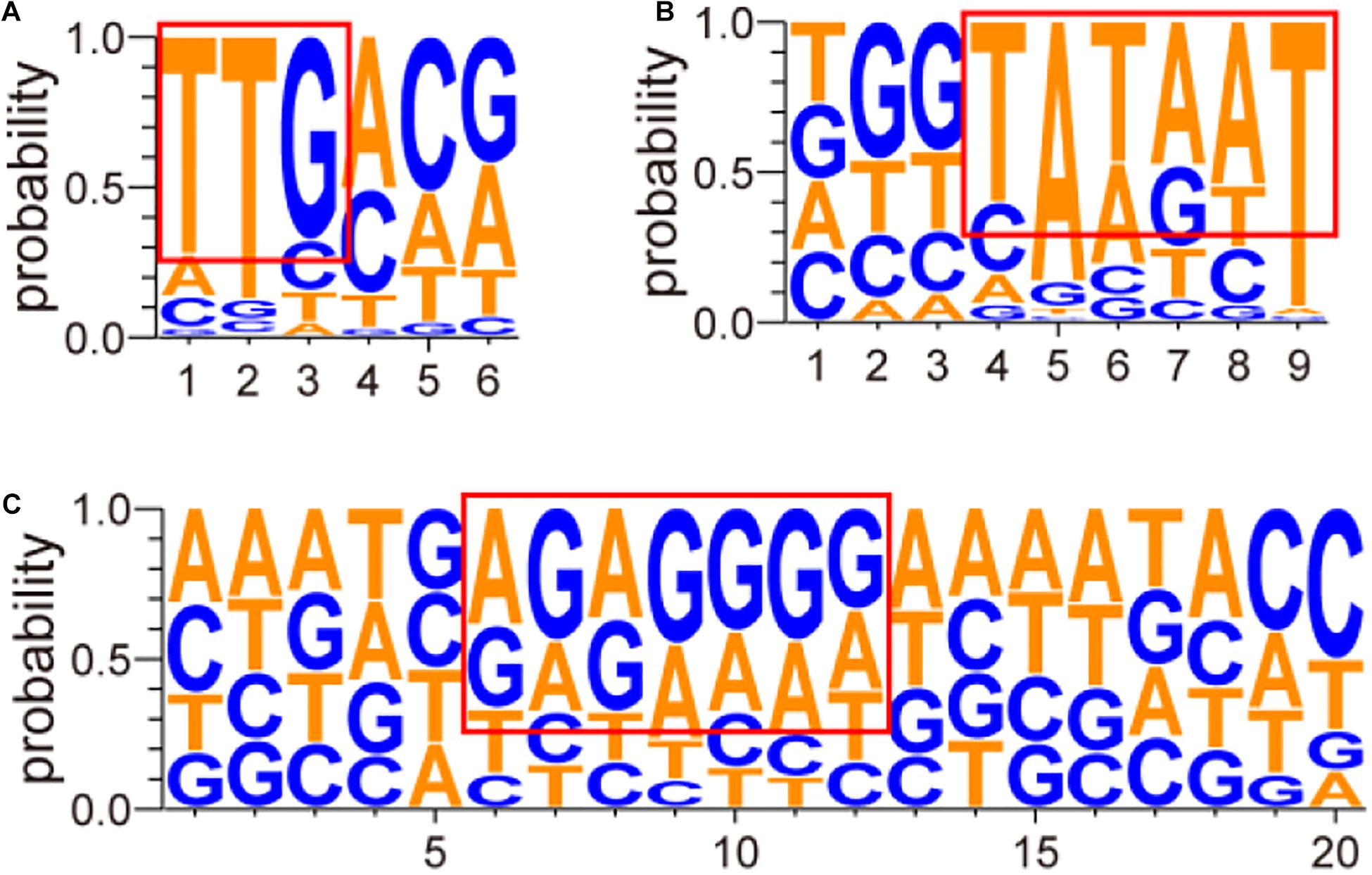
Figure 4. Sequence analysis of conserved nucleotides of screened promoters. The sequence analysis of conserved nucleotides of screened promoters was conducted using Softberry (Salamov and Solovyevand, 2011) and Neural Network Promoter Prediction (Reese, 2001). The abscissa axis shows the positions of nucleotides in different promoters. The 20 nucleotides before the initiation codon “ATG” or “GTG” were selected for the prediction of RBS. The ordinate axis shows the probability of each nucleotide. In the figure, (A) stands for the prediction result of the conserved region near position −35, (B) stands for the prediction result of the conserved region near position −10, and (C) stands for the prediction result of the conserved RBS.
Strain G. oxydans has potential in the biosynthesis of 2-KLG from D-sorbitol (Wang et al., 2018). It has been reported that SDH was an essential dehydrogenase in the conversion of L-sorbose to form 2-KLG in G. oxydans (Hoshino et al., 1990; Saito et al., 1998). In our previous study, it was also found that the rate-limiting step of the fermentation is the enzyme activity of SDH (Chen et al., 2019). In this study, a group of gradient promoters was selected to overexpress SDH in G. oxydans WSH-003. As shown in Figure 5, when weak promoters were used, the strains produced almost no 2-KLG; when medium-strength promoters were used, the strains could only synthesize 2-KLG with yields lower than 2.0 g/L; when strong promoters were applied, the strains could synthesize 2-KLG with yields of up to 3.7 g/L. The highest conversion yields achieved about 7% in mole number using strong promoters, almost twofold higher than that of those using medium-strength promoters. In addition, all the strains achieved similar biomass (Figure 5). Taken together, it can be concluded that the titer of 2-KLG increased with the enhancement of promoters or, in other words, 2-KLG production was positively related to the expression of SDH. These results demonstrated that the activity of SDH was indeed a rate-limiting step in the fermentation of 2-KLG.
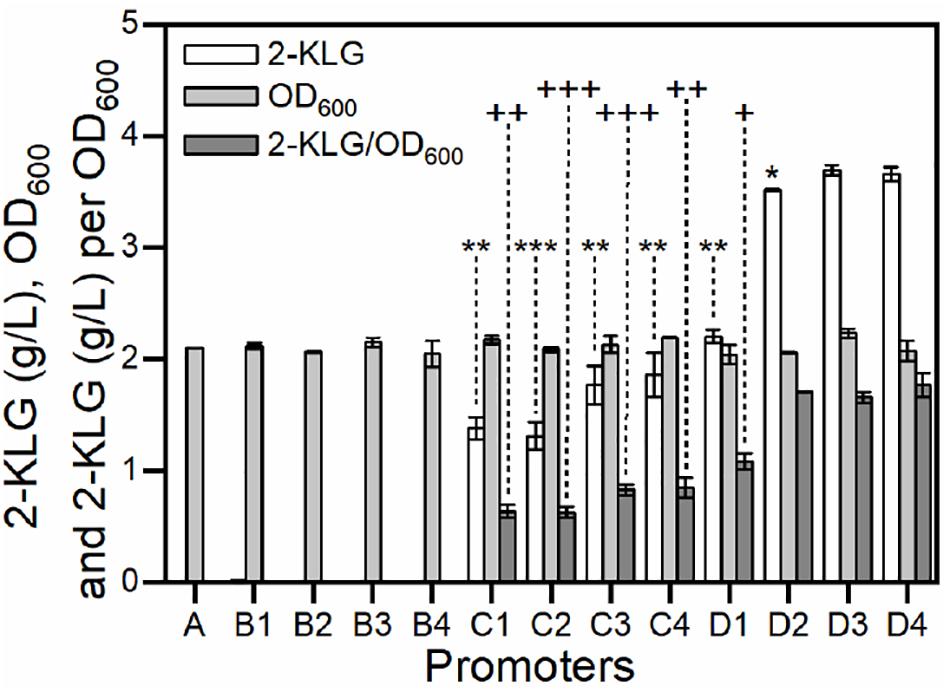
Figure 5. Effect of over-expressing SDH by gradient promoters on the growth of G. oxydans and the yield of 2-KLG. The abscissa axis shows different promoters controlling the expression of SDH in G. oxydans. (A) negative control; (B1–B4) weak promoters P1142, P0991, P1295, and P0327; (C1–C4) medium-strength promoters P0647, P0205, P0804, and P1653; (D1–D4) strong promoters P3022, P2038, P2057, and P2703. The ordinate axis shows the titer of 2-KLG (g/L), OD600 values and the titer of 2-KLG per OD600. *P < 0.05 2-KLG titer compared with D3 by two-tailed t test; **P < 0.01 2-KLG titer compared with D3 by two-tailed t test; ***P < 0.001 2-KLG titer compared with D3 by two-tailed t test; D3 and D4 are not statistically significant in 2-KLG titer with p = 0.0667. + P < 0.05 2-KLG titer per OD600 compared with D3 by two-tailed t test; ++P < 0.01 2-KLG titer per OD600 compared with D3 by two-tailed t test; +++P < 0.001 2-KLG titer per OD600 compared with D3 by two-tailed t test; D2 and D3 are not statistically significant in 2-KLG titer per OD600 with p = 0.227; D4 and D3 are not statistically significant in 2-KLG titer per OD600 with p = 0.083.
Gluconobacter oxydans is an excellent host to produce 2-KLG, which is an essential precursor of vitamin C (Saito et al., 1998; Gao et al., 2014; Wang et al., 2018). With the help of gradient promoters screened, a series of 2-KLG-producing strains have been obtained. These 2-KLG-producing strains showed the highest titer when the strongest promoters were used, while almost no production when weak promoters were employed. It was consistent with the study of Saito et al. (1997) that the productivity of 2-KLG could be improved by optimizing promoters. The degradation of 2-KLG by a class of aldo-keto reductases was reported in some G. oxydans strains (Sugisawa et al., 1990; Saito et al., 1997) and Aspergillus niger (Kuivanen et al., 2017). That may be the reason that G. oxydans WSH-003 could not accumulate 2-KLG when the expression level of sorbose dehydrogenase was low. These results may explain why many G. oxydans strains possess the entire set of 2-KLG biosynthesis genes, but only a few strains produce 2-KLG naturally (Wang et al., 2018).
In this study, a group of promoters with different strengths was obtained based on RNA-Seq data of whole transcripts. The relative strength of these promoters covered a range of about 28 times, from 400 to 11,000, while reported promoters covered about 10 times, from 400 to 4000. Among them, the activity of the newly discovered strongest promoter P2703 was approximately threefold that of the reported strong promoters P264 and Pdnak. Besides, two promoters P0943 and P3022 showed high activity in both E. coli and G. oxydans, revealing great potential in the construction of a shuttle expression system. The promoter region and the transcription start site of the screened promoters were also analyzed. The two strong shuttle promoters P3022 and P0943 had an excellent linear discriminant function (LDF) value, which may be the reason why these two promoters had high activities in both E. coli and G. oxydans.
In recent years, researchers have conducted many studies on the promoters of prokaryotes, especially model microorganisms such as E. coli (Schuller et al., 2020), Bacillus subtilis (Castillo-Hair et al., 2019), and Corynebacterium glutamicum (Dostalova et al., 2019). Based on these studies, researchers could have a detailed knowledge of these promoters’ structures and transcription factors. In this study, we found a “TTGnnn” region, with a highly conserved “TTG,” nearing position −35 and a “TATAAT” region nearing position −10, which was in accordance with the results in many other bacteria like E. coli. However, the “TATnnT” region nearing position −10 was not observed in the strong promoters of G. oxydans (Supplementary Table 4). This result is in agreement with the study of Kranz, and the main reason may be that the promoter motif is recognized by alternative sigma factors except σ70 (Kranz et al., 2018). The higher frequency of “A” or “G” at transcription initiation sites supported the theory that purine nucleotides are related to the increased transcription initiation rates (Mendoza-Vargas et al., 2009). Consistent with a prediction by Kranz et al. (2018) the conserved RBS motif “AGGAg” was also found in the strong promoters of G. oxydans.
With the development of synthetic biology, many methods can be applied to improve the strength of promoters, for example, randomization of the non-conserved region of the promoters (Siegl et al., 2013), error-prone PCR (Swagatika et al., 2019), hybrid or cascade promoters (Zhou et al., 2017), the design of RBS by RBS Calculator (Salis, 2011), and the use of a promoter library based on machine learning (Zhao et al., 2020). On the other hand, many other promoters like shuttle promoters and inducible promoters are also crucial in protein engineering and metabolic engineering. A strong shuttle promoter, Pbs, for B. subtilis, E. coli, and Saccharomyces cerevisiae was constructed by Yang et al. (2018). Three broad-spectrum promoters (Pbs1, Pbs2, and Pbs3) with different strengths, were generated by random mutation and characterized. In a recent study, a newly tunable L-arabinose-inducible PBAD promoter was discovered to be useful in G. oxydans 621H, and the activity of this promoter was affected by the pH of the medium (Fricke et al., 2020). In summary, the identification of gradient promoters in this study expanded the toolbox of available promoters, and these promoters revealed promising prospects in metabolic engineering of G. oxydans for high-value products. With further research, more serviceable promoters of G. oxydans are expected to be discovered and constructed.
The data presented in the study are deposited in online repositories: https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN18147321.
YC, LL, and SY performed the experiments and data analysis. YC and JZ wrote the manuscript and conceived the study. JL, JZ, and JC coordinated the project. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2019YFA0904900), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (32021005), and the National Science Fund for Excellent Young Scholars (21822806).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.673844/full#supplementary-material
Battling, S., Wohlers, K., Igwe, C., Kranz, A., Pesch, M., Wirtz, A., et al. (2020). Novel plasmid-free Gluconobacter oxydans strains for production of the natural sweetener 5-ketofructose. Microb. Cell Fact. 19, 1–15.
Blank, M., and Schweiger, P. (2018). Surface display for metabolic engineering of industrially important acetic acid bacteria. Peerj 6, e4626. doi: 10.7717/peerj.4626
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Castillo-Hair, S. M., Baerman, E. A., Fujita, M., Igoshin, O. A., and Tabor, J. J. (2019). Optogenetic control of Bacillus subtilis gene expression. Nat. Commun. 10, 1–11. doi: 10.1128/mmbr.57.1.1-33.1993
Chen, Y., Liu, L., Shan, X., Du, G., Zhou, J., and Chen, J. (2019). High-throughput screening of a 2-keto-L-gulonic acid-producing Gluconobacter oxydans strain based on related dehydrogenases. Front. Bioeng. Biotechnol. 7:385.
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
De La Morena, S., Wojtusik, M., Santos, V. E., and Garcia-Ochoa, F. (2020). Kinetic modeling of dihydroxyacetone production from glycerol by Gluconobacter oxydans ATCC 621 resting cells: Effect of fluid dynamics conditions. Catalysts 10, 101. doi: 10.3390/catal10010101
De Muynck, C., Pereira, C., Naessens, M., Parmentier, S., Soetaert, W., and Vandamme, E. (2007). The genus Gluconobacter oxydans: comprehensive overview of biochemistry and biotechnological applications. Crit. Rev. Biotechnol. 27, 147. doi: 10.1080/07388550701503584
De Wulf, P., Soetaert, W., and Vandamme, E. J. (2000). Optimized synthesis of L-sorbose by C5-dehydrogenation of D-sorbitol with Gluconobacter oxydans. Biotechnol. Bioeng. 69, 339–343. doi: 10.1002/1097-0290(20000805)69:3<339::aid-bit12>3.0.co;2-e
Deppenmeier, U., Hoffmeister, M., and Prust, C. (2002). Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60, 233. doi: 10.1007/s00253-002-1114-5
Dostalova, H., Busche, T., Holátko, J., Rucka, L., Štěpánek, V., Barvik, I., et al. (2019). Overlap of promoter recognition specificity of stress response sigma factors SigD and SigH in Corynebacterium glutamicum ATCC 13032. Front. Microbiol. 9:3287.
Fang, J., Wan, H., Zeng, W., Li, J., Chen, J., and Zhou, J. (2020). Transcriptome analysis of Gluconobacter oxydans WSH-003 exposed to elevated 2-keto-L-gulonic acid reveals the responses to osmotic and oxidative stress. Appl. Biochem. Biotechnol. 193, 128–141. doi: 10.1007/s12010-020-03405-8
Fricke, P. M., Link, T., Gätgens, J., Sonntag, C., Otto, M., Bott, M., et al. (2020). A tunable L-arabinose-inducible expression plasmid for the acetic acid bacterium Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 104, 9267–9282. doi: 10.1007/s00253-020-10905-4
Gao, H., Li, J., Sivakumar, D., Kim, T.-S., Patel, S. K., Kalia, V. C., et al. (2019). NADH oxidase from Lactobacillus reuteri: a versatile enzyme for oxidized cofactor regeneration. Int. J. Biol. Macromol. 123, 629–636. doi: 10.1016/j.ijbiomac.2018.11.096
Gao, L., Hu, Y., Liu, J., Du, G., Zhou, J., and Chen, J. (2014). Stepwise metabolic engineering of Gluconobacter oxydans WSH-003 for the direct production of 2-keto-L-gulonic acid from D-sorbitol. Metab. Eng. 24, 30–37. doi: 10.1016/j.ymben.2014.04.003
Gao, L., Zhou, J., Liu, J., Du, G., and Chen, J. (2012). Draft genome sequence of Gluconobacter oxydans WSH-003, a strain that is extremely tolerant of saccharides and alditols. J. Bacteriol. 194, 4455–4456. doi: 10.1128/jb.00837-12
Hahn, T., Torkler, S., Van Der Bolt, R., Gammel, N., Hesse, M., Möller, A., et al. (2020). Determining different impact factors on the xylonic acid production using Gluconobacter oxydans DSM 2343. Process Biochem. 94, 172–179. doi: 10.1016/j.procbio.2020.04.011
Hoffmann, J. J., Hövels, M., Kosciow, K., and Deppenmeier, U. (2020). Synthesis of the alternative sweetener 5-ketofructose from sucrose by fructose dehydrogenase and invertase producing Gluconobacter strains. J. Biotechnol. 307, 164–174. doi: 10.1016/j.jbiotec.2019.11.001
Hoshino, T., Sugisawa, T., Tazoe, M., Shinjoh, M., and Fujiwara, A. (1990). Metabolic pathway for 2-keto-L-gulonic acid formation in Gluconobacter melanogenus IFO 3293. Agric Biol Chem. 54, 1211–1218. doi: 10.1271/bbb1961.54.1211
Hövels, M., Kosciow, K., Kniewel, J., Jakob, F., and Deppenmeier, U. (2020). High yield production of levan-type fructans by Gluconobacter japonicus LMG 1417. Int. J. Biol. Macromol. 164, 295–303. doi: 10.1016/j.ijbiomac.2020.07.105
Hu, Y., Wan, H., Li, J., and Zhou, J. (2015). Enhanced production of L-sorbose in an industrial Gluconobacter oxydans strain by identification of a strong promoter based on proteomics analysis. J. Ind. Microbiol. Biotechnol. 42, 1039–1047. doi: 10.1007/s10295-015-1624-7
Kallnik, V., Meyer, M., Deppenmeier, U., and Schweiger, P. (2010). Construction of expression vectors for protein production in Gluconobacter oxydans. J. Biotechnol. 150, 460–465. doi: 10.1016/j.jbiotec.2010.10.069
Khan, N., Yeung, E., Farris, Y., Fansler, S. J., and Bernstein, H. C. (2020). A broad-host-range event detector: expanding and quantifying performance between Escherichia coli and Pseudomonas species. Synth. Biol. 5, ysaa002.
Kim, T.-S., Patel, S. K., Selvaraj, C., Jung, W.-S., Pan, C.-H., Kang, Y. C., et al. (2016). A highly efficient sorbitol dehydrogenase from Gluconobacter oxydans G624 and improvement of its stability through immobilization. Sci. Rep. 6, 1–11.
Kranz, A., Busche, T., Vogel, A., Usadel, B., Kalinowski, J., Bott, M., et al. (2018). RNAseq analysis of α-proteobacterium Gluconobacter oxydans 621H. BMC genomics 19:24.
Kuivanen, J., Arvas, M., and Richard, P. (2017). Clustered genes encoding 2-keto-L-gulonate reductase and L-idonate 5-dehydrogenase in the novel fungal D-glucuronic acid pathway. Front. Microbiol. 8:225.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Meth. 9, 357. doi: 10.1038/nmeth.1923
Lee, S. H., Kim, M.-S., Jung, H. C., Lee, J., Lee, J.-H., Lee, H. S., et al. (2015). Screening of a novel strong promoter by RNA sequencing and its application to H2 production in a hyperthermophilic archaeon. Appl. Microbiol. Biotechnol. 99, 4085–4092. doi: 10.1007/s00253-015-6444-1
Li, K., Mao, X., Liu, L., Lin, J., Sun, M., Wei, D., et al. (2016). Overexpression of membrane-bound gluconate-2-dehydrogenase to enhance the production of 2-keto-D-gluconic acid by Gluconobacter oxydans. Microb. Cell Fact. 15, 121.
Li, N., Zeng, W., Xu, S., and Zhou, J. (2020). Obtaining a series of native gradient promoter-5’-UTR sequences in Corynebacterium glutamicum ATCC 13032. Microb. Cell Fact. 19, 1–11.
Liao, Y., Huang, L., Wang, B., Zhou, F., and Pan, L. (2015). The global transcriptional landscape of Bacillus amyloliquefaciens XH7 and high-throughput screening of strong promoters based on RNA-seq data. Gene 571, 252–262. doi: 10.1016/j.gene.2015.06.066
Mendoza-Vargas, A., Olvera, L., Olvera, M., Grande, R., Vega-Alvarado, L., Taboada, B., et al. (2009). Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. doi: 10.1371/journal.pone.0007526
Merfort, M., Herrmann, U., Bringer-Meyer, S., and Sahm, H. (2006). High-yield 5-keto-D-gluconic acid formation is mediated by soluble and membrane-bound gluconate-5-dehydrogenases of Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 73, 443–451. doi: 10.1007/s00253-006-0467-6
Mientus, M., Kostner, D., Peters, B., Liebl, W., and Ehrenreich, A. (2017). Characterization of membrane-bound dehydrogenases of Gluconobacter oxydans 621H using a new system for their functional expression. Appl. Microbiol. Biotechnol. 101, 3189–3200. doi: 10.1007/s00253-016-8069-4
Nishikura-Imamura, S., Matsutani, M., Insomphun, C., Vangnai, A. S., Toyama, H., Yakushi, T., et al. (2014). Overexpression of a type II 3-dehydroquinate dehydratase enhances the biotransformation of quinate to 3-dehydroshikimate in Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 98, 2955–2963. doi: 10.1007/s00253-013-5439-z
Poulsen, L. D., and Vinther, J. (2018). RNA-Seq for bacterial gene expression. Curr. Protoc. Nucleic Acid Chem. 73, e55.
Prust, C., Hoffmeister, M., Liesegang, H., Wiezer, A., Fricke, W. F., Ehrenreich, A., et al. (2005). Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 23, 195–200. doi: 10.1038/nbt1062
Reese, M. G. (2001). Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26, 51–56. doi: 10.1016/s0097-8485(01)00099-7
Saito, Y., Ishii, Y., Hayashi, H., Imao, Y., Akashi, T., Yoshikawa, K., et al. (1997). Cloning of genes coding for L-sorbose and L-sorbosone dehydrogenases from Gluconobacter oxydans and microbial production of 2-keto-L-gulonate, a precursor of L-ascorbic acid, in a recombinant G. oxydans strain. Appl. Environ. Microbiol. 63, 454–460. doi: 10.1128/aem.63.2.454-460.1997
Saito, Y., Ishii, Y., Hayashi, H., Yoshikawa, K., Noguchi, Y., Yoshida, S., et al. (1998). Direct fermentation of 2-keto-L-gulonic acid in recombinant Gluconobacter oxydans. Biotechnol. Bioeng. 58, 309–315. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<309::aid-bit30>3.0.co;2-4
Salamov, V. S. A., and Solovyevand, A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its applications in agriculture, biomedicine and environmental studies, ed. R. W. Li (New York: Nova Science Publishers), 61–78.
Salis, H. M. (2011). The ribosome binding site calculator. Methods Enzymol. 498, 19–42. doi: 10.1016/b978-0-12-385120-8.00002-4
Schedel, M. (2000). Regioselective oxidation of aminosorbitol with Gluconobacter oxydans, key reaction in the industrial 1-deoxynojirimycin synthesis. Biotechnology 8, 295–308. doi: 10.1002/9783527620913.ch7
Schneider, T. D., and Stephens, R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100. doi: 10.1093/nar/18.20.6097
Schuller, A., Cserjan-Puschmann, M., Tauer, C., Jarmer, J., Wagenknecht, M., Reinisch, D., et al. (2020). Escherichia coli σ70 promoters allow expression rate control at the cellular level in genome-integrated expression systems. Microb. Cell. Fact. 19, 1–11.
Selvaraj, C., Krishnasamy, G., Jagtap, S. S., Patel, S. K., Dhiman, S. S., Kim, T.-S., et al. (2016). Structural insights into the binding mode of D-sorbitol with sorbitol dehydrogenase using QM-polarized ligand docking and molecular dynamics simulations. Biochem. Eng. J. 114, 244–256. doi: 10.1016/j.bej.2016.07.008
Shan, X., Liu, L., Zeng, W., Chen, J., and Zhou, J. (2020). High throughput screening platform for a FAD-dependent L-sorbose dehydrogenase. Front. Bioeng. Biotechnol. 8:194.
Shen, Y., Zhou, X., and Xu, Y. (2020). Enhancement of Gluconobacter oxydans resistance to lignocellulosic-derived inhibitors in xylonic acid production by overexpressing thioredoxin. Appl. Biochem. Biotechnol. 191, 1072–1083. doi: 10.1007/s12010-020-03253-6
Siegl, T., Tokovenko, B., Myronovskyi, M., and Luzhetskyy, A. (2013). Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 19, 98–106. doi: 10.1016/j.ymben.2013.07.006
Stark, R., Grzelak, M., and Hadfield, J. (2019). RNA sequencing: the teenage years. Nat. Rev. Genet. 20, 631–656. doi: 10.1038/s41576-019-0150-2
Sugisawa, T., Hoshino, T., Masuda, S., Nomura, S., Setoguchi, Y., Tazoe, M., et al. (1990). Microbial production of 2-keto-L-gulonic acid from L-sorbose and D-sorbitol by Gluconobacter melanogenus. Agric. Biol. Chem. 54, 1201–1209. doi: 10.1271/bbb1961.54.1201
Swagatika, P., Nikhil, K., Barkha, R., Pashupathi, M., Parthasarathi, B., and Ajay, K. (2019). Effect of mutation resulted from error prone PCR on the strength of promoter activity. J. Exp. Zool. India 22, 981–986.
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Wan, H., Xia, Y., Li, J., Kang, Z., and Zhou, J. (2017). Identification of transporter proteins for PQQ-secretion pathways by transcriptomics and proteomics analysis in Gluconobacter oxydans WSH-003. Front. Chem. Sci. Eng. 11:72–88. doi: 10.1007/s11705-016-1580-4
Wang, P., Zeng, W., Xu, S., Du, G., Zhou, J., and Chen, J. (2018). Current challenges facing one-step production of L-ascorbic acid. Biotechnol. Adv. 36, 1882–1899. doi: 10.1016/j.biotechadv.2018.07.006
Yang, S., Liu, Q., Zhang, Y., Du, G., Chen, J., and Kang, Z. (2018). Construction and characterization of broad-spectrum promoters for synthetic biology. ACS Synth. Biol. 7, 287–291. doi: 10.1021/acssynbio.7b00258
Yuan, J. F., Wu, M. B., Lin, J. P., and Yang, L. R. (2016). Combinatorial metabolic engineering of industrial Gluconobacter oxydans DSM2343 for boosting 5-keto-D-gluconic acid accumulation. BMC Biotechnol. 16:42.
Zhang, L., Lin, J., Ma, Y., Wei, D., and Sun, M. (2010). Construction of a novel shuttle vector for use in Gluconobacter oxydans. Mol. Biotechnol. 46, 227–233. doi: 10.1007/s12033-010-9293-2
Zhao, M., Zhou, S., Wu, L., and Deng, Y. (2020). Machine learning-based promoter strength prediction derived from a fine-tuned synthetic promoter library in Escherichia coli. bioRxiv
Zhou, S., Ding, R., Chen, J., Du, G., Li, H., and Zhou, J. (2017). Obtaining a panel of cascade promoter-5′-UTR complexes in Escherichia coli. ACS Synth. Biol. 6, 1065–1075. doi: 10.1021/acssynbio.7b00006
Keywords: 2-keto-L-gulonic acid, Gluconobacter oxydans, promoters, L-sorbose, sorbose dehydrogenase
Citation: Chen Y, Liu L, Yu S, Li J, Zhou J and Chen J (2021) Identification of Gradient Promoters of Gluconobacter oxydans and Their Applications in the Biosynthesis of 2-Keto-L-Gulonic Acid. Front. Bioeng. Biotechnol. 9:673844. doi: 10.3389/fbioe.2021.673844
Received: 28 February 2021; Accepted: 22 March 2021;
Published: 09 April 2021.
Edited by:
Dawei Zhang, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, ChinaReviewed by:
Sanjay Kumar Singh Patel, Konkuk University, South KoreaCopyright © 2021 Chen, Liu, Yu, Li, Zhou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwen Zhou, emhvdWp3MTk4MkBqaWFuZ25hbi5lZHUuY24=; Jian Chen, amNoZW5AamlhbmduYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.