
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol., 01 July 2021
Sec. Synthetic Biology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.673270
This article is part of the Research TopicRecent Advances in Application of Synthetic Biology for Production of Bioactive CompoundsView all 11 articles
Isoflavonoids are well-known plant secondary metabolites that have gained importance in recent time due to their multiple nutraceutical and pharmaceutical applications. In plants, isoflavonoids play a role in plant defense and can confer the host plant a competitive advantage to survive and flourish under environmental challenges. In animals, isoflavonoids have been found to interact with multiple signaling pathways and have demonstrated estrogenic, antioxidant and anti-oncologic activities in vivo. The activity of isoflavonoids in the estrogen pathways is such that the class has also been collectively called phytoestrogens. Over 2,400 isoflavonoids, predominantly from legumes, have been identified so far. The biosynthetic pathways of several key isoflavonoids have been established, and the genes and regulatory components involved in the biosynthesis have been characterized. The biosynthesis and accumulation of isoflavonoids in plants are regulated by multiple complex environmental and genetic factors and interactions. Due to this complexity of secondary metabolism regulation, the export and engineering of isoflavonoid biosynthetic pathways into non-endogenous plants are difficult, and instead, the microorganisms Saccharomyces cerevisiae and Escherichia coli have been adapted and engineered for heterologous isoflavonoid synthesis. However, the current ex-planta production approaches have been limited due to slow enzyme kinetics and traditionally laborious genetic engineering methods and require further optimization and development to address the required titers, reaction rates and yield for commercial application. With recent progress in metabolic engineering and the availability of advanced synthetic biology tools, it is envisaged that highly efficient heterologous hosts will soon be engineered to fulfill the growing market demand.
Living organisms like plants, fungi and unicellular prokaryotes and eukaryotes produce a myriad of chemicals, broadly classified as primary metabolites and secondary metabolites. Primary metabolites are fundamental compounds of life as they are involved in vital cellular processes such as respiration and photosynthesis. Secondary metabolites are a specialized class of chemicals, and they are usually produced under specific conditions and give producing organisms an additional advantage to survive, compete or attack other organisms (Nabavi et al., 2020). Secondary metabolites are usually divided into three classes (alkaloids, terpenoids, and phenylpropanoids) based on their chemical structure and precursor primary metabolite. The phenylpropanoid is the largest class of plant secondary metabolites, which play an important role in plant growth and development (Vogt, 2010). The phenylpropanoid pathway has different branches that lead to different groups of compounds including chalcones, flavanones, isoflavonoids, and anthocyanins (Novelli et al., 2019). The structure and function of most of these groups have been well researched and documented in the literature.
The isoflavonoids are a large group of plant secondary metabolites and possess a 3-phenylchroman skeleton, which is biogenetically derived from the 2-phenylchroman skeleton of the parent flavonoid (Figure 1). Isoflavonoids are predominantly present in Papilionoideae, a subfamily of Leguminosae (Dixon and Sumner, 2003). More than 2,400 isoflavonoids from over 300 plants have been identified so far (Veitch, 2007, 2009, 2013; Al-Maharik, 2019). Isoflavonoids play multiple roles in host plant, and their role in plant defense and plant–rhizobia relationships is the most significant (Larose et al., 2002). Due to their significance for the host plant, biosynthetic pathways involved in the synthesis and accumulation of many isoflavonoids have been explored. Following that, several attempts have been made to increase the content of isoflavonoids in endogenous as well as in related plants. However, due to the complexity of plant secondary metabolism, no significant improvement has been achieved.
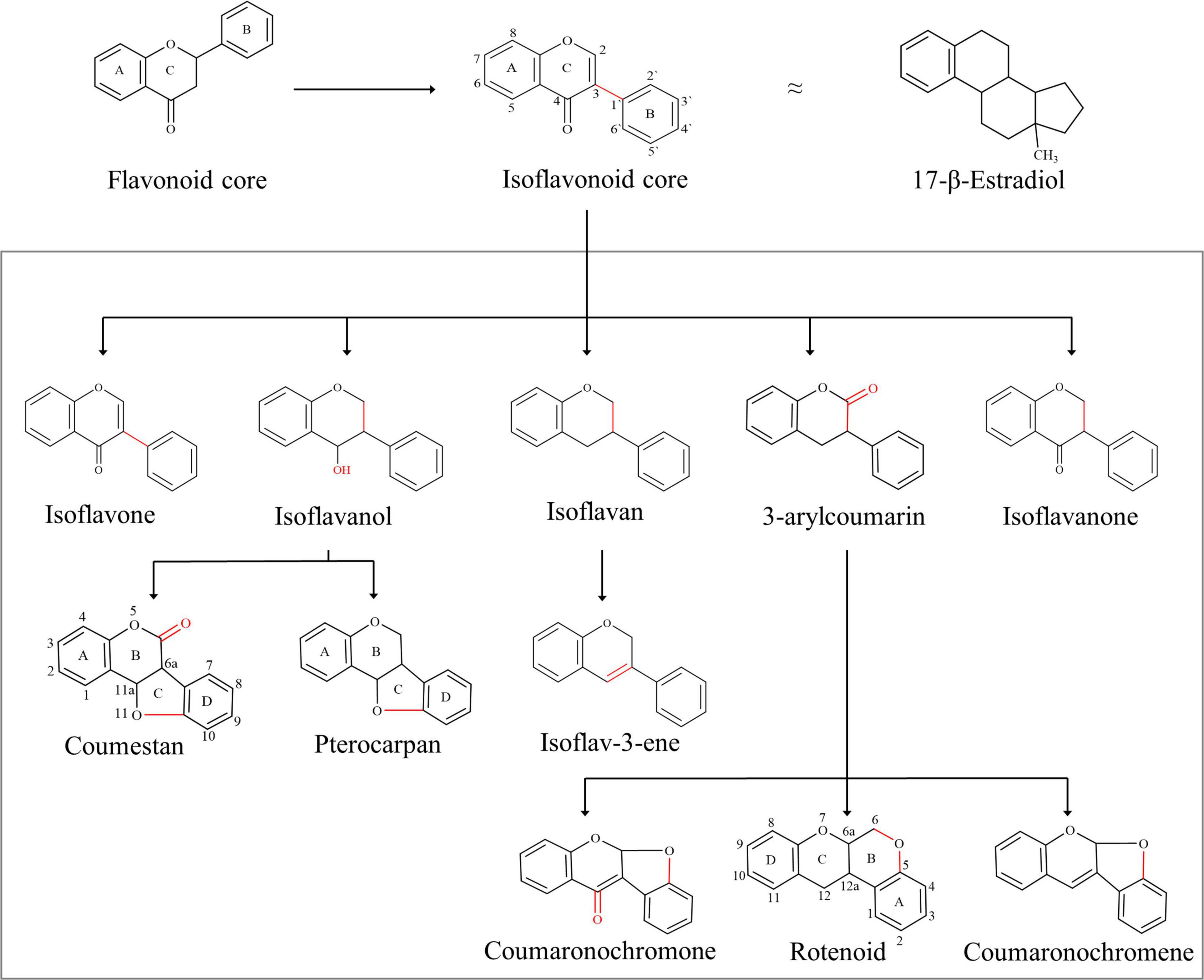
Figure 1. Basic skeleton of isoflavonoids: isoflavonoids are structurally different to flavonoids, with the B-ring migration from position 2 to 3, which in turn leads to the structural similarities to estrogen, e.g., 17β-estradiol. Isoflavonoid diversity is regulated by simple functional additions such as hydroxyl, which in turn can generate additional rings into the backbone, e.g., pterocarpan and coumestan. The addition of ketones can also generate additional rings, for example, rotenoid and coumaronochromone.
Isoflavonoids have played a distinctive role in the history of disease prevalence across continents. It is believed that the difference in the prevalence of cancer across continents is linked with the preference for soy foods (Ko, 2014). Soybean products are rich in organic chemicals that are structurally similar to 17β-estradiol, a human sex hormone (Figure 1). Due to this structural similarity, isoflavonoids play an important role in cellular signaling pathways and control multiple functions in humans and are commonly known as phytoestrogens (Prasad et al., 2010). Together with this, isoflavonoids are commonly used in cosmetics, nutraceuticals, pharmaceuticals, food, and beverage industry; however, the pharmaceutical sector holds the largest share in the market due to potential applications of isoflavonoids in chronic and cardiovascular diseases. The market size of isoflavonoids was over US$ 13.5 billion in 2018 and is estimated to reach US$ 30 billion by 2025 (Ahuja and Mamtani, 2019). Isoflavonoids are presently extracted from plants; however, alternative production platforms are also being explored for their sustainable production to maintain a constant supply in the growing market.
Due to a range of potential applications, the demand for isoflavonoids is growing in recent times. However, the issue of traditional extraction and low yield in plants along with recent climate change and competition to use cultivatable land have questioned their availability for the general public. Therefore, the present paper is aimed to discuss the biosynthetic pathways and potential applications of isoflavonoids (as a subclass of flavonoids) generally but specifically about seven key isoflavonoids: daidzein, formononetin, pisatin, medicarpin, coumestrol, genistein, and biochanin-A with a special focus on their biosynthesis in heterologous hosts.
Isoflavonoids are commonly present in low amounts in seeds and roots of the Leguminosae/Fabaceae family including several commonly consumed plants like barley, broccoli, cauliflower, fava beans, lupine, kudzu, and soy (Prasad et al., 2010; Table 1). Traces of isoflavonoids are also present in red wine and in other plants like alfalfa, red clover and linseed (Pilsakova et al., 2010). Quite interestingly, isoflavonoids have also been identified from at least 59 non-leguminous plant families (i.e., Iridaceae, Rosaceae, and Liliaceae), as it is commonly believed that isoflavonoids’ biosynthetic machinery is not widely distributed in plant families except legumes (Lapčík, 2007).
With increasing climate and environmental pressures, the potential utilization of isoflavonoids in planta to enhance plant resistance against herbivore insects and to improve the interactions of the plant with the rhizobiome has resulted in increased interest and research (Dillon et al., 2017).
Isoflavonoids produce a spectrum of benefits for the host plant (Figure 2). Isoflavonoids play an important role in plant defense, as they possess a range of antimicrobial activities (commonly analyzed in vitro) (Dixon, 1999). They are famous as plant defensive chemicals and are active against vertebrates, molluscs, herbivorous insects, and microorganisms (Dakora and Phillips, 1996; Nwachukwu et al., 2013). For example, the well-known isoflavonoid pterocarpans, maackiain, and pisatin play an important role as phytoalexins in the interaction between Nectria haematococca and the host plant Pisum sativum (garden pea) (Wasmann and VanEtten, 1996; Enkerli et al., 1998). Both of these pterocarpans are targets of fungal virulence factors and detoxification enzymes, which indicates their importance for the host plant. Recently, Dillon and colleagues have shown that UV-B-induced accumulation of genistein enhances resistance of field-grown soybean plants against Anticarsia gemmatalis neonates (Dillon et al., 2017). A 30% reduction in survival and 45% reduction in mass gain of larvae was documented, and the authors have concluded that UV-B-induced accumulation of isoflavonoids increases the resistance of plants against A. gemmatalis (Dillon et al., 2017). An overview of UV-B-based induction of isoflavonoids is described in section “Regulation of Isoflavonoid Biosynthesis in Plants.”
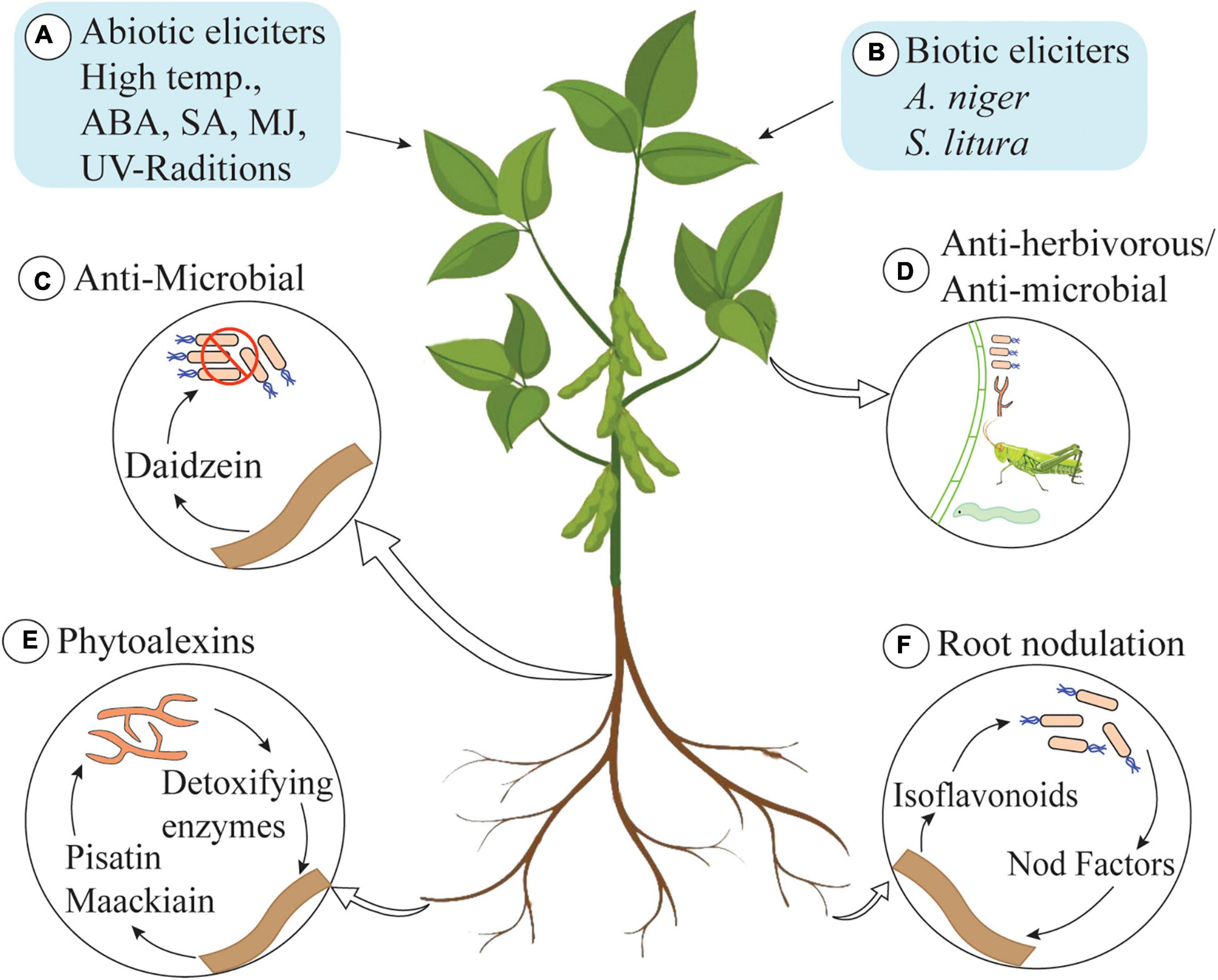
Figure 2. Role of isoflavonoids in plants. (A) Abiotic elicitors: high temperature, abscisic acid (ABA), salicylic acid (SA), methyl jasmonate (MJ), and UV-radiations. (B) Biotic elicitors: Aspergillus niger and Spodoptera litura trigger isoflavonoid biosynthesis in plants. (C) Daidzein secreted by roots kills bacteria in rhizosphere and shapes the rhizosphere communities. (D) Isoflavonoids also possess anti-herbivorous and antimicrobial activities. (E) Pisatin and maachiain kill pathogenic fungi in rhizosphere (some fungi produce detoxifying enzymes to degrade pisatin and maachiain to cause infection in roots of host plant). (F) Some isoflavonoids induce nodulation genes in receptive bacterial species and play an important role in root-nodule formation.
Isoflavonoids are not only active inside the cell but also play a beneficial role in the rhizosphere. The role of isoflavonoids in the induction of nodulation genes and as allelopathic agents has also been documented (Dixon, 1999). Daidzein, secreted by soybean roots, acts as a signaling molecule for nodulation and alters the structure and functioning of rhizosphere communities (Okutani et al., 2020). In addition to this, isoflavonoids play a role in the induction of transcription of genes involved in the production of the Nod factor. These Nod factors are rhizobial signaling molecules that make plants receptive to symbiotic root infection. Some isoflavonoids are very specific and only induce the production of Nod factors in compatible hosts, thus playing an important role in host selection (Aoki et al., 2000). Isoflavonoids are also involved in developing mutualistic interaction with compatible fungal species. The role of isoflavonoid in initiating the spore germination, hyphal growth and root colonization as well as the formation of arbuscule inside the root cell has been documented (Larose et al., 2002).
Several epidemiological studies have shown that an isoflavonoid-rich diet is associated with a low risk of chronic diseases like menopausal, diabetes, cancer, and cardiovascular diseases (Kozłowska and Szostak-Wȩgierek, 2017). Due to chemical similarity with 17β-estradiol, isoflavonoids can bind with estrogen receptors (ERs) such as ER-α and ER-β (Chen et al., 2018). Due to this affinity, isoflavonoids interfere with cellular signaling mechanisms and play an important role in cellular growth and protection (Figure 3). Isoflavonoid aglycones (without glucose) are sometimes biologically more active and available than glycones, as glucose moiety has a strong effect on their function and absorbance in the human gastrointestinal tract (Lee et al., 2018). For example, in its un-glycosylated form, the affinity of genistein is comparable with that of 17β-estradiol, but in its glycosylated forms, its affinity is up to 100–500 times less (Kuiper et al., 1997; Breinholt and Larsen, 1998).
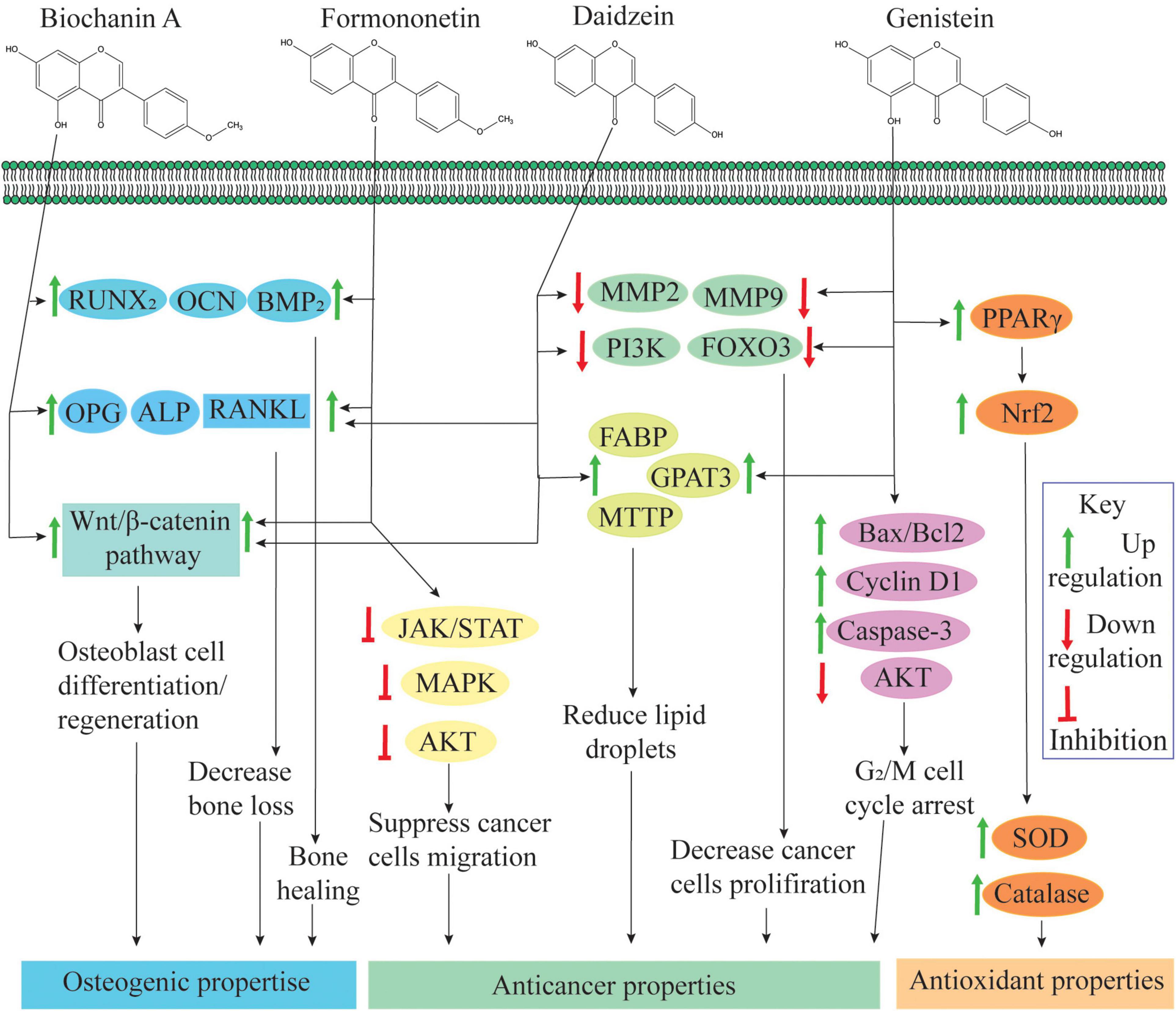
Figure 3. Role of isoflavonoids in human health: biochanin-A and formononetin are involved in bone healing and regeneration effects by upregulating runt-related transcription factor 2 (RUNX2), osteocalcin (OCN) and bone morphogenetic protein 2 (BMP2) expression at the injury site (Singh et al., 2017). Biochanin-A, formononetin, and daidzein also regulate osteoprotegerin (OPG), alkaline phosphatase (ALP), and receptor activator of nuclear factor κβ ligand (RANKL) expression; and these compounds are actively involved in osteogenic activities (Zakłos-Szyda et al., 2020). Additionally, an isoflavone mixture (biochanin-A, formononetin, and daidzein) promotes osteoblast cell differentiation and proliferation through the activation of the Wnt/β-catenin pathway (Chen et al., 2018). Additionally, formononetin inactivates signaling pathways, namely, Janus kinase/signal transducers, and activators of transcription (JAK/STAT) pathway, protein kinase B (PKB or AKT) pathway and mitogen-activated protein kinase (extracellular signal regulated kinase 1/2) [MAPK (ERK1/2)] pathways and suppresses cell migration, invasion and angiogenesis (Qi et al., 2016; Park et al., 2018). Daidzein and genistein can inhibit expression of matrix metalloproteinase-2/9 (MMP2/9), phosphatidylinositol-3-kinase (PI3K) and forkhead box O-3 (FOXO3) and reduce the cancer cell proliferation. Through activation of fatty acid-binding protein (FABP), glycerol-3-phosphate acyltransferase 3 (GPAT3) and microsomal triglyceride transfer protein (MTTP), genistein and daidzein reduce lipid droplet accumulation. Both of these situations induce apoptosis in cancer cells (as reviewed by Hsiao et al., 2020). Genistein activates the expression of Bcl-2-associated X-protein (Bax/Bcl-2), cyclin D1 and caspase-3 pathways and suppressed PI3K/AKT phosphorylation, which results in genistein-induced G2/M cell cycle arrest (Shafiee et al., 2016). Genistein activates peroxisome proliferator-activated receptor gamma (PPARγ) and enhances expression of superoxide dismutase (SOD) and catalase via Nrf2 activation (Chan et al., 2018).
The application of isoflavonoids in human health is a diverse topic, which is not in the scope of the present review. Therefore, in the following paragraphs, a short and precise overview of their health applications has been covered. Interested readers are request to consult recent review papers for further details (Zaheer and Humayoun Akhtar, 2017; Das et al., 2020; Hu et al., 2020; Liu et al., 2020).
The applications of isoflavonoids as phytoestrogens are one of the most exciting areas of interest in clinical research and nutrition. The declining level of estrogen hormone in aging women is a main cause of osteoporosis, and soy isoflavonoids can substitute the natural estrogen and control bone loss (Chen et al., 2018; Zakłos-Szyda et al., 2020). A meta-analysis has shown that soy isoflavonoid intake for 6 months has a beneficial effect on bone mineral density (BMD), especially on the lumbar spine (Wei et al., 2012). That is why the prevalence of osteoporosis is low in the Chinese and Japanese populations as compared with that in European and Americans. The beneficial role of isoflavonoids is explained due to their molecular similarity with natural estrogen and binding affinity with ERs, especially with ER-β. In addition to this, isoflavonoids intake has positive effects on learning and memory expression, and isoflavonoids are also involved in controlling hot flashes in menopause women (Henderson et al., 2000; Li et al., 2015). Several isoflavonoids including genistein and daidzein as well their derivative have been identified and characterized for their beneficial estrogenic properties.
The role of isoflavonoids as antioxidants is well established; and it is sometimes believed that the antioxidant potential of isoflavonoids is comparable with that of the well-known antioxidant vitamin E (Djuric et al., 2001). Antioxidant properties are achieved either by the regulation of gene expression of antioxidant enzymes, for example, catalase, or by inhibiting the secondary oxidant production like hydrogen peroxide (Mortensen et al., 2009). Genistein and daidzein are well-known metal chelator and radical scavenger; and due to the presence of three hydroxyl groups, the former is better than the latter (Han et al., 2009). Genistein increases the production of superoxide dismutase (SOD), which scavenges free radicals (Kuriyama et al., 2013). Conversion of daidzein to equol is an important physiological phenomenon, as equol has 100-fold higher ER binding affinity and a greater antioxidant ability than daidzein (Zaheer and Humayoun Akhtar, 2017). Isoflavonoids support disease prevention like type 2 diabetes and health maintenance by repressing the oxidative stress (Umeno et al., 2016). However, it is largely unclear to which extent isoflavonoids mediate antioxidant activities in vivo, which is difficult to access, and their antioxidant activities are mostly accessed in vitro.
Isoflavones are also well-known for their anticancer potential. Their role in the increase of prostacyclin level, activation of the endothelial level of nitric oxide synthase, inhibition of cell proliferation and DNA synthesis, relaxation of vessels and reduction of plaque, and maintenance of circulatory systems has also been documented (Cano et al., 2010; Tay et al., 2019). Isoflavonoids also interact with epigenetic modifications and are involved in the hypermethylation of tumor suppressor genes, and the underlying mechanism of methylation and acetylation of histones in breast cancer cell lines has also been revealed (Dagdemir et al., 2013).
Genistein, daidzein, formononetin, and, to some extent, coumestrol are well-studied isoflavonoids due to their anticancer potential. Genistein is a strong anticancer candidate isoflavonoid compound with a 50% inhibitory concentration (IC50) value of 37.5 μM against human topoisomerase II (Mizushina et al., 2013). Genistein is known to inhibit protein tyrosine kinase (PTK) and DNA topoisomerases I and II and, therefore, affects a range of cellular activities specifically in carcinogenesis and other neurodegenerative diseases (Kuriyama et al., 2013). Genistein and daidzein can induce cell cycle arrest at S, G2/M and G1 stages in various cancer cells (Bossard et al., 2012; Adjakly et al., 2013; Shafiee et al., 2016). In a recent study, the authors have concluded that genistein and daidzein can play an important role in cancer cell metastasis, tumorigenesis and stem-like properties, and they have potential as alternative therapies for ovarian cancer patients (Chan et al., 2018).
The IC50 value of formononetin ranges from 10 to 300 μM when tested against various cancer cell lines. Formononetin is also able to efficiently inhibit tumor growth in vivo, and it is effective against many types of tumors including breast, bone, colon, nasopharyngeal and multiple myeloma cells (Qi et al., 2016; Kim C. et al., 2018; Park et al., 2018). In most of the studies, 1–200 μM (0.3–53.7 μg/ml) of concentration of formononetin was tested, and a variable response has been observed on different cell lines (Tay et al., 2019). Coumestrol is also an important anticancer isoflavonoid candidate molecule with an IC50 value of 228 nM tested against casein kinase 2 (CK2) (Liu S. et al., 2013). Selective reduction in CK2 activity has been seen for coumestrol in a dose-dependent manner in various cancer cell lines (Liu S. et al., 2013; Park et al., 2015; Kim et al., 2017). In silico modeling has suggested that coumestrol binds to ATP binding pocket of haspin kinase to suppress its activity and results in inhibition of cancer cell proliferation (Kim et al., 2017).
Isoflavonoids are a diverse and distinctive subclass of flavonoids, and despite their limited distribution in the plant kingdom, isoflavonoids are structurally very diverse (Stobiecki and Kachlicki, 2006; Figure 1). The number and complexity of substitution on the basic 3-phenylchroman skeleton along with the different levels of oxidation and the presence of additional heterocyclic rings are responsible for such an outstanding diversity. Isoflavonoids are further divided into several groups, which are shown in Figure 1 (Dixon and Steele, 1999).
The ring system of isoflavones is derived from two different pathways: the A-ring is derived from the acetate pathway, whereas the B- and C-rings are formed from the shikimate pathway, resulting in a basic C6-C3-C6 skeleton (Ververidis et al., 2007). The basic skeleton is then further decorated with various rounds of glycosylation, methylation and hydroxylation reactions performed by several enzymes and enzyme complexes. This versatile decoration of the basic isoflavonoid skeleton is responsible for the enormous diversity of isoflavonoids. In general, the most common hydroxylation sites of isoflavonoids are 5,7,2′,3′-, and 4′-C, and common C- and O-glycosylation and methylation sites are 6,7, 8-, and 4′-C (Stobiecki and Kachlicki, 2006). Naturally, isoflavonoids are present in a glycosylated form in a plant cell, and the dominant glycosidic form is β-D-glycoside. Other glycosylated forms like 6 ″-O-acetyl-glycoside and 6 ″-O-malonyl-glycoside are also possible; however, the aglycone form is sometimes biologically more active (Ko, 2014). Pterocarpans are an interesting group of isoflavonoids due to the presence of the fourth ring, which is formed due to fusion of 4-C keto group with 6′-C; and due to this fusion, the ring system of pterocarpans is renamed (Whitten et al., 1997).
The isoflavonoids are synthesized via the phenylpropanoid pathway, utilizing flavonoid intermediates. First, the aromatic amino acids (phenylalanine) are transformed into the p-coumaroyl CoA by a set of three enzymes: phenylalanine ammonia lyase (PAL), trans-cinnamate-4-hydroxylase (C4H) and 4-coumaroyl CoA lyase (4CL). Some plant species have a promiscuous PAL, which is also able to incorporate tyrosine in the pathway (Rosler et al., 1997). The next set of three important enzymes—chalcone synthase (CHS), chalcone isomerase (CHI), and chalcone reductase (CHR)—is responsible for producing naringenin and liquiritigenin from p-coumaroyl CoA. A detailed overview of flavonoid biosynthesis has been recently published, and readers are requested to consult Nabavi et al. (2020) and references therein for further details. Both naringenin and liquiritigenin are important flavanones, which are intermediates for various other flavonoid subgroups such as anthocyanins, proanthocyanidins, flavonols, and flavones as well as precursors for isoflavonoids.
Leguminous plants produce isoflavonoids via two different routes, which, however, share many of their chemical reactions and biogenetic machinery. Migration of B-ring from the C-2 position to C-3 position is the first committed and unique step in isoflavonoid biosynthesis, which is catalyzed by isoflavone synthase (IFS), a cytochrome P450 class CYP93C enzyme (Steele et al., 1999; Jung et al., 2000; Supplementary Table 1). The immediate product of this reaction is 2,7,4′-trihydroxyisoflavanone, which is an unstable compound and dehydrated to corresponding isoflavanone, i.e., genistein or daidzein, either spontaneously or with the action of another enzyme, 2-hydroxyisoflavanone dehydratase (HIDH) (Steele et al., 1999; Akashi et al., 2005). Daidzein, formononetin, genistein, biochanin-A (isoflavones), pisatin, medicarpin (pterocarpans), and coumestrol (coumestans) are key isoflavonoids that are well-known for their potential pharmaceutical applications (Du et al., 2010). Biosynthesis of key isoflavonoids is discussed in detail in the following paragraphs and shown in Figure 4 and Supplementary Figures 1–4.
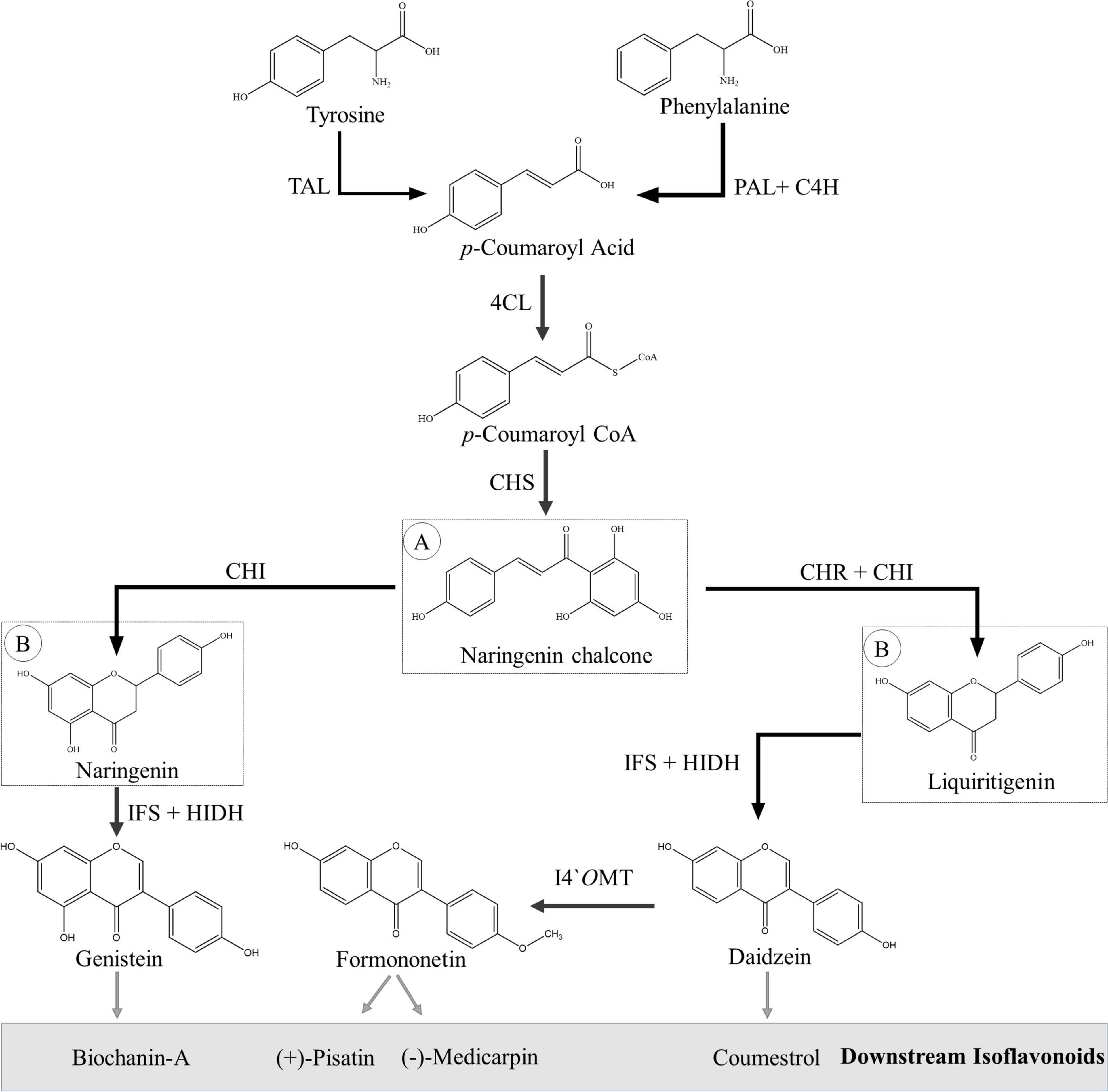
Figure 4. Biosynthesis of basic isoflavonoids from amino acids (phenylalanine and tyrosine). (A) Flavonoid precursor molecule. (B) Isoflavonoid precursor molecules. Abbreviations, 4CL, 4-coumarate CoA ligase; C4H, trans-cinnamate 4-hydroxylase; CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; HIDH, 2-hydroxyisoflavanone dehydratase; I4′OMT, isoflavone 4′-O-methyltransferase; IFS, isoflavone synthase; PAL, phenylalanine ammonia lyase; TAL, tyrosine ammonia lyase.
Formononetin is an important isoflavone that is synthesized from daidzein is a single-step reaction catalyzed by isoflavanone 4′-O-methyltransferase (I4′OMT). I4′OMT, first identified from Medicago truncatula, transfer a methyl group from S-adenosyl-L-methionine (SAM) to the 4′-C position of daidzein (Liu et al., 2006). Other carbons of the basic skeleton can also be methylated; for example, in alfalfa, 7-C is methylated to produce isoformononetin by isoflavone-7-O-methyltransferase (I7OMT) (Zubieta et al., 2001). Sometimes, both 4′-C and 7-C sites are methylated, and the resulting product is known as dimethyldaidzein (Preedy, 2012).
Pisatin is an important phytoalexin that belongs to the pterocarpan group of isoflavonoids; and like coumestans, these compounds have two asymmetric carbons, C-6a and C-11a (Slade et al., 2005). Only dextrorotatory pterocarpans [(+)-pterocarpans] possess the antimicrobial activity and are produced in a few plant species such as peanut (Arachis hypogaea) (Strange et al., 1985). Pisatin is the first chemically identified (+)-pterocarpan that is exclusively synthesized by pea (P. sativum) (Cruickshank and Perrin, 1960). Starting from formononetin, the first chemical reaction in (+)-pisatin biosynthesis pathway is catalyzed by isoflavone 3′-hydroxylase (I3′H), a P450 class CYP81E9 enzyme that adds OH group at 3′-C of formononetin to form calycosin (Supplementary Figure 1). Calycosin is then converted to pseudobaptigenin by the action of pseudobaptigenin synthase (PBS) (Clemens and Barz, 1996; Liu et al., 2003). The next step is the formation of 2′,7-dihydroxy-4′,5′-methylenedioxyisoflavone (DMD) by another P450 class CYP81E1/E7 enzyme known as isoflavone 2′-hydroxylase (I2′H) (Akashi et al., 1998; Liu et al., 2003). The next chemical reaction is catalyzed by isoflavone reductase (IFR), a unique enzyme of the pathway that introduces chirality in pterocarpan biosynthesis that converts DMD to (3R)-sophorol (Tiemann et al., 1987; Uchida et al., 2017). The next step to IFR is the formation of (3R,4R)-2′-hydroxyisoflavanol from (3R)-sophorol by the help of 2′-hydroxyisoflavanone 4-reductase (I4′R). Pea I4′R is also known as sophorol reductase (SOR), as it specifically converts (3R)-sophorol to (3R,4R)-7,2′-dihydroxy-4′5′-methylenedioxyisoflavanol [cis-(−)-DMDI] (DiCenzo and VanEtten, 2006). RNA-mediated downregulation of IFS and SOR genes in pea resulted in decreased accumulation of (+)-pisatin, which indicates that (+)-pisatin synthesis proceeds through (3R)-sophorol and cis-(−)-DMDI intermediates (Kaimoyo and VanEtten, 2008). The next step to SOR is the catalyzation of isoflav-3-enes synthase (I3S), identified very recently, which converts cis-(−)-DMDI to 7,2′-dihydroxy-4′,5′-methylenedioxyisoflav-3-ene (DMDIF) (Uchida et al., 2020). The enzyme involved in the next step, conversion of achiral DMDIF to (+)-6a-hydroxymaackiain (+)-6a-(HMK), is not yet identified; however, it is believed that (+)-6a-(HMK) is the direct precursor of (+)-pisatin. The final methylation reaction is catalyzed by (+)-6a-hydroxymaackiain 3-O-methyltransferase (HMM), which converts (+)-6a-(HMK) to (+)-pisatin (Wu et al., 1997).
Medicarpin is another important pterocarpan that is formed from formononetin (Supplementary Figure 2). I2′H and IFR catalyze the initial chemical reaction in medicarpin biosynthesis (Paiva et al., 1991). I2′H performs oxidation reaction at 2′-C and produces 2′-hydroxy formononetin, which is then reduced to (−)-vestitone by IFR (Tiemann et al., 1987). Two reactions are catalyzed by vestitone reductase (VR) and 7,2′-dihydroxy-4′-methoxyisoflavanol dehydratase (DMID); and (−)-vestitone is reduced and dehydrated to (−)-medicarpin, a major phytoalexin of alfalfa (Medicago sativa) (Guo et al., 1994).
Coumestrol belongs to the coumestans group of isoflavonoid; and like pterocarpans, these compounds also have two asymmetric carbons, C-6a and C-11a; however, only cis-configurations are sterically possible and present in nature (Whitten et al., 1997). The biosynthetic pathway of coumestrol synthesis is not completely understood; however, few steps have been genetically tested, and the remaining are predicted based on differential gene expression and clustering analysis (Ha et al., 2019; Supplementary Figure 3). The pathway starts from daidzein, and the first two chemical reactions involve the conversion of daidzein to 2′-hydroxydaidzein to (3R)-2′-hydroxydihydrodaidzein catalyzed by I2′H and IFR, respectively (Dewick et al., 1970; Berlin et al., 1972). Dehydration of (3R)-2′-hydroxydihydrodaidzein to 3,9-dihydroxyterocarp-6a-en and following chemical reactions up to coumestrol biosynthesis are NAD(P)-dependent redox reactions catalyzed by unidentified NAD(P)-linked oxidoreductases. Overall, seven genes are predicted to be involved in coumestrol biosynthesis starting from daidzein (Ha et al., 2019). These genes are supposed to encode NAD(P)-linked oxidoreductases, which are responsible for catalyzing NAD(P)-dependent oxidation reactions for coumestrol biosynthesis (Ha et al., 2019).
Genistein and biochanin-A (isoflavones) are also well-known isoflavonoids and are synthesized through the same pathway that starts from naringenin (second route of isoflavonoid biosynthesis) (Figure 4 and Supplementary Figure 4). IFS is responsible for the migration of B-ring from the C-2 position to C-3 position, as mentioned earlier; and the resulting product is 2-hydroxy-2,3-dihydrogenistein (Jung et al., 2000). Like 2,7,4′-trihydroxyisoflavanone, 2-hydroxygenistein is an unstable compound and dehydrated to genistein either spontaneously or with the action of another enzyme, HIDH (Akashi et al., 2005). Genistein is then methylated at the 4′-C position by I4′OMT, and biochanin-A is formed (Liu et al., 2006).
The isoflavonoid biosynthesis pathways are exceptionally complicated, and the overall accumulation of isoflavonoids inside a plant cell depends not only on pathway-specific enzymes but also on their interaction with other enzymes (Burbulis and Winkel-Shirley, 1999). Genes involved in isoflavone synthesis have shown a functional differentiation, which is because of two recent genome-duplication events: a soybean lineage-specific duplication (13 million years ago) and an early legume duplication (59 million years ago) (Schmutz et al., 2010; Chu et al., 2014). In the following paragraph, several factors that influence isoflavonoid biosynthesis in the host plant and the underlying molecular mechanisms are discussed.
Abiotic and biotic factors are key regulators of isoflavonoid pathway genes because anything that upregulated the expression of CHS and IFS will have a strong effect on the overall synthesis and accumulation (Dhaubhadel et al., 2003, 2007; Cheng et al., 2008). This is so because CHS and IFS are important enzymes in isoflavonoid biosynthesis, as the farmer directs the C-flow from the phenylpropanoid pathway to flavonoids, and the latter diverts C-flow from the flavonoid pathway to isoflavonoids. In one study, the authors have reported that the accumulation of isoflavonoids and the expression of four key genes (CHS7, CHS8, IFS1, and IFS2) were increased in soybean plants under high temperature stress (Chennupati et al., 2012). However, scientific evidence to explain a direct positive correlation between temperature, gene expression and accumulation of isoflavonoid content is very few. The correlation between the expression of CHS and IFS genes, isoflavonoid accumulation and effect of biotic and abiotic stresses in soybean has also been investigated (Chen et al., 2009; Devi et al., 2020). In biotic factors, Aspergillus niger has been tested as an elicitor of isoflavonoid biosynthesis at 0.1% concentration and a nearly fourfold increase in IFS1 expression, and 5.9-fold increase in isoflavonoid accumulation has been documented as compared with control plants (Devi et al., 2020). Recently, Murakami and colleagues have demonstrated that the content of daidzein and formononetin is increased following the herbivory of Spodoptera litura and foliar applications of S. litura oral sections (Murakami et al., 2014).
The effect of phytohormones as elicitors for isoflavonoid biosynthesis has also been tested, as plant hormones are extensively studied as signaling molecules involved in defense response to environmental signals in plants (Creelman and Mullet, 1997; Draper, 1997). In one study, effect of salicylic acid (SA) and methyl jasmonate (MJ) was analyzed, and it was noted that SA was more active than MJ. A fivefold increase in the expression of IFS2, onefold increase in the expression of CHS8 and a 4.5-fold increase in isoflavonoid contents were observed as compared with control when SA was applied at the concentration of 10 μM. Similarly, when MJ was applied at the same concentration, a maximum of onefold increase in the expression of IFS1 and 3.75-fold increase in isoflavonoid accumulation were seen over control (Devi et al., 2020). In another recent study, an overall correlation between abscisic acid (ABA) and UV-B-induced isoflavonoid accumulation and its molecular basis have been investigated. ABA along with guanosine-3′,5′-cyclic monophosphate (cGMP) can upregulate the expression of CHS and IFS genes and ultimately result in higher accumulation of isoflavonoids under UV-B treatment in soybean. ABA causes inhibition of type 2C protein phosphate (PP2C) and activation of SNF1-related protein kinase (SnRK) and upregulate the expression of CHS and IFS and finally results in higher accumulation of isoflavonoids in the plant cell (Jiao and Gu, 2019). Overall, the role of the IFS enzyme is more critical than that of CHS, as plants always try to accumulate transcript of IFS in higher concentrations than CHS. Therefore, it is speculated that either the IFS enzyme has translational regulation or it is enzymatically very slow and has a short lifespan.
Some transcription factors (TFs) of the MYB class involved in isoflavonoid biosynthesis have been identified. The first interesting candidate is R1-type MYB TF GmMYB176, which regulates the expression of CHS8 and thus controls the overall flavonoid/isoflavonoid contents in plants (Yi et al., 2010). Similarly, the R2R3-type MYB TFs GmMYB39 and GmMYB100 are reported to downregulate the expression of structural genes of the isoflavonoids pathway, thus negatively controlling isoflavonoid biosynthesis in plants. Recently, a soybean TF, GmMYB29, has been characterized for its positive role in isoflavonoid biosynthesis. Comparative genomic analyses have shown that GmMYB29 has maintained the highly conserved R2R3 domain and small amino acid motif in the C-terminal region, which is related to stress resistance in plants (Chu et al., 2017). The expression pattern of GmMYB29 is similar to that of IFS2, which supports the hypothesis that the GmMYB29 is a regulator of the IFS2 gene (Höll et al., 2013). A positive correlation between the expression of GmMYB29 and the accumulation of isoflavonoids in different tissues of plants has also been documented (Liu X. et al., 2013; Yan et al., 2015). It is said that the soybean genome has 4,343 predicted TF encoding genes, which are roughly equal to 6.5% of the total number of genes in the plant (Doerge, 2002). Therefore, it seems that more TF involved in isoflavonoid biosynthesis and regulation will be identified and characterized in the future.
Gene expression is also regulated at the post-transcriptional level, which is largely mediated by two small RNA classes: microRNA (miRNA) and short interfering RNA (siRNA) (Khraiwesh et al., 2010). SiRNA-based regulation of the flavonoid pathway was first reported by Tuteja et al. (2009). The authors have reported tissue-specific (seed coat) silencing of the CHS by siRNA in soybean. In Arabidopsis, miRNAs like miR156, miR163, miR393 and miR828 are reported to be involved in the regulation of synthesis of secondary metabolites (Bulgakov and Avramenko, 2015; Gupta et al., 2017a). Additionally, miR156-SPL (Squamosa Promoter Binding Protein like) target pair destabilizes WD40-bHLH-MYB TF and negatively regulates anthocyanin biosynthesis in plants (Gou et al., 2011). The role of miRNA in the secondary metabolism of several medicinal plants like Catharanthus roseus, Papaver somniferum, and Picrorhiza kurroa have also been reported (Hao et al., 2012; Pani and Mahapatra, 2013; Boke et al., 2015; Vashisht et al., 2015). As most of the isoflavone biosynthesis and accumulation occur in the developing seed, five new miRNA and their target genes that were predicted to be involved in isoflavonoid biosynthesis have been identified (Gupta et al., 2017b). Interestingly, expression correlation analysis of Gma-miRNA26/28 and their corresponding targets 4CL and I7′OMT genes have shown a perfect negative correlation across all stages and genotypes studied so far. It means that decreasing the expression of Gma-miRNA26 has resulted in increase of the expression of 4CL, which could potentially divert the flux toward the synthesis of phenylpropanoid pathways and also resulted in increased accumulation of total isoflavone in the respective plant (Gupta et al., 2017b).
Isoflavonoids are naturally mostly produced in legumes or pea family; however, bulk production of isoflavonoid faces some challenges due to their low content in parent plants. Therefore, alternative cost-effective production platforms are required to meet the growing demand and to ensure availability throughout the year. Model plants like Nicotiana benthamiana are considered useful transient expression hosts, as necessary cofactors and substrate pool are likely to be maintained in planta (Cravens et al., 2019). However, genetic manipulations, even for model plants, are difficult and slow as compared with microorganisms, and thus, microbial hosts are usually preferred.
Microorganisms are excellent production hosts for plant natural products (PNPs) due to low genetic complexity, ease in genetic manipulation, availability of genetic tool kit and genetic tractability. During the last couple of decades, researchers have engineered artificial isoflavonoid biosynthesis pathways in Saccharomyces cerevisiae and Escherichia coli (Cress et al., 2013; Table 2). At least seven enzymes (PAL/TAL, 4CL, CHS, CHI, CHR, IFS, and IFD) are required for the de novo synthesis of parent isoflavonoids: daidzein and genistein. The expression and functionality of plant P450 class enzymes are not optimal in heterologous hosts; therefore, the heterologous synthesis of isoflavonoids is challenging.
Among the many production chassis available, S. cerevisiae is most commonly used for heterologous PNP synthesis, as being eukaryote, it has most of the cellular compartments found in the plant cell. Availability of genetic tool kit, high rate of genetic recombination, ease in genomic manipulation and integration along with generally recognised as safe (GRAS) status are some of the additional benefits for using yeast as a heterologous host (Cravens et al., 2019).
Initial attempts to synthesize isoflavonoids in yeast were focused to convert a flavonoid precursor into the corresponding isoflavonoid. Akashi et al. were the first to express the IFS gene from licorice in yeast and have successfully synthesized genistein from naringenin (Akashi et al., 1999). Kim et al. have reported that an engineered yeast strain can produce up to 20 mg/L of genistein when the necessary IFS/CPR is expressed and naringenin is added in the medium (Kim et al., 2005). De novo synthesis (construction of the complete pathway in the engineered microbial host) of isoflavonoids has also been achieved (Trantas et al., 2009; Rodriguez et al., 2017). An engineered yeast strain, overexpressing seven heterologous enzymes, was able to produce genistein from different precursors added in the growth medium. The final yield of genistein was 7.7 mg/L (28.5 μM), 0.14 mg/L (0.52 μM), and 0.1 mM (0.4 μM) when 0.5 mM of naringenin, 1 mM of p-coumaric acid and 10 mM of phenylalanine were added in the media, respectively (Trantas et al., 2009). An interesting case was the synthesis of quercetin, which was not detected in the medium when phenylalanine was used as a precursor (eight heterologous enzyme reactions), but the engineered strain was able to synthesize 0.26 mg/L (0.9 μM) of quercetin when p-coumaric acid was added in the medium (six heterologous enzyme reactions) (Trantas et al., 2009). Recently, with further advancement in knowledge, the synthesis of quercetin has been achieved (eight heterologous enzyme reactions); however, it is speculated that de novo synthesis of other isoflavonoids is difficult and requires multiple rounds of genetic engineering (Rodriguez et al., 2017). Employing yeast as a production host for the heterologous synthesis of isoflavonoids has distinctive advantages in the functional expression of plant P450 class enzymes; however, further knowledge and optimization are still needed for the synthesis of key isoflavonoids.
E. coli has also been extensively used for heterologous biosynthesis of natural products due to the availability of genetic tools, ease in the engineering of native biochemical pathways, simple cultivation techniques and rapid growth (Yang et al., 2020). However, E. coli was not the first choice of genetic engineers for the heterologous biosynthesis of isoflavonoids due to issues in the expression of IFS and other plant P450 class enzymes.
The functional expression of IFS was the first bottleneck for the synthesis of isoflavonoids in prokaryotic hosts; therefore, efforts were put forward to express plant P450 enzymes in E. coli. Kim et al. (2009) were the first to engineer IFS of red clover by in-frame fusion with a CPR from rice, and the resulting chimeric protein was able to synthesize genistein (up to 15.1 mg/L) from naringenin. In the same year, Leonard et al. have also expressed an engineered chimeric IFS in E. coli and have successfully converted naringenin and liquiritigenin into genistein and daidzein, respectively. The IFS gene from Glycine max and CPR from C. roseus were used, and the membrane-spanning regions were replaced with a mammalian leader sequence (Leonard et al., 2009). Co-cultivation of S. cerevisiae and E. coli strategy was also exploited to boost the final yield of the desired isoflavonoid. In one study, when a naringenin-producing E. coli strain was co-cultured with IFS expressing S. cerevisiae strain, up to 6 mg/L of genistein was detected in the medium (Katsuyama et al., 2007). In recent years, significant progress has been made in heterologous product synthesis, and many PNPs are now being produced and commercialized. Some of these developments are discussed in section “Advance Genetic Engineering Approaches for ex-planta Isoflavonoids Biosynthesis.”
Significant advancement in biotechnology and synthetic biology has been made during the last couple of decades, and heterologous biosynthesis of many PNPs has been successfully achieved. As discussed in section “Heterologous Biosynthesis of Isoflavonoids,” many attempts have also been made to synthesize isoflavonoids in the microbial host, as heterologous synthesis has several advantages over natural and organic synthesis. Some of these are cost-effectiveness, environmentally friendly, on-demand production and being easy to operate, and it is also a desired technology in the green economy initiative. Therefore, in the following section, some advancements in synthetic biology and metabolic engineering are discussed, which will be helpful to produce industrially acceptable titre, rate and yield (TRY) of heterologous synthesis of most of the key isoflavonoids.
Most of the enzymes involved in the synthesis of isoflavonoids are yet to be characterized; thus, identification and characterization of specific and highly efficient enzymes are a key requirement for the successful heterologous biosynthesis of isoflavonoids. Increasing availability of genetic information is helping enzyme identification and characterization efforts, and genome sequencing projects will speed up the process (One Thousand Plant Transcriptomes Initiative, 2019). If genome sequences are available, it becomes very easy to identify or discover new enzymes by comparing plants that produce or lack a specific compound, which will then help us to explain which enzyme might be the candidate for the biosynthesis of that compound. Following the gene identification, approaches like codon optimization, promoter and other regulatory sequence selections and, finally, protein engineering can be used to improve the enzyme activity and expression level (Lee et al., 2016). Therefore, engineering of the heterologous host that can produce an isoflavonoid of interest at industrially acceptable yield is not easy and straightforward.
As mentioned in Subsection “Isoflavonoid biosynthesis in plants,” enzymes involved in the synthesis of key isoflavonoids are identified but not well characterized, and suboptimal expression of an enzyme results in bottlenecks in a diversion of C-flux from central metabolism to a final product (Supplementary Table 1). Enzyme engineering can help to address some of the issues of low expression and/or low activity of plant-origin enzymes in a heterologous host such as issues of incorrect folding, feedback inhibition and suboptimal pH (Cravens et al., 2019). Enzyme localization by either synthetic or RNA-based scaffolds is a powerful strategy under enzyme engineering approaches. An interesting example in this regard is the use of a synthetic scaffold to cluster a group of three enzymes of the mevalonate pathway, and 77-fold increase in the final product was observed (Dueber et al., 2009). Together with this, protein fusion can effectively reduce pathway competition by bringing the active sites of enzymes near each other and facilitate product channeling. A truncated flavonoid 3′-hydroxylase (tF3′H), a plant P450 class enzymes and a truncated P450 reductase (tCPR) were expressed together as a fusion protein, and successful synthesis of eriodictyol was achieved in E. coli (Zhu et al., 2014). Directed evolution is another approach that can help to increase enzyme activity, stability and substrate specificity. Directed evolution becomes even easier if the relationship between enzyme structure, sequence and function are relatively well established.
The chemical space accessed by the heterologous pathway can be expanded to a range of new products by adding additional enzymes in the pathway. In this way, it is easy to transform natural products into their halogenated, hydroxylated, methylated and glycosylated forms by using an enzyme that can accept that natural product as a substrate. Additionally, protein engineering techniques can help to engineer existing enzymes to accept the molecule of interest, as is demonstrated for halogenase enzyme (Payne et al., 2015). The possibility to synthesize novel derivatives is another interesting opportunity. Once a pathway has been established, it is easy to add, replace or remove any pathway enzyme to produce novel derivatives.
Techniques used to construct metabolic pathway in the heterologous host are extensively reviewed in the literature and includes methods for multigene genomic integration, gene editing and combinatorial enzyme expression (Li et al., 2019). Recent advances in genome engineering make it possible to establish heterologous pathways in previously un-cultivatable organisms. The introduction of the CRISPR/Cas9 system for genome engineering has revolutionized metabolic engineering, as the engineering efficiency of CRISPR/Cas9 sometimes reaches 100% in E. coli (Cho et al., 2018). CRISPR/Cas9 variants such as catalytically dead CRISPR-associated protein 9 (dCas9) are even more useful, as they are helpful to control gene expression and to divert metabolic C-flow toward the product of interest. However, approaches to engineer a metabolic pathway in a heterologous host are not yet clear and straightforward.
Modular pathway engineering approaches are helping the researchers to get a higher titer of the product of interest. This approach involves the engineering of a parent strain to increase substrate supply, improvement in the overall flux of the selected pathway and elimination or downregulation of side products. The first interesting example in this regard is the development of a yeast strain for high p-coumaric acid production, as it is an important intermediate in the isoflavonoid pathway. Rodriguez and colleagues have developed a yeast strain capable of producing 1.9 g/L of p-coumaric acid through a combination of six genetic changes in the central metabolism (Rodriguez et al., 2015). As discussed in section “Heterologous Biosynthesis of Isoflavonoids,” the expression of CHS and IFS is not optimal, and to optimize the overall flux of the pathway, researchers have concluded that the optimal copy number for CHS and IFS is 5 and 2, respectively (Lv et al., 2019). Following further optimization of pH and carbon/nitrogen ratio (C/N), the engineered strain was capable of producing 252.4 mg/L of naringenin from glucose in a shake flask culture (Lv et al., 2019). Side product formation, i.e., phloretic acid formation via reduction of p-coumaroyl CoA, is also a hurdle in getting a commercially acceptable yield, as it results in metabolic C loss. Researchers have finally identified the enzyme, enoyl reductase Tsc13, responsible for phloretic acid formation. However, Tsc13 is an essential enzyme and cannot be deleted. Therefore, yeast TSC13 is replaced with a plant TSC13, and the unwanted side reaction is eliminated while retaining the natural function of Tsc13 as such (Lehka et al., 2017). Thus, significant progress has been undertaken to engineer a strain able to produce a commercially acceptable yield of an isoflavonoid molecule of interest.
The development of bio-foundries is considered revolutionary progress in pathway construction and validation (Chao et al., 2017). A bio-foundry is a collection of wet lab robotics and software developed to systematize the construction, assembly and testing of pathway parts in the host strain. Until now, bio-foundry-based approaches are used to construct only a short pathway (< 5 enzymes) or already validated pathways (Casini et al., 2018). It is not clear when enzyme discovery and characterization will be automated for long pathways (> 5 enzymatic pathway), as most of the approaches are custom-made up to now; however, the potential is huge, and once optimized, such automation will revolutionize metabolic engineering projects.
It is now possible to transform and express over 25 genes in a single strain as demonstrated by Li et al. (2018). However, it is demonstrated that when the number of genes required for the biosynthesis of a compound is increased, the performance of a heterologous system is decreased, and strain optimization becomes difficult and laborious (Trantas et al., 2009). To address such issues, co-culture approach, in which two or more organisms expressing complementary genes are grown together, is an interesting and useful strategy.
The co-culture approach (growing two or more organisms having different modules of the same pathway together) can help metabolic engineering in multiple ways. The effectiveness of co-culturing approach is the most obvious in the strain re-optimization process, as it becomes easy to identify and manipulate the issue of heterologous expression in a strain expression in only a part/module of the complete pathway as compared with traditional monoculture approach (Jones and Koffas, 2016). In a traditional monoculture approach, an extension of an established heterologous pathway requires additional genes to be transformed and expressed in the previously engineered strain. This might destabilize the parent strain in a genetic or fermentation perspective and demands further rounds of optimization. Genetic re-optimization is laborious work, and sometimes, it becomes very difficult to regain the previously achieved fluxes (Wu et al., 2016). However, in poly-culture approaches, previous genetic optimizations are preserved on the one hand, and establishment/optimization of a new module into a new strain are easy (as compared with already engineered strain) on the other hand; and finally, few fermentation optimization steps are required to adjust the new strain in co-culture (Li et al., 2018). Together with this, co-culturing can also help to address the issue of the expression of plant P450 class enzymes in prokaryotic hosts, and in this way, the potential of prokaryotic systems can be used for the synthesis of isoflavonoids of interest. For example, a tyrosine-producing E. coli and naringenin producing yeast strain (using D-xylose as a C source) were co-cultured for naringenin biosynthesis. The optimized co-culture was able to produce up to 21.16 mg/L of naringenin from simple sugars, and the final titer was eightfolds higher than the monoculture of the engineered yeast strain (Zhang et al., 2017).
Detection and quantification of heterologous products in the microbial host by using genetic biosensors is an exciting area of research and advantages offered by co-culture opportunities have increased their importance (de Frias et al., 2018). For example, a naringenin-sensitive TF FdeR (from Herbaspirillum seropedicae) when combined with a green fluorescent protein (GFP) can serve as genetically encoded biosensor and helps in the detection and reporting of intracellular naringenin level (Siedler et al., 2014). Another interesting example is the development of the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) method, which can activate the expression of downstream genes following activation by naringenin. A nearly threefold increase in the expression of reporter genes in E. coli strain expressing the riboswitch was noted when 200 mg/L of naringenin was added in the media (Jang et al., 2017). In another study, an RNA riboswitch-based biosensor module was used to control the growth of naringenin-responsive E. coli strains grown together in a co-culture system. A positive correlation was seen between the naringenin production (by one strain) and the expression of a reporter gene (in the second strain) (Xiu et al., 2017). The authors have concluded that a naringenin-responsive biosensor has helped the second strain to control metabolic burden, and it also allowed the authors to do module-specific strain optimization comparatively easily and efficiently.
The isoflavonoids are an important group of plant secondary metabolites that play multiple significant roles in plants as well as in humans. The diversity of isoflavonoids, as well as other PNPs, is a result of an ongoing process of evolution that has generated rich and diverse enzyme sets and will continue to do so in the future. Genome sequencing projects for the identification of candidate genes and TFs will continue to produce a wealth of knowledge for isoflavonoid biosynthesis and engineering in plants.
In plants, the isoflavonoid-mediated natural plant defense mechanisms are a potential tool to address pre-harvest crop losses due to pests and diseases, along with reducing the use of toxic and expensive pesticides. Therefore, an attractive avenue for further research is to engineer beneficial isoflavonoid biosynthesis pathways into non-endogenous commercial crop plants. However, this approach has technical and social limitations, in terms of the complex interactions and interdependencies of functional genes with added challenges from the public perception and regulatory requirements for the use of genetically modified organisms.
The widespread availability of isoflavonoids for use in agricultural, nutraceutical and pharmaceutical applications is limited due to low yield in plants; therefore, alternative approaches such as ex-planta biosynthesis synthetic methods are being investigated. Most of the reports published so far have used E. coli and S. cerevisiae as heterologous hosts; however, interest in using other available microbial chassis is growing. Therefore, the identification and engineering of enzymes and other genetic components, as well as the exploration of new hosts, will set the direction of future research in the heterologous synthesis of isoflavonoids. Developments in few key areas mentioned in Box 1 will pave the way of successful biosynthesis of isoflavonoids in the near future.
BOX 1. Emerging developments in synthetic biology approaches for heterologous flavonoid biosynthesis.
| Current challenges | Emerging developments |
| Characterization of flavonoid biosynthetic pathways and gene identification | Genome sequencing projects and machine learning approaches will help in ✓ Identification of microbial homologous of plant enzymes ✓ Annotation and characterization of genes from plant hosts |
| Slow enzyme kinetics | Protein engineering and directed evolution will help in ✓ Enhancing and optimizing enzyme activity and stability in prokaryotic systems ✓ Engineering binding epitopes for enhanced pathway partner protein interactions |
| Limitations in metabolic engineering | CRISPR/Cas9-based genomic integration approaches will help in ✓ Integration of natural product biosynthetic pathways into non-endogenous microorganisms ✓ Manipulation of microbial chassis ✓ Use of inexpensive microbial feed stock to reduce synthesis cost |
| Co-culturing approaches will help to ✓ Manage gene expression, reduce metabolic burden and allow rapid testing of new biosynthetic pathways |
|
The successful development of commercially scalable ex-planta production platforms for isoflavonoids will have the further benefit of opening exciting avenues toward the biosynthesis and exploitation of alternative PNPs. The realization of these efforts will pave the way toward future economically sustainable, socially beneficial, green-modular bioindustries.
MS, SRS, and PK conceived, designed, and wrote the initial draft of this review article. SS and PK reviewed and edited the manuscript. All authors have read and approved the contents of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
MS acknowledges the University of Western Australia for providing international fee scholarship and University Postgraduate Award. PK is supported by the University of Western Australia with additional research funding from Ex Planta Pty Ltd. toward Bio-Synthesis of Isoflavones—Formononetin (FMN) Proof of Concept project. We gratefully acknowledge the computational resources and support from the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.673270/full#supplementary-material
Adjakly, M., Ngollo, M., Boiteux, J.-P., Bignon, Y.-J., Guy, L., and Bernard-Gallon, D. (2013). Genistein and daidzein: different molecular effects on prostate cancer. Anticancer Res. 33, 39–44.
Ahuja, K., and Mamtani, K. (2019). Industry Analysis Report, Regional Outlook, Application Development Potential, Price Trends, Competitive Market Share & Forecast, 2019–2025. Selbyville, DE: Global Market Insights.
Akashi, T., Aoki, T., and Ayabe, S. (1998). CYP81E1, a cytochrome P450 cDNA of licorice (Glycyrrhiza echinata L.), encodes Isoflavone 2′-hydroxylase. Biochem. Biophys. Res. Commun. 251, 67–70. doi: 10.1006/bbrc.1998.9414
Akashi, T., Aoki, T., and Ayabe, S. (1999). Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 121, 821–828. doi: 10.1104/pp.121.3.821
Akashi, T., Aoki, T., and Ayabe, S. (2005). Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol. 137, 882–891. doi: 10.1104/pp.104.056747
Al-Maharik, N. (2019). Isolation of naturally occurring novel isoflavonoids: an update. Nat. Prod. Rep. 36, 1156–1195. doi: 10.1039/c8np00069g
Aoki, T., Akashi, T., and Ayabe, S. (2000). Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J. Plant Res. 113:475. doi: 10.1007/pl00013958
Berlin, J., Dewick, P. M., Barz, W., and Grisebach, H. (1972). Biosynthesis of coumestrol in Phaseolus aureus. Phytochemistry 11, 1689–1693. doi: 10.1016/0031-9422(72)85020-9
Boke, H., Ozhuner, E., Turktas, M., Parmaksiz, I., Ozcan, S., and Unver, T. (2015). Regulation of the alkaloid biosynthesis by mi RNA in opium poppy. Plant Biotechnol. J. 13, 409–420. doi: 10.1111/pbi.12346
Bossard, C., Busson, M., Vindrieux, D., Gaudin, F., Machelon, V., Brigitte, M., et al. (2012). Potential role of estrogen receptor beta as a tumor suppressor of epithelial ovarian cancer. PLoS One 7:e44787. doi: 10.1371/journal.pone.0044787
Breinholt, V., and Larsen, J. C. (1998). Detection of weak estrogenic flavonoids using a recombinant yeast strain and a modified MCF7 cell proliferation assay. Chem. Res. Toxicol. 11, 622–629. doi: 10.1021/tx970170y
Bulgakov, V. P., and Avramenko, T. V. (2015). New opportunities for the regulation of secondary metabolism in plants: focus on microRNAs. Biotechnol. Lett. 37, 1719–1727. doi: 10.1007/s10529-015-1863-8
Burbulis, I. E., and Winkel-Shirley, B. (1999). Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. 96, 12929–12934. doi: 10.1073/pnas.96.22.12929
Cano, A., and García-Pérez, M. Á, and Tarín, J. J. (2010). Isoflavones and cardiovascular disease. Maturitas 67, 219–226. doi: 10.1016/j.maturitas.2010.07.015
Casini, A., Chang, F.-Y., Eluere, R., King, A. M., Young, E. M., Dudley, Q. M., et al. (2018). A pressure test to make 10 molecules in 90 days: external evaluation of methods to engineer biology. J. Am. Chem. Soc. 140, 4302–4316. doi: 10.1021/jacs.7b13292
Chan, K. K., Siu, M. K., Jiang, Y., Wang, J., Leung, T. H., and Ngan, H. Y. (2018). Estrogen receptor modulators genistein, daidzein and ERB-041 inhibit cell migration, invasion, proliferation and sphere formation via modulation of FAK and PI3K/AKT signaling in ovarian cancer. Cancer Cell Int. 18, 1–14. doi: 10.3892/etm.2020.9238
Chao, R., Mishra, S., Si, T., and Zhao, H. (2017). Engineering biological systems using automated biofoundries. Metab. Eng. 42, 98–108. doi: 10.1016/j.ymben.2017.06.003
Chen, H., Seguin, P., Archambault, A., Constan, L., and Jabaji, S. (2009). Gene expression and isoflavone concentrations in soybean sprouts treated with chitosan. Crop Sci. 49, 224–236. doi: 10.2135/cropsci2007.09.0536
Chen, L., Teng, H., Jia, Z., Battino, M., Miron, A., Yu, Z., et al. (2018). Intracellular signaling pathways of inflammation modulated by dietary flavonoids: the most recent evidence. Crit. Rev. Food Sci. Nutr. 58, 2908–2924. doi: 10.1080/10408398.2017.1345853
Cheng, H., Yu, O., and Yu, D. (2008). Polymorphisms of IFS1 and IFS2 gene are associated with isoflavone concentrations in soybean seeds. Plant Sci. 175, 505–512. doi: 10.1016/j.plantsci.2008.05.020
Chennupati, P., Seguin, P., Chamoun, R., and Jabaji, S. (2012). Effects of high-temperature stress on soybean isoflavone concentration and expression of key genes involved in isoflavone synthesis. J. Agric. Food Chem. 60, 12421–12427. doi: 10.1021/jf3036319
Cho, C.-H., Jung, Y. S., Nam, T. G., Rha, C.-S., Ko, M.-J., Jang, D., et al. (2020). pH-adjusted solvent extraction and reversed-phase HPLC quantification of isoflavones from soybean (Glycine max (L.) Merr.). J. Food Sci. 85, 673–681. doi: 10.1111/1750-3841.15051
Cho, S., Shin, J., and Cho, B.-K. (2018). Applications of CRISPR/Cas system to bacterial metabolic engineering. Int. J. Mol. Sci. 19:1089. doi: 10.3390/ijms19041089
Chu, S., Wang, J., Cheng, H., Yang, Q., and Yu, D. (2014). Evolutionary study of the isoflavonoid pathway based on multiple copies analysis in soybean. BMC Genet. 15:76. doi: 10.1186/1471-2156-15-76
Chu, S., Wang, J., Zhu, Y., Liu, S., Zhou, X., Zhang, H., et al. (2017). An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PLoS Genet. 13:e1006770. doi: 10.1371/journal.pgen.1006770
Clemens, S., and Barz, W. (1996). Cytochrome P450-dependent methylenedioxy bridge formation in Cicer arietinum. Phytochemistry 41, 457–460. doi: 10.1016/0031-9422(95)00618-4
Cravens, A., Payne, J., and Smolke, C. D. (2019). Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 10:2142.
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Biol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
Cress, B. F., Linhardt, R. J., and Koffas, M. A. (2013). Isoflavonoid production by genetically engineered microorganisms. Nat. Prod. Phytochem. Bot. Metab. Alkaloids Phenolics Terpenes 2, 1647–1681. doi: 10.1007/978-3-642-22144-6_53
Cruickshank, I. A. M., and Perrin, D. R. (1960). Isolation of a phytoalexin from Pisum sativum L. Nature 187, 799–800. doi: 10.1038/187799b0
Dagdemir, A., Durif, J., Ngollo, M., Bignon, Y.-J., and Bernard-Gallon, D. (2013). Histone lysine trimethylation or acetylation can be modulated by phytoestrogen, estrogen or anti-HDAC in breast cancer cell lines. Epigenomics 5, 51–63. doi: 10.2217/epi.12.74
Dakora, F. D., and Phillips, D. A. (1996). Diverse functions of isoflavonoids in legumes transcend anti-microbial definitions of phytoalexins. Physiol. Mol. Plant Pathol. 49, 1–20. doi: 10.1006/pmpp.1996.0035
Das, S., Sharangi, A. B., Egbuna, C., Jeevanandam, J., Ezzat, S. M., Adetunji, C. O., et al. (2020). “Health benefits of isoflavones found exclusively of plants of the fabaceae family,” in Functional Foods and Nutraceuticals, eds C. Egbuna and G. Dable Tupas (Cham: Springer), 473–508. doi: 10.1007/978-3-030-42319-3_22
de Frias, U. A., Pereira, G. K. B., Guazzaroni, M.-E., and Silva-Rocha, R. (2018). Boosting secondary metabolite production and discovery through the engineering of novel microbial biosensors. BioMed Res. Int. 2018:7021826.
de Lima, P. F., Colombo, C. A., Chiorato, A. F., Yamaguchi, L. F., Kato, M. J., and Carbonell, S. A. M. (2014). Occurrence of isoflavonoids in Brazilian common bean germplasm (Phaseolus vulgaris L.). J. Agri. Food Chem. 62, 9699–9704. doi: 10.1021/jf5033312
Devi, M. A., Kumar, G., and Giridhar, P. (2020). Effect of biotic and abiotic elicitors on isoflavone biosynthesis during seed development and in suspension cultures of soybean (Glycine max L.). 3 Biotech 10:98.
Dewick, P. M., Barz, W., and Grisebach, H. (1970). Biosynthesis of coumestrol in Phaseolus aureus. Phytochemistry 9, 775–783. doi: 10.1016/s0031-9422(00)85180-8
Dhaubhadel, S., Gijzen, M., Moy, P., and Farhangkhoee, M. (2007). Transcriptome analysis reveals a critical role of CHS7 and CHS8 genes for isoflavonoid synthesis in soybean seeds. Plant Physiol. 143, 326–338. doi: 10.1104/pp.106.086306
Dhaubhadel, S., McGarvey, B. D., Williams, R., and Gijzen, M. (2003). Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol. Biol. 53, 733–743. doi: 10.1023/b:plan.0000023666.30358.ae
DiCenzo, G. L., and VanEtten, H. D. (2006). Studies on the late steps of (+) pisatin biosynthesis: evidence for (−) enantiomeric intermediates. Phytochemistry 67, 675–683. doi: 10.1016/j.phytochem.2005.12.027
Dillon, F. M., Chludil, H. D., and Zavala, J. A. (2017). Solar UV-B radiation modulates chemical defenses against Anticarsia gemmatalis larvae in leaves of field-grown soybean. Phytochemistry 141, 27–36. doi: 10.1016/j.phytochem.2017.05.006
Dixon, R. A. (1999). “Isoflavonoids: biochemistry, molecular bilolgy, and biological functions,” in Comprehensive Natural Products Chemistry. Polyketides and Other Secondary Metabolites Including Fatty Acids and Their Derivative, Vol. 1, ed. U. Sankawa (Amsterdam: Elsevier), 773–823. doi: 10.1016/b978-0-08-091283-7.00030-8
Dixon, R. A., and Steele, C. L. (1999). Flavonoids and isoflavonoids–a gold mine for metabolic engineering. Trends Plant Sci. 4, 394–400. doi: 10.1016/s1360-1385(99)01471-5
Dixon, R. A., and Sumner, L. W. (2003). Legume natural products: understanding and manipulating complex pathways for human and animal health. Plant Physiol. 131, 878–885. doi: 10.1104/pp.102.017319
Djuric, Z., Chen, G., Doerge, D. R., Heilbrun, L. K., and Kucuk, O. (2001). Effect of soy isoflavone supplementation on markers of oxidative stress in men and women. Cancer Lett. 172, 1–6. doi: 10.1016/s0304-3835(01)00627-9
Doerge, R. W. (2002). Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 3, 43–52. doi: 10.1038/nrg703
Draper, J. (1997). Salicylate, superoxide synthesis and cell suicide in plant defence. Trends Plant Sci. 2, 162–165. doi: 10.1016/s1360-1385(97)01030-3
Du, H., Huang, Y., and Tang, Y. (2010). Genetic and metabolic engineering of isoflavonoid biosynthesis. Appl. Microbiol. Biotechnol. 86, 1293–1312. doi: 10.1007/s00253-010-2512-8
Dueber, J. E., Wu, G. C., Malmirchegini, G. R., Moon, T. S., Petzold, C. J., Ullal, A. V., et al. (2009). Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27:753. doi: 10.1038/nbt.1557
Enkerli, J., Bhatt, G., and Covert, S. F. (1998). Maackiain detoxification contributes to the virulence of Nectria haematococca MP VI on chickpea. Mol. Plant Microbe Interact. 11, 317–326. doi: 10.1094/mpmi.1998.11.4.317
Fuentes-Herrera, P. B., Delgado-Alvarado, A., Herrera-Cabrera, B. E., Luna-Guevara, M. L., and Olvera-Hernández, J. I. (2020). Quantification of isoflavones in stems of faba bean (Vicia faba L.). Rev. Fac. Cienc. Agrar. 52, 43–51.
Gou, J.-Y., Felippes, F. F., Liu, C.-J., Weigel, D., and Wang, J.-W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Guo, L., Dixon, R. A., and Paiva, N. L. (1994). Conversion of vestitone to medicarpin in alfalfa (Medicago sativa L.) is catalyzed by two independent enzymes. Identification, purification, and characterization of vestitone reductase and 7, 2′-dihydroxy-4′-methoxyisoflavanol dehydratase. J. Biol. Chem. 269, 22372–22378. doi: 10.1016/s0021-9258(17)31799-4
Gupta, O. P., Karkute, S. G., Banerjee, S., Meena, N. L., and Dahuja, A. (2017a). Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Front. Plant Sci. 8:374. doi: 10.3389/fpls.2017.00374
Gupta, O. P., Nigam, D., Dahuja, A., Kumar, S., Vinutha, T., Sachdev, A., et al. (2017b). Regulation of isoflavone biosynthesis by miRNAs in two contrasting soybean genotypes at different seed developmental stages. Front. Plant Sci. 8:567. doi: 10.3389/fpls.2017.00567
Ha, J., Kang, Y.-G., Lee, T., Kim, M., Yoon, M. Y., Lee, E., et al. (2019). Comprehensive RNA sequencing and co-expression network analysis to complete the biosynthetic pathway of coumestrol, a phytoestrogen. Sci. Rep. 9:1934.
Han, R.-M., Tian, Y.-X., Liu, Y., Chen, C.-H., Ai, X.-C., Zhang, J.-P., et al. (2009). Comparison of flavonoids and isoflavonoids as antioxidants. J. Agric. Food Chem. 57, 3780–3785. doi: 10.1021/jf803850p
Hao, D.-C., Yang, L., Xiao, P.-G., and Liu, M. (2012). Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis. Physiol. Plant. 146, 388–403. doi: 10.1111/j.1399-3054.2012.01668.x
Henderson, V. W., Paganini-Hill, A., Miller, B. L., Elble, R. J., Reyes, P. F., Shoupe, D., et al. (2000). Estrogen for Alzheimer’s disease in women: randomized, double-blind, placebo-controlled trial. Neurology 54, 295–295.
Höll, J., Vannozzi, A., Czemmel, S., D’Onofrio, C., Walker, A. R., Rausch, T., et al. (2013). The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25, 4135–4149. doi: 10.1105/tpc.113.117127
Hsiao, Y.-H., Ho, C.-T., and Pan, M.-H. (2020). Bioavailability and health benefits of major isoflavone aglycones and their metabolites. J. Funct. Foods 74:104164. doi: 10.1016/j.jff.2020.104164
Hu, C., Wong, W.-T., Wu, R., and Lai, W.-F. (2020). Biochemistry and use of soybean isoflavones in functional food development. Crit. Rev. Food Sci. Nutr. 60, 2098–2112. doi: 10.1080/10408398.2019.1630598
One Thousand Plant Transcriptomes Initiative (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679. doi: 10.1038/s41586-019-1693-2
Jang, S., Jang, S., Xiu, Y., Kang, T. J., Lee, S.-H., Koffas, M. A., et al. (2017). Development of artificial riboswitches for monitoring of naringenin in vivo. ACS Synth. Biol. 6, 2077–2085. doi: 10.1021/acssynbio.7b00128
Jiao, C., and Gu, Z. (2019). Cyclic GMP mediates abscisic acid-stimulated isoflavone synthesis in soybean sprouts. Food Chem. 275, 439–445. doi: 10.1016/j.foodchem.2018.09.071
Jones, J. A., and Koffas, M. A. G. (2016). Optimizing metabolic pathways for the improved production of natural products. Methods Enzymol. 575, 179–193. doi: 10.1016/bs.mie.2016.02.010
Jung, W., Yu, O., Lau, S.-M. C., O’Keefe, D. P., Odell, J., Fader, G., et al. (2000). Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18, 208–212. doi: 10.1038/72671
Kaimoyo, E., and VanEtten, H. D. (2008). Inactivation of pea genes by RNAi supports the involvement of two similar O-methyltransferases in the biosynthesis of (+)-pisatin and of chiral intermediates with a configuration opposite that found in (+)-pisatin. Phytochemistry 69, 76–87. doi: 10.1016/j.phytochem.2007.06.013
Katsuyama, Y., Miyahisa, I., Funa, N., and Horinouchi, S. (2007). One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 73, 1143–1149. doi: 10.1007/s00253-006-0568-2
Khraiwesh, B., Arif, M. A., Seumel, G. I., Ossowski, S., Weigel, D., Reski, R., et al. (2010). Transcriptional control of gene expression by microRNAs. Cell 140, 111–122. doi: 10.1016/j.cell.2009.12.023
Kim, B.-G. (2020). Biological synthesis of genistein in Escherichia coli. J. Microbiol. Biotechnol. 30, 770–776. doi: 10.4014/jmb.1911.11009
Kim, C., Lee, S.-G., Yang, W. M., Arfuso, F., Um, J.-Y., Kumar, A. P., et al. (2018). Formononetin-induced oxidative stress abrogates the activation of STAT3/5 signaling axis and suppresses the tumor growth in multiple myeloma preclinical model. Cancer Lett. 431, 123–141. doi: 10.1016/j.canlet.2018.05.038
Kim, D. H., Kim, B. G., Lee, H. J., Lim, Y., Hur, H. G., and Ahn, J.-H. (2005). Enhancement of isoflavone synthase activity by co-expression of P450 reductase from rice. Biotechnol. Lett. 27, 1291–1294. doi: 10.1007/s10529-005-0221-7
Kim, D.-H., Kim, B.-G., Jung, N.-R., and Ahn, J.-H. (2009). Production of genistein from naringenin using Escherichia coli containing isoflavone synthase-cytochrome P450 reductase fusion protein. J. Microbiol. Biotechnol. 19, 1612–1616. doi: 10.4014/jmb.0905.05043
Kim, J.-E., Lee, S.-Y., Jang, M., Choi, H.-K., Kim, J. H., Chen, H., et al. (2017). Coumestrol epigenetically suppresses cancer cell proliferation: coumestrol is a natural haspin kinase inhibitor. Int. J. Mol. Sci. 18:2228. doi: 10.3390/ijms18102228
Kim, K.-M., Park, J.-S., Choi, H., Kim, M.-S., Seo, J.-H., Pandey, R. P., et al. (2018). Biosynthesis of novel daidzein derivatives using Bacillus amyloliquefaciens whole cells. Biocatal. Biotransformation 36, 469–475. doi: 10.1080/10242422.2018.1461212
Ko, K.-P. (2014). Isoflavones: chemistry, analysis, functions and effects on health and cancer. Asian Pac. J. Cancer Prev. 15, 7001–7010. doi: 10.7314/apjcp.2014.15.17.7001
Koirala, N., Pandey, R. P., Thuan, N. H., Ghimire, G. P., Jung, H. J., Oh, T.-J., et al. (2019). Metabolic engineering of Escherichia coli for the production of isoflavonoid-4′-O-methoxides and their biological activities. Biotechnol. Appl. Biochem. 66, 484–493. doi: 10.1002/bab.1452
Kozłowska, A., and Szostak-Wȩgierek, D. (2017). “Flavonoids–food sources, health benefits, and mechanisms involved,” in Bioactive Molecules in Food, eds J.-M. Mérillon and K. G. Ramawat (Cham: Springer), 1–27. doi: 10.1007/978-3-319-54528-8_54-1
Kuiper, G. G., Carlsson, B. O., Grandien, K. A. J., Enmark, E., Häggblad, J., Nilsson, S., et al. (1997). Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138, 863–870. doi: 10.1210/endo.138.3.4979
Kuriyama, I., Takahashi, Y., Yoshida, H., and Mizushina, Y. (2013). Inhibitory effect of isoflavones from processed soybeans on human dna topoisomerase II activity. J. Plant Biochem. Physiol. 1, 106–112.
Lapčík, O. (2007). Isoflavonoids in non-leguminous taxa: a rarity or a rule? Phytochemistry 68, 2909–2916. doi: 10.1016/j.phytochem.2007.08.006
Larose, G., Chênevert, R., Moutoglis, P., Gagné, S., Piché, Y., and Vierheilig, H. (2002). Flavonoid levels in roots of Medicago sativa are modulated by the developmental stage of the symbiosis and the root colonizing arbuscular mycorrhizal fungus. J. Plant Physiol. 159, 1329–1339. doi: 10.1078/0176-1617-00896
Lee, D.-H., Kim, M. J., Park, S.-H., Song, E.-J., Nam, Y.-D., Ahn, J., et al. (2018). Bioavailability of isoflavone metabolites after korean fermented soybean paste (doenjang) ingestion in estrogen-deficient Rats. J. Food Sci. 83, 2212–2221. doi: 10.1111/1750-3841.14214
Lee, H., Kim, B.-G., and Ahn, J.-H. (2014). Production of bioactive hydroxyflavones by using monooxygenase from Saccharothrix espanaensis. J. Biotechnol. 176, 11–17. doi: 10.1016/j.jbiotec.2014.02.002
Lee, P.-G., Kim, J., Kim, E.-J., Jung, E., Pandey, B. P., and Kim, B.-G. (2016). P212A mutant of dihydrodaidzein reductase enhances (S)-equol production and enantioselectivity in a recombinant Escherichia coli whole-cell reaction system. Appl. Environ. Microbiol. 82, 1992–2002. doi: 10.1128/aem.03584-15
Lehka, B. J., Eichenberger, M., Bjørn-Yoshimoto, W. E., Vanegas, K. G., Buijs, N., Jensen, N. B., et al. (2017). Improving heterologous production of phenylpropanoids in Saccharomyces cerevisiae by tackling an unwanted side reaction of Tsc13, an endogenous double-bond reductase. FEMS Yeast Res. 17:fox004. doi: 10.1093/femsyr/fox004
Leonard, E., and Koffas, M. A. (2007). Engineering of artificial plant cytochrome P450 enzymes for synthesis of isoflavones by Escherichia coli. Appl. Environ. Microbiol. 73, 7246–7251. doi: 10.1128/aem.01411-07
Leonard, E., Runguphan, W., O’connor, S., and Prather, K. J. (2009). Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat. Chem. Biol. 5:292. doi: 10.1038/nchembio.160
Li, J., Li, Z., Li, C., Gou, J., and Zhang, Y. (2014). Molecular cloning and characterization of an isoflavone 7-O-glucosyltransferase from Pueraria lobata. Plant Cell Rep. 33, 1173–1185. doi: 10.1007/s00299-014-1606-7
Li, L., Liu, X., Wei, K., Lu, Y., and Jiang, W. (2019). Synthetic biology approaches for chromosomal integration of genes and pathways in industrial microbial systems. Biotechnol. Adv. 37, 730–745. doi: 10.1016/j.biotechadv.2019.04.002
Li, L., Lv, Y., Xu, L., and Zheng, Q. (2015). Quantitative efficacy of soy isoflavones on menopausal hot flashes. Br. J. Clin. Pharmacol. 79, 593–604. doi: 10.1111/bcp.12533
Li, Y., Li, S., Thodey, K., Trenchard, I., Cravens, A., and Smolke, C. D. (2018). Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U.S.A. 115, E3922–E3931.
Liu, C.-J., Deavours, B. E., Richard, S. B., Ferrer, J.-L., Blount, J. W., Huhman, D., et al. (2006). Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell 18, 3656–3669. doi: 10.1105/tpc.106.041376
Liu, C.-J., Huhman, D., Sumner, L. W., and Dixon, R. A. (2003). Regiospecific hydroxylation of isoflavones by cytochrome p450 81E enzymes from Medicago truncatula. Plant J. 36, 471–484. doi: 10.1046/j.1365-313x.2003.01893.x
Liu, L., Chen, X., Hao, L., Zhang, G., Jin, Z., Li, C., et al. (2020). Traditional fermented soybean products: processing, flavor formation, nutritional and biological activities. Crit. Rev. Food Sci. Nutr. 13, 1–19. doi: 10.1080/10408398.2020.1848792
Liu, S., Hsieh, D., Yang, Y.-L., Xu, Z., Peto, C., Jablons, D. M., et al. (2013). Coumestrol from the national cancer Institute’s natural product library is a novel inhibitor of protein kinase CK2. BMC Pharmacol. Toxicol. 14:36. doi: 10.1186/2050-6511-14-36
Liu, X., Yuan, L., Xu, L., Xu, Z., Huang, Y., He, X., et al. (2013). Over-expression of GmMYB39 leads to an inhibition of the isoflavonoid biosynthesis in soybean (Glycine max. L). Plant Biotechnol. Rep. 7, 445–455. doi: 10.1007/s11816-013-0283-2
Lv, Y., Marsafari, M., Koffas, M., Zhou, J., and Xu, P. (2019). Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth. Biol. 8, 2514–2523. doi: 10.1021/acssynbio.9b00193
Mazur, W. (1998). 11 Phytoestrogen content in foods. Baillieres Clin. Endocrinol. Metab. 12, 729–742. doi: 10.1016/S0950-351X(98)80013-X
Mazur, W. M., Duke, J. A., Wähälä, K., Rasku, S., and Adlercreutz, H. (1998). Isoflavonoids and lignans in legumes: nutritional and health aspects in humans. J. Nutr. Biochem. 9, 193–200. doi: 10.1016/S0955-2863(97)00184-8
Megías, C., Cortés-Giraldo, I., Alaiz, M., Vioque, J., and Girón-Calle, J. (2016). Isoflavones in chickpea (Cicer arietinum) protein concentrates. J. Funct. Foods 21, 186–192. doi: 10.1016/j.jff.2015.12.012
Mizushina, Y., Shiomi, K., Kuriyama, I., Takahashi, Y., and Yoshida, H. (2013). Inhibitory effects of a major soy isoflavone, genistein, on human DNA topoisomerase II activity and cancer cell proliferation. Int. J. Oncol. 43, 1117–1124. doi: 10.3892/ijo.2013.2032
Mortensen, A., Kulling, S. E., Schwartz, H., Rowland, I., Ruefer, C. E., Rimbach, G., et al. (2009). Analytical and compositional aspects of isoflavones in food and their biological effects. Mol. Nutr. Food Res. 53, S266–S309.
Murakami, S., Nakata, R., Aboshi, T., Yoshinaga, N., Teraishi, M., Okumoto, Y., et al. (2014). Insect-induced daidzein, formononetin and their conjugates in soybean leaves. Metabolites 4, 532–546. doi: 10.3390/metabo4030532
Nabavi, S. M., Šamec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., et al. (2020). Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnol. Adv. 38:107316. doi: 10.1016/j.biotechadv.2018.11.005
Novelli, S., Gismondi, A., Di Marco, G., Canuti, L., Nanni, V., and Canini, A. (2019). Plant defense factors involved in Olea europaea resistance against Xylella fastidiosa infection. J. Plant Res. 132, 439–455. doi: 10.1007/s10265-019-01108-8
Nwachukwu, I. D., Luciano, F. B., and Udenigwe, C. C. (2013). The inducible soybean glyceollin phytoalexins with multifunctional health-promoting properties. Food Res. Int. 54, 1208–1216. doi: 10.1016/j.foodres.2013.01.024
Okutani, F., Hamamoto, S., Aoki, Y., Nakayasu, M., Nihei, N., Nishimura, T., et al. (2020). Rhizosphere modelling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant Cell Environ. 43, 1036–1046. doi: 10.1111/pce.13708
Paiva, N. L., Edwards, R., Sun, Y., Hrazdina, G., and Dixon, R. A. (1991). Stress responses in alfalfa (Medicago sativa L.) 11. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol. Biol. 17, 653–667. doi: 10.1007/bf00037051
Pani, A., and Mahapatra, R. K. (2013). Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genom. Data 1, 2–6. doi: 10.1016/j.gdata.2013.06.001
Park, G., Baek, S., Kim, J.-E., Lim, T., Lee, C. C., Yang, H., et al. (2015). Flt3 is a target of coumestrol in protecting against UVB-induced skin photoaging. Biochem. Pharmacol. 98, 473–483. doi: 10.1016/j.bcp.2015.08.104
Park, S., Bazer, F. W., Lim, W., and Song, G. (2018). The O-methylated isoflavone, formononetin, inhibits human ovarian cancer cell proliferation by sub G0/G1 cell phase arrest through PI3K/AKT and ERK1/2 inactivation. J. Cell. Biochem. 119, 7377–7387. doi: 10.1002/jcb.27041
Payne, J. T., Poor, C. B., and Lewis, J. C. (2015). Directed evolution of RebH for site-selective halogenation of large biologically active molecules. Angew. Chem. 127, 4300–4304. doi: 10.1002/ange.201411901
Pilsakova, L., Riecanský, I., and Jagla, F. (2010). The physiological actions of isoflavone phytoestrogens. Physiol. Res. 59:651. doi: 10.33549/physiolres.931902
Prasad, S., Phromnoi, K., Yadav, V. R., Chaturvedi, M. M., and Aggarwal, B. B. (2010). Targeting inflammatory pathways by flavonoids for prevention and treatment of cancer. Planta Med. 76, 1044–1063. doi: 10.1055/s-0030-1250111
Preedy, V. R. (2012). Isoflavones: Chemistry, Analysis, Function and Effects. London: Royal Society of Chemistry.
Qi, C., Xie, M., Liang, J., Li, H., Li, Z., Shi, S., et al. (2016). Formononetin targets the MAPK and PI3K/Akt pathways to induce apoptosis in human nasopharyngeal carcinoma cells in vitro and in vivo. Int. J. Clin. Exp. Med. 9, 1180–1189.
Rodriguez, A., Kildegaard, K. R., Li, M., Borodina, I., and Nielsen, J. (2015). Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 31, 181–188. doi: 10.1016/j.ymben.2015.08.003
Rodriguez, A., Strucko, T., Stahlhut, S. G., Kristensen, M., Svenssen, D. K., Forster, J., et al. (2017). Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour. Technol. 245, 1645–1654. doi: 10.1016/j.biortech.2017.06.043
Rosler, J., Krekel, F., Amrhein, N., and Schmid, J. (1997). Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113, 175–179. doi: 10.1104/pp.113.1.175
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183.
Shafiee, G., Saidijam, M., Tavilani, H., Ghasemkhani, N., and Khodadadi, I. (2016). Genistein induces apoptosis and inhibits proliferation of HT29 colon cancer cells. Int. J. Mol. Cell. Med. 5:178.
Siedler, S., Stahlhut, S. G., Malla, S., Maury, J., and Neves, A. R. (2014). Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab. Eng. 21, 2–8. doi: 10.1016/j.ymben.2013.10.011
Silva, L. R., Pereira, M. J., Azevedo, J., Gonçalves, R. F., Valentão, P., de Pinho, P. G., et al. (2013). Glycine max (L.) Merr., Vigna radiata L. and Medicago sativa L. sprouts: A natural source of bioactive compounds. Food Res. Inter. 50, 167–175. doi: 10.1016/j.foodres.2012.10.025
Singh, K. B., Dixit, M., Dev, K., Maurya, R., and Singh, D. (2017). Formononetin, a methoxy isoflavone, enhances bone regeneration in a mouse model of cortical bone defect. Br. J. Nutr. 117, 1511–1522. doi: 10.1017/S0007114517001556
Slade, D., Ferreira, D., and Marais, J. P. (2005). Circular dichroism, a powerful tool for the assessment of absolute configuration of flavonoids. Phytochemistry 66, 2177–2215. doi: 10.1016/j.phytochem.2005.02.002
Steele, C. L., Gijzen, M., Qutob, D., and Dixon, R. A. (1999). Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch. Biochem. Biophys. 367, 146–150. doi: 10.1006/abbi.1999.1238
Stobiecki, M., and Kachlicki, P. (2006).“Isolation and identification of flavonoids,” in The Science of Flavonoids, ed. E. Grotewold (Cham: Springer), 47–69. doi: 10.1007/0-387-28822-8_2
Strange, R. N., Ingham, J. L., Cole, D. L., Cavill, M. E., Edwards, C., Cooksey, C. J., et al. (1985). Isolation of the phytoalexin medicarpin from leaflets of Arachis hypogaea and related species of the tribe Aeschynomeneae. Z. Naturforsch. C 40, 313–316. doi: 10.1515/znc-1985-5-605
Tay, K.-C., Tan, L. T.-H., Chan, C. K., Hong, S. L., Chan, K.-G., Yap, W. H., et al. (2019). Formononetin: a review of its anticancer potentials and mechanisms. Front. Pharmacol. 10:820. doi: 10.3389/fphar.2019.00820
Tiemann, K., Hinderer, W., and Barz, W. (1987). Isolation of NADPH: isoflavone oxidoreductase, a new enzyme of pterocarpan phytoalexin biosynthesis in cell suspension cultures of Cicer arietinum. FEBS Lett. 213, 324–328. doi: 10.1016/0014-5793(87)81515-6
Trantas, E., Panopoulos, N., and Ververidis, F. (2009). Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab. Eng. 11, 355–366. doi: 10.1016/j.ymben.2009.07.004
Tuteja, J. H., Zabala, G., Varala, K., Hudson, M., and Vodkin, L. O. (2009). Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. Plant Cell 21, 3063–3077. doi: 10.1105/tpc.109.069856
Uchida, K., Akashi, T., and Aoki, T. (2017). The missing link in leguminous pterocarpan biosynthesis is a dirigent domain-containing protein with isoflavanol dehydratase activity. Plant Cell Physiol. 58, 398–408. doi: 10.1093/pcp/pcw213
Uchida, K., Aoki, T., Suzuki, H., and Akashi, T. (2020). Molecular cloning and biochemical characterization of isoflav-3-ene synthase, a key enzyme of the biosyntheses of (+)-pisatin and coumestrol. Plant Biotechnol. 37, 301–310. doi: 10.5511/plantbiotechnology.20.0421a
Umeno, A., Horie, M., Murotomi, K., Nakajima, Y., and Yoshida, Y. (2016). Antioxidative and antidiabetic effects of natural polyphenols and isoflavones. Molecules 21:708. doi: 10.3390/molecules21060708
Vashisht, I., Mishra, P., Pal, T., Chanumolu, S., Singh, T. R., and Chauhan, R. S. (2015). Mining NGS transcriptomes for miRNAs and dissecting their role in regulating growth, development, and secondary metabolites production in different organs of a medicinal herb, Picrorhiza kurroa. Planta 241, 1255–1268. doi: 10.1007/s00425-015-2255-y
Veitch, N. C. (2007). Isoflavonoids of the leguminosae. Nat. Prod. Rep. 24, 417–464. doi: 10.1039/b511238a
Veitch, N. C. (2009). Isoflavonoids of the leguminosae. Nat. Prod. Rep. 26, 776–802. doi: 10.1039/b616809b
Veitch, N. C. (2013). Isoflavonoids of the leguminosae. Nat. Prod. Rep. 30, 988–1027. doi: 10.1039/c3np70024k
Ververidis, F., Trantas, E., Douglas, C., Vollmer, G., Kretzschmar, G., and Panopoulos, N. (2007). Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: chemical diversity, impacts on plant biology and human health. Biotechnol. J. Healthcare Nutr. Technol. 2, 1214–1234. doi: 10.1002/biot.200700084
Wasmann, C. C., and VanEtten, H. D. (1996). Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol. Plant Microbe Interact. 9, 793–803. doi: 10.1094/mpmi-9-0793
Wei, P., Liu, M., Chen, Y., and Chen, D.-C. (2012). Systematic review of soy isoflavone supplements on osteoporosis in women. Asian Pac. J. Trop. Med. 5, 243–248. doi: 10.1016/s1995-7645(12)60033-9
Wei, Z., Zu, Y., Fu, Y., Wang, W., Luo, M., Zhao, C., et al. (2013). Ionic liquids-based microwave-assisted extraction of active components from pigeon pea leaves for quantitative analysis. Sep. Purif. Technol. 102, 75–81. doi: 10.1016/j.seppur.2012.09.031
Whitten, P. L., Kudo, S., and Okubo, K. K. (1997). “Isoflavonoids,” in Handbook of Plant and Fungal Toxicants, ed. J. P. Felix D’Mello (Boca Raton, FL: CRC Press), 117–137.
Wu, G., Yan, Q., Jones, J. A., Tang, Y. J., Fong, S. S., and Koffas, M. A. (2016). Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 34, 652–664. doi: 10.1016/j.tibtech.2016.02.010
Wu, Q., Preisig, C. L., and VanEtten, H. D. (1997). Isolation of the cDNAs encoding (+) 6a-hydroxymaackiain 3-O-methyltransferase, the terminal step for the synthesis of the phytoalexin pisatin in Pisum satvium. Plant Mol. Biol. 35, 551–560.
Xiu, Y., Jang, S., Jones, J. A., Zill, N. A., Linhardt, R. J., Yuan, Q., et al. (2017). Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures. Biotechnol. Bioeng. 114, 2235–2244. doi: 10.1002/bit.26340
Yan, J., Wang, B., Zhong, Y., Yao, L., Cheng, L., and Wu, T. (2015). The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 89, 35–48. doi: 10.1007/s11103-015-0349-3
Yang, D., Park, S. Y., Park, Y. S., Eun, H., and Lee, S. Y. (2020). Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 38, 745–765. doi: 10.1016/j.tibtech.2019.11.007
Yi, J., Derynck, M. R., Li, X., Telmer, P., Marsolais, F., and Dhaubhadel, S. (2010). A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 62, 1019–1034.
Zaheer, K., and Humayoun Akhtar, M. (2017). An updated review of dietary isoflavones: nutrition, processing, bioavailability and impacts on human health. Crit. Rev. Food Sci. Nutr. 57, 1280–1293. doi: 10.1080/10408398.2014.989958
Zakłos-Szyda, M., Budryn, G., Grzelczyk, J., Pérez-Sánchez, H., and Żyżelewicz, D. (2020). Evaluation of isoflavones as bone resorption inhibitors upon interactions with receptor activator of nuclear factor-κB ligand (RANKL). Molecules 25:206. doi: 10.3390/molecules25010206
Zhang, W., Liu, H., Li, X., Liu, D., Dong, X.-T., Li, F.-F., et al. (2017). Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae. Eng. Life Sci. 17, 1021–1029. doi: 10.1002/elsc.201700039
Zhu, S., Wu, J., Du, G., Zhou, J., and Chen, J. (2014). Efficient synthesis of eriodictyol from L-tyrosine in Escherichia coli. Appl. Environ. Microbiol. 80, 3072–3080. doi: 10.1128/aem.03986-13
Keywords: synthetic biology, heterologous synthesis, green bioindustries, isoflavonoids, plant secondary metabolites
Citation: Sajid M, Stone SR and Kaur P (2021) Recent Advances in Heterologous Synthesis Paving Way for Future Green-Modular Bioindustries: A Review With Special Reference to Isoflavonoids. Front. Bioeng. Biotechnol. 9:673270. doi: 10.3389/fbioe.2021.673270
Received: 27 February 2021; Accepted: 27 May 2021;
Published: 01 July 2021.
Edited by:
Luan Luong Chu, Phenikaa University, VietnamReviewed by:
Jingyu Wang, Westlake Institute for Advanced Study (WIAS), ChinaCopyright © 2021 Sajid, Stone and Kaur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Parwinder Kaur, cGFyd2luZGVyLmthdXJAdXdhLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.