- State Key Laboratory of Biobased Material and Green Papermaking, School of Bioengineering, Qilu University of Technology (Shandong Academy of Sciences), Jinan, China
Introduction
As the typical species of the genus Serratia, Serratia marcescens is a rod-shaped, facultatively anaerobic, Gram-negative bacterium of the family Enterobacteriaceae (Hejazi and Falkiner, 1997; Matsushita et al., 2009; Gaultier et al., 2018). It has been reported as an opportunistic human pathogen that may cause the hospital-acquired infections (Ferreira et al., 2020). Remarkably, S. marcescens has the ability to produce a series of valuable products, including prodigiosin, chitinase, protease, lipase, nuclease, bacteriocin, surfactant, and wetting agent. The prodigiosin is a bioactive secondary metabolite with many pharmaceutical values such as antimicrobial, algicidal, anticancer, antimalarial, anti-inflammatory, anti-diabetic, and immunomodulatory effects (Atsushi et al., 2014; Darshan and Manonmani, 2015; Arivizhivendhan et al., 2018). Although it could be produced by several bacterial species from the genera Serratia, Pseudoalteromonas, Vibrio, and so on (Lee et al., 2011; Elkenawy et al., 2017), the genus Serratia is well-known as the main prodigiosin producing strains (Li et al., 2015).
Considering the potential applications, S. marcescens has attracted great attentions from many researchers Due to advances in high-throughput sequencing technologies, more and more sequencing projects have been set up and researchers could better understand the function, environmental adaptation and potential application of bacteria. There have been 682 genomes of S. marcescens reported in NCBI (https://www.ncbi.nlm.nih.gov/genome/?term=Serratiamarcescens). However, most of these strains were isolated from the intestines and ecological niches, such as air, soil, water, plants and animals. Here, we firstly isolated strain ZPG19 from the compost generated by aerobic composting of Flammulina velutipes residue collected in Dezhou, Shandong Province, China (37.45°N, 116.37°E). We sequenced and characterized its complete genomes in order to provide a promising resource to conduct the biosynthesis analysis and the molecular investigations of genus Serratia.
Materials and Methods
Genomic DNA Isolation
Strain ZPG19 of S. marcescens was cultured for 24 h in LB media (Tryptone 10 g/L, Yeast extract 5 g/L, NaCl10 g/L). Then genomic DNA was extracted with TIANamp Bacteria DNA Kit (TIANGEN Biotech Co., Ltd, Beijing, China) following the manufacturer's instructions. The quality and quantity of purified genomic DNA were determined by NanoDrop 2000 (Thermo Scientific, MA, USA).
Genome Sequencing, Assembly, and Annotation
The genome of the strain ZPG19 was sequenced at Personal (Shanghai Personal Biotechnology Co., Ltd, China) using two different technologies: Illumina NovaSeq with 400 bp library and the PacBio Sequel with a 20-kb library. The adapters of the 3' end were removed using AdapterRemoval (Schubert et al., 2016). Raw reads were quality filtered and error corrected with SOAPEC (kmer = 17) (Luo et al., 2012). De novo assembly of the read sequences was carried out using the hierarchical genome assembly process workflow (Chin et al., 2016). The annotation of the sequences was carried out using a modified version of the Prokka annotation pipeline, which incorporated Prodigal 2.60, Aragorn, and RNAmmer 1.2 for the prediction of open reading frames, tRNAs, and rRNAs, respectively (Seemann, 2014; Yabe and Fukushima, 2020). The prediction of other ncRNAs was mainly obtained by comparing with Rfam database (Kalfari et al., 2018).
Genome Comparison
We selected the whole sequenced genomes of five S. marcescens strains isolated from different habitats for comparative genomic analysis. Chromosomal genome comparison among strain ZPG19 and these five fully sequenced genomes was carried out by using progressive Mauve genome aligner with Geneious software at the default settings (Kearse et al., 2012).
Direct Link to Deposited Data and Information to Users
The BioProject designations for S. marcescens strain ZPG19 are PRJNA665610. And the raw sequences have been deposited in GenBank under the accession numbers SRR12714697 in September 2020. Strain ZPG19 of S. marcescens is available from the China Center for Type Culture Collection (CTCC) under accession numbers M2019645.
Interpretation of Data Set
General Genome Features
We obtained 8,126,164 raw reads covering a total of 1,212,079,667 bp with 230× genome coverage for strain ZPG19. The complete chromosome contained a circular molecule of 5,269,270 bp with 59.49% G+C content. A total of 5,169 genes were predicted including 4,934 coding DNA sequences (CDSs), 95 tRNA genes, 22 rRNA genes and 118 other non-coding RNA genes. In this work, no plasmid was found. The detailed genomic information of ZPG19 and five S. marcescens strains was list in Table 1.
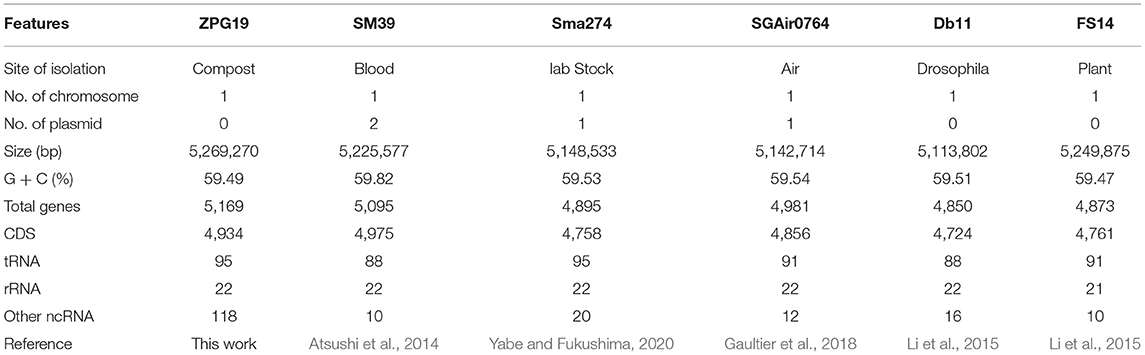
Table 1. Detailed information of Chromosomal genomes from S. marcescens Strain ZPG19 and five reported S. marcescens strains.
Genome Comparison
The whole-genome sizes, GC contents and gene contents of the six Serratia strains were comparable with a slight difference (Table 1). At the same time, the number of genes increased with the enlargement of genome size. The number of plasmids varied in six Serratia strains. SM39 carried two plasmids, Sma274 and SGAir0764 carried one plasmid, respectively. While the other three Serratia strains analyzed had no plasmid.
A global alignment of genome sequences from six Serratia strains was performed using Mauve software and the results showed that the synteny between S. marcescens was not very conserved (Figure 1). Gene rearrangements were commonly observed along the whole stretch of the six chromosomal genomes which was consistent with previous reports (Li et al., 2015).
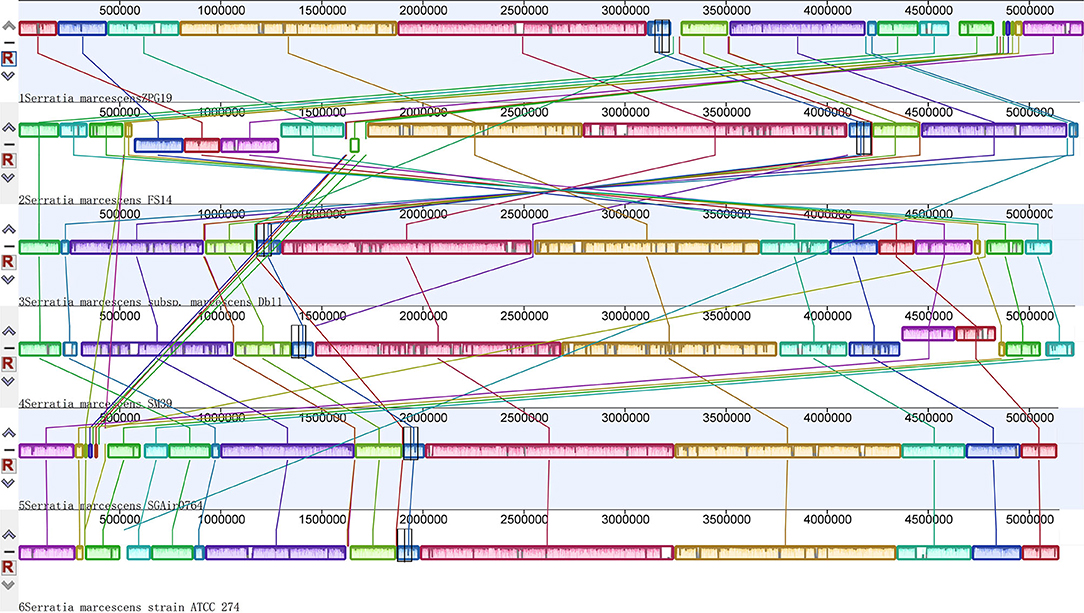
Figure 1. Global multiplealignments of chromosomes from six Serratia strains using progressive MAUVE with default parameters. Colored blocks indicated the genome sequences that aligned to part of another genome and was possibly homologous and internally free from genomic rearrangement (locally collinear blocks). White regions represented sequences that did not aligned and probably contained sequences specific to a particular genome. Blocks below the center line showed regions that aligned in the reverse orientation. The names of the strains were listed at the bottom of the blocks.
Prodigiosin-Producing Enzymes
We have confirmed that the strain ZPG19 could produce prodigiosin via HPLC-MS method. Here, the prodigiosin-producing inventory of ZPG19 was identified including genes for 3-oxoacyl-[acyl-carrier protein] reductase (fabG), [acyl-carrier-protein] S-malonyltransferase (fabD) and pig cluster. The pig cluster contained 14 candidate genes which arranged pigA through to pigN flanked by cueR and copA (Figure 2). We have putatively assigned the products of one gene to the condensing enzymes. Ten genes that encoded proteins required for the biosynthesis of the monopyrroles. Three genes encoding proteins required for the biosynthesis of 4-methoxy-2,2-bipyrrole-5-carboxyaldehyde. This observation is congruent with S. marcescens strain FS14 and S. plymuthica strain AS13 in earlier studies (Li et al., 2015). The order of the genes was conserved among these Serratia species and the corresponding 14 predicted proteins were similar in size and share significant amino acid.
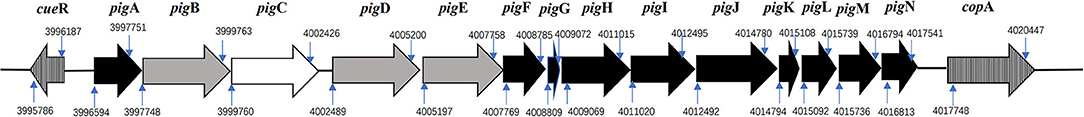
Figure 2. The pig cluster of prodigiosin biosynthesis in strain ZPG19. Genes were symbolized by arrows. The white arrows denoted genes involving in condensing enzymes; black arrows represented genes encoding proteins required for the biosynthesis of the monopyrroles; gray arrows genes encoding proteins required for the biosynthesis of 4-methoxy-2,2-bipyrrole-5-carboxyaldehyde; vertical striped arrows denoted flanking genes cueR and copA; The numbers in small blue arrows indicated the start positions and end positions of each gene.
In conclusion, the complete genome of S. marcescens strain ZPG19 was sequenced and assembled into one chromosome. Comparative genome and sequence analyses showed that rearrangements occurred in six Serratia strains. Genes fabG, fabD, and pig cluster responsible for prodigiosin production were detected in this work.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Wet lab execution was performed by XL. Data processing and handling was performed by XT. The manuscript was written by JingZ. Project planning was conducted by JieZ. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the Foundation of Qilu University of Technology of Cultivating Subject for Biology and Biochemistry (No. 202019) and The Shandong Province Agricultural Major Application Technology Innovation Project (No. 20182130106).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.665077/full#supplementary-material
References
Arivizhivendhan, K. V., Mahesh, M., Boopathy, R., Swarnalatha, S., Regina Mary, R., and Sekaran, G. (2018). Antioxidant and antimicrobial activity of bioactive prodigiosin produces from Serratia marcescens using agricultural waste as a substrate. J. Food Sci. Technol. 55, 2661–2670. doi: 10.1007/s13197-018-3188-9
Atsushi, I., Yutaka, N., Elizabeth, P., Tadasuke, O., Yoshitoshi, O., Keisuke, K., et al. (2014). Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome biology and evolution. Genome Biol. Evol. 6, 2096–2110. doi: 10.1093/gbe/evu160
Chin, C. S., Peluso, P., Sedlazeck, F. J., Nattestad, M., Concepcion, G. T., Clum, A., et al. (2016). Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054. doi: 10.1038/nmeth.4035
Darshan, N., and Manonmani, H. K. (2015). Prodigiosin and its potential applications. J. Food Sci. Technol. 52, 5393–5407. doi: 10.1007/s13197-015-1740-4
Elkenawy, N. M., Yassin, A. S., Elhifnawy, H. N., and Amin, M. A. (2017). Optimization of prodigiosin production by Serratia marcescens using crude glycerol and enhancing production using gammaradiation. Biotechnol Rep. 14, 47–53. doi: 10.1016/j.btre.2017.04.001
Ferreira, L. C., Maul, J. E., Viana, M., Sousa, T., Azevedo, V., Roberts, D. P., et al. (2020). Complete genome sequence of the biocontrol agent Serratia marcescens strain n4–5 uncovers an assembly artefact. Braz. J. Microbiol. 52, 245–250. doi: 10.1007/s42770-020-00382-2
Gaultier, N. E., Junqueira, A. C. M., Uchida, A., Purbojati, R. W., Houghton, J. N. I., Chénard, C., et al. (2018). Complete genome sequence of the bacterium Serratia marcescens SGAir0764, isolated from Singapore air. Genome Announc. 6:e00637-18. doi: 10.1128/genomeA.00637-18
Hejazi, A., and Falkiner, F. R. (1997). Serratia marcescens. J. Med. Microbiol. 46, 903–912. doi: 10.1099/00222615-46-11-903
Kalfari, I., Argasinska, J., Quinones-Olvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., et al. (2018). Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 4, D335–D342. doi: 10.1093/nar/gkx1038
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28,1647–1649. doi: 10.1093/bioinformatics/bts199
Lee, J. S., Kim, Y. S., Park, S., Kim, J., Kang, S. J., Lee, M. H., et al. (2011). Exceptional production of both prodigiosin and cycloprodigiosin as major metabolic constituents by a novel marine bacterium, Zooshikella rubidus S1-1. Appl. Environ. Microbiol. 77, 4967–4973. doi: 10.1128/AEM.01986-10
Li, P. P., Kwok, A. H. Y., Jiang, J. W., Ran, T. T., Xu, D. Q, Wang, W. W., et al. (2015). Comparative genome analyses of Serratia marcescens FS14 reveals its high antagonistic potential. PLoS ONE 10:e0123061. doi: 10.1371/journal.pone.0123061
Luo, R. B., Liu, B. H., Xie, Y. L., and Li, Z. Y. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Giga Science. 1:18. doi: 10.1186/2047-217X-1-18
Matsushita, K., Uchiyama, J., Kato, S., Ujihara, T., Hoshiba, H., Sugihara, S., et al. (2009). Morphological and genetic analysis of three bacteriophages of Serratia marcescens isolated from environmental water. FEMS Microbiol. Lett. 291, 201–208. doi: 10.1111/j.1574-6968.2008.01455.x
Schubert, M., Lindgreen, S., and Orlando, L. (2016). AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9:88. doi: 10.1186/s13104-016-1900-2
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Keywords: Serratia marcescens, strain ZPG19, prodigiosin, genomic sequence, comparative genomics
Citation: Li X, Tan X, Zhang J and Zhang J (2021) Complete Genome Sequences of One Prodigiosin-Producing Serratia marcescens Strain ZPG19. Front. Bioeng. Biotechnol. 9:665077. doi: 10.3389/fbioe.2021.665077
Received: 07 February 2021; Accepted: 06 April 2021;
Published: 11 May 2021.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Fengjie Cui, Jiangsu University, ChinaSumit Kumar, Indian Institute of Technology Delhi, India
Donatella Cimini, University of Campania Luigi Vanvitelli, Italy
Farshad Darvishi, Alzahra University, Iran
Copyright © 2021 Li, Tan, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zhang, aGVsbGVuemhhbmdqQDE2My5jb20=; Jie Zhang, emhhbmdqaWVAcWx1LmVkdS5jbg==
 Xue Li
Xue Li Xinfeng Tan
Xinfeng Tan Jing Zhang
Jing Zhang Jie Zhang
Jie Zhang