- 1Shenzhen Key Laboratory of Marine Bioresources and Eco-environmental Science, Shenzhen Engineering Laboratory for Marine Algal Biotechnology, Guangdong Provincial Key Laboratory for Plant Epigenetics, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province, College of Physics and Optoelectronic Engineering, Shenzhen University, Shenzhen, China
- 3College of Chemistry and Environmental Engineering, Shenzhen University, Shenzhen, China
Euglena gracilis, a green microalga known as a potential candidate for jet fuel producers and new functional food resources, is highly tolerant to antibiotics, heavy metals, and other environmental stresses. Its cells contain many high-value products, including vitamins, amino acids, pigments, unsaturated fatty acids, and carbohydrate paramylon as metabolites, which change contents in response to various extracellular environments. However, mechanism insights into the cellular metabolic response of Euglena to different toxic chemicals and adverse environmental stresses were very limited. We extensively investigated the changes of cell biomass, pigments, lipids, and paramylon of E. gracilis under several environmental stresses, such as heavy metal CdCl2, antibiotics paromomycin, and nutrient deprivation. In addition, global metabolomics by Ultra-high-performance liquid chromatography tandem mass spectrometry (UHPLC–MS/MS) was applied to study other metabolites and potential regulatory mechanisms behind the differential accumulation of major high-valued metabolites. This study collects a comprehensive update on the biology of E. gracilis for various metabolic responses to stress conditions, and it will be of great value for Euglena cultivation and high-value [154mm][10mm]Q7metabolite production.
Introduction
Euglena gracilis, which began to appear 500 million years ago, is a species of unicellular organisms that live mostly in freshwater (Goto and Beneragama, 2010) and has no cell walls, giving it dual characteristics of plants and animals, which enable this microalga to adopt photosynthetic, heterotrophic, and mixotrophic conditions during long-term evolution (Edmunds, 1965; Zakryś et al., 2017). During this long evolution, Euglena became a highly adaptable microorganism that survived diverse and extreme conditions on the earth, such as high UV radiation, acid mine water, man-made antibiotics, heavy metal pollution, and nutrient deprivation (Ferreira et al., 2007; Moreno-Sánchez et al., 2017).
Among microalgae, E. gracilis is well-known as a producer of polyunsaturated fatty acids (PUFAs) (Schwarzhans et al., 2015), vitamin E (Takeyama et al., 2015), chlorophyll a and b, several types of carotenoid pigments (Tanno et al., 2020), and polysaccharose paramylon (β-1,3-glucan) (Ivusic and Santek, 2015). Nitrogen deficiency (as nitrogen deprivation, N–) is one of the most common environmental stresses to enhance lipid (biofuels) accumulation in microalgae (Peccia et al., 2013). An increase of total lipids in stationary growth phase cells in comparison to exponential growth phase cells in N deprived, mixotrophic E. gracilis cultures had been reported (Regnault et al., 1990). And a decrease in chlorophyll production by E. gracilis was observed during a short-term exposure to N– (García-Ferris et al., 1996). A long-term N limitation on lipid, protein, and pigment production of E. gracilis in photoheterotrophic cultures indicated that N– could increase total fatty acid production, with lower protein contents and pigment production (Tossavainen et al., 2019). The symptoms of N deficiency are well-documented, but the underlying molecular mechanisms are largely unknown in microalgae (Tossavainen et al., 2019).
Euglena gracilis is a highly promising group of microorganisms to be used in the bio-remediation of heavy metal-polluted aerobic and anaerobic acidic aquatic environments. Considering the current data and the fact that E. gracilis has an innate tolerance to relatively high concentrations of heavy metals, as demonstrated by the minimum inhibitory concentrations (MICs) obtained, that is, CdCl2 (50 μM) induced diminution in cell growth (Castro-Guerrero et al., 2008) this organism has the potential for the management of particular heavy metal contamination of the environment (Khatiwada et al., 2020). In E. gracilis, CdCl2 exposure can induce morphological alteration, linked to reactive oxygen stress (Watanabe and Suzuki, 2002). So far, the connection between Cd, paramylon, and lipid accumulation in E. gracilis is still unclear.
Euglena gracilis was reported to be highly tolerant to various antibiotics (Shao et al., 2018). When bacteria and other microalgal cells were significantly inhibited by antibiotics at the low concentration of 20–50 μg/ml (50 μg/ml Kanamycin Chlamydomonas; Bateman and Purton, 2000), E. gracilis cells still grew well. In Euglena, rapamycin induced the reduction of chlorophyll and the accumulation of neutral lipids without deterring its cell proliferation. In another green microalga of Chlamydomonas, however, rapamycin induced serious growth inhibition as reported elsewhere (Mukaida et al., 2016). Various antibiotics were used to bleach Euglena with concentrations as high as 200 μg/ml (Ebringer, 1964). Based on our preliminary experiments, paromomycin (PRM) could not inhibit E. gracilis cell growth at concentrations ranging from 10 to 50 μg/ml. Since streptomycin significantly bleached Euglena cells by deleting part of the chloroplast genome, we did not choose streptomycin for further investigation in this study. Currently, no report of PRM against E. gracilis has been listed, and the antibiotic resistance mechanism of E. gracilis is still unclear.
The research on different high-value metabolites and their potential control mechanisms under various environmental stresses are especially important and interesting. However, because of the absence of a high-dimension genome of E. gracilis (Ebenezer et al., 2017) and with limited previous OMICS studies, there is no definite conclusion on molecular mechanisms involved in responses to different environmental stresses.
With the advent of the post-genome era, various OMICS technologies, such as proteomics and metabolomics, appeared one after the other, in which metabolomics was the first to develop and the most widely applied technology (Johnson et al., 2016). Metabolomics has been an important branch of system biology in recent years. It can help to understand the biological process more directly and effectively, and the study of metabolites can also help to analyze complex sample traits (Johnson et al., 2016). We used the metabolomic approach to investigate the effects of different inoculum sizes on a green microalga Chlorella growth (Lu et al., 2012), salt stress in a model cyanobacterium (Wang et al., 2014), astaxanthin induction in Haematococcus (Su et al., 2014), antioxidant butylated hydroxyanisole on lipid accumulation in Crypthecodinium cohnii (Sui et al., 2014), and obtaining better understanding on the metabolic response mechanism of microalgae with different environments.
In this study, we explored global metabolomics to analyze metabolite changes under various conditions in E. gracilis for the first time, that is, heavy metals, antibiotics, and nutrient deficiency, to explore the mechanism of tolerance, acclimatization, and high-value product accumulations. The results show that pigments, lipids, and carbohydrates (paramylon) can be altered significantly under different stresses. Further, comparative metabolomics revealed some common and unique metabolic pathways and modules regulated by all selected stresses and individual stress, respectively.
Materials and Methods
Strains and Culture Conditions
Euglena gracilis CCAP 224/5Z was obtained from the Culture Collection of Algae and Protozoa (https://www.ccap.ac.uk/). The microalgal cells were grown in the EM medium [1.8 g/L NH4Cl, 0.6 g/L KH2PO4, 0.6 g/L MgSO4, 60 mg/L urea, 0.02 g/L CaCl2, 0.48 mg/L Na2EDTA, 2 mg/L Fe2(SO4)3, 60 μl HCl, 0.01 mg/L Vb1, 0.0005 mg/L Vb12, 20 mg/L CuSO4·5H2O, 0.4 g/L ZnSO4·7H2O, 1.3 g/L Co(NH3)·H2O, and 1.6 g/L MnCl2·4H2O] in 6 ml culture volumes of a 6-well plate under a light intensity of ~100 μmol/m2/s in an illuminating incubator without shaking at 26°C until microalgal cells reached the stationary phase (Afiukwa and Ogbonna, 2007; Yanming et al., 2018).
Cultivation and Stress Treatments
Euglena gracilis cells were cultured for 6 days in the EM medium (10 ml EtOH in 100 ml EM medium), then 1 × 106 cells/ml were centrifuged at 5,000 × g for 3 min and transferred into an equal volume of the EM medium as the 1:10 dilution was applied to each treatment well in a 12-well plate. Treatments were applied, including supplementation with PRM (25 μg/ml), CdCl2 (Cd) (0.5 mM), or nitrogen deprivation (N–) (without NH4Cl nor urea). After 6 days, stationary phase cell samples were collected and used for future experiments.
Cell Growth, Chlorophyll, Paramylon, and Total Lipids
Cell growth was measured by counting cell numbers with a microscope in a 0.1-ml counting chamber. The total algal chlorophyll was extracted with 95% ethanol, and the content was spectrophotometrically assayed according to the method (Harris, 2009; Smith, 2009). The paramylon of 5 × 107-108 cells was extracted and measured as described previously (Sugiyama et al., 2009; Nakazawa et al., 2017; Guo et al., 2020). The total lipids of 107-108 cells were measured using the oven-drying method (Yang et al., 2018).
UHPLC–MS/MS Metabolite Analysis
For the LC–MS/MS analysis, samples took after 6 days of incubation were first centrifuged at 5,000 × g for 5 min at 4°C to harvest ~106-107 cells. Metabolite extraction was followed as per previously reported protocol (Zhang et al., 2020). Cell pellets were then transported to a 1.5-ml Eppendorf microcentrifuge tube (Hamburg, Germany). After adding 1,000 μl extract solvent (acetonitrile:methanol:water, 2:2:1, containing internal standard), the samples were vortexed for 30 s using a XW-80A vortex mixer (Kylin-Bell Lab Instruments Co., Ltd., Haimen, China), homogenized at 45 Hz for 4 min using a tissue grinding machine (JXFSTPRP-24, Shanghai Jingxin Industrial Development Co., Ltd., Shanghai, China), and sonicated for 5 min in an ice-water bath using an ultrasonic cell-crushing device (Fangao Microelectronics Co., Ltd., Shenzhen, China). The homogenate and sonicate circle was repeated three times, followed by incubation at −20°C for 1 h and centrifugation at 12,000 rpm 4°C for 15 min. The LC–MS/MS analyses were performed using a ultra-high-performance liquid chromatography (UHPLC) system (1290, Agilent Technologies, Waldbronn, Germany) with a UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm) coupled to Q Exactive mass spectrometer (Orbitrap MS, Thermo Fisher Scientific, San Jose, CA, USA). Mobile phase A was 0.1% formic acid in water for positive mode, and 5 mmol/L ammonium acetate in water for negative mode, and the mobile phase B was acetonitrile. The elution gradient was set as follows: 0–1.0 min, 1% B; 1.0–8.0 min, 1–99% B; 8.0–10.0 min, 99% B; 10.0–10.1 min, 99–1% B; 10.1–12 min, 1% B. The flow rate was 0.5 ml/min. The injected volume was 2 μl. QE mass spectrometer was used for its ability to acquire MS/MS spectra information-dependent acquisition (IDA) mode in the control of the acquisition software (Xcalibur 4.0.27, Thermo Fisher Scientific, San Jose, CA, USA). In this mode, the acquisition software continuously evaluates the full-scan MS spectrum. The electrospray ionization (ESI) source conditions were set as following: sheath gas flow rate as 45 arb, Aux gas flow rate as 15 arb, capillary temperature 400°C, full MS resolution as 70,000, MS/MS resolution as 17,500, collision energy as 20/40/60 eV in normalized collision energy (NCE) mode and spray voltage as 4.0 kV (positive) or −3.6 kV (negative), respectively (Periannan, 2003).
Compound identification from a non-targeted metabolite database were converted to the mzXML format using ProteoWizard and processed with an in-house program, which was developed using R and based on XCMS, for peak detection, extraction, alignment, and integration. Then an in-house MS2 database (BiotreeDB) was applied in metabolite annotation. The cutoff for annotation was set at 0.3. (Smith et al., 2006). Identified peaks were normalized to the peak intensity of ribitol. The pathway enrichment analysis on the metabolite dataset based on Kyoto Encyclopedia of Genes and Genomes (KEGG) was performed in Metaboanalyst 3.01 (Xia et al., 2015). Related assistance was performed at Shanghai Biotree Biotech Co., Ltd. (Shanghai, China).
Statistical Analysis
Significant differences in growth, chlorophyll content, biomass, and paramylon content were tested using the Dunnett's t-test. All data were obtained and averaged from at least three independent experiments, and standard errors were calculated and displayed as error bars. And after raw data profiles were processed, the Student's t-test was used for the univariate analysis. Principal component analysis (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA) were used for multivariate analysis. Variable importance in projection (VIP) score was combined with p-value to screen significant differential metabolites, and then the significant differential metabolites were qualitatively analyzed based on the relevant KEGG pathway and existing materials (Mangalam et al., 2013).
Results
Cell Growth, Chlorophyll, Paramylon, and Lipids
Cell growth differed greatly under different environmental stresses (Table 1). The PRM treatment caused a slight decrease (7.17%, p > 0.05), whereas CdCl2 (Cd) and nitrogen deprivation (N–) treatments significantly inhibited the growth of E. gracilis by 7.79% (p < 0.05) and 11.95% (p < 0.05), respectively.
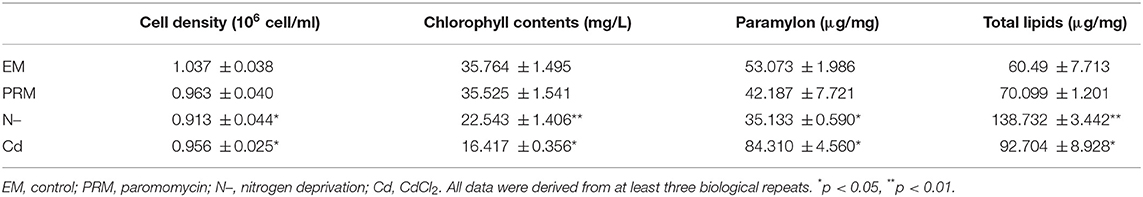
Table 1. Physiological parameter, cell density, chlorophyll contents, paramylon, and total lipids determined under different environmental stresses.
Photosynthesis systems and chlorophyll in microalgae are sensitive to environmental stress. The addition of Cd, and N– decreased the chlorophyll content of E. gracilis significantly (Table 1), whereas PRM had no significant effect. Chlorophyll content decreased by 55% (p < 0.05) and 38% (p < 0.001) in cells treated with Cd and N–, respectively.
Similarly, there were obvious differences in the paramylon content of E. gracilis under different environmental treatments. PRM did not affect paramylon content; Cd caused a significant increase (171% of the control, p < 0.05), whereas N– showed an obvious reduction (66% of the control, p < 0.05) (Table 1).
Many environmental stresses could alter the metabolism, cellular membrane structure, and lipids. In this study, significant changes of total lipids were also detected in both Cd and N– treatments, with 53 and 129% increases compared to EM control, respectively. PRM caused no significant change in total lipid content in E. gracilis.
Comparative Metabolomics of Euglena Under Different Environmental Stresses
General Features of Metabolomics
In this study, the metabolic plasticity of E. gracilis under different stresses was elucidated using comparative metabolomic analysis. UHPLC–MS/MS revealed a total of 10,892 negative ion mode (NEG) and 11,003 positive ion mode (POS) peaks. Furthermore, 254 NEG and 583 POS metabolites, respectively, were annotated by the KEGG database (Supplementary Table 1).
The PCA analysis showed that all four groups, EM, PRM, N–, and Cd, of metabolites could be distinguished (Figure 1), indicating that the metabolic changes to stress were significantly different from each other. Among them, two groups of metabolites (EM and PRM) were relatively closer, indicating that smaller differences had occurred in the metabolic alterations of microalgae cells treated with PRM. Furthermore, the metabolites (N– and Cd) of two groups of microalgae cells were clearly distinguished between each other, and far from EM and PRM groups (Figure 1), suggesting that the metabolic pathways of the microalgae cells changed dramatically under N– and Cd.
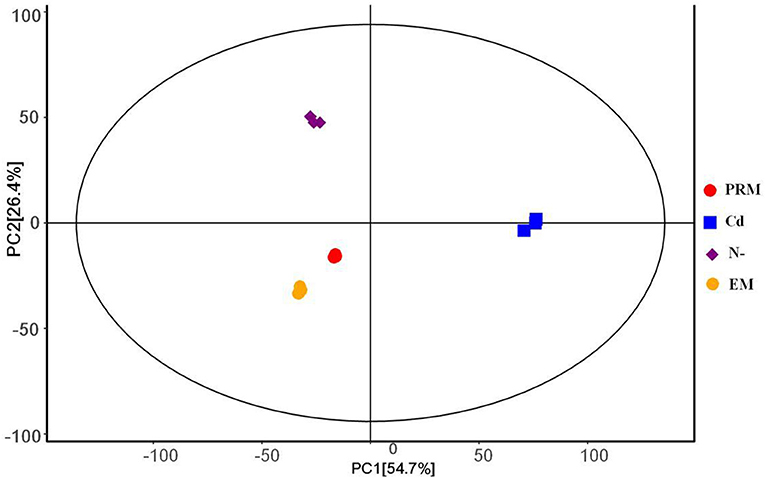
Figure 1. PCA score plot under different treatment groups based on metabolites, each with 3 biological replicates. PRM, paromomycin; Cd, CdCl2 addition; N–, nitrogen deprivation; EM, control.
According to the criteria of fold change (>2.0, or <0.5, and p < 0.05), 401 out of 426 (POS) and 170 of 196 (NEG) candidate metabolites were significantly changed under different treatments (Figure 2).
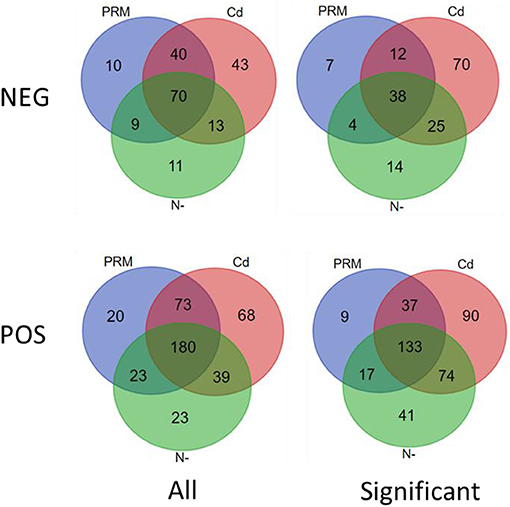
Figure 2. Venn diagram of all (All) and significantly changed (significant) metabolites under stresses (PRM, paromomycin; Cd, CdCl2; N–, nitrogen deprivation), under negative (NEG) and positive (POS) ion modes.
In total, numbers of significantly decreased metabolites were much more than increased ones in all stress treatment groups, for instance, 177 vs. 19 (PRM) and 313 vs. 21 (Cd) under the POS mode, whereas similar trends were also observed under the NEG mode, for instance, 52 vs. 8 (PRM) and 108 vs. 36 (Cd) (Table 2). These stresses inhibited Euglena cell growth by reducing the metabolism levels of cellular activity. Among stresses, Cd seems to cause more dramatic metabolic changes, with the most significantly changed metabolites as total 334 and 166 under POS and NEG mode, respectively, in contrast to N– group (179, 80) and PRM (196, 60) compared to the EM control.
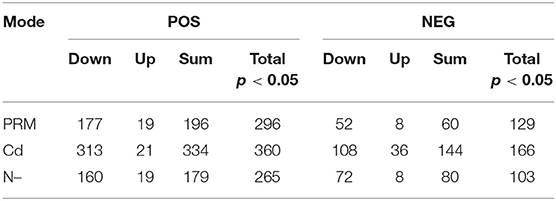
Table 2. Summary of metabolomics: differential metabolites under different stresses (PRM, paromomycin; Cd, CdCl2; N–, nitrogen deprivation) and under negative (POS) and positive (NEG) ion modes.
TOP Changed Metabolites Under Stresses
Based on fold changes increased and decreased, TOP 10 metabolites under different stresses were analyzed (Supplementary Table 2). Interestingly, metabolites as organic oxygen compounds, organic acids, and derivatives (mostly amino acids) were deceased, and lipids and lipid-like molecules increased in PRM treatment. Under Cd, TOP changed metabolites include lipids and organoheterocyclic compounds and increased polyketides and benzenoids. While as expected, N– treatment induced significant lipids like PC (16:0/P-16:0; 24:0/14:0), PS (18:1/15:0; 15:0/16:1), DG (20:3), and reduced organic acids (mostly single amino acids as Tyr, Asp, and Arg, and two peptides like Arg-Ser, Thr-Arg, and Arg-Phe).
KEGG Pathway Enrichment
The KEGG-enriched metabolome bubble view showed all matched pathways according to the p-values from pathway enrichment analysis and the pathway impact values from pathway topology analysis (Figure 3). The candidate metabolites with p < 0.05 were enriched by one or several metabolic pathways under three stresses, such as Aminoacyl-tRNA biosynthesis, Purine metabolism, Nitrogen metabolism, “Taurine and hypotaurine metabolism,” Butanoate metabolism, “Alanine, aspartate and glutamate metabolism,” “Pantothenate and coenzyme A (CoA) biosynthesis,” and Pyrimidine metabolism (Figure 3; for more details see Supplementary Table 3 KEGG list only).
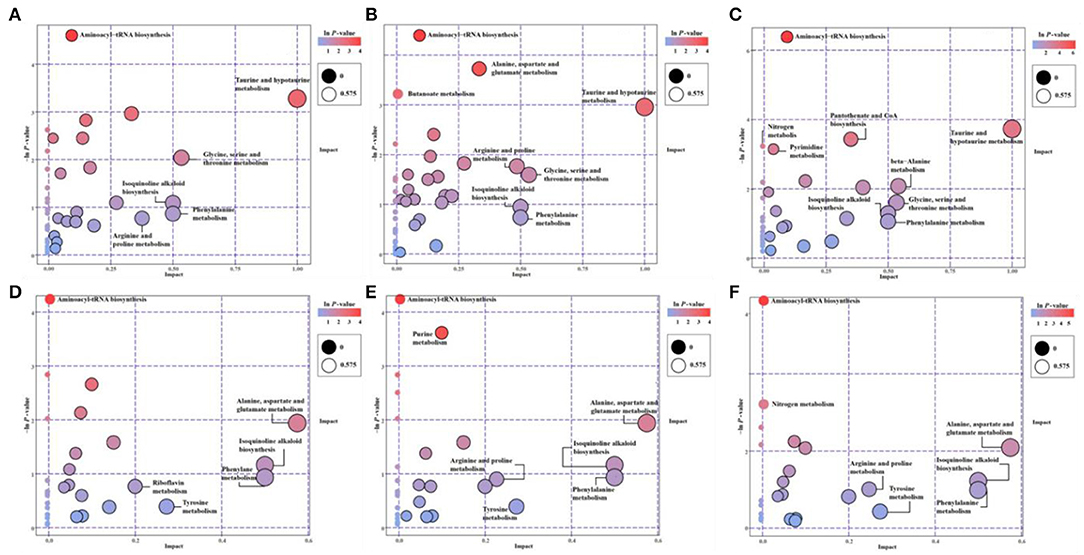
Figure 3. The pathway enrichment and topology analysis of candidate metabolites from E. gracilis under different environmental stresses. (A–C) NEG modes; (D–F) POS modes. (A,D) PRM-NEG, PRM-POS; (B,E) Cd-NEG, Cd-POS; (C,F) N-NEG, N-POS. “Pathway Impact” on the x-axis represents the impact of these enriched pathways computed via topology analysis. “–log10(p)” on the y-axis refers to the negative natural logarithmic value of the original p-value from a statistical analysis of the pathway difference between the groups of stresses vs. the EM control. The redder the color, the higher the –log10(p) and pathway impact value, the more significant the enrichment degree.
Under both POS and NEG modes, the aminoacyl-tRNA biosynthesis showed up under all stresses, which indicated that this metabolic pathway might be a key stress response of Euglena cells to different environmental stresses, such as nutrient deprivation, heavy metal, and antibiotics.
Overall, microalgae cell metabolic pathways under PRM conditions were primarily involved in the biosynthesis of Aminoacyl-tRNA, and Taurine and hypotaurine. With higher p-values (p > 0.05), some highly impacted pathways involved in amino acids, such as “Ala, Asp, and Glu metabolism,” Phe metabolism, Tyrosine metabolism, “Glycine, Ser, and Thr metabolism,” “Arginine and Proline metabolism,” Isoquinoline alkaloid biosynthesis, and Riboflavin metabolism.
Relatively more significantly enriched metabolic pathways were enriched when cells were treated with Cd. In addition to the biosynthesis of aminoacyl-tRNA, 3 more pathways, Purine metabolism, “Ala, Asp, and Glu metabolism,” and Butanoate metabolism, showed up in the Cd treatment. Similarly, all less significant pathways (p > 0.05) enriched in PRM were on the Cd pathway list (Figure 3), except for the Riboflavin metabolism.
Compared with PRM and Cd, N— treatment enriched the most significant metabolic pathways, including Aminoacyl-tRNA biosynthesis, “Taurine and hypotaurine metabolism,” “Pantothenate and CoA biosynthesis,” Nitrogen metabolism, and Pyrimidine metabolism. The late three pathways were N– group-specific, and nitrogen metabolism as the well-known pathways changed when nitrogen nutrient was absent.
Network Analyses of Metabolomics
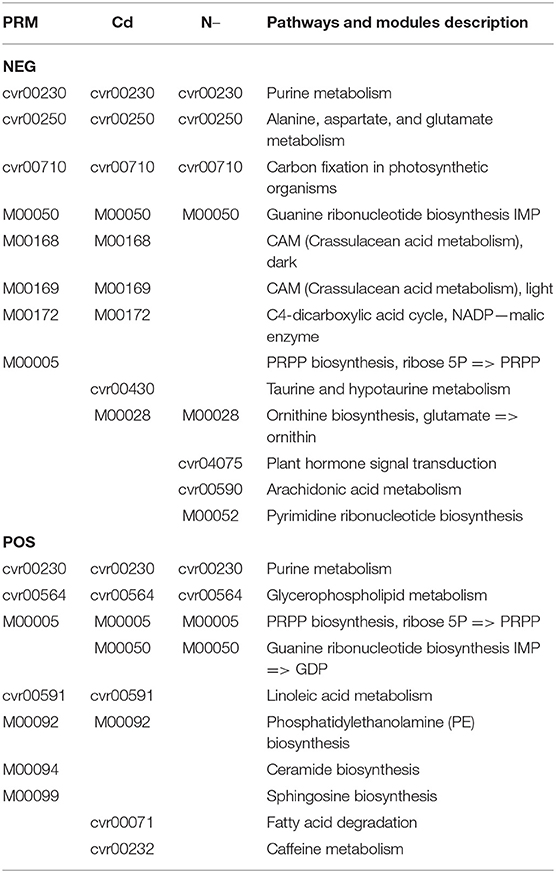
Based on the metabolomic KEGG and network analyses, we also obtained groups of metabolic pathways and modules that play core roles in the metabolic networking (Table 3 network plot). Several pathways and modules were plotted in all stress groups simultaneously, including Purine metabolism (module cvr00230), “Ala, Asp, and Glu metabolism” (cvr00250), Carbon fixation in photosynthetic organisms (cvr00710), Guanine ribonucleotide biosynthesis (M00050), Glycerophospholipid metabolism (cvr00564), and PRPP biosynthesis (M00005). M00005 is a module on KEGG, The KEGG MODULE database consists of KEGG modules identified by M numbers and KEGG reaction modules identified by RM numbers, which are manually defined functional units of gene sets and reaction sets, respectively. KEGG modules are further divided into pathway modules and signature modules as shown https://www.genome.jp/entry/M00005, and the PRPP: 5-Phosphoribosyl-1-Pyrophosphate.
On both PRM and Cd network lists, we found Crassulacean acid metabolism (CAM), dark (M00168) and light (M00169); C4-dicarboxylic acid cycle, nicotinamide adenine dinucleotide phosphate (NADP)-malic enzyme (M00172), Linoleic acid metabolism (cvr00591), and Phosphatidylethanolamine (PE) biosynthesis (M00092). However, on both Cd and N– lists, only two networks were detected as Guanine ribonucleotide biosynthesis (M00050) and Ornithine biosynthesis (M00028).
Several specific networks were only detected in one individual stress. For examples, Ceramide biosynthesis (M00094) and Sphingosine biosynthesis (M00090) in the PRM group; Fatty acid degradation (cvr00071) and caffeine metabolism (cvr00232) in the Cd group; Plant hormone signal transduction (cvr04075), Arachidonic acid (ARA) metabolism (cvr00590), and Pyrimidine ribonucleotide biosynthesis (M00052) under N– group.
Discussion
Euglena is highly adaptable to changing environments, some adverse stresses like UVB (Takahashi et al., 2006), heavy metals (Moreno-Sánchez et al., 2017), and antibiotics (Ebringer, 1964). In this study, microalgal cells showed tolerance to antibiotics, PRM, without significant cell growth inhibition, chlorophyll reduction, and paramylon and lipid alterations. The inhibition of chlorophyll formation in Euglena by antibiotics was reported previously (Linnane and Stewart, 1967). This may be caused by different antibiotics and different tolerant concentrations of E. gracilis cells. In the Cr(VI) treatments, a reduction of chlorophyll was observed, the ratio of proteins to paramylon content was augmented, and total lipid content was increased (Rocchetta et al., 2012). In contrast, the other two stresses, N– and Cd, showed obvious effects on above-mentioned physiological parameters and major macromolecules, such as polysaccharide (paramylon) and lipids.
This is the first time to apply global metabolomics to study the cellular response of E. gracilis to environmental stresses, such as antibiotics, heavy metals, and nutrient deprivation. Many metabolites and metabolic pathways, instead of standard physiological parameters, and macromolecules, were detected significantly altered by these stresses and shed light on the mechanisms of tolerance or toxic responses in E. gracilis. PRM did not cause much change of pigment, carbohydrates, and lipids in E. gracilis cells. However, based on metabolomics, hundreds of metabolites and several important metabolic pathways were detected significantly disturbed by PRM, indicating the sensitivity of metabolomics when studying the microalgal cellular response to changing environments. Metabolomics easily differentiates the groups of control, PRM, N–, and Cd treatments, even between control and PRM, by which cell growth, chlorophyll content, paramylon, and lipid changes could not distinguish significantly.
In this study, we detected paramylon degradation and lipids accumulation under N–. In most microalgae, lipids were induced to high accumulation under nitrogen starvation (Chen et al., 2017). There is a report showing induction of carbohydrates and no lipid accumulation with no nitrogen source in Euglena with NH4Cl as the only nitrogen source (Coleman et al., 1988). However, there is a major difference compared to the reference above since we added urea and NH4Cl as nitrogen sources. Thus, N– in this study means both NH4Cl and urea depletion and may cause differential accumulation (reduction) of carbohydrates and lipids in our study.
Based on hundreds of changed metabolites, significantly increased or decreased TOP 10 metabolites may suggest the major changes under different treatments. Even with no significantly changed lipids and paramylon under the PRM treatment, amino acids were found to decrease while lipids and lipid-like molecules increase. PRM is a broad-spectrum aminoglycoside antibiotic produced by Streptomyces rimosus var. paromomycinus (Davidson et al., 2009). The in vitro and in vivo antibacterial action of PRM closely parallels that of neomycin. PRM bind specifically to the RNA oligonucleotide at the A site of bacterial 30S ribosomes thereby causing misreading and premature termination of translation and leading to inhibition of protein synthesis followed by cell death (PubChem CID 165580). As reported, some antibiotics, such as chloramphenicol and other antibiotics that specifically inhibit the synthesis of proteins in mitochondria and bacteria, would also selectively inhibit protein synthesis by chloroplast (Linnane and Stewart, 1967). PRM did not reduce chlorophyll biosynthesis in E. gracilis cells at the concentration of 25 μg/ml, suggesting its inhibition pathways may not involve the synthesis of chloroplast proteins. Cd treatment in this study resulted in chlorophyll degradation, paramylon, and lipid accumulation. Under Cd treatment, metal-causing reactive oxygen species (ROS) changed the lipids and organoheterocyclic compounds, and N– increased lipids and decreased protein contents in Euglena cells. These TOP changed metabolites showed similar patterns as previous reports (Moreno-Sánchez et al., 2017; Tossavainen et al., 2019).
The KEGG enrichment provides metabolic pathways through which cells are regulated under specific treatments. The only common pathway enriched in all treatments is Aminoacyl-tRNA biosynthesis, suggesting this may be the common/universal stress responding pathway in Euglena. Aminoacyl-tRNA species are prevalent amino acid donors in the cell due to their critical role as substrates for ribosomal protein biosynthesis. It has become clear that aminoacyl-tRNAs are also utilized in an ever-growing number of other non-ribosomal biosynthetic pathways, particularly in bacteria, although some pathways also exist in eukaryotes (Shepherd and Ibba, 2013). Much has been uncovered in recent years about many novel functions, disease connection, and inter pathway connection of tRNA synthetases (Park et al., 2008; Zhou et al., 2020). PRM might inhibit protein synthesis by binding specifically to the RNA oligonucleotide at the site of bacterial, or microalgal mitochondrial and 30S ribosomes of chloroplast, and thus alter aminoacyl-tRNA biosynthesis. Nitrogen deprivation involves the degradation of carbohydrates, paramylon in E. gracilis, and constructive proteins, and accumulation of membrane lipids (Richter et al., 2015), and at the same time, specific enzymes/macromolecules have to be transformed or de novo synthesized via aminoacyl-tRNA biosynthesis pathway. Similarly, heavy metals are created ROS through lipid peroxidation and DNA damage (Rodriguez-Zavala et al., 2007; Khatiwada et al., 2020). Then the cells are induced to produce antioxidant chemicals and enzymes, such as superoxide dismutase and DNA repair proteins from protein biosynthesis with aminoacyl-tRNA biosynthesis pathway. However, whether the aminoacyl-tRNA biosynthesis pathway could be a novel biomarker for environmental stresses has yet to be worked out in the future with more investigations.
There are more pathways enriched under individual stresses according to our global metabolomics, reflecting different metabolic responses by different stresses.
In Cd treatment, “Purine metabolism,” “Alanine, aspartate, and glutamate metabolism,” and “Butanoate metabolism” pathways were detected significantly changed. Purine nucleotides are essential for many biochemical processes like energy transfer, metabolic regulation, and synthesis of DNA and RNA (Watanabe et al., 2014). Alanine, aspartate, and glutamate are derived from intermediates of central metabolism, mostly the citric acid cycle, in one or two steps. While the pathways are short, the importance and complexity of the functions of these amino acids befit their proximity to central metabolism (Limami et al., 2008). Butanoate metabolism describes the metabolic fate of several short-chain fatty acids or short-chain alcohols. Many of these molecules are eventually used in the production of ketone bodies, the creation of short-chain lipids, or as precursors to the citrate cycle (pubchem.ncbi.nlm.nih.gov/pathway/PathBank:SMP0000073). Cd degraded chlorophyll and reduced the activity of photosynthesis of microalgal cells; however, short-term (6 days) Cd treatment also increased the contents of paramylon and lipids (Mendoza-Cozatl et al., 2002; Einicker-Lamas et al., 2003). Thus, we speculated that the mutual transformation of three major macromolecules, proteins, carbohydrates, and lipids may respond to heavy metals. In Cd treatment, decreasing amino acid contents and significantly enriched protein synthesis pathways with enhanced carbohydrates and lipid production were detected, indicating the mutual transformation of proteins into paramylon and lipids in E. gracilis.
When microalgal cells were treated with PRM, the “Taurine and hypotaurine metabolism” pathway was also enriched. Taurine, an active substance that regulates the normal physiological activities of cells, is associated with maintaining the osmotic pressure balance in cells, plus strengthening cell membranes and enhancing antioxidant capacity (Tevatia et al., 2015, 2019). Taurine is synthesized in large amounts by animals; however, plants, green microalgae, and fungi could only synthesize minute amounts (Tevatia et al., 2019). E. gracilis enriched this metabolic pathway where the metabolite was located under PRM treatment. Thus, our results indicated that the metabolic pathway played an important role in regulating antibiotics tolerance of E. gracilis, as it stabilized the cell membrane and maintained cellular functions.
Nitrogen deprivation also enriched the taurine and hypotaurine compounds, suggesting this pathway may also involve in nutrient deprivation stress perhaps just by using taurine and its relative amino acid compounds as alternative nitrogen resources for biosynthesis of new enzymes for maintaining basic cell survival. Under N– treatment, KEGG pathway enrichment also collected other pathways such as “Nitrogen metabolism,” “Pantothenate and CoA biosynthesis,” and “Pyrimidine metabolism.”
Nitrogen makes up only a small fraction of plant and microalgal dry weight, but N compounds are extremely important physiologically. For instance, structural and storage proteins, enzymes, amino acids and amides, nucleic acids, and plant hormones, all contain N. The substituted purine and pyrimidine bases that constitute nucleic acids, nucleotides, and nicotinamide nucleotides also contain N (Pallardy, 2008). Transcriptome analysis revealed that genes involved in metabolism, plant hormone signal transduction (e.g., abscisic acid, auxin, and jasmonate), transporter activity, and oxidative stress responses were rapidly regulated by N– (based on our unpublished transcriptomic data in N– Euglena).
Pantothenate is vitamin B5 and is the key precursor for the biosynthesis of CoA, a universal and essential cofactor involved in a myriad of metabolic reactions, including the synthesis of phospholipids, the synthesis and degradation of fatty acids, and the operation of the tricarboxylic acid cycle (Leonardi and Jackowski, 2007). Pyrimidines metabolism pathway suggests the cells use the nitrogen from nucleic acids under N–. Although both pyrimidines and purines are components in nucleic acids, they are made in different ways. Likewise, products of pyrimidine degradation are more water-soluble than are the products of purine degradation (Wan et al., 2019).
In summary, the KEGG pathway enrichment showed that both common and specific metabolic pathways were employed to respond against different environmental stresses by E. gracilis cells.
Based on network plot analyses, we also targeted some specific metabolic pathways and modules under different stresses, especially on photosynthesis and carbon fixation, ROS, membrane lipids, and amino acid metabolisms.
Photosynthesis is a global sensor of environmental stress in green plants, microalgae, and cyanobacteria (Biswal et al., 2011). Photosynthesis and photosynthetic pigments in E. gracilis were reported as sensitive endpoints for the toxicity evaluation of liquid detergents (Azizullah et al., 2014). Carbon fixation is the process by which inorganic carbon is added to an organic molecule, and it occurs during the light-independent reaction of photosynthesis with the help of chlorophyll. This pathway is showed as a core responsive pathway in all three stresses in metabolic networking analyses, indicating photosynthesis and carbon fixation involves in the alternations caused by antibiotics, heavy metals, and N–. Cd and N– treatments significantly reduced chlorophyll contents with lower photosynthetic activity, whereas PRM did not decrease chlorophyll significantly. Further experiments with the photosynthetic activity will clarify this observation.
It is very interesting to locate the CAM pathway in PRM and Cd groups. CAM (also known as CAM photosynthesis) is a carbon fixation pathway that evolved in some plants as an adaptation to arid conditions. In CAM, the carboxylation (C3 + C1) occurs at night, and the (C4 – C3) decarboxylation occurs during the day (Libik-Konieczny et al., 2019; Gilman and Edwards, 2020). The C3 + C1 mechanism has been found in the green Ulvophyceae benthic macroalga Udotea flabellum and the planktonic diatom Thalassiosira weissflogii (Cheng et al., 2019). The photosynthetic metabolism (C3, C4, or CAM) seems to affect plant sensitivity to environmental stresses. CAM is a metabolic strategy allowing plants to maintain photosynthesis under stress conditions (Libik-Konieczny et al., 2019). Although N– also showed a significant reduction of chlorophyll contents as Cd treatment; however, only PRM and Cd triggered CAM metabolism changes. Heterotrophic CO2 fixation by E. gracilis was reported in Levedahl (1966). A previous study indicated that E. gracilis can remove Cd under anaerobic conditions, which might be advantageous for heavy metal removal in sediment from polluted water bodies or bioreactors, where the O2 concentration is particularly low (Geovanni Santiago-Martinez et al., 2015). Perhaps under PRM and Cd treatments, the damage of photosynthesis might be compensated partially by CAM metabolism, which needs further verification shortly.
Finally, Euglena appears to exemplify a strategy for survival and adaptation to various environmental conditions during the evolutionary process of euglenoids (Geovanni Santiago-Martinez et al., 2015; Ishikawa et al., 2017). Among various environmental stresses, heavy metal accumulation and resistance in Euglena are widely studied. Under Cd treatment, mitochondria and chloroplasts were altered in shape, and thylakoid arrangement was changed, and osmiophilic plastoglobuli was increased (Duret et al., 1986). Cd causes cellular damage to Euglena, including DNA strand breaks and intracellular membrane damage, as a result of reactive oxygen stress (Watanabe and Suzuki, 2002). Enhanced alternative oxidase and antioxidant enzymes under Cd stress in Euglena were reported (Castro-Guerrero et al., 2008). The data suggested that Cd was compartmentalized into chloroplasts in a process that may involve the transport of free Cd and the participation of thiol-peptides in E gracilis (Mendoza-Cozatl et al., 2002). The chloroplasts appear to have a major role in the tolerance and accumulation of Cd in E. gracilis (Khatiwada et al., 2020). The variety of biochemical mechanisms evolved in E. gracilis to resist, accumulate, and remove heavy metals from the environment are summarized as follows: adsorption to the external cell pellicle, intracellular binding by glutathione and glutathione polymers, and their further compartmentalization as heavy metal complexes into chloroplasts and mitochondria, polyphosphate biosynthesis, and secretion of organic acids (Moreno-Sánchez et al., 2017).
Alternation of membrane lipids and PUFA pathways was also reported in all stresses. Lipid metabolism is crucial for the survival and propagation of the species since lipids are an essential cellular component for maintaining homeostasis in the presence of environmental stressors. Reactions in glycerolipid and glycerophospholipid metabolism occur at the membrane, which plays an important role in signaling and response to environmental stress. PE is one of the most abundant membrane phospholipids, in which metabolism was significantly changed under PRM and Cd. In both prokaryotes and eukaryotes, PE plays important role in various membrane functions. It is suggested that the synthesis of these membrane lipids was beneficial to increase the peristalsis and swimming ability of E. gracilis, which protected the integrity of the cell, improving the viability of the cell under these stresses.
Under the PRM treatment, ceramide biosynthesis and sphingosine metabolism were also disturbed. Ceramide biosynthesis begins in the endoplasmic reticulum with the condensation of palmitoyl-CoA and serine, catalyzed by the multimeric enzyme serine palmitoyltransferase to produce 3-ketosphinganine (Chaurasia and Summers, 2015). Sphingosine is a long-chain unsaturated amino alcohol C18H37O2N that is found especially in cell membranes and is a primary constituent of sphingolipids. Sphingosine itself increases the permeability of phospholipid membranes. With no cell walls in E. gracilis unlike other microalgae, the external cell pellicle and the cell membrane are the first front-line against the changing aquatic environment. Thus, distortions and alterations of membrane lipids of Euglena have to be sensitive and elastic in the environment.
As the major compound of membrane lipids, PUFAs are also very sensitive to environmental stresses at qualitative and quantitative levels. Halter et al. (2012) found that under adverse conditions, such as the presence of heavy metal arsenic, the content of unsaturated fatty acids in the cell membrane had significantly increased. In this study, linoleic acid (LA) metabolism was highlighted in groups of PRM and Cd (Halter et al., 2012). LA [18:2 (n−6)] is an omega-6 fatty acid and helps stimulate skin and hair growth, maintain bone health, regulate metabolism, and to maintain the reproductive system (Lee et al., 2018). Under abiotic stress by salt or ultraviolet light, the production of lutein and PUFAs were observed by the acidophilic eukaryotic microalga Coccomyxa onubensis (Bermejo et al., 2018). Thus, increasing the fluidity and hydrophobicity of the cell membrane by increasing PUFA allowed E. gracilis adaptability in the surrounding environments, such as PRM and Cd. Similar PUFA accumulation was also reported in freshwater microalgae Chlamydomonas mexicana and Scenedesmus obliquus grown under salt stress (Salama et al., 2013). Furthermore, it was determined that there is a relationship between the effects of Cd-induced oxidative stress on PUFAs and their degradation products and is also reflected on the increase in tolerance to Cd toxicity in plant cells (Pavlík et al., 2017). Specifically to N–, ARA metabolism reacted significantly. ARA is an integral constituent of the biological cell membrane, conferring it with fluidity and flexibility, so necessary for the function of all cells. Free ARA modulates the function of ion channels, several receptors, and enzymes via activation as well as inhibition (Tallima and El Ridi, 2018). Similarly, the green oleaginous microalga Lobosphaera incisa accumulates triacylglycerols enriched in the long-chain PUFA, such as ARA under nitrogen deprivation. This is consistent with the enhanced lipid accumulation under N– stress in E. gracilis.
Conclusions
This study demonstrated that metabolomics is sensitive and accurate enough to elucidate cellular responses of E. gracilis to different environmental stresses, even with no significantly changed physiological parameters, such as cell numbers, pigments, paramylon, and lipids under PRM treatment. Based on global metabolomics, a common metabolic pathway responding to multiple environmental stresses, aminoacyl-tRNA biosynthesis, was firstly targeted in E. gracilis. The other shared metabolic pathways and modules under selected stress include Purine metabolism, carbon fixation, Ala, Asp, and Glu metabolism, Glycerophospholipid metabolism, and PRPP biosynthesis. We observed the well-known changed metabolic pathways involved in stresses, like ROS in heavy metals, N metabolism during N deprivation, and protein biosynthesis inhibition with PRM. Some unique modules were individually detected under different stresses, like ceramide and sphingosine biosynthesis under PRM, ARA metabolism in N–, and fatty acid degradation and caffeine metabolism with Cd treatment. Under stresses, without cell walls, membrane lipids and PUFA in E. gracilis cells were altered to resist, acclimatize to, and remove the adverse factors like Cd and reactive oxidative species. All data suggest that the mutual interaction and transformation of macromolecular metabolites as proteins, pigments, lipids, and paramylon are regulated delicately to resist, tolerate, and/or remove these environmental stresses.
Data Availability Statement
The data presented in the study were deposited in the MetaboLights repository, with the accession number MTBLS2878.
Author Contributions
JW and ZH contributed to the conception and design of the study. JH, CL, MD, and XZ conducted the experiments. JH, CL, AL, and XZ involved in the statistical analysis. JW and JH wrote the first draft of the manuscript. AL and JW wrote the sections of the manuscript. JW and CL contributed to manuscript revision. All authors read and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0902500), the Natural Science Foundation of Guangdong Province, China (2014A030313562), the Guangxi Innovation-Driven Development Special Fund (Gui Ke AA18242047), Shenzhen Basic Research Projects (JCYJ20180507182405562), and Grant Plan for Demonstration City Project for Marine Economic Development in Shenzhen (No. 86). They are used for the design of the study, data collection, data analysis, and writing the manuscript, respectively.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge support from the Instrumental Analysis Center of Shenzhen University (LiHu Campus). They are obliged to Biotechnology Co., Ltd. Shanghai, China for the suggestions of metabolomic analysis. They would like to thank TopEdit (http://www.topeditsci.com) for English language editing of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.662655/full#supplementary-material
Abbreviations
ARA, arachidonic acid; CAM, Crassulacean acid metabolism; CoA, coenzyme A; E. gracilis, Eugleaa gracilis; KEGG, Kyoto Encyclopedia of Genes and Genomes; OPLS-DA, orthogonal projections to latent structures discriminant analysis; PCA, principal component analysis; PE, phosphatidylethanolamine; PRM, paromomycin; PUFAs, polyunsaturated fatty acids; ROS, reactive oxygen species; VIP, variable importance in projection.
Footnotes
References
Afiukwa, C. A., and Ogbonna, J. C. (2007). Effects of mixed substrates on growth and vitamin production by Euglena gracilis. Afr. J. Biotechnol. 6:2417. doi: 10.5897/AJB2007.000-2417
Azizullah, A., Richter, P., and Hder, D. P. (2014). Photosynthesis and photosynthetic pigments in the flagellate Euglena gracilis- As sensitive endpoints for toxicity evaluation of liquid detergents. J. Photochem. Photobiol. B. Biol. 133, 18–26. doi: 10.1016/j.jphotobiol.2014.02.011
Bateman, J. M., and Purton, S. (2000). Tools for chloroplast transformation in Chlamydomonas: expression vectors and a new dominant selectable marker. Mol. Gene. Genet. Mgg 263, 404–410. doi: 10.1007/s004380051184
Bermejo, E., Ruiz-Domínguez, M. C., Cuaresma, M., Vaquero, I., Ramos-Merchante, A., Vega, J. M., et al. (2018). Production of lutein, and polyunsaturated fatty acids by the acidophilic eukaryotic microalga Coccomyxa onubensis under abiotic stress by salt or ultraviolet light - ScienceDirect. J. Biosci. Bioeng. 125, 669–675. doi: 10.1016/j.jbiosc.2017.12.025
Biswal, B., Joshi, P. N., Raval, M. K., and Biswal, U. C. (2011). Photosynthesis, a global sensor of environmental stress in green plants: Stress signalling and adaptation. Curr. Sci. 101, 47–56.
Castro-Guerrero, N. A., Rodríguez-Zavala, J. S., Marín-Hernández, A., Rodríguez-Enríquez, S., and Moreno-Sánchez, R. (2008). Enhanced alternative oxidase and antioxidant enzymes under Cd2+ stress in Euglena. J. Bioenerget. Biomembr. 40:227. doi: 10.1007/s10863-007-9098-6
Chaurasia, B., and Summers, S. A. (2015). Ceramides - lipotoxic inducers of metabolic disorders. Trends Endocrinol. Metabo. 26, 538–550. doi: 10.1016/j.tem.2015.07.006
Chen, B., Wan, C., Mehmood, M. A., Chang, J. S., Bai, F., and Zhao, X. (2017). Manipulating environmental stresses and stress tolerance of microalgae for enhanced production of lipid and value-added products-A review. Bioresour. Technol. 244, 1198–1206. doi: 10.1016/j.biortech.2017.05.170
Cheng, J., Zhu, Y., Zhang, Z., and Yang, W. (2019). Modification and improvement of microalgae strains for strengthening CO2 fixation from coal-fired flue gas in power plants. Bioresour. Technol. 291:121850. doi: 10.1016/j.biortech.2019.121850
Coleman, L. W., Barry, H. R., Steven, D., and Schwartzbach. (1988). Environmental control of carbohydrate and lipid synthesis in Euglena. Plant Cell Physiol. 29, 423–432
Davidson, R. N., Boer, M. D., and Ritmeijer, K. (2009). Paromomycin. Transact. R. Soc. Trop. Med. Hygiene 103, 653–660. doi: 10.1016/j.trstmh.2008.09.008
Duret, S., Bonaly, J., Bariaud, A., Vannereau, A., and Mestre, J. C. (1986). Cadmium-induced ultrastructural changes in Euglena cells. Environ. Res. 39, 96–103. doi: 10.1016/S0013-9351(86)80011-1
Ebenezer, T. G. E., Carrington, M., Lebert, M., Kelly, S., and Field, M. C. (2017). Euglena gracilis Genome and Transcriptome: Organelles, Nuclear Genome Assembly Strategies and Initial Features. London: Springer International Publishing. doi: 10.1007/978-3-319-54910-1_7
Ebringer, L. (1964). Bleaching of euglenas by antibiotics–a specific form of antagonism in actinomycetes. Folia Microbiol. 9, 249–255. doi: 10.1007/BF02875844
Edmunds, L. N. (1965). Studies on synchronously dividing cultures of Euglena gracilis Klebs (strain Z). II. Patterns of biosynthesis during the cell cycle. J. Cell. Compar. Physiol. 66, 159–181. doi: 10.1002/jcp.1030660205
Einicker-Lamas, M., Morales, M. M., Miranda, K., Garcia-Abreu, J., Oliveira, A. J. F., and Oliveira, F. L. S. S.M. (2003). P-glycoprotein-like protein contributes to cadmium resistance in Euglena gracilis. J. Compar. Physiol. B 173, 559–564. doi: 10.1007/s00360-003-0365-5
Ferreira, V. S., Rocchetta, I., Conforti, V., Bench, S., Feldman, R., and Levin, M. J. (2007). Gene expression patterns in Euglena gracilis: Insights into the cellular response to environmental stress. Gene 389, 136–145. doi: 10.1016/j.gene.2006.10.023
García-Ferris, C., Ríos, A. D. L., Ascaso, C., and Moreno, J. (1996). Correlated biochemical and ultrastructural changes in nitrogen-starved Euglena gracilis. J. Phycol. 32, 953–963. doi: 10.1111/j.0022-3646.1996.00953.x
Geovanni Santiago-Martinez, M., Lira-Silva, E., Encalada, R., Pineda, E., Carlos Gallardo-Perez, J., Zepeda-Rodriguez, A., et al. (2015). Cadmium removal by Euglena gracilis is enhanced under anaerobic growth conditions. J. Hazard. Mater. 288, 104–112. doi: 10.1016/j.jhazmat.2015.02.027
Gilman, I. S., and Edwards, E. J. (2020). Crassulacean acid metabolism. Curr. Biol. 30:R57–R62. doi: 10.1016/j.cub.2019.11.073
Goto, K., and Beneragama, C. K. (2010). Circadian clocks and antiaging: do non-aging microalgae like Euglena reveal anything? Ageing Res. Rev. 9, 91–100. doi: 10.1016/j.arr.2009.09.003
Guo, Q., Bi, D., Wu, M., Yu, B., Hu, L., Liu, C., et al. (2020). Immune activation of murine RAW264.7 macrophages by sonicated and alkalized paramylon from Euglena gracilis. BMC Microbiol. 20:171. doi: 10.1186/s12866-020-01782-y
Halter, D., Casiot, C., Heipieper, H. J., Plewniak, F., Marchal, M., Simon, S., et al. (2012). Surface properties and intracellular speciation revealed an original adaptive mechanism to arsenic in the acid mine drainage bio-indicator Euglena mutabilis. Appl. Microbiol. Biotechnol. 93, 1735–1744. doi: 10.1007/s00253-011-3493-y
Harris, E. H. (2009). The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, 2nd Edn. San Diego, CA: Academic Press, 119–158, 267–274
Ishikawa, T., Tamaki, S., Maruta, T., and Shigeoka, S. (2017). Biochemistry and physiology of reactive oxygen species in Euglena. Adv. Experi. Med. Biol. 979, 47–64. doi: 10.1007/978-3-319-54910-1_4
Ivusic, F., and Santek, B. (2015). Optimization of complex medium composition for heterotrophic cultivation of Euglena gracilis and paramylon production. Bioprocess Biosyst. Eng. 38, 1103–1112. doi: 10.1007/s00449-015-1353-3
Johnson, C. H., Ivanisevic, J., and Siuzdak, G. (2016). Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17:451. doi: 10.1038/nrm.2016.25
Khatiwada, B., Hasan, M. T., Sun, A., Kamath, K. S., Mirzaei, M., Sunna, A., et al. (2020). Probing the role of the chloroplasts in heavy metal tolerance and accumulation in Euglena gracilis. Microorganisms 8:115. doi: 10.3390/microorganisms8010115
Lee, M.-C., Park, J. C., and Lee, J.-S. (2018). Effects of environmental stressors on lipid metabolism in aquatic invertebrates. Aquatic Toxicol. 200, 83–92. doi: 10.1016/j.aquatox.2018.04.016
Leonardi, R., and Jackowski, S. (2007). Biosynthesis of pantothenic acid and coenzyme A. Ecosal Plus 2:4. doi: 10.1128/ecosal.3.6.3.4
Levedahl, B. H. (1966). Heterotrophic CO2 fixation by a bleached Euglena. Exp. Cell Res. 44, 393–402. doi: 10.1016/0014-4827(66)90445-9
Libik-Konieczny, M., Kuzniak, E., Surówka, E., Slesak, I., Nosek, M, and Miszalski, Z. (2019). Crassulacean acid metabolism and its role in plant acclimatization to abiotic stresses and defence against pathogens. Prog. Botany 81, 277–306. doi: 10.1007/124_2019_33
Limami, A. M., Glévarec, G., Ricoult, C., Cliquet, J. B., and Planchet, E. (2008). Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J. Exp. Bot. 59, 2325–2335. doi: 10.1093/jxb/ern102
Linnane, A. W., and Stewart, P. R. (1967). The inhibition of chlorophyll formation in Euglena by antibiotics which inhibit bacterial and mitochondrial protein synthesis. Biochem. Biophys. Res. Commun. 27, 511–516. doi: 10.1016/S0006-291X(67)80016-0
Lu, S., Wang, J., Niu, Y., Yang, J., Zhou, J., and Yuan, Y. (2012). Metabolic profiling reveals growth related FAME productivity and quality of Chlorella sorokiniana with different inoculum sizes. Biotechnol. Bioeng. 109, 1651–1662. doi: 10.1002/bit.24447
Mangalam, A., Poisson, L., Nemutlu, E., Datta, I., and Giri, S. (2013). Profile of circulatory metabolites in a relapsing-remitting animal model of multiple sclerosis using global metabolomics. J. Clin. Cell. Immunol. 4, 1–12. doi: 10.4172/2155-9899.1000150
Mendoza-Cozatl, D., Devars, S., Loza-Tavera, H., and Moreno-Sánchez, R. (2002). Cadmium accumulation in the chloroplast of Euglena gracilis. Physiol. Plant. 115, 276–283. doi: 10.1034/j.1399-3054.2002.1150214.x
Moreno-Sánchez, R., Rodríguez-Enríquez, S., Jasso-Chávez, R., Saavedra, E., and García-García, J. D. (2017). Biochemistry and physiology of heavy metal resistance and accumulation in euglena. Adv. Exp. Med. Biol. 979, 91–121. doi: 10.1007/978-3-319-54910-1_6
Mukaida, S., Ogawa, T., Ohishi, K., Tanizawa, Y., Ohta, D., and Arita, M. (2016). The effect of rapamycin on biodiesel-producing protist Euglena gracilis. Biosci. Biotechnol. Biochem. 80, 1223–1229. doi: 10.1080/09168451.2016.1141040
Nakazawa, M., Hayashi, R., Takenaka, S., Inui, H., Ishikawa, T., Ueda, M., et al. (2017). Physiological functions of pyruvate:NADP+ oxidoreductase and 2-oxoglutarate decarboxylase in Euglena gracilis under aerobic and anaerobic conditions. Biosci. Biotechnol. Biochem. 81, 1386–1393. doi: 10.1080/09168451.2017.1318696
Park, S. G., Schimmel, P., and Kim, S. (2008). Aminoacyl tRNA synthetases and their connections to disease. Proc. Nat. Acad. Sci. U.S.A. 105, 11043–11049. doi: 10.1073/pnas.0802862105
Pavlík, M., Zemanová, V., Pavlíková, D., Kyjaková, P., and Hlavsa, T. (2017). Regulation of odd-numbered fatty acid content plays an important part in the metabolism of the hyperaccumulator Noccaea spp. adapted to oxidative stress. J. Plant Physiol. 208, 94–101. doi: 10.1016/j.jplph.2016.09.014
Peccia, J., Haznedaroglu, B., Gutierrez, J., and Zimmerman, J. B. (2013). Nitrogen supply is an important driver of sustainable microalgae biofuel production. Trends Biotechnol. 31, 134–138. doi: 10.1016/j.tibtech.2013.01.010
Periannan, A. (2003). Quantitative estimation of ebastine in formulations by HPTLC. Planar chromatography. JPC 2:16. doi: 10.1556/JPC.16.2003.2.16
Regnault, A., Piton, F., and Calvayrac, R. (1990). Growth, proteins and chlorophyll in Euglena adapted to various C/N balances. Phytochemistry 29, 3711–3715. doi: 10.1016/0031-9422(90)85318-A
Richter, P. R., Liu, Y., An, Y., Li, X., Nasir, A., Strauch, S. M., et al. (2015). Amino acids as possible alternative nitrogen source for growth of Euglena gracilis Z in life support systems. Life Sci. Space Res. 4:1. doi: 10.1016/j.lssr.2014.11.001
Rocchetta, I., Mazzuca, M., Conforti, V., Balzaretti, V., and Molina, M. D. C. R.D. (2012). Chromium induced stress conditions in heterotrophic and auxotrophic strains of Euglena gracilis. Ecotoxicol. Environ. Saf. 84, 147–154. doi: 10.1016/j.ecoenv.2012.07.020
Rodriguez-Zavala, J. S., Garcia-Garcia, J. D., Ortiz-Cruz, M. A., and Moreno-Sanchez, R. (2007). Molecular mechanisms of resistance to heavy metals in the protist Euglena gracilis. J. Environ. Sci. Health Part A Toxic. 42, 1365–1378. doi: 10.1080/10934520701480326
Salama, E. S., Kim, H. C., Abou-Shanab, R. A. I., Ji, M. K., Oh, Y. K., Kim, S. H., et al. (2013). Biomass, lipid content, and fatty acid composition of freshwater Chlamydomonas mexicana and Scenedesmus obliquus grown under salt stress. Bioprocess Biosyst. Eng. 36, 827–833. doi: 10.1007/s00449-013-0919-1
Schwarzhans, J. P., Cholewa, D., Grimm, P., Beshay, U., Risse, J. M., Friehs, K., et al. (2015). Dependency of the fatty acid composition of Euglena gracilis on growth phase and culture conditions. J. Appl. Phycol. 27, 1389–1399. doi: 10.1007/s10811-014-0458-4
Shao, Q., Jiang, Y., Lei, A., Hu, Z., and Wang, J. (2018). Advances in genetic transformation techniques of Euglena. Acta Hydrobiol. Sinica. 42, 207–214. doi: 10.7541/2018.081
Shepherd, J., and Ibba, M. (2013). Direction of aminoacylated transfer RNAs into antibiotic synthesis and peptidoglycan-mediated antibiotic resistance. FEBS Lett. 587, 2895–2904. doi: 10.1016/j.febslet.2013.07.036
Smith, C. A., Want, E. J., O'Maille, G., Abagyan, R., and Siuzdak, G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787. doi: 10.1021/ac051437y
Smith, E. F. (2009). Physiology of Woody Plants. San Diego, CA: Academic Press. doi: 10.1016/B978-0-12-370873-1.00044-7
Su, Y., Wang, J., Shi, M., Niu, X., and Zhang, W. (2014). Metabolomic and network analysis of astaxanthin-producing Haematococcus pluvialis under various stress conditions. Bioresour. Technol. 170, 522–529. doi: 10.1016/j.biortech.2014.08.018
Sugiyama, A., Suzuki, K., Mitra, S., Arashida, R., Yoshida, E., Nakano, R., et al. (2009). Hepatoprotective effects of paramylon, a β-1, 3-D-glucan isolated from Euglena gracilis Z, on acute liver injury induced by carbon tetrachloride in rats. J. Vet. Med. Sci. 71, 885–890. doi: 10.1292/jvms.71.885
Sui, X., Niu, X., Shi, M., Pei, G., Li, J., Chen, L., et al. (2014). Metabolomic analysis reveals mechanism of antioxidant butylated hydroxyanisole on lipid accumulation in Crypthecodinium cohnii. J. Agricult. Food Chem. 62, 12477–12484. doi: 10.1021/jf503671m
Takahashi, A., Shibata, N., Nishikawa, S., Ohnishi, K., Ishioka, N., and Ohnishi, T. (2006). UV-B light induces an adaptive response to UV-C exposure via photoreactivation activity in Euglena gracilis. Photochem. Photobiol. Sci. Offi. J. Eur. Photochem. Assoc. 5, 467–471. doi: 10.1039/b601971d
Takeyama, H., Kanamaru, A., Yoshino, Y., Kakuta, H., and Matsunaga, T. (2015). Production of antioxidant vitamins, beta-carotene, vitamin C, and vitamin E, by two-step culture of Euglena gracilis Z. Biotechnol. Bioeng. 53:185–190. doi: 10.1002/(SICI)1097-0290(19970120)53:2<185::AID-BIT8>3.0.CO;2-K
Tallima, H., and El Ridi, R. (2018). Arachidonic acid: physiological roles and potential health benefits - A review. J. Adv. Res. 11, 33–41. doi: 10.1016/j.jare.2017.11.004
Tanno, Y., Kato, S., Takahashi, S., Tamaki, S., and Shinomura, T. (2020). Light dependent accumulation of β-carotene enhances photo-acclimation of Euglena gracilis. J. Photochem. Photobiol. B. Biol. 209:111950. doi: 10.1016/j.jphotobiol.2020.111950
Tevatia, R., Allen, J., Rudrappa, D., White, D., Clemente, T. E., Cerutti, H., et al. (2015). The taurine biosynthetic pathway of microalgae. Algal Res. 9, 21–26. doi: 10.1016/j.algal.2015.02.012
Tevatia, R., Payne, S., Allen, J., White, D., and Blum, P. (2019). A synthetic cdo/csad taurine pathway in the green unicellular alga Chlamydomonas reinhardtii. Algal Res. 40:101491. doi: 10.1016/j.algal.2019.101491
Tossavainen, M., Ilyass, U., Ollilainen, V., Valkonen, K., and Romantschuk, M. (2019). Influence of long term nitrogen limitation on lipid, protein and pigment production of Euglena gracilis in photoheterotrophic cultures. PeerJ 7:6624. doi: 10.7717/peerj.6624
Wan, Q. L., Meng, X., Fu, X., Chen, B., Yang, J., Yang, H., et al. (2019). Intermediate metabolites of the pyrimidine metabolism pathway extend the lifespan of C. elegans through regulating reproductive signals. Aging 11:102033. doi: 10.18632/aging.102033
Wang, J., Zhang, X., Shi, M., Gao, L., Niu, X., Te, R., et al. (2014). Metabolomic analysis of the salt-sensitive mutants reveals changes in amino acid and fatty acid composition important to long-term salt stress in Synechocystis sp. PCC 6803. Funct. Integrat. Genomics 14, 431–440. doi: 10.1007/s10142-014-0370-7
Watanabe, M., and Suzuki, T. (2002). Involvement of reactive oxygen stress in cadmium-induced cellular damage in Euglena gracilis. Compar. Biochem. Physiol. Toxicol. Pharmacol. Cbp 131, 491–500. doi: 10.1016/S1532-0456(02)00036-4
Watanabe, S., Matsumoto, M., Hakomori, Y., Takagi, H., Shimada, H., and Sakamoto, A. (2014). The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ. 37, 1022–1036. doi: 10.1111/pce.12218
Xia, J., Sinelnikov, I. V., Han, B., and Wishart, D. S. (2015). MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 43, 251–257. doi: 10.1093/nar/gkv380
Yang, L., Chen, J., Qin, S., Zeng, M., and Wang, J. (2018). Growth and lipid accumulation by different nutrients in the microalga Chlamydomonas reinhardtii. Biotechnol. Biofuels. 11:40. doi: 10.1186/s13068-018-1041-z
Yanming, W., Tuulikki, S. N. L., Heiko, R., Wiebe, M. G., and Adrianna, I. (2018). Euglena gracilis growth and cell composition under different temperature, light and trophic conditions. PLoS ONE. 13:e0195329. doi: 10.1371/journal.pone.0195329
Zakryś, B., Milanowski, R., and Karnkowska, A. (2017). Evolutionary origin of Euglena. Oxygen Transport Tissue. 979, 3–17. doi: 10.1007/978-3-319-54910-1_1
Zhang, T., Zhang, S., Chen, L., Ding, H., Wu, P., Zhang, G., et al. (2020). UHPLC-MS/MS-Based nontargeted metabolomics analysis reveals biomarkers related to the freshness of chilled chicken. Foods 9:1326. doi: 10.3390/foods9091326
Keywords: Euglena, metabolomics, antibiotics, heavy metals, nutrient deprivation, environmental stresses
Citation: He J, Liu C, Du M, Zhou X, Hu Z, Lei A and Wang J (2021) Metabolic Responses of a Model Green Microalga Euglena gracilis to Different Environmental Stresses. Front. Bioeng. Biotechnol. 9:662655. doi: 10.3389/fbioe.2021.662655
Received: 01 February 2021; Accepted: 10 June 2021;
Published: 20 July 2021.
Edited by:
Wei Xiong, National Renewable Energy Laboratory (DOE), United StatesCopyright © 2021 He, Liu, Du, Zhou, Hu, Lei and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangxin Wang, anh3YW5nQHN6dS5lZHUuY24=
 Jiayi He
Jiayi He ChenChen Liu
ChenChen Liu Mengzhe Du3
Mengzhe Du3 Zhangli Hu
Zhangli Hu Anping Lei
Anping Lei Jiangxin Wang
Jiangxin Wang