
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 17 June 2021
Sec. Bioprocess Engineering
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.650247
This article is part of the Research TopicEndophytes and Their Biotechnological ApplicationsView all 9 articles
 Rajreepa Talukdar1
Rajreepa Talukdar1 Srichandan Padhi2
Srichandan Padhi2 Amit K. Rai2
Amit K. Rai2 Marco Masi3
Marco Masi3 Antonio Evidente3
Antonio Evidente3 Dhruva Kumar Jha1
Dhruva Kumar Jha1 Alessio Cimmino3
Alessio Cimmino3 Kumananda Tayung1*
Kumananda Tayung1*An endophytic fungus isolated from healthy leaf tissues of Houttuynia cordata Thunb., an ethnomedicinal plant of North East India, showed a considerable amount of antimicrobial activity. The ethyl acetate extract of the fungal culture filtrates displayed promising antimicrobial activity against a panel of clinically significant pathogens including Candida albicans, Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa. Bioassay guided purification of the organic extract using column and thin layer chromatography yielded a pure homogenous compound which was identified using spectroscopic methods (essentially by 1H NMR and MS) as tyrosol, a well-known phenylethanoid present in several natural sources. Besides, molecular docking studies against tyrosyl tRNA synthetases (TyrRS) of S. aureus (PDB ID: 1JIL) and E. coli (PDB ID: 1VBM), and CYP45014α-lanosterol demethylase (CYP51) of C. albicans (PDB ID: 5FSA) revealed tyrosol has a strong binding affinity with the enzyme active site residues. The fungus was identified as Colletotrichum sp. and characterized by its genomic ITS rDNA and ITS2 sequences. Phylogenetic analyses showed clustering of our isolate with Colletotrichum coccodes. Species of Colletotrichum are also reported to be plant pathogens. Therefore, to confirm the endophytic lifestyle of the isolate, ITS2 RNA secondary structure study was undertaken. The result indicated our isolate exhibited differences in the folding pattern as well as in motif structures when compared to those of pathogenic C. coccodes. The findings indicated that endophytic fungi harboring H. cordata could be explored as a potent source of antimicrobial agents.
Exploring the secondary metabolites produced by microorganisms colonizing unusual or unique environmental settings have turned on new possibilities toward discovering novel compounds of therapeutic interest and developing them as lead drugs. Endophytic fungi associated with the plants used in the traditional medicines are of special interest as these live in between the intercellular spaces of healthy tissues and cause hardly any infection while also producing a plethora of novel chemical entities with diverse pharmacological and biotechnological applications (Strobel and Daisy, 2003). Furthermore, the symbiotic association of these endophytes with the host plants indicates their compounds to be less toxic to the cell and does not kill the eukaryotic host system (Shubin et al., 2014; Chutulo and Chalannavar, 2018). The total number of fungal species including the endophytes is estimated to be 1.5 million, of which only 6–7% has been described till now, while the remaining still waiting to be introduced to the existing world of microbes. Besides, it is also estimated that 51% of bioactive substances isolated from these endophytic fungi were previously unknown (Hawksworth, 2001; Kharwar et al., 2011). On the other hand, bioinformatics approaches such as molecular docking have emerged as a usual and essential component of drug discovery and design pipeline. It assigns and employs a variety of conformation search strategies to the receptor and ligands, and predicts possible binding affinity by establishing chemical interactions between them (Ruyck et al., 2016; Padhi et al., 2020).
Northeast India, a major part of the Indo-Burma Belt, is recognized as a global biodiversity hotspot owing to its unique habitat and rich diversity of endemic flora and fauna. This region is a treasure-house of a large number of medicinal plants which are being used by various ethnic tribes for the treatment of various ailments. However, a repertoire of them remains untapped despite their tremendous potential in traditional medicines. Herein, an endophytic fungus identified as Colletotrichum sp. was isolated from the healthy leaf tissues of Houttuynia cordata Thunb., a potential ethnomedicinal plant being used for the treatment of stomach related disorders. The organic extract of its culture filtrates displayed considerable antimicrobial activity against some clinically significant human test pathogens. The isolate was also characterized in terms of ITS rDNA sequence and ITS2 secondary structures. The bioactive extract which was purified by chromatographic and spectroscopic techniques yielded a pure compound identified as tyrosol. Molecular docking studies revealed that the compound possesses a high binding affinity toward specific bacterial and fungal enzymes responsible for their growth and pathogenesis.
The fungus was isolated from healthy leaf tissues of H. cordata collected from Sonapur (26° 06′ 57.5″ N 91° 58′ 44.8″ E), Assam, India predominantly inhabited by tribal communities. A voucher specimen of the plant species has been deposited at the herbaria of Department of Botany, Gauhati University with accession number 18538 upon identification. Briefly, for isolation of endophytic fungi, healthy leaves were washed thoroughly with tap water followed by washing with 10% mild bio-detergent and finally with double distilled water to remove the surface debris. Surface sterilization of the leaves was carried out following standard protocol with slight modifications (Jena and Tayung, 2013). The leaves were dipped sequentially in 70% ethanol (3 min), followed by 0.5% sodium hypochlorite (NaOCl) (2 min), and then rinsed thoroughly with sterile distilled water (1 min). Finally, the leaves were dried over sterile blotting paper inside the laminar airflow chamber. Surface sterilized leaves measuring about 0.5 mm in diameter were then punched out using a sterile puncture and were inoculated onto Petri plates containing potato dextrose agar (PDA) medium supplemented with streptomycin sulfate (50 μg/ml). The plates with the leaf fragments were then incubated at 25 ± 2°C for 2 weeks and observed once a day for the growth of mycelia. Hyphal tips growing out of the leaf fragments were transferred to PDA slants, sub-cultured, and stored at 4°C. The isolates were identified based on their morphological and microscopic characters referring to standard identification manuals (Gilman, 1971; Barnett and Hunter, 1996; Damm et al., 2012). The fungus used in the present study was one of the several endophytic fungal isolates obtained from the surface-sterilized leaf fragments which showed considerable antimicrobial activity during the preliminary antimicrobial assay.
The fungus was cultivated in potato dextrose broth (PDB) and incubated at 28°C in BOD shaking incubator for 2 weeks at 120 rpm. After the specified incubation period, the culture broth was filtered through sterile Whatman filter paper to remove the mycelia mat. The filtrate so obtained was collected and fungal metabolites were extracted using an equal volume of ethyl acetate (EtOAc) through vigorous shaking for 10–15 min in a separating funnel. The solvent was then evaporated in a rotary evaporator and the organic extract thus obtained was weighed. The antimicrobial activity of the extracts was determined by Agar cup diffusion assay against some clinically significant test pathogens. The test organisms included three bacterial pathogens, namely, Staphylococcus aureus (MTCC 737), Pseudomonas aeruginosa (MTCC 424), and Escherichia coli (MTCC 443), and one pathogenic fungus, Candida albicans (MTCC 227). The test organisms were procured from the Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (CSIR-IMTECH), Chandigarh, India. The test organisms were activated by cultivating them on freshly prepared nutrient agar (NA) and sabouraud dextrose agar (SDA) media, respectively for bacterial and fungal pathogens. Meanwhile, sterile NA and SDA plates were prepared and the plates were inoculated with 0.2 ml of overnight grown bacterial and fungal cultures containing 1.0 × 106 cells. A lawn culture was prepared on each plate by evenly spreading the inoculums with the help of a sterile cotton swab. Agar cups were prepared in the plates by scooping out the solid medium with a sterile cork borer (7 mm in diameter). The cups were then filled in with 100 μl of the EtOAc organic extracts dissolved in 10% dimethyl sulfoxide (DMSO) at a concentration of 1 mg/ml and incubated respectively at 37 ± 1°C for 24 h for bacterial and at 28°C ± 1°C for 48 h for fungal pathogens. The assay was carried out in triplicates and antimicrobial activity was determined by the appearance of clear zones of inhibition against the test organism around the agar cups. 10% DMSO was used as the negative control for the assay.
The minimum inhibitory concentration (MIC) of the organic extract was determined by micro-broth dilution assay in a sterile 96-well plate (Padhi and Tayung, 2013) against P. aeruginosa, S. aureus, E. coli, and C. albicans. A two-fold dilution of the extracts with the concentration ranging from 1,000 to 62.5 μg/ml was made. The wells were filled with 90 μl of each test bacterial and fungal suspension (approx. 106 CFU/ml). Test extracts (10 μl) of different concentrations were added into each well to make upto a final volume of 100 μl. Medium containing 10% DMSO was used as the negative control. After incubation at 37°C for 24 h for bacteria and 48 h for fungal pathogens, a solution (10 μl) of triphenyltetrazolium chloride (TTC) was added to each well as microbial growth indicator, and the microplates were incubated for an additional 30 min. MIC was determined as the lowest concentration of the test extract at which no pink color appeared.
The organic extract obtained from Colletotrichum coccodes HCS3 culture filtrates (99.9 mg) was purified on a silica gel (0.063−0.200 mm, Kieselgel 60, Merck) column eluted with chloroform and isopropanol (CHCl3-i-PrOH 9:1) and a specific volume of eluent (5 ml) was collected in vials. The fraction contained in each vial was subjected to thin-layer chromatography (TLC). The plates were visualized by exposure to UV radiation or spraying first with 10% sulfuric acid (H2SO4) in methanol (MeOH) and then with 5% phosphomolybdic acid in ethanol (EtOH), followed by heating at 110°C for 10 min on a hot plate. The vials containing related fractions were clubbed into one vial affording seven homogeneous fractions. The fractions were concentrated in a rotary evaporator, weighed, and evaluated for antimicrobial activity at 1 mg/ml against the test organisms using an agar well diffusion assay. The active fractions were further purified by analytical and preparative TLC eluted with CHCl3-i-PrOH (9:1) yielding a pure and homogenous compound. To elucidate the structure of this pure compound and also to check the purity, 1H NMR spectra were recorded in deuterated chloroform (CDCl3) at 500 MHz on a Varian instrument. The same solvent (CDCl3) was used as an internal standard. Electrospray Ionization/Mass Spectrometry(ESI/MS) and liquid chromatography (LC)/MS analyses were performed using the LC/MS time-of- light (TOF) system (Agilent 6230B, HPLC 1260 Infinity) column Phenomenex LUNA [C18 (2), 5 μm, 150 mm × 4.6 mm]. Analytical and preparative TLC was carried out on silica gel (0.25 and 0.5 mm, respectively, F254, Kieselgel 60, Merck) and reversed-phase (0.20 mm, F254, Kieselgel 60 RP-18, Merck) plates.
Based on the antimicrobial activity of the EtOAc extract, molecular docking was carried out to examine the possible binding affinity of the isolated compound toward TyrRS of S. aureus (1JIL) and E. coli (1VBM), and CYP51 of C. albicans (5FSA). The 3D crystal structures of these enzymes were retrieved from Protein Data Bank (PDB) and optimized (removal of unwanted water molecules, heteroatoms, and addition of polar hydrogens, missing amino acids) using the “prepare protein” protocol of Discovery Studio (DS) Client v20.1.0.19.295. Similarly, the 2D structure of the metabolite was optimized using the “prepare ligand” protocol. The active site of the enzymes was selected as the binding site for the ligand and a docking experiment was performed using the DS CDOCKER program which is an implementation of a CHARMm based docking tool (Gagnon et al., 2014). Docking optimization was carried out with a root mean square threshold (RMSD) 0.5 Å and pose cluster radius 0.5 to ensure the docked poses are diverse. The pose with the highest negative interaction energy was selected as the best binding conformation. The entire experiment was carried out following the procedure mentioned by Padhi et al. (2021).
Species confirmation of the endophytic isolate was determined by ITS rDNA sequence analysis. The fungal strain was cultured on PDB and a small amount of the mycelia was suspended in 40 μl MQ water. Genomic DNA was isolated by the CTAB method (Clarke, 2009). A portion of the genomic DNA was diluted upto 50 ng/μl for use in PCR. The nuclear ribosomal DNA and ITS region of the isolate were amplified using the universal primers ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The PCR was set up using the following components: 2.5 μl buffer (109), 1.5 μl MgCl2 (25 mM), 2.5 μl dNTPs (2 mM), 0.2 μl Promega Taq (5 U/μl), 1.0 μl each of forward and reverse primers (5 pm/μl) and 6.0 μl DNA from the diluted extract. The PCR condition was run with an initial denaturation at 94°C for 3 min. Denaturation, annealing, and extension were done at 96°C for 10 s, 55°C for 10 s, and 72°C for 30 s, respectively, in 45 cycles. The final extension was done at 72°C for 10 min and held at 4°C. After the PCR cycle, 2 μl of the product was used to check on 1% agarose gel. DNA sequencing was performed using an ABI 3730 sequencer. The forward and reverse sequence reads thus obtained were assembled to generate the contig using CAP3, a bioinformatics tool used for the assembling of genomic DNA reads (Huang and Madan, 1999). The annotated ITS rDNA contig was submitted to GenBank and an accession number was obtained.
A homology search and analysis for the ITS rDNA sequence as obtained above was performed with the Basic Local Alignment Search Tool (BLAST). Based on the results, 995 ITS rDNA sequences belonging to Colletotrichum spp. and having similarity with the isolate were randomly selected and retrieved from GenBank. The sequences were further filter searched and altogether 83 sequences with complete ITS rDNA and 30 sequences with ITS2 which were trimmed from the ITS rDNA using ITS2 secondary structure database (Koetschan et al., 2010) were selected for phylogenetic study. For phylogenetic analysis, multiple sequence alignments were performed using CLUSTALW software utilizing default settings, and trees were generated by the character state Maximum Parsimony (MP) method using MEGA 6.0 (Tamura et al., 2013). The robustness of the tree was assessed by bootstrap analysis with 1,000 replications. Based on the phylogenetic analysis, selected ITS2 sequences belonging to C. coccodes of varied lifestyles (endophytic and pathogenic) were used to generate the RNA secondary structure using the mfold web server (Zuker, 2003). The structure prediction was performed with a temperature of 37°C; ionic conditions: 1 M NaCl, no divalent ions; the maximum number of nucleotides in a bulge or loop: 30; maximum asymmetry of an interior/bulge loop: 30; percentage sub-optimality number: 5 and upper bound on the number of computed foldings: 50. The structure chosen from different output files was the one with the highest negative free energy if various similar structures were obtained. The consensus ITS2 secondary structures were compared among different lifestyles and differences in their folding patterns as well as in number and types of motifs were investigated.
The isolate described in this study was designated as strain HCS3 and was one among the several isolates that showed antimicrobial activity against the test pathogens used. The isolate was identified as Colletotrichum sp. based on the colony morphology and microscopic reproductive structures. The isolate was grown on PDA and malt extract agar (MEA) for identification. Colonies on PDA were circular, white initially with an orange tint that turns gray with age and darker at the center with black spots. Aerial mycelia white to grayish, cottony without visible conidial masses. The reverse side was white to pale orange in color. Growth on PDA was slow showing a diameter of about 55 mm in 7 days at 25°C (Figures 1A,B). While colonies on MEA were white with an orange tint at first but turned gray to dark blackish later. Colonies showed concentric ring-like growth both on front and reverse sides. Aerial mycelia were dark gray, sparse, and turned darker with age with conidial masses. The growth rate on MEA was faster showing a diameter of 67 mm in 7 days at 25°C (Figures 1C,D). Conidia measures 20–22 × 5–7 μm and is aseptate, smooth-walled, hyaline, and cylindrical with rounded ends, often with conspicuous papillate basal scars, guttulate, and slightly constricted centrally (Figure 2).
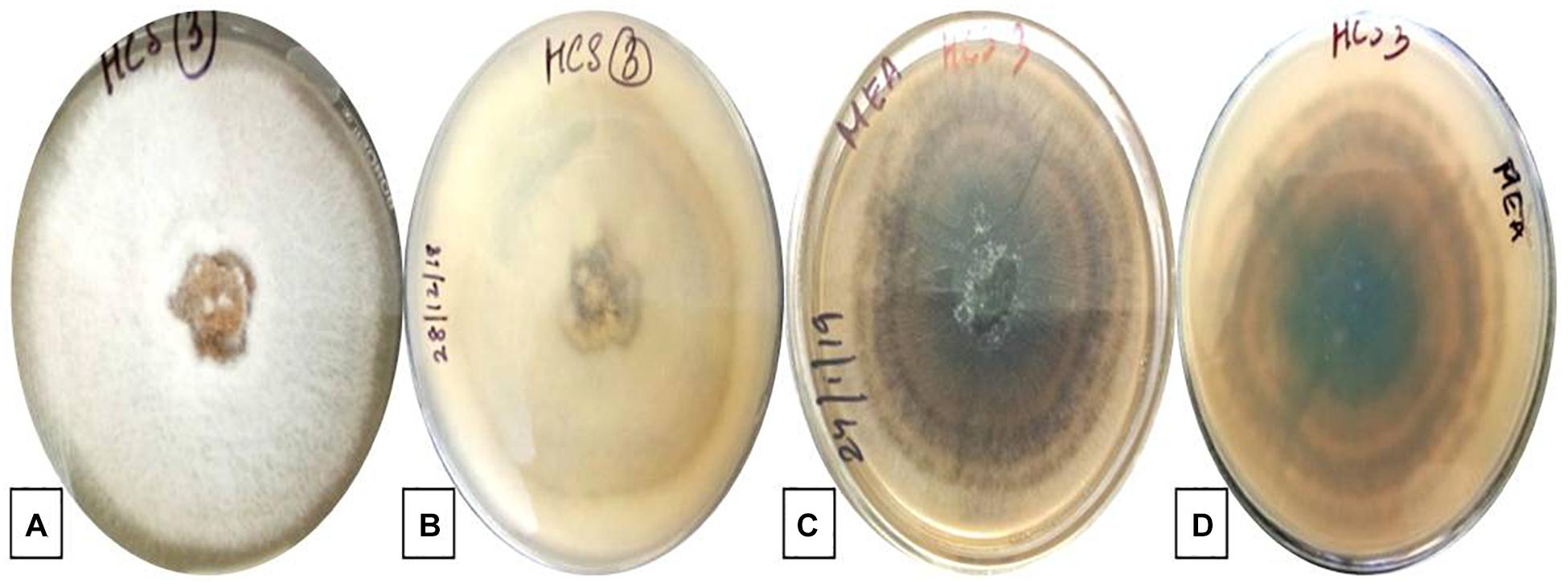
Figure 1. Colony morphology of Colletotrichum coccodes. (HCS3) on PDA: (A) front view, (B) reverse view and on MEA: (C) front view, (D) reverse view.
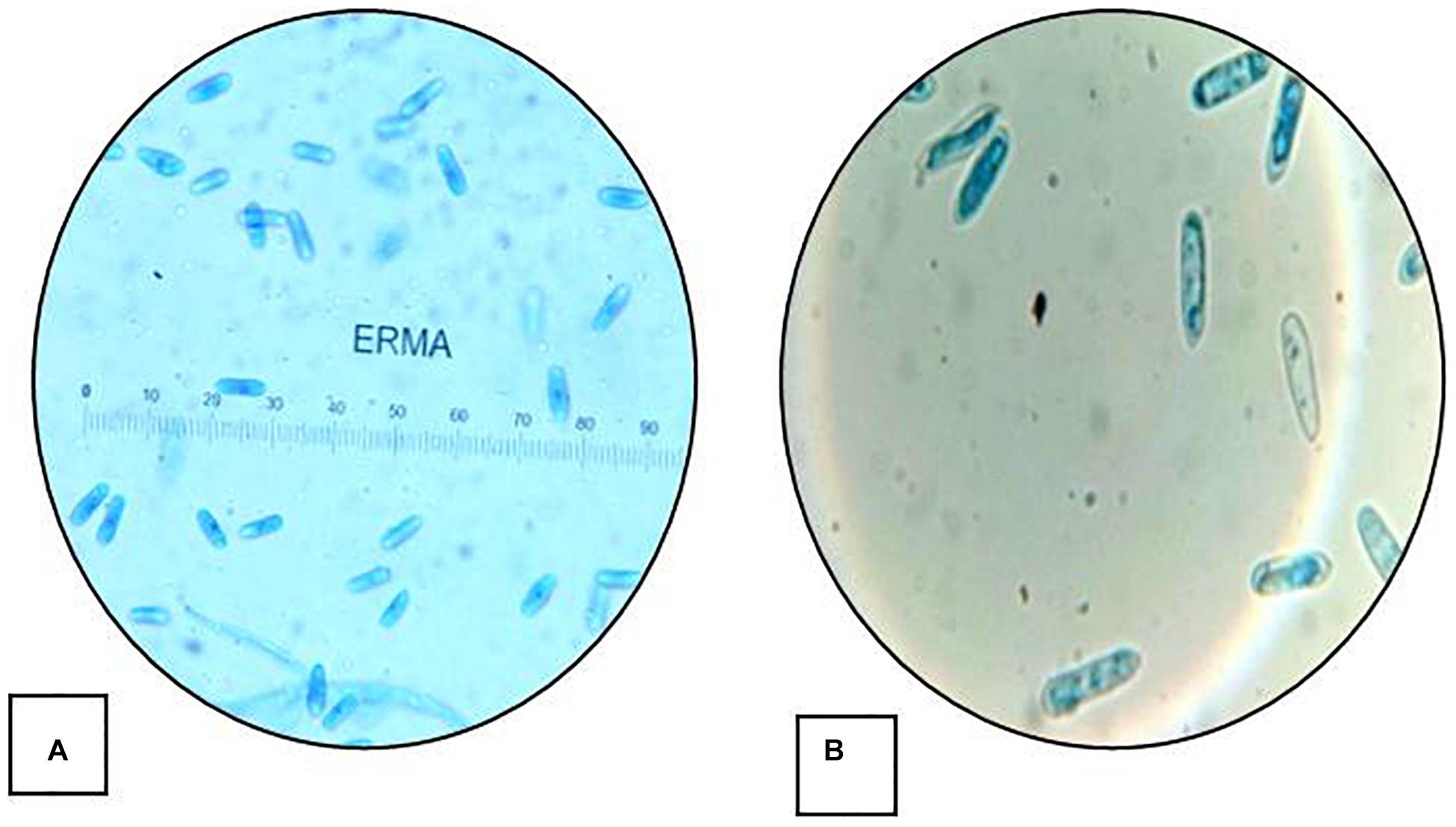
Figure 2. Conidial morphology Colletotrichum coccodes. (HCS3): (A) conidia (20–22 μm × 5–7 μm) (40×) and (B) conidia (100×).
The results showed the organic extract exhibited considerable antimicrobial activity against all the test pathogens used. However, maximum inhibition was observed against S. aureus followed by E. coli and C. albicans (Figure 3A). MIC of the organic extract was determined by micro-broth dilution assay in sterile 96-well plate against all the test pathogens, P. aeruginosa, S. aureus, E. coli, and C. albicans. Different concentration of the organic extract such as 1,000, 500, 250, 125, and 62.5 μg/ml was used to determine the MIC. The wells were filled with 90 μl of each test bacterial and fungal suspension with 10 μl of different concentrations of the test extract. The MIC for S. aureus, E. coli and C. albicans was found to be 125 μg/ml and that for P. aeruginosa was 250 μg/ml (Figure 3B).
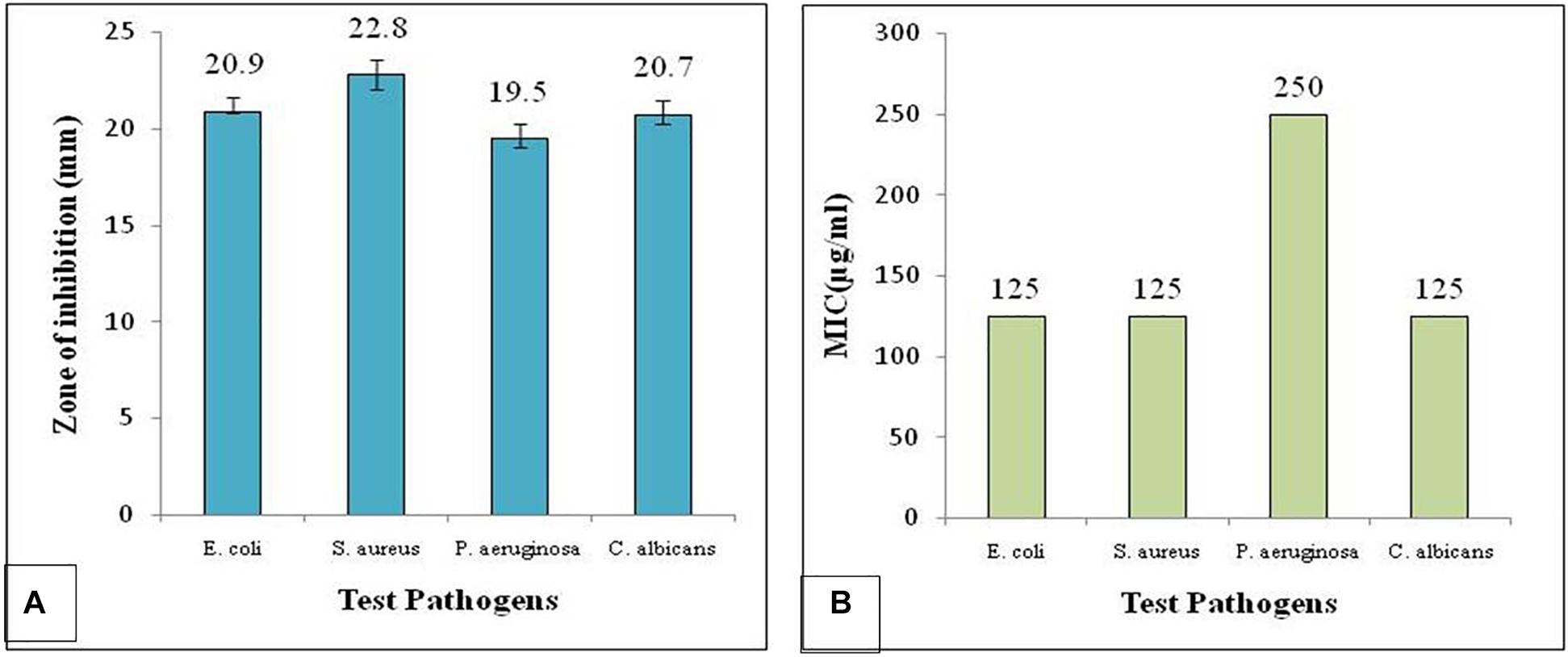
Figure 3. Antimicrobial activity of the Colletotrichum coccodes (EtOAc extract) showing (A) zone of inhibition and (B) MIC against test pathogens.
The EtOAc extract was purified on column chromatography and yielded 7 homogenous fractions namely F1 (3.5 mg), F2 (4.0 mg), F3 (7.6 mg), F4 (2.6 mg), F5 (1.8 mg), F6 (1.5 mg), F7 (2.1 mg), and MeOH washes (37.4 mg). Among all, the fraction F3 was observed to be having substantial activity against the test pathogens, and the residue was further purified on TLC to yield a pure and homogenous solid (2.3 mg). 1H NMR spectrum of this compound showed signal system characteristics for a p-substituted benzene derivative and a hydroxyethyl residue. The compound was identified as “tyrosol” (Figure 4) by comparing its spectroscopic properties with those available in the literature. The identification was further validated and confirmed by ESI-MS spectrum recorded in a positive modality which showed the dimer sodiated [2M + Na]+, and sodiated [M + Na]+ adduct ions at m/z: 299 and 161, respectively. The spectroscopic descriptions of tyrosol are given below;
Tyrosol: 1H NMR (500 MHz), δ: 7.10 (d, J = 8.2 Hz, H-2 and H-6), 6.79 (d, J = 8.2 Hz, H-3 and H-5), 3.82 (t, J = 6.6 Hz, H2-8), 2.80 (t, J = 6.6 Hz, H2-7); ESIMS (+), m/z: 299 [2M + Na]+, 161 [M + Na]+.
Molecular docking was carried out to understand the binding interaction and affinity of tyrosol toward the enzymes TyrRS of S. aureus and E. coli, and CYP51 of C. albicans. The isolated compound showed varied affinity toward the active sites of different enzymes used. The affinity was highest against TyrRS of S. aureus (CDOCKER energy −26.8457 kcal/mol; interaction energy −30.7030 kcal/mol) followed by that of E. coli (CDOCKER energy −24.8513 kcal/mol; interaction energy −29.3093 kcal/mol). The binding affinity of the tyrosol was also observed to be substantial against CYP51 of C. albicans. The binding between tyrosol and TyrRS of S. aureus was represented by two covalent C–H and three non-covalent (2 H-bond and 1 Pi-Alkyl) interactions. Similarly, the interaction between tyrosol and TyrRS of E. coli was characterized by 1 covalent C–H and 6 non-covalent ones including 1 Pi-alkyl and 5 H-bonds. However, the test ligand tyrosol was found to be bonded to the C. albicans CYP51 active site with the support of five non-covalent bonds including 3 H-bond, 1 Pi-Alkyl, and 1 Pi-Pi T shaped bonds. The CDOCKER energy and interaction energy for this binding was respectively computed to be −20.7595 and −24.2725 kcal/mol. The types of interaction, bond distance, and amino acids involved between tyrosol and the target enzymes are given in Tables 1, 2.
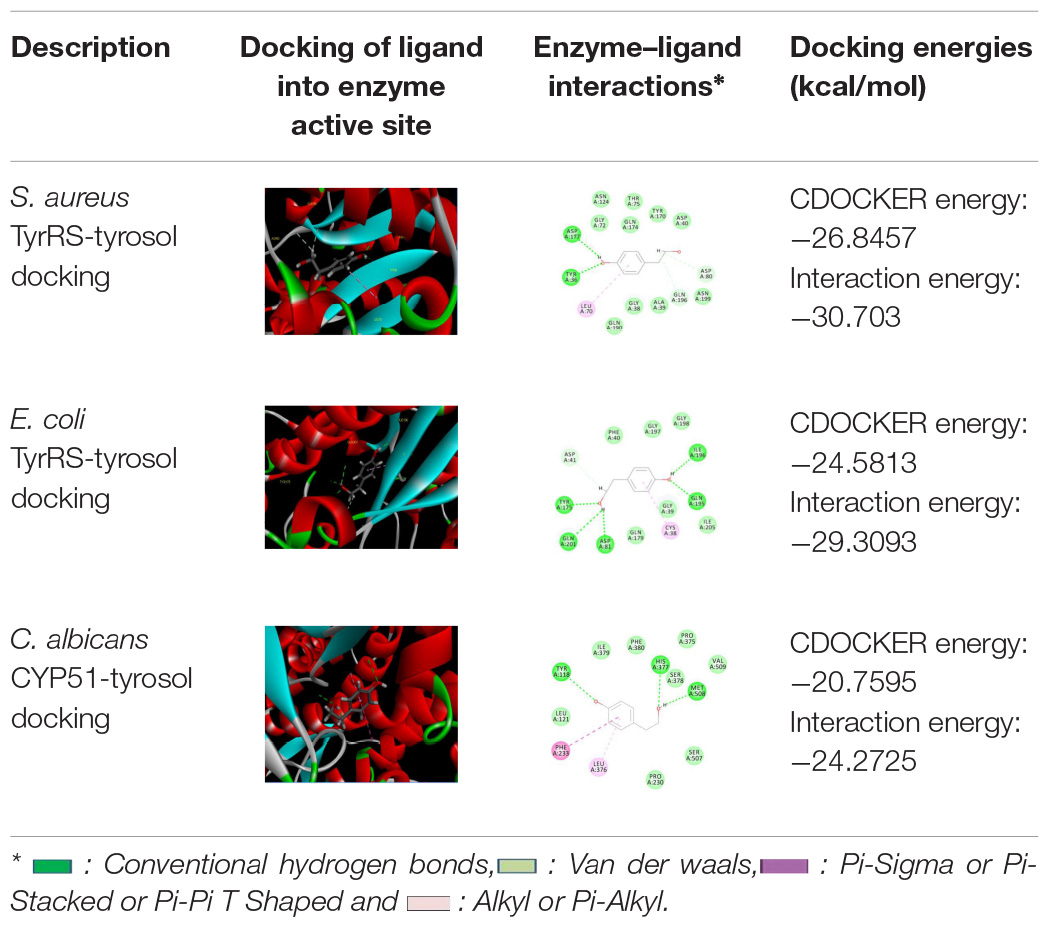
Table 1. Molecular docking of tyrosol into the active sites of TyrRS (S. aureus and E. coli) and CYP51 (C. albicans).
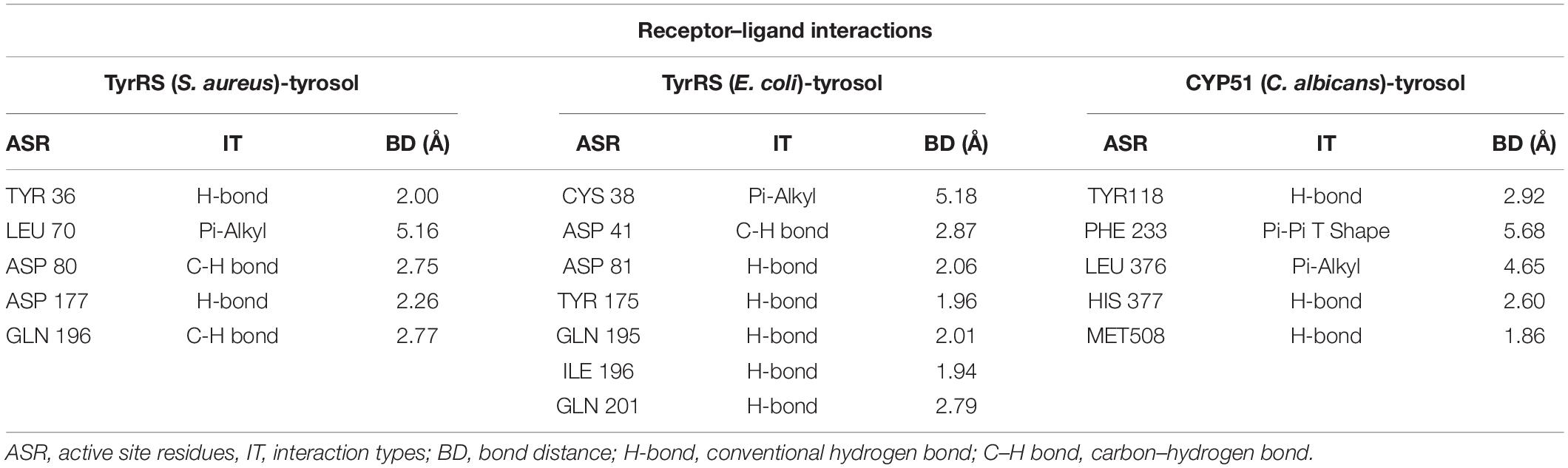
Table 2. Receptor-ligand interactions between tyrosol and TyrRS (S. aureus and E. coli) and CYP51 (C. albicans).
The genomic DNA of the isolate was isolated and the region for ITS rDNA was amplified and sequenced using universal primers ITS4 and ITS5. The contig thus obtained from assembling the forward and reverse reads were deposited to the GenBank with the accession MN128230.1. A BLAST homology searches ITS rDNA was carried out against the NCBI non-redundant nucleotide (nr) database and the results showed our isolate resembling that of Colletotrichum coccodes (MF076580.1) with Maximum Identity 98.25%; Query Coverage 91% and E-value 0.0. Based on this, a total of 995 ITS rDNA sequences belonging to different Colletotrichum spp., were randomly retrieved from GenBank and screened for the presence of complete ITS rDNA region (18S rDNA-ITS1-5.8S-ITS2-rDNA-28S rDNA). Phylogenetic analysis was performed using selected 83 sequences and the tree showed clustering of our isolate under the clade C. coccodes (Figure 5A). Further, the sequences were trimmed for the ITS2, and as such a final tree was constructed using 30 ITS2 sequences. The MP phylogenetic tree displayed our isolate shared a close affinity toward C. coccodes which is supported by a bootstrap value 92 (Figure 5B).
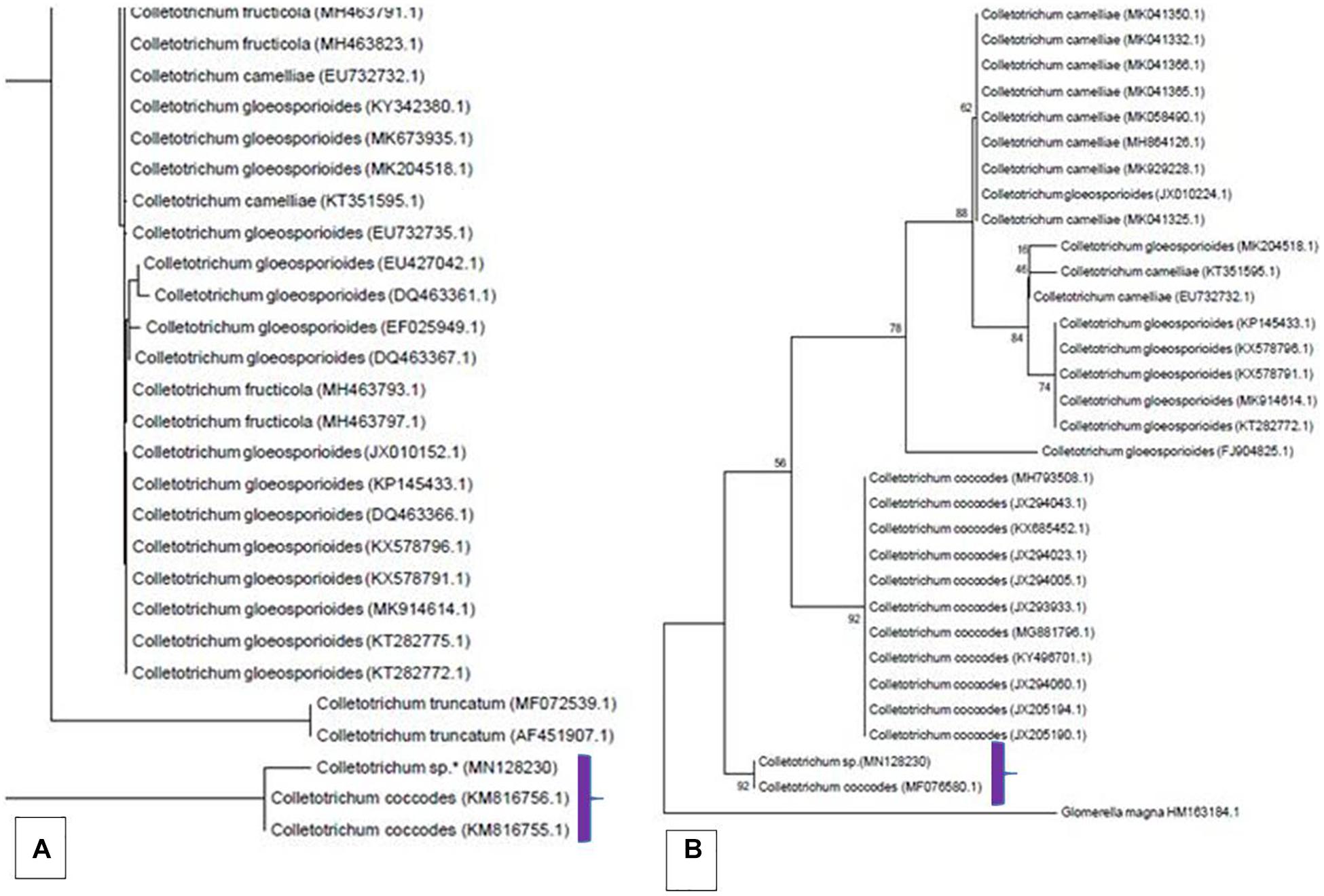
Figure 5. Snapshots of phylogenetic tree generated using Maximum Parsimony (MP) method showing clustering of our isolate under the clade Colletotrichum coccodes. Trees were constructed using ITS rDNA (A) and ITS2 (B) sequences.
Prediction of secondary structures was carried out using selected 12 ITS2 sequences belonging to C. coccodes (11 pathogenic and 1 endophytic) and was compared to that of our isolate. Briefly, an ITS2 consensus structure for the pathogenic C. coccodes was generated and a comparative assessment between the pathogenic, endophytic lifestyles and our isolate was made. The consensus structure for pathogenic C. coccodes consisted of a conserved core bulge of which radiating three major helices (H1, H2, and H3), however, the ITS2 secondary structure for the endophytic C. coccodes as well as for that belong to our endophytic isolate comprised of one incomplete (H1) and four complete helices (H2, H3, H4, and H5) (Figure 6). The results displayed the pathogenic and the endophytic ITS2 structures exhibited different folding patterns and dissimilar structural motifs. Our isolate shared extreme similarity with that of endophytic lifestyle both in terms of ITS2 folding pattern as well as identical structural motifs. The details about different structural features among ITS2 structures of different lifestyles of C. coccodes are presented in Table 3.
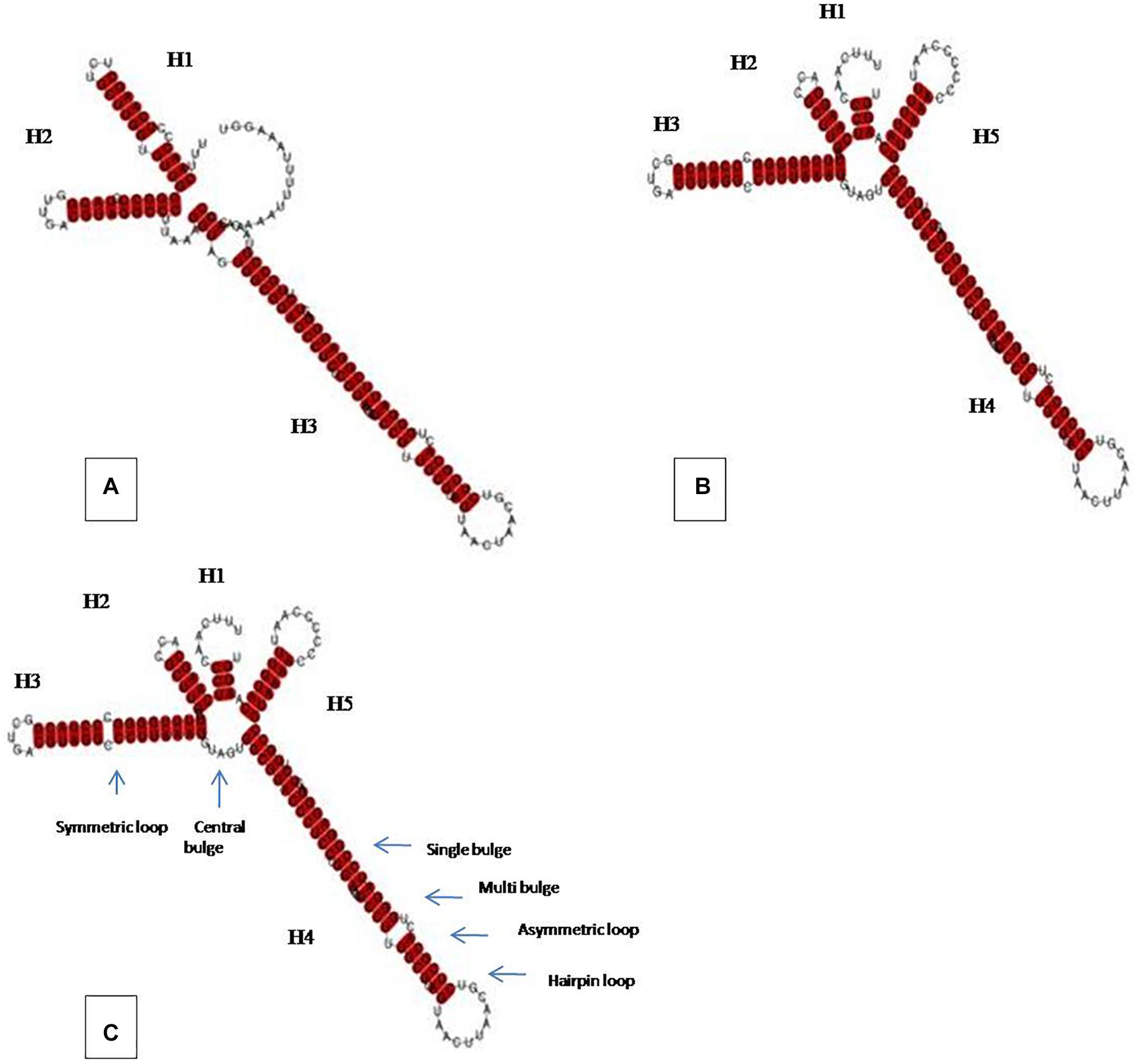
Figure 6. ITS2 secondary structures of C. coccodes among different life styles showing helices emerging from a central bulge, (A) pathogenic, (B) endophytic and (C) own isolate (MN128230.1). Motifs in the structures are indicated by arrow.
Endophytic fungi which live in close association with vascular plants without causing any obvious symptoms are an important component of plant-micro ecosystems. They confer profound impacts on host plants as mutualists by promoting their growth and tolerance to diseases (Abdel-Azeem et al., 2019; Kaur, 2020). There is growing interest among researchers to isolate and characterize the endophytic fungi inhabiting medicinal plants as they have been producing a plethora of chemical entities of biotechnological and pharmaceutical significance. Besides, some of them have also the ability to produce the same bioactive metabolites as their host plants (Tan and Zou, 2001; Schulz et al., 2002; Gouda et al., 2016). In the present study, endophytic fungi colonizing leaf tissues of H. cordata Thunb., an ethnomedicinal plant of North East India was investigated. Among several other endophytic isolates, a fungus identified as Colletrotrichum sp. (HCS3), showed considerable antimicrobial activity against some clinically important pathogenic bacteria and fungi. The organic extract exhibited strong inhibition against S. aureus, E. coli, P. aeruginosa, and C. albicans with MIC range 125–250 μg/ml. The occurrence of Colletotrichum spp. as an endophyte is not rare; it has been isolated from several medicinal plants with potent biological activity against several drug-resistant pathogens (Lu et al., 2000; Arivudainambi et al., 2011; Sharma et al., 2017; Kim and Shim, 2019).
Molecular tools including phylogenetic analyses that aid along with the traditional techniques have been proven to be successful for correct identification of fungi and have further revolutionized the fungal classification (Sarwar et al., 2019). Moreover, the ITS region which consists of a highly conserved 5.8S rRNA and variable regions ITS1 and, ITS2, is one of the widely used phylogenetic markers for fungal species identification (Tamura et al., 2004; Xu and Adamowicz, 2016). Further, the spacer regions such as ITS1 and ITS2 are reportedly useful in detecting and identifying fungal pathogens from clinical specimens and environmental samples. Also, recent reports revealed the sequence variability in ITS2 is more apposite for phylogenetic reconstruction and species differentiation in eukaryotes as well as in fungi (Iwen et al., 2002). The resolution of the phylogenetic and evolutionary prediction can be improved further if structural features are taken into account as RNA secondary structures include morphological information that is not found in the sequence (Marinho et al., 2011; Heeg and Wolf, 2015; Zhang et al., 2020). Undeniably, ITS2 secondary structure plays a critical part in rRNA processing and formation of functional 60S subunit. Despite considerable variation in its nucleotide sequences, a typical eukaryotic ITS2 secondary structure (also known as four-helix or ring-pin model) consists of four helices that are maintained through several common and shared structure motifs including internal loops, hairpin loops, and bulges (Schultz et al., 2005; Kapoor et al., 2018). These structure motifs impart stability to the ITS2 structure and are reportedly susceptible to polymorphisms that further give rise to non-canonical G–U pairs within the conserved regions of double-stranded helices (Cote and Peculis, 2001; Freire et al., 2012). Above all, closely related species share very similar folding patterns of ITS2 with nearly identical structure motifs (Rampersad, 2014; Verma et al., 2020). Homology search and phylogenetic analysis using ITS as well as ITS2 sequence of the isolate (HCS3) revealed it to be Colletotrichum coccodes as the trees displayed clustering of our isolate under C. coccodes. Furthermore, a comparison of ITS2 secondary structure features revealed close affinity among pathogenic and endophytic C. coccodes including significant differences in the folding pattern and motif structures. Similar observations have also been put forth by several studies (Padhi and Tayung, 2013; Padhi et al., 2016) which suggested ITS2 structure could be used as a phylogenetic marker to establish similarities and variations between different lifestyles of the same species.
The EtOAc-derived organic extract which showed promising activity in inhibiting the growth of test pathogens was further purified to yield a potent phenolic compound identified to be tyrosol based on its NMR and ESIMS spectra. Tyrosol is a well-known secondary metabolite produced by both plants (Capasso et al., 1992) and fungi including Diplodia seriata (Venkatasubbaiah and Chilton, 1990), Alternaria tagetica (Gamboa-Angulo et al., 2001), Neofusicoccum parvum (Evidente et al., 2010), and Neofusicoccum australe (Andolfi et al., 2012), Lasiodiplodia spp. (Cimmino et al., 2017), Diaporthella cryptica (Cimmino et al., 2018). Moreover, several studies have also reported tyrosol from the bioactive extracts of endophytic Colletotrichum (Colletotrichum gleosporoides, Colletotrichum crassipes) (Anisha and Radhakrishnan, 2017; Masi et al., 2018; Plotnikov and Plotnikova, 2020). However, the report of its isolation from C. coccodes is limited. Interestingly, in the present study tyrosol was isolated from an endophytic C. coccodes extract having significant antimicrobial activity. This suggests the potential of endophytic C. coccodes colonizing H. cordata as producers of bioactive metabolites.
Natural products are often considered for their rich gallows which let them acting as ligands for various clinically relevant receptors and enzymes. The position of enzyme inhibitors as candidate drugs is very cosmic because of their expediency in treating several ailments. Searching inhibitors of enzymes responsible for the growth and maintenance of microbes (for example, enzymes involved in DNA replication, protein translation, cell wall synthesis, etc.) have become a familiar intent for antimicrobial drug discovery (Pereira et al., 2017; Belete, 2019). Tyrosyl tRNA synthetases (TyrRS) belonging to enzyme class aminoacyl tRNA synthetase have been playing an essential role in the faithful translation and protein synthesis in the bacterial cell. Several studies on natural products have targeted and validated inhibition of this enzyme for novel and effective therapeutics development against clinically significant bacterial pathogens such as E. coli, S. aureus, and Mycobacterium tuberculosis (Xiao et al., 2011; Zhu et al., 2015; Skupinska et al., 2017; Sun et al., 2017). Similarly, “lanosterol-14α-demethylase” which is a cytochrome P450 enzyme, has been reported to be having supreme significance in the biosynthesis of sterols in eukaryotic cells. The enzyme catalyzes the conversion of lanosterol to ergosterol, an important component of the fungal cell membrane, depletion of which leads to growth inhibition or disruption of cells. Because of this fact this enzyme has long been considered as a target of interest for most antifungal drug discovery against Aspergillus niger, Cryptococcus albidus, and especially against C. albicans (Kelly et al., 1990; Hargrove et al., 2017; Stana et al., 2017; Rana et al., 2019). Furthermore, the determination of crystal structures of these enzymes has led to the evolution of computational approaches which have been extremely advantageous in screening a large number of small molecules against specific targets of interest. Molecular docking examines and evaluates the affinity of probable drug candidates toward the binding cavities of target proteins or enzymes by establishing chemical connections of various strengths (Alam and Khan, 2018; Padhi et al., 2020). Tyrosol has earlier been reported as an inhibitor of several enzymes including α-glucosidase, cytochrome oxidase through in silico and in vitro investigations (Zhang et al., 2019; Yadav et al., 2020). Amini et al. (2017) reported the possible antibacterial action of tyrosol as it inhibited ATPase activity in E. coli. Nevertheless, reports describing the antimicrobial action mechanism of tyrosol are truly limited. In this study, the purified compound tyrosol was screened against the active sites of TyrRS of S. aureus and E. coli, and CYP51 of C. albicans using CDOCKER docking program. It was observed that the compound made significant contacts with the active site amino acids of all the target enzymes. As revealed from the energy parameters, tyrosol has the highest affinity toward S. aureus TyrRs followed by that of E. coli. Similarly, the binding affinity of tyrosol was also found to be substantial with C. albicans CYP51. The non-covalent bonds formed during any receptor-ligand interactions use to participate in a major role in pharmaceutical drug designing and development. The H-bonds usually confer structural stability and rigidity to the protein-ligand complexes. Moreover, there exist Pi-Sigma interactions (Pi-anion, Pi-cation, and Pi-alkyl) and salt bridges that reportedly maximize the binding affinity between protein and ligands in a physiological environment (Rahman et al., 2016; Aljoundi et al., 2020). In the present study, the interaction between tyrosol and target enzymes (TyrRS and CYP51) witnessed some non-covalent forces inclusively H-bonds and Pi-Sigma. This proposes that the antimicrobial activity of the organic extract obtained from C. coccodes culture filtrates could be linked to the presence of tyrosol which in turn might hinder the normal growth of the test bacteria and fungus by inhibiting the activity of the target enzymes. The findings further suggest that endophytic C. coccodes inhabiting medicinal plants like H. cordata Thunb. could be exploited as a potential producer of antimicrobial agents. Moreover, the in silico findings can be used in combination with in vitro enzymatic assays to investigate the efficacy of tyrosol as a potent enzyme inhibitor and an effective antimicrobial agent.
In this study, bioactive extract of an endophytic C. coccodes isolated from the leaves of a medicinal plant H. cordata Thunb. was characterized to yield a pure metabolite tyrosol. Molecular docking revealed tyrosol has a strong affinity toward bacterial tyrosyl tRNA synthetase and fungal CYP45014α-lanosterol demethylase, suggesting its possible antimicrobial action mechanism. The isolate was characterized by ITS rDNA and ITS2 sequence, and ITS2 structure was used to differentiate our isolate from the same species of different lifestyles. The findings of the study could be useful in exploring the bioactive potential of fungal endophytes colonizing plants of ethnomedicinal importance. Further, ITS2 could be used as a potential marker for species differentiation among dissimilar lifestyles. To the best of our knowledge, tyrosol was isolated from C. coccodes for the first time.
The datasets presented in the study can be found at NCBI GenBank with the following URL: https://www.ncbi.nlm.nih.gov/nuccore/MN128230.
RT, KT, and DJ designed, carried out the experimental works, and drafted the manuscript. SP and AR contributed to the molecular docking and phylogeny. AC, MM, and AE did purification and characterization of the metabolite. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful to the Head, Department of Botany, Gauhati University and Director, Institute of Bioresources and Sustainable Development, India for providing the necessary facilities to carry out the work.
Abdel-Azeem, A., Azeem, M. A., and Khalil, W. (2019). “Endophytic fungi as a new source of antirheumatoid metabolites,” in Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases, 2nd Edn, eds R. R. Watson and V. R. Preedy (Cambridge, MA: Academic Press), 335–384.
Alam, S., and Khan, F. (2018). Virtual screening, docking, ADMET and system pharmacology studies on Garcinia caged xanthone derivatives for anticancer activity. Sci. Rep. 8:5524.
Aljoundi, A., Bjij, I., El Rashedy, A., and Soliman, M. E. S. (2020). Covalent versus non-covalent enzyme inhibition: which route should we take? A justification of the good and bad from molecular modelling perspective. Protein J. 39, 97–105. doi: 10.1007/s10930-020-09884-2
Amini, A., Liu, M., and Ahmad, Z. (2017). Understanding the link between antimicrobial properties of dietary olive phenolics and bacterial ATP synthase. Int. J. Biol. Macromol. 101, 153–164. doi: 10.1016/j.ijbiomac.2017.03.087
Andolfi, A., Maddau, L., Cimmino, A., Linaldeddu, B. T., Franceschini, A., Serra, S., et al. (2012). Cyclobotryoxide, a phytotoxic metabolite produced by the plurivorous pathogen Neofusicoccum australe. J. Nat. Prod. 75, 1785–1791. doi: 10.1021/np300512m
Anisha, C., and Radhakrishnan, E. K. (2017). Metabolite analysis of endophytic fungi from cultivars of Zingiber officinale Rosc. identifies myriad of bioactive compounds including tyrosol. 3 Biotech. 7:146.
Arivudainambi, U. S. E., Anand, T. D., Shanmugaiah, V., Karunakaran, C., and Rajenrdan, A. (2011). Novel bioactive metabolites producing endophytic fungus Colletotrichum gloeosporioides against multidrug resistant Staphylococcus aureus. FEMS. Immunol. Med.Microbiol. 61, 340–345. doi: 10.1111/j.1574-695x.2011.00780.x
Barnett, H. L., and Hunter, B. B. (1996). Illustrated Genera of Imperfect Fungi, 4th Edn, St. Paul Minnesota: APS Press.
Belete, T. M. (2019). Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Hum. Microb. J. 11:100052. doi: 10.1016/j.humic.2019.01.001
Capasso, R., Cristinzio, G., Evidente, A., and Scognamiglio, F. (1992). Isolation, spectroscopy and selective phytotoxic effects of polyphenols from vegetable waste waters. Phytochemistry 31, 4125–4128. doi: 10.1016/0031-9422(92)80426-f
Chutulo, E. C., and Chalannavar, R. K. (2018). Endophytic mycoflora and their bioactive compounds from Azadirachta indica: a comprehensive review. J. Fungi 4:42. doi: 10.3390/jof4020042
Cimmino, A., Cinelli, T., Masi, M., Reveglia, P., da Silva, M. A., Mugnai, L., et al. (2017). Phytotoxic lipophilic metabolites produced by grapevine strains of Lasiodiplodia species in Brazil. J. Agric. Food Chem. 65, 1102–1107. doi: 10.1021/acs.jafc.6b04906
Cimmino, A., Nocera, P., Linaldeddu, B. T., Masi, M., Gorecki, M., Pescitelli, G., et al. (2018). Phytotoxic metabolites produced by Diaporthella cryptica, the causal agent of hazelnut branch canker. J. Agric. Food. Chem. 66, 3435–3442. doi: 10.1021/acs.jafc.8b00256
Clarke, J. D. (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc. 3:pdb.rot5177.
Cote, C. A., and Peculis, B. A. (2001). Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res. 29, 2106–2116. doi: 10.1093/nar/29.10.2106
Damm, U., Johnston, P. R., and Weir, B. S. (2012). The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73, 115–180. doi: 10.3114/sim0011
Evidente, A., Punzo, B., Andolfi, A., Cimmino, A., Melck, D., and Luque, J. (2010). Lipophilic phytotoxins produced by Neofusicoccum parvum, a grapevine canker agent. Phytopathol. Mediterr. 49, 74–79.
Freire, M. C. M., Roméria da Silva, M., Zhang, X., Almeida, A. M. R., Stacey, G., and de Oliveira, L. O. (2012). Nucleotide polymorphism in the 58S nrDNA gene and internal transcribed spacers in Phakopsora pachyrhizi viewed from structural models. Fungal Genet. Biol. 49, 95–100. doi: 10.1016/j.fgb.2011.12.010
Gagnon, J. A., Valen, E., Thyme, S. B., Huang, P., Ahkmetova, L., Pauli, A., et al. (2014). Efficient mutagenesis by Cas9 Protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9:e98186. doi: 10.1371/journal.pone.0098186
Gamboa-Angulo, M. M., Garcıá-Sosa, K., Alejos-González, F., Escalante-Erosa, F., Delgado-Lamas, G., and Peña-Rodríguez, L. M. (2001). Tagetolone and tagetenolone: two phytotoxic polyketides from Alternaria tagetica. J. Agric. Food Chem. 49, 1228–1232. doi: 10.1021/jf000872k
Gouda, S., Das, G., Sen, S. K., Shin, H. S., and Patra, J. K. (2016). Endophytes: a treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 7:1538. doi: 10.3389/fmicb.2016.01538
Hargrove, T. Y., Friggeri, L., Wawrzak, Z., Qi, A., Hoekstra, W. J., Schotzinger, R. J., et al. (2017). Structural analyses of Candida albicans sterol 14-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 292, 6728–6743. doi: 10.1074/jbc.m117.778308
Hawksworth, D. L. (2001). The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105, 1422–1432. doi: 10.1017/s0953756201004725
Heeg, J. S., and Wolf, M. (2015). ITS2 and 18S rDNA sequence-structure phylogeny of Chlorella and allies (Chlorophyta, Trebouxiophyceae, Chlorellaceae). Plant Gene 4, 20–28. doi: 10.1016/j.plgene.2015.08.001
Huang, X., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877. doi: 10.1101/gr.9.9.868
Iwen, P. C., Hinrichs, S. H., and Rupp, M. E. (2002). Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 40, 87–109. doi: 10.1080/mmy.40.1.87.109
Jena, S. K., and Tayung, K. (2013). Endophytic fungal communities associated with two ethno-medicinal plants of Similipal biosphere reserve, India and their antimicrobial prospective. J. App. Pharm. Sci. 3, S7–S12.
Kapoor, N., Gambhir, L., and Saxena, S. (2018). Secondary structure prediction of ITS rRNA region and molecular phylogeny: an integrated approach for the precise speciation of Muscodor species. Ann. Microbiol. 68, 763–772. doi: 10.1007/s13213-018-1381-8
Kaur, T. (2020). “Fungal endophyte-host plant interactions: role in sustainable agriculture,” in Sustainable Crop Production, eds M. Hassanuzaman, M. C. M. Filho, M. Fujita, and T. A. R. Nogueira (London: Intech Open), 1–18.
Kelly, S. L., Kenna, S., Bligh, H. F. J., Watson, P. F., Stansfield, I., Ellis, S. W., et al. (1990). “Lanosterol to Ergosterol- Enzymology, inhibition and genetics,” in Biochemistry of Cell Walls and Membranes in Fungi, eds P. J. Kuhn, A. P. J. Trinci, M. J. Jung, M. W. Goosey, and L. G. Copping (Berlin: Springer), 223–243. doi: 10.1007/978-3-642-74215-6_15
Kharwar, R. N., Mishra, A., Gond, S. K., Stierle, A., and Stierle, D. (2011). Anticancer compounds derived from fungal endophytes: their importance and future challenges. Nat. Prod. Rep. 28, 1208–1228. doi: 10.1039/c1np00008j
Kim, J. W., and Shim, S. H. (2019). The fungus Colletotrichum as a source for bioactive secondary metabolites. Arch. Pharm. Res. 42, 735–753. doi: 10.1007/s12272-019-01142-z
Koetschan, C., Forster, F., Keller, A., Schleicher, T., Ruderisch, B., Schwarz, R., et al. (2010). The ITS2 Database III- sequences and structures for phylogeny. Nucleic Acids Res. 38, D275–D279.
Lu, H., Zou, W. X., Meng, J. C., Hu, J., and Tan, R. X. (2000). New bioactive metabolites produced by Colletotrichum sp., an endophytic fungus in Artemisia annua. Plant Sci. 151, 67–73. doi: 10.1016/s0168-9452(99)00199-5
Marinho, M. A. T., Junqueira, A. C. M., and Azeredo-Espin, A. M. L. (2011). Evaluation of the internal transcribed spacer 2 (ITS2) as a molecular marker for phylogenetic inference using sequence and secondary structure information in blow flies (Diptera: Calliphoridae). Genetica 139, 1189–1207. doi: 10.1007/s10709-011-9621-x
Masi, M., Maddau, L., Linaldeddu, B. T., Scanu, B., Evidente, A., and Cimmino, A. (2018). Bioactive metabolites from pathogenic and endophytic fungi of forest trees. Curr. Med. Chem. 25, 208–252. doi: 10.2174/0929867324666170314145159
Padhi, S., Masi, M., Chourasia, R., Rajashekar, Y., Rai, A. K., and Evidente, A. (2021). ADMET profile and virtual screening of plant and microbial natural metabolites as SARS-CoV-2 S1 glycoprotein receptor binding domain and main protease inhibitors. Eur. J. Pharmacol. 890:173648. doi: 10.1016/j.ejphar.2020.173648
Padhi, S., Masi, M., Panda, S. K., Luyten, W., Cimmino, A., Evidente, A., et al. (2020). Antimicrobial secondary metabolites of an endolichenicAspergillusniger isolated from lichen thallus of Parmotremaravum. Nat. Prod. Res. 34, 2573–2580. doi: 10.1080/14786419.2018.1544982
Padhi, S., Panda, M. K., Das, D., and Tayung, K. (2016). ITS2 RNA secondary structure analysis reveals close affinity between endophytic and pathogenic fungi: a case study in Fusarium species. Ann. Microbiol. 66, 625–633. doi: 10.1007/s13213-015-1142-x
Padhi, S., and Tayung, K. (2013). Antimicrobial activity and molecular characterization of an endophytic fungus, Quambalaria sp. isolated from Ipomoea carnea. Ann. Microbiol. 63, 793–800. doi: 10.1007/s13213-012-0534-4
Pereira, J. J., Pereira, A. P., Jandú, J. J., da Paz, J. A., Crovella, S., dos Santos Correia, M. T., et al. (2017). Commiphoraleptophloeos phytochemical and antimicrobial characterization. Front. Microbial. 8:52. doi: 10.3389/fmicb.2017.00052
Plotnikov, M. B., and Plotnikova, T. M. (2020). Tyrosol as a neuroprotector: strong effects of a “weak” antioxidant. Curr. Neuropharmacol. 19, 34–48. doi: 10.2174/1570159X18666200507082311
Rahman, A., Hoque, M. M., Khan, M. A., Sarwar, M. G., and Halim, M. A. (2016). Non-covalent interactions involving halogenated derivatives of capecitabine and thymidylate synthase: a computational approach. Springer Plus 5:146.
Rampersad, S. N. (2014). ITS1, 5.8S and ITS2 secondary structure modelling for intra-specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex. Springer Plus 3:684.
Rana, R., Sharma, R., and Kumar, A. (2019). Repurposing of fluvastatin against Candida albicans CYP450 lanosterol 14 α-demethylase, a target enzyme for antifungal therapy: an in silico and In vitro Study. Curr. Mol. Med. 19:506. doi: 10.2174/1566524019666190520094644
Ruyck, J., Brysbaert, G., Blossey, R., and Lensink, M. F. (2016). Molecular docking as a popular tool in drug design, an in silico travel. Adv. Appl. Bioinform. 9, 1–11. doi: 10.2147/aabc.s105289
Sarwar, S., Firdous, Q., and Khalid, A. N. (2019). “Importance of molecular and phylogenetic analyses for identification of basidiomycetes,” in Recent Advances in Phylogenetics, ed. Y. Zubaida (London: IntechOpen), doi: 10.5772/intechopen.80671
Schultz, J., Maisel, S., Gerlach, D., Müller, T., and Wolf, M. (2005). A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11, 361–364. doi: 10.1261/rna.7204505
Schulz, B., Boyle, C., Draeger, S., Rommert, A. K., and Krohn, K. (2002). Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol. Res. 106, 996–1004. doi: 10.1017/s0953756202006342
Sharma, V. K., Kumar, J., Singh, D. K., Mishra, A., Verma, S. K., Gond, S. K., et al. (2017). Induction of cryptic and bioactive metabolites through natural dietary components in an endophytic fungus Colletotrichum gloeosporioides (Penz.) Sacc. Front. Microbiol. 8:1126. doi: 10.3389/fmicb.2017.01126
Shubin, L., Juan, H., Ren-Chao, Z., Shi-Ru, X., and Yuan-Xiao, J. (2014). Fungal endophytes of Alpinia officinarum rhizomes: insights on diversity and variation across growth years, growth sites, and the inner active chemical concentration. PLoS One 9:e115289. doi: 10.1371/journal.pone.0115289
Skupinska, M., Stȩpniak, P., Łȩtowska, I., Rychlewski, L., Barciszewska, M., and Barciszewski, J. (2017). Natural compounds as inhibitors of Tyrosyl-tRNA Synthetase. Microb. Drug Resist. 23, 308–320. doi: 10.1089/mdr.2015.0272
Stana, A., Vodnar, D. C., Tamaian, R., Pîrnău, A., Vlase, L., Ionu?, I., et al. (2017). Design, synthesis and antifungal activity evaluation of new Thiazolin-4-ones as Potential Lanosterol 14α-Demethylase Inhibitors. Int. J. Mol. Sci. 18:177. doi: 10.3390/ijms18010177
Strobel, G., and Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67, 491–502. doi: 10.1128/mmbr.67.4.491-502.2003
Sun, J., Lv, P. C., and Zhu, H. L. (2017). Tyrosyl-tRNA synthetase inhibitors: a patent review. Expert. Opin. Ther. Pat. 27, 557–564. doi: 10.1080/13543776.2017.1273350
Tamura, K., Masatoshi, N., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Boil. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, R. X., and Zou, W. X. (2001). Endophytes: a rich source of functional metabolites (1987 to 2000). Nat. Prod. Rep. 18, 448–459. doi: 10.1039/b100918o
Venkatasubbaiah, P., and Chilton, W. S. (1990). Phytotoxins of Botryosphaeria obtusa. J. Nat. Prod. 53, 1628–1630. doi: 10.1021/np50072a044
Verma, C., Mishra, G., and Omkar (2020). Widespread inspection and comparative analysis of ITS secondary structure conservation and covariation of Coccinellidae. Int. J. Trop. Insect Sci. 40, 587–597. doi: 10.1007/s42690-020-00105-y
Xiao, Z. P., Ma, T. W., Liao, M. L., Feng, Y. T., Peng, X. C., Li, J. L., et al. (2011). Tyrosyl-tRNA synthetase inhibitors as antibacterial agents: Synthesis, molecular docking and structure-activity relationship analysis of 3-aryl-4-arylaminofuran-2(5H)-ones. Eur. J. Med. Chem. 46, 4904–4914. doi: 10.1016/j.ejmech.2011.07.047
Xu, J., and Adamowicz, S. (2016). Fungal DNA barcoding. Genome 59, 913–932. doi: 10.1139/gen-2016-0046
Yadav, T. C., Kumar, N., Raj, U., Goel, N., Vardawaj, P. K., Prasad, R., et al. (2020). Exploration of interaction mechanism of tyrosol as a potent anti-inflammatory agent. J. Biomol. Struct. Dyn. 38, 382–397. doi: 10.1080/07391102.2019.1575283
Zhang, J., Nugrahaningrum, D. A., Marcelina, O., Ariyanti, A. D., Wang, G., Liu, C., et al. (2019). Tyrosol facilitates neovascularization by enhancing skeletal muscle cells viability and paracrine function in diabetic hindlimb ischemia mice. Front. Pharmacol. 10:909. doi: 10.3389/fphar.2019.00909
Zhang, W., Tian, W., Gao, Z., Wang, G., and Zhao, H. (2020). Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. U.S.A. 117, 14857–14863. doi: 10.1073/pnas.2009637117
Zhu, N., Lin, Y., Li, D., Gao, N., Liu, C., You, X., et al. (2015). Identification of an anti-TB compound targeting the tyrosyl-tRNA synthetase. J. Antimicrob. Chemother. 70, 2287–2294. doi: 10.1093/jac/dkv110
Keywords: Houttuynia cordata, Colletotrichum coccodes, antimicrobial activity, tyrosol, ITS2 RNA secondary structure
Citation: Talukdar R, Padhi S, Rai AK, Masi M, Evidente A, Jha DK, Cimmino A and Tayung K (2021) Isolation and Characterization of an Endophytic Fungus Colletotrichum coccodes Producing Tyrosol From Houttuynia cordata Thunb. Using ITS2 RNA Secondary Structure and Molecular Docking Study. Front. Bioeng. Biotechnol. 9:650247. doi: 10.3389/fbioe.2021.650247
Received: 06 January 2021; Accepted: 17 May 2021;
Published: 17 June 2021.
Edited by:
Adeline Su Yien Ting, Monash University Malaysia, MalaysiaReviewed by:
Piyush Pandey, Assam University, IndiaCopyright © 2021 Talukdar, Padhi, Rai, Masi, Evidente, Jha, Cimmino and Tayung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kumananda Tayung, a3VtYW5hbmRAZ2F1aGF0aS5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.